- 1Department of Breast Surgery, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Program of Innovative Cancer Therapeutics, Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 3Key Laboratory of Combined Multi-Organ Transplantation, Ministry of Public Health, Key Laboratory of Organ Transplantation, Zhejiang University, Hangzhou, China
Pseudogenes, abundant in the human genome, are traditionally considered as non-functional “junk genes.” However, recent studies have revealed that pseudogenes act as key regulators at DNA, RNA or protein level in diverse human disorders (including cancer), among which pseudogene-derived long non-coding RNA (lncRNA) transcripts are extensively investigated and has been reported to be frequently dysregulated in various types of human cancer. Growing evidence demonstrates that pseudogene-derived lncRNAs play important roles in cancer initiation and progression by serving as competing endogenous RNAs (ceRNAs) through competitively binding to shared microRNAs (miRNAs), thus affecting both their cognate genes and unrelated genes. Herein, we retrospect those current findings about expression, functions and potential ceRNA mechanisms of pseudogene-derived lncRNAs in human cancer, which may provide us with some crucial clues in developing potential targets for cancer therapy in the future.
Background
Cancer, a major killer of human health, constitutes an enormous burden on society in both more and less economically developed countries. According to GLOBOCAN statistics, about 14.1 million new cancer cases were diagnosed and 8.2 million cancer-associated deaths occurred in 2012 all over the world (Torre et al., 2015). Cancer is a complex disease. Multiple lines of evidence suggest that numerous risk elements, containing genetic and environmental factors, account for cancer initiation and development, among which the involvement of mis-regulation of non-coding RNAs (ncRNAs) in cancer has aroused extensive attention during the past few decades.
Based on length, ncRNAs can be divided into several subtypes, such as microRNA (miRNA), long non-coding RNA (lncRNA) as well as circular RNA (circRNA). ncRNAs, especially miRNA, have been solely studied until a famous hypothesis, namely competing endogenous RNA (ceRNA) mechanism, which was proposed by the team of Salmena et al. (2011). ceRNA hypothesis represents a hidden RNA language that messenger RNA (mRNA) can cross talk with lncRNA or circRNA by competitively binding to shared miRNAs, and eventually influence physiological and pathological processes (Ou et al., 2019; Wang W. et al., 2019; Zhang et al., 2020). After the emergence of ceRNA hypothesis, ncRNA-related research has been developed rapidly.
Jacq et al. (1977) introduced a new word “pseudogene” when they discovered a copy of the 5S rRNA gene in Xenopus laevis. The copy with 5′-end truncation and 14-bp mismatches makes it non-functional. From then on, a large number of pseudogenes have been gradually found in a variety of prokaryotes and eukaryotes, including homo species. For a long time, pseudogenes are considered as non-functional “junk genes,” “relics of evolution” or “genomic fossil.”
In recent years, with the huge advancement of in-deep research, multilayered functions of pseudogene DNAs, RNAs or proteins have been reported in various types of human cancer (Xiao-Jie et al., 2015), among which pseudogene-derived RNA transcripts are the most investigated. Pseudogene-expressed RNAs are key components of lncRNAs. Similar as common lncRNAs, pseudogene-derived lncRNAs can also function as critical modulators in initiation and development of human cancer by ceRNA mechanism via sponging miRNAs, which is supported by increasing findings. Therefore, in this work, we highlight recent findings regarding the expression, function and miRNA sponging mechanism of pseudogene-derived lncRNAs in diverse types of human cancer.
Origination and Classification of Pseudogenes and Pseudogene-Derived lncRNAs
Based on the origination form from its ancestral gene, pseudogenes can be classified into three types: (1) processed pseudogenes or retrotransposed pseudogenes deriving from retrotransposition of processed mRNA back into the genome; (2) unprocessed pseudogenes or duplicated pseudogenes deriving from unfaithful gene duplications; and (3) unitary pseudogenes deriving from gene mutations (Xiao-Jie et al., 2015). lncRNAs are divided into several categories according to genomic organization and relation to coding genes, such as long intergenic non-coding RNAs, antisense RNAs, sense overlapping RNAs, sense intronic RNAs, enhancer RNAs as well as pseudogene-expressed lncRNAs (Grander and Johnsson, 2016). Although only few of pseudogenes can be transcribed, all the three types of pseudogenes may transcribe and are called transcribed pseudogenes or pseudogene-derived transcripts. However, compared with other members of lncRNAs, transcribed pseudogenes-derived lncRNAs have not been paid attention previously. Recent studies have demonstrated that transcribed pseudogene-derived lncRNAs play important roles in multiple biological processes, such as cell proliferation, cell cycle, cell migration and cell death (Lai et al., 2019; Oliveira-Mateos et al., 2019; Varesio et al., 2019).
Competing Endogenous RNA Mechanism of Pseudogene-Derived lncRNA
Previous evidences have suggested that pseudogene-expressed RNAs could function as antisense RNAs or endo-siRNAs (Korneev et al., 1999; Watanabe et al., 2008). Besides, recent studies have also found that pseudogene-expressed RNAs serve as sponges of miRNAs and thus exert biological roles. To better understand the miRNA sponge mechanism of pseudogene-derived lncRNAs in cancer, competing endogenous RNA (ceRNA) mechanism proposed by Salmena et al. (2011) should be introduced. In this hypothesis, messenger RNA, lncRNA and circRNA can “talk” to each other by binding to shared miRNAs using miRNA response elements (MREs). Dysregulation of lncRNAs, pseudogenes and circRNAs leads to alteration of abundance of miRNAs, thus affecting their inhibition of downstream target expression. This mechanism also applies to pseudogene-derived transcripts. To date, ceRNA mechanism is validated to participate in initiation and progression of human cancer when its dysregulated (Qu et al., 2015; Yang C. et al., 2016). Based on ceRNA mechanism, researchers and scholars have discovered a variety of potential cancer-associated pseudogenes using in silico analysis. For example, Wei Y. et al. (2017) identified three pseudogene-involved ceRNA triples in lung adenocarcinoma, including NKAPP1-miR-21-5p-PRDM11, MSTO2P-miR-29c-3p-EZH2 and RPLP0P2-miR-29c-3p-EZH2; Jiang et al. (2018) screened several prostate cancer-related pseudogenes by establishing pseudogene-miRNA-gene triple ceRNA regulatory network. Lab experiments also confirmed the involvement of pseudogene-mediated ceRNA mechanism in cancer development. For instance, HMGA1 pseudogenes, HMGA1P6 and HMGA1P7, were reported to serve as candidate proto-oncogenic ceRNAs (Esposito et al., 2014); HMGA1P7 was also found to sustain overexpression of H19 and lgf2 by acting as decoy for miR-15, miR-16, miR-214, and miR-761 (De Martino et al., 2016). Karreth et al. (2015) suggested that BRAF pseudogenes BRAF-RS1 and BRAFP1 functioned as ceRNAs to elevate BRAF expression and activate MAPK signaling, thereby eliciting their roles in lymphoma.
Expression and Functions of Pseudogene-Derived lncRNAs in Human Cancer
Dysregulation of pseudogenes and their transcripts has been implicated into initiation and/or progression of human disorders, including cancer. Among pseudogene-derived lncRNAs, some act as tumor promotors, facilitating cancer development, whereas the other function as tumor suppressors, inhibiting cancer progression. In this part, we summarized the upregulated oncogenic pseudogene-derived lncRNAs and downregulated tumor suppressive pseudogene-derived lncRNAs in diverse types of human cancer (Figure 1).

Figure 1. Overview of reported upregulated and downregulated pseudogene-derived lncRNAs in diverse types of human cancer.
Upregulated Oncogenic Pseudogene-Derived lncRNAs
Liver Cancer
As is known to all, hepatitis B virus (HBV) infection is closely linked to occurrence of hepatocellular carcinoma (HCC). PCNAP1 was reported to boost HBV replication, thus enhancing tumor growth of HCC (Feng et al., 2019). lncRNA PTTG3P was markedly upregulated in HCC samples compared with normal samples and its overexpression promoted growth and metastasis of HCC by increasing its cognate gene PTTG1 and activating PI3K/AKT signaling pathway (Huang J.L. et al., 2018). p53 signaling is a widely acknowledged tumor suppressive pathway in multiple human cancers, including HCC (Masutomi et al., 2001). PDIA3P1, upregulated in HCC tissues compared with paired normal adjacent tissues, could increase proliferation, migration and invasion, and decrease apoptosis of HCC by suppressing p53 signaling pathway (Kong et al., 2017). UBE2CP3 was significantly upregulated in HCC and was discovered to function as an oncogene by increasing tumor metastasis through inducing epithelial-mesenchymal transition (Cao et al., 2017). Lin et al. (2018) also confirmed the oncogenic role of UBE2CP3 in HCC, facilitating angiogenesis via activation of ERK1/2/HIF-1alpha/VEGFA signaling. Another highly expressed pseudogene-derived lncRNA, PDPK2P, also exhibited these oncogenic effects on HCC by PDK1/AKT/caspase 3 signaling pathway (Pan et al., 2019). OCT4-pg4, acting as a natural sponging of miR-145 to upregulating OCT4, exerted an oncogenic role in hepatocarcinogenesis by promoting growth (Wang et al., 2013). HCC is notorious for its high aggressiveness and easy to relapse. A recent study conducted by the team of Wang Q demonstrated that ANXA2P2 was highly expressed in HCC and promoted an aggressive phenotype of HCC (Wang Q.S. et al., 2019). And Wang M.Y. et al. (2019) confirmed that RACGAP1P facilitated early recurrence of HCC by activating RACGAP1/Rho/ERK signaling axis. Dysregulation of pseudogenes and their transcripts accounts for the development of chemo-resistance and radio-resistance of HCC which greatly reduces the efficacy of chemotherapy and radiotherapy. For example, Xie et al. (2019) suggested that PDIAP3 caused doxorubicin resistance of HCC by targeting miR-125/124-TRAF6/NF-KB signaling; Zhou Y. et al. (2019) found that knockdown of SUMO1P3 markedly enhanced radio-sensitivity in HCC.
Breast Cancer
CYP4Z2P, an oncogenic pseudogene of CYP4Z1, has been widely investigated in breast cancer. The 3′UTR of CYP4Z2P was found to promote angiogenesis of breast cancer (Zheng et al., 2014, 2015). The ceRNA network of CYP4Z2P and its parental gene CYP4Z1 exerted an anti-apoptotic function in breast cancer (Li C. et al., 2017). Tamoxifen is an effective therapy for estrogen receptor (ER)-positive breast cancer. Zheng et al. (2016) discovered that the ceRNA network of CYP4Z2P and CYP4Z1 conferred tamoxifen resistance of breast cancer. The ceRNA network, activated by transcriptional factor six2, was also responsible for maintaining the stemness of breast cancer (Zheng L. et al., 2019). In addition to CYP4Z2P, some pseudogene-derived lncRNAs were also found to function as oncogenes in breast cancer. For example, Liu et al. (2017a) confirmed that SUMO1P3 promoted proliferation, migration and invasion of breast cancer; Barrow et al. (2019) found that CRYbetaB2P1 was upregulated in breast cancer and facilitated progression of breast cancer by increasing CRYbetaB2 expression; and Wang R. et al. (2018) demonstrated that FTH1P3 led to paclitaxel resistance of breast cancer. PTTG3P was markedly upregulated in breast cancer samples compared with normal controls and its upregulated linked to poor prognosis of patients with breast cancer (Lou et al., 2019).
Gastric Cancer
NANOGP8 expression was significantly elevated in gastric cancer and knockdown of NANOGP8 resulted in decreased proliferation and promoted apoptosis of gastric cancer cells (Li et al., 2019). Ma X. et al. (2018) also suggested that NANOGP8 was a positive regulator of gastric cancer stem cells, associated with epithelial-mesenchymal transition, stemness, cancer stem cell markers as well as Wnt signaling pathway. DUXAP8, a 2107 nt RNA derived from pseudogene, was obviously upregulated in gastric cancer and significantly associated with greater tumor size, advanced clinical stage and lymphatic metastasis (Ma et al., 2017). Overexpression of DUXAP8 promoted cell proliferation and migration of gastric cancer by epigenetically inhibiting PLEKHO1 expression. Another DUXA-associated pseudogene, DUXAP10, also exerted its oncogenic roles in gastric cancer by transcriptionally repressing LATS1 expression and maintaining the stability of beta-catenin mRNA (Xu et al., 2018). POU5F1B, a processed pseudogene highly homologous to OCT4, was frequently amplified in gastric cancer cell lines and clinical specimens when compared with their corresponding controls (Hayashi et al., 2015). Furthermore, after POU5F1B overexpression in gastric cancer, a variety of growth factors were induced and an aggressive phenotype was exhibited. Ni et al. (2017) showed that MSTO2P was markedly associated with lymphatic metastasis and distal metastasis and promoted growth, colony formation, migration and invasion of gastric cancer. High expression PTTG3P linked to large tumor size, increased tumor invasiveness and served as an unfavorable prognostic biomarker (Weng, 2017). Besides, the abilities of gastric cancer proliferation and invasion were enhanced after ectopic expression of PTTG3P. Wang X. et al. (2019) found that DUSP5P1, directly induced by C8orf76, fueled gastric tumorigenicity and metastasis by activating MAPK signaling pathway. SUMO1P3 was significantly upregulated in gastric cancer and correlated with tumor size, differentiation, lymphatic metastasis and invasion (Mei et al., 2013). ROC curve analysis also indicated that high expression level of SUMO1P3 might be a potential diagnostic biomarker for gastric cancer. However, its functions in gastric cancer remain unclear and need to be further investigated.
Colorectal Cancer
High SUMO1P3 level was reported to be associated with advanced histological stages, metastasis, angiogenesis and poor prognosis of colon cancer patients (Zhang L.M. et al., 2017). Inhibition of SUMO1P3 repressed proliferation, migration, invasion and pro-angiogenesis of colon cancer cells in vitro, and reduced growth, liver metastasis and vascularization of colon cancer in vivo by decreasing cyclin D1, vimentin and VEGFA but increasing E-cadherin expression. FLT1P1markedly promoted cell proliferation in vitro and xenograft tumor growth in vivo (Ye et al., 2015). TPTE2P1 levels in colorectal cancer tissues were higher than that in adjacent normal tissues, and its upregulation was markedly associated with poor patients’ survival (Dai et al., 2019). The authors also indicated that silencing expression of TPTE2P1 resulted in cell cycle arrest at S phase and caused cell apoptosis in colorectal cancer. Moreover, the tumor suppressive effects of TPTE2P1 on colorectal cancer was observed in the in vivo experiment. DUXAP8 was remarkably overexpressed in colorectal cancer and DUXAP8 knockdown led to inhibited proliferation, migration and invasion and enhanced apoptosis (Du et al., 2019). Lian et al. (2017) suggested that another DUXA-associated pseudogene-derived lncRNA, DUXAP10, was also evidently upregulated in colorectal cancer and positively correlated with advanced pathological stages, larger tumor sizes and lymph node metastasis. DUXAP10 promoted cell proliferation and cell cycle progression and blocked cell apoptosis by epigenetically silencing expression of p21 and PTEN.
Lung Cancer
SUMO1P3 expression was markedly increased in non-small cell lung cancer tissues and cells compared with corresponding normal controls and in metastatic lymph node specimens in comparison to primary tumor tissue specimens (Zhang et al., 2019). The authors also confirmed high SUMO1P3 expression was correlated with late clinical stage, lymph node metastasis, distant metastasis and poor differentiated degree. Moreover, SUMO1P3 enhanced cell migration and invasion of non-small cell lung cancer. Sun et al. (2017) reported that DUXAP8, upregulated in non-small cell lung cancer and an unfavorable prognostic biomarker, significantly facilitated cell growth, migration and invasion, and impaired apoptosis both in vitro and in vivo by epigenetically silencing EGR1 and RHOB. DUXAP10 expression was identified to be overexpressed in non-small cell lung cancer tissues and cell lines and its upregulation was correlated with poor prognosis of patients with non-small cell lung cancer (Wei C.C. et al., 2017). Functional and mechanistic experiments suggested that DUXAP10 exerted oncogenic roles in non-small cell lung cancer by binding to LSD1 and epigenetic silencing expression of LATS2 and RRAD.
Brain Cancer
Glioma and glioblastoma are two main types of brain cancer. Pseudogene-derived lncRNAs are found to facilitate development and progression of glioma and glioblastoma. For example, Zhang et al. (2018) FTH1P3 was reported to be upregulated in glioma tissues and high-grade glioma tissues when compared with normal brain tissues and low-grade glioma tissues, respectively. Overexpression of FTH1P3 enhanced glioma cell proliferation and inhibited apoptosis by regulating miR-224-5p/TPD52 pathway. Another study by Xu et al. (2019) demonstrated that LGMNP1 was significantly increased in glioblastoma cells after radiation and its overexpression conferred radio-therapy resistance in glioblastoma by reducing DNA damage and apoptotic population.
Endometrial Cancer
The expression level of OGFRP1 was significantly upregulated in endometrial cancer (Lv et al., 2019). Lv et al. (2019) also experimentally confirmed that knockdown of OGFRP1 suppressed the malignant behaviors of endometrial cancer, including suppressed cell viability, enhanced cell apoptosis and inhibited cell migration and invasion. OCT4-pg5 was found to be aberrantly activated in endometrial cancer samples compared with benign endometrium samples, and increased expression of OCT4-pg5 enhanced proliferation by promoting OCT4/PI3K/AKT/CCND1 signaling (Bai et al., 2015).
Esophageal Squamous Cell Carcinoma
Feng et al. (2017) reported that PHBP1, markedly upregulated in esophageal squamous cell carcinoma and positively correlated with clinical advanced stage, facilitated proliferation, colony formation and xenograft tumor growth in vitro and in vivo by inducing cell cycle arrest. PTTG3P expression was found to increase in esophageal squamous cell carcinoma tissues and cell lines (Zhang and Shi, 2019). Enhanced expression of PTTG3P greatly stimulated migration and invasion of esophageal squamous cell carcinoma through upregulating expression levels of PTTG1 and PTTG2 (Zhang and Shi, 2019). Yang L. et al. (2018) demonstrated that FTH1P3 was notably upregulated in esophageal squamous cell carcinoma and knockdown of FTH1P3 significantly inhibited proliferation, migration and invasion of esophageal squamous cell carcinoma cells by regulating SP1/NF-kB signaling. The study conducted Wang Z. et al. (2018) certified an obvious increased expression of DUXAP10 in esophageal squamous cell carcinoma. They also showed that DUXAP10 was positively correlated with poor prognosis and epigenetically silenced p21 by recruiting EZH2 to the promoter of p21, thereby promoting cell proliferation and metastasis.
Renal Cell Carcinoma
Chen et al. (2019) demonstrated that DUXAP8 and DUXAP9 were significantly upregulated in renal cell carcinoma, promoted tumor growth and served as two unfavorable prognostic biomarkers for patients with renal cell carcinoma. DUXAP8 was also discovered to enhance progression of renal cell carcinoma (Huang T. et al., 2018). Androgen receptor plays key roles in development of renal cell carcinoma. Wang K. et al. (2019) found androgen receptor could decrease the expression of ASS1P3, a pseudogene of ASS1, and thus facilitate cell growth of renal cell carcinoma.
Pancreatic Cancer
The oncogenic function of SUMO1P3 in the development of pancreatic cancer was reported by Tian et al. (2018). They found that SUMO1P3 expression was markedly elevated in pancreatic cancer tissues when compared with normal controls. Functional experiments revealed the enhanced effect of SUMO1P3 on proliferation, migration and invasion of pancreatic cancer. By epigenetically silencing CDKN1A an KLF2, DUXAP8, upregulated in pancreatic cancer, promoted growth of pancreatic cancer (Lian et al., 2018b). The team of Lian et al. (2018a) demonstrated that DUXAP10 expression was higher in renal cell carcinoma patients with an advanced TNM stage and positive lymph node metastasis. They also confirmed that knockdown of DUXAP10 could result in inhibited proliferation, migration, invasion and enhanced apoptosis in renal cell carcinoma through interacting with RNA-binding proteins, EZH2 and LSD1. Depletion of ZFP91P significantly decreased cell proliferation and migration capacities of pancreatic cancer by altering beta-catenin and vimentin expression (Huang et al., 2016).
Other Types of Cancer
SUMO1P3 was increased in bladder cancer and its high expression predicted poor prognosis of patients with bladder cancer (Zhan et al., 2016). Furthermore, after knockdown of SUMO1P3, bladder cancer exhibited cell proliferation and migration inhibition and apoptosis induction (Zhan et al., 2016).
PTTG3P, a pseudogene of PTTG1 which is upregulated in many types of cancer, was found to promote cervical cancer growth and metastasis by enhancing PTTG1 expression (Guo et al., 2019). Recently, Liu et al. (2017c) showed that RSU1P2 was increased in cervical cancer and boosted the malignant phenotype of cervical cancer. POU5F1B was also reported to be upregulated in cervical cancer tissues and cell lines by Yu et al. (2019). The authors confirmed that interference with expression of POU5F1B significantly suppressed cell proliferation, migration and invasion, and induced apoptosis in cervical cancer (Yu et al., 2019).
Gallbladder is a rare malignant tumor with poor prognosis all over the world. Two pseudogene-derived lncRNAs, TPTE2P1 and Loc344887, have been documented to play oncogenic roles in development of gallbladder cancer (Lv et al., 2015; Wu et al., 2017). Depletion of TPTE2P1 significantly blocked epithelial-mesenchymal transition, migration and invasion of gallbladder cancer (Lv et al., 2015). Loc344887 was elevated in gallbladder cancer, was positively associated with larger tumor size, and facilitated cell proliferation, cell cycle progression, migration and invasion (Wu et al., 2017).
Uchino et al. (2012) suggested that NANOGP8 overexpression remarkably increased cell proliferation whereas its inhibition suppressed the proliferation in human gastrointestinal cancer cells.
Yuan et al. (2019) confirmed FTH1P3 was significantly upregulated in laryngeal squamous cell cancer and positively linked to the poor differentiation, high T classification, positive lymph node metastasis and advanced clinical stage. The authors also demonstrated that FTH1P3 overexpression resulted in enhancement of cell proliferation, migration and invasion and inhibition of cell apoptosis in laryngeal squamous cell cancer (Yuan et al., 2019).
FTH1P3 expression was also increased in uveal melanoma and elevated expression of FTH1P3 promoted cell proliferation, cell cycle and migration of uveal melanoma (Azzariti et al., 2017).
The group of Zang (2017) suggested that FTH1P3 facilitated cell proliferation and colony formation in oral squamous cell carcinoma by upregulating the expression of fizzled 5.
Yang X. et al. (2018) demonstrated that PDIA3P was highly expressed in multiple myeloma, was correlated with poor prognosis of patients with multiple myeloma, and induced growth and drug resistance of multiple myeloma.
Two pseudogene-derived lncRNAs, HMGA1P6 and HMGA1P7, contributed to increase its cognate gene HMGA1 level in human pituitary tumor and then to pituitary tumorigenesis (Esposito et al., 2015).
Downregulated Tumor Suppressive Pseudogene-Derived lncRNAs
Liver Cancer
psiPPM1K, a retrotranscript pseudogene, was significantly reduced in HCC surgical specimens (Chan et al., 2013). Two endo-siRNAs originated from psiPPM1K are found to inhibit oncogenic cell growth of HCC. Low expression of lncRNA PTENP1 in HCC has been reported (Chen et al., 2015). A similar result was also identified by another study conducted by Qian et al. (2017). They found that PTENP1 suppress HCC migration and invasion through enhancing PTEN signaling by interacting with miR-193a-3p. INTS6P1 upregulated its cognate gene INTS6 through competitively binding to miR-17-5p, thereby functioning its tumor suppressive roles in HCC (Peng et al., 2015). Wang et al. (2015) indicated that AOC4P inhibited metastasis of HCC by enhancing degradation of vimentin and suppressing epithelial-mesenchymal transition. MT1DP, directly inhibited by YAP and Runx2, resulted in reduced cell proliferation and colony formation in soft agar, but increased apoptosis in liver cancers (Yu W. et al., 2014).
Breast Cancer
PTENP1 was reported as a tumor suppressor in development and progression of breast cancer. For instance, a recent study conducted by Tang et al. (2017a) showed that PTENP1 suppressed proliferation and migration of breast cancer by inhibiting AKT and MAPK signaling pathways; another study performed by Gao et al. (2019) demonstrated that PETNP1 increased expression of PTEN, thus mediating proliferation, invasion and drug resistance of breast cancer by activation of PI3K/AKT pathway. Connnexin 43 pseudogene, PsiCx43, increased sensitivity to cytotoxic chemotherapy of breast cancer (Bier et al., 2009). SEPW1P RNA was also validated to participate in PIWI-interacting RNA-36712-mediating suppression of breast cancer progression (Tan et al., 2019).
Gastric Cancer
PTENP1 has been demonstrated to function as a tumor suppressor in several cancer cells, containing gastric cancer. For example, Guo et al. (2016) found that PTENP1 was frequently downregulated in gastric cancer, and suppressed proliferation, migration, invasion and promoted apoptosis of gastric cancer through increasing PTEN expression; Zhang R. et al. (2017) also confirmed the tumor suppressive roles of PTENP1 in gastric cancer by decoying miR-106b and miR-93 and thus upregulating PTEN expression. Apart from PTENP1, some pseudogene-derived lncRNAs were also demonstrated to be downregulated in gastric cancer and play tumor suppressive roles in gastric cancer progression. SFTA1P, which is 693 nt long, was obviously decreased in gastric cancer tissues compared with the adjacent normal tissues (Ma H. et al., 2018). Increased expression of SFTA1P suppressed proliferation, migration and invasion of gastric cancer by partially inhibiting p53 signaling. Downregulation of KRT19P3 in gastric cancer cells and tissues was confirmed by Zheng J. et al. (2019). Functional experiments suggested that enforced expression of KRT19P3 significantly blocked cell proliferation and metastasis both in vitro and in vivo. KRT19P3-mediated enhancement of COPS7A protein stability accounted for its roles in suppressing gastric cancer.
Colorectal Cancer
Increasing evidence have supported the involvement of pseudogene-derived lncRNAs in the suppression of pathogenesis of cancer, including colorectal cancer. For example, Chen et al. (2016) confirmed that CTNNAP1 was significantly downregulated in colorectal cancer. The degree of CTNNAP1 dysregulation was markedly correlated with TNM stage. Furthermore, overexpression of CTNNAP1 could suppressed cell proliferation and tumor growth in vitro and in vivo via induction of G0/G1 cell cycle arrest. Sulforaphane, an anticancer agent, exerts its effects partially by inducing Nrf2-dependent signaling. The investigation performed by the group of Johnson G demonstrated that silencing of NMRAL2P could protect against sulforaphane-mediated inhibition of cell growth, colony formation and migration in colon cancer (Johnson et al., 2017).
Lung Cancer
By combination of bioinformatics analysis and experimental validation, Zhang H. et al. (2017) suggested that SFTA1P was downregulated in both lung adenocarcinoma and lung squamous cell carcinoma and inhibited tumor growth, migration and invasion. The downregulation of SFTA1P in lung squamous cell carcinoma was also reported by the team of Li L. et al. (2017). They found elevated SFTA1P induced apoptosis and enhanced the sensitivity to cisplatin of lung squamous cell carcinoma. Intriguingly, low expression of SFTA1P was only correlated with lung adenocarcinoma patients’ poor prognosis but not with lung squamous cell carcinoma patients’ prognosis (Zhang H. et al., 2017). Besides, Shang et al. (2019) confirmed that CHIAP2 expression was markedly decreased in lung adenocarcinoma. After upregulation of CHIAP2, the impaired proliferation and invasion of lung adenocarcinoma were observed.
Brain Cancer
PTENP1 is a tumor suppressive pseudogene-derived lncRNA reported in multiple types of cancer as well as brain glioma. Hu et al. (2019) reported that expression of PTENP1 was decreased in glioma tissues compared with normal brain tissues and forced expression of PTENP1 inhibited cell proliferation and migration in vitro by inducing p21 expression and blocking p38 signaling pathway. In addition, Yang C. et al. (2019) first identified HERC2P2 as a potential tumor suppressor in glioma by constructing a ceRNA network based on the Chinese Glioma Genome Atlas database. Subsequently, they also validated that HERC2P2 overexpression attenuated migration and colony formation abilities of glioma in vitro and inhibited glioma xenograft growth in vivo.
Esophageal Squamous Cell Carcinoma
The tumor suppressive role of PTENP1 in esophageal squamous cell carcinoma was reported by Gong et al. (2017). They found a downregulated expression of PTENP1 in esophageal squamous cell carcinoma compared with the corresponding adjacent normal tissues and overexpression of PTENP1 inhibited tumor proliferation (esophageal squamous cell carcinoma). Liu et al. (2018) suggested TUSC2P was decreased in esophageal squamous cell carcinoma and its downregulation linked to unfavorable prognosis of patients with esophageal squamous cell carcinoma. The inhibitory effects of TUSC2P on proliferation and invasion of esophageal squamous cell carcinoma were also experimentally validated in the study (Liu et al., 2018). TUSC2P-mediated upregulation of TUSC2 was partially responsible for the tumor suppressive roles of TUSC2P in esophageal squamous cell carcinoma (Liu et al., 2018).
Other Types of Cancer
PTENP1 was downregulated in head and neck squamous cell carcinoma and its low expression correlated with poor patients’ prognosis (Liu et al., 2017b). Upregulation of PTENP1 led to inhibited proliferation, colony formation and migration in vitro and suppressed growth in vivo in head and neck squamous cell carcinoma (Liu et al., 2017b).
PTENP1 was reported to act as a tumor suppressor in clear-cell renal cell carcinoma by Yu G. et al. (2014). They found that PTENP1 was downregulated in clear-cell renal cell carcinoma and overexpression of PTENP1 reduced cell proliferation, migration, invasion in vitro and tumor growth and metastasis in vivo (Yu G. et al., 2014).
Gao et al. (2017) suggested that PTENP1, protecting PTEN transcripts from being inhibited by miR-21, suppressed proliferation and colony formation of oral squamous cell carcinoma.
ceRNA Mechanism Contributes to the Roles of Pseudogene-Derived lncRNAs in Human Cancer
Over the past decades, pseudogenes have been documented to play crucial roles in diverse cancer types in DNA, RNA and protein levels (Panagopoulos et al., 2008; Xiao-Jie et al., 2015). Considering study progresses, this work mainly reviewed the miRNA sponging function of pseudogene-derived lncRNAs in human cancer (Table 1).
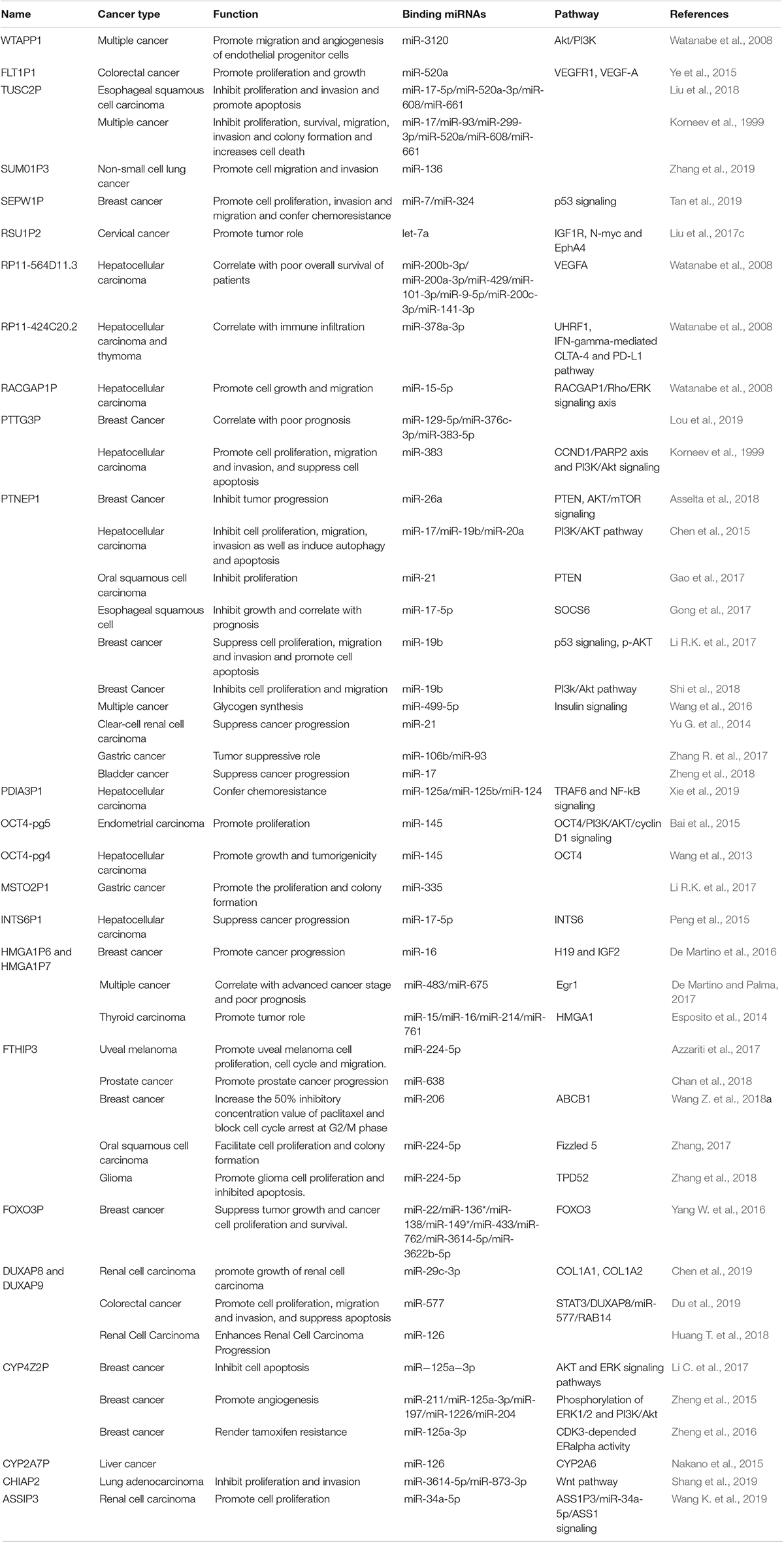
Table 1. Summary of dysregulated pseudogenes and its molecular mechanism in diverse types of cancer.
PTENP1
PTENP1 functions as a ceRNA and suppress tumor progression in multi-cancers, including breast cancer, HCC, oral squamous cell carcinoma, esophageal squamous cell carcinoma, clear-cell renal cell carcinoma, gastric cancer and bladder cancer. In breast cancer, PTENP1 was found to stimulates expression of PTEN transcript and inhibits cell proliferation and migration through decoying miR-19b (Li R.K. et al., 2017; Shi et al., 2018). Further, leads to p53 upregulation and p-AKT downregulation. In HCC, long non-coding RNA PTENP1 via its ceRNA interaction with miR-17, miR-19b and miR-20a, and finally suppressed cell growth through inhibited oncogenic PI3K/AKT pathway (Chen et al., 2015). Additionally, PTENP1 could mediated miR-21 expression and decreased the proliferation ability of oral squamous cell carcinoma and clear-cell renal cell carcinoma (Yu G. et al., 2014; Gao et al., 2017). Meanwhile, PTENP1 could suppress bladder cancer progression by regulating miR-17 as well as esophageal squamous cell carcinoma (Gong et al., 2017; Zheng et al., 2018). What’s more, PTENP1 functions to modulate PTEN expression by sponging miR-93 and miR-106b, plays a tumor suppressive role in gastric cancer (Zhang R. et al., 2017). Not only in cancer, PTEN also contributing to insulin resistance through binding miR-499-5p directly (Wang et al., 2016).
FTH1P3
Increasing evidence has demonstrated that FTH1P3 functions as a ceRNA and participates in tumor initiation and development. For example, FTH1P3 facilitated progression of glioma (Zhang et al., 2018), uveal melanoma (Azzariti et al., 2017), and oral squamous cell carcinoma (Zhang, 2017). Furthermore, FTH1P3 induced paclitaxel resistance by regulating miR-206/ABCB1 axis in breast cancer (Wang R. et al., 2018). Chan et al. (2018) also suggested a FTH1 gene: pseudogene: microRNA modulated tumorigenesis in prostate cancer.
FLT1P1
FLT1 encodes a member of the vascular endothelial growth factor receptor family. FLT1P1 is a bidirectionally transcribed pseudogene of FLT1. Ye et al. (2015) confirmed that FLT1P1 antisense transcript suppressed VEGFA by interaction with miR-520a and inhibited tumor cell proliferation and xenograft tumor growth.
CYP4Z2P
CYP4Z2P and CYP4Z1 promoted angiogenesis of breast cancer by commonly targeting miR-211, miR-197, miR-1226, miR-125a, and miR-204 (Zheng et al., 2015). Moreover, the ceRNA network between CYP4Z1 and CYP4Z2P led to progression of breast cancer, including suppressed apoptosis (Li C. et al., 2017) and induced tamoxifen resistance (Zheng et al., 2016).
DUXAP8 and DUXAP9
To date, two DUXA-associated pseudogene-derived lncRNAs, DUXAP8 and DUXAP9, have been discovered to act as ceRNAs to enhance cancer development. For example, Chen et al. (2019) demonstrated that DUXAP8 and DUXAP9 enhanced growth of renal cell carcinoma by binding to miR-29c-3p and leading to upregulation of COL1A1 and COL1A2; Huang T. et al. (2018) suggested that DUXAP8 facilitated progression of renal cell carcinoma by sponging miR-126. DUXAP8, upregulated by STAT3, also fueled migration and invasion of colorectal cancer by functioning as a ceRNA for miR-577 to regulate RAB14 (Du et al., 2019).
HMGA1P6 and HMGA1P7
Esposito et al. (2014) for the first time, reported HMGA1 pseudogenes as candidate proto-oncogenic ceRNAs, including HMGA1P6 and HMGA1P7. HMGA1P7 was found to increase H19 and lgf2 expression by acting as decoy for miR-15, miR-16, miR-214, and miR-761 (De Martino et al., 2016). De Martino and Palma (2017) also showed that HMGA1P7 upregulated miR-483 and miR-675 through a ceRNA mechanism with Egr1.
OCT4-pg4 and OCT4-pg5
Two OCT4-associated pseudogene-derived lncRNAs, OCT4-pg4 and OCT4-pg5, have been reported to act as ceRNAs, thus involving in cancer initiation and progression (Wang et al., 2013; Bai et al., 2015). OCT4-pg4 promoted growth and tumorigenicity of HCC by growing OCT4 expression through competing for miR-145 (Wang et al., 2013). OCT4-pg5 also upregulated OCT4 by sponging miR-145 and thus facilitated cell proliferation of endometrial carcinoma (Bai et al., 2015).
PTTG3P
Pseudogene PTTG3P was found to be closely related with poor prognosis in HCC and breast cancer. Lou et al. (2019) suggested that PTTG3P expression positively associated with PTTG1 expression and may function by sponging miR-129-5p, miR-383-5p, and miR-376c-3p. Recently, Zhou Q. et al. (2019) reported that downregulation of PTTG3P promoted apoptosis and decreased proliferation, invasion and migration of HCC cells via increasing the expression levels of miR-383 targets, PARP2 and CCND1.
TUSC2P
Liu et al. (2018) indicated that TUSC2P could suppress proliferation, invasion and accelerate apoptosis of esophageal squamous cell carcinoma in vivo and in vitro through modulating expression of miR-17-5p, miR-520a-3p, miR-608, and miR-661. Besides, it also could serve as a prognostic factor for patients with esophageal squamous cell carcinoma (Liu et al., 2018). In addition, TUSC2P was founded to act as a key tumor-inhibitor that could inhibits cell proliferation, migration and invasion in 4T1 and MDA-MB-231 cells (Rutnam et al., 2014). What’s more, TUSC2P promotes those functions by binding miR-17, miR-93, miR-299-3p, miR-520a, miR-608, and miR-661 according to the research conducted by Rutnam et al. (2014).
ASS1P3
ASS1P3, a pseudogene of ASS1, promoted cell proliferation by functioning as a miRNA decoy for miR-34a-5p in renal cell carcinoma reported by the team of Wang K. et al. (2019).
CHIAP2
Pseudogene-derived lncRNA, CHIAP2, suppressed lung adenocarcinoma cell proliferation and invasion by modulation of miR-3614-5p and miR-873-3p-mediated inhibition of NFATC2 and GSK3B (Shang et al., 2019).
CYP2A7P
CYP2A7P was discovered to affect expression of its cognate gene CYP2A6 by functioning as a decoy for miR-126 in human liver (Nakano et al., 2015). However, the effect of CYP2A7P/miR-126/CYP2A6 axis in human cancer containing liver cancer is still not determined.
FOXO3P
FOXO3P inhibited tumor growth and angiogenesis by activating FOXO3 activity (Yang W. et al., 2016). By luciferase reporter assay, the authors confirmed that FOXO3P exerted its roles through sponging several miRNAs, including miR-22, miR-136∗, miR-138, miR-149∗, miR-433, miR-762, miR-3614-5p, and miR-3622b-5p (Yang W. et al., 2016).
GBAP1
GBA encodes lysosomal glucocerebrosidase and its mutations are associated with Parkinson’s disease. Recently, Tang et al. (2017b) demonstrated that GBAP1, a pseudogene of GBA, acted as a ceRNA for GBA by competing with miR-22-3p.
INTS6P1
INTS6P1, a tumor suppressive pseudogene-derived lncRNA, exerted its roles in HCC by competitively binding to oncogenic miR-17-5p and thus upregulating its cognate gene INTS6 (Peng et al., 2015). Lui et al. (2017) suggested that INTS6 suppressed growth of HCC through Wnt pathway. Taken together, INTS6P1-miR-17-5p-INTS6-Wnt signaling pathway may be a potential therapeutic target in treating HCC.
MSTO2P1
MSTO2P1 functioned as an oncogenic molecule, including enhanced growth, colony formation, migration and invasion in cervical cancer as mentioned above (Li R.K. et al., 2017). The mechanistic investigation revealed that MSTO2P1 exhibited these effects by regulating expression of miR-335 which has been well documented as a potential suppressor of gastric cancer progression (Sandoval-Borquez et al., 2017).
PDIA3P1
hMTR4, which promotes RNA degradation, could bind to PDIA3P1, and this interaction was disrupted by Dox treatment (Xie et al., 2019). miR-125a/b and miR-124 directly targeted TRAF6, however, PDIA3P1 bound to miR-125a/b and miR-124 and relieved their suppression on TRAF6, thereby causing activation of NF-kB pathway (Xie et al., 2019). The novel hMTR4-PDIA3P1-miR-125/miR-124-TRAF6 axis may play a key role in chemoresistance of HCC.
RACGAP1P
Wang M.Y. et al. (2019) lately confirmed that pseudogene RACGAP1 is an indispensable molecule for the malignant progression of HCC cells, which was dependent on RACGAP1/Rho/ERK signaling axis.
RP11-424C20.2
UHRF1 is a parental gene of pseudogene RP11-424C20.2, it is significantly related with immune infiltration (Yang J. et al., 2019). RP11-424C20.2, a ceRNA, could upregulate UHRF1 expression via sponging miR-378a-3p. This axis regulated immune escape of THYM and LIHC by PD-L1 and IFN-gamma-mediated CLTA-4 pathway.
RP11-564D11.3
According to bioinformatic analysis, Song et al. (2019) found that high expression of RP11-564D11.3 was obviously associated with poor overall survival of multitype cancer patients, such as HCC, mesothelioma, kidney diseases, lung adenocarcinoma, paraganglioma, and pheochromocytoma. Furthermore, seven miRNAs binding with RP11-564D11.3 were predicted, including miR-200b-3p, miR-200a-3p, miR-429, miR-101-3p, miR-9-5p, miR-200c-3p, and miR-141-3p.
RSU1P2
Pseudogene-derived LncRNA, RSU1P2, was founded to be a tumor-promoting molecule in cervical cancer by against let-7a, next regulated N-myc, IGF1R and EphA4 expression (Liu et al., 2017c).
SEPW1P
A recent study reported that piRNA-36712 restrained proliferation, invasion, migration and paclitaxel and doxorubicin resistance of breast cancer, while SEPW1P decreased piRNA-36712 expression by regulating miRNA-7 and miRNA-324, then inhibited P53, p21 and E-cadherin, but upregulated Slug (Tan et al., 2019).
SUMO1P3
Recent research found that the expression level of SUMO1P3 was significantly upregulated in non-small cell lung cancer cell lines and cancer tissues. Furthermore, SUMO1P3 promotes NSCLC cells invasion and migration by binding and regulating miR-136 (Zhang et al., 2019).
WTAPP1
WTAPP1, a molecular decoy for miR-3120-5p, promoted cell migration, invasion and tube formation both in vitro and in vivo by increasing MMP1 expression and activating PI3K/Akt/mTOR signaling (Li et al., 2018).
Potential Dysregulated Mechanisms of Pseudogene-Derived lncRNAs in Human Cancer
As we mentioned above, a variety of pseudogene-derived lncRNAs have been reported to be frequently dysregulated in various types of human cancer, and a lot attention has been paid to explore the function and action mechanisms of these identified pseudogene-derived lncRNAs in cancer. Only few potential dysregulated mechanisms of pseudogene-derived lncRNA have been reported (Figure 2). Several transcriptional factors are discovered to link to the mis-regulation of pseudogene-derived lncRNAs. For instance, Du et al. (2019) revealed that DUXAP8 upregulation in colorectal cancer was induced by STAT3; SIX2 played a positive role in CYP4Z2P expression in breast cancer (Zheng L. et al., 2019). Some studies demonstrated that the methylation level of pseudogenes linked to the alteration of pseudogene-derived lncRNAs’ expression (Kovalenko et al., 2013, 2018). Hormone and hormone receptor correlated with mis-regulation of pseudogene-derived lncRNAs. For example, Lu et al. (2006) suggested that KLK31P was stimulated by androgen in prostate cancer; the team of Wang K. et al. (2019) found that androgen receptor negatively modulated ASS1P3 expression in renal cell carcinoma. Emerging evidence showed that RNA stability-associated proteins participated in modulation of expression of pseudogene-derived lncRNAs. Human homolog of mRNA transport mutant 4 (hMTR4) led to PDIA3P1 degradation and caused its expression downregulation in HCC (Xie et al., 2019). A recent study confirmed that baculovirus mediated expression of PTENP1 in HCC (Chen et al., 2015). Besides, exosome also linked to the aberrant expression of pseudogene-derived lncRNA in cancer. Zheng et al. (2018) revealed that PTENP1, transmitted by exosome, significantly suppressed progression of bladder cancer. However, investigations into the mechanisms of pseudogene-derived lncRNAs’ upregulation or downregulation remain inadequate. More possible mechanisms causing deregulation of pseudogene-derived lncRNA in cancer need to be explored in the future.
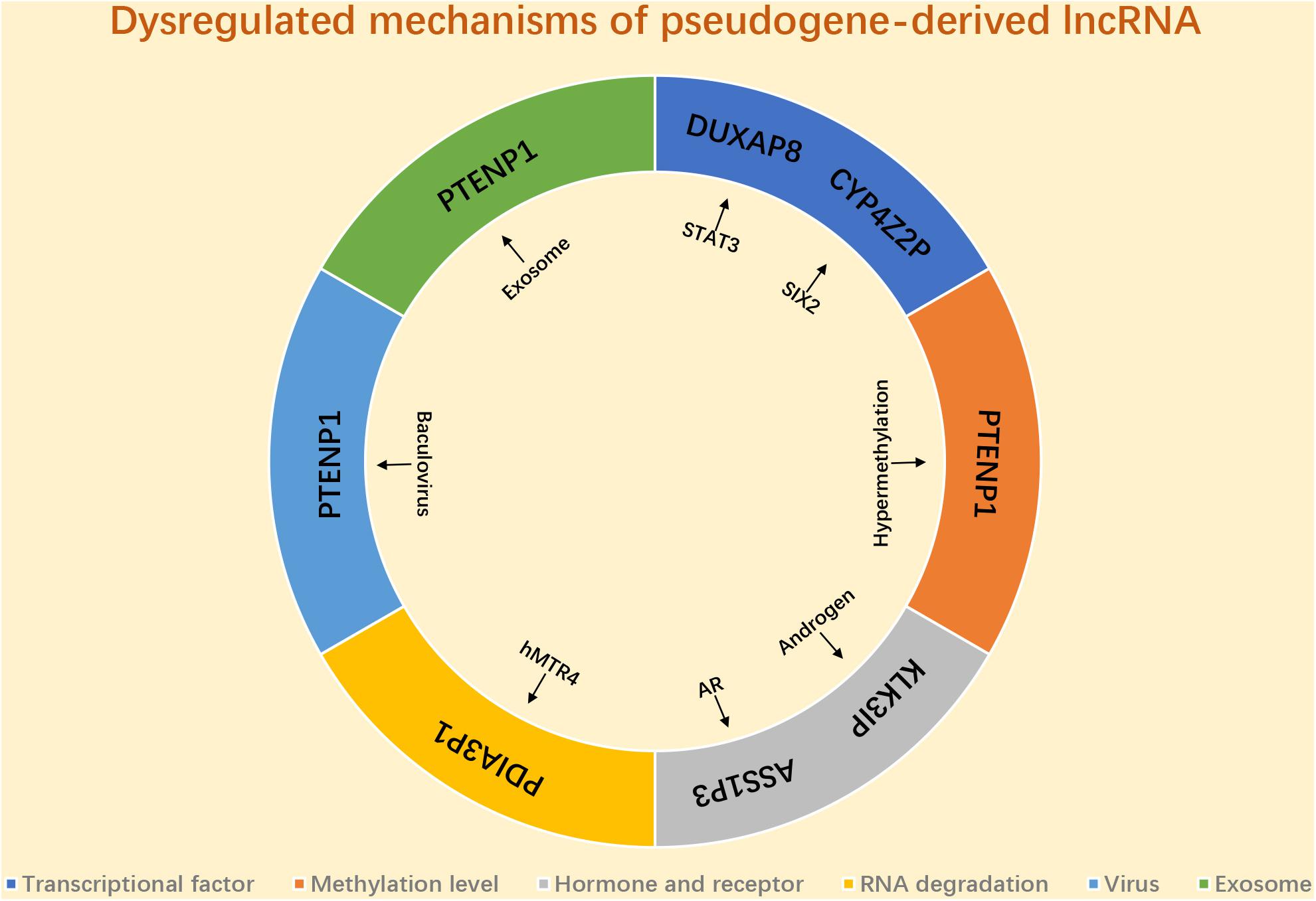
Figure 2. The potential mechanism responsible for dysregulation of pseudogene-derived lncRNA in cancer.
Conclusion and Future Perspectives
Pseudogenes are regarded as non-functional “Genomic evolutional junks” for a long time. During the past few years, researchers have reported that a variety of pseudogenes possess transcribed activities, and some of pseudogene-derived RNA transcripts are validated to play as key regulators in diverse biological processes (Wen et al., 2012). Expression and/or function dysregulation of these RNA transcripts account for the occurrence of multiple human disorders, containing cancer (Pink et al., 2011; Poliseno, 2012). Therefore, performing an all-round overview regarding the expression, function and molecular mechanisms of pseudogene-derived lncRNAs in human cancers may be helpful for developing effective anti-cancer measures.
Benefiting from the advancement of high-throughput sequencing technology and development of bioinformatics analysis, a growing number of pseudogene-derived lncRNAs have been identified, annotated and functionally predicted (Kalyana-Sundaram et al., 2012; Han et al., 2014). As reported, pseudogene-derived lncRNAs are frequently mis-regulated in diverse diseases. But, to date, the role of majority of pseudogene-derived lncRNAs remains unknown. Low expression abundance and high tissue specificity may hamper the interpretation of their functions. The beginning of functional studies for most of pseudogene-derived lncRNAs is usually the cognate genes of pseudogenes. The high homology of pseudogene-derived lncRNAs and transcripts from their cognate genes may be another drag force to impede investigation progression of pseudogene-derived lncRNAs’ functions. Of note, the influence of pseudogene-expressed transcripts needs to be excluded when researchers are performing the functional investigations for transcripts from cognate genes.
ceRNA hypothesis, proposed by Salmena et al. (2018) presents a new language among messenger RNAs, lncRNAs and circRNAs by using miRNA response elements. Pseudogene-derived lncRNA transcripts make up a part of lncRNAs. To the best of our knowledge, no studies regarding pseudogene-derived circRNAs have been reported although Dong et al. (2016) proposed that circRNAs can be retrotranscribed and finally inserted back into the host genome as processed pseudogenes. With the deepening of research, pseudogene-derived circRNA transcripts may be gradually discovered in the future.
To date, most of studies regarding molecular action mechanisms of pseudogene-derived lncRNAs mainly focused on the ceRNA hypothesis by competing shared miRNAs with cognate genes or non-cognate genes (Wei Y. et al., 2017; Wu et al., 2018). As a subtype of lncRNA, pseudogene-derived lncRNA may also exert its regulatory effects in cancer by other mechanisms, such as binding to transcription factors, producing miRNA/piRNA and encoding proteins (Bhan et al., 2017; Renganathan and Felley-Bosco, 2017). However, these fields are still inadequate. Moreover, most of individual transcripts can’t eventually compromise miRNA activity (Thomson and Dinger, 2016). In the future, more corresponding work should be performed. Of course, the current findings about roles and mechanisms of known pseudogene-derived lncRNAs need to be further precisely validated by basic lab experiments and large clinical trials. Despite all this, finding novel pseudogene-derived lncRNAs as well as circRNAs, identifying their roles in different cancer types and subtypes and developing these pseudogene-derived transcripts-based strategies for diagnosis, therapy and prognosis exhibit very promising in overcoming cancer.
Author Contributions
WL and PF designed this work. WL collected the reference and drafted the manuscript. BD polished the language. PF critically revised the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ceRNA, competing endogenous RNA; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; lncRNA, long non-coding RNA; miRNA, microRNA; MRE, miRNA response element; ncRNA, non-coding RNA.
References
Asselta, R., Yndestad, S., Austreid, E., Skaftnesmo, K. O., Lonning, P. E., and Eikesdal, H. P. (2018). Divergent activity of the pseudogene PTENP1 in ER-positive and negative breast cancer. Mol. Cancer Res. 16, 78–89. doi: 10.1158/1541-7786.MCR-17-0207
Azzariti, A., Arra, C., Fusco, A., Esposito, F., and Zheng, X. (2017). Long non-coding RNA FTH1P3 facilitates uveal melanoma cell growth and invasion through miR-224-5p. PLoS One 12:e0184746. doi: 10.1371/journal.pone.0184746
Bai, M., Yuan, M., Liao, H., Chen, J., Xie, B., Yan, D., et al. (2015). OCT4 pseudogene 5 upregulates OCT4 expression to promote proliferation by competing with miR-145 in endometrial carcinoma. Oncol. Rep. 33, 1745–1752. doi: 10.3892/or.2015.3763
Barrow, M. A., Martin, M. E., Coffey, A., Andrews, P. L., Jones, G. S., Reaves, D. K., et al. (2019). A functional role for the cancer disparity-linked genes, CRYbetaB2 and CRYbetaB2P1, in the promotion of breast cancer. Breast Cancer Res. 21:105. doi: 10.1186/s13058-019-1191-3
Bhan, A., Soleimani, M., and Mandal, S. S. (2017). Long noncoding RNA and cancer: a new paradigm. Cancer Res. 77, 3965–3981. doi: 10.1158/0008-5472.CAN-16-2634
Bier, A., Oviedo-Landaverde, I., Zhao, J., Mamane, Y., Kandouz, M., and Batist, G. (2009). Connexin43 pseudogene in breast cancer cells offers a novel therapeutic target. Mol. Cancer Ther. 8, 786–793. doi: 10.1158/1535-7163.MCT-08-0930
Cao, S. W., Huang, J. L., Chen, J., Hu, Y. W., Hu, X. M., Ren, T. Y., et al. (2017). Long non-coding RNA UBE2CP3 promotes tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget 8, 65370–65385. doi: 10.18632/oncotarget.18524
Chan, J. J., Kwok, Z. H., Chew, X. H., Zhang, B., Liu, C., Soong, T. W., et al. (2018). A FTH1 gene:pseudogene:microRNA network regulates tumorigenesis in prostate cancer. Nucleic Acids Res. 46, 1998–2011. doi: 10.1093/nar/gkx1248
Chan, W. L., Yuo, C. Y., Yang, W. K., Hung, S. Y., Chang, Y. S., Chiu, C. C., et al. (2013). Transcribed pseudogene ψPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma. Nucleic Acids Res. 41, 3734–3747. doi: 10.1093/nar/gkt047
Chen, C. L., Tseng, Y. W., Wu, J. C., Chen, G. Y., Lin, K. C., Hwang, S. M., et al. (2015). Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials 44, 71–81. doi: 10.1016/j.biomaterials.2014.12.023
Chen, J., Lou, W., Ding, B., and Wang, X. (2019). Overexpressed pseudogenes, DUXAP8 and DUXAP9, promote growth of renal cell carcinoma and serve as unfavorable prognostic biomarkers. Oncogene 11, 5666–5688. doi: 10.18632/aging.102152
Chen, X., Zhu, H., Wu, X., Xie, X., Huang, G., Xu, X., et al. (2016). Downregulated pseudogene CTNNAP1 promote tumor growth in human cancer by downregulating its cognate gene CTNNA1 expression. Oncotarget 7, 55518–55528. doi: 10.18632/oncotarget.10833
Dai, X., Xie, Y., Dong, M., Zhao, J., Yu, H., Zhou, B., et al. (2019). The long noncoding RNA TPTE2P1 promotes the viability of colorectal cancer cells. J. Cell. Biochem. 120, 5268–5276. doi: 10.1002/jcb.27801
De Martino, M., Forzati, F., Marfella, M., Pellecchia, S., Arra, C., Terracciano, L., et al. (2016). HMGA1P7-pseudogene regulates H19 and Igf2 expression by a competitive endogenous RNA mechanism. Sci. Rep. 6:37622. doi: 10.1038/srep37622
De Martino, M., and Palma, G. (2017). The HMGA1 pseudogene 7 induces miR-483 and miR-675 upregulation by activating Egr1 through a ceRNA mechanism. Genes 8:330. doi: 10.3390/genes8110330
Dong, R., Zhang, X. O., Zhang, Y., Ma, X. K., Chen, L. L., and Yang, L. (2016). CircRNA-derived pseudogenes. Cell Res. 26, 747–750. doi: 10.1038/cr.2016.42
Du, C., Wang, H. X., Chen, P., and Chen, C. H. (2019). STAT3-induced upregulation of lncRNA DUXAP8 functions as ceRNA for miR-577 to promote the migration and invasion in colorectal cancer through the regulation of RAB14. Eur. Rev. Med. Pharmacol. Sci. 23, 6105–6118. doi: 10.26355/eurrev_201907_18424
Esposito, F., De Martino, M., D’Angelo, D., Mussnich, P., Raverot, G., Jaffrain-Rea, M. L., et al. (2015). HMGA1-pseudogene expression is induced in human pituitary tumors. Cell Cycle 14, 1471–1475. doi: 10.1080/15384101.2015.1021520
Esposito, F., De Martino, M., Petti, M. G., Forzati, F., Tornincasa, M., Federico, A., et al. (2014). HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget 5, 8341–8354.
Feng, F., Qiu, B., Zang, R., Song, P., and Gao, S. (2017). Pseudogene PHBP1 promotes esophageal squamous cell carcinoma proliferation by increasing its cognate gene PHB expression. Oncotarget 8, 29091–29100. doi: 10.18632/oncotarget.16196
Feng, J., Yang, G., Liu, Y., Gao, Y., Zhao, M., Bu, Y., et al. (2019). LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics 9, 5227–5245. doi: 10.7150/thno.34273
Gao, L., Ren, W., Zhang, L., Li, S., Kong, X., Zhang, H., et al. (2017). PTENp1, a natural sponge of miR-21, mediates PTEN expression to inhibit the proliferation of oral squamous cell carcinoma. Mol. Carcinog. 56, 1322–1334. doi: 10.1002/mc.22594
Gao, X., Qin, T., Mao, J., Zhang, J., Fan, S., Lu, Y., et al. (2019). PTENP1/miR-20a/PTEN axis contributes to breast cancer progression by regulating PTEN via PI3K/AKT pathway. J. Exp. Clin. Cancer Res. 38:256. doi: 10.1186/s13046-019-1260-6
Gong, T., Zheng, S., Huang, S., Fu, S., Zhang, X., Pan, S., et al. (2017). PTENP1 inhibits the growth of esophageal squamous cell carcinoma by regulating SOCS6 expression and correlates with disease prognosis. Mol. Carcinog. 56, 2610–2619. doi: 10.1002/mc.22705
Grander, D., and Johnsson, P. (2016). Pseudogene-expressed RNAs: emerging roles in gene regulation and disease. Curr. Top. Microbiol. Immunol. 394, 111–126. doi: 10.1007/82_2015_442
Guo, X., Deng, L., Deng, K., Wang, H., Shan, T., Zhou, H., et al. (2016). Pseudogene PTENP1 suppresses gastric cancer progression by modulating PTEN. Anticancer Agents Med. Chem. 16, 456–464. doi: 10.2174/1871520615666150507121407
Guo, X. C., Li, L., Gao, Z. H., Zhou, H. W., Li, J., and Wang, Q. Q. (2019). The long non-coding RNA PTTG3P promotes growth and metastasis of cervical cancer through PTTG1. Aging 11, 1333–1341. doi: 10.18632/aging.101830
Han, L., Yuan, Y., Zheng, S., Yang, Y., Li, J., Edgerton, M. E., et al. (2014). The pan-cancer analysis of pseudogene expression reveals biologically and clinically relevant tumour subtypes. Nat. Commun. 5:3963. doi: 10.1038/ncomms4963
Hayashi, H., Arao, T., Togashi, Y., Kato, H., Fujita, Y., De Velasco, M. A., et al. (2015). The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene 34, 199–208. doi: 10.1038/onc.2013.547
Hu, S., Xu, L., Li, L., Luo, D., Zhao, H., Li, D., et al. (2019). Overexpression of lncRNA PTENP1 suppresses glioma cell proliferation and metastasis in vitro. Onco Targets Ther. 12, 147–156. doi: 10.2147/OTT.S182537
Huang, J. L., Cao, S. W., Ou, Q. S., Yang, B., Zheng, S. H., Tang, J., et al. (2018). The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol. Cancer 17:93. doi: 10.1186/s12943-018-0841-x
Huang, T., Wang, X., Yang, X., Ji, J., Wang, Q., Yue, X., et al. (2018). Long non-coding RNA DUXAP8 enhances renal cell carcinoma progression via downregulating miR-126. Med. Sci. Monit. 24, 7340–7347. doi: 10.12659/MSM.910054
Huang, W., Li, N., Hu, J., and Wang, L. (2016). Inhibitory effect of RNA-mediated knockdown of zinc finger protein 91 pseudogene on pancreatic cancer cell growth and invasion. Oncol. Lett. 12, 1343–1348. doi: 10.3892/ol.2016.4794
Jacq, C., Miller, J. R., and Brownlee, G. G. (1977). A pseudogene structure in 5S DNA of Xenopus laevis. Cell 12, 109–120. doi: 10.1016/0092-8674(77)90189-1
Jiang, T., Guo, J., Hu, Z., Zhao, M., Gu, Z., and Miao, S. (2018). Identification of potential prostate cancer-related pseudogenes based on competitive endogenous RNA network hypothesis. Med. Sci. Monit. 24, 4213–4239. doi: 10.12659/MSM.910886
Johnson, G. S., Li, J., Beaver, L. M., Dashwood, W. M., Sun, D., Rajendran, P., et al. (2017). A functional pseudogene, NMRAL2P, is regulated by Nrf2 and serves as a coactivator of NQO1 in sulforaphane-treated colon cancer cells. Mol. Nutr. Food Res. 61:1600769. doi: 10.1002/mnfr.201600769
Kalyana-Sundaram, S., Kumar-Sinha, C., Shankar, S., Robinson, D. R., Wu, Y. M., Cao, X., et al. (2012). Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 149, 1622–1634. doi: 10.1016/j.cell.2012.04.041
Karreth, F. A., Reschke, M., Ruocco, A., Ng, C., Chapuy, B., Leopold, V., et al. (2015). The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 161, 319–332. doi: 10.1016/j.cell.2015.02.043
Kong, Y., Zhang, L., Huang, Y., He, T., Zhang, L., Zhao, X., et al. (2017). Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 407, 76–83. doi: 10.1016/j.canlet.2017.07.031
Korneev, S. A., Park, J. H., and O’Shea, M. (1999). Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 19, 7711–7720. doi: 10.1523/jneurosci.19-18-07711.1999
Kovalenko, T. F., Morozova, K. V., Ozolinya, L. A., Lapina, I. A., and Patrushev, L. I. (2018). The PTENP1 pseudogene, unlike the PTEN gene, is methylated in normal endometrium, as well as in endometrial hyperplasias and carcinomas in middle-aged and elderly females. Acta Naturae 10, 43–50.
Kovalenko, T. F., Sorokina, A. V., Ozolinia, L. A., and Patrushev, L. I. (2013). [Pseudogene PTENP1 5’-region methylation in endometrial cancer and hyperplasias]. Bioorg. Khim. 39, 445–453.
Lai, Y., Li, J., Zhong, L., He, X., Si, X., Sun, Y., et al. (2019). The pseudogene PTENP1 regulates smooth muscle cells as a competing endogenous RNA. Clin. Sci. 133, 1439–1455. doi: 10.1042/CS20190156
Li, C., Zheng, L., Xin, Y., Tan, Z., Zhang, Y., Meng, X., et al. (2017). The competing endogenous RNA network of CYP4Z1 and pseudogene CYP4Z2P exerts an anti-apoptotic function in breast cancer. FEBS Lett. 591, 991–1000. doi: 10.1002/1873-3468.12608
Li, H., Zhu, H., Zhou, Y., Wang, H., Niu, Z., Shen, Y., et al. (2017). Long non-coding RNA MSTO2P promotes the proliferation and colony formation in gastric cancer by indirectly regulating miR-335 expression. Tumour Biol. 39:1010428317705506. doi: 10.1177/1010428317705506
Li, L., Feng, R., Fei, S., Cao, J., Zhu, Q., Ji, G., et al. (2019). NANOGP8 expression regulates gastric cancer cell progression by transactivating DBC1 in gastric cancer MKN-45 cells. Oncol. Lett. 17, 555–563. doi: 10.3892/ol.2018.9595
Li, L., Yin, J. Y., He, F. Z., Huang, M. S., Zhu, T., Gao, Y. F., et al. (2017). Long noncoding RNA SFTA1P promoted apoptosis and increased cisplatin chemosensitivity via regulating the hnRNP-U-GADD45A axis in lung squamous cell carcinoma. Oncotarget 8, 97476–97489. doi: 10.18632/oncotarget.22138
Li, R. K., Gao, J., Guo, L. H., Huang, G. Q., and Luo, W. H. (2017). PTENP1 acts as a ceRNA to regulate PTEN by sponging miR-19b and explores the biological role of PTENP1 in breast cancer. Cancer Gene Ther. 24, 309–315. doi: 10.1038/cgt.2017.29
Li, W. D., Zhou, D. M., Sun, L. L., Xiao, L., Liu, Z., Zhou, M., et al. (2018). LncRNA WTAPP1 promotes migration and angiogenesis of endothelial progenitor cells via MMP1 through MicroRNA 3120 and Akt/PI3K/autophagy pathways. Stem Cells 36, 1863–1874. doi: 10.1002/stem.2904
Lian, Y., Xiao, C., Yan, C., Chen, D., Huang, Q., Fan, Y., et al. (2018a). Knockdown of pseudogene derived from lncRNA DUXAP10 inhibits cell proliferation, migration, invasion, and promotes apoptosis in pancreatic cancer. J. Cell. Biochem. 119, 3671–3682. doi: 10.1002/jcb.26578
Lian, Y., Yang, J., Lian, Y., Xiao, C., Hu, X., and Xu, H. (2018b). DUXAP8, a pseudogene derived lncRNA, promotes growth of pancreatic carcinoma cells by epigenetically silencing CDKN1A and KLF2. Cancer Commun. 38:64. doi: 10.1186/s40880-018-0333-9
Lian, Y., Xu, Y., Xiao, C., Xia, R., Gong, H., Yang, P., et al. (2017). The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN. Sci. Rep. 7:7312. doi: 10.1038/s41598-017-07954-7
Lin, J., Cao, S., Wang, Y., Hu, Y., Liu, H., Li, J., et al. (2018). Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1alpha/VEGFA signalling in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 37:113. doi: 10.1186/s13046-018-0727-1
Liu, F., Gong, R., He, B., Chen, F., and Hu, Z. (2018). TUSC2P suppresses the tumor function of esophageal squamous cell carcinoma by regulating TUSC2 expression and correlates with disease prognosis. BMC Cancer 18:894. doi: 10.1186/s12885-018-4804-9
Liu, J., Song, Z., Feng, C., Lu, Y., Zhou, Y., Lin, Y., et al. (2017a). The long non-coding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR-320a. Am. J. Transl. Res. 9, 5594–5602.
Liu, J., Xing, Y., Xu, L., Chen, W., Cao, W., and Zhang, C. (2017b). Decreased expression of pseudogene PTENP1 promotes malignant behaviours and is associated with the poor survival of patients with HNSCC. Sci. Rep. 7:41179. doi: 10.1038/srep41179
Liu, Q., Guo, X., Que, S., Yang, X., Fan, H., Liu, M., et al. (2017c). LncRNA RSU1P2 contributes to tumorigenesis by acting as a ceRNA against let-7a in cervical cancer cells. Oncotarget 8, 43768–43781. doi: 10.18632/oncotarget.10844
Lou, W., Ding, B., and Fan, W. (2019). High expression of pseudogene PTTG3P indicates a poor prognosis in human breast cancer. Mol. Ther. Oncolytics 14, 15–26. doi: 10.1016/j.omto.2019.03.006
Lu, W., Zhou, D., Glusman, G., Utleg, A. G., White, J. T., Nelson, P. S., et al. (2006). KLK31P is a novel androgen regulated and transcribed pseudogene of kallikreins that is expressed at lower levels in prostate cancer cells than in normal prostate cells. Prostate 66, 936–944. doi: 10.1002/pros.20382
Lui, K. Y., Zhao, H., Qiu, C., Li, C., Zhang, Z., Peng, H., et al. (2017). Integrator complex subunit 6 (INTS6) inhibits hepatocellular carcinoma growth by Wnt pathway and serve as a prognostic marker. BMC Cancer 17:644. doi: 10.1186/s12885-017-3628-3
Lv, W., Wang, L., Lu, J., Mu, J., Liu, Y., and Dong, P. (2015). Downregulation of TPTE2P1 inhibits migration and invasion of gallbladder cancer cells. Chem. Biol. Drug Des. 86, 656–662. doi: 10.1111/cbdd.12533
Lv, Y., Chen, S., Wu, J., Lin, R., Zhou, L., Chen, G., et al. (2019). Upregulation of long non-coding RNA OGFRP1 facilitates endometrial cancer by regulating miR-124-3p/SIRT1 axis and by activating PI3K/AKT/GSK-3beta pathway. Artif. Cells Nanomed. Biotechnol. 47, 2083–2090. doi: 10.1080/21691401.2019.1617727
Ma, H., Ma, T., Chen, M., Zou, Z., and Zhang, Z. (2018). The pseudogene-derived long non-coding RNA SFTA1P suppresses cell proliferation, migration, and invasion in gastric cancer. Biosci. Rep. 38:BSR20171193. doi: 10.1042/BSR20171193
Ma, H. W., Xie, M., Sun, M., Chen, T. Y., Jin, R. R., Ma, T. S., et al. (2017). The pseudogene derived long noncoding RNA DUXAP8 promotes gastric cancer cell proliferation and migration via epigenetically silencing PLEKHO1 expression. Oncotarget 8, 52211–52224. doi: 10.18632/oncotarget.11075
Ma, X., Wang, B., Wang, X., Luo, Y., and Fan, W. (2018). NANOGP8 is the key regulator of stemness, EMT, Wnt pathway, chemoresistance, and other malignant phenotypes in gastric cancer cells. PLoS One 13:e0192436. doi: 10.1371/journal.pone.0192436
Masutomi, K., Kaneko, S., and Kobayashi, K. (2001). [Tumor suppressor gene, p53, and hepatocarcinogenesis]. Nihon Rinsho 59(Suppl. 6), 138–141.
Mei, D., Song, H., Wang, K., Lou, Y., Sun, W., Liu, Z., et al. (2013). Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med. Oncol. 30:709. doi: 10.1007/s12032-013-0709-2
Nakano, M., Fukushima, Y., Yokota, S., Fukami, T., Takamiya, M., Aoki, Y., et al. (2015). CYP2A7 pseudogene transcript affects CYP2A6 expression in human liver by acting as a decoy for miR-126. Drug Metab. Dispos. 43, 703–712. doi: 10.1124/dmd.115.063255
Ni, S., Wang, Y., Xu, M., Zhang, Q., Yang, Y., Wu, Y., et al. (2017). Long non-coding RNA MSTO2P promotes the proliferation and colony formation in gastric cancer by indirectly regulating miR-335 expression. Tumour Biol. 39:1010428317705506.
Oliveira-Mateos, C., Sanchez-Castillo, A., and Soler, M. (2019). The transcribed pseudogene RPSAP52 enhances the oncofetal HMGA2-IGF2BP2-RAS axis through LIN28B-dependent and independent let-7 inhibition. Nat. Commun. 10:3979. doi: 10.1038/s41467-019-11910-6
Ou, R., Lv, J., Zhang, Q., Lin, F., Zhu, L., Huang, F., et al. (2019). circAMOTL1 motivates AMOTL1 expression to facilitate cervical cancer growth. Mol. Ther. Nucleic Acids 19, 50–60. doi: 10.1016/j.omtn.2019.09.022
Pan, W., Li, W., Zhao, J., Huang, Z., Zhao, J., Chen, S., et al. (2019). lncRNA-PDPK2P promotes hepatocellular carcinoma progression through the PDK1/AKT/Caspase 3 pathway. Mol. Oncol. 13, 2246–2258. doi: 10.1002/1878-0261.12553
Panagopoulos, I., Moller, E., Collin, A., and Mertens, F. (2008). The POU5F1P1 pseudogene encodes a putative protein similar to POU5F1 isoform 1. Oncol. Rep. 20, 1029–1033.
Peng, H., Ishida, M., Li, L., Saito, A., Kamiya, A., Hamilton, J. P., et al. (2015). Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget 6, 5666–5677.
Pink, R. C., Wicks, K., Caley, D. P., Punch, E. K., Jacobs, L., and Carter, D. R. (2011). Pseudogenes: pseudo-functional or key regulators in health and disease? RNA 17, 792–798. doi: 10.1261/rna.2658311
Poliseno, L. (2012). Pseudogenes: newly discovered players in human cancer. Sci. Signal. 5:re5. doi: 10.1126/scisignal.2002858
Qian, Y. Y., Li, K., Liu, Q. Y., and Liu, Z. S. (2017). Long non-coding RNA PTENP1 interacts with miR-193a-3p to suppress cell migration and invasion through the PTEN pathway in hepatocellular carcinoma. Oncotarget 8, 107859–107869. doi: 10.18632/oncotarget.22305
Qu, J., Li, M., Zhong, W., and Hu, C. (2015). Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int. J. Clin. Exp. Med. 8, 17110–17116.
Renganathan, A., and Felley-Bosco, E. (2017). Long noncoding RNAs in cancer and therapeutic potential. Adv. Exp. Med. Biol. 1008, 199–222. doi: 10.1007/978-981-10-5203-3_7
Rutnam, Z. J., Du, W. W., Yang, W., Yang, X., and Yang, B. B. (2014). The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat. Commun. 5:2914. doi: 10.1038/ncomms3914
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Sandoval-Borquez, A., Polakovicova, I., Carrasco-Veliz, N., Lobos-Gonzalez, L., Riquelme, I., Carrasco-Avino, G., et al. (2017). MicroRNA-335-5p is a potential suppressor of metastasis and invasion in gastric cancer. Clin. Epigenetics 9:114. doi: 10.1186/s13148-017-0413-8
Shang, J., Wang, Z., Chen, W., Yang, Z., Zheng, L., Wang, S., et al. (2019). Pseudogene CHIAP2 inhibits proliferation and invasion of lung adenocarcinoma cells by means of the WNT pathway. J. Cell. Physiol. 234, 13735–13746. doi: 10.1002/jcp.28053
Shi, X., Tang, X., and Su, L. (2018). Overexpression of long noncoding RNA PTENP1 inhibits cell proliferation and migration via suppression of miR-19b in breast cancer cells. Oncol. Res. 26, 869–878. doi: 10.3727/096504017X15123838050075
Song, H., Yang, J., Zhang, Y., Zhou, J., Li, Y., and Hao, X. (2019). Integrated analysis of pseudogene RP11-564D11.3 expression and its potential roles in hepatocellular carcinoma. Epigenomics 11, 267–280. doi: 10.2217/epi-2018-0152
Sun, M., Nie, F. Q., Zang, C., Wang, Y., Hou, J., Wei, C., et al. (2017). The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol. Ther. 25, 739–751. doi: 10.1016/j.ymthe.2016.12.018
Tan, L., Mai, D., Zhang, B., Jiang, X., Zhang, J., Bai, R., et al. (2019). PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer 18:9. doi: 10.1186/s12943-019-0940-3
Tang, H., Zhao, X., Sun, Y., Jiang, Y., Liu, Y., Chen, S., et al. (2017a). Long non-coding RNA PTENP1 inhibits proliferation and migration of breast cancer cells via AKT and MAPK signaling pathways. Oncol. Lett. 14, 4659–4662. doi: 10.3892/ol.2017.6823
Tang, H., Zhao, X., Sun, Y., Jiang, Y., Liu, Y., Straniero, L., et al. (2017b). The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p. Sci. Rep. 7:12702. doi: 10.1038/s41598-017-12973-5
Thomson, D. W., and Dinger, M. E. (2016). Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17, 272–283. doi: 10.1038/nrg.2016.20
Tian, C., Jin, Y., and Shi, S. (2018). Long non-coding RNA SUMO1P3 may promote cell proliferation, migration, and invasion of pancreatic cancer via EMT signaling pathway. Oncol. Lett. 16, 6109–6115. doi: 10.3892/ol.2018.9378
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Uchino, K., Hirano, G., Hirahashi, M., Isobe, T., Shirakawa, T., Kusaba, H., et al. (2012). Human Nanog pseudogene8 promotes the proliferation of gastrointestinal cancer cells. Exp. Cell Res. 318, 1799–1807. doi: 10.1016/j.yexcr.2012.04.011
Varesio, L. M., Willett, J. W., Fiebig, A., and Crosson, S. (2019). A carbonic anhydrase pseudogene sensitizes select Brucella lineages to low CO2 tension. J. Bacteriol. 201:e00509-19. doi: 10.1128/JB.00509-19
Wang, K., Sun, Y., Guo, C., Liu, T., Fei, X., and Chang, C. (2019). Androgen receptor regulates ASS1P3/miR-34a-5p/ASS1 signaling to promote renal cell carcinoma cell growth. Cell Death Dis. 10:339. doi: 10.1038/s41419-019-1330-x
Wang, L., Guo, Z. Y., Zhang, R., Xin, B., Chen, R., Zhao, J., et al. (2013). Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis 34, 1773–1781. doi: 10.1093/carcin/bgt139
Wang, L., Zhang, N., Wang, Z., Ai, D. M., Cao, Z. Y., and Pan, H. P. (2016). Pseudogene PTENP1 functions as a competing endogenous RNA (ceRNA) to regulate PTEN expression by sponging miR-499-5p. Biochemistry 81, 739–747. doi: 10.1134/S0006297916070105
Wang, M. Y., Chen, D. P., Qi, B., Li, M. Y., Zhu, Y. Y., Yin, W. J., et al. (2019). Pseudogene RACGAP1P activates RACGAP1/Rho/ERK signalling axis as a competing endogenous RNA to promote hepatocellular carcinoma early recurrence. Cell Death Dis. 10:426. doi: 10.1038/s41419-019-1666-2
Wang, Q. S., Shi, L. L., Sun, F., Zhang, Y. F., Chen, R. W., and Yang, S. L. (2019). High expression of ANXA2 pseudogene ANXA2P2 promotes an aggressive phenotype in hepatocellular carcinoma. Dis. Markers 2019:9267046. doi: 10.1155/2019/9267046
Wang, R., Zhang, T., Yang, Z., Jiang, C., and Seng, J. (2018). Long non-coding RNA FTH1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB1. J. Cell. Mol. Med. 22, 4068–4075. doi: 10.1111/jcmm.13679
Wang, T. H., Lin, Y. S., Chen, Y., Yeh, C. T., Huang, Y. L., Hsieh, T. H., et al. (2015). Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget 6, 23342–23357.
Wang, W., Lou, W., Ding, B., Yang, B., Lu, H., Kong, Q., et al. (2019). A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging 11, 2610–2627. doi: 10.18632/aging.101933
Wang, X., Liang, Q., Zhang, L., Gou, H., Li, Z., Chen, H., et al. (2019). C8orf76 promotes gastric tumorigenicity and metastasis by directly inducing lncRNA DUSP5P1 and associates with patient outcomes. Clin. Cancer Res. 25, 3128–3140. doi: 10.1158/1078-0432.CCR-18-2804
Wang, Z., Ren, B., Huang, J., Yin, R., Jiang, F., and Zhang, Q. (2018). LncRNA DUXAP10 modulates cell proliferation in esophageal squamous cell carcinoma through epigenetically silencing p21. Cancer Biol. Ther. 19, 998–1005. doi: 10.1080/15384047.2018.1470723
Watanabe, T., Totoki, Y., Toyoda, A., Kaneda, M., Kuramochi-Miyagawa, S., Obata, Y., et al. (2008). Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543. doi: 10.1038/nature06908
Wei, C. C., Nie, F. Q., Jiang, L. L., Chen, Q. N., Chen, Z. Y., Chen, X., et al. (2017). The pseudogene DUXAP10 promotes an aggressive phenotype through binding with LSD1 and repressing LATS2 and RRAD in non small cell lung cancer. Oncotarget 8, 5233–5246. doi: 10.18632/oncotarget.14125
Wei, Y., Chang, Z., Wu, C., Zhu, Y., Li, K., and Xu, Y. (2017). Identification of potential cancer-related pseudogenes in lung adenocarcinoma based on ceRNA hypothesis. Oncotarget 8, 59036–59047. doi: 10.18632/oncotarget.19933
Wen, Y. Z., Zheng, L. L., Qu, L. H., Ayala, F. J., and Lun, Z. R. (2012). Pseudogenes are not pseudo any more. RNA Biol. 9, 27–32. doi: 10.4161/rna.9.1.18277
Weng, W. (2017). PTTG3P promotes gastric tumour cell proliferation and invasion and is an indicator of poor prognosis. J. Cell. Mol. Med. 21, 3360–3371. doi: 10.1111/jcmm.13239
Wu, C., Wei, Y., Zhu, Y., Li, K., Zhu, Y., Zhao, Y., et al. (2018). Identification of cancer-related potential biomarkers based on lncRNA-pseudogene-mRNA competitive networks. FEBS Lett. 592, 973–986. doi: 10.1002/1873-3468.13011
Wu, X. C., Wang, S. H., Ou, H. H., Zhu, B., Zhu, Y., Zhang, Q., et al. (2017). The NmrA-like family domain containing 1 pseudogene Loc344887 is amplified in gallbladder cancer and promotes epithelial-mesenchymal transition. Chem. Biol. Drug Des. 90, 456–463. doi: 10.1111/cbdd.12967
Xiao-Jie, L., Ai-Mei, G., Li-Juan, J., and Jiang, X. (2015). Pseudogene in cancer: real functions and promising signature. J. Med. Genet. 52, 17–24. doi: 10.1136/jmedgenet-2014-102785
Xie, C., Zhang, L. Z., Chen, Z. L., Zhong, W. J., Fang, J. H., Zhu, Y., et al. (2019). A novel hMTR4-PDIA3P1-miR-125/124-TRAF6 regulatory axis and its function in NF-K B signaling and chemoresistance. Hepatology doi: 10.1002/hep.30931 [Epub ahead of print].
Xu, H., Chen, B., Xing, J., Wei, Z., Liu, C., Qiu, Y., et al. (2019). Upregulation of LGMNP1 confers radiotherapy resistance in glioblastoma. Oncol. Rep. 41, 3435–3443. doi: 10.3892/or.2019.7128
Xu, Y., Yu, X., Wei, C., Nie, F., Huang, M., and Sun, M. (2018). Over-expression of oncigenic pesudogene DUXAP10 promotes cell proliferation and invasion by regulating LATS1 and beta-catenin in gastric cancer. J. Exp. Clin. Cancer res. 37:13. doi: 10.1186/s13046-018-0684-8
Yang, C., Wang, L., Sun, J., Zhou, J. H., Tan, Y. L., Wang, Y. F., et al. (2019). Identification of long non-coding RNA HERC2P2 as a tumor suppressor in glioma. Carcinogenesis 40, 956–964. doi: 10.1093/carcin/bgz043
Yang, C., Wu, D., Gao, L., Liu, X., Jin, Y., Wang, D., et al. (2016). Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget 7, 13479–13490. doi: 10.18632/oncotarget.7266
Yang, J., Zhang, Y., and Song, H. (2019). A disparate role of RP11-424C20.2/UHRF1 axis through control of tumor immune escape in liver hepatocellular carcinoma and thymoma. Aging 11, 6422–6439. doi: 10.18632/aging.102197
Yang, L., Sun, K., Chu, J., Qu, Y., Zhao, X., Yin, H., et al. (2018). Long non-coding RNA FTH1P3 regulated metastasis and invasion of esophageal squamous cell carcinoma through SP1/NF-kB pathway. Biomed. Pharmacother. 106, 1570–1577. doi: 10.1016/j.biopha.2018.07.129
Yang, W., Du, W. W., Li, X., Yee, A. J., and Yang, B. B. (2016). Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 35, 3919–3931. doi: 10.1038/onc.2015.460
Yang, X., Ye, H., He, M., Zhou, X., Sun, N., Guo, W., et al. (2018). LncRNA PDIA3P interacts with c-Myc to regulate cell proliferation via induction of pentose phosphate pathway in multiple myeloma. Biochem. Biophys. Res. Commun. 498, 207–213. doi: 10.1016/j.bbrc.2018.02.211
Ye, X., Fan, F., Bhattacharya, R., Bellister, S., Boulbes, D. R., Wang, R., et al. (2015). VEGFR-1 pseudogene expression and regulatory function in human colorectal cancer cells. Mol. Cancer Res. 13, 1274–1282. doi: 10.1158/1541-7786.MCR-15-0061
Yu, G., Yao, W., Gumireddy, K., Li, A., Wang, J., Xiao, W., et al. (2014). Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Mol. Cancer Ther. 13, 3086–3097. doi: 10.1158/1535-7163.MCT-14-0245
Yu, J., Zhang, J., Zhou, L., Li, H., Deng, Z. Q., and Meng, B. (2019). The octamer-binding transcription factor 4 (OCT4) pseudogene, POU domain class 5 transcription factor 1B (POU5F1B), is upregulated in cervical cancer and down-regulation inhibits cell proliferation and migration and induces apoptosis in cervical cancer cell lines. Med. Sci. Monit. 25, 1204–1213. doi: 10.12659/MSM.912109
Yu, W., Qiao, Y., Tang, X., Ma, L., Wang, Y., Zhang, X., et al. (2014). Tumor suppressor long non-coding RNA, MT1DP is negatively regulated by YAP and Runx2 to inhibit FoxA1 in liver cancer cells. Cell. Signal. 26, 2961–2968. doi: 10.1016/j.cellsig.2014.09.011
Yuan, H., Jiang, H., Wang, Y., and Dong, Y. (2019). Increased expression of lncRNA FTH1P3 predicts a poor prognosis and promotes aggressive phenotypes of laryngeal squamous cell carcinoma. Biosci. Rep. 39:BSR20181644. doi: 10.1042/BSR20181644
Zhan, Y., Liu, Y., Wang, C., Lin, J., Chen, M., Chen, X., et al. (2016). Increased expression of SUMO1P3 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Oncotarget 7, 16038–16048. doi: 10.18632/oncotarget.6946
Zhang, C. Z. (2017). Long non-coding RNA FTH1P3 facilitates oral squamous cell carcinoma progression by acting as a molecular sponge of miR-224-5p to modulate fizzled 5 expression. Gene 607, 47–55. doi: 10.1016/j.gene.2017.01.009
Zhang, H., Xiong, Y., Xia, R., Wei, C., Shi, X., and Nie, F. (2017). The pseudogene-derived long noncoding RNA SFTA1P is down-regulated and suppresses cell migration and invasion in lung adenocarcinoma. Tumour Biol. 39:1010428317691418. doi: 10.1177/1010428317691418
Zhang, L. M., Wang, P., Liu, X. M., and Zhang, Y. J. (2017). LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am. J. Transl. Res. 9, 5461–5472.
Zhang, R., Guo, Y., Ma, Z., Ma, G., Xue, Q., Li, F., et al. (2017). Long non-coding RNA PTENP1 functions as a ceRNA to modulate PTEN level by decoying miR-106b and miR-93 in gastric cancer. Oncotarget 8, 26079–26089. doi: 10.18632/oncotarget.15317
Zhang, Y., Han, T., Li, J., Cai, H., Xu, J., Chen, L., et al. (2020). Comprehensive analysis of the regulatory network of differentially expressed mRNAs, lncRNAs and circRNAs in gastric cancer. Biomed. Pharmacother. 122:109686. doi: 10.1016/j.biopha.2019.109686
Zhang, Y., Li, Y., Han, L., Zhang, P., and Sun, S. (2019). SUMO1P3 is associated clinical progression and facilitates cell migration and invasion through regulating miR-136 in non-small cell lung cancer. Biomed. Pharmacother. 113:108686. doi: 10.1016/j.biopha.2019.108686
Zhang, Y., Li, Y., Wang, J., and Lei, P. (2018). Long non-coding RNA ferritin heavy polypeptide 1 pseudogene 3 controls glioma cell proliferation and apoptosis via regulation of the microRNA2245p/tumor protein D52 axis. Mol. Med. Rep. 18, 4239–4246. doi: 10.3892/mmr.2018.9491
Zhang, Z., and Shi, Z. (2019). The pseudogene PTTG3P promotes cell migration and invasion in esophageal squamous cell carcinoma. Open Med. 14, 516–522. doi: 10.1515/med-2019-0057
Zheng, J., Zhang, H., Ma, R., Liu, H., and Gao, P. (2019). Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-kappaB pathway in gastric cancer. Oncogene 38, 7073–7088. doi: 10.1038/s41388-019-0934-z
Zheng, L., Guo, Q., Xiang, C., Liu, S., Jiang, Y., Gao, L., et al. (2019). Transcriptional factor six2 promotes the competitive endogenous RNA network between CYP4Z1 and pseudogene CYP4Z2P responsible for maintaining the stemness of breast cancer cells. J. Hematol. Oncol. 12:23.
Zheng, L., Li, X., Gu, Y., Lv, X., and Xi, T. (2015). The 3’UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res. Treat. 150, 105–118. doi: 10.1007/s10549-015-3298-2
Zheng, L., Li, X., Gu, Y., Ma, Y., and Xi, T. (2014). Pseudogene CYP4Z2P 3’UTR promotes angiogenesis in breast cancer. Biochem. Biophys. Res. Commun. 453, 545–551. doi: 10.1016/j.bbrc.2014.09.112
Zheng, L., Li, X., Meng, X., Chou, J., Hu, J., Zhang, F., et al. (2016). Competing endogenous RNA networks of CYP4Z1 and pseudogene CYP4Z2P confer tamoxifen resistance in breast cancer. Mol. Cell. Endocrinol. 427, 133–142. doi: 10.1016/j.mce.2016.03.012
Zheng, R., Du, M., Wang, X., Xu, W., Liang, J., Wang, W., et al. (2018). Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol. Cancer 17:143. doi: 10.1186/s12943-018-0880-3
Zhou, Q., Zhang, W., Wang, Z., and Liu, S. (2019). Long non-coding RNA PTTG3P functions as an oncogene by sponging miR-383 and up-regulating CCND1 and PARP2 in hepatocellular carcinoma. BMC Cancer 19:731. doi: 10.1186/s12885-019-5936-2
Keywords: pseudogene, microRNA (miRNA), long non-coding RNA (lncRNA), circular RNA (circRNA), competing endogenous RNA (ceRNA), cancer
Citation: Lou W, Ding B and Fu P (2020) Pseudogene-Derived lncRNAs and Their miRNA Sponging Mechanism in Human Cancer. Front. Cell Dev. Biol. 8:85. doi: 10.3389/fcell.2020.00085
Received: 06 November 2019; Accepted: 30 January 2020;
Published: 28 February 2020.
Edited by:
Nejat Dalay, Istanbul University, TurkeyReviewed by:
Apiwat Mutirangura, Chulalongkorn University, ThailandMatthew Wong, Children’s Cancer Institute, Australia
Copyright © 2020 Lou, Ding and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiyang Lou, MTE3MTgyNjRAemp1LmVkdS5jbg==; Peifen Fu, UGVpZmVuX0Z1QDE2My5jb20=
 Weiyang Lou
Weiyang Lou Bisha Ding2,3
Bisha Ding2,3