
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 23 October 2018
Sec. Membrane Traffic and Organelle Dynamics
Volume 6 - 2018 | https://doi.org/10.3389/fcell.2018.00146
This article is part of the Research Topic Autophagy: from Big Data to Physiological Significance View all 16 articles
During the last decade, autophagy has been pointed out as a central process in cellular homeostasis with the consequent implication in most cellular settings and human diseases pathology. At present, there is significant data available about molecular mechanisms that regulate autophagy. Nevertheless, autophagy pathway itself and its importance in different cellular aspects are still not completely clear. In this article, we are focused in four main aspects: (a) Induction of Autophagy: Autophagy is an evolutionarily conserved mechanism induced by nutrient starvation or lack of growth factors. In higher eukaryotes, autophagy is a cell response to stress which starts as a consequence of organelle damage, such as oxidative species and other stress conditions. (b) Initiation of Autophagy; The two major actors in this signaling process are mTOR and AMPK. These multitasking protein complexes are capable to summarize the whole environmental, nutritional, and energetic status of the cell and promote the autophagy induction by means of the ULK1-Complex, that is the first member in the autophagy initiation. (c) ULK1-Complex: This is a highly regulated complex responsible for the initiation of autophagosome formation. We review the post-transductional modifications of this complex, considering the targets of ULK1. (d)The mechanisms involved in autophagosome formation. In this section we discuss the main events that lead to the initial structures in autophagy. The BECN1-Complex with PI3K activity and the proper recognition of PI3P are one of these. Also, the transmembrane proteins, such as VMP1 and ATG9, are critically involved. The membrane origin and the cellular localization of autophagosome biogenesis will be also considered. Hence, in this article we present an overview of the current knowledge of the molecular mechanisms involved in the initial steps of mammalian cell autophagosome biogenesis.
There are three types of autophagy, processes where cytoplasmic components are delivered to lysosomes for degradation: microautophagy/endosomal microautophagy (Li et al., 2012; Galluzzi et al., 2017), chaperone-mediated autophagy (CMA) (Cuervo and Wong, 2014; Kaushik and Cuervo, 2018) and macroautophagy (hereafter mentioned as autophagy). This is the engulfment of cytoplasmic contents by a double membrane vesicle, named autophagosome. The outer membrane of the autophagosome eventually fuses with the lysosome, where the inner vesicle is delivered (Figure 1). Here we present a brief overview of the mechanisms involved in the initial steps of mammalian cell autophagosome biogenesis.
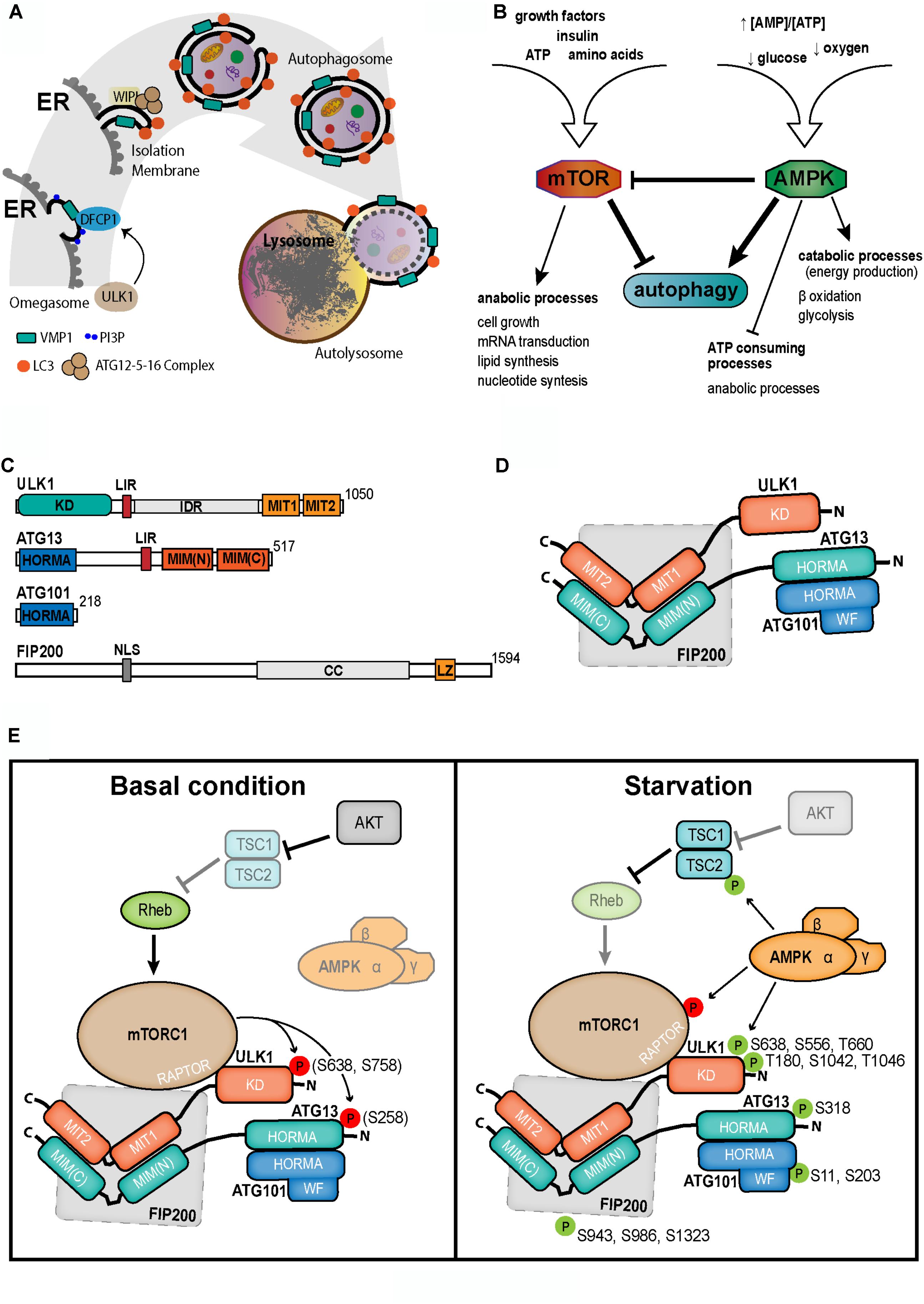
FIGURE 1. (A) Schematic overview of autophagy. UKL1 activation leads to autophagosome biogenesis. On the ER surface, the transmembrane protein VMP1 recruits a PI3K complex. The consequent PI3P subdomain is recognized by DFCP1 on the omegasome structure. Then, in the isolation membrane, WIPI proteins recruit the ATG5-ATG12-ATG16 complex which in turn make possible the lipidation of LC3 on the membrane. The formation of autophagosome, a double membrane vesicle, allows the carrying of cargo to lysosome. Eventually, cargo is degraded in the resulted autophagolysosome. ER, endoplasmic reticulum; PI3K, phosphatidylinositol 3-kinase; PI3P, phosphatidylinositol (3,4,5) triphosphate (PI3P). (B) Diagram of interrelationship among the cellular energetic and metabolic regulators, mTOR and AMPK, and the autophagy. (C) Representative scheme of the ULK1 complex proteins. Upper right number in each scheme shows the length of the amino acid chain. Described domains are showed for each protein. (D) Possible structure and interrelationship among the ULK1 complex proteins, suggested from available data. KD, kinase domain; LIR, LC3-interacting region; IDR, intrinsically disordered region; MIT, microtubule interacting and trafficking domain; HORMA, HOP1, REV7, and MAD2 domain; MIM, MIT-interacting motif; NLS, nuclear localization signal; CC, coil-coil region; LZ, leucine zipper; WF, WF finger motif. (E) Regulation of the autophagy initiation complex ULK1 by mTOR and AMPK at basal (left) and starvation (right) conditions.
The main task of autophagy is to deal against poor nutrient environments. In superior eukaryote cells, mTOR, which is a serine/threonine kinase, checks the presence of growth factors and nutrients. In presence of amino acids (mainly leucine, glutamine and arginine), mTORC1 maintains the autophagy inhibition. When nutrients are no longer available, the inhibition of mTORC1 releases the ‘brake’ and autophagy is eventually induced (Carroll et al., 2016). Growth factors negatively regulate the autophagy by activation of mTOR. Activation of the insulin receptor induces the phosphorylation of TSC2, avoiding the TSC1/2 complex formation and the mTORC1 inhibition (Haeusler et al., 2018). Other growth factors induce the RAS pathway, which activates the ERK1/2 dimer that inhibits the TSC1/2 complex and phosphorylates RAPTOR activating mTORC1 and suppressing autophagy (Carriere et al., 2011).
AMPK is a key serine/threonine kinase that is activated in low energy conditions (Egan et al., 2011). Then, AMPK activates the autophagosome formation by mean of direct and indirect ways. Furthermore, AMPK can be activated by CaMKKB in the ER-overloaded response (Hoyer-Hansen et al., 2007). The unfolded protein response, by mean of IRE1α, PERK and ATF6, is also an autophagy triggering event, enhancing LC3 conjugation (Ding et al., 2007; Kouroku et al., 2007).
During quick and intense oxygen fluctuations, autophagy is induced by mTORC1-dependent pathways and/or by ER stress. (Papandreou et al., 2008; Rouschop et al., 2010). In moderate but chronic hypoxia, autophagy is triggered mainly by HIF1α and PKCδ-JNK1 pathways (Mazure and Pouyssegur, 2010). HIF1α is the major transcription factor involved in cell response to hypoxia (Brocato et al., 2014). Among the genes transcribed by HIF1α is BNIP3 which disrupts the Bcl2-BECN1 interaction releasing BECN1 to be part of the autophagy process (Zhang et al., 2008), and VMP1, which interacts with BECN1 and is required for autophagosome formation (Ropolo et al., 2007; Rodriguez et al., 2017). Regarding to the PKCδ pathway, this kinase activates JNK1 that in turn phosphorylates Bcl2 to release it from BECN1 (Pattingre et al., 2009).
Oxidative stress induces autophagy in order to recycle damaged mitochondria (and other damaged organelles), and eliminate proteins aggregates (Ureshino et al., 2014). NRF2 is bound to antioxidant response elements promoting the transcription of p62, a cargo receptor for autophagy (Puissant et al., 2012). FOXO3 induces the expression of LC3 (an ATG protein that is described below) and BNIP3 (Mahalingaiah and Singh, 2014). Finally, ROS inhibit ATG4-mediated LC3 delipidation, that takes place immediately after formation of the autolysosome, conferring stability to LC3 and favoring its recruitment to the autophagosome (Scherz-Shouval et al., 2007).
Independently of the induction agent, in canonical autophagy, the initiation of autophagosome biogenesis is managed by the kinases mTOR and AMPK. In fact, through the association with RAPTOR, DEPTOR, PRAS40 and mLST8, mTOR constitutes the complex 1 [mTORC1]. At basal conditions, mTORC1 is stimulated by the small GTPase Rheb. In turn, mTOR triggers cell growth and diverse anabolic processes such as lipids, proteins and nucleotides synthesis (Lamb et al., 2013; Klionsky and Schulman, 2014). On the other hand, active mTORC1 abolishes most of catabolic processes including the autophagy (Lamb et al., 2013; Klionsky and Schulman, 2014; Figure 1B). Therefore, mTOR inhibits autophagy, by several phosphorylations on the first complex of the pathway (see further), when optimal nutrients concentration is available.
During starvation, Rheb is inhibited by the TSC1/2 heterodimer removing the activation stimulus on mTOR (Huang and Manning, 2008). This inhibition of mTORC1 decreases its influence on autophagy and as a consequence, the mechanism of autophagosome biogenesis is triggered (Carroll et al., 2016; Figure 1E). Moreover, the inactivation of mTORC1 allows that the dephosphorylated TFEB translocates to the nucleus (Puertollano et al., 2018) where it induces the transcription of ATG genes, such as UVRAG, WIPI, MAPLC3B, SQSTM1, Vps11, Vps18, and ATG9B. TFEB also promotes the lysosomal function in the cell (Settembre et al., 2011).
AMPK is a heterotrimeric complex composed by a catalytic α subunit and two regulatory subunits, β and γ (Egan et al., 2011). Since AMPK is activated in low energy conditions, this kinase inhibits anabolic processes, and induces catabolic pathways, such as autophagy (Egan et al., 2011; Zhang et al., 2013; Figure 1B). AMP binding allows LKB1 to phosphorylate AMPK (Thr172) (Xiao et al., 2007; Zhang et al., 2013), which in turn directly and indirectly activates the autophagosome formation as is explained in the next sections.
ULK1 is so far the first complex in the core molecular machinery involved in the biogenesis of autophagosomes. This complex is composed by the serin/threonin protein kinase ULK1, ATG13, FIP200, and ATG101. Activated ULK1 is capable of triggering series of phosphorylations that enable the nucleation process and autophagosome biogenesis. At N-terminal ULK1 is the kinase domain followed by a disordered region that is postulated as highly regulated. On the opposite side, there are two MIT domains in tandem that compose a globular structure (Noda and Fujioka, 2015). ULK1 structure was characterized in complex with ATG13. On the C-terminal of ATG13 there are two MIT-interacting motifs in a helical region for recognition-interaction with the ULK1 MIT domains (Noda and Fujioka, 2015; Qi et al., 2015). Additionally, both proteins, ULK1 and ATG13, have a LIR domain for interaction with LC3 family members. ATG101, the smallest member of the complex, is essential for autophagy (Mercer et al., 2009). ATG101 is almost fully composed by a HORMA domain with direct interaction with the HORMA domain at the N-terminus of ATG13. ATG101 stabilizes ATG13 and ULK1 (Mercer et al., 2009; Suzuki et al., 2015) and seems to recruit downstream molecules through its WF finger motif (Suzuki et al., 2015). The last member of ULK1-complex is FIP200, that is the largest molecule involved in this complex (Hara et al., 2008; Figures 1C,D).
ULK1 complex is regulated by the two major key proteins related to nutritional and energetic sensing, mTOR and AMPK (He and Klionsky, 2009). Under growth factors stimulation and nutrient availability, the activated mTORC1 interacts with ULK1 through RAPTOR and phosphorylates several sites of ULK1 (Ser757/5637 in mouse, Ser758 in human) (Alers et al., 2012) and Atg13 (Ser258 in mouse) subunits (Kim et al., 2011; Puente et al., 2016). Then, ULK1 complex remains inactivated and autophagy repressed. AMPK induces ULK1-mediated autophagy by three strategies: 1- AMPK phosphorylates TSC2 at Ser1345 enhancing the activity of this mTORC1 inhibitor (Inoki et al., 2003). 2- AMPK is able to inhibit mTORC1 activity directly by phosphorylation of Raptor in Ser792/722 (Gwinn et al., 2008; Egan et al., 2011). 3- AMPK interacts with and phosphorylates ULK1 in Ser317/777 for its activation (Kim et al., 2011; Figure 1E).
Another pathway for ULK1 autophagy activation has been proposed: AMBRA1 may act as a bridge between ULK1 and the ubiquitin ligase E3 TRAF6 (Nazio et al., 2013; Grumati and Dikic, 2018). TRAF6-mediated poly ubiquitination, K63 type branched ubiquitin, potentiates autophagy activation by promoting stabilization and self-association of ULK1. This event initiates a positive loop, where ULK1 phosphorylates AMBRA1 enhancing TRAF6-mediated ULK1 ubiquitination (Nazio et al., 2013; Grumati and Dikic, 2018). Further, growth factors withdrawal might induce the activation of TIP60 by GSK3-mediated phosphorylation at Ser86. TIP60 is an acetyltransferase that induces the activation of ULK1 by acetylation of Lys162/606 enhancing the triggering of autophagy (Lin et al., 2012).
Once activated, ULK1 is able to phosphorylate several substrates. Among them, there are two initial complexes, the ULK1 complex itself and the PI3KC3 complex 1 (PI3KC3-C1). In the first complex, ULK1 phosphorylates to itself (Thr180/1046, Ser1042) (Bach et al., 2011), and the other members of the complex, Atg13 (Ser318/203), FIP200 (Ser943/986/1323) and ATG101 (Ser11/203) (Lin and Hurley, 2016; Orhon and Reggiori, 2017; Figure 1E). In the second complex, ULK1 potentiates the PI3K activity of the catalytic subunit Vps34, by the phosphorylation of two members of the complex, BECN1 (Ser14) and ATG14L (Ser29), resulting in the increment of PI3P production (Russell et al., 2013). Following to ULK1 complex activation, the transmembrane protein VMP1 interacts with the BH3 domain of BECN1 through its ATG domain, recruiting the PI3KC3-C1 to the autophagosomal membrane (Molejon et al., 2013).
There are two main PI3KC3 complexes in autophagosome biogenesis. The complex 1 is composed by BECN1, ATG14L, Vps15 and Vps34, which is a key component in autophagosome initiation. The other complex, PI3KC3-C2, is related to autophagosome maturation and endosomal trafficking and is composed by the same members except for the regulatory protein ATG14L which is replaced by UVRAG. Structurally, the PI3KC3-C1 is stabilized in pairs, BECN1/ATG14L and Vps15/Vps34 (Stjepanovic et al., 2017). Upon autophagy induction, BECN1 recruitment induces the complex assembly, through the adaptor ATG14L, where the WD domain of Vps15 organizes the proteins into the complex allowing the activity of Vps34 (Stjepanovic et al., 2017). Moreover, the KAP1-mediated SUMOylation of Vps34 enhances the interaction of this protein with the rest of the complex (Yang et al., 2013). As it was commented before, ULK1-mediated phosphorylation of BECN1, ATG14L and Vps34 potentiates PI3K activity in this complex. The tumor suppressor DAPK, a calcium/calmodulin serine/threonine kinase, also contributes to the PI3KC3-C1 recruitment to the autophagosome membrane. This kinase phosphorylates BECN1 on its BH3 domain interfering with the BECN1-Bcl-xL association and releasing BECN1 (Zalckvar et al., 2009). This effect is reaffirmed by TRAF6 which ubiquitinates BECN1 on the same region (Shi and Kehrl, 2010). Recently, it has been proposed that Vps34 activity may be switched on/off by an EP300-dependent acetylation/deacetylation on K771, as another regulation of the PI3KC3-C1 (Su and Liu, 2017; Su et al., 2017).
The cascade of subsequent activations of ULK1 and PI3KC3-C1 complex members is limited by a series of degradative processes. The deubiquitinase A20 (DUB A20) controls BECN1 participation on autophagosome formation by elimination of poly ubiquitin chain in the BH3 domain placed by ATF6 E3 ligase. Beyond that regulation, the E3 ligases NEDD4 and NEDD4L induce degradation of key members in ULK1, and Vps34 complexes respectively (Platta et al., 2012; Nazio et al., 2016). BECN1 is poly ubiquitinated with K11-linked ubiquitin chain by NEDD4 to be eliminated in the proteasome. Similar activity is carried out by NEDD4L on ULK1 targeting this protein with K27- and K29-linked ubiquitin chains. In both cases, the proteasome-mediated elimination of those proteins causes the destabilization of its respective complexes. In a redundant way of labeling for degradation, the poly ubiquitination with K48-linked ubiquitin chains on ULK1, BECN1, and Vps34 is catalyzed by the complex CUL3-KHLH20 (Liu and Chen, 2016).
The local enrichment of PI3P in ER-subdomains acts as the signal for the nucleation of several autophagy-related proteins in a structure named omegasome that resembles the Greek letter omega (Ktistakis and Tooze, 2016). The first protein which recognizes the PI3P is DFCP1. DFCP1 possesses a diffuse pattern over the ER, mitochondria and Golgi but it is rapidly mobilized to the PI3P spots by the recognition of this phospholipid with the two FYVE motifs of its structure. Although it is a marker of omegasome, little is known about its role during the initial steps of autophagosome biogenesis. Additionally, the DFCP1 depletion does not seem to interfere with the progression of autophagy.
The rising omegasome leads to extension of a sack-like structure named isolation membrane or phagophore. WIPI2b, a member of the PROPPIN family, recognizes the local PI3P by the FRRG motif of its WD40-repeat β-propeller on the isolation membrane (Nascimbeni et al., 2017a). The process continues with two ubiquitin like systems: ATG12 and LC3. Cytoplasmic ATG12 is covalently attached to a C-terminal glycine of ATG5. This catalytic reaction resembles the ubiquitination process where ATG7 and ATG10 are subrogated to E1 and E2 enzymes, respectively (Klionsky and Schulman, 2014). ATG5-ATG12 complex is highly important, since it functions as E3 enzyme for LC3 conjugation to phosphatidylethanolamine (PE) on the autophagosomal membrane. This process seems to be mediated by ATG16L, which is composed by a WD40-repeat β-propeller domain localized in the C-terminal sequence. At N-terminal sequences, ATG16L possesses a binding domain that allows the interaction with ATG5 to eventually form the ATG12-ATG5-ATG16L complex (Wilson et al., 2014). The middle sequence of ATG16L expands a coil-coil (cc) dimerization domain that induces the formation of ATG16L dimers (Wilson et al., 2014). Then, WIPI2b is recognized by a region of ATG16L, between the cc-dimerization domain and the WD40-repeated β-propeller domain. Consequently, the ATG12-ATG5-ATG16L complex is recruited to the isolation membrane. LC3 plays a central role in autophagy being involved in vesicle elongation, maturation, fusion of autophagosome-lysosome and even as an adaptor to cargo recognition (Nakatogawa et al., 2007; Lee and Lee, 2016). LC3 shows a diffuse pattern distributed over the cytoplasm and into the nucleus (known as LC3-I) in basal conditions. Upon autophagy triggering, LC3 is deacetylated in the nucleus by SIRT1 (Huang et al., 2015) and is cleaved in cytoplasm by ATG4B, which eliminates the C-terminal arginine residue to expose a glycine (Satoo et al., 2009; Maruyama and Noda, 2017). In an ubiquitin-like reaction, the exposed glycine is combined to form a thioester bound, first with ATG7 (E1-like enzyme) and then with ATG3 (the E2-like enzyme) (Satoo et al., 2009; Maruyama and Noda, 2017). ATG3 is recognized by ATG12 of the ATG12-ATG5-ATG16L complex which has been already recruited to isolation membrane through WIPI2b. The ATG12-ATG5-ATG16L complex functions as the E3 enzyme leading the formation of an amide bound with the amine headgroup of PE (Noda et al., 2013; Otomo et al., 2013; Dooley et al., 2015). The lipidated LC3 (LC3-II) is present at the isolation membrane and on the autophagosome, in both sides of the membrane. The arrival of autophagosome to the lysosome is a fusion dependent mechanism of the HOPS complex, through STX17 (Jiang et al., 2014), and RAB7 (Gutierrez et al., 2004). Since LC3 is present in both membranes of autophagosome, once exposed to lysosomal hydrolases, there is a pool of LC3 that is degraded with cargo. However, the LC3 localized in the external membrane is cleaved from the PE, by ATG4B, and then recycled. (Noda et al., 2013; Otomo et al., 2013; Dooley et al., 2015).
Furthermore, of which is explained above, autophagy is able to follow unconventional pathways. ER-stress or glucose influx after starvation in NIH3T3, can induce autophagy independent of mTOR inhibition and where AMPK activation is not essential (Corona Velazquez and Jackson, 2018). Moreover, the glucose influx in mouse embryonic fibroblast can trigger autophagy independent of ULK1/2. Starved chicken DT40 cells show an autophagy dependent of ATG13-FIP200 interaction but independent of ULK1. Similar behavior is observed in some viral infection, such as coronaviruses, HBV or Poliovirus, which induce a non-degradative ULK1-independent form of autophagy. Even more interesting is that the oleate fatty acid can induce an autophagy mechanism that lacks of PI3P synthesis, since it cannot be inhibited by knocking-down of BECN1, Vps34, or ATG14. These examples suggest that autophagy is flexible and the pathways in autophagosome biogenesis may adapt to different situations depending on the inductor and the biological context (Corona Velazquez and Jackson, 2018).
It is accepted that the initial structure related to autophagy is located on the ER. The data suggest that ULK1 complex translocates to phosphatidylinositol-enriched ER-subdomains and then, the membrane structure is fed by ATG9A-containing vesicles (Nishimura et al., 2017). Then, autophagosomes are formed in highly active ER-subdomains where lipidic interchange between ER and other cytoplasmic organelles occurs.
Two sites of autophagosome biogenesis have been recently demonstrated: The ER-plasma membrane contact site (ER-PM) and the ER-Mitochondria contact site (Hamasaki et al., 2013; Nascimbeni et al., 2017b). VMP1 is a key player in the biogenesis of autophagosomes that remains in the autophagosomal membrane (Grasso et al., 2011). VMP1-BECN1 interaction allows the recruitment of PI3KC3-C1 to the ER-PM contact site by the interaction with the proteins Esyt 1, 2, and 3 (Nascimbeni et al., 2017b). Moreover, VMP1 was suggested to also regulate the ER-mitochondria contact site during autophagy and to be involved in the release of the initial autophagosome vesicle by activation of SERCA pump (Tabara and Escalante, 2016; Zhao et al., 2017). The transmembrane protein ATG9A is in Golgi and endosomal system, in early and late endosomes with a minimal percentage of recycling ones (Feng and Klionsky, 2017). In starvation, the TRAPPIII complex, related to ER-Golgi vesicular trafficking, mobilizes ATG9A vesicles to the sites of nascent autophagosomes (Shirahama-Noda et al., 2013). The adaptor protein AP-4 is required for this event, since it mediates the trafficking of ATG9A from trans-Golgi network to the site of autophagosomes maturation (Mattera et al., 2017). This event would potentiate the expansion of the isolation membrane. Nevertheless, the contribution of this membrane by the ATG9A vesicles is not enough to explain the growth of the membrane itself. Moreover, ATG9 seems to take a distinctive role in different systems. In contrast to mammals, yeast ATG9 has a fundamental role at very early steps in the pre-autophagosomal structure. On the other hand, in plants, the depletions of Arabidopsis ATG9 still allows formation of autophagosomal structures supplemented with ATG8 (LC3 ortholog) suggesting divergent regulation and mechanisms of this types of vesicles (Zhuang et al., 2017).
Ribosomes-free regions specialized in ER-Golgi communication are present in the rough ER. Vesicles arise targeted to the Golgi from these areas, described as ER-exit sites (ERES). These vesicles are supplemented by the proteins Sar1, Sec23, Sec24, Sec13 and Sec31, that constitute the COPII coat (Zahoor and Farhan, 2018). Before reaching Golgi, the COPII-coated vesicles go through an intermediated structure named ER-Golgi intermediate compartment (ERGIC) (Ben-Tekaya et al., 2005). The function of these structures is not completely understood, but they might participate in the autophagosome biogenesis. An impairment of these compartments causes an autophagy downregulation (Karanasios et al., 2016; Zahoor and Farhan, 2018).
Data suggest that the bulk contribution for the growth of the autophagosome membrane comes from the ER-Golgi vesicular trafficking. During starvation, the FIP200-CTAGES5 interaction induces the remodeling and enlargement of ERES positives for Sec12 (Ge et al., 2017). This allows the production of COPII-coated vesicles that are released to contribute to autophagosome formation. Moreover, ULK1 phosphorylates Sec23A, a member of the COPII multiprotein complex. This event is related to morphological variations on ERES during starvation and might turn the secretory machinery from anabolic to catabolic state.
A recent work shows a previously unexpected key role of Rab11A-positive membranes in autophagosome biogenesis (Puri et al., 2018). They demonstrated that WIPI2 relies, beyond the recognition of PI3P, in the interaction with Rab11A for recruitment of ATG16L. Also, the authors suggest a model where isolation membrane is represented by Rab11A-positive membrane, likely to be recycling endosomes. In this context, Rab11A-positive membranes constitute the platform for autophagosome formation initial steps.
The initial molecular steps in autophagosome biogenesis are determined by three mains complexes: ULK1 complex; PI3KC3-C1; and ATG16L1–ATG5–ATG12 which eventually favors LC3 lipidation in the growing isolation membrane. LC3 family seems to play a relevant role in cargo recognition, autophagosome closure and fusion with lysosomes. However, while the initial molecular steps seem to be essential and well-known in canonical autophagy, the subsequent events in mammalian autophagosome biogenesis are less characterized. Moreover, the wide spectrum of autophagy-related events and the number of molecules involved (Table 1) leads to the concept that different pathways might account for diverse types of autophagy and may reveal different functions of autophagy in physiological and pathological cellular processes. Furthermore, the meaning of different origins and composition of the autophagosomal membrane, such as those supplied by ATG9A and COP-II vesicles (Feng and Klionsky, 2017), are still not fully understood.
Moreover, autophagosome biogenesis is regulated by a variety of signaling pathways through posttranslational modification, such as phosphorylations, ubiquitinations, SUMOylations and acetylation, that may account for diverse conditions, functions or selectivity. Furthermore, this molecular regulation, that are eminently druggable, may be relevant in the development of therapeutic strategies of autophagy modulation for complex pathologies such as cancer (Galluzzi et al., 2015) or neurodegenerative diseases (Zare-Shahabadi et al., 2015).
Although there are many aspects still unclear on mammalian autophagosome biogenesis, future findings that shed light on this sophisticated intracellular process can be taken for granted.
DG did the literature search, wrote the first draft of the manuscript and designed all the figures. FR wrote a session of the first draft of the manuscript and assisted with the edited version. MV edited and added to the draft of the manuscript and figures and revised the final version of the manuscript.
This work was supported by grants from the University of Buenos Aires (UBACyT) The National Council for Scientific Research and Technology (CONICET-PIP) and the National Agency for Scientific and Technological Promotion (PICT).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AMBRA1, Activating molecule in BECN1-regulated autophagy protein 1; AMPK, AMP-activated Kinase; AP-4, Adaptor protein 4; ATF6, Activating transcription factor 6; ATG, Autophagy related gen or protein; Bcl-2, B-cell lymphoma 2; BECN1, Coiled-Coil Moesin-Like BCL2-Interacting Protein; BH3, Bcl-2 homology 3; BiP, Binding immunoglobulin protein; BNIP3, BCL2 interacting protein 3; CaMKKB, Calcium/calmodulin-dependent protein kinase kinase 2; COPII, Coat complex protein II; CTAGES5, Cutaneous T-cell linphoma-associated antigen 5; CUL3, Cullin-3; DAPK, Death-associated protein kinase; DEPTOR, DEP domain containing mTOR-interacting protein; DFCP1, Double FYVE containing protein 1; EP300, Histone acetyltransferase p300; ERK1/2, Mitogen-activated protein kinase; Esyt, Extended synaptotagmin; FIP200, FAK family-interacting protein of 200 kDa (also known as RB1CC1); FOXO3, Forkhead box protein O3; GSK3, Glycogen synthase kinase 3; HIF1α, Hypoxia-inducible factor 1 alpha; HORMA, Hop/Rev7/Mad2 domain; IDR, Intrinsically disordered region; IRE1α, Inositol-requiring enzyme 1 alpha; JNK, c-Jun N-terminal kinases; KAP1, E3 SUMO-protein ligase TRIM28; KHLH20, Kelch-like protein 20; LC3, Microtubule-associated proteins 1A/1B light chain 3B (also known as MAP1LC3B); LIR, LC3-interacting region; LKB1, Serine/threonine-protein kinase STKB1; MIT, Microtubule interacting and trafficking domain; mLST8, mammalian Letal with SEC13 protein 8; NEDD4, Neural precursor cell expressed developmentally down-regulated protein 4; NEDD4L, Neural precursor cell expressed developmentally downregulated gene 4-like; NRF2, Nuclear factor erythroid 2-related factor 2; PDK1, 3-phosphoinositide-dependent protein kinase 1; PERK, Proline-rich receptor-like protein kinase; PI3K, Phosphatidylinositol 3-kinase; PI3P, Phosphatidylinositol 3-phosphate; PKCδ, Protein kinase C delta type; PRAS40, Proline-rich Akt substrate of 40 kDa; PROPPIN, β-propeller that bind polyphosphoinositides; RAB, Ras-related protein; RAPTOR, Regulatory-associated protein of mTOR; Rheb, Ras homolog enriched in brain; ROS, Reactive oxygen species; SERCA, sarco/endoplasmic reticulum Ca2+-ATPase; SIRT1, Sirtuin 1; SQSTM1, Sequestosome 1 (also known as p62); STX17, Sintaxin 17; SUMO, Small ubiquitin-like modifier; TFEB, Transcription factor EB; TIP60, 60 KDa Tat-Interactive Protein; mTOR, mammalian Target of Rapamycin; TRAF6, Tumor necrosis factor receptor (TNFR)-associated factor 6; TRAPPIII, transport protein particle (TRAPP) III complex; TSC1/2, Tuberous sclerosis 1/2; ULK1, unc-51-like kinase 1; UVRAG, UV radiation resistance associated protein; VMP1, Vacuole membrane protein 1; Vps, Vacuolar protein sorting; WIPI, WD repeat domain phosphoinositide-interacting protein.
Alers, S., Loffler, A. S., Wesselborg, S., and Stork, B. (2012). Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 32, 2–11. doi: 10.1128/MCB.06159-11
Bach, M., Larance, M., James, D. E., and Ramm, G. (2011). The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem. J. 440, 283–291. doi: 10.1042/BJ20101894
Ben-Tekaya, H., Miura, K., Pepperkok, R., and Hauri, H. P. (2005). Live imaging of bidirectional traffic from the ERGIC. J. Cell Sci. 118(Pt 2), 357–367. doi: 10.1242/jcs.01615
Brocato, J., Chervona, Y., and Costa, M. (2014). Molecular responses to hypoxia-inducible factor 1alpha and beyond. Mol. Pharmacol. 85, 651–657. doi: 10.1124/mol.113.089623
Carriere, A., Romeo, Y., Acosta-Jaquez, H. A., Moreau, J., Bonneil, E., Thibault, P., et al. (2011). ERK1/2 phosphorylate raptor to promote ras-dependent activation of mTOR complex 1 (mTORC1). J. Biol. Chem. 286, 567–577. doi: 10.1074/jbc.M110.159046
Carroll, B., Maetzel, D., Maddocks, O. D., Otten, G., Ratcliff, M., Smith, G. R., et al. (2016). Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. eLife 5:e11058. doi: 10.7554/eLife.11058
Corona Velazquez, A. F., and Jackson, W. T. (2018). So many roads: the multi-faceted regulation of autophagy induction. Mol. Cell. Biol. doi: 10.1128/MCB.00303-18 [Epub ahead of print].
Cuervo, A. M., and Wong, E. (2014). Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 24, 92–104. doi: 10.1038/cr.2013.153
Ding, W. X., Ni, H. M., Gao, W., Hou, Y. F., Melan, M. A., Chen, X., et al. (2007). Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 282, 4702–4710. doi: 10.1074/jbc.M609267200
Dooley, H. C., Wilson, M. I., and Tooze, S. A. (2015). WIPI2B links PtdIns3P to LC3 lipidation through binding ATG16L1. Autophagy 11, 190–191. doi: 10.1080/15548627.2014.996029
Egan, D. F., Shackelford, D. B., Mihaylova, M. M., Gelino, S., Kohnz, R. A., Mair, W., et al. (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461. doi: 10.1126/science.1196371
Feng, Y., and Klionsky, D. J. (2017). Autophagic membrane delivery through ATG9. Cell Res. 27, 161–162. doi: 10.1038/cr.2017.4
Galluzzi, L., Baehrecke, E. H., Ballabio, A., Boya, P., Bravo-San Pedro, J. M., Cecconi, F., et al. (2017). Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836. doi: 10.15252/embj.201796697
Galluzzi, L., Pietrocola, F., Bravo-San Pedro, J. M., Amaravadi, R. K., Baehrecke, E. H., Cecconi, F., et al. (2015). Autophagy in malignant transformation and cancer progression. EMBO J. 34, 856–880. doi: 10.15252/embj.201490784
Ge, L., Zhang, M., Kenny, S. J., Liu, D., Maeda, M., Saito, K., et al. (2017). Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 18, 1586–1603. doi: 10.15252/embr.201744559
Grasso, D., Ropolo, A., Lo Re, A., Boggio, V., Molejon, M. I., Iovanna, J. L., et al. (2011). Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J. Biol. Chem. 286, 8308–8324. doi: 10.1074/jbc.M110.197301
Grumati, P., and Dikic, I. (2018). Ubiquitin signaling and autophagy. J. Biol. Chem. 293, 5404–5413. doi: 10.1074/jbc.TM117.000117
Gutierrez, M. G., Munafo, D. B., Beron, W., and Colombo, M. I. (2004). Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 117(Pt 13), 2687–2697. doi: 10.1242/jcs.01114
Gwinn, D. M., Shackelford, D. B., Egan, D. F., Mihaylova, M. M., Mery, A., Vasquez, D. S., et al. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226. doi: 10.1016/j.molcel.2008.03.003
Haeusler, R. A., McGraw, T. E., and Accili, D. (2018). Biochemical and cellular properties of insulin receptor signalling. Nat. Rev. Mol. Cell Biol. 19, 31–44. doi: 10.1038/nrm.2017.89
Hamasaki, M., Furuta, N., Matsuda, A., Nezu, A., Yamamoto, A., Fujita, N., et al. (2013). Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393. doi: 10.1038/nature11910
Hara, T., Takamura, A., Kishi, C., Iemura, S., Natsume, T., Guan, J. L., et al. (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510. doi: 10.1083/jcb.200712064
He, C., and Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. doi: 10.1146/annurev-genet-102808-114910
Hoyer-Hansen, M., Bastholm, L., Szyniarowski, P., Campanella, M., Szabadkai, G., Farkas, T., et al. (2007). Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol. Cell 25, 193–205. doi: 10.1016/j.molcel.2006.12.009
Huang, J., and Manning, B. D. (2008). The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 412, 179–190. doi: 10.1042/BJ20080281
Huang, R., Xu, Y., Wan, W., Shou, X., Qian, J., You, Z., et al. (2015). Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466. doi: 10.1016/j.molcel.2014.12.013
Inoki, K., Zhu, T., and Guan, K. L. (2003). TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590. doi: 10.1016/S0092-8674(03)00929-2
Jiang, P., Nishimura, T., Sakamaki, Y., Itakura, E., Hatta, T., Natsume, T., et al. (2014). The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell 25, 1327–1337. doi: 10.1091/mbc.E13-08-0447
Karanasios, E., Walker, S. A., Okkenhaug, H., Manifava, M., Hummel, E., Zimmermann, H., et al. (2016). Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7:12420. doi: 10.1038/ncomms12420
Kaushik, S., and Cuervo, A. M. (2018). The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381. doi: 10.1038/s41580-018-0001-6
Kim, J., Kundu, M., Viollet, B., and Guan, K. L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. doi: 10.1038/ncb2152
Klionsky, D. J., and Schulman, B. A. (2014). Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 21, 336–345. doi: 10.1038/nsmb.2787
Kouroku, Y., Fujita, E., Tanida, I., Ueno, T., Isoai, A., Kumagai, H., et al. (2007). ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14, 230–239. doi: 10.1038/sj.cdd.4401984
Ktistakis, N. T., and Tooze, S. A. (2016). Digesting the expanding mechanisms of autophagy. Trends Cell Biol. 26, 624–635. doi: 10.1016/j.tcb.2016.03.006
Lamb, C. A., Yoshimori, T., and Tooze, S. A. (2013). The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774. doi: 10.1038/nrm3696
Lee, Y. K., and Lee, J. A. (2016). Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 49, 424–430. doi: 10.5483/BMBRep.2016.49.8.081
Li, W. W., Li, J., and Bao, J. K. (2012). Microautophagy: lesser-known self-eating. Cell. Mol. Life Sci. 69, 1125–1136. doi: 10.1007/s00018-011-0865-5
Lin, M. G., and Hurley, J. H. (2016). Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 39, 61–68. doi: 10.1016/j.ceb.2016.02.010
Lin, S. Y., Li, T. Y., Liu, Q., Zhang, C., Li, X., Chen, Y., et al. (2012). GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336, 477–481. doi: 10.1126/science.1217032
Liu, C. C., and Chen, R. H. (2016). KLHL20 links the ubiquitin-proteasome system to autophagy termination. Autophagy 12, 890–891. doi: 10.1080/15548627.2016.1157243
Mahalingaiah, P. K., and Singh, K. P. (2014). Chronic oxidative stress increases growth and tumorigenic potential of MCF-7 breast cancer cells. PLoS One 9:e87371. doi: 10.1371/journal.pone.0087371
Maruyama, T., and Noda, N. N. (2017). Autophagy-regulating protease Atg4: structure, function, regulation and inhibition. J. Antibiot. doi: 10.1038/ja.2017.104 [Epub ahead of print].
Mattera, R., Park, S. Y., De Pace, R., Guardia, C. M., and Bonifacino, J. S. (2017). AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc. Natl. Acad. Sci. U.S.A. 114, E10697–E10706. doi: 10.1073/pnas.1717327114
Mazure, N. M., and Pouyssegur, J. (2010). Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Cell Biol. 22, 177–180. doi: 10.1016/j.ceb.2009.11.015
Mercer, C. A., Kaliappan, A., and Dennis, P. B. (2009). A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5, 649–662. doi: 10.4161/auto.5.5.8249
Molejon, M. I., Ropolo, A., Re, A. L., Boggio, V., and Vaccaro, M. I. (2013). The VMP1-Beclin 1 interaction regulates autophagy induction. Sci. Rep. 3:1055. doi: 10.1038/srep01055
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178. doi: 10.1016/j.cell.2007.05.021
Nascimbeni, A. C., Codogno, P., and Morel, E. (2017a). Local detection of PtdIns3P at autophagosome biogenesis membrane platforms. Autophagy 13, 1602–1612. doi: 10.1080/15548627.2017.1341465
Nascimbeni, A. C., Giordano, F., Dupont, N., Grasso, D., Vaccaro, M. I., Codogno, P., et al. (2017b). ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 36, 2018–2033. doi: 10.15252/embj.201797006
Nazio, F., Carinci, M., Valacca, C., Bielli, P., Strappazzon, F., Antonioli, M., et al. (2016). Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J. Cell Biol. 215, 841–856. doi: 10.1083/jcb.201605089
Nazio, F., Strappazzon, F., Antonioli, M., Bielli, P., Cianfanelli, V., Bordi, M., et al. (2013). mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15, 406–416. doi: 10.1038/ncb2708
Nishimura, T., Tamura, N., Kono, N., Shimanaka, Y., Arai, H., Yamamoto, H., et al. (2017). Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 36, 1719–1735. doi: 10.15252/embj.201695189
Noda, N. N., and Fujioka, Y. (2015). Atg1 family kinases in autophagy initiation. Cell. Mol. Life Sci. 72, 3083–3096. doi: 10.1007/s00018-015-1917-z
Noda, N. N., Fujioka, Y., Hanada, T., Ohsumi, Y., and Inagaki, F. (2013). Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 14, 206–211. doi: 10.1038/embor.2012.208
Orhon, I., and Reggiori, F. (2017). Assays to monitor autophagy progression in cell cultures. Cells 6:E20. doi: 10.3390/cells6030020
Otomo, C., Metlagel, Z., Takaesu, G., and Otomo, T. (2013). Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 20, 59–66. doi: 10.1038/nsmb.2431
Papandreou, I., Lim, A. L., Laderoute, K., and Denko, N. C. (2008). Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 15, 1572–1581. doi: 10.1038/cdd.2008.84
Pattingre, S., Bauvy, C., Carpentier, S., Levade, T., Levine, B., and Codogno, P. (2009). Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J. Biol. Chem. 284, 2719–2728. doi: 10.1074/jbc.M805920200
Platta, H. W., Abrahamsen, H., Thoresen, S. B., and Stenmark, H. (2012). Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 441, 399–406. doi: 10.1042/BJ20111424
Puente, C., Hendrickson, R. C., and Jiang, X. (2016). Nutrient-regulated phosphorylation of ATG13 inhibits starvation-induced autophagy. J. Biol. Chem. 291, 6026–6035. doi: 10.1074/jbc.M115.689646
Puertollano, R., Ferguson, S. M., Brugarolas, J., and Ballabio, A. (2018). The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 37:e98804. doi: 10.15252/embj.201798804
Puissant, A., Fenouille, N., and Auberger, P. (2012). When autophagy meets cancer through p62/SQSTM1. Am. J. Cancer Res. 2, 397–413.
Puri, C., Vicinanza, M., Ashkenazi, A., Gratian, M. J., Zhang, Q., Bento, C. F., et al. (2018). The RAB11A-positive compartment is a primary platform for autophagosome assembly mediated by WIPI2 recognition of PI3P-RAB11A. Dev. Cell 45, 114.e8–131.e8. doi: 10.1016/j.devcel.2018.03.008
Qi, S., Kim, D. J., Stjepanovic, G., and Hurley, J. H. (2015). Structure of the human Atg13-Atg101 HORMA heterodimer: an interaction hub within the ULK1 complex. Structure 23, 1848–1857. doi: 10.1016/j.str.2015.07.011
Rodriguez, M. E., Catrinacio, C., Ropolo, A., Rivarola, V. A., and Vaccaro, M. I. (2017). A novel HIF-1alpha/VMP1-autophagic pathway induces resistance to photodynamic therapy in colon cancer cells. Photochem. Photobiol. Sci. 16, 1631–1642. doi: 10.1039/c7pp00161d
Ropolo, A., Grasso, D., Pardo, R., Sacchetti, M. L., Archange, C., Lo Re, A., et al. (2007). The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 282, 37124–37133. doi: 10.1074/jbc.M706956200
Rouschop, K. M., van den Beucken, T., Dubois, L., Niessen, H., Bussink, J., Savelkouls, K., et al. (2010). The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 120, 127–141. doi: 10.1172/JCI40027
Russell, R. C., Tian, Y., Yuan, H., Park, H. W., Chang, Y. Y., Kim, J., et al. (2013). ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750. doi: 10.1038/ncb2757
Satoo, K., Noda, N. N., Kumeta, H., Fujioka, Y., Mizushima, N., Ohsumi, Y., et al. (2009). The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28, 1341–1350. doi: 10.1038/emboj.2009.80
Scherz-Shouval, R., Shvets, E., Fass, E., Shorer, H., Gil, L., and Elazar, Z. (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760. doi: 10.1038/sj.emboj.7601623
Settembre, C., Di Malta, C., Polito, V. A., Garcia Arencibia, M., Vetrini, F., Erdin, S., et al. (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433. doi: 10.1126/science.1204592
Shi, C. S., and Kehrl, J. H. (2010). TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 3:ra42. doi: 10.1126/scisignal.2000751
Shirahama-Noda, K., Kira, S., Yoshimori, T., and Noda, T. (2013). TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J. Cell Sci. 126(Pt 21), 4963–4973. doi: 10.1242/jcs.131318
Stjepanovic, G., Baskaran, S., Lin, M. G., and Hurley, J. H. (2017). Unveiling the role of VPS34 kinase domain dynamics in regulation of the autophagic PI3K complex. Mol. Cell. Oncol. 4:e1367873. doi: 10.1080/23723556.2017.1367873
Su, H., and Liu, W. (2017). PIK3C3/VPS34 control by acetylation. Autophagy 14, 1086–1087. doi: 10.1080/15548627.2017.1385676
Su, H., Yang, F., Wang, Q., Shen, Q., Huang, J., Peng, C., et al. (2017). VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol. Cell 67, 907.e7–921.e7. doi: 10.1016/j.molcel.2017.07.024
Suzuki, H., Kaizuka, T., Mizushima, N., and Noda, N. N. (2015). Structure of the Atg101-Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat. Struct. Mol. Biol. 22, 572–580. doi: 10.1038/nsmb.3036
Tabara, L. C., and Escalante, R. (2016). VMP1 establishes ER-microdomains that regulate membrane contact sites and autophagy. PLoS One 11:e0166499. doi: 10.1371/journal.pone.0166499
Ureshino, R. P., Rocha, K. K., Lopes, G. S., Bincoletto, C., and Smaili, S. S. (2014). Calcium signaling alterations, oxidative stress, and autophagy in aging. Antioxid. Redox. Signal. 21, 123–137. doi: 10.1089/ars.2013.5777
Wilson, M. I., Dooley, H. C., and Tooze, S. A. (2014). WIPI2b and Atg16L1: setting the stage for autophagosome formation. Biochem. Soc. Trans. 42, 1327–1334. doi: 10.1042/BST20140177
Xiao, B., Heath, R., Saiu, P., Leiper, F. C., Leone, P., Jing, C., et al. (2007). Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature 449, 496–500. doi: 10.1038/nature06161
Yang, Y., Fiskus, W., Yong, B., Atadja, P., Takahashi, Y., Pandita, T. K., et al. (2013). Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. U.S.A. 110, 6841–6846. doi: 10.1073/pnas.1217692110
Zahoor, M., and Farhan, H. (2018). Crosstalk of autophagy and the secretory pathway and its role in diseases. Int. Rev. Cell Mol. Biol. 337, 153–184. doi: 10.1016/bs.ircmb.2017.12.004
Zalckvar, E., Berissi, H., Mizrachy, L., Idelchuk, Y., Koren, I., Eisenstein, M., et al. (2009). DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 10, 285–292. doi: 10.1038/embor.2008.246
Zare-Shahabadi, A., Masliah, E., Johnson, G. V., and Rezaei, N. (2015). Autophagy in Alzheimer’s disease. Rev. Neurosci. 26, 385–395. doi: 10.1515/revneuro-2014-0076
Zhang, H., Bosch-Marce, M., Shimoda, L. A., Tan, Y. S., Baek, J. H., Wesley, J. B., et al. (2008). Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 283, 10892–10903. doi: 10.1074/jbc.M800102200
Zhang, Y. L., Guo, H., Zhang, C. S., Lin, S. Y., Yin, Z., Peng, Y., et al. (2013). AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell Metab. 18, 546–555. doi: 10.1016/j.cmet.2013.09.005
Zhao, Y. G., Chen, Y., Miao, G., Zhao, H., Qu, W., Li, D., et al. (2017). The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell 67, 974.e6–989.e6. doi: 10.1016/j.molcel.2017.08.005
Keywords: autophagy regulation, mTOR, AMPK, ULK1, VMP1
Citation: Grasso D, Renna FJ and Vaccaro MI (2018) Initial Steps in Mammalian Autophagosome Biogenesis. Front. Cell Dev. Biol. 6:146. doi: 10.3389/fcell.2018.00146
Received: 31 July 2018; Accepted: 08 October 2018;
Published: 23 October 2018.
Edited by:
Yanzhuang Wang, University of Michigan, United StatesReviewed by:
Ming Zhu, University of California, San Diego, United StatesCopyright © 2018 Grasso, Renna and Vaccaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Ines Vaccaro, bXZhY2Nhcm9AZmZ5Yi51YmEuYXI=; bWFyaWEudmFjY2Fyb0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.