- Biological and Environmental Science and Engineering Division, KAUST Environmental Epigenetics Program, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
The raising worldwide prevalence of Type 1 and Type 2 diabetes mellitus (T1DM and T2DM) solicits the derivation of in vitro methods yielding mature and fully functional β-cells to be used in regenerative medicine. Several protocols to differentiate human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) into human pancreatic β-like cells have recently been developed. These methods, coupled with a bioengineering approach using biocompatible encapsulating devices, have recently led to experimental clinical trials showing great promises to ultimately end the battle of diabetic patients for managing hyperglycemia. However, in vitro differentiation protocols face the challenge of achieving homogenous population of mono-hormonal insulin-secreting mature β-cells. Major epigenetic events such as DNA methylation, post-translational modification of histones and non-coding RNAs expression, orchestrate physiological endocrine pancreas specification into α-, β-, γ-, and δ-cells, both in vivo and in vitro. The dysregulation of such epigenetic processes is associated to multiple pancreatic disorders including diabetes. Understanding the epigenomic and transcriptomic landscape underlying endocrine pancreas development could, therefore, improve in vitro differentiation methods. In this review, we summarize the most effective protocols for in vitro differentiation of hESCs/hiPSCs toward pancreatic β-cells and we discuss the current limitations in the derivation of functional glucose-responsive, insulin-releasing β-cells. Moreover, we focus on the main transcriptional and epigenetic events leading to pancreatic specification and on the applicative potential of novel epigenetic drugs for the establishment of innovative pharmacological therapeutic approaches.
Introduction
Insulin-dependent T1DM is characterized by selective autoimmune depletion of pancreatic β-cells resulting in deficient insulin release (Katsarou et al., 2017). Current pharmacological treatments mostly rely on the use of exogenous insulin to control glycemia (Pickup, 2012). The transplantation of cadaveric islets in combination with immunosuppressive therapies has been proven to reestablish normoglycemia up to several years but is limited by the scarcity of donor tissues (Bellin et al., 2012). Therefore, the availability of hESC-derived β-cells in combination with immune-protective devices could provide an innovative therapeutic approach for the treatment of the disease (Agulnick et al., 2015; Vegas et al., 2016). T2DM is a metabolic syndrome characterized by impaired glucose regulation resulting from insulin resistance and progressive loss of β-cells functionality. T2DM pharmacological protocols mainly target the endogenous response to glucose stimulation by increasing insulin sensitivity, insulin secretion, and glucose uptake (Marín-Peñalver et al., 2016). However, the efficacy of such treatments is progressively reduced due to eventual pancreatic β-cells failure (Kahn, 1994; Miller and Nguyen, 2014) and exogenous insulin administration is required. Thus, both T1DM and T2DM patients would benefit from regenerative medicine applications aimed at reconstituting physiological β-cells mass and function.
The adult human pancreas comprises two distinct endocrine and exocrine functional compartments. Acinar and ductal cells of the exocrine area occupy about 95% of the pancreas parenchyma, while islets of endocrine cells randomly scattered throughout the organ constitute the remaining 5% (Paris et al., 2004). These regions, known as the islets of Langerhans, contain a heterogeneous population of endocrine cells with specialized functions: α-, β-, γ-, δ-, and 𝜀-cells secreting glucagon (GCG), insulin (INS), pancreatic polypeptide (PPY), somatostatin (SST), and ghrelin (GHR) hormones, respectively. The breakthrough of single-cell RNA-Seq technology analysis has allowed the study of islet biology and its subpopulations at single cell level (Baron et al., 2016; Wang et al., 2016), thus opening the road to a deep understanding of pancreatic physiopathology at the molecular level. Moreover, the combination of techniques such as imaging mass spectrometry (IMS) (Jansson et al., 2016), mass cytometry (Spitzer and Nolan, 2016; Wang et al., 2016) and single cell RNA-Seq profiling of subpopulations of human and mouse islets (Muraro et al., 2016; Zeng et al., 2017), confirmed proteomic and transcriptomic signatures related to glucose sensing, uptake, and insulin releasing, but also revealed an unexpected heterogeneity in the transcriptomic pattern of individual β-cells and highlighted main molecular changes in the profiles of healthy and diabetic patients. Recent findings have identified up to five different β-cell subpopulations with diverging expression of subsets of genes implicated in insulin resistance, sensitivity, and diabetes [e.g., the adipokine RBP4, the free fatty acid receptor FFAR4 and the transcription factors (TFs) ID1 and ID3], oxidative metabolism and endoplasmic reticulum stress (Baron et al., 2016; Muraro et al., 2016; Segerstolpe et al., 2016). Despite these advances, further knowledge may come from a thorough analysis of the transcriptomic and proteomic signatures of different cells within the Langerhans islets and the generation of innovative organoid-based differentiation protocols mimicking the physiological three-dimensional (3D) architecture of pancreatic islets.
Human pluripotent stem cells (hPSCs) can serve as an inexhaustible source of differentiated cells due to their unique features of unlimited expansion and specification into virtually all somatic cells including pancreatic β-cells. However, to date, reproducing in vitro physiological conditions resulting in sustained β-cells maturation has been largely unsuccessful. Over the last decade, the increased understanding of the mechanisms regulating pancreatic differentiation has allowed the derivation of protocols to direct hPSCs toward insulin-producing β-like cells. While the available protocols efficiently yield differentiated intermediates, including definitive endoderm (DE), primitive gut tube-like (PGT), and pancreatic progenitors (PPs), they fail to generate functional, mono-hormonal β-cells. Consequently, the final stages of in vitro differentiation protocols result in a mixture of immature poly-hormonal endocrine cells featuring α-, β-, γ-, and δ-cells characteristics. Here, we dissect the temporally activated molecular events regulating pancreas specification in vivo and in vitro and discuss the role played by epigenetic modifiers in driving pancreas organogenesis and pathogenesis. Moreover, we explore the potential benefits of the implementation of drugs acting on epigenetic modifiers, also known as “epidrugs,” to optimize β-cells differentiation in vitro (Zullo et al., 2017).
Differentiation of Hesc and Hipsc Into Pancreatic β-Cells: Closing the Gaps
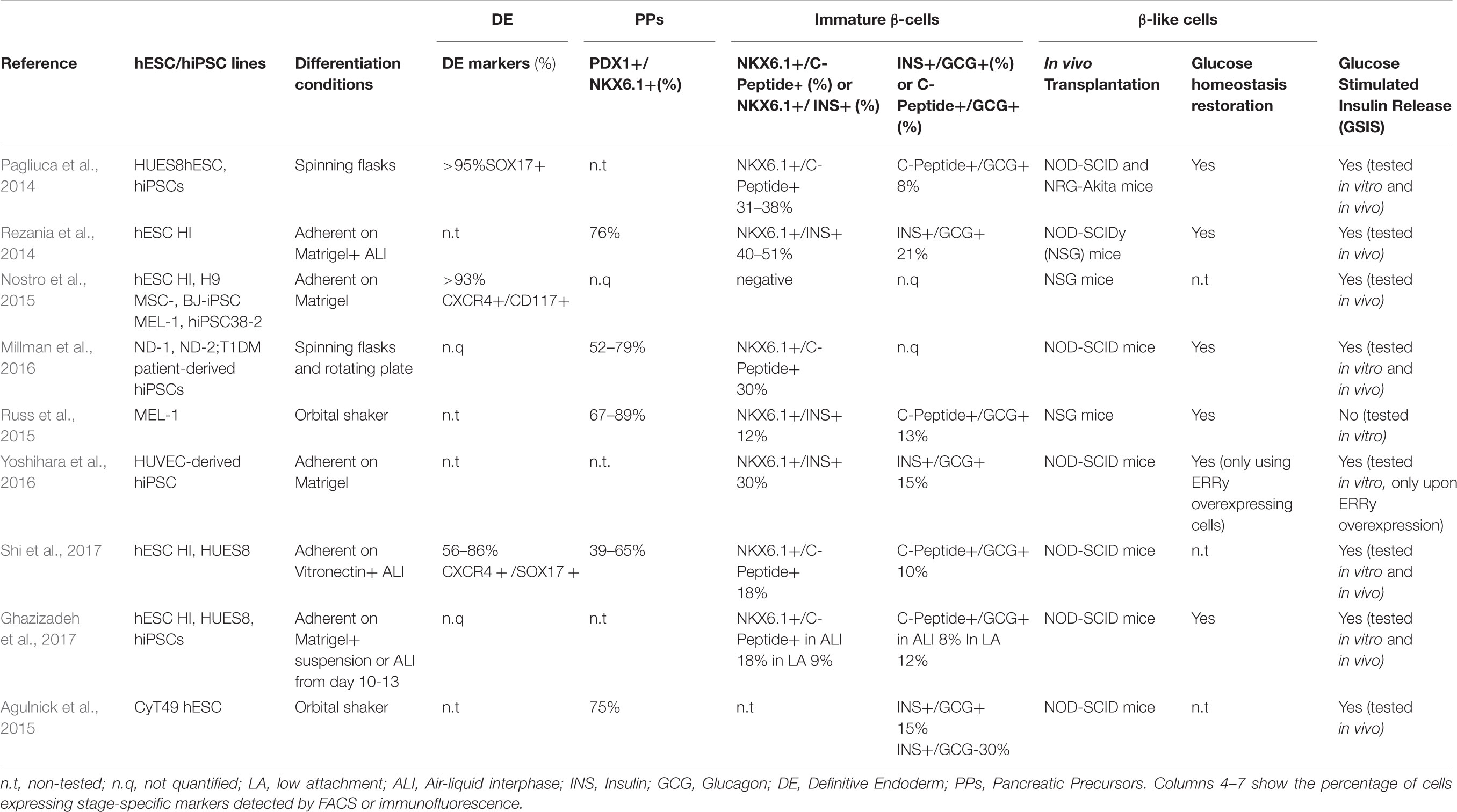
Ontogenetic studies performed in rodents elucidated the sequential developmental steps that, following gastrulation and endodermal specification, give rise to pancreatic endocrine progenitors. The endodermal germinal layer undergoes an intricate patterning of invaginations leading to the formation of the PGT organized into foregut, midgut, and hindgut. The commitment of foregut tube endoderm cells toward PPs is driven by orchestrated signaling mediated by transforming growth factor-β (TGF-β) proteins of the activin/nodal families, bone morphogenic protein (BMP), retinoic acid (RA) and AKT/PI3K pathways (D’Amour et al., 2005; Gadue and Gordon, 2006). Formation of pre-pancreatic endoderm is subsequently marked by the induction of the pancreatic and duodenal homeobox 1 (PDX1) TF, a master regulator expressed by all common progenitors of the mature pancreas (Herrera et al., 2002). Subsequently, pancreagenesis progresses as PDX1+ precursors proliferate and sprout to form ventral and dorsal buddings, that in turn fuse to form a single organ. Induction of the TF NKX6.1 restricts PDX1+ cells fate to endocrine and ductal cells (Schaffer et al., 2010) and drives the expression of key β-cell specification genes (Pedersen et al., 2005; Wescott et al., 2009; Taylor et al., 2013; Shih et al., 2015). At this stage, subsequent waves of proliferation produce a dense epithelial bud, where Neurogenin-3 (NGN3) positive pre-endocrine cells emerge and differentiate into proto-islet structures expressing ISL LIM Homeobox 1 (ISL1), Neuronal Differentiation 1 (NEUROD1), and Insulinoma associated protein 1 (INSM1) (Rosa and German, 2004; Gierl, 2006; Jia et al., 2015). From these late progenitors, mono-hormonal INS+, GCG+, PPY+, SST+, and GHR+ cell subtypes arise. Leveraging this knowledge, several groups developed multistep in vitro protocols to generate mono-hormonal glucose-responsive and insulin-releasing β-cells from hPSCs (Rezania et al., 2014; Millman et al., 2016; Table 1). However, several technical and biological limitations have prevented the establishment of a robust and standardized differentiation protocol: first, differentiation efficiency varies across different hPSCs lines and might be affected by growth rate, cell density, and batch-to-batch inconsistency of Matrigel and serum derivatives (e.g., knockout serum replacement, and KSR) often used in in vitro methods (Blauwkamp et al., 2012; Zeng et al., 2016; Shi et al., 2017; Tchieu et al., 2017); second, the in vivo transplantation required for the final maturation of the in vitro-derived β-like cells is intrinsically linked to biological variability; third, the amount of insulin released and the response to glucose stimuli are not yet comparable to bona fide β-cells (e.g., cadaveric islets used as gold standard positive control), suggesting faults in glucose sensing and insulin secretion machinery (Pagliuca et al., 2014; Rezania et al., 2014).
During in vitro specification, three key stages can be considered informative check-points to predict the efficacy of the differentiation protocols:
Specification of hPSCs Into DE and PGT
Induction of DE cells from hPSCs was initially achieved by combining Activin A (AA) and WNT3 recombinant proteins, activating the nodal/activin and β-catenin pathways, respectively (D’Amour et al., 2006). More recently, the glycogen synthase kinase 3β (GSK3-β) inhibitor and specific activator of the canonical Wnt/β-catenin pathway CHIR99021 was shown to induce DE more efficiently in combination with AA (Rezania et al., 2014). Lately, novel small molecules, including several ROCK inhibitors identified through a high-throughput screening approach, have further improved the differentiation of hPSCs toward DE and PPs (Korostylev et al., 2017). The molecular mechanism responsible for the increased formation of endoderm intermediates upon ROCK inhibition remains elusive, but similar results are observed using high density cultures (Toyoda et al., 2017) and 3D aggregation conditions (Takeuchi et al., 2014). This parallelism suggests that sustained reshaping of the cellular cytoskeleton could promote pancreatic specification in vitro. Consequently, an increased awareness that mechanical forces, cell–cell, and cell–substrate interactions boost pancreatic differentiation, encouraged scientists to implement pioneering expedients including rotating-bioreactors (Agulnick et al., 2015), microgravity chambers (Tanaka et al., 2013), hydrogel micro-well platforms (Bernard et al., 2012), and biomimetic scaffolds (Wang et al., 2017) which resulted in improved differentiation efficiency. However, current protocols yield about 85% of cells expressing the DE markers FOXA2, SOX17, and CXCR4 (Nostro et al., 2015; Zhu et al., 2016; Shi et al., 2017), but fail to produce fully homogenous populations of DE cells and require additional studies and optimization. According to several published protocols, further differentiation of DE cells into PGT can be achieved by the use of culture media containing fibroblast growth factor 10 (FGF10) or keratinocytes growth factor (KGF) (Kroon et al., 2008; Pagliuca et al., 2014). PGT specification leads to the formation of a compact epithelium expressing hepatocyte nuclear factors HNF4α, HNF1β, and HNF6 (Sampaziotis et al., 2017).
Derivation of PDX1+ NKX6.1+ Pancreatic Progenitors
The derivation of PDX1+ progenitors from PGT cells is achieved by using RA in combination with BMP, sonic hedgehog (SHH) signaling inhibitors, and either FGF10 or KGF (Guo et al., 2013; Hua et al., 2013). Additionally, low concentrations of Protein kinase C (PKC) agonists such as (2S,5S)-(E,E)-8-(5-(4-(trifluoromethyl)phenyl)-2,4-pentadienoylamino)benzolactam (TPB) and Phorbol 12,13-dibutyrate (PDBu), further stimulate PDX1 induction (Rezania et al., 2014; Zhu et al., 2016). PDX1+ cells must progressively acquire expression of the master regulator NKX6.1 to further progress toward endocrine commitment into β-cells. This is achieved by the use of endothelial growth factor (EGF), nicotinamide and Noggin (Nostro et al., 2015). It has been proposed that a subtle and short-term stimulation of BMP and SHH signaling is crucial to commit toward a mono-hormonal endocrine fate characterized by the expression of NGN3 in PDX1+/NKX6.1+ precursors, from which NKX2.2+ and subsequently C-Peptide+/INS+ cells emerge (Jennings et al., 2013; Russ et al., 2015). Conversely, extended stimulation promotes NGN3 expression in a subpopulation of PDX1+/NKX6.1- cells and lead to C-Peptide+/GCG+ poly-hormonal cells (Gu et al., 2002; Nostro et al., 2015; Russ et al., 2015).
Specification of PPs Into β-Like Cells
Existing in vitro protocols promote the generation of immature poly-hormonal cells (e.g., GCG+/INS+; SST+/INS+; and PPY+/INS+) characterized by defective glucose-sensing insulin-releasing characteristics (Table 1; D’Amour et al., 2006; Kroon et al., 2008; Rezania et al., 2012). The immature state of the in vitro-derived β-like cells recapitulates features of the immature fetal pancreas, whose functional maturation occurs postnatally during the transition from weaning to increased carbohydrate intake, and is likely triggered by the transition from glycolysis to oxidative phosphorylation (Aguayo-Mazzucato et al., 2013; Stolovich-Rain et al., 2015). In line with these observations, the exogenous expression of a transcriptional regulator of mitochondrial oxidative metabolism such as the estrogen-related receptor gamma (ERRγ) ameliorates in vitro maturation and glucose stimulated insulin secretion (GSIS) of iPSC-derived β-like cells (Yoshihara et al., 2016). Consistently, an independent study demonstrated that cellular homeostasis and metabolism are influenced by oxygen tension and that hyperoxic cell culture conditions improve differentiation toward endocrine β-cells (Hakim et al., 2014). While the optimal in vitro culture conditions to further specify poly-hormonal cells into single-signature β-cells remain elusive, xenotransplantation of these cells into diabetic mice is sufficient to drive the final maturation into INS+ cells expressing specific bona fide markers of β-cells and to reestablish normoglycemia (Pagliuca et al., 2014; Rezania et al., 2014; Dhawan et al., 2015; Russ et al., 2015; Millman et al., 2016). The encouraging data obtained in diabetic mice and the generation of biocompatible and immune-protective devices have paved the way to approve clinical trials for T1DM patients (ViaCyte Inc., Clinical trial number: NCT03162926). The main goal of these trials is to assess the safety, tolerability, and efficacy of the treatment related to tumorigenic risk, scarce immune-tolerance, and inefficient vascularization.
Epigenetics and in vitro Differentiation
Seminal work from Sander’s group thoroughly characterized the epigenetic landscape of endodermal progenitors and pancreatic β-cells precursors derived from hESCs (Xie et al., 2013). During endodermal differentiation, hPSCs undergo dynamic epigenetic remodeling processes regulated by Polycomb group proteins (PcG) and the histone H3 lysine 27 (H3K27) demethylase KDM6B. Specifically, differentiation of hPSCs into DE is accompanied by the resolution of bivalent domains associated with critical DE marker genes such as SOX17, EOMES, and GATA6 whose induction is KDM6B-dependent (Xie et al., 2013). As differentiation toward later stages of pancreatic specification proceeds, DE markers become silenced in a PcG-dependent manner and late markers are progressively induced. Later work from the same laboratory demonstrated that the enhancers associated with lineage-specific TFs that regulate pancreas development (including PDX1, SOX9, and PROX1) are in a “poised” state (H3K4me1 only) at the PGT stage. This process primes the PGT progenitors to respond to subsequent differentiation cues and is regulated by the TFs FOXA1 and FOXA2 (Wang et al., 2015). Subsequently, lineage-specific and H3K4me1-marked enhancers are terminally activated by acquiring H3K27 acetylation (H3K4me1/H3K27ac) (Wang et al., 2015). Over the last decade, several groups have tested multiple epidrugs to optimize β-cell differentiation in vitro (Table 2). In 2009, Melton’s group identified two compounds, IDE1 and IDE2, from a library of HDACs inhibitors, that drive efficient differentiation of mouse ESCs into DE through induction of the endodermal marker Sox17 (Borowiak et al., 2009). The class-I HDAC inhibitors sodium butyrate and valproic acid have also been used in combination with AA to prime hESCs toward DE (Hay et al., 2008; Kondo et al., 2014), while the demethylating agent 5′-Azadeoxycytidine (AZA) has been shown to stimulate NGN3 expression (Lefebvre et al., 2010). Thus, the increased knowledge on the epigenetic and transcriptional landscape of PPs and mature β-cells coupled to a systematic high-throughput screening of epidrugs at each step of differentiation offers new opportunities for the optimization of the current protocols.
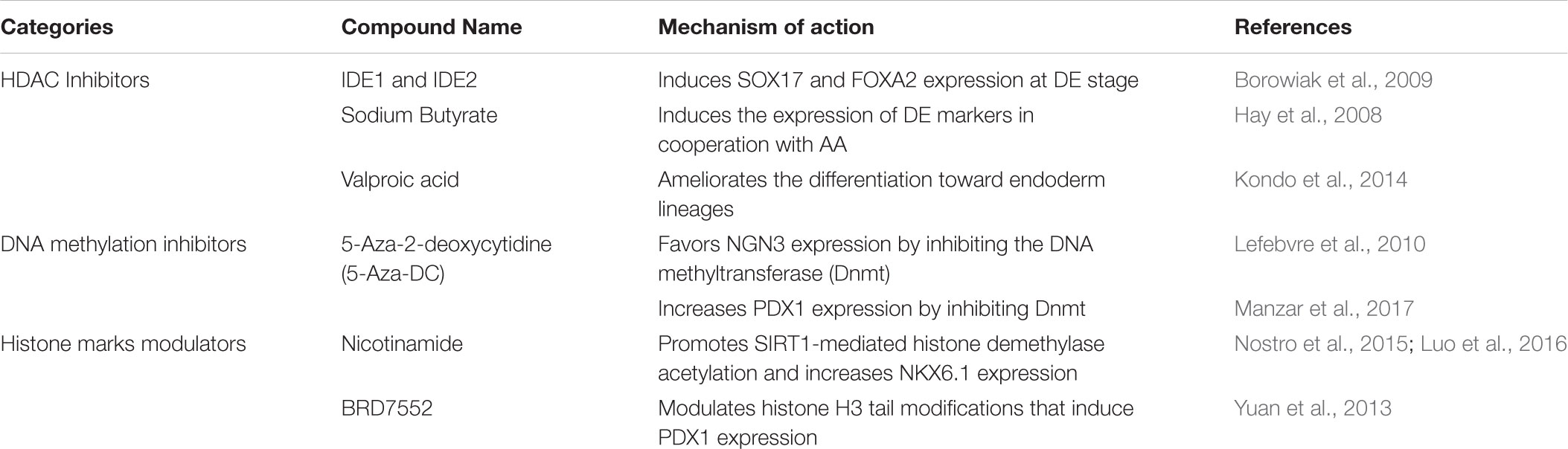
TABLE 2. Epigenetic modulators implemented in protocols for in vitro differentiation into β-like cells.
Transcriptional and Epigenetic (Mis)Regulation of the Endocrine Pancreas
Environmental variables such as lifestyle, diet, and active exercise impact the overall health status of individuals through complex homeostatic mechanisms that involve transcriptional and epigenetic remodeling (Ling and Groop, 2009; Jerram et al., 2017). These variables could be particularly relevant in concomitance with genetic predispositions including haploinsufficiency of key genes regulating pancreatic development and function (Seidman and Seidman, 2002). For example, heterozygous loss of function (LoF) mutations of the pancreatic TFs HNF1α, HNF4α and HNF1β are associated to the maturity-onset diabetes of the young (MODY) (Ryffel, 2001), and the wide spectrum of phenotypes associated to these mutations could result from the integration with non-genetic variables. Work from Huangfu’s laboratory has recently dissected the role played by the pancreatic TFs GATA4 and GATA6 during stepwise differentiation of hESCs toward β-cells. They demonstrated that GATA6 expression is crucial to convert hESCs into DE, while GATA4 is essential for the specification of DE-PGT into PPs through a mechanism not reproducible in in vivo studies performed on mouse models (Shi et al., 2017).
Several reports have highlighted the pivotal role played by key epigenetic modifiers such as Dnmt1, PRC1/PRC2, and MLLs complexes during pancreatic development and how their dysregulation may lead to diabetes progression. For instance, β-cell specific Dnmt3A-KO mice show aberrant expression of major developmental metabolic genes, such as herokinase 1 (hk1) and lactate dehydrogenase A (ldhA), resulting in defective GSIS in the postnatal life (Dhawan et al., 2015). The DNA methylation state of promoters of genes regulating pancreatic specification such as INS, is also important for regulating β-cell mass and functions during aging (Avrahami et al., 2015). Dhawan et al. demonstrated that β-cells-specific DNA methyltransferase Dnmt1 inactivation leads to loss of β-cell mass and concomitant trans-differentiation into α-cells (Dhawan et al., 2011). This plasticity is mediated by the reactivation of the α-cell-restricted TF Arx and simultaneous repression of the β-cell-restricted TF Pax4. These data are coherent with a symmetrical study by Collombat et al. (2009) in which the overexpression of Pax4 drives conversion of α-cells into β-cells (Collombat et al., 2009). Moreover, Talchai et al. showed that β-cell-specific Foxo1 deletion in mice leads to reduced β-cell mass due to induced β-cells dedifferentiation and reactivation of TFs markers of endocrine progenitors (Talchai et al., 2012). More evidence demonstrated that the histone acetyltransferase (HAT) p300 and the histone methyltransferase (HMT) Set9 are responsible for the glucose-dependent insulin gene activation (Andrali et al., 2008). Moreover, combining immunostaining techniques, deep epigenome mapping, and single-cell transcriptomics Lu and colleagues identified shared aberrant chromatin signatures in T2DM patients and diabetic mouse models (Lu et al., 2018). Reduced levels of mature β-cells markers such as PDX1, MAFA, NKX6.1, NKX2.2, and UCN3, are accompanied by a marked H3K27me3 nuclear signal depletion in diabetic versus control islets, a change linked to the loss of PCR2 functions. Analyzing islets obtained from a β-cell restricted KO mouse strain for the embryonic ectoderm development PRC2 subunit (Eed-KO mice), the authors demonstrated a simultaneous shutdown of mature β-cells TFs (Neurod1, NKX6.1, and Mafa), the upregulation of earlier PP markers (i.e., Gata6, Gata4, Onecut2, and Foxa1) and increased levels of the dedifferentiation marker Gli2. These data indicate that PRC2 maintains global silencing in terminally differentiated β-cells and that PCR2-insufficiency contributes to the onset of T2DM (Lu et al., 2018). The transcriptomic and epigenomic analysis of healthy and T2DM pancreatic tissues has recently identified epigenomic and transcriptomic signatures associated to diabetes including impaired distribution of DNA methylation and histone modification marks at key genes regulating β-pancreatic cell maturation (Dayeh et al., 2014; Olsson et al., 2014; Rui et al., 2016). Overall, the accumulated evidence suggests that active epigenetic remodeling is occurring during progression of T2DM and that epigenetic memory-related mechanisms may influence the outcome of the pharmacological treatment to restore normo-glycemia.
Concluding Remarks and Future Perspectives
The increasing prevalence of T1DM and T2DM demands derivation of novel regenerative medicine approaches aimed at reconstituting functional β-cells. Before the ground-breaking invention of the somatic cell reprogramming technique (Takahashi and Yamanaka, 2006; Takahashi et al., 2007), the study of diabetes largely relied on the use of bioptic tissues obtained from post-mortem healthy or diabetic pancreatic tissues and mouse models. The availability of hESC and hiPSC overcame these limitations allowing the study of the onset and progression of diseases “in a dish” and providing an unprecedented cellular in vitro drug discovery platform (Tiscornia et al., 2011; Soldner and Jaenisch, 2012). The derivation of diabetic hPSC-derived β-cells offers a novel tool for the study of the transcriptional and epigenetic dysregulation resulting from the integration of genetic backgrounds and environmental variables. In fact, aberrant deposition of epigenetic marks is a well-known molecular event associated to diabetes progression and epigenetic memory-related mechanisms may influence the outcome of the pharmacological treatments to restore normo-glycemia. For example, T1DM has been linked to dysregulated epigenetic signatures at critical genes involved in self-immunity and regulation of glucose homeostasis (Jayaraman et al., 2013; Zullo et al., 2017) Notably, inhibitors targeting both HATs and histone deacetylases (HDACs) are currently used in T2DM clinical trials (Sommese et al., 2017) and Class-II HDACs inhibitors are particularly promising since they simultaneously exert anti-inflammatory effect, amelioration of insulin sensitivity and enhanced β-cells maturation. The availability of next generation sequencing techniques coupled with single-cell multi-omics analysis offers the unprecedented possibility to obtain a comprehensive understanding of the epigenetic and transcriptional mechanisms regulating pancreagenesis. This could lead to personalized epidrugs–based pharmacological treatments and to improved culture conditions for the production of hPSC-derived β-cells in vitro. Finally, increased epigenetic knowledge would allow exploration of alternative therapeutic approaches such as the conversion of fibroblasts and α-cells to-β cells, a process that can be promoted by the use of 5-AZA-DC (Table 2; Lefebvre et al., 2010; Katz et al., 2013; Pennarossa and Brevini, 2013).
Author Contributions
VA and AA conceived the work and wrote the manuscript.
Funding
This work was supported by KAUST baseline research fund to AA.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the KAUST Environmental Epigenetics Program members, especially Prof. Christian Froekjaer Jensen and Dr. Roberta Pennucci for their thoughtful suggestions and discussions. We regret that several reports could not be cited due to space constraints.
References
Aguayo-Mazzucato, C., Zavacki, A. M., and Diabetes, A. M. (2013). Thyroid hormone promotes postnatal rat pancreatic β-cell development and glucose-responsive insulin secretion through MAFA. Am. Diabetes Assoc. 62,1569–1580. doi: 10.2337/db12-0849/-/DC1
Agulnick, A. D., Ambruzs, D. M., Moorman, M. A., Bhoumik, A., Cesario, R. M., Payne, J. K., et al. (2015). Insulin-Producing endocrine cells differentiated In Vitro from human embryonic stem cells function in macroencapsulation devices In Vivo. Stem Cells Transl. Med. 4, 1214–1222. doi: 10.5966/sctm.2015-0079
Andrali, S. S., Sampley, M. L., Vanderford, N. L., and Ozcan, S. (2008). Glucose regulation of insulin gene expression in pancreatic beta-cells. Biochem. J. 415, 1–10. doi: 10.1042/BJ20081029
Avrahami, D., Li, C., Zhang, J., Schug, J., Avrahami, R., Rao, S., et al. (2015). Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved β cell function. Cell Metabolism 22, 619–632. doi: 10.1016/j.cmet.2015.07.025
Baron, M., Veres, A., Wolock, S. L., Faust, A. L., Gaujoux, R., Vetere, A., et al. (2016). A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 1–35. doi: 10.1016/j.cels.2016.08.011
Bellin, M., Marchetto, M. C., Gage, F. H., and Mummery, C. L. (2012). Induced pluripotent stem cells: the new patient? Nat. Rev. Mol. Cell Biol. 13, 713–726. doi: 10.1038/nrm3448
Bernard, A. B., Lin, C.-C., and Anseth, K. S. (2012). A microwell cell culture platform for the aggregation of pancreatic β-Cells. Tissue Eng. Part C Methods 18, 583–592. doi: 10.1089/ten.tec.2011.0504
Blauwkamp, T. A., Nigam, S., Ardehali, R., Weissman, I. L., and Nusse, R. (2012). Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat. Commun. 3:1070. doi: 10.1038/ncomms2064
Borowiak, M., Maehr, R., Chen, S., Chen, A. E., Tang, W., Fox, J. L., et al. (2009). Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Stem Cell 4, 348–358. doi: 10.1016/j.stem.2009.01.014
Collombat, P., Xu, X., Ravassard, P., Sosa-Pineda, B., Dussaud, S., Billestrup, N., et al. (2009). The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into α and subsequently β Cells. Cell 138, 449–462. doi: 10.1016/j.cell.2009.05.035
D’Amour, K. A., Agulnick, A. D., Eliazer, S., Kelly, O. G., Kroon, E., and Baetge, E. E. (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534–1541. doi: 10.1038/nbt1163
D’Amour, K. A., Bang, A. G., Eliazer, S., Kelly, O. G., Agulnick, A. D., Smart, N. G., et al. (2006). Production of pancreatic hormone–expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 24, 1392–1401. doi: 10.1038/nbt1259
Dayeh, T., Volkov, P., Salö, S., Hall, E., Nilsson, E., Olsson, A. H., et al. (2014). Genome-Wide DNA methylation analysis of human pancreatic islets from Type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 10:e1004160. doi: 10.1371/journal.pgen.1004160
Dhawan, S., Georgia, S., Tschen, S.-I., Fan, G., and Bhushan, A. (2011). Pancreatic β Cell identity is maintained by DNA methylation-mediated repression of Arx. Dev. Cell 20, 419–429. doi: 10.1016/j.devcel.2011.03.012
Dhawan, S., Tschen, S.-I., Zeng, C., Guo, T., Hebrok, M., Matveyenko, A., et al. (2015). DNA methylation directs functional maturation of pancreatic β cells. J. Clin. Invest. 125, 2851–2860. doi: 10.1172/JCI79956
Gadue, P., and Gordon, M. K. (2006). Wnt and TGF-β signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 16806–16811.
Ghazizadeh, Z., Kao, D.-I., Amin, S., Cook, B., Rao, S., Zhou, T., et al. (2017). ROCKII inhibition promotes the maturation of human pancreatic beta-like cells. Nat. Commun. 8, 1–12. doi: 10.1038/s41467-017-00129-y
Gierl, M. S. (2006). The Zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic beta cells and intestinal endocrine cells. Genes Dev. 20, 2465–2478. doi: 10.1101/gad.381806
Gu, G., Dubauskaite, J., and Melton, D. A. (2002). Direct evidence for the pancreatic lineage: NGN3 + cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447–2457.
Guo, S., Dai, C., Guo, M., Taylor, B., Harmon, J. S., Sander, M., et al. (2013). Inactivation of specific β cell transcription factors in type 2 diabetes. J. Clin. Invest. 123, 3305–3316. doi: 10.1172/JCI65390
Hakim, F., Kaitsuka, T., Raeed, J. M., Wei, F.-Y., Shiraki, N., Akagi, T., et al. (2014). High oxygen condition facilitates the differentiation of mouse and human pluripotent stem cells into pancreatic progenitors and insulin-producing cells. J. Biol. Chem. 289, 9623–9638. doi: 10.1074/jbc.M113.524363
Hay, D. C., Zhao, D., Fletcher, J., Hewitt, Z. A., McLean, D., Urruticoechea-Uriguen, A., et al. (2008). Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development In Vivo. Stem Cells 26, 894–902. doi: 10.1634/stemcells.2007-0718
Herrera, P. L., Nepote, V., and Delacour, A. (2002). Pancreatic cell lineage analyses in mice. Endocrine 19, 267–278.
Hua, H., Shang, L., Martinez, H., Freeby, M., Gallagher, M. P., Ludwig, T., et al. (2013). iPSC-derived β cells model diabetes due to glucokinase deficiency. J. Clin. Invest. 123, 3146–3153. doi: 10.1172/JCI67638
Jansson, E. T., Comi, T. J., Rubakhin, S. S., and Sweedler, J. V. (2016). Single cell peptide heterogeneity of rat islets of langerhans. ACS Chem. Biol. 11, 2588–2595. doi: 10.1021/acschembio.6b00602
Jayaraman, S., Patel, A., Jayaraman, A., Patel, V., Holterman, M., and Prabhakar, B. (2013). Transcriptome analysis of epigenetically modulated genome indicates signature genes in manifestation of type 1 diabetes and its prevention in NOD mice. PLoS One 8:e55074. doi: 10.1371/journal.pone.0055074
Jennings, R. E., Berry, A. A., Kirkwood-Wilson, R., Roberts, N. A., Hearn, T., Salisbury, R. J., et al. (2013). Development of the human pancreas from foregut to endocrine commitment. Diabetes Metab. Res. Rev. 62, 3514–3522. doi: 10.2337/db12-1479
Jerram, S. T., Dang, M. N., and Leslie, R. D. (2017). The role of epigenetics in Type 1 diabetes. Curr. Diab. Rep. 17, 1–11. doi: 10.1007/s11892-017-0916-x
Jia, S., Ivanov, A., Blasevic, D., Muller, T., Purfurst, B., Sun, W., et al. (2015). Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic -cell function. EMBO J. 34, 1417–1433. doi: 10.15252/embj.201490819
Kahn, C. R. (1994). Banting Lecture. Insulin action, diabetogenes, and the cause of type II diabetes. Diabetes 43, 1066–1084. doi: 10.2337/diab.43.8.1066
Katsarou, A., Gudbjörnsdottir, S., Rawshani, A., Dabelea, D., Bonifacio, E., Anderson, B. J., et al. (2017). Type 1 diabetes mellitus. Nat. Rev. Dis. Primers 3:17016. doi: 10.1038/nrdp.2017.16
Katz, L. S., Geras-Raaka, E., and Gershengorn, M. C. (2013). Reprogramming adult human dermal fibroblasts to islet-like cells by epigenetic modification coupled to transcription factor modulation. Stem Cells Dev. 22, 2551–2560. doi: 10.1089/scd.2013.0134
Kondo, Y., Iwao, T., Yoshihashi, S., Mimori, K., Ogihara, R., Nagata, K., et al. (2014). Histone deacetylase inhibitor valproic acid promotes the differentiation of human induced pluripotent stem cells into hepatocyte-like cells. PLoS One 9:e104010. doi: 10.1371/journal.pone.0104010
Korostylev, A., Mahaddalkar, P. U., Keminer, O., Hadian, K., Schorpp, K., Gribbon, P., et al. (2017). A high-content small molecule screen identifies novel inducers of definitive endoderm. Mol. Metabolism 6, 640–650. doi: 10.1016/j.molmet.2017.04.009
Kroon, E., Martinson, L. A., Kadoya, K., Bang, A. G., Kelly, O. G., Eliazer, S., et al. (2008). Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 26, 443–452. doi: 10.1038/nbt1393
Lefebvre, B., Belaich, S., Longue, J., Vandewalle, B., Oberholzer, J., Gmyr, V., et al. (2010). 5′-AZA induces Ngn3 expression and endocrine differentiation in the PANC-1 human ductal cell line. Biochem. Biophys. Res. Commun. 391, 305–309. doi: 10.1016/j.bbrc.2009.11.054
Ling, C., and Groop, L. (2009). Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes Metab. Res. Rev. 58,2718–2725. doi: 10.2337/db09-1003
Lu, T. T.-H., Heyne, S., Dror, E., Casas, E., Leonhardt, L., Boenke, T., et al. (2018). The polycomb-dependent epigenome controls β cell dysfunction, dedifferentiation, and diabetes. Cell Metabolism 27, 1294.e7–1308.e7. doi: 10.1016/j.cmet.2018.04.013
Luo, H., Shenoy, A. K., Li, X., Jin, Y., Jin, L., Cai, Q., et al. (2016). MOF acetylates the histone demethylase LSD1 to suppress epithelial-to-mesenchymal transition. CellReports 15, 1–27. doi: 10.1016/j.celrep.2016.05.050
Manzar, G. S., Kim, E.-M., and Zavazava, N. (2017). Demethylation of induced pluripotent stem cells from type 1 diabetic patients enhances differentiation into functional pancreatic β cells. J. Biol. Chem. 292, 14066–14079. doi: 10.1074/jbc.M117.784280
Marín-Peñalver, J. J., Martín-Timón, I., and del Cañizo-Gómez, F. J. (2016). Management of hospitalized type 2 diabetes mellitus patients. J. Transl. Int. Med. 4, 155–161. doi: 10.1515/jtim-2016-0027
Miller, B. R., and Nguyen, H. (2014). New and emerging drugs and targets for Type 2 diabetes. Am. Health drug Benefits 7, 452–463.
Millman, J. R., Xie, C., Van Dervort, A., Gürtler, M., Pagliuca, F. W., and Melton, D. A. (2016). Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat. Commun. 7, 11463–11468. doi: 10.1038/ncomms11463
Muraro, M. J., Dharmadhikari, G., Grün, D., Groen, N., Dielen, T., Jansen, E., et al. (2016). A single-cell transcriptome atlas of the human pancreas. Cell Systems 3, 385.e3–394.e3. doi: 10.1016/j.cels.2016.09.002
Nostro, M. C., Sarangi, F., Yang, C., Holland, A., Elefanty, A. G., Stanley, E. G., et al. (2015). Efficient generation of NKX6-1 + pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Rep. 4, 591–604. doi: 10.1016/j.stemcr.2015.02.017
Olsson, A. H., Volkov, P., Bacos, K., Dayeh, T., Hall, E., Nilsson, E. A., et al. (2014). Genome-Wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 10:e1004735. doi: 10.1371/journal.pgen.1004735
Pagliuca, F. W., Millman, J. R., Gürtler, M., Segel, M., Van Dervort, A., Ryu, J. H., et al. (2014). Generation of functional human pancreatic β cells In Vitro. Cell 159, 428–439. doi: 10.1016/j.cell.2014.09.040
Paris, M., Tourrel-Cuzin, C., Plachot, C., and Ktorza, A. (2004). Review: pancreatic β-Cell neogenesis revisited. Exp. Diabesity Res. 5, 111–121. doi: 10.1080/15438600490455079
Pedersen, J. K., Nelson, S. B., Jorgensen, M. C., Henseleit, K. D., Fujitani, Y., Wright, C. V. E., et al. (2005). Endodermal expression of Nkx6 genes depends differentially on Pdx1. Dev. Biol. 288, 487–501. doi: 10.1016/j.ydbio.2005.10.001
Pennarossa, G., and Brevini, T. A. L. (2013). Brief demethylation step allows the conversion of adult human skin fibroblasts into insulin-secreting cells. Proc. Natl. Acad. Sci. U.S.A. 110, 1–6. doi: 10.1073/pnas.1220637110
Pickup, J. C. (2012). Insulin-Pump therapy for type 1 diabetes mellitus. N. Engl. J. Med. 366, 1616–1624. doi: 10.1056/NEJMct1113948
Rezania, A., Bruin, J. E., Arora, P., Rubin, A., Batushansky, I., Asadi, A., et al. (2014). Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133. doi: 10.1038/nbt.3033
Rezania, A., Bruin, J. E., Riedel, M. J., Mojibian, M., and Diabetes, A. A. (2012). Maturation of human embryonic stem cell–derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Am. Diabetes Assoc. 61, 2016–2029. doi: 10.2337/db11-1711/-/DC1
Rosa, G., and German, M. (2004). Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc. Natl. Acad. Sci. U.S.A.101, 1–6.
Rui, J., Deng, S., Lebastchi, J., Clark, P. L., Usmani-Brown, S., and Herold, K. C. (2016). Methylation of insulin DNA in response to proinflammatory cytokines during the progression of autoimmune diabetes in NOD mice. Diabetologia 59, 1021–1029. doi: 10.1007/s00125-016-3897-4
Russ, H. A., Parent, A. V., Ringler, J. J., Hennings, T. G., Nair, G. G., Shveygert, M., et al. (2015). Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 34, 1759–1772. doi: 10.15252/embj.201591058
Ryffel, G. U. (2001). Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 27, 11–29.
Sampaziotis, F., de Brito, M. C., Geti, I., Bertero, A., Hannan, N. R., and Vallier, L. (2017). Directed differentiation of human induced pluripotent stem cells into functional cholangiocyte-like cells. Nat. Protoc. 12, 814–827. doi: 10.1038/nprot.2017.011
Schaffer, A. E., Freude, K. K., Nelson, S. B., and Sander, M. (2010). Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev. Cell 18, 1022–1029. doi: 10.1016/j.devcel.2010.05.015
Segerstolpe, A., Palasantza, A., Eliasson, P., Andersson, E.-M., Andréasson, A.-C., Sun, X., et al. (2016). Single-Cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism 24, 593–607. doi: 10.1016/j.cmet.2016.08.020
Seidman, J. G., and Seidman, C. (2002). Transcription factor haploinsufficiency: when half a loaf is not enough. J. Clin. Invest. 109, 451–455. doi: 10.1172/JCI15043
Shi, Z.-D., Lee, K., Yang, D., Amin, S., Verma, N., Li, Q. V., et al. (2017). Genome editing in hPSCs reveals GATA6 haploinsufficiency and a genetic interaction with GATA4 in human pancreatic development. Stem Cell 20, 1–21. doi: 10.1016/j.stem.2017.01.001
Shih, H. P., Seymour, P. A., Patel, N. A., Xie, R., Wang, A., Liu, P. P., et al. (2015). A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Reports 13, 326–336. doi: 10.1016/j.celrep.2015.08.082
Soldner, F., and Jaenisch, R. (2012). Medicine. iPSC disease modeling. Science 338, 1155–1156. doi: 10.1126/science.1227682
Sommese, L., Zullo, A., Mancini, F. P., Fabbricini, R., Soricelli, A., and Napoli, C. (2017). Clinical relevance of epigenetics in the onset and management of type 2 diabetes mellitus. Epigenetics 12, 1–15. doi: 10.1080/15592294.2016.1278097
Spitzer, M. H., and Nolan, G. P. (2016). Mass cytometry: single cells. many features. Cell 165, 780–791. doi: 10.1016/j.cell.2016.04.019
Stolovich-Rain, M., Enk, J., Vikesa, J., Nielsen, F. C., Saada, A., Glaser, B., et al. (2015). Weaning triggers a maturation step of pancreatic β cells. Dev. Cell 32, 535–545. doi: 10.1016/j.devcel.2015.01.002
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. doi: 10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Takeuchi, H., Nakatsuji, N., and Suemori, H. (2014). Endodermal differentiation of human pluripotent stem cells to insulin-producing cells in 3D culture. Sci. Rep. 4, 230–239. doi: 10.1038/srep04488
Talchai, C., Xuan, S., Lin, H. V., Sussel, L., and Accili, D. (2012). Pancreatic β Cell dedifferentiation as a mechanism of diabetic β cell failure. Cell 150, 1223–1234. doi: 10.1016/j.cell.2012.07.029
Tanaka, H., Tanaka, S., Sekine, K., Kita, S., Okamura, A., Takebe, T., et al. (2013). The generation of pancreatic β-cell spheroids in a simulated microgravity culture system. Biomaterials 34, 5785–5791. doi: 10.1016/j.biomaterials.2013.04.003
Taylor, B. L., Liu, F.-F., and Sander, M. (2013). Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Reports 4, 1262–1275. doi: 10.1016/j.celrep.2013.08.010
Tchieu, J., Zimmer, B., Fattahi, F., Amin, S., Zeltner, N., Chen, S., et al. (2017). A Modular platform for differentiation of human pscs into all major ectodermal lineages. Stem Cell 21, 399.e7–410.e7. doi: 10.1016/j.stem.2017.08.015
Tiscornia, G., Vivas, E. L., and Belmonte, J. C. I. U. A. (2011). Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat. Med. 17, 1570–1575. doi: 10.1038/nm.2504
Toyoda, T., Kimura, A., Tanaka, H., Ameku, T., Mima, A., Hirose, Y., et al. (2017). Rho-Associated kinases and non-muscle myosin IIs inhibit the differentiation of human iPSCs to pancreatic endoderm. Stem Cell Reports 9, 1–21. doi: 10.1016/j.stemcr.2017.07.005
Vegas, A. J., Veiseh, O., Gürtler, M., Millman, J. R., Pagliuca, F. W., Bader, A. R., et al. (2016). Long-term glycemic control using polymer-encapsulated human stem cell–derived beta cells in immune-competent mice. Nat. Med. 22, 306–311. doi: 10.1038/nm.4030
Wang, A., Yue, F., Li, Y., Xie, R., Harper, T., Patel, N. A., et al. (2015). Epigenetic priming of enhancers predicts developmental competence of hESC-Derived endodermal lineage intermediates. Stem Cell 16, 386–399. doi: 10.1016/j.stem.2015.02.013
Wang, W., Jin, S., and Ye, K. (2017). Development of islet organoids from H9 human embryonic stem cells in biomimetic 3D Scaffolds. Stem Cells Dev. 26, 394–404. doi: 10.1089/scd.2016.0115
Wang, Y. J., Golson, M. L., Schug, J., Traum, D., Liu, C., Vivek, K., et al. (2016). Single-Cell mass cytometry analysis of the human endocrine pancreas. Cell Metabolism 24, 616–626. doi: 10.1016/j.cmet.2016.09.007
Wescott, M. P., Rovira, M., and Reichert, M. (2009). Pancreatic ductal morphogenesis and the Pdx1 homeodomain transcription factor. Am. Soc. Cell Biol. 20, 4838–4844. doi: 10.1091/mbc.E09
Xie, R., Everett, L. J., Lim, H.-W., Patel, N. A., Schug, J., Kroon, E., et al. (2013). Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Stem Cell 12,224–237. doi: 10.1016/j.stem.2012.11.023
Yoshihara, E., Wei, Z., Lin, C. S., Fang, S., Ahmadian, M., Kida, Y., et al. (2016). ERRγ is required for the metabolic maturation of therapeutically functional glucose-responsive β Cells. Cell Metabolism 23, 622–634. doi: 10.1016/j.cmet.2016.03.005
Yuan, Y., Hartland, K., Boskovic, Z., Wang, Y., Walpita, D., Lysy, P. A., et al. (2013). A small-molecule inducer of PDX1 expression identified by high-throughput screening. Chem. Biol. 20, 1513–1522. doi: 10.1016/j.chembiol.2013.10.013
Zeng, C., Mulas, F., Sui, Y., Guan, T., Miller, N., Tan, Y., et al. (2017). Pseudotemporal ordering of single cells reveals metabolic control of postnatal β cell proliferation. Cell Metabolism 25, 1160.e11–1175.e11. doi: 10.1016/j.cmet.2017.04.014
Zeng, H., Guo, M., Zhou, T., Tan, L., Chong, C. N., Zhang, T., et al. (2016). An isogenic human ESC platform for functional evaluation of genome-wide-association-study- identified diabetes genes and drug discovery. Stem Cell 19, 326–340. doi: 10.1016/j.stem.2016.07.002
Zhu, Z., Li, Q. V., Lee, K., Rosen, B. P., González, F., Soh, C.-L., et al. (2016). Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Stem Cell 18, 755–768. doi: 10.1016/j.stem.2016.03.015
Keywords: β-cells, epigenetics, diabetes, in vitro differentiation, regenerative medicine
Citation: Astro V and Adamo A (2018) Epigenetic Control of Endocrine Pancreas Differentiation in vitro: Current Knowledge and Future Perspectives. Front. Cell Dev. Biol. 6:141. doi: 10.3389/fcell.2018.00141
Received: 20 July 2018; Accepted: 04 October 2018;
Published: 25 October 2018.
Edited by:
Beatrice Bodega, Istituto Nazionale Genetica Molecolare (INGM), ItalyReviewed by:
Zong Wei, Salk Institute for Biological Studies, United StatesFrancesco Rusconi, Università degli Studi di Milano, Italy
Copyright © 2018 Astro and Adamo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio Adamo, YW50b25pby5hZGFtb0BrYXVzdC5lZHUuc2E=
 Veronica Astro
Veronica Astro Antonio Adamo
Antonio Adamo