
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Cell Dev. Biol. , 15 October 2018
Sec. Stem Cell Research
Volume 6 - 2018 | https://doi.org/10.3389/fcell.2018.00124
This article is part of the Research Topic Perspectives in Zebrafish Research View all 16 articles
Hematopoiesis is an optimal system for studying stem cell maintenance and lineage differentiation under physiological and pathological conditions. In vertebrate organisms, billions of differentiated hematopoietic cells need to be continuously produced to replenish the blood cell pool. Disruptions in this process have immediate consequences for oxygen transport, responses against pathogens, maintenance of hemostasis and vascular integrity. Zebrafish is a widely used and well-established model for studying the hematopoietic system. Several new hematopoietic regulators were identified in genetic and chemical screens using the zebrafish model. Moreover, zebrafish enables in vivo imaging of hematopoietic stem cell generation and differentiation during embryogenesis, and adulthood. Finally, zebrafish has been used to model hematopoietic diseases. Recent technological advances in single-cell transcriptome analysis, epigenetic regulation, proteomics, metabolomics, and processing of large data sets promise to transform the current understanding of normal, abnormal, and malignant hematopoiesis. In this perspective, we discuss how the zebrafish model has proven beneficial for studying physiological and pathological hematopoiesis and how these novel technologies are transforming the field.
Over the past three decades, zebrafish has been established as an important model to study various biological processes during development and homeostasis, including hematopoiesis. Many attractive features underpin the success of zebrafish as a model for vertebrate hematopoiesis. Cell-intrinsic and -extrinsic signaling mechanisms in hematopoiesis are well conserved between zebrafish and mammals, with the exception of a few hematopoietic niche components (Liao et al., 1998; Murayama et al., 2006; Bertrand and Traver, 2009; Paik and Zon, 2010; Goessling and North, 2011; Zhang and Liu, 2011; Zhang et al., 2013; Frame et al., 2017; Nik et al., 2017; Gore et al., 2018). Moreover, zebrafish embryos are small and transparent so they are ideal for imaging and easy to manipulate, at low cost. Additionally, genetic manipulation is easy and population studies can be easily performed in zebrafish. Thus, zebrafish have become invaluable vertebrate models for robust large-scale genetic screens (Mullins et al., 1994; Driever et al., 1996; Amsterdam et al., 1999) and, more recently, high-throughput chemical compound screens (North et al., 2007; Yeh et al., 2009). However, there are certain disadvantages in the zebrafish model. For example, zebrafish is not a mammal, but rather a poikilothermic animal in which the development of embryos occurs outside of the animal body and without placenta. That may lead to many metabolic and other differences between zebrafish and mammals, including drug action and utilization. Finally, the zebrafish genome is duplicated and thus many genes have paralogs and homologs that make the otherwise easy genetic manipulation complicated (Glasauer and Neuhauss, 2014).
Embryonic hematopoiesis in zebrafish is a multistep process occurring in a spatially restricted manner in three distinct waves. During the intraembryonic primitive wave, the medial and anterior lateral mesoderm give rise to erythroid and myeloid cells, respectively. Erythro-myeloid progenitors (EMPs) form in the posterior blood island (PBI) during a transient intermediate wave. Finally, during the definitive wave, hematopoietic stem cells (HSC) with multilineage capacity originate in the aorta-gonad-mesonephros (AGM) region. The HSCs then translocate to and expand in the caudal hematopoietic tissue (CHT), which is followed by the colonization of the kidney and the thymus (Figure 1). Interestingly, it was recently discovered that HSC-independent T-cells can originate from the AGM and PBI during the embryonic and larval stages of development (Tian et al., 2017).
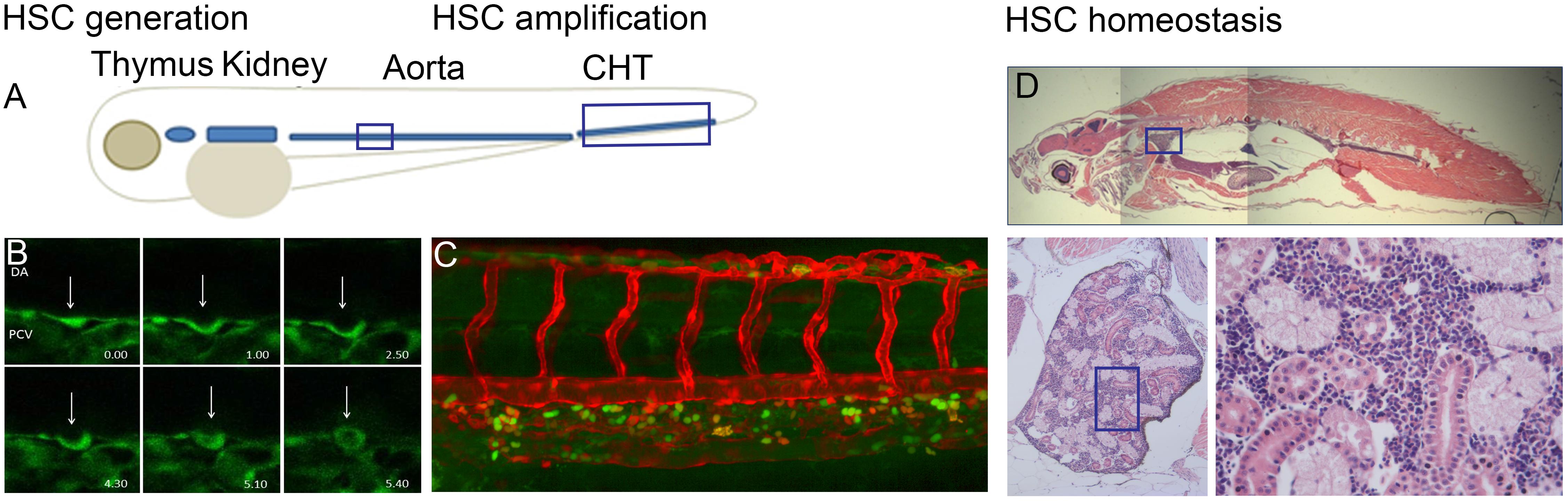
FIGURE 1. (A) Overview of definitive hematopoietic sites in the developing embryo where HSPCs are born from hemogenic endothelial cells of the dorsal aorta (DA). HSPCs are amplified in the caudal hematopoietic tissue (CHT) and migrate to the kidney and thymus. (B) Endothelial-to-hematopoietic transition (EHT) event imaged in a Tg(fli:GFP) embryo between 32 and 40 hpf. (C) CHT region in Tg(flt:RFP)/Tg(CD41:GFP) embryo at 56 hpf indicating erythroid myeloid progenitors (EMP) in green and definitive HSPCs in yellow as they originate from the artery and retain RFP at this timepoint. (D) Sagittal section through an adult zebrafish where the head kidney is indicated with enlargement showing the hematopoietic cells in between the kidney tubules.
Zebrafish has been extensively used for modeling human hematopoietic disease, including anemia, thrombocytopenia, bone marrow failure syndromes, leukemia, and lymphoma (Taylor and Zon, 2011; Kwan and North, 2017; Potts and Bowman, 2017; Gore et al., 2018). The first transplantable leukemia modeled in zebrafish was T-cell acute lymphoblastic leukemia (T-ALL), which was induced by T cell-specific c-Myc overexpression (Langenau et al., 2003). Thereafter, several models of myelodysplastic syndromes and myeloproliferative neoplasms have been described (Le et al., 2007; He et al., 2014; Gjini et al., 2015; Peng et al., 2015).
Although for many years zebrafish was mainly used to study embryonic and larval developmental hematopoiesis, recent technological advances have transformed the field. In this perspective, we will briefly discuss how research using zebrafish genetic models in combination with chemical screens, high-end imaging, and genome-wide molecular, metabolics and proteomics approaches has contributed to our understanding of hematopoiesis.
The transparency and accessibility of zebrafish embryos was pivotal to collect evidence showing that hematopoietic stem and progenitor cells (HSPCs) emerge from the ventral wall of the dorsal aorta in vivo (Bertrand et al., 2010; Kissa and Herbomel, 2010). Moreover, high-end imaging techniques in zebrafish embryos uncovered the mechanisms of thymus development (Hess and Boehm, 2012) and revealed that HSPCs are amplified and interact with endothelial cells in the CHT (Tamplin et al., 2015). Multiple signaling pathways and cell-interactions affect HSPC emergence. For instance, inflammatory signaling provided by neutrophils is required for HSC generation (Espin-Palazon et al., 2014; Li et al., 2014; Sawamiphak et al., 2014; He et al., 2015). These unique properties of zebrafish allowed to uncover the role of macrophages in HPSC mobilization and definitive hematopoiesis (Travnickova et al., 2015). Where transient and rapid cell-interactions occur, light-sheet microscopy, SPIM (selective plane illumination) or spinning disk microscopy can be used to visualize these processes in vivo in embryos and adults, because these systems record time-lapse 3D fluorescent images 100–1000x faster than conventional confocal microscopy (Inoue and Inoue, 1996; Huisken et al., 2004; Arrenberg et al., 2010). Moreover, transparent adult zebrafish models (White et al., 2008) have enabled the imaging of adult hematopoiesis, thereby opening the way to research exploring different HSC niche components and HSC-niche interactions in the adult kidney marrow, thymus, and spleen.
The development of single-cell RNA-sequencing (scRNA-seq) revolutionized the way we understand hematopoiesis. As most cellular compartments have a certain degree of heterogeneity, with bulk RNA-seq one cannot distinguish between a small transcriptional difference in many cells, and a large transcriptional difference in a few cells. Several insightful reviews describe the different methods used for single-cell RNA-seq (Kolodziejczyk et al., 2015; Ziegenhain et al., 2017; Dal Molin and Di Camillo, 2018).
In zebrafish, one of the first methods used to characterize the transcriptome of single cells was massive parallel qPCR, where up to 96 transcripts could be analyzed in great sequencing depth using the Fluidigm system. This method revealed two distinct sub-populations of HSPCs in the CD41-GFP low-expressing stem cell compartment of the adult kidney marrow. Moreover, by using this technique and genetic ablation of T cells, a previously uncharacterized hematopoietic cytotoxic T/NK cell population in zebrafish was uncovered (Moore et al., 2016).
Recent technological advances in scRNA-seq have enabled analyses without restriction to specific transcripts. A re-examination of the CD41-GFPlow population revealed four HSPC sub-populations with different cellular characteristics and potential novel markers for HSCs were uncovered. Importantly, some cells in these subpopulations expressed the thrombocyte differentiation program long before they would have been characterized as thrombocytes, showing that there is an early lineage bias (Guo et al., 2013; Buenrostro et al., 2015; Paul et al., 2015; Drissen et al., 2016; Grover et al., 2016; Nestorowa et al., 2016; Olsson et al., 2016; Alberti-Servera et al., 2017; Velten et al., 2017; Villani et al., 2017; Buenrostro et al., 2018; Dahlin et al., 2018). In addition, scRNA-seq analyses in various transgenic lines revealed that ribosomal genes and lineage regulators control hematopoietic differentiation (Athanasiadis et al., 2017) and uncovered several novel hematopoietic populations, including two new types of NK cells (Tang et al., 2017). Finally, elegant comparative evolutionary studies on LCK-GFP transgenic zebrafish and mammals showed that membrane proteins are less conserved in NK cells than in T cells (Carmona et al., 2017). In the pathological context, scRNA-seq analysis of Myc-induced T-ALLs demonstrated that few cells expressed an immature stem cell program, suggesting that only a small proportion of leukemia cells promote the disease. This is remarkable as a single transgenic approach was used to initiate leukemogenesis and all leukemia cells overexpress Myc (Moore et al., 2016).
Zebrafish has traditionally been utilized to lineage-trace differentiation during embryonic stages by labeling single cells with dyes and following them throughout development, until the dye fades or dilutes. However, the recent development of complex genetic models has removed this time restriction and enabled lineage tracing from the embryo into adulthood. For instance, HSPCs generated from the hemogenic endothelium of the aorta have been lineage-traced by using the multicolor transgenic labeling system “blood bow” (Henninger et al., 2017) in combination with high-end imaging and fluorescence-activated cell sorting (FACS). Additionally, labeling with CRISPR/Cas9 scarring in embryos and tracing of unique hematopoietic clones into adulthood has revealed that the hematopoietic system is only generated from a handful of cells present at dome stage (Alemany et al., 2018). This study claimed that all clones contribute to all blood lineages, a subject that is controversial in mammalian studies (Yamamoto et al., 2013; Notta et al., 2016; Pei et al., 2017).
A different approach for lineage-tracing cells consists of performing high-throughput scRNAseq at various developmental stages and then mapping similarities in transcriptional profiles across a pseudo timescale of differentiation (Macosko et al., 2015). By using this method in early embryogenesis, two independent studies have described gradually divergent differentiation patterns for specific lineages and uncovered signaling networks required for zebrafish development (Farrell et al., 2018; Wagner et al., 2018).
Future studies combining scRNAseq with lineage tracing will be paramount to advance our understanding of the developmental origins of hematopoietic populations. However, this approach has the important caveat that scRNA-seq does not provide topographic information for each individual cell. To overcome this limitation, the Van Oudernaarden and Bakkers laboratories have developed RNA-tomography (TOMOSEQ), a method that combines traditional histological techniques with low-input RNA sequencing and mathematical image reconstruction (Junker et al., 2014).
Zebrafish is an ideal vertebrate model system to conduct bio-reactive compound screens (Zon and Peterson, 2005; Cusick et al., 2012; Tamplin et al., 2012; Veinotte et al., 2014; Rennekamp and Peterson, 2015; Deveau et al., 2017). The animals are small-sized and lay hundreds of eggs that develop very rapidly, thereby allowing the monitoring of compound activity and biotoxicity in vivo across development. Such screens have led to the identification of prostaglandin E2 as a compound that increases HSC production (North et al., 2007). Prostaglandin E2 is currently being investigated for HSC expansion applications in human and non-human primates (Goessling et al., 2011; Cutler et al., 2013).
Important insights into the molecular regulation of T-ALL came from zebrafish studies where immature T cells served as models for T-ALL cells. By screening small molecules for an effect on immature T cells using LCK-GFP transgenic zebrafish, a novel compound, 1H-indole-3-carbaldehyde quinolin-8-yl-hydrazone, named Lenaldekar, was identified with the potential to specifically attack T-ALL cells (Ridges et al., 2012). Lenaldekar also has a potential effect against autoimmune diseases such as multiple sclerosis, as they are caused by an off-target activity of T cells (Cusick et al., 2012). Currently there are ongoing clinical trials to study the effectiveness of this promising compound. These examples highlight the power of zebrafish models for screening novel chemical compounds affecting normal, abnormal or malignant hematopoiesis (Shafizadeh et al., 2004; Yeh et al., 2009; Paik et al., 2010; Gutierrez et al., 2014; Arulmozhivarman et al., 2016).
Future studies addressing malignancy heterogeneity may combine chemical screens with scRNAseq to identify therapy-resistant cells and explore the mechanisms underpinning resistance to treatment in individual cells, a fundamental unresolved question in the cancer research field.
Several acute myeloid leukemia (AML) predisposition syndromes are caused by innate mutations in transcription factors that affect embryonic hematopoiesis, such as Gata2 and Runx1 (Babushok et al., 2016), suggesting that perturbations in embryonic hematopoiesis affect the adult HSC compartment. As the effects of alterations in embryonic hematopoiesis can be easily monitored in zebrafish throughout development, as well as during adulthood, this is an excellent system to study AML predisposition syndromes. Until the recent development of targeted gene editing, manipulating the zebrafish genome to create specific mutations for making knockout and knockin animals was challenging. Although TILLING (Targeting Induced Local Lesions in Genomes) was a significant advancement, this is a costly method that requires thousands of fish to search for a STOP codon in the gene of interest. Moreover, TILLING is rather limiting as it does not allow to induce specific mutations (Wienholds et al., 2003; Draper et al., 2004). Targeting the zebrafish genome with zinc-finger nucleases was the beginning of a new era in zebrafish biology, as selected genes could finally be specifically targeted for genome editing (Amacher, 2008; Foley et al., 2009). Shortly after this technology was introduced, TALENS (Dahlem et al., 2012; Hwang et al., 2014; Huang et al., 2016; Liu et al., 2016), and more recently, CRISPR/Cas9 (Hruscha et al., 2013; Irion et al., 2014; Shah et al., 2015; Li et al., 2016; Liu et al., 2017) were developed. Whilst it is relatively easy to generate knockouts and large deletions with these gene-targeting techniques, making knockin animals remains challenging. Nevertheless, several laboratories have successfully created knockin animals by using CRISPR/Cas9 and co-injecting a repair template to facilitate homology-directed repair (Hruscha et al., 2013; Auer et al., 2014; Albadri et al., 2017; Kesavan et al., 2017). Additionally, Cre/lox, Flp/FRT, and ΦC31 systems are also currently being used in zebrafish for precise genome editing (Mosimann et al., 2013; Felker and Mosimann, 2016; Carney and Mosimann, 2018). Importantly, tissue-specific expression of Cas9 in the hematopoietic system can be performed in zebrafish to enable conditional manipulation of hematopoietic cells (Ablain et al., 2015). A major caveat in both perturbing the zebrafish genome and comparing the zebrafish with the mammalian transcriptome in the context of clinical translation, is, as previously mentioned, the Teleost genome duplication (Glasauer and Neuhauss, 2014). As a result most genes are present twice with (partially) redundant biological roles. This means that for a complete perturbation of a mammalian gene, the zebrafish counterparts have to be removed both, complicating genetic crossings and analyses.
Chromatin conformation is essential for controlling gene expression, and deregulation of this process may cause malignant transformation (Groschel et al., 2014). Zebrafish is an excellent system to explore the mechanisms underlying chromatin regulation and to evaluate the effects of chromatin-modifying drugs in vivo. Gene regulatory elements can be identified in zebrafish using chromatin immunoprecipitation combined with sequencing (ChIP-seq), however, the technique is limited by the low number of zebrafish-specific antibodies currently available and the large amount of input material required (Havis et al., 2006; Trompouki et al., 2011; Bogdanovic et al., 2013). ChIP-seq has been mostly used in early zebrafish embryos (Paik et al., 2010; Vastenhouw et al., 2010; Bogdanovic et al., 2012; Xu et al., 2012; Winata et al., 2013; Nelson et al., 2017; Meier et al., 2018). Antibodies against histone marks, which are highly conserved between species, have been successfully utilized in zebrafish erythrocytes to describe the potential locus control region (LCR) regulating globin expression (Ganis et al., 2012). Moreover, given the functional conservation of these genes, zebrafish is useful to functionally validate enhancers identified in mouse and/or human models (Tijssen et al., 2011; Chiang et al., 2017).
Other techniques for identifying gene regulatory elements are based on the detection of open chromatin, for instance, assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq). ATAC-seq requires much less input material than ChIP-seq, and has even been used successfully with single cells (Fernandez-Minan et al., 2016; Doganli et al., 2017). This method allowed the identification of endothelial enhancers (Quillien et al., 2017) and revealed the role of cohesin in rearranging the genomic architecture during the transition of maternal to zygotic transcription in early embryos (Meier et al., 2018). Combining scRNA-seq with ATAC-seq and immune phenotypic analysis is a powerful approach to integrate our understanding of lineage differentiation with the regulatory elements involved in that process (Buenrostro et al., 2018). DNA methylation studies can also be used to understand chromatin accessibility, although more material is needed in these methods. Methylation experiments have been conducted in zebrafish albeit not specifically in the hematopoietic system (Lee et al., 2015; Kaaij et al., 2016). Since many tissue-specific fluorescent lines exist in zebrafish, future research should aim to identify enhancers and promoters in specific cell types, rather than using whole embryos.
Despite the advantages of ATAC-seq and methylation analyses, these approaches cannot offer the same information as ChIP-seq. Thus, improved ChIP-seq protocols, such as the high sensitivity indexing-first chromatin immunoprecipitation approach (iChIP) developed in Ido Amit’s laboratory, should be adapted to zebrafish (Gury-BenAri et al., 2016). Moreover, it would be important to unravel chromatin interactions in active enhancer and promoter regions during hematopoiesis. However, although chromatin conformation has been studied in early zebrafish embryos (Gomez-Marin et al., 2015; Fernandez-Minan et al., 2016), to date no studies have addressed this question specifically in zebrafish hematopoiesis. It is important to mention the combined effort of many groups to collate all available genome-wide data in zebrafish in the DANIO-CODE Data Coordination Center1 (Tan et al., 2016). This recently launched database will provide an easy access to high-quality genome data to all scientists.
In the era of genome-wide technology, gene expression studies should be complemented with proteomic studies, as transcriptional and translational outcomes can sometimes differ. Additionally, the extension of these analyses to metabolomics may uncover another layer of regulation critical for hematopoiesis. Indeed, it was recently shown that dormant stem cell populations have low metabolic activity, and this is required to maintain the hematopoietic system during aging and periods of intense stress (Cabezas-Wallscheid et al., 2017). Although proteomics and metabolomics methods have not yet been extensively explored in zebrafish, particularly in the hematopoietic system, some studies have reported differences between transcript and protein levels in multiple genes by using proteomic analyses either in whole zebrafish embryos or in specific cell populations during regeneration (Alli Shaik et al., 2014; Baral et al., 2014; Rabinowitz et al., 2017). Metabolomics has proven useful to understand the neurological damage resulting from chemical perturbations in zebrafish embryos (Ong et al., 2009; Rabinowitz et al., 2017; Roy et al., 2017). Finally, as mass spectrometry analyses are constantly improving, the sensitivity of these methods will likely overcome the current problem of heterogeneous and low cell-number populations.
The zebrafish has become an invaluable model system for understanding how HSCs form and are maintained, and how hematopoietic cell differentiation is regulated during embryogenesis and in adulthood. The unique advantages offered by this model system over traditional mouse models regarding the use in chemical screens and the accessibility during embryonic stages allowing easy manipulation and visualization and tracing into adult stages, in combination with recent new technologies (Figure 2), have opened the way for novel exciting hypotheses on the mechanisms promoting hematopoietic diseases, the role of the niche in normal and malignant hematopoiesis, and the effect of chemical compounds on malignant cells. The high conservation between the zebrafish and human hematopoietic systems means that discoveries in fish may have strong translational potential and important clinical implications for the treatment of hematopoietic diseases.
EdP and ET conceived and wrote this manuscript.
EdP was supported by EHA junior non-clinical research fellowship and by KWF/Alpe’dHuzes (SK10321). ET was supported by the Max Planck Society, a Marie Curie Career Integration Grant (631432 Bloody Signals), the Deutsche Forschungsgemeinschaft, Research Training Group GRK2344 “MeInBio – BioInMe,” and by The Fritz Thyssen Stiftung (Az 10.17.1.026MN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. I. P. Touw for careful reading of the manuscript, Dr. van Royen, E. Gioacchino, J. Peulen for graphical contributions and Dr. T. Clapes for producing the second figure.
Ablain, J., Durand, E. M., Yang, S., Zhou, Y., and Zon, L. I. (2015). A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 32, 756–764. doi: 10.1016/j.devcel.2015.01.032
Albadri, S., Del Bene, F., and Revenu, C. (2017). Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods 121-122, 77–85. doi: 10.1016/j.ymeth.2017.03.005
Alberti-Servera, L., von Muenchow, L., Tsapogas, P., Capoferri, G., Eschbach, K., Beisel, C., et al. (2017). Single-cell RNA sequencing reveals developmental heterogeneity among early lymphoid progenitors. EMBO J. 36, 3619–3633. doi: 10.15252/embj.201797105
Alemany, A., Florescu, M., Baron, C. S., Peterson-Maduro, J., and van Oudenaarden, A. (2018). Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112. doi: 10.1038/nature25969
Alli Shaik, A., Wee, S., Li, R. H., Li, Z., Carney, T. J., Mathavan, S., et al. (2014). Functional mapping of the zebrafish early embryo proteome and transcriptome. J. Proteome Res. 13, 5536–5550. doi: 10.1021/pr5005136
Amacher, S. L. (2008). Emerging gene knockout technology in zebrafish: zinc-finger nucleases. Brief. Funct. Genomic. Proteomic. 7, 460–464. doi: 10.1093/bfgp/eln043
Amsterdam, A., Burgess, S., Golling, G., Chen, W., Sun, Z., Townsend, K., et al. (1999). A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 13, 2713–2724. doi: 10.1101/gad.13.20.2713
Arrenberg, A. B., Stainier, D. Y., Baier, H., and Huisken, J. (2010). Optogenetic control of cardiac function. Science 330, 971–974. doi: 10.1126/science.1195929
Arulmozhivarman, G., Stoter, M., Bickle, M., Krater, M., Wobus, M., Ehninger, G., et al. (2016). In vivo chemical screen in zebrafish embryos identifies regulators of hematopoiesis using a semiautomated imaging assay. J. Biomol. Screen. 21, 956–964. doi: 10.1177/1087057116644163
Athanasiadis, E. I., Botthof, J. G., Andres, H., Ferreira, L., Lio, P., and Cvejic, A. (2017). Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat. Commun. 8:2045. doi: 10.1038/s41467-017-02305-6
Auer, T. O., Duroure, K., De Cian, A., Concordet, J. P., and Del Bene, F. (2014). Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24, 142–153. doi: 10.1101/gr.161638.113
Babushok, D. V., Bessler, M., and Olson, T. S. (2016). Genetic predisposition to myelodysplastic syndrome and acute myeloid leukemia in children and young adults. Leuk. Lymphoma 57, 520–536. doi: 10.3109/10428194.2015.1115041
Baral, R., Ngounou Wetie, A. G., Darie, C. C., and Wallace, K. N. (2014). Mass spectrometry for proteomics-based investigation using the zebrafish vertebrate model system. Adv. Exp. Med. Biol. 806, 331–340. doi: 10.1007/978-3-319-06068-2_15
Bertrand, J. Y., Chi, N. C., Santoso, B., Teng, S., Stainier, D. Y., and Traver, D. (2010). Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111. doi: 10.1038/nature08738
Bertrand, J. Y., and Traver, D. (2009). Hematopoietic cell development in the zebrafish embryo. Curr. Opin. Hematol. 16, 243–248. doi: 10.1097/MOH.0b013e32832c05e4
Bogdanovic, O., Fernandez-Minan, A., Tena, J. J., de la Calle-Mustienes, E., and Gomez-Skarmeta, J. L. (2013). The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods 62, 207–215. doi: 10.1016/j.ymeth.2013.04.011
Bogdanovic, O., Fernandez-Minan, A., Tena, J. J., de la Calle-Mustienes, E., Hidalgo, C., van Kruysbergen, I., et al. (2012). Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 22, 2043–2053. doi: 10.1101/gr.134833.111
Buenrostro, J. D., Corces, M. R., Lareau, C. A., Wu, B., Schep, A. N., Aryee, M. J., et al. (2018). Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173, 1535–1548.e16. doi: 10.1016/j.cell.2018.03.074
Buenrostro, J. D., Wu, B., Litzenburger, U. M., Ruff, D., Gonzales, M. L., Snyder, M. P., et al. (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. doi: 10.1038/nature14590
Cabezas-Wallscheid, N., Buettner, F., Sommerkamp, P., Klimmeck, D., Ladel, L., Thalheimer, F. B., et al. (2017). Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell 169, 807–823.e19. doi: 10.1016/j.cell.2017.04.018
Carmona, S. J., Teichmann, S. A., Ferreira, L., Macaulay, I. C., Stubbington, M. J., Cvejic, A., et al. (2017). Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 27, 451–461. doi: 10.1101/gr.207704.116
Carney, T. J., and Mosimann, C. (2018). Switch and trace: recombinase genetics in zebrafish. Trends Genet. 34, 362–378. doi: 10.1016/j.tig.2018.01.004
Chiang, I. K., Fritzsche, M., Pichol-Thievend, C., Neal, A., Holmes, K., Lagendijk, A., et al. (2017). SoxF factors induce Notch1 expression via direct transcriptional regulation during early arterial development. Development 144, 2629–2639. doi: 10.1242/dev.146241
Cusick, M. F., Libbey, J. E., Trede, N. S., Eckels, D. D., and Fujinami, R. S. (2012). Human T cell expansion and experimental autoimmune encephalomyelitis inhibited by Lenaldekar, a small molecule discovered in a zebrafish screen. J. Neuroimmunol. 244, 35–44. doi: 10.1016/j.jneuroim.2011.12.024
Cutler, C., Multani, P., Robbins, D., Kim, H. T., Le, T., Hoggatt, J., et al. (2013). Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood 122, 3074–3081. doi: 10.1182/blood-2013-05-503177
Dahlem, T. J., Hoshijima, K., Jurynec, M. J., Gunther, D., Starker, C. G., Locke, A. S., et al. (2012). Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8:e1002861. doi: 10.1371/journal.pgen.1002861
Dahlin, J. S., Hamey, F. K., Pijuan-Sala, B., Shepherd, M., Lau, W. W. Y., Nestorowa, S., et al. (2018). A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood 131, e1–e11. doi: 10.1182/blood-2017-12-821413
Dal Molin, A., and Di Camillo, B. (2018). How to design a single-cell RNA-sequencing experiment: pitfalls, challenges and perspectives. Brief. Bioinform. doi: 10.1093/bib/bby007 [Epub ahead of print].
Deveau, A. P., Bentley, V. L., and Berman, J. N. (2017). Using zebrafish models of leukemia to streamline drug screening and discovery. Exp. Hematol. 45, 1–9. doi: 10.1016/j.exphem.2016.09.012
Doganli, C., Sandoval, M., Thomas, S., and Hart, D. (2017). Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-Seq) protocol for zebrafish embryos. Methods Mol. Biol. 1507, 59–66. doi: 10.1007/978-1-4939-6518-2_5
Draper, B. W., McCallum, C. M., Stout, J. L., Slade, A. J., and Moens, C. B. (2004). A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods Cell Biol. 77, 91–112. doi: 10.1016/S0091-679X(04)77005-3
Driever, W., Solnica-Krezel, L., Schier, A. F., Neuhauss, S. C., Malicki, J., Stemple, D. L., et al. (1996). A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37–46.
Drissen, R., Buza-Vidas, N., Woll, P., Thongjuea, S., Gambardella, A., Giustacchini, A., et al. (2016). Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat. Immunol. 17, 666–676. doi: 10.1038/ni.3412
Espin-Palazon, R., Stachura, D. L., Campbell, C. A., Garcia-Moreno, D., Del Cid, N., Kim, A. D., et al. (2014). Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell 159, 1070–1085. doi: 10.1016/j.cell.2014.10.031
Farrell, J. A., Wang, Y., Riesenfeld, S. J., Shekhar, K., Regev, A., and Schier, A. F. (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360:eaar3131. doi: 10.1126/science.aar3131
Felker, A., and Mosimann, C. (2016). Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. Methods Cell Biol. 135, 219–244. doi: 10.1016/bs.mcb.2016.01.009
Fernandez-Minan, A., Bessa, J., Tena, J. J., and Gomez-Skarmeta, J. L. (2016). Assay for transposase-accessible chromatin and circularized chromosome conformation capture, two methods to explore the regulatory landscapes of genes in zebrafish. Methods Cell Biol. 135, 413–430. doi: 10.1016/bs.mcb.2016.02.008
Foley, J. E., Maeder, M. L., Pearlberg, J., Joung, J. K., Peterson, R. T., and Yeh, J. R. (2009). Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat. Protoc. 4, 1855–1867. doi: 10.1038/nprot.2009.209
Frame, J. M., Lim, S. E., and North, T. E. (2017). Hematopoietic stem cell development: using the zebrafish to identify extrinsic and intrinsic mechanisms regulating hematopoiesis. Methods Cell Biol. 138, 165–192. doi: 10.1016/bs.mcb.2016.08.004
Ganis, J. J., Hsia, N., Trompouki, E., de Jong, J. L., DiBiase, A., Lambert, J. S., et al. (2012). Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev. Biol. 366, 185–194. doi: 10.1016/j.ydbio.2012.03.021
Gjini, E., Mansour, M. R., Sander, J. D., Moritz, N., Nguyen, A. T., Kesarsing, M., et al. (2015). A zebrafish model of myelodysplastic syndrome produced through tet2 genomic editing. Mol. Cell. Biol. 35, 789–804. doi: 10.1128/MCB.00971-14
Glasauer, S. M., and Neuhauss, S. C. (2014). Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics 289, 1045–1060. doi: 10.1007/s00438-014-0889-2
Goessling, W., Allen, R. S., Guan, X., Jin, P., Uchida, N., Dovey, M., et al. (2011). Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell 8, 445–458. doi: 10.1016/j.stem.2011.02.003
Goessling, W., and North, T. E. (2011). Hematopoietic stem cell development: using the zebrafish to identify the signaling networks and physical forces regulating hematopoiesis. Methods Cell Biol. 105, 117–136. doi: 10.1016/B978-0-12-381320-6.00005-9
Gomez-Marin, C., Tena, J. J., Acemel, R. D., Lopez-Mayorga, M., Naranjo, S., de la Calle-Mustienes, E., et al. (2015). Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc. Natl. Acad. Sci. U.S.A. 112, 7542–7547. doi: 10.1073/pnas.1505463112
Gore, A. V., Pillay, L. M., Venero Galanternik, M., and Weinstein, B. M. (2018). The zebrafish: a fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 7:e312. doi: 10.1002/wdev.312
Groschel, S., Sanders, M. A., Hoogenboezem, R., de Wit, E., Bouwman, B. A. M., Erpelinck, C., et al. (2014). A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157, 369–381. doi: 10.1016/j.cell.2014.02.019
Grover, A., Sanjuan-Pla, A., Thongjuea, S., Carrelha, J., Giustacchini, A., Gambardella, A., et al. (2016). Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 7:11075. doi: 10.1038/ncomms11075
Guo, G., Luc, S., Marco, E., Lin, T. W., Peng, C., Kerenyi, M. A., et al. (2013). Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 13, 492–505. doi: 10.1016/j.stem.2013.07.017
Gury-BenAri, M., Thaiss, C. A., Serafini, N., Winter, D. R., Giladi, A., Lara-Astiaso, D., et al. (2016). The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246.e13. doi: 10.1016/j.cell.2016.07.043
Gutierrez, A., Pan, L., Groen, R. W., Baleydier, F., Kentsis, A., Marineau, J., et al. (2014). Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. J. Clin. Invest. 124, 644–655. doi: 10.1172/JCI65093
Havis, E., Anselme, I., and Schneider-Maunoury, S. (2006). Whole embryo chromatin immunoprecipitation protocol for the in vivo study of zebrafish development. Biotechniques 40, 34, 36, 38 passim. doi: 10.2144/000112098
He, B. L., Shi, X., Man, C. H., Ma, A. C., Ekker, S. C., Chow, H. C., et al. (2014). Functions of flt3 in zebrafish hematopoiesis and its relevance to human acute myeloid leukemia. Blood 123, 2518–2529. doi: 10.1182/blood-2013-02-486688
He, Q., Zhang, C., Wang, L., Zhang, P., Ma, D., Lv, J., et al. (2015). Inflammatory signaling regulates hematopoietic stem and progenitor cell emergence in vertebrates. Blood 125, 1098–1106. doi: 10.1182/blood-2014-09-601542
Henninger, J., Santoso, B., Hans, S., Durand, E., Moore, J., Mosimann, C., et al. (2017). Clonal fate mapping quantifies the number of haematopoietic stem cells that arise during development. Nat. Cell Biol. 19, 17–27. doi: 10.1038/ncb3444
Hess, I., and Boehm, T. (2012). Intravital imaging of thymopoiesis reveals dynamic lympho-epithelial interactions. Immunity 36, 298–309. doi: 10.1016/j.immuni.2011.12.016
Hruscha, A., Krawitz, P., Rechenberg, A., Heinrich, V., Hecht, J., Haass, C., et al. (2013). Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140, 4982–4987. doi: 10.1242/dev.099085
Huang, P., Xiao, A., Tong, X., Lin, S., and Zhang, B. (2016). Targeted mutagenesis in zebrafish by TALENs. Methods Mol. Biol. 1338, 191–206. doi: 10.1007/978-1-4939-2932-0_15
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J., and Stelzer, E. H. (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009. doi: 10.1126/science.1100035
Hwang, W. Y., Peterson, R. T., and Yeh, J. R. (2014). Methods for targeted mutagenesis in zebrafish using TALENs. Methods 69, 76–84. doi: 10.1016/j.ymeth.2014.04.009
Inoue, S., and Inoue, T. (1996). Digital unsharp masking reveals fine detail in images obtained with new spinning-disk confocal microscope. Biol. Bull. 191, 269–270. doi: 10.1086/BBLv191n2p269
Irion, U., Krauss, J., and Nusslein-Volhard, C. (2014). Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141, 4827–4830. doi: 10.1242/dev.115584
Junker, J. P., Noel, E. S., Guryev, V., Peterson, K. A., Shah, G., Huisken, J., et al. (2014). Genome-wide RNA Tomography in the zebrafish embryo. Cell 159, 662–675. doi: 10.1016/j.cell.2014.09.038
Kaaij, L. J., Mokry, M., Zhou, M., Musheev, M., Geeven, G., Melquiond, A. S., et al. (2016). Enhancers reside in a unique epigenetic environment during early zebrafish development. Genome Biol. 17:146. doi: 10.1186/s13059-016-1013-1
Kesavan, G., Chekuru, A., Machate, A., and Brand, M. (2017). CRISPR/Cas9-mediated zebrafish knock-in as a novel strategy to study midbrain-hindbrain boundary development. Front. Neuroanat. 11:52. doi: 10.3389/fnana.2017.00052
Kissa, K., and Herbomel, P. (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115. doi: 10.1038/nature08761
Kolodziejczyk, A. A., Kim, J. K., Svensson, V., Marioni, J. C., and Teichmann, S. A. (2015). The technology and biology of single-cell RNA sequencing. Mol. Cell. 58, 610–620. doi: 10.1016/j.molcel.2015.04.005
Kwan, W., and North, T. E. (2017). Netting novel regulators of hematopoiesis and hematologic malignancies in zebrafish. Curr. Top. Dev. Biol. 124, 125–160. doi: 10.1016/bs.ctdb.2016.11.005
Langenau, D. M., Traver, D., Ferrando, A. A., Kutok, J. L., Aster, J. C., Kanki, J. P., et al. (2003). Myc-induced T cell leukemia in transgenic zebrafish. Science 299, 887–890. doi: 10.1126/science.1080280
Le, X., Langenau, D. M., Keefe, M. D., Kutok, J. L., Neuberg, D. S., and Zon, L. I. (2007). Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 9410–9415. doi: 10.1073/pnas.0611302104
Lee, H. J., Lowdon, R. F., Maricque, B., Zhang, B., Stevens, M., Li, D., et al. (2015). Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat. Commun. 6:6315. doi: 10.1038/ncomms7315
Li, M., Zhao, L., Page-McCaw, P. S., and Chen, W. (2016). Zebrafish genome engineering using the CRISPR-Cas9 system. Trends Genet. 32, 815–827. doi: 10.1016/j.tig.2016.10.005
Li, Y., Esain, V., Teng, L., Xu, J., Kwan, W., Frost, I. M., et al. (2014). Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 28, 2597–2612. doi: 10.1101/gad.253302.114
Liao, E. C., Paw, B. H., Oates, A. C., Pratt, S. J., Postlethwait, J. H., and Zon, L. I. (1998). SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 12, 621–626. doi: 10.1101/gad.12.5.621
Liu, J., Zhou, Y., Qi, X., Chen, J., Chen, W., Qiu, G., et al. (2017). CRISPR/Cas9 in zebrafish: an efficient combination for human genetic diseases modeling. Hum. Genet. 136, 1–12. doi: 10.1007/s00439-016-1739-6
Liu, Y., Zhao, H., and Cheng, C. H. (2016). Mutagenesis in Xenopus and Zebrafish using TALENs. Methods Mol. Biol. 1338, 207–227. doi: 10.1007/978-1-4939-2932-0_16
Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. doi: 10.1016/j.cell.2015.05.002
Meier, M., Grant, J., Dowdle, A., Thomas, A., Gerton, J., Collas, P., et al. (2018). Cohesin facilitates zygotic genome activation in zebrafish. Development 145:dev156521. doi: 10.1242/dev.156521
Moore, J. C., Tang, Q., Yordan, N. T., Moore, F. E., Garcia, E. G., Lobbardi, R., et al. (2016). Single-cell imaging of normal and malignant cell engraftment into optically clear prkdc-null SCID zebrafish. J. Exp. Med. 213, 2575–2589. doi: 10.1084/jem.20160378
Mosimann, C., Puller, A. C., Lawson, K. L., Tschopp, P., Amsterdam, A., and Zon, L. I. (2013). Site-directed zebrafish transgenesis into single landing sites with the phiC31 integrase system. Dev. Dyn. 242, 949–963. doi: 10.1002/dvdy.23989
Mullins, M. C., Hammerschmidt, M., Haffter, P., and Nusslein-Volhard, C. (1994). Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr. Biol. 4, 189–202. doi: 10.1016/S0960-9822(00)00048-8
Murayama, E., Kissa, K., Zapata, A., Mordelet, E., Briolat, V., Lin, H. F., et al. (2006). Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 25, 963–975. doi: 10.1016/j.immuni.2006.10.015
Nelson, A. C., Cutty, S. J., Gasiunas, S. N., Deplae, I., Stemple, D. L., and Wardle, F. C. (2017). In vivo regulation of the zebrafish endoderm progenitor niche by T-box transcription factors. Cell Rep. 19, 2782–2795. doi: 10.1016/j.celrep.2017.06.011
Nestorowa, S., Hamey, F. K., Pijuan Sala, B., Diamanti, E., Shepherd, M., Laurenti, E., et al. (2016). A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–e31. doi: 10.1182/blood-2016-05-716480
Nik, S., Weinreb, J. T., and Bowman, T. V. (2017). Developmental HSC microenvironments: lessons from zebrafish. Adv. Exp. Med. Biol. 1041, 33–53. doi: 10.1007/978-3-319-69194-7_4
North, T. E., Goessling, W., Walkley, C. R., Lengerke, C., Kopani, K. R., Lord, A. M., et al. (2007). Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011. doi: 10.1038/nature05883
Notta, F., Zandi, S., Takayama, N., Dobson, S., Gan, O. I., Wilson, G., et al. (2016). Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351:aab2116. doi: 10.1126/science.aab2116
Olsson, A., Venkatasubramanian, M., Chaudhri, V. K., Aronow, B. J., Salomonis, N., Singh, H., et al. (2016). Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702. doi: 10.1038/nature19348
Ong, E. S., Chor, C. F., Zou, L., and Ong, C. N. (2009). A multi-analytical approach for metabolomic profiling of zebrafish (Danio rerio) livers. Mol. Biosyst. 5, 288–298. doi: 10.1039/b811850g
Paik, E. J., de Jong, J. L., Pugach, E., Opara, P., and Zon, L. I. (2010). A chemical genetic screen in zebrafish for pathways interacting with cdx4 in primitive hematopoiesis. Zebrafish 7, 61–68. doi: 10.1089/zeb.2009.0643
Paik, E. J., and Zon, L. I. (2010). Hematopoietic development in the zebrafish. Int. J. Dev. Biol. 54, 1127–1137. doi: 10.1387/ijdb.093042ep
Paul, F., Arkin, Y., Giladi, A., Jaitin, D. A., Kenigsberg, E., Keren-Shaul, H., et al. (2015). Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677. doi: 10.1016/j.cell.2015.11.013
Pei, W., Feyerabend, T. B., Rossler, J., Wang, X., Postrach, D., Busch, K., et al. (2017). Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460. doi: 10.1038/nature23653
Peng, X., Dong, M., Ma, L., Jia, X. E., Mao, J., Jin, C., et al. (2015). A point mutation of zebrafish c-cbl gene in the ring finger domain produces a phenotype mimicking human myeloproliferative disease. Leukemia 29, 2355–2365. doi: 10.1038/leu.2015.154
Potts, K. S., and Bowman, T. V. (2017). Modeling myeloid malignancies using zebrafish. Front. Oncol. 7:297. doi: 10.3389/fonc.2017.00297
Quillien, A., Abdalla, M., Yu, J., Ou, J., Zhu, L. J., and Lawson, N. D. (2017). Robust identification of developmentally active endothelial enhancers in zebrafish using FANS-assisted ATAC-Seq. Cell Rep. 20, 709–720. doi: 10.1016/j.celrep.2017.06.070
Rabinowitz, J. S., Robitaille, A. M., Wang, Y., Ray, C. A., Thummel, R., Gu, H., et al. (2017). Transcriptomic, proteomic, and metabolomic landscape of positional memory in the caudal fin of zebrafish. Proc. Natl. Acad. Sci. U.S.A. 114, E717–E726. doi: 10.1073/pnas.1620755114
Rennekamp, A. J., and Peterson, R. T. (2015). 15 years of zebrafish chemical screening. Curr. Opin. Chem. Biol. 24, 58–70. doi: 10.1016/j.cbpa.2014.10.025
Ridges, S., Heaton, W. L., Joshi, D., Choi, H., Eiring, A., Batchelor, L., et al. (2012). Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood 119, 5621–5631. doi: 10.1182/blood-2011-12-398818
Roy, U., Conklin, L., Schiller, J., Matysik, J., Berry, J. P., and Alia, A. (2017). Metabolic profiling of zebrafish (Danio rerio) embryos by NMR spectroscopy reveals multifaceted toxicity of beta-methylamino-L-alanine (BMAA). Sci. Rep. 7:17305. doi: 10.1038/s41598-017-17409-8
Sawamiphak, S., Kontarakis, Z., and Stainier, D. Y. (2014). Interferon gamma signaling positively regulates hematopoietic stem cell emergence. Dev. Cell 31, 640–653. doi: 10.1016/j.devcel.2014.11.007
Shafizadeh, E., Peterson, R. T., and Lin, S. (2004). Induction of reversible hemolytic anemia in living zebrafish using a novel small molecule. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 138, 245–249.
Shah, A. N., Davey, C. F., Whitebirch, A. C., Miller, A. C., and Moens, C. B. (2015). Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods 12, 535–540. doi: 10.1038/nmeth.3360
Tamplin, O. J., Durand, E. M., Carr, L. A., Childs, S. J., Hagedorn, E. J., Li, P., et al. (2015). Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252. doi: 10.1016/j.cell.2014.12.032
Tamplin, O. J., White, R. M., Jing, L., Kaufman, C. K., Lacadie, S. A., Li, P., et al. (2012). Small molecule screening in zebrafish: swimming in potential drug therapies. Wiley Interdiscip. Rev. Dev. Biol. 1, 459–468. doi: 10.1002/wdev.37
Tan, H., Onichtchouk, D., and Winata, C. (2016). DANIO-CODE: toward an encyclopedia of DNA elements in zebrafish. Zebrafish 13, 54–60. doi: 10.1089/zeb.2015.1179
Tang, Q., Iyer, S., Lobbardi, R., Moore, J. C., Chen, H., Lareau, C., et al. (2017). Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 214, 2875–2887. doi: 10.1084/jem.20170976
Taylor, A. M., and Zon, L. I. (2011). Modeling diamond blackfan anemia in the zebrafish. Semin. Hematol. 48, 81–88. doi: 10.1053/j.seminhematol.2011.02.002
Tian, Y., Xu, J., Feng, S., He, S., Zhao, S., Zhu, L., et al. (2017). The first wave of T lymphopoiesis in zebrafish arises from aorta endothelium independent of hematopoietic stem cells. J. Exp. Med. 214, 3347–3360. doi: 10.1084/jem.20170488
Tijssen, M. R., Cvejic, A., Joshi, A., Hannah, R. L., Ferreira, R., Forrai, A., et al. (2011). Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell 20, 597–609. doi: 10.1016/j.devcel.2011.04.008
Travnickova, J., Tran Chau, V., Julien, E., Mateos-Langerak, J., Gonzalez, C., Lelievre, E., et al. (2015). Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat. Commun. 6:6227. doi: 10.1038/ncomms7227
Trompouki, E., Bowman, T. V., Dibiase, A., Zhou, Y., and Zon, L. I. (2011). Chromatin immunoprecipitation in adult zebrafish red cells. Methods Cell Biol. 104, 341–352. doi: 10.1016/B978-0-12-374814-0.00019-7
Vastenhouw, N. L., Zhang, Y., Woods, I. G., Imam, F., Regev, A., Liu, X. S., et al. (2010). Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926. doi: 10.1038/nature08866
Veinotte, C. J., Dellaire, G., and Berman, J. N. (2014). Hooking the big one: the potential of zebrafish xenotransplantation to reform cancer drug screening in the genomic era. Dis. Model. Mech. 7, 745–754. doi: 10.1242/dmm.015784
Velten, L., Haas, S. F., Raffel, S., Blaszkiewicz, S., Islam, S., Hennig, B. P., et al. (2017). Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281. doi: 10.1038/ncb3493
Villani, A. C., Satija, R., Reynolds, G., Sarkizova, S., Shekhar, K., Fletcher, J., et al. (2017). Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356:eaah4573. doi: 10.1126/science.aah4573
Wagner, D. E., Weinreb, C., Collins, Z. M., Briggs, J. A., Megason, S. G., and Klein, A. M. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. doi: 10.1126/science.aar4362
White, R. M., Sessa, A., Burke, C., Bowman, T., LeBlanc, J., Ceol, C., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189. doi: 10.1016/j.stem.2007.11.002
Wienholds, E., van Eeden, F., Kosters, M., Mudde, J., Plasterk, R. H., and Cuppen, E. (2003). Efficient target-selected mutagenesis in zebrafish. Genome Res. 13, 2700–2707. doi: 10.1101/gr.1725103
Winata, C. L., Kondrychyn, I., Kumar, V., Srinivasan, K. G., Orlov, Y., Ravishankar, A., et al. (2013). Genome wide analysis reveals Zic3 interaction with distal regulatory elements of stage specific developmental genes in zebrafish. PLoS Genet. 9:e1003852. doi: 10.1371/journal.pgen.1003852
Xu, C., Fan, Z. P., Muller, P., Fogley, R., DiBiase, A., Trompouki, E., et al. (2012). Nanog-like regulates endoderm formation through the Mxtx2-Nodal pathway. Dev. Cell 22, 625–638. doi: 10.1016/j.devcel.2012.01.003
Yamamoto, R., Morita, Y., Ooehara, J., Hamanaka, S., Onodera, M., Rudolph, K. L., et al. (2013). Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 154, 1112–1126. doi: 10.1016/j.cell.2013.08.007
Yeh, J. R., Munson, K. M., Elagib, K. E., Goldfarb, A. N., Sweetser, D. A., and Peterson, R. T. (2009). Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat. Chem. Biol. 5, 236–243. doi: 10.1038/nchembio.147
Zhang, C., Patient, R., and Liu, F. (2013). Hematopoietic stem cell development and regulatory signaling in zebrafish. Biochim. Biophys. Acta 1830, 2370–2374. doi: 10.1016/j.bbagen.2012.06.008
Zhang, P., and Liu, F. (2011). In vivo imaging of hematopoietic stem cell development in the zebrafish. Front. Med. 5:239–247. doi: 10.1007/s11684-011-0123-0
Ziegenhain, C., Vieth, B., Parekh, S., Reinius, B., Guillaumet-Adkins, A., Smets, M., et al. (2017). Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 65, 631–643e4. doi: 10.1016/j.molcel.2017.01.023
Keywords: zebrafish, hematopoiesis, next generation sequencing, hematopoietic (stem) cells, technology
Citation: de Pater E and Trompouki E (2018) Bloody Zebrafish: Novel Methods in Normal and Malignant Hematopoiesis. Front. Cell Dev. Biol. 6:124. doi: 10.3389/fcell.2018.00124
Received: 16 July 2018; Accepted: 10 September 2018;
Published: 15 October 2018.
Edited by:
Masatake Osawa, Gifu University, JapanReviewed by:
Veronica Ramos-Mejia, Centro Pfizer-Universidad de Granada-Junta de Andalucía de Genómica e Investigación Oncológica (GENYO), SpainCopyright © 2018 de Pater and Trompouki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emma de Pater, ZS5kZXBhdGVyQGVyYXNtdXNtYy5ubA== Eirini Trompouki, dHJvbXBvdWtpQGllLWZyZWlidXJnLm1wZy5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.