
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Cell Dev. Biol. , 27 September 2018
Sec. Signaling
Volume 6 - 2018 | https://doi.org/10.3389/fcell.2018.00121
This article is a correction to:
Cystathionine β-Synthase Is Necessary for Axis Development in Vivo
 Shubhangi Prabhudesai1†
Shubhangi Prabhudesai1† Chris Koceja1†
Chris Koceja1† Anindya Dey2†
Anindya Dey2† Shahram Eisa-Beygi3
Shahram Eisa-Beygi3 Noah R. Leigh4
Noah R. Leigh4 Resham Bhattacharya2,5*
Resham Bhattacharya2,5* Priyabrata Mukherjee2,5,6,7*
Priyabrata Mukherjee2,5,6,7* Ramani Ramchandran1,3,8*
Ramani Ramchandran1,3,8*A Corrigendum on
Cystathionine β-Synthase Is Necessary for Axis Development in vivo
by Prabhudesai, S., Koceja, C., Dey, A., Eisa-Beygi, S., Leigh, N. R., Bhattacharya, R., et al. (2018) Front. Cell Dev. Biol. 6:14. doi: 10.3389/fcell.2018.00014
In the original article, there was an error in Figure 2C as published. The sequence of the cbsa splice 1 (CBSA-S1) morpholino was incorrectly typed as
TACCTGCACAAAGTGAACACACAACCA
The correct sequence is
TACCTGCACAAAGTGAACACAACCA
The name of the morpholino was changed in the figure from cbsa splice (CBSA-S1) to cbsa splice 1 (CBSA-S1) to match the legend.
The corrected Figure 2 appears below. The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way.
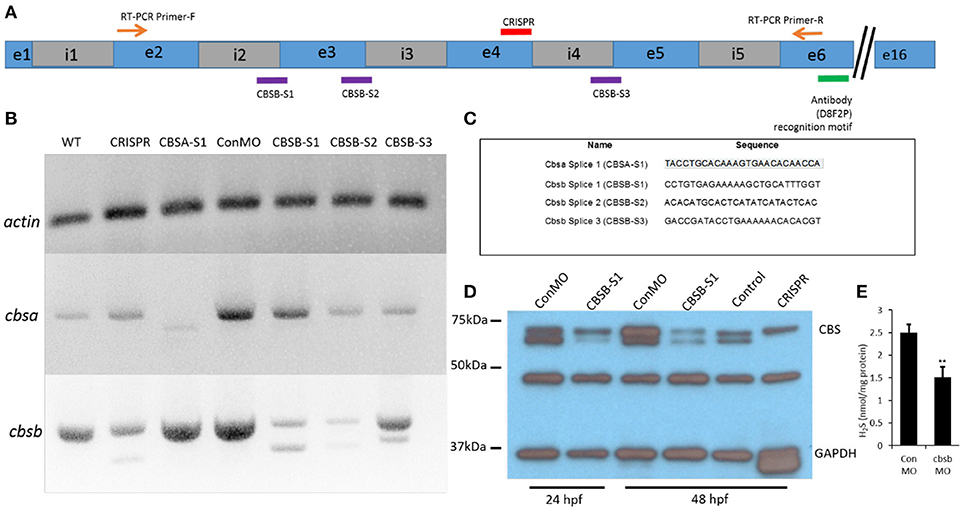
Figure 2. Loss-of-function efficacy studies. Panel (A) shows a “partial” cartoon representation of the cbsb genomic site with the location of the MO sites (purple rectangles) at appropriate intron (i) and exon (e) junctions, CRISPR-targeted site (red rectangle), site of RT-PCR forward (F) and reverse (R) primers, and the antibody recognition site. Panel (B) shows RT-PCR for three genes (cbsb, cbsa, actin) in total RNA from injected embryos (~24 hpf) (left to right): wild type (WT) control and cbsb CRISPR-injected fish, cbsa splice 1 (CBSA-S1), control morpholino (ConMO), cbsb splice1 (CBSB-S1), cbsb splice 2 (CBSB-S2), cbsb splice 3 (CBSB-S3). Panel (C) shows the sequence of the morpholinos used in this study. Panel (D) shows CBS and GAPDH western blots for ConMO, CBSB-S1 at 24 and 48 hpf along with control and cbsb CRISPR fish at 48 hpf. Panel (E) shows the comparison between CBSB-S1 MO and ConMO-injected embryos for hydrogen sulfide production. n = 3 for both groups (data from three experiments). Twenty embryos in each group in each experiment. **P < 0.01.
The original article has been updated.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Keywords: zebrafish, CRISPR, small molecules, methionine, homcystinuria, hydrogen sulfide, morpholino
Citation: Prabhudesai S, Koceja C, Dey A, Eisa-Beygi S, Leigh NR, Bhattacharya R, Mukherjee P and Ramchandran R (2018) Corrigendum: Cystathionine β-Synthase Is Necessary for Axis Development in vivo. Front. Cell Dev. Biol. 6:121. doi: 10.3389/fcell.2018.00121
Received: 13 August 2018; Accepted: 05 September 2018;
Published: 27 September 2018.
Edited and reviewed by: Gregory Kelly, University of Western Ontario, Canada
Copyright © 2018 Prabhudesai, Koceja, Dey, Eisa-Beygi, Leigh, Bhattacharya, Mukherjee and Ramchandran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Resham Bhattacharya, UmVzaGFtLUJoYXR0YWNoYXJ5YUBPVUhTQy5lZHU=
Priyabrata Mukherjee, UHJpeWFicmF0YS1NdWtoZXJqZWVAT1VIU0MuZWR1
Ramani Ramchandran, cnJhbWNoYW5AbWN3LmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.