- 1Department of Otorhinolaryngology Head and Neck Surgery, University of Maryland School of Medicine, Baltimore, MD, United States
- 2Section on Sensory Cell Development and Function, National Institute on Deafness and Other Communication Disorders, Bethesda, MD, United States
- 3Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD, United States
- 4Department of Anatomy and Neurobiology, University of Maryland School of Medicine, Baltimore, MD, United States
A Corrigendum on
Transcriptomic Profiling of Zebrafish Hair Cells Using RiboTag
by Matern, M. S., Beirl, A., Ogawa, Y., Song, Y., Paladugu, N., Kindt, K. S., et al. (2018). Front. Cell Dev. Biol. 6:47. doi: 10.3389/fcell.2018.00047
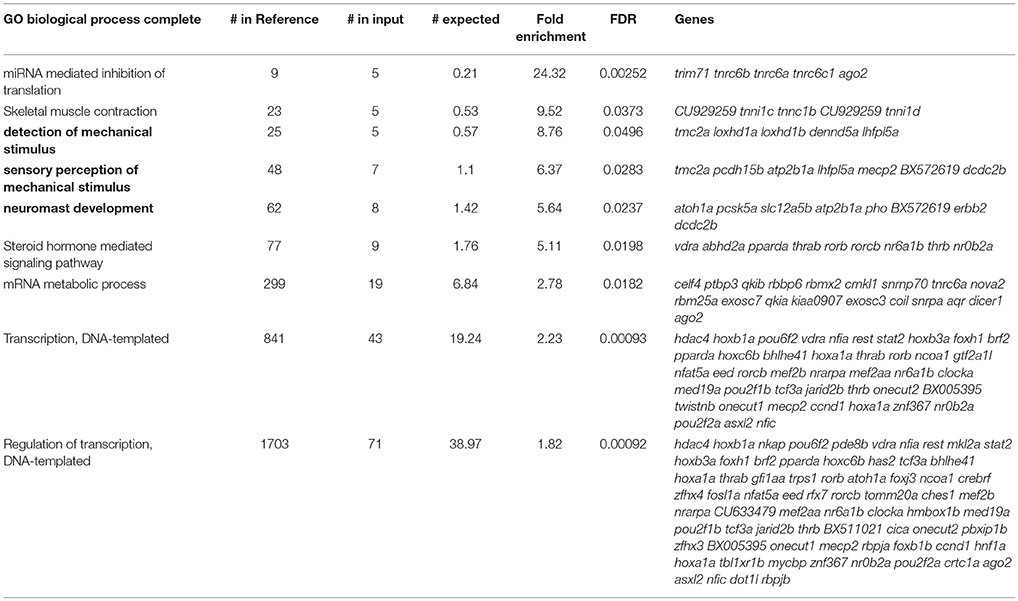
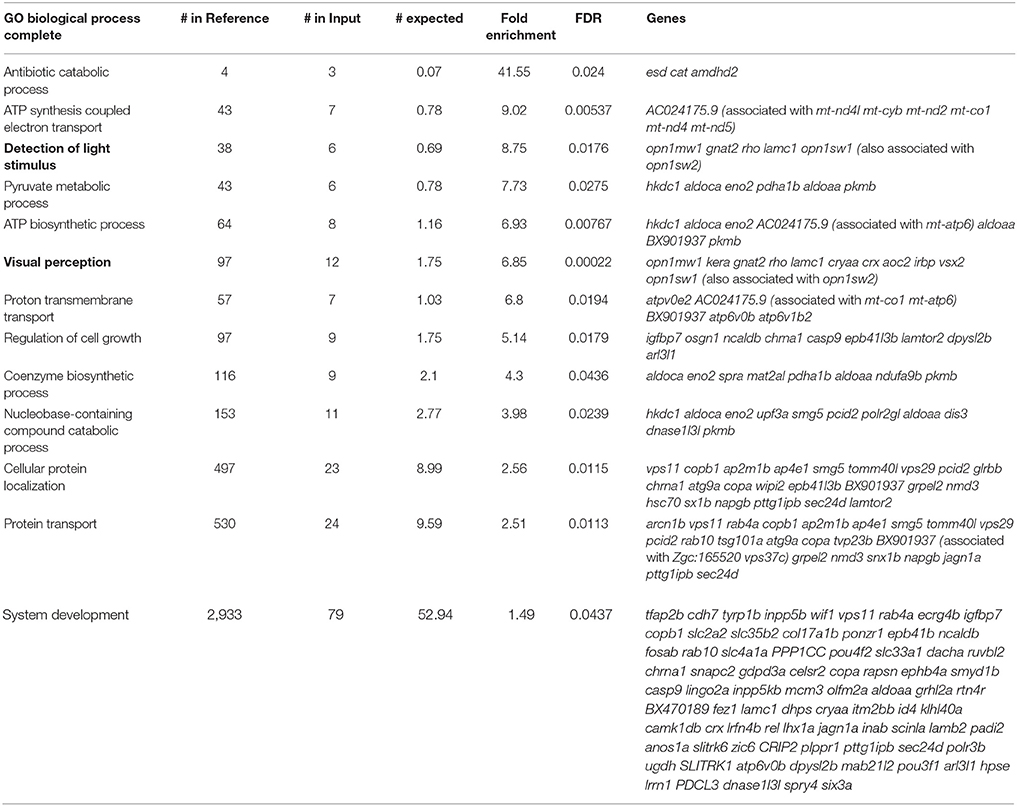
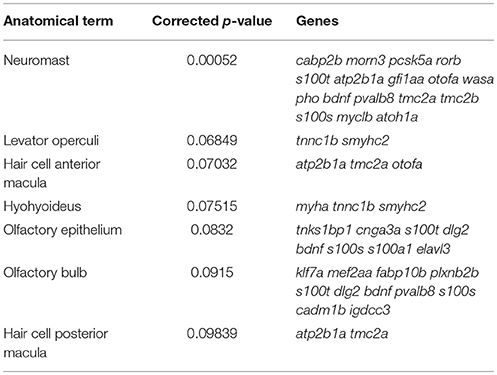
Although the reported fold change values and statistics were accurate, there was a mistake in the CPM values shown in Supplementary Dataset 1 in the original article. This mistake has been corrected and it has resulted in a slight change in the number of transcripts meeting the CPM cutoff for expression (n = 17,164), enrichment (n = 2,379), and depletion (n = 2,258). These corrected numbers of expressed, enriched and depleted transcripts are now reflected in the text, as well as in Figure 3a. The gene ontology and ZEOGS analyses have been redone and changes have been made to Tables 1–3, and Supplementary Table 2. The genes chosen for validation now fall within the top 100 enriched transcripts rather than the top 50; therefore, Supplementary Table 3 has been changed to show the top 100 enriched transcripts. The scientific conclusions made from these analyses have not changed in any way.
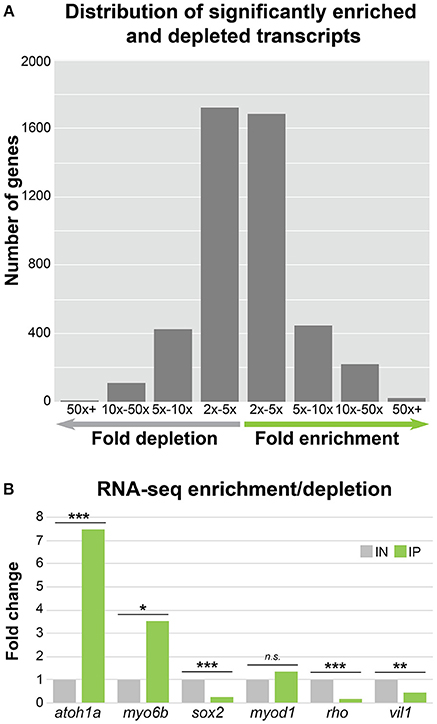
Figure 3. HC ribosome immunoprecipitation can reliably detect HC enriched and depleted transcripts by RNA-Seq. (A) Bar graph showing the distribution of significantly enriched and depleted transcripts in the IN vs. IP samples binned by fold change range. For genes with depleted transcripts in the IP samples, the number of genes per bin is as follows: 2x − 5x = 1726, 5x − 10x = 422, 5x − 10x = 109, and 50x + = 1. For IP enriched gene transcripts, the number of genes per bin is as follows: 2x − 5x = 1685, 5x − 10x = 450, 5x − 10x = 223, and 50x + = 21. (B) RNA-Seq fold change enrichment and depletion of HC expressed and non-expressed genes replicates the results obtained by RT-qPCR (Figure 2C). Statistical significance was assessed using DEseq (see Methods). *FDR < 0.05, **FDR < 0.01, ***FDR < 0.001.
A fourth input sample, which was included in the original analysis, was missing from Supplementary Dataset 1 and Supplementary Table 1 in the original article. This sample has now been added to both the files. Sentence one in the subsection “RNA sequencing and informatics” under “Materials and Methods”, now reads “RNA from 5 dpf Tg (myo6b:RiboTag) zebrafish IN and IP samples was submitted in biological quadruplicates for RNA-Seq at the UMSOM Institute for Genome Sciences.” The following sentence has also been added: “One IP sample had a high intergenic content suggestive of DNA contamination and was excluded from the analysis.” Finally, sentence two in paragraph one of the subsection “RNA sequencing of IP and IN samples from Tg(myo6b:RiboTag) zebrafish” under “Results”, now reads “We therefore performed RNA-Seq on IP and IN samples in at least three biological replicates from 5 dpf Tg(myo6b:RiboTag) zebrafish”.
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way.
The original article has been updated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2018.00084/full#supplementary-material
Keywords: inner ear, hair cells, zebrafish, RiboTag, RNA-Seq
Citation: Matern MS, Beirl A, Ogawa Y, Song Y, Paladugu N, Kindt KS and Hertzano R (2018) Corrigendum: Transcriptomic Profiling of Zebrafish Hair Cells Using RiboTag. Front. Cell Dev. Biol. 6:84. doi: 10.3389/fcell.2018.00084
Received: 06 June 2018; Accepted: 17 July 2018;
Published: 27 August 2018.
Edited and reviewed by: Kelvin Y. Kwan, Rutgers University, The State University of New Jersey, United States
Copyright © 2018 Matern, Beirl, Ogawa, Song, Paladugu, Kindt and Hertzano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ronna Hertzano, cmhlcnR6YW5vQHNvbS51bWFyeWxhbmQuZWR1
 Maggie S. Matern
Maggie S. Matern Alisha Beirl
Alisha Beirl Yoko Ogawa1
Yoko Ogawa1 Ronna Hertzano
Ronna Hertzano