- 1School of Life Sciences, Queen's Medical Centre, University of Nottingham, Nottingham, United Kingdom
- 2Division of Cancer and Stem Cells, Centre for Biomolecular Sciences, School of Medicine, University of Nottingham, Nottingham, United Kingdom
5-methylcytosine (5mC) is the best understood DNA modification and is generally believed to be associated with repression of gene expression. Over the last decade, sequentially oxidized forms of 5mC (oxi-mCs) have been discovered within the genomes of vertebrates. Their discovery was accompanied by that of the ten-eleven translocation (TET) methylcytosine dioxygenases, the enzymes that catalyze the formation of the oxi-mCs. Although a number of studies performed on different vertebrate models and embryonic stem cells demonstrated that both TET enzymes and oxi-mCs are likely to be important for several developmental processes it is currently unclear whether their developmental roles are conserved among vertebrates. Here, we summarize recent developments in this field suggesting that biological roles of TETs/oxi-mCs may significantly differ between mice and zebrafish. Thus, although the role of TET proteins in late organogenesis has been documented for both these systems; unlike in mice the enzymatic oxidation of 5mC does not seem to be involved in zygotic reprogramming or gastrulation in zebrafish. Our analysis may provide an insight into the general principles of epigenetic regulation of animal development and cellular differentiation.
DNA Methylation and Plasticity of Gene Expression in Embryonic Development
Embryonic development and organogenesis are epigenetic processes directed by intricate networks of transcriptional regulators (Boyer et al., 2005; Chambers et al., 2007). During embryogenesis, the transcriptional programmes of individual cells are influenced by cell intrinsic factors and stochastic events (Tang et al., 2011; Moris et al., 2016). Cells also need to be responsive to molecular signals originating from their microenvironment (Rossant and Tam, 2009; Shi and Wu, 2009). The plasticity of gene expression allows distinct populations of cells to arise within the embryo (Tam and Loebel, 2007). The regulation of gene expression during development is remarkably complex and heavily reliant upon epigenetic mechanisms, such as histone and DNA modifications (Reik, 2007).
Eukaryotic DNA can be methylated at cytosine residues, by DNA methyltransferases (DNMTs) (Bird, 2002). The methylation of a gene's cis-regulatory elements is generally considered to inhibit its transcription (Suzuki and Bird, 2008) and, remarkably, the impairment of DNA methylation regulation has been implicated in the pathologies of many developmental diseases (Robertson, 2005). Most metazoan DNA methylation (99.98% in human somatic cells, Jang et al., 2017) takes place at CpG dinucleotide sites within the genome. CpG sites represent approximately 7% of the mouse genome (Smith et al., 2009) and 5.3% of the zebrafish genome (Chatterjee et al., 2013). Regions of the genome with high CpG density, so-called CpG islands, generally overlap with mapped transcription start sites of identified gene promoters (Carninci et al., 2006; Potok et al., 2013). Importantly, DNA methylation is reversible (Wu and Zhang, 2014) and 5mC deposition and removal pathways operate during several key stages of vertebrate embryogenesis (Smith and Meissner, 2013).
The TET Enzymes and 5mC Oxidized Derivatives (Oxi-mCs)
Over the past 8 years, renewed interest in active demethylation and the role of 5mC in development has been fueled by the discovery of a novel function for the ten-eleven translocation (TET) enzymes (Tahiliani et al., 2009; Ito et al., 2010). These enzymes catalyze the iterative oxidation of 5mC to: 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009; Ito et al., 2011), from here on collectively referred to as oxi-mCs. The discovery that Thymine DNA glycosylase (TDG) can excise 5fC and 5caC, leaving an abasic site to be re-filled by non-modified cytosine, allowed a pathway for TET-mediated active demethylation to be posited (He et al., 2011; Maiti and Drohat, 2011). Figure 1 illustrates the steps involved in active demethylation. A comprehensive discussion of currently proposed demethylation mechanisms is provided in the excellent review of Wu and Zhang (Wu and Zhang, 2017).
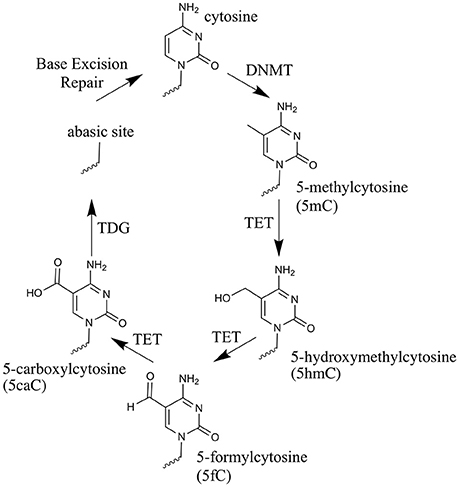
Figure 1. The cytosine oxidation cycle. The chemical structures of cytosine, 5-methylcytosine and each oxi-mC within DNA. Their synthesis route is depicted by arrows. The enzyme catalyzing each reaction and/or the physical process taking place, is labeled next to the arrow.
5hmC was first detected in viruses, using a combination of UV-spectroscopy and paper chromatography (Wyatt and Cohen, 1953), but the first reported detection of this modification in animals (Penn et al., 1972) could not be reproduced. In fact, the generation of 5hmC was interpreted as an artifact caused by the method for hydrolysing the DNA during sample preparation (Kothari and Shankar, 1976). Using thin-layer chromatography and the purified DNA from lysates of Purkinje neurons, the existence of 5hmC was proven in 2009 and it was shown to be a substantial component of mammalian genomic DNA (Kriaucionis and Heintz, 2009). This discovery was accompanied by the discovery that TET1 could oxidize 5mC to 5hmC (Tahiliani et al., 2009). Two other oxidized derivatives of 5mC; 5fC and 5caC were later identified (Ito et al., 2011).
Subsequent studies have since demonstrated the necessity for the pre-existence of 5mC for 5hmC formation. The knockdown of Dnmt expression results in the disappearance of 5hmC from the genome (Ficz et al., 2011; Williams et al., 2011). The direct generation of 5hmC from non-methylated DNA has not yet been reported, either in vitro or in vivo (Szwagierczak et al., 2010; Ficz et al., 2011).
Whilst participating as intermediates in an active demethylation pathway, evidence of a bona fide epigenetic function of oxi-mCs has also been presented. 5mC, 5hmC, 5fC, and 5caC all appear to have an individual set of binding proteins (or “readers”) that associate with them in a cell type-specific manner (Iurlaro et al., 2013; Spruijt et al., 2013). To show this, pulldowns of proteins crosslinked to DNA fragments specifically containing 5mC, 5hmC, 5fC, and 5caC on nuclear lysates from mouse embryonic stem cells (mESCs), mouse neuronal precursor cells (NPCs) and mouse adult brain cells were performed (Spruijt et al., 2013). Although the exclusion of indirectly bound factors from the analysis was not possible, the constituents of bound complexes were found to differ between both the oxi-5mCs and across some of the cell types analyzed in this study (Spruijt et al., 2013). Whilst most of the complex components were associated with DNA demethylation, indicative of oxi-mCs acting as intermediates in this process, transcription factors such as MeCP2, found to bind 5hmC, were also detected in these experiments (Spruijt et al., 2013). MeCP2 had previously been shown to interact with 5hmC in neuronal cells where 5hmC was mainly found associated with actively expressed genes (Mellén et al., 2012). This together with the finding that 5fC preferentially localizes to poised enhancers in mESCs suggests that oxidation of 5mC itself may serve a regulatory purpose (Song et al., 2013).
Key Regulators of Mouse and Zebrafish DNA Methylation
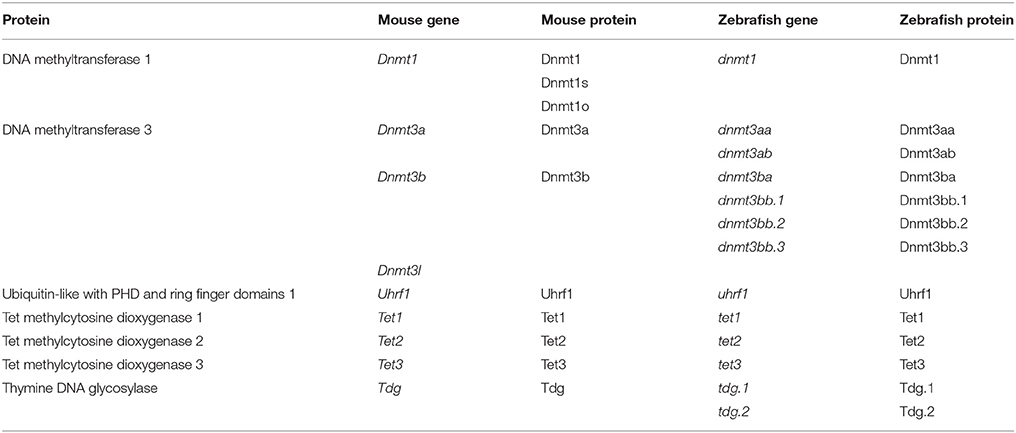
In vertebrates, DNMT1 maintains existing methylation patterns (Stein et al., 1982; Yoder et al., 1997). There are three main splice variants of Dnmt1 expressed during mouse development—Dnmt1 (Li et al., 1992), the oocyte specific Dnmt1o and the somatic Dnmt1s (Howell et al., 2001; Cirio et al., 2008; Hirasawa et al., 2008). Expression of Dnmt1 occurs after embryonic day 7 (E7) (Carlson et al., 1992; Mertineit et al., 1998). Dnmt1o is maternally deposited and the protein active in the zygote (Hirasawa et al., 2008). DNMT1s activity occurs primarily after the first cell division (Cirio et al., 2008), with limited activity likely in the zygote (Hirasawa et al., 2008). DNMT1 activity is believed to be guided by interaction with UHRF1 (also known as NP95) (Bostick et al., 2007; Sharif et al., 2007). UHRF1 also binds to H3K9me3, which marks heterochromatin (Bannister et al., 2001; Lachner et al., 2001; Peters et al., 2001), thus, UHRF1 directs DNMT1 to preserve the long-term silencing of genes (Fang et al., 2016; Harrison et al., 2016; Iurlaro et al., 2017). The de novo methylation of DNA is conducted by two enzymes in mice; DNMT3a and DNMT3b (Li et al., 1998; Okano et al., 1999). DNMT3a is first seen in the developing embryo at E7.5 (Okano et al., 1999), most strongly within extraembryonic tissue, with weak expression seen in the mesoderm (Okano et al., 1999). From E8.5, expression becomes ubiquitous (Okano et al., 1999). DNMT3b is first observed at E7.5 (Okano et al., 1999), most strongly within ectodermal tissue (Okano et al., 1999). DNMT3l is non-catalytic, but recruits DNMT3a and DNMT3b to nucleosomes devoid of H3K4 methylation (Saitou et al., 2012) and is expressed during gametogenesis (Bourc'his et al., 2001). Based upon sequence complementarity, zebrafish carry two likely orthologs to mouse Dnmt3a—dnmt3aa and dnmt3ab, which are conserved in other fish species (Goll and Halpern, 2011), with the orthologs to Dnmt3b likely being dnmt3ba, dnmt3bb.1, dnmt3bb.2, and dnmt3bb.3 (Goll and Halpern, 2011). Orthologs to mouse Dnmt1 and Uhrf1 in zebrafish have also been identified (Mhanni et al., 2001; Goll and Halpern, 2011; Kent et al., 2016). Table 1 provides a list of the epigenetic players in mouse and zebrafish.
In the following text, we will describe the early developmental differences in the dynamics of the methylome and hydroxymethylome in mice and zebrafish. The divergent phenotypes arising from the loss of DNMT, UHRF1 and TET in these two animals highlight the inter-species variation of the role for DNA methylation and hydroxymethylation during vertebrate embryogenesis.
Dnmt and Uhrf1 Knockout Phenotypes in Mice
The homozygous knockout of Dnmt1 causes lethality at E11 and these embryos display overall methylation levels of 30% compared to wildtype controls (Li et al., 1992) (A list of relevant mouse knockout phenotypes is given in Table 2). The presence of 5mC in these embryos was suggestive of other factors participating in its maintenance. These factors were subsequently identified to be DNMT1o (Mertineit et al., 1998) and DNMT1s (Howell et al., 2001). When DNMT1o deposition is prevented, embryonic imprints are largely maintained (Hirasawa et al., 2008), though embryos die at late gestation (Howell et al., 2001). This incomplete erasure is likely due to the presence of zygotic DNMT1s (Hirasawa et al., 2008). When DNMT1o is present in DNMT1s depleted embryos (Kurihara et al., 2008), maternal imprints are partially maintained (Kurihara et al., 2008). The methylome of the inactive maternal X-chromosome is protected by STELLA (Nakamura et al., 2007). The homozygous knockout of Uhrf1 expression results in mid-gestational lethality (Sharif et al., 2007) that phenocopies the homozygous loss of Dnmt1 (Li et al., 1992).
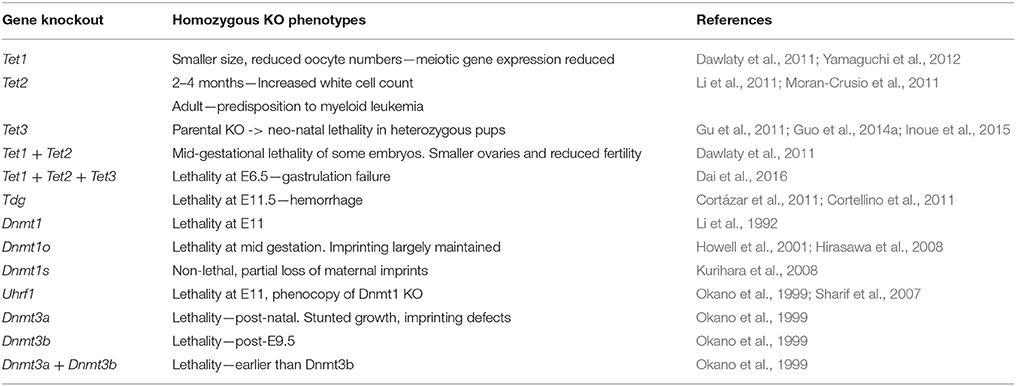
Table 2. Phenotypes resulting from the homozygous knockout of cytosine methylation and oxidation pathway genes in the mouse.
Mice heterozygous for either Dnmt3a or Dnmt3b knockout alleles are normal and fertile (Okano et al., 1999). The homozygous knockout of Dnmt3a is also not embryonically lethal, but results in imprinting defects, stunted growth and premature, post-natal death (Okano et al., 1999). DNMT3b−/− mutant mice do not survive beyond mid-gestation and die after E9.5 (Okano et al., 1999). This timing is indicative of defects in extraembryonic tissue development (Copp, 1995). The combined knockout of Dnmt3a and Dnmt3b results in slightly earlier lethality than in DNMT3b-null mice (Okano et al., 1999).
In primordial germ cells (PGCs), DNMT1 is responsible for the maintenance of DNA methylation during their early maturation (Hargan-Calvopina et al., 2016). The germline-specific deletion of Dnmt1 leads to premature meiotic gene expression and premature erasure of imprinting, causing infertility (Hargan-Calvopina et al., 2016). In PGCs, the loss of Dnmt3l blocks the re-establishment of methylation in maternally imprinted genes (Bourc'his et al., 2001). This loss results in the mid-gestational lethality of heterozygous progeny derived from outcrossing female mice that carry germline deletion of Dnmt3l (Bourc'his et al., 2001). These progeny display biallelic expression of maternally imprinted genes (Bourc'his et al., 2001), with the mid-gestational lethality being indicative of defects in placental formation (Copp, 1995). These Dnmt3l defects are re-capitulated in an analogous experiment, using females carrying the germline specific knockout of Dnmt3a instead (Kaneda et al., 2004). Curiously, the paternal germline-specific knockout of Dnmt3a also results in defects to gametogenesis, characterized by the absence of methylation at certain imprinted loci and sterility (Kaneda et al., 2004). DNMT3b appears to play no role in the development of the germline (Kaneda et al., 2004).
Methods for Studying Methylomic and Hydroxymethylomic Changes
The three most common methods employed for detecting 5mC and oxi-mCs in mice are immunofluorescent staining, bisulfite sequencing and mass spectrometry, but each of these methodologies have limitations (reviewed in Wu and Zhang, 2017). Only mass spectrometry is capable of directly quantifying 5mC across the whole developmental span (Amouroux et al., 2016; Okamoto et al., 2016), but is not currently sensitive enough to quantify oxi-mCs during early development (Okamoto et al., 2016). In zebrafish, the large clutch size makes these problems less pronounced, with direct 5mC and 5hmC quantification being possible using mass spectrometry across the whole development span (Kamstra et al., 2015). In adult organisms, the study of inter-tissue 5mC and 5hmC variation is much more informative than overall changes (Kriaucionis and Heintz, 2009; Globisch et al., 2010; Kamstra et al., 2015). Fortunately, in adult tissue, quantification of 5mC and 5hmC is much easier than in embryos (Kriaucionis and Heintz, 2009; Globisch et al., 2010).
The Developmental Profile of the Mouse Methylome
The early developmental methylomic profile (i.e., pre-organogenesis) within mice, which is illustrated in Figure 2A, has been well defined and re-programming phases coincide with particular development stages (see these excellent reviews, Saitou et al., 2012; Iurlaro et al., 2017). Methylomic re-programming begins at fertilization. At this stage, the sperm and oocyte combine their respective haploid methylomes to form the zygotic methylome. Initial reprogramming of the parental pronuclei takes place rapidly and the first cell division then occurs (Gu et al., 2011; Guo et al., 2014a). Blastomeres divide and differentiation commences in the morula, from which the blastocyst forms. The blastocyst further differentiates into the epiblast and gastrulation occurs. Germline development begins with a population of epiblast cells being epigenetically specified to develop as PGCs (Lawson et al., 1999). Post-gastrulation, organogenesis begins and the embryo develops to adulthood.
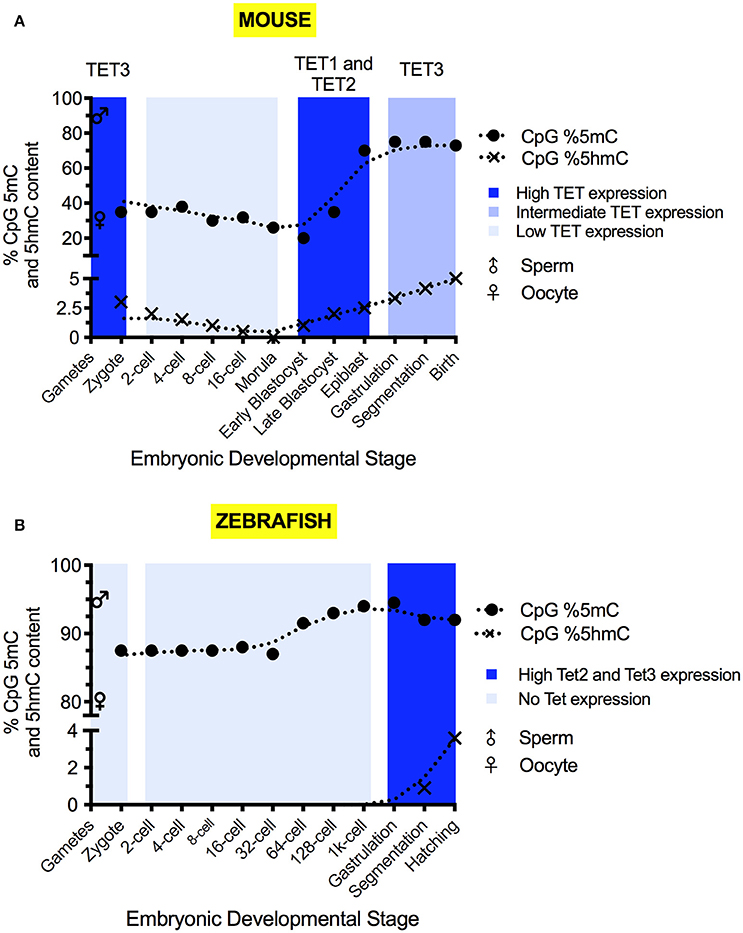
Figure 2. The 5mC and 5hmC content of the mouse (A) and zebrafish (B) during development. 5mC and 5hmC content are expressed as % of genomic CpG sites. Solid circles represent the overall genomic CpG 5mC status (Jiang et al., 2013; Amouroux et al., 2016; Okamoto et al., 2016; Guo et al., 2017), with crosses representing the overall genomic CpG 5hmC status (Pastor et al., 2013; Wu and Zhang, 2014; Kamstra et al., 2015; Amouroux et al., 2016). Sex symbols indicate the 5mC content of sperm or oocytes. Blue panels represent the general expression level of TET mRNA at particular stages (Yamaguchi et al., 2012; Ge et al., 2014; Amouroux et al., 2016; Bogdanović et al., 2016). X-axes labels indicate the developmental stage of the embryo, with shared positions being analogous between species.
Sperm carries a highly methylated genome, with 80–90% of CpG sites marked by 5mC (Amouroux et al., 2016; Guo et al., 2017). The oocyte is comparatively hypomethylated with approximately 30% of CpG sites marked (Amouroux et al., 2016; Guo et al., 2017). Discrepancies between the reported magnitude of methylomic changes exist due to differences of quantitation method used and the cellular pools analyzed (Amouroux et al., 2016; Guo et al., 2017). By the first cell division, paternal 5mC levels have declined to 30–40% of the sperm (Amouroux et al., 2016; Guo et al., 2017), whilst the maternal methylome remains relatively stable, with 5mC declining by no more than 10% (Okamoto et al., 2016; Guo et al., 2017). Overall zygotic methylation levels decrease by roughly 20% from fertilization to the first cell division (Guo et al., 2017) and, in addition to global demethylation of 5mC occurring, it has been shown that limited de novo methylation also takes place in the zygote (Amouroux et al., 2016). Between the 2-cell stage and 8-cell stage, minimal DNA demethylation takes place (Okamoto et al., 2016; Guo et al., 2017).
After the 8-cell stage, 5mC levels decline (Okamoto et al., 2016). Differentiation begins in the morula, with separation of the trophectoderm and the inner cell mass (ICM) allowing implantation to commence (Wang and Dey, 2006). In humans, the two cell layers are distinguished by slight differences to global 5mC levels (Guo et al., 2014b). Embryonic cells have lost their totipotency by blastocyst formation (Wang and Dey, 2006) and 5mC levels reach their developmental minimum outside of the germline (Smith et al., 2012; Kobayashi et al., 2013; Okamoto et al., 2016; Guo et al., 2017). Only 20% of blastocyst CpG sites are methylated (Okamoto et al., 2016; Guo et al., 2017), suggesting that low 5mC levels in mice are required for naïve pluripotency to enable both gastrulation and germline development.
The naïve pluripotent cells of the blastocyst's ICM differentiate into the epiblast in which re-methylation raises 5mC levels to 70% of CpG sites (Smith et al., 2012; Auclair et al., 2014). The epiblast generates the embryonic soma and the germline. PGC formation is epigenetically induced by a BMP signal released from the extraembryonic ectoderm (Lawson et al., 1999). The three embryonic germ layers arise as the primitive streak appears and marks the onset of gastrulation (Tam and Loebel, 2007). In somatic cells, 5mC continues to accumulate post-epiblast, and adult tissues typically display high levels of 5mC (Globisch et al., 2010).
Sexual reproduction requires the formation of gametes. These develop from PGCs. PGC formation begins at approximately E6.5, when BMP4 signaling from the extraembryonic ectoderm (Lawson et al., 1999) induces the transcription of Blimp1 and Prdm14 within a small number of epiblast cells (Saitou et al., 2012). These transcription factors repress the transcription of Dnmt3a, Dnmt3b, Dnmt3l and cause the repression of UHRF1 activity (Kagiwada et al., 2012; Iurlaro et al., 2017). The methylation dynamics of germline development have been well explored in this review (Messerschmidt et al., 2014). The re-establishment of totipotency in gametes requires erasure of genomic imprints (Hackett et al., 2013; Yamaguchi et al., 2013b), re-activation of the silenced X-chromosome (Chuva de Sousa Lopes et al., 2008) and reset of the somatic methylome within the PGC precursors (Saitou et al., 2012). This demethylation is complete prior to PGC sexual specification (Messerschmidt et al., 2014). In mice, germ cell specification and maturation is an epigenetic process, involving two main stages of demethylation (Seisenberger et al., 2012).
The first stage of PGC demethylation commences upon their specification and results in a drop of 5mC levels from approximately 70% of CpG sites to 25% of CpG sites (Seisenberger et al., 2012) (please see Figure 3 for an illustration). This demethylation initiates the re-establishment of totipotency via the reversal of the somatic cell methylation patterns which had developed post-gastrulation (Seki et al., 2005; Kurimoto et al., 2008). During this stage DNMT1 maintains the methylation, specifically, of imprinted regions and meiotic genes (Hargan-Calvopina et al., 2016). DNMT1−/− PGCs display premature expression of key meiotic factors for both sperm and oocyte development, such as Stra8, γH2AX, and Tex12, as well as significantly reduced methylation at imprinting control regions (ICRs), for genes such as H19 and Snrpn (Hargan-Calvopina et al., 2016).
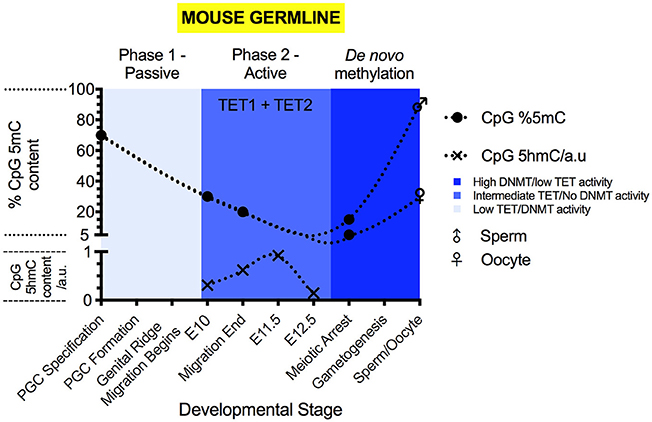
Figure 3. The 5mC and 5hmC content of the mouse germline during development. 5mC content is expressed as % of genomic CpG sites, while 5hmC is expressed as relative change to genomic 5hmC status. Solid circles represent the overall genomic CpG 5mC status (Seisenberger et al., 2012; Guo et al., 2017; Iurlaro et al., 2017), with crosses representing the relative genomic CpG 5hmC status (Hackett et al., 2013; Yamaguchi et al., 2013a). Sex symbols indicate the 5mC content of sperm or oocytes. Blue panels represent the general activities of TET and DNMT enzymes at particular stages (Kagiwada et al., 2012; Hackett et al., 2013). Above each colored panel, the coinciding methylation phase is labeled. X-axes labels indicate the developmental stage of the embryo.
Active demethylation and passive dilution of 5mC, localized to discrete genomic locations, drives the second phase of PGC methylomic reprogramming (E9.5-13.5) (Seisenberger et al., 2012; Yamaguchi et al., 2012; Hackett et al., 2013), after which totipotency is re-established. (Figure 3) The timing of demethylation during the second stage of reprogramming is not simultaneous at all sites (Hackett et al., 2013). Initiation between loci can differ by as much as 24 h (Hackett et al., 2013), suggesting that demethylation of imprints and genes involved in gametogenesis is progressive and controlled. However, areas of the genome are able to escape from the near complete erasure of 5mC in PGCs prior to their sexual specification and re-methylation (Lane et al., 2003; Borgel et al., 2010; Iurlaro et al., 2017). These “escapees” (Iurlaro et al., 2017) are typically repeat elements and of the IAPLTR1 subclass (Hackett et al., 2013), though evidence suggests that some parasitic elements are also demethylated (Wang et al., 2014), for currently obscure reasons.
Once demethylation is complete, sexual specification can proceed (Messerschmidt et al., 2014). In E13.5 female PGCs, the lowest level of 5mC content is observed, at 2.2% of CpG sites (Hackett et al., 2013). From the initiation of sexual specification onward, de novo methylation proceeds, leading to re-establishment of parental imprints in the gametes (Hata et al., 2002; Seisenberger et al., 2012). Eventually, sperm carry 5mC at 80–90% of CpG sites and oocytes carry 30% (Amouroux et al., 2016; Guo et al., 2017).
The Developmental Profile of the Mouse Hydroxymethylome
5hmC is detectable at all stages of pre-implantational development (Ruzov et al., 2011) (Figure 2A). In the zygote, 5hmC is detectable at the mid-pronuclear stage (Ruzov et al., 2011; Amouroux et al., 2016) and increases in abundance up to the first cell division (Gu et al., 2011; Iqbal et al., 2011; Ruzov et al., 2011; Pastor et al., 2013; Amouroux et al., 2016). This increase accompanies the demethylation of the paternal genome seen over time in the zygote (Amouroux et al., 2016; Okamoto et al., 2016; Guo et al., 2017). Zygotic 5fC and 5caC formation transiently accompany 5hmC accumulation (Inoue et al., 2011; Wang et al., 2014; Amouroux et al., 2016). The embryonic levels of 5fC and 5caC are substantially reduced by the four-cell stage (Wang et al., 2014).
Changes to 5hmC levels after the first cell division are low until the morula forms, after which 5hmC content increases and peaks in the blastocyst ICM (Ruzov et al., 2011). As the ICM differentiates and gastrulation occurs, 5hmC becomes localized to particular tissue types and putative stem cell niches (Ruzov et al., 2011).
During PGC maturation, 5hmC is not present during the first phase of demethylation (Hackett et al., 2013; Yamaguchi et al., 2013a) (Figure 3). During the second phase of PGC demethylation, starting at E9.5, 5hmC becomes detectable (Hackett et al., 2013; Yamaguchi et al., 2013a) and the decrease in 5mC abundance is mirrored by increases to 5hmC levels. At E11.5, 5hmC content begins to decline progressively (Hackett et al., 2013; Yamaguchi et al., 2013a), reaching a minimum at E13.5 (Hackett et al., 2013; Yamaguchi et al., 2013a). During the second phase of demethylation in PGCs, 5hmC enrichment can be found at all genetic elements outside of the small number of constitutively methylated sites.
5hmC quantification in adult mice has been restricted to discrete tissues (Kriaucionis and Heintz, 2009; Globisch et al., 2010). LC-MS data from adult tissues reveals that 5hmC content is highly variable, ranging from approximately 0.35–0.7% of genomic cytosine within the CNS, particularly enriched in neurons (Kriaucionis and Heintz, 2009), to being practically undetectable in testes (Globisch et al., 2010). It is believed that neurons contain the highest amount of 5hmC within terminally differentiated cells (Globisch et al., 2010). Why this variation exists is unclear, but is suggestive of roles for 5hmC and TET in regulating the development of discrete adult tissue (Spruijt et al., 2013), or that oxi-mCs can operate as stable, bona fide epigenetic marks (Spruijt et al., 2013).
Tet and Tdg mRNA Expression Patterns
Tet3 mRNA is maternally deposited and protein is expressed in the zygote (Gu et al., 2011) (Figure 2A), before undergoing rapid degradation or cytoplasmic sequestration (Gu et al., 2011; Iqbal et al., 2011). Tet3 mRNA is then re-expressed post mid-gestation (Yamaguchi et al., 2012). The zygotic expression of Tet1 and Tet2 mRNA is very low (Iqbal et al., 2011; Amouroux et al., 2016). Significant Tet1 mRNA expression occurs after the zygotic genome activation (ZGA) and peaks in the ICM (Amouroux et al., 2016). Significant Tet2 mRNA expression is mainly confined to the blastocyst and the somatic tissue post mid-gestation (Yamaguchi et al., 2012; Amouroux et al., 2016; Bogdanović et al., 2016).
In PGCs, Tet1 mRNA expression peaks during the second phase of their methylomic reprogramming (Yamaguchi et al., 2012; Hackett et al., 2013) (Figure 3). PGCs display co-expression of Tet1 and Tet2 mRNA (Yamaguchi et al., 2012), however, Tet2 mRNA expression peaks in the germline much later (Yamaguchi et al., 2012). The peak of Tet2 mRNA expression coincides with the sudden onset of Tet3 mRNA expression within oocytes (Gu et al., 2011; Yamaguchi et al., 2012).
During embryonic development, prior to the blastocyst, Tdg mRNA is barely detectable (Tang et al., 2011). In the ICM and, especially in mESCs, Tdg mRNA expression is comparatively high (Tang et al., 2011), while embryonic expression levels post-gastrulation have not been reported. Tdg mRNA is expressed at high levels within PGCs (Kagiwada et al., 2012) and is present from early to late stages of their maturation (Kagiwada et al., 2012).
Tet and Tdg Knockout Phenotypes in Mice
To study the role of TET1 in mouse development, chimeric males transplanted at blastocyst stage with Tet1+/− mESCs were bred to WT females to produce Tet1+/− progeny. These mice were phenotypically normal and fertile. Homozygous mice derived from a heterozygous incross, although smaller in size, turn out to be viable, showing that TET1 is not essential for ontogenesis (Table 2). When these mice are crossed to WT mice, however, litter sizes were smaller than expected, suggesting reduced fertility of Tet1−/− males and females, and a role for TET1 in male and female germline development (Dawlaty et al., 2011).
An independently generated Tet1 gene-trap line that displayed the same overall phenotype was used to further study the role of TET1 in the male and female germ line (Yamaguchi et al., 2012). Adult females homozygous for the gene trap allele had smaller ovaries with reduced germ cell numbers. Oocyte numbers were already reduced in the Tet1−/− female fetus at E16.5. Fetal ovaries showed increased numbers of apoptotic cells that had failed to progress through the meiotic prophase. Analysis of the methylation profile of the female PGCs at E13.5 revealed that while genome wide demethylation was unaffected, a subset of meiotic genes displayed hypermethylation and reduced expression (Yamaguchi et al., 2012). Unlike the Tet1 knockout ovaries, Tet1 knockout testes were functionally normal. However, embryos derived from crosses of Tet1−/− males with WT females showed heterogeneous phenotypes (Yamaguchi et al., 2013b). All displayed placental growth defects that resulted in variable embryonic and postnatal growth retardation. The placental defects were reminiscent of abnormalities observed in mice with a mutation in the imprinted gene Peg10 (Ono et al., 2006). RNA-Seq experiments performed on E9.5 embryos demonstrated that imprinted genes were indeed dysregulated in the progeny (Yamaguchi et al., 2013b). In E13.5 PGCs of the Tet1−/− homozygous males, imprinted genes displayed hypermethylation that is maintained in the sperm (Yamaguchi et al., 2013b). Altogether these data show that TET1 expression in developing PGCs is not required for global PGC genome demethylation, but plays an important role in (a) the activation of meiotic genes in the developing oocytes and (b) in the erasure of imprints in the germ cells.
Tet2−/− mice display no gestational developmental defects, have normal morphology and are fertile (Li et al., 2011) (Table 2). Incrosses of Tet2−/− mice produce normal litter sizes and morphologically normal embryos (Li et al., 2011). While heterozygous and homozygous Tet2 mutants appear macroscopically normal, analysis of their blood at 2–4 months revealed increased numbers of white blood cells. In mice carrying hematopoietic stem cell (HSC)-specific depletion of Tet2, an increased rate of self-renewal, elevated myeloid progenitor proliferation and a predisposition to myeloid leukemia was observed (Moran-Crusio et al., 2011). These defects were cell-autonomous (Moran-Crusio et al., 2011). Interestingly, the phenotype of Tet1 and Tet2 double homozygous mice (Dawlaty et al., 2013) was reminiscent of Tet1−/− homozygous mice (Yamaguchi et al., 2012, 2013b).
The expression of Tet3 in oocytes and zygotes (Figure 2A) suggests a role for Tet3 at the earliest stages of mouse embryonic development. To study this role, germline-specific knockouts were generated (Table 3). When female mice carrying the germline-specific Tet3 knockout alleles were outcrossed to wildtype mice, the litter size was smaller than expected (Gu et al., 2011). In the zygote, 5mC levels were maintained and 5hmC levels failed to rise in the male and female pronuclei (Gu et al., 2011; Guo et al., 2014a). This coincided with a high rate of developmental failure at mid-gestation stages (Gu et al., 2011), which, however, since has been shown to be dependent on the genetic background (Inoue et al., 2015). Irrespective of this background, the Tet3+/− progeny displayed a considerable degree of neonatal lethality, as well as growth defects which were also observed in Tet3+/− mice that were derived from a cross of Tet3+/− males with wildtype females, suggesting that the neonatal lethality is due to haploinsufficiency rather than the lack of 5mC oxidation at zygotic stages (Inoue et al., 2015). Tet3 mutants all died neonatally (Gu et al., 2011). Altogether, these data suggest important roles for TET3 at different stages of embryonic development that require further investigation.

Table 3. Phenotypes resulting from the homozygous knockout of cytosine methylation and oxidation pathway genes in the mouse germline.
To investigate whether functional redundancy masked roles of TET proteins in embryonic development in the single and Tet1/Tet2 double knockouts, triple knockout embryos were generated by deleting the three Tet genes in the maternal and paternal germline (Table 2). The mice carrying triple Tet+/− sperm and oocytes were crossed to generate triple Tet−/− progeny. These embryos initially developed normally, but then failed to progress beyond gastrulation, with a phenotype being observable at E6.5 (Dai et al., 2016). The phenotype consists of buckling of the epiblast layer in the primitive streak region, defective migration of mesodermal cells and a lack of head folds in the nascent fetus (Dai et al., 2016). In spite of these defects, the expression of early gastrulation markers is still seen (Dai et al., 2016). The definitive endoderm and axial mesoderm are specified, but fail to mature (Dai et al., 2016). The paraxial mesoderm is absent and neuroectodermal development is abnormal (Dai et al., 2016). These observations indicate that gastrulation was initiated, but had failed to progress (Dai et al., 2016). This phenotype was caused by hyperactive NODAL signaling due to the transcriptional silencing of Lefty1 and Lefty2 (Dai et al., 2016). The Lefty genes encode inhibitors of NODAL signaling (Meno et al., 1998, 1999). The silencing of the Lefty genes correlates with elevated DNA methylation at their cis-regulatory element in the absence of all three TET proteins (Dai et al., 2016). Normal Lefty expression and the phenotype of the embryos were rescued by the presence of a single copy of WT Tet3 or by the knockout of Dnmt3 (Dai et al., 2016).
TDG is essential for mouse embryonic development (Cortázar et al., 2011; Cortellino et al., 2011) (Table 2). While Tdg+/− embryos and adult mice are normal, Tdg−/− mice develop internal hemorrhage and necrosis (Cortázar et al., 2011; Cortellino et al., 2011). The phenotype manifests at E10.5 and leads to lethality at E11.5, for unknown reasons (Wu and Zhang, 2017). The analysis of gene expression in mouse embryonic fibroblasts (MEFs) isolated from E9.5 Tdg−/− mouse embryos suggests a role for TDG in transcriptional gene regulation (Cortázar et al., 2011; Cortellino et al., 2011). Tdg−/− MEFs, as well as Tdg−/− ES cells induced to differentiate into neural progenitor cells show reduced expression of numerous developmental genes (Cortázar et al., 2011). The downregulation of these genes in differentiating cells coincides with the presence of repressive histone modifications and the methylation of CpG sites in their promoter regions (Cortázar et al., 2011; Cortellino et al., 2011). These data suggest that TDG acts in concert with other partners like the TET proteins, the deaminase AID and histone modifiers, like CBP/p300 and MLL to maintain activating marks on developmental gene promoters to prevent their Polycomb- and DNMT3a/b-mediated repression during differentiation (Cortázar et al., 2011; Cortellino et al., 2011).
Zebrafish Embryonic Development and the Role of DNA Methylation
In zebrafish, external fertilization of the egg is followed by external embryonic development. The embryo's yolk provides the nutrition and gas diffusion through the skin is sufficient for gas exchange. Extraembryonic membranes are not formed, and the embryos undergo an extended period of cell cleavages before zygotic genome activation occurs at approximately the 1,000 (1k) cell stage (Kimmel et al., 1995; Pálfy et al., 2017). Unlike in mice, PGCs are not induced, but predetermined by the maternal deposition of germ plasm in the oocyte (Raz, 2003). Therefore, blastodermal cells of the 1k stage embryo that lack germ plasm do not need to be programmed to be able give rise to cells of the germline. Thus, naïve pluripotent cells do not exist. These differences in embryology are reflected in the methylome and hydroxymethylome of zebrafish, as will be explained below.
DNA methylation plays an important role during zebrafish development. The loss of zygotic expression of Dnmt1 in dnmt1−/− mutants causes an embryonic phenotype that manifests from 84 hpf and leads to larval death by 8 dpf. Embryos exhibit a small-sized pancreas, liver and eyes, as well as dysmorphic branchial arches (Anderson et al., 2009). Within the exocrine pancreas, acinar cells are specified, but fail to proliferate and eventually undergo apoptosis (Anderson et al., 2009). A smaller liver is also observed upon zygotic loss of uhrf1 expression (Sadler et al., 2007; Jacob et al., 2015). uhrf1−/− mutants show defects in liver outgrowth and liver regeneration. Hepatocytes undergo re-replication of their DNA, fail to divide and undergo apoptosis. Uhrf1 and dnmt1 mutants also display defects in lens development (Tittle et al., 2011) and intestinal barrier function (Marjoram et al., 2015). Dnmt1 mutants also struggle to maintain haematopoietic progenitor cells (Liu et al., 2015). Maternal supply of Dnmt1 and Uhrf1 is likely to mask earlier roles of DNA methylation in the zygotic mutants.
Translation-blocking morpholinos that target maternal and zygotic mRNA show that depletion of maternal and zygotic Uhrf1 protein causes pre-gastrulation lethality, suggesting an essential role for maternal Uhrf1 protein at ZGA (Chu et al., 2012; Kent et al., 2016). By contrast, Dnmt1 morphants display late differentiation defects in the intestine, the exocrine pancreas and the retina (Rai et al., 2006). The presence of 6 orthologs of mouse Dnmt3 in zebrafish makes loss of function studies difficult as the role of individual genes may be masked by functional redundancy (Goll and Halpern, 2011). Nevertheless, dnmt3bb.1 mutants have been shown to have very specific defects in haematopoietic progenitor maintenance (Gore et al., 2016). Morpholino-mediated knockdown of Dnmt3bb.2 causes neurogenic defects in the brain and the retina (Rai et al., 2010). Inhibition of all Dnmt activity, using 5-azacytidine, lead to defects in somite development, notochord and muscle, as well as to trunk shortening (Martin et al., 1999; Goll and Halpern, 2011), however the compound's instability in water complicates data analysis and interpretation (Goll and Halpern, 2011). The different embryonic phenotypes caused by 5-azacytidine treatment and Uhrf1, Dnmt1 and Dnmt3 morpholino-injection highlights that more work is required to gain a better understanding of the role of DNA methylation during early zebrafish embryogenesis.
Developmental Dynamics of the Zebrafish Methylome and Hydroxymethylome
Bisulfite sequencing of genomic DNA isolated from sperm, oocytes and early stage embryos revealed that 91–95% of CpG dinucleotides are methylated in sperm and 75–80% are methylated in oocytes (Jiang et al., 2013; Potok et al., 2013) (Figure 2B). There is very little methylation in non-CpG sites (Potok et al., 2013). As the zygote undergoes cell divisions, DNA methylation is reduced and reaches a minimum at the 64-cell stage. Subsequent re-methylation is gradual and takes CpG methylation levels back to that of sperm by the time the embryo reaches sphere stage (Potok et al., 2013). By the time of ZGA, the oocyte methylome has been reprogrammed to match the sperm (Jiang et al., 2013; Potok et al., 2013). Close inspection of the methylation in CpG islands at promoter sites revealed that the methylome is static for large numbers of genes (Potok et al., 2013). Constitutively hypomethylated TSSs are found in genes involved in basal cellular processes, such as metabolism, transcription and early development (Potok et al., 2013). Shared, constitutively hypermethylated promoters are associated with genes related to later developmental processes, such as neurogenesis (Potok et al., 2013). A small number of CpG islands display dynamic methylation patterns (Potok et al., 2013). A subset of those are hypermethylated in the maternal genome and are subsequently demethylated prior to the midblastula transition (MBT) (Potok et al., 2013). These include the TSSs of genes with roles in germ cell and embryonic development (Potok et al., 2013). Other genes have hypomethylated TSSs in the oocyte that are methylated upon zygotic gene activation. These include genes involved in oogenesis and late development (Potok et al., 2013). Comparison of methylomic and transcriptomic data revealed that genes with hypermethylated promoter sequences were silent while genes with hypomethylated TSSs were more likely to be expressed. Some hypomethylated genes that are not expressed are likely to be under the additional epigenetic control, for example by histones (Jiang et al., 2013; Potok et al., 2013). The hox genes are hypomethylated in sperm, but packaged in H3K4me3 and H3K27me3-bivalently marked chromatin and, therefore, poised, but not active (Jiang et al., 2013; Potok et al., 2013). Outside of promoters, differentially methylated regions are also found insides genes. These are suspected to include enhancer sequences (Potok et al., 2013). Thus, the dynamic methylomic changes that occur on the maternal genome adjust the maternal genome to the paternal genome. This is in preparation for the onset of biallelic zygotic gene transcription. The methylomic changes on the maternal genome do not require the presence of the paternal template. They occur even in maternal haploids generated by parthenogenesis, when eggs are triggered to develop by fertilization with UV-treated and therefore genome-inactivated sperm (Potok et al., 2013). The absence of widespread demethylation of the zebrafish genome between fertilization and ZGA likely reflects differences in embryonic development compared to mice. The early zebrafish cells neither need to give rise to extraembryonic tissue nor do they need to generate PGCs. The lack of imprinting (Macleod et al., 1999; Potok et al., 2013) means that the global, active demethylation occurring prior to gametogenesis in mice is not required in zebrafish (Yamaguchi et al., 2013b). PGCs are also pre-determined by the inheritance of germ plasm and set aside at the beginning of embryonic development (Raz, 2003). Thus, blastomeres lacking germ plasm neither require the ability to generate PGCs nor need to be programmed for naïve pluripotency, which is linked to genome demethylation. This allows the zebrafish to pre-set the zygotic methylome in the gametes, with minor amendments to the maternal methylome being performed during the early cleavage period.
From MBT onwards, the embryonic methylome begins to diverge from the sperm (Jiang et al., 2013). The methylomic changes that occur during this period are much more substantial than those seen prior to MBT (Jiang et al., 2013; Potok et al., 2013). A comparison of the methylome of sphere stage embryos with that of differentiated muscle cells reveals thousands of differentially methylated sequences. Most of these are likely to be intronic enhancers and many of them are hypermethylated in the differentiated muscle cells (Potok et al., 2013). Single-base resolution methylome maps of zebrafish embryos at different stages of development also pinpoint thousands of differentially methylated regions of the genome (Lee et al., 2015). Many of these lie outside of promoters, CpG islands and nearby shores. These sequences are enriched for evolutionary conserved sequences or have previously been associated with histone modification and transcription factor binding signatures, suggesting that they constitute cis-regulatory elements. Twenty of the elements have been tested in stable transgenic zebrafish and shown to act as tissue-specific enhancers (Lee et al., 2015). Thus, differential methylation is another indicator for active enhancer sequences. Overall, the use of DNA methylation in the regulation of promoter and enhancer activity is conserved in fish and mouse.
The changes in the methylation profiles seen after MBT coincide with an increase in 5hmC levels (Lee et al., 2015) (Figure 2B)—indicative of active demethylation occurring. 5hmC only becomes observable at 12 hpf (Kamstra et al., 2015) and, unlike in mice, 5hmC does not correlate positively with pluripotency (Ruzov et al., 2011; Almeida et al., 2012). Embryonic 5hmC content peaks at 96 hpf with 0.23% of genomic cytosine, and the rise is concomitant with a decline in 5mC content (Kamstra et al., 2015). 5hmC presence is maintained into adulthood, displaying inter-tissue enrichment variation akin to mammals (Globisch et al., 2010; Kamstra et al., 2015). As in mammals (Globisch et al., 2010), 5hmC is most abundant in the brain, reaching 0.5% of cytosine content (Kamstra et al., 2015) and the reasons for this enrichment are likewise unknown.
Expression Profiles of Tet and Tdg mRNA and Knockout Phenotypes in Zebrafish
The zebrafish genome contains paralogs to Tdg in the mouse, these are tdg.1 and tdg.2 (Yates et al., 2016, ENSEMBL release 90). Their expression patterns have not been published. Neither loss of function nor gain of function data has been published.
The expression of the tet genes and the presence of 5hmC are virtually undetectable prior to 24 hpf (Almeida et al., 2012; Ge et al., 2014; Kamstra et al., 2015; Bogdanović et al., 2016), suggesting they make no contribution to pre-organogenic zebrafish development. The generally low expression of tet1 mRNA compared to tet2 and tet3 (Bogdanović et al., 2016) indicates that Tet2 and Tet3 make the dominant contribution to 5mC oxidation in zebrafish. Why the level of tet1 expression is low remains to be determined.
Individual tet gene deletion does not cause a noticeable reduction to 5hmC levels at embryonic stages. All tet mutants are viable and fertile (Li et al., 2015) (Table 4). The lack of 5hmC reduction in zebrafish suggests functional redundancy and that compensatory increases in expression to the remaining tet genes may occur. The latter has not been explored. In double tet2−/− tet3−/− fish, a reduction in the 5hmC level is observed at 5 dpf and embryonic abnormalities are visible (Li et al., 2015). The phenotype includes the loss of HSC formation on day 1 (Li et al., 2015). From day 1 of development, abnormalities in eye development, brain morphology and pigmentation are visible, and defects in HSC production are detectable (Li et al., 2015). The tet2−/− and tet3−/− double knockout zebrafish die during the larval period (Li et al., 2015). Triple tet mutants show a further reduction of 5mC levels, but present the same phenotype (Li et al., 2015). There are no gastrulation defects. Thus, the role that mouse Tet proteins have in the activation of lefty1 expression and the control of the Nodal morphogen gradient prior to gastrulation does not seem to be conserved in the zebrafish embryo. By contrast, a late role for Tet2 in haematopoietic cells is conserved from fish to man. 24-month-old Tet2 mutant zebrafish display a myelodysplastic syndrome that is also observed in human patients and mouse models with TET2 mutation in their haematopoietic cells (Gjini et al., 2015). Altogether, the phenotypes of the tet2 single and tet2/tet3 double mutant zebrafish suggest that Tet proteins play very specific roles during lineage specification and differentiation in zebrafish. Current data suggest that these roles may or may not be conserved between mammals and fish.
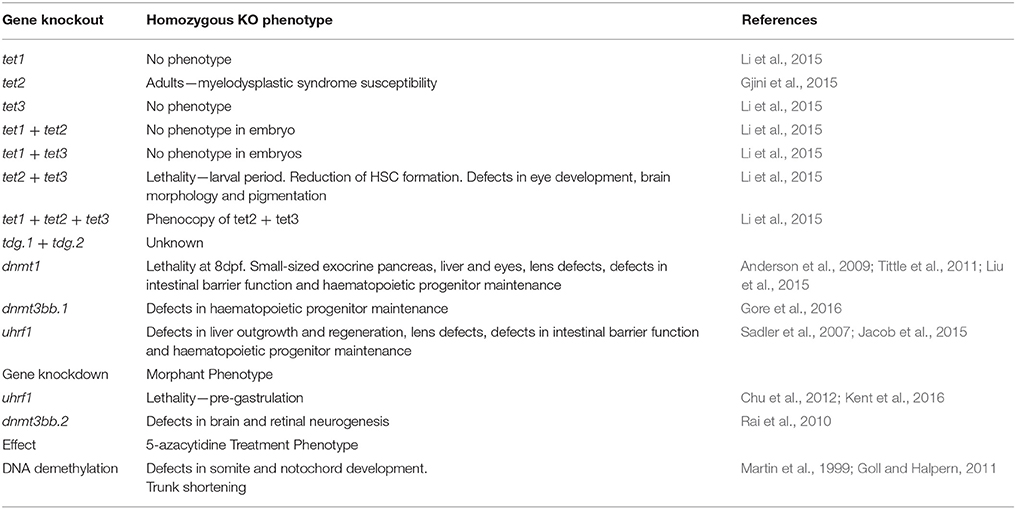
Table 4. Phenotypes resulting from the loss of function of cytosine methylation and oxidation pathway genes in the zebrafish.
Concluding Remarks
During vertebrate embryonic development, cellular differentiation relies on the differential expression of thousands of genes. Epigenetic mechanisms, including DNA methylation, act on cis-regulatory elements to control their expression. DNA methylation usually leads to the repression of gene expression. Zebrafish and mouse use essentially the same enzymatic machinery to set, maintain, modify and remove these methylation marks. The way how they use this machinery early in embryogenesis, however, differs considerably and reflects important differences in their embryogenesis. The mammalian embryo (a) has the immediate need to generate extraembryonic membranes for its survival, (b) uses imprinting to prevent parthenogenesis, and (c) needs to generate naïve pluripotent cells in the blastocyst that are able give rise to cells of the germline. Zebrafish do not have any of these requirements. Their yolk provides the nutrition, while gas exchange is achieved through the skin, as embryos develop externally. Zebrafish have not evolved a mechanism to prevent parthenogenesis through imprinting. Furthermore, zebrafish PGCs are predetermined by germ-plasm and are not induced through epigenetic mechanisms. Therefore, unlike in the mouse, the roles of zebrafish Tet proteins are restricted to the regulation of specific processes occurring during lineage specification and differentiation. The few examples we know suggest that their involvement in a particular process may or may not be conserved between mammals and fish.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the Medical Research Council (MR/J000841/1 and G0601134 to MG) and by the BBSRC (BB/N005759/1 to AR). PJ has been supported by the BBSRC [DTP fellowship (BB/J014508/1)] and the University of Nottingham.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Almeida, R. D., Loose, M., Sottile, V., Matsa, E., Denning, C., Young, L., et al. (2012). 5-hydroxymethyl-cytosine enrichment of non-committed cells is not a universal feature of vertebrate development. Epigenetics 7, 383–389. doi: 10.4161/epi.19375
Amouroux, R., Nashun, B., Shirane, K., Nakagawa, S., Hill, P. W. S., D'Souza, Z., et al. (2016). De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18, 225–233. doi: 10.1038/ncb3296
Anderson, R. M., Bosch, J. A., Goll, M. G., Hesselson, D., Dong, P. D. S., Shin, D., et al. (2009). Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev. Biol. 334, 213–223. doi: 10.1016/j.ydbio.2009.07.017
Auclair, G., Guibert, S., Bender, A., and Weber, M. (2014). Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol. 15:545. doi: 10.1186/s13059-014-0545-5
Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., et al. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124. doi: 10.1038/35065138
Bird, A. (2002). DNA methylation patterns and epigenetic memory DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. doi: 10.1101/gad.947102
Bogdanović, O., Smits, A. H., de la Calle Mustienes, E., Tena, J. J., Ford, E., Williams, R., et al. (2016). Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat. Genet. 48, 417–426. doi: 10.1038/ng.3522
Borgel, J., Guibert, S., Li, Y., Chiba, H., Schübeler, D., Sasaki, H., et al. (2010). Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42, 1093–1100. doi: 10.1038/ng.708
Bostick, M., Kim, J. K., Estève, P. O., Clark, A., Pradhan, S., and Jacobsen, S. E. (2007). UHRF1 plays a role in maintaining dna methylation in mammalian cells. Science 317, 1760–1764. doi: 10.1126/science.1147939
Bourc'his, D., Xu, G.-L., Lin, C.-S., Bollman, B., and Bestor, T. H. (2001). Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539. doi: 10.1126/science.1065848
Boyer, L. A., Lee, T. I., Cole, M. F., Johnstone, S. E., Levine, S. S., Zucker, J. P., et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. doi: 10.1016/j.cell.2005.08.020
Carlson, L. L., Page, A. W., and Bestor, T. H. (1992). Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 6, 2536–2541. doi: 10.1101/gad.6.12b.2536
Carninci, P., Sandelin, A., Lenhard, B., Katayama, S., Shimokawa, K., Ponjavic, J., et al. (2006). Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626–635. doi: 10.1038/ng1789
Chambers, I., Silva, J., Colby, D., Nichols, J., Nijmeijer, B., Robertson, M., et al. (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234. doi: 10.1038/nature06403
Chatterjee, A., Ozaki, Y., Stockwell, P. A., Horsfield, J. A., Morison, I. M., and Nakagawa, S. (2013). Mapping the zebrafish brain methylome using reduced representation bisulfite sequencing. Epigenetics 8, 979–989. doi: 10.4161/epi.25797
Chu, J., Loughlin, E. A., Gaur, N. A., SenBanerjee, S., Jacob, V., Monson, C., et al. (2012). UHRF1 phosphorylation by cyclin A2/cyclin-dependent kinase 2 is required for zebrafish embryogenesis. Mol. Biol. Cell 23, 59–70. doi: 10.1091/mbc.E11-06-0487
Chuva de Sousa Lopes, S. M., Hayashi, K., Shovlin, T. C., Mifsud, W., Surani, M. A., McLaren, A., et al. (2008). X chromosome activity in mouse xx primordial germ cells. PLoS Genet. 4:e30. doi: 10.1371/journal.pgen.0040030
Cirio, M. C., Ratnam, S., Ding, F., Reinhart, B., Navara, C., and Chaillet, J. R. (2008). Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev. Biol. 8:9. doi: 10.1186/1471-213X-8-9
Copp, A. J. (1995). Death before birth: clues from gene knockouts and mutations. Trends Genet. 11, 87–93. doi: 10.1016/S0168-9525(00)89008-3
Cortázar, D., Kunz, C., Selfridge, J., Lettieri, T., Saito, Y., MacDougall, E., et al. (2011). Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423. doi: 10.1038/nature09672
Cortellino, S., Xu, J., Sannai, M., Moore, R., Caretti, E., Cigliano, A., et al. (2011). Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79. doi: 10.1016/j.cell.2011.06.020
Dai, H.-Q., Wang, B.-A., Yang, L., Chen, J.-J., Zhu, G.-C., Sun, M.-L., et al. (2016). TET-mediated DNA demethylation controls gastrulation by regulating Lefty–Nodal signalling. Nature 538, 528–532. doi: 10.1038/nature20095
Dawlaty, M. M., Breiling, A., Le, T., Raddatz, G., Barrasa, M. I., Cheng, A. W., et al. (2013). Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323. doi: 10.1016/j.devcel.2012.12.015
Dawlaty, M. M., Ganz, K., Powell, B. E., Hu, Y.-C., Markoulaki, S., Cheng, A. W., et al. (2011). Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9, 166–175. doi: 10.1016/j.stem.2011.07.010
Fang, J., Cheng, J., Wang, J., Zhang, Q., Liu, M., Gong, R., et al. (2016). Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat. Commun. 7:11197. doi: 10.1038/ncomms11197
Ficz, G., Branco, M. R., Seisenberger, S., Santos, F., Krueger, F., Hore, T. A., et al. (2011). Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402. doi: 10.1038/nature10008
Ge, L., Zhang, R.-P., Wan, F., Guo, D.-Y., Wang, P., Xiang, L.-X., et al. (2014). TET2 plays an essential role in erythropoiesis by regulating lineage-specific genes via dna oxidative demethylation in a Zebrafish Model. Mol. Cell. Biol. 34, 989–1002. doi: 10.1128/MCB.01061-13
Gjini, E., Mansour, M. R., Sander, J. D., Moritz, N., Nguyen, A. T., Kesarsing, M., et al. (2015). A Zebrafish Model of myelodysplastic syndrome produced through tet2 genomic editing. Mol. Cell. Biol. 35, 789–804. doi: 10.1128/MCB.00971-14
Globisch, D., Münzel, M., Müller, M., Michalakis, S., Wagner, M., Koch, S., et al. (2010). Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE 5:e15367. doi: 10.1371/journal.pone.0015367
Goll, M. G., and Halpern, M. E. (2011). DNA methylation in zebrafish. Prog. Mol. Biol. Transl. Sci. 101, 193–218. doi: 10.1016/B978-0-12-387685-0.00005-6
Gore, A. V., Athans, B., Iben, J. R., Johnson, K., Russanova, V., Castranova, D., et al. (2016). Epigenetic regulation of hematopoiesis by DNA methylation. Elife 5, 1–25. doi: 10.7554/eLife.11813
Gu, T.-P., Guo, F., Yang, H., Wu, H.-P., Xu, G.-F., Liu, W., et al. (2011). The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610. doi: 10.1038/nature10443
Guo, F., Li, L., Li, J., Wu, X., Hu, B., Zhu, P., et al. (2017). Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 27, 967–988. doi: 10.1038/cr.2017.82
Guo, F., Li, X., Liang, D., Li, T., Zhu, P., Guo, H., et al. (2014a). Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–458. doi: 10.1016/j.stem.2014.08.003
Guo, H., Zhu, P., Yan, L., Li, R., Hu, B., Lian, Y., et al. (2014b). The DNA methylation landscape of human early embryos. Nature 511, 606–610. doi: 10.1038/nature13544
Hackett, J. A., Sengupta, R., Zylicz, J. J., Murakami, K., Lee, C., Down, T. A., et al. (2013). Germline DNA demethylation dynamics and imprint erasure through 5-Hydroxymethylcytosine. Science 339, 448–452. doi: 10.1126/science.1229277
Hargan-Calvopina, J., Taylor, S., Cook, H., Hu, Z., Lee, S. A., Yen, M.-R., et al. (2016). Stage-Specific demethylation in primordial germ cells safeguards against precocious differentiation. Dev. Cell 39, 75–86. doi: 10.1016/j.devcel.2016.07.019
Harrison, J. S., Cornett, E. M., Goldfarb, D., Darosa, P. A., Li, Z. M., Yan, F., et al. (2016). Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. Elife 5, 1–24. doi: 10.7554/eLife.17101
Hata, K., Okano, M., Lei, H., and Li, E. (2002). Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993.
He, Y.-F., Li, B.-Z., Li, Z., Liu, P., Wang, Y., Tang, Q., et al. (2011). Tet-Mediated formation of 5-Carboxylcytosine and its excision by TDG in mammalian, D. N. A. Science 333, 1303–1307. doi: 10.1126/science.1210944
Hirasawa, R., Chiba, H., Kaneda, M., Tajima, S., Li, E., Jaenisch, R., et al. (2008). Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 22, 1607–1616. doi: 10.1101/gad.1667008
Howell, C. Y., Bestor, T. H., Ding, F., Latham, K. E., Mertineit, C., Trasler, J. M., et al. (2001). Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104, 829–838. doi: 10.1016/S0092-8674(01)00280-X
Inoue, A., Shen, L., Dai, Q., He, C., and Zhang, Y. (2011). Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 21, 1670–1676. doi: 10.1038/cr.2011.189
Inoue, A., Shen, L., Matoba, S., and Zhang, Y. (2015). Haploinsufficiency, but not defective paternal 5mC oxidation, accounts for the developmental defects of maternal Tet3 knockouts. Cell Rep. 10, 463–470. doi: 10.1016/j.celrep.2014.12.049
Iqbal, K., Jin, S.-G., Pfeifer, G. P., and Szabó, P. E. (2011). Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U. S. A. 108, 3642–3647. doi: 10.1073/pnas.1014033108
Ito, S., D'Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi: 10.1038/nature09303
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi: 10.1126/science.1210597
Iurlaro, M., Ficz, G., Oxley, D., Raiber, E.-A., Bachman, M., Booth, M. J., et al. (2013). A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 14:R119. doi: 10.1186/gb-2013-14-10-r119
Iurlaro, M., von Meyenn, F., and Reik, W. (2017). DNA methylation homeostasis in human and mouse development. Curr. Opin. Genet. Dev. 43, 101–109. doi: 10.1016/j.gde.2017.02.003
Jacob, V., Chernyavskaya, Y., Chen, X., Tan, P. S., Kent, B., Hoshida, Y., et al. (2015). DNA hypomethylation induces a DNA replication-associated cell cycle arrest to block hepatic outgrowth in uhrf1 mutant zebrafish embryos. Development 142, 510–521. doi: 10.1242/dev.115980
Jang, H. S., Shin, W. J., Lee, J. E., and Do, J. T. (2017). CpG and Non-CpG methylation in epigenetic gene regulation and brain function. Genes (Basel). 8:148. doi: 10.3390/genes8060148
Jiang, L., Zhang, J., Wang, J. J., Wang, L., Zhang, L., Li, G., et al. (2013). Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153, 773–784. doi: 10.1016/j.cell.2013.04.041
Kagiwada, S., Kurimoto, K., Hirota, T., Yamaji, M., and Saitou, M. (2012). Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 32, 340–353. doi: 10.1038/emboj.2012.331
Kamstra, J. H., Løken, M., Aleström, P., and Legler, J. (2015). Dynamics of DNA Hydroxymethylation in zebrafish. Zebrafish 12, 230–237. doi: 10.1089/zeb.2014.1033
Kaneda, M., Okano, M., Hata, K., Sado, T., Tsujimoto, N., Li, E., et al. (2004). Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903. doi: 10.1038/nature02633
Kent, B., Magnani, E., Walsh, M. J., and Sadler, K. C. (2016). UHRF1 regulation of Dnmt1 is required for pre-gastrula zebrafish development. Dev. Biol. 412, 99–113. doi: 10.1016/j.ydbio.2016.01.036
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kobayashi, H., Sakurai, T., Miura, F., Imai, M., Mochiduki, K., Yanagisawa, E., et al. (2013). High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 23, 616–627. doi: 10.1101/gr.148023.112
Kothari, R. M., and Shankar, V. (1976). 5-methylcytosine content in the vertebrate deoxyribonucleic acids: species specificity. J. Mol. Evol. 7, 325–329. doi: 10.1007/BF01743628
Kriaucionis, S., and Heintz, N. (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. doi: 10.1126/science.1169786
Kurihara, Y., Kawamura, Y., Uchijima, Y., Amamo, T., Kobayashi, H., Asano, T., et al. (2008). Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev. Biol. 313, 335–346. doi: 10.1016/j.ydbio.2007.10.033
Kurimoto, K., Yamaji, M., Seki, Y., and Saitou, M. (2008). Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle 7, 3514–3518. doi: 10.4161/cc.7.22.6979
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120. doi: 10.1038/35065132
Lane, N., Dean, W., Erhardt, S., Hajkova, P., Surani, A., Walter, J., et al. (2003). Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35, 88–93. doi: 10.1002/gene.10168
Lawson, K. A., Dunn, N. R., Roelen, B. A. J., Zeinstra, L. M., Davis, A. M., Wright, C. V. E., et al. (1999). Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13, 424–436. doi: 10.1101/gad.13.4.424
Lee, H. J., Lowdon, R. F., Maricque, B., Zhang, B., Stevens, M., Li, D., et al. (2015). Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat. Commun. 6:6315. doi: 10.1038/ncomms7315
Li, C., Lan, Y., Schwartz-Orbach, L., Korol, E., Tahiliani, M., Evans, T., et al. (2015). Overlapping requirements for Tet2 and Tet3 in normal development and Hematopoietic stem cell emergence. Cell Rep. 12, 1133–1143. doi: 10.1016/j.celrep.2015.07.025
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. doi: 10.1016/0092-8674(92)90611-F
Li, E., Okano, M., and Xie, S. (1998). Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219–220. doi: 10.1038/890
Li, Z., Cai, X., Cai, C.-L., Wang, J., Zhang, W., Petersen, B. E., et al. (2011). Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118, 4509–4518. doi: 10.1182/blood-2010-12-325241
Liu, X., Jia, X., Yuan, H., Ma, K., Chen, Y., Jin, Y., et al. (2015). DNA methyltransferase 1 functions through C/ebpa to maintain hematopoietic stem and progenitor cells in zebrafish. J. Hematol. Oncol. 8:15. doi: 10.1186/s13045-015-0115-7
Macleod, D., Clark, V. H., and Bird, A. (1999). Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat. Genet. 23, 139–140. doi: 10.1038/13767
Maiti, A., and Drohat, A. C. (2011). Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338. doi: 10.1074/jbc.C111.284620
Marjoram, L., Alvers, A., Deerhake, M. E., Bagwell, J., Mankiewicz, J., Cocchiaro, J. L., et al. (2015). Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proc. Natl. Acad. Sci.U.S.A. 112, 2770–2775. doi: 10.1073/pnas.1424089112
Martin, C. C., Laforest, L., Akimenko, M. A., and Ekker, M. (1999). A role for DNA methylation in gastrulation and somite patterning. Dev. Biol. 206, 189–205. doi: 10.1006/dbio.1998.9105
Mellén, M., Ayata, P., Dewell, S., Kriaucionis, S., and Heintz, N. (2012). MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430. doi: 10.1016/j.cell.2012.11.022
Meno, C., Gritsman, K., Ohishi, S., Ohfuji, Y., Heckscher, E., Mochida, K., et al. (1999). Mouse lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol. Cell 4, 287–298. doi: 10.1016/S1097-2765(00)80331-7
Meno, C., Shimono, A., Saijoh, Y., Yashiro, K., Mochida, K., Ohishi, S., et al. (1998). Lefty-1 is required for Left-Right determination as a regulator of Lefty-2 and nodal. Cell 94, 287–297. doi: 10.1016/S0092-8674(00)81472-5
Mertineit, C., Yoder, J., Taketo, T., Laird, D. W., Trasler, J. M., and Bestor, T. H. (1998). Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development 125, 889–897.
Messerschmidt, D. M., Knowles, B. B., and Solter, D. (2014). DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812–828. doi: 10.1101/gad.234294.113
Mhanni, A. A., Yoder, J. A., Dubesky, C., and McGowan, R. A. (2001). Cloning and sequence analysis of a zebrafish cDNA encoding DNA (cytosine-5)-methyltransferase-1. Genesis 30, 213–219. doi: 10.1002/gene.1067
Moran-Crusio, K., Reavie, L., Shih, A., Abdel-Wahab, O., Ndiaye-Lobry, D., Lobry, C., et al. (2011). Tet2 Loss Leads to increased Hematopoietic stem cell Self-Renewal and myeloid transformation. Cancer Cell 20, 11–24. doi: 10.1016/j.ccr.2011.06.001
Moris, N., Pina, C., and Arias, A. M. (2016). Transition states and cell fate decisions in epigenetic landscapes. Nat. Rev. Genet. 17, 693–703. doi: 10.1038/nrg.2016.98
Nakamura, T., Arai, Y., Umehara, H., Masuhara, M., Kimura, T., Taniguchi, H., et al. (2007). PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9, 64–71. doi: 10.1038/ncb1519
Okamoto, Y., Yoshida, N., Suzuki, T., Shimozawa, N., Asami, M., Matsuda, T., et al. (2016). DNA methylation dynamics in mouse preimplantation embryos revealed by mass spectrometry. Sci. Rep. 6:19134. doi: 10.1038/srep19134
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. doi: 10.1016/S0092-8674(00)81656-6
Ono, R., Nakamura, K., Inoue, K., Naruse, M., Usami, T., Wakisaka-Saito, N., et al. (2006). Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106. doi: 10.1038/ng1699
Pálfy, M., Joseph, S. R., and Vastenhouw, N. L. (2017). The timing of zygotic genome activation. Curr. Opin. Genet. Dev. 43, 53–60. doi: 10.1016/j.gde.2016.12.001
Pastor, W. A., Aravind, L., and Rao, A. (2013). TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356. doi: 10.1038/nrm3589
Penn, N. W., Suwalski, R., O'Riley, C., Bojanowski, K., and Yura, R. (1972). The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 126, 781–790.
Peters, A. H. F. M., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schöfer, C., et al. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337. doi: 10.1016/S0092-8674(01)00542-6
Potok, M. E., Nix, D. A., Parnell, T. J., and Cairns, B. R. (2013). Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772. doi: 10.1016/j.cell.2013.04.030
Rai, K., Jafri, I. F., Chidester, S., James, S. R., Karpf, A. R., Cairns, B. R., et al. (2010). Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J. Biol. Chem. 285, 4110–4121. doi: 10.1074/jbc.M109.073676
Rai, K., Nadauld, L. D., Chidester, S., Manos, E. J., James, S. R., Karpf, A. R., et al. (2006). Zebra Fish Dnmt1 and Suv39h1 regulate Organ-Specific terminal differentiation during development. Mol. Cell. Biol. 26, 7077–7085. doi: 10.1128/MCB.00312-06
Raz, E. (2003). Primordial germ-cell development: the zebrafish perspective. Nat. Rev. Genet. 4, 690–700. doi: 10.1038/nrg1154
Reik, W. (2007). Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432. doi: 10.1038/nature05918
Robertson, K. D. (2005). DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610. doi: 10.1038/nrg1655
Rossant, J., and Tam, P. P. L. (2009). Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701–713. doi: 10.1242/dev.017178
Ruzov, A., Tsenkina, Y., Serio, A., Dudnakova, T., Fletcher, J., Bai, Y., et al. (2011). Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 21, 1332–1342. doi: 10.1038/cr.2011.113
Sadler, K. C., Krahn, K. N., Gaur, N., and Ukomadu, C. (2007). Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc. Natl. Acad. Sci. U.S.A. 104, 1570–1575. doi: 10.1073/pnas.0610774104
Saitou, M., Kagiwada, S., and Kurimoto, K. (2012). Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139, 15–31. doi: 10.1242/dev.050849
Seisenberger, S., Andrews, S., Krueger, F., Arand, J., Walter, J., Santos, F., et al. (2012). The dynamics of Genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862. doi: 10.1016/j.molcel.2012.11.001
Seki, Y., Hayashi, K., Itoh, K., Mizugaki, M., Saitou, M., and Matsui, Y. (2005). Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 278, 440–458. doi: 10.1016/j.ydbio.2004.11.025
Sharif, J., Muto, M., Takebayashi, S., Suetake, I., Iwamatsu, A., Endo, T. A., et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. doi: 10.1038/nature06397
Shi, L., and Wu, J. (2009). Epigenetic regulation in mammalian preimplantation embryo development. Reprod. Biol. Endocrinol. 7:59. doi: 10.1186/1477-7827-7-59
Smith, Z. D., Chan, M. M., Mikkelsen, T. S., Gu, H., Gnirke, A., Regev, A., et al. (2012). A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344. doi: 10.1038/nature10960
Smith, Z. D., Gu, H., Bock, C., Gnirke, A., and Meissner, A. (2009). High-throughput bisulfite sequencing in mammalian genomes. Methods 48, 226–232. doi: 10.1016/j.ymeth.2009.05.003
Smith, Z. D., and Meissner, A. (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220. doi: 10.1038/nrg3354
Song, C. X., Szulwach, K. E., Dai, Q., Fu, Y., Mao, S. Q., Lin, L., et al. (2013). Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691. doi: 10.1016/j.cell.2013.04.001
Spruijt, C. G., Gnerlich, F., Smits, A. H., Pfaffeneder, T., Jansen, P. W. T. C., Bauer, C., et al. (2013). Dynamic readers for 5-(Hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159. doi: 10.1016/j.cell.2013.02.004
Stein, R., Gruenbaum, Y., Pollack, Y., Razin, A., and Cedar, H. (1982). Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc. Natl. Acad. Sci. U.S.A. 79, 61–65. doi: 10.1073/pnas.79.1.61
Suzuki, M. M., and Bird, A. (2008). DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476. doi: 10.1038/nrg2341
Szwagierczak, A., Bultmann, S., Schmidt, C. S., Spada, F., and Leonhardt, H. (2010). Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 38:e181. doi: 10.1093/nar/gkq684
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. doi: 10.1126/science.1170116
Tam, P. P. L., and Loebel, D. A. F. (2007). Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8, 368–381. doi: 10.1038/nrg2084
Tang, F., Barbacioru, C., Nordman, E., Bao, S., Lee, C., Wang, X., et al. (2011). Deterministic and stochastic allele specific gene expression in single mouse blastomeres. PLoS ONE 6:e21208. doi: 10.1371/journal.pone.0021208
Tittle, R. K., Sze, R., Ng, A., Nuckels, R. J., Swartz, M. E., Anderson, R. M., et al. (2011). Uhrf1 and Dnmt1 are required for development and maintenance of the zebrafish lens. Dev. Biol. 350, 50–63. doi: 10.1016/j.ydbio.2010.11.009
Wang, H., and Dey, S. K. (2006). Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet. 7, 185–199. doi: 10.1038/nrg1808
Wang, L., Zhang, J., Duan, J., Gao, X., Zhu, W., Lu, X., et al. (2014). Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991. doi: 10.1016/j.cell.2014.04.017
Williams, K., Christensen, J., Pedersen, M. T., Johansen, J. V., Cloos, P. A. C., Rappsilber, J., et al. (2011). TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348. doi: 10.1038/nature10066
Wu, H., and Zhang, Y. (2014). Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68. doi: 10.1016/j.cell.2013.12.019
Wu, X., and Zhang, Y. (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18, 517–534. doi: 10.1038/nrg.2017.33
Wyatt, G. R., and Cohen, S. S. (1953). The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem. J. 55, 774–82.
Yamaguchi, S., Hong, K., Liu, R., Inoue, A., Shen, L., Zhang, K., et al. (2013a). Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 23, 329–339. doi: 10.1038/cr.2013.22
Yamaguchi, S., Hong, K., Liu, R., Shen, L., Inoue, A., Diep, D., et al. (2012). Tet1 controls meiosis by regulating meiotic gene expression. Nature 492, 443–447. doi: 10.1038/nature11709
Yamaguchi, S., Shen, L., Liu, Y., Sendler, D., and Zhang, Y. (2013b). Role of Tet1 in erasure of genomic imprinting. Nature 504, 460–464. doi: 10.1038/nature12805
Yates, A., Akanni, W., Amode, M. R., Barrell, D., Billis, K., Carvalho-Silva, D., et al. (2016). Ensembl 2016. Nucleic Acids Res. 44, D710–D716. doi: 10.1093/nar/gkv1157
Keywords: DNA methylation, epigenetics, 5mC, 5hmC, TET, zebrafish, mouse, gene expression
Citation: Jessop P, Ruzov A and Gering M (2018) Developmental Functions of the Dynamic DNA Methylome and Hydroxymethylome in the Mouse and Zebrafish: Similarities and Differences. Front. Cell Dev. Biol. 6:27. doi: 10.3389/fcell.2018.00027
Received: 02 December 2017; Accepted: 05 March 2018;
Published: 20 March 2018.
Edited by:
Kyoko Yokomori, University of California, Irvine, United StatesReviewed by:
Yves Renaudineau, Université de Bretagne Occidentale, FranceShicheng Guo, Marshfield Clinic Research Institute, United States
Copyright © 2018 Jessop, Ruzov and Gering. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexey Ruzov, YWxleGV5LnJ1em92QG5vdHRpbmdoYW0uYWMudWs=
Martin Gering, bWFydGluLmdlcmluZ0Bub3R0aW5naGFtLmFjLnVr
 Peter Jessop
Peter Jessop Alexey Ruzov
Alexey Ruzov Martin Gering
Martin Gering