- Department of Biology, Syracuse University, Syracuse, NY, USA
Polycystic kidney disease (PKD) proteins are trans-membrane proteins that have crucial roles in many aspects of vertebrate development and physiology, including the development of many organs as well as left–right patterning and taste. They can be divided into structurally-distinct PKD1-like and PKD2-like proteins and usually one PKD1-like protein forms a heteromeric polycystin complex with a PKD2-like protein. For example, PKD1 forms a complex with PKD2 and mutations in either of these proteins cause Autosomal Dominant Polycystic Kidney Disease (ADPKD), which is the most frequent potentially-lethal single-gene disorder in humans. Here, we identify the complete family of pkd genes in zebrafish and other teleosts. We describe the genomic locations and sequences of all seven genes: pkd1, pkd1b, pkd1l1, pkd1l2a, pkd1l2b, pkd2, and pkd2l1. pkd1l2a/pkd1l2b are likely to be ohnologs of pkd1l2, preserved from the whole genome duplication that occurred at the base of the teleosts. However, in contrast to mammals and cartilaginous and holostei fish, teleosts lack pkd2l2, and pkdrej genes, suggesting that these have been lost in the teleost lineage. In addition, teleost, and holostei fish have only a partial pkd1l3 sequence, suggesting that this gene may be in the process of being lost in the ray-finned fish lineage. We also provide the first comprehensive description of the expression of zebrafish pkd genes during development. In most structures we detect expression of one pkd1-like gene and one pkd2-like gene, consistent with these genes encoding a heteromeric protein complex. For example, we found that pkd2 and pkd1l1 are expressed in Kupffer's vesicle and pkd1 and pkd2 are expressed in the developing pronephros. In the spinal cord, we show that pkd1l2a and pkd2l1 are co-expressed in KA cells. We also identify potential co-expression of pkd1b and pkd2 in the floor-plate. Interestingly, and in contrast to mouse, we observe expression of all seven pkd genes in regions that may correspond to taste receptors. Taken together, these results provide a crucial catalog of pkd genes in an important model system for elucidating cell and developmental processes and modeling human diseases and the most comprehensive analysis of embryonic pkd gene expression in any vertebrate.
Introduction
Polycystic kidney disease (PKD) proteins are trans-membrane proteins that share a conserved polycystin-cation-channel domain, located in their last six trans-membrane domains. These proteins are crucially important for human health as they have essential functions in many aspects of vertebrate development and physiology (Delmas, 2004b; Venkatachalam and Montell, 2007; Zhou, 2009; Semmo et al., 2014). Most notably, this gene/protein family is named after PKD because mutations in either PKD1 or PKD2 account for all of the known forms of Autosomal Dominant Polycystic Kidney Disease (ADPKD), which is the most common genetic cause and the fourth most common cause of kidney failure. ADPKD affects one in 400–1000 individuals, across all ethnic groups, which also makes it the most frequent potentially-lethal single-gene disorder in humans (Dalgaard, 1957; Iglesias et al., 1983; Reeders et al., 1985; Levy and Feingold, 2000; Sutters and Germino, 2003; Zhou, 2009). In PKD, large epithelial-lined cysts develop and fill with fluid. This causes abnormally enlarged kidneys and the cysts compress normal renal tissue, destroying it and impairing normal kidney function. This usually results in chronic renal failure by middle age. In addition, cysts can also form in the liver, pancreas, spleen, ovaries, large bowel, brain, and heart and patients often have cardiovascular defects (Grantham, 1993; Wu and Somlo, 2000; Delmas, 2004b; Harris and Torres, 2009; Zhou, 2009; Cornec-Le Gall et al., 2013; Paul et al., 2014; Semmo et al., 2014). Mice heterozygous for a mutation in Pkd2 also develop kidney cysts and renal failure and die as young adults (Wu and Somlo, 2000). In contrast, mice that have homozygous mutations in Pkd2 or Pkd1 die before birth, probably due to cardiac failure caused by incorrect heart development (Wu and Somlo, 2000; Boulter et al., 2001). In addition, these embryos have defects in their kidneys and pancreas (Lu et al., 1997, 2001; Kim et al., 2000; Wu et al., 2000; Boulter et al., 2001). Pkd1 homozygous mutants also have skeletal defects (Boulter et al., 2001; Lu et al., 2001) and Pkd2 and Pkd1l1 are required for left-right patterning/asymmetry and the correct localization of several organs (Pennekamp et al., 2002; McGrath et al., 2003; Field et al., 2011; Kamura et al., 2011; Yoshiba et al., 2012; Yuan et al., 2015).
While less is known about the functions of the other Pkd genes, about 50% of mice homozygous for a mutation in Pkd2l1 have heterotaxy (intestinal malrotation; Delling et al., 2013) and up-regulation of Pkd1l2 in mouse causes profound neuromuscular defects (Mackenzie et al., 2009). Pkdrej is expressed in sperm suggesting that it may have a role in male fertility (Veldhuisen et al., 1999; Butscheid et al., 2006) and there is in vitro evidence that a complex of PKD1L3 and PKD2L1 may function as sour-taste receptors (Huang et al., 2006; Ishimaru et al., 2006), although this may not be the case in vivo, at least in mouse, as analysis of a mouse Pkd1l3 mutant found no significant defect in taste reception (Nelson et al., 2010).
In humans and mouse there are eight PKD genes: PKDREJ, PKD1, PKD1L1, PKD1L2, PKD1L3, PKD2, PKD2L1, and PKD2L2 (Zhou, 2009). These can be divided into two main sub-groups. PKD1-like (also called polycystin-1) genes (PKDREJ, PKD1, PKD1L1, PKD1L2, PKD1L3) are large multi-exon genes, encoding proteins of 1700–4300 amino acids. For example, human and mouse PKD1 each have 46 exons encoding about 4300 amino acids (Li et al., 2003). PKD1-like proteins have 11 trans-membrane domains, a large extracellular N-terminal domain and a short intracellular C-terminal tail with a G-protein binding site and, in some cases, a coiled-coil domain. The N-terminal domain typically contains several repeats of an Ig-fold-containing domain called the PKD domain, a lipoxygenase homology/polycystin-lipoxygenase-atoxin PLAT/LH2 domain, a G-protein-coupled receptor proteolytic site (GPS) and a receptor egg jelly (REJ) domain (Delmas, 2004b; Zhou, 2009; Hofherr and Kottgen, 2011; Semmo et al., 2014). In contrast, PKD2-like (also called TRPP) proteins (PKD2, PKD2L1 and PKD2L2) are shorter, <1000 amino acids in each case (Veldhuisen et al., 1999; Li et al., 2003; Zhou, 2009; Semmo et al., 2014). These proteins are non-selective cation-channel proteins with six trans-membrane domains, an intracellular N-terminal domain and an intracellular C-terminal domain that sometimes contains a coiled-coil domain (Delmas, 2004b; Venkatachalam and Montell, 2007; Zhou, 2009; Hofherr and Kottgen, 2011; Semmo et al., 2014). PKD2-like proteins are part of the transient receptor potential (TRP) channel superfamily (Delmas, 2004b; Ishimaru et al., 2006; Owsianik et al., 2006; Ramsey et al., 2006; Venkatachalam and Montell, 2007; Zhou, 2009; Nilius and Owsianik, 2011; Semmo et al., 2014). TRP proteins all have six trans-membrane domains with a pore domain between the 5th and 6th domains and they have crucial roles in many different sensory functions including detection of mechanical, chemical, and thermal stimuli (Montell, 2005; Owsianik et al., 2006; Ramsey et al., 2006; Venkatachalam and Montell, 2007; Damann et al., 2008; Nilius and Owsianik, 2011; Venkatachalam et al., 2014). Interestingly, it has been proposed that the PKD2-like/TRPP proteins may be the most evolutionary ancient of all of the TRP proteins as they are found not just in vertebrates and invertebrates but also in yeast (Palmer et al., 2005; Venkatachalam and Montell, 2007; Semmo et al., 2014).
PKD1-like and PKD2-like proteins form heteromeric polycystin-receptor-channel complexes and, in at least some cases, physical interaction between these proteins is crucial for correct membrane localization of the resulting complex as well as correct physiological function (Li et al., 2003; Murakami et al., 2005; Ishimaru et al., 2006; Giamarchi et al., 2010; Field et al., 2011; Semmo et al., 2014). Consistent with this, mutations in partner proteins usually produce almost identical phenotypes both in humans and model organisms (e.g., Barr and Sternberg, 1999; Sutters and Germino, 2003; Field et al., 2011). For example, PKD2 and PKD1L1 physically interact and mutations in either of these genes cause defects in left-right patterning (Field et al., 2011). Similarly, PKD1 complexes with PKD2 and mutations in either of these genes cause ADPKD (Qian et al., 1997; Tsiokas et al., 1997; Yu et al., 2009; Zhu et al., 2011).
PKD heteromeric complexes are thought to form receptor-mediated non-selective cation-channels that are often located in primary cilia. For example, PKD1 and PKD2 form a non-selective cation-channel located in primary cilia of renal epithelial cells, that is thought to transduce extracellular stimuli such as fluid flow, possibly through altering general intracellular calcium signals (Hanaoka et al., 2000; Nauli et al., 2003; Delmas, 2004a; Delmas et al., 2004; Zhou, 2009 although see Delling et al., 2016, which challenges this model) or by altering local ciliary calcium concentrations (Delling et al., 2013; DeCaen et al., 2016). Similarly, PKD2 and PKD1L1 may form a calcium channel in the primary cilia of cells in the node, which could help establish left/right asymmetry during early stages of development, by sensing and transducing the left-biased signal (Pennekamp et al., 2002; McGrath et al., 2003; Field et al., 2011; Kamura et al., 2011; Yoshiba et al., 2012, although again see Delling et al., 2016, which challenges this model).
Given the importance of PKD genes to many different aspects of vertebrate embryonic development and physiology, it is crucial that we know where all of these genes are expressed. This may help us to identify other potential functions and interacting partners for this family of proteins. Zebrafish is a powerful model system for elucidating developmental and cell biological processes and for modeling and studying human diseases (e.g., Hostetter et al., 2003; Huang et al., 2014; Avagyan and Zon, 2016; Bournele and Beis, 2016; Brown et al., 2016; Carneiro et al., 2016; Griffin et al., 2016; Harrison et al., 2016; Kozol et al., 2016; Myllymaki et al., 2016; Poureetezadi and Wingert, 2016; Song et al., 2016; Wager et al., 2016; Wojciechowska et al., 2016; Zon, 2016). Consistent with this, all of the evidence so far suggests that zebrafish pkd genes function in ways that are highly conserved with their mammalian orthologs. For example, pkd2 expression is enriched in the developing zebrafish pronephros (Bisgrove et al., 2005; Schottenfeld et al., 2007), Pkd2 protein is present in zebrafish kidney epithelial cells (Obara et al., 2006), and knock-down of pkd2 function causes cyst formation in the zebrafish pronephros (Sun et al., 2004; Obara et al., 2006; Streets et al., 2006; Fu et al., 2008; Chang et al., 2011; Arif Pavel et al., 2016). In addition, pkd2 is expressed in Kupffer's vesicle (KV) during early zebrafish embryogenesis (Bisgrove et al., 2005; Schottenfeld et al., 2007; Roxo-Rosa et al., 2015). The KV is a transient organ that forms during late gastrulation stages from dorsal forerunner cells that coalesce near the caudal end of the zebrafish embryo, and it is required to set up left/right asymmetry (Essner et al., 2005; Kramer-Zucker et al., 2005; Sampaio et al., 2014; Smith et al., 2014). Consistent with this, knock-down of Pkd2 function in zebrafish causes disturbed left-right patterning/asymmetry and randomization of heart and gut looping (Bisgrove et al., 2005; Schottenfeld et al., 2007) and zebrafish pkd2 mutants have impaired cardiac function (Paavola et al., 2013). Similarly, knock-down of Pkd1 causes cyst formation in the liver (Tietz Bogert et al., 2013). In addition, studies in zebrafish have identified novel functions for Pkd proteins, such as helping to integrate mechanosensory feedback into locomotor neural circuits (Bohm et al., 2016).
Despite the importance of pkd genes, when we started this study only three pkd genes had been described in zebrafish, pkd1, pkd1b, and pkd2, although analyses of pkd2l1 were also published more recently (Sun et al., 2004; Bisgrove et al., 2005; Obara et al., 2006; Streets et al., 2006; Schottenfeld et al., 2007; Feng et al., 2008; Fu et al., 2008; Francescatto et al., 2010; Giamarchi et al., 2010; Hurd et al., 2010; Mangos et al., 2010; Chang et al., 2011; Fogelgren et al., 2011; Merrick et al., 2012; Graham et al., 2013; Paavola et al., 2013; Tietz Bogert et al., 2013; Coxam et al., 2014; Djenoune et al., 2014; Fidelin and Wyart, 2014; Goetz et al., 2014; Quan et al., 2015; Roxo-Rosa et al., 2015; Yuan et al., 2015; Arif Pavel et al., 2016; Bohm et al., 2016). It was also unclear whether zebrafish have duplicate copies (ohnologs) of any of the Pkd genes found in mammals, from the genome duplication event at the base of the teleosts (Amores et al., 1998; Postlethwait et al., 1998; Force et al., 1999; Postlethwait, 2007). Therefore, we decided to identify the full complement of zebrafish pkd genes. Using bioinformatics and RT-PCR-based cloning we have identified seven zebrafish pkd genes: pkd1, pkd1b, pkd1l1, pkd1l2a, pkd1l2b, pkd2, and pkd2l1. We have also identified what may be a remnant of pkd1l3 that lacks the polycystin-cation-channel domain sequence that is conserved in all other pkd genes. Therefore, we do not consider this a bona-fide pkd gene. In this paper we identify the sequences and genomic locations of all of these genes. We also confirm that no additional pkd genes exist in three other teleosts: medaka, stickleback or green spotted pufferfish. In addition, we describe the expression of each of the seven zebrafish pkd genes during embryonic and larval development. Taken together, we provide the first description of the complete family of zebrafish pkd genes and the most comprehensive analysis of embryonic pkd gene expression in any vertebrate.
Materials and Methods
Ethics Approval
All zebrafish experiments in this research were approved by the Syracuse University IACUC committee.
Zebrafish Husbandry and Fish Lines
Zebrafish (Danio rerio) were maintained on a 14-h light/10-h dark cycle at 28.5°C. Embryos were obtained from natural paired and/or grouped spawnings of wild-type (WT; AB, TL, or AB/TL hybrid) or mindbomb (mibta52b; Jiang et al., 1996) or Tg(−8.1gata1:gata1-EGFP) (Kobayashi et al., 2001) fish. Embryos were staged in hours post fertilization at 28.5°C (h) or days post fertilization (dpf) according to Kimmel et al. (1995).
Identification of pkd Genes
Initially we searched NCBI, http://www.ZFIN.org and Ensembl for zebrafish pkd genes. We then blasted nucleotide sequences for these genes against the zebrafish genome using Tblastn on Ensembl http://www.ensembl.org/Danio_rerio/Tools/Blast?db=core. We identified polycystin-cation-channel domains and performed a Tblastn with these peptide sequences using default parameters at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=tblastn&PAGE_TYPE=BlastSearch&BLAST_SPEC=OGP__7955__9557&LINK_LOC=blasttab&LAST_PAGE=blastn).
Protein sequences were obtained from mapped mRNA transcripts using the Translate tool at the ExPASy Bioinformatics Resource Portal: http://web.expasy.org/translate/. To compare and analyze protein structures, protein domains were identified by searching against the Pfam protein database at EMBL-EBI: http://pfam.xfam.org (Finn et al., 2015).
To amplify in situ hybridization probe templates and confirm particular open reading frames we created zebrafish cDNA from 27 h WT zebrafish embryos. Total RNA was extracted by homogenizing 50–100 mg of embryos in 1 mL of TRIzol reagent (Ambion, 15596-026). RNA integrity (2:1 ratio of 28S:18S rRNA bands) and quality (A260/A280 ratio of ~2.0) was confirmed using agarose gel electrophoresis and spectrophotometry respectively. cDNA was synthesized using Bio-Rad iScript Reverse Transcription Supermix kit (Bio-Rad, 170-8891).
To map and confirm open reading frames, PCRs were performed using 5 μl of cDNA template in a 50 μl reaction, with Phusion High-Fidelity DNA Polymerase (NEB, M0530L) and mapping primers listed in Supplementary Table 1. Reaction conditions were 98.0°C for 30 s, followed by 30 cycles of: 98.0°C for 10 s, Annealing (see Supplementary Table 1 for temperatures) for 20 s, and extension at 72.0°C (see Supplementary Table 1 for extension times). A final extension step was performed for 5 min at 72.0°C.
For pkd1, pkd1l2a, and pkd1l2b we also performed inverse PCR to identify missing 5′ sequence, as described in Lewis et al. (1999), with the following modifications. One microgram of total RNA extracted from 27 h WT zebrafish embryos (see above) was incubated with 10 μM of gene specific primer (see Supplementary Table 2) and 1 mM each of dNTPs in a final volume of 10 μl for 5 min at 65°C. Two-hundred units of M-MuLV Reverse Transcriptase (NEB, M0253S) and eight units of Protector RNase Inhibitor (Roche, 03335399001) were then added and first strand cDNA synthesized by incubating for 1 h at 42°C. Second strand cDNA synthesis was performed immediately as described in Lewis et al. (1999), but the reaction was incubated for 4 h at 14°C, followed by 10 min at 70°C, before adding five units of T4 DNA Polymerase (NEB, M0203S) and incubating for 10 min at 37°C. Circularization was performed as described in Lewis et al. (1999), with the exception that RNA ligase was omitted and purification was performed using Amicon Ultra-0.5 Centrifugal Filter Units with Ultracel-30 Membrane (Millipore Sigma, UFC503024). Reaction products were diluted to a final volume of 500 μl using nuclease-free water and filtered by centrifuging for 10 min at 14000 × g, before eluting by inverting filter and centrifuging for 2 min at 1000 × g. Five microliters of purified, circularized product was used in a 50 μl PCR with Phusion High-Fidelity DNA Polymerase (NEB, M0530L). Reaction conditions were: 98.0°C for 30 s, followed by 35 cycles of: 98.0°C for 10 s, Annealing—(see Supplementary Table 1 for temperatures) for 20 s and Extension (see Supplementary Table 1 for extension times) at 72.0°C. A final extension step was performed for 5 min at 72.0°C.
For pkd1 and pkd1l2b, nested PCR was performed. The first round of PCR was performed as described above, using the respective Nested_Set 1 primers (Supplementary Table 1). This product was diluted 1:10 in nuclease-free water and 2.5 μl of that dilution used as a template in the second round PCR, using Nested_Set 2 primers (Supplementary Table 1).
In all cases, PCR products were verified on a 1% agarose TAE gel and then purified using EZ-10 Spin Column PCR Products Purification kit (Bio Basic Inc, BS664). Purified PCR products were sequenced using the PCR primers (Supplementary Table 1) to prime the reactions and the resulting sequences blasted against zebrafish genome assembly GRCz10 using Tblastn and default parameters on Ensembl (http://www.ensembl.org/Danio_rerio/Tools/Blast?db=core).
Our mapped mRNA transcript sequences for zebrafish pkd1, pkd1l2a, and pkd1l2b have been submitted to NCBI [KY074550 (pkd1), KY074551 (pkd1l2a), and KY074552 (pkd1l2b)].
Phylogenetic Analyses
The peptide sequence for the polycystin-cation-channel domain was identified using Pfam (http://pfam.xfam.org/; Finn et al., 2015) and isolated, when present, from all of the pkd genes in zebrafish (Danio rerio, dre), green spotted pufferfish (Tetraodon nigroviridis, tni), medaka (Oryzias latipes, ola), stickleback (Gasterosteus aculeatus, gac), spotted gar (Lepisosteus oculatus, loc), elephant shark (Callorhinchus milii, cmi), fly (Drosophila melanogaster, dme), human (Homo sapiens, hsa), and mouse (Mus musculus, mmu). The following genome assemblies were used: zebrafish—GRCz10, green spotted pufferfish—TETRAODON 8.0, medaka—HdrR and stickleback—BROAD S1. For spotted gar, elephant shark, fly, human, and mouse proteins, the polycystin-cation-channel domain sequences were isolated from the longest protein isoforms available in Ensembl genomes LepOcu1 (GCA_000242695.1), ESHARK1, BDGP6 (GCA_000001215.4), GRCh38.p7 (GCA_000001405.22), and GRCm38.p4 (GCA_000001635.6), respectively. Protein sequence alignment was performed using Clustal Omega server at EMBL-EBI (Version 1.2.3) and default parameters: http://www.ebi.ac.uk/Tools/msa/clustalo/ (Goujon et al., 2010; Sievers et al., 2011; McWilliam et al., 2013). Phylogenetic trees for the PKD1-like and PKD2-like families were generated using regions of the polycystin-cation-channel domain contained in all of the proteins (see Supplementary Figures 1, 2). We used both the neighbor-joining (NJ) method [plotted using Phylodendron software (version 0.8d) http://iubio.bio.indiana.edu/treeapp/treeprint-form.html] and the maximum likelihood method, implementing a WAG substitution model, performed using PhyML (v3.1/3.0 aLRT) accessed at the Phylogeny.Fr web interface (http://www.phylogeny.fr/index.cgi; Dereeper et al., 2008, 2010). The maximum likelihood analyses are presented here (Figure 3).
Syntenic Analyses
Having identified genomic loci of teleost pkd genes through Tblastn analysis (see above), we compared these to PKD loci in human and mouse genomes using location-based displays in Ensembl to identify any conserved synteny.
In situ Hybridization and Immunohistochemistry
Embryos were fixed in 4% paraformaldehyde and single in situ hybridization or fluorescent in situ hybridization plus immunohistochemistry experiments were performed as previously described (Concordet et al., 1996; Batista et al., 2008). Embryos older than 24 h were often incubated in 0.003% 1-phenyl-2-thiourea (PTU) to prevent pigment formation. For fluorescent in situ hybridization + immunohistochemistry, after detection of the in situ hybridization reaction using TSA Kit #5, with HRP, Goat anti-mouse IgG and Alexa Fluor 594 Tyramide (ThermoFisher Scientific, T20915), embryos were washed 8 × 15 min in PBST and incubated in Image-iT FX Signal Enhancer (ThermoFisher Scientific, I36933) for 30 min at room temperature. Immunohistochemistry was performed using a chicken polyclonal anti-GFP primary antibody (Abcam, Ab13970, 1:500) and a Goat anti-chicken IgY (H+L), Alexa Fluor 488 secondary antibody (ThermoFisher Scientific, A-11039, 1:1000). Probes for in situ hybridization experiments were prepared using PCR-based DNA templates from 27 h cDNA, made as described above, and primers listed in Supplementary Table 3. Primers for all zebrafish pkd genes, except pkd1 Primer Set 1, were designed using the following parameter ranges: nucleotide length—21 bases (minimum)-28 bases (maximum), tm—58°C (minimum)−65°C (maximum) and GC content—45% (minimum)-60% (maximum) with Primer3 web version 4.0.0 at http://bioinfo.ut.ee/primer3/ (Koressaar and Remm, 2007; Untergasser et al., 2012). All reverse primers include the sequence for the T3 RNA Polymerase minimal promoter: ATTAACCCTCACTAAAGGGA. This sequence is shown in bold and underlined in the reverse primers listed in Supplementary Table 3. To avoid cross-reactivity, whenever possible, riboprobes were designed against 3′ UTR or coding sequence lacking all of the conserved protein domains shown in Figure 2. pkd1 Set 1 primers used to make the zebrafish pkd1 riboprobe were identical to those described by Coxam et al. (2014). These primers generated a 580 bp PCR product and the resulting RNA probe revealed specific embryonic expression. However, this region of the pkd1 transcript was no longer included in the annotation of the pkd1 gene in Ensembl GRCz10 (Figure 1A). Therefore, we also generated an alternative pkd1 in situ probe that binds 3′ to the Coxam riboprobe, in a region included in the newer Ensembl transcript (pkd1 Set 2 primers—see Supplementary Table 3, Figure 1A). This probe produced identical, albeit weaker, expression to the first (Coxam) riboprobe (data not shown). The stronger Coxam riboprobe was therefore used throughout this study.
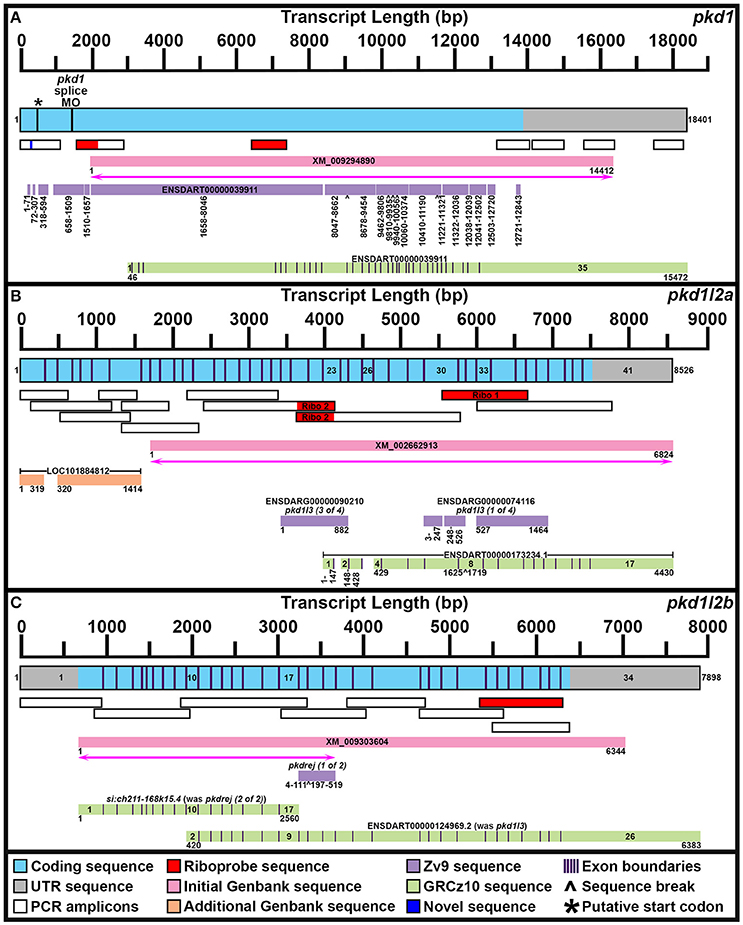
Figure 1. Mapping pkd1, pkd1l2a, and pkd1l2b mRNA Transcripts. Summary of mRNA transcript mapping results for pkd1 (A), pkd1l2a (B), and pkd1l2b (C). Approximate length in base pairs (bp) is indicated by scale at top of each panel. Mapped transcripts are shown in next row of each panel. Coding sequence is blue and UTR is gray. Numbers flanking these mapped transcripts indicate nucleotide positions. Black vertical lines (coding sequence box, A) indicate putative start codon, and morpholino sequence positions. Purple vertical lines (coding sequence boxes, A–C) indicate exon boundaries, where known. Mapped PCR amplicons generated in this study are indicated with white boxes. Red indicates riboprobe sequences used in this study. Dark blue indicates novel sequence identified in this study but not currently present in Ensembl GRCz10 genome. Genbank reference sequences used at beginning of this study are shown as pink boxes. Magenta lines with double arrows beneath these indicate regions of sequence homology identified at start of this project. Genbank reference sequences identified during this study are shown as orange boxes. Ensembl Zv9 transcript sequences are shown as lilac boxes. Ensembl GRCz10 transcript sequences are shown as green boxes. Numbers beneath sequences show nucleotide positions. ∧, break in aligned sequence. Thin purple vertical lines in green boxes indicate exon boundaries, where known. Key exons for interpreting mapping results are numbered. (A) Our newly mapped pkd1 transcript contains all but 173 bases of the older Zv9 ENSDART00000039911 transcript (lilac boxes) as well as all but the first 45 nucleotides of the current GRCz10 ENSDART00000039911 transcript (green boxes). We have also identified additional 5′ sequence and missing regions of coding sequence. The GRCz10 ENSDART00000039911 transcript corresponds to nucleotides 2975–18401 of our mapped transcript. The Zv9 ENSDART00000039911 transcript corresponds to nucleotides 218–13798 of our mapped transcript but contains some gaps (nucleotides 292–357, 430–597, 875–925, 1778–1786, 1935–1973, 8363–8410, 9804–9811, 10715–10725, 11608–11640, 12356, 12359–12389, 12852–12890, and 13109–13675 of our mapped transcript). Inverse PCR identified 5′ transcript sequence along with a novel stretch of nucleotides (292–357) absent from GRCz10 Ensembl genome (shown in dark blue). Nucleotides 2659–12720 of the Zv9 transcript are almost 100% identical to nucleotides 46–10179 of the GRCz10 transcript and these regions align with nucleotides 2975–13108 of our mapped transcript. Nucleotides 10180–10746 of the GRCz10 transcript share no homology with the Zv9 transcript, but correspond to nucleotides 13109–13675 of our mapped transcript. Nucleotides 12721–12843 of the Zv9 transcript share 100% homology with nucleotides 10747–10869 of the GRCz10 transcript and correspond to nucleotides 13676–13798 of our mapped transcript. The coding sequence of the GRCz10 transcript terminates 30 nucleotides downstream of the Zv9 transcript and is followed by 4573 bp of unique 3′ UTR sequence. Using RT-PCR we have confirmed that our mapped transcript utilizes the same stop codon and 3′ UTR sequence. Specifically, we have confirmed that nucleotides 10192–11136, 11173–12085, 12598–13484, and 14524–15376 of 3′ UTR sequence in the GRCz10 transcript are transcribed and map to nucleotides 13121–14065, 14102–15014, 15527–16413, and 17453–18395 of our mapped transcript, respectively. Our inverse PCR revealed 217 nucleotides of coding sequence upstream of the Zv9 transcript and 66 nucleotides of novel coding sequence between nucleotides 71 and 72 of the Zv9 transcript. In total, this produces a 18401 bp transcript that encodes a 4608 amino acid protein and we have deposited this sequence in NCBI (NCBI accession number KY074550). This sequence lacks a start methionine. *indicates in-frame methionine at 527–529 nucleotides. However, if this is the start codon, the resulting protein would lack the leucine rich repeat domain, encoded by the 175 amino acids in-frame upstream of this methionine, that is present in mouse, human and stickleback PKD1. There is a putative in-frame start codon a further 54 nucleotides (18 amino acids) upstream of our present transcript, which we think is more likely to be the true start codon. The location of the splice-blocking morpholino sequence used by Mangos et al. (2010) that resulted in kidney cysts in some animals is also indicated (nucleotides 1187–1197 of the Zv9 transcript). (B) Our current transcript for pkd1l2a encompasses both LOC101884812 and XM_002662913 and contains additional exons not present in either of these sequences. The start of the current ENSDART00000173234.1 transcript coincides with the start of exon 23 in our longer transcript, but the first exon of ENSDART00000173234.1 is shorter than exon 23 in our transcript. Exons 2–3, 4–7, and 9–17 of ENSDART00000173234.1 are identical to exons 24–25, 27–30, and 33–41 of our transcript. Exon 26 of our transcript is absent in ENSDART00000173234.1 and exons 31–32 and intron 31–32 exist as a single exon, exon 8, in ENSDART00000173234.1. (C) Our current transcript for pkd1l2b contains both the si:ch211-168k15.4 and ENSDARG00000101214 (ENSDART00000124969.2) sequences, utilizing a start codon 4 bases upstream of exon 1 in the current si:ch211-168k15.4 annotation, and transitioning between exon 16 of si:ch211-168k15.4 immediately into exon 9 of ENSDART00000124969.2. Nucleotides 683–7026 of our new 7898 bp mRNA transcript align perfectly with the original XM_009303604 6344 bp reference sequence. The start codon was identified in this study along with novel 5′ UTR sequence.
To confirm genomic structure of pkd1l2a, two separate riboprobes were generated and tested (Supplementary Table 3, Figure 1B). These were generated against two adjacent genes that have been retired in Ensembl GRCz10 but that we show here encompass different parts of pkd1l2a. The pkd1l2a riboprobe generated with primer Set 1 is the most 3′ of the two probes. It was designed against ENSDARG00000074116 and it partially overlaps the current Ensembl pkd1l2a transcript. In contrast, the pkd1l2a riboprobe generated with primer Set 2 was designed against ENSDARG00000090210 and is immediately 5′ to the current Ensembl pkd1l2a transcript. Whilst both probes produced the same expression patterns, the latter probe was weaker in putative taste buds and therefore the probe designed against ENSDARG00000074116 (primer set 1) was used for all of the studies in this paper.
Each 50 μL probe reaction PCR contained 5 μL cDNA and one unit of Phusion High-Fidelity DNA Polymerase (NEB, M0530L). PCR conditions were: 94°C for 3 min followed by 35 cycles of 94°C for 30 s, 56.5°C for 30 s, 72°C for 1.5 min and then a final extension step of 72°C for 10 min. PCR products were purified by phenol:chloroform extraction. in situ hybridization probes were made using 1 μg purified PCR product, T3 RNA Polymerase (Roche, 11031171001) and DIG RNA Labeling Mix (Roche, 11277073910).
Imaging
Embryos 24 h and older were deyolked in 70% glycerol/30% sterile water using mounting pins. For lateral and dorsal views of the embryo, whole embryos were mounted in 70% glycerol in coverslip sandwiches (24 × 60 mm coverslips; VWR, 48393-106), with 2–4 coverslips (22 × 22 mm; VWR, 16004-094) on either side of the sample to avoid sample compression. For ventral views of putative taste receptors, the trunk was dissected with a razor blade and the head carefully inverted on to a 24 × 60 mm coverslip and a similar coverslip sandwich made. For lateral views of eyes, they were dissected from forebrain using mounting pins and mounted as for whole embryos, but using only 1–2 coverslips each side of the specimen. Cross-sections were cut by hand using a razor blade mounted in a 12 cm blade holder (World Precision Instruments, Cat. #14134). Differential interference contrast (DIC) pictures were taken using an AxioCam MRc5 camera mounted on a Zeiss Axio Imager M1 compound microscope. A Zeiss LSM 710 confocal microscope was used to image embryos mounted in DABCO (1,4-Diazabicyclo[2.2.2]octane, Sigma, D-2522, 2% w/v solution in 80% sterile glycerol) for fluorescent double-labeling experiments. Images were processed using Adobe Photoshop software (Adobe, Inc) and Image J software (Abràmoff et al., 2004).
Cell Counts and Statistics
In all cases, cells counts are for both sides of a five-somite length of the spinal cord adjacent to somites 6–10. Values are an average of five embryos. Results were analyzed using the student's t-test; Error bars indicate standard error of the mean.
Results
Zebrafish Have Seven pkd Genes
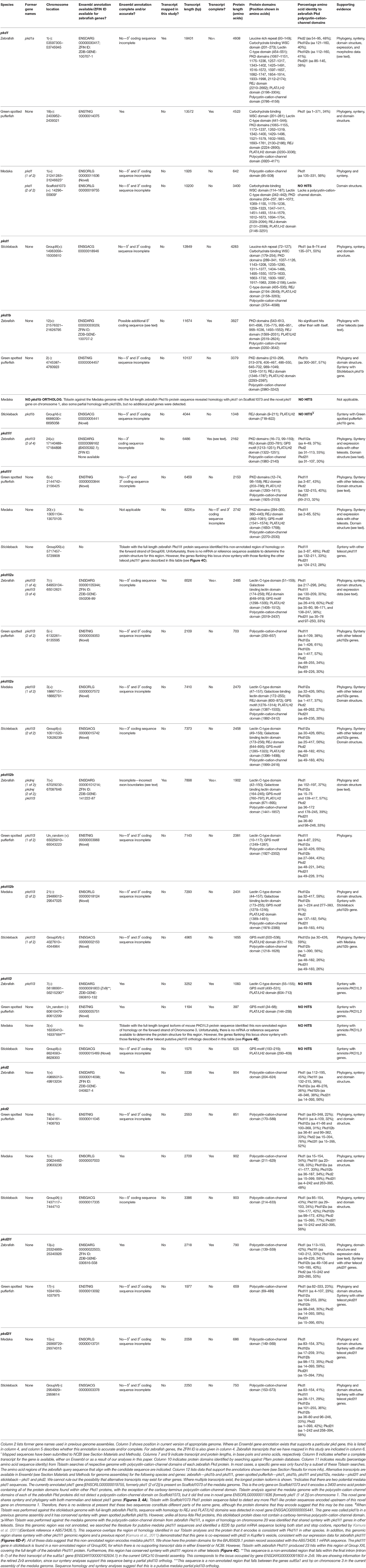
To establish the full complement of zebrafish pkd genes we initially searched several online resources. We found NCBI nucleotide reference sequences for six genes: XM_009294890 (called pkd1), XM_009297371 (called pkd1l1), XM_009303604 (called pkd1l2), XM_002662913 (called pkd1l3), DQ175629 (called pkd2), and XM_690312 (called pkd2l1) and an additional gene, called pkd1b, on the zebrafish database website, ZFIN (Note: some of these records have since been retired as a result of standard genome-annotation processing and our data suggest that some of these names are not correct). To identify additional potential pkd genes, we blasted each of these sequences against zebrafish genome assembly Ensembl Zv9 using Tblastn. We also performed a textual search for pkd genes on Ensembl. Using these methods, we identified 10 potential pkd genes [called at that time pkd1, pkd1b, pkd1l3, pkd1l3 (1 of 4), pkd1l3 (2 of 4), pkd1l3 (3 of 4), pkdrej (1 of 2), pkdrej (2 of 2), pkd2, and pkd2l1]. We examined each of these, in order to determine which of them were indeed bona-fide pkd genes. These analyses identified seven pkd genes as described below.
pkd1
When we commenced our bioinformatic analyses of zebrafish pkd genes, the reference sequence XM_009294890 aligned in Ensembl Zv9 with a 12843 bp transcript, ENSDART00000039911 (associated with gene ENSDARG00000030417 on chromosome 1) called pkd1, that lacked both start and stop codons (Figure 1A; Table 1). However, this annotation changed in the current genome assembly, GRCz10, which contains a revised 15472 bp ENSDART00000039911 transcript that lacks the first 2607 nucleotides present in the older transcript. Supporting the older 5′ sequence, Coxam et al. (2014) showed enriched expression in zebrafish trunk at late embryonic stages using an in situ hybridization riboprobe designed against nucleotides 1307–1838 of the older transcript. We have also amplified this region from zebrafish cDNA and, in our hands, a riboprobe designed against this region is strongly expressed in embryonic pronephros, consistent with pkd1 expression in other animals (see expression analyses below). This expression is identical, although stronger, to the expression that we see when we use an alternative riboprobe designed against a more 3′ region included in the newer Ensembl transcript (see Section Materials and Methods and Figure 1A). In addition, Mangos et al. (2010) describe a splice-blocking pkd1 morpholino aligned with the older transcript (Figure 1A), that induced kidney cysts in some animals, consistent with Pkd1 function in other animals. Taken together, these data suggest that the current Ensembl annotation is incorrect and that at least some pkd1 transcripts include parts of the older upstream sequence.
Therefore, we used inverse PCR and overlapping PCR amplicons, to identify/map the correct pkd1 sequence (see Section Materials and Methods, Figure 1A and Table 1). All but 173 bases of the older ENSDART00000039911 transcript (Zv9) are present in our newly mapped pkd1 transcript as are all but the first 45 nucleotides of the current ENSDART00000039911 transcript (GRCz10). However, compared to our new transcript, there are some gaps in the Zv9 transcript, and we have also identified novel 5′ sequence that is not present in either transcript nor the 5′ genomic sequence in GRCz10, suggesting that there may be sequence missing from chromosome 1 in Ensembl. Taken together, our data suggest that a transcript of at least 18401 bp exists that encodes a 4608 amino acid protein and we have deposited this sequence in NCBI (accession number KY074550). This sequence lacks a start methionine. There is a methionine codon in-frame at 527–529 nucleotides (Figure 1A), but the region upstream of this methionine encodes a leucine-rich-repeat domain which is conserved in mouse, human and stickleback PKD1. Therefore, we think that this methionine is unlikely to be the start codon. We consider that the start codon is more likely to be a putative in-frame methionine 54 nucleotides upstream of our present transcript. We are confident that this gene is pkd1, given our synteny and phylogeny analyses discussed below (see also Table 1).
pkd1b
In contrast to pkd1, since the inception of this study the annotation of ENSDARG00000033029, the gene called pkd1b in our initial bioinformatic searches, has remained unchanged within Ensembl. The current longest pkd1b transcript, ENSDART00000153412.2, encodes a 3817 amino acid protein (Table 1, Figure 2). This Ensembl sequence is strongly supported by the reference sequence XM_017358624.1, which has the Gene ID 565697 (https://www.ncbi.nlm.nih.gov/gene/), with the exception that this reference sequence encodes an additional 73 amino acids at the amino terminus. Therefore, we cannot rule out the possibility that the pkd1b transcript might be longer than that currently shown in Ensembl. However, this would not affect the predicted domain structure of Pkd1b protein, as the additional 73 amino acids do not contain any additional predicted protein domains (Figure 2). We are confident that this gene is pkd1b based on the protein domains that it encodes (Figure 2) and our phylogeny analyses discussed below (see also Table 1). However, it is worth noting that, despite its name and the absence of this gene in mammals, we do not think that this gene is a teleost duplicate of pkd1 (see Section Discussion below).
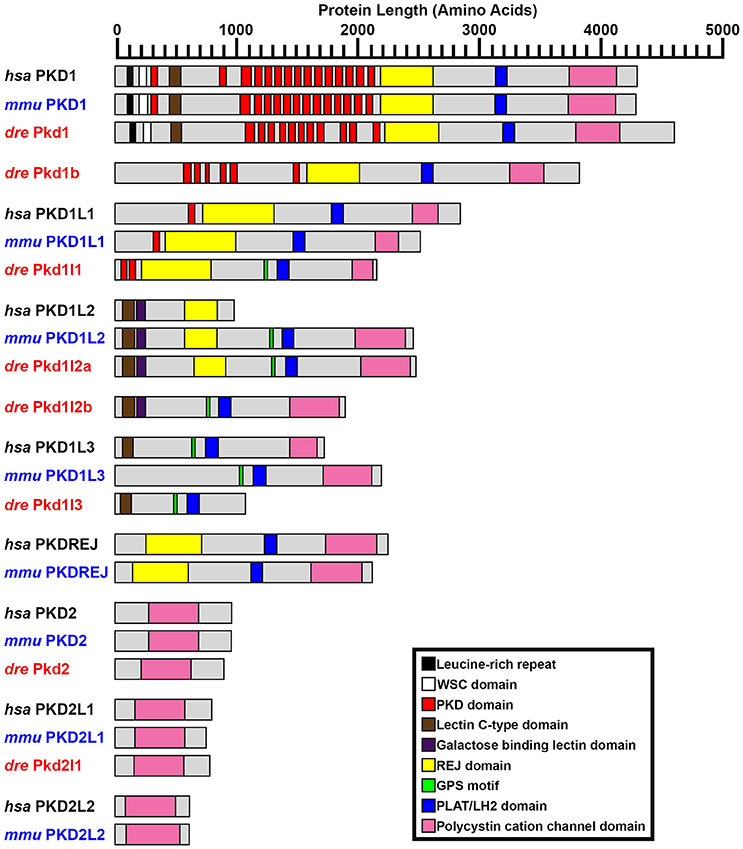
Figure 2. PKD protein domains. Schematics of protein domains identified in all eight human (Homo sapiens, hsa) and mouse (Mus musculus, mmu) and seven zebrafish (Danio rerio, dre) PKD proteins. The zebrafish putative partial pkd1l3 ortholog is also shown. Approximate protein length is indicated by scale at top. Where multiple transcripts exist in Ensembl, the longest protein isoform is shown. In all three species, PKD1 is the longest PKD protein and the only protein to contain a leucine-rich repeat and carbohydrate-binding WSC domain in the amino-terminus. Pkd1b is not present in mammals. Zebrafish Pkd1b resembles Pkd1 with multiple PKD domain repeats in the amino-terminus and REJ, PLAT/LH2, and polycystin-cation-channel domains in the carboxy-terminus. In all three species, PKD1L1 contains a shorter polycystin-cation-channel domain, approximately half the size of that in other Pkd proteins. Unlike mammals, zebrafish Pkd1l1 also contains a GPS motif upstream of the PLAT/LH2 domain. The 5′ coding sequence of mouse Pkd1l1 gene is presently incomplete. PKD1L2 is unusual in humans in that, according to information on Ensembl, longer transcripts represent polymorphic pseudogenes that have acquired mutations, preventing them from being expressed as functional proteins. As a result, the current version of human PKD1L2 is half the size of mouse and zebrafish Pkd1l2 and lacks the polycystin-cation-channel domain characteristic of PKD proteins. If this is correct, then this suggests that human PKD1L2 is no longer a bona-fide PKD gene. Mouse PKD1L2 and Zebrafish Pkd1l2a and Pkd1l2b have identical domain structures, with the exception that Pkd1l2b lacks the REJ domain. PKD1L3 protein structure differs slightly between mammals. Human PKD1L3 has a Lectin C-type domain in the amino-terminus and mouse PKD1L3 does not. In addition, the polycystin-cation-channel domain in mouse PKD1L3 is 411 amino acids long, compared to only 237 amino acids in human PKD1L3. We have identified a putative partial pkd1l3 ortholog in zebrafish, but the sequence lacks the polycystin-cation-channel domain, so we do not consider it a bona-fide pkd gene. Pkdrej and Pkd2l2 are not present in zebrafish. The only currently identified domain in PKD2, PKD2L1, and PKD2L2 proteins is the polycystin-cation-channel domain.
pkd1l1
The reference sequence XM_009297371, called pkd1l1 in our initial analyses, aligns with the forward strand of chromosome 24. In Ensembl Zv9 this region contained a small ORF of 225 amino acids, called pkd1l3 (2 of 4). In GRCz10, this region of homology is now called BX005392.1 (gene ENSDARG00000099162). The longest transcript at this locus is ENSDART00000169516.1, which encodes a protein of 2153 amino acids. Whilst there is no stop codon in either ENSDART00000169516.1 or an additional shorter transcript associated with BX005392.1, there is a putative stop codon in-frame 27 bp downstream in the 3′ flanking sequence of ENSDART00000169516.1. To assess whether this locus might encode a pkd gene, we generated two alternative riboprobes, one designed against exons 39–43 and the other against exons 46–49. As described below, both of these riboprobes labeled putative taste receptors and we also saw expression in dorsal forerunner cells / Kupffer's vesicle similar to pkd1l1 expression in medaka (Kamura et al., 2011). The structure of this protein, including its shorter polycystin-cation-channel domain and fewer PKD domains is consistent with that of PKD1L1 in other vertebrates (Figure 2). In addition, our synteny and phylogenetic analyses described below also suggest that this gene is pkd1l1.
pkd1l2a
The XM_002662913 (used to be called pkd1l3) reference sequence aligns with the reverse strand of chromosome 7. In an early release of Zv9, this region contained two adjacent novel genes, ENSDARG00000074116 and ENSDARG00000090210, encoding proteins of 488 and 294 amino acids respectively (Figure 1B). Temporarily these genes received the annotations pkd1l3 (1 of 4) and pkd1l3 (3 of 4), before being retired from the final release of Zv9. Given the proximity of these genes to one another on the same chromosome and their consecutive alignment with the XM_002662913 reference sequence, we hypothesized that these genes might constitute different parts of one longer gene. Consistent with this, riboprobes generated against each gene produced identical expression patterns in zebrafish ventral spinal cord and putative taste receptors (Figure 1B; and see expression analyses below).
In GRCz10 this locus is now called pkd1l2a. A single 17-exon transcript, ENSDART00000173234.1, is predicted to encode an 1124 amino acid protein. However, the 5′ sequence of this transcript only partially overlaps the 3′ sequence of ENSDARG00000090210 and is, therefore, likely to be incomplete (Figure 1B). It is also likely to contain inaccuracies because sequence present in our ENSDARG00000074116 riboprobe is annotated as being intronic in ENSDART00000173234.1.
To identify the correct gene sequence, we used the XM_002662913 sequence, which aligns with both of the retired pkd1l3 genes as well as sequence upstream of them, to map the mRNA transcript using RT-PCR. Since a preliminary protein domain search of the reference sequence suggested we might be missing amino-terminus sequence, we used the RefSeq GFF3 annotation import track in GRCz10 to identify a putative locus, LOC101884812, immediately upstream (Figure 1B). A protein domain search of this sequence using the Pfam protein database identified lectin and galactose-binding-lectin domains typically found in the amino-terminus of PKD1L2 proteins (Figure 2). We confirmed that these regions were transcribed using RT-PCR (Figure 1B). We also performed inverse PCR to identify any 5′ coding sequence that might be further upstream (Figure 1B). Using these approaches we identified an 8526 bp mRNA transcript, comprising 41 exons, both 5′ and 3′ UTR sequences and encoding a protein of 2485 amino acids that is identical in domain structure and length to mouse PKD1L2 (Figure 2, Table 1). This transcript encompasses both LOC101884812 and XM_002662913 and contains additional exons not present in either of these sequences (Figure 1B). There is also considerable overlap between our transcript and the current ENSDART00000173234.1 transcript (Figure 1B). The structure of the protein encoded by our mapped transcript, including its lack of a PKD domain and the presence of a galactose-binding-lectin domain, is consistent with PKD1L2 proteins in other animals (Figure 2). Our synteny and phylogenetic analyses (see below) also suggest that this is a pkd1l2 gene. Our pkd1l2a transcript has been deposited at NCBI (accession number KY074551).
pkd1l2b
In Zv9 the XM_009303604 reference sequence partially aligned with pkd1l2 (ENSDARG00000088121) on the forward strand of chromosome 7 (1–2985 bp) and to a non-coding region on the forward strand of Zv9_Scaffold3511 (1782–6344 bp), suggesting that the pkd1l2 annotation on chromosome 7 might be incomplete (Figure 1C). Consistent with this, the 5′ coding sequence of the longest ENSDARG00000088121 transcript, ENSDART00000156286, lacked a start codon and only encoded a protein of 859 amino acids. This gene was subsequently renamed pkdrej (2 of 2) in a later Zv9 release. Interestingly, in that same genome release an additional gene, pkdrej (1 of 2), encoding a protein of 124 amino acids, was reported immediately downstream of pkdrej (2 of 2). Our PFAM protein domain analysis revealed that these “Pkdrej” proteins contained classic features of Pkd proteins (Lectin C-type domain, galactose-binding-lectin domain, and GPS motif [Pkdrej (2 of 2)] and PLAT/LH2 domain [Pkdrej (1 of 2)]. However, neither protein contained the REJ domain, present in amniote PKDREJ proteins (Figure 2), nor the polycystin-cation-channel domain present in all other Pkd proteins. By the first release of GRCz10, pkdrej (2 of 2) had become si:ch211-168k15.4 and parts of pkdrej (1 of 2) had been included in the largest transcript of a new gene, ENSDARG00000101214, called pkd1l3, although in the most recent release of GRCz10 (version 86.10), ENSDARG00000101214 is named pkd1l2. These two genes overlap each other (Figure 1C). Exons 10–16 and the first 231 bases of exon 17 of si:ch211-168k1.5.4 are identical to exons 2–9 of ENSDARG00000101214. However, the si:ch211-168k15.4 transcript utilizes a stop codon present in intron 9–10 of ENSDARG00000101214, and the transcript for ENSDARG00000101214 (ENSDART00000124969.2) utilizes a start codon present in exon 10 of si:ch211-168k15.4. Since the protein encoded by ENSDART00000124969.2 is 1442 amino acids long and contains a polycystin-cation-channel domain, we tested whether this transcript and si:ch211-168k15.4 might actually be part of a larger pkd gene using RT-PCR. Since the 5′ coding sequence is incomplete in si:ch211-168k15.4 we also performed inverse PCR to identify any 5′ coding sequence that might be further upstream (Table 1). These analyses identified a longer, combined 7898 bp transcript that contains both the si:ch211-168k15.4 and ENSDARG00000101214 sequences, utilizing a start codon 4 bases upstream of exon 1 in the current si:ch211-168k15.4 annotation, and transitioning between exon 16 of si:ch211-168k15.4 immediately into exon 9 of ENSDART00000124969.2. Whilst the identification of the start codon is unique to this study and we have also identified novel 3′ UTR sequence, nucleotides 683–7026 of our new 7898 bp mRNA transcript align perfectly with the original XM_009303604 6344 bp reference sequence. The resulting 1902 amino acid protein has very similar domains to Pkd1l2a (Table 1, Figure 2) and its polycystin-cation-channel domain is most similar to that of Pkd1l2a, with which it has >60% identity, more than 30% higher than with any other zebrafish Pkd protein (Table 2). The lack of a PKD domain and the presence of a galactose-binding-lectin domain are also consistent with PKD1L2 proteins in other animals, with the exception that, unlike zebrafish Pkd1l2a and amniote PKD1L2, this protein is missing a REJ domain. Our synteny and phylogenetic analyses, discussed below, also suggest that this is a pkd1l2 gene. Therefore, we are confident that this gene is pkd1l2b and we have deposited the transcript sequence at NCBI (accession number KY074552).
pkd2
During this study the annotation of the gene called pkd2 remained unchanged within Ensembl. The pkd2 mRNA reference sequence identified at the start of this study, DQ175629.1, aligns with the 14 exons present in the current Ensembl pkd2 transcript, ENSDART00000020412.7, although the latter contains additional UTR sequence. This suggests that pkd2 (ENSDARG00000014098) encodes a 904 amino acid protein. Our synteny and phylogeny analyses (see below) confirm that this gene is pkd2. Similar to PKD2 proteins in other vertebrates, the main conserved domain in the encoded protein is the polycystin-cation-channel domain (Figure 2, Table 1).
pkd2l1
The annotation of the gene called pkd2l1 also remained unchanged within Ensembl during this study. The pkd2l1 mRNA reference sequence XM_690312 aligns with 100% homology to exons 1–9, 11–12, and 14–15 of the current pkd2l1 transcript ENSDART00000145948.1, which contains the additional exons 10 and 13. This suggests that pkd2l1 generates a 790 amino acid protein and is encoded by ENSDARG00000022503. Consistent with this, our in situ hybridization riboprobe (see expression analyses below), which was designed against the 3′ coding and UTR sequence present in ENSDART00000145948.1 (nucleotides 2062–2614), generated data similar to expression reported using a riboprobe designed against more upstream sequence (nucleotides 1148–2022; Djenoune et al., 2014). As for zebrafish Pkd2, and PKD2 family proteins in other vertebrates, the main conserved domain in this protein is the polycystin-cation-channel domain (Figure 2, Table 1). Our synteny and phylogeny analyses (see below) also suggest that this gene is pkd2l1.
We also performed additional bioinformatics searches using a newer version of the zebrafish genome released during our study, GRCz10, to test if there were any additional potential pkd genes. For this, we identified peptide sequences for the polycystin-cation-channel domain in each of the zebrafish pkd genes and performed Tblastn analyses with each of these sequences against this newer version of the genome (see Section Materials and Methods). We used the polycystin-cation-channel domain because this is the domain that defines Pkd proteins (Figure 2). When we did this, several of the domains identified other already-identified pkd genes (Table 1), but no new pkd genes were identified.
Since the zebrafish pkd genes that we had identified included only one set of potential teleost-duplicates or ohnologs, (just pkd1l2a/pkd1l2b as we do not think that pkd1 and pkd1b are teleost duplicates because these genes exist in both cartilaginous and holostei fish as described below), we further investigated whether teleost ohnologs of any additional pkd genes might exist by searching for pkd genes in medaka, stickleback and green spotted pufferfish. We performed a textual search for pkd genes, and also blasted the polycystin-cation-channel domain for all seven zebrafish Pkd proteins, against each of these genomes. We identified the same complement of seven genes in both stickleback and green spotted pufferfish and six genes in medaka (pkd1b was missing; Table 1). For each zebrafish Pkd protein, the polycystin-cation-channel domain had greatest homology with the gene that our phylogeny and synteny analyses suggest is its closest ortholog in each of the other teleost genomes (Table 1). Whilst Pkd1l2b in green spotted pufferfish and stickleback have slightly higher sequence homology with the polycystin-cation-channel domain of zebrafish Pkd1l2a than Pkd1l2b, Pkd1l2a in both of these teleosts shows even higher sequence homology with zebrafish Pkd1l2a. Therefore, these data, together with our phylogeny and synteny analyses, suggest that we have correctly classified the genes that encode these proteins (Table 1). We found no evidence in any of the teleosts examined for additional duplicate (ohnolog) pkd genes.
Interestingly, our Tblastn analyses with full-length zebrafish Pkd1b identified a small region of homology in each of the teleost genomes with the PLAT/LH2 region of Pkd1b (data not shown). Visual inspection of each of these loci revealed conserved synteny with the region surrounding amniote PKD1L3 genes (Figure 4E). To assess whether these teleost genomes might contain pkd1l3 orthologs, we performed Tblastn analyses with full-length mouse PKD1L3 (Supplementary Table 4). These identified the same loci. The putative pkd1l3 locus is not annotated in either zebrafish or medaka genomes, although a transcript is present in a previous zebrafish genome assembly (ENSDARG00000091803 in Zv9, Table 1). In the green spotted pufferfish and stickleback genomes, this locus contains a novel gene. Consistent with our Tblastn analyses with polycystin-cation-channel sequences, we have not detected these sequences encoding this domain within any of these loci. Given that the polycystin-cation-channel domain is the one domain that is present in all PKD proteins, we do not consider these sequences bona-fide pkd genes and therefore have not analyzed them further in this study.
Given that we found sequences with homology to part of the pkd1l3 gene, to confirm that pkd212 and pkdrej are absent in teleosts we performed Tblastn with full-length mouse PKD2L2 and PKDREJ against all of the teleost genomes discussed above. Both of these analyses only produced alignments with already identified Pkd proteins. Therefore, we are confident that there are no pkd212 or pkdrej genes in these teleost genomes.
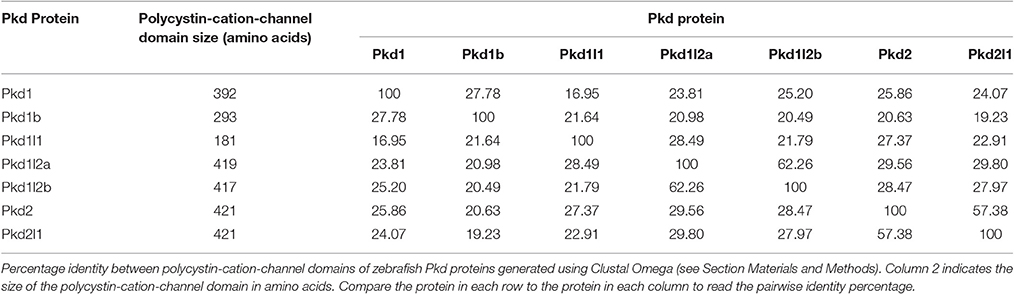
To investigate potential relationships between the zebrafish pkd genes we aligned the polycystin-cation-channel domains for each of the Pkd proteins and determined the percentage identity of this domain between each of them. Pkd1l2a and Pkd1l2b have the highest identity (62%; Table 2), which is consistent with them being recently duplicated genes. Pkd2l1 and Pkd2 also have a high degree of identity at 57%. However, all of the other pair-wise comparisons have <30% identity at the amino acid level.
To investigate the evolution of zebrafish pkd genes we identified all of the PKD genes in both spotted gar (Lepisosteus oculatus; a holostei fish) and elephant shark (Callorhinchus milii; a cartilaginous fish). In both of these species we found 8 pkd genes: pkd1, pkd1b, pkd1l1, pkd1l2, pkdrej, pkd2, pkd2l1, and pkd2l2 (Figure 3, Supplementary Table 5). Unlike mammals, spotted gar, and elephant shark both have a pkd1b gene. In contrast, similar to mammals and unlike teleosts they only have one pkd1l2 gene (Supplementary Table 5). However, like teleosts, spotted gar only has a partial pkd1l3 sequence that lacks the polycystin-cation-channel domain but is located in a region with conserved synteny with other pkd1l3 regions (data not shown). It is currently less clear whether a pkd1l3 gene exists in elephant shark. In the mammalian, teleost and holostei genomes that we have examined, PKD1L3 is always located close to a gene called DHODH (Figure 4E). dhodh is located on Scaffold_12 of the elephant shark current genome, but we did not find any evidence for a pkd gene nearby (data not shown), although we cannot rule out the possibility that this gene exists elsewhere in the genome.
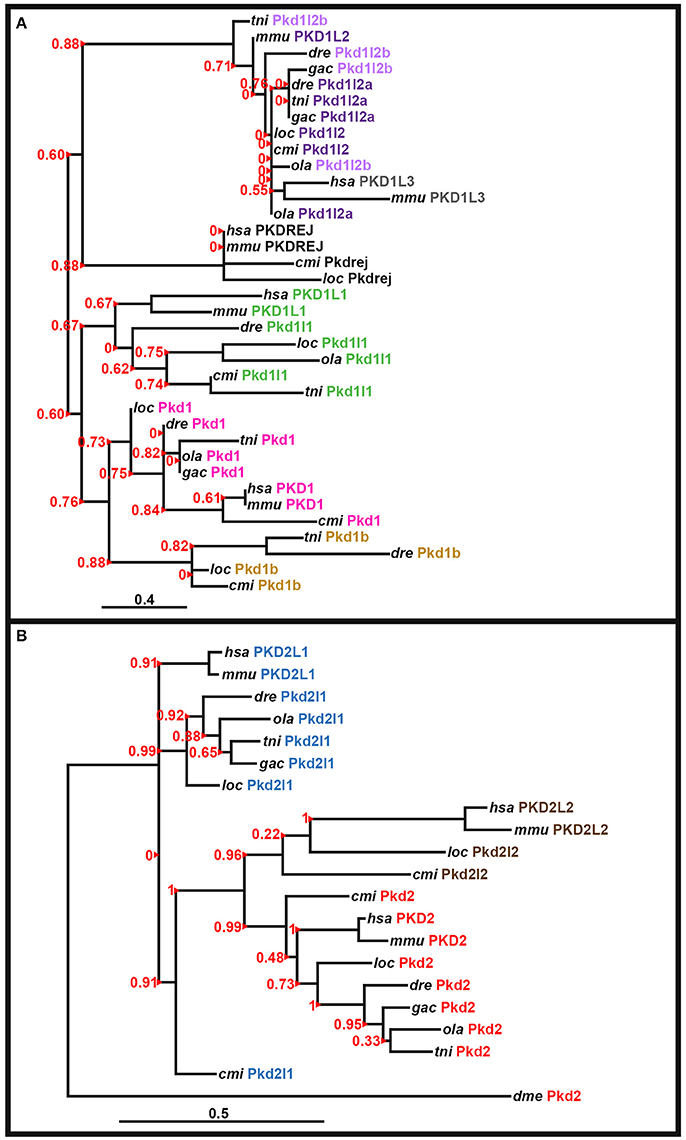
Figure 3. Phylogenetic analysis of PKD proteins. Phylogenetic analysis of human (Homo sapiens, hsa), mouse (Mus musculus, mmu), spotted gar (Lepisosteus oculatus, loc), elephant shark (Callorhinchus milii, cmi), zebrafish (Danio rerio, dre), medaka (Oryzias latipes, ola), green spotted pufferfish (Tetraodon nigroviridis, tni), and stickleback (Gasterosteus aculeatus, gac) PKD1-like proteins (A) and PKD2-like proteins with the Drosophila melanogaster (dme) Pkd2 protein as an outgroup (B). In both cases a region of the polycystin-cation-channel domain that was present in all of the proteins was used (see Section Materials and Methods and Supplementary Figures 1, 2). Both analyses used a maximum likelihood method, with WAG substitution, performed using PhyML (v3.1/3.0 Alrt; see Section Materials and Methods). Human PKD1L2, stickleback Pkd1b and teleost, and spotted gar Pkd1l3 proteins are not included as they lack the polycystin-cation-channel domain. We did not include an invertebrate protein in the analysis of PKD1-like proteins as the evolution of this gene family seems to be more complex and while vertebrate PKD1-like proteins do have some homology to invertebrate proteins, this homology is limited and we did not identify an invertebrate protein that had good support for being a clear outgroup for this family. aLRT Sh-like branch support values are shown in red to the left of each branch. Red arrowheads indicate the branch that each value corresponds to. Scale bar = 0.4 nucleotide substitutions per site (A), 0.5 nucleotide substitutions per site (B).

Figure 4. Conserved synteny around zebrafish pkd1-like genes. Examination of syntenic relationships between pkd and neighboring genes in genomic regions associated with zebrafish pkd1 family genes. Species is indicated on left and chromosomes on right. Un-Random (tni), unordered random sequences that have yet to be assigned to a chromosome. hsa, human (Homo sapiens); mmu, mouse (Mus musculus); dre, zebrafish (Danio rerio); ola, medaka (Oryzias latipes); tni, green spotted pufferfish (Tetraodon nigroviridis); and gac, stickleback (Gasterosteus aculeatus). Pkd genes are indicated in bold red text. Schematics are not to scale. For ease of comparison, gene clusters are shown in the same orientation, even though in some cases, gene organization is as shown, but on the opposite strand of the chromosome. Schematics only include annotated coding genes. Antisense processed transcripts and ribosomal and long-non-coding RNA loci are not included. Colors indicate homologous genes within an individual panel. So, for example, pink genes in pkd1 (A) are homologous to each other (they are all SLC9A3R2 despite their slightly different positions) but they are not homologous to pink genes in the pkd1l1 panel. However, gray (novel) genes in (A) are an exception, as these three genes are not homologous to each other. We did not find a pkd1b gene in medaka, and none of the genes flanking pkd1b in green spotted pufferfish and stickleback are found near the zebrafish pkd1b gene (B). The PKD1L1 locus is syntenic within but not between amniotes and teleosts (C). Zebrafish pkd1l2a is the only teleost gene to share synteny with both the aminote and other teleost PKD1L2 loci (D). Only stickleback and medaka pkd1l2b genes share any synteny among the pkd1l2b genes (D). As in amniotes, all teleost putative partial pkd1l3 orthologs are flanked by dhodh genes (E).
To confirm orthologous relationships between teleost and mammalian PKD1-family and PKD2-family genes we performed phylogenetic analyses of human (Homo sapiens), mouse (Mus musculus), spotted gar (Lepisosteus oculatus), elephant shark (Callorhinchus milii), zebrafish (Danio rerio), medaka (Oryzias latipes), green spotted pufferfish (Tetraodon nigroviridis), and stickleback (Gasterosteus aculeatus) PKD1-like and PKD2-like proteins. We used the region of the polycystin-cation-channel domain that was present in all of the proteins and both Neighbor-Joining (NJ) and maximum likelihood methods (see Section Materials and Methods, Supplementary Figures 1–2, Figure 3). In the resulting phylogenetic trees (Figure 3; data not shown), all of the zebrafish proteins cluster with the expected proteins from other species, suggesting that their annotations are correct. Consistent with Pkd1l2a and Pkd1l2b being teleost duplicates of PKD1L2 proteins in other vertebrates, all of the PKD1L2 proteins cluster together. Interestingly, in the maximum likelihood tree, mammalian PKD1L3 genes are also contained in this cluster, although this was not the case in the NJ analysis (Figure 3 and data not shown). However, we are confident that none of the teleost pkd1l2 genes are pkd1l3 genes as we have also found partial pkd1l3 genes in teleosts as discussed above.
To further test whether we had correctly identified orthologous relationships, we also examined the genomic regions around each of the zebrafish pkd genes and their proposed orthologs in other vertebrates for conserved syntenic relationships with other neighboring genes. We found that the pkd1 genomic locus contains both a NTHL1 and TSC2 gene in humans, mouse and all four teleost species (Figure 4A). In contrast, whilst the genomic regions around green spotted pufferfish and stickleback pkd1b share synteny with each other, none of the genes in this region are found near zebrafish pkd1b (Figure 4B). The PKD1L1 locus has considerable shared synteny between human and mouse and between teleosts, but the teleost loci don't have any obvious shared synteny with the amniote loci (Figure 4C). In contrast, PKD1L2 is located near a GCSH and a BCO1 gene in both mammals and at least one teleost and there are other genes found in common near most of the teleost pkd1l2a genes (Figure 4D).
The PKD2 locus, like the PKD1 locus, also has some conserved synteny between different vertebrates (Figure 5A). This genomic region contains an ABCG2 and PPM1K gene in humans, zebrafish, green spotted pufferfish, medaka, and stickleback. While the mouse Pkd2 locus doesn't seem to contain these genes—it shares three other genes with the human locus. The Pkd2l1 locus also has considerable conserved synteny between different teleost genomes and between mouse and human. In addition, a couple of genes are present in both mammals and at least one teleost: CHUK (present in mammals and zebrafish) and a SEC gene—SEC31B (present in mammals) and sec23lp (present in zebrafish and stickleback; Figure 5B).
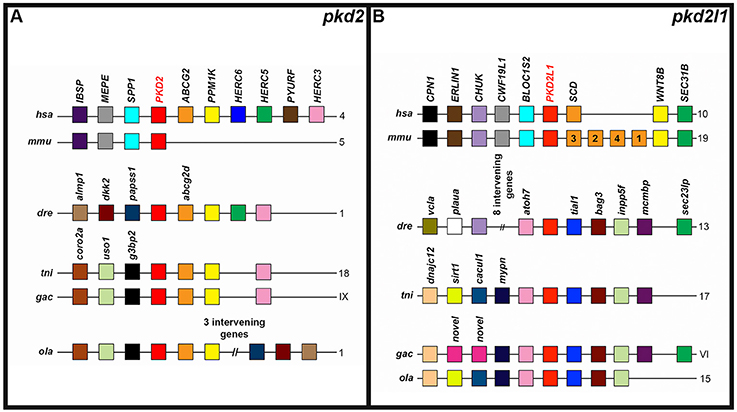
Figure 5. Conserved synteny around zebrafish pkd2-like genes. Examination of syntenic relationships between pkd and neighboring genes in genomic regions associated with zebrafish pkd2 family genes. Species is indicated on left and chromosomes on right. hsa, human (Homo sapiens); mmu, mouse (Mus musculus); dre, zebrafish (Danio rerio); ola, medaka (Oryzias latipes); tni, green spotted pufferfish (Tetraodon nigroviridis); and gac, stickleback (Gasterosteus aculeatus). Pkd genes are indicated in bold red text. Schematics are not to scale. For ease of comparison, gene clusters are shown in the same orientation, even though in some cases, gene organization is as shown, but on the opposite strand of the chromosome. Schematics only include annotated coding genes. Antisense processed transcripts and ribosomal and long-non-coding RNA loci are not included. Colors only indicate homologous genes within an individual panel. So, for example, pink genes in the pkd2 panel are not homologous to pink genes in the pkd2l1 panel. The teleost pkd2 genes share synteny with human but not mouse PKD2 (A). The teleosts share considerable synteny at the pkd2l1 locus, but only zebrafish and stickleback pkd2l1 genes share any synteny with amniotes (B).
Based on all of these analyses, we are convinced that the genes that we have identified are indeed pkd1, pkd1b, pkd1l1, pkd1l2a, pkd1l2b, pkd2, and pkd2l1 and that these are the only bona-fide pkd genes in zebrafish and the other teleosts that we have examined.
Expression of pkd Genes during Zebrafish Development
As described in the introduction, pkd genes have important developmental functions in many different tissues and in at least some of these locations, PKD1-like and PKD2-like proteins act in a heteromeric complex. However, before we started this study the expression patterns, and hence the potential functions and binding partners, were not known for several of the zebrafish pkd genes. Therefore, we performed a comprehensive expression analysis for all seven genes at developmental stages from 8 h to 5 dpf. We identified expression of specific subsets of these genes in many different tissues/structures as described below.
Dorsal Forerunner Cell and Kupffer's Vesicle Expression
As previous studies have established that pkd2 is expressed in Kupffer's vesicle (KV) during early zebrafish embryogenesis (Bisgrove et al., 2005; Schottenfeld et al., 2007; Roxo-Rosa et al., 2015), we investigated expression of all seven zebrafish pkd genes in dorsal forerunner cells/KV at 8.3, 10, and 12 h (Figure 6). We found no expression of pkd1, pkd1b, pkd1l2a, pkd1l2b, and pkd2l1 at any of these stages (Figures 6J–S), with the exception that there is some spinal cord expression of pkd1b at 12 h (arrows in Figure 6M). In contrast, pkd1l1 was expressed in a cluster of dorsal forerunner cells at 8.3 h (Figure 6D), that later condense to form the KV at 10 and 12 h (Figures 6E–F). pkd2 expression was not detected at 8.3 h (Figure 6G), but was present in a group of cells in the KV region at 10 h, resolving in to a ring of cells surrounding the KV by 12 h (Figures 6H–I).
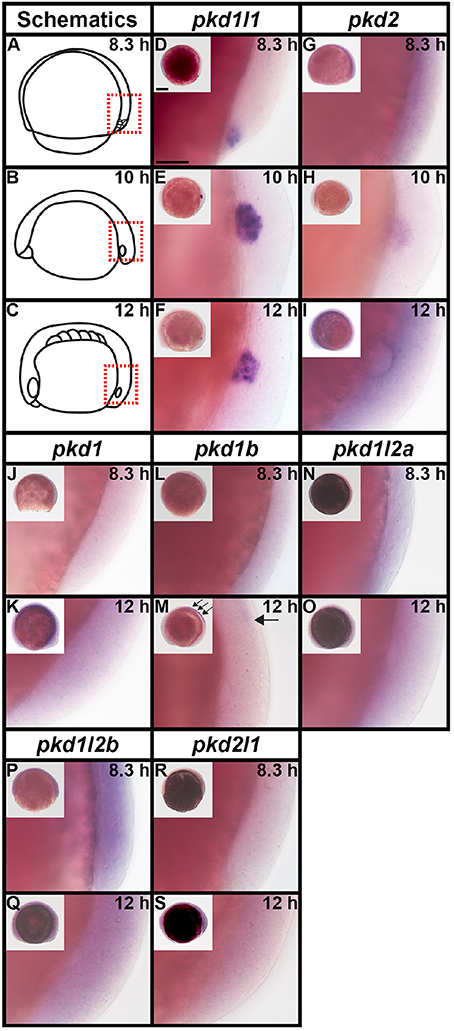
Figure 6. pkd2 and pkd1l1 are expressed in Kupffer's vesicle. Lateral expression of pkd genes at 8.3, 10, and 12 h. Region shown in main panel at each stage is indicated by red dotted boxes in schematics (A–C). Inset images in (D–S) show whole-mount view of embryo, dorsal forerunner cells/KV located in bottom right-hand corner. There is no expression of pkd1, pkd1b, pkd1l2a, pkd1l2b, or pkd2l1 in dorsal forerunner cells or KV at any of these stages. The boundary of the KV cavity is faintly visible in M as a slightly different focal plane has been shown to include spinal cord expression. However, pkd1b is not expressed in the margin of KV. Arrows in (M) indicate caudal limit of spinal cord expression of pkd1b. pkd1l1 is expressed in the KV region at all three stages (D–F) and pkd2 is expressed at 10 and 12 h but not 8.3 h (G–I). Scale bar (D) = 50 μm, (D–S) main panels and 200 μm, inset panels.
Pronephros/Kidney Expression
It has already been reported that pkd2 expression is enriched in developing zebrafish pronephros (Bisgrove et al., 2005; Schottenfeld et al., 2007), and Pkd2 protein is present in zebrafish kidney epithelial cells (Obara et al., 2006). However, before this study, it was unknown if any other pkd genes are expressed in the developing zebrafish pronephros. Therefore, we examined expression in WT embryos (8.3, 10, 12, 24, 27, 30, 36, and 48 h, 3, 4, and 5 dpf). We found no expression of pkd1b, pkd1l1, pkd1l2a, pkd1l2b, or pkd2l1 in the developing pronephros at any of these stages (Figures 6–9 and data not shown). In contrast, while pkd1 and pkd2 are not expressed in presumptive pronephric mesoderm at 8.3, 10, or 12 h (Figures 6G–K), they are both expressed in the pronephros by 24 h, although the expression of pkd1 is much stronger than that of pkd2 (Figures 7A,K, 9O,P,A',B'). The expression of pkd1 in the pronephros persists until 3 dpf. We also found that weak pkd2 expression persists in the pronephros during the pharyngula period (arrows in 27 and 36 h) but it was not enriched above the otherwise ubiquitous expression of pkd2 by 2 dpf (Figures 10A–D and data not shown).
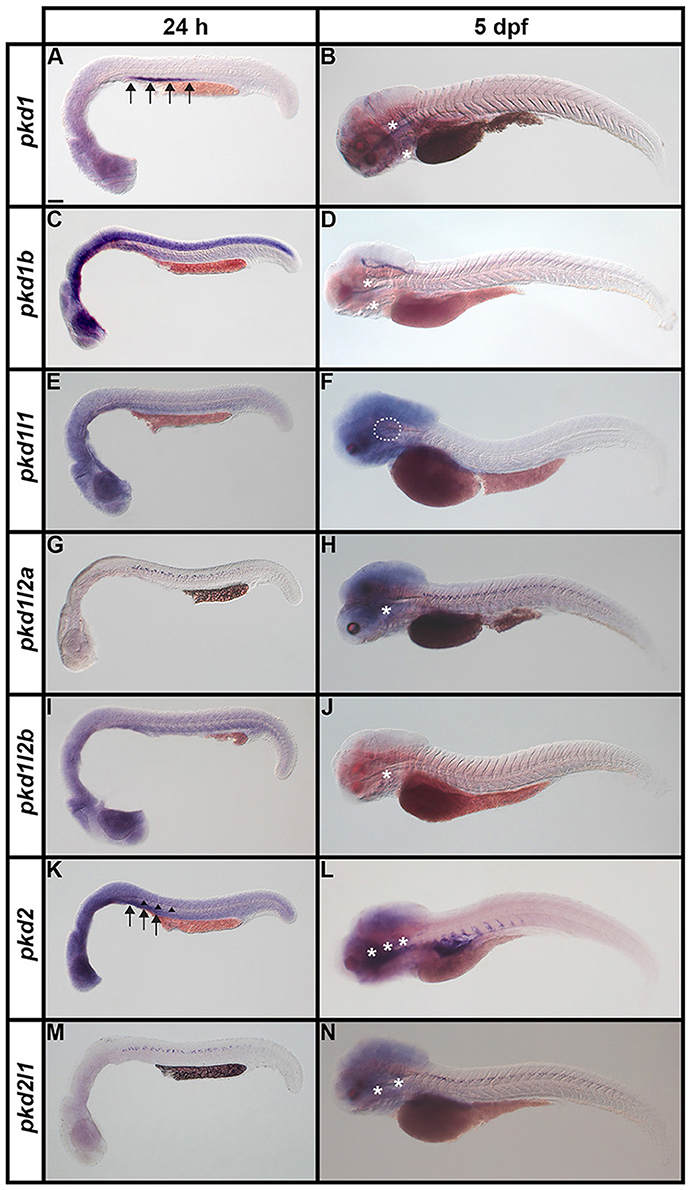
Figure 7. Expression of pkd genes in zebrafish embryos and larvae. Lateral views of whole embryo expression of pkd genes at 24 h and 5 dpf. Rostral left, dorsal up. (A,B) pkd1 is strongly expressed in the pronephros at 24 h (arrows, A) but not at 5 dpf. By 5 dpf, pkd1 expression persists only in the putative taste receptors (white asterisks, B). (C,D) pkd1b is broadly expressed throughout the dorsal-ventral hindbrain and spinal cord, and in the caudal-most midbrain at 24 h. By 5 dpf, strong expression persists in the floor plate in the midbrain and hindbrain, whilst weaker expression persists in putative taste receptors of the pharynx (white asterisks, D). (E,F) pkd1l1 is not expressed at 24 h but is detected in the ear at 5 dpf (white dotted line, F). (G,H) pkd1l2a is expressed in cells in the ventral-most spinal cord at 24 h. This expression persists at 5 dpf, as does expression in putative taste receptors (white asterisk, H). (I,J) pkd1l2b expression is not detected at 24 h and persists only weakly in the pharyngeal cartilage at 5 dpf (white asterisk, J). (K,L) pkd2 is expressed in the pronephros (arrows, K) and perhaps very weakly in the floor plate at 24 h (arrowheads, K). By 5 dpf, pkd2 expression is restricted to the ventral region of the rostral somites and putative taste receptors (white asterisks, L). (M,N) Like pkd1l2a, pkd2l1 is also expressed in cells in the ventral-most spinal cord at 24 h. This expression also persists at 5 dpf, together with weak expression in putative taste receptors (white asterisks, N). Low level diffuse staining in the brain in (A,C,F,H,L,N) and more widely in (E,I,K) is probably background staining. These embryos were stained for longer periods in order to try and detect any weak, but specific, expression in the spinal cord. As a consequence of this, the brain, which contains large ventricles which sometimes trap RNA riboprobes, often has background staining (see Section Discussion). Scale bar (A) = 100 μm.
Spinal Cord Expression
Zebrafish pkd2l1 was recently shown to be expressed in a unique population of spinal cord cells, called Kolmer Agduhr (KA) cells or cerebrospinal fluid-contacting neurons (CSF-cNs; Djenoune et al., 2014). In addition, pkd1b has been reported as being expressed broadly in the spinal cord at late somitogenesis stages but restricted to medial floor plate ependymal cells by 3.5 dpf (Mangos et al., 2010) and low level expression of pkd2 has been observed in the floor plate during somitogenesis stages (Bisgrove et al., 2005; Schottenfeld et al., 2007). Therefore, we were very interested in investigating expression of pkd genes in spinal cord cells, particularly to see if we could identify a potential partner for Pkd2l1 in KA cells and/or additional Pkd proteins expressed in the floor plate.
We examined spinal cord expression of all 7 pkd genes in WT embryos at 12, 24, 27, 30, 36, and 48 h, and 3, 4, and 5 dpf. In addition, we examined expression in mindbomb mutants at 24 h. mindbomb encodes an E3-ubiquitin-ligase that is required for efficient Notch signaling. In mindbomb mutants Notch signaling is lost and, as a result, the vast majority of spinal cord progenitor cells precociously differentiate as early-forming populations of spinal cord neurons at the expense of later forming neurons and glia (Jiang et al., 1996; Schier et al., 1996; Itoh et al., 2003; Park and Appel, 2003; Batista et al., 2008). Therefore, comparing expression of genes in the spinal cords of mindbomb mutants and WT embryos enables us to distinguish between progenitor domain expression (which should be lost) and post-mitotic expression (which is often, although not always, expanded). In addition, if a gene is expressed very weakly in post-mitotic spinal cord cells, its expression is usually easier to observe in mindbomb mutants, where the expression is often expanded and stronger (Batista et al., 2008). Therefore, examining expression in mindbomb mutants also helps us to be more confident about whether a gene is expressed in the spinal cord or not.
Both WT and mindbomb mutant expression analyses suggest that pkd1, pkd1l1, and pkd1l2b are not expressed in zebrafish spinal cord (Figures 7A,B,E,F,I,J, 8S–A', 9M–X and Supplementary Figures 3G–U). It is likely that pkd2 is also not expressed in spinal cord, other than very weakly in floor plate (Figures 7K,L, 8B'–D', 9Y–B' and Supplementary Figures 3V–Z). In contrast, pkd1b was expressed broadly in the spinal cord and pkd1l2a and pkd2l1 were both expressed in post-mitotic cells in ventral spinal cord (Figures 7–9, 11, see more detailed descriptions below).
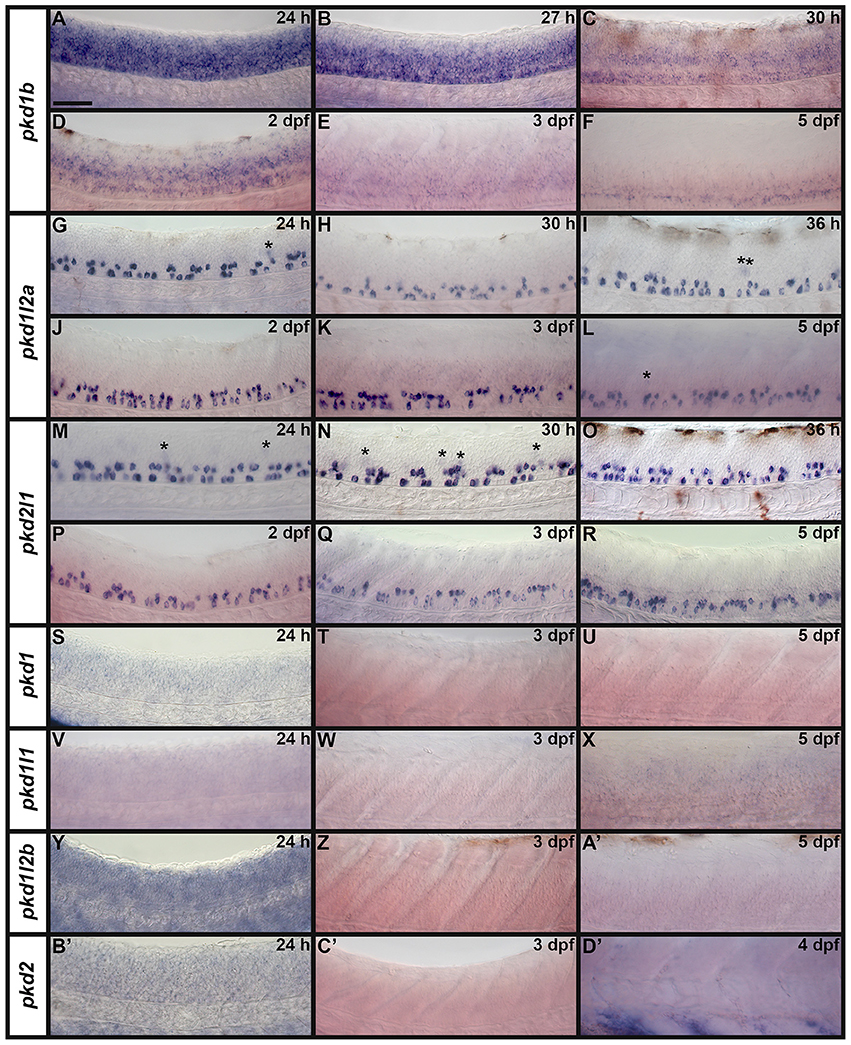
Figure 8. Spinal cord expression of zebrafish pkd genes. Lateral views showing expression of pkd genes at 1–5 dpf. Rostral left, dorsal up. (A–F) pkd1b is expressed broadly in the spinal cord. pkd1l2a (G–L) and pkd2l1 (M–R) are both expressed in two rows of cells in the ventral spinal cord and occasionally weakly in more dorsal cells (asterisk). (S–U) pkd1, (V–X) pkd1l1, (Y–A') pkd1l2b, and (B'–D') pkd2 are not expressed in spinal cord. Some of these embryos have background expression as we stained them for long periods of time to try and detect any weak, but specific, expression. Expression of pkd2 is visible in the rostral ventral somites (D'). Scale bar (A) = 50 μm.
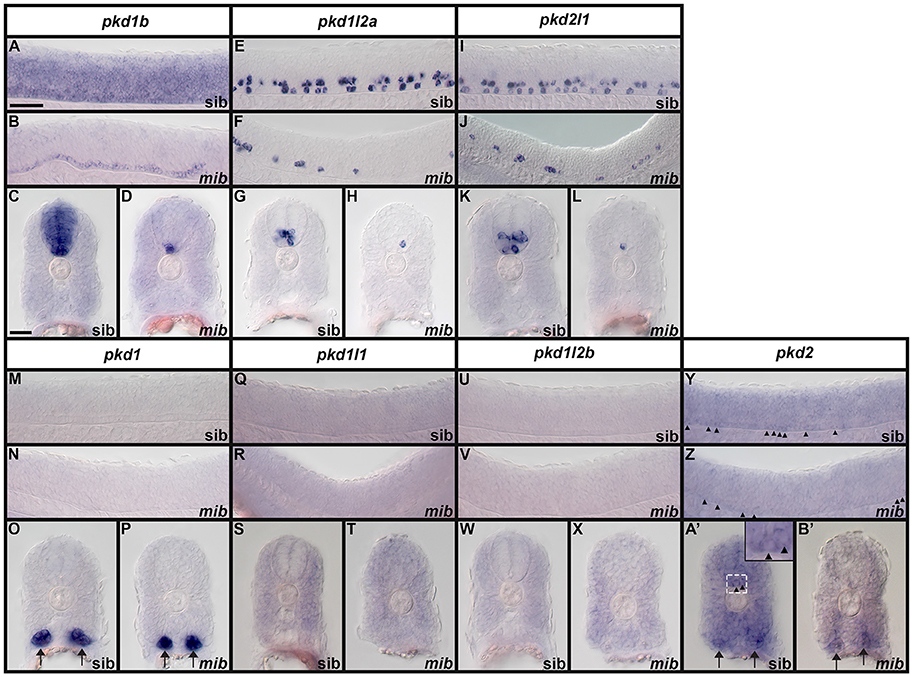
Figure 9. Expression of zebrafish pkd genes in mindbomb mutants. Lateral views (A,B,E,F,I,J,M,N,Q,R,U,V,Y,Z) and cross-sections (C,D,G,H,K,L,O,P,S,T,W,X,A',B') of pkd expression in the trunk of mindbomb mutants and sibling embryos with WT phenotypes. Dorsal is up. In lateral views, rostral is left and only the spinal cord region is shown. Arrows (O,P,A',B') indicate pronephros expression. Arrowheads (Y,Z,A' and higher magnification inset in A') indicate weak expression of pkd2 in the floor plate of the spinal cord. The focal plane in B' does not include labeled floor plate cells. Scale bar (A) = 50 μm (lateral views, A,B,E,F,I,J,M,N,Q,R,U,V,Y,Z); Scale Bar (C) = 30 μm (cross-sections, C,D,G,H,K,L,O,P,S,T,W,X,A',B') and 10 μm (inset in A').
pkd1b is expressed very broadly throughout the dorsal/ventral extent of the spinal cord at 24 h in what appears to be mainly progenitor cells (Figures 8A, 9A,C). By 27 h, expression in the most dorsal part of the spinal cord has reduced. By 30 h the expression has resolved into two broad domains in the ventral spinal cord, one just above the notochord and one in the middle of the dorsal/ventral axis. This continues until 5 dpf, at which point expression in the more dorsal domain is much weaker (Figures 8B–F). Consistent with pkd1b being expressed by progenitor cells, most of its spinal expression is lost in mindbomb mutants, with the exception of the floor plate expression, which remains. This suggests that pkd1b may be expressed in floor plate in addition to being broadly expressed in spinal cord progenitor cells (Figures 9B,D). Consistent with this, we observe pkd1b expression in the floor plate of the hindbrain from 24 h until at least 5 dpf (insets in Figures 12 E'–G').
pkd1l2a and pkd2l1 have very similar spinal expression patterns. Both genes are already expressed in two rows of ventral cells by 24 h (Figures 8G,M) as well as being expressed more weakly in occasional more dorsal cells (Figures 8, 11). This expression also extends into caudal hindbrain (insets in Figures 12M–O, Y–A'). Unusually for post-mitotic spinal cord cells, but consistent with KA cells, these cells are located medially in ventral spinal cord, in positions where they can contact the CSF-containing central canal (Figures 11A,B, 9E,G,I,K). The expression of both of these genes continues throughout all of the stages that we examined, although the two rows become less distinct at later stages and pkd2l1 expression seems to become weaker in the more ventral row of cells by 4 dpf (Figures 8H–L,N–R). To confirm that these genes are both expressed by KA cells, we performed double labels with Tg(−8.1gata1:gata1-EGFP) zebrafish which express GFP in both KA and V2b cells in the spinal cord (Kobayashi et al., 2001; Batista et al., 2008). Consistent with the previous report (Djenoune et al., 2014), we find that pkd2l1 is expressed in all ventral KA″ cells and more dorsal KA′ cells. In addition, we show for the first time that zebrafish pkd1l2a is also expressed in both of these cell types (Figures 11C–I').
Ear Expression
We also detected expression of two pkd genes in specific territories of the ear at 4–5 dpf (Figures 10E–H). We observed pkd1l1 and pkd2l1 expression in the ectoderm of the inner ear that supports the posterior canal and posterior crista at 4 dpf. pkd1l1 is also weakly expressed in the utricular otolith. By 5 dpf, pkd1l1 expression persists in the utricular otolith and the underlying utricular macula. It is also expressed in the neighboring ectoderm flanking the lateral canal and lateral crista (Figures 10E,F). In contrast, the expression of pkd2l1 persists in the tissue surrounding the posterior canal and posterior crista (Figures 10G,H).
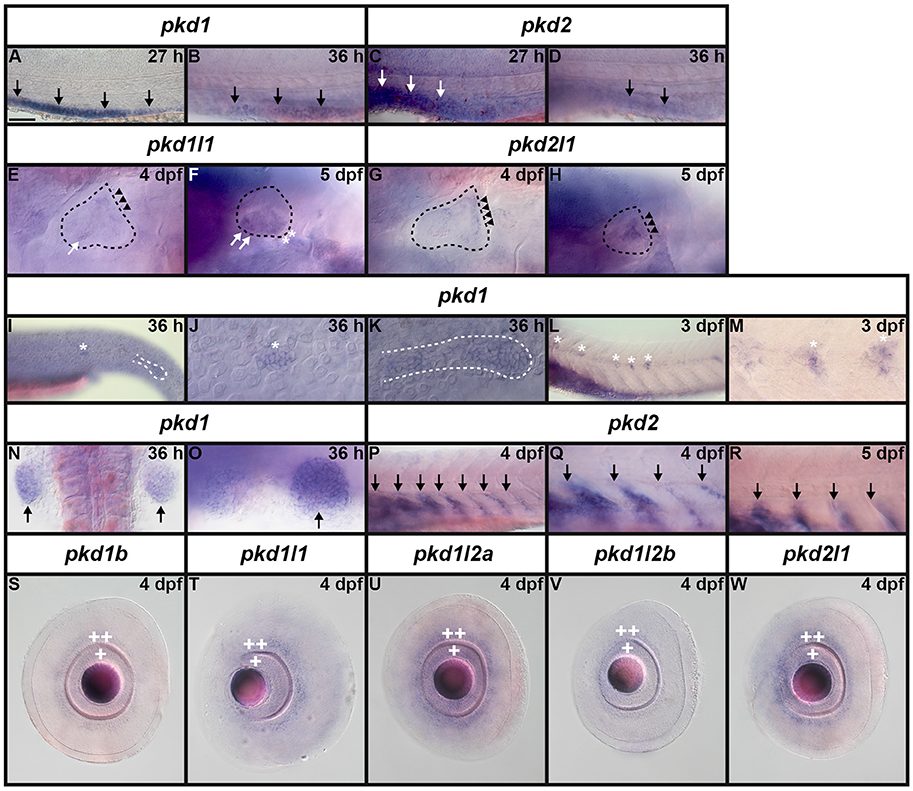
Figure 10. Expression of pkd genes in kidney, somites and sensory organs. (A–D) Lateral view of pkd1 (A,B) or pkd2 (C,D) expression in pronephros (black and white arrows) at 27 and 36 h. Rostral left, dorsal up. pkd1 is strongly expressed in pronephros at 27 h (A). Expression starts to decline at 36 h (B). Expression of pkd2 is weak in pronephros at 27 h (C) and is reduced even further by 36 h (D). (E–H) Lateral expression of pkd genes in the ear at 4–5 dpf. Dotted line shows ear boundary. Weak expression of pkd1l1 (E) and pkd2l1 (G) is first detected at 4 dpf in the inner ear ectoderm that supports the posterior canal and posterior crista (black arrowheads). pkd1l1 is also weakly expressed in the utricular otolith (white arrows). By 5 dpf, pkd1l1 expression persists in the utricular otolith and the underlying utricular macula (white arrows). It is also expressed in neighboring ectoderm flanking the lateral canal and lateral crista (white asterisks; F). At 5 dpf the expression of pkd2l1 persists in tissue surrounding the posterior canal and posterior crista (black arrowheads; H). (I–M) Lateral view of pkd1 expression in neuromasts (white asterisks) and lateral line primordium (white dotted line) at 36 h and 3 dpf. Rostral left, dorsal up. Weak expression of pkd1 in neuromasts and lateral line primordium is first detected at 36 h [I, higher magnification of the neuromasts (J) and lateral line primordium (K)]. By 3 dpf expression persists in neuromasts (L and higher magnification view, M). pkd1 is also expressed in pectoral fin buds (black arrows) at 36 h [dorsal view, rostral top (N), and lateral view—rostral left, dorsal up (O)]. (P–R) Lateral expression of pkd2 in rostral somites at 4 and 5 dpf. Rostral left, dorsal up. pkd2 is first expressed in the ventral half of each rostral somite at 4 dpf (black arrows in P, higher magnification in Q) and persists at 5 dpf (black arrows in R). (S–W) Lateral expression of pkd genes in the eye at 4 dpf. Rostral left, dorsal up. pkd1b, pkd1l1, pkd1l2a, pkd1l2b, and pkd2l1 are expressed in the ganglion cell layer (adjacent to lens, single white cross) and amacrine cells (outer cell layer immediately adjacent to ganglion cell layer, double white cross) of the eye at 4 dpf. The expression of pkd1b (S) and pkd1l2b is weak (V) and the expression of pkd1l1 (T), pkd1l2a (U), and pkd2l1 (W) is stronger. Only the expression of pkd1l2b persists in these cell layers at 5 dpf (data not shown). Scale bar (A) = 23 μm (J,K,M); 42 μm (E–H,O); 50 μm (A–D,Q,R); 55 μm (N); 62.5 μm (S–W); and 100 μm (I,L,P).
Lateral Line and Neuromasts
Interestingly, we also detected expression of pkd1, and only pkd1, in the neuromasts (asterisks, Figures 10I,J,L,M) and lateral line primordium (white dotted line, Figures 10I,K). This expression was first apparent at 36 h and it persists in the neuromasts until 3 dpf, but is lost by 4 dpf.
Fin Buds
Similarly, out of all 7 pkd genes, we only detected expression of pkd1 in the pectoral fin buds. pkd1 is expressed in these structures at 36, 48 h, and 3 dpf, but we could not detect expression above background by 4 dpf (arrows in Figures 10N,O and data not shown). At 36 and 48 h, pkd1 expression is ubiquitous throughout these structures but as reported previously (Mangos et al., 2010) expression is restricted to the base of the fin by 3 dpf.
Somite Expression
We also only detected expression of one pkd gene in somites. pkd2 is first expressed in the ventral half of each rostral somite at 4 dpf and this expression persists at 5 dpf (Figure 7L, arrows in Figures 10P–R).
Eye Expression
We detected transient expression of five different pkd genes in the retina. pkd1b, pkd1l1, pkd1l2a, pkd1l2b, and pkd2l1 are expressed in the ganglion cell layer (adjacent to the lens, single cross) and the amacrine cells (outer cell layer immediately adjacent to the ganglion cell layer, double cross) of the eye at 4 dpf. The expression of pkd1b and pkd1l2b is weak, but the expression of pkd1l1, pkd1l2a, and pkd2l1 is stronger (Figures 10S–W). However, only pkd1l2b expression persists at 5 dpf (data not shown).
Taste Bud Expression
In other vertebrates, PKD1L3 and PKD2L1 have been proposed to function as sour taste receptors (Huang et al., 2006; Ishimaru et al., 2006, although also see Nelson et al., 2010; Hofherr and Kottgen and references therein). In zebrafish, the taste buds first form at about 3 dpf on the lips/jaw and pharyngeal arches and then at 4 dpf they also form on the mouth and oro-pharyngeal cavity (Hansen et al., 2002; Kapsimali et al., 2011). Consistent with possible expression in the taste buds, we observe dynamic expression of all seven zebrafish pkd genes in the pharyngeal and/or jaw regions at these stages.
Mangos et al. (2010) reported expression of pkd1 in the pharyngeal arches and jaw forming regions at both 52 and 72 h and our data agrees with this. We see strong expression in the pharyngeal arches by 48 h (data not shown) and this expression remains strong at 3 dpf on both the ceratobranchial arches (the more caudal parts of the pharyngeal arches, posterior to the eyes, arrowheads Figures 12D, 13A), and also more anteriorly on the hyosymplectic cartilage (medio-lateral and just posterior to the eyes, derived from the second pharyngeal arch, arrowheads Figures 12D, 13A). Expression is decreased at 4 and 5 dpf (arrowheads Figures 12A–F, 13A–C). Consistent with an earlier report (Bisgrove et al., 2005), we also find pkd2 expression in the pharyngeal arches at 3 dpf (arrowheads Figures 12G,J). In addition, we demonstrate here that strong expression of pkd2 persists in the pharyngeal arches at 4 dpf (arrowheads Figures 12H,K). By 5 dpf, there is also some expression on the pharyngeal walls and the expression in the pharyngeal cartilage has reduced caudally and remains in only the rostral-most pharyngeal cartilage (arrowheads Figures 12I,L, 13D–F).
pkd1l2a and pkd1l2b are also expressed in putative taste buds, although their expression patterns differ from one another and from those of pkd1 and pkd2. We observed pkd1l2a expression on the pharyngeal walls and cartilage at all stages examined. In contrast to pkd1, which is expressed not only across the entire surface of the ceratobranchial arches but also on the medio-lateral hyosymplectic cartilage at 3 dpf, pkd1l2a is expressed only laterally on the ceratobranchial arches and in very few cells medially, in the pharyngeal cavity between the eyes. By 4 dpf, pkd1l2a expression is strongest on the ceratohyal cartilage, which is the most rostral and medial component of the pharyngeal cartilage and this expression persists at 5 dpf (arrowheads Figures 12M–R, 13G–I). In contrast to pkd1l2a, we did not detect expression of pkd1l2b on the pharyngeal walls. Instead it is expressed in a few medial cells of the ceratobranchial arches (caudal-most pharyngeal arches) at 3 dpf. This expression spreads laterally by 4 dpf, and is expanded further by 5 dpf such that it is adjacent and caudal to that of pkd1l2a at these stages (arrowheads Figures 12S–X, 13J–L). At 3 dpf the expression of pkd2l1 partially overlaps that of pkd1l2a and pkd1l2b across the ceratobranchial arches, although, like pkd1l2b, pkd2l1 is not expressed on the pharyngeal walls, it is only expressed on the pharyngeal cartilage. This expression of pkd2l1 increases and expands laterally across the ceratobranchial arches over the next 2 days (arrowheads Figures 12Y,Z,A'–D', 13M–O).
We also see weak expression of pkd1b in the pharynx, probably on the pharyngeal walls at 2, 3, 4, and 5 dpf. By 4 dpf, and persisting at 5 dpf, this gene is also expressed in a few locations on the pharyngeal cartilage (arrowheads Figures 12E'–J', 13P–R). pkd1l1 is only expressed very transiently in these regions. There is no obvious expression on either the pharyngeal cartilage or the pharyngeal walls at either 3 or 5 dpf. However, there is a transient pulse of expression on both the pharyngeal walls and parts of the pharyngeal cartilage at 4 dpf (arrowheads Figures 12K'–P', 13S–U).
Discussion
In this paper, we identify seven zebrafish pkd genes and confirm their orthologous relationships with Pkd genes in other vertebrates using phylogenetic and syntenic analyses. Our bioinformatics analyses strongly suggest that this is the full complement of pkd genes in zebrafish and other teleosts and we are confident in our classification/naming of all of these genes, based on our synteny and phylogenetic analyses. The only possible exceptions are zebrafish pkd1l2a and pkd1l2b. The gene that we have called pkd1l2b in zebrafish has much more shared synteny with pkd1l2a genes in other teleosts than the gene that we have called zebrafish pkd1l2a. However, our phylogenetic analysis suggests that our assignments of pkd1l2a and pkd1l2b in zebrafish are correct. In addition, the other teleost Pkd1l2a proteins share higher homology with the zebrafish Pkd1l2a polycystin-cation-channel domain than with the Pkd1l2b polycystin-cation-channel domain. Finally, all of the teleost Pkd1l2a proteins, including zebrafish, contain a REJ domain (with the exception of green spotted pufferfish for which we only have a very short protein sequence available) and none of the teleost Pkd1l2b proteins, including zebrafish, contain a REJ domain.
Interestingly, our data suggest that only two pkd ohnologs from the whole genome duplication that occurred at the base of the teleosts have been retained: all four teleost genomes that we analyzed have both a pkd1l2a and a pkd1l2b gene, whereas we have not found Pkd1l2b in other vertebrates. In contrast, we found orthologs of all of the other teleost pkd genes in at least some other vertebrate lineages.
This limited number of teleost duplicate genes or ohnologs is accompanied by the lack of some pkd genes that exist in mammals. Our systematic bioinformatic analyses of all four teleost genomes, demonstrates that they lack pkd212 and pkdrej genes. Given that both of these genes are present in the cartilaginous fish, elephant shark (Callorhinchus milii) and the holostei fish, spotted gar (Lepisosteus oculatus), it is likely that these two genes have been lost in the teleost lineage. In addition, while we have found putative partial pkd1l3 genes in teleosts, these sequences lack the region that encodes the polycystin-cation-channel domain that is present in all other PKD genes, suggesting that they are no longer bona-fide PKD genes. The spotted gar genome also contains a partial pkd1l3 sequence that is missing this polycystin-cation-channel domain region but has conserved synteny with pkd1l3 genomic regions in other species. This suggests that pkd1l3 may be in the process of being lost in the ray-finned fish lineage. In contrast, it is likely that the Pkd1b gene has been lost somewhere in the lobe-finned fish lineage as elephant shark, spotted gar, zebrafish, green spotted pufferfish and stickleback genomes all contain this gene, but mouse and human genomes do not (Figures 2, 3; Supplementary Table 5).
Interestingly, our data suggest that pkd1b has also undergone relatively rapid evolution in the teleost lineage. This gene appears to be absent in medaka. In addition, stickleback Pkd1b lacks the polycystin-cation-channel domain, suggesting that it is no longer a bona-fide Pkd protein and the polycystin-cation-channel domain is, unusually, not very highly conserved between zebrafish and pufferfish Pkd1b.
It is not clear what the functional significance is of teleosts having fewer pkd genes than mammals. It is possible that PKD proteins have additional functions in mammals compared to teleosts, but as not much is currently known about the functions of PKD1L3, PKD2L2, and PKDREJ it is difficult to test this idea. PKD1L3 is implicated in detection of sour-taste in mammals (Huang et al., 2006; Ishimaru et al., 2006) although this may not be the case in vivo in mouse, as analysis of a mouse mutant in Pkd1l3 detected no defects in taste reception (Nelson et al., 2010; Chen et al., 2011; Hofherr and Kottgen, 2011 and references therein). However, as we find expression of all seven zebrafish pkd genes in potential taste-bud regions, taste detection is unlikely to be an amniote-specific PKD function. PKDREJ expression has only been detected in sperm (Butscheid et al., 2006; Sutton et al., 2008; Hofherr and Kottgen, 2011) and PKD2L2 is expressed most prominently in mouse testis and oocytes (Veldhuisen et al., 1999; Guo et al., 2000; Taft et al., 2002; Chen et al., 2008; Hofherr and Kottgen, 2011), so it is possible that PKD proteins have a function in germ cells in mammals but not teleosts.
Prior to the start of this project, expression data for zebrafish pkd genes was limited to pkd1 (previously called pkd1a), pkd1b and pkd2 (Bisgrove et al., 2005; Obara et al., 2006; Schottenfeld et al., 2007; Mangos et al., 2010; Coxam et al., 2014). In addition, expression of pkd2l1 in spinal cord KA cells was described during this study (Djenoune et al., 2014). In this paper we confirm and extend these earlier expression analyses and we also provide completely novel information on the expression of pkd1l1, pkd1l2a, and pkd1l2b. In general, our results are consistent both with earlier studies in zebrafish and with studies of PKD gene expression in mammals. The main difference is that, for some of the pkd genes, previous reports suggest that they are expressed ubiquitously, whereas we see more specific expression patterns. It is always difficult to distinguish between low-level generalized expression and background staining, so we cannot rule out that there is also low-level ubiquitous expression in these cases.
In most structures we only detect expression of one pkd1-like gene and one pkd2-like gene, suggesting that these genes encode a heteromeric Pkd1/Pkd2 complex in each case. For example, in the dorsal forerunner cells/KV only pkd2 and pkd1l1 are expressed (Figure 6). This is consistent with medaka (Kamura et al., 2011) and mammals, where these two genes are expressed in the equivalent structure, the node (Field et al., 2011; Barratt et al., 2014). This suggests that the expression of these two genes in these related structures, and presumably their function in left/right patterning, is highly conserved between teleosts and mammals. However, the expression of these two genes is not identical in zebrafish. pkd1l1 is expressed before pkd2 and while pkd1l1 is expressed in a condensed group of cells at 12 h, pkd2 expression resolves into a hollow ring. These expression patterns appear to be at least partially complementary, with the pkd2 expression surrounding the pkd1l1 expression (Figures 6D–I). This suggests that the proteins encoded by these genes may only physically interact in a subset of the cells expressing each gene. Interestingly, a recent report suggests that in the mouse node an extracellular domain of PKD1L1 may be required for left-right patterning and that PKD1L1 may be an upstream genetic repressor of PKD2 in this organ (Grimes et al., 2016), which could explain the mainly complementary expression patterns of these two genes in zebrafish.
Similarly, in the developing pronephros we only detect expression of pkd1 and pkd2. This is again, consistent with other vertebrates and with the fact that mutations in these two PKD genes, and only these two PKD genes, cause ADPKD in humans. While pkd2 expression in the zebrafish pronephros was published previously (Bisgrove et al., 2005; Obara et al., 2006; Schottenfeld et al., 2007), this is the first report of pkd1 expression in the developing pronephros. Both pkd1 and pkd2 are expressed in the pronephros by 24 h, although the expression of pkd1 is much stronger than that of pkd2 (Figures 7A,K, 9O,P,A',B'). The expression of pkd1 persists until 3 dpf, although expression of pkd2 is no longer enriched above its otherwise ubiquitous expression by 2 dpf. In contrast, Obara et al. (2006) observed expression of Pkd2 protein in the pronephros at 2 dpf by immunohistochemistry, suggesting that enrichment of Pkd2 protein expression may persist longer than that of the RNA.
In the spinal cord, we confirm the recently reported expression of pkd2l1 in zebrafish KA cells (Figures 7M,N, 8M–R, 9I,K, 11B,C,G–I'). This expression is also conserved in mouse (Huang et al., 2006; Orts-Del'immagine et al., 2012; Petracca et al., 2016). Excitingly, we identify the potential partner of Pkd2l1 in these specialized neurons as Pkd1l2a (Figures 7G,H, 8G–L, 9E,G, 11A,C–F'). This is also consistent with a very recent report in mouse (Petracca et al., 2016). Our data suggests that all KA′ and KA″ cells express both pkd1l2a and pkd2l1 (Figure 11). We also see expression of both of these genes in a very small number of more dorsal and lateral spinal neurons that are likely to be V2b neurons (Figures 8G,I,L–N, 11A,B). This is consistent with the previous report from Djenoune et al. (2014) as they also saw a small number of dorsal cells that did not contact the central canal and might, therefore, be cells other than KAs.
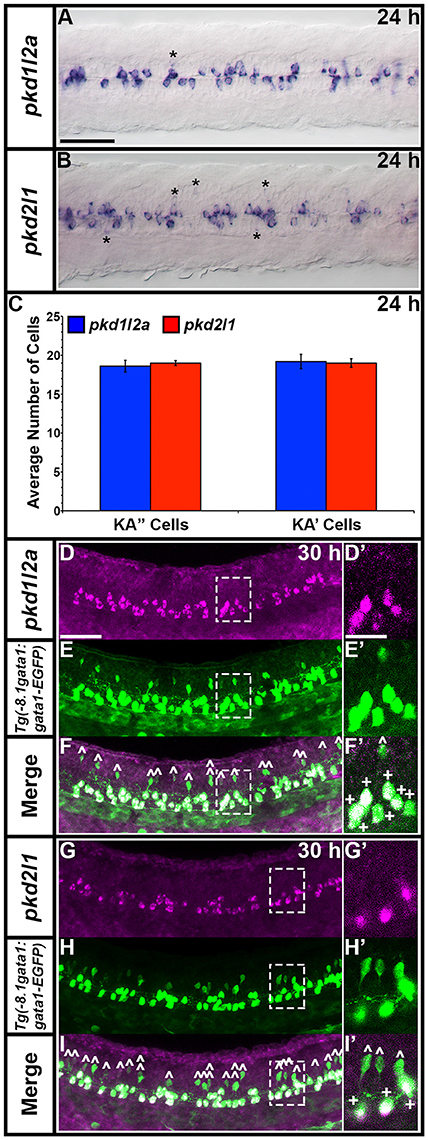
Figure 11. pkd1l2a and pkd2l1 are co-expressed in zebrafish KA cells. Dorsal view of pkd1l2a (A) and pkd2l1 (B) expression in 24 h spinal cord. Rostral left. Most of the labeled cells are KA cells that abut the central canal. Asterisks indicate expression in occasional weak, more lateral cells (that correspond to the more dorsal cells indicated in Figures 8G–R). Prolonged staining sometimes reveals additional weak, lateral cells (data not shown). (C) Average number of cells (y-axis) expressing pkd1l2a (blue) and pkd2l1 (red) in KA″ and KA′ cells (x-axis) at 24 h in WT spinal cord region adjacent to somites 6–10 (n = 5). Error bars indicate standard error of the mean. There is no statistical difference between the number of pkd1l2a and pkd2l1-expressing KA″ (p = 0.6419) and KA′ (p = 0.8571) cells respectively (Student's t-test). These data do not include occasional non-KA lateral cells (2 cells each in 2/5 pkd2l1-labeled embryos; 0 cells in 5 pkd1l2a-labeled embryos). (D–F',G–I') Lateral views of zebrafish spinal cord at 30 h. Anterior left, dorsal top. In situ hybridization (purple) for pkd1l2a (D,D') and pkd2l1 (G,G'), EGFP immunohistochemistry (green) in Tg(–8.1gata1:gata1-EGFP) embryos (E,E',H,H') and merged views (F,F',I,I'). (D'–I') Magnified single confocal plane of white dotted box region. 100% of pkd1l2a and pkd2l1-expressing KA cells co-express Tg(–8.1gata1:gata1-EGFP) and 100% of GFP-positive Tg(–8.1gata1:gata1-EGFP) KA cells co-express either pkd1l2a (F, indicated with + in F') or pkd2l1 (I, indicated with + in I'). No GFP-positive Tg(–8.1gata1:gata1-EGFP) dorsal V2b cells co-express either pkd1l2a (white ˆ in F,F') or pkd2l1 (white ˆ in I,I'). Double-labeled cells are not indicated in (D–I) main panels as they are so numerous. Scale bar (A) = 50 μm (A,B). Scale bar (D) = 50 μm (D–I) and 20 μm (D'–I').
We also observe broad expression of pkd1b in the spinal cord (Figures 8A–F), consistent with an earlier report (Mangos et al., 2010) and weak expression of pkd2 in the floor plate (Figures 9Y,Z,A'). Our mindbomb mutant results (Figure 9) suggest that the broad expression of pkd1b is in progenitor cells and that this gene is also expressed in the floor plate. This suggests that a Pkd2/Pkd1b heterocomplex may have a function in the floor plate. In mouse, Pkd1 is also strongly expressed in the spinal cord (Guillaume et al., 1999), suggesting that there may be a conserved role for Pkd1 proteins in this region of the CNS. Interestingly, mouse PKD1 expression also seems to be enriched in the floor plate region (Guillaume et al., 1999). In contrast, only very low to undetectable spinal cord expression of Pkd2 has been reported in mouse (Guillaume and Trudel, 2000). Given our results in zebrafish, it would be interesting to examine more closely if there is any enrichment of PKD2 in the mouse floor plate.
In contrast to the spinal cord, and also to some previous reports (e.g., Bisgrove et al., 2005; Mangos et al., 2010; Coxam et al., 2014), with the exceptions of pkd2l1 and pkd1l2a expression in KA cells, which extends into the caudal hindbrain (Djenoune et al., 2014; Figures 7G,H,M,N, insets in Figures 12M–O,Y,Z,A') and pkd1b expression in the floor plate in the hindbrain (Figures 7C,D, insets in Figures 12E'–G') we do not see substantial expression of any of the pkd genes in the zebrafish brain at the stages that we examined. Due to the tubular structure of the brain, it is prone to probe trapping, particularly in the folds between the forebrain and midbrain and midbrain/hindbrain boundary, which often causes background expression. While we cannot rule out that some of the other genes are expressed broadly or ubiquitously in the brain, we think that any staining is more likely to be background rather than specific expression.
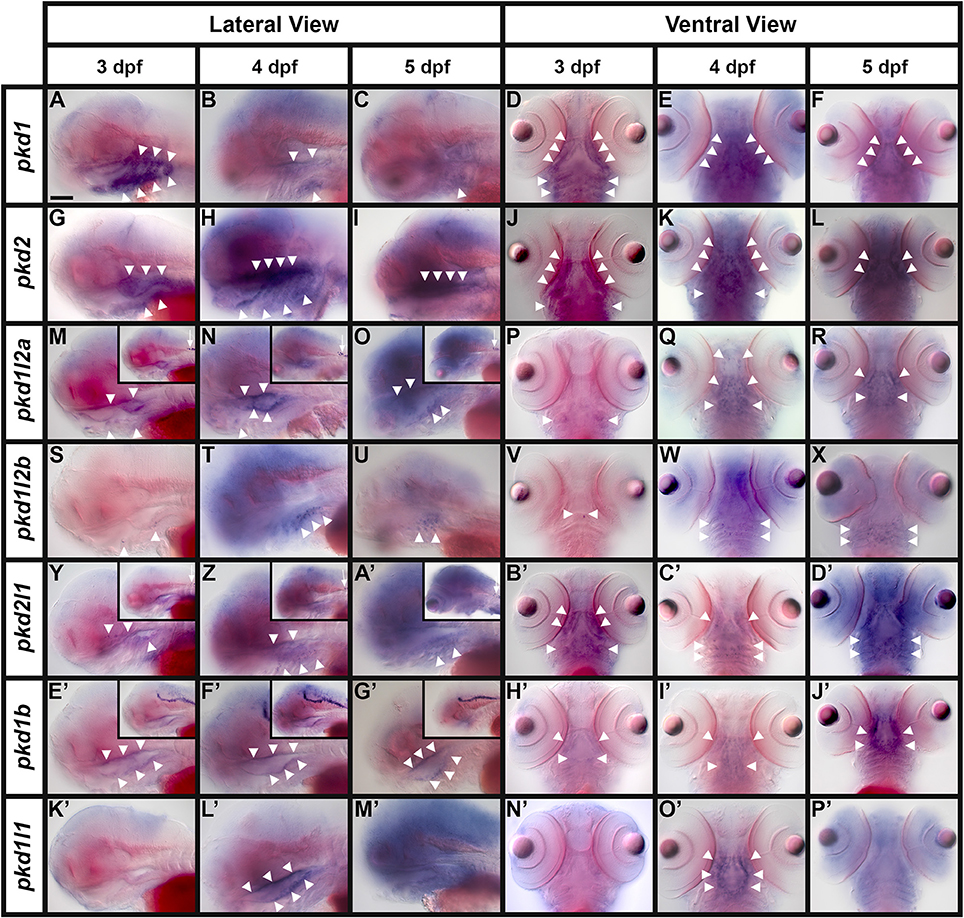
Figure 12. Expression of pkd genes in taste bud regions. Lateral (A–C,G–I,M–O,S–U,Y,Z,A',E'–G',K'–M') and ventral (D–F,J–L,P–R,V–X, B'–D',H'–J',N'–P') views of pkd gene expression at 3, 4, and 5 dpf. Rostral is left Rostral is left (A–C,G–I,M–O,S–U,Y,Z,A',E'–G',K'–M') and top (D–F,J–L,P–R,V–X,B'–D',H'–J',N'–P'). In most of the lateral views, the eyes are out of focus. Insets in (M–O) and (Y–A') show expression of pkd1l2a and pkd2l1 in KA cells in the rostral spinal cord (small white arrows). Insets in (E'–G') show expression of pkd1b in the floor plate of the midbrain and hindbrain. White arrowheads indicate the locations of pharyngeal expression. Scale bar (A) = 100 μm.
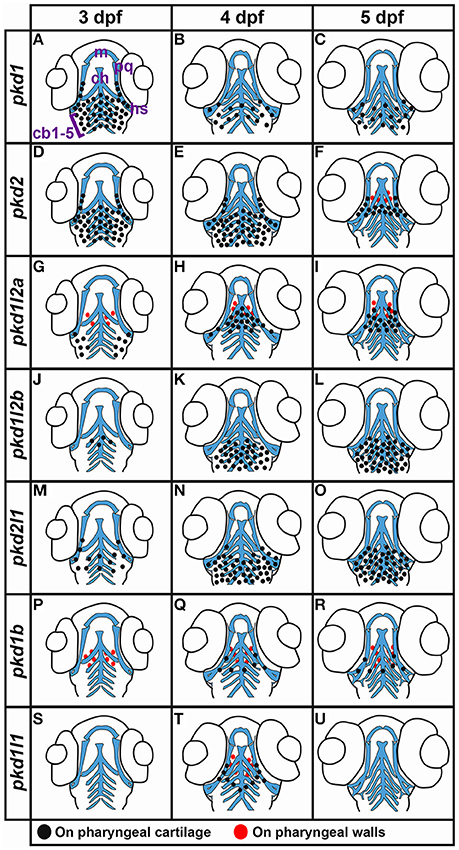
Figure 13. Summary of pkd expression in ventral pharyngeal regions. (A–U) Schematics showing ventral views of zebrafish pharyngeal regions summarizing expression of pkd genes at 3, 4, and 5 dpf. Black dots indicate expression on pharyngeal cartilage and red dots indicate expression on pharyngeal walls. Cartilage schematics are modified from Schilling and Kimmel (1997), Knight et al. (2003), and Edmunds et al. (2016). Locations of the major cartilaginous elements are shown in panel (A). m, Meckel's cartilage (ventral component of lower jaw); ch, ceratohyal cartilage (derived from the second pharyngeal arch); pq, palatoquadrate cartilage (derived from the first pharyngeal arch, forms the dorsal mandibular cartilage); hs, hyosymplectic cartilage (also derived from the second pharyngeal arch); cb1–5, ceratobranchial cartilage 1–5 (forms the ventral branchial or gill arches).
Our experiments also detected expression of pkd1l1 and pkd2l1 in the ear, and of pkd1b, pkd1l1, pkd1l2a, pkd1l2b, and pkd2l1 in the retina, suggesting that these genes may be important for development of these crucial sensory organs. In addition, as mentioned above, we observe dynamic expression of all seven zebrafish pkd genes in regions that may correspond to taste receptors. This is in contrast to mouse, where only Pkd1l3 and Pkd1l2 have been described as being expressed in these cell types (Huang et al., 2006; LopezJimenez et al., 2006). Notably, LopezJimenez and colleagues did not observe any expression of Pkd1, Pkd2, or Pkd1l1 in mouse taste receptors (LopezJimenez et al., 2006). It will be interesting in future studies to confirm whether all of the pkd expression in zebrafish pharyngeal regions corresponds to taste receptors and which pkd genes are co-expressed in particular subsets of these cells. If, indeed, pkd gene expression demarcates different types of receptor cells, these additional pkd expression domains in zebrafish might be consistent with suggestions that teleosts have relatively large numbers of different taste receptors (Okada, 2015).
We also observe expression of pkd1 in neuromasts, although interestingly, we did not find expression of either of the pkd2-like genes in these cells. Given that zebrafish neuromasts are deposited by the migrating lateral line primordium, it is intriguing that the large extracellular domain of Pkd1 has been implicated in both cell migration and adhesion (Streets et al., 2003; Castelli et al., 2015).
In addition, we found expression of pkd2, and only pkd2, in the rostral-ventral somites (Figures 8D', 10P–R), suggesting that this gene is expressed in a subset of muscle cells. Interestingly, in Drosophila, Pkd2 is required for optimal contractility of smooth muscle cells (Gao et al., 2004), suggesting that it may have a similar role in zebrafish muscle. Potentially consistent with this idea, there is evidence that Pkd2 physically interacts with Tropomyosin I (Li et al., 2003a) and skeletal isoforms of Troponin I (Li et al., 2003b).
Finally, we also observe expression of pkd1 in the pectoral fins, as previously reported (Mangos et al., 2010). However, in contrast to this paper, we do not detect expression of pkd1 in the caudal notochord or around the KV. Unfortunately the authors of this paper do not report which region of pkd1 they used to make their RNA in situ hybridization probe, but as the sequence that their ATG start codon morpholino is designed against is not included in our new pkd1 transcript, it is possible that their probe contained sequence that recognized other genes in addition to pkd1.
In conclusion, in this paper we identify and provide a comprehensive description of the zebrafish family of pkd genes and their expression domains during embryogenesis and larval stages. Our data suggest that different pairs of Pkd1-like and Pkd2-like proteins have specific functions during vertebrate development. In particular, that Pkd2 and Pkd1l1 may be required for setting up correct left-right patterning, that Pkd1 and Pkd2 may be required for correct kidney development, that Pkd1l1 and Pkd2l1 may be required for ear development and that Pkd1l2a and Pkd2l1 may be required for correct function of spinal KA cells. We also identify expression of all seven pkd genes in potential taste bud-forming regions and of five pkd genes in the retina. Interestingly, we also find expression of single pkd1 or pkd2 genes in certain structures, such as pkd1 in neuromasts and pectoral fins and pkd2 in somites, suggesting that proteins encoded by these genes may also have functions independent of Pkd1/Pkd2 heterocomplexes. Given the importance of PKD proteins for many different aspects of vertebrate development and physiology, this description of the full complement of zebrafish pkd genes and their expression in different tissues and organs is a fundamental first step to characterizing the functional roles of these biologically vital proteins.
Author Contributions
SE performed most of the experiments and analyses in the paper, including all the bioinformatics analyses, the synteny, and phylogenetic analyses, most of the cloning (including all of the inverse PCR), some of the in situ hybridizations and photography, PC performed and photographed some in situ hybridization experiments, AS also performed some in situ hybridization experiments, SB performed the double-labeling experiments and took some of the photographs, KL and SE designed and directed the study and wrote the paper. All authors read and approved the final manuscript.
Funding
This work was funded by HFSP grant RGP063 and NIH NINDS R21NS073979 to KL.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Ginny Grieb, Jessica Bouchard, and several SU undergraduate fish husbandry workers for help with maintaining zebrafish lines. We would also like to thank Andy Prendergast for help with identifying initial pkd reference sequences, Richard Bates for assistance with an in situ hybridization experiment, Steve Dorus and Jannice Friedman for help with phylogenetic analyses and José L. Juárez-Morales and Jeff Amack for comments on earlier drafts of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcell.2017.00005/full#supplementary-material
References
Abràmoff, M. D., Magalhães, P. J., and Ram, S. J. (2004). Image processing with imageJ. Biophotonics Int. 11, 36–41.
Amores, A., Force, A., Yan, Y. L., Joly, L., Amemiya, C., Fritz, A., et al. (1998). Zebrafish hox clusters and vertebrate genome evolution. Science 282, 1711–1714. doi: 10.1126/science.282.5394.1711
Arif Pavel, M., Lv, C., Ng, C., Yang, L., Kashyap, P., Lam, C., et al. (2016). Function and regulation of TRPP2 ion channel revealed by a gain-of-function mutant. Proc. Natl. Acad. Sci. U.S.A. 113, E2363–E2372. doi: 10.1073/pnas.1517066113
Avagyan, S., and Zon, L. I. (2016). Fish to learn: insights into blood development and blood disorders from zebrafish hematopoiesis. Hum. Gene Ther. 27, 287–294. doi: 10.1089/hum.2016.024
Barr, M. M., and Sternberg, P. W. (1999). A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature 401, 386–389. doi: 10.1038/43913
Barratt, K. S., Glanville-Jones, H. C., and Arkell, R. M. (2014). The Zic2 gene directs the formation and function of node cilia to control cardiac situs. Genesis 52, 626–635. doi: 10.1002/dvg.22767
Batista, M. F., Jacobstein, J., and Lewis, K. E. (2008). Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev. Biol. 322, 263–275. doi: 10.1016/j.ydbio.2008.07.015
Bisgrove, B. W., Snarr, B. S., Emrazian, A., and Yost, H. J. (2005). Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer's vesicle are required for specification of the zebrafish left-right axis. Dev. Biol. 287, 274–288. doi: 10.1016/j.ydbio.2005.08.047
Bohm, U. L., Prendergast, A., Djenoune, L., Nunes Figueiredo, S., Gomez, J., Stokes, C., et al. (2016). CSF-contacting neurons regulate locomotion by relaying mechanical stimuli to spinal circuits. Nat. Commun. 7:10866. doi: 10.1038/ncomms10866
Boulter, C., Mulroy, S., Webb, S., Fleming, S., Brindle, K., and Sandford, R. (2001). Cardiovascular, skeletal, and renal defects in mice with a targeted disruption of the Pkd1 gene. Proc. Natl. Acad. Sci. U.S.A. 98, 12174–12179. doi: 10.1073/pnas.211191098
Bournele, D., and Beis, D. (2016). Zebrafish models of cardiovascular disease. Heart Fail. Rev. 21, 803–813. doi: 10.1007/s10741-016-9579-y
Brown, D. R., Samsa, L. A., Qian, L., and Liu, J. (2016). Advances in the study of heart development and disease using zebrafish. J. Cardiovasc. Dev. Dis. 3:13. doi: 10.3390/jcdd3020013
Butscheid, Y., Chubanov, V., Steger, K., Meyer, D., Dietrich, A., and Gudermann, T. (2006). Polycystic kidney disease and receptor for egg jelly is a plasma membrane protein of mouse sperm head. Mol. Reprod. Dev. 73, 350–360. doi: 10.1002/mrd.20410
Carneiro, M. C., de Castro, I. P., and Ferreira, M. G. (2016). Telomeres in aging and disease: lessons from zebrafish. Dis. Model. Mech. 9, 737–748. doi: 10.1242/dmm.025130
Castelli, M., De Pascalis, C., Distefano, G., Ducano, N., Oldani, A., Lanzetti, L., et al. (2015). Regulation of the microtubular cytoskeleton by Polycystin-1 favors focal adhesions turnover to modulate cell adhesion and migration. BMC Cell Biol. 16:15. doi: 10.1186/s12860-015-0059-3
Chang, M. Y., Lu, J. K., Tian, Y. C., Chen, Y. C., Hung, C. C., Huang, Y. H., et al. (2011). Inhibition of the P2X7 receptor reduces cystogenesis in PKD. J. Am. Soc. Nephrol. 22, 1696–1706. doi: 10.1681/ASN.2010070728
Chen, D., Li, P., Guo, W., Ye, F., Wu, J., Wei, D., et al. (2011). Molecular evolution of candidate sour taste receptor gene PKD1L3 in mammals. Genome 54, 890–897. doi: 10.1139/g11-057
Chen, Y., Zhang, Z., Lv, X. Y., Wang, Y. D., Hu, Z. G., Sun, H., et al. (2008). Expression of Pkd2l2 in testis is implicated in spermatogenesis. Biol. Pharm. Bull. 31, 1496–1500. doi: 10.1248/bpb.31.1496
Concordet, J. P., Lewis, K. E., Moore, J. W., Goodrich, L. V., Johnson, R. L., Scott, M. P., et al. (1996). Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development 122, 2835–2846.
Cornec-Le Gall, E., Audrezet, M. P., Chen, J. M., Hourmant, M., Morin, M. P., Perrichot, R., et al. (2013). Type of PKD1 mutation influences renal outcome in ADPKD. J. Am. Soc. Nephrol. 24, 1006–1013. doi: 10.1681/ASN.2012070650
Coxam, B., Sabine, A., Bower, N. I., Smith, K. A., Pichol-Thievend, C., Skoczylas, R., et al. (2014). Pkd1 regulates lymphatic vascular morphogenesis during development. Cell Rep. 7, 623–633. doi: 10.1016/j.celrep.2014.03.063
Dalgaard, O. Z. (1957). Bilateral polycystic disease of the kidneys; a follow-up of two hundred and eighty-four patients and their families. Acta Med. Scand. Suppl. 328, 1–255.
Damann, N., Voets, T., and Nilius, B. (2008). TRPs in our senses. Curr. Biol. 18, R880–R889. doi: 10.1016/j.cub.2008.07.063
DeCaen, P. G., Liu, X., Abiria, S., and Clapham, D. E. (2016). Atypical calcium regulation of the PKD2-L1 polycystin ion channel. eLife 5:e13413. doi: 10.7554/eLife.13413
Delling, M., DeCaen, P. G., Doerner, J. F., Febvay, S., and Clapham, D. E. (2013). Primary cilia are specialized calcium signalling organelles. Nature 504, 311–314. doi: 10.1038/nature12833
Delling, M., Indzhykulian, A. A., Liu, X., Li, Y., Xie, T., Corey, D. P., et al. (2016). Primary cilia are not calcium-responsive mechanosensors. Nature 531, 656–660. doi: 10.1038/nature17426
Delmas, P. (2004a). The gating of polycystin signaling complex. Biol. Res. 37, 681–691. doi: 10.4067/S0716-97602004000400026
Delmas, P. (2004b). Polycystins: from mechanosensation to gene regulation. Cell 118, 145–148. doi: 10.1016/j.cell.2004.07.007
Delmas, P., Nauli, S. M., Li, X., Coste, B., Osorio, N., Crest, M., et al. (2004). Gating of the polycystin ion channel signaling complex in neurons and kidney cells. FASEB J. 18, 740–742. doi: 10.1096/fj.03-0319fje
Dereeper, A., Audic, S., Claverie, J. M., and Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. doi: 10.1186/1471-2148-10-8
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Djenoune, L., Khabou, H., Joubert, F., Quan, F. B., Nunes Figueiredo, S., Bodineau, L., et al. (2014). Investigation of spinal cerebrospinal fluid-contacting neurons expressing PKD2L1: evidence for a conserved system from fish to primates. Front. Neuroanat. 8:26. doi: 10.3389/fnana.2014.00026
Edmunds, R. C., Su, B., Balhoff, J. P., Eames, B. F., Dahdul, W. M., Lapp, H., et al. (2016). Phenoscape: identifying candidate genes for evolutionary phenotypes. Mol. Biol. Evol. 33, 13–24. doi: 10.1093/molbev/msv223
Essner, J. J., Amack, J. D., Nyholm, M. K., Harris, E. B., and Yost, H. J. (2005). Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132, 1247–1260. doi: 10.1242/dev.01663
Feng, S., Okenka, G. M., Bai, C. X., Streets, A. J., Newby, L. J., DeChant, B. T., et al. (2008). Identification and functional characterization of an N-terminal oligomerization domain for polycystin-2. J. Biol. Chem. 283, 28471–28479. doi: 10.1074/jbc.M803834200
Fidelin, K., and Wyart, C. (2014). Inhibition and motor control in the developing zebrafish spinal cord. Curr. Opin. Neurobiol. 26, 103–109. doi: 10.1016/j.conb.2013.12.016
Field, S., Riley, K. L., Grimes, D. T., Hilton, H., Simon, M., Powles-Glover, N., et al. (2011). Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development 138, 1131–1142. doi: 10.1242/dev.058149
Finn, R. D., Clements, J., Arndt, W., Miller, B. L., Wheeler, T. J., Schreiber, F., et al. (2015). HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38. doi: 10.1093/nar/gkv397
Fogelgren, B., Lin, S. Y., Zuo, X., Jaffe, K. M., Park, K. M., Reichert, R. J., et al. (2011). The exocyst protein Sec10 interacts with Polycystin-2 and knockdown causes PKD-phenotypes. PLoS Genet. 7:e1001361. doi: 10.1371/journal.pgen.1001361
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y.-L., and Postlethwait, J. H. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545.
Francescatto, L., Rothschild, S. C., Myers, A. L., and Tombes, R. M. (2010). The activation of membrane targeted CaMK-II in the zebrafish Kupffer's vesicle is required for left-right asymmetry. Development 137, 2753–2762. doi: 10.1242/dev.049627
Fu, X., Wang, Y., Schetle, N., Gao, H., Putz, M., von Gersdorff, G., et al. (2008). The subcellular localization of TRPP2 modulates its function. J. Am. Soc. Nephrol. 19, 1342–1351. doi: 10.1681/ASN.2007070730
Gao, Z., Joseph, E., Ruden, D. M., and Lu, X. (2004). Drosophila Pkd2 is haploid-insufficient for mediating optimal smooth muscle contractility. J. Biol. Chem. 279, 14225–14231. doi: 10.1074/jbc.M312223200
Giamarchi, A., Feng, S., Rodat-Despoix, L., Xu, Y., Bubenshchikova, E., Newby, L. J., et al. (2010). A polycystin-2 (TRPP2) dimerization domain essential for the function of heteromeric polycystin complexes. EMBO J. 29, 1176–1191. doi: 10.1038/emboj.2010.18
Goetz, J. G., Steed, E., Ferreira, R. R., Roth, S., Ramspacher, C., Boselli, F., et al. (2014). Endothelial cilia mediate low flow sensing during zebrafish vascular development. Cell Rep. 6, 799–808. doi: 10.1016/j.celrep.2014.01.032
Goujon, M., McWilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J., et al. (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids. Res. 38, W695–W699. doi: 10.1093/nar/gkq313
Graham, D. M., Huang, L., Robinson, K. R., and Messerli, M. A. (2013). Epidermal keratinocyte polarity and motility require Ca(2)(+) influx through TRPV1. J. Cell Sci. 126(Pt 20), 4602–4613. doi: 10.1242/jcs.122192
Grantham, J. J. (1993). Polycystic kidney disease: hereditary and acquired. Adv. Intern. Med. 38, 409–420.
Griffin, A., Krasniak, C., and Baraban, S. C. (2016). Advancing epilepsy treatment through personalized genetic zebrafish models. Prog. Brain Res. 226, 195–207. doi: 10.1016/bs.pbr.2016.03.012
Grimes, D. T., Keynton, J. L., Buenavista, M. T., Jin, X., Patel, S. H., Kyosuke, S., et al. (2016). Genetic analysis reveals a hierarchy of interactions between polycystin-encoding genes and genes controlling cilia function during left-right determination. PLoS Genet. 12:e1006070. doi: 10.1371/journal.pgen.1006070
Guillaume, R., D'Agati, V., Daoust, M., and Trudel, M. (1999). Murine Pkd1 is a developmentally regulated gene from morula to adulthood: role in tissue condensation and patterning. Dev. Dyn. 214, 337–348. doi: 10.1002/(SICI)1097-0177(199904)214:4<337::AID-AJA6>3.0.CO;2-O
Guillaume, R., and Trudel, M. (2000). Distinct and common developmental expression patterns of the murine Pkd2 and Pkd1 genes. Mech. Dev. 93, 179–183. doi: 10.1016/S0925-4773(00)00257-4
Guo, L., Schreiber, T. H., Weremowicz, S., Morton, C. C., Lee, C., and Zhou, J. (2000). Identification and characterization of a novel polycystin family member, polycystin-L2, in mouse and human: sequence, expression, alternative splicing, and chromosomal localization. Genomics 64, 241–251. doi: 10.1006/geno.2000.6131
Hanaoka, K., Qian, F., Boletta, A., Bhunia, A. K., Piontek, K., Tsiokas, L., et al. (2000). Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature 408, 990–994. doi: 10.1038/35050128
Hansen, A., Reutter, K., and Zeiske, E. (2002). Taste bud development in the zebrafish, Danio rerio. Dev. Dyn. 223, 483–496. doi: 10.1002/dvdy.10074
Harris, P. C., and Torres, V. E. (2009). Polycystic kidney disease. Annu. Rev. Med. 60, 321–337. doi: 10.1146/annurev.med.60.101707.125712
Harrison, N. R., Laroche, F. J., Gutierrez, A., and Feng, H. (2016). Zebrafish models of human leukemia: technological advances and mechanistic insights. Adv. Exp. Med. Biol. 916, 335–369. doi: 10.1007/978-3-319-30654-4_15
Hofherr, A., and Kottgen, M. (2011). TRPP channels and polycystins. Adv. Exp. Med. Biol. 704, 287–313. doi: 10.1007/978-94-007-0265-3_16
Hostetter, C. L., Sullivan-Brown, J. L., and Burdine, R. D. (2003). Zebrafish pronephros: a model for understanding cystic kidney disease. Dev. Dyn. 228, 514–522. doi: 10.1002/dvdy.10371
Huang, A. L., Chen, X., Hoon, M. A., Chandrashekar, J., Guo, W., Trankner, D., et al. (2006). The cells and logic for mammalian sour taste detection. Nature 442, 934–938. doi: 10.1038/nature05084
Huang, L., Xiao, A., Wecker, A., McBride, D. A., Choi, S. Y., Zhou, W., et al. (2014). A possible zebrafish model of polycystic kidney disease: knockdown of wnt5a causes cysts in zebrafish kidneys. J. Vis. Exp. 94:e52156. doi: 10.3791/52156
Hurd, T., Zhou, W., Jenkins, P., Liu, C. J., Swaroop, A., Khanna, H., et al. (2010). The retinitis pigmentosa protein RP2 interacts with polycystin 2 and regulates cilia-mediated vertebrate development. Hum. Mol. Genet. 19, 4330–4344. doi: 10.1093/hmg/ddq355
Iglesias, C. G., Torres, V. E., Offord, K. P., Holley, K. E., Beard, C. M., and Kurland, L. T. (1983). Epidemiology of adult polycystic kidney disease, Olmsted County, Minnesota: 1935-1980. Am. J. Kidney Dis. 2, 630–639. doi: 10.1016/S0272-6386(83)80044-4
Ishimaru, Y., Inada, H., Kubota, M., Zhuang, H., Tominaga, M., and Matsunami, H. (2006). Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 12569–12574. doi: 10.1073/pnas.0602702103
Itoh, M., Kim, C., Palardy, G., Oda, T., Jiang, Y., Maust, D., et al. (2003). Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell. 4, 67–82. doi: 10.1016/S1534-5807(02)00409-4
Jiang, Y. J., Brand, M., Heisenberg, C. P., Beuchle, D., Furutani-Seiki, M., Kelsh, R. N., et al. (1996). Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development 123, 205–216.
Kamura, K., Kobayashi, D., Uehara, Y., Koshida, S., Iijima, N., Kudo, A., et al. (2011). Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Development 138, 1121–1129. doi: 10.1242/dev.058271
Kapsimali, M., Kaushik, A. L., Gibon, G., Dirian, L., Ernest, S., and Rosa, F. M. (2011). Fgf signaling controls pharyngeal taste bud formation through miR-200 and Delta-Notch activity. Development 138, 3473–3484. doi: 10.1242/dev.058669
Kim, K., Drummond, I., Ibraghimov-Beskrovnaya, O., Klinger, K., and Arnaout, M. A. (2000). Polycystin 1 is required for the structural integrity of blood vessels. Proc. Natl. Acad. Sci. U.S.A. 97, 1731–1736. doi: 10.1073/pnas.040550097
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Knight, R. D., Nair, S., Nelson, S. S., Afshar, A., Javidan, Y., Geisler, R., et al. (2003). lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development 130, 5755–5768. doi: 10.1242/dev.00575
Kobayashi, M., Nishikawa, K., and Yamamoto, M. (2001). Hematopoietic regulatory domain of gata1 gene is positively regulated by GATA1 protein in zebrafish embryos. Development 128, 2341–2350.
Koressaar, T., and Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291. doi: 10.1093/bioinformatics/btm091
Kozol, R. A., Abrams, A. J., James, D. M., Buglo, E., Yan, Q., and Dallman, J. E. (2016). Function over form: modeling groups of inherited neurological conditions in zebrafish. Front. Mol. Neurosci. 9:55. doi: 10.3389/fnmol.2016.00055
Kramer-Zucker, A. G., Olale, F., Haycraft, C. J., Yoder, B. K., Schier, A. F., and Drummond, I. A. (2005). Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development 132, 1907–1921. doi: 10.1242/dev.01772
Levy, M., and Feingold, J. (2000). Estimating prevalence in single-gene kidney diseases progressing to renal failure. Kidney Int. 58, 925–943. doi: 10.1046/j.1523-1755.2000.00250.x
Lewis, K. E., Concordet, J. P., and Ingham, P. W. (1999). Characterisation of a second patched gene in the zebrafish Danio rerio and the differential response of patched genes to Hedgehog signalling. Dev. Biol. 208, 14–29. doi: 10.1006/dbio.1998.9169
Li, A., Tian, X., Sung, S. W., and Somlo, S. (2003). Identification of two novel polycystic kidney disease-1-like genes in human and mouse genomes. Genomics 81, 596–608. doi: 10.1016/S0888-7543(03)00048-X
Li, Q., Dai, Y., Guo, L., Liu, Y., Hao, C., Wu, G., et al. (2003a). Polycystin-2 associates with tropomyosin-1, an actin microfilament component. J. Mol. Biol. 325, 949–962. doi: 10.1016/S0022-2836(02)01333-5
Li, Q., Shen, P. Y., Wu, G., and Chen, X. Z. (2003b). Polycystin-2 interacts with troponin I, an angiogenesis inhibitor. Biochemistry 42, 450–457. doi: 10.1021/bi0267792
LopezJimenez, N. D., Cavenagh, M. M., Sainz, E., Cruz-Ithier, M. A., Battey, J. F., and Sullivan, S. L. (2006). Two members of the TRPP family of ion channels, Pkd1l3 and Pkd2l1, are co-expressed in a subset of taste receptor cells. J. Neurochem. 98, 68–77. doi: 10.1111/j.1471-4159.2006.03842.x
Lu, W., Peissel, B., Babakhanlou, H., Pavlova, A., Geng, L., Fan, X., et al. (1997). Perinatal lethality with kidney and pancreas defects in mice with a targetted Pkd1 mutation. Nat. Genet. 17, 179–181. doi: 10.1038/ng1097-179
Lu, W., Shen, X., Pavlova, A., Lakkis, M., Ward, C. J., Pritchard, L., et al. (2001). Comparison of Pkd1-targeted mutants reveals that loss of polycystin-1 causes cystogenesis and bone defects. Hum. Mol. Genet. 10, 2385–2396. doi: 10.1093/hmg/10.21.2385
Mackenzie, F. E., Romero, R., Williams, D., Gillingwater, T., Hilton, H., Dick, J., et al. (2009). Upregulation of PKD1L2 provokes a complex neuromuscular disease in the mouse. Hum. Mol. Genet. 18, 3553–3566. doi: 10.1093/hmg/ddp304
Mangos, S., Lam, P. Y., Zhao, A., Liu, Y., Mudumana, S., Vasilyev, A., et al. (2010). The ADPKD genes pkd1a/b and pkd2 regulate extracellular matrix formation. Dis. Model. Mech. 3, 354–365. doi: 10.1242/dmm.003194
McGrath, J., Somlo, S., Makova, S., Tian, X., and Brueckner, M. (2003). Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 114, 61–73. doi: 10.1016/S0092-8674(03)00511-7
McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., et al. (2013). Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600. doi: 10.1093/nar/gkt376
Merrick, D., Chapin, H., Baggs, J. E., Yu, Z., Somlo, S., Sun, Z., et al. (2012). The gamma-secretase cleavage product of polycystin-1 regulates TCF and CHOP-mediated transcriptional activation through a p300-dependent mechanism. Dev. Cell 22, 197–210. doi: 10.1016/j.devcel.2011.10.028
Montell, C. (2005). The TRP superfamily of cation channels. Sci. STKE 2005:re3. doi: 10.1126/stke.2722005re3
Murakami, M., Ohba, T., Xu, F., Shida, S., Satoh, E., Ono, K., et al. (2005). Genomic organization and functional analysis of murine PKD2L1. J. Biol. Chem. 280, 5626–5635. doi: 10.1074/jbc.M411496200
Myllymaki, H., Bauerlein, C. A., and Ramet, M. (2016). The zebrafish breathes new life into the study of tuberculosis. Front. Immunol. 7:196. doi: 10.3389/fimmu.2016.00196
Nauli, S. M., Alenghat, F. J., Luo, Y., Williams, E., Vassilev, P., Li, X., et al. (2003). Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137. doi: 10.1038/ng1076
Nelson, T. M., Lopezjimenez, N. D., Tessarollo, L., Inoue, M., Bachmanov, A. A., and Sullivan, S. L. (2010). Taste function in mice with a targeted mutation of the pkd1l3 gene. Chem. Senses 35, 565–577. doi: 10.1093/chemse/bjq070
Nilius, B., and Owsianik, G. (2011). The transient receptor potential family of ion channels. Genome Biol. 12:218. doi: 10.1186/gb-2011-12-3-218
Obara, T., Mangos, S., Liu, Y., Zhao, J., Wiessner, S., Kramer-Zucker, A. G., et al. (2006). Polycystin-2 immunolocalization and function in zebrafish. J. Am. Soc. Nephrol. 17, 2706–2718. doi: 10.1681/ASN.2006040412
Okada, S. (2015). The taste system of small fish species. Biosci. Biotechnol. Biochem. 79, 1039–1043. doi: 10.1080/09168451.2015.1023251
Orts-Del'immagine, A., Wanaverbecq, N., Tardivel, C., Tillement, V., Dallaporta, M., and Trouslard, J. (2012). Properties of subependymal cerebrospinal fluid contacting neurones in the dorsal vagal complex of the mouse brainstem. J. Physiol. 590, 3719–3741. doi: 10.1113/jphysiol.2012.227959
Owsianik, G., D'Hoedt, D., Voets, T., and Nilius, B. (2006). Structure-function relationship of the TRP channel superfamily. Rev. Physiol. Biochem. Pharmacol. 156, 61–90. doi: 10.1007/s10254-005-0006-0
Paavola, J., Schliffke, S., Rossetti, S., Kuo, I. Y., Yuan, S., Sun, Z., et al. (2013). Polycystin-2 mutations lead to impaired calcium cycling in the heart and predispose to dilated cardiomyopathy. J. Mol. Cell. Cardiol. 58, 199–208. doi: 10.1016/j.yjmcc.2013.01.015
Palmer, C. P., Aydar, E., and Djamgoz, M. B. (2005). A microbial TRP-like polycystic-kidney-disease-related ion channel gene. Biochem. J. 387(Pt 1), 211–219. doi: 10.1042/BJ20041710
Park, H. C., and Appel, B. (2003). Delta-notch signaling regulates oligodendrocyte specification. Development 130, 3747–3755. doi: 10.1242/dev.00576
Paul, B. M., Consugar, M. B., Ryan Lee, M., Sundsbak, J. L., Heyer, C. M., Rossetti, S., et al. (2014). Evidence of a third ADPKD locus is not supported by re-analysis of designated PKD3 families. Kidney Int. 85, 383–392. doi: 10.1038/ki.2013.227
Pennekamp, P., Karcher, C., Fischer, A., Schweickert, A., Skryabin, B., Horst, J., et al. (2002). The ion channel polycystin-2 is required for left-right axis determination in mice. Curr. Biol. 12, 938–943. doi: 10.1016/S0960-9822(02)00869-2
Petracca, Y. L., Sartoretti, M. M., Di Bella, D. J., Marin-Burgin, A., Carcagno, A. L., Schinder, A. F., et al. (2016). The late and dual origin of cerebrospinal fluid-contacting neurons in the mouse spinal cord. Development 143, 880–891. doi: 10.1242/dev.129254
Postlethwait, J. H. (2007). The zebrafish genome in context: ohnologs gone missing. J. Exp. Zool. B Mol. Dev. Evol. 308, 563–577. doi: 10.1002/jez.b.21137
Postlethwait, J., Yan, Y., Gates, M., Horne, S., Amores, A., Brownlie, A., et al. (1998). Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 18, 345–349. doi: 10.1038/ng0498-345
Poureetezadi, S. J., and Wingert, R. A. (2016). Little fish, big catch: zebrafish as a model for kidney disease. Kidney Int. 89, 1204–1210. doi: 10.1016/j.kint.2016.01.031
Qian, F., Germino, F. J., Cai, Y., Zhang, X., Somlo, S., and Germino, G. G. (1997). PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat. Genet. 16, 179–183. doi: 10.1038/ng0697-179
Quan, F. B., Dubessy, C., Galant, S., Kenigfest, N. B., Djenoune, L., Leprince, J., et al. (2015). Comparative distribution and in vitro activities of the urotensin II-related peptides URP1 and URP2 in zebrafish: evidence for their colocalization in spinal cerebrospinal fluid-contacting neurons. PLoS ONE 10:e0119290. doi: 10.1371/journal.pone.0119290
Ramsey, I. S., Delling, M., and Clapham, D. E. (2006). An introduction to TRP channels. Annu. Rev. Physiol. 68, 619–647. doi: 10.1146/annurev.physiol.68.040204.100431
Reeders, S. T., Breuning, M. H., Davies, K. E., Nicholls, R. D., Jarman, A. P., Higgs, D. R., et al. (1985). A highly polymorphic DNA marker linked to adult polycystic kidney disease on chromosome 16. Nature 317, 542–544. doi: 10.1038/317542a0
Roxo-Rosa, M., Jacinto, R., Sampaio, P., and Lopes, S. S. (2015). The zebrafish Kupffer's vesicle as a model system for the molecular mechanisms by which the lack of Polycystin-2 leads to stimulation of CFTR. Biol. Open 4, 1356–1366. doi: 10.1242/bio.014076
Sampaio, P., Ferreira, R. R., Guerrero, A., Pintado, P., Tavares, B., Amaro, J., et al. (2014). Left-right organizer flow dynamics: how much cilia activity reliably yields laterality? Dev. Cell 29, 716–728. doi: 10.1016/j.devcel.2014.04.030
Schier, A. F., Neuhauss, S. C., Harvey, M., Malicki, J., Solnica, K. L., Stainier, D. Y., et al. (1996). Mutations affecting the development of the embryonic zebrafish brain. Development 123, 165–178.
Schilling, T., and Kimmel, C. (1997). Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development 124, 2945–2960.
Schottenfeld, J., Sullivan-Brown, J., and Burdine, R. D. (2007). Zebrafish curly up encodes a Pkd2 ortholog that restricts left-side-specific expression of southpaw. Development 134, 1605–1615. doi: 10.1242/dev.02827
Semmo, M., Kottgen, M., and Hofherr, A. (2014). The TRPP subfamily and polycystin-1 proteins. Handb. Exp. Pharmacol. 222, 675–711. doi: 10.1007/978-3-642-54215-2_27
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Smith, D. J., Montenegro-Johnson, T. D., and Lopes, S. S. (2014). Organized chaos in Kupffer's vesicle: how a heterogeneous structure achieves consistent left-right patterning. Bioarchitecture 4, 119–125. doi: 10.4161/19490992.2014.956593
Song, Z., Zhang, X., Jia, S., Yelick, P. C., and Zhao, C. (2016). Zebrafish as a model for human ciliopathies. J. Genet. Genomics 43, 107–120. doi: 10.1016/j.jgg.2016.02.001
Streets, A. J., Moon, D. J., Kane, M. E., Obara, T., and Ong, A. C. (2006). Identification of an N-terminal glycogen synthase kinase 3 phosphorylation site which regulates the functional localization of polycystin-2 in vivo and in vitro. Hum. Mol. Genet. 15, 1465–1473. doi: 10.1093/hmg/ddl070
Streets, A. J., Newby, L. J., O'Hare, M. J., Bukanov, N. O., Ibraghimov-Beskrovnaya, O., and Ong, A. C. (2003). Functional analysis of PKD1 transgenic lines reveals a direct role for polycystin-1 in mediating cell-cell adhesion. J. Am. Soc. Nephrol. 14, 1804–1815. doi: 10.1097/01.ASN.0000076075.49819.9B
Sun, Z., Amsterdam, A., Pazour, G. J., Cole, D. G., Miller, M. S., and Hopkins, N. (2004). A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131, 4085–4093. doi: 10.1242/dev.01240
Sutters, M., and Germino, G. G. (2003). Autosomal dominant polycystic kidney disease: molecular genetics and pathophysiology. J. Lab. Clin. Med. 141, 91–101. doi: 10.1067/mlc.2003.13
Sutton, K. A., Jungnickel, M. K., and Florman, H. M. (2008). A polycystin-1 controls postcopulatory reproductive selection in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 8661–8666. doi: 10.1073/pnas.0800603105
Taft, R. A., Denegre, J. M., Pendola, F. L., and Eppig, J. J. (2002). Identification of genes encoding mouse oocyte secretory and transmembrane proteins by a signal sequence trap. Biol. Reprod. 67, 953–960. doi: 10.1095/biolreprod.102.005546
Tietz Bogert, P. S., Huang, B. Q., Gradilone, S. A., Masyuk, T. V., Moulder, G. L., Ekker, S. C., et al. (2013). The zebrafish as a model to study polycystic liver disease. Zebrafish 10, 211–217. doi: 10.1089/zeb.2012.0825
Tsiokas, L., Kim, E., Arnould, T., Sukhatme, V. P., and Walz, G. (1997). Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc. Natl. Acad. Sci. U.S.A. 94, 6965–6970. doi: 10.1073/pnas.94.13.6965
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Veldhuisen, B., Spruit, L., Dauwerse, H. G., Breuning, M. H., and Peters, D. J. (1999). Genes homologous to the autosomal dominant polycystic kidney disease genes (PKD1 and PKD2). Eur. J. Hum. Genet. 7, 860–872. doi: 10.1038/sj.ejhg.5200383
Venkatachalam, K., Luo, J., and Montell, C. (2014). Evolutionarily conserved, multitasking TRP channels: lessons from worms and flies. Handb. Exp. Pharmacol. 223, 937–962. doi: 10.1007/978-3-319-05161-1_9
Venkatachalam, K., and Montell, C. (2007). TRP channels. Annu. Rev. Biochem. 76, 387–417. doi: 10.1146/annurev.biochem.75.103004.142819
Wager, K., Zdebik, A. A., Fu, S., Cooper, J. D., Harvey, R. J., and Russell, C. (2016). Neurodegeneration and Epilepsy in a Zebrafish Model of CLN3 Disease (Batten Disease). PLoS ONE 11:e0157365. doi: 10.1371/journal.pone.0157365
Wojciechowska, S., van Rooijen, E., Ceol, C., Patton, E. E., and White, R. M. (2016). Generation and analysis of zebrafish melanoma models. Methods Cell Biol. 134, 531–549. doi: 10.1016/bs.mcb.2016.03.008
Wu, G., Markowitz, G. S., Li, L., D'Agati, V. D., Factor, S. M., Geng, L., et al. (2000). Cardiac defects and renal failure in mice with targeted mutations in Pkd2. Nat. Genet. 24, 75–78. doi: 10.1038/71724
Wu, G., and Somlo, S. (2000). Molecular genetics and mechanism of autosomal dominant polycystic kidney disease. Mol. Genet. Metab. 69, 1–15. doi: 10.1006/mgme.1999.2943
Yoshiba, S., Shiratori, H., Kuo, I. Y., Kawasumi, A., Shinohara, K., Nonaka, S., et al. (2012). Cilia at the node of mouse embryos sense fluid flow for left-right determination via Pkd2. Science 338, 226–231. doi: 10.1126/science.1222538
Yu, Y., Ulbrich, M. H., Li, M. H., Buraei, Z., Chen, X. Z., Ong, A. C., et al. (2009). Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc. Natl. Acad. Sci. U.S.A. 106, 11558–11563. doi: 10.1073/pnas.0903684106
Yuan, S., Zhao, L., Brueckner, M., and Sun, Z. (2015). Intraciliary calcium oscillations initiate vertebrate left-right asymmetry. Curr. Biol. 25, 556–567. doi: 10.1016/j.cub.2014.12.051
Zhou, J. (2009). Polycystins and primary cilia: primers for cell cycle progression. Annu. Rev. Physiol. 71, 83–113. doi: 10.1146/annurev.physiol.70.113006.100621
Zhu, J., Yu, Y., Ulbrich, M. H., Li, M. H., Isacoff, E. Y., Honig, B., et al. (2011). Structural model of the TRPP2/PKD1 C-terminal coiled-coil complex produced by a combined computational and experimental approach. Proc. Natl. Acad. Sci. U.S.A. 108, 10133–10138. doi: 10.1073/pnas.1017669108
Keywords: PKD, polycystin, TRP proteins, spinal cord, taste buds, node, dorsal forerunner cells, kidney
Citation: England SJ, Campbell PC, Banerjee S, Swanson AJ and Lewis KE (2017) Identification and Expression Analysis of the Complete Family of Zebrafish pkd Genes. Front. Cell Dev. Biol. 5:5. doi: 10.3389/fcell.2017.00005
Received: 19 November 2016; Accepted: 19 January 2017;
Published: 21 February 2017.
Edited by:
David Ellard Keith Ferrier, University of St Andrews, UKReviewed by:
Paolo Sordino, Stazione Zoologica Anton Dohrn, ItalyTokiharu Takahashi, The University of Manchester, UK
Copyright © 2017 England, Campbell, Banerjee, Swanson and Lewis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katharine E. Lewis, a2VsZXdpMDJAc3lyLmVkdQ==
 Samantha J. England
Samantha J. England Paul C. Campbell
Paul C. Campbell Santanu Banerjee
Santanu Banerjee Annika J. Swanson
Annika J. Swanson Katharine E. Lewis
Katharine E. Lewis