
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 26 November 2015
Sec. Membrane Traffic and Organelle Dynamics
Volume 3 - 2015 | https://doi.org/10.3389/fcell.2015.00074
This article is part of the Research Topic GTPases regulate mitotic cell logistics View all 7 articles
Over the last two decades, the small GTPase Ran has emerged as a central regulator of both mitosis and meiosis, particularly in the generation, maintenance, and regulation of the microtubule (MT)-based bipolar spindle. Ran-regulated pathways in mitosis bear many similarities to the well-characterized functions of Ran in nuclear transport and, as with transport, the majority of these mitotic effects are mediated through affecting the physical interaction between karyopherins and Spindle Assembly Factors (SAFs)—a loose term describing proteins or protein complexes involved in spindle assembly through promoting nucleation, stabilization, and/or depolymerization of MTs, through anchoring MTs to specific structures such as centrosomes, chromatin or kinetochores, or through sliding MTs along each other to generate the force required to achieve bipolarity. As such, the Ran-mediated pathway represents a crucial functional module within the wider spindle assembly landscape. Research into mitosis using the model organism Drosophila melanogaster has contributed substantially to our understanding of centrosome and spindle function. However, in comparison to mammalian systems, very little is known about the contribution of Ran-mediated pathways in Drosophila mitosis. This article sets out to summarize our understanding of the roles of the Ran pathway components in Drosophila mitosis, focusing on the syncytial blastoderm embryo, arguing that it can provide important insights into the conserved functions on Ran during spindle formation.
With a fast generation time of 9 days at 25°C, relative ease of genetic manipulation, and a fully sequenced genome (Adams et al., 2000), Drosophila is a powerful model organism for studying basic biological processes such as mitosis. Drosophila tissues are an easily obtainable source material with which to investigate different types of cell division, including asymmetric cell division of neuroblasts, polarized mitosis of the ovarian epithelium, meiosis in the testes and oocytes, and syncytial mitosis in early embryos. In addition, Drosophila cell lines are available for in vitro cell culture techniques. Although Drosophila differs from vertebrate mitosis in that it undergoes semi-open mitosis (Figure 1A), where the nuclear envelope only fully breaks down at the spindle poles and where nuclear pores only fully dissociate at metaphase (Fuchs et al., 1983; Stafstrom and Staehelin, 1984), the overarching pathways involved in spindle formation are very similar.
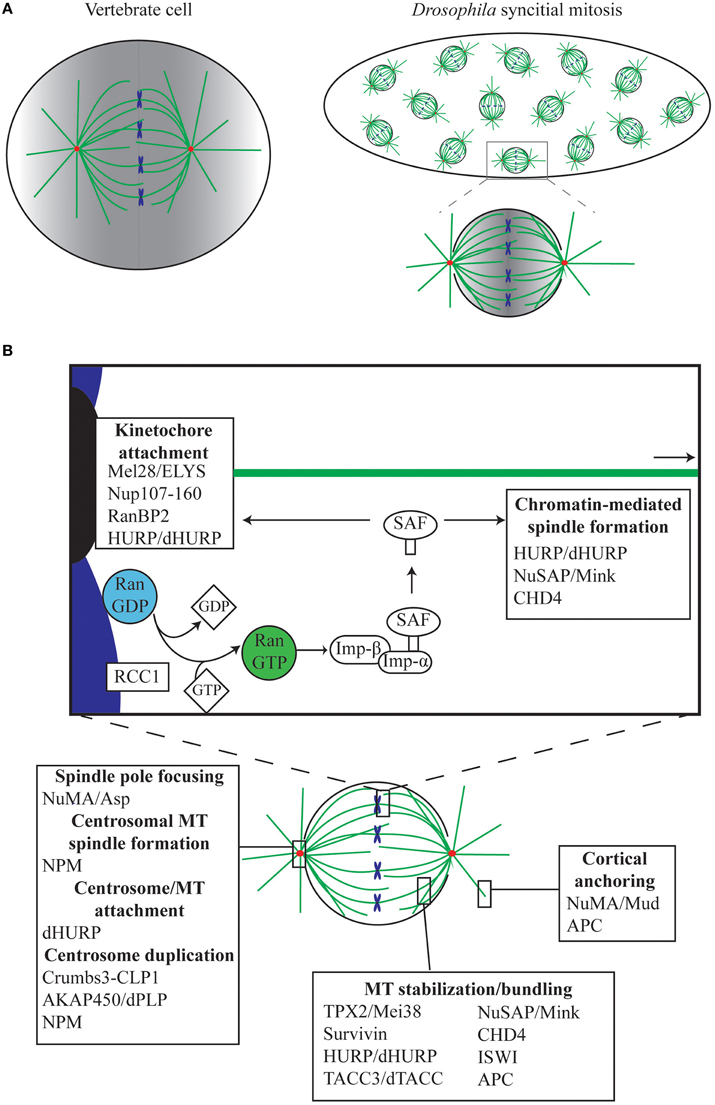
Figure 1. The role of Ran in mitosis. (A) The Drosophila syncytial embryo as a tool for understanding mitosis. In the Drosophila early embryo, the first 13 rounds of mitosis occur rapidly and take place in a shared cytoplasm. Unlike vertebrate cells, which undergo open mitosis and disassemble the nuclear envelope during mitosis, Drosophila undergoes semi-open mitosis, only disassembling the nuclear envelope at the spindle poles. Red, centrosomes; green, MTs; blue, chromosomes. In both vertebrates and Drosophila, Ran.GTP is generated in the vicinity of the chromatin, resulting in a gradient (shown in gray), which is strongest around the chromosomes and weakest at the poles and cortex. (B) Ran mediates mitotic functions via release of Spindle Assembly Factors (SAFs). During mitosis, Ran.GTP is generated around the chromosomes by the chromatin-bound RanGEF RCC1, facilitating the release of SAFs, which are otherwise sequestered by Importins (Imp-β/Imp-α). SAFs have critical roles in, amongst other things, MT anchoring to the kinetochores and centrosomes, in spindle growth from the chromatin, in MT bundling and stabilization, and in anchoring of astral MTs to the cell cortex.
Of the various tissues available, the Drosophila early embryo exhibits some additional advantages in teasing out fundamental concepts of spindle assembly. Following fertilization, the embryo undergoes 13 rounds of synchronous mitoses within a common cytoplasm (Figure 1A), before proceeding to cellularization. In contrast to the much longer time required for a full cell cycle in most animal cells, these syncytial cycles traverse through sequential S and M phases without intervening growth phases, with each round completed within 10–25 min (Foe and Alberts, 1983). During mitotic cycles 10–13 the nuclei are positioned close to the cortex of the embryo, and are therefore easy to image using confocal microscopy. Importantly, mitosis in the syncytium depends largely on maternally-supplied proteins, with the majority of transcription and translation silenced until cycle 14 (Foe and Alberts, 1983; Sullivan and Theurkauf, 1995; De Renzis et al., 2007; Lécuyer et al., 2007; Benoit et al., 2009). This, in conjunction with the shared cytoplasm and the large size of the embryo in relation to normal somatic cells (~150 μm long), means that the effects on spindle assembly of disrupting single proteins or protein complexes can be easily observed via microinjection-based methods (Brust-Mascher and Scholey, 2009; Conduit et al., 2015). Furthermore, embryonic mitoses appear to be much more tolerant to subtle perturbation than mammalian somatic cells, as syncytial nuclei continue to cycle, attempting to form spindles and segregate chromosomes, even in the presence of centrosome, spindle, or chromosome abnormalities (Hayward et al., 2014). However, although the syncytial embryo and Drosophila in general has contributed substantially to our understanding of canonical centrosome-driven spindle formation, very little is actually known in the fly about chromatin-driven spindle self-organization in which Ran plays such a central role. This review will outline the current understanding of the Ran pathway in Drosophila, and how it relates to the Ran pathway in other metazoans.
Ran was first discovered as a substrate for the RCC1 Guanine nucleotide Exchange Factor (GEF; Bischoff and Ponstingl, 1991), and has been predominantly characterized as a central player in nuclear transport, shuttling proteins and mRNA into and out of the nucleus. It is a ~25 kDa guanosine triphosphatase (GTPase) related to the Ras superfamily of GTPases, and can exist in the active guanosine triphosphate-bound (GTP) state or the inactive guanosine diphosphate-bound (GDP) state. Although Ran has inherent GTPase activity, Ran binding proteins (RanBPs) and Ran GTPase activating protein (RanGAP) are essential for effective GTP hydrolysis to take place in a cellular context (Bischoff et al., 1994; Bischoff and Görlich, 1997). Nuclear transport has been well-characterized (Figure 2) and excellent reviews already exist (Stewart, 2007; Cautain et al., 2015). While these functions are outside the main scope of this review, it is important to consider the similarities between the role of Ran in nuclear transport and in mitosis.
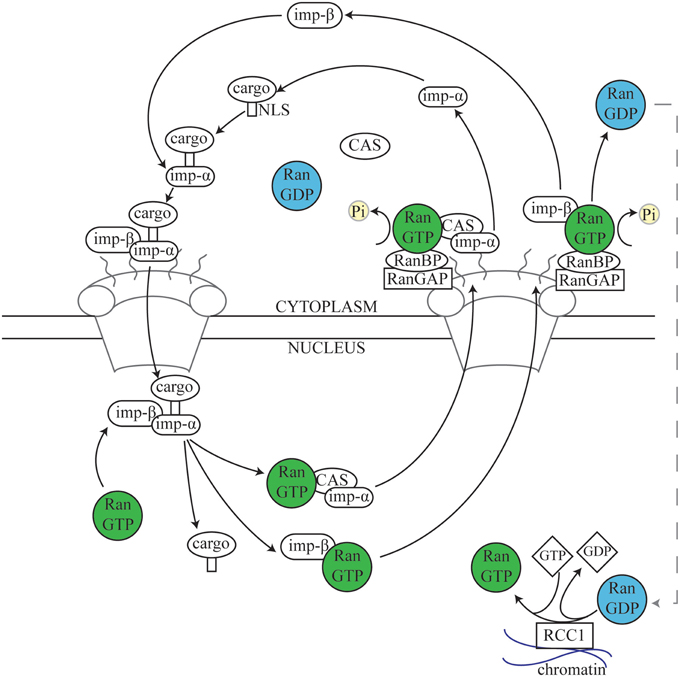
Figure 2. The nuclear transport cycle. During nuclear import, Importins in the cytoplasm recognize and bind to the Nuclear Localization Signal (NLS) of a target protein. The complex of cargo plus the Importin α/β dimer docks at the cytoplasmic side of the nuclear pore and is transported into the nucleus. Subsequently, Ran.GTP in the nucleus binds to Importin β, resulting in disassembly of the complex and releasing the cargo into the nuclear space. Importin α is then recycled back to the cytosol by the Exportin Cellular Apoptosis Susceptibility (Cas) protein via normal Ran-dependent export pathways (Kutay et al., 1997; Tekotte et al., 2002), while the Importin β/Ran.GTP complex is transported separately (Kose et al., 1999). Upon reaching the cytosolic side of the nuclear envelope, Ran.GTPase Activating Protein 1 (RanGAP1) and Ran Binding Proteins (RanBPs) stimulate Ran-dependent GTP hydrolysis, causing release of Importin β from Ran (Kutay et al., 1997; Lounsbury and Macara, 1997; Seewald et al., 2003). RanGAP1 is unable to directly affect Ran.GTP complexed with either importins or exportins, and instead acts via one of several Ran binding proteins (RanBPs) (Bischoff and Görlich, 1997; Floer et al., 1997; Kutay et al., 1997; Lounsbury and Macara, 1997). Nuclear export follows a similar process; Exportins such as chromosome region maintenance 1 (Crm1) recognize and bind to Nuclear Export Signals (NESs) on target proteins. Exportin, Ran.GTP, and cargo form a complex which passes out of the nucleus through the nuclear pore and, as described above (not shown) (Fornerod and Ohno, 2002; Kuersten et al., 2002). Cytosolic Ran.GDP is transported into the nucleus, where the chromatin-bound Ran guanine exchange factor (RanGEF) RCC1 re-generates a pool of Ran.GTP (Ohtsubo et al., 1989; Bischoff and Ponstingl, 1991; Klebe et al., 1995). Thus, the spatial restriction of RanGAP1, RanBPs, and RCC1 results in a large cytoplasmic pool of Ran.GDP and a large nuclear pool of Ran.GTP (Bischoff et al., 1994; Klebe et al., 1995).
The current model explaining how the Ran signal transduction pathway contributes to spindle assembly was originally conceptualized from observations in Xenopus embryo extracts (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999); indeed, the majority of studies to date on Ran function during mitosis have been carried out in Xenopus and verified in mammalian cell lines. Both systems utilize open mitosis, where the nuclear envelope breaks down fully and cytoplasmic and nuclear contents—including Ran—mingle during mitosis (Hutchins et al., 2009). RCC1 binds to chromatin (Ohtsubo et al., 1989; England et al., 2010), and through its interaction with Histones (Nemergut et al., 2001; Makde et al., 2010) and DNA (Chen et al., 2007), generates a locally high concentration of Ran.GTP (Bischoff and Ponstingl, 1991). As Ran.GTP diffuses away from the chromatin, RanGAP induces GTP hydrolysis, and Ran.GTP is converted to Ran.GDP (Kalab and Heald, 2008); an activity enhanced by RanBP1 (Seewald et al., 2003). As a result, a Ran.GTP gradient is formed throughout the cytoplasm with the highest concentration around the chromosomes. Ran.GTP acts to release a set of proteins termed Spindle Assembly Factors (SAFs) from their interaction with the Importin family of proteins [see Section The Role of Karyopherins (Importins and Exportins) in Drosophila Mitosis]. As these SAFs are locally released around the chromatin, it is in this region of the cell that they promote MT polymerization, stabilization and organization (Figure 1). The classic model of mitotic Ran function suggests that the Ran gradient has a particularly crucial role in generating MTs, and therefore spindles, independently of the classic MT organizing centers, centrosomes (Gruss et al., 2001; Wiese et al., 2001). For example, a bipolar spindle array can be generated from just chromatin-coated beads (Heald et al., 1996), or from beads coated with RCC1 in Xenopus embryo extract (Halpin et al., 2011), while activating Ran at the plasma membrane through specific targeting of a modified RCC1 results in ectopic MT aster generation (Zonis and Wilde, 2011). However, as described below, the localization of Ran to additional cellular locations during mitosis, and variations in the requirement of a Ran gradient in the generation of a mitotic spindle, suggests a more complex role for Ran than a simple gradient model suggests.
The clear dominance of centrosome-driven MT nucleation during Drosophila embryonic spindle formation, and differences in the presence or spatio-temporal localization of Ran pathway components between Drosophila and vertebrate mitosis, has left the role of Ran in Drosophila unclear. For example, in addition to localizing in a gradient around mitotic chromatin, Ran.GTP has also been described at both human mitotic centrosomes (Keryer et al., 2003), and mouse meiotic MTs (Cao et al., 2005), while in Drosophila embryos, Ran is found only throughout the spindle region; albeit in a diffuse gradient (Trieselmann and Wilde, 2002; Moutinho-Pereira et al., 2013). Similarly, the absence of a mitotic phenotype upon RNAi-mediated knock-down of Drosophila RCC1, either in the presence or absence of centrosomes, has led some to propose that the Ran.GTP gradient is unnecessary in flies (Moutinho-Pereira et al., 2013). Furthermore, there is no apparent homolog of RanBP1, which is essential for Ran function in human cells (Bischoff and Görlich, 1997; Floer et al., 1997; Fornerod and Ohno, 2002; Kuersten et al., 2002; Seewald et al., 2003), in Drosophila. Finally, whereas RanGAP localizes to spindles and kinetochores in mammalian cells (Joseph et al., 2002), in Drosophila early embryos it is predominantly found at the nuclear envelope, and occasionally at peripheral spindle MTs (Trieselmann and Wilde, 2002).
However, these apparent differences need not preclude a functional role for Ran.GTP in Drosophila spindle formation. The absence of an RNAi-induced RCC1 phenotype can be explained by incomplete knockdown. Indeed, although RCC1 knockdown in HeLa cells has been shown to result in reduced Ran.GTP generation around chromatin, it has no effect on mitotic spindle assembly either (Bamba et al., 2002; Kaláb et al., 2006). Similarly, the differences in localization of Ran pathway components between Drosophila and other cells may reflect detection issues or system specialisms. For example, given the syncytial nature of the Drosophila early embryo, the presence of RanGAP at the nuclear envelope may in fact, create an effective barrier, which restricts Ran-mediated spindle assembly around each individual mass of chromatin, rather than allowing diffusion throughout the entire embryo. Moreover, a biochemical association between Ran and MTs has been demonstrated by mass spectrometry (Hughes et al., 2008), suggesting a functional relationship between the Ran pathway and spindle formation.
Until very recently, the strongest evidence supporting a role for Ran in Drosophila mitosis came from studies by the Wilde lab (Trieselmann and Wilde, 2002; Silverman-Gavrila and Wilde, 2006). Microinjecting syncytial embryos with dominant negative Ran (T24N) resulted in a range of phenotypes ranging from the severe, such as complete failure to generate MTs or very few MTs, to more moderate, such as spindle fusion and spindle pole disorganization. Moreover, injection of Ran inhibitors such as Importin α/β and human RanBP1 reproduced these less severe phenotypes. These results indicate that the Ran pathway does have a MT-related role in the embryo (Silverman-Gavrila and Wilde, 2006). However, the dominance of centrosome-driven spindle formation in this tissue made it difficult to address the precise role of Ran in generation of MTs from chromatin in Drosophila. Recently, this barrier has been overcome by exploiting the temperature dependent nature of MT polymerization. When Drosophila embryos that have built a bipolar spindle using centrosomes are cooled to 4°C, their MTs depolymerize. Upon return to room temperature, a dramatic shift in the temporal and spatial nucleation, stabilization and sorting of MTs occurs (Hayward et al., 2014); instead of MTs re-initiating from the centrosomes, the mitotic spindle is organized almost exclusively from MTs generated around chromosomes (Hayward and Wakefield, 2014; Hayward et al., 2014). When dominant negative Ran (T24N) is injected into these cold-treated Drosophila embryos and spindle formation re-initiated following the temperature shift, chromatin-dependent MT generation is completely abolished (Hayward and Wakefield, 2014). Thus, it seems highly likely that the Ran pathway does function in Drosophila similarly to vertebrate systems.
Ran.GTP largely elicits its cellular functions through affecting the interactions between a class of proteins, termed Karyopherins, and their targets (Chook and Blobel, 2001; Mosammaparast and Pemberton, 2004). Karyopherins can be broadly divided into two classes—those involved in nuclear import (Importins), and those involved in nuclear export (Exportins). While Exportins bind Nuclear Export Signals (NESs), Importins bind Nuclear Localization Signals (NLSs). Together, these proteins therefore coordinate the transport of cargo in and out of nucleus (Figure 2). A sub-set of NLS-containing Importin cargoes, those that affect MT function during mitosis, are termed SAFs. During interphase these SAFs are sequestered in the nucleus by Importins, preventing interaction with interphase MTs. However, during open mitosis, the nuclear envelope breaks down, resulting in an influx of Tubulin dimer to the nucleus. The SAFs are then able to interact with Tubulin or with nascent MTs nucleated in the nuclear space, promoting spindle assembly (Figure 1B).
The complex regulation of Ran-mediated Importin-SAF interactions during both interphase and mitosis is derived from the specificity of the NLS-Importin interaction. The most well characterized example is the Importin β, Karyopherin β1, which can recognize targets either independently (Palmeri and Malim, 1999) or in association with an adapter protein—one of several variants of Importin α (Goldfarb et al., 2004). Each Importin α variant imparts specificity for different subsets of cargo (Pumroy and Cingolani, 2015). Moreover, many other variants of Karyopherin β exist, which recognize different NLSs and do not require an Importin α adaptor (Chook and Süel, 2011). Karyopherin β2 is one such example, which has also been implicated in mitotic regulation, through recognition of distinct NLS motifs (Lau et al., 2009; Bernis et al., 2014).
In all, over 20 different Karyopherins exist, with the vast majority having Drosophila homologs (Quan et al., 2008; Table 1). Even though only partial nuclear envelope breakdown occurs during most Drosophila mitoses, (Fuchs et al., 1983; Stafstrom and Staehelin, 1984; Harel et al., 1989; Katsani et al., 2008), the importance of nuclear import/export of proteins on mitotic spindle formation is clear. Abrogation of Importin α/β binding to NLSs, through injection of NLS peptides, results in monopolar, unfocused, narrow, and large barrel-shaped spindles, while co-injection of NLS peptide with dominant-negative Ran rescues spindle assembly, confirming that these phenotypes are due to Importin binding to NLSs (Virágh et al., 2012).
The most widely studied Importin in flies, Importin β1/Ketel, localizes predominantly to the nuclear envelope throughout the cell cycle of the early embryo (Trieselmann and Wilde, 2002) and, biochemically, is able to bind MTs (Tirian et al., 2003; Hughes et al., 2008). However, a point mutation which locks Importin β1/Ketel in a Ran.GDP binding conformation has no effect on nuclear import or spindle formation, though it does affect nuclear envelope reformation (Timinszky et al., 2002), suggesting other Importin β proteins contribute to SAF release during mitosis. Indeed, Importin 5/Karyβ3 has been identified as a MT-associated protein (Hughes et al., 2008) and as an interactor of the SAF Drosophila Hepatoma Up-Regulated Protein (dHURP), alongside Importin β1/Ketel (Hayward and Wakefield, 2014). Thus, it is conceivable that Importin 5/Karyβ3 may sequester a specific subset of SAFs that are involved in embryonic spindle formation. Of the other Karyopherin β or α proteins present in Drosophila, nothing is known with regards to potential mitotic SAF-related roles. Indeed, even in other experimental systems, the vast potential array of Karyopherin combinations currently precludes a comprehensive understanding of the role of Importin-SAF release on mitotic spindle formation.
The list of Ran-dependent SAFs has steadily increased since the first targets, TPX2 and NuMA, were identified in 2001 (Gruss et al., 2001; Wiese et al., 2001). These SAFs, primarily characterized in humans and Xenopus, play diverse roles and contribute to many aspects of spindle formation and function. Unsurprisingly, given the localization of Ran.GTP around chromatin, the majority of these have key roles in chromosome-mediated spindle self-assembly. However, several also function in the nucleus during interphase, including TPX2 (Neumayer and Nguyen, 2014), CHD4 (O'shaughnessy and Hendrich, 2013), NuMA (Ohata et al., 2013; Vidi et al., 2014), ISWI (Aydin et al., 2014; Vidi et al., 2014), Bard1 (Jasin, 2002), NuSAP (Kotian et al., 2014), Adenomatous Polyposis Coli (Apc) (Jaiswal and Narayan, 2008), TACC3 (Ha et al., 2015), and Survivin (Chakravarti et al., 2004; Jiang et al., 2009; Reichert et al., 2011); all of which have been implicated in the DNA damage response. Of the 29 Ran-dependent SAFs considered here, 25 have annotated homologs in Drosophila (Table 2). Many of these have cellular roles related to that of their vertebrate counterparts, and yet very few have been functionally verified as being Ran dependent. It is therefore likely that the overarching biochemical pathways and processes of Ran-mediated spindle assembly are conserved, though with some re-wiring unique to Drosophila. For the purposes of this review, we have separated SAFs into two groups, motor (Table 2) and non-motor SAFs (Table 3). In the following sections, we summarize the known functions of these proteins in Ran-dependent mitotic pathways and compare their roles in Drosophila and non-Drosophila models.
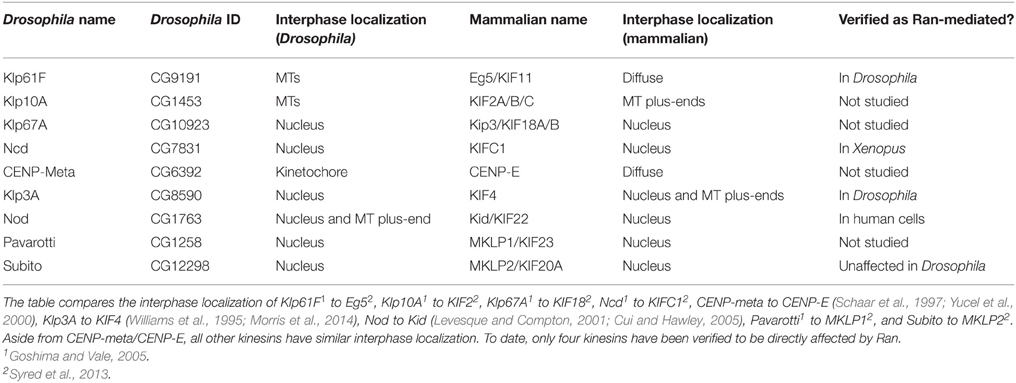
Table 2. Comparison of Drosophila Kinesins identified as mitotically relevant and their human homologs.
Although TPX2 was one of the first Ran-dependent SAFs to be identified, examination of the Drosophila genome by standard homology-based methods, over a period of a decade failed to identify a TPX2 homolog. More recent analysis found that the protein encoded by the mei38/short spindles 1 (ssp1) locus shares significant homology to parts of the human TPX2 protein, though lacking both the Aurora A and Kinesin-5 binding domains (Goshima, 2011; Hayward et al., 2014). Based on current Flybase nomenclature, we will refer to this protein as Mei38/dTPX2, but acknowledge the uncertainty that comes with attributing standardized names to divergent gene products from different species. In vertebrates, TPX2 has multiple mitotic roles; it can directly nucleate and bundle MTs (Brunet et al., 2004), it facilitates autophosphorylation of the mitotic kinase Aurora A and protects it from dephosphorylation (Dodson and Bayliss, 2012; Zorba et al., 2014), and it targets Xklp2/kinesin-12/KIF15 to spindle poles (Wittmann et al., 1998). TPX2 is also required for localization of KIF11/Eg5 to spindle MTs and facilitates its functions in kinetochore MT formation and spindle pole organization (Ma et al., 2011). In contrast, Mei38/dTPX2 does not bind Aurora A and a mei38 null mutation does not affect Aurora A or the localization of its effectors in Drosophila embryos (Hayward et al., 2014). Moreover, as Drosophila lack a clear Xklp2/kinesin-12/KIF15 homolog (Wickstead et al., 2010), and as Mei38/dTPX2 lacks a kinesin-5 binding domain (Goshima, 2011), it appears likely that Mei38/dTPX2 does not possess kinesin-directed activities. However, Mei38/dTPX2 has MT bundling capacity in vitro (Goshima, 2011) and mitotic spindles lacking Mei38/dTPX2 are significantly shorter than wild type (Hayward et al., 2014; Helmke and Heald, 2014; Fu et al., 2015), suggesting that at least some MT-related functions of vertebrate TPX2 (Hayward et al., 2014; Helmke and Heald, 2014; Fu et al., 2015) are shared with Mei38/dTPX2. Indeed, although Drosophila lacking Mei38/dTPX2 are viable and fertile, the protein does have a crucial role during self-organization of the mitotic spindle. Upon chromatin-derived spindle self-assembly driven by cold treatment and regrowth, the Drosophila embryonic spindles are unstable and tend toward collapse (Hayward et al., 2014). Taken together, the current evidence suggests that Mei38 may well represent a divergent TPX2-like protein, where additional proteins (e.g., see Section Ran.GTP and Mitotic Kinases) compensate for the loss of the key functions maintained in both Xenopus and humans.
Similarly, there is no true Drosophila homolog of the conserved SAF, Nuclear Mitotic Apparatus (NuMA). NuMA was initially described as an Importin β-inhibited protein promoting MT aster formation (Wiese et al., 2001) and has two clear roles during spindle formation. First, it is transported to MT minus-ends by the Dynein-Dynactin complex, where it crosslinks MTs and focuses the spindle poles (Gaglio et al., 1995; Merdes et al., 1996, 2000; Compton, 1998; Radulescu and Cleveland, 2010). Second, it plays a role in anchoring of astral MTs to the cell cortex, correctly orienting mitotic spindles during both symmetric (Silk et al., 2009; Kotak et al., 2012) and asymmetric cell division (Morin and Bellaïche, 2011; Peyre et al., 2011). In Drosophila, these two functions appear to be carried out by two distinct proteins. The Mushroom Body Defect (Mud) protein is the closest homolog to NuMA as assessed by primary structural homology (Bowman et al., 2006), while the Abnormal Spindle protein (Asp) also possesses some limited homology to NuMA domains (Saunders et al., 1997). Both proteins localize to the spindle poles during syncytial mitosis (Saunders et al., 1997; Yu et al., 2006). However, while Mud does not appear to have a role in focusing spindle poles, the absence of Asp leads to splayed spindle poles, free centrosomes and downstream mitotic and meiotic abnormalities (do Carmo Avides et al., 2001; Wakefield et al., 2001). Conversely, loss of Mud specifically affects MT-cortex interactions (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). In vertebrates, the membrane-anchored heterotrimeric G-proteins (mainly Gαi1, Gαi2, and Gαi3) interact with the leucine-glycine-asparagine repeat protein (Lgn) (Siderovski et al., 1999), which in turn interacts with NuMA and, though this, Dynein to tether MTs (Du et al., 2001; Bergstralh and St Johnston, 2014). This NuMA-facilitated anchoring is inhibited in the central region of the cell surrounding the metaphase plate, where the concentration of Ran.GTP is high (Kiyomitsu and Cheeseman, 2012; Bird et al., 2013), and as such is key to the orientation of the bipolar spindle (Morin et al., 2007; Konno et al., 2008; Zheng et al., 2010). In Drosophila, Mud binds to the homolog of Lgn, Partner of Inscrutable (Pins), and Gαi1 (Siller et al., 2006). The requirement of Ran in Mud function is indirect. Targeting of Mud and Pins to the cortex requires an additional protein, Canoe/Afadin (Wee et al., 2011), and only occurs when Canoe is bound to Ran.GTP; it is theorized that Canoe brings Ran.GTP to the cortex, and the localized high concentration of cortical Ran.GTP prevents Importin β from inhibiting Mud (Wee et al., 2011). This is in sharp contrast to the situation in vertebrates, where NuMA-mediated anchoring is inhibited by Ran.GTP (Kiyomitsu and Cheeseman, 2012; Bird et al., 2013); this discrepancy outlines the incompleteness of our knowledge of Ran function in spindle pole positioning.
Hepatoma Up-Regulated Protein (HURP) is a MT nucleating and stabilizing SAF, important in both centrosome and chromatin driven spindle assembly (Koffa et al., 2006). Crucially, its Drosophila homolog, Mars/dHURP, is one of the few Drosophila SAFs for which there is evidence for a Ran-dependent function (Cesario and McKim, 2011). In vertebrates, HURP plays a crucial role in kinetochore-fiber stabilization (Koffa et al., 2006; Sillje et al., 2006; Wong and Fang, 2006). During mitosis HURP is phosphorylated by Aurora A, which protects it from degradation (Yu et al., 2005), allowing its association with MT plus-ends and facilitating kinetochore-MT attachments (Wu et al., 2013). While the dependence of dHURP by Aurora A has not yet been investigated, its role in kinetochore-fiber stabilization and kinetochore-MT attachment appears to be conserved (Yang and Fan, 2008). dHURP is also required for MT attachment to centrosomes (Zhang et al., 2009) and for localization of γ-tubulin to the spindle (Yang and Fan, 2008), loss of which results in decreased spindle density. Moreover, in both Drosophila and vertebrate systems, HURP has been shown to be critical for chromatin-mediated MT generation (Wong and Fang, 2006; Hayward et al., 2014). Therefore, while certain functions have not yet been verified between vertebrate and Drosophila systems, dHURP is an excellent candidate for further studies of Ran-mediated spindle assembly in Drosophila.
The Transforming Acidic Coiled Coil (TACC) family of proteins, which includes TACC3 in humans, exist in a complex with TOG/XMAP215 to promote MT polymerization (Fox et al., 2014; Gutiérrez-Caballero et al., 2015), and Clathrin to promote kinetochore-MT bundling (Fu et al., 2010; Hubner et al., 2010; Lin et al., 2010; Booth et al., 2011). It therefore stabilizes kinetochore MTs (Booth et al., 2011) centrosomal MTs (Barros et al., 2005) and the mitotic spindle, generally (Gergely et al., 2000a). Although not commonly regarded as a Ran-dependent SAF, TACC3 immunoprecipitates with Importin β, and their interaction is reduced in the presence of constitutively active RanL43E (Albee et al., 2006); further, in human cell lines, TACC3 is sequestered to the nucleus during interphase, presumably by Importins (Gergely et al., 2000a). In flies, the interaction between the TACC3 and TOG homologs (dTACC and Msps respectively) is conserved. dTACC, however, lacks an apparent NLS and does not localize to the nucleus, but concentrates at centrosomes during embryonic interphase (Gergely et al., 2000b). This might indicate that dTACC is not Importin-mediated. However, dTACC localization is disrupted in the presence of dominant negative Ran (T24N) during Drosophila female meiosis (Cesario and McKim, 2011). Since TACC3 and dTACC both localize to spindle MTs during mitosis (Barros et al., 2005; Kinoshita et al., 2005), and are functionally conserved (Gergely et al., 2000a), Drosophila may be a useful tool to explore the mechanistic details between Ran and TACC.
During interphase, the Nucleolar and Spindle Associated Protein (NuSAP) plays a role in the DNA damage response (Kotian et al., 2014). However, at the onset of mitosis, NuSAP is released around chromatin by Ran.GTP, binding to DNA and both stabilizing and crosslinking MTs in order to promote spindle formation from the chromosomes (Raemaekers et al., 2003; Ribbeck et al., 2006, 2007). Cells either deficient in or overexpressing NuSAP show defects in cell division (Raemaekers et al., 2003; Ribbeck et al., 2006; Li et al., 2007) and knockout of NuSAP in mice is lethal at the early embryonic stage (Vanden Bosch et al., 2010). The Drosophila homolog of NuSAP, Mink, was recently identified as a MAP in mitotic, but not interphase, S2 cells (Syred et al., 2013). While the role of Ran in Mink function has not yet been investigated, it is nuclear during interphase and therefore is likely to be bound by Importins, and has a role in crosslinking and stabilizing MTs in Drosophila S2 cells (Syred et al., 2013).
Somewhat surprisingly, three nucleic acid binding proteins have been identified as Ran-dependent SAFs. The chromatin remodeling factors, Imitation Switch (ISWI) and Chromodomain-Helicase-DNA-binding 4 (CHD4), both associate with MTs in a Ran.GTP-dependent manner in Xenopus (Yokoyama et al., 2009, 2013). Both localize to chromatin during interphase but relocalize to the spindle during mitosis in Xenopus egg extracts, and both play roles in spindle assembly and stability. ISWI can nucleate and bundle MTs in Xenopus embryo extracts (Yokoyama et al., 2009), while CHD4 stabilizes, but does not nucleate, MTs (Yokoyama et al., 2013). Encouragingly, both have Drosophila homologs which appear to function similarly to their vertebrate counterparts (Yokoyama et al., 2009, 2013). CHD4 acts in early mitosis to promote spindle assembly from the chromatin, and depletion in either HeLa cells or Drosophila S2 cells results in reduced spindle density and disorganization, with resultant chromosome mis-segregation (Yokoyama et al., 2013). ISWI stabilizes the spindle prior to anaphase, and depletion of this protein in Xenopus egg extracts or Drosophila S2 cells produces defective and disintegrating spindles (Yokoyama et al., 2009). In addition, Rae1, an mRNA export protein normally found in the nucleus, binds directly to Importin β (Blower et al., 2005), suggesting that its activity is spatially and temporally coordinated by Ran. Indeed, it appears to act by increasing the ability of NuMA to focus and bundle MTs and perturbation of this interaction results in spindle defects (Wong et al., 2006). Interestingly, Rae1 acts as part of a large ribonucleoprotein complex, which includes Nup98 and the Ran-dependent SAF, TACC3 (Blower et al., 2005), though the modes of activation and regulation of this complex are unclear. However, as yet, nothing is known of Rae1 mitotic MT function in Drosophila.
Two additional MT effectors, APC and Microspherule Protein 1 (MCRS1) have been identified as Ran-dependent SAFs in vertebrates (Dikovskaya et al., 2010; Meunier and Vernos, 2011), and have Drosophila homologs. APC is involved in a multitude of cellular functions, including Wnt signaling, transcription, DNA damage response, cell adhesion, and mitosis (Bahmanyar et al., 2009; Lui et al., 2012). In both Drosophila and vertebrates, there are two APC genes; apc and apc2. In vertebrates, loss of APC results in embryonic lethality, while APC2 knock-outs are viable. In Drosophila, APC and APC2 appear to function redundantly in Wnt/Wingless signaling though the distribution of each differs in Drosophila tissues (Ahmed et al., 2002). APC has multiple mitotic roles in vertebrates. It can bind either directly or indirectly to MTs and plays roles in nucleation, bundling, and MT dynamics (Deka et al., 1998; Banks and Heald, 2004; Dikovskaya et al., 2004), affecting processes such as kinetochore attachment and MT anchoring at the centrosomes (Bahmanyar et al., 2009). However, in Drosophila, it is APC2 that appears to play a more crucial role in mitosis. Early embryos derived from hypomorphic apc2 alleles show defects in centrosome separation and spindle pole positioning and subsequent loss of nuclei from the embryonic cortex in a process called “nuclear fall-out” (McCartney et al., 2001; Buttrick et al., 2008). This is thought to occur through loss of stable attachments between astral microtubules plus ends to the cell cortex, a process mediated via catenins (McCartney et al., 2001; Buttrick et al., 2008). Moreover, RNAi of apc2 in Drosophila also interferes with oriented cell division in the embryonic epidermis, suggesting a wider role for APC2 in regulating MT-cortex interactions (Lu et al., 2001). It is unclear whether this function is conserved in vertebrate APC or APC2, though APC loss in mammalian cell lines results in mis-orientation of the spindle (Green et al., 2005). In vertebrates, APC protein function is mediated by Ran; vertebrate APC interacts with Importin β, and this interaction is reduced by constitutively-active Ran-Q69L (Dikovskaya et al., 2010). Further, Importin β inhibits the ability of APC to nucleate MTs from pure Tubulin, and to bundle Taxol stabilized MTs (Dikovskaya et al., 2010). However, whether Ran also regulates Drosophila APC or APC2 function is as yet unknown. Since apc2 mutant embryos present such a distinctive centrosome and cortical phenotype in the Drosophila syncytial embryo (see above), inhibition of Ran function in these mutants, under both normal cycling and cold-treatment/re-growth conditions, presents an ideal opportunity to test this hypothesis. Much less is known about the Drosophila homolog of MCRS1. In vertebrates, MCRS1 plays a protective role at the minus ends of MTs generated from the chromosomes, preventing depolymerization (Meunier and Vernos, 2011). Interestingly, although the Drosophila homolog, Rcd5, has been implicated in transcription during interphase (Andersen et al., 2010), it was originally identified in a genome-wide screen for proteins that affected the recruitment of the MT effector, Centrosomin (Cnn), to centrosomes in mitotic S2 cells—hence its name, Reduction in Cnn dots 5 (Dobbelaere et al., 2008). However, it remains unclear whether Rcd5 has a mitotic role, or whether it is Ran-regulated.
One further Ran-dependent SAF with a sequence homolog in Drosophila, is the cell polarity protein Crumbs3 (Crumbs in Drosophila) (Pocha and Knust, 2013). Although generally regarded as a transmembrane protein, the role of Crumbs3 in mitosis is dependent upon a specific splice variant with a distinct C-terminus, Crumbs3-CLPI. Crumbs3-CLPI appears to play a key role in centrosomal regulation, and depletion in mammalian cells leads to a variety of phenotypes including supernumerary centrosomes, multipolar spindle formation and multinuclear cells (Fan et al., 2007). Crumbs3-CLPI is dependent upon Ran.GTP for localization to centrosomes (Fan et al., 2007), and may be recruited by a centrosomal pool of Ran.GTP. However, the Drosophila genome does not appear to possess such a distinct splice variant and so whether Crumbs is able to moonlight as a Ran effector in flies is currently unknown.
Finally, there are also three Ran-dependent SAFs identified in vertebrates that appear to have no sequence homologs in Drosophila. These are Bard1, which associates with BRCA1 to localize TPX2 to spindle poles (Joukov et al., 2006); RHAMM, which interacts with TPX2 and γ-Tubulin at centrosomes (Groen et al., 2004); and Xnf7, which bundles MT and protects them from depolymerization (Maresca et al., 2005). These proteins feature domains and coiled-coil motifs that confound conventional search methods, and as such it is difficult to determine whether they are indeed absent in Drosophila or still remain to be identified. Bard1, at least, has been identified in the genome of the honeybee Apis mellifera and therefore may have been lost or truncated in Drosophila and other Diptera, similarly to dTPX2/Mei38. Alternatively, RHAMM and Xnf7 may represent vertebrate-specific proteins; analysis of the evolutionary patterns of these proteins across a range of opisthokont organisms would answer this question.
MT motor proteins play essential roles in regulating MT dynamics and generating the forces involved in centrosome movement, and chromosome alignment and segregation. The minus-end directed motor cytoplasmic Dynein is important throughout mitosis, with roles in centrosome separation (Vaisberg et al., 1993; Robinson et al., 1999; Raaijmakers et al., 2012), spindle pole focusing in conjunction with NuMA (Radulescu and Cleveland, 2010), and shedding of the Rod-Zw10-Zwilch (RZZ) complex from kinetochores polewards along kinetochore-MT bundles (Barisic and Geley, 2011). Kinesin-like proteins (Klps) include both plus end- and minus end- directed motors which can transport cargo to specific cellular locations, crosslink and slide MTs to generate forces for centrosome or chromosome separation, attach MTs to kinetochores, push MTs away from chromatin, or alter MT dynamics; see Cross and McAinsh (2014) for a recent review.
Of the 25 kinesins identified in Drosophila, depletion of 9 results in mitotic defects (Table 2; Yucel et al., 2000; Giunta et al., 2002; Goshima and Vale, 2005; Cesario et al., 2006). These 9 mitotically relevant kinesins have homologs in humans and five—Klp3A (KIF4), Klp67A (KIF18), Ncd (KIFC1), Pavarotti (KIF23), and Nod (Kid/KIF22)—are sequestered to the nucleus in both Drosophila interphase cells (Cui et al., 2005; Goshima and Vale, 2005) and human cell lines (Mazumdar et al., 2011; Syred et al., 2013). Expression of Klp67A, Ncd, or Pavarotti without their NLS results in MT network disruption (Goshima and Vale, 2005), highlighting the importance of sequestering them away from interphase MTs. However, whether Ran plays a role in the mitotic function of these kinesins beyond sequestration during interphase in Drosophila is largely unknown.
Ran has, however, been shown to regulate two kinesins in Drosophila; Klp61F (KIF11/Kinesin-5/Eg5) and Klp3A (Kinesin-4/KIF4), though it is not clear how this impacts upon spindle assembly. Klp61F shows reduced binding affinity to MTs in the presence of dominant negative Ran (T24N) (Silverman-Gavrila and Wilde, 2006), a property that is conserved in its Xenopus homolog, Eg5 (Wilde et al., 2001). Notably, in both Drosophila and vertebrates, Klp61F/KIF11 is excluded from the nucleus during interphase, indicating that Ran is able to mediate the protein's activity in the absence of an NLS through an unknown mechanism (Silverman-Gavrila and Wilde, 2006). Less is known about Klp3A, though upon injection of dominant negative Ran (T24N) into Drosophila syncytial embryos, Klp3A localization to MTs is lost, and instead re-localizes to chromosomes (Silverman-Gavrila and Wilde, 2006).
Outside of Drosophila, Ran has been shown to affect the regulation of the minus-end directed motor KIFC1 (also known as HSET or XCTK2) (Ems-McClung et al., 2004; Weaver et al., 2015), and the chromokinesin, Kid (KIF22) (Trieselmann et al., 2003). Importin α/β binds the non-motor tail of KIFC1 and inhibits its MT crosslinking activity (Ems-McClung et al., 2004). Therefore, the Ran.GTP gradient effectively limits the MT sliding activity of KIFC1 to the regions near the chromosomes (Weaver et al., 2015); indeed, expressing mutant forms of KIFC1 which cannot be inhibited by Importin α/β causes spindles to become elongated (Cai et al., 2009). The chromokinesin Kid/KIF22 plays a key role in generating the forces that align and maintain chromosomes at the metaphase plate (Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001). This activity relies upon Kid's motor functions, but Importin α/β is able to directly inhibit its MT binding ability (Trieselmann et al., 2003). This inhibition of MT binding ability effectively targets Kid to chromosomes, where Kid is released from Importin α/β in the presence of high Ran.GTP (Tahara et al., 2008). While Drosophila has homologs of both KIF1C and Kid, Ncd and Nod respectively, the involvement of Ran in the function of these proteins has not yet been determined.
Kinases play a central role in mitosis, from cell cycle control to spindle assembly, and in cytokinesis. The classic mitotic kinases include the Cyclin Dependent Kinase, CDK1, the Polo-like kinases (PLKs), the Aurora kinases A and B, and the NIMA (never in mitosis in Aspergillus nidulans)-related kinases (NEKs) (Ma and Poon, 2011; Bayliss et al., 2012). It is therefore unsurprising that at least some of these kinases are regulated by, or themselves regulate, Ran. To date, there is evidence linking CDK1, Plk1, and Aurora A to Ran-dependent pathways in mitosis.
CDK1 is the master regulator of mitotic entry (Malumbres, 2014), and plays a role in Ran-mediated spindle assembly by phosphorylating RCC1 (Hutchins et al., 2004). RCC1 contains an N-terminal NLS, which mediates nuclear localization (Nemergut and Macara, 2000; Talcott and Moore, 2000). During interphase, the interaction of RCC1 with chromatin is highly dynamic, cycling between chromatin and nucleoplasm; the interaction of RCC1 with chromatin is coupled to its ability to phosphorylate Ran and phosphorylation dissociates RCC1 from chromatin (Li et al., 2003). During mitosis, phosphorylation of the NLS of RCC1 by CDK1 is required to maintain the dynamic nature of this RCC1-chromatin interaction, preventing RCC1 inhibition by Importins (Hutchins et al., 2004; Li and Zheng, 2004). This phosphorylation event is therefore essential for generation of the Ran.GTP gradient around the chromatin, and disruption results in abnormal spindle formation and chromosome mis-segregation (Li and Zheng, 2004). Again, there are unfortunately no published studies investigating the relationship between Drosophila CDK1 and Ran pathway components.
Plk1 regulates, amongst other things, mitotic entry, centrosome maturation, spindle assembly, and cytokinesis (Zitouni et al., 2014). During interphase in human cells, Plk1 localizes to both the cytoplasm and the nucleus, though disruption of the NLS blocks mitotic entry, suggesting constant shuttling in and out of the nucleus impacts cell cycle progression (Taniguchi et al., 2002). During mitosis, Plk1 phosphorylates Ran, and while the precise functional significance of this is unknown, a Plk1 phospho-mimetic mutation in Ran results in abnormal spindle morphology (Feng et al., 2006). Plk1 also phosphorylates RanBP1 (Hwang et al., 2011), and this phosphorylation is crucial for proper spatial regulation of the Ran.GTP gradient (Zhang et al., 2014). Together, this suggests that Plk1 functions upstream of Ran-dependent mitotic pathways. However, nothing is yet known of the relationship between the Drosophila homolog of Plk1, Polo, and Ran.
There is, however, some evidence linking Aurora kinases and Ran in Drosophila. In vertebrate systems, the Ran-dependent SAF, TPX2, is at the heart of Aurora A localization and activation. However, as described in Section Non-motor SAFs, Drosophila TPX2 (Mei38) lacks the Aurora A interaction motif and Aurora A does not bind to dTPX2 (Goshima, 2011; Hayward et al., 2014), neither does loss of the protein affect the localization of Aurora A effectors (Hayward et al., 2014). However, injection of dominant negative Ran into Drosophila early embryos inhibits both the centrosomal and spindle association of Aurora A, demonstrating a clear regulatory link between the two (Silverman-Gavrila and Wilde, 2006). How this is achieved is unclear, but there are two potential pathways. Firstly, HURP plays a role in Aurora A targeting in Xenopus (Koffa et al., 2006). In Drosophila dHURP has been demonstrated to be regulated by Ran.GTP (Cesario and McKim, 2011), and loss of either dHURP or injection of dominant negative Ran into cold-treated mitotic embryos results in complete cessation of chromatin-induced spindle formation (Hayward and Wakefield, 2014; Hayward et al., 2014). Therefore, it is possible that dHURP has evolved to regulate Aurora A in the absence of full-length TPX2. Alternatively, there may be an as-yet unknown link between Ran, Aurora A and other regulators such as Aurora Borealis (Bora) and Ajuba. In Drosophila, Bora activates Aurora A (Hutterer et al., 2006) and localization of Aurora A relies on Ajuba (Sabino et al., 2011). Bora is a good candidate for a Ran-regulated SAF, as it is retained in the nucleus during interphase (Hutterer et al., 2006), indicating the presence of a NLS. Both Bora and Ajuba have vertebrate homologs with roles in mitosis (Hirota et al., 2003; Hutterer et al., 2006), though Ran dependency has not been investigated. It may be that in vertebrates the TPX2 and Bora/Ajuba pathways function redundantly in Ran-dependent mitotic pathways, or it may be that, due to the limited functionality of dTPX2 in relation to human TPX2, the Bora/Ajuba pathway and/or dHURP have been co-opted to compensate for TPX2-mediated Aurora A activation.
Aurora B is an obligate part of the hetero-tetrameric Chromosomal Passenger Complex (CPC) together with INCENP, Borealin, and Survivin. Ran.GTP has been shown to directly bind Survivin in vertebrates (Xia et al., 2008), but surprisingly, dominant-negative Ran elicits no effect on CPC kinase activity (Kelly et al., 2007) suggesting Ran activity may be dispensable for CPC activity. Instead, the significance of the Ran-Survivin interaction appears deliver TPX2 to MTs (Xia et al., 2008). Whether Ran also interacts with Survivin in Drosophila is unknown; the truncation of dTPX2/Mei38 may abrogate binding. However, as the MT bundling activities of dTPX2/Mei38 appear to be conserved in Drosophila, a potential role for Ran/Survivin in loading dTPX2 on to MTs, at least during mitosis, may still be required.
The interactions between kinases and Ran function in mitosis are most likely broader than described above. For example, the Cyclin Dependent Kinase, CDK11, is involved in multiple aspects of cell cycle regulation, such as centrosome maturation and separation (Petretti et al., 2006) sister chromatid cohesion (Hu et al., 2007; Rakkaa et al., 2014) and cytokinesis (Wilker et al., 2007; Franck et al., 2011) and, in Xenopus, a long isoform of this kinase, p110, has been demonstrated to be regulated by Ran (Yokoyama et al., 2008). Although the Drosophila homolog of CDK11, Pitslre, has been identified as a regulator of Rho activity during cytokinesis (Gregory et al., 2007), any relationship between it and Ran has yet to be investigated.
The Ran gradient model, in which RCC1 is sequestered exclusively at mitotic chromosomes, suggests that only low concentrations of Ran.GTP are present around the centrosomes and thus that Ran would play a minor part in centrosome function. However, it has been demonstrated that a pool of Ran remains associated with centrosomes throughout the cell cycle (Keryer et al., 2003) and proteins linked with Ran function at centrosomes are beginning to emerge. In human cell lines, Ran is maintained at the centrosome by association with the scaffolding protein AKAP450 (Keryer et al., 2003). It is difficult to tease apart the nature of the relationship between the two proteins during mitosis, as AKAP450 is also responsible for anchoring of several other molecules, including the kinases Casein Kinase 1 (CK1) and Cyclin E-Cdk2 to the centrosome (Nishimura et al., 2005). In addition, AKAP450 can anchor the MT nucleating complex γ-TuRC to the centrosome, promoting MT nucleation (Takahashi et al., 2002). Therefore, while loss of AKAP450 from the centrosome leads to a failure in centrosome duplication and defects in mitosis, it is unclear whether these effects are due to Ran, to CK1, to γ-TuRC or to other anchored proteins. The situation is even more complicated in Drosophila, where the closest identified homolog of AKAP450 is the Pericentrin-like protein, dPLP. dPLP is the only large fly protein with a PACT (Pericentrin/AKAP450 centrosomal targeting) domain in Drosophila (Martinez-Campos et al., 2004) and, as such, it is not entirely clear whether it is a homolog of pericentrin or AKAP450 or represents a functional composite of both. What is clear is that dPLP affects centrosome duplication (Dobbelaere et al., 2008) and has an important role in the structural maintenance of centrosomes, though this seems to be primarily in interphase (Müller et al., 2010; Lerit et al., 2015; Richens et al., 2015). Whether this role is due to direct effects of dPLP or whether, like AKAP450, it acts as an anchoring protein for other kinases, and whether Ran is in any way involved, all remains to be seen.
A second centrosomal protein that has a clear functional relationship to the Ran pathway is Nucleophosmin (NPM/B23). Originally described as a nucleolar phosphoprotein (Okuda et al., 2000; Tarapore et al., 2002), NPM contains both an NLS and an NES, the latter of which is recognized by the Exportin Crm1, which shuttles the protein to centrosomes (Wang et al., 2005). Crm1 is required to maintain NPM at centrosomes (Shinmura et al., 2005; Wang et al., 2005) and NPM dissociation from centrosomes following phosphorylation by CDK2/Cyclin E triggers centrosome duplication at the onset of mitosis in mammalian cells (Okuda et al., 2000; Tokuyama et al., 2001). NPM is also an activator of Aurora A kinase at the centrosome at G2/M, suggesting potential cross-talk between the Ran and Aurora A pathways. Although there is a Nucleophosmin homolog in Drosophila, as yet there are no studies on its centrosomal or mitotic functions.
Most metazoan model organisms undergo mitosis having completely disassembled their nuclear envelope and their nuclear pore complexes. However, in Drosophila and C. elegans, which only disassemble the nuclear envelope at the spindle pole regions (Fuchs et al., 1983; Stafstrom and Staehelin, 1984; Harel et al., 1989; Katsani et al., 2008) nuclear pore disassembly is gradual, and is only complete at metaphase (Stafstrom and Staehelin, 1984).
The core of the nuclear pore complex is the Nup107–160 complex, consisting of Nup 37, Nup 43, Nup 85, Nup 96, Nup 107, Nup 133, Nup 160, She 1, and Sec 13 (Belgareh et al., 2001; Vasu et al., 2001; Harel et al., 2003; Loïodice et al., 2004). During interphase, this complex acts as a scaffold for the rest of the nuclear pore (Szymborska et al., 2013). During mitosis, however, Nup107–160 relocalizes to chromatin and recruits the MT nucleating complex γ-TuRC to the kinetochore (Mishra et al., 2010); a process which is Ran-dependent in Xenopus embryo extracts (Franz et al., 2007). A further nuclear pore complex subunit, Mel28/ELYS, has been identified as the initial building block for the nuclear pore; in vertebrates it is required for recruitment of the Nup107–160 complex to the reassembling nuclear pore (Rasala et al., 2006; Franz et al., 2007), and for recruitment of the same complex to the chromatin during mitosis (Galy et al., 2006; Rasala et al., 2006). However, Mel28/ELYS can directly recruit γ-TuRC to MT nucleation sites on the spindle, and this process is both independent of the Nup107–160 complex and essential for Ran-dependent spindle assembly (Yokoyama et al., 2014). Since, in Drosophila, the nuclear pores disassemble only gradually, the role of Nup107–160 in kinetochore attachment to MTs is unclear. Moreover, no research has, as yet, been undertaken on the Mel28/ELYS homolog in Drosophila, CG14215. However, in C. elegans, which also utilizes semi-open mitosis, Mel28 likewise localizes to the chromatin during mitosis and recruits the Nup107–160 complex (Galy et al., 2006).
Nup358/RanBP2 is also another nuclear pore complex protein, which during interphase is involved in nuclear transport (Figure 2; Wu et al., 1995; Yokoyama et al., 1995; Walther et al., 2002). During mitosis it associates with MTs, especially the spindle poles and kinetochores, and RNAi knockdown in HeLa cells results in metaphase catastrophe (Hashizume et al., 2013), and MT-kinetochore attachment defects (Salina et al., 2003). Although there is only minimal information on mitotic functions of Nup358/RanBP2 in Drosophila, it has been identified as a MAP, as a mitotic centrosomal protein and as an Ndc80 interactor using mass spectrometry (Przewloka et al., 2007; Hughes et al., 2008; Müller et al., 2010).
Following chromosome segregation, nuclear envelopes reform in order to generate two daughter nuclei. In Xenopus, karyopherins play an essential role, recruiting nuclear envelope vesicles to chromatin (Zhang et al., 2002b). As with other Karyopherin functions, this process is dependent on Ran, and Ran immobilized on beads is able to drive nuclear envelope reformation in Xenopus embryo extracts (Zhang and Clarke, 2000; Zhang et al., 2002a). The presence of Ran itself, however, is not sufficient for nuclear reformation to occur; addition of non-hydrolysable GTP inhibits this process, suggesting the GTPase cycle is important (Hetzer et al., 2000). Similarly to spindle assembly, it is theorized that, at late anaphase/early telophase, Ran.GTP releases nuclear envelope components around chromatin (Hetzer, 2010). Although the nuclear envelope does not fully disassemble in Drosophila, Ran is clearly involved in the dynamics of nuclear assembly. Injection of a mutant form of Importin β with reduced Ran.GTP binding affinity into Drosophila embryos stops Lamin from accumulating at the nuclear envelope (Timinszky et al., 2002), while addition of the same mutant to Drosophila embryo extracts inhibits nuclear envelope formation (Tirian et al., 2003). Therefore, although additional evidence is needed, it can be reasonably assumed Ran plays a similar role in nuclear envelope reformation following mitosis for both Drosophila and vertebrate systems.
The idea of the spindle matrix, a persistent nuclear-derived cellular structure in the vicinity of the spindle, was first conceived in 1984 (Pickett-Heaps et al., 1984). The spindle matrix is thought to provide a physical framework upon which MTs or SAFs attach (Tsai et al., 2006; Johansen and Johansen, 2007; Zheng, 2010). While it is somewhat poorly defined, the perduring spindle-like localization of certain proteins following MT depolymerization and the presences of forces within the spindle region following laser-ablation of MTs suggest such a structure exists and has a role in spindle assembly (Johansen and Johansen, 2009).
The current literature suggests two distinct, yet possibly related, spindle matrices. In vertebrates, at the onset of nuclear envelope breakdown, Lamin B assembles into a matrix-like network that can retain SAFs after MT depolymerization (Tsai et al., 2006; Zheng, 2010). RNAi knockdown of the Drosophila homolog Lamin Dm0 produces monopolar spindles with low MT density, suggesting it has some similar function to the Lamin B derived spindle matrix (Goshima et al., 2007). In addition, it has recently been demonstrated that the conserved MT-associated protein, BuGZ, undergoes liquid-liquid phase transitions in vitro, driven by low complexity hydrophobic residues. This aggregation promotes spindle matrix assembly, driving MT concentration and spindle formation (Jiang et al., 2015). Intriguingly, in Drosophila, four proteins—Skeletor, Chromator, Megator, and East—have each been shown to localize as matrix components, where disruption of any of these results in disruption of the matrix and spindle defects (Johansen and Johansen, 2009). All contain low complexity FG repeats, and have been hypothesized to generate a gel-like colloid (Johansen and Johansen, 2009). While a vertebrate homolog of Megator exists [the Translocated Promotor Region (Tpr) protein] (Lince-Faria et al., 2009), the others appear to be Drosophila-specific (Johansen and Johansen, 2009). Irrespective of their precise molecular composition, there is evidence that both vertebrate and Drosophila spindle matrices are Ran-dependent; in Xenopus embryo extracts, Ran is required for assembly of Lamin B into the spindle matrix (Tsai et al., 2006) while injection of dominant-negative Ran into Drosophila embryos results in disruption of Skeletor localization (Silverman-Gavrila and Wilde, 2006). Thus, it seems likely that Ran plays a direct role in spindle matrix formation.
The robustness of bipolar spindle assembly is enhanced by Augmin, a conserved hetero-octomeric protein complex which binds to pre-existing MTs within the spindle, recruiting the MT nucleating complex γ-TuRC to generate further new MTs (Goshima et al., 2007, 2008; Hughes et al., 2008; Lawo et al., 2009; Uehara et al., 2009). The ability of Xenopus Augmin to generate branched MTs has recently been shown to be greatly enhanced in the presence of constitutively active RanQ69L and/or the Ran-mediated SAF TPX2 (Petry et al., 2013), strongly suggesting that complex activity is Ran-mediated. Further, inhibition of Augmin activity through injection of interfering antibodies into Drosophila embryos completely abolishes chromatin-driven MT nucleation following cold treatment (Hayward et al., 2014). Although it is possible that at least some aspects of this chromatin-driven pathway are Ran-independent, injection of dominant negative Ran (T24N) into cold-treated Drosophila embryos phenocopies Augmin loss (Hayward and Wakefield, 2014). Furthermore, RNAi against Drosophila Augmin in S2 cells has been shown to attenuate kinetochore-MT attachment, a Ran-dependent process (Bucciarelli et al., 2009). Together, this evidence suggests that Ran and Augmin act in the same pathways of chromatin-mediated spindle assembly and kinetochore-MT attachment, although the details have yet to be determined.
There is some evidence that Ran plays a role in furrow formation and cytokinesis. Although cytokinesis does not occur in Drosophila syncytial divisions, pseudocleavage furrows form prior to the onset of anaphase, separating the dividing chromosomes from neighboring spindles and preventing nuclear fusion (Sullivan et al., 1990). These pseudocleavage furrows resemble cytokinetic furrows, and share many of the same components (Mazumdar and Mazumdar, 2002); thus, the mechanisms that regulate them are likely to be similar. In one of the first demonstrations of a role for Ran in furrow formation, injection of dominant-negative Ran (T24N) or Importin α into syncytial embryos was found to significantly reduce pseudocleavage furrow generation, correlating with a loss of Anillin and Peanut recruitment—proteins which are important for furrow stability (Silverman-Gavrila et al., 2008). These events occur distantly to the chromosomes, in metaphase, in a region where Ran.GTP is believed to be at a low concentration. However, there is evidence for a localized population of Ran.GTP at the cortex in humans (Wee et al., 2011) as discussed in Section Non-motor SAFs, which could drive an additional cortical Ran gradient independently of chromosomes. These results are somewhat confounded by a recent study using a temperature sensitive RCC1 cell line, also in humans, which suggests that chromosomally-derived Ran.GTP in anaphase is actually responsible for reducing the amount of cortical Anillin during asymmetric cell divisions (Kiyomitsu and Cheeseman, 2013). Moreover, the ability of Importin β2 to bind Anillin has been shown not to affect mitosis or cytokinesis; rather it required to sequester the protein in the nucleus in interphase, preventing abnormal cellular architecture (Chen et al., 2015). Although this could reflect differences between distinct populations of Anillin regulated through distinct Importins (see Section The Core Ran Pathway in Drosophila Embryos), such an explanation does not explain the discrepancy between a cortically-derived Ran.GTP gradient which may act positively upon Anillin and a chromosomally-derived Ran.GTP gradient that negatively regulates Anillin localization.
There is also indirect evidence for a second target of Ran during cytokinesis—the kinesin, KIF14. KIF14 is known to interact with the MT bundling protein PRC1, localizing to the central spindle in anaphase, to co-ordinate cytokinesis (Gruneberg et al., 2006). The germline-specific paralog of KIF14 in Xenopus, Nuclear and Meiotic Actin Bundling Kinesin (NabKin), which is required for cytokinesis during the second meiotic cycle, has recently been shown to be negatively regulated by Importin β and activated by Ran.GTP, strongly suggesting it as a canonical, but anaphase specific, SAF (Samwer et al., 2013). Interestingly, KIF14 in Drosophila is encoded by Klp38B, which has been shown to localize to condensed chromosomes and regulate spindle formation (Molina et al., 1997; Ruden et al., 1997). However, as a precise role in post-metaphase events has not been described, whether Klp38B is a conserved Ran target involved in central spindle formation and/or cytokinesis remains only a possibility.
Finally, as eluded to in Ran-dependent SAFs, there is evidence, albeit limited and again contradictory, that CDK11, which has a clear role in cytokinesis (Wilker et al., 2007; Franck et al., 2011), is regulated through Ran. Given the role of the Drosophila homolog, Pitslre, as a regulator of Rho activity during cytokinesis (Gregory et al., 2007), it is tempting to speculate that more Ran-dependent proteins with roles in cytokinesis remain to be discovered.
Originally conceptualized as a molecular and chemical “GPS” for SAFs (Kalab and Heald, 2008), through the restriction of RCC1 to chromosomes, it is now becoming apparent that the spatially and temporally controlled generation of Ran.GTP in mitosis regulates events from centrosome duplication to bipolar spindle maintenance to reformation of the nuclear envelope post-mitosis. Although this helps to explain the pleiotropic effects of interfering with Ran function in mitosis, the many targets of the pathway hinder a detailed understanding of the molecular basis of these phenotypes. As such, we still do not have a complete understanding of how this signal coordinates a cellular response. Indeed, several important questions remain unresolved. First, is a Ran.GTP gradient essential for robust spindle formation or maintenance? Clearly levels of the RanGEF RCC1 may be greatly reduced without affecting spindle assembly, suggesting that different systems may tolerate a reduction in Ran.GTP to a greater or lesser degree and further supporting the notion that the robustness of mitotic spindle formation is achieved through the presence of multiple functional “modules,” whose input is tuned in each individual cell type and organism (Duncan and Wakefield, 2011). However, without RCC1 null mutants in multiple species, it is difficult to be absolutely clear about the universality of this pathway. Second, do most or all SAFs retain functionality in the interphase nucleus? As SAFs are translocated to the nucleus during interphase, the high concentration of Ran.GTP in the nucleus should result in their release from Importin complexes. Therefore, unless additional regulatory events such as mitotic phosphorylation are required, these SAFs ought to be active in the nucleus outside of mitosis. In support of this, several SAFs have, indeed, been shown to function in the nucleus during interphase (Jasin, 2002; Jaiswal and Narayan, 2008; Jiang et al., 2009; Reichert et al., 2011; Ohata et al., 2013; O'shaughnessy and Hendrich, 2013; Aydin et al., 2014; Neumayer et al., 2014; Vidi et al., 2014; Ha et al., 2015). Moreover, removal of NLSs from SAFs can result in interphase MT abnormalities (e.g., Chen et al., 2015). Such sequestration of already-active proteins in the nucleus prior to mitosis may therefore provide a way for cells to generate MTs quickly as soon as NEB occurs—a notion supported by the observation that dHURP immediately and preferentially localizes to those centrosomally-nucleated MTs facing the nucleus within the nucleus upon mitotic entry (Hayward et al., 2014). Thirdly, the ability of different Importins to sequester different SAFs and localize differentially suggests a complex and co-ordinated regulation of Ran.GTP in mitosis. Moreover, as a final curve-ball, it appears that Ran may not be the only GTPase whose activity releases SAFs. Very recently, it has been shown that, in humans, the RCC1-like protein, TD60/RCC2, acts as a GEF for the GTPase RalA. Loss of TD60, or expression of dominant-negative Ral into cells, results in multiple spindle defects (Papini et al., 2015).
In summary, the list of GTPase-dependent SAFs and their specific roles in mitosis, continues to grow. There are, therefore, many future avenues to explore, in order to fully appreciate the coordinated response of the cell to the dynamic Importin-SAF-Ran relationship. Drosophila, with its many experimental advantages, may yet provide one of the best systems in which to drive forward our understanding.
The article was researched and written by JC, edited by AB and JW and revised by all authors. Figures were produced by AB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
JC was funded by the BBSRC grant BB/K017837/1 to JW.
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Ahmed, Y., Nouri, A., and Wieschaus, E. (2002). Drosophila Apc1 and Apc2 regulate Wingless transduction throughout development. Development 129, 1751–1762.
Albee, A. J., Tao, W., and Wiese, C. (2006). Phosphorylation of maskin by Aurora-A is regulated by RanGTP and importin beta. J. Biol. Chem. 281, 38293–38301. doi: 10.1074/jbc.M607203200
Andersen, D. S., Raja, S. J., Colombani, J., Shaw, R. L., Langton, P. F., Akhtar, A., et al. (2010). Drosophila MCRS2 associates with RNA polymerase II complexes to regulate transcription. Mol. Cell. Biol. 30, 4744–4755. doi: 10.1128/MCB.01586-09
Antonio, C., Ferby, I., Wilhelm, H., Jones, M., Karsenti, E., Nebreda, A. R., et al. (2000). Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102, 425–435. doi: 10.1016/S0092-8674(00)00048-9
Aydin, Ö. Z., Vermeulen, W., and Lans, H. (2014). ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle 13, 3016–3025. doi: 10.4161/15384101.2014.956551
Bahmanyar, S., Nelson, W. J., and Barth, A. I. (2009). Role of APC and its binding partners in regulating microtubules in mitosis. Adv. Exp. Med. Biol. 656, 65–74. doi: 10.1007/978-1-4419-1145-2_6
Bamba, C., Bobinnec, Y., Fukuda, M., and Nishida, E. (2002). The GTPase Ran regulates chromosome positioning and nuclear envelope assembly in vivo. Curr. Biol. 12, 503–507. doi: 10.1016/S0960-9822(02)00741-8
Banks, J. D., and Heald, R. (2004). Adenomatous polyposis coli associates with the microtubule-destabilizing protein XMCAK. Curr. Biol. 14, 2033–2038. doi: 10.1016/j.cub.2004.10.049
Barisic, M., and Geley, S. (2011). Spindly switch controls anaphase: spindly and RZZ functions in chromosome attachment and mitotic checkpoint control. Cell Cycle 10, 449–456. doi: 10.4161/cc.10.3.14759
Barros, T. P., Kinoshita, K., Hyman, A. A., and Raff, J. W. (2005). Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J. Cell Biol. 170, 1039–1046. doi: 10.1083/jcb.200504097
Bayliss, R., Fry, A., Haq, T., and Yeoh, S. (2012). On the molecular mechanisms of mitotic kinase activation. Open Biol. 2:120136. doi: 10.1098/rsob.120136
Belgareh, N., Rabut, G., Baï, S. W., van Overbeek, M., Beaudouin, J., Daigle, N., et al. (2001). An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154, 1147–1160. doi: 10.1083/jcb.200101081
Benoit, B., He, C. H., Zhang, F., Votruba, S. M., Tadros, W., Westwood, J. T., et al. (2009). An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 136, 923–932. doi: 10.1242/dev.031815
Bergstralh, D. T., and St Johnston, D. (2014). Spindle orientation: what if it goes wrong? Semin. Cell Dev. Biol. 34, 140–145. doi: 10.1016/j.semcdb.2014.06.014
Bernis, C., Swift-Taylor, B., Nord, M., Carmona, S., Chook, Y. M., and Forbes, D. J. (2014). Transportin acts to regulate mitotic assembly events by target binding rather than Ran sequestration. Mol. Biol. Cell 25, 992–1009. doi: 10.1091/mbc.E13-08-0506
Bird, S. L., Heald, R., and Weis, K. (2013). RanGTP and CLASP1 cooperate to position the mitotic spindle. Mol. Biol. Cell 24, 2506–2514. doi: 10.1091/mbc.E13-03-0150
Bischoff, F. R., and Görlich, D. (1997). RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 419, 249–254. doi: 10.1016/S0014-5793(97)01467-1
Bischoff, F. R., Klebe, C., Kretschmer, J., Wittinghofer, A., and Ponstingl, H. (1994). RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. U.S.A. 91, 2587–2591. doi: 10.1073/pnas.91.7.2587
Bischoff, F. R., and Ponstingl, H. (1991). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80–82. doi: 10.1038/354080a0
Blower, M. D., Nachury, M., Heald, R., and Weis, K. (2005). A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121, 223–234. doi: 10.1016/j.cell.2005.02.016
Booth, D. G., Hood, F. E., Prior, I. A., and Royle, S. J. (2011). A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 30, 906–919. doi: 10.1038/emboj.2011.15
Bowman, S. K., Neumüller, R. A., Novatchkova, M., Du, Q., and Knoblich, J. A. (2006). The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 10, 731–742. doi: 10.1016/j.devcel.2006.05.005
Brunet, S., Sardon, T., Zimmerman, T., Wittmann, T., Pepperkok, R., Karsenti, E., et al. (2004). Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell 15, 5318–5328. doi: 10.1091/mbc.E04-05-0385
Brust-Mascher, I., and Scholey, J. M. (2009). Microinjection techniques for studying mitosis in the Drosophila melanogaster syncytial embryo. J. Vis. Exp. 15:e1382. doi: 10.3791/1382
Bucciarelli, E., Pellacani, C., Naim, V., Palena, A., Gatti, M., and Somma, M. P. (2009). Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and gamma-tubulin to promote kinetochore-driven MT formation. Curr. Biol. 19, 1839–1845. doi: 10.1016/j.cub.2009.09.043
Buttrick, G. J., Beaumont, L. M., Leitch, J., Yau, C., Hughes, J. R., and Wakefield, J. G. (2008). Akt regulates centrosome migration and spindle orientation in the early Drosophila melanogaster embryo. J. Cell Biol. 180, 537–548. doi: 10.1083/jcb.200705085
Cai, S., Weaver, L. N., Ems-McClung, S. C., and Walczak, C. E. (2009). Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol. Biol. Cell 20, 1348–1359. doi: 10.1091/mbc.E08-09-0971
Cao, Y. K., Zhong, Z. S., Chen, D. Y., Zhang, G. X., Schatten, H., and Sun, Q. Y. (2005). Cell cycle-dependent localization and possible roles of the small GTPase Ran in mouse oocyte maturation, fertilization and early cleavage. Reproduction 130, 431–440. doi: 10.1530/rep.1.00391
Carazo-Salas, R. E., Guarguaglini, G., Gruss, O. J., Segref, A., Karsenti, E., and Mattaj, I. W. (1999). Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181. doi: 10.1038/22133
Cautain, B., Hill, R., de Pedro, N., and Link, W. (2015). Components and regulation of nuclear transport processes. FEBS J. 282, 445–462. doi: 10.1111/febs.13163
Cesario, J., and McKim, K. S. (2011). RanGTP is required for meiotic spindle organization and the initiation of embryonic development in Drosophila. J. Cell Sci. 124, 3797–3810. doi: 10.1242/jcs.084855
Cesario, J. M., Jang, J. K., Redding, B., Shah, N., Rahman, T., and McKim, K. S. (2006). Kinesin 6 family member Subito participates in mitotic spindle assembly and interacts with mitotic regulators. J. Cell Sci. 119, 4770–4780. doi: 10.1242/jcs.03235
Chakravarti, A., Zhai, G. G., Zhang, M., Malhotra, R., Latham, D. E., Delaney, M. A., et al. (2004). Survivin enhances radiation resistance in primary human glioblastoma cells via caspase-independent mechanisms. Oncogene 23, 7494–7506. doi: 10.1038/sj.onc.1208049
Chen, A., Akhshi, T. K., Lavoie, B. D., and Wilde, A. (2015). Importin b2 mediates the spatio-temporal regulation of anillin through a noncanonical nuclear localization signal. J. Biol. Chem. 290, 13500–13509. doi: 10.1074/jbc.M115.649160
Chen, T., Muratore, T. L., Schaner-Tooley, C. E., Shabanowitz, J., Hunt, D. F., and Macara, I. G. (2007). N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol. 9, 596–603. doi: 10.1038/ncb1572
Chook, Y., and Blobel, G. (2001). Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715. doi: 10.1016/S0959-440X(01)00264-0
Chook, Y. M., and Süel, K. E. (2011). Nuclear import by karyopherin-betas: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606. doi: 10.1016/j.bbamcr.2010.10.014
Conduit, P. T., Hayward, D., and Wakefield, J. G. (2015). Microinjection techniques for studying centrosome function in Drosophila melanogaster syncytial embryos. Methods Cell Biol. 129, 229–249. doi: 10.1016/bs.mcb.2015.03.007
Cross, R. A., and McAinsh, A. (2014). Prime movers: the mechanochemistry of mitotic kinesins. Nat. Rev. Mol. Cell Biol. 15, 257–271. doi: 10.1038/nrm3768
Cui, W., and Hawley, R. S. (2005). The HhH2/NDD domain of the Drosophila Nod chromokinesin-like protein is required for binding to chromosomes in the oocyte nucleus. Genetics 171, 1823–1835. doi: 10.1534/genetics.105.047464
Cui, W., Sproul, L. R., Gustafson, S. M., Matthies, H. J., Gilbert, S. P., and Hawley, R. S. (2005). Drosophila Nod protein binds preferentially to the plus ends of microtubules and promotes microtubule polymerization in vitro. Mol. Biol. Cell 16, 5400–5409. doi: 10.1091/mbc.E05-06-0582
De Renzis, S., Elemento, O., Tavazoie, S., and Wieschaus, E. F. (2007). Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5:e117. doi: 10.1371/journal.pbio.0050117
Deka, J., Kuhlmann, J., and Müller, O. (1998). A domain within the tumor suppressor protein APC shows very similar biochemical properties as the microtubule-associated protein tau. Eur. J. Biochem. 253, 591–597. doi: 10.1046/j.1432-1327.1998.2530591.x
Dikovskaya, D., Li, Z., Newton, I. P., Davidson, I., Hutchins, J. R., Kalab, P., et al. (2010). Microtubule assembly by the Apc protein is regulated by importin-beta—RanGTP. J. Cell Sci. 123, 736–746. doi: 10.1242/jcs.060806
Dikovskaya, D., Newton, I. P., and Näthke, I. S. (2004). The adenomatous polyposis coli protein is required for the formation of robust spindles formed in CSF Xenopus extracts. Mol. Biol. Cell 15, 2978–2991. doi: 10.1091/mbc.E03-08-0613
do Carmo Avides, M., Tavares, A., and Glover, D. M. (2001). Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat. Cell Biol. 3, 421–424. doi: 10.1038/35070110
Dobbelaere, J., Josué, F., Suijkerbuijk, S., Baum, B., Tapon, N., and Raff, J. (2008). A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6:e224. doi: 10.1371/journal.pbio.0060224
Dodson, C. A., and Bayliss, R. (2012). Activation of Aurora-A kinase by protein partner binding and phosphorylation are independent and synergistic. J. Biol. Chem. 287, 1150–1157. doi: 10.1074/jbc.M111.312090
Du, Q., Stukenberg, P. T., and Macara, I. G. (2001). A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 3, 1069–1075. doi: 10.1038/ncb1201-1069
Duncan, T., and Wakefield, J. G. (2011). 50 ways to build a spindle: the complexity of microtubule generation during mitosis. Chromosome Res. 19, 321–333. doi: 10.1007/s10577-011-9205-8
Ems-McClung, S. C., Zheng, Y., and Walczak, C. E. (2004). Importin alpha/beta and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell 15, 46–57. doi: 10.1091/mbc.E03-07-0454
England, J. R., Huang, J., Jennings, M. J., Makde, R. D., and Tan, S. (2010). RCC1 uses a conformationally diverse loop region to interact with the nucleosome: a model for the RCC1-nucleosome complex. J. Mol. Biol. 398, 518–529. doi: 10.1016/j.jmb.2010.03.037
Fan, S., Fogg, V., Wang, Q., Chen, X. W., Liu, C. J., and Margolis, B. (2007). A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J. Cell Biol. 178, 387–398. doi: 10.1083/jcb.200609096
Feng, Y., Yuan, J. H., Maloid, S. C., Fisher, R., Copeland, T. D., Longo, D. L., et al. (2006). Polo-like kinase 1-mediated phosphorylation of the GTP-binding protein Ran is important for bipolar spindle formation. Biochem. Biophys. Res. Commun. 349, 144–152. doi: 10.1016/j.bbrc.2006.08.028
Floer, M., Blobel, G., and Rexach, M. (1997). Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J. Biol. Chem. 272, 19538–19546. doi: 10.1074/jbc.272.31.19538
Foe, V. E., and Alberts, B. M. (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31–70.
Fornerod, M., and Ohno, M. (2002). Exportin-mediated nuclear export of proteins and ribonucleoproteins. Results Probl. Cell Differ. 35, 67–91. doi: 10.1007/978-3-540-44603-3_4
Fox, J. C., Howard, A. E., Currie, J. D., Rogers, S. L., and Slep, K. C. (2014). The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol. Biol. Cell 25, 2375–2392. doi: 10.1091/mbc.E13-08-0501
Franck, N., Montembault, E., Romé, P., Pascal, A., Cremet, J. Y., and Giet, R. (2011). CDK11(p58) is required for centriole duplication and Plk4 recruitment to mitotic centrosomes. PLoS ONE 6:e14600. doi: 10.1371/journal.pone.0014600
Franz, C., Walczak, R., Yavuz, S., Santarella, R., Gentzel, M., Askjaer, P., et al. (2007). MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 8, 165–172. doi: 10.1038/sj.embor.7400889
Fu, J., Bian, M., Xin, G., Deng, Z., Luo, J., Guo, X., et al. (2015). TPX2 phosphorylation maintains metaphase spindle length by regulating microtubule flux. J. Cell Biol. 210, 373–383. doi: 10.1083/jcb.201412109
Fu, W., Tao, W., Zheng, P., Fu, J., Bian, M., Jiang, Q., et al. (2010). Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J. Cell Sci. 123, 3645–3651. doi: 10.1242/jcs.075911
Fuchs, J. P., Giloh, H., Kuo, C. H., Saumweber, H., and Sedat, J. (1983). Nuclear structure: determination of the fate of the nuclear envelope in Drosophila during mitosis using monoclonal antibodies. J. Cell Sci. 64, 331–349.
Funabiki, H., and Murray, A. W. (2000). The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102, 411–424. doi: 10.1016/S0092-8674(00)00047-7
Gaglio, T., Saredi, A., and Compton, D. A. (1995). NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 131, 693–708. doi: 10.1083/jcb.131.3.693
Galy, V., Askjaer, P., Franz, C., Lopez-Iglesias, C., and Mattaj, I. W. (2006). MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol 16, 1748–1756. doi: 10.1016/j.cub.2006.06.067
Gergely, F., Karlsson, C., Still, I., Cowell, J., Kilmartin, J., and Raff, J. W. (2000a). The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. U.S.A. 97, 14352–14357. doi: 10.1073/pnas.97.26.14352
Gergely, F., Kidd, D., Jeffers, K., Wakefield, J. G., and Raff, J. W. (2000b). D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 19, 241–252. doi: 10.1093/emboj/19.2.241
Giunta, K. L., Jang, J. K., Manheim, E. A., Subramanian, G., and McKim, K. S. (2002). subito encodes a kinesin-like protein required for meiotic spindle pole formation in Drosophila melanogaster. Genetics 160, 1489–1501.
Goldfarb, D. S., Corbett, A. H., Mason, D. A., Harreman, M. T., and Adam, S. A. (2004). Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514. doi: 10.1016/j.tcb.2004.07.016
Goshima, G. (2011). Identification of a TPX2-like microtubule-associated protein in Drosophila. PLoS ONE 6:e28120. doi: 10.1371/journal.pone.0028120
Goshima, G., Mayer, M., Zhang, N., Stuurman, N., and Vale, R. D. (2008). Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421–429. doi: 10.1083/jcb.200711053
Goshima, G., and Vale, R. D. (2005). Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell 16, 3896–3907. doi: 10.1091/mbc.E05-02-0118
Goshima, G., Wollman, R., Goodwin, S. S., Zhang, N., Scholey, J. M., Vale, R. D., et al. (2007). Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316, 417–421. doi: 10.1126/science.1141314
Green, R. A., Wollman, R., and Kaplan, K. B. (2005). APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol. Biol. Cell 16, 4609–4622. doi: 10.1091/mbc.E05-03-0259
Gregory, S. L., Shandala, T., O'Keefe, L., Jones, L., Murray, M. J., and Saint, R. (2007). A Drosophila overexpression screen for modifiers of Rho signalling in cytokinesis. Fly (Austin.) 1, 13–22. doi: 10.4161/fly.3806
Groen, A. C., Cameron, L. A., Coughlin, M., Miyamoto, D. T., Mitchison, T. J., and Ohi, R. (2004). XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol. 14, 1801–1811. doi: 10.1016/j.cub.2004.10.002
Gruneberg, U., Neef, R., Li, X., Chan, E. H., Chalamalasetty, R. B., Nigg, E. A., et al. (2006). KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 172, 363–372. doi: 10.1083/jcb.200511061
Gruss, O. J., Carazo-Salas, R. E., Schatz, C. A., Guarguaglini, G., Kast, J., Wilm, M., et al. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83–93. doi: 10.1016/S0092-8674(01)00193-3
Gutiérrez-Caballero, C., Burgess, S. G., Bayliss, R., and Royle, S. J. (2015). TACC3-ch-TOG track the growing tips of microtubules independently of clathrin and Aurora-A phosphorylation. Biol. Open 4, 170–179. doi: 10.1242/bio.201410843
Ha, G. H., Kim, J. L., Petersson, A., Oh, S., Denning, M. F., Patel, T., et al. (2015). TACC3 deregulates the DNA damage response and confers sensitivity to radiation and PARP inhibition. Oncogene 34, 1667–1678. doi: 10.1038/onc.2014.105
Halpin, D., Kalab, P., Wang, J., Weis, K., and Heald, R. (2011). Mitotic spindle assembly around RCC1-coated beads in Xenopus egg extracts. PLoS Biol. 9:e1001225. doi: 10.1371/journal.pbio.1001225
Harel, A., Chan, R. C., Lachish-Zalait, A., Zimmerman, E., Elbaum, M., and Forbes, D. J. (2003). Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell 14, 4387–4396. doi: 10.1091/mbc.E03-05-0275
Harel, A., Zlotkin, E., Nainudel-Epszteyn, S., Feinstein, N., Fisher, P. A., and Gruenbaum, Y. (1989). Persistence of major nuclear envelope antigens in an envelope-like structure during mitosis in Drosophila melanogaster embryos. J. Cell Sci. 94(Pt 3), 463–470.
Hashizume, C., Kobayashi, A., and Wong, R. W. (2013). Down-modulation of nucleoporin RanBP2/Nup358 impaired chromosomal alignment and induced mitotic catastrophe. Cell Death Dis. 4, e854. doi: 10.1038/cddis.2013.370
Hayward, D., Metz, J., Pellacani, C., and Wakefield, J. G. (2014). Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev. Cell 28, 81–93. doi: 10.1016/j.devcel.2013.12.001
Hayward, D., and Wakefield, J. G. (2014). Chromatin-mediated microtubule nucleation in Drosophila syncytial embryos. Commun. Integr. Biol. 7:e28512. doi: 10.4161/cib.28512
Heald, R., Tournebize, R., Blank, T., Sandaltzopoulos, R., Becker, P., Hyman, A., et al. (1996). Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382, 420–425. doi: 10.1038/382420a0
Helmke, K. J., and Heald, R. (2014). TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol. 206, 385–393. doi: 10.1083/jcb.201401014
Hetzer, M., Bilbao-Cortés, D., Walther, T. C., Gruss, O. J., and Mattaj, I. W. (2000). GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell 5, 1013–1024. doi: 10.1016/S1097-2765(00)80266-X
Hetzer, M. W. (2010). The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2:a000539. doi: 10.1101/cshperspect.a000539
Hirota, T., Kunitoku, N., Sasayama, T., Marumoto, T., Zhang, D., Nitta, M., et al. (2003). Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114, 585–598. doi: 10.1016/S0092-8674(03)00642-1
Hu, D., Valentine, M., Kidd, V. J., and Lahti, J. M. (2007). CDK11(p58) is required for the maintenance of sister chromatid cohesion. J. Cell Sci. 120, 2424–2434. doi: 10.1242/jcs.007963
Hubner, N. C., Bird, A. W., Cox, J., Splettstoesser, B., Bandilla, P., Poser, I., et al. (2010). Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754. doi: 10.1083/jcb.200911091
Hughes, J. R., Meireles, A. M., Fisher, K. H., Garcia, A., Antrobus, P. R., Wainman, A., et al. (2008). A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol. 6:e98. doi: 10.1371/journal.pbio.0060098
Hutchins, J. R., Moore, W. J., and Clarke, P. R. (2009). Dynamic localisation of Ran GTPase during the cell cycle. BMC Cell Biol. 10:66. doi: 10.1186/1471-2121-10-66
Hutchins, J. R., Moore, W. J., Hood, F. E., Wilson, J. S., Andrews, P. D., Swedlow, J. R., et al. (2004). Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr. Biol. 14, 1099–1104. doi: 10.1016/j.cub.2004.05.021
Hutterer, A., Berdnik, D., Wirtz-Peitz, F., Zigman, M., Schleiffer, A., and Knoblich, J. A. (2006). Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev. Cell 11, 147–157. doi: 10.1016/j.devcel.2006.06.002
Hwang, H. I., Ji, J. H., and Jang, Y. J. (2011). Phosphorylation of Ran-binding protein-1 by Polo-like kinase-1 is required for interaction with Ran and early mitotic progression. J. Biol. Chem. 286, 33012–33020. doi: 10.1074/jbc.M111.255620
Izumi, Y., Ohta, N., Hisata, K., Raabe, T., and Matsuzaki, F. (2006). Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8, 586–593. doi: 10.1038/ncb1409
Jaiswal, A. S., and Narayan, S. (2008). A novel function of adenomatous polyposis coli (APC) in regulating DNA repair. Cancer Lett. 271, 272–280. doi: 10.1016/j.canlet.2008.06.024
Jasin, M. (2002). Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 21, 8981–8993. doi: 10.1038/sj.onc.1206176
Jiang, G., Ren, B., Xu, L., Song, S., Zhu, C., and Ye, F. (2009). Survivin may enhance DNA double-strand break repair capability by up-regulating Ku70 in human KB cells. Anticancer Res. 29, 223–228.
Jiang, H., Wang, S., Huang, Y., He, X., Cui, H., Zhu, X., et al. (2015). Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 163, 108–122. doi: 10.1016/j.cell.2015.08.010
Johansen, J., and Johansen, K. M. (2009). The spindle matrix through the cell cycle in Drosophila. Fly (Austin.) 3, 213–220. doi: 10.4161/fly.3.3.9340
Johansen, K. M., and Johansen, J. (2007). Cell and molecular biology of the spindle matrix. Int. Rev. Cytol. 263, 155–206. doi: 10.1016/S0074-7696(07)63004-6
Joseph, J., Tan, S. H., Karpova, T. S., McNally, J. G., and Dasso, M. (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156, 595–602. doi: 10.1083/jcb.200110109
Joukov, V., Groen, A. C., Prokhorova, T., Gerson, R., White, E., Rodriguez, A., et al. (2006). The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 127, 539–552. doi: 10.1016/j.cell.2006.08.053
Kalab, P., and Heald, R. (2008). The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 121, 1577–1586. doi: 10.1242/jcs.005959
Kaláb, P., Pralle, A., Isacoff, E. Y., Heald, R., and Weis, K. (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701. doi: 10.1038/nature04589
Kalab, P., Pu, R. T., and Dasso, M. (1999). The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9, 481–484. doi: 10.1016/S0960-9822(99)80213-9
Katsani, K. R., Karess, R. E., Dostatni, N., and Doye, V. (2008). In vivo dynamics of Drosophila nuclear envelope components. Mol. Biol. Cell 19, 3652–3666. doi: 10.1091/mbc.E07-11-1162
Kelly, A. E., Sampath, S. C., Maniar, T. A., Woo, E. M., Chait, B. T., and Funabiki, H. (2007). Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell 12, 31–43. doi: 10.1016/j.devcel.2006.11.001
Keryer, G., Di Fiore, B., Celati, C., Lechtreck, K. F., Mogensen, M., Delouvee, A., et al. (2003). Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol. Biol. Cell 14, 4260–4271. doi: 10.1091/mbc.E02-11-0773
Kinoshita, K., Noetzel, T. L., Pelletier, L., Mechtler, K., Drechsel, D. N., Schwager, A., et al. (2005). Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 170, 1047–1055. doi: 10.1083/jcb.200503023
Kiyomitsu, T., and Cheeseman, I. M. (2012). Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14, 311–317. doi: 10.1038/ncb2440
Kiyomitsu, T., and Cheeseman, I. M. (2013). Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 154, 391–402. doi: 10.1016/j.cell.2013.06.010
Klebe, C., Bischoff, F. R., Ponstingl, H., and Wittinghofer, A. (1995). Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34, 639–647. doi: 10.1021/bi00002a031
Koffa, M. D., Casanova, C. M., Santarella, R., Köcher, T., Wilm, M., and Mattaj, I. W. (2006). HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 16, 743–754. doi: 10.1016/j.cub.2006.03.056
Konno, D., Shioi, G., Shitamukai, A., Mori, A., Kiyonari, H., Miyata, T., et al. (2008). Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 10, 93–101. doi: 10.1038/ncb1673
Kose, S., Imamoto, N., Tachibana, T., Yoshida, M., and Yoneda, Y. (1999). Beta-subunit of nuclear pore-targeting complex (importin-beta) can be exported from the nucleus in a Ran-independent manner. J. Biol. Chem. 274, 3946–3952. doi: 10.1074/jbc.274.7.3946
Kotak, S., Busso, C., and Gönczy, P. (2012). Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199, 97–110. doi: 10.1083/jcb.201203166
Kotian, S., Banerjee, T., Lockhart, A., Huang, K., Catalyurek, U. V., and Parvin, J. D. (2014). NUSAP1 influences the DNA damage response by controlling BRCA1 protein levels. Cancer Biol. Ther. 15, 533–543. doi: 10.4161/cbt.28019
Kuersten, S., Arts, G. J., Walther, T. C., Englmeier, L., and Mattaj, I. W. (2002). Steady-state nuclear localization of exportin-t involves RanGTP binding and two distinct nuclear pore complex interaction domains. Mol. Cell. Biol. 22, 5708–5720. doi: 10.1128/MCB.22.16.5708-5720.2002
Kutay, U., Bischoff, F. R., Kostka, S., Kraft, R., and Görlich, D. (1997). Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061–1071. doi: 10.1016/S0092-8674(00)80372-4
Lau, C. K., Delmar, V. A., Chan, R. C., Phung, Q., Bernis, C., Fichtman, B., et al. (2009). Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol. Biol. Cell 20, 4043–4058. doi: 10.1091/mbc.E09-02-0152
Lawo, S., Bashkurov, M., Mullin, M., Ferreria, M. G., Kittler, R., Habermann, B., et al. (2009). HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19, 816–826. doi: 10.1016/j.cub.2009.04.033
Lécuyer, E., Yoshida, H., Parthasarathy, N., Alm, C., Babak, T., Cerovina, T., et al. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187. doi: 10.1016/j.cell.2007.08.003
Lerit, D. A., Jordan, H. A., Poulton, J. S., Fagerstrom, C. J., Galletta, B. J., Peifer, M., et al. (2015). Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J. Cell Biol. 210, 79–97. doi: 10.1083/jcb.201503117
Levesque, A. A., and Compton, D. A. (2001). The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154, 1135–1146. doi: 10.1083/jcb.200106093
Li, H. Y., Wirtz, D., and Zheng, Y. (2003). A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 160, 635–644. doi: 10.1083/jcb.200211004
Li, H. Y., and Zheng, Y. (2004). Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 18, 512–527. doi: 10.1101/gad.1177304
Li, L., Zhou, Y., Sun, L., Xing, G., Tian, C., Sun, J., et al. (2007). NuSAP is degraded by APC/C-Cdh1 and its overexpression results in mitotic arrest dependent of its microtubules' affinity. Cell. Signal. 19, 2046–2055. doi: 10.1016/j.cellsig.2007.05.017
Lin, C. H., Hu, C. K., and Shih, H. M. (2010). Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J. Cell Biol. 189, 1097–1105. doi: 10.1083/jcb.200911120
Lince-Faria, M., Maffini, S., Orr, B., Ding, Y., Cláudia, F., Sunkel, C. E., et al. (2009). Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184, 647–657. doi: 10.1083/jcb.200811012
Loïodice, I., Alves, A., Rabut, G., Van Overbeek, M., Ellenberg, J., Sibarita, J. B., et al. (2004). The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell 15, 3333–3344. doi: 10.1091/mbc.E03-12-0878
Lounsbury, K. M., and Macara, I. G. (1997). Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin β and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin β. J. Biol. Chem. 272, 551–555. doi: 10.1074/jbc.272.1.551
Lu, B., Roegiers, F., Jan, L. Y., and Jan, Y. N. (2001). Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature 409, 522–525. doi: 10.1038/35054077
Lui, C., Mills, K., Brocardo, M. G., Sharma, M., and Henderson, B. R. (2012). APC as a mobile scaffold: regulation and function at the nucleus, centrosomes, and mitochondria. IUBMB Life 64, 209–214. doi: 10.1002/iub.599
Ma, H. T., and Poon, R. Y. (2011). How protein kinases co-ordinate mitosis in animal cells. Biochem. J. 435, 17–31. doi: 10.1042/BJ20100284
Ma, N., Titus, J., Gable, A., Ross, J. L., and Wadsworth, P. (2011). TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J. Cell Biol. 195, 87–98. doi: 10.1083/jcb.201106149
Makde, R. D., England, J. R., Yennawar, H. P., and Tan, S. (2010). Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566. doi: 10.1038/nature09321
Maresca, T. J., Niederstrasser, H., Weis, K., and Heald, R. (2005). Xnf7 contributes to spindle integrity through its microtubule-bundling activity. Curr. Biol. 15, 1755–1761. doi: 10.1016/j.cub.2005.08.049
Martinez-Campos, M., Basto, R., Baker, J., Kernan, M., and Raff, J. W. (2004). The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165, 673–683. doi: 10.1083/jcb.200402130
Mason, D. A., Stage, D. E., and Goldfarb, D. S. (2009). Evolution of the metazoan-specific importin alpha gene family. J. Mol. Evol. 68, 351–365. doi: 10.1007/s00239-009-9215-8
Mazumdar, A., and Mazumdar, M. (2002). How one becomes many: blastoderm cellularization in Drosophila melanogaster. Bioessays 24, 1012–1022. doi: 10.1002/bies.10184
Mazumdar, M., Sung, M. H., and Misteli, T. (2011). Chromatin maintenance by a molecular motor protein. Nucleus 2, 591–600. doi: 10.4161/nucl.2.6.18044
McCartney, B. M., McEwen, D. G., Grevengoed, E., Maddox, P., Bejsovec, A., and Peifer, M. (2001). Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nat. Cell Biol. 3, 933–938. doi: 10.1038/ncb1001-933
Merdes, A., Heald, R., Samejima, K., Earnshaw, W. C., and Cleveland, D. W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851–862. doi: 10.1083/jcb.149.4.851
Merdes, A., Ramyar, K., Vechio, J. D., and Cleveland, D. W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447–458. doi: 10.1016/S0092-8674(00)81365-3
Meunier, S., and Vernos, I. (2011). K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat. Cell Biol. 13, 1406–1414. doi: 10.1038/ncb2372
Mishra, R. K., Chakraborty, P., Arnaoutov, A., Fontoura, B. M., and Dasso, M. (2010). The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12, 164–169. doi: 10.1038/ncb2016
Molina, I., Baars, S., Brill, J. A., Hales, K. G., Fuller, M. T., and Ripoll, P. (1997). A chromatin-associated kinesin-related protein required for normal mitotic chromosome segregation in Drosophila. J. Cell Biol. 139, 1361–1371. doi: 10.1083/jcb.139.6.1361
Morin, X., and Bellaiche, Y. (2011). Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 21, 102–119. doi: 10.1016/j.devcel.2011.06.012
Morin, X., Jaouen, F., and Durbec, P. (2007). Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat. Neurosci. 10, 1440–1448. doi: 10.1038/nn1984
Morris, E. J., Nader, G. P., Ramalingam, N., Bartolini, F., and Gundersen, G. G. (2014). Kif4 interacts with EB1 and stabilizes microtubules downstream of Rho-mDia in migrating fibroblasts. PLoS ONE 9:e91568. doi: 10.1371/journal.pone.0091568
Mosammaparast, N., and Pemberton, L. F. (2004). Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556. doi: 10.1016/j.tcb.2004.09.004
Moutinho-Pereira, S., Stuurman, N., Afonso, O., Hornsveld, M., Aguiar, P., Goshima, G., et al. (2013). Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc. Natl. Acad. Sci. U.S.A. 110, 19808–19813. doi: 10.1073/pnas.1320013110
Müller, H., Schmidt, D., Steinbrink, S., Mirgorodskaya, E., Lehmann, V., Habermann, K., et al. (2010). Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 29, 3344–3357. doi: 10.1038/emboj.2010.210
Nemergut, M. E., and Macara, I. G. (2000). Nuclear import of the ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol. 149, 835–850. doi: 10.1083/jcb.149.4.835
Nemergut, M. E., Mizzen, C. A., Stukenberg, T., Allis, C. D., and Macara, I. G. (2001). Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292, 1540–1543. doi: 10.1126/science.292.5521.1540
Neumayer, G., Belzil, C., Gruss, O. J., and Nguyen, M. D. (2014). TPX2: of spindle assembly, DNA damage response, and cancer. Cell. Mol. Life Sci. 71, 3027–3047. doi: 10.1007/s00018-014-1582-7
Neumayer, G., and Nguyen, M. D. (2014). TPX2 impacts acetylation of histone H4 at lysine 16: implications for DNA damage response. PLoS ONE 9:e110994. doi: 10.1371/journal.pone.0110994
Nishimura, T., Takahashi, M., Kim, H. S., Mukai, H., and Ono, Y. (2005). Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E-cdk2 complex. Genes Cells 10, 75–86. doi: 10.1111/j.1365-2443.2005.00816.x
Ohata, H., Miyazaki, M., Otomo, R., Matsushima-Hibiya, Y., Otsubo, C., Nagase, T., et al. (2013). NuMA is required for the selective induction of p53 target genes. Mol. Cell. Biol. 33, 2447–2457. doi: 10.1128/MCB.01221-12
Ohba, T., Nakamura, M., Nishitani, H., and Nishimoto, T. (1999). Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284, 1356–1358. doi: 10.1126/science.284.5418.1356
Ohtsubo, M., Okazaki, H., and Nishimoto, T. (1989). The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 109, 1389–1397. doi: 10.1083/jcb.109.4.1389
Okuda, M., Horn, H. F., Tarapore, P., Tokuyama, Y., Smulian, A. G., Chan, P. K., et al. (2000). Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103, 127–140. doi: 10.1016/S0092-8674(00)00093-3
O'shaughnessy, A., and Hendrich, B. (2013). CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem. Soc. Trans. 41, 777–782. doi: 10.1042/BST20130027
Palmeri, D., and Malim, M. H. (1999). Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 19, 1218–1225. doi: 10.1128/MCB.19.2.1218
Papini, D., Langemeyer, L., Abad, M. A., Kerr, A., Samejima, I., Eyers, P. A., et al. (2015). TD-60 links RalA GTPase function to the CPC in mitosis. Nat. Commun. 6, 7678. doi: 10.1038/ncomms8678
Petretti, C., Savoian, M., Montembault, E., Glover, D. M., Prigent, C., and Giet, R. (2006). The PITSLRE/CDK11p58 protein kinase promotes centrosome maturation and bipolar spindle formation. EMBO Rep. 7, 418–424. doi: 10.1038/sj.embor.7400639
Petry, S., Groen, A. C., Ishihara, K., Mitchison, T. J., and Vale, R. D. (2013). Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152, 768–777. doi: 10.1016/j.cell.2012.12.044
Peyre, E., Jaouen, F., Saadaoui, M., Haren, L., Merdes, A., Durbec, P., et al. (2011). A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J. Cell Biol. 193, 141–154. doi: 10.1083/jcb.201101039
Pickett-Heaps, J., Spurck, T., and Tippit, D. (1984). Chromosome motion and the spindle matrix. J. Cell Biol. 99, 137s–143s. doi: 10.1083/jcb.99.1.137s
Pocha, S. M., and Knust, E. (2013). Complexities of Crumbs function and regulation in tissue morphogenesis. Curr. Biol. 23, R289–R293. doi: 10.1016/j.cub.2013.03.001
Przewloka, M. R., Zhang, W., Costa, P., Archambault, V., D'Avino, P. P., Lilley, K. S., et al. (2007). Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE 2:e478. doi: 10.1371/journal.pone.0000478
Pumroy, R. A., and Cingolani, G. (2015). Diversification of importin-alpha isoforms in cellular trafficking and disease states. Biochem. J. 466, 13–28. doi: 10.1042/BJ20141186
Quan, Y., Ji, Z. L., Wang, X., Tartakoff, A. M., and Tao, T. (2008). Evolutionary and transcriptional analysis of karyopherin beta superfamily proteins. Mol. Cell. Proteomics 7, 1254–1269. doi: 10.1074/mcp.M700511-MCP200
Raaijmakers, J. A., van Heesbeen, R. G., Meaders, J. L., Geers, E. F., Fernandez-Garcia, B., Medema, R. H., et al. (2012). Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 31, 4179–4190. doi: 10.1038/emboj.2012.272
Radulescu, A. E., and Cleveland, D. W. (2010). NuMA after 30 years: the matrix revisited. Trends Cell Biol. 20, 214–222. doi: 10.1016/j.tcb.2010.01.003
Raemaekers, T., Ribbeck, K., Beaudouin, J., Annaert, W., Van Camp, M., Stockmans, I., et al. (2003). NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 162, 1017–1029. doi: 10.1083/jcb.200302129
Rakkaa, T., Escudé, C., Giet, R., Magnaghi-Jaulin, L., and Jaulin, C. (2014). CDK11(p58) kinase activity is required to protect sister chromatid cohesion at centromeres in mitosis. Chromosome Res. 22, 267–276. doi: 10.1007/s10577-013-9400-x
Rasala, B. A., Orjalo, A. V., Shen, Z., Briggs, S., and Forbes, D. J. (2006). ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. U.S.A. 103, 17801–17806. doi: 10.1073/pnas.0608484103
Reichert, S., Rödel, C., Mirsch, J., Harter, P. N., Tomicic, M. T., Mittelbronn, M., et al. (2011). Survivin inhibition and DNA double-strand break repair: a molecular mechanism to overcome radioresistance in glioblastoma. Radiother. Oncol. 101, 51–58. doi: 10.1016/j.radonc.2011.06.037
Ribbeck, K., Groen, A. C., Santarella, R., Bohnsack, M. T., Raemaekers, T., Kocher, T., et al. (2006). NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell 17, 2646–2660. doi: 10.1091/mbc.E05-12-1178
Ribbeck, K., Raemaekers, T., Carmeliet, G., and Mattaj, I. W. (2007). A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol. 17, 230–236. doi: 10.1016/j.cub.2006.11.050
Richens, J. H., Barros, T. P., Lucas, E. P., Peel, N., Pinto, D. M., Wainman, A., et al. (2015). The Drosophila Pericentrin-like-protein (PLP) cooperates with Cnn to maintain the integrity of the outer PCM. Biol. Open 4, 1052–1061. doi: 10.1242/bio.012914
Robinson, J. T., Wojcik, E. J., Sanders, M. A., McGrail, M., and Hays, T. S. (1999). Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146, 597–608. doi: 10.1083/jcb.146.3.597
Ruden, D. M., Cui, W., Sollars, V., and Alterman, M. (1997). A Drosophila kinesin-like protein, Klp38B, functions during meiosis, mitosis and segmentation. Dev. Biol. 191, 284–296. doi: 10.1006/dbio.1997.8726
Sabino, D., Brown, N. H., and Basto, R. (2011). Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. J. Cell Sci. 124, 1156–1166. doi: 10.1242/jcs.076711
Salina, D., Enarson, P., Rattner, J. B., and Burke, B. (2003). Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162, 991–1001. doi: 10.1083/jcb.200304080
Samwer, M., Dehne, H. J., Spira, F., Kollmar, M., Gerlich, D. W., Urlaub, H., et al. (2013). The nuclear F-actin interactome of Xenopus oocytes reveals an actin-bundling kinesin that is essential for meiotic cytokinesis. EMBO J. 32, 1886–1902. doi: 10.1038/emboj.2013.108
Saunders, R. D., Avides, M. C., Howard, T., Gonzalez, C., and Glover, D. M. (1997). The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 137, 881–890. doi: 10.1083/jcb.137.4.881
Schaar, B. T., Chan, G. K., Maddox, P., Salmon, E. D., and Yen, T. J. (1997). CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139, 1373–1382. doi: 10.1083/jcb.139.6.1373
Seewald, M. J., Kraemer, A., Farkasovsky, M., Körner, C., Wittinghofer, A., and Vetter, I. R. (2003). Biochemical characterization of the Ran-RanBP1-RanGAP system: are RanBP proteins and the acidic tail of RanGAP required for the Ran-RanGAP GTPase reaction? Mol. Cell. Biol. 23, 8124–8136. doi: 10.1128/MCB.23.22.8124-8136.2003
Shinmura, K., Tarapore, P., Tokuyama, Y., George, K. R., and Fukasawa, K. (2005). Characterization of centrosomal association of nucleophosmin/B23 linked to Crm1 activity. FEBS Lett. 579, 6621–6634. doi: 10.1016/j.febslet.2005.10.057
Siderovski, D. P., Diversé-Pierluissi, M., and De Vries, L. (1999). The GoLoco motif: a Galphai/o binding motif and potential guanine-nucleotide exchange factor. Trends Biochem. Sci. 24, 340–341. doi: 10.1016/S0968-0004(99)01441-3
Silk, A. D., Holland, A. J., and Cleveland, D. W. (2009). Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J. Cell Biol. 184, 677–690. doi: 10.1083/jcb.200810091
Siller, K. H., Cabernard, C., and Doe, C. Q. (2006). The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8, 594–600. doi: 10.1038/ncb1412
Silljé, H. H., Nagel, S., Körner, R., and Nigg, E. A. (2006). HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16, 731–742. doi: 10.1016/j.cub.2006.02.070
Silverman-Gavrila, R. V., and Wilde, A. (2006). Ran is required before metaphase for spindle assembly and chromosome alignment and after metaphase for chromosome segregation and spindle midbody organization. Mol. Biol. Cell 17, 2069–2080. doi: 10.1091/mbc.E05-10-0991
Silverman-Gavrila, R. V., Hales, K. G., and Wilde, A. (2008). Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol. Biol. Cell 19, 3735–3744. doi: 10.1091/mbc.E08-01-0049
Stafstrom, J. P., and Staehelin, L. A. (1984). Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo. Eur. J. Cell Biol. 34, 179–189.
Stewart, M. (2007). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208. doi: 10.1038/nrm2114
Sullivan, W., Minden, J. S., and Alberts, B. M. (1990). daughterless-abo-like, a Drosophila maternal-effect mutation that exhibits abnormal centrosome separation during the late blastoderm divisions. Development 110, 311–323.
Sullivan, W., and Theurkauf, W. E. (1995). The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr. Opin. Cell Biol. 7, 18–22. doi: 10.1016/0955-0674(95)80040-9
Syred, H. M., Welburn, J., Rappsilber, J., and Ohkura, H. (2013). Cell cycle regulation of microtubule interactomes: multi-layered regulation is critical for the interphase/mitosis transition. Mol. Cell. Proteomics 12, 3135–3147. doi: 10.1074/mcp.M113.028563
Szymborska, A., de Marco, A., Daigle, N., Cordes, V. C., Briggs, J. A., and Ellenberg, J. (2013). Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658. doi: 10.1126/science.1240672
Tahara, K., Takagi, M., Ohsugi, M., Sone, T., Nishiumi, F., Maeshima, K., et al. (2008). Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J. Cell Biol. 180, 493–506. doi: 10.1083/jcb.200708003
Takahashi, M., Yamagiwa, A., Nishimura, T., Mukai, H., and Ono, Y. (2002). Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell 13, 3235–3245. doi: 10.1091/mbc.E02-02-0112
Talcott, B., and Moore, M. S. (2000). The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin alpha3/Qip. J. Biol. Chem. 275, 10099–10104. doi: 10.1074/jbc.275.14.10099
Taniguchi, E., Toyoshima-Morimoto, F., and Nishida, E. (2002). Nuclear translocation of plk1 mediated by its bipartite nuclear localization signal. J. Biol. Chem. 277, 48884–48888. doi: 10.1074/jbc.M206307200
Tarapore, P., Okuda, M., and Fukasawa, K. (2002). A mammalian in vitro centriole duplication system: evidence for involvement of CDK2/cyclin E and nucleophosmin/B23 in centrosome duplication. Cell Cycle 1, 75–81. doi: 10.4161/cc.1.1.103
Tekotte, H., Berdnik, D., Török, T., Buszczak, M., Jones, L. M., Cooley, L., et al. (2002). Dcas is required for importin-alpha3 nuclear export and mechano-sensory organ cell fate specification in Drosophila. Dev. Biol. 244, 396–406. doi: 10.1006/dbio.2002.0612
Timinszky, G., Tirián, L., Nagy, F. T., Tóth, G., Perczel, A., Kiss-László, Z., et al. (2002). The importin-beta P446L dominant-negative mutant protein loses RanGTP binding ability and blocks the formation of intact nuclear envelope. J. Cell Sci. 115, 1675–1687.
Tirian, L., Timinszky, G., and Szabad, J. (2003). P446L-importin-beta inhibits nuclear envelope assembly by sequestering nuclear envelope assembly factors to the microtubules. Eur. J. Cell Biol. 82, 351–359. doi: 10.1078/0171-9335-00324
Tokuyama, Y., Horn, H. F., Kawamura, K., Tarapore, P., and Fukasawa, K. (2001). Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J. Biol. Chem. 276, 21529–21537. doi: 10.1074/jbc.M100014200
Trieselmann, N., Armstrong, S., Rauw, J., and Wilde, A. (2003). Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci. 116, 4791–4798. doi: 10.1242/jcs.00798
Trieselmann, N., and Wilde, A. (2002). Ran localizes around the microtubule spindle in vivo during mitosis in Drosophila embryos. Curr. Biol. 12, 1124–1129. doi: 10.1016/S0960-9822(02)00934-X
Tsai, M. Y., Wang, S., Heidinger, J. M., Shumaker, D. K., Adam, S. A., Goldman, R. D., et al. (2006). A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science 311, 1887–1893. doi: 10.1126/science.1122771
Uehara, R., Nozawa, R. S., Tomioka, A., Petry, S., Vale, R. D., Obuse, C., et al. (2009). The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. U.S.A. 106, 6998–7003. doi: 10.1073/pnas.0901587106
Vaisberg, E. A., Koonce, M. P., and McIntosh, J. R. (1993). Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 123, 849–858. doi: 10.1083/jcb.123.4.849
Vanden Bosch, A., Raemaekers, T., Denayer, S., Torrekens, S., Smets, N., Moermans, K., et al. (2010). NuSAP is essential for chromatin-induced spindle formation during early embryogenesis. J. Cell Sci. 123, 3244–3255. doi: 10.1242/jcs.063875
Vasu, S., Shah, S., Orjalo, A., Park, M., Fischer, W. H., and Forbes, D. J. (2001). Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155, 339–354. doi: 10.1083/jcb.200108007
Vidi, P. A., Liu, J., Salles, D., Jayaraman, S., Dorfman, G., Gray, M., et al. (2014). NuMA promotes homologous recombination repair by regulating the accumulation of the ISWI ATPase SNF2h at DNA breaks. Nucleic Acids Res. 42, 6365–6379. doi: 10.1093/nar/gku296
Virágh, E., Gorjánácz, M., Török, I., Eichhorn, T., Kallakuri, S., Szlanka, T., et al. (2012). Specific cooperation between Imp-alpha2 and Imp-beta/Ketel in spindle assembly during Drosophila early nuclear divisions. G3 (Bethesda) 2, 1–14. doi: 10.1534/g3.111.001073
Wakefield, J. G., Bonaccorsi, S., and Gatti, M. (2001). The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J. Cell Biol. 153, 637–648. doi: 10.1083/jcb.153.4.637
Walther, T. C., Pickersgill, H. S., Cordes, V. C., Goldberg, M. W., Allen, T. D., Mattaj, I. W., et al. (2002). The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158, 63–77. doi: 10.1083/jcb.200202088
Wang, W., Budhu, A., Forgues, M., and Wang, X. W. (2005). Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 7, 823–830. doi: 10.1038/ncb1282
Weaver, L. N., Ems-McClung, S. C., Chen, S. H., Yang, G., Shaw, S. L., and Walczak, C. E. (2015). The Ran-GTP gradient spatially regulates XCTK2 in the spindle. Curr. Biol. 25, 1509–1514. doi: 10.1016/j.cub.2015.04.015
Wee, B., Johnston, C. A., Prehoda, K. E., and Doe, C. Q. (2011). Canoe binds RanGTP to promote Pins(TPR)/Mud-mediated spindle orientation. J. Cell Biol. 195, 369–376. doi: 10.1083/jcb.201102130
Wickstead, B., Gull, K., and Richards, T. A. (2010). Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol. Biol. 10:110. doi: 10.1186/1471-2148-10-110
Wiese, C., Wilde, A., Moore, M. S., Adam, S. A., Merdes, A., and Zheng, Y. (2001). Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science 291, 653–656. doi: 10.1126/science.1057661
Wilde, A., Lizarraga, S. B., Zhang, L., Wiese, C., Gliksman, N. R., Walczak, C. E., et al. (2001). Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 3, 221–227. doi: 10.1038/35060000
Wilde, A., and Zheng, Y. (1999). Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359–1362. doi: 10.1126/science.284.5418.1359
Wilker, E. W., van Vugt, M. A., Artim, S. A., Huang, P. H., Petersen, C. P., Reinhardt, H. C., et al. (2007). 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature 446, 329–332. doi: 10.1038/nature05584
Williams, B. C., Riedy, M. F., Williams, E. V., Gatti, M., and Goldberg, M. L. (1995). The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 129, 709–723. doi: 10.1083/jcb.129.3.709
Wittmann, T., Boleti, H., Antony, C., Karsenti, E., and Vernos, I. (1998). Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J. Cell Biol. 143, 673–685. doi: 10.1083/jcb.143.3.673
Wong, J., and Fang, G. (2006). HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 173, 879–891. doi: 10.1083/jcb.200511132
Wong, R. W., Blobel, G., and Coutavas, E. (2006). Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. U.S.A. 103, 19783–19787. doi: 10.1073/pnas.0609582104
Wu, J. M., Chen, C. T., Coumar, M. S., Lin, W. H., Chen, Z. J., Hsu, J. T., et al. (2013). Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules. Proc. Natl. Acad. Sci. U.S.A. 110, E1779–E1787. doi: 10.1073/pnas.1220523110
Wu, J., Matunis, M. J., Kraemer, D., Blobel, G., and Coutavas, E. (1995). Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270, 14209–14213. doi: 10.1074/jbc.270.23.14209
Xia, F., Canovas, P. M., Guadagno, T. M., and Altieri, D. C. (2008). A survivin-ran complex regulates spindle formation in tumor cells. Mol. Cell. Biol. 28, 5299–5311. doi: 10.1128/MCB.02039-07
Yang, C. P., and Fan, S. S. (2008). Drosophila mars is required for organizing kinetochore microtubules during mitosis. Exp. Cell Res. 314, 3209–3220. doi: 10.1016/j.yexcr.2008.08.004
Yokoyama, H., Gruss, O. J., Rybina, S., Caudron, M., Schelder, M., Wilm, M., et al. (2008). Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate. J. Cell Biol. 180, 867–875. doi: 10.1083/jcb.200706189
Yokoyama, H., Koch, B., Walczak, R., Ciray-Duygu, F., Gonzalez-Sanchez, J. C., Devos, D. P., et al. (2014). The nucleoporin MEL-28 promotes RanGTP-dependent gamma-tubulin recruitment and microtubule nucleation in mitotic spindle formation. Nat. Commun. 5, 3270. doi: 10.1038/ncomms4270
Yokoyama, H., Nakos, K., Santarella-Mellwig, R., Rybina, S., Krijgsveld, J., Koffa, M. D., et al. (2013). CHD4 is a RanGTP-dependent MAP that stabilizes microtubules and regulates bipolar spindle formation. Curr. Biol. 23, 2443–2451. doi: 10.1016/j.cub.2013.09.062
Yokoyama, H., Rybina, S., Santarella-Mellwig, R., Mattaj, I. W., and Karsenti, E. (2009). ISWI is a RanGTP-dependent MAP required for chromosome segregation. J. Cell Biol. 187, 813–829. doi: 10.1083/jcb.200906020
Yokoyama, N., Hayashi, N., Seki, T., Panté, N., Ohba, T., Nishii, K., et al. (1995). A giant nucleopore protein that binds Ran/TC4. Nature 376, 184–188. doi: 10.1038/376184a0
Yu, C. T., Hsu, J. M., Lee, Y. C., Tsou, A. P., Chou, C. K., and Huang, C. Y. (2005). Phosphorylation and stabilization of HURP by Aurora-A: implication of HURP as a transforming target of Aurora-A. Mol. Cell. Biol. 25, 5789–5800. doi: 10.1128/MCB.25.14.5789-5800.2005
Yu, J. X., Guan, Z., and Nash, H. A. (2006). The mushroom body defect gene product is an essential component of the meiosis II spindle apparatus in Drosophila oocytes. Genetics 173, 243–253. doi: 10.1534/genetics.105.051557
Yucel, J. K., Marszalek, J. D., McIntosh, J. R., Goldstein, L. S., Cleveland, D. W., and Philp, A. V. (2000). CENP-meta, an essential kinetochore kinesin required for the maintenance of metaphase chromosome alignment in Drosophila. J. Cell Biol. 150, 1–11. doi: 10.1083/jcb.150.1.1
Zhang, C., and Clarke, P. R. (2000). Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288, 1429–1432. doi: 10.1126/science.288.5470.1429
Zhang, C., Goldberg, M. W., Moore, W. J., Allen, T. D., and Clarke, P. R. (2002a). Concentration of Ran on chromatin induces decondensation, nuclear envelope formation and nuclear pore complex assembly. Eur. J. Cell Biol. 81, 623–633. doi: 10.1078/0171-9335-00288
Zhang, C., Hutchins, J. R., Muhlhausser, P., Kutay, U., and Clarke, P. R. (2002b). Role of importin-beta in the control of nuclear envelope assembly by Ran. Curr. Biol. 12, 498–502. doi: 10.1016/S0960-9822(02)00714-5
Zhang, G., Breuer, M., Förster, A., Egger-Adam, D., and Wodarz, A. (2009). Mars, a Drosophila protein related to vertebrate HURP, is required for the attachment of centrosomes to the mitotic spindle during syncytial nuclear divisions. J. Cell Sci. 122, 535–545. doi: 10.1242/jcs.040352
Zhang, M. S., Arnaoutov, A., and Dasso, M. (2014). RanBP1 governs spindle assembly by defining mitotic Ran-GTP production. Dev. Cell 31, 393–404. doi: 10.1016/j.devcel.2014.10.014
Zheng, Y. (2010). A membranous spindle matrix orchestrates cell division. Nat. Rev. Mol. Cell Biol. 11, 529–535. doi: 10.1038/nrm2919
Zheng, Z., Zhu, H., Wan, Q., Liu, J., Xiao, Z., Siderovski, D. P., et al. (2010). LGN regulates mitotic spindle orientation during epithelial morphogenesis. J. Cell Biol. 189, 275–288. doi: 10.1083/jcb.200910021
Zitouni, S., Nabais, C., Jana, S. C., Guerrero, A., and Bettencourt-Dias, M. (2014). Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15, 433–452. doi: 10.1038/nrm3819
Zonis, J., and Wilde, A. (2011). The site of RanGTP generation can act as an organizational cue for mitotic microtubules. Biol. Cell 103, 421–434. doi: 10.1042/BC20100135
Keywords: mitosis, RanGTP, microtubules, mitotic spindle, Drosophila melanogaster
Citation: Chen JWC, Barker AR and Wakefield JG (2015) The Ran Pathway in Drosophila melanogaster Mitosis. Front. Cell Dev. Biol. 3:74. doi: 10.3389/fcell.2015.00074
Received: 04 September 2015; Accepted: 09 November 2015;
Published: 26 November 2015.
Edited by:
Patrizia Lavia, Institute of Molecular Biology and Pathology - CNR, ItalyReviewed by:
Andrew Wilde, University of Toronto, CanadaCopyright © 2015 Chen, Barker and Wakefield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James G. Wakefield, ai5nLndha2VmaWVsZEBleGV0ZXIuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.