- 1InnoSyn B. V., Geleen, Netherlands
- 2Novo Nordisk A/S, Bagsværd, Denmark
We here report four biocatalytic approaches for the synthesis of the protected amino acid building block α-benzyl L-glutamate. Screenings of these routes to identify active and selective enzymes were conducted, and major hits were confirmed in retest reactions. In the first approach, N-Boc L-glutamic acid is mono-benzylesterified by the protease Alcalase with 81% yield; and in the other three approaches, a biocatalytic γ-selective hydrolysis of α,γ-dibenzyl L-glutamate, a selective amide hydrolysis of α-benzyl L-glutamine, and a selective lactam hydrolysis of alpha-benzyl L-pyroglutamate is performed with up to 71% yield.
1 Introduction
Amino acids and their protected derivatives are important building blocks in peptide synthesis. α-Benzyl L-glutamate is a simple, yet challenging target in this group since any preparation from the naturally occurring L-glutamic acid will require discrimination between the α- and γ-carboxylic acid functionalities, either during synthesis or during subsequent purification steps. The structural element gamma-glutamate is a moiety in many modern drugs (Kurtzhals et al., 2023), and its synthetic introduction would typically require an α-protected glutamate building block. While the versatile acid-labile tert-butyl protecting group is often applied, some synthetic schemes require an alternative, and the benzyl ester may be used since it is orthogonal to the tert-butyl ester and can be cleaved either under aqueous basic conditions or by hydrogenolysis.
Chemical approaches to α-benzyl L-glutamate rely on the (moderate) difference in the reactivity of the α- and γ-carboxylic acid functionalities, both as a nucleophilic or electrophilic moiety, and in all cases, the amino functionality needs to be protected. For instance, N-carbobenzoxy-L-glutamic acid can be converted to its cyclic anhydride and reacted with benzyl alcohol, giving the α-benzyl ester in moderate selectivity, which can be isolated as its dicyclohexylamine salt in 59% yield (Klieger et al., 1962) after which the carbobenzoxy group can be removed by HBr/acetic acid. The yield of such chemical processes is low, and they have poor sustainability due to the large amount of toxic waste, large consumption of organic solvents, and low yield. For these reasons and due to the many solid handling procedures, the costs of these chemical production processes are high. Hence, there is a need for a selective, cost-efficient, and sustainable process.
Higher selectivity, yield, and potential sustainability could be accomplished by using enzymes as biocatalysts. Enzymes are typically applied because of their usually high chemo-, regio-, and/or enantio-selectivities, which typically make the chemistry of the protection group superfluous. In chemical and pharmaceutical industries, enzymes are applied to increase the step efficiency of multi-step chemical processes, reduce the amounts of reagents and organic solvents, and sometimes enable the use of alternative starting materials which may be cheaper, may originate from renewable resources, or just be more readily available, thereby typically improving the economic and the environmental footprint of the overall process (Alcántara et al., 2022). Consequently, a number of industrial chemo-enzymatic processes have been developed and implemented (Wu et al., 2021; France et al., 2023).
In this study, we have explored several possibilities to obtain the target α-benzyl L-glutamate via a biocatalytic route from simple starting materials by exploiting the propensity of enzymes for high chemo- or regio-selectivity. We describe the results obtained during the screening of four biocatalytic approaches to the target product α-benzyl L-glutamate (Scheme 1).
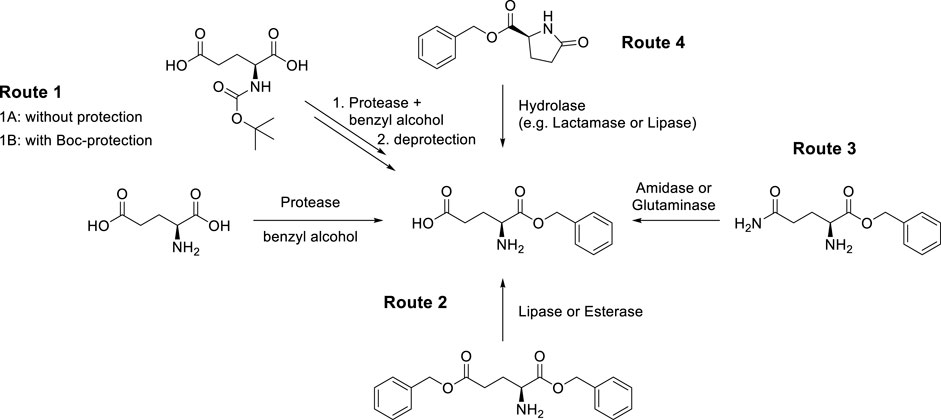
SCHEME 1. Devised and investigated enzymatic routes to α-benzyl L-glutamate enabled by different types of hydrolase enzymes.
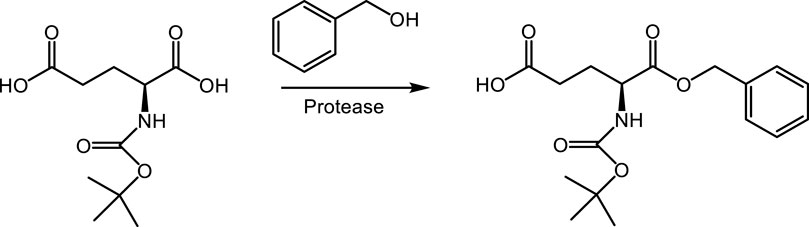
The first approach (route 1) exploits the α-selectivity of proteases to form the α-benzyl-ester of L-glutamic acid in benzyl alcohol as the (co-) solvent, either with (1B) or without (1A) an N-protecting group. Application of an N-protecting group will require a subsequent chemical step for removal. α-selective enzymatic esterifications of L-glutamic and L-aspartic acid have previously been reported by Nuijens et al. (2009), but not with benzyl alcohol.
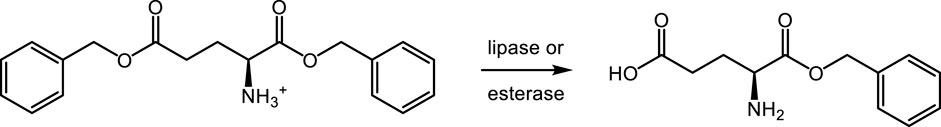
The second approach (route 2) applies lipases or esterases for γ-selective hydrolysis of dibenzyl L-glutamate.
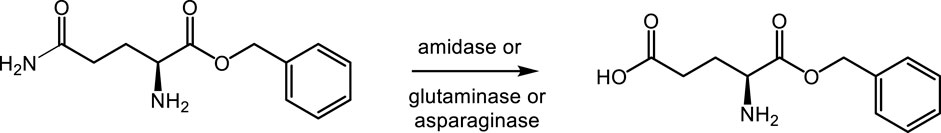
The third approach (route 3) applies amidases, glutaminases, or asparaginases for γ-selective amide hydrolysis of α-benzyl L-glutamine. Glutaminases are well-known to convert glutamine into glutamic acid not only in the amino acid form but also in tripeptides, e.g., Leu-Gln-Gly (Brown et al., 2008; Ito et al., 2013). Asparaginases can act as glutaminases, but until now they have not been reported to act on ester derivatives of L-asparagine or L-glutamine (Orabi et al., 2019).
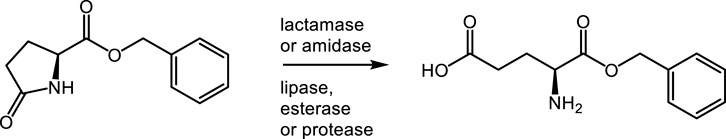
The fourth approach (route 4) applies lactamases, oxoprolinases, amidases, or other hydrolytic enzymes to hydrolyze the lactam functionality in α-benzyl L-pyroglutamate. Oxoprolinases (EC 3.5.2.9), also known as pyroglutamate hydrolases, catalyze the hydrolysis of pyroglutamate by forming glutamate. Although most of these enzymes require the consumption of the high-energy co-substrate ATP (Niehaus et al., 2017), there are also ATP-independent sub-classes described (Nishimura et al., 1999).
A range of enzymes from commercial suppliers and in-house recombinant production in E. coli were screened for all four routes.
2 Experimental
2.1 Materials
2.1.1 Chemicals
N-Boc L-glutamic acid, α-benzyl-N-Boc L-glutamic acid, and α,γ-dibenzyl L-glutamate.HCl were obtained from TCI EUROPE (Zwijndrecht, Belgium). α-Benzyl L-glutamine was sourced from eNovation Chemicals LLC (Bridgewater, United States). α-Benzyl L-pyroglutamate was obtained from Biosynth (Compton, United Kingdom). Other standard laboratory chemicals and solvents were sourced from commercial suppliers in highest and chromatography grades.
2.1.2 Enzymes
Immobilized proteases Alcalase, Savinase, Esperase, and Protex 6L were obtained from Chiral Vision (Den Hoorn, Netherlands). Papain was obtained from NAGASE (Düsseldorf, Germany). Esterase from Bacillus subtilis was sourced from abcr (Karlsruhe, Germany). Liquid and immobilized Candida antarctica lipase B (Lipozyme and Novozym 435) as well as liquid and powder formulation of Alcalase were sourced from Novozymes (Bagsværd, Denmark). Glutaryl acylase and penicillin G amidase were obtained from Boehringer Mannheim (Penzberg, Germany). Protease Promod 24L was obtained from Biocatalysts (Cardiff, United Kingdom). Lipase obtained from Pseudomonas alcaligenes and recombinant pig liver esterases (PLEs) are products of InnoSyn.
2.2 Analytical method
HPLC analysis was performed using an Inertsil C18 (250 mm × 3.0 mm, 5 µm particle size) column at a column temperature of 40°C with a run time of 20 min at an injection volume of 0.5 or 1 μL. For detection, a VWD detector at 210 nm was used. For separation, a gradient of mobile phases A (H2O + 0.1% (v/v) methane sulfonic acid (MSA)) and B (acetonitrile +0.1% (v/v) MSA) at a flow rate of 0.5 mL/min was applied. The reactions with N-Boc L-glutamic acid were analyzed with the following gradient of the mobile phases: 0 min: 95/5 A/B; 2 min: 95/5 A/B; 12 min: 10/90 A/B; 17 min: 10/90 A/B; 17.1 min: 95/5 A/B. For all other reactions, a different gradient of the same mobile phases was used: 0 min: 90/10 A/B; 12 min: 10/90 A/B; 12.5 min: 5/95 A/B; 15.5 min: 5/95 A/B; 16 min: 90/10 A/B.
2.3 Recombinant enzyme production
Chemically competent Escherichia coli K12 and BL21 strains were transformed with pBR322-derived expression vectors, including the genes of interest. Recombinant clones were selected on agar plates supplemented with 50 μg/mL neomycin. Recombinant protein production was performed in shake flasks containing 100 mL LB medium (10 g/L tryptone, 5 g/L yeast extract, and 7 g/L NaCl) at 28°C. Enzyme production was induced by addition of 0.02% L-rhamnose in the middle of the exponential growth phase (OD620 = 0.4–0.8). After overnight incubation, cell pellets were harvested, and cell-free extracts were prepared by sonication and removal of cell debris by centrifugation.
2.4 Enzyme screenings
To 96-well microtiter plates or individual HPLC vials were added buffer, co-solvent, substrate solution, and enzyme. The plates/vials were shaken on an orbital shaker with a speed of 250 rpm at the indicated temperature overnight, before the reactions were quenched, and samples were withdrawn, diluted, and analyzed by HPLC. As controls, biological and chemical blank reactions were included in all screenings. The chemical blanks consisted of reactions in which of all reaction components, only the enzyme was left out. The missing weight or volume of the enzyme was compensated by the addition of the respective weight or volume of the buffer in which the reaction was performed. As biological blanks, reactions with cell-free extracts of E. coli strains not overexpressing a recombinant enzyme or Pichia pastoris culture supernatants not containing a recombinantly expressed and secreted enzyme were performed. In the tables showing the results of the screening hits, only the results with the biological blanks are presented, which corresponded to the E. coli or P. pastoris strain of the listed recombinantly produced enzyme(s). In the cases where several replicates of the corresponding blanks were available, the average of the blank reaction values is given.
2.4.1 α-selective benzyl esterification of N-Boc L-glutamic acid
N-Boc L-glutamic acid (17 g/L) was incubated in an overnight reaction at 50°C with 182 mg of the enzyme formulation in dry benzyl alcohol at a 1-mL reaction volume. After dilution in acetonitrile, the reaction mixture was analyzed by HPLC, as described in Section 2.2. 14 and proteases were screened (immobilized or powder).
2.4.2 α-selective benzyl esterification of L-glutamic acid
A measure of 17 mg of L-glutamic acid or L-glutamic acid.HCl was incubated with 1 mL of dry benzyl alcohol or methanol, respectively, each in the presence of 180 mg of Alcalase. Each reaction was carried out with or without molecular sieves (20 mg, 3 Å pore size) and incubated overnight at 50°C. Reactions were worked up by 10-fold dilution with water for glutamic acid analysis or with a tetrahydrofuran (THF)/water mixture (90/10 v/v%) for product analysis and analyzed by HPLC, as described in Section 2.2.
2.4.3 γ-selective hydrolysis of dibenzyl L-glutamate, pH 7
Approximately 7 mg α,γ-dibenzyl L-glutamate in 50 µL toluene was mixed with 100 µL aqueous potassium phosphate (KPi) buffer pH 7.5 and 100 µL enzyme solution to a final substrate concentration of 90 mM (∼30 g/L) and a final volume of 250 μL at pH 7.0. Approximately 120 individual lipase, esterase, and protease enzymes were screened. Reactions were incubated overnight at 28°C on an orbital shaker with a shaking speed of 250 rpm. Enzymes were denaturated by the addition of an organic solvent [THF/H2O (90/10 v/v%)] and removed from the solution by centrifugation for 20 min at 5,000 rpm and at room temperature. The remaining supernatant was analyzed via HPLC, as described in Section 2.2.
2.4.4 γ-selective hydrolysis of dibenzyl L-glutamate, pH 5
α,γ-Dibenzyl L-glutamate.HCl in 100 µL water was mixed with 50 µL aqueous KPi buffer pH 7.5 containing glacial acetic acid and 100 µL enzyme solution to a final substrate concentration of 90 mM (∼30 g/L) and a final volume of 250 μL at pH 5. Approximately 120 individual lipase, esterase, and protease enzymes were screened. Reactions were incubated overnight at 28°C on an orbital shaker with a shaking speed of 250 rpm. Enzymes were denaturated by the addition of an organic solvent (THF/H2O (90/10 v/v%)) and removed from the solution by centrifugation for 20 min at 5,000 rpm and at room temperature. The remaining supernatant was analyzed via HPLC, as described in Section 2.2. For the calculation of the resulting α-benzyl L-glutamate yield, the area of the peak—not corresponding to the desired product, but co-eluting at the same retention time—detected in the used quenching solution was subtracted from the area detected in the enzymatic and the control/blank reactions.
2.4.5 γ-selective amide hydrolysis of α-benzyl L-glutamine
Stock solutions (33 mM) of α-benzyl L-glutamine in 100 mM Na-acetate buffers of pH 4.0, 4.5, and 4.8 were prepared. A measure of 150 μL of the stock solutions was mixed in HPLC vials with 100 μL of the enzyme solution (pH ∼ 7.5), resulting in a final concentration of 20 mM, a final volume of 250 μL, and a pH of 4.9, 5.5, and 6.0. Reaction mixtures were incubated overnight at 20°C on an orbital shaker with a shaking speed of 250 rpm. Subsequently, the enzymes were precipitated by the addition of an acetonitrile/water (1:1) mixture and removed by centrifugation. The remaining supernatant was analyzed via HPLC, as described in Section 2.2. Glutaminases and asparaginases were screened at all three pH values. Amidases were screened either in vials or in 96-well microtiter plates at pH 5.5.
2.4.6 Selective lactam hydrolysis of α-benzyl L-pyroglutamate without ATP
Stock solutions (33 mM) of α-benzyl L-pyroglutamate in 100 mM Na-acetate buffers pH 4.5 and 4.8 were prepared. A measure of 150 μL of the stock solutions was mixed in HPLC vials with 100 μL of the enzyme solution (pH ∼7.5), resulting in a final concentration of 20 mM, a final volume of 250 μL, and pH of 5.5 and 6.0. Reaction mixtures were incubated overnight at 20°C on a shaker with a shaking speed of 250 rpm. Subsequently, the enzymes were precipitated by the addition of an acetonitrile/water (1:1) mixture and removed by centrifugation. The remaining supernatant was analyzed via HPLC, as described in Section 2.2. ATP-independent oxoprolinases and γ-lactamases were screened at pH 5.5 and 6.0. Approximately 120 individual lipase, esterase, and protease enzymes were screened in 96-well microtiter plates at pH 5.5.
2.4.7 Selective lactam hydrolysis of α-benzyl L-pyroglutamate with ATP
Reactions were conducted similar to the protocol described by Niehaus et al. (2017). Final reaction mixtures consisted of 5 mM α-benzyl L-pyroglutamate, 5 mM ATP, 5 mM MgCl2, 100 mM KCl, buffer (Na-acetate or potassium phosphate, depending on the pH), and 100 μL enzyme solution in a total volume of 250 μL. Reaction mixtures were incubated overnight at 20°C on an orbital shaker with a shaking speed of 250 rpm. Subsequently, the enzymes were precipitated by the addition of an acetonitrile/water (1:1) mixture and removed by centrifugation. The remaining supernatant was analyzed via HPLC, as described in Section 2.2. ATP-dependent oxoprolinases were tested at pH 4.9, 5.5, and 6.0.
2.5 Retests
To confirm some of the screening results, the α-selective benzyl esterification of N-Boc L-glutamic acid with Alcalase and the γ-selective hydrolysis of dibenzyl L-glutamate at pH 5 retests were performed at a 10-mL reaction volume scale. The reaction conditions, as described under Sections 2.4.1 and 2.4.4, were linearly scaled up in each of the parameters. Except for the reaction volume and the size of the glass test vials, the conditions of these experiments were kept identical to the original screening conditions.
2.6 Modeling and docking
For the docking of N-Boc-L-glutamate and L-glutamate to Alcalase/subtilisin Carlsberg, respectively, in the presence of benzyl alcohol, the experimental 3D structure of subtilisin Carlsberg with PDB accession number 1sbc was used. To obtain a 3D model of pig liver esterase isoform 4 (PLE-4), the published experimental crystal structure of PLE isoform 5 (PLE-5) with PDB accession number 5fv4 was manually mutated in the four positions that PLE-5 and PLE-4 differ, namely, R87G, F285P, F286L, and A287T. Subsequently, dibenzyl L-glutamate was docked in two different confirmations that either lead to the hydrolysis of the α-benzyl group or the γ-benzyl group. For the docking of α-benzyl L-glutamine and L-glutamine to the 3D structures of glutaminase 1 from E. coli (Glsa1) and for that of glutaminase 2 from B. subtilis (Glsa2), the experimental 3D structure of Glsa1 with PDB accession number 1u60 and an AlphaFold2 (version 2.3.2, release date 05 April 2023)-generated 3D structure of Glsa2 (UniProt accession no. O07637, probability score pLDDT of 95.71), respectively, were used. To obtain a 3D model of the oxoprolinases of Alcaligenes faecalis N-38A (UniProt accession no. Q9FDT5) and Alcaligenes aquatilis (RefSeq accession no. WP_108728566), AlphaFold2 (version 2.3.2, release date 05 April 2023, score pLDDT score of 90.81 and 90.85, respectively) was used. Subsequently, α-benzyl L-pyroglutamate and L-pyroglutamate were docked to the AlphaFold2-generated 3D structures. Dockings were performed with AutoDock Vina (version 1.1.2., Trott and Olsen (2010)). Modeling and visualizations were done with PyMOL and Chimera.
3 Results and discussions
3.1 α-selective benzyl esterification of N-Boc L-glutamic acid (route 1)
The screening of 14 commercial proteases for conversion of N-Boc-L-glutamic acid to N-Boc-L-glutamic acid α-benzyl ester (Scheme 2) revealed eight hits with detectable product formation (Table 1). A wide range of yields were observed under the screening conditions, and immobilized Alcalase provided the highest yield of 81%. The activity of this enzyme was confirmed in a retest.
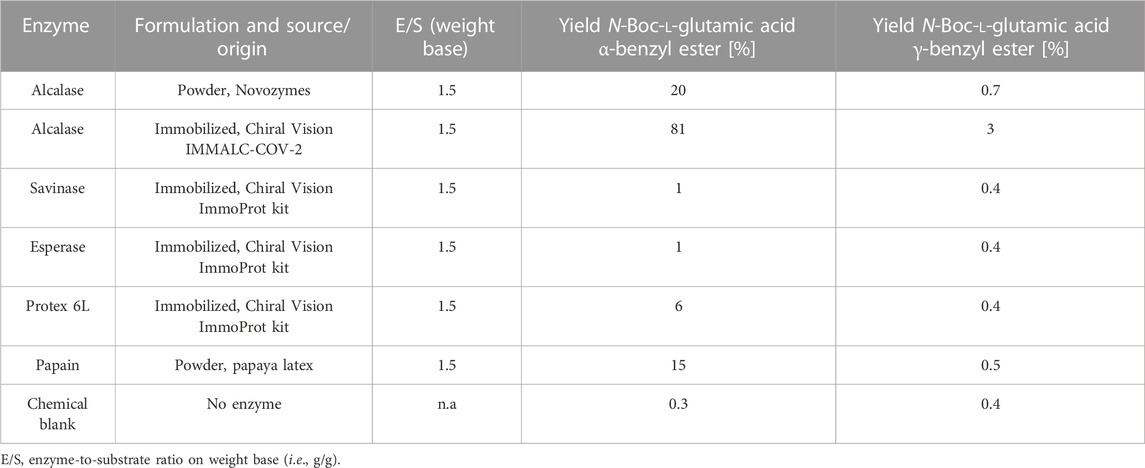
TABLE 1. Hits from the protease screening in the α-selective benzyl esterification of N-Boc L-glutamic acid. (HPLC yields).
Testing Alcalase—the best enzyme in α-selective benzyl esterification of N-Boc-L-glutamic acid—for the conversion of L-glutamic acid or its HCl salt in benzyl alcohol did not result in any target product formation. Attempts to form the corresponding methyl ester in the methanol solvent did not reveal any hits either. The absence of enzymatic esterification products is probably due to the insolubility of L-glutamic acid and its HCl salt in benzyl alcohol and methanol in combination with a very unfavorable reaction equilibrium. We believe the N-protecting group is required to solubilize the amino acid in benzyl alcohol and to increase the pKa of the α-carboxylic acid moiety so that the thermodynamic equilibrium is more on the side of the benzyl ester.
N-Boc-L-glutamate and L-glutamate were docked to the 3D structure of Alcalase (subtilisin Carlsberg, PDB no. 1sbc) in the presence of benzyl alcohol which acted as the reaction medium and substrate for esterification. The docking showed that both amino acid derivatives are bound in similar positions and distances to the catalytic triad consisting of Ser221, His664, and Asp32 (Figure 1). The distance of the catalytic Ser221 to the closest carboxylate oxygen atom of N-Boc-L-glutamate and L-glutamate is 3.6 and 3.7 Å, respectively, and therefore almost identical. Equally, benzyl alcohol occupies similar positions in both dockings. It is therefore unlikely that a difference in binding efficiency causes the absence of esterification of L-glutamate and benzyl alcohol, underlining that probably their low solubility in each other and the very unfavorable equilibrium are the reasons for the absence of esterification of L-glutamate and benzyl alcohol.
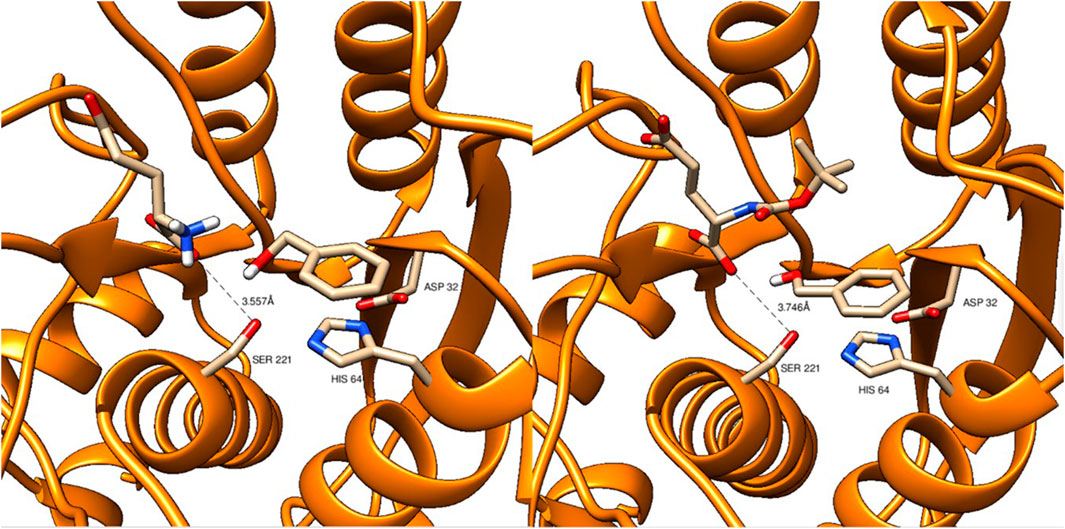
FIGURE 1. Docking of L-glutamate (left panel) and N-Boc-L-glutamate (right panel), both together with benzyl alcohol to the 3D structure of subtilisin Carlsberg (PDB no. 1sbc).
The screening was performed with N-tert-butyloxycarbonyl (N-Boc) protected L-glutamic acid, but we believe a wider choice of protecting groups, for instance, other carbamate types such as benzyloxycarbonyl (Z) or 9-fluorenylmethoxycarbonyl (Fmoc), or amide types such as acetyl, trifluoro-acetyl, formyl, or benzoyl can be utilized.
3.2 γ-selective hydrolysis of dibenzyl L-glutamate (route 2)
The initial screening of lipase, esterase, and protease libraries (in total 128 enzymes) for selective hydrolysis of dibenzyl L-glutamate (Scheme 3) was conducted in a biphasic toluene–water mixture, due to the low solubility of dibenzyl L-glutamate free base in water at pH 7. The screening identified a series of active enzymes, but with the undesired selectivity, giving γ-benzyl glutamate as the predominant product (results not shown).
A second screening was performed in a monophasic aqueous system at a lower pH of approximately 5, where the HCl salt of the substrate was readily soluble. Again, many active enzymes forming γ-benzyl glutamate were found, but now also five highly active enzymes were identified, which, besides γ-benzyl L-glutamate, formed α-benzyl L-glutamate in relevant yields of 11%–64% (Table 2). The enzyme PLE-4, recombinantly produced in E. coli, favored the formation of the desired α-benzyl L-glutamate, 64% vs. γ-benzyl L-glutamate, 16%. The activity of this enzyme was confirmed in a retest under identical conditions as the screening itself on the 10-mL scale.
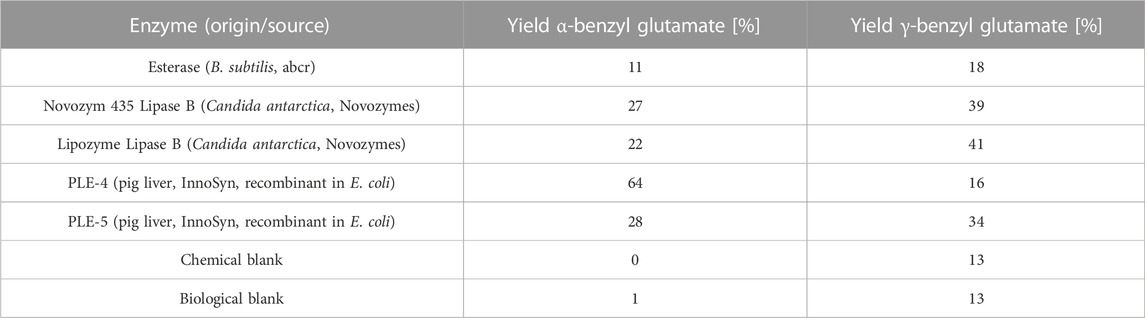
TABLE 2. Hits from the aqueous lipase/esterase screening of the hydrolysis of dibenzyl L-glutamate at pH 5.0 (HPLC yields).
We hypothesize that at a lower pH, the protonated amine is the predominant species allowing for better discrimination between the two esters due to the positive charge being closer to the α-benzyl ester moiety. The retests at the 10-mL scale showed that the final levels of the reaction products were already obtained within the reaction time of 1 h at 28°C. By conducting the PLE-4 reaction at the 10-mL scale and at a lower reaction temperature, we observed an increase of α/γ-selectivity toward α-benzyl L-glutamate with respect to γ-benzyl L-glutamate. It seems that at this lower temperature, the relative reaction rates of the two competing hydrolysis reactions are changed in favor of the formation of the desired α-isomer as the non-selective chemical (hydrolysis) reactions are typically more suppressed than enzyme-enhanced reactions.
Enzyme engineering of amino acid residues surrounding the binding pocket may give variants with improved substrate acceptance, leading to increased activity and selectivity due to improved accommodation of the substrate in the right position. To obtain a better understanding of the mechanisms of α/γ-selectivity, dibenzyl L-glutamate was docked to the 3D model of PLE-4 in conformations, leading to the hydrolysis of the α-benzyl group and the γ-benzyl group producing γ-benzyl L-glutamate or α-benzyl L-glutamate, respectively. The docking indicated the E208 binds the α-amino group when hydrolysis would yield α-benzyl glutamate (Figure 2), while in case of the hydrolysis yielding γ-benzyl glutamate, the α-amino group would be in a hydrophobic environment without any hydrogen bonding. At lower pH and therefore higher degree of protonation of the α-amino group, the salt bridge to E208 is probably stronger than at neutral pH. On the contrary, at neutral pH, the accommodation of the α-amino group in the hydrophobic pocket close to G125 and G126 has a higher probability than at pH 5.
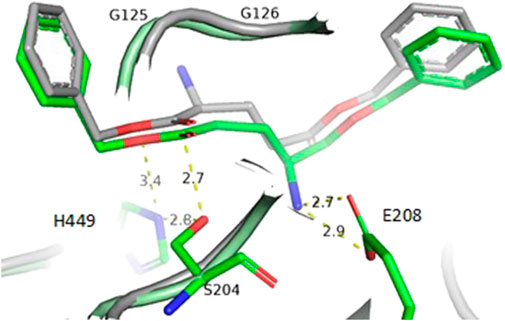
FIGURE 2. Superimposition of PLE-4 3D-models with dibenzyl L-glutamate docked in conformations, leading to the hydrolysis of the α-benzyl group (gray) and the γ-benzyl group (green) producing γ-benzyl L-glutamate or α-benzyl L-glutamate, respectively.
3.3 γ-selective amide hydrolysis of α-benzyl L-glutamine (route 3)
Glutaminases, asparaginases, and amidases (in total 107 enzymes) were screened for γ-selective hydrolysis of α-benzyl L-glutamine (Scheme 4). This substrate, however, shows significant chemical hydrolysis of the benzyl ester functionality at pH above 5.5. Therefore, the screening was conducted at pH 5.5. pH values below 6 are not favorable for most of the asparaginases and glutaminases, so these were also tested at pH 4.9 and 6.0. In this manner, the preferred conditions for substrate and enzyme stability were accounted for.
Four enzymes were identified that gave a yield between 1.5% and 3.8% of the α-benzyl L-glutamate product (Table 3). The highest activity was found with glutaminase 1 of E. coli (UniProt accession no. P77454) at pH 4.9 and 5.5, while less activity of this enzyme could be detected at pH 6 (1.1%). Glutaminase 2 of B. subtilis (UniProt accession no. O07637), on the contrary, showed the highest activity at pH 6 (2.3%), less at pH 5.5 (1.5%), and no activity at pH 4.9. Furthermore, an immobilized glutaryl acylase and a penicillin-G amidase showed the formation of α-benzyl L-glutamate. Benzyl alcohol was detected as a byproduct in all reactions due to chemical hydrolysis of the α-benzyl ester and not due to enzymatic activity, as confirmed by control reactions without the enzyme. The enzymatic reactions thus appear highly selective for hydrolysis of the amine functionality over the ester functionality. Only the activity of glutaminase 2 of B. subtilis could be confirmed during a retest on 10-ml scale.
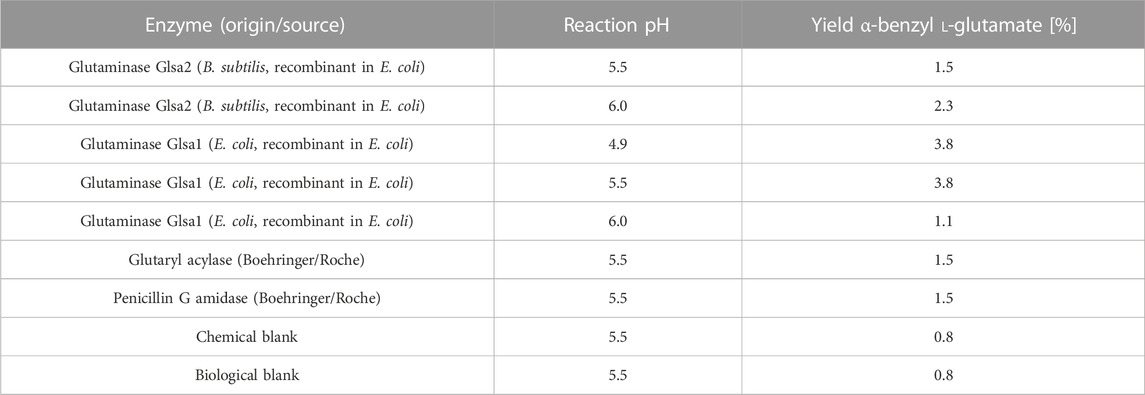
TABLE 3. Hits from the screening of glutaminases, asparaginases, and amidases for the γ-amide hydrolysis of α-benzyl L-glutamine (HPLC yields).
The physiological substrate L-glutamine and the non-physiological target substrate α-benzyl L-glutamine were docked into the experimental and AlphaFold2-generated 3D structures of glutaminase 1 from E. coli (PDB accession no. 1u60) and glutaminase 2 from B. subtilis, respectively. The docking resulted in very similar docking poses of the amide nitrogen atom to the hydroxyl group of the catalytic triad serine residues (Ser66 for Glsa1 and Ser65 for Glsa2, Figure 3). The distances between these were relatively large but not larger for the non-physiological substrate α-benzyl L-glutamine than for L-glutamine, therefore not explaining the low conversion obtained. Probably, the low pH values that had to be applied because of substrate instability at neutral pH values, which are the pH optima of the glutaminases (Brown et al., 2008), led to the low conversions of α-benzyl L-glutamine.
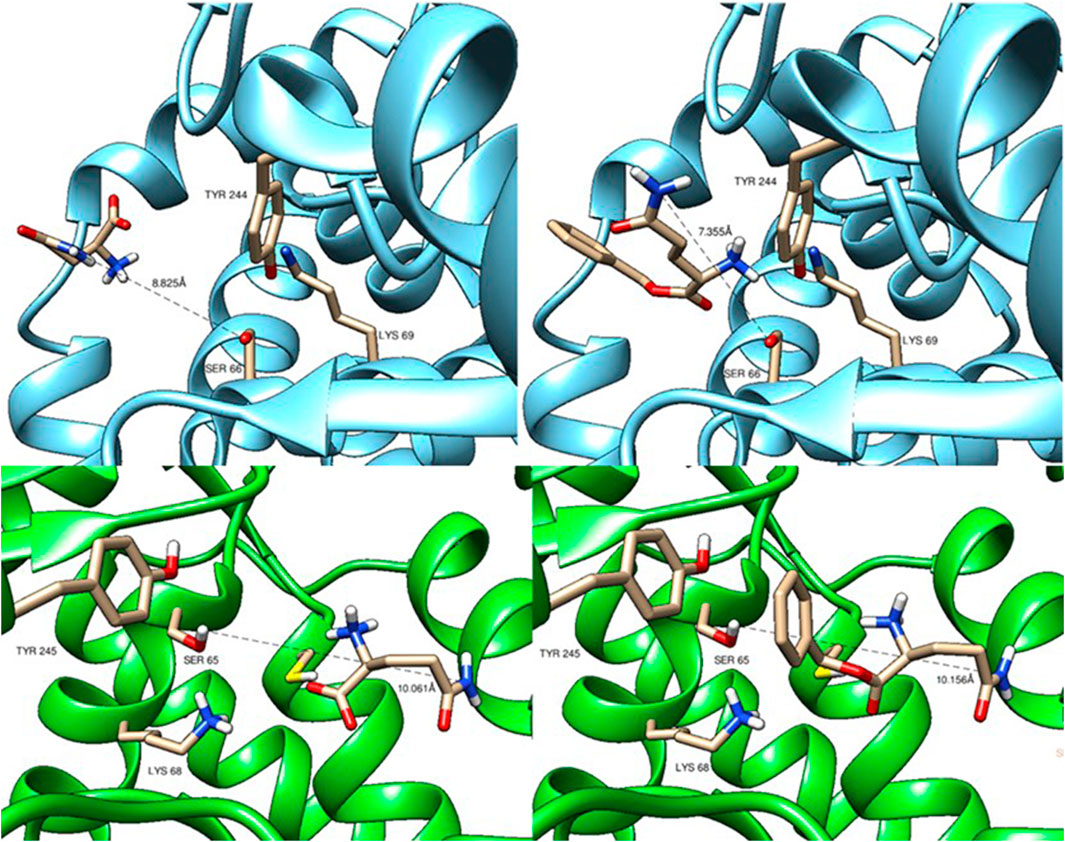
FIGURE 3. Docking of L-glutamine (left panels) and α-benzyl-L -glutamine (right panels) to the 3D structures of glutaminase 1 from Escherichia coli (upper panels) and glutaminase 2 from Bacillus subtilis (lower panels).
More elaborate modelling and enzyme engineering of the enzymes may give variants with improved substrate acceptance and increased activity via improved binding of α-benzyl L-glutamine in the active site of the enzyme. It would also be relevant if the enzymes could be modified to be more active and/or stable at pH values lower than 5.5 or preferably lower than 5.0, as this would allow suppression of the chemical hydrolysis of the benzyl ester.
3.4 Selective lactam hydrolysis of α-benzyl L-pyroglutamate (route 4)
Oxoprolinases, γ-lactamases, esterases, lipases, amidases, and proteases (in total 147 enzymes) were screened for their ability to selectively hydrolyze the lactam in α-benzyl L-pyroglutamate (Scheme 5). Five enzymes forming the desired α-benzyl L-glutamate were identified (Table 4). The oxoprolinases of Alcaligenes faecalis N-38A (UniProt accession no. Q9FDT5) and Alcaligenes aquatilis (RefSeq accession no. WP_108728566) gave 2.7% and 1.3% of the product α-benzyl L-glutamate, respectively, and Alcalase 2.4L gave 1.8%. Retesting these five enzymes in individual reactions under the same conditions as the screening confirmed the activity of Promod 24L (1.2% yield) and Alcalase 2.4L (10.1% yield), showing considerable improvement compared to the screening result.
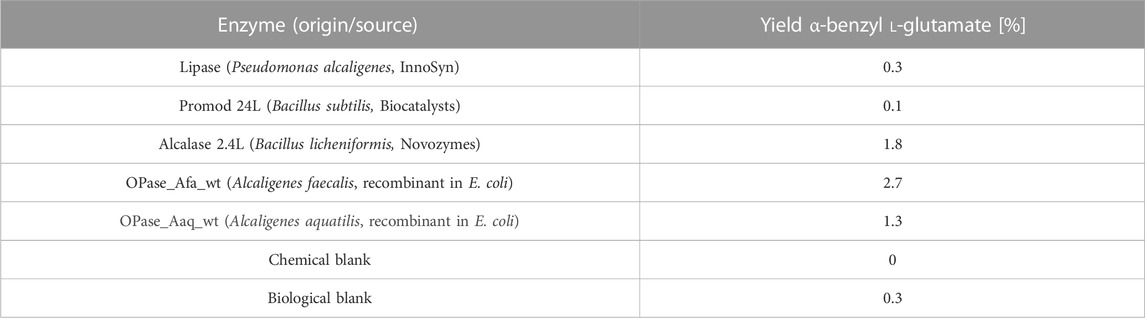
TABLE 4. Hits of the screening of oxoprolinases, γ-lactamases, esterases, lipases, amidases, and proteases for the lactam hydrolysis of α-benzyl L-pyroglutamate (HPLC yields). All enzymes in this table are ATP-independent and had thus been tested without ATP.
For these enzymes, enzyme engineering may provide variants with improved affinity for α-benzyl L-glutamate and activity in lactam hydrolysis. However, the ATP-independent oxoprolinases from A. faecalis and A. aquatilis share very little sequence similarity to any other known enzyme class and no similarity to ATP-dependent oxoprolinases, for which experimental 3D structures have been obtained. Based on the AlphaFold2-generated 3D structures of the A. faecalis and A. aquatilis oxoprolinases, we could not identify a catalytic triad that would define the active site of these enzymes. For the A. aquatilis oxoprolinase, the closest serine residue was found to be Ser183 at a distance of 4.6–4.8 Å to the lactam nitrogen atom of both L-pyroglutamate and α-benzyl-L-pyroglutamate (Figure 4). The A. faecalis oxoprolinase is 91% identical on the amino acid level, and not surprisingly the binding poses and distances of the two substrates and the distances to the potential active site Ser183 were very similar, thereby not providing an obvious explanation for the observed low conversions of the target substrate.
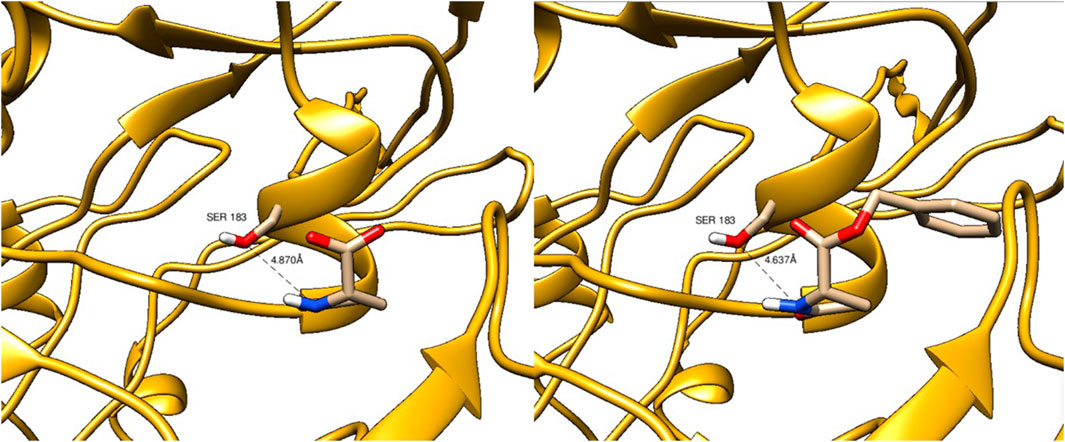
FIGURE 4. Docking of L-pyroglutamate (left panel) and α-benzyl-L-pyroglutamate (right panel) to the 3D structure of oxoprolinase from Alcaligenes aquatilis.
4 Conclusion
In this study, four different enzymatic routes were investigated for the synthesis of α-benzyl L-glutamate from simple starting materials. Active enzyme hits were identified for all routes with generally good selectivities, even though in one case, concurrent chemical hydrolysis leads to formation of the undesired byproduct, glutamine (route 3). All of these reactions have their specific advantages and challenges (Table 5).
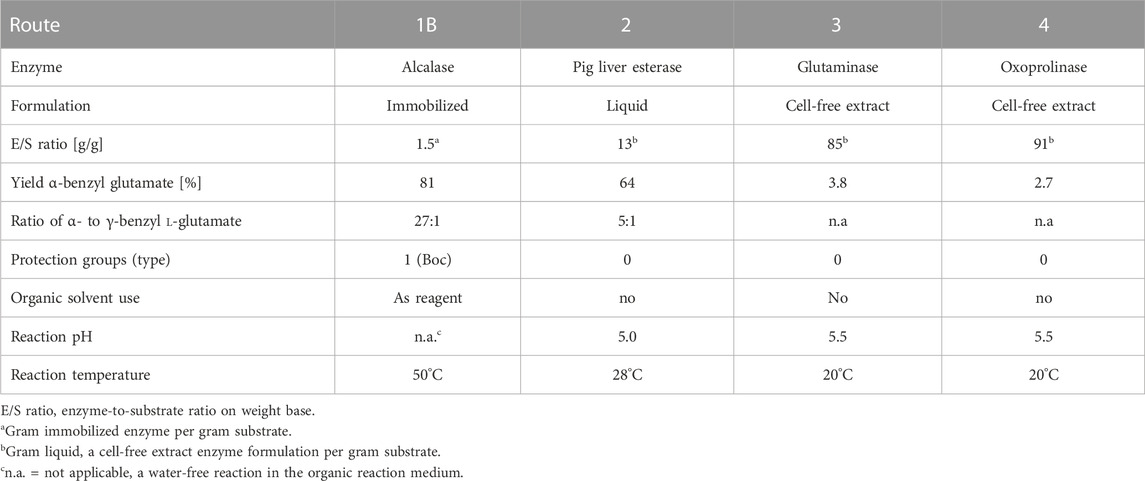
TABLE 5. Characteristics of the different enzymatic routes toward α-benzyl L-glutamate investigated in this study. Yields as analytical yields.
All of the enzymatic routes have in common that no organic solvent is required as the reaction medium. The benzyl alcohol used in Routes 1A and 1B serves as a substrate and solvent at the same time. Although Route 1A has proven to be unfeasible, its alternative 1B is characterized by the need for a Boc-protection of the L-glutamate’s amine functionality. Its key advantage is its high selectivity and efficiency (on screening scale) and the use of the cheap enzyme Alcalase which is available from several commercial suppliers.
In route 2, the γ-benzyl group does not function as a protecting group in the original sense, but is “lost” after the enzymatic hydrolysis, while the α-benzyl group remains as an integral part of the product. On the small/screening scale, the regio-selectivity of the PLE reaction was not complete, and the yields were lower than in route 1B.
The hydrolysis of α-benzyl L-pyroglutamate by oxoprolinases, γ-lactamases, or other hydrolase enzymes in route 4 has proven to be more challenging than the amide hydrolysis of α-benzyl L-glutamine by glutaminases or amidases in route 3, but both routes have the advantage of not requiring any protecting group.
Activities obtained during screenings also include low levels, but it is expected that a general optimization study of the biocatalytic reaction conditions will facilitate improved activities and yields. Optimization of the biocatalysts via enzyme engineering may also lead to improved performance in activity and selectivity. It is further possible that enzyme immobilization will increase enzyme stability, ease of enzyme removal, and allow for re-use of the enzyme. Finally, the here reported biocatalytic route scouting and its experimental validation on screening scale has given a direction to the development of an efficient biocatalytic route to benzyl L-glutamate.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
PQ: conceptualization, project administration, resources, supervision, writing–original draft, and writing–review and editing. LJ: data curation, investigation, visualization, writing–original draft, and writing–review and editing. MM: conceptualization, investigation, and writing–original draft. LV: investigation and writing–original draft. VP: writing–review and editing, data curation, investigation, visualization. J-MvdL: writing–review and editing, methodology, investigation, visualization. LN: conceptualization, supervision, writing–original draft, and writing–review and editing. MS: conceptualization, supervision, writing–original draft, and writing–review and editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. The authors declare that this study received funding from Novo Nordisk A/S. The funder had the following involvement in the study: funder’s employee LN contributed to conceptualization, supervision, writing–original draft, and writing–review and editing. Otherwise the funder did not have a further involvement in the study.
Acknowledgments
The authors wish to thank Math Boesten and Frédéric Tayar for their expert analytical support during the course of the project.
Conflict of interest
The authors are employees of InnoSyn B. V. (PQ, LJ, MM, LV, VP, J-MvdL and MS) or Novo Nordisk A/S (LN), respectively. The research project, which was conducted at InnoSyn B. V. (Geleen, Netherlands), was funded by Novo Nordisk A/S (Bagsværd, Denmark). For conducting the research, the authors did not receive any additional payment.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alcántara, A. R., Domínguez de María, P., Littlechild, J. A., Schürmann, M., Sheldon, R. A., and Wohlgemuth, R. (2022). Biocatalysis as key to sustainable industrial chemistry. ChemSusChem 15, e202102709. doi:10.1002/cssc.202102709
Brown, G., Singer, A., Proudfoot, M., Skarina, T., Kim, Y., Chang, C., et al. (2008). Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis. Biochemistry 47 (21), 5724–5735. doi:10.1021/bi800097h
France, S. P., Lewis, R. D., and Martinez, C. A. (2023). The evolving nature of Biocatalysis in pharmaceutical research and development. JACS Au 3, 715–735. doi:10.1021/jacsau.2c00712
Ito, K., Hanya, Y., and Koyama, Y. (2013). Purification and characterization of a glutaminase enzyme accounting for the majority of glutaminase activity in Aspergillus sojae under solid-state culture. Appl. Microbiol. Biotechnol. 97, 8581–8590. doi:10.1007/s00253-013-4693-4
Klieger, E., and Gibian, H. (1962). Über Peptidsynthesen, X. Vereinfachte Darstellung und Reaktionen von Carbobenzoxy-l-glutaminsäure-α-halbestern. Justus Liebigs Ann. Chem. 655, 195–210. doi:10.1002/jlac.19626550124
Kurtzhals, P., Østergaard, S., Nishimura, E., and Kjeldsen, T. (2023). Derivatization with fatty acids in peptide and protein drug discovery. Nat. Rev. Drug Discov. 22 (1), 59–80. doi:10.1038/s41573-022-00529-w
Niehaus, T. D., Elbadawi-Sidhu, M., Crécy-Lagard de, V., Fiehn, O., and Hanson, A. D. (2017). Discovery of a widespread prokaryotic 5-oxoprolinase that was hiding in plain sight. J. Biol. Chem. 292 (39), 16360–16367. doi:10.1074/jbc.m117.805028
Nishimura, A., Ozaki, Y., Oyama, H., Shin, T., and Murao, S. (1999). Purification and characterization of a novel 5-oxoprolinase (without ATP-hydrolyzing activity) from Alcaligenes faecalis N-38A. Appl. Environ. Microbiol. 65 (2), 712–717. doi:10.1128/aem.65.2.712-717.1999
Nuijens, T., Cusan, C., Kruijtzer, J. A. W., Rijkers, D. T. S., Liskamp, R. M. J., and Quaedflieg, P. (2009). Versatile selective α-carboxylic acid esterification of N-protected amino acids and peptides by Alcalase. Synthesis 5, 809–814. doi:10.1055/s-0028-1083362
Orabi, H. M., El-Fakharany, E. M., Abdelkhalek, E. S., and Sidkey, N. M. (2019). L-asparaginase and L-glutaminase: sources, production, and applications in medicine and industry. J. Microbiol. Biotechnol. Food Sci. 9 (2), 179–190. doi:10.15414/jmbfs.2019.9.2.179-190
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Keywords: α-benzyl L-glutamate, L-glutamic acid, enzymatic, sustainable, screening
Citation: Quaedflieg PJLM, Jente LMH, Müller M, Vermote L, Plesciuc V, van der Laan J-M, Nielsen L and Schürmann M (2024) Biocatalytic route scouting and enzyme screening toward the synthesis of α-benzyl L-glutamate. Front. Catal. 3:1285074. doi: 10.3389/fctls.2023.1285074
Received: 29 August 2023; Accepted: 05 December 2023;
Published: 31 January 2024.
Edited by:
Frank Hollmann, Delft University of Technology, NetherlandsReviewed by:
Davide Tessaro, Polytechnic University of Milan, ItalyAna I. Benítez-Mateos, ETH Zürich, Switzerland
Copyright © 2024 Quaedflieg, Jente, Müller, Vermote, Plesciuc, van der Laan, Nielsen and Schürmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Schürmann, bWFydGluLnNjaHVlcm1hbm5AaW5ub3N5bi5jb20=
 Peter J. L. M. Quaedflieg
Peter J. L. M. Quaedflieg Lisanne M. H. Jente1
Lisanne M. H. Jente1 Lone Nielsen
Lone Nielsen Martin Schürmann
Martin Schürmann