
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Catal. , 20 December 2021
Sec. Biocatalysis
Volume 1 - 2021 | https://doi.org/10.3389/fctls.2021.803416
 Simone A. De Rose1
Simone A. De Rose1 William Finnigan1†
William Finnigan1† Nicholas J. Harmer2
Nicholas J. Harmer2 Jennifer A. Littlechild1*
Jennifer A. Littlechild1* The HotSolute consortium
The HotSolute consortiumOsmolytes protect microbial cells against temperature, osmolarity and other stresses. The osmolyte cyclic 2,3-diphosphoglycerate, originally isolated from the thermophilic archaeon Methanothermus fervidus, naturally protects cellular proteins under extreme conditions. The biosynthetic pathway for cyclic 2,3-diphosphoglycerate has been introduced into the thermophilic bacterium Thermus thermophilus. The two enzymes in this synthetic pathway, 2-phosphoglycerate kinase and cyclic diphosphoglycerate synthetase, were incorporated into a newly designed modular BioBricks vector. The expression of this two-enzyme cascade resulted in the whole cell production of cyclic 2,3 diphosphoglycerate. In vivo production of cyclic 2,3-diphosphoglycerate was confirmed by mass spectrometry to a concentration up to 650 µM. This study demonstrates the feasibility of using this well studied thermophilic bacterium as a host in a whole-cell factory approach to produce cyclic 2,3 diphosphoglycerate. This raises the potential for commercialisation of cDPG for cosmetic and healthcare applications. Our work demonstrates the application of Thermus thermophilus as an alternative host for other high value small organic molecules of industrial interest.
Osmolytes are small organic molecules that are accumulated within cells as a response to stress conditions (Yancey, 2001; Littlechild et al., 2013). They act to increase the thermodynamic stability of biological macromolecules in extremophilic microorganisms without compromising their native functional activities (Yancey et al., 1982). The concentration of osmolytes in the cell can range from millimolar to 1–2 M in response to the extracellular osmolarity. They accumulate to up to 25% of dry cell weight in microorganisms exposed to stressful environmental conditions (Raddadi et al., 2015; Faria et al., 2018). As some osmolytes are tolerated by the cells over this wide concentration range, these compounds were also termed compatible solutes (Brown, 1976).
Extremolytes are organic osmolytes from extremophilic microorganisms that are adapted to grow at high or low temperature, high salinity and other environmental extremes (Lippert and Galinski, 1992; Rothschild and Mancinelli, 2001). These molecules have many applications in several different industrial sectors. These include additives to improve the stability of high value molecules such as enzymes, drugs, and antibodies (Iyer and Ananthanarayan, 2008; Tani et al., 2015; Hamon et al., 2017; Castro-Ochoa et al., 2019), food and cosmetic product ingredients (Buenger and Driller, 2004; Graf et al., 2008; Marini et al., 2014), and as potential therapeutics for several neurodegenerative disorders (Jorge et al., 2016).
In the past decade, there has been a surge in the successful biotechnological production of extremolytes, and most prominently ectoine and hydroxyectoine (Czech et al., 2018). These compounds are produced at large scale (5 tons per year). The world leader for ectoine manufacturing is the German company Bitop AG, who relies on a non-GMO Halomonas elongate strain that has the ability to export these extremolytes from the cells in a process referred to as “bacterial milking” (Sauer and Galinski, 1998). Ectoine accumulates inside H. elongata cells under high-salt growth conditions (15% (w/v) NaCl). It is released into the growth medium by osmotic downshock (3% (w/v) NaCl). More recently, ‘superleaky’ mutants of H. elongata have been isolated that excrete ectoine during high-salt growth (Kunte et al., 2014; Becker and Wittmann, 2020).
Another extremolyte is cyclic 2,3-diphosphoglycerate (cDPG). cDPG has been exclusively found in the hyperthermophilic archaeal methanogens such as Methanothermus fervidus, Methanopyrus kandleri, and Methanothermobacter thermoautotrophicus, at concentrations from 0.3–1.1 M (Hensel and König 1988; Ciulla et al., 1994; Matussek et al., 1998). cDPG is biosynthesised by a two-step pathway from the glycolytic intermediate 2-phosphoglycerate (2PG). The process requires two enzymes, 2-phosphoglycerate kinase (2PGK) that forms 2,3-di-phosphoglycerate (2,3DPG) from 2PG and cyclic di-phosphoglycerate synthase (cDPGS) that cyclises 2,3DPG to form the extremolyte cDPG (Lehmacher et al., 1990) (Scheme 1).
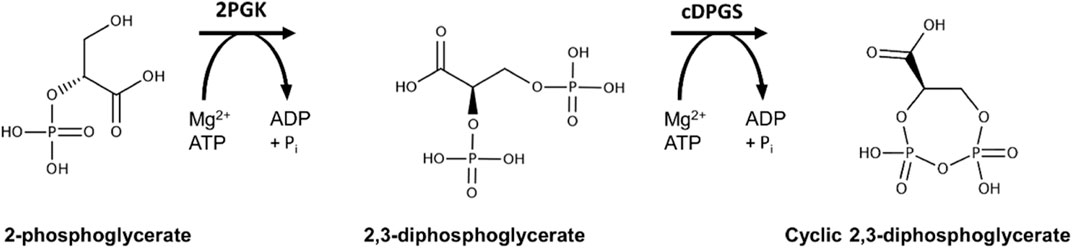
SCHEME 1. Reaction scheme for production of cyclic 2,3-diphosphoglycerate using the enzymes 2PGK and cDPGS derived from the archaeon Methanococcus fervidus.
In the native archaeal methanogen species cDPG biosynthesis is triggered by increasing the growth temperature (Lehmacher et al., 1990) and the accumulation of this extremolyte in the cells is correlated with high temperature environments. The concentration increases from 70 mM in M. thermoautotrophicum (65°C), to 300 mM in M. fervidus (84°C), and 1 M in M. kandleri (98°C) (Shima et al., 1998). One other role for cDPG has been suggested to be as a phosphate and energy storage compound (Sastry et al., 1992; Van Alebeek et al., 1994; Lentzen and Schwarz, 2006). Because the cDPGS reaction is exergonic at cellular concentrations, cDPG accumulation is favoured thermodynamically until this reaction reaches equilibrium (Shima et al., 1998). cDPG appears to play a role in the thermoprotection of proteins, and increased thermostability has been demonstrated for several model enzymes (Hensel and Jakob, 1993; Borges et al., 2002). Furthermore, cDPG protects plasmid DNA against oxidative damage by hydroxyl radicals (Lentzen and Schwarz, 2006). It can also function as a superoxide scavenger, with one third of the activity of the strong antioxidant ascorbic acid (Valentão et al., 2002).
The growth of these archaeal methanogens at a scale to obtain sufficient quantities of the cDPG extremolyte would be extremely challenging since these organisms are fastidious strict anaerobes that use C1 and C2 compounds as terminal electron acceptors (Thauer et al., 2008; Jabłoński et al., 2015). Their cultivation requires sophisticated and specialised equipment (Stetter et al., 1981). Attempts to use Escherichia coli as a host to bio-synthesise cDPG have had limited success, with a maximum production of 8.8 µmol per milligram of expressed protein (Moritz, 2003).
Transfer of the cDPG pathway (Scheme 1) into an aerobic, easy to grow thermophilic organism would allow the industrial production of cDPG from cheap feedstocks. In addition, it would maintain the optimum temperature for the enzymes of the cDPG pathway. The use of a so called “thermophilic cell factory” offers additional advantages such as a lower risk of contamination, the potential for the recovery of volatile products, higher mass transfer rates, improved solubility of some substrates, and the potential for integration with high temperature chemical processes (Frock and Kelly, 2012). Thermus thermophilus is an ideal candidate for this purpose, as it offers fast growth rates, high cell yields, and natural competency which significantly simplifies the genetic manipulation required (Cava et al., 2009; Verdú, et al., 2019). For these reasons, a wealth of genetic tools have already been established for T. thermophilus, including plasmids (Tamakoshi et al., 1999; Moreno et al., 2003; Fujino et al., 2020), promoters (Moreno et al., 2005; Cava et al., 2009), and selection markers (Lasa et al., 1992; Nakamura et al., 2005). Several studies have already demonstrated that T. thermophilus could potentially be used as a cell factory (Doi et al., 2009; Aulitto et al., 2017). These include early studies by the group of Tairo Oshima who demonstrated the expression of an isopropyl malate dehydrogenase chimeric enzyme in T. thermophilus (Kotsuka et al., 1996). Furthermore, 2PG is produced by the central metabolism of T. thermophilus as part of the Embden-Meyerhof-Parnas pathway from which the cDPG pathway branches (Swarup et al., 2014; Cordova et al., 2016).
This study reports the first example of using T. thermophilus as a whole-cell host to produce the valuable extremolyte cDPG. The enzymes 2PGK and cDPGS have been recombinantly expressed in T. thermophilus using a flexible plasmid design allowing production of cDPG to a level compatible with an industrial application. These results validate the concept of using a thermophilic host for in vivo biosynthesis of a variety of high value small molecules.
A new E. coli to T. thermophilus shuttle vector (pBBTth) was designed to facilitate the use of the BioBrick cloning method (http://hdl.handle.net/1721.1/21168). This vector combines origins of replication for E. coli and T. thermophilus, and a thermostable selectable marker from pMKE2 to allow selection in either E. coli or T. thermophilus (Supplementary Figure S4). The BioBricks cloning site and reporter gene from pSB1C3 (Supplementary Figure S3B) were added to facilitate the cloning steps in E. coli. The vector was designed for the addition of two Biobricks to form the final vector, removing the RFP reporter gene. More than two pieces could be combined if desired using the Gibson (Thomas et al., 2015) or Golden Gate (Engler and Marillonnet, 2014) cloning methods.
Four T. thermophilus promoters were identified, covering a range of strengths (Supplementary Table S1; Prof. José Berenguer and Prof. Aurelio Hidalgo, Universidad Autónoma de Madrid, and personal communication). To quantify promoter strength, the super-folder GFP (sfGFP), which functions at 70°C (Cava et al., 2008), or mRFP, were cloned downstream of each promoter in pBBTth. T. thermophilus was transformed with these constructs as well as controls with no promoter and grown in suitable minimal media (Averhoff, 2006). Both sfGFP and mRFP were codon optimized for expression in T. thermophilus and featured a C-terminal FLAG tag. Expression of both proteins was faintly visible on SDS-PAGE and was clearly detected by an anti-FLAG Western blot (Figure 1). No mRFP fluorescence was detected above background. This is likely due to the instability of the mRFP fluorophore at high temperature (Deepankumar et al., 2015). Expression of thermostable sfGFP was quantified from cultures grown in minimal media (Figure 2). As expected, PslpA showed strong expression (Luisa Faraldo et al., 1991; Verdú, et al., 2019). However, Prmp and Pnqo (Cava et al., 2008) both showed comparable expression while Pnar (Hidalgo et al., 2004), even when not induced, and showed expression at two-thirds of this level. Surprisingly, constructs without a promoter showed some expression, and likely from read-through from the selectable marker. These results confirmed that multiple T. thermophilus promoters can express recombinant proteins, and that none are sufficiently strong to cause cellular toxicity.
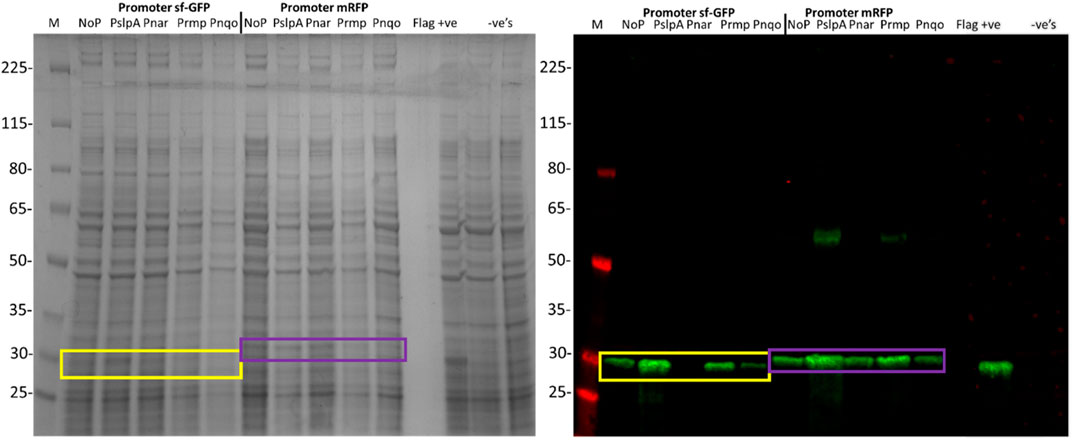
FIGURE 1. SDS-PAGE and Western blot of the total cell lysate from T. thermophilus expressing codon optimised sfGFP and mRFP, under the control of PslpA, Pnar, Prmp, Pnqo, and without a promoter. T. thermophilus containing empty pBBTthK and without a plasmid were included as negative controls. A T. thermophilus cell lysate expressing a 29 kDa FLAG tagged protein (pBBTth-Archaeoglobus fulgidus esterase) was included as a positive control. The markers are the Spectra™ Multicolor Broad Range Protein Ladder (Thermo #11842124). The Western blot was probed using a chicken anti-FLAG primary antibody and a goat anti-chicken IRDye 800CW secondary antibody. The expected sizes of sfGFP (26.8 kDa) and mRFP (28 kDa) are shown by the yellow and purple boxes respectively. The larger MW band in some mRFP lanes (∼56 kDa) likely reflects a detergent resistant dimer of mRFP.
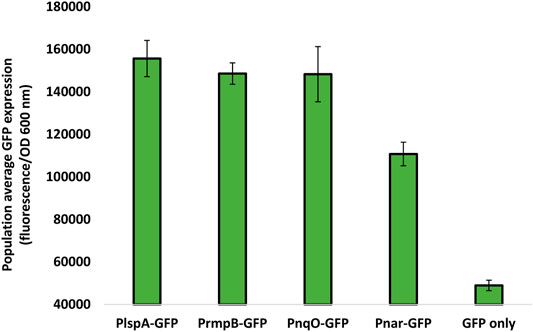
FIGURE 2. Population average sfGFP fluorescence as a measure of promoter strength sfGFP expression was measured by reading the fluorescence and OD600 nm of T. thermophilus cultures grown in minimal media. 200 μl aliquots was taken in triplicate from three shake flasks for each culture into a 96-well microtiter plate for reading using a plate reader. Population average GFP expression was calculated as the fluorescence recorded divided by OD600 nm. A blank was also measured using T. thermophilus cultures with no sfGFP expression and subtracted from all readings. Error bars represent standard error. Data are the mean of three biological replicates, each of which contained three technical replicates.
The 2PGK and cDPGS enzymes from M. fervidus were codon-optimised for T. thermophilus and cloned into pBBTth to form a new enzymatic cascade in T. thermophilus. Expression of Mf2PGK and MfcDPGS was directed using a polycistronic construct, with 6*His tags added to both proteins to allow confirmation of expression. Constructs were prepared with each of the four promoters identified above. The T. thermophilus was transformed with each construct and grown for 24 h. After approximately 3.5 h cultures containing constructs featuring the Pnar promoter were placed in a sealed and para-filmed 50 ml Falcon tube with the addition of 40 mM potassium nitrate. This was incubated at 70°C with no shaking to achieve anaerobic conditions that would trigger Pnar induction (Moreno et al., 2003; Hidalgo et al., 2004). cDPGs expression was detected from the three constitutive promoters (Supplementary Figure S1). In contrast, no expression was detected for 2PGK with any of the promoters. New ribosome binding sites were designed using the Salis RBS calculator (Salis, 2011) (Supplementary Table S3). The predicted expression strength correlated with the observed expression level with the 2PGK expression being detected only after using the Mf-2PGK RBS2 which had a predicted translation initiation rate of 236,855 compared to the previous Mf-2PGK RBS that was only 90,966. However, it is difficult to exclude the influence of other factors. Although the RBS calculator can be used for the design of functional RBSs for T. thermophilus good 2PGK expression was only observed with the new RBS2 and PslpA promoter (Fig. S2) but not with the other promoters (data not shown).
T. thermophilus cells expressing cDPGS and 2PGK showed significantly different growth in culture, when compared to cells transformed with empty pBBTth (Figure 3). As recently reviewed by Zeng and colleagues, synthetic pathways compete with their host for the cellular resources required for their respective functions. This resource competition can lead to poor performance and unexpected behaviour of the designed systems (Zeng et al., 2021). In our case there was a longer lag phase delay, of approximately 4 h and a lower final OD600 nm compared to cells with empty pBBTth. However, the exponential growth rate is almost unaffected. This might be due to the energetic burden of producing 2PGK and cDPGS, or it might reflect the diversion of resources from central metabolism to produce 2,3DPG and cDPG. This was tested by repeating the experiment in stress conditions (addition of high salt concentrations; Figure 4). In contrast to un-supplemented media, there was little effect shown from expressing the cDPG pathway in media supplemented with 150 mM NaCl, whilst cells transformed with the empty pBBTth grew slower. Supplementation of the media with 300 mM NaCl dramatically slowed the growth of all strains. The T. thermophilus cells expressing the cDPG pathway grew significantly better than the control, reaching a final OD600 nm of 0.64 ± 0.04 compared to the 0.28 ± 0.02 for the control (p < 0.001). These results strongly suggest that the cells with the synthetic pathway added grow slower in the base media because they are synthesising cDPG to bio-protective levels at the cost of the growth rate. Furthermore, although there are no reports of cDPG production being modulated by salt stress, cDPG can still have a protective/beneficial function in osmotic stress conditions as reported for other extremolytes such as ectoine and hydroxyectoine (Czech et al., 2018). Indeed, cDPG at pH 8.0 can counter balance potassium ions in a 1–3.3 ratio. This makes it a good extremolyte to respond to osmotic stress (Hensel and König, 1988; Shima et al., 1998).
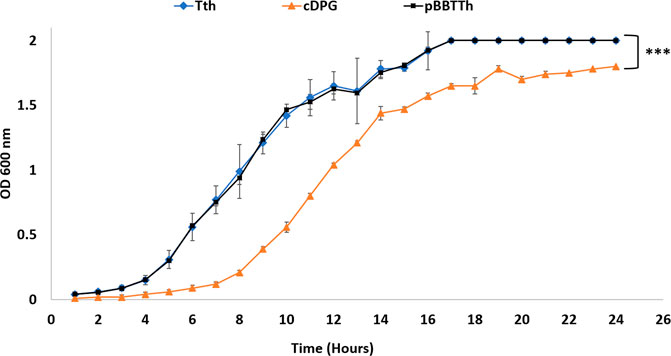
FIGURE 3. Growth curve of T. thermophilus cells transformed with different plasmids over a 24-h period. Blue diamond: Untransformed T. thermophilus cells (growth without kanamycin), black square: T. thermophilus cells transformed with empty pBBTth vector, and green triangle: T. thermophilus cells transformed with pBBTth-PslpA-cDPGS-2PGK plasmid. Error bars represent standard error. Data are the mean of three biological replicates. The entire growth curves were analysed by ordinary one-way ANOVA using GraphPad Prism v9.2. Statistical differences were considered significant if the p value was less than 0.05 (denoted by stars *p < 0.05; **p < 0.005; ***p < 0.001).
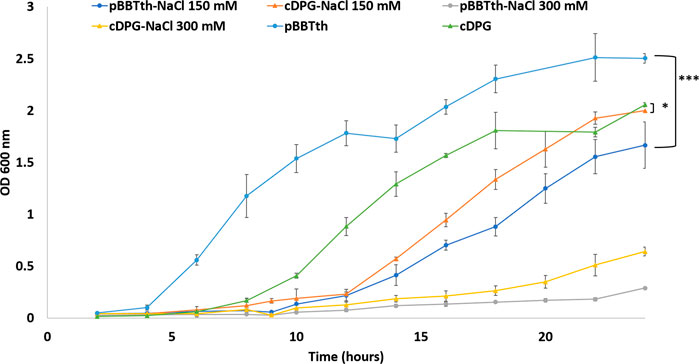
FIGURE 4. Growth curve of T. thermophilus showing the OD at 600 nm of cells transformed with empty pBBTth vector (circle) or pBBTth-PslpA-cDPGS-2PGK (triangle) in the presence of 0, 150, or 300 mM NaCl over a 24-h period. The entire growth curves were analysed by ordinary one-way ANOVA using GraphPad Prism v9.2. Statistical differences were considered significant if the p value was less than 0.05 (denoted by stars *p < 0.05; **p < 0.005; ***p < 0.001).
The cDPG production by T. thermophilus was determined over a 24-h period by HPLC-MS (Figure 5, 6A). As expected, wild-type T. thermophilus showed no cDPG production. The T. thermophilus expressing the 2PGK-cDPGS synthetic operon produced cDPG, with production peaking at 650 µM after 16 h. This corresponds to the late stage of exponential growth. As the cells enter stationary phase (24 h) there is a decline in cDPG concentration, consistent with the hypothesis that cDPG acts as an energy store (Sastry et al., 1992; Van Alebeek et al., 1994; Lentzen and Schwarz, 2006). The concentration of the substrate 2PG was also monitored (Figure 6B) showing that the 2PG concentration also peaks at late log phase. Notably, the 2PG concentration is not significantly lower in the cells expressing the cDPG pathway. This indicates that T. thermophilus can boost 2PG production to meet the requirement for cDPG biosynthesis, while maintaining 2PG concentration to approximately the same level as the control. No cDPG was detected in the culture medium without cell lysis (data not shown). This demonstrates that T. thermophilus can accumulate extremolytes in the cell up to 650 µM.
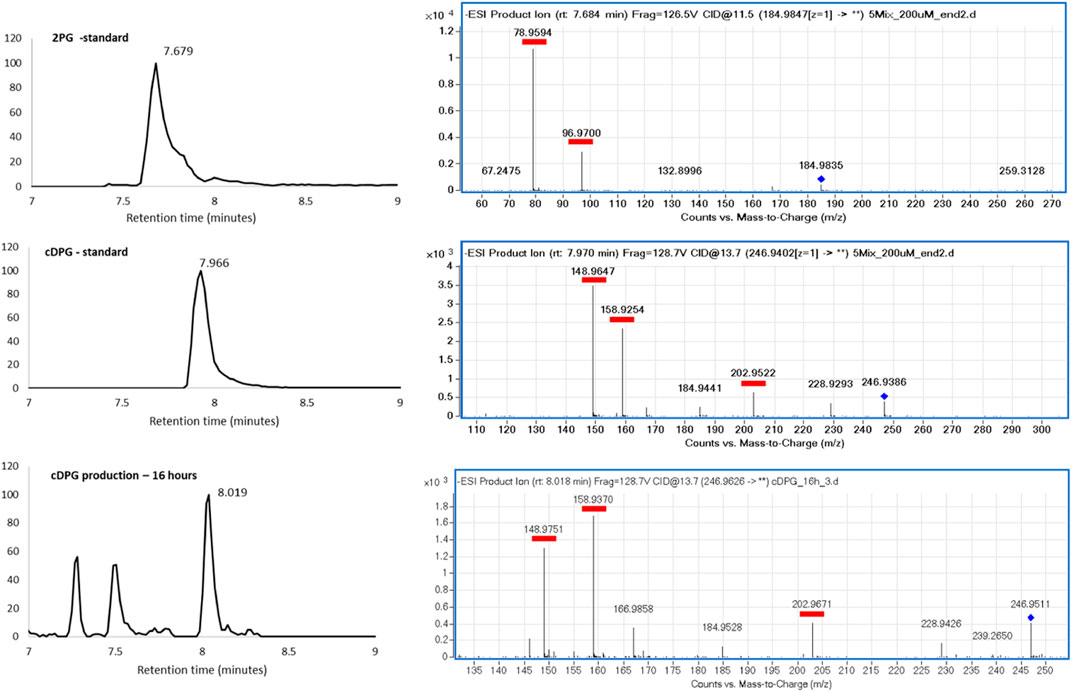
FIGURE 5. HPLC-MS analysis of 2PG and cDPG in standard and reaction. (Left) Extracted ion chromatograms. (Right) MS fragmentation patterns, with the whole molecule mass indicated by the blue diamond, and the fragments underlined in red.
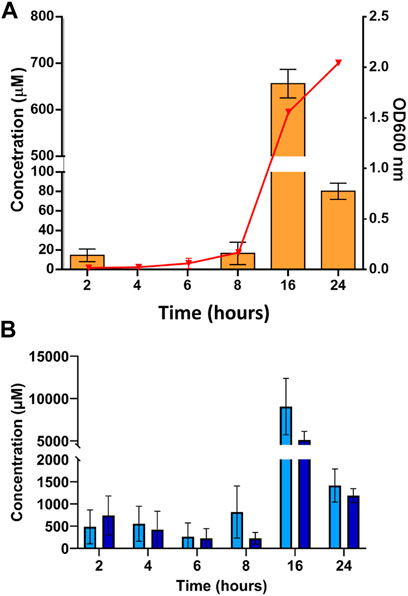
FIGURE 6. Whole cell production of the extremolyte cDPG in T. thermophilus cell cultures performed in small-scale shake flasks. T. thermophilus from an overnight starter culture was added to 50 ml of TBM media supplemented with 30 μg/ml kanamycin to an OD 600 nm of 0.0019. Cultures were grown in a 250 ml baffled flask and growth followed at OD600 nm. (A) The concentration of cDPG was monitored by HPLC-MS over 24 h of cell growth of T. thermophilus transformed with an empty pBBTth plasmid (light green) or with the pBBTth-PslpA-cDPGS-2PGK plasmid (orange), the red triangle overlay shows the growth curve of T. thermophilus cells transformed with pBBTth-PslpA-cDPGS-2PGK 24-h period. (B) concentration of 2PG as detected by HPLC-MS over 24 h of T. thermophilus cell growth transformed with empty pBBTth (light blue), and T. thermophilus cells transformed with pBBTth-PslpA-cDPGS-2PGK plasmid (dark blue). Error bars represent standard error. Data are the mean of three biological replicates.
The concentration of cDPG produced at the 16 h point is sufficiently high to make industrial production feasible in principle. To realise this opportunity there are three key challenges to overcome. Firstly, the cDPG concentration decreases after the exponential phase of cell growth at 16 h (dropping from ∼650 to 80 µM), indicating that T. thermophilus after reaching stationary phase likely utilises cDPG as an energy source as described in other studies (Sastry et al., 1992; Van Alebeek et al., 1994; Shima et al., 1998; Lentzen and Schwarz, 2006). Changing the expression strategy to a fed-batch system (Hewitt and Nienow, 2007) could reduce the cDPG from being used as an energy source and help to reach a higher OD600 nm value. Secondly, an optimised protocol for purification of cDPG from the cell culture is required. An ideal strategy would be similar to the “bacterial milking” described for ectoine. T. thermophilus naturally produces two other extremolytes, mannosylglycerate and trehalose (Nunes et al., 1995; Silva et al., 2005). A T. thermophilus trehalose deficient strain has been reported to have increased mannosylglycerate production under salt stress condition (Silva et al., 2003). The use of a trehalose and mannosylglycerate deficient strain could be beneficial to increase cDPG accumulation levels in response to salt stress. Finally, the data reported here are at a laboratory scale. Scale up to an industrial fermentation setting would likely require re-optimisation of the parameters at each stage of scale-up (Yang, 2014; Delvigne et al., 2017).
Here, we demonstrate for the first time a two-step whole-cell reaction performed in the thermophilic bacteria T. thermophilus. Two thermostable enzymes from M. fervidus have been cloned into a single plasmid and over-expressed under the control of the strong PslpA promoter. The production of the extremolyte cDPG was monitored with HPLC-MS, and the maximum concentration detected was approximately 650 µM compared to previous in vivo studies performed in E. coli which showed a maximum production of 8.8 µmol (Moritz, 2003). The whole-cell production of cDPG is associated with a slowdown of T. thermophilus growth. The introduction of synthetic pathways often results in growth defects because cellular resources are diverted away from biomass production (Zeng et al., 2021) and require additional energy (2 ATP for cDPG). Despite the slower growth of T. thermophilus, these results represent a major improvement when compared to the extraction of cDPG from cultivated methanogenic archaea which are strict anaerobes and are extremely difficult to grow.
Recently a new inducible promoter for the silica-induced protein (Sip) was used to express a thermostable β-galactosidase in T. thermophilus with a yield of 27 mg/L of culture (Fujino et al., 2016, 2020). This inducible promoter could be used in further experiments with T. thermophilus expressing the cDPG biosynthesis pathway for the industrial production of the extremolyte cDPG. There are key cost benefits of production in vivo when compared to a cell free system since the provision of substrate and cofactor is provided by the host cell.
T. thermophilus HB27 was kindly gifted by Prof Jose Berenguer (Universidad Autónoma de Madrid, Madrid, and Spain). Pure cDPG for use as a standard for the HPLC-MS was kindly donated by Prof Bettina Siebers (University of Duisburg-Essen, Essen, and Germany).
The pBBTTh vector was designed to obtain a T. thermophilus vector that could be used with the BioBricks cloning system (http://hdl.handle.net/10871/27323). Two vectors were selected as templates for the construction of the new vector. pMKE2 (Supplementary Figure S3A) is a commercially available vector capable of replication and selection in both E. coli and T. thermophilus, and carries the Pnar promoter (Moreno et al., 2005). pSB1C3 (Fig. S3B), is an established BioBricks vector for E. coli with a BioBricks cloning site flanked by two insulating terminator regions (Shetty et al., 2008). The BioBricks cloning site contains a stuffer gene for the expression of mRFP under the control of a lac promoter which helps identify successful cloning attempts. Fragments from the two vectors were designed with overhangs for Gibson assembly, amplified by PCR and then digested with DpnI. Gibson assembly was performed using the Gibson Assembly Cloning Kit (New England Biolabs #E5510S), following the manufacturer’s recommendations. The resulting plasmid was confirmed by restriction digest with XbaI and PstI giving bands at the expected sizes after agarose gel electrophoresis.
The Mf-2PGK and Mf-cDPGS genes were codon optimised based on the codon bias of T. thermophilus using the GeneArt, Gene Optimizer algorithm with a C-Terminal 6x His-Tag added. Ribosome binding sites (RBS) (Supplementary Table S3) were designed using the RBS calculator (Salis, 2011). Synthetic genes were ordered from GeneArt (Thermofisher, Waltham, MA, and United States).
PslpA and Pnar were cloned as BioBricks from pMKE2 by PCR, while Prmp and Pnqo were cloned from T. thermophilus genomic DNA. To ensure promoters from the T. thermophilus genome were captured, 200 bp and 150 bp regions upstream of the rmpB and nqo genes respectively were used.
Assembly parts were amplified by PCR using the Accuprime GC rich polymerase (Thermofisher, Waltham, MA, United States) and subcloned into pBBTTh (Supplementary Table S1). Primers for the assembly were designed using the NEBuilder assembly tool (Supplementary Table S2). PCR products were assembled with the NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolabs #E5520S) following the manufacturer instructions. The PslpA, Pnar, Prmp, and Pnqo promoters were used for all constructs. Constructs were assembled in E. coli with several steps; firstly, the genes for Mf-2PGK and Mf-cDPGS were cloned separately in pBBTth under the control of each promoter. After verification of successful cloning by colony PCR the entire synthetic operon was assembled (Figure 7) and verified by Sanger sequencing.
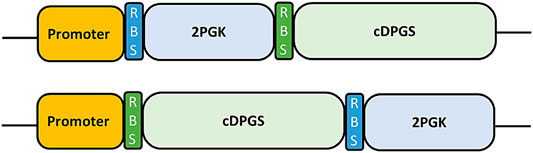
FIGURE 7. Schematic representation of the synthetic operon parts as originally designed (Top) and in the final construct (Bottom). Promoter: A promoter part, either PslpA, Pnar, Prmp or Pnqo. RBS: A custom RBS, designed using the RBS calculator (Salis, 2011). 2PGK and cDPGS: Codon optimized genes for expression.
The final polycistronic constructs were transformed in T. thermophilus by incubating 20 μg of plasmid DNA to 500 μl of T. thermophilus culture in 10 ml breather tubes for four hours at 70°C, 200 rpm. Cultures were plated on Thermus broth media (TBM) agar (Koyama et al., 1986; Averhoff, 2006) with 30 μg/ml kanamycin. Plates were incubated at 70°C overnight. Colonies were picked and grown overnight in TBM supplemented with 30 μg/ml kanamycin at 70°C and 200 rpm. Negative controls of T. thermophilus with an empty pBBTth plasmid, and without a plasmid were also set up (without kanamycin added where no plasmid was used).
Overnight cultures of T. thermophilus transformed with plasmids for the expression of sfGFP from the range of promoters were set up in TBM as standard. Minimal media cultures were inoculated with 500 μl of the overnight culture in 30 ml minimal media with 30 μg/ml kanamycin. Cultures were incubated at 60°C, 200 rpm. 200 μl samples were taken in triplicate into a 96-well microtiter plate for OD600 nm and fluorescence readings taken using an M200 Pro plate reader (Tecan, Mannedorf, and Switzerland). At approximately OD600 nm 0.15 the population average sfGFP expression was determined allowing the quantification of the gene expression from each promoter. Fluorescence settings were as follows: Excitation wavelength 450 nm, excitation bandwidth 9 nm, emission wavelength 508 nm, emission bandwidth 20 nm, gain 120, 10 flashes, 10 μs integration time, lag time 0 μs, settle time 0 μs, and an automatically calculated Z-position of 18 268 μm. Population average GFP fluorescence was calculated (blank subtracted fluorescence divided by blank subtracted OD600 nm). Standard deviations for each reading were combined and shown as error bars.
A single colony of T. thermophilus HB27 containing pBBTTh-cDPG was inoculated into 5 ml TBM supplemented with 30 μg/ml kanamycin and cultured at 65°C, 200 rpm, for 16 h. This seed culture was transferred to 50 ml TBM in a 250 ml baffled flask and grown for up to 48 h at 65°C, 200 rpm, and growth followed using OD600 nm. Samples from the expression culture were taken every 4 h for measurement of the OD. After 24 h samples were taken for SDS-PAGE and Western blot analysis. The SDS-PAGE gel was run on ExpressPAGE or SurePAGE™ (Genscript) Bis-Tris 4–20% gels at 140 V for 50 min. Gels were blotted onto an Amersham™ Protan™ 0.45 µM nitrocellulose membrane (GE Healthcare) with a Thermo Scientific™ Pierce™ G2 Fast Blotter. The Western blot was performed using a iBind system (Thermo Fischer, Waltam, MA, United States ) using a mouse Anti-His Tag (Sigma-Aldritch, St Louis, MI, United States #05–949) primary antibody at a 1:1,000 dilution and goat Anti-Mouse IRDye 680RD (Li-cor, Lincon, NE, United States , #926–68070) secondary antibody at a 1:2000 dilution. Either Spectra™ Multicolor Broad Range Protein Ladder (4–225 kDa; Thermo #11842124) or Blue Plus® IV Protein Marker (10–180 kDa; Generon, Slough, United Kingdom) was used for these analyses.
T. thermophilus starter cultures were grown in 5 ml of TBM media (supplemented with 30 μg/ml kanamycin where appropriate) and shaken at 180 r.p.m. overnight at 65°C. Cells were diluted in fresh TBM to an optical density (OD)600 nm of 1.9. From these cultures, lower initial density populations were appropriately diluted with media in a 1:1000 ratio.
Experiments were performed in triplicates using 50 ml Falcon tubes filled with 5 ml of media supplemented with 30 μg/ml kanamycin and/or 150 mM or 300 mM NaCl were appropriate. Cultures were incubated at 65°C at 180 rpm with an incline of 45°. Growth curves were recorded by measuring the OD600 nm over a 24 h period.
Growth curve parameters were estimated as described by Reding-Roman and colleagues except they used c.f.u. instead of OD to measure population density (N) from growth data using the logistic equation:
where N(t) is density at time t and L is lag and denotes the time taken to reach the mid-point of the sigmoid. K is the carrying capacity and r is the growth rate (Reding-Roman et al., 2017).
Statistical tests and data analysis were performed using Graphpad prism v9.2. The parameters associated with the logistic equation were estimated by fitting the equation to growth data using the Nonlinear fit option in Graphpad prism v9.2. Growth curve comparisons were performed with an ordinary one-way ANOVA. Multiple comparisons were made with general linear models (GLMs) with post-hoc Tukey multi-comparison of means (honest significant differences method).
500 μl samples were taken in triplicate at different time points (2 h/4 h/8 h/16 h/24 h). These were freeze-thawed three times (−20–60°C) and added to 1 ml of ice-cold acetonitrile. Samples were incubated for 10 min on ice and then centrifuged at a maximum speed of 16 000 g for 10 min. The supernatant was collected and transferred into a HPLC vial. Samples were kept at 4°C and analysed by HPLC within 24 h.
Metabolite profiling was performed using a QToF 6,520 Mass spectrometer (Agilent Technologies, Palo Alto, United States) coupled to a 1,200 series Rapid Resolution HPLC system. 5 µl of sample extract was loaded onto an Agilent Infinity Lab Poroshell 120 HILIC-Z 2.7 µm, 2.1 × 250 mm analytical column. For detection using negative ion mode, mobile phase A comprised 100% water with 10 mM ammonium acetate and 5 µM medronic acid and mobile phase B was 90% (v/v) acetonitrile with 10 mM ammonium acetate and 5 µM medronic acid. All solvents were of LC-MS grade.
The following gradient was used: 0 min–95% B; 5 min–65% B; 10 min–50% B; 11 min–95% B; 15 min–95% B followed by 1 min post time. The flow rate was 0.25 ml min−1 and the column temperature was held at 25°C for the duration. The source conditions for electrospray ionisation were as follows: gas temperature was 325°C with a drying gas flow rate of 9 L min−1 and a nebuliser pressure of 35 psi. The capillary voltage was 3.5 kV, the fragmentor voltage was 115 V and skimmer 70 V. Scanning was performed using the auto MS/MS function at 5 scans sec−1 for precursor ion surveying and 4 scans sec−1 for MS/MS, with a sloped collision energy of 3.5 V/100 Da with an offset of 5 V.
MassHunter qualitative analysis (version B.07.00) was used to identify potentially interesting compounds with similar masses to the sugar phosphate of interest and consistent fragmentation profile (Supplementary Figure S5). MassHunter quantitative analysis (version B.07.01) was used to compare compounds (peak areas) between samples.
List of the HotSolute consortium collaborators: Bettina Siebers, YmV0dGluYS5zaWViZXJzQHVuaS1kdWUuZGU=; Christopher Bräsen, Y2hyaXN0b3BoZXIuYnJhZXNlbkB1bmktZHVlLmRl; Christina Stracke, Y2hyaXN0aW5hLnN0cmFja2VAdW5pLWR1ZS5kZQ==; Benjamin Meyer, YmVuamFtaW4ubWV5ZXJAdW5pLWR1ZS5kZQ==; Misha N. Isupov, bWlzdXBvdkBleGV0ZXIuYWMudWs=; Nicholas J. Harmer, Ti5KLkhhcm1lckBleGV0ZXIuYWMudWs=; Simone Antonio De Rose, Uy5BLkRlLVJvc2VAZXhldGVyLmFjLnVr; Jennifer Ann Littlechild, Si5BLkxpdHRsZWNoaWxkQGV4ZXRlci5hYy51aw==; Elizaveta Bonch-Osmolovskaya, ZWxpemF2ZXRhLmJvQGdtYWlsLmNvbQ==; Sergey Gavrilov, c25nYXZyaWxvdkBnbWFpbC5jb20=; Ilya Kublanov, a3VibGFub3YuaWx5YUBnbWFpbC5jb20=; Daniela Monti, ZGFuaWVsYS5tb250aUBzY2l0ZWMuY25yLml0; Erica Ferrandi, ZXJpY2EuZmVycmFuZGlAc2NpdGVjLmNuci5pdA==; Eleonora Dore, ZWxlb25vcmEuZG9yZUBzY2l0ZWMuY25yLml0; Felix Müller, ZmVsaXgubXVlbGxlckBldm9uaWsuY29t; Jacky Snoep, amFja3kuc25vZXBAbWFjLmNvbQ==.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author. Data has been deposited with the University of Exeter ORE https://doi.org/10.24378/exe.3683.
WF designed and assembled the pBBTth vector. WF and SD characterised the promoters. SD cloned and expressed the cDPG pathway genes and performed all the bio-production experiments. SD and NH analysed the data. JL and NH coordinated the work. JL and the HotSolute consortium obtained funding for this work as part of a ERA-CoBioTech grant. SD wrote the initial draft of the manuscript with further contributions from all authors.
Funds for open access publication fees from University of Exeter, Library BBSRC component of ERA-Biotech grant HotSolute.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This work has been carried out as part of the HotSolute project under by the ERA-CoBiotech EU programme and financed by local countries funding agencies. The project aimed to identify new thermophilic bacterial and archaeal chassis for extremolyte production. The authors would like to thank the United Kingdom BBSRC-funded grant BB/R02166X/1 and the University of Exeter for support. The authors would also like to thank Dr Deborah Salmon for her help and support in running the HPLC-MS experiments and for the data analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fctls.2021.803416/full#supplementary-material
Aulitto, M., Fusco, S., Fiorentino, G., Limauro, D., Pedone, E., Bartolucci, S., et al. (2017). Thermus thermophilus as Source of Thermozymes for Biotechnological Applications: Homologous Expression and Biochemical Characterization of an α-galactosidase. Microb. Cel. Fact. 16, 1–10. doi:10.1186/S12934-017-0638-4
Averhoff, B. (2006). 12 Genetic Systems for Thermus. Methods Microbiol. 35, 279–308. doi:10.1016/S0580-9517(08)70015-1
Becker, J., and Wittmann, C. (2020). Microbial Production of Extremolytes - High-Value Active Ingredients for Nutrition, Health Care, and Well-Being. Curr. Opin. Biotechnol. 65, 118–128. doi:10.1016/j.copbio.2020.02.010
Borges, N., Ramos, A., Raven, N., Sharp, R., and Santos, H. (2002). Comparative Study of the Thermostabilizing Properties of Mannosylglycerate and Other Compatible Solutes on Model Enzymes. Extremophiles 6, 209–216. doi:10.1007/s007920100236
Brown, A. D. (1976). Microbial Water Stress. Bacteriol. Rev. 40, 803–846. doi:10.1128/mmbr.40.4.803-846.197610.1128/br.40.4.803-846.1976
Buenger, J., and Driller, H. (2004). Ectoin: An Effective Natural Substance to Prevent UVA-Induced Premature Photoaging. Skin Pharmacol. Physiol. 17, 232–237. doi:10.1159/000080216
Castro-Ochoa, K. F., Vargas-Robles, H., Chánez-Paredes, S., Felipe-López, A., Cabrera-Silva, R. I., Shibayama, M., et al. (2019). Homoectoine Protects against Colitis by Preventing a Claudin Switch in Epithelial Tight Junctions. Dig. Dis. Sci. 64, 409–420. doi:10.1007/s10620-018-5309-8
Cava, F., De Pedro, M. A., Blas-Galindo, E., Waldo, G. S., Westblade, L. F., and Berenguer, J. (2008). Expression and Use of Superfolder green Fluorescent Protein at High Temperatures In Vivo: A Tool to Study Extreme Thermophile Biology. Environ. Microbiol. 10, 605–613. doi:10.1111/j.1462-2920.2007.01482.x
Cava, F., Hidalgo, A., and Berenguer, J. (2009). Thermus thermophilus as Biological Model. Extremophiles 13, 213–231. doi:10.1007/s00792-009-0226-6
Ciulla, R. A., Burggraf, S., Stetter, K. O., and Roberts, M. F. (1994). Occurrence and Role of Di- Myo -Inositol-1,1′-Phosphate in Methanococcus igneus. Appl. Environ. Microbiol. 60, 3660–3664. (Accessed May 26, 2017). doi:10.1128/aem.60.10.3660-3664.1994
Cordova, L. T., Lu, J., Cipolla, R. M., Sandoval, N. R., Long, C. P., and Antoniewicz, M. R. (2016). Co-utilization of Glucose and Xylose by Evolved Thermus thermophilus LC113 Strain Elucidated by 13 C Metabolic Flux Analysis and Whole Genome Sequencing. Metab. Eng. 37, 63–71. doi:10.1016/j.ymben.2016.05.001
Czech, L., Hermann, L., Stöveken, N., Richter, A., Höppner, A., Smits, S., et al. (2018). Role of the Extremolytes Ectoine and Hydroxyectoine as Stress Protectants and Nutrients: Genetics, Phylogenomics, Biochemistry, and Structural Analysis. Genes 9, 177–258. doi:10.3390/genes9040177
Deepankumar, K., Nadarajan, S. P., Bae, D.-H., Baek, K.-H., Choi, K.-Y., and Yun, H. (2015). Temperature Sensing Using Red Fluorescent Protein. Biotechnol. Bioproc. E 20, 67–72. doi:10.1007/s12257-014-0456-z
Delvigne, F., Takors, R., Mudde, R., van Gulik, W., and Noorman, H. (2017). Bioprocess Scale-Up/down as Integrative Enabling Technology: from Fluid Mechanics to Systems Biology and beyond. Microb. Biotechnol. 10, 1267–1274. doi:10.1111/1751-7915.12803
Doi, K., Fujino, Y., Inagaki, F., Kawatsu, R., Tahara, M., Ohshima, T., et al. (2009). Stimulation of Expression of a Silica-Induced Protein (Sip) in Thermus thermophilus by Supersaturated Silicic Acid. Appl. Environ. Microbiol. 75, 2406–2413. doi:10.1128/AEM.02387-08
Engler, C., and Marillonnet, S. (2014). Golden Gate Cloning. Methods Mol. Biol. 1116, 119–131. doi:10.1007/978-1-62703-764-8_9
Faraldo, M. L., De Pedro, M. A., and Berenguer, J. (1991). Cloning and Expression in Escherichia coli of the Structural Gene Coding for the Monomeric Protein of the S Layer of Thermus thermophilus HB8. J. Bacteriol. 173, 5346–5351. doi:10.1128/jb.173.17.5346-5351.1991
Faria, C., Borges, N., Rocha, I., and Santos, H. (2018). Production of Mannosylglycerate in Saccharomyces cerevisiae by Metabolic Engineering and Bioprocess Optimization. Microb. Cel. Fact. 17, 1–11. doi:10.1186/s12934-018-1023-7
Frock, A. D., and Kelly, R. M. (2012). Extreme Thermophiles: Moving beyond Single-Enzyme Biocatalysis. Curr. Opin. Chem. Eng. 1, 363–372. doi:10.1016/J.COCHE.2012.07.003
Fujino, Y., Goda, S., Suematsu, Y., and Doi, K. (2020). Development of a New Gene Expression Vector for Thermus thermophilus Using a Silica-Inducible Promoter. Microb. Cel. Fact. 19, 1–12. doi:10.1186/s12934-020-01385-2
Fujino, Y., Nagayoshi, Y., Iwase, M., Yokoyama, T., Ohshima, T., and Doi, K. (2016). Silica-Induced Protein (Sip) in Thermophilic Bacterium Thermus thermophilus Responds to Low Iron Availability. Appl. Environ. Microbiol. 82, 3198–3207. doi:10.1128/AEM.04027-15
Graf, R., Anzali, S., Buenger, J., Pfluecker, F., and Driller, H. (2008). The Multifunctional Role of Ectoine as a Natural Cell Protectant. Clin. Dermatol. 26, 326–333. doi:10.1016/j.clindermatol.2008.01.002
Hamon, N., Mouline, C. C., and Travert, M. (2017). Synthesis of Mannosylglycerate Derivatives as Immunostimulating Agents. Eur. J. Org. Chem. 2017, 4803–4819. doi:10.1002/ejoc.201700682
Hensel, R., and Jakob, I. (1993). Stability of Glyceraldehyde-3-Phosphate Dehydrogenases from Hyperthermophilic Archaea at High Temperature. Syst. Appl. Microbiol. 16, 742–745. doi:10.1016/S0723-2020(11)80348-6
Hensel, R., and Kã¶nig, H. (1988). Thermoadaptation of Methanogenic Bacteria by Intracellular Ion Concentration. FEMS Microbiol. Lett. 49, 75–79. doi:10.1111/j.1574-6968.1988.tb02685.x
Hewitt, C. J., and Nienow, A. W. (2007). The Scale‐Up of Microbial Batch and Fed‐Batch Fermentation Processes. Adv. Appl. Microbiol. 62, 105–135. doi:10.1016/S0065-2164(07)62005-X
Hidalgo, A., Betancor, L., Moreno, R., Zafra, O., Cava, F., Fernández-Lafuente, R., et al. (2004). Thermus thermophilus as a Cell Factory for the Production of a Thermophilic Mn-dependent Catalase Which Fails to Be Synthesized in an Active Form in Escherichia coli. Appl. Environ. Microbiol. 70, 3839–3844. doi:10.1128/AEM.70.7.3839-3844.2004
Iyer, P. V., and Ananthanarayan, L. (2008). Enzyme Stability and Stabilization-Aqueous and Non-aqueous Environment. Process Biochem. 43, 1019–1032. doi:10.1016/j.procbio.2008.06.004
Jabłoński, S., Rodowicz, P., and Łukaszewicz, M. (2015). Methanogenic Archaea Database Containing Physiological and Biochemical Characteristics. Int. J. Syst. Evol. Microbiol. 65, 1360–1368. doi:10.1099/IJS.0.000065
Jorge, C. D., Borges, N., Bagyan, I., Bilstein, A., and Santos, H. (2016). Potential Applications of Stress Solutes from Extremophiles in Protein Folding Diseases and Healthcare. Extremophiles 20, 251–259. doi:10.1007/s00792-016-0828-8
Kotsuka, T., Akanuma, S., Tomuro, M., Yamagishi, A., and Oshima, T. (1996). Further Stabilization of 3-isopropylmalate Dehydrogenase of an Extreme Thermophile, Thermus thermophilus, by a Suppressor Mutation Method. J. Bacteriol. 178, 723–727. doi:10.1128/jb.178.3.723-727.1996
Koyama, Y., Hoshino, T., Tomizuka, N., and Furukawa, K. (1986). Genetic Transformation of the Extreme Thermophile Thermus thermophilus and of Other Thermus Spp. J. Bacteriol. 166, 338–340. doi:10.1128/jb.166.1.338-340.1986
Kunte, H., Lentzen, G., and Galinski, E. (2014). Industrial Production of the Cell Protectant Ectoine: Protection Mechanisms, Processes, and Products. Cbiot. 3, 10–25. doi:10.2174/22115501113026660037
Lasa, I., Castón, J. R., Fernández-Herrero, L. A., Pedro, M. A., and Berenguer, J. (1992). Insertional Mutagenesis in the Extreme Thermophilic Eubacteria Thermus thermophilus HB8. Mol. Microbiol. 6, 1555–1564. doi:10.1111/j.1365-2958.1992.tb00877.x
Lehmacher, A., Vogt, A.-B., and Hensel, R. (1990). Biosynthesis of Cyclic 2,3-diphosphoglycerate. FEBS Lett. 272, 94–98. doi:10.1016/0014-5793(90)80456-S
Lentzen, G., and Schwarz, T. (2006). Extremolytes: Natural Compounds from Extremophiles for Versatile Applications. Appl. Microbiol. Biotechnol. 72, 623–634. doi:10.1007/s00253-006-0553-9
Lippert, K., and Galinski, E. (1992). Enzyme Stabilization Be Ectoine-type Compatible Solutes: protection against Heating, Freezing and Drying. Appl. Microbiol. Biotechnol. 37, 61–65. doi:10.1007/BF00174204
Littlechild, J., Novak, H., James, P., and Sayer, C. (2013). “Mechanisms of thermal Stability Adopted by Thermophilic Proteins and Their Use in white Biotechnology,” in Thermophilic Microbes in Environmental and Industrial Biotechnology: Biotechnology of Thermophiles (Springer Netherlands), 481–507. doi:10.1007/978-94-007-5899-5_19
Marini, A., Reinelt, K., Krutmann, J., and Bilstein, A. (2014). Ectoine-containing Cream in the Treatment of Mild to Moderate Atopic Dermatitis: A Randomised, Comparator-Controlled, Intra-individual Double-Blind, Multi-center Trial. Skin Pharmacol. Physiol. 27, 57–65. doi:10.1159/000351381
Matussek, K., Moritz, P., Brunner, N., Eckerskorn, C., and Hensel, R. (1998). Cloning, Sequencing, and Expression of the Gene Encoding Cyclic 2,3-Diphosphoglycerate Synthetase, the Key Enzyme of Cyclic 2,3-Diphosphoglycerate Metabolism in Methanothermus fervidus. J. Bacteriol. 180, 5997–6004. (Accessed April 3, 2020). doi:10.1128/jb.180.22.5997-6004.1998
Moreno, R., Haro, A., Castellanos, A., and Berenguer, J. (2005). High-level Overproduction of His-Tagged Tth DNA Polymerase in Thermus thermophilus. Appl. Environ. Microbiol. 71, 591–593. doi:10.1128/AEM.71.1.591-593.2005
Moreno, R., Zafra, O., Cava, F., and Berenguer, J. (2003). Development of a Gene Expression Vector for Thermus thermophilus Based on the Promoter of the Respiratory Nitrate Reductase. Plasmid 49, 2–8. doi:10.1016/S0147-619X(02)00146-4
Moritz, P. (2003). Die Bedeutung der cyclischen 2, 3-Diphosphoglycerat Synthetase für die temperaturabhängige Regulation der intrazellulären Konzentration von cyclischem 2, 3-Diphosphoglycerat im hyperthermophilen Archaeum Methanothermus fervidus. PhD thesis, University of Duisburg-Essen.
Nakamura, A., Takakura, Y., Kobayashi, H., and Hoshino, T. (2005). In Vivo directed Evolution for Thermostabilization of Escherichia coli Hygromycin B Phosphotransferase and the Use of the Gene as a Selection Marker in the Host-Vector System of Thermus thermophilus. J. Biosci. Bioeng. 100, 158–163. doi:10.1263/JBB.100.158
Nunes, O. C., Manaia, C. M., Da Costa, M. S., and Santos, H. (1995). Compatible Solutes in the Thermophilic Bacteria Rhodothermus marinus and “Thermus thermophilus”. Appl. Environ. Microbiol. 61, 2351–2357. doi:10.1128/aem.61.6.2351-2357.1995
Raddadi, N., Cherif, A., Daffonchio, D., Neifar, M., and Fava, F. (2015). Biotechnological Applications of Extremophiles, Extremozymes and Extremolytes. Appl. Microbiol. Biotechnol. 99, 7907–7913. doi:10.1007/s00253-015-6874-9
Reding-Roman, C., Hewlett, M., Duxbury, S., Gori, F., Gudelj, I., and Beardmore, R. (2017). The Unconstrained Evolution of Fast and Efficient Antibiotic-Resistant Bacterial Genomes. Nat. Ecol. Evol. 1, 1–11. doi:10.1038/s41559-016-0050
Rothschild, L. J., and Mancinelli, R. L. (2001). Life in Extreme Environments. Nature 409, 1092–1101. doi:10.1038/35059215
Salis, H. M. (2011). “The Ribosome Binding Site Calculator,” in Methods In Enzymology (Academic Press), 19–42. doi:10.1016/B978-0-12-385120-8.00002-4
Sastry, M. V. K., Robertson, D. E., Moynihan, J. A., and Roberts, M. F. (1992). Enzymatic Degradation of Cyclic 2,3-diphosphoglycerate to 2,3-diphosphoglycerate in Methanobacterium thermoautotrophicum. Biochemistry 31, 2926–2935. doi:10.1021/bi00126a012
Sauer, T., and Galinski, E. A. (1998). Bacterial Milking: A Novel Bioprocess for Production of Compatible Solutes. Biotechnol. Bioeng. 57, 306–313. doi:10.1002/(SICI)1097-0290(19980205)57:3<306:AID-BIT7>3.0
Shetty, R. P., Endy, D., and Knight, T. F. (2008). Engineering BioBrick Vectors from BioBrick Parts. J. Biol. Eng. 2, 1. doi:10.1186/1754-1611-2-5
Shima, S., Hérault, D. A., Berkessel, A., and Thauer, R. K. (1998). Activation and Thermostabilization Effects of Cyclic 2,3-diphosphoglycerate on Enzymes from the Hyperthermophilic Methanopyrus kandleri. Arch. Microbiol. 170, 469–472. doi:10.1007/s002030050669
Silva, Z., Alarico, S., Nobre, A., Horlacher, R., Marugg, J., Boos, W., et al. (2003). Osmotic Adaptation of Thermus thermophilus RQ-1: Lesson from a Mutant Deficient in Synthesis of Trehalose. J. Bacteriol. 185, 5943–5952. doi:10.1128/jb.185.20.5943-5952.2003
Silva, Z. l., Alarico, S., and Da Costa, M. S. (2005). Trehalose Biosynthesis in Thermus thermophilus RQ-1: Biochemical Properties of the Trehalose-6-Phosphate Synthase and Trehalose-6-Phosphate Phosphatase. Extremophiles 9, 29–36. doi:10.1007/s00792-004-0421-4
Stetter, K. O., Thomm, M., Winter, J., Wildgruber, G., Huber, H., Zillig, W., et al. (1981). Methanothermus fervidus, Sp. nov., a Novel Extremely Thermophilic Methanogen Isolated from an Icelandic Hot spring. Zentralblatt für Bakteriologie Mikrobiologie Hygiene: Abt. Originale C: Allgemeine, Angewandte Ökologische Mikrobiologie 2, 166–178. doi:10.1016/s0721-9571(81)80038-5
Swarup, A., Lu, J., DeWoody, K. C., and Antoniewicz, M. R. (2014). Metabolic Network Reconstruction, Growth Characterization and 13C-Metabolic Flux Analysis of the Extremophile Thermus thermophilus HB8. Metab. Eng. 24, 173–180. doi:10.1016/j.ymben.2014.05.013
Tamakoshi, M., Yaoi, T., Oshima, T., and Yamagishi, A. (1999). An Efficient Gene Replacement and Deletion System for an Extreme thermophile, Thermus. thermophilus. FEMS Microbiol. Lett. 173, 431–437. doi:10.1111/J.1574-6968.1999.TB13535.X
Tani, Y., Miyake, R., Yukami, R., Dekishima, Y., China, H., Saito, S., et al. (2015). Functional Expression of L-Lysine α-oxidase from Scomber japonicus in Escherichia coli for One-Pot Synthesis of L-Pipecolic Acid from DL-Lysine. Appl. Microbiol. Biotechnol. 99, 5045–5054. doi:10.1007/s00253-014-6308-0
Thauer, R. K., Kaster, A.-K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic Archaea: Ecologically Relevant Differences in Energy Conservation. Nat. Rev. Microbiol. 6 (6), 579–591. doi:10.1038/nrmicro1931
Thomas, S., Maynard, N. D., and Gill, J. (2015). DNA Library Construction Using Gibson Assembly. Nat. Methods 12, i–ii. doi:10.1038/nmeth.f.384
Valentão, P., Fernandes, E., Carvalho, F., Andrade, P. B., Seabra, R. M., and Bastos, M. L. (2002). Antioxidant Activity of Hypericum androsaemum Infusion: Scavenging Activity against Superoxide Radical, Hydroxyl Radical and Hypochlorous Acid. Biol. Pharm. Bull. 25, 1320–1323. doi:10.1248/bpb.25.1320
Van Alebeek, G.-J. W. M., Tafazzul, G., Kreuwels, M. J. J., Keltjens, J. T., and Vogels, G. D. (1994). Cyclic 2,3-diphosphoglycerate Metabolism in Methanobacterium thermoautotrophicum (Strain AH): Characterization of the Synthetase Reaction. Available at: https://link.springer.com/content/pdf/10.1007%2FBF00314474.pdf (Accessed October 10, 2018).
Verdú, C., Sanchez, E., Ortega, C., Hidalgo, A., Berenguer, J., and Mencía, M. (2019). A Modular Vector Toolkit with a Tailored Set of Thermosensors to Regulate Gene Expression in Thermus thermophilus. ACS Omega 4, 14626–14632. doi:10.1021/acsomega.9b02107
Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D., and Somero, G. N. (1982). Living with Water Stress: Evolution of Osmolyte Systems. Science 217, 1214–1222. doi:10.1126/science.7112124
Yancey, P. H. (2001). Water Stress, Osmolytes and Proteins. Am. Zool. 41, 699–709. doi:10.1093/icb/41.4.699
Yang, X. (2014). Scale-Up of Microbial Fermentation Process. Man. Ind. Microbiol. Biotechnol., 669–675. doi:10.1128/9781555816827.CH47
Keywords: industrial application, compatible solute, thermophiles, enzyme cascade
Citation: De Rose SA, Finnigan W, Harmer NJ, Littlechild JA and The HotSolute consortium (2021) Production of the Extremolyte Cyclic 2,3-Diphosphoglycerate Using Thermus thermophilus as a Whole-Cell Factory. Front. Catal. 1:803416. doi: 10.3389/fctls.2021.803416
Received: 27 October 2021; Accepted: 19 November 2021;
Published: 20 December 2021.
Edited by:
Wuyuan Zhang, Tianjin Institute of Industrial Biotechnology (CAS), ChinaReviewed by:
Ram Karan, King Abdullah University of Science and Technology, Saudi ArabiaCopyright © 2021 De Rose, Finnigan, Harmer, Littlechild and The HotSolute consortium. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer A. Littlechild, Si5BLkxpdHRsZWNoaWxkQGV4ZXRlci5hYy51aw==
†Present address: Manchester Institute of Biotechnology, University of Manchester, United Kingdom
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.