- 1School of Chemical and Biological Engineering, Seoul National University, Seoul, South Korea
- 2Institute of Molecular Biology and Genetics, Seoul National University, Seoul, South Korea
- 3Department of Chemical Engineering, Pusan National University, Busan, South Korea
- 4Department of Bioscience and Biotechnology, Konkuk University, Seoul, South Korea
- 5Bioengineering Institute, Seoul National University, Seoul, South Korea
- 6Institute of Engineering Research, Seoul National University, Seoul, South Korea
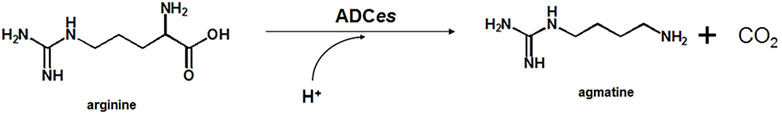
Agmatine, involved in various modulatory actions in cellular mechanisms, is produced from arginine (Arg) by decarboxylation reaction using arginine decarboxylase (ADC, EC 4.1.1.19). The major obstacle of using wild-type Escherichia coli ADC (ADCes) in agmatine production is its sharp activity loss and instability at alkaline pH. Here, to overcome this problem, a new disulfide bond was rationally introduced in the decameric interface region of the enzyme. Among the mutants generated, W16C/D43C increased both thermostability and activity. The half-life (T1/2) of W16C/D43C at pH 8.0 and 60°C was 560 min, which was 280-fold longer than that of the wild-type, and the specific activity at pH 8.0 also increased 2.1-fold. Site-saturation mutagenesis was subsequently performed at the active site residues of ADCes using the disulfide-bond mutant (W16C/D43C) as a template. The best variant W16C/D43C/I258A displayed a 4.4-fold increase in the catalytic efficiency when compared with the wild-type. The final mutant (W16C/D43C/I258A) was successfully applied to in vitro synthesis of agmatine with an improved yield and productivity (>89.0% yield based on 100 mM of Arg within 5 h).
Introduction
Agmatine ((4-aminobutyl)guanidine) is a critical arginine metabolite synthesized through ADC reaction, and it has various actions in the body. It is a potential candidate for therapeutic use in various diseases, including neurotrauma diseases, mood disorders, diabetes mellitus, cognitive disorders, and cancer (Aricioglu et al., 2003; Roth et al., 2004; Wade et al., 2009; Keynan et al., 2010; Haenisch et al., 2011; Song et al., 2014; Rahangdale et al., 2021). An interesting recent study evaluated the pharmacological effects of treating agmatine on the offspring of rats exposed to alcohol in the prenatal period. The results showed potential for treating cognitive impairment and other neurological complications associated with fetal alcohol spectrum disorders (Aglawe et al., 2021). Agmatine has also been shown to have significant hepatic and renal protective functions in cholestatic animals. This mechanism of action of agmatine may be related to its antioxidant properties (Ommati et al., 2020). The practical function of agmatine as a diverse drug candidate in the body is also related to its interference with imidazoline receptors (Zomkowski et al., 2002; Neis et al., 2016). Based on the promising results of agmatine against various illnesses, the molecule may be utilized for potential therapeutic targets (Piletz et al., 2013).
ADCes is a robust biocatalyst that produces agmatine from the decarboxylation of Arg. Studies have shown that ADCes have excellent characteristics, such as high specific activity and high substrate selectivity, compared to ADCs from other origins (Blethen et al., 1968; Morris and Boeker, 1983). The major problem using ADCes for the synthesis of agmatine is that the activity and stability of the enzyme vary greatly depending on pH conditions. ADCes uses pyridoxal 5-phosphate (PLP) as a cofactor to decarboxylate the α-carboxyl group of Arg and consumes a proton (Figure 1). As a result, the reaction pH increases during the generation of agmatine and significantly affects ADCes structural stability and activity. Under acidic conditions, the five homodimers of ADCes spontaneously associate as a decamer, becoming an active form, whereas the structure dissociates as an inactive homodimer form under neutral or alkaline conditions. Even though it is not sure why the homodimer structures are inactive, ADCes gain their active functionality depending on the pH (Andrell et al., 2009; Kanjee et al., 2011). Thus, it is desirable to overcome this problem via protein engineering to apply the enzyme for stable agmatine production.
Various diamines from renewable carbon sources are becoming essential in building a sustainable chemical industry, and decarboxylases are frequently applied for such purposes (Chae et al., 2020). However, similar to ADCes, various decarboxylases such as lysine decarboxylase (Kanjee et al., 2011), glutamate decarboxylase (Capitani et al., 2003), and ornithine decarboxylase often tend to lose their activities depending on the reaction pH. Even though buffers and pH titers can be used to maintain the pH of the decarboxylation reaction at the required pH, the use of additives in mass production adds additional costs, dilutes the concentration of the target product, and complicates the purification process. Thus, if the enzyme is active even in alkaline conditions without the additives, it can reduce the final production cost, which is advantageous for the industrial-scale process. For this reason, several studies aimed to improve various decarboxylase activities at alkaline pH (Guirard and Snell, 1980; Thu Ho et al., 2013; Hong et al., 2017). Even though there is a previous study producing agmatine using ADC, recombinant wild-type ADCs are used in the reaction (Sun et al., 2015), and no attempt was undertaken to alter the properties of the enzyme for industrial applications.
Random mutagenesis, often used in directed evolution to improve the desired properties of a target protein, creates a wide variety of libraries covering the entire protein sequence. Intensive labor is required to find desirable mutants within a limited time (Porter et al., 2016). Thus, to reduce the library size efficiently and screen effective mutants, advanced bioinformatics and computational tools are often used to create smart libraries. Through rigorous computational methods, calculating the conservative scores based on multiple sequence alignments to analyze putative positive mutation sites called “hot spots” (Hazes and Dijkstra, 1988; Craig and Dombkowski, 2013), or homology modeling based on the sequence similarity of proteins are some frequently applied technical examples to help select critical residues in the enzyme (Choi et al., 2014a; Choi et al., 2014b; Choi et al., 2016; Jung et al., 2018). Subsequently, protein engineering techniques such as multiple site-saturation mutagenesis (SSM)) (Reetz and Carballeira, 2007; Sun et al., 2016) or site-directed mutagenesis (SDM) (Bornscheuer and Pohl, 2001) can be applied to the selected residues to generate mutation libraries.
In this study, to improve ADCes enzyme activity and stability at alkaline pH, we selected residues to perform mutations by computationally analyzing the enzyme’s structural changes and active sites. Substantial improvements in thermal stability and specific activity at alkaline pH were achieved by introducing a new disulfide pair in the decameric interface of the ADCes homodimer subunit. Our results provide a general mutation strategy for enhancing enzyme stability by disulfide bond formation on oligomeric state proteins and subsequently enhancing their specific activity via active site mutations.
Materials and Methods
Construction of ADCes in Expression Vector
DNA manipulations were performed according to the method of Sambrook, etc. (Sambrook et al., 2006). E.coli DH5α was used for DNA manipulation, and E.coli BL21 (DE3) was used as a host strain for gene expression. Escherichia coli BL21 (DE3) genomic DNA was used as the source for adiA gene, which encodes protein ADCes (GenBank accession: CAQ34466). To express C-terminal His-tagged fusion ADCes, the coding region of adiA was amplified by PCR using forward primer 5′- TATAATCATATGATGAAAGTATTAATTGTTGAA -3′ and reverse primer 5′- ATATACTCGAGCGCTTTCACGCACAT -3’. The PCR product was digested with restriction enzymes (NdeI/XhoI) and inserted into the corresponding restriction enzyme sites in pET24ma vector (Lee et al., 2010) as reported previously (Hong, 2017).
Disulfide Bond Design of ADCes
ADCes PDB structure (2VYC) was analyzed using DbD v2.12, which predicts the appropriate pair of residues capable of disulfide bonding (Craig and Dombkowski, 2013). Predicted pairs were identified using Pymol, and mutation was performed by only selecting disulfide residue pairs in the interfacial region. The primer sequences for the three disulfide-linked mutants W16C/D42C, W16C/D43C, and W16C/D42C/D43C are shown in Supplementary Table S1.
Site-Saturation Mutagenesis
To screen for positive mutants in selected residues of ADCes, NNK codons were used for site-saturated mutagenesis (Hong et al., 2018). The primers corresponding to Supplementary Table S1 were used for PCR to perform alanine scanning and site-saturated mutation. The gene libraries were treated with DpnI for 1 h at 37°C, and 10 μl of the library plasmids were transformed into E. coli BL21 (DE3). To screen mutants from a single residue, ∼200 colonies were examined to ensure a mutation coverage of 95% or greater. The mutant libraries were cultured in 96 deep-well plates for 20 h, and after centrifuging the cultured cells, the supernatant was discarded, and the relative activity of mutants was measured.
Expression and Purification of ADCes Variants
According to the previously reported method, the expression and purification of ADCes variants were carried out (Hong, 2017). IPTG (0.2 mM) was added to the culture when the OD600 reached 0.8, and the cells were harvested after 18 h induction at 18°C. His6-tagged fusion proteins were purified at 4°C with Ni-NTA agarose resin from Qiagen (Hilden, Germany). Elution containing the purified His6-tagged ADCes variants was concentrated by ultrafiltration using Centriplus YM-30 (Millipore, Bedford, MA) with a molecular weight cut-off of 100 kDa.
Temperature Effect on ADCes Variants
The protein melting temperature (Tm) of the variants were compared by SYPRO Orange excitation/emission wavelength profile (Niesen et al., 2007). 1000X SYPRO Orange was dissolved in Tris-HCl (50 mM, pH 8.0) buffer, and 20 μl solution was added to the 0.5 μg purified enzyme. Light cycler 480 II System (Roche Applied Science) was used for the detection of fluorescence changes at specific time points. By the excitation/emission curve, Δ Tm was calculated (Hong, 2017).
The thermal activity of ADC was evaluated by measuring the enzyme activity at various temperatures. The purified enzyme variants were incubated for 1 h at 33 to 65°C, and then cooled on ice for 10 min. The residual enzyme activity was then measured.
To confirm thermal stability under acidic or alkaline conditions, the purified enzyme was placed in 50 mM citrate buffer, pH 5.6, or 50 mM sodium phosphate buffer, pH 8.0, respectively. The samples were then placed in 60°C water bath. Residual activity was calculated based on the ADC activities of non-incubated enzymes. All reactions were done in triplicate.
Enzyme Assay
The relative reaction rates of the mutant libraries were quantitatively measured by a colorimetric method using cresol red (pKa = 8.3) (Hong, 2017). The decarboxylase reaction was measured using 5 mM pH8.0 sodium phosphate buffer, 4 mM L-Arg, 0.2 mM PLP and 0.2 mM indicator. For high-throughput library screening, the same method was applied, and the cultured mutants were examined in 96-well plates. The increase in absorbance at 574 nm was recorded using a UV spectrophotometer (BMG LABTECH, Germany), and the activity was determined via the ΔOD on the basis of the solution containing all substances other than the enzyme.
To determine the kcat and KM values, 200 μl reaction was performed in 96-well plates using purified enzymes. The reactivity was confirmed at 45°C by increasing Arg concentrations from 0.5 to 20 mM in pH 8.0 potassium phosphate buffer. kcat and KM were calculated with the nonlinear regression analysis of the Michaelis-Menten equation by GraphPad Prism 7 (https://www.graphpad.com/scientific-software/prism/) software.
To determine enzyme activity at various pH, a reaction solution was obtained using the following 60 mM buffers: sodium citrate (pH 5.0–6.0); sodium phosphate (pH 7.0–8.0); and Tris-HCl (pH 8.8). 30 mM L-Arg, 0.1 mM PLP, and 7.5 μg of purified enzyme were added to the reaction mixture. Enzyme activity was measured using HPLC.
Batch Reaction for Agmatine Production
To compare agmatine production over time, 150 μg of the purified enzyme was added to the 500 μl reaction mixture of 100 mM Arg and 0.2 mM PLP. Reactions were carried out at 45°C and quantitated by HPLC after certain time intervals. All the reactions were done in triplicate.
Analysis of Substrate and Products
Quantitative analysis of Arg and agmatine was performed using Younglin HPLC system with IonoSpher 5 C, G100x3 Column (Agilent, United States). The flow rate of eluant consisting of 0.6 g L−1 citrate, 4 g L−1 tartaric acid, 1.4 g L−1 ethylene diamine, 5% methanol in 95% water was 0.9 ml min−1 at 40°C. Refractive index (RI) detection was used for the detection of substrate and product. Retention times for Arg and agmatine were 1.5 and 4.1 min, respectively.
Molecular Modeling Studies
The ADC mutants were modeled by introducing the mutant residues into the reported wild-type structure (PDB 2VYC) using SWISS-MODEL (Bordoli et al., 2009). The template and W16C/D43C/I258A ADC mutant had three amino acid differences, resulting in a sequence identity of nearly 100%. The docking model of substrate Arg into the ADC active site was generated around 5 Å of the PLP site using AutoDock Vina 1.1.2 (Trott and Olson, 2010).
Results
Design and Selection of ADCes Disulfide Bridge Variants
ADCes have been reported to be activated depending on pH due to its conformation changes (Andrell et al., 2009). To confirm the pH dependent activity of ADCes, the specific activity was measured in a pH range between 5.0 and 8.0. The highest specific activity of the ADCes was observed at pH 5.0, and the activity decreased sharply when the pH increased. Little activity remaining at pH above 7.0, which is in good agreement with the reported activity profile (Kanjee et al., 2011) (Supplementary Figure S1).
To rationally create a focused, smart library for selecting ADCes mutants with enhanced structural stability at alkaline pH, insertion of disulfide bonds at the decameric interface was attempted. A web-based tool, “Disulfide by Design (DbD) v2.12”, was used for in silico design of new disulfide bridges using the 3D structure of ADCes (PDB 2VYC). The computational algorithm predicted the distance between the two Cβs of a Cys pair when a particular target residue is mutated to cysteine (Cys) and verified possible disulfide bonds (Craig and Dombkowski, 2013). Out of 83 disulfide pair candidates, five pairs were situated on the decamer interface region. Among the five putative disulfide bond pairs, we narrowed down the putative mutation targets to the residues involved in the pentameric inner ring formation. Thus, the N-terminal wing domain interface in helix α′ 1 and helix α′ 2 regions were selected and counted as ‘hot-spot’ (Andrell et al., 2009) (Figure 2A). From the two putative disulfide pair candidates in the hot-spot regions, W16C was predicted to form a disulfide bond with D42C or D43C, respectively. W16 is located in helix α′ 1, whereas D42 and D43 residues are in helix α′ 2 of another homodimer subunit (Figure 2B). To examine the activity and stability changes of ADCes through Cys substitution, two mutants were generated: W16C/D42C and W16C/D43C.
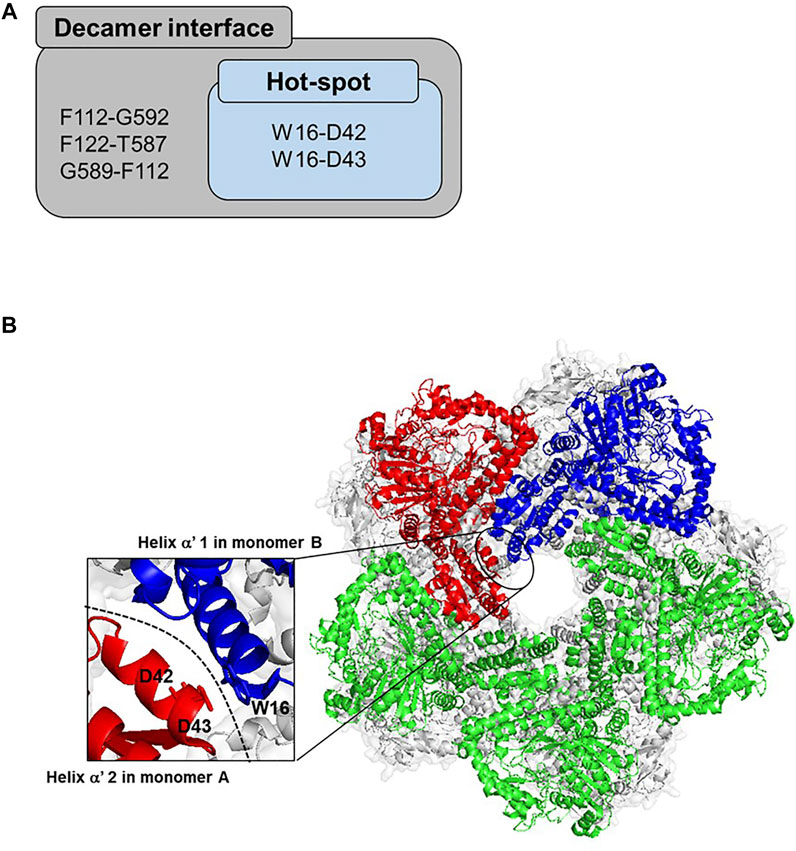
FIGURE 2. Proposed disulfide bridges in the interface of ADCes. (A) The potential residue pairs for disulfide formation from DbD V1.20. Out of the five predicted pairs located in the decamer interface, only two pairs are in the “hot-spot”, the intrinsic interface only for decameric structures. (B) The “hot-spot” decameric interface region of ADCes (PDB 2VYC). The basic unit of ADCes is composed of five homodimers that combine as a decamer. To define the monomers, monomer A is colored in red and monomer B is colored in blue. The green structure represents the other monomers in the pentameric ring among the decameric monomers. There were two disulfide bond pairs predicted in the interface. W16 in helix α′ 1 in monomer B was predicted to form a disulfide bond with D42 and D43 in helix α′ 2 in monomer A, respectively.
The enzyme activities of the two resulting mutants were measured at pH 8.0 using their cell extracts. As given in Supplementary Figure S2, the relative activities of the variants were measured using ADCes activity as control. Among the two mutants examined, only W16C/D43C mutation was active towards the substrate. When protein expression levels were compared, the protein expression level of the W16C/D42C mutant was similar to that of the wild-type, but this mutant lost activity (Supplementary Figure S3); thus, only W16C/D43C mutant was chosen for further enzyme characterization.
Enzymatic Activity of Purified W16C/D43C Mutant at Alkaline pHs
To better understand and precisely compare the W16C/D43C mutant activity, enzyme reactions were conducted in different alkaline pH buffers using the purified enzymes. At pH 8.0, the specific activity of ADCes and W16C/D43C mutant were 3.01 ± 0.10 and 6.28 ± 0.12 μmol/min/mg, respectively, whereas at pH 8.8, the specific activity of ADCes and W16C/D43C mutant were 2.73 ± 0.11 and 5.05 ± 0.6 μmol/min/mg, respectively. One thing to note is that W16C/D43C mutant showed almost two-fold enhanced specific activity (i.e., 2.1 and 1.9 folds at pH 8.0 and 8.8, respectively) at alkaline conditions compared to the wild-type (Figure 3). The purified enzyme reaction at pH 8.0 showed higher activity enhancement than the cell extract (Supplementary Figure S2).
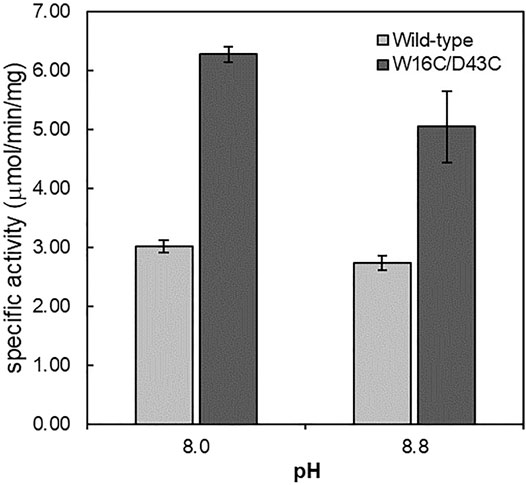
FIGURE 3. Specific activity of ADCes and W16C/D43C in alkaline pH. Each purified enzyme was tested against 30 mM concentration Arg. Values are expressed as μmol per min per mg of purified enzyme. Experimental points represent mean values of triple replicates. Error bars represent the standard deviations.
Determination of the Thermostability of ADCes Variants
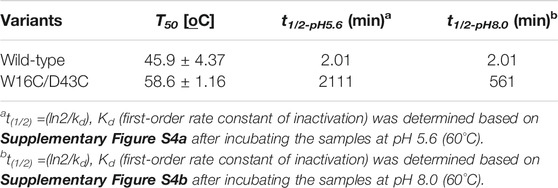
The thermostability of wild-type and W16C/D43C was compared by incubating the purified enzymes at 60°C, as explained in the Material and Methods section. To examine the thermostability at different pH, the samples were incubated at pH 5.6 (Figure 4A) and pH 8.0 (Figure 4B), respectively. The wild-type activity dropped to almost zero within 10 min at both pH 5.6 and 8.0 conditions; However, W16C/D43C maintained activity compared to wild-type at both pHs. When the enzymes were heated at pH 5.6, W16C/D43C was exceptionally stable, and nearly 97% of the initial activity remained after 10 min incubation. The dramatic thermostability enhancement was calculated by the half-life (t1/2-pH5.6) as shown in Supplementary Figure S4a. The t1/2-pH5.6 of W16C/D43C mutant was 2100 min, 1050-fold as that of ADCes at 60°C. In the case of pH 8.0 (Supplementary Figure S4b), the t1/2-pH8.0 of W16C/D43C mutant was 560 min, almost 280-fold as that of the wild-type indicating that the dramatic thermostability enhancement of W16C/D43C mutant was maintained under both acidic and alkaline conditions.
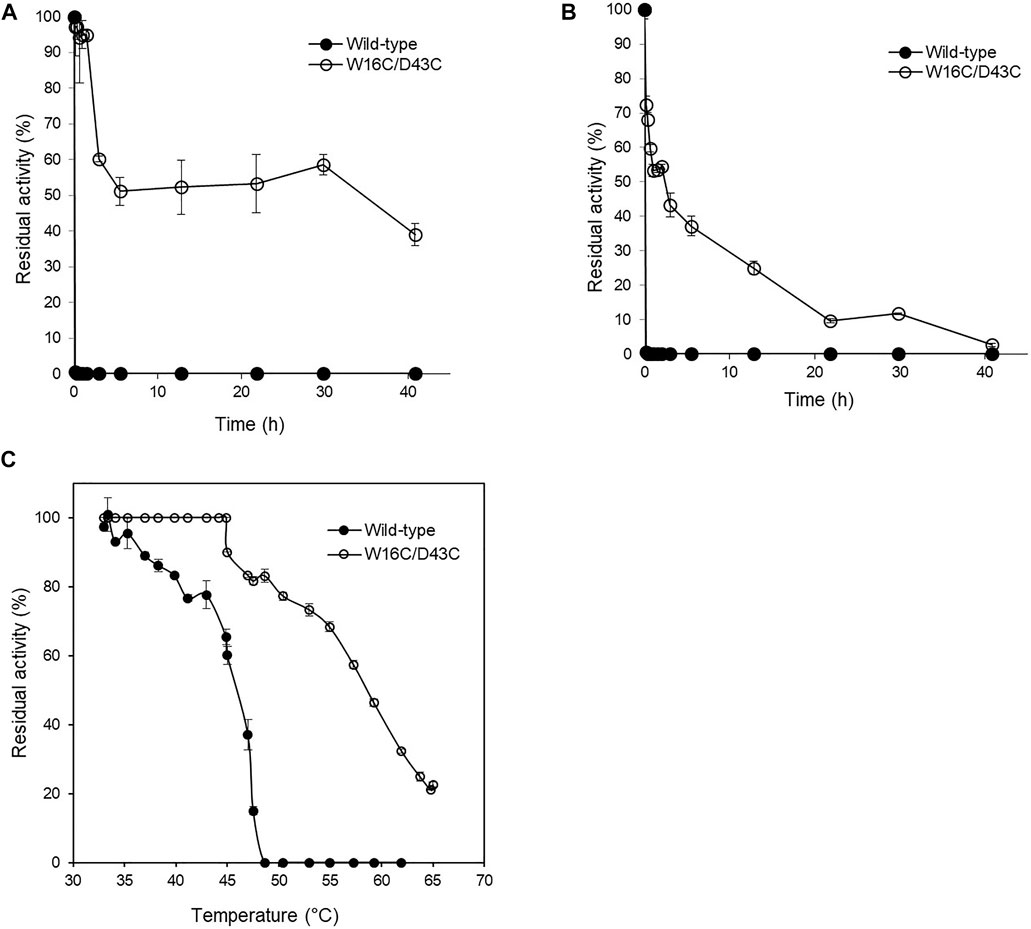
FIGURE 4. Thermal stability of W16C/D43C mutant compared to ADCes. The half-life of W16C/D43C mutant was compared to that of ADCes by incubating the purified samples at 60°C and measuring the remaining activity. The samples were respectively incubated at (A) pH5.6 using 50 mM citric buffer and (B) pH 8.0 using 50 mM sodium phosphate buffer. (C) To determine T50, the remaining enzyme activities were measured after the enzyme variants were incubated from 33°C to 65°C for 1 h. The closed circle represents ADCes, and the open circle represents W16C/D43C. Experimental points represent mean values of triplicate measurements. Error bars represent the standard deviations.
T50, the temperature at which any enzyme loses 50% of its activity after 1 h incubation, was also measured (Figure 4C), and the W16C/D43C mutant displayed 12.7 ± 3.2°C higher T50 than ADCes. All of the examined thermostability values were summarized in Table 1, verifying that the disulfide bond mutation substantially enhanced the ADC’s stability.
Site-Saturation Mutagenesis in the Active Site Residues
In order to further increase the activity of W16C/D43C disulfide bond mutant, single SSMs at the active site residues of ADCes were subsequently performed. Candidate residues for mutagenesis were selected based on ADCes -Arg docking simulations and the previously reported agmatine aldimine structure at ADCes active site (Andrell et al., 2009). Among the residues directly contacting the cofactor PLP or the substrate Arg, the residues interacting with the phosphate group of PLP and carboxylic group of Arg were carefully examined. Among 13 residues, seven residues near the phosphate group of PLP and six residues near the carboxylic group of Arg were chosen (Figure 5A). To minimize the experiments for SSM, alanine scanning was performed in these residues before SSM. When the corresponding amino acids of the selected residues were substituted to Ala, only the two mutants, C254A and I258A, maintained more than 60% of their original activities towards Arg (Figures 5B,C). The activity loss of most of Ala substituted mutants was somewhat expected because the examined residues were highly conserved based on the conservation analysis using HotSpot Wizard 2.0 (Bendl et al., 2016) (Figure 5A).
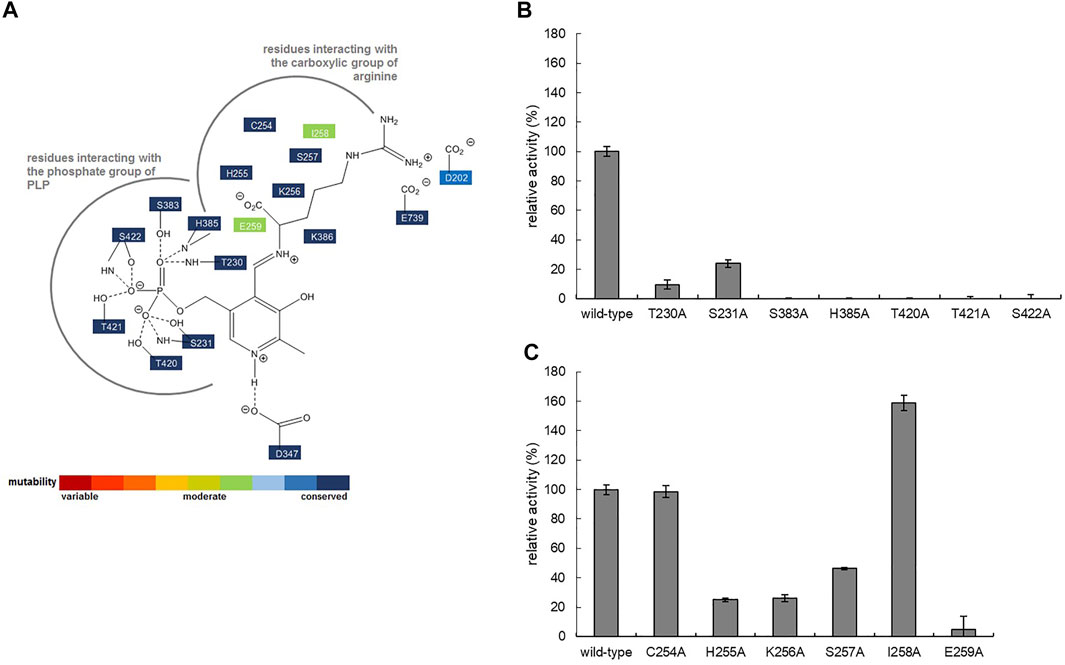
FIGURE 5. (A) Schematic diagram showing the interactions stabilizing PLP in the active site of ADCes (PDB2VYC). The thick grey line shows the pocket interacting with the substrate Arg. The degree of conservation is indicated by different colors and is based on the result of HotSpot Wizard. Relative activities of alanine-substituted mutant enzymes for ADCes residues interacting with the (B) phosphate group of PLP and (C) carboxylic group of Arg.
C254 and I258 were subjected to SSM using the disulfide bond mutant (W16C/D43C) template. Degenerate NNK codons were used for the library construction, and at least 172 mutant clones were examined to ensure a mutation coverage of 95% or more for a single SSM (Nov 2013). 200 colonies were selected from each residue library from the agar plates, and, in total, 400 clone activities were compared through whole-cell cultures in 96 well plates at pH 8.0. Among the mutants, 12 colonies with higher activity than the template W16C/D43C were identified. However, only one mutant displayed more than 1.4-fold increased activity, and its mutation site was identified as W16C/D43C/I258A. Interestingly, the best mutant from the SSM library at position I258 was the same mutant created by alanine scanning.
Characterization of ADCes Variants
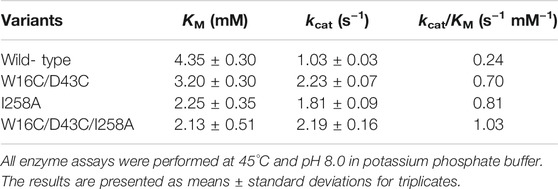
To investigate the mutational effect on the kinetic values, the kinetic parameters of W16C/D43C, I258A, and the final mutant W16C/D43C/I258A of ADC were determined after purification at pH 8.0, as shown in Table 2. Compared with ADCes, all the three variants displayed an increase in kcat and a decrease in KM value, resulting in enhanced catalytic efficiency (kcat/KM). In particular, the final mutant W16C/D43C/I258A showed enhanced kcat value for Arg (2.1 fold increase) and showed decreased KM value (2.0 fold decrease), resulting in a significant kcat/KM value (4.4 fold increase) compared to that of the wild-type.
In order to confirm the influence of the additional active site mutation (W16C/D43C/I258A) on the disulfide mutant (W16C/D43C), the melting temperatures (Tm) of ADCes variants were measured using differential scanning fluorometry (DSF) (Supplementary Figure S5). The disulfide bond W16C/D43C mutant exhibited 18°C higher Tm than that of ADCes. However, I258A mutation alone harmed the stability of ADCes and displayed 2.4°C lower Tm value. Nonetheless, when the single mutation (I258A) was combined with the disulfide mutation (W16C/D43C), the melting temperature of the final mutant (W16C/D43C/I258A) was 81.7 ± 0.01°C, which was 16.0°C higher than the wild-type. The final mutant W16C/D43C/I258A provided dramatic structural stability against the single I258A mutation.
Batch Reaction of ADCes Variants
Finally, the time course of agmatine production was monitored by HPLC with RI detector using the purified enzyme variants (150 μg) together with 100 mM Arg and 0.2 mM PLP. The starting pH of the batch reaction for agmatine production using Arg monohydrochloride as substrate was ca. 6.7. As shown in Figure 6, the examined three mutants were identified to be superior to ADCes in converting Arg into agmatine throughout the whole reaction. I258A and W16C/D43C/I258A mutants exhibited an initial conversion rate of up to 1.4- and 1.5-fold, respectively, within 20 min compared to the wild type. In terms of the final yield, W16C/D43C and W16C/D43C/I258A mutants with the disulfide bond mutation showed 1.8- and 2-fold increases of agmatine yield compared to the wild-type within 5 h, respectively. As a result, due to the improved stability and the activity, W16C/D43C/I258A mutant showed the highest agmatine yield, which is ca. 2-fold improvement (>89% yield within 5 h) in total compared to that of the wild-type (>43% yield within 5 h).
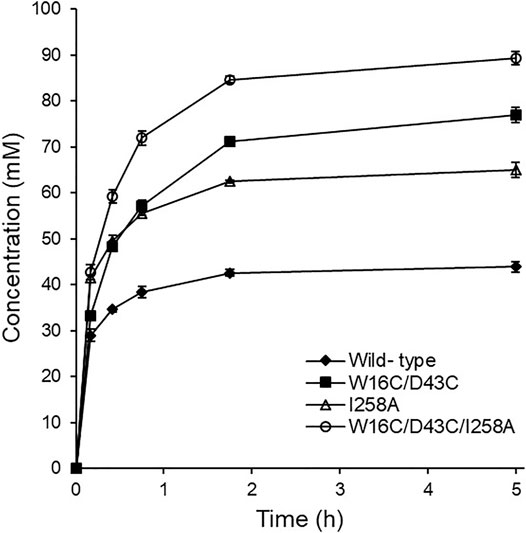
FIGURE 6. Time course agmatine production. Agmatine production was compared at 45°C using the purified enzymes of ADCes, W16C/D43C, I258A, and W16C/D43C/I258A. The reaction mixture contained 100 mM Arg, 0.2 mM of PLP, and 150 μg purified enzymes.
Discussion
Intracellular ADCes activate according to the acid stress response of E. coli cells, and by its decarboxylation mechanism, consumes a proton in the cytoplasm. Consumption of the protons in acidic conditions allows the cells to maintain a neutral pH suitable for cell growth. However, ADCes are no longer active when the pH becomes neutral or alkaline. This phenomenon occurs to avoid excessive degradation of Arg in the cells (Andrell et al., 2009). However, the acid response of ADCes causes problems when applied for industrial agmatine production because the pH increases rapidly due to the high concentration of substrate used in the reaction process.
ADCes belongs to the prokaryotic ornithine decarboxylase (pODC) subclass of Fold Type 1 PLP-dependent decarboxylase, composed of five domains: an N-terminal wing domain, linker domain, PLP-binding domain, the aspartate aminotransferase-like small domain followed by a C-terminal domain (Andrell et al., 2009). The N-terminal wing domain is vital in creating the decamer structure. Through hydrogen bondings or electrostatic interactions in this domain, ADCes forms an active decamer from the inactive homodimers depending on the pH. In particular, the first two α-helices present in the ADCes wing domain are essential for creating the pentameric structure of the enzyme (Andrell et al., 2009). Thus, these specific interface regions were selected to generate disulfide bridge mutants.
When creating SSM libraries, residues close to the active site and substrate-binding pocket were selected. We were particularly interested in the residues interacting with the phosphate group of cofactor PLP, which are essential in binding the ligand to PLP-dependent enzymes (Denesyuk et al., 2002). The residues interacting with the phosphate group of PLP were examined primarily. Due to the extremely high conservation scores of the examined seven residues (Figure 5A), all the Ala substituted mutants lost their original activities. According to a reported study of other amino acid decarboxylases, the carbon dioxide releasing process from the α-carbon of the substrate is the rate-limiting step of decarboxylation reactions (Kluger and Rathgeber, 2008); thus, the residues near the carboxyl group of Arg where its dynamic interaction occurs were selected for further Ala scanning. Although mutating the substrate interacting residues is very likely to reduce the original activity of the enzyme (Toscano et al., 2007), interestingly enough, a mutant with improved activity was discovered.
A few reports analyzed ADC catalytic efficiency from various origins. In one of the studies, the ADC from G. forsetii was evaluated after analyzing the phylogenetic distribution of various forms of ADC (Burrell et al., 2010). The kcat/KM value of the G. forsetii ADC at pH 8.5 was 0.22 mM-1s-1. Considering that the mutant W16C/D43C/I258A in this study exhibited a value of 1.03 mM-1s-1 kcat/KM at pH 8.0, it is not easy to compare the activities accurately because of the slightly different alkaline pH conditions used for the evaluation. However, roughly speaking, our final W16C/D43C/I258A mutant is approximately 4.7-fold more active than the G. forsetii ADC at alkaline conditions. From another recent study, the catalytic efficiency of ADC from Shewanella algae was characterized (Pei et al., 2021). Even though the kcat (12.62 ± 0.68 s−1) for this variant was comparatively high, because of its high KM (14.55 ± 1.45 mM), when the overall kcat/KM value was compared to the final W16C/D43C/I258A mutant of this study it had lower catalytic efficiency. Compared with the papers reported so far, the W16C/D43C/I258A mutant developed in our article has higher catalytic activity at alkaline pH.
The increased activity of the W16C/D43C/I258A mutant showed that the catalytic activity of an enzyme could be improved by subtle changes in the amino acid side chains, as reported (Taylor and Vaisman, 2010). To better understand the cause of increased activity, the structure of the final mutant W16C/D43C/I258A was modeled and compared with that of the wild type (Figure 7). For ADCes, the distance between the sec-butyl side chain of I258 and the Cα atom of Arg and the distance between the sec-butyl side chain of I258 and the guanidino group of Arg were determined as 9.6 Å and 6.3 Å, respectively. On the other hand, in the W16C/D43C/I258A mutant case, the distances were identified as 10.1 Å and 7.1 Å, respectively. Based on the protein modeling results of the I258A mutant, when the bulky hydrophobic side chain of Ile was replaced with the more diminutive amino acid Ala, this may have minimized the steric hindrance, thus improving substrate binding to the active site. However, it is difficult to explain the affinity change due to the long structural distance between the substrate Arg and the Ala of I258A amino acid residue (>6 Å) when docking simulation is performed. Therefore, the docking simulation result cannot suggest the exact cause of the productivity improvement due to the interaction change caused by the mutation of the enzyme. If the protein structure of the W16C/D43C/I258A mutation is resolved through further studies, a more accurate explanation will be possible. Further studies such as molecular dynamics simulations could also explain the cause of kcat increase and KM decrease in the mutant.
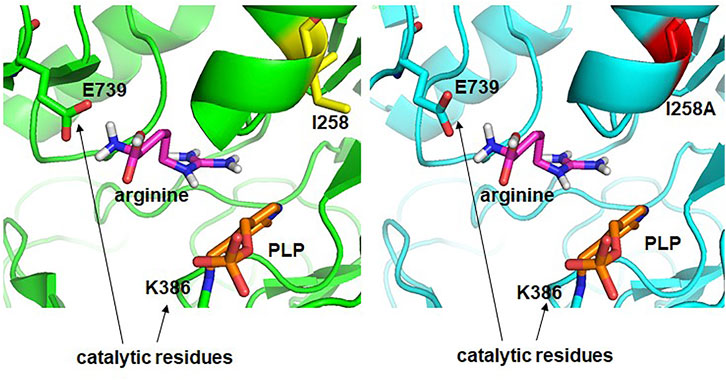
FIGURE 7. Structural comparison of active sites of ADCes (left) and W16C/D43C/I258A mutant (right). Arg is placed in the active site through simulation. I258 in ADCes and the Ala in I258A mutant are shown in yellow and red, respectively.
The starting pH of the batch reaction for agmatine production using Arg monohydrochloride as substrate was ca. 6.7. After 5 h, the pH of the reaction mixture using the ADCes reached around 8.1, whereas the mutant reached near 8.4. The higher final pH of the reaction using the mutant indicates that the W16C/D43C/I258A mutant’s reaction consumes more protons which means more agmatine was produced than the wild-type at alkaline conditions. The stability enhancement of the enzyme could be advantageous in the process development of agmatine production. Although the water solubility of Arg is high at 20°C, i.e., 149 g/L, the substrate solubility could be further increased at a higher temperature, and more significant amounts of agmatine production could be achieved using the mutant.
Conclusion
In conclusion, we rationally selected mutational target sites of ADCes for disulfide bond formation based on its PDB structure (2VYC). We chose the best performers by creating smart, focused libraries with SSM. Protein-ligand docking simulations for this mutant showed that when the hydrophobic side chain of Ile at position 258 was replaced by the smaller amino acid Ala, the space of the active site to which the substrate binds was expanded. Due to the enhanced activity and stability at alkaline pH, the final mutant W16C/D43C/I258A showed better agmatine production throughout the reaction than the wild-type.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
All authors contributed to the conception and the main idea of the work. EH drafted the main text, figures, and tables. B-GK supervised the work and provided the comments and additional scientific information. S-GL and HY also reviewed and revised the text. All authors read and approved the final version of the work to be published.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1A6A3A01012771) and (NRF-2019R1I1A1A01059843). This work was supported by the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) (Project Number: KMDF_PR_20200901_0151). This work was supported by the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) (Project Number: KMDF_PR_21A0301L1-11). The Institute of Engineering Research at Seoul National University provided research facilities for this work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fctls.2021.774512/full#supplementary-material
References
Aglawe, M. M., Kale, M. B., Rahangdale, S. R., Kotagale, N. R., Umekar, M. J., and Taksande, B. G. (2021). Agmatine Improves the Behavioral and Cognitive Impairments Associated with Chronic Gestational Ethanol Exposure in Rats. Brain Res. Bull. 167, 37–47. doi:10.1016/j.brainresbull.2020.11.015
Andréll, J., Hicks, M. G., Palmer, T., Carpenter, E. P., Iwata, S., and Maher, M. J. (2009). Crystal Structure of the Acid-Induced Arginine Decarboxylase from Escherichia coli: Reversible Decamer Assembly Controls Enzyme Activity. Biochemistry 48, 3915–3927. doi:10.1021/bi900075d
Aricioglu, F., Korcegez, E., Bozkurt, A., and Ozyalcin, S. (2003). Effect of Agmatine on Acute and Mononeuropathic Pain. Ann. N.Y Acad. Sci. 1009, 106–115. doi:10.1196/annals.1304.010
Bendl, J., Stourac, J., Sebestova, E., Vavra, O., Musil, M., Brezovsky, J., et al. (2016). HotSpot Wizard 2.0: Automated Design of Site-Specific Mutations and Smart Libraries in Protein Engineering. Nucleic Acids Res. 44, W479–W487. doi:10.1093/nar/gkw416
Blethen, S. L., Boeker, E. A., and Snell, E. E. (1968). Arginine Decarboxylase from Escherichia coli. J. Biol. Chem. 243, 1671–1677. doi:10.1016/s0021-9258(18)93498-8
Bordoli, L., Kiefer, F., Arnold, K., Benkert, P., Battey, J., and Schwede, T. (2009). Protein Structure Homology Modeling Using SWISS-MODEL Workspace. Nat. Protoc. 4, 1–13. doi:10.1038/nprot.2008.197
Bornscheuer, U. T., and Pohl, M. (2001). Improved Biocatalysts by Directed Evolution and Rational Protein Design. Curr. Opin. Chem. Biol. 5, 137–143. doi:10.1016/s1367-5931(00)00182-4
Burrell, M., Hanfrey, C. C., Murray, E. J., Stanley-Wall, N. R., and Michael, A. J. (2010). Evolution and Multiplicity of Arginine Decarboxylases in Polyamine Biosynthesis and Essential Role in Bacillus Subtilis Biofilm Formation. J. Biol. Chem. 285, 39224–39238. doi:10.1074/jbc.m110.163154
Capitani, G., De Biase, D., Aurizi, C., Gut, H., Bossa, F., and Grutter, M. G. (2003). Crystal Structure and Functional Analysis of Escherichia coli Glutamate Decarboxylase. EMBO J. 22, 4027–4037. doi:10.1093/emboj/cdg403
Chae, T. U., Ahn, J. H., Ko, Y.-S., Kim, J. W., Lee, J. A., Lee, E. H., et al. (2020). Metabolic Engineering for the Production of Dicarboxylic Acids and Diamines. Metab. Eng. 58, 2–16. doi:10.1016/j.ymben.2019.03.005
Choi, H., Kyeong, H.-H., Choi, J. M., and Kim, H.-S. (2014a). Rational Design of Ornithine Decarboxylase with High Catalytic Activity for the Production of Putrescine. Appl. Microbiol. Biotechnol. 98, 7483–7490. doi:10.1007/s00253-014-5669-8
Choi, Y. H., Kim, J. H., Park, J. H., Lee, N., Kim, D.-H., Jang, K.-S., et al. (2014b). Protein Engineering of α2,3/2,6-Sialyltransferase to Improve the Yield and Productivity of In Vitro Sialyllactose Synthesis. Glycobiology 24, 159–169. doi:10.1093/glycob/cwt092
Choi, Y. H., Kim, J. H., Park, B. S., and Kim, B.-G. (2016). Solubilization and Iterative Saturation Mutagenesis of α1,3-Fucosyltransferase fromHelicobacter Pylorito Enhance its Catalytic Efficiency. Biotechnol. Bioeng. 113, 1666–1675. doi:10.1002/bit.25944
Craig, D. B., and Dombkowski, A. A. (2013). Disulfide by Design 2.0: a Web-Based Tool for Disulfide Engineering in Proteins. BMC Bioinf. 14, 346. doi:10.1186/1471-2105-14-346
Denesyuk, A. I., Denessiouk, K. A., Korpela, T., and Johnson, M. S. (2002). Functional Attributes of the Phosphate Group Binding Cup of Pyridoxal Phosphate-dependent Enzymes. J. Mol. Biol. 316, 155–172. doi:10.1006/jmbi.2001.5310
Guirard, B. M., and Snell, E. E. (1980). Purification and Properties of Ornithine Decarboxylase from Lactobacillus Sp. 30a. J. Biol. Chem. 255, 5960–5964. doi:10.1016/s0021-9258(19)70724-8
Haenisch, B., Bönisch, H., Cichon, S., Allam, J.-P., Novak, N., and Molderings, G. J. (2011). Effects of Exogenous Agmatine in Human Leukemia HMC-1 and HL-60 Cells on Proliferation, Polyamine Metabolism and Cell Cycle. Leuk. Res. 35, 1248–1253. doi:10.1016/j.leukres.2010.12.023
Hazes, B., and Dijkstra, B. W. (1988). Model Building of Disulfide Bonds in Proteins with Known Three-Dimensional Structure. Protein Eng. Des. Sel. 2, 119–125. doi:10.1093/protein/2.2.119
Hong, E. Y., Lee, S. G., Park, B. J., Lee, J. M., Yun, H., and Kim, B. G. (2017). Simultaneously Enhancing the Stability and Catalytic Activity of Multimeric Lysine Decarboxylase CadA by Engineering Interface Regions for Enzymatic Production of Cadaverine at High Concentration of Lysine. Biotechnol. J. 12, 1700278. doi:10.1002/biot.201700278
Hong, E. Y., Kim, J. Y., Upadhyay, R., Park, B. J., Lee, J. M., and Kim, B.-G. (2018). Rational Engineering of Ornithine Decarboxylase with Greater Selectivity for Ornithine over Lysine through Protein Network Analysis. J. Biotechnol. 281, 175–182. doi:10.1016/j.jbiotec.2018.07.020
Hong, E. Y. (2017). Study on Diamine Production by Protein Engineering of Amino Acid Decarboxylases. Seoul National University. Doctoral dissertation Doctoral dissertation.
Jung, E., Park, B. G., Yoo, H.-W., Kim, J., Choi, K.-Y., and Kim, B.-G. (2018). Semi-rational Engineering of CYP153A35 to Enhance ω-hydroxylation Activity toward Palmitic Acid. Appl. Microbiol. Biotechnol. 102, 269–277. doi:10.1007/s00253-017-8584-y
Kanjee, U., Gutsche, I., Ramachandran, S., and Houry, W. A. (2011). The Enzymatic Activities of the Escherichia coli Basic Aliphatic Amino Acid Decarboxylases Exhibit a pH Zone of Inhibition. Biochemistry 50, 9388–9398. doi:10.1021/bi201161k
Keynan, O., Mirovsky, Y., Dekel, S., Gilad, V. H., and Gilad, G. M. (2010). Safety and Efficacy of Dietary Agmatine Sulfate in Lumbar Disc-Associated Radiculopathy. An Open-Label, Dose-Escalating Study Followed by a Randomized, Double-Blind, Placebo-Controlled Trial. Pain Med. 11, 356–368. doi:10.1111/j.1526-4637.2010.00808.x
Kluger, R., and Rathgeber, S. (2008). Catalyzing Separation of Carbon Dioxide in Thiamin Diphosphate-Promoted Decarboxylation. FEBS J. 275, 6089–6100. doi:10.1111/j.1742-4658.2008.06739.x
Lee, J.-H., Chung, S.-W., Lee, H.-J., Jang, K.-S., Lee, S.-G., and Kim, B.-G. (2010). Optimization of the Enzymatic One Pot Reaction for the Synthesis of Uridine 5′-diphosphogalactose. Bioproc. Biosyst. Eng. 33, 71–78. doi:10.1007/s00449-009-0365-2
Morris, D. R., and Boeker, E. A. (1983). [18] Biosynthetic and Biodegradative Ornithine and Arginine Decarboxylases from Escherichia coli. Methods Enzymol. 94, 125–134. doi:10.1016/s0076-6879(83)94020-x
Neis, V. B., Bettio, L. E. B., Moretti, M., Rosa, P. B., Ribeiro, C. M., Freitas, A. E., et al. (2016). Acute Agmatine Administration, Similar to Ketamine, Reverses Depressive-like Behavior Induced by Chronic Unpredictable Stress in Mice. Pharmacol. Biochem. Behav. 150-151, 108–114. doi:10.1016/j.pbb.2016.10.004
Niesen, F. H., Berglund, H., and Vedadi, M. (2007). The Use of Differential Scanning Fluorimetry to Detect Ligand Interactions that Promote Protein Stability. Nat. Protoc. 2, 2212–2221. doi:10.1038/nprot.2007.321
Nov, Y. (2013). Fitness Loss and Library Size Determination in Saturation Mutagenesis. PLoS One 8, e68069. doi:10.1371/journal.pone.0068069
Ommati, M. M., Farshad, O., Mousavi, K., Taghavi, R., Farajvajari, S., Azarpira, N., et al. (2020). Agmatine Alleviates Hepatic and Renal Injury in a Rat Model of Obstructive Jaundice. PharmaNutrition 13, 100212. doi:10.1016/j.phanu.2020.100212
Pei, X. D., Lu, L. H., Yue, S. Y., Li, Y., Liu, X. L., Li, F., et al. (2021). Characterization of a Novel Shewanella Algae Arginine Decarboxylase Expressed in Escherichia coli. Mol. Biotechnol. doi:10.1007/s12033-021-00397-6
Piletz, J. E., Aricioglu, F., Cheng, J.-T., Fairbanks, C. A., Gilad, V. H., Haenisch, B., et al. (2013). Agmatine: Clinical Applications after 100 Years in Translation. Drug Discov. Today 18, 880–893. doi:10.1016/j.drudis.2013.05.017
Porter, J. L., Rusli, R. A., and Ollis, D. L. (2016). Directed Evolution of Enzymes for Industrial Biocatalysis. Chembiochem 17, 197–203. doi:10.1002/cbic.201500280
Rahangdale, S., Fating, R., Gajbhiye, M., Kapse, M., Inamdar, N., Kotagale, N., et al. (2021). Involvement of Agmatine in Antidepressant-like Effect of HMG-CoA Reductase Inhibitors in Mice. Eur. J. Pharmacol. 892, 173739. doi:10.1016/j.ejphar.2020.173739
Reetz, M. T., and Carballeira, J. D. (2007). Iterative Saturation Mutagenesis (ISM) for Rapid Directed Evolution of Functional Enzymes. Nat. Protoc. 2, 891–903. doi:10.1038/nprot.2007.72
Roth, B. L., Sheffler, D. J., and Kroeze, W. K. (2004). Magic Shotguns versus Magic Bullets: Selectively Non-selective Drugs for Mood Disorders and Schizophrenia. Nat. Rev. Drug Discov. 3, 353–359. doi:10.1038/nrd1346
Sambrook, J., Russell, D. W., and Sambrook, J. (2006). The Condensed Protocols from Molecular Cloning : A Laboratory Manual. Cold Spring Harbor, N.Y. Cold Spring Harbor Laboratory Press.
Song, J., Hur, B. E., Bokara, K. K., Yang, W., Cho, H. J., Park, K. A., et al. (2014). Agmatine Improves Cognitive Dysfunction and Prevents Cell Death in a Streptozotocin-Induced Alzheimer Rat Model. Yonsei Med. J. 55, 689–699. doi:10.3349/ymj.2014.55.3.689
Sun, X., Song, W., and Liu, L. (2015). Enzymatic Production of Agmatine by Recombinant Arginine Decarboxylase. J. Mol. Catal. B: Enzym. 121, 1–8. doi:10.1016/j.molcatb.2015.06.008
Sun, Z., Lonsdale, R., Wu, L., Li, G., Li, A., Wang, J., et al. (2016). Structure-Guided Triple-Code Saturation Mutagenesis: Efficient Tuning of the Stereoselectivity of an Epoxide Hydrolase. ACS Catal. 6, 1590–1597. doi:10.1021/acscatal.5b02751
Taylor, T. J., and Vaisman, I. I. (2010). Discrimination of Thermophilic and Mesophilic Proteins. BMC Struct. Biol. 10 Suppl 1 (Suppl. 1), S5. doi:10.1186/1472-6807-10-S1-S5
Thu Ho, N. A., Hou, C. Y., Kim, W. H., and Kang, T. J. (2013). Expanding the Active pH Range of Escherichia coli Glutamate Decarboxylase by Breaking the Cooperativeness. J. Biosci. Bioeng. 115, 154–158. doi:10.1016/j.jbiosc.2012.09.002
Toscano, M. D., Woycechowsky, K. J., and Hilvert, D. (2007). Minimalist Active-Site Redesign: Teaching Old Enzymes New Tricks. Angew. Chem. Int. Ed. 46, 3212–3236. doi:10.1002/anie.200604205
Trott, O., and Olson, A. J. (2010). AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 31, 455–461. doi:10.1002/jcc.21334
Wade, C. L., Eskridge, L. L., Nguyen, H. O. X., Kitto, K. F., Stone, L. S., Wilcox, G., et al. (2009). Immunoneutralization of Agmatine Sensitizes Mice to μ-Opioid Receptor Tolerance. J. Pharmacol. Exp. Ther. 331, 539–546. doi:10.1124/jpet.109.155424
Keywords: disulfide bridge, enzyme catalysis, Escherichia coli arginine decarboxylase, protein design, site-saturation mutagenesis
Citation: Hong EY, Lee S-G, Yun H and Kim B-G (2021) Improving the Stability and Activity of Arginine Decarboxylase at Alkaline pH for the Production of Agmatine. Front. Catal. 1:774512. doi: 10.3389/fctls.2021.774512
Received: 12 September 2021; Accepted: 02 November 2021;
Published: 23 November 2021.
Edited by:
Gonzalo De Gonzalo, Sevilla University, SpainReviewed by:
Rosli Md Illias, Universiti Teknologi Malaysia, MalaysiaRolandas Meskys, Vilnius University, Lithuania
Copyright © 2021 Hong, Lee, Yun and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Gee Kim, Ynl1bmdraW1Ac251LmFjLmty
 Eun Young Hong1,2
Eun Young Hong1,2 Hyungdon Yun
Hyungdon Yun Byung-Gee Kim
Byung-Gee Kim