- 1Division of Cardiac Surgery, Department of Surgery, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 2Department of Pediatrics, Division of Endocrinology, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 3Division of Cardiovascular Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States
- 4Department of Physiology, Center for Metabolism and Obesity Research, Johns Hopkins University School of Medicine, Baltimore, MD, United States
The exact role of subcutaneous adipose tissue in the interplay between type 2 diabetes (T2D) and coronary artery disease (CAD) is yet to be determined. A prospective cohort study of adult patients with and without T2D undergoing CABG was performed. Subcutaneous adipose tissue was collected during the procedure and RNA seq analysis was performed. A total of 741 differentially expressed genes (DEGs) were identified (332 up- and 409 down-regulated in the T2D group). Our results demonstrated that pathways related to apoptosis and immune response were significantly dysregulated in the adipose tissue of T2D subjects. The main molecular pathways involved were CXCR, NOTCH, STAT, NFKB1 and FGFR pathways, which have a well-documented role in diabetes and CAD. SPI1 and MTF1 were two novel upstream transcription factors identified which have been suggested to be involved in the inflammatory cascade and insulin regulation in diabetes. Three miRNAs were differentially expressed between the two groups (miR-27a, miR-335 and miR-146). These preliminary results provide fertile ground for further research of potential targets for patients with diabetes and coronary artery disease.
Introduction
Coronary artery disease (CAD) is the most common cause of mortality in patients with type 2 diabetes (T2D). Despite the well-documented association between obesity and T2D, and the adverse effects of visceral adipose tissue (AT), subcutaneous adipose tissue (ScAT) is believed to have beneficial paracrine and endocrine effects on insulin resistance (1). However, its exact role in the interplay between T2D and CAD is yet to be determined (2). Herein, we sought to investigate changes in the transcriptomic profile of the ScAT associated with T2D in patients with advanced CAD undergoing coronary artery bypass grafting (CABG), compared to individuals without T2D.
Method
This is a prospective cohort of adult patients with and without T2D undergoing CABG at the Johns Hopkins Hospital (JHH) approved by JHH Institutional Review Board (IRB00318696). Exclusion criteria included immunocompromised patients and patients with type 1 diabetes, active cancer, inflammatory bowel disease or previous cardiac surgery. Patient demographics, comorbidities, medications and preoperative blood work were collected. ScAT samples collected from the sternotomy site at the beginning of the case were immediately frozen and stored in liquid nitrogen. RNA seq was performed as previously described (3). Genes with adjusted False discovery rate (FDR) <0.05 and log2-fold change (>1) were characterized as differentially expressed genes (DEGs) for each comparison. MetaCore (v20.2), Appyter (v0.2.6) and GSEA were used for functional enrichment and pathway analysis. STRING (12.0) was used for protein-protein interaction networks. Mutliomic analysis to visualize the miRNA-gene networks was performed in miRNet (2.0).
Results
A total of 6 patients were enrolled, 3 with T2D (mean age 60.3 years, mean BMI 29.26 kg/m2, all with 4-vessel disease, and 2 with HTN, 1 with hyperlipidemia) and 3 without T2D (mean age 65 years, mean BMI 37.07 kg/m2, all with HTN and hyperlipidemia, and 4-vessel disease). To ascertain the global transcriptomic changes associated with T2D in the ScAT of these patients, we performed RNA sequencing and gene ontology (GO) pathway analysis. A total of 741 differentially expressed genes (DEGs) were identified (332 up- and 409 down-regulated in the T2D group) (Figure 1A). Principal component analysis (PCA) indicated that the transcriptomic profiles of the two groups were separate and discrete (Figure 1B). Cumulative GO analysis of 6 different databanks showed that the top upregulated pathways in T2D included leptin influence on immune response, inflammatory response to chemokines, leukocyte aggregation, neutrophil activation, CXC receptor binding and heat sock factor 1 (HSF-1) activation (Figure 1C). GSEA enrichment analysis identified apoptosis, inflammation and complement activation to be significantly upregulated in the T2D group (Figure 1D). Transcription factors (TF) PU.1 (SPI1), erythroid transcription factor (GATA), nuclear factor kappa B subunit 1 (NFKB1), signal transducer and activator of transcription factor 3 (STAT3), E74 like ETS transcription factor 4 (ELF4) were found to be upstream regulators of the activated pathways among others (Figure 1E). The network of the most significantly enriched pathways in T2D and their subsequent subclusters are better depicted in Figure 1F indicating the presence of neutrophil degranulation, inflammatory and immune response. Further GO analysis of the top downregulated targets showed that NOTCH signaling and fibroblast growth factor receptor (FGFR) signaling pathways were significantly downregulated in the diabetic patients, while the top predicted upstream regulators for those pathways were found to be the microphthalmia transcription factor 1 (MTF1) -a regulator of beta pancreatic cells- and the fibroblast growth factor receptor 3b (FGFR3b) -a receptor found to be dysregulated in visceral obesity (data not shown). Protein-protein interaction network analysis of the enriched targets indicated that the top 5 clusters are associated with innate immune response, oxygen carrier activity, chemokine activity, regulation of mitosis and cellular heat acclimation (Figure 1G). Multiomic analysis of the top dysregulated miRNAs and proteins in the diabetes group showed that 3 mRNAs, miR-335, miR-27a and miR-146a formed networks with targets associated with pathways of aging, apoptosis and cell death; while the associated disease processes included heart failure, diabetes and different types of cancer. The transcription factors regulating those miRNAs include NFKB1, STAT5 and MYC (Figure 1H).
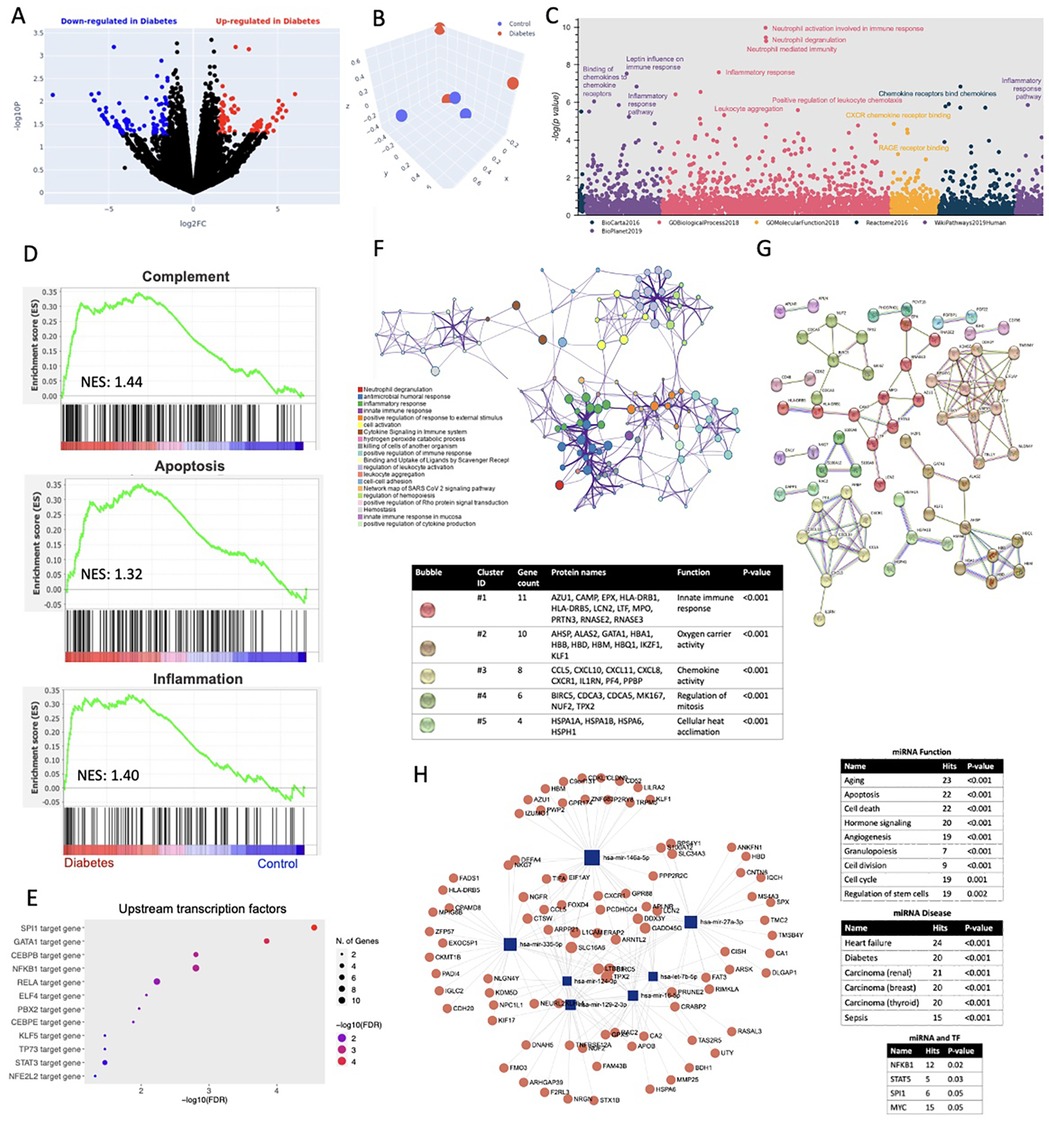
Figure 1. (A) Volcano plot indicating the upregulated (red) and downregulated (blue) genes in the diabetes group compared to control. (B) Principal component analysis (PCA) for control (blue) and diabetes (red) groups. (C) Manhattan plot of six databases indicating the most significant Gene Ontology (GO) molecular functions when comparing diabetes to control groups. (D) Enrichment plot analysis indicating upregulated pathways in the diabetes group compared to control. NES: Normalized enrichment score. (E) Forest plot indicating significantly upregulated transcription factors in the diabetes group compared to control. FDR: False discovery rate. (F) Network analysis of enriched pathways associated with diabetes in our study population. (G) Protein string interaction network analysis showing the most significant clusters when comparing diabetes group to control. (H) miRNA and transcription factor (TF) interaction analysis network.
Discussion
This is a pilot study investigating the transcriptomic shifts of ScAT associated with T2D in patients with advanced CAD requiring surgical revascularization. It is reported that ScAT potentially exerts a protective role against glucose homeostasis dysregulation. Our results showed that pathways related to apoptosis and immune response were significantly dysregulated in the adipose tissue of T2D subjects. The association between these pathways and the pathogenesis of CAD and T2D through modulation of insulin resistance and low-grade inflammation among others has been previously studied. The main molecular pathways involved in our study, namely CXCR, NOTCH, STAT, NFKB1 and FGFR pathways, have a well-documented role in diabetes and CAD (3). SPI1 and MTF1 were two novel upstream ΤFs identified which have been suggested to be involved in the inflammatory cascade and insulin regulation in diabetes (4). Moreover, three miRNAs were differentially expressed in the adipose tissue of diabetic patients. Interestingly, miR-27a has been previously described as a negative regulator of adipogenesis and is increased by white adipose tissue inflamation (5). However, the role of miR-335 and miR-146 has not been explored beyond inflammation and oncogenesis suggesting a possible link of those miRNAs to targets regulating the inflammatory response and secretion of diabetic adipocytes. Our findings suggest that ScAT is an active paracrine organ which is involved in the low-grade inflammation associated with CAD and is modulated by T2D. Thus, development of drugs that can target these miRNAs could potentially decrease the proinflammatory environment promoted by T2D and CAD and lead to decreased risk for comorbidities and complications.
The extrapolation of our results may be limited by the relatively small sample size which did not allow for subgroup analyses based on further comorbidities or medications. The nature of the current study is exploratory and was not focused on solid comparisons between the two patient populations. Additionally, these are patients with advanced CAD and their transcriptome may differ from those with earlier stages of the disease.
Our next steps are focused on different sources of adipose tissue including perivascular and epicardial fat since their proximity to the heart may unveil new pathways involved in CAD and insulin hemostasis.
Data availability statement
The RNA sequencing data analysed in this study was obtained from Novogene Corporation Inc, the following restrictions apply: data are deleted 30 calendar days after release to the customer. Requests to access the other datasets, including DEGs should be directed to ID,ZG91bGFtaXMuaUBnbWFpbC5jb20=.
Ethics statement
The studies involving humans were approved by Johns Hopkins Hospital Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
ID: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. BP: Data curation, Investigation, Methodology, Writing – review & editing. AT: Conceptualization, Data curation, Formal Analysis, Investigation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. JP: Conceptualization, Investigation, Methodology, Supervision, Validation, Writing – review & editing. GW: Conceptualization, Investigation, Methodology, Resources, Supervision, Validation, Writing – review & editing. AK: Conceptualization, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing. RW: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. ID received funding from the Physician Scientist Training Program of Johns Hopkins University School of Medicine.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Carruthers NJ, Strieder-Barboza C, Caruso JA, Flesher CG, Baker NA, Kerk SA, et al. The human type 2 diabetes-specific visceral adipose tissue proteome and transcriptome in obesity. Sci Rep. (2021) 11:1. doi: 10.1038/s41598-021-96995-0
2. Van Der Kolk BW, Kalafati M, Adriaens M, Van Greevenbroek MMJ, Vogelzangs N, Saris WHM, et al. Subcutaneous adipose tissue and systemic inflammation are associated with peripheral but not hepatic insulin resistance in humans. Diabetes. (2019) 68:2247–58. doi: 10.2337/db19-0560
3. Saxena A, Mathur N, Tiwari P, Mathur SK. Whole transcriptome RNA-Seq reveals key regulatory factors involved in type 2 diabetes pathology in peripheral fat of Asian Indians. Sci Rep. (2021) 11:10632. doi: 10.1038/s41598-021-90148-z
4. Mazur MA, Winkler M, Ganić E, Colberg JK, Johansson JK, Bennet H, et al. Microphthalmia transcription factor regulates pancreatic β-cell function. Diabetes. (2013) 62:2834–42. doi: 10.2337/db12-1464
Keywords: coronary artery disease, diabetes, subcutaneous fat, transcriptome, RNA seq
Citation: Doulamis IP, Pan B, Tzani A, Plutzky J, Wong GW, Kilic A and Wolf RM (2025) Transcriptomic signatures of subcutaneous adipose tissue in patients with diabetes and coronary artery disease: a pilot study. Front. Cardiovasc. Med. 12:1524605. doi: 10.3389/fcvm.2025.1524605
Received: 7 November 2024; Accepted: 9 January 2025;
Published: 5 February 2025.
Edited by:
Turgay Celik, Wake Forest Baptist Medical Center, United StatesReviewed by:
Mahesh Appari, University of Oxford, United KingdomYi Guo, The Quzhou Affiliated Hospital of Wenzhou Medical University, China
Copyright: © 2025 Doulamis, Pan, Tzani, Plutzky, Wong, Kilic and Wolf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ilias P. Doulamis, ZG91bGFtaXMuaUBnbWFpbC5jb20=
 Ilias P. Doulamis
Ilias P. Doulamis Bernard Pan2
Bernard Pan2 G. William Wong
G. William Wong Risa M. Wolf
Risa M. Wolf