
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 25 July 2024
Sec. Cardiovascular Pharmacology and Drug Discovery
Volume 11 - 2024 | https://doi.org/10.3389/fcvm.2024.1417701
This article is part of the Research Topic Exploring Molecular Mechanisms and Novel Diagnostics in Cardiovascular Disease Treatment View all 8 articles
Background: The relationship between human immunodeficiency virus (HIV) infection and pulmonary arterial hypertension (PAH) has garnered significant scrutiny. Individuals with HIV infection have a higher risk of developing PAH. However, the specific mechanism of HIV-associated PAH remains unclear. Our study aims at investigating the shared biomarkers in HIV infection and PAH and predicting the potential therapeutic target for HIV-associated PAH.
Methods: Data for HIV infection and PAH were downloaded from Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) analysis was performed to detect shared genes in HIV infection and PAH. Enrichment analysis was conducted to identify the function of common DEGs. Protein-protein interaction (PPI) analysis was used to detect key genes. These crucial genes were subsequently verified by RT-qPCR. Finally, candidate drugs were identified by using the Drug Signatures Database (DSigDB).
Results: Nineteen common DEGs were identified in HIV infection and PAH. Enrichment analysis exhibited that the functions of these genes were mainly enriched in inflammatory responses, mainly including cellular immunity and interaction between viral proteins and cytokines. By constructing PPI networks, we identified the key gene CC-type chemokine ligand 5 (CCL5), and we verified that CCL5 was highly expressed in hypoxia induced human pulmonary artery endothelial cells (hPAECs) and human pulmonary artery smooth muscle cells (hPASMCs). In addition, we predicted 10 potential drugs targeting CCL5 by Autodock Vina.
Conclusion: This study revealed that CCL5 might be a common biomarker of HIV infection and PAH and provided a new therapeutic target for HIV-associated PAH. However, further clinical validation is still indispensable.
HIV is a retrovirus that includes two types: HIV-1 and HIV-2 (1). HIV infection is a serious public health problem that has caused approximately 40 million deaths worldwide (2). Acquired immunodeficiency syndrome (AIDS) is the final outcome of HIV infection, and people diagnosed with AIDS often die from serious infections or cancer because of their highly weakened immune systems. In recent years, with the application of antiviral drugs, the life span of HIV infected people has been greatly extended (3). Thus, the chronic complications of HIV infection have attracted more attention. Previous studies indicated that HIV infection was linked to a higher risk of cardiovascular disease, which may be connected to persistent inflammation (4). Among them, HIV-associated PAH has received great attention because of its poor prognosis. A previous study indicated that PAH was a long-term complication of HIV infection, and individuals with HIV-associated PAH had a worse survival rate compared with HIV-positive patients without PAH (5). The prevalence of PAH is 0.5% in HIV-positive individuals, which is much greater than that in HIV-negative individuals (6).
As a subtype of pulmonary hypertension, pulmonary arterial hypertension is distinguished by pulmonary arterial remodeling. Right heart failure is the usual cause of death for PAH patients, causing a huge disease burden (7). Studies elucidating the mechanism of the relationship between HIV infection and PAH are very limited. DNA damage response and chronic inflammation may contribute to the development of HIV-associated PAH (8, 9). Besides, previous studies showed that increased levels of asymmetric dimethylarginine (10) and smooth muscle cell proliferation (11) also played key roles in HIV-associated PAH. In order to detect novel therapeutic targets for HIV-associated PAH, studying the common mechanism between PAH and HIV infection is very crucial.
Nowadays, we are able to better understand diseases owing to advancements in sequencing technology and bioinformatics analysis. In this study, we identified the common biomarkers between HIV infection and PAH and predicted potential drugs for HIV-associated PAH.
Datasets on HIV infection and pulmonary arterial hypertension were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) (12). For discovery cohorts, dataset GSE37250 contained 274 HIV-positive and 263 HIV-negative whole blood samples (13), and dataset GSE117261 included lung tissue samples from 58 PAH patients and 25 controls (14). For validation cohorts, dataset GSE30310 involved 48 HIV-positive and 19 HIV-negative peripheral blood mononuclear cell samples (15), and dataset GSE53408 comprised lung tissue samples from 12 PAH patients and 11 healthy controls (16). All the data were derived from human species. Details of these datasets can be found in Supplementary Table S1. Figure 1 (by Figdraw) exhibits the workflow of this study.
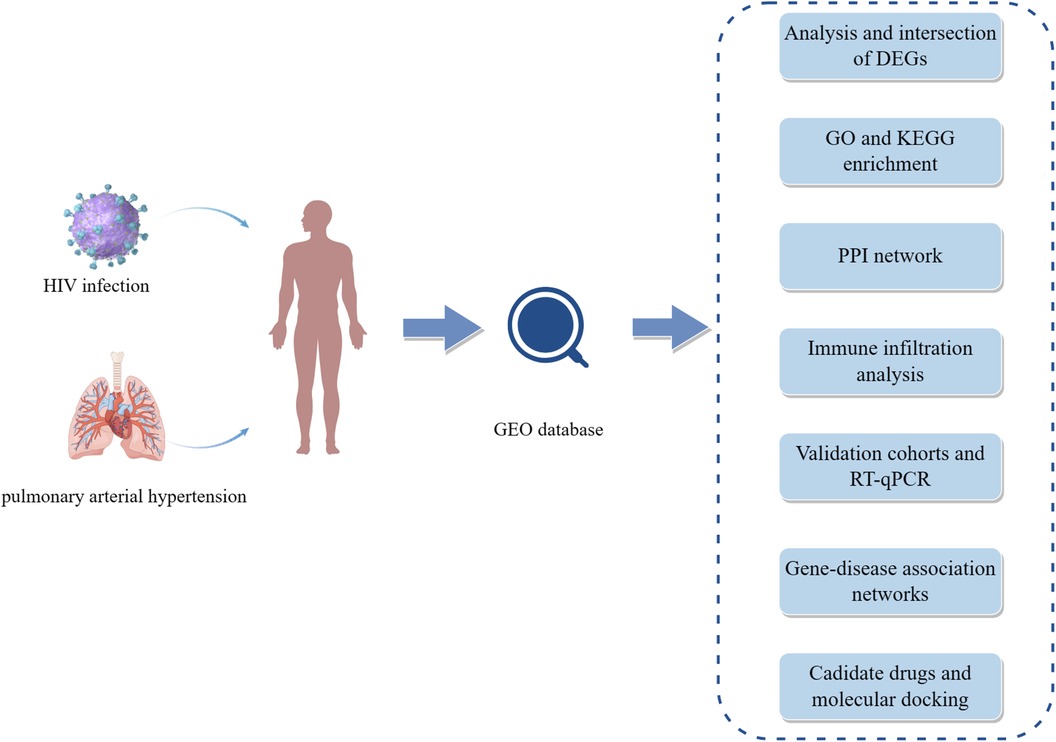
Figure 1 The workflow of this study. HIV, human immunodeficiency virus; GEO, gene expression omnibus; DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; PPI, protein-protein interaction; RT-qPCR, quantitative real-time polymerase chain reaction.
DEGs analysis was performed by using “limma” package. For dataset GSE37250, the cutoff criteria was |Log2 fold change| >0.1 and adjusted P-value <0.001 (17). For dataset GSE117261, the cutoff criteria was |Log2 fold change| >0.585 and adjusted P-value <0.05 (18). The above results were visualized by heatmaps and volcano plots. The common DEGs of the two diseases were obtained by using “venn” package (19).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses are commonly used to find the biological function of gene sets (20, 21). We conducted function enrichment analysis of common DEGs by using “ClusterProfiler” package. P-value <0.05 was considered significant.
Using the STRING database (version 12.0; https://cn.string-db.org/) (22), we built a PPI network of common DEGs. The minimum required interaction score was 0.150, and unconnected nodes were hidden. Besides, the PPI network was visualized by Cytoscape (version 3.10.1) (23). Subsequently, Molecular Complex Detection (MCODE) (24) was used to identify functional modules, and the default parameters were set as follows: degree cutoff = 2, node score cutoff = 0.2, K-core = 2, and max depth = 100. Finally, the top ten genes were identified by CytoHubba plugin according to Degree algorithm (25).
Single sample gene set enrichment analysis (ssGSEA) algorithm was utilized to calculate enrichment scores between distinct groups by “GSVA” R package (26, 27). Next, the abundance of 28 types of immune cells in each group was visualized. Plus, we explored the linkage between hub genes and immune cells by Spearman's rank correlation analysis.
hPAECs were purchased from ScienCell (Shanghai, China, Cat. No. 3100). hPASMCs were obtained from Procell (Wuhan, China, Cat. No. CP-H243). They were cultured in endothelial cell medium (ScienCell, Cat. No. 1001) and smooth muscle cell medium (Procell, Cat. No. CM-H243), respectively, at 37°C with 5% CO2. Hypoxia induced hPAECs and hPASMCs were incubated with 2% O2, 5% CO2 and 93% N2 at 37°C for 24 h in anoxic incubators.
RNA was extracted from cells by Trizol reagent (Invitrogen, USA, Cat. No. 15596026). Next, the purity of the RNA was determined using NanoDrop 2000 (Thermo, USA). Then, the PrimeScript®RT Reagent Kit with gDNA Eraser (Takara, Japan, Cat. No. RR047A) was used for reverse transcription in T100 Thermal Cycler (Biorad, USA). Finally, qPCR was executed employing the SYBR®Premix Ex TaqII kit (Takara, Japan, Cat. No. RR820A) in qPCR instrument (Applied Biosystems 7500, USA). The primer sequences are displayed in Supplementary Table S2.
DisGeNET is one of the largest collections of genes and variants linked to human disease (28). We constructed gene-disease association networks filtering by disease class infections and cardiovascular diseases, respectively. The above process was completed in DisGeNET Cytoscape App (version 7.3.0).
The top ten candidate drugs with the best statistical significance were obtained from DSigDB database (29) by Enrichr (https://maayanlab.cloud/Enrichr/) (30) (Supplementary Figure S1). Small molecule drug structures were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Structures of CCL5 (PDB code: 5DNF) were accessed from the PDB database (https://www.rcsb.org/). Autodock Vina software (31) and Pymol (32) were used for validation of the docking protocol and molecular docking. Firstly, the co-crystalline ligand (beta-D-glucopyranose) of CCL5 was extracted and saved. Then the co-crystalline ligand was re-docked with CCL5. The root mean square deviation (RMSD) between the conformation of the redocked ligands and the conformation of the original crystal structure was calculated to validate the docking protocol (33). Finally, the molecular docking process between ten drugs and CCL5 was carried out.
R software (v. 4.3.0) and GraphPad Prism (v.9.5.1) were employed in statistical analysis. Correlation between CCL5 and immune cells was determined by Spearman correlation analysis. P-value <0.05 was considered statistically significant.
2216 DEGs and 307 DEGs were identified in datasets GSE37250 and GSE117261, respectively (Supplementary Table S3). Figures 2A,B showed the DEGs of HIV infection, and Figures 2C,D showed the DEGs of PAH. Further, the common DEGs were identified, and 12 up-regulated and 7 down-regulated genes were detected in HIV infection and PAH (Figures 2E,F).
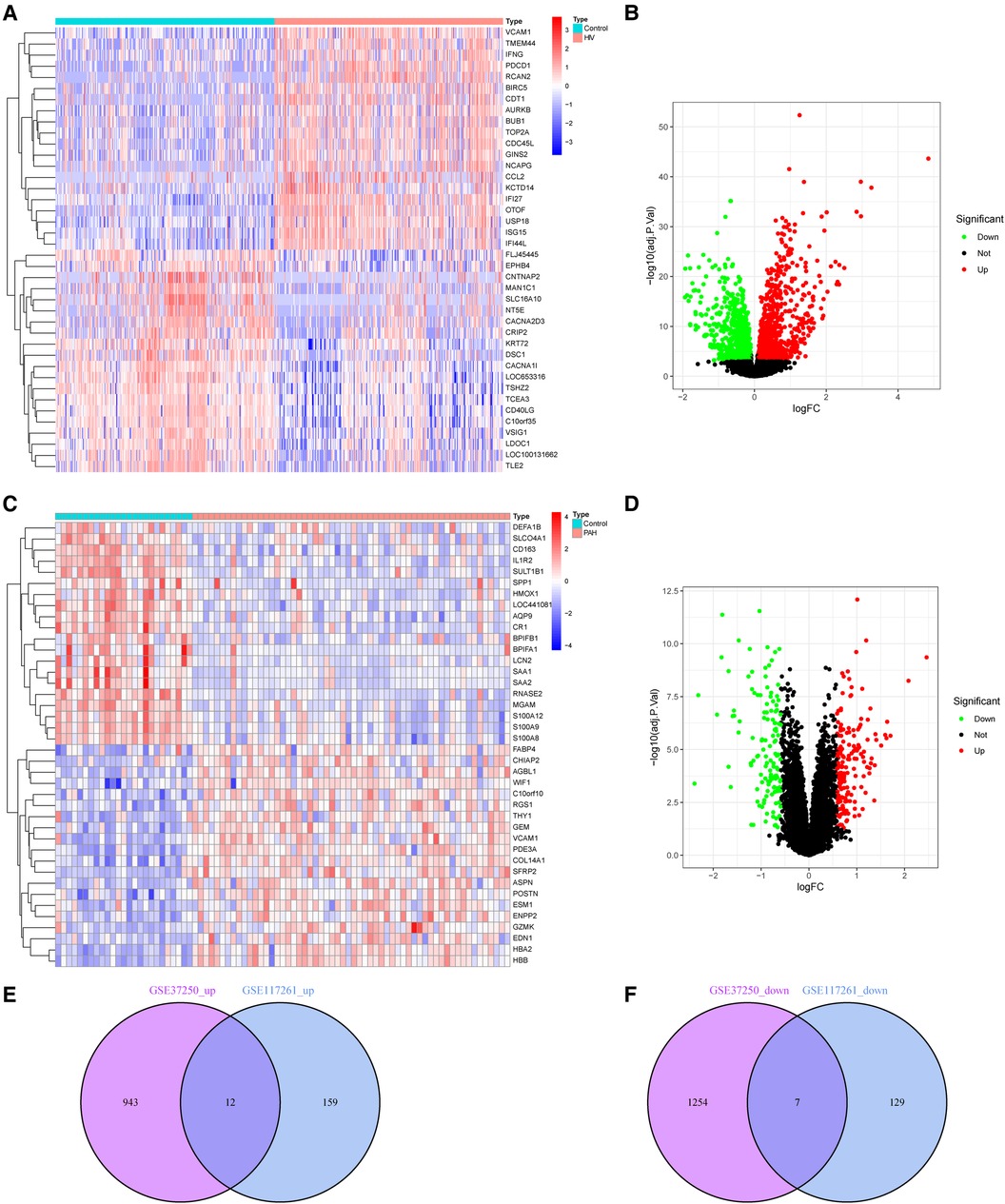
Figure 2 DEGs in HIV infection and PAH group. (A,B) Heatmap and volcano plot of DEGs in HIV infection. (C,D) Heatmap and volcano plot of DEGs in PAH. (E,F) The intersection of DEGs between HIV infection and PAH. DEGs, differentially expressed genes; HIV, human immunodeficiency virus; PAH, pulmonary arterial hypertension.
To investigate the possible common mechanism between HIV infection and PAH, GO and KEGG enrichment analyses were performed. According to GO analysis, these shared genes were mostly associated with positive regulation of mononuclear cell migration, cellular extravasation, positive regulation of T cell proliferation (biological process), fibrillar center (cellular component), and CCR chemokine receptor binding (molecular function) (Figure 3A). Besides, KEGG enrichment indicated that these genes were mainly related to viral protein interaction with cytokine and cytokine receptor (Figure 3B).
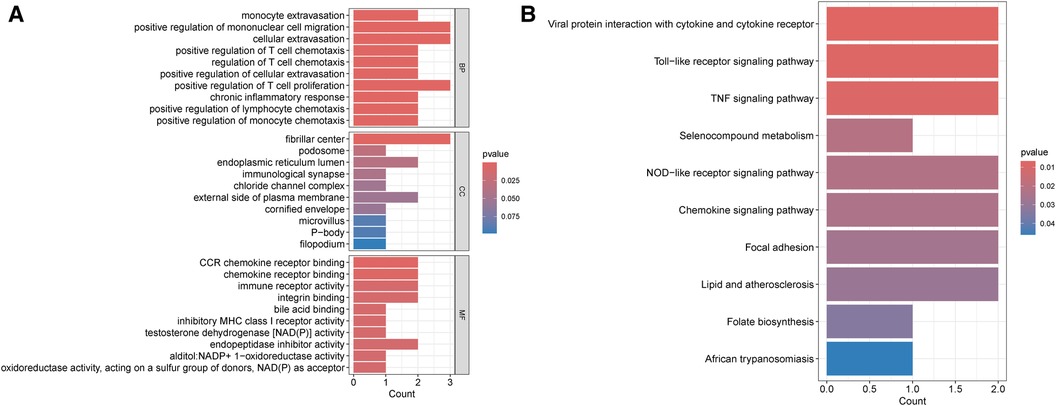
Figure 3 Enrichment analysis of common DEGs between HIV infection and PAH. (A) GO enrichment analysis of common DEGs between HIV infection and PAH. (B) KEGG enrichment analysis of common DEGs between HIV infection and PAH. DEGs, differentially expressed genes; HIV, human immunodeficiency virus; PAH, pulmonary arterial hypertension; GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; BP, biological process; CC, cellular component; MF, molecular function.
As shown in Figures 4A,B, a PPI network was constructed and visualized. Next, we extracted one closely related gene cluster module using the MCODE plug-in (Figure 4C). Subsequently, the top 10 candidate hub genes were screened by the Degree algorithm. As shown in Figure 4D, the top three genes were CCL5, GZMA, and CCR2. They were closely linked.
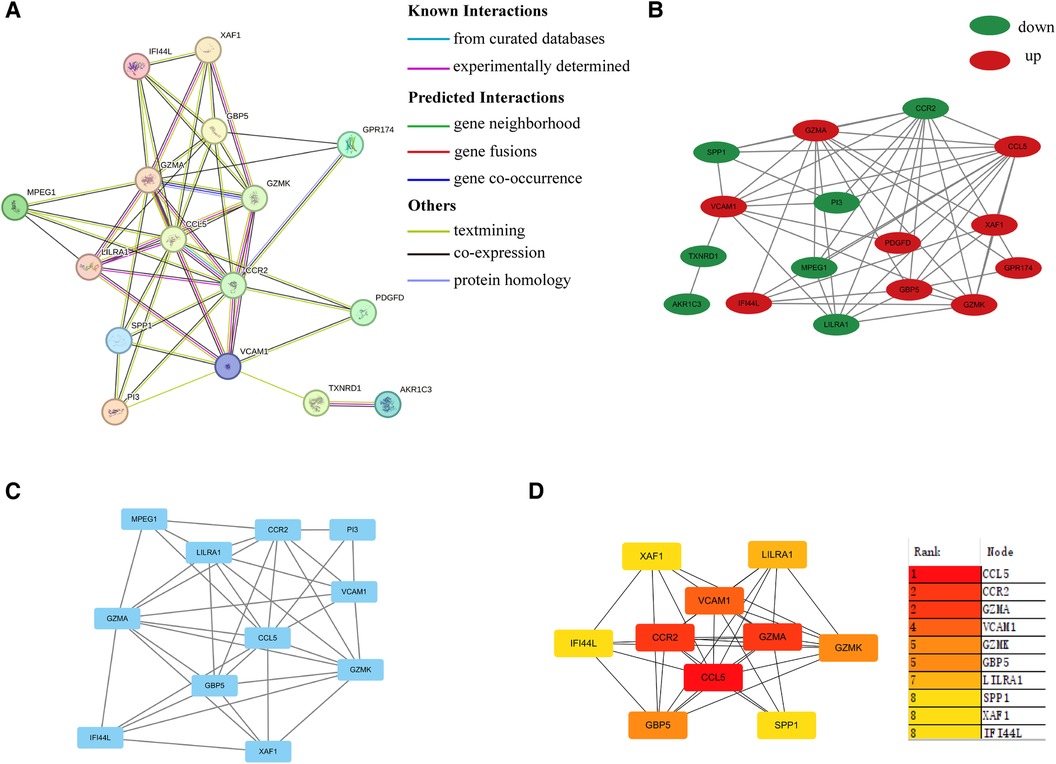
Figure 4 PPI network. (A) PPI network of common DEGs between HIV infection and PAH. (B) PPI network visualized by Cytoscape. Up-regulated and down-regulated genes were marked in red and green, respectively. (C) Sub-cluster of PPI network by MCODE. (D) Top ten hub genes according to Degree algorithm. PPI, protein-protein interaction; DEGs, differentially expressed genes.
The expression levels of CCL5, GZMA, and CCR2 were analyzed in validation cohorts GSE30310 and GSE53408. Only CCL5 showed significant differences in both validation groups (Figures 5E,G). For dataset GSE30310, there was no significant difference in CCR2 (Supplementary Figure S2), and GZMA was not detected. Similarly, for dataset GSE53408, the expression level of GZMA was significantly increased in the PAH group (Supplementary Figure S3), but CCR2 was not detected. Therefore, CCL5 was identified as a hub gene in HIV infection and PAH.
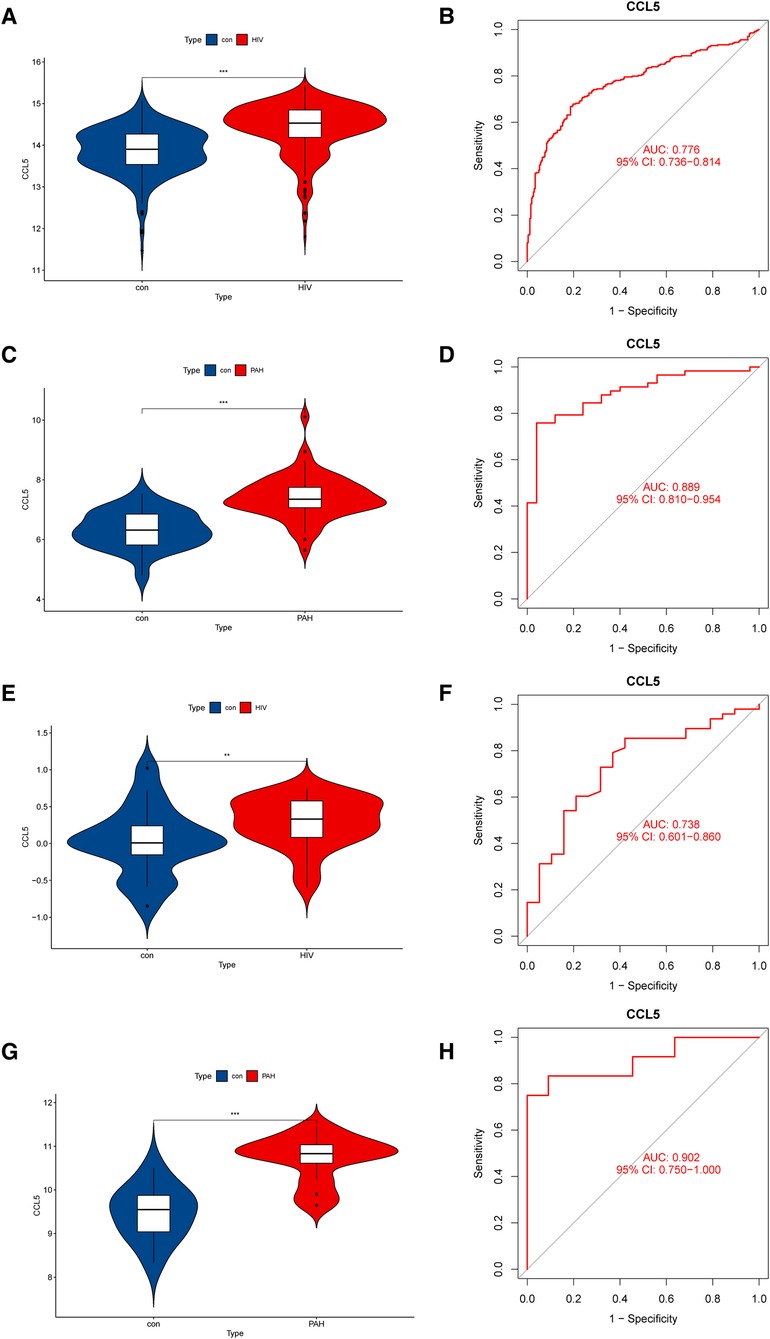
Figure 5 Expression levels and diagnostic values of CCL5. (A) Expression level of CCL5 in the discovery cohorts for HIV infection (GSE37250). (B) ROC curve of CCL5 in the discovery cohorts for HIV infection (GSE37250). (C) Expression level of CCL5 in the discovery cohorts for PAH (GSE117261). (D) ROC curve of CCL5 in the discovery cohorts for PAH (GSE117261). (E) Expression level of CCL5 in the validation cohorts for HIV infection (GSE30310). (F) ROC curve of CCL5 in the validation cohorts for HIV (GSE30310). (G) Expression level of CCL5 in the validation cohorts for PAH (GSE53408). (H) ROC curve of CCL5 in the validation cohorts for PAH (GSE53408). HIV, human immunodeficiency virus; PAH, pulmonary arterial hypertension; ROC, receiver operating characteristic; CCL5, CC-type chemokine ligand 5.
To make our study more credible, we evaluated the expression levels and diagnostic values of CCL5 in both the discovery cohort and the validation cohort. In the discovery cohort, CCL5 was upregulated in HIV infection group (Figure 5A) and PAH group (Figure 5C). The receiver operating characteristic curves (ROC curves) showed certain diagnostic values of CCL5 (Figures 5B,D). Similarly, in the validation cohort, CCL5 was upregulated in the disease group with certain diagnostic values (Figures 5E–H).
Immune infiltration analysis showed the roles that immune cells played in HIV infection and PAH. In the HIV infection group, the proportion of activated CD4 T cells, activated CD8 T cells and activated dendritic cells increased, and there was a decline in the percentage of effector memory CD4 T cells, monocytes, and neutrophils (Figures 6A,B). In the PAH group, the proportion of activated B cells, activated CD8 T cells and eosinophils increased, and the proportion of activated dendritic cells, neutrophils, and regulatory T cells decreased (Figures 6C,D). Correlation analysis showed a correlation between CCL5 and immune cells. In dataset GSE37250, CCL5 was positively related to effector memory CD4 T cells, activated CD4 T cells, and activated CD8 T cells (Figure 6E). CCL5 was also positively correlated with the above immune cells in dataset GSE117261 (Figure 6F).
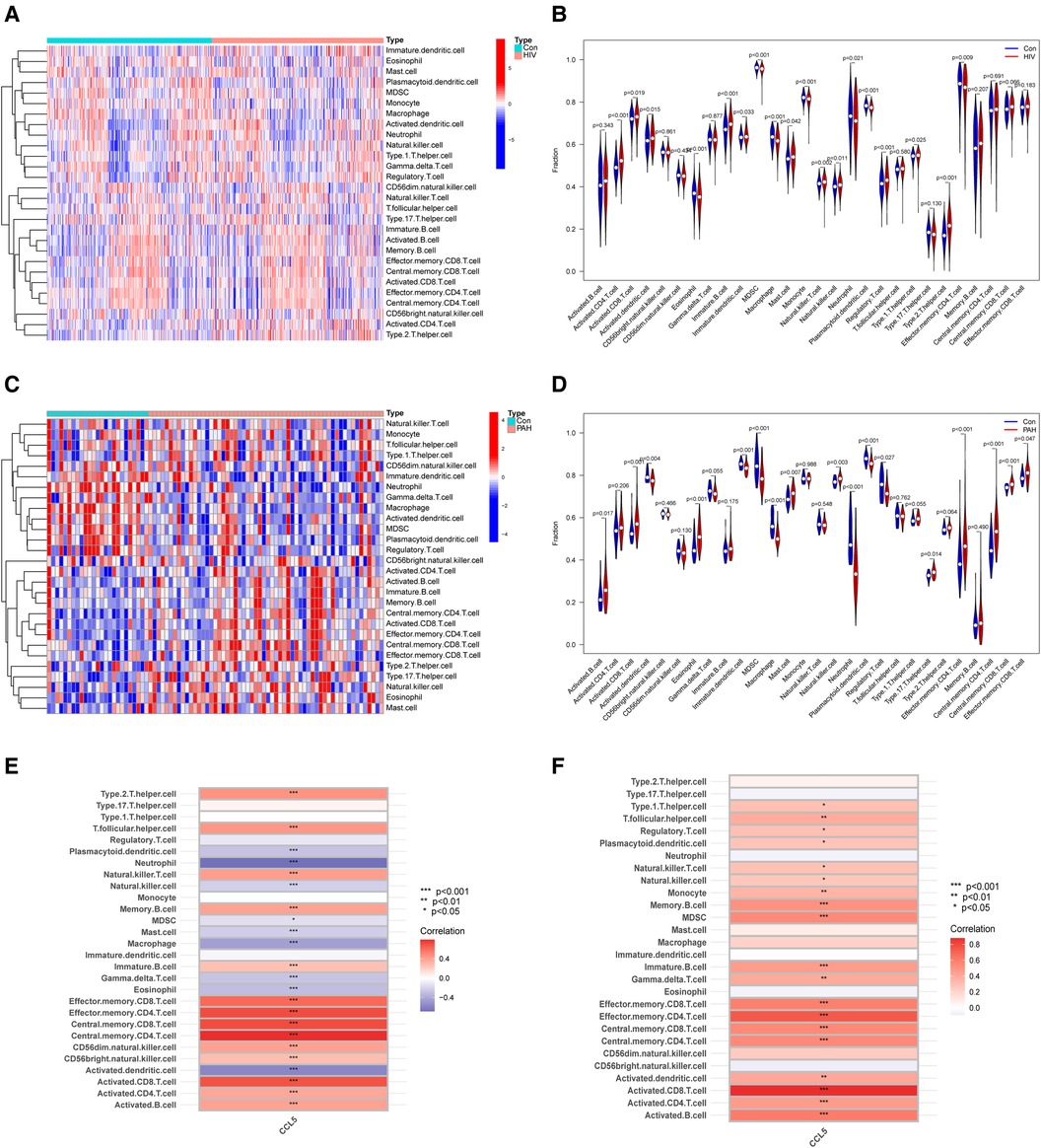
Figure 6 Immune infiltration analysis. (A) Heatmap of immune cells between control and HIV infection groups. (B) Violin plot of immune cells fraction between control and HIV infection groups. (C) Heatmap of immune cells between control and PAH groups. (D) Violin plot of immune cells fraction between control and PAH groups. (E) Correlation analysis of immune cell and CCL5 in GSE37250. (F) Correlation analysis of immune cell and CCL5 in GSE117261. HIV, human immunodeficiency virus; PAH, pulmonary arterial hypertension; CCL5, CC-type chemokine ligand 5.
The results of the PCR indicated that the expression of CCL5 was increased significantly in hypoxia-induced hPAECs and hPASMCs (Figures 7A,B).
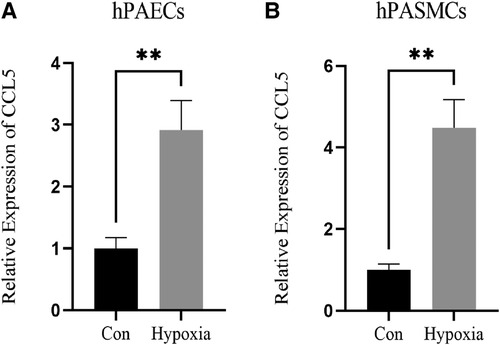
Figure 7 Expression of CCL5 in hPAECs and hPASMCs. (A) Expression level of CCL5 between control and hypoxia groups in hPAECs. (B) Expression level of CCL5 between control and hypoxia groups in hPASMCs. hPAECs, human pulmonary artery endothelial cells; hPASMCs, human pulmonary artery smooth muscle cells. n = 3, *P < 0.05, **P < 0.01.
Figure 8A showed the association between CCL5 and infections, including HIV infection. In addition, Figure 8B showed the association between CCL5 and cardiovascular disease, including PAH.
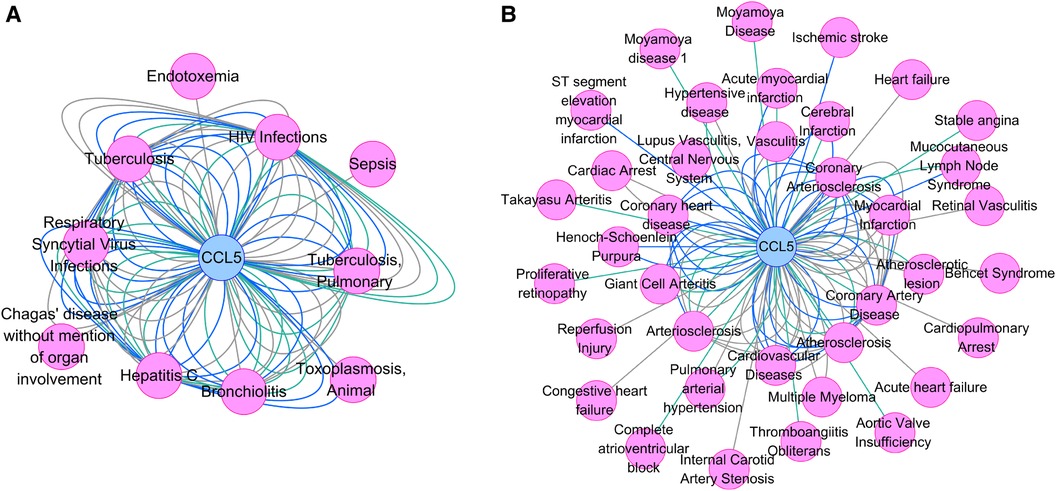
Figure 8 Gene-disease association network. (A) The association between CCL5 and infections. (B) The association between CCL5 and cardiovascular diseases. CCL5, CC-type chemokine ligand 5.
The native co-crystallized ligand of CCL5 was extracted and re-docked by Autodock (Supplementary Figure S4). The calculated RMSD value was 0.266Å, which was less than 2Å, indicating that the docking protocol was valid. Besides, molecular docking was performed between the ten potential drugs and CCL5. All of the anticipated binding sites had binding energies of less than −5 kcal/mol (Supplementary Table S4), suggesting that ligands can spontaneously attach to the receptor molecule. The binding modes and binding interactions between candidate drugs (ligands) and CCL5 were displayed in Figure 9. Ligands were located in the active pocket. Except for ticlopidine, the other ligands were bound to amino acid residues around the active pocket by forming hydrogen bonds, indicating that they formed stable conformations.
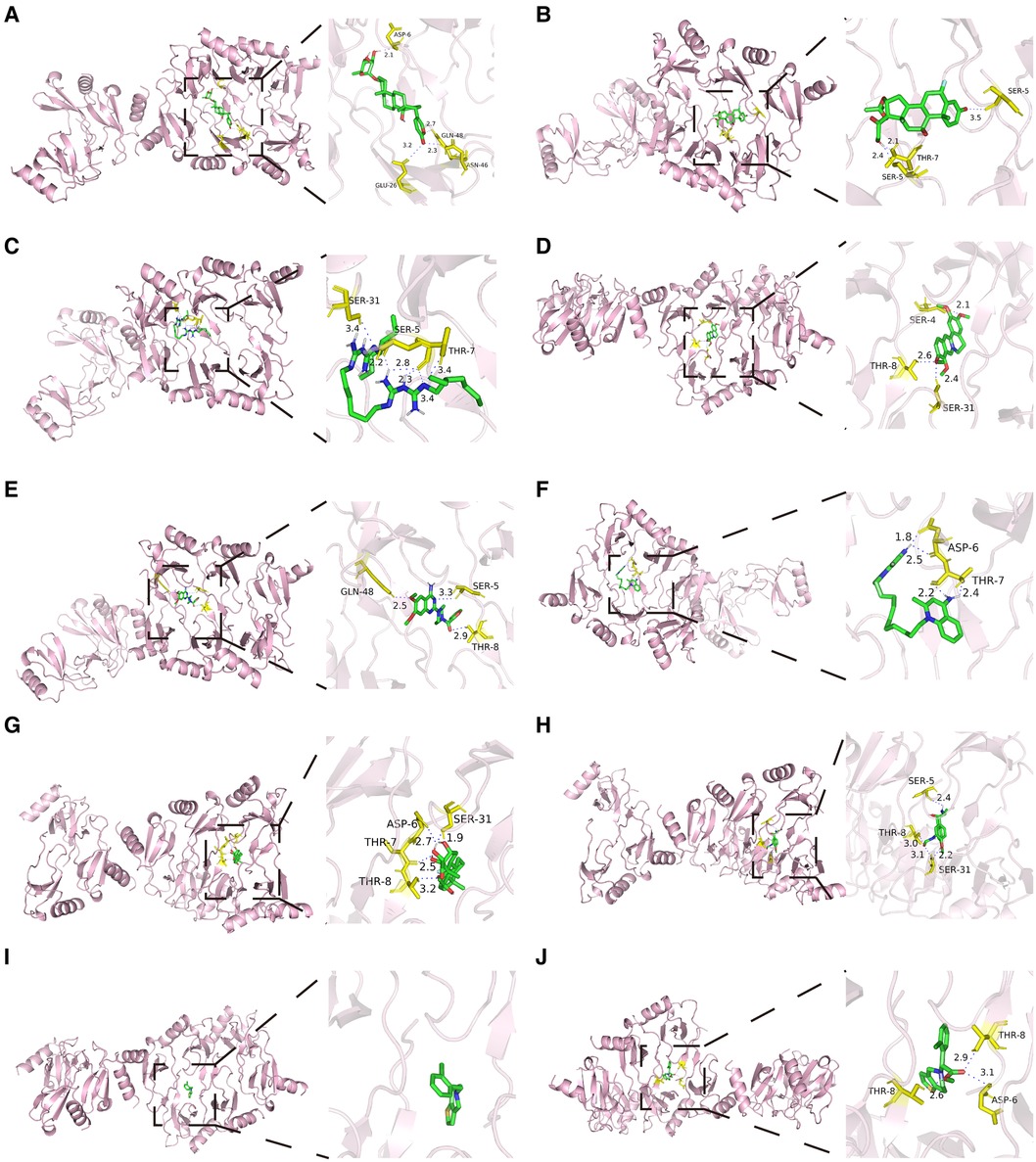
Figure 9 Molecular docking results of ten potential drugs and CCL5. Hydrogen bonds were displayed in blue dashed lines. Amino acid residues were labeled yellow. (A) Proscillaridin. (B) Flunisolide. (C) Alexidine. (D) Palmatine. (E) Prazosin. (F) Dequalinium. (G) Triamcinolone. (H) Formoterol. (I) Ticlopidine. (J) Clopidogrelum. CCL5, CC-type chemokine ligand 5.
This study aimed to explore the shared biomarkers between HIV infection and PAH and seek potential therapeutic targets for HIV-associated PAH. We found that CCL5 may be a key gene in HIV-associated PAH, and several potential drugs were identified. Over the past few decades, it has been recognized that there is a correlation between HIV infection and PAH. Kim and Factor reported the first case report of a man with both HIV infection and pulmonary arterial hypertension in 1987 (34). Shortly thereafter, more similar cases were reported, which revealed a possible correlation between HIV infection and PAH (35). HIV-positive individuals have a 1,000-fold higher risk of PAH than the general population (36). So far, the pathophysiological mechanism of PAH in HIV-infected people has not been clarified, and it may be related to the HIV-viral proteins GP120, TAT, and NEF. HIV-viral proteins lead to pulmonary artery endothelial damage through inflammatory responses, which further leads to pulmonary artery remodeling and pulmonary hypertension (37). However, its mechanism at the gene level needs to be further revealed.
In this study, we performed DEGs analysis on the HIV infection dataset and the PAH dataset, respectively. By intersecting the above two DEGs sets, 19 common DEGs were obtained. Functional enrichment analyses were conducted to investigate the biological functions of the above genes. GO analysis showed that the common DEGs were mainly enriched in positive regulation of mononuclear cell migration, positive regulation of T cell proliferation, and CCR chemokine receptor binding, which are mainly involved in inflammatory responses. It is consistent with previous studies. Immune activation and chronic inflammatory responses play important roles in HIV infection. HIV targets monocyte-derived dendritic cells, monocyte-derived macrophages, and CD4 T cells with the help of chemokine receptors to activate a series of complex signaling pathways that cause the long-term activation of inflammatory responses (38, 39). Similarly, in PAH, chemokines recruit various immune cells in lung tissue, including monocytes and T cells, and promote pulmonary artery hypertension by producing a series of inflammatory factors (40). Furthermore, KEGG enrichment analysis revealed that the shared genes were mainly enriched in viral protein interaction with cytokine and cytokine receptor. This is somewhat in line with previous studies. HIV-associated PAH might possess the following pathogenesis: HIV viral proteins bind to receptors on lung endothelial cells, promoting cytokine release and inflammation, thus resulting in vascular remodeling and pulmonary hypertension (35). Therefore, we can conclude that immune activation and inflammatory responses play an important role in HIV infection and PAH.
In order to further identify the key genes, we constructed a PPI network. Moreover, it was further confirmed in the validation cohort that CCL5 was highly expressed in HIV infection and PAH, enhancing the reliability of our study.
CCL5 (CC-type chemokine ligand 5), also known as RANTES (Regulated on Activation, Normal T Cell Expressed and Secreted), is mainly secreted by activated T cells. Numerous previous investigations demonstrated that CCL5 played a crucial role in human diseases, including solid tumors (41), autoimmune diseases (42), metabolic diseases (43), etc. Similarly, CCL5 is essential for HIV infection and PAH, although the exact mechanism remains to be further studied.
CCL5 was identified as an HIV-suppressive factor produced by CD8 T cells according to a previous study (44). A clinical study showed that the expression levels of CCL5 in HIV-infected people was increased compared with HIV-negative controls (45). A meta-analysis showed that the expression level of CCL5 was negatively associated with the risk of HIV infection. CCL5 can competitively bind to CCR5 or promote the internalization of CCR5, thereby preventing the entry and replication of HIV (46). The expression level of CCL5 in individuals with HIV infection may be influenced by many factors. In HIV-positive individuals, the increased cell frequency of memory-like NK cells (47), virtual memory CD8 T cells (48), and co-infection with the human T cell lymphotropic virus (HTLV) (49) may elevate the expression of CCL5, playing a role in anti-HIV and delaying AIDS. Consistent with previous studies, we discovered that CCL5 expression was markedly elevated in the HIV infection group, which may be connected to the activation of cellular immune responses caused by viral infection.
In addition, CCL5 expression was positively correlated with the risk of PAH. A recent study showed that CCL5 expression levels were significantly elevated in the pulmonary endarterectomy tissue of patients with chronic thromboembolic pulmonary hypertension compared to healthy controls, and CCL5 may lead to pulmonary hypertension by promoting fibroblast migration (50). A study indicated that CCL5-CCR5 pathway was activated in PAH, thus promoting macrophage recruitment and pulmonary-artery smooth muscle cells proliferation (51). Besides, another study indicated that CCL5 promoted platelet activation, thus leading to endothelial cell injury and vascular remodeling in PAH (52). Furthermore, a study showed that CCL5 deficiency could reverse hypoxia-induced pulmonary hypertension by restoring bone morphogenetic protein receptor 2 (BMPR2) signaling (53). Consistent with previous studies, our study indicated that the expression level of CCL5 was increased in transcriptome data. To make our study more rigorous, in vitro experiments were conducted. Hypoxia-induced hPAECs and hPASMCs are commonly used to construct models of PAH. Our study indicated that CCL5 was highly expressed in hypoxia-induced hPAECs and hPASMCs, suggesting that CCL5 may promote the pathogenesis of PAH. It was consistent with our findings in the transcriptome data. In a word, although CCL5 plays a beneficial role in HIV infection, elevated levels of CCL5 promote the progression of PAH. Therefore, we can reasonably speculate that CCL5 is a key gene in HIV-associated PAH.
Regulating the expression of CCL5 may be a target for the treatment of HIV-associated PAH in the future. Previous studies have revealed several drugs that can regulate the expression of CCL5. A study showed that dimethylfumarate downregulated the secretion of CCL5 by inhibiting NF-KB (54). Similarly, Chen et al.'s study indicated that traditional Chinese medicines Shuanghuanglian and Qingkailing inhibited the expression of CCL5 by suppressing NF-kB (55). Likewise, Terminalia chebula Retz. Extract (56), desipramine, and atomoxetine (57) have been proven to downregulate the expression level of CCL5. Therefore, we speculated that the above drugs might play a role in the treatment of HIV-associated PAH by inhibiting the expression level of CCL5.
Furthermore, several drugs that target CCL5 were predicted using the DSigDB, including proscillaridin, flunisolide, alexidine, palmatine, prazosin, dequalinium, etc. Proscillaridin is a cardiac glycoside that can alleviate heart failure by inhibiting the Na+/K + pump (58), and it can also promote tumor cell apoptosis, thus playing an anti-tumor role (59). Flunisolide is a corticosteroid that is used in asthma and rhinitis by reducing inflammatory responses (60). These drugs may delay the progression of HIV-associated PAH by reducing the inflammatory responses. However, studies on the application of the above candidate drugs in HIV-related PAH are very limited. Our study provides a new idea for drug development in HIV-related PAH in the future. Due to the fact that the effect of the candidate drugs in vivo cannot be predicted by molecular docking, further studies are needed.
There are some advantages in our study. Firstly, previous studies on the common mechanisms of HIV infection and PAH are very limited. This was the first study to explore shared biomarkers between HIV infection and PAH by using bioinformatics methods, and the key role of inflammatory responses in HIV infection and PAH was identified by enrichment analysis and immune infiltration analysis, providing new diagnostic and treatment targets for HIV patients with PAH. Besides, the above findings were verified in validation datasets and in vitro experiments, which strengthened the persuasiveness of this study. Lastly, candidate drugs were predicted by molecular docking, which provided new ideas for drug development in the future.
Nevertheless, there are some shortcomings in our study. First of all, although our study was intended to explore the shared genes of HIV infection and PAH, we were unable to obtain transcriptome data from patients with both HIV infection and PAH. It is necessary to collect clinical samples for further analysis in the future. Secondly, our study did not clarify the specific mechanism by which CCL5 promotes the development of PAH, and further animal or cell experiments are required. Lastly, the specific mechanisms of interactions of 10 potential drugs with CCL5 are unknown, and further studies are required.
In conclusion, our study identified shared biomarkers in HIV infection and PAH. We illustrated that immune responses might be a key step in HIV infection and PAH, and CCL5 was a key gene in HIV-associated PAH. Finally, ten candidate drugs were predicted for HIV-associated PAH.
Publicly available datasets were analyzed in this study. This data can be found here: GEO database (https://www.ncbi.nlm.nih.gov/geo/) under the accession numbers GSE37250, GSE117261, GSE30310, and GSE53408.
MY: Formal Analysis, Writing – original draft. WB: Writing – review & editing. ZZ: Writing – review & editing, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
The current study was financed by Shenzhen Nanshan District Health Science and Technology Project under grant No. 2020108.
We appreciate all authors in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2024.1417701/full#supplementary-material
HIV, human immunodeficiency virus; PAH, pulmonary arterial hypertension; GEO, gene expression omnibus; DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; PPI, protein-protein interaction; CCL5, CC-type chemokine ligand 5; hPAECs, human pulmonary artery endothelial cells; hPASMCs, human pulmonary artery smooth muscle cells; RT-qPCR, quantitative real-time polymerase chain reaction; AIDS, Acquired Immunodeficiency Syndrome; MCODE, molecular complex detection; ssGSEA, single sample gene set enrichment analysis; DSigDB, drug signatures database; RMSD, root mean square deviation; RANTES, regulated on activation, normal T cell expressed and secreted; CCR5, C-C chemokine receptor type 5.
2. Bekker LG, Beyrer C, Mgodi N, Lewin SR, Delany-Moretlwe S, Taiwo B, et al. HIV infection. Nat Rev Dis Primers. (2023) 9:42. doi: 10.1038/s41572-023-00452-3
3. Deeks SG, Overbaugh J, Phillips A, Buchbinder S. HIV infection. Nat Rev Dis Primers. (2015) 1:15035. doi: 10.1038/nrdp.2015.35
4. Hemkens LG, Bucher HC. HIV infection and cardiovascular disease. Eur Heart J. (2014) 35:1373–81. doi: 10.1093/eurheartj/eht528
5. Sitbon O. HIV-related pulmonary arterial hypertension: clinical presentation and management. AIDS. (2008) 22(Suppl 3):S55–62. doi: 10.1097/01.aids.0000327517.62665.ec
6. Basyal B, Jarrett H, Barnett CF. Pulmonary hypertension in HIV. Can J Cardiol. (2019) 35:288–98. doi: 10.1016/j.cjca.2019.01.005
7. Delcroix M, Howard L. Pulmonary arterial hypertension: the burden of disease and impact on quality of life. Eur Respir Rev. (2015) 24:621–9. doi: 10.1183/16000617.0063-2015
8. Simenauer A, Nozik-Grayck E, Cota-Gomez A. The DNA damage response and HIV-associated pulmonary arterial hypertension. Int J Mol Sci. (2020) 21(9):3305. doi: 10.3390/ijms21093305
9. Feinstein MJ, Hsue PY, Benjamin LA, Bloomfield GS, Currier JS, Freiberg MS, et al. Characteristics, prevention, and management of cardiovascular disease in people living with HIV: a scientific statement from the American Heart Association. Circulation. (2019) 140:e98–124. doi: 10.1161/CIR.0000000000000695
10. Parikh RV, Scherzer R, Nitta EM, Leone A, Hur S, Mistry V, et al. Increased levels of asymmetric dimethylarginine are associated with pulmonary arterial hypertension in HIV infection. AIDS. (2014) 28:511–9. doi: 10.1097/QAD.0000000000000124
11. Guo ML, Kook YH, Shannon CE, Buch S. Notch3/VEGF-A axis is involved in TAT-mediated proliferation of pulmonary artery smooth muscle cells: implications for HIV-associated PAH. Cell Death Discov. (2018) 4:22. doi: 10.1038/s41420-018-0087-9
12. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. (2012) 41:D991–5. doi: 10.1093/nar/gks1193
13. Cattamanchi A, Kaforou M, Wright VJ, Oni T, French N, Anderson ST, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. (2013) 10:e1001538. doi: 10.1371/journal.pmed.1001538
14. Stearman RS, Bui QM, Speyer G, Handen A, Cornelius AR, Graham BB, et al. Systems analysis of the human pulmonary arterial hypertension lung transcriptome. Am J Respir Cell Mol Biol. (2019) 60:637–49. doi: 10.1165/rcmb.2018-0368OC
15. Morse CG, Voss JG, Rakocevic G, McLaughlin M, Vinton CL, Huber C, et al. HIV infection and antiretroviral therapy have divergent effects on mitochondria in adipose tissue. J Infect Dis. (2012) 205:1778–87. doi: 10.1093/infdis/jis101
16. Zhao YD, Yun HZH, Peng J, Yin L, Chu L, Wu L, et al. De novo synthesize of bile acids in pulmonary arterial hypertension lung. Metabolomics. (2014) 10:1169–75. doi: 10.1007/s11306-014-0653-y
17. Yan C, Niu Y, Wang X. Blood transcriptome analysis revealed the crosstalk between COVID-19 and HIV. Front Immunol. (2022) 13:1008653. doi: 10.3389/fimmu.2022.1008653
18. Chen W, Yang Q, Hu L, Wang M, Yang Z, Zeng X, et al. Shared diagnostic genes and potential mechanism between PCOS and recurrent implantation failure revealed by integrated transcriptomic analysis and machine learning. Front Immunol. (2023) 14:e1175384. doi: 10.3389/fimmu.2023.1175384
19. Chen Z, Wang W, Jue H, Hua Y. Bioinformatics and system biology approach to identify potential common pathogenesis for COVID-19 infection and osteoarthritis. Sci Rep. (2023) 13(1):9330. doi: 10.1038/s41598-023-32555-y
20. The Gene Ontology, C. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. (2019) 47:D330–8. doi: 10.1093/nar/gky1055
21. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. (2000) 28:27–30. doi: 10.1093/nar/28.1.27
22. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. (2023) 51:D638–46. doi: 10.1093/nar/gkac1000
23. Otasek D, Morris JH, Boucas J, Pico AR, Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. (2019) 20:185. doi: 10.1186/s13059-019-1758-4
24. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. (2003) 4:2. doi: 10.1186/1471-2105-4-2
25. Guo Q, Zhu Q, Zhang T, Qu Q, Cheang I, Liao S, et al. Integrated bioinformatic analysis reveals immune molecular markers and potential drugs for diabetic cardiomyopathy. Front Endocrinol (Lausanne). (2022) 13:933635. doi: 10.3389/fendo.2022.933635
26. Yao M, Wang S, Han Y, Zhao H, Yin Y, Zhang Y, et al. Micro-inflammation related gene signatures are associated with clinical features and immune status of fibromyalgia. J Transl Med. (2023) 21:594. doi: 10.1186/s12967-023-04477-w
27. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMCBioinformatics. (2013) 14:7. doi: 10.1186/1471-2105-14-7
28. Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. (2017) 45:D833–9. doi: 10.1093/nar/gkw943
29. Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee S, et al. DSigDB: drug signatures database for gene set analysis. Bioinformatics. (2015) 31:3069–71. doi: 10.1093/bioinformatics/btv313
30. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. (2016) 44:W90–7. doi: 10.1093/nar/gkw377
31. Trott O, Olson AJ. Autodock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. (2010) 31:455–61. doi: 10.1002/jcc.21334
32. Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. (2010) 24:417–22. doi: 10.1007/s10822-010-9352-6
33. Kwofie SK, Asiedu SO, Koranteng R, Quarshie E, Tiburu EK, Miller WA 3rd, et al., Computer-aided identification of potential inhibitors against necator americanus glutathione S-transferase 3. Inform Med Unlocked. (2022) 30:100957. doi: 10.1016/j.imu.2022.100957
34. Kim KK, Factor SM. Membranoproliferative glomerulonephritis and plexogenic pulmonary arteriopathy in a homosexual man with acquired immunodeficiency syndrome. Hum Pathol. (1987) 18:1293–6. doi: 10.1016/S0046-8177(87)80417-3
35. Butrous G. Human immunodeficiency virus-associated pulmonary arterial hypertension: considerations for pulmonary vascular diseases in the developing world. Circulation. (2015) 131:1361–70. doi: 10.1161/CIRCULATIONAHA.114.006978
36. Triplette M, Crothers K, Attia EF. Non-infectious pulmonary diseases and HIV. Curr HIV/AIDS Rep. (2016) 13:140–8. doi: 10.1007/s11904-016-0313-0
37. Correale M, Palmiotti GA, Lo Storto MM, Montrone D, Foschino Barbaro MP, Di Biase M, et al. HIV-associated pulmonary arterial hypertension: from bedside to the future. Eur J Clin Invest. (2015) 45:515–28. doi: 10.1111/eci.12427
38. Silvin A, Manel N. Innate immune sensing of HIV infection. Curr Opin Immunol. (2015) 32:54–60. doi: 10.1016/j.coi.2014.12.003
39. Lv T, Cao W, Li T. HIV-related immune activation and inflammation: current understanding and strategies. J Immunol Res. (2021) 2021:7316456. doi: 10.1155/2021/7316456
40. Yan Q, Liu S, Sun Y, Chen C, Yang Y, Yang S, et al. CC chemokines modulate immune responses in pulmonary hypertension. J Adv Res. (2023). doi: 10.1016/j.jare.2023.10.015
41. Dangaj D, Bruand M, Grimm AJ, Ronet C, Barras D, Duttagupta PA, et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. (2019) 35:885–900.e810. doi: 10.1016/j.ccell.2019.05.004
42. Hinrichs AC, Blokland SLM, Kruize AA, Lafeber FPJ, Leavis HL, van Roon JAG. CCL5 release by CCR9+ CD8T cells: a potential contributor to immunopathology of primary Sjogren’s syndrome. Front Immunol. (2022) 13:887972. doi: 10.3389/fimmu.2022.887972
43. Ooms M, Strom A, Strassburger K, Menart B, Leslie RD, Schloot NC. Increased spontaneous CCL2 (MCP-1) and CCL5 (RANTES) secretion in vitro in LADA compared to type 1 diabetes and type 2 diabetes: action LADA 14. Diabetes Metab Res Rev. (2021) 37:e3431. doi: 10.1002/dmrr.3431
44. Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. (1995) 270:1811–5. doi: 10.1126/science.270.5243.1811
45. Ullum H, Cozzi Lepri A, Victor J, Aladdin H, Phillips AN, Gerstoft J, et al. Production of beta-chemokines in human immunodeficiency virus (HIV) infection: evidence that high levels of macrophage inflammatory protein-1beta are associated with a decreased risk of HIV disease progression. J Infect Dis. (1998) 177:331–6. doi: 10.1086/514192
46. Silva MJA, Marinho RL, Dos Santos PAS, Dos Santos CS, Ribeiro LR, Rodrigues YC, et al. The association between CCL5/RANTES SNPs and susceptibility to HIV-1 infection: a meta-analysis. Viruses. (2023) 15(9):1958. doi: 10.3390/v15091958
47. Florez-Alvarez L, Hernandez JC, Zapata W. NK cells in HIV-1 infection: from basic science to vaccine strategies. Front Immunol. (2018) 9:2290. doi: 10.3389/fimmu.2018.02290
48. Hu W, Li YJ, Zhen C, Wang YY, Huang HH, Zou J, et al. CCL5-Secreting virtual memory CD8+ T cells inversely associate with viral reservoir size in HIV-1-infected individuals on antiretroviral therapy. Front Immunol. (2022) 13:897569. doi: 10.3389/fimmu.2022.897569
49. Oo Z, Barrios CS, Castillo L, Beilke MA. High levels of CC-chemokine expression and downregulated levels of CCR5 during HIV-1/HTLV-1 and HIV-1/HTLV-2 coinfections. J Med Virol. (2015) 87:790–7. doi: 10.1002/jmv.24070
50. Zabini D, Heinemann A, Foris V, Nagaraj C, Nierlich P, Balint Z, et al. Comprehensive analysis of inflammatory markers in chronic thromboembolic pulmonary hypertension patients. Eur Respir J. (2014) 44:951–62. doi: 10.1183/09031936.00145013
51. Abid S, Marcos E, Parpaleix A, Amsellem V, Breau M, Houssaini A, et al. CCR2/CCR5-mediated macrophage-smooth muscle cell crosstalk in pulmonary hypertension. Eur Respir J. (2019) 54(4):1802308. doi: 10.1183/13993003.02308-2018
52. Delaney C, Davizon-Castillo P, Allawzi A, Posey J, Gandjeva A, Neeves K, et al. Platelet activation contributes to hypoxia-induced inflammation. Am J Physiol Lung Cell Mol Physiol. (2021) 320:L413–21. doi: 10.1152/ajplung.00519.2020
53. Nie X, Tan J, Dai Y, Liu Y, Zou J, Sun J, et al. CCL5 deficiency rescues pulmonary vascular dysfunction, and reverses pulmonary hypertension via caveolin-1-dependent BMPR2 activation. J Mol Cell Cardiol. (2018) 116:41–56. doi: 10.1016/j.yjmcc.2018.01.016
54. Seidel P, Roth M. Anti-inflammatory dimethylfumarate: a potential new therapy for asthma? Mediators Inflamm. (2013) 2013:875403. doi: 10.1155/2013/875403
55. Chen X, Howard OMZ, Yang X, Wang L, Oppenheim JJ, Krakauer T. Effects of Shuanghuanglian and Qingkailing, two multi-components of traditional Chinese medicinal preparations, on human leukocyte function. Life Sci. (2002) 70:2897–913. doi: 10.1016/S0024-3205(02)01541-2
56. Kim HJ, Song HK, Park SH, Jang S, Park KS, Song KH, et al. Terminalia Chebula Retz. Extract ameliorates the symptoms of atopic dermatitis by regulating anti-inflammatory factors in vivo and suppressing STAT1/3 and NF-kB signaling in vitro. Phytomedicine. (2022) 104:154318. doi: 10.1016/j.phymed.2022.154318
57. O’Sullivan JB, Ryan KM, Harkin A, Connor TJ. Noradrenaline reuptake inhibitors inhibit expression of chemokines IP-10 and RANTES and cell adhesion molecules VCAM-1 and ICAM-1 in the CNS following a systemic inflammatory challenge. J Neuroimmunol. (2010) 220:34–42. doi: 10.1016/j.jneuroim.2009.12.007
58. Da Costa EM, Armaos G, McInnes G, Beaudry A, Moquin-Beaudry G, Bertrand-Lehouillier V, et al. Heart failure drug proscillaridin A targets MYC overexpressing leukemia through global loss of lysine acetylation. J Exp Clin Cancer Res. (2019) 38:251. doi: 10.1186/s13046-019-1242-8
59. Li RZ, Fan XX, Duan FG, Jiang ZB, Pan HD, Luo LX, et al. Proscillaridin A induces apoptosis and suppresses non-small-cell lung cancer tumor growth via calcium-induced DR4 upregulation. Cell Death Dis. (2018) 9:696. doi: 10.1038/s41419-018-0733-4
Keywords: HIV infection, PAH, DEGs, CCL5, molecular docking
Citation: Yang M, Bi W and Zhang Z (2024) Identification and validation of CCL5 as a key gene in HIV infection and pulmonary arterial hypertension. Front. Cardiovasc. Med. 11: 1417701. doi: 10.3389/fcvm.2024.1417701
Received: 15 April 2024; Accepted: 16 July 2024;
Published: 25 July 2024.
Edited by:
Jianguo Sun, China Pharmaceutical University, ChinaReviewed by:
Jun-Feng Cao, Southwest Jiaotong University, China© 2024 Yang, Bi and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijie Zhang, ZHJ6aGlqaWV6aGFuZ0AxNjMuY29t
†ORCID:
Mengyue Yang
orcid.org/0009-0007-6266-1014
Wen Bi
orcid.org/0000-0002-5613-6412
Zhijie Zhang
orcid.org/0009-0003-0417-282X
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.