
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med., 29 July 2024
Sec. Cardiac Rhythmology
Volume 11 - 2024 | https://doi.org/10.3389/fcvm.2024.1411784
Background: Clinical observations and epidemiological studies suggest a potential linkage between gastroesophageal reflux disease (GERD) and arrhythmias, yet the underlying mechanism remains elusive. This study investigates the causal relationship between GERD and four types of arrhythmia through a genetic lens, employing Mendelian randomization analysis to elucidate the directionality of these associations.
Methods: Selected single nucleotide polymorphisms (SNPs) from genome-wide association study (GWAS) data were utilized as instrumental variables. The inverse variance weighting (IVW) method, MR-Egger regression analysis, and the weighted median method were employed in two-sample Mendelian randomization analysis. Horizontal pleiotropy was detected and corrected using the MR-PRESSO test and MR-Egger regression. The stability and reliability of the Mendelian randomization results were assessed using the leave-one-out method, Cochran's Q test, and funnel plots. The causal relationship between GERD and four types of arrhythmias was evaluated using the odds ratio (OR).
Results: IVW results indicated that GERD could increase the risk of arrhythmias. A one standard deviation increases in the logarithmically transformed GERD score resulted in a 34% increase in the risk of arrhythmia (OR = 1.34; 95% CI 1.19–1.51; p = 1.66E-06). No significant correlation was found between GERD and other arrhythmias.
Conclusion: A causal relationship exists between GERD and arrhythmias, suggesting that GERD increases the risk of developing these arrhythmias.
Arrhythmia, the most prevalent cardiovascular disorders, are primarily observed in pathological states, encompassing both cardiac and non-cardiac conditions. Non-cardiac triggers of arrhythmias include hyperthyroidism, anemia, infection, and gastroesophageal reflux disease (GERD). These conditions are categorized into various types, such as atrial fibrillation, atrial flutter, supraventricular tachycardia, ventricular fibrillation, ventricular flutter, and conduction blocks. Cardiac arrhythmias affect over 700,000 people in England, with the most prevalent type, atrial fibrillation (AF), affecting up to 1% of the population and accounting for 1% of the entire NHS budget (1, 2). In the United States, the projected increase in arrhythmia cases to 16 million by 2050 is expected to result in considerable morbidity and mortality (3). This highlights the urgent need for strategies to prevent arrhythmia.Several arrhythmias we included in this study: paroxysmal tachycardia (paroxysmal tachycardia is regarded by clinicians as vegetative dystonia with crises and centres of overexcitatiou in different departments of the conduction apparatus of the heart.) (4), right bundle branch block, left bundle branch block, conduction disorders (RBBB, LBBB and conduction disorders: A block in the bundle branch of the electrical conduction system of the heart) (5).
GERD is characterized by the backflow of stomach contents into the esophagus, leading to discomfort and potential complications. Common symptoms include heartburn, characterized by a burning sensation beneath the sternum that ascends from the upper abdomen to the neck. GERD is highly prevalent, affecting approximately 20% of adults in the Western world (6). Population-based studies indicate its widespread presence in communities, with prevalence rates ranging from 10% to 30% (7).
Numerous prior studies have demonstrated that risk factors associated with arrhythmia, such as hypertension, diabetes, smoking, and dyslipidemia, elevate the risk of arrhythmias (8). Consequently, addressing these risk factors can alleviate the economic burden on patients, reduce the incidence of arrhythmias, and assist clinicians in adopting innovative treatment approaches (9). Several studies suggest a potential link between gastroesophageal reflux and arrhythmias, although the underlying mechanisms, possibly related to vagal tone and parasympathetic reflexes, remain unclear (10–12). Roemheld, in the last century, identified a correlation between the upper digestive tract and cardiovascular diseases, coining the term “gastrocardiac syndrome”. He demonstrated that esophageal stimulation could induce arrhythmias (13). Over the years, the investigation into the causal relationship between digestive and cardiovascular diseases has intensified. An observational studies indicate that esophageal acid stimulation can initiate a cardiac autonomic reflex, leading to arrhythmias (14). Previous research reveals that sensory input from the gut or other internal organs can influence heart function. For instance, rat studies have shown that bloating can affect cardiovascular regulation through the activation of neuronal circuits of excitation and inhibition (15). However, in humans, the effects of gastrointestinal stimulation on cardiac autonomic nervous function remain elusive. Tougas and colleagues investigated the impact of esophageal stimulation on cardiac autonomic function, applying mechanical or electrical stimulation to the esophagus and observing its effect on heart rate, thereby testing the hypothesis of a vagal afferent response (16). In summary, a potential correlation exists between GERD and arrhythmias, and this paper aims to determine whether a causal relationship exists between them through rigorous scientific methods.
Mendelian randomization (MR) is an analytical method employed to assess the observed causal relationships between modifiable risk factors and clinically relevant outcomes, adhering to the Mendelian laws of genetics, which dictate the random distribution of allele genes. MR analysis leverages genetic variation linked to an exposure as an instrumental variable to deduce causality with the outcome. Should a causal link exist between the exposure and outcome, the instrumental variable associated with the exposure will proportionally influence the outcome, thereby substantially reducing confounding and reverse causation (17).The objective of Mendelian randomization is to evaluate causal hypotheses within non-experimental data (18). To estimate the causal relationships between GERD and arrhythmias, we utilized pooled level data from several large consortia in a two-sample MR (TSMR) study.
As illustrated in Figure 1, an MR study was designed to systematically explore the association between gastroesophageal reflux and arrhythmias risk. All studies included in our analysis received approval from their respective academic ethics review boards, with each participant providing written informed consent. The institutional review boards granted the necessary approvals. Notably, the current study, being a reanalysis of publicly available GWAS data, did not require additional ethical approval. This study adhered to the STROBE-MR (Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization) guidelines (Supplementary Table S4).
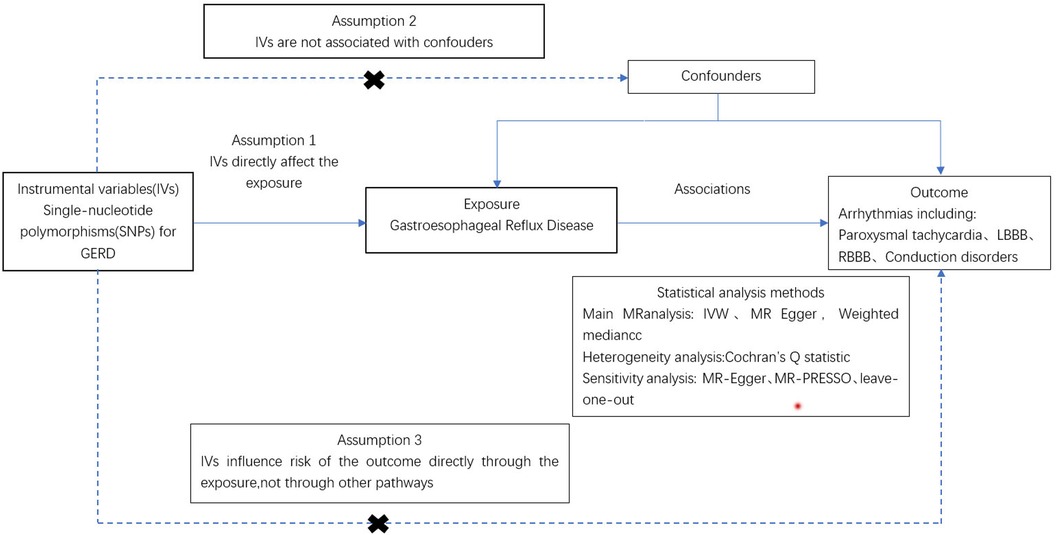
Figure 1 Procedure for an MR analysis of causal associations gastroesophageal reflux disease and risk for arrhythmia.
In this study, a comprehensive MR analysis was conducted to elucidate the causal relationship between GERD and arrhythmias (paroxysmal tachycardia, right bundle branch block, left bundle branch block, and conduction disorders), All the patients met the international diagnostic criteria for ICD. The GWAS data for GERD were sourced from the GWAS Catalog (ebi-a-GCST90000514), encompassing 129,080 cases and 473,524 controls of European ancestry (19),as the data related to GERD is derived from the GWAS database, the classification of GERD remains unknown to us. Genetic instrumental variables for the four arrhythmias were obtained from the R10 database of the FinnGen Biobank (20). Details are provided in Supplementary Table S1. To adhere to the two-sample MR study's assumption of independent samples for exposure and outcomes, we excluded GWAS pooled measurements of arrhythmias phenotypes with significant overlap with the GERD samples. The selected arrhythmias ensured all participants were of European descent, thereby reducing the risk of pleiotropic bias across the analyses (21). This study aimed to investigate the causal link between genetic predisposition to GERD and arrhythmias risk.
MR analysis is predicated on three fundamental conditions: (1) the genetic variant is strongly associated with the exposure; (2) the genetic variant is not influenced by confounding factors; (3) the genetic variant is not affected by the outcome (21–23). Initially, a genome-wide significance threshold of 5 × 10−8 was applied to identify SNPs significantly associated with GERD (24). Secondly, to ensure the independence of SNPs, we required that those associated with exposure should not be in linkage disequilibrium (LD), setting the LD threshold at r2 = 0.001 distance greater than 10,000 KB (25). Furthermore, the F-statistic was calculated for the IVs to assess the presence of weak instrumental bias. The F-value was determined using the equation F = Beta2/Se2, where Beta represents the allele effect value, and Se represents the estimated standard error of Beta. To prevent any potential bias caused by weak IVs, only IVs with F > 10 were retained for further analysis (26). Lastly, SNPs meeting these criteria were verified using the Phenoscanner database (www.phenoscanner.medschl.cam.ac.uk) on March 10, 2024, to exclude any that might be associated with a confounding phenotype (27). SNPS not linked to phenotypes affecting the results were retained for further analysis.
In this study, the inverse variance weighting (IVW) method served as the primary approach to estimate the causal effect. Within two-sample MR Analysis, IVW is regarded as a potent method for causality detection (28). Supplementary analyses were conducted using MR-Egger (29) and the weighted median methods (30). A series of sensitivity analyses were then performed. Initially, to mitigate the impact of heterogeneity on causal effect, Cochran's Q test was employed to assess heterogeneity. If p > 0.05, indicating that the influence of heterogeneity on the causal effect is negligible, the fixed effect model was applied. Conversely, the random effect model was utilized to minimize the impact of heterogeneity on the causal effect (Supplementary Table S5) (31). Subsequently, the MR-Egger intercept test was employed to detect horizontal pleiotropy (29). Additionally, the MR-PRESSO outlier test was utilized to identify and adjust for outliers among SNPs, thereby correcting for horizontal pleiotropy (32). To assess the robustness of the findings, a residual sensitivity analysis was conducted. The R programming language (version 4.3.1) facilitated the statistical analysis, employing the “TwoSampleMR” and “MRPRESSO” packages for MR analysis and the “forest plot” package for visualization. A p-value < 0.05 was deemed statistically significant.
In this analysis, 79 SNPS associated with GERD and arrhythmias were identified. Allelic frequency palindromes within these SNPs were excluded, specifically rs2145318, rs2358016, rs9517313, and rs957345. The F statistic was calculated for each SNP, all demonstrating F values greater than 10, indicative of no weak instrumental bias. For detailed F values, refer to Supplementary Table S2. All selected SNPs were then verified using the Phenoscanner V2 database, resulting in no additional SNP exclusions. Ultimately, these 79 SNPs were employed as instrumental variables for GERD, with detailed information available in Supplementary Table.
Three methods (MR Egger, weighted median, and IVW) were employed to assess the causal relationship between GERD and arrhythmias. The IVW method, serving as the primary analytical approach, identified GERD as an independent risk factor for paroxysmal tachycardia (OR = 1.34; 95% CI 1.19–1.51; p = 1.66E-06), as depicted in Figure 2. Our analysis did not reveal significant causal associations between GERD and other arrhythmias, such as conduction disorders, right bundle branch block (RBBB), and left bundle branch block (LBBB), as detailed in Supplementary Table S3.
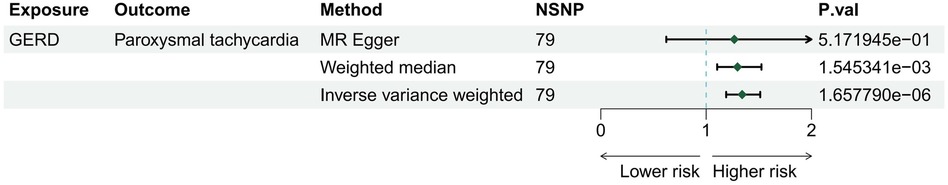
Figure 2 The risk association between gastroesophageal reflux disease and risk for arrhythmia in a forest plot.
These findings indicate that GERD may elevate the risk of paroxysmal tachycardia, both epidemiologically and genetically, whereas no causal links were established between GERD and LBBB, RBBB, or conduction disorders. Scatter plots illustrating the estimated effect sizes for gene-predicted GERD and paroxysmal tachycardia are presented in Figure 3.
Furthermore, MR-Egger intercept and MR-PRESSO tests were conducted to detect pleiotropy, with P-values > 0.05 suggesting no pleiotropic interference. A leave-one-out analysis was also performed, sequentially excluding each SNP to verify the consistency of the causality results, which confirmed the robustness of the MR analysis findings (Supplementary Figure).
GERD, a prevalent condition of the digestive system, is notably linked to various cardiovascular diseases due to the unique anatomical proximity of the esophagus to the heart. Our MR analysis investigated the causal relationship between GERD and four arrhythmias, revealing a significant association between GERD and paroxysmal tachycardia. GERD could increase the risk of arrhythmias. A one standard deviation increases in the logarithmically transformed GERD score resulted in a 34% increase in the risk of arrhythmia. Several epidemiological studies have highlighted GERD's pivotal role in triggering and promoting arrhythmias. Huang et al. conducted a population-based study and demonstrated a higher incidence of future arrhythmias in patients with GERD compared to controls (HR of 1.31, 95% CI of 1.06–1.61, P = 0.013) as part of a nationwide cohort in Taiwan (33). Similarly, Kunz et al. reported the relative risk for developing arrhythmias in GERD patients compared with controls during healthcare encounters in the USA (HR of 1.39, 95% CI of 1.33-1.45) (34). However, it is important to note that this study did not observe an association between GERD and left bundle branch block(LBBB), right bundle branch block(RBBB), or conduction disorders. Despite some observational studies suggesting a link between GERD and arrhythmias (35), many predisposing factors for arrhythmias are similar to those associated with GERD, including increasing age, sleep apnea, obesity, and diabetes, making it challenging to distinguish GERD as a predisposing factor independent of common confounding factors The actual incidence of patients with both GERD and arrhythmias is reported to be 5% (36). Current studies, either too small or retrospective, fail to show a clear association between GERD and arrhythmias. In a meta-analysis involving 82 882 GERD patients and 29 671 patients without GERD, there was no statistically significant increase in the risk of arrhythmias (37). Therefore, we speculate that the lack of association between GERD and LBBB, RBBB, and conduction disorders may be related to the small size of study statistics or the presence of common confounding factors.
Clinical observational studies have consistently demonstrated a significant link between GERD and certain arrhythmias (35). Another research found a causal connection between gastroesophageal reflux and supraventricular tachycardia (38). Electrophysiological disturbances are crucial for the onset of paroxysmal tachycardia, with clinical evidence underscoring inflammation as a key factor in cardiac electrophysiological abnormalities (39). GERD triggers an increase in interleukin-8 mRNA (IL-8 mRNA) expression in esophageal mucosal cells, leading to neutrophil infiltration and an inflammatory response (40). This inflammation can cause myocardial damage and influence the electrophysiological remodeling of the myocardium by altering myocardial membrane potential, affecting cardiac ion channels, and impacting the action potential (39), ultimately resulting in paroxysmal tachycardia. In summary, we hypothesize that GERD, through local acidic irritation, prompts an inflammatory reaction in adjacent tissues, influencing systemic inflammation levels and, consequently, the heart's conduction system.
In addition, gastric reflux through esophageal acid stimulation can diminish coronary blood perfusion, subsequently elevating the risk of myocardial ischemia (41). We hypothesize that autonomic excitation is associated with sympathetic inhibition (42). Post-myocardial ischemia nerve growth is a significant arrhythmias contributor. Nerve growth factor, by binding to the tyrosine protein kinase A receptor on the superior cervical ganglion surface, activates the extracellular regulatory protein kinase (ERK1/2), catalyzes serine phosphorylation at S727 in STAT3, and activates STAT3 along with downstream signaling molecules to facilitate nerve growth (43). Tougas et al. observed that autonomic balance was disrupted following esophageal acid injection (44), suggesting that esophageal acid signals could influence cardiac electrical rhythm, potentially leading to arrhythmias. Additionally, according to one study: the autonomic nervous system directly impacts arrhythmia, where the vagus nerve and parasympathetic preganglionic fibers converge on the fat pad between the superior vena cava and the aorta to form a ganglionic plexus, thereby innervating the sinoatrial and atrioventricular nodes (45).
The strengths of our two-sample bidirectional MR study include the following: Firstly, the MR analysis method allows for the emulation of randomized controlled trials within an observational setting, providing higher-level experimental evidence through a cost-effective approach. Moreover, the MR methodology effectively circumvents reverse causation and confounding variables. Secondly, all instrumental variables (IVs) employed in the MR analysis were meticulously selected to ensure result accuracy. Thirdly, due to data being sourced from distinct tissues, there was minimal overlap between exposures and outcomes in the samples. Finally, our analysis's findings could have significant implications for healthcare policy, as elucidating a causal link between GERD and paroxysmal tachycardia may influence public health strategies concerning prevention and treatment.
Our study possesses several limitations. Firstly, the GWAS datasets utilized were exclusively derived from European populations, restricting the generalizability and applicability of our findings to diverse populations. Hence, further validation with a broader and more inclusive database is necessary. Secondly, the inherent constraints of the GWAS datasets necessitate additional basic research to substantiate causality and explore potential pathological mechanisms, pivotal for clinical interventions. Thirdly, the outcome data from GERD did not delineate the frequency and severity of GERD episodes, warranting further investigation into the relationship between GERD severity and the risk of paroxysmal tachycardia and other arrhythmias.
In summary, our research underscores a potential causal link between GERD and arrhythmias. While the underlying mechanisms connecting GERD and arrhythmia remain elusive, GERD significantly contributes to arrhythmias risk assessment. Considering the causal association between GERD and arrhythmias, enhancing arrhythmias prevention strategies in individuals with GERD is advisable.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
JL: Data curation, Methodology, Validation, Writing – original draft, Writing – review & editing, Conceptualization, Formal Analysis, Funding acquisition, Investigation, Project administration, Resources, Software, Supervision, Visualization. LT: Writing – original draft, Writing – review & editing. JY: Writing – review & editing. YL: Writing – review & editing. XY: Writing – review & editing. CH: Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We thank all investigators for making available their GWAS data. We also want to acknowledge the participants and investigators of the FinnGen study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2024.1411784/full#supplementary-material
1. Huikuri HV, Castellanos A, Myerburg RJ. Sudden death due to cardiac arrhythmias. N Engl J Med. (2001) 345(20):1473–82. doi: 10.1056/NEJMra000650
2. Marchesi JR, Russell NJ, White GF, House WA. Effects of surfactant adsorption and biodegradability on the distribution of bacteria between sediments and water in a freshwater microcosm. Appl Environ Microbiol. (1991) 57(9):2507–13. doi: 10.1128/aem.57.9.2507-2513.1991
3. Tousoulis D. A new era in arrhythmia management: from medical treatment to invasive strategies. Hellenic J Cardiol. (2016) 57(5):301–3. doi: 10.1016/j.hjc.2017.01.017
4. Subbotnik SI, Feinberg YS, Spielberg PI. Electroencephalography in paroxysmal tachycardia. Electroencephalogr Clin Neurophysiol. (1955) 7(4):577–84. doi: 10.1016/0013-4694(55)90084-5
5. Josephson ME. Clinical Cardiac Electrophysiology: Techniques and Interpretations. 2nd. Philadelphia: Lea & Febiger (1993).
6. Maret-Ouda J, Markar SR, Lagergren J. Gastroesophageal reflux disease. JAMA. (2020) 324(24):2565. doi: 10.1001/jama.2020.21573
7. Davis PJ, Davis FB. Nongenomic actions of thyroid hormone on the heart. Thyroid. (2002) 12(6):459–66. doi: 10.1089/105072502760143827
8. Mariani MV, Pierucci N, Trivigno S, Cipollone P, Piro A, Chimenti C, et al. Probability score to predict spontaneous conversion to sinus rhythm in patients with symptomatic atrial fibrillation when less could be more? J Clin Med. (2024) 13(5):3–4. doi: 10.3390/jcm13051470
9. Mariani MV, Pierucci N, Piro A, Trivigno S, Chimenti C, Galardo G, et al. Incidence and determinants of spontaneous cardioversion of early onset symptomatic atrial fibrillation. Medicina (Kaunas). (2022) 58(11):10. doi: 10.3390/medicina58111513
10. Floria M, Bărboi O, Grecu M, Cijevschi PC, Balan G, Drug VL. Atrial fibrillation and sympathovagal balance in patients with gastroesophageal reflux disease. Turk J Gastroenterol. (2017) 28(2):88–93. doi: 10.5152/tjg.2017.16540
11. Floria M, Drug VL. Atrial fibrillation and gastroesophageal reflux disease: from the cardiologist perspective. World J Gastroenterol. (2015) 21(10):3154–6. doi: 10.3748/wjg.v21.i10.3154
12. Gesualdo M, Scicchitano P, Carbonara S, Ricci G, Principi M, Ierardi E, et al. The association between cardiac and gastrointestinal disorders: causal or casual link? J Cardiovasc Med (Hagerstown). (2016) 17(5):330–8. doi: 10.2459/JCM.0000000000000351
13. Jervell O, Lødøen O. The gastrocardiac syndrome. Acta Med Scand Suppl. (1952) 266:595–9. doi: 10.1111/j.0954-6820.1952.tb13409.x
14. Cuomo R, De Giorgi F, Adinolfi L, Sarnelli G, Loffredo F, Efficie E, et al. Oesophageal acid exposure and altered neurocardiac function in patients with GERD and idiopathic cardiac dysrhythmias. Aliment Pharmacol Ther. (2006) 24(2):361–70. doi: 10.1111/j.1365-2036.2006.02987.x
15. Sabbatini M, Molinari C, Grossini E, Mary DA, Vacca G, Cannas M. The pattern of c-fos immunoreactivity in the hindbrain of the rat following stomach distension. Exp Brain Res. (2004) 157(3):315–23. doi: 10.1007/s00221-004-1845-x
16. Tougas G, Kamath M, Watteel G, Fitzpatrick D, Fallen EL, Hunt RH, et al. Modulation of neurocardiac function by oesophageal stimulation in humans. Clin Sci (Lond). (1997) 92(2):167–74. doi: 10.1042/cs0920167
17. Chen X, Kong J, Pan J, Huang K, Zhou W, Diao X, et al. Kidney damage causally affects the brain cortical structure: a Mendelian randomization study. EBioMedicine. (2021) 72:103592. doi: 10.1016/j.ebiom.2021.103592
18. Larsson SC, Butterworth AS, Burgess S. Mendelian randomization for cardiovascular diseases: principles and applications. Eur Heart J. (2023) 44(47):4913–24. doi: 10.1093/eurheartj/ehad736
19. Ong JS, An J, Han X, Law MH, Nandakumar P, Schumacher J, et al. Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett’s oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut. (2022) 71(6):1053–61. doi: 10.1136/gutjnl-2020-323906
20. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. Finngen provides genetic insights from a well-phenotyped isolated population. Nature. (2023) 613(7944):508–18. doi: 10.1038/s41586-022-05473-8
21. Burgess S, Davey SG, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. (2019) 4:186. doi: 10.12688/wellcomeopenres.15555.1
22. Davey SG, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. (2014) 23(R1):R89–98. doi: 10.1093/hmg/ddu328
23. Rasooly D, Patel CJ. Conducting a reproducible Mendelian randomization analysis using the R analytic statistical environment. Curr Protoc Hum Genet. (2019) 101(1):e82. doi: 10.1002/cphg.82
24. Li S, Chen M, Zhang Q, Fang M, Xiong W, Bai L. Ankylosing spondylitis and glaucoma in European population: a Mendelian randomization study. Front Immunol. (2023) 14:1120742. doi: 10.3389/fimmu.2023.1120742
25. Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. (2001) 69(1):1–14. doi: 10.1086/321275
26. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. (2011) 40(3):740–52. doi: 10.1093/ije/dyq151
27. Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. Phenoscanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. (2019) 35(22):4851–3. doi: 10.1093/bioinformatics/btz469
28. Burgess S, Scott RA, Timpson NJ, Davey SG, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. (2015) 30(7):543–52. doi: 10.1007/s10654-015-0011-z
29. Bowden J, Davey SG, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. (2015) 44(2):512–25. doi: 10.1093/ije/dyv080
30. Bowden J, Davey SG, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40(4):304–14. doi: 10.1002/gepi.21965
31. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. (2013) 37(7):658–65. doi: 10.1002/gepi.21758
32. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50(5):693–8. doi: 10.1038/s41588-018-0099-7
33. Huang CC, Chan WL, Luo JC, Chen YC, Chen TJ, Chung CM, et al. Gastroesophageal reflux disease and atrial fibrillation: a nationwide population-based study. PLoS One. (2012) 7(10):e47575. doi: 10.1371/journal.pone.0047575
34. Kunz JS, Hemann B, Atwood JE, Jackson J, Wu T, Hamm C, et al. Is there a link between gastroesophageal reflux disease and atrial fibrillation? Clin Cardiol. (2009) 32(10):584–7. doi: 10.1002/clc.20660
35. Gerson LB, Friday K, Triadafilopoulos G. Potential relationship between gastroesophageal reflux disease and atrial arrhythmias. J Clin Gastroenterol. (2006) 40(9):828–32. doi: 10.1097/01.mcg.0000225571.42890.a5
36. Armaganijan L, Patel D, Lopes RD, Morillo CA, Araújo RR, Munhoz FP, et al. Gastroesophageal reflux and atrial fibrillation: is there any correlation? Expert Rev Cardiovasc Ther. (2012) 10(3):317–22. doi: 10.1586/erc.11.198
37. Xu L, Zhang Y, Xie J, Liu Y, Xu L. Association between gastroesophageal reflux disease and atrial fibrillation: a systematic review and meta-analysis. Rev Esp Enferm Dig. (2019) 111(11):874–9. doi: 10.17235/reed.2019.5389/2017
38. Fuertes Á, Alshweki A, Pérez-Muñuzuri A, Couce ML. Supraventricular tachycardia in newborns and its association with gastroesophageal reflux disease. An Pediatr (Barc). (2017) 87(4):206–10. doi: 10.1016/j.anpedi.2016.10.006
39. Lewek J, Kaczmarek K, Cygankiewicz I, Wranicz JK, Ptaszynski P. Inflammation and arrhythmias: potential mechanisms and clinical implications. Expert Rev Cardiovasc Ther. (2014) 12(9):1077–85. doi: 10.1586/14779072.2014.942286
40. Yoshida N. Inflammation and oxidative stress in gastroesophageal reflux disease. J Clin Biochem Nutr. (2007) 40(1):13–23. doi: 10.3164/jcbn.40.13
41. Dobrzycki S, Skrodzka D, Musiał WJ, Go M, Korecki J, Gugała K, et al. Relationship between gastroesophageal reflux disease and myocardial ischemia. Effect of reflux on temporary activity of autonomic nervous system. Rocz Akad Med Bialymst. (2004) 49:93–7. PMID: 15631321.
42. Bortolotti M, Pandolfo N, Miglioli M. Abnormal esophagocardiac reflex in patients with non-cardiac chest pain. Dis Esophagus. (2001) 14(1):57–9. doi: 10.1111/j.1442-2050.2001.00150.x
43. Pellegrino MJ, Habecker BA. STAT3 integrates cytokine and neurotrophin signals to promote sympathetic axon regeneration. Mol Cell Neurosci. (2013) 56:272–82. doi: 10.1016/j.mcn.2013.06.005
44. Tougas G, Spaziani R, Hollerbach S, Djuric V, Pang C, Upton AR, et al. Cardiac autonomic function and oesophageal acid sensitivity in patients with non-cardiac chest pain. Gut. (2001) 49(5):706–12. doi: 10.1136/gut.49.5.706
Keywords: gastroesophageal reflux disease, Mendelian randomization, paroxysmal tachycardia, arrhythmia, GWAS
Citation: Liang J, Tang L, Yang J, Li Y, Yang X and Hou C (2024) Gastroesophageal reflux disease and risk for arrhythmias: a Mendelian randomization analysis. Front. Cardiovasc. Med. 11: 1411784. doi: 10.3389/fcvm.2024.1411784
Received: 3 April 2024; Accepted: 2 July 2024;
Published: 29 July 2024.
Edited by:
Rui Providencia, University College London, United KingdomReviewed by:
Vincenzo Mirco La Fazia, Texas Cardiac Arrhythmia Institute, United States© 2024 Liang, Tang, Yang, Li, Yang and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: ChiJun Hou, NDY5MTAyNDQzQHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.