
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Cardiovasc. Med. , 01 August 2024
Sec. Cardiovascular Genetics and Systems Medicine
Volume 11 - 2024 | https://doi.org/10.3389/fcvm.2024.1339701
This article is part of the Research Topic Precision medicine in cardiovascular disease View all 4 articles
Background: At present, no consensus is reached among articles that investigate the relationship of paraoxonase 1(PON1) -108C/T polymorphism with susceptibility of coronary heart disease (CHD) so far. In this regard, the present meta-analysis was conducted to comprehensively review existing articles related to the relationship of PON1 -108C/T polymorphism with CHD susceptibility. It was preregistered in the International Platform of Registered Systematic Review and Meta-Analysis Protocols (INPLASY)-INPLASY202430117.
Methods: Articles that explored the relationship between PON1 -108C/T polymorphism and CHD incidence were searched from electronic databases according to our preset study selection criteria. Thereafter, we adopted stata 12.0 software to analyze our screened studies. At the same time, odds ratios (ORs) and related 95% confidence intervals (95% CIs) were determined for evaluating association strength.
Results: At last, this meta-analysis selected altogether 13 case-control studies that involved 2,979 cases and 2,887 control subjects. We found that the PON1 -108C/T polymorphism displayed marked relationship with CHD susceptibility (T vs. C: OR = 1.24, 95% CI 1.07–1.45; CT vs. CC: OR = 1.33, 95% CI 1.17–1.52; TT vs. CC: OR = 1.51, 95% CI 1.09–2.09; Recessive model: OR = 1.16, 95% CI 0.93–1.45; Dominant model: OR = 1.45, 95% CI 1.16–1.81). Moreover, subgroup analysis showed that race and sample size had no impact on the results. Bioinformatics analysis showed that -108C>T polymorphism was relation to PON1 gene expression (https://gtexportal.org/home/).
Conclusions: The PON1 -108T allele is identified as the possible low-penetrant risk factor of CHD, as suggested by our present meta-analysis.
Systematic Review Registration: https://inplasy.com/inplasy-2024-3-0117/, Identifier INPLASY202430117.
Coronary heart disease (CHD) represents a leading cause resulting in disability and mortality worldwide, which also becomes a main public health issue (1). Although our knowledge of CHD has advanced considerably, the detailed pathogenesis of CHD remains unclear. A series of modifiable and non-modifiable risk factors can influence the formation of atherosclerotic plaque, including age, gender, smoking habit, intake of alcohol, physical inactivity, overweight/obesity, abnormal lipid profile, high blood pressure, as well as family history of CHD (2, 3). Paraoxonase (PON) family includes PON1, PON2, and PON3. PONl and PON3 belong to secretory enzymes, which are mainly expressed in liver and kidney tissues, and are closely bound to high-density lipoprotein in serum. PON2 belongs to intracellular enzymes and can be widely expressed in various tissue cells. In PON family, domestic and foreign scholars have conducted the most comprehensive and in-depth research on PON1. PON1 is the HDL-related Ca2+ dependent glycoprotein (lactonase), which presents antioxidant and anti-atherogenic features through paraoxon hydrolysis (4).
PON1 is an HDL-associated esterase that hydrolyses lipoperoxides. It acts as a protective factor against oxidative modifications of LDL, indicating that it possibly exerts an important part in preventing atherosclerotic process. This has led to the hypothesis that the lower the PON1 activity is, the higher will be the accumulation of oxidized LDL and risk of CHD. Recent meta-analysis showed that PON-1 activity is significantly lower in CAD patients. In human being, PON1 locates within q21.3–q22.1 in chromosome 7 long arm. The PON1 gene has nearly 200 single nucleotide polymorphisms (SNPs). Notably, the PON1 -108 polymorphism promoter region [c.-108C>T (C: cytosine T: thymine); dbSNP: rs705379; OMIM database: +1688200003] greatly affects PON1 function. Besides, in certain cellular systems, the -108C/T polymorphism of this gene takes up 23% expression of PON1, while -108TT constructs decreased PON1 level relative to -108CC constructs.
Many articles are conducted to analyze the association of PON1 -108C>T polymorphisms with CHD susceptibility, however, no consensus results are obtained so far. According to a previous systematic review, the PON1 -108C>T may be potential CHD risk factors. Due to three missing eligible studies, this meta-analysis did not include all potentially relevant studies. In addition, previous meta-analysis did not use subgroup analysis and bioinformatics analysis. To evaluate the impact of PON1 -108C>T polymorphism on CHD clinical outcomes, we conducted the current updated meta-analysis.
This meta-analysis was implemented according to PRISMA checklist (Attachment 1). With the guidance of Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols guidelines we has completed our meta-analysis protocol, and has been registered in the International Platform of Registered Systematic Review and Meta-analysis Protocols (INPLASY) with the number is INPLASY202430117. Web of Science, PubMed, CNKI and Embase were systemically retrieved to enroll eligible articles published before June 2020. The keywords shown below were used, including “paraoxonase 1”, “paraoxonase-1”, “PON 1”, “PON-1”, “variant”, “variation”, “mutation”, “SNP”, “polymorphism”, “atherosclerosis”, “arteriosclerosis”, “coronary heart disease”, “coronary artery disease”, “acute coronary syndrome”, “myocardialinfarction”. Besides, reference lists were manually searched to prevent omitting any eligible study.
We selected studies based on the following criteria: (a) case-control articles evaluating the relationship of PON1 -108C>T with CHD susceptibility; (b) studies conducted by enrolling irrelevant subjects; (c) studies with enough data on ORs and 95% CIs. At the same time, reports, reviews, letters and comments were eliminated from this study. For duplicates, the one that had the greatest sample size was adopted for the present meta-analysis.
Two independent investigators collected data and were also responsible for result checking. Data extracted were as follows, first author, publication year, area, case/control numbers, frequencies of genotypes among cases/controls, along with Hardy-Weinberg equilibrium (HWE) evidence among controls. Any disagreement between them was settled down by mutual negotiation.
HWE among controls of every article was assessed through chi-square test, with P < 0.05 indicating significance of disequilibrium. Besides, ORs and 95%CIs were adopted for estimating the association strength of PON1 -108C>T polymorphism with CHD risk through allele model (T vs. C), heterozygote comparison (CT vs. CC), homozygote comparison (TT vs. CC), recessive model (TT vs. CT + CC) and dominant model (TT + CT vs. CC). In addition, chi-square test was conducted to quantify the heterogeneity influence, with I2 > 50% suggesting that there was heterogeneity among different studies, as a result, a random effects model should be selected; else, the fixed effects model should be used. We also conducted sensitivity for evaluating result stability through eliminating a study each time for determining its impact on combined ORs. Furthermore, Begg's funnel plot was employed to investigate the bias of publication, with P < 0.05 indicating the presence of significant bias of publication. STATA12.0 (Stata Corporation, College Station, Texas) was utilized for meta-analysis.
Using the GTEx expression quantitative trait locus (eQTL) data, -108C>T polymorphism was examined in relation to PON1 gene expression (https://gtexportal.org/home/).
Figure 1 presents the study screening procedure. Altogether 526 relevant papers were identified from Web of Science, PubMed, CNKI and Embase. Finally, we enrolled 13 case-control studies into the present meta-analysis (5–14). These studies were published between 2000 and 2020. The detailed information of the included papers was demonstrated in Table 1. Of these, four studies focused on Caucasians, nine studies focused on Asians. The genetic distributions of controls conformed to HWE among the enrolled studies, with the only exception of Ahmad et al. The general features of the eight researches were summarized in Table 2.
Table 1 and Figure 2 display the association of PON1 -108C>T polymorphism with CHD incidence. It was observed that, PON1 -108T polymorphism showed significant correlation with CHD incidence (T vs. C: OR = 1.24, 95% CI 1.07–1.45; CT vs. CC: OR = 1.33, 95% CI 1.17–1.52; TT vs. CC: OR = 1.51, 95% CI 1.09–2.09; Recessive model: OR = 1.16, 95% CI 0.93–1.45; Dominant model: OR = 1.45, 95% CI 1.16–1.81). In addition, significant association was found from subgroup analyses based on HWE, sample size and race. Sensitivity analysis was also conducted, which suggested that none of the enrolled studies had significant influence on the combined ORs, indicating our result stability. The cumulative meta-analyses showed a increasing trend in the estimated risk effect, which showed PON1 -108 polymorphism is associated with HCC risk, and the results were stable (Figure 3).
Sensitivity analysis was performed by omitting each study in turn to evaluate the influence of single studies on the overall estimation (Figure 4). The corresponding pooled OR and significant results did not change materially, indicating that our results were statistically robust.
We visualized the funnel plot to analyze the potential bias of publication, and no obvious bias of publication was detected (Figure 5), which indicated that the present meta-analysis had low bias of publication.
The GTEx portal shows a significant correlation between PON1 -108C>T polymorphism and heart PON1 gene expression. In detail, the T allele of PON1 -108C>T polymorphism significantly correlated with decreased PON1 expression (Figure 6).
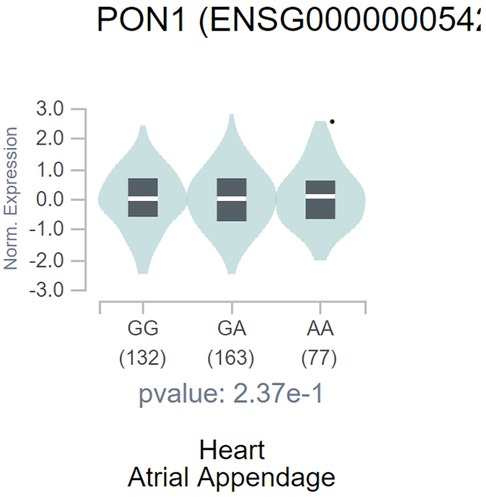
Figure 6. Violin plot shows the correlation between PON1 -108C/T polymorphism with PON1 expression. The figure was from the GTEx.
CHD represents a disorder that involves multiple factors, and its etiology remains largely unclear at present. It has been increasingly suggested that CHD results from the complicated interactions among genetic and environmental factors. PON1, an enzyme related to HDL-C, can resist the peroxidation of LDL-C. The decreased PON1 activity has been suggested to induce an increased incidence of coronary atherosclerosis (12). Besides, -108TT has been previously reported to down-regulate the PON1 level relative to the -108CC constructs (7). In this regard, we predicted that PON1 -108T might participate in CHD incidence. PON1 -108C>T polymorphism is previously suggested to be related to CHD incidence, yet no consistent findings are obtained. For clarifying those heterogeneous results, the present meta-analysis was conducted aiming to examine the association between PON1 -108C>T polymorphism and CHD susceptibility.
This meta-analysis enrolled 13 case-control studies that involved 2,979 patients along with 2,887 controls, which aimed to examine the association between PON1 -108C>T polymorphism and CHD susceptibility. As far as we know, the present meta-analysis was the first to comprehensively assess the correlation between PON1 -108C>T polymorphism and CHD susceptibility. Overall, PON1 -108C>T polymorphism was significantly associated with CHD incidence among all the studied populations. Due to different living environments and genetic backgrounds, subgroup analysis based on race was also conducted, which indicated that PON1 -108C>T polymorphism showed obvious relation with CHD susceptibility among the Asian and Caucasian populations. After that, subgroup analysis based on sample size showed that the sample size had no impact on the results. Furthermore, we also conducted sensitivity analysis, which revealed that this polymorphism was markedly correlated with CHD susceptibility. There was no obvious evidence regarding the bias of publication detected. Due to the limited sample size, our results must be further investigated.
This study did not illustrate the related mechanism of such relation. Previous study and our bioinformatics analysis found that -108T allele decreased PON1 level (8). PON1 is believed to promote the anti-inflammatory and anti-oxidant functions of lipoproteins (11). Ultimately, decreased PON1 led to the occurrence and progression of CHD. Moreover, Bouman et al. demonstrated that PON1 is the rate-limiting enzyme in the second step of clopidogrel bioactivation, in which 2-oxo-clopidogrel is hydrolyzed into the active thiol molecule. An analysis of 112 individuals revealed that the PON1 -108C>T polymorphism is the major factor affecting clopidogrel bioactivation (14).
Certain limitations must be noted in this meta-analysis. Firstly, this meta-analysis was carried out according to the non-adjusted ORs, since ORs were not available in each study and the available ORs were unadjusted relative to identical possible confounding factors, like exposure, age and sex. The insufficient information for data analysis possibly induced severe confounding bias. Secondly, the insufficient raw materials prevented us from better evaluating the possible associations between genes and the environment or between genes. Thirdly, only few studies and participants were involved in certain subgroups-based meta-analysis.
To sum up, this meta-analysis indicates the relationship of PON1 -108C>T polymorphism with CHD susceptibility. More articles that estimate the impacts of interactions between genes and between genes and the environment are needed to further and comprehensively understand the correlation between PON1 -108C>T polymorphism and CHD susceptibility.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
JL: Writing – original draft. PW: Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Timmis A, Townsend N, Gale C, Grobbee R, Maniadakis N, Flather M, et al. European society of cardiology: cardiovascular disease statistics 2017. Eur Heart J. (2018) 39:508–79. doi: 10.1093/eurheartj/ehx628
2. Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S. Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ Res. (2016) 118:535–46. doi: 10.1161/CIRCRESAHA.115.307611
3. Sekuri C, Cam FS, Ercan E, Tengiz I, Sagcan A, Eser E, et al. Renin-angiotensin system gene polymorphisms and premature coronary heart disease. J Renin Angiotensin Aldosterone Syst. (2005) 6:38–42. doi: 10.3317/jraas.2005.005
4. Huo X, Guo Y, Zhang Y, Li J, Wen X, Liu J. Paraoxonase 1 gene (Q192R) polymorphism confers susceptibility to coronary artery disease in type 2 diabetes patients: evidence from case-control studies. Egypt Heart J. (2012) 2:29–37.
5. Huo X, Guo Y, Zhang Y, Li J, Wen X, Liu J. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
6. Ahmad I, Narang R, Venkatraman A, Das N. Frequency distribution of the single-nucleotide -108C/T polymorphism at the promoter region of the PON1 gene in Asian Indians and its relationship with coronary artery disease. J Community Genet. (2011) 2:27–32. doi: 10.1007/s12687-011-0037-1
7. Blatter Garin MC, Moren X, James RW. Paraoxonase-1 and serum concentrations of HDL-cholesterol and apoA-I. J Lipid Res. (2006) 47:515–20. doi: 10.1194/jlr.M500281-JLR200
8. Gupta N, Singh S, Maturu VN, Sharma YP, Gill KD. Paraoxonase 1 (PON1) polymorphisms, haplotypes and activity in predicting cad risk in North-West Indian Punjabis. PLoS One. (2011) 6:e17805. doi: 10.1371/journal.pone.0017805
9. Gupta N, Binu KBK, Singh S, Maturu NV, Sharma YP, Bhansali A, et al. Low serum PON1 activity: an independent risk factor for coronary artery disease in north-west Indian type 2 diabetics. Gene. (2012) 498:13–9. doi: 10.1016/j.gene.2012.01.091
10. Huo L, Zhou X, Zheng F. Relationship between a polymorphism in the promoter region of Chinese Hans paraoxonase gene and coronary heart diseases. Basic Med Sci Clin. (2002) 22:534–7.
11. James RW, Leviev I, Ruiz J, Passa P, Froguel P, Garin MC. Promoter polymorphism T(-107)C of the paraoxonase PON1 gene is a risk factor for coronary heart disease in type 2 diabetic patients. Diabetes. (2000) 49:1390–3. doi: 10.2337/diabetes.49.8.1390
12. Saeed M, Perwaiz Iqbal M, Yousuf FA, Perveen S, Shafiq M, Sajid J, et al. Interactions and associations of paraoxonase gene cluster polymorphisms with myocardial infarction in a Pakistani population. Clin Genet. (2007) 71:238–44. doi: 10.1111/j.1399-0004.2007.00753.x
13. Wang X, Fan Z, Huang J, Su S, Yu Q, Zhao J, et al. Extensive association analysis between polymorphisms of PON gene cluster with coronary heart disease in Chinese Han population. Arterioscler Thromb Vasc Biol. (2003) 23:328–34. doi: 10.1161/01.ATV.0000051702.38086.C1
Keywords: coronary heart disease, -108C/T, risk, meta-analysis, PON1
Citation: Liao J and Wang P (2024) Association between paraoxonase 1 -108C/T polymorphism and coronary heart disease: an updated meta-analysis. Front. Cardiovasc. Med. 11: 1339701. doi: 10.3389/fcvm.2024.1339701
Received: 16 November 2023; Accepted: 17 June 2024;
Published: 1 August 2024.
Edited by:
Anupam Basu, The University of Burdwan, IndiaReviewed by:
Jerzy Beltowski, Medical University of Lublin, Poland© 2024 Liao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengcheng Wang, d2FuZ3BlbmdjaGVuZ21lZEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.