- 1Department of Cardiology and Institutes for Systems Genetics, Frontiers Science Center for Disease-Related Molecular Network, West China Hospital, Sichuan University, Chengdu, China
- 2Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, China
- 3Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
Myocardial infarction (MI) is a prevalent cardiovascular disease characterized by myocardial necrosis resulting from coronary artery ischemia and hypoxia, which can lead to severe complications such as arrhythmia, cardiac rupture, heart failure, and sudden death. Despite being a research hotspot, the etiological mechanism of MI remains unclear. The emergence and widespread use of omics technologies, including genomics, transcriptomics, proteomics, metabolomics, and other omics, have provided new opportunities for exploring the molecular mechanism of MI and identifying a large number of disease biomarkers. However, a single-omics approach has limitations in understanding the complex biological pathways of diseases. The multi-omics approach can reveal the interaction network among molecules at various levels and overcome the limitations of the single-omics approaches. This review focuses on the omics studies of MI, including genomics, epigenomics, transcriptomics, proteomics, metabolomics, and other omics. The exploration extended into the domain of multi-omics integrative analysis, accompanied by a compilation of diverse online resources, databases, and tools conducive to these investigations. Additionally, we discussed the role and prospects of multi-omics approaches in personalized medicine, highlighting the potential for improving diagnosis, treatment, and prognosis of MI.
1. Introduction
Cardiovascular disease (CVD) is a leading cause of mortality globally, responsible for nearly half of all deaths. Among various types of CVD, myocardial infarction (MI), is the main cause of cardiovascular death and one of the most common types of coronary artery disease (CAD) (1). In recent years, MI have been occurring in increasingly younger individuals due to changes in lifestyle, unclear circadian rhythms, and increased social pressure (2, 3). MI is characterized by irreversible myocardial necrosis caused by coronary artery ischemia and hypoxia. After MI occurs, a large number of fibroblasts replace necrotic cardiomyocytes, leading to ventricular remodeling such as myocardial fibrosis and cardiac hypertrophy (4). These changes can ultimately lead to adverse events such as cardiac rupture, heart failure (HF), and sudden death due to insufficient cardiac motility (5–7). Although methods such as bypass grafting, percutaneous coronary intervention (PCI), and antithrombotic drugs are available for MI treatment, they can only reduce the severity of CAD to a certain extent. Due to the complexity and diversity of the disease, these treatments cannot reverse myocardial necrosis and ventricular remodeling caused by ischemia and hypoxia (4). Therefore, it remains the focus of research to elucidate the diverse molecular mechanism of MI and to find efficient markers, which is of great significance to improve the diagnosis, treatment effect and prognosis of MI.
Since the turn of the century, the completion and deepening of the Human Genome Project (HGP) (8) and the Encyclopedia of DNA Elements (ENCODE) (9) have established a robust foundation for personalized medicine research and the investigation of the pathogenesis of complex diseases. In recent years, the wide application of high-throughput technology and high-resolution mass spectrometry (HRMS) has not only increased the amount of biological data on the molecular mechanisms of diseases but also expanded the dimension of such data. This has encouraged researchers to explore the molecular mechanisms of diseases using multi-omics technologies and methods. Diseases, including MI, are now understood more comprehensively at various molecular levels, including genomics, epigenetics, transcriptomics, proteomics, metabolomics, etc.
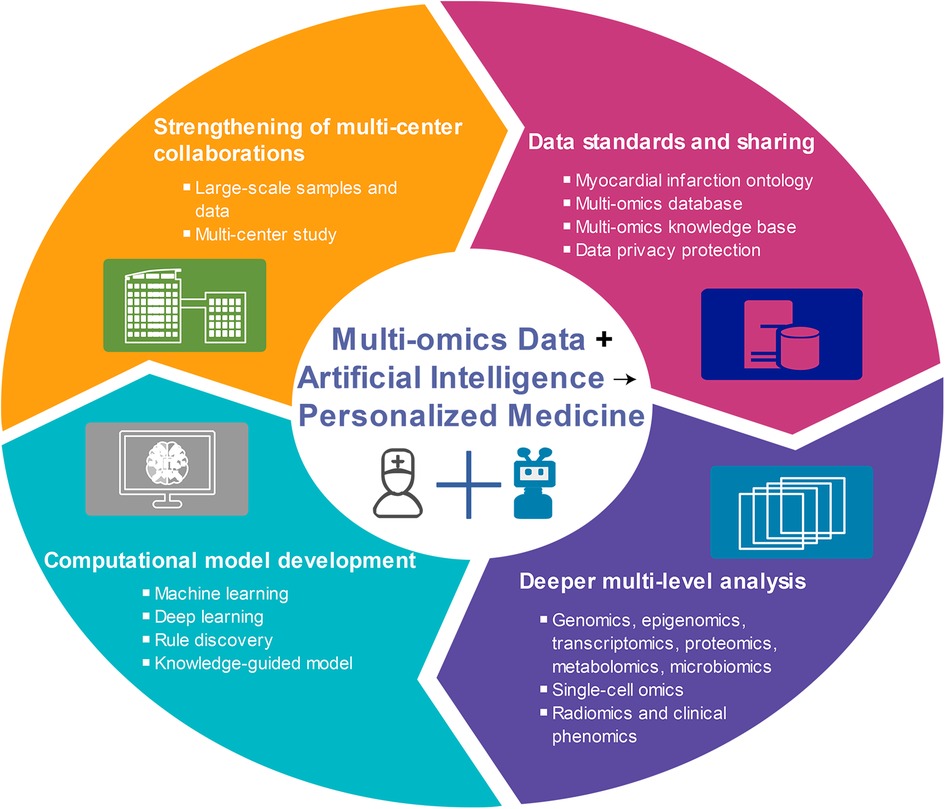
MI is a complex disease caused by environmental and genetic factors, and its various subtypes exhibit different pathogenesis and prognoses (10). While some progress has been made in single-omics studies of MI over the past few decades, the pathogenesis of MI remains unclear. The development of multi-omics methods is expected to shed light on the molecular pathogenesis and differences among various MI subtypes, identify biomarkers with diagnostic, therapeutic, and prognostic values, and ultimately enable the prediction, prevention, and personalized treatment of MI. Therefore, this study summarized the research progress made in MI research across various omics fields, including genomics, epigenomics, transcriptomics, proteomics, metabolomics, and others. In particular, we emphasized the significance and potential of multi-omics approaches in realizing personalized medicine for MI (Figure 1).
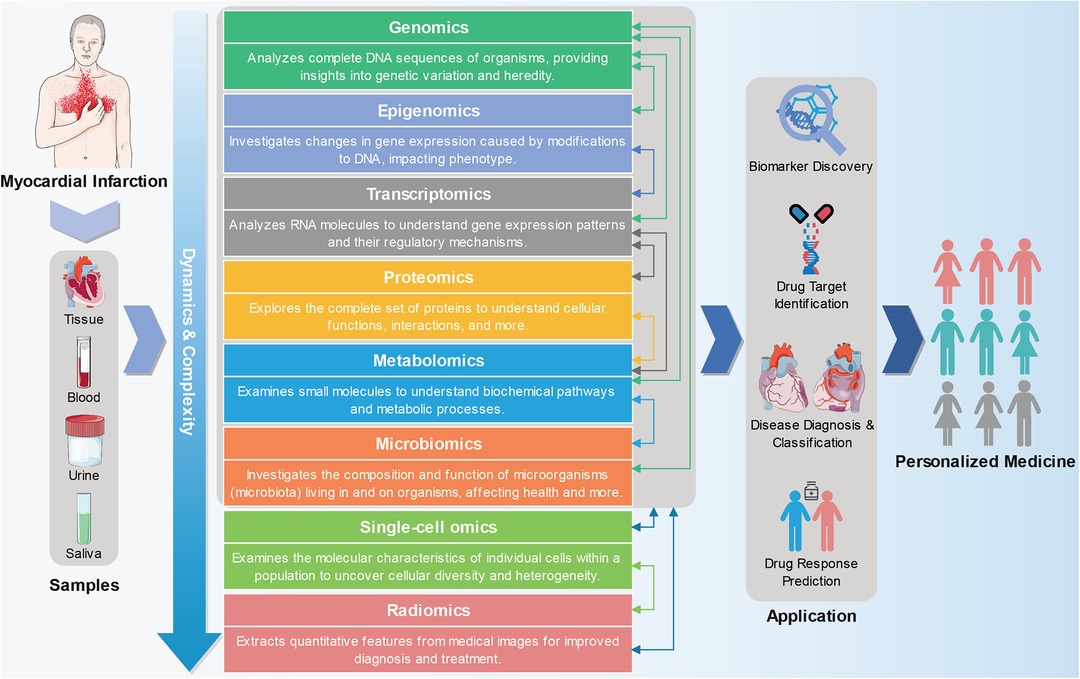
Figure 1. From multi-omics approaches to personalized medicine in myocardial infarction. Genomics, epigenomics, transcriptomics, proteomics, metabolomics, and other omics technologies can be used to detect and analyze tissue or body fluid (such as blood, urine, and saliva) samples from MI patients. Omics and multi-omics approaches offer a wide range of applications in advancing precision medicine, encompassing biomarker discovery, drug target identification, patient stratification, and predicting drug responses. The part of the figure is drawn with the pictures of Servier Medical Art (https://smart.servier.com).
2. Single-omics approaches
2.1. Genomics
Genomics is a field of study that encompasses the systematic analysis of all genes in an organism. The development of genome sequencing technologies, including Sanger sequencing (11), DNA microarray (12), and next-generation sequencing (NGS) (13). Of these technologies, DNA microarrays and NGS are widely used for studying gene mutations, which can help identify candidate genes associated with diseases and analyze genotype sensitivity to drugs. Gene mutations can be broadly categorized into three types: single nucleotide variations (SNVs), insertions/deletions (In/Dels), and copy number variations (CNVs). These types of mutations can contribute to the development of various diseases, including MI. Therefore, genomics plays an important role in elucidating the genetic basis of diseases and developing personalized medicine.
Researchers have identified numerous gene associated with an increased risk of MI, offering crucial insights into a deeper understanding of the genetic basis of this heart disease. Among these, genes in the 9p21.3 region have been identified as important genetic risk factors for MI. Variants in this genetic region, including CDKN2B/CDKN2B-AS1 rs1333049 (14, 15), MTAP rs7027989 (16), and ANRIL rs9632884 (17), were significantly associated with an elevated risk of MI. The C risk allele and CC genotype of rs1333049 were both linked to a higher risk of not only MI but also other CVDs (14, 15, 18, 19). The renin-angiotensin system is crucial regulating blood pressure and the development of CAD. Studies have found that polymorphisms in renin-angiotensin system-related genes, including AGT rs4762 (20), AGTR1 rs5186 (21), AGTR2 rs11091046 (22), KLK1 rs5517 (23), ACE rs1799752 (24), as well as ACE2 rs4646142 and rs1978124 (25), were associated with the risk of MI. The ACE gene insert/delete (I/D, rs1799752) polymorphism is the most extensively studied. This polymorphism refers to the existence or deletion of an Alu repeat sequence of 287 base pairs in intron 16 of the ACE gene, which can alter the activity of the angiotensin-converting enzyme (ACE) protein and lead to enhanced plaque vulnerability, ulceration, and thrombosis, ultimately leading to MI (26). Numerous studies have shown that the DD genotype of the ACE gene is not only a risk factor for MI (23, 27–29) but also related to the poor prognosis (30–32). The coagulation and fibrinolytic systems play pivotal roles in thrombus formation and dissolution. Abnormal mutations in specific coagulation factors, such as fibrinogen (FGA, FGB, FGG) (33), F2 (34), F5 (34), F7 (35), VWF (36), as well as genes in the fibrinolytic system like PLAT (37), and SERPINE1 (38), can lead to abnormal thrombosis or the formation of thrombi that are challenging to dissolve, subsequently increasing the risk of MI. Furthermore, genetic variants related to lipid metabolism and the inflammatory response are closely associated with the risk of MI, such as APOE rs7412 (39), CETP rs429358 (39), LPL rs328 (40), IL-6 rs1800795 (41), and TNF rs1800629 (42). These genetic variations can influence multiple biological processes, including cholesterol metabolism, vascular inflammation, plaque formation, and more, thereby increasing the risk of MI (43–45). The identification of these genetic risk factors not only helps uncover the genetic basis of MI but also offers new opportunities for disease prediction and prevention. Based on known genetic risk factors, researchers have developed genetic risk scoring systems that can estimate an individual's risk of MI (46–48). This personalized risk assessment aids medical professionals in better identifying high-risk patients and initiating appropriate interventions, including lifestyle modifications and medications, to reduce the risk of MI.
Genomics technology can be employed to identify genetic markers that can predict an individual's response to drugs. Clopidogrel is an antiplatelet drug commonly used for anticoagulant therapy in patients with MI. However, some individuals respond poorly to clopidogrel, which may be related to CYP2C19 loss-of-function mutations that slow drug metabolism and thus reduce the drug's efficacy (49, 50). Warfarin is a commonly used anticoagulant drug for treating patients with heart disease. Individual warfarin dosage requirements vary based on polymorphisms *1, *2 and *3 for CYP2C9, −1639G > A for VKORC1 allowing for a more accurate determination of the drug dosage to prevent issues such as bleeding or insufficient clotting (51, 52). Angiotensin modulators refer to a class of drugs that interact with the renin-angiotensin-aldosterone system in the body to regulate blood pressure and fluid balance, including ACE inhibitors and angiotensin II receptor blockers (ARBs). The ACE I/D polymorphism, AGT rs7079, and AGTR1 haplotypes have been proven to be associated with variable responses to angiotensin modulators, impacting both neurological outcomes and blood pressure variations (53, 54). Orbofiban is a medication that belongs to a class of drugs known as platelet aggregation inhibitors or antiplatelet agents. MI patients with the T allele of the GNB3 gene were more likely to experience bleeding when given orbofiban, indicating that genetics can influence the risk of bleeding with antiplatelet drugs (55). These genetic markers can help doctors better select appropriate drugs and dosages to increase the effectiveness of treatment and reduce the risk of adverse reactions.
2.2. Epigenomics
Epigenomics is a specialized branch of genomics that focuses on the comprehensive analysis of epigenetic changes at the genomic level, including DNA methylation, histone modification, and chromatin structure, across the entire genome of an organism or a specific cell type (56). These epigenetic changes play a critical role in the development and progression of human diseases, including MI. By identifying the specific epigenetic changes associated with MI, researchers can identify potential therapeutic targets and develop personalized treatment strategies for patients.
DNA methylation is an epigenetic marker that can be inherited, and it involves the transfer of a methyl group to the cytosine 5 carbon site through DNA methyltransferase. This process typically occurs in the promoter region and serves to inhibit gene transcription. Abnormal hypermethylation, which can lead to transcriptional silencing, is often associated with diseases and can be used as a biomarker (57). Recent studies have found that DNA methylation plays an important role in the development of MI. DNA methylation patterns differ between patients with MI and healthy individuals (58). These aberrant methylation patterns can result in altered gene expression, influencing the onset and progression of MI. GPR15 gene may contribute to MI by influencing inflammation and angiogenesis-related pathways (59, 60), with DNA hypomethylation in this gene associated with an increased risk of early-onset MI due to the upregulation of GPR15 expression levels (61). ABO blood types are linked to the risk of MI, with non-blood type O individuals having a higher risk (62, 63). The DNA methylation of the ABO gene promoter plays a pivotal role in regulating ABO gene expression (64), where increased methylation status of the ABO gene promoter was associated with an elevated risk of AMI by suppressing ABO mRNA expression (65, 66). An epigenome-wide association study identified 34 differentially methylated CpG sites associated with MI. These CpGs may contribute to MI through influencing the expression of genes related to inflammatory and lipid metabolism, including MPO, SERPINA1, NISCH, DLEU1, ZFPM1, and others (67). Furthermore, biomarkers derived from DNA methylation data, such as GrimAgeAccel, PhenoAgeAccel, EEAA, and DNAmRS, can predict the risk of MI. Among these, GrimAgeAccel has proven to be the most effective tool for evaluating MI risk (68).
DNA methylation is an emerging and exciting research field in the treatment of MI, which is still in the early stages of exploration and development. DNA methylation can affect the expression and activity of mitochondria, antioxidant and apoptosis genes, thereby leading to ischemia-reperfusion (I/R) injury (69). I/R injury plays a key role in the process of MI (70), so a deep understanding of the role of DNA methylation in this process is crucial for developing targeted therapeutic strategies. Some studies have begun to explore drug intervention in DNA methylation to improve the recovery of patients with MI. These drugs can affect the methylation status of specific genes and are expected to promote cardiac repair and regeneration after MI. DNA methyltransferase-1 (DNMT1) has been identified as a key player in cardiac fibrosis by regulating miR-133b methylation, affecting myofibroblast activation and CTGF expression. 5-Azacytidine can counteract DNMT1-induced miR-133b methylation, delaying myocardial fibrosis (71). Additionally, APAF1 gene is a key player in the regulation of apoptosis, a fundamental process in cell biology (72). DNMT1 has also been identified as playing a role in the methylation of the APAF1 promoter, silencing the APAF1 gene. This mechanism reinforces the protective effect of sevoflurane against cardiomyocyte injury induced by hypoxia/reoxygenation (73). Long non-coding RNA (lncRNA) ZFAS1 has been demonstrated to be upregulated during cardiac I/R injury, affecting cardiac function by influencing the methylation level of the Notch1 gene. Nicotinamide mononucleotide could enhance Notch1 expression, leading to improved cardiomyocyte survival and cardiac function (74). Furthermore, lifestyle factors, such as diet, exercise, and smoking cessation, have been demonstrated to influence DNA methylation (75–77). Maintaining a healthy lifestyle during MI recovery can potentially enhance cardiovascular health by impacting DNA methylation. Healthcare professionals play a crucial role in mitigating the risk of MI by educating patients about making healthy lifestyle choices.
Histones are essential for maintaining the shape and structure of chromatin, and the tail region of histones undergoes various post-translational modifications such as methylation, acetylation, and phosphorylation. These modifications can lead to changes in chromatin conformation, ultimately affecting the transcription of important genes (78, 79). Histone methylation, particularly trimethylation of histone H3 on lysine-4 (H3K4me3), plays a crucial role in MI (80). JMJD3 demethylase was found to exacerbate cardiac fibrosis by reducing H3K27me3 at the beta-catenin promoter in activated cardiac fibroblasts, worsening cardiac fibrosis. Inhibiting JMJD3 may be a potential therapy for cardiac fibrosis (81). Another study showed that Salvia miltiorrhiza and Carthamus tinctorius extract (SCE) effectively reduced myocardial fibrosis and inflammation by inhibiting H3K4me3 and H3K36 trimethylation (H3K36me3) at the Smad3 promoter in cardiac fibroblasts, leading to decreased Smad3 transcription (82). Additionally, riboflavin has been shown to protect myocardium from damage by regulating phospholipid metabolism and H3K4me2 (83). Histone acetyltransferases (HATs) and histone deacetylases (HDACs) play crucial roles in regulating histone acetylation, which in turn affects inflammation, myocardial function, and cardiac repair following MI (84, 85). After a MI, miRNA-134-5p goes up, inhibiting histone H3K14 acetylation by lowering lysine acetyltransferase 7 expression. This reduces antioxidant enzyme activity, raising oxidative stress and promoting harmful heart remodeling (86, 87). Eicosapentaenoic acid, docosahexaenoic acid, and ecklonia stolonifera Okamura extract were found to inhibit p300 HAT activity. This inhibition reduced histone acetylation and regulates gene expression, ultimately suppressing cardiomyocyte hypertrophy and preventing HF development (88, 89). Sodium caprylate has been shown to enhance cardiac recovery after MI by acting on HAT Kat2a, increasing histone acetylation levels, activating antioxidant gene expression, and reducing cardiomyocyte apoptosis (90). Studies have shown that inhibition of HDAC1 (91), HDAC3 (92), HDAC4 (93), HDAC5 (94), HDAC6 (95), and HDAC9 (96) has a protective effect on cardiomyocyte apoptosis, left ventricular remodeling (LVR), cardiac disfunction, and cardiac fibrosis after MI. This signifies the potential therapeutic value of targeting these specific HDACs to enhance post-MI cardiac recovery and mitigate adverse remodeling processes. Therefore, studying histone modifications is expected to offer valuable insights for developing new treatment strategies to alleviate MI and promote patient recovery.
2.3. Transcriptomics
Transcriptomics refers to the study of all RNA transcripts within a specific species, including both mRNA and non-coding RNA (ncRNA). Gene transcription is known to be spatiotemporal specific, meaning that gene expression can vary between different tissues or at different stages of the same tissue. The analysis of transcription plays a critical role in identifying the structure and function of the genome, decoding the genetic network underlying diseases, and searching for sensitive molecular biomarkers for diseases, drugs, and pathogens (97, 98).
Transcriptomic studies have identified many genes associated with MI, including genes that are up- or down-regulated after MI. These differentially expressed genes can reveal changes in cell signaling pathways after infarction and possible therapeutic targets. Sheng et al. (99) identified 552 differentially expressed genes and 23 differentially expressed lncRNAs of MI by analyzing the gene expression profile of circulating endothelial cells. They observed that inflammation-related genes such as NR4A2, IRAK3, NFIL3, IL1R2, CLEC4E and BCL3AMI were highly up-regulated, indicating that inflammation is an important feature of MI. Another study employed a bioinformatic analysis to pinpoint eight key immuno-inflammation-related genes, namely SH2D1B, ADM, PI3, MMP9, NRG1, CBLB, RORA, and FASLG, which have been identified as potential biomarkers for AMI (100). Zhuo et al. (101) constructed an MI-related lncRNA-miRNA-mRNA network using RNA sequencing data and identified lncRNA SNHG8, hsa-miR-411-5p, SOCS3 and ICAM1 were key nodes in the network. Their findings demonstrated that lncRNA SNHG8 not only emerged as a risk factor for MI but also exhibited substantial diagnostic potential. Furthermore, extensive transcriptomic studies have revealed a significant correlation between the abnormal expression of inflammation-related genes, such as VEGFA (102), TNF (103), IL6 (103, 104), IL6R (104), PTGS2 (105), immune response-related genes including CDKN2B (106), CDKN1C (107), TLR2 (108), TLR4 (108), apoptosis and proliferation-related genes like F3 (109), BTG2 (110), TXNIP (111), SAMSN1 (112), lipid metabolism-related genes such as PPARGC1A (113), ACSL1 (114), ABCG1 (115), SULT2B1 (116), and extracellular matrix (ECM) and collagen-related genes, such as MMP2 (117), MMP9 (117), LTBP4 (118), TNXB (118), and the occurrence and development of MI. These findings offer crucial insights into a deeper comprehension of the mechanisms underlying MI and hold promise for the development of more precise approaches for preventing and treating heart disease in the future.
NcRNAs, including microRNAs (miRNAs), lncRNAs and circular RNAs (circRNAs), have a significant attention in the field of MI research. They not only contribute to a deeper understanding of the disease's pathogenesis but also serve as valuable biomarkers, diagnostic tools, prognostic indicators, and potential therapeutic targets, offering new hope for managing MI patients (119–123). Numerous studies have demonstrated significant changes in the expression profiles of ncRNAs in both cardiac tissue and body fluids after MI. Some ncRNAs are upregulated, while others are downregulated. These changes are involved in the regulation of crucial pathological processes such as inflammation, apoptosis, and fibrosis, aiding in a more profound comprehension of the molecular basis of this disease (121, 123–125). Furthermore, many ncRNAs have been identified as biomarkers that can be utilized for MI diagnosis. These include miRNAs such as miRNA-1-3p (126, 127), miRNA-208a-3p (127, 128), miRNA-499a-5p (127), miRNA-486-5p (129), miRNA-21-5p (129, 130), as well as lncRNAs such as N1LR (131), SNHG1 (131), HIF1A-AS2 (132), TTTY15 (133), HULC (133), and circRNAs like cZNF292 (134), circTMEM165 (135), circUBAC2 (135), circZNF609 (135), circANKRD12 (135), circSLC8A1 (135), and among others. Additionally, ncRNAs are also employed to assess patients' prognosis, assist in determining treatment strategies, and predict disease progression. They are associated with patients' cardiac function, the risk of recurrent MI, and the occurrence of adverse events (124, 136–140). This information can offer valuable assistance to healthcare professionals in making well-informed decisions regarding treatment and predicting patient outcomes.
Transcriptome research plays a crucial role in treating MI. By studying how genes are active in heart tissue or cells, transcriptome research uncovers how drugs affect the body's molecular responses. This helps us understand treatment effects and mechanisms better, leading to more personalized therapies. In rat studies, it was indicated that photobiomodulation therapy can intervene in the activation of cardiac fibrosis after MI by altering gene activities and miRNAs in heart (141, 142). Su et al. (143) analyzed gene and ncRNA expression to uncover potential molecular mechanisms linked to the varying effectiveness of acubitril/valsartan treatment in patients with HF after AMI. These type of studies offers valuable insights into a deeper comprehension of drug mechanisms of action and resistance. Transcriptome research also aids in identifying potential biomarkers that can be utilized to assess treatment outcomes, monitor treatment progression, and predict patient responses to therapy. For instance, plasma levels of miR-223 and miR-126 have demonstrated potential as predictive biomarkers of dual antiplatelet therapy response and prognosis in patients with ST-segment elevation MI (STEMI) (144). Furthermore, abundant research suggests that miRNA hold promising potential in treating MI by regulating gene expression and crucial cellular processes. Key therapeutic candidates include miRNA-21 (145–147), miRNA-192-5p (148), and miRNA-432-5p (148), involved in inflammation and fibrosis; miRNA-499 (149), implicated in endothelial injury; miRNA-126 (150, 151), contributing to angiogenesis; miRNA-133 (152), influencing cardiac function; and miRNA-208a (153), linked to fibrosis. Intervening with these miRNAs could fundamentally change MI treatment by adjusting core processes and facilitating cardiac recovery.
2.4. Proteomics
Proteomics studies all proteins in tissues or cells, including protein expression levels, post-translational modifications, and protein-protein interactions to the study structure and location of protein and protein-protein interactions, providing a direct basis for clarifying the nature of life phenomena. Commonly used proteomics technologies include mass spectrometry (MS), two-dimensional gel electrophoresis, protein microarrays, protein-protein interaction assays, and imaging techniques like fluorescence and electron microscopy (154). Proteomics has played a crucial role in improving our understanding of the molecular mechanisms underlying MI and identifying potential protein markers of this disease.
Proteomics has unveiled dynamic fluctuations in protein expression during MI. By analyzing proteins in myocardial samples obtained from MI patients and animal models, scientists have identified a variety of proteins associated with MI, including inflammatory mediators, apoptosis-related proteins, and cardiac contractile proteins, shedding light on multiple key pathways involved in the development of MI (155–159). For example, Das et al. (160) used Orbitrap MS to identify 38 up-regulated and 26 down-regulated proteins in MI patients, most of which were related to chronic inflammation, atherosclerosis, and cholesterol reverse transport. Pan et al. (161) used ITRAQ (Isobaric tags for relative and absolute quantification) combined with LC-MS/MS (Liquid chromatography-tandem mass spectrometer) technologies to identify 95 MI differential expression proteins related to carbon metabolism, toll-like receptor signal pathway and hypertrophic cardiomyopathy. The proteomics approach has led to the discovery of an increasing number of diagnostic biomarkers, such as plasminogen (162), complement C8 beta chain (162), coagulation factor II (162), alpha-1 acid glycoprotein 2 (163), corticosteroid-binding globulin (163), serotransferrin (163), lactate dehydrogenase (164), creatine kinase (164), haptoglobin (165), etc. In a mouse model, researchers conducted proteomic analysis and identified cardiac myosin binding protein-C (cMyBP-C) as a potential up-regulated biomarker for MI (166). The potential of cMyBP-C as a sensitive cardiac-specific biomarker of MI was further confirmed by measuring its increased levels in the plasma of MI patients through enzyme-linked immunosorbent assay (ELISA) (167). Furthermore, proteomic approaches have identified many potential biomarkers for predicting MI prognosis by revealing distinct protein expression patterns. By examining the distinct protein expression patterns associated with MI, researchers have been able to discover valuable biomarkers that can provide insights into various outcomes, including HF (168), pulmonary hypertension (169), LVR (170), chronic kidney disease (171), and long-term outcomes (171). Liu et al. (168) found NF-κB signaling-related proteins linked to HF after MI. Another study also identified 50 proteins during MI patient hospitalization for predicting long-term HF occurrence (172). These proteins hold promise as potential markers for diagnosing HF after MI. LVR is a prevalent complication following MI, and researchers have identified several potential protein biomarkers associated with this condition. These biomarkers include apolipoprotein A1, immunoglobulin A, interleukin-17E, tissue inhibitor of metalloproteinases-1, urokinase-type plasminogen activator, midkine, proprotein convertase subtilisin/kexin type 6, among others (170, 173, 174). Furthermore, proteomic studies can provide useful information in assessing left ventricular ejection fraction (LVEF) and infarct size after MI (175).
Currently, established cardiac biomarkers such as cardiac troponin (cTn), creatine kinase (CK), creatine kinase MB (CK-MB), copeptin, and heart-type fatty acid binding protein (H-FABP) have been validated as valuable for the diagnosis and prognosis of MI (176, 177). Among these, cTn is acknowledged as a biomarker for assessing the risk of acute coronary syndrome (ACS), including cardiac troponin T (cTnT) and cardiac troponin I (cTnI) (178). Furthermore, protein biomarkers can provide significant diagnostic information for MI, which is crucial for follow-up treatment. For instance, myeloid-related protein 8/14 (MRP-8/14) was found to have higher concentrations, and high-sensitivity cardiac troponin I (hs-cTnI) to have lower concentrations in type 2 MI compared to type 1 MI (179). Pandey et al. (180) demonstrated that the levels of cTnT and CK-MB were significantly higher in patients with type 1 MI than in those with type 2 MI, and cTnT increased disproportionately with CK-MB in type 2 MI patients. Additionally, coagulation factor VII levels were significantly higher in patients with silent MI than in those with clinical MI in another study (181).
Proteomics analysis can be employed to unravel the complex mechanisms underlying the therapeutic effects of various interventions in the context of MI. Within the domain of MI and its treatment, proteomic studies have illuminated the cardioprotective effects of different compounds. For example, a proteomic study demonstrated that the cardioprotective effects of Shexiang Baoxin pill (SBP) and Suxiao Jiuxin pill (SJP) in MI rats are achieved by modulating pathways associated with focal adhesion, and platform activation (182). Additionally, SBP's cardioprotective mechanisms were also found to involve energy metabolism within cardiac tissue (183). Wang et al. (184) employed proteomics to delve into the therapeutic effects of Salviae Miltiorrhizae and Cortex Moutan extract post-MI in rats. Their findings underscored the significance of metabolism, oxidative stress, and cytoskeleton modulation in these effects. Proteomics has provided insights into the regulatory protein targets impacted by specific treatments, offering valuable knowledge for advancing therapeutic strategies. Through the utilization of the SOMAScan aptamer-proteomics platform, George et al. (185) identified a cluster of five proteins whose regulation is influenced by the antagonistic effects of IL-6. These identified proteins were speculated to potentially play a role in mediating the therapeutic effects of tocilizumab in cases of non-ST segment elevation MI (NSTEMI). Furthermore, proteomic investigations have unveiled the positive influence of physical exercise on cardiac remodeling after MI. These benefits are attributed to increased anti-oxidant levels, diminished ion channel expression, favorable adaptations in energy metabolism, dampened inflammation, and alterations in ECM organization (186, 187).
2.5. Metabolomics
Metabolomics is a research method used to quantitatively analyze all metabolites in an organism and investigate the relationship between metabolites and physiological and pathological changes. This method employs various technologies, such as nuclear magnetic resonance (NMR) spectroscopy, MS, gas chromatography (GC), liquid chromatography (LC), capillary electrophoresis (CE), Fourier transform infrared (FTIR) spectroscopy, and Raman spectroscopy to study the small molecules produced by metabolic processes in biological fluids, cells, and tissues (188). Metabolomics studies have identified several dysregulated metabolites during MI, including lipids, amino acids, energy metabolites, and oxidative stress markers (189–191).
Research has revealed notable distinctions in metabolite profiles between patients with MI and healthy individuals or other populations (192, 193). Some metabolites, such as 12,13-diHOME, noradrenaline, tryptophan, and cysteic acid, were significantly elevated in patients with MI, while some antioxidants like glutathione were decreased (194–196). Changes in these metabolites may serve as potential biomarkers of MI. Abnormal amino acid metabolism is one of the early indicators of MI. Research has shown that amino acids and their metabolites such as tryptophan, carnitine, l-homocysteine sulfinic acid, kynurenine, and cysteic acid, significantly increase in the serum after MI, while amino acids like leucine, isoleucine, l-proline, l-alanine, glycine, l-cysteine, and l-cysteine sulfinic acid decrease (190, 195, 197). These changes in amino acid concentrations can serve as biomarkers for MI, aiding in early diagnosis and monitoring of patients' conditions. Lipid metabolism also undergoes significant changes in MI. Triglycerides, low-density lipoprotein cholesterol (LDL-C), non-high-density lipoprotein cholesterol (non-HDL-C), remnant cholesterol, and total cholesterol levels significantly increase after MI, while high-density lipoprotein cholesterol (HDL-C) decreases (198–201). This lipid metabolism abnormality is closely associated with the development of coronary artery atherosclerosis and can be used to predict the risk of MI. MI results in an insufficient energy supply to myocardial cells. Therefore, metabolomics research places a significant emphasis on metabolites related to energy metabolism. Examples of such metabolites include lactic acid (202), fatty acids (203), phosphates (204, 205), and creatine (206), all of which experience alterations following MI and can serve as biomarkers for this condition. Additionally, MI is characterized by disruptions in glucose metabolism. Various studies have observed substantial changes in metabolites related to glucose metabolism, including glucose and lactate, following a MI (207, 208). These changes reflect a shift in the myocardial cells' energy dependence. The measurement of these metabolites can provide valuable insights into the metabolic status of patients with MI.
Metabolomic studies have revealed a series of metabolites related to the prognosis of MI, including amino acids, lipids, and sugar metabolites. These metabolites can be used to predict adverse outcomes in patients with MI, such as major adverse cardiovascular events (MACEs), HF, death, etc. (209–211). For example, recent research has identified a significant positive correlation between heightened levels of various metabolites, such as phenylacetyl glutamine, indoxyl sulfate, deoxycholic acid, trimethylamine N-oxide, trimethyllysine, dimethylarginines, and MACEs, in individuals suffering from MI (212–214). Increased plasma kynurenine levels have been found to be positively associated with the occurrence of STEMI and its adverse outcomes (190). These findings provide valuable insights for healthcare professionals, enabling them to better assess a patient's risk and implement appropriate interventions. After a MI, the body rapidly activates its inflammatory response, resulting in increased levels of markers like interleukins and C-reactive protein, which can damage cardiac tissue (215). Metabolomic studies reveal the intricate interactions between these inflammatory molecules and metabolic compounds, providing a comprehensive understanding of inflammation's metabolic effects (216–218). Additionally, metabolic alterations have been shown to reflect energy deficit, acidosis, oxidative stress, ion imbalance, and cardiac injury following MI (191). Analyzing changes in metabolic markers allows us to assess the extent of myocardial tissue damage and predict patient prognosis.
Metabolomic studies provide valuable insights into the treatment of MI. Firstly, through metabolomics techniques, doctors can gain a better understanding of the metabolic profiles of each patient, thereby adjusting drug selection and dosages to enhance treatment effectiveness. Xia et al. (189) conducted research into the distinct metabolic changes in diabetic patients experiencing AMI. They uncovered essential metabolites linked to compromised mitochondrial function, impaired glucose utilization, and heightened inflammation. These finding sheds light on personalized therapeutic strategies for cases involving diabetes-associated AMI, representing a promising avenue for targeted interventions. Additionally, metabolomics can also contribute to the discovery of new drugs that may treat MI by intervening in metabolic pathways. Metabolomic studies have been employed to elucidate the therapeutic impacts of drugs on MI, such as the hydroethanolic extract of Cucumis sativus L. seeds (219), colchicine (220), SJP (221), and SBP (222). These studies offer a comprehensive insight into the mechanisms underlying drug treatments for MI. They hold the potential to guide the development of more effective treatments and provide valuable insights into evaluating the safety and potential side effects of drugs. Fan et al. (223) used metabolomics to reveal the therapeutic potential and synergistic mechanism of total saponins and flavonoids in notoginseng-safflower (NS-SF) in treating MI, emphasizing its superior efficacy compared to individual components and highlighting their combined regulation of key metabolic pathways in MI treatment. This study provides valuable insights for the clinical development of NS-SF as a potential treatment for CVDs. Furthermore, metabolomic studies can shed light on the impact of specific interventions in patients with MI. For instance, ketone ester supplementation has been shown to attenuate cardiac inflammation and enhance cardiac energy in a porcine model of AMI (224). Another metabolomic investigation has indicated that the combination of smoking and a high-fat diet may exacerbate cardiac dysfunction following MI, resulting in substantial disruptions in metabolic pathways related to inflammation, energy metabolism, and excessive oxidative stress (225). Therefore, gaining a deeper understanding of how different dietary patterns, supplements, and dietary modifications affect metabolic pathways could prove instrumental in enhancing the recovery and treatment outcomes of MI patients.
2.6. Microbiomics
Microbiomics is an interdisciplinary field that combines microbiology, macrotranscriptomics, macroproteomics, metabolomics, chemistry, culturomics, ecology, phylogeny, and systems biology to investigate the composition, diversity, and function of microorganisms (226, 227). The primary sequencing methods used in microbiomics include marker gene sequencing (e.g., 16S amplicon sequencing), macrogenome sequencing, and macrotranscriptome sequencing (228).
The human body contains over 2,000 types of microorganisms that interact in complex ways with the host (229). Disruptions in microbial ecology can promote atherosclerosis and CVD through inflammation, arterial fibrosis, and dyslipidemia (230–233). Firstly, the microbial community within the human body is closely related to the pathogenesis of MI. Research has shown that an imbalance in the gut microbiota is a key factor contributing to the development of CVDs (234). This imbalance can lead to an increase in harmful microorganisms while simultaneously decreasing the presence of beneficial microorganisms, thereby promoting an increase in inflammatory responses and disturbances in lipid metabolism. These factors may result in atherosclerosis, ultimately leading to the occurrence of MI (235). For example, short-chain fatty acids (SCFAs) are metabolites produced when intestinal microorganisms ferment dietary fibers such as cellulose. Research shows that SCFAs can have anti-inflammatory and antioxidant effects, which contribute to the maintenance of cardiovascular health (236). Nevertheless, an imbalance in gut microbes may lead to reduced SCFA production, thereby diminishing this protective effect and potentially promoting the development of MI (237). Additionally, a study has shown that the gut microbiota may impact the occurrence and development of AMI through the SCFA pathway (238). Trimethylamine oxide (TMAO) is a compound produced through intestinal microbial metabolism and is commonly associated with the consumption of fish, red meat, and other foods rich in choline and L-carnitine (239). Studies have demonstrated that elevated levels of TMAO are linked to the promotion of plaque formation, inflammatory responses, and an increased risk of cardiac events (240, 241). TMAO may also exacerbate the occurrence and progression of MI by damaging arterial endothelial function and inducing atherosclerosis. Furthermore, the gut microbiota differed significantly not only between MI patients and healthy individuals but also among subgroups of MI patients (238). Similarly, blood microbiota was also associated with MI. A study found that the structure and abundance of blood microbiota differed between MI patients and healthy individuals (242). These microbiome changes may become potential biomarkers, providing new avenues for early diagnosis and risk assessment of MI.
Microbiomics has made significant progress in MI prognosis research. In patients with STEMI, intestinal bacterial translocation was positively correlated with systemic inflammation and adverse cardiovascular events. Treatment with antibiotics to eliminate intestinal bacterial translocation alleviated systemic inflammation and myocardial cell damage in MI mice (243). Butyrate, a SCFA produced by the microbiota, can suppress inflammation and prevent myocardial hypertrophy. Research has demonstrated its capacity to enhance cardiac function and promote sympathetic nerve remodeling after MI in rats (244). Conversely, elevated plasma levels of TMAO independently correlated with a high risk of plaque rupture in STEMI patients (245). Additionally, studies have consistently indicated that high TMAO levels are positively associated with MACEs, atrial fibrillation, and coronary atherosclerotic burden (209, 213, 246–248). TMAO holds promise as a valuable biomarker for predicting poor prognosis and improving risk assessment and management in MI patients. Considering these findings, microbiomics opens up new diagnostic and therapeutic avenues for MI patients. By restoring balance to the intestinal microecology, it is anticipated to reduce chronic inflammation levels, enhance immune function, lower the risk of recurrent MI, and ultimately improve patient survival rates.
Microbiome research has made exciting advancements in MI treatment, offering innovative perspectives for novel therapeutic strategies. Firstly, research has established a strong link between the gut microbiome and chronic inflammation, a significant contributor to MI (235, 249). By modulating the intestinal microecology, it holds the potential to alleviate chronic inflammation in patients and support post-MI recovery. For instance, studies have demonstrated that supplementing with probiotics or prebiotics can restore a healthier balance of intestinal flora, effectively reducing inflammation and facilitating recovery following MI (250–252). This provides direction for the development of new biologics or dietary interventions. Secondely, studies have shown that gut microbiota remodeling through certain interventions, such as Lactobacillus johnsonii, dapagliflozin, and flavonoids, can result in enhanced cardiac function after AMI (253–255). SCFAs have been found to improve cardiomyocyte function and heart contractility (244, 256). Therefore, by increasing the intake of dietary fiber and promoting the production of SCFAs, it is expected to improve the cardiac function of patients with MI. Furthermore, exercise intervention can influence the gut microbiota to improve cardiac function after MI (257, 258). Thirdly, the microbiome can influence drug metabolism and effects. Studies have shown that the efficacy of some drugs, such as antibiotics (259), immunosuppressive drugs (260, 261), and anticoagulants (262), in the treatment of MI may be affected by the microbiome. Therefore, by adjusting the microbiome, drug efficacy can be optimized and therapeutic effects improved.
2.7. Single-cell omics
Cells are the fundamental building blocks of all living organisms, and exploring the phenotype and function of individual cells can lead to a deeper understanding of biological activities, helping to comprehend the causes, progression, and treatment of medical conditions such as MI. The use of single-cell omics has greatly advanced our understanding of cell heterogeneity, with technologies such as single-cell RNA sequencing (scRNA-seq), single-cell DNA sequencing (scDNA-seq), single-cell ATAC sequencing (scATAC-seq), and cytometry providing unprecedented levels of detail when examining individual cells (263, 264). Single-omics analysis unveils cell population diversity and functional disparities, opening a new dimension for studying diseases like MI.
Single-cell genomics has provided unprecedented insights into the responses of distinct cell types within cardiac tissue during MI. MI involves a cascade of complex biological events, including myocardial cell necrosis, infiltration of inflammatory cells, and fibroblast proliferation, among others. Traditional research methods struggle to capture the nuances and changes within these cell types. Fortunately, single-cell genomics technology empowers researchers to decode the gene expression patterns of individual cells. Through scRNA-seq of cardiac samples from MI patients, researchers have identified various cell types, including myocardial cells, immune cells, fibroblasts, and endothelial cells, and have explored their dynamic changes during MI (265–268). This comprehension of cellular diversity is pivotal in the study of cardiac development, injury responses, and the progression of cardiac diseases. In a study by Qian et al. (269), utilizing the scRNA-seq method, the transcriptomes of peripheral blood mononuclear cells in AMI patients were analyzed, revealing a total of 27 cell clusters. This investigation unveiled that peripheral immune cells in patients with plaque rupture exhibited marked pro-inflammatory characteristics, while plaque erosion was associated with intermediate monocyte expansion, neutrophil activation, and granule release. These single-cell analysis techniques enable scientists to delve deeper into the distinctive roles of various cell types in MI pathology. Research has consistently shown that myocardial cell heterogeneity is closely linked to cardiac remodeling, angiogenesis, and inflammation following MI (267, 270–274). A recent study has revealed a highly enriched regulatory T cell (Treg) subtype within the myocardium of MI mice, which facilitates cardiac damage repair post-MI (275). Furthermore, in an analysis of myocardial cells from a pig AMI model, a cell cluster potentially related to cardiac injury was identified. This cluster regulates the cardiac injury response by upregulating genes such as TBX5, TBX20, ERBB4, and GRK5 (276). These research findings underscore the significance of myocardial cell heterogeneity and potential mechanisms in the treatment of MI and cardiac injury repair. Furthermore, scRNA-seq is a valuable tool for investigating communication between different types of heart cells. Understanding the network of interactions among cardiac cells that constitute the heart is crucial for comprehending cardiac homeostasis and the progression of diseases (277). Skelly et al. (278) delved into the analysis of non-muscle cells in the mouse heart using scRNA-seq technology, uncovering the diversity and sex differences among cardiac cells. This research provides essential insights for the study of cardiac development and diseases, deepening our understanding of cardiac cell composition and function. Another study conducted a focused study on the cellular composition of the mouse left ventricle. Through scRNA-seq, they identified various types of cardiac cells and elucidated their functions. This study not only offers potential therapeutic targets for CVDs but also unveils an extensive network of interconnected communication among cells within the left ventricle (279). Through a comprehensive exploration of the functions of different cell types and their interactions, we enhance our understanding of MI's pathogenesis and provide invaluable insights for future treatment strategies.
Single-cell genomics provides an opportunity to discover new biomarkers. By examining individual cells, particularly their gene expression patterns within different cell subpopulations, researchers have successfully identified a range of potential biomarkers associated with MI. For example, Zhang et al. (280) used scRNA-seq technology to reveal the potential roles that IL1B and TLR2 may play in the diagnosis of MI, which are closely related to various infiltrating immune cells. Another study also identified a series of immune cell-related genes, including FOS, DUSP1, CXCL8, and NFKBIA, which can not only differentiate between AMI and coronary heart disease (CHD) but also predict the risk of HF in AMI patients (281). Furthermore, single-cell studies have revealed some monocyte-related genes, including CUX1, CTSD, ADD3, PRKAR1A, and SDCBP, which can be used to predict the risk of developing HF after AMI (282, 283). These findings provide new clues for early diagnosis and prognosis assessment of MI, with the potential to improve patient treatment and management. Additionally, single-cell genomics also helps uncover individual differences among MI patients. Each patient's cell composition and gene expression may vary, suggesting that personalized treatment strategies may hold promise. Single-cell research can provide a basis for developing personalized treatment plans, thereby enhancing treatment effectiveness.
2.8. Radiomics
Radiomics is a methodology that enables the precise characterization of pathological observations detected in radiological imaging by essentially converting images into data. Its research stages typically include data selection, medical imaging, feature extraction, exploratory analysis, and modeling (284). A The advancements in technologies such as magnetic resonance imaging (MRI), computed tomography (CT), echocardiography, nuclear imaging, and others have greatly supported accurate diagnosis and treatment in the context of MI.
MRI stands as a high-resolution non-invasive technique employed to gain intricate insights into heart structure and function, including parameters such as ventricular wall thickness, myocardial function, and volume. Recent research indicates that MRI-based radiomics, particularly in non-contrast MRI, holds great potential in predicting MI, promising to revolutionize the management and treatment of heart diseases. A study by Smith et al. (285) demonstrated the reliability of machine learning-based radiomic features extracted from non-contrast MRI in distinguishing between MI and normal tissue, offering a novel avenue for clinical diagnosis. Another study revealed that radiomic analysis using non-contrast MRI can predict adverse LVR following STEMI, thereby enhancing the accuracy of patient prognosis assessment (286). Furthermore, the integration of native T1 and extracellular volume (ECV) mapping within MRI technology, along with radiomic analysis, elevates the accuracy of predicting heart function recovery and microvascular damage. Ma et al. (287) indicated that radiomic analysis using non-contrast CMR T1 mapping can play an important role in the diagnosing AMI and the predicting myocardial function recovery. This method not only enhances the accuracy of detecting microvascular obstruction but is also expected to improve long-term predictions of myocardial contraction function, providing valuable tools for the clinical management of cardiac diseases. Additionally, radiomics based on non-contrast-enhanced T1 mapping exhibits the capacity to predict MACEs in STEMI patients, facilitating patient risk stratification (288). Chen et al. (289) unveiled that radiomic texture analysis-based ECV score mapping can differentiate between reversible and irreversible myocardial injuries in STEMI patients while predicting LVR, potentially assuming a significant role in clinical applications. Late gadolinium enhancement (LGE) is a commonly used MRI technique for detecting myocardial abnormalities. Di Noto et al. (290) found that radiomic features of LGE can accurately differentiate between MI and myocarditis, offering a promising tool for non-invasive diagnosis. Furthermore, combining LGE-based radiomics with machine learning can predict MACEs in STEMI patients (291). Therefore, MRI-based radiomics and machine learning have opened new horizons in CVD research. They can be seamlessly integrated with MRI and clinical information to enhance model accuracy and predictive performance.
CT plays a significant role in the diagnosis and evaluation of MI. Coronary CT angiography (CCTA) enables the non-invasive assessment of coronary artery narrowing and obstruction, aiding doctors in determining the patient's coronary condition. Pericardial adipose tissue (PCAT) is the fat surrounding the heart within the pericardial sac, often relevant in cardiovascular research and clinical contexts due to its potential impact on heart health (292). A recent study has discovered that the use of CCTA for the radiomic analysis of PCAT can effectively differentiate between patients with AMI and those with stable coronary arterial disease (CAD) (293). This research highlights the potential role of radiology and machine learning in cardiac disease image diagnosis. Specifically, the combination of clinical characteristics, PCAT attenuation, and radiomic parameters can enhance the accuracy of identifying AMI patients. Additionally, Si et al. (294) indicate that radiomics analysis of CCTA-based PCAT radiological features excels at distinguishing between AMI and unstable angina pectoris (UA). They found that the application of PCAT's radiomics characteristics and the fat attenuation index can improve performance in recognizing AMI, offering a promising new approach for non-invasive CHD diagnosis. Another study has also proven that radiomics features based on CCTA-derived PCAT can be used to distinguish between NSTEMI and UA. However, this study also noted certain limitations in the radiological model of epicardial adipose tissue for this task (295). Therefore, the radiomic features of PCAT based on CCTA can be used for the differential diagnosis of MI.
Therefore, radiomics plays an increasingly vital role in the diagnosis, treatment planning, and overall patient management of MI. The ongoing development and enhancement of these technologies hold the promise of delivering more precise and personalized diagnostic and treatment options for MI patients, ultimately contributing to improved recovery and survival rates. However, it's imperative to address challenges related to data privacy, standardization, and clinical integration to effectively introduce these innovations into routine clinical practice and, ultimately, to the benefit of patients.
3. From single-omics to multi-omics integrative analyses: toward the era of MI personalized medicine
3.1. Necessity of multi-omics integration in personalized medicine
As depicted in Figure 1, diverse omics exhibit unique characteristics when delving into the intricacies within organisms. The different omics provide insights into different layers of biological information. Furthermore, there exists a strong interconnectedness among these omics. Genomics concentrates on an organism's genetic information, which generally remain relatively static and stable. Conversely, other omics domains like epigenomics, transcriptomics, proteomics, and metabolomics offer dynamic insights into gene expression, protein activity, and metabolite levels (296–298). Progressing from genomics to epigenomics, and subsequently to transcriptomics, proteomics, metabolomics, and other omics, the complexity and dynamism of information progressively intensify, more effectively showcasing a wide range of disease phenotypes and contributing to the comprehension of disease origins and progression mechanisms (299). Nevertheless, as complexity deepens, data analysis becomes increasingly intricate.
Furthermore, each omics approach has its own advantages and disadvantages (Table 1). Single-omics studies offer the advantage of conducting in-depth examinations of specific biological or molecular mechanisms. There studies provide highly focused information that allows researchers to delve deeply into specific genes, proteins or molecular mechanisms, revealing microscopic level details and mechanisms. This focus makes single-omics studies particularly useful when addressing specific biological questions and research goals, such as identifying disease-causing genes, analyzing protein function, or studying a specific metabolic pathway (300). Additionally, single-omics studies usually have high experimental controllability, facilitating the acquisition of reproducible results and offering relatively low costs. This makes single-omics studies a powerful tool for studying specific biological questions or conducting preliminary explorations. However, the disadvantage of single-omics studies is their narrow focus, and they often fail to provide insights into the interactions and integration of the overall biological system. Life science often involves multi-level and complex interactions, so relying solely on data from a single research field may not fully understand the integrity and comprehensiveness of biological systems (301–303). Furthermore, each omics approach has its own limitations, including technical constraints, experimental setups, and data analysis.
Therefore, a pressing need has arisen to transition from single-omics to multi-omics integrative approaches in current biomedical research. Although conventional single-omics techniques retain significance within specific fields, they encounter challenges in fully capturing the intricacies and diversity present within organisms. Particularly, when delving deeply into complex maladies like MI and the analysis of individual variabilities, single-omics approaches exhibit inherent limitations. The etiology of MI is exceptionally intricate, potentially giving rise to abnormalities across multiple levels. Genetic variations and irregularities in gene expression may hold pivotal roles in the progression of the disease. Genetic mutations could render individuals more susceptible to cardiac ailments, thereby amplifying the risk of MI (304). In parallel, aberrant gene expression could disrupt the functionality of the cardiovascular system, further hastening the onset of MI (305, 306). However, beyond genetics, subsequent alterations could serve as triggers for MI, encompassing factors such as environmental influences, lifestyles, and epigenetics (123, 307–310). This underscores that MI constitutes a multi-tiered and complex process, with deviations at each omics level potentially influencing the eventual outcome. Consequently, when confronted with the intricacies of MI, the integration of multi-omics approaches becomes pivotal, not only furnishing additional evidence but also propelling deep phenotyping analysis and the advancement of personalized medicine.
The integration of multi-omics approaches refers to the practice of combining and analyzing data from various omics technologies, such as genomics, epigenomics, transcriptomics, proteomics, and metabolomics. These approaches enable a more comprehensive understanding of biological systems, yielding profound insights into complex biological processes and diseases. By merging molecular omics with clinical phenotype omics data, we can delve into the underlying causes of MI, unravel the complex connections between genes and phenotypes, and create a detailed patient information landscape. This not only enhances the accuracy of disease diagnosis but also empowers the creation of tailor-made treatment protocols and the real-time tracking of patient responses to treatment. Thus, the shift from single-omics methods to multi-omics integrative approaches has evolved into an indispensable trend in contemporary biomedical research. This transformation paves the way for broader possibilities in future medical research and clinical practice, ushering in a new era of personalized medicine for MI.
3.2. Advancements in multi-omics integration research for MI
In recent years, researchers have adopted a strategy of integrating two or more “omics” approaches to conduct in-depth research on the molecular mechanisms and potential biomarkers of MI. These studies help us better understand the pathogenesis of MI and provide new clues for early diagnosis and treatment. Table 2 displays some MI research involving multi-omics integration.
Multi-omics integration analysis is a powerful tool for exploring the relationships between molecules at various levels in MI and identifying potential biomarkers and key molecules. The integrated research of genomics and transcriptomics is used to study the relationship between genetic variations and changes in transcriptional expression, which is conducive to clarifying the potential molecular mechanisms of MI. Chen et al. (311) revealed that genetic variations significantly reduce the transcriptional activity of the SIRT5 gene promoter. This alteration could result in changes in SIRT5 levels, thereby increasing the risk of AMI. SIRT5 overexpression has been demonstrated to provide protection against cardiac dysfunction induced by pressure overload, while simultaneously suppressing adverse metabolic and fibrotic pathways associated with HF (342). Similarly, other studies have uncovered that genetic variations in ATG7 (312), GATA6 (313), and VEGFR-1 (314) can significantly impact the transcriptional activity of these genes, further influencing the risk of MI. Furthermore, Haase et al. (61) employed a combination of genomics, epigenomics, and transcriptomics to reveal that elevated GPR15 expression is associated with early-onset MI, potentially mediating the adverse effects of smoking on MI risk. Additionally, they found that DNA hypomethylation and a nucleotide polymorphism (rs2230344) of GPR15 are also linked to MI risk. Since rs2230344 is located in close proximity to GPR15 DNA methylation sites (343), it may influence DNA methylation, leading to increased GPR15 levels. The integrated research of genomics and metabolomics is employed to explore the intricate connections between genetic variations and alterations in metabolite profiles. 25-hydroxy vitamin D (25OHD) serves as an intermediary product in the body's vitamin D metabolism, participating in lipid metabolism. Its deficiency may increase the risk of MI (344). Sedky et al. (315) revealed that genetic variations in the CYP2R1 gene can modulate serum 25OHD levels, thereby impacting MI risk. Lipid metabolism abnormalities are intricately linked to the risk of MI, as elevated cholesterol levels and imbalanced lipid profiles can contribute to atherosclerosis and heighten the likelihood of MI (345). Studies have indicated that genetic variations in the LRP8 (rs10788952 and rs7546246) (316), CETP (rs708272) (317), NRIL (rs9632884) and MALAT1 (rs320040) (17) are closely associated with MI risk by exerting their influence on lipid metabolism. These research findings underscore the potential significance of genetic variations in the pathogenesis of MI, offering promising avenues for future disease treatment and personalized medicine.
The integrated research in epigenomics and transcriptomics is utilized to investigate the impact of epigenomic alterations on gene expression and to identify key molecular signatures and potential mechanism. A recent study has revealed that substantial alterations in DNA methylation and gene expression take place in the early phases of AMI, particularly within the initial 6 h following AMI. Additionally, the study has identified promising epigenetic-based biomarkers for early clinical diagnosis and potential therapeutic targets for AMI, which include Ptpn6, Csf1r, Col6a1, Cyba, and Map3k14 (305). Wang et al. (318) unveiled the critical roles of histone modifications such as histone H3 lysine 27 acetylation (H3K27ac), histone H3 lysine 9 acetylation (H3K9ac), and H3K4me3 in the early stages of MI. They identified that at least 195 genes were upregulated and associated with one of these modifications, impacting various key biological processes, including cardiac cell development, inflammation, vascularization, and metabolism. Additionally, they discovered that enhancers rich in H3K27ac may play a crucial role in early vascular responses. In another study, researchers discovered that knocking down DYRK1A enhanced the expression of numerous genes involved in cell proliferation, thereby activating the cell cycle of cardiac cells and promoting cardiac repair following MI. Throughout this process, epigenetic modifications, particularly H3K4me3 and H3K27ac modifications, played a crucial role, involving proteins such as WD repeat-containing protein 82 and lysine acetyltransferase 6A. Therefore, by regulating these epigenetic modifications and protein activity, it may be possible to promote more effective cardiac cell repair, potentially leading to improved recovery and cardiac function in MI patients (338). Spleen focus forming virus proviral integration oncogene (SPI1) belongs to the ETS family of transcription factors and participates in a wide range of cellular processes, including inflammation and cell apoptosis (346, 347). Liu et al. (320) revealed that the upregulation of SPI1 gene expression, driven by reduced DNA methylation, leads to heightened inflammation and cardiac damage through the activation of the TLR4/NFκB pathway, further exacerbating MI development. This research sheds light on the pivotal role of SPI1 in the pathological process of MI and establishes a foundation for exploring new intervention strategies. Aldehyde dehydrogenase 2 (ALDH2) downregulation is related to MI severity, and epigenetic changes may contribute to its downregulation (348). Researchers found that ALDH2 mRNA and protein downregulation after MI was partly due to CpG hypermethylation in the upstream ALDH2 gene promoter, as revealed by multi-omics analysis in the rat MI model (337). The integration of epigenomics and transcriptomics provides important insights into the mechanisms linking smoking and MI. A recent study discovered that smoking can induce DNA demethylation at the F2RL3 site, potentially increasing the expression level of the PAR4 gene. This alteration may lead to heightened platelet reactivity, thereby escalating the risk of MI in smokers (319). Furthermore, Ward-Caviness et al. (339) correlated epigenetic fingerprint sites with cis-gene expression and integrated them into the gene expression metabolomics network. They found that DNA methylation changes in MI were associated with alterations in branched-chain amino acid metabolism. This discovery suggests that epigenetic changes may play a vital role in metabolic regulation after MI. This has significant implications for our understanding of the metabolic regulation of MI and for identifying potential therapeutic pathways. Collectively, these studies underscore the regulatory role of epigenomics in the pathogenesis of MI and provide valuable insights and targets for understanding and treating this condition.
Liu et al. (324) conducted an analysis of transcriptomes and proteomes in myocardial tissue at various time points following AMI. They noted that the earliest significant changes occurred at 6 h after AMI, and pyroptosis was activated at 24 h after AMI. Additionally, they highlighted the potential of the pyroptosis inhibitor VX-765 as a promising drug target and identified the protein WIPI1 (WD repeat domain, phosphoinositide interacting 1) as a valuable early diagnostic biomarker for AMI. Another study also revealed transcriptomic and proteomic changes associated with MI, with a particular emphasis on changes in immune, cell cycle, and ECM-related pathways. In addition, the study identified potential blood markers that could be used for the diagnosis or treatment monitoring of MI. Wu et al. (321) employed an integrated approach combining transcriptomics and proteomics to successfully identify seven key genes associated with AMI, with Coro1a exhibiting upregulation in both omics levels. This pivotal discovery not only presents potential new biomarkers for early AMI diagnosis but also offers hope for enhanced treatment and patient management strategies. Notably, the Coro1a gene, a member of the Coronin family, primarily functions within the immune system, and its aberrations can lead to immune system dysfunction and other health complications (349). Furthermore, a recent study that integrated transcriptomics and proteomics has unveiled tissue remodeling signatures specific to NSTEMI. In conjunction with the elevation of inflammation and fibrosis markers, the ischemic regions of NSTEMI displayed unique patterns of complex galactosylated and sialylated N-glycans within cellular membranes and the ECM (325). These findings offer valuable insights into potential drug therapies aimed at addressing fibrous remodeling in NSTEMI, presenting novel perspectives for future therapeutic strategies.
Integrated analysis of metabolomics and other omics has been used to uncover the protective mechanisms of drugs against MI. Sodium salvianolic acid A (SAAS) is a novel drug derived from a traditional Chinese medicine Salvia miltiorrhiza. It is currently undergoing Phase I clinical trials in China for the treatment of CHD and stable angina. Through proteomics and metabolomics analyses, Jia et al. (326) revealed that SASS possesses the remarkable ability to counteract a multitude of metabolic alterations induced by MI. SAAS exerts this protective effect by selectively binding to specific proteins integral to metabolic pathways, thereby effectively safeguarding rats from the detrimental consequences of MI. Furthermore, Jia et al. (322) utilized transcriptomic and proteomic analyses to reveal the cardioprotective effects of SAAS in MI by regulating pathways such as the actin cytoskeleton, phagosomes, focal adhesions, and others. These studies offer valuable insights into SAAS treatment for MI and pave the way for innovative research directions in future treatment strategies. Nutmeg-5 is an ancient and classic formula in traditional Mongolian medicine composed of five traditional Chinese medicines, widely used in the treatment of MI. Yan et al. (327) further elucidated the mechanisms underlying Nutmeg-5's protective efficacy against MI by employing proteomics and metabolomics analyses. Their findings revealed that Nutmeg-5 achieves its protective effect on MI through the inhibition of ECM-receptor interaction pathways and TGF-β1/Smad2 signaling transduction, which was achieved by regulating plasma metabolites. Acid ceramidase (AC) serves as the primary enzyme responsible for catalyzing the hydrolysis of ceramide, thereby producing free fatty acids and sphingosine. Recent research has highlighted the promising potential of AC gene therapy in alleviating pulmonary arterial hypertension with right heart dysfunction (350). In a study by Hadas et al. (340), utilizing a comprehensive approach encompassing transcriptomics, proteomics, and metabolomics, it was revealed that overexpressing AC via modified mRNA (modRNA) delivery can lower ceramide levels, promote cell survival, and offer cardiac protection after MI by improving heart function and reducing scar size. This discovery underscores the therapeutic potential of AC modRNA in ischemic heart disease. Zhang et al. (331) utilized metabolomics and metagenomics analyses to reveal that gut microbiota dysbiosis is a key contributing factor to high platelet activity in STEMI patients receiving ticagrelor treatment after PCI. This finding underscores the importance of gut microbiota in treating MI patients and offers new potential strategies for improving platelet activity. Liao et al. (333) used metabolomics and microbiomics analyses and discovered that the cardiac protective effects of SJP in AMI may be linked to its impact on gut microbiota and host fatty acid metabolism, especially its regulation of eicosanoids. This study offers valuable insights into the mechanism of this traditional Chinese medicine formulation and provides new avenues for AMI treatment and prevention. Furthermore, multi-omics analysis can also be used to reveal the protective effect of exercise on MI. Jiang et al. (93) conducted an integrated analysis of transcriptomics, proteomics, and metabolomics, shedding light on the beneficial effects of exercise in rats suffering from HF. Their investigation revealed that exercise not only enhances heart function but also improves glucose metabolism in HF rats through the activation of the AMPK-HDAC4-MEF2a pathway, inhibition of HDAC4 activity, and enhancement of GLUT1 expression. These discoveries provide novel and valuable insights into the potential treatment of HF following MI.
With the advancement of single-cell and multi-omics technologies, single-cell analysis has entered the multi-omics era. Multi-omics single-cell analysis integrates multiple types of omics information, such as genomics, epigenomics, transcriptomics, proteomics, metabolomics, or spatial state, simultaneously at single-cell resolution. This provides a more comprehensive understanding of cellular states and fates, which is of great significance for the development of precision medicine for MI. Kuppe et al. (335) developed a multi-omics map of MI using single-cell gene expression, chromatin accessibility, and spatial transcriptomic profiling. They created a comprehensive molecular map of human cardiac remodeling post-MI, revealing insights into cell-type shifts, transcriptome and epigenome changes, and tissue reorganization. These findings have significant implications for understanding cardiac diseases and potential therapeutic approaches. Kim et al. (334), through the use of single-cell transcriptomics and spatial transcriptomics techniques, have unveiled the heterogeneity of macrophages in the post-MI heart and identifies Trem2hi macrophages as potential therapeutic targets. Additionally, they found that the injection of soluble form of triggering receptor expressed on myeloid cells 2 (Trem2) during the subacute phase of MI improved myocardial function and remodeling, offering promise for novel MI treatments. In another study, a comprehensive approach involving multi-omic single-cell gene expression analysis, epitope mapping, and chromatin accessibility profiling identified a specific subset of fibroblasts in human cardiac disease that contributes to tissue fibrosis. This study found that their emergence is driven by interactions with C-C chemokine receptor type 2 (CCR2) macrophages via interleukin 1 beta (IL-1β) signaling. These discoveries emphasize the broader therapeutic potential of targeting inflammation as a strategy to combat tissue fibrosis and restore normal organ function (336).
Furthermore, multi-omics data integration analysis enables the development of biomarkers for early diagnosis and personalized medicine of MI. Lim et al. (341) conducted a comprehensive investigation utilizing metabolomics, lipidomics, glycomics, and metallomics approaches to construct intricate multi-omics maps of interconnected biomolecules, significantly advancing our comprehension of MI. Their research not only unveiled the potential role of glycerophospholipids in immune regulation mediated by N-glycans but also underscored the critical importance of sarcoplasmic reticulum Ca-ATPase (SRCA) in CVD. Additionally, they elucidated the contribution of phosphatidylethanolamines to SRCA function. These findings provide pivotal insights into the molecular mechanisms of MI. Furthermore, their multi-omics classifier exhibited exceptional performance in distinguishing AMI cases from healthy subjects, achieving an impressive AUC (Area Under the Curve) of 0.953. Dong et al. (332) employed an integrated approach, combining metabolomics and metagenomics analyses, and successfully identified four gut microbiota species (Alistipes, Streptococcus, Lactobacillus, Faecalibacterium), three critical fecal metabolites (formic acid, threonine, tyrosine), and two urinary metabolites (urea, lactulose) associated with MI. The combination of these factors yielded a set of potential, valuable, and non-invasive biomarkers for distinguishing AMI from stable CAD, with an AUC of 0.932. These findings provide a valuable predictive tool with significant clinical applications. Chan et al. (329) used proteomics and single-cell transcriptomics methods to identify several potential protein biomarkers for diagnosing HF in patients following MI. These biomarkers, including N-terminal B-type natriuretic peptide, cTnT, angiopoietin-2, thrombospondin-2, latent transforming growth factor-beta binding protein-4, and follistatin-related protein-3, hold promise for early HF diagnosis and treatment. Furthermore, the study elucidated the sources of these biomarkers, with NPPB and TNNT2 displaying the highest gene expression levels in cardiac muscle cells. In summary, integrating and analyzing multi-omics data holds significant promise in advancing early diagnosis and personalized medicine for MI. By analyzing multi-omics data, we can gain a more comprehensive understanding of this severe CVD, identify potential biomarkers and treatment targets, and thereby provide more accurate diagnoses, prognosis assessments, and personalized treatment plans.
4. Databases and online tools for multi-omics integrative analyses
4.1. Databases and knowledge bases
Numerous omics databases and knowledge bases offer researchers invaluable resources to integrate and analyze diverse biological data across various molecular levels, encompassing genomics, transcriptomics, proteomics, metabolomics, and more. These resources facilitate in-depth exploration of interactions between genes, proteins, metabolites, and offer comprehensive insights into biological pathways, disease mechanisms, and beyond. Table 3 presents a series of databases and knowledge bases available for integrating multi-omics data of MI. Among these, the CVD Knowledge Portal (CVDKP) (351), MI Knowledge Base (MIKB) (10), and HeartBioPortal (352) concentrate on multi-omics data pertaining to CVDs and MI, providing robust support for cardiovascular research. The Genome-Wide Association Study (GWAS) Catalog standardizes genomic association study data, streamlining analyses of genetic variations linked to traits (353). ArrayExpress serves as a repository for gene expression data, furnishing rich resources for the study of gene regulation and expression patterns (354). The GTEx project (355) directs attention toward tissue-specific gene expression, offering precious resources for comprehending gene functions in distinct tissues. Furthermore, the European Nucleotide Archive (ENA) (356) functions as a repository for nucleic acid sequencing data, playing a critical role in genomics and transcriptomics research by furnishing extensive datasets.
ProteomeXchange consortium (357), PeptideAtlas repository (358), Proteomics Identifications Database (PRIDE) (359), Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) (360), and Human Protein Atlas (362) supply ample proteomics and MS data, aiding researchers in probing protein structure and function. Human Cell Atlas (366), Single Cell Portal (SCP) (367), and Database of Deeply Integrated Single-Cell Omics data (DISCO) (368) zero in on single-cell genomics, providing pivotal platforms to uncover cellular heterogeneity and dynamic changes. Metabolomics WorkBench (363), MetaboLights (364), and Global Natural Products Social Molecular Networking (GNPS) (365) concentrate on exploring metabolomics data, underpinning research on metabolic pathways and bioactive molecules.
The National Center for Biotechnology Information (NCBI) (369) and the European Molecular Biology Laboratory European Bioinformatics Institute (EMBL-EBI) (370) stand as pivotal bioinformatics resource centers, pivotal in integrating, storing, and disseminating biological data, tools, and knowledge, thereby supporting interdisciplinary multi-omics analyses and in-depth exploration within the global life science research community. Search Tool for the Retrieval of Interaction Gene/Proteins (STRING) (371), Reactome (372), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (373) provide researchers with the means to delve deeply into protein interactions, pathways, and functional annotations, furnishing essential insights for investigating molecular mechanisms. The presence of these databases lends strong support to interdisciplinary research, propelling the cross-application and analysis of multi-omics data, yielding invaluable insights for life science research, and further advancing our comprehension of the intricate nature of biological systems.
4.2. Analysis tools
Integrative analysis of multi-omics data is a complex and vital task, involving combining information from different omics layers to uncover deeper biological insights. Researchers can use various tools, categorized into three main types: toolkits, desktop applications, and web-based tools. Toolkits are designed for skilled programmers and typically require installation and setup. They offer extensive functions and flexibility, allowing researchers to customize analysis workflows and settings. Common toolkits include Bioconductor (R language) (374), mixOmics (R language) (375), Seurat (R language) (376), scikit-learn (Python) (377), and scGREAT (Python) (378). However, using toolkits requires programming and data skills, making it challenging for non-experts. Users also need their own computing and storage resources. Desktop applications provide graphical interfaces, enabling non-programmers to conduct multi-omics analysis. For example, Taverna (379) and KNIME (380) offer graphical user interface-based workflow design for integrating diverse omics data. However, desktop apps might have limitations in analysis due to computing and data constraints.
Web-based online tools provide a convenient solution for conducting integrative analysis of multi-omics data. These tools don't require installation and can be accessed directly through web browsers. They typically feature user-friendly interfaces that guide users step-by-step through the analysis process using graphical interfaces, eliminating the need for programming skills. However, these online tools might have limitations due to server computational resources and data privacy concerns, potentially restricting their usability for large-scale data analysis. Users could encounter constraints related to computational resources, affecting the speed of analysis. Furthermore, as the analysis workflows are predefined, customization options for analyses could be limited. Table 4 shows several integrative multi-omics online tools along with their descriptions. For example, LDexpress (381) specializes in integrating population-specific linkage disequilibrium data and tissue-specific gene expression information to investigate the impact of germ cell lineage variations on gene expression and disease associations. Paitomics (382) integrates transcriptomics and metabolomics data, Quickomics (383) combines transcriptomics and proteomics data, and GeneTrail (384) encompasses genomics, transcriptomics, miRNAomics, proteomics, epigenetics, and single-cell data integration. TIMEOR (385) integrates genomics, transcriptomics, and proteomics data to unveil temporal changes in gene regulatory networks. Omics Integrator (386) uses network optimization algorithms to integrate proteomics, gene expression, and epigenetics data. Mergeomics (387) is used to combine various types of omics data to uncover disease pathways and potential drugs. These tools offer researchers a wide range of choices to explore and interpret multi-omics data, leading to profound insights into biological processes more easily. However, when choosing an appropriate tool, users should carefully consider factors like data scale, available computational resources, privacy requirements, and analysis objectives.
5. Conclusion and perspectives
MI is a complex CVD, and its occurrence and development are associated with pathological changes in multiple biological systems. Single-omic approaches, such as genomics, epigenomics, transcriptomics, proteomics, and metabolomics, have greatly advanced our understanding of the molecular mechanisms of MI. However, single-omics approaches possess limitations, as they fail to fully account for the complex interactions among molecules across various levels within the context of the disease. By integrating data from various omics approaches, multi-omics research goes beyond the limitations of single omics methods and addresses gaps in information, revealing a comprehensive understanding of the causes of MI from different perspectives. This approach helps us better understand the complete range of molecular changes in MI, which in turn assists in identifying important biomarkers for diagnosis and treatment. Importantly, multi-omics integration is not only effective in revealing disease mechanisms but also has significant potential in the field of drug therapy. Analyzing data across different omics layers allows us to gain deeper insights into how drugs work in treating MI. This aids in developing more personalized and precise treatment strategies, optimizing treatment effectiveness, and minimizing the risk of adverse reactions. As a result, multi-omics integration not only helps uncover the complex molecular mechanisms of MI but also strongly supports the advancement of precision medicine and individualized treatment approaches. This approach offers new perspectives and possibilities for better understanding, prevention, and treatment of this serious condition.
Currently, multi-omics studies on MI are predominantly focused on individual omics layers, lacking comprehensive integration This limitation hinders the realization of personalized medicine in MI. Furthermore, the diversity of data and the lack of standardization make cross-study comparisons and integrated analyses challenging. Simultaneously, the high-dimensionality of multi-omics data requires more advanced algorithms and models to uncover underlying correlations. To advance precise medical research on MI, future directions in multi-omics studies should emphasize the following aspects, as illustrated in Figure 2: Firstly, conducting large-scale, multicenter, standardized clinical studies is essential to gather extensive and diverse multi-omics data on MI. This approach enhances statistical power, robustness, and replicability of research outcomes. Secondly, the establishment of unified data standards and sharing platforms is crucial. This includes constructing a MI ontology, databases, and knowledge repositories. Such practices ensure data quality and consistency, enhancing comparability across different studies. Sharing platforms foster collaboration and information exchange among researchers, driving continuous progress in multi-omics studies of MI (398, 399). Thirdly, future research should delve deeper into multi-omics analyses. Beyond genomics, transcriptomics, proteomics, and metabolomics, other omics layers such as epigenomics, microbiomics, and clinical phenomics could be explored. Integrating data from these diverse layers can provide comprehensive information, offering stronger support for accurate MI diagnosis and individualized treatment. Fourthly, the development of more advanced computational models and algorithms is necessary to effectively handle large-scale multi-omics data. Technologies like machine learning and artificial intelligence can be widely applied to analyze and interpret multi-omics data. This aids in discovering potential biomarkers, predicting disease risks, and providing guidance for developing personalized medicine approaches for MI. Finally, medical advancement is progressing towards intelligence, especially in the realm of precision medicine. Given the intricate nature of genes and molecules, conventional manual analysis techniques fall short of meeting requirements. The forthcoming medical paradigm will lean on intelligent models fueled by data and guided by knowledge, facilitating swifter and more precise analysis of intricate multi-omics data. These models will support physicians in rendering accurate diagnoses and treatment determinations. Particularly in the case of complex ailments like MI, the implementation of intelligent models can assist medical professionals in enhancing their comprehension of disease mechanisms and devising tailored treatment approaches.
Author contributions
BS and CZ: contributed to conception and initial outline of the review. CZ, TT, EW, YuZ, MH, RW, and CB: wrote sections of the manuscript. BS, CZ, JW, and YiZ: contributed to manuscript content and language editing. All authors contributed to the conception of the work, manuscript revision, and approved the submitted version. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32270690 and 32070671).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Li Y, Cao GY, Jing WZ, Liu J, Liu M. Global trends and regional differences in incidence and mortality of cardiovascular disease, 1990–2019: findings from 2019 global burden of disease study. Eur J Prev Cardiol. (2023) 30(3):276–86. doi: 10.1093/eurjpc/zwac285
2. Tsao CW, Aday AW, Almarzooq ZI, Anderson CAM, Arora P, Avery CL, et al. Heart disease and stroke statistics-2023 update: a report from the American heart association. Circulation. (2023) 147(8):e93–e621. doi: 10.1161/cir.0000000000001123
3. Desai R, Mishra V, Chhina AK, Jain A, Vyas A, Allamneni R, et al. Cardiovascular disease risk factors and outcomes of acute myocardial infarction in young adults: evidence from 2 nationwide cohorts in the United States a decade apart. Curr Probl Cardiol. (2023) 48(9):101747. doi: 10.1016/j.cpcardiol.2023.101747
4. Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA, et al. Fourth universal definition of myocardial infarction. Circulation. (2018) 138(20):e618–e51. doi: 10.1161/cir.0000000000000617
5. Oualha D, Ben Abderrahim S, Ben Abdeljelil N, BelHadj M, Ben Jomâa S, Saadi S, et al. Cardiac rupture during acute myocardial infarction: autopsy study (2004–2020). Ann Cardiol Angeiol. (2023) 72(3):101601. doi: 10.1016/j.ancard.2023.101601
6. Harrington J, Butler J. Heart failure after myocardial infarction: glass emptier than full. Eur J Heart Fail. (2023) 25(8):1225–7. doi: 10.1002/ejhf.2961
7. Koivunen M, Tynkkynen J, Oksala N, Eskola M, Hernesniemi J. Incidence of sudden cardiac arrest and sudden cardiac death after unstable angina pectoris and myocardial infarction. Am Heart J. (2023) 257:9–19. doi: 10.1016/j.ahj.2022.11.009
8. Green ED, Watson JD, Collins FS. Human genome project: twenty-five years of big biology. Nature. (2015) 526(7571):29–31. doi: 10.1038/526029a
9. Consortium EP. The ENCODE (ENCyclopedia of DNA elements) project. Science. (2004) 306(5696):636–40. doi: 10.1126/science.1105136
10. Zhan C, Zhang Y, Liu X, Wu R, Zhang K, Shi W, et al. MIKB: a manually curated and comprehensive knowledge base for myocardial infarction. Comput Struct Biotechnol J. (2021) 19:6098–107. doi: 10.1016/j.csbj.2021.11.011
11. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. (1977) 74(12):5463–7. doi: 10.1073/pnas.74.12.5463
12. Bumgarner R. Overview of DNA microarrays: types, applications, and their future. Curr Protoc Mol Biol. (2013) Chapter 22:Unit 22.1. doi: 10.1002/0471142727.mb2201s101
13. van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. (2014) 30(9):418–26. doi: 10.1016/j.tig.2014.07.001
14. Leu HB, Chung CM, Chen JW, Pan WH. The Mediterranean diet reduces the genetic risk of chromosome 9p21 for myocardial infarction in an Asian population community cohort. Sci Rep. (2019) 9(1):18405. doi: 10.1038/s41598-019-54938-w
15. Tibaut M, Naji F, Petrovič D. Association of myocardial infarction with CDKN2B antisense RNA 1 (CDKN2B-AS1) rs1333049 polymorphism in slovenian subjects with type 2 diabetes mellitus. Genes. (2022) 13(3):526. doi: 10.3390/genes13030526
16. Zhang Q, Cheng SS, Yan J, Chen XD, Wang XF, Yang XC. Association between methylthioadenosine phosphorylase gene single nucleotide polymorphisms and myocardial infarction in Chinese Han ethnicity. Zhonghua Liu Xing Bing Xue Za Zhi. (2010) 31(1):83–6.20302706
17. Li Y, Zhang D, Zhang Y, Xu X, Bi L, Zhang M, et al. Association of lncRNA polymorphisms with triglyceride and total cholesterol levels among myocardial infarction patients in Chinese population. Gene. (2020) 724:143684. doi: 10.1016/j.gene.2019.02.085
18. Li R, Zhang X, Yin W, Wang Y, Liu Y. Common genetic variants on chromosome 9p21 confers risk of ischemic stroke: a large-scale genetic association study. Cell Mol Biol. (2021) 67(2):132–7. doi: 10.14715/cmb/2021.67.2.20
19. Mobeen Zafar M, Saqlain M, Mehmood Raja A, Arzoo Shaiq P, Javaid Asad M, Nawaz Shah MK, et al. 9p21 locus polymorphism is a strong predictor of metabolic syndrome and cardiometabolic risk phenotypes regardless of coronary heart disease. Genes. (2022) 13(12):2226. doi: 10.3390/genes13122226
20. Li YY, Wang H, Wang H, Zhang YY. Myocardial infarction and AGT p.Thr174Met polymorphism: a meta-analysis of 7657 subjects. Cardiovasc Ther. (2021) 2021:6667934. doi: 10.1155/2021/6667934
21. Ozturk O, Ozturk U, Nergiz S, Karahan MZ. The relationship between angiotensin-II type 1 receptor gene polymorphism and repolarization rarameters after a first anterior acute myocardial infarction. Korean Circ J. (2016) 46(6):791–7. doi: 10.4070/kcj.2016.46.6.791
22. Aoki S, Mukae S, Itoh S, Sato R, Nishio K, Ueda H, et al. Genetic background in patients with acute myocardial infarction. Jpn Heart J. (2001) 42(1):15–28. doi: 10.1536/jhj.42.15
23. Dai SH, Li JF, Feng JB, Li RJ, Li CB, Li Z, et al. Association of serum levels of AngII, KLK1, and ACE/KLK1 polymorphisms with acute myocardial infarction induced by coronary artery stenosis. Jo Renin Angiotensin Aldosterone Syst. (2016) 17(2):1470320316655037. doi: 10.1177/1470320316655037
24. Damar İ H, Eroz R. The association of hereditary prothrombotic risk factors with ST-elevation myocardial infarction. Medeni Med J. (2020) 35(4):295–303. doi: 10.5222/mmj.2020.67366
25. Yang W, Huang W, Su S, Li B, Zhao W, Chen S, et al. Association study of ACE2 (angiotensin I-converting enzyme 2) gene polymorphisms with coronary heart disease and myocardial infarction in a Chinese Han population. Clin Sci. (2006) 111(5):333–40. doi: 10.1042/cs20060020
26. Cambien F, Costerousse O, Tiret L, Poirier O, Lecerf L, Gonzales MF, et al. Plasma level and gene polymorphism of angiotensin-converting enzyme in relation to myocardial infarction. Circulation. (1994) 90(2):669–76. doi: 10.1161/01.cir.90.2.669
27. Hmimech W, Idrissi HH, Diakite B, Korchi F, Baghdadi D, Tahri Joutey Hassani Idrissi H, et al. Impact of I/D polymorphism of angiotensin-converting enzyme (ACE) gene on myocardial infarction susceptibility among young Moroccan patients. BMC Res Notes. (2017) 10(1):763. doi: 10.1186/s13104-017-3039-1
28. Dai S, Ding M, Liang N, Li Z, Li D, Guan L, et al. Associations of ACE I/D polymorphism with the levels of ACE, kallikrein, angiotensin II and interleukin-6 in STEMI patients. Sci Rep. (2019) 9(1):19719. doi: 10.1038/s41598-019-56263-8
29. Žaliaduonytė-Pekšienė D, Lesauskaitė V, Liutkevičienė R, Tamakauskas V, Kviesulaitis V, Šinkūnaitė-Maršalkienė G, et al. Association of the genetic and traditional risk factors of ischaemic heart disease with STEMI and NSTEMI development. J Renin Angiotensin Aldosterone Syst. (2017) 18(4):1470320317739987. doi: 10.1177/1470320317739987
30. Chen Y, Dong S, He M, Qi T, Zhu W. Angiotensin-converting enzyme insertion/deletion polymorphism and risk of myocardial infarction in an updated meta-analysis based on 34993 participants. Gene. (2013) 522(2):196–205. doi: 10.1016/j.gene.2013.03.076
31. Karahan Z, Ugurlu M, Ucaman B, Veysel Ulug A, Kaya I, Cevik K, et al. Association between ACE gene polymorphism and QT dispersion in patients with acute myocardial infarction. Open Cardiovasc Med J. (2016) 10:117–21. doi: 10.2174/1874192401610010117
32. Martínez-Quintana E, Chirino R, Nieto-Lago V, Pérez-Jiménez P, López-Ríos L, Rodríguez-González F. Prognostic value of ACE I/D, AT1R A1166C, PAI-I 4G/5G and GPIIIa a1/a2 polymorphisms in myocardial infarction. Cardiol J. (2014) 21(3):229–37. doi: 10.5603/CJ.a2013.0107
33. Siegerink B, Rosendaal FR, Algra A. Genetic variation in fibrinogen; its relationship to fibrinogen levels and the risk of myocardial infarction and ischemic stroke. J ThrombHaemost. (2009) 7(3):385–90. doi: 10.1111/j.1538-7836.2008.03266.x
34. Rallidis LS, Gialeraki A, Tsirebolos G, Tsalavoutas S, Rallidi M, Iliodromitis E. Prothrombotic genetic risk factors in patients with very early ST-segment elevation myocardial infarction. J Thromb Thrombolysis. (2017) 44(2):267–73. doi: 10.1007/s11239-017-1520-2
35. Huang H, Jia S, Chen S, Sha Y, Ma A, Ma X, et al. The coagulation factor VII gene polymorphisms in patients with myocardial infarction in Ningxia Hui and Han populations. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2009) 26(6):653–8. doi: 10.3760/cma.j.issn.1003-9406.2009.06.010
36. Schneppenheim R, Hellermann N, Brehm MA, Klemm U, Obser T, Huck V, et al. The von Willebrand factor Tyr2561 allele is a gain-of-function variant and a risk factor for early myocardial infarction. Blood. (2019) 133(4):356–65. doi: 10.1182/blood-2018-04-843425
37. Abd El-Aziz TA, Rezk NA. Relation of PAI-1 and TPA genes polymorphisms to acute myocardial infarction and its outcomes in Egyptian patients. Cell Biochem Biophys. (2015) 71(1):227–34. doi: 10.1007/s12013-014-0188-x
38. Osmak GJ, Sidko AR, Kiselev IS, Favorova OO. Age-dependent approach to search for genetic variants associated with myocardial infarction. Mol Biol. (2020) 54(4):699–704. doi: 10.31857/s0026898420040138
39. Semaev S, Shakhtshneider E, Shcherbakova L, Orlov P, Ivanoshchuk D, Malyutina S, et al. Association of common variants of APOE, CETP, and the 9p21.3 chromosomal region with the risk of myocardial infarction: a prospective study. Int J Mol Sci. (2023) 24(13):10908. doi: 10.3390/ijms241310908
40. He K, Zhu Z, Chen Y. Lipoprotein lipase gene polymorphisms are associated with myocardial infarction risk: a meta-analysis. Genet Test Mol Biomarkers. (2021) 25(6):434–44. doi: 10.1089/gtmb.2021.0042
41. Hashad IM, Nosseir H, Shaban GM, Abdel Rahman MF, Gad MZ. Is there a correlation between -174(G/C) polymorphism of IL-6 gene and the incidence of acute myocardial infarction? J Genet Eng Biotechnol. (2021) 19(1):139. doi: 10.1186/s43141-021-00243-x
42. Hua XP, Qian J, Cao CB, Xie J, Zeng XT, Zhang ZJ. Association between TNF-α rs1800629 polymorphism and the risk of myocardial infarction: a meta-analysis. Genet Mol Res. (2016) 15(3). doi: 10.4238/gmr.15037292
43. Pinheiro-Júnior S, Pinhel MA, Nakazone MA, Pinheiro A, Amorim GF, Florim GM, et al. Effect of genetic variants related to lipid metabolism as risk factors for cholelithiasis after bariatric surgery in Brazilian population. Obes Surg. (2012) 22(4):623–33. doi: 10.1007/s11695-012-0590-7
44. Levstek T, Podkrajšek N, Rehberger Likozar A, Šebeštjen M, Trebušak Podkrajšek K. The influence of treatment with PCSK9 inhibitors and variants in the CRP (rs1800947), TNFA (rs1800629), and IL6 (rs1800795) genes on the corresponding inflammatory markers in patients with very high lipoprotein(a) levels. J Cardiovasc Dev Dis. (2022) 9(5):127. doi: 10.3390/jcdd9050127
45. Ellman N, Keswell D, Collins M, Tootla M, Goedecke JH. Ethnic differences in the association between lipid metabolism genes and lipid levels in black and white South African women. Atherosclerosis. (2015) 240(2):311–7. doi: 10.1016/j.atherosclerosis.2015.03.027
46. Qi L, Ma J, Qi Q, Hartiala J, Allayee H, Campos H. Genetic risk score and risk of myocardial infarction in Hispanics. Circulation. (2011) 123(4):374–80. doi: 10.1161/circulationaha.110.976613
47. Patel RS, Sun YV, Hartiala J, Veledar E, Su S, Sher S, et al. Association of a genetic risk score with prevalent and incident myocardial infarction in subjects undergoing coronary angiography. Circ Cardiovasc Genet. (2012) 5(4):441–9. doi: 10.1161/circgenetics.111.960229
48. Wu Z, Sheng H, Su X, Gao X, Lu L, Jin W. Mediating effect of diabetes mellitus on the association between chromosome 9p21.3 locus and myocardial infarction risk: a case-control study in Shanghai, China. Front Endocrinol. (2018) 9:362. doi: 10.3389/fendo.2018.00362
49. Yadav AK, Chakkumkollath AK, Helna A, Birla S, Thimmulappa RK, Shambu SK, et al. Substantiation of a clopidogrel metabolism-associated gene (CYP2C19) variation among healthy individuals. Indian Heart J. (2023) 75(5):343–6. doi: 10.1016/j.ihj.2023.05.005
50. Ma Q, Chen GZ, Zhang YH, Zhang L, Huang LA. Clinical outcomes and predictive model of platelet reactivity to clopidogrel after acute ischemic vascular events. Chin Med J. (2019) 132(9):1053–62. doi: 10.1097/cm9.0000000000000210
51. de Freitas Campos EI, Gomes KB, Ribeiro DD, Puurunen MK, Oliveira Magalhães Mourão A, Ferreira IG, et al. Influence of polymorphisms in CYP2C9, VKORC1, MDR1 and APOE genes on the warfarin maintenance dose in Brazilian patients. Pharmacogenomics. (2023) 24(13):701–12. doi: 10.2217/pgs-2023-0099
52. Liu TY, Hsu HY, You YS, Hsieh YW, Lin TC, Peng CW, et al. Efficacy of warfarin therapy guided by pharmacogenetics: a real-world investigation among Han Taiwanese. Clin Ther. (2023) 45(7):662–70. doi: 10.1016/j.clinthera.2023.04.006
53. Oliveira FF, Almeida SS, Chen ES, Smith MC, Bertolucci PHF. Pharmacogenetics of angiotensin modulators according to APOE-ɛ4 alleles and the ACE insertion/deletion polymorphism in Alzheimer's Disease. Acta Neuropsychiatr. (2023):1–16. doi: 10.1017/neu.2023.38. [Epub ahead of print]37605989
54. Su X, Lee L, Li X, Lv J, Hu Y, Zhan S, et al. Association between angiotensinogen, angiotensin II receptor genes, and blood pressure response to an angiotensin-converting enzyme inhibitor. Circulation. (2007) 115(6):725–32. doi: 10.1161/circulationaha.106.642058
55. Maree AO, Vangjeli C, Jneid H, Ryan J, Cox D, Cannon CP, et al. G-protein beta3 subunit polymorphism and bleeding in the orbofiban in patients with unstable coronary syndromes-thrombolysis in myocardial infarction 16 trial. J Thromb Haemost. (2010) 8(5):934–41. doi: 10.1111/j.1538-7836.2010.03775.x
56. Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. (2010) 28(10):1057–68. doi: 10.1038/nbt.1685
57. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. (2013) 38(1):23–38. doi: 10.1038/npp.2012.112
58. Agha G, Mendelson MM, Ward-Caviness CK, Joehanes R, Huan T, Gondalia R, et al. Blood leukocyte DNA methylation predicts risk of future myocardial infarction and coronary heart disease. Circulation. (2019) 140(8):645–57. doi: 10.1161/circulationaha.118.039357
59. Haase T, Müller C, Krause J, Röthemeier C, Stenzig J, Kunze S, et al. Novel DNA methylation sites influence GPR15 expression in relation to smoking. Biomolecules. (2018) 8(3):74. doi: 10.3390/biom8030074
60. Pan B, Wang X, Nishioka C, Honda G, Yokoyama A, Zeng L, et al. G-protein coupled receptor 15 mediates angiogenesis and cytoprotective function of thrombomodulin. Sci Rep. (2017) 7(1):692. doi: 10.1038/s41598-017-00781-w
61. Haase T, Müller C, Stoffers B, Kirn P, Waldenberger M, Kaiser FJ, et al. G protein-coupled receptor 15 expression is associated with myocardial infarction. Int J Mol Sci. (2022) 24(1):180. doi: 10.3390/ijms24010180
62. Lilova Z, Hassan F, Riaz M, Ironside J, Ken-Dror G, Han T, et al. Blood group and ischemic stroke, myocardial infarction, and peripheral vascular disease: a meta-analysis of over 145,000 cases and 2,000,000 controls. J Stroke Cerebrovasc Dis. (2023) 32(8):107215. doi: 10.1016/j.jstrokecerebrovasdis.2023.107215
63. Pang H, Zong Z, Hao L, Cao Q. ABO blood group influences risk of venous thromboembolism and myocardial infarction. J Thromb Thrombolysis. (2020) 50(2):430–8. doi: 10.1007/s11239-019-02012-7
64. Kominato Y, Hata Y, Takizawa H, Tsuchiya T, Tsukada J, Yamamoto F. Expression of human histo-blood group ABO genes is dependent upon DNA methylation of the promoter region. J Biol Chem. (1999) 274(52):37240–50. doi: 10.1074/jbc.274.52.37240
65. Yousuf FA, Kazmi K, Iqbal J, Ahmed N, Iqbal MP. Higher DNA methylation of ABO gene promoter is associated with acute myocardial infarction in a hospital-based population in Karachi. Pak J Med Sci. (2020) 36(3):505–10. doi: 10.12669/pjms.36.3.1406
66. Shao M, Lyu XP, Yang QK, Zhu WT, Song J, Kong YK, et al. Effects of DNA methylation on ABO gene expression in leukemia. Zhonghua Xue Ye Xue Za Zhi. (2016) 37(9):795–9. doi: 10.3760/cma.j.issn.0253-2727.2016.09.013
67. Fernández-Sanlés A, Sayols-Baixeras S, Subirana I, Sentí M, Pérez-Fernández S, de Castro Moura M, et al. DNA methylation biomarkers of myocardial infarction and cardiovascular disease. Clin Epigenetics. (2021) 13(1):86. doi: 10.1186/s13148-021-01078-6
68. Wang C, Ni W, Yao Y, Just A, Heiss J, Wei Y, et al. DNA methylation-based biomarkers of age acceleration and all-cause death, myocardial infarction, stroke, and cancer in two cohorts: the NAS, and KORA F4. EBioMedicine. (2021) 63:103151. doi: 10.1016/j.ebiom.2020.103151
69. Boovarahan SR, AlAsmari AF, Ali N, Khan R, Kurian GA. Targeting DNA methylation can reduce cardiac injury associated with ischemia reperfusion: one step closer to clinical translation with blood-borne assessment. Front Cardiovasc Med. (2022) 9:1021909. doi: 10.3389/fcvm.2022.1021909
70. Zou Y, Li L, Li Y, Chen S, Xie X, Jin X, et al. Restoring cardiac functions after myocardial infarction-ischemia/reperfusion via an exosome anchoring conductive hydrogel. ACS Appl Mater Interfaces. (2021) 13(48):56892–908. doi: 10.1021/acsami.1c16481
71. Zhang S, Xie H, Du Y, Wang B, Lan B, Wang H. DNMT1-induced miR-133b suppression via methylation promotes myocardial fibrosis after myocardial infarction. Gen Physiol Biophys. (2023) 42(5):417–29. doi: 10.4149/gpb_2023018
72. Shaikh NI, Sethi RS. Impairment of apoptosis pathway via Apaf1 downregulation during chlorpyrifos and/or cypermethrin induced lung damage. Anim Biotechnol. (2023) 34(3):738–45. doi: 10.1080/10495398.2021.1981918
73. Jin G, Zheng J, Zhang Y, Yang Z, Chen Y, Huang C. LncRNA UCA1 epigenetically suppresses APAF1 expression to mediate the protective effect of sevoflurane against myocardial ischemia-reperfusion injury. Funct Integr Genomics. (2022) 22(5):965–75. doi: 10.1007/s10142-022-00874-4
74. Li M, Jiao L, Shao Y, Li H, Sun L, Yu Q, et al. LncRNA-ZFAS1 promotes myocardial ischemia-reperfusion injury through DNA methylation-mediated Notch1 down-regulation in mice. JACC Basic Transl Sci. (2022) 7(9):880–95. doi: 10.1016/j.jacbts.2022.06.004
75. Fernández-Carrión R, Sorlí JV, Asensio EM, Pascual EC, Portolés O, Alvarez-Sala A, et al. DNA-methylation signatures of tobacco smoking in a high cardiovascular risk population: modulation by the Mediterranean diet. Int J Environ Res Public Health. (2023) 20(4):3635. doi: 10.3390/ijerph20043635
76. Shan M, Li S, Zhang Y, Chen Y, Zhou Y, Shi L. Maternal exercise upregulates the DNA methylation of Agtr1a to enhance vascular function in offspring of hypertensive rats. Hypertens Res. (2023) 46(3):654–66. doi: 10.1038/s41440-022-01124-7
77. Gillman AS, Gardiner CK, Koljack CE, Bryan AD. Body mass index, diet, and exercise: testing possible linkages to breast cancer risk via DNA methylation. Breast Cancer Res Treat. (2018) 168(1):241–8. doi: 10.1007/s10549-017-4573-1
78. Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, et al. The structural basis of modified nucleosome recognition by 53BP1. Nature. (2016) 536(7614):100–3. doi: 10.1038/nature18951
79. Taberlay PC, Achinger-Kawecka J, Lun AT, Buske FA, Sabir K, Gould CM, et al. Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations. Genome Res. (2016) 26(6):719–31. doi: 10.1101/gr.201517.115
80. Yuan J, Peng H, Mo B, Yin C, Fang G, Li Y, et al. Inhibition of Wdr5 attenuates ang-II-induced fibroblast-to-myofibroblast transition in cardiac fibrosis by regulating Mdm2/P53/P21 pathway. Biomolecules. (2022) 12(11):1574. doi: 10.3390/biom12111574
81. Long F, Wang Q, Yang D, Zhu M, Wang J, Zhu Y, et al. Targeting JMJD3 histone demethylase mediates cardiac fibrosis and cardiac function following myocardial infarction. Biochem Biophys Res Commun. (2020) 528(4):671–7. doi: 10.1016/j.bbrc.2020.05.115
82. Yang J, Wang B, Li N, Zhou Q, Zhou W, Zhan Z. Salvia miltiorrhiza and Carthamus tinctorius extract prevents cardiac fibrosis and dysfunction after myocardial infarction by epigenetically inhibiting smad3 expression. Evid Based Complement Alternat Med. (2019) 2019:6479136. doi: 10.1155/2019/6479136
83. Wang P, Fan F, Li X, Sun X, Ma L, Wu J, et al. Riboflavin attenuates myocardial injury via LSD1-mediated crosstalk between phospholipid metabolism and histone methylation in mice with experimental myocardial infarction. J Mol Cell Cardiol. (2018) 115:115–29. doi: 10.1016/j.yjmcc.2018.01.006
84. Cao W, Feng Z, Zhu D, Li S, Du M, Ye S, et al. The role of PGK1 in promoting ischemia/reperfusion injury-induced microglial M1 polarization and inflammation by regulating glycolysis. NeuroMol Med. (2023) 25(2):301–11. doi: 10.1007/s12017-023-08736-3
85. Kang H. Regulation of acetylation states by nutrients in the inhibition of vascular inflammation and atherosclerosis. Int J Mol Sci. (2023) 24(11):9338. doi: 10.3390/ijms24119338
86. Li X, Wei C, Zhang Z, Jin Q, Xiao X. MiR-134-5p regulates myocardial apoptosis and angiogenesis by directly targeting KDM2A after myocardial infarction. Int Heart J. (2020) 61(4):815–21. doi: 10.1536/ihj.19-468
87. Hao S, Sui X, Wang J, Zhang J, Pei Y, Guo L, et al. Secretory products from epicardial adipose tissue induce adverse myocardial remodeling after myocardial infarction by promoting reactive oxygen species accumulation. Cell Death Dis. (2021) 12(9):848. doi: 10.1038/s41419-021-04111-x
88. Sunagawa Y, Katayama A, Funamoto M, Shimizu K, Shimizu S, Sari N, et al. The polyunsaturated fatty acids, EPA and DHA, ameliorate myocardial infarction-induced heart failure by inhibiting p300-HAT activity in rats. J Nutr Biochem. (2022) 106:109031. doi: 10.1016/j.jnutbio.2022.109031
89. Katagiri T, Sunagawa Y, Maekawa T, Funamoto M, Shimizu S, Shimizu K, et al. Ecklonia stolonifera Okamura extract suppresses myocardial infarction-induced left ventricular systolic dysfunction by inhibiting p300-HAT activity. Nutrients. (2022) 14(3):580. doi: 10.3390/nu14030580
90. Lei I, Tian S, Gao W, Liu L, Guo Y, Tang P, et al. Acetyl-CoA production by specific metabolites promotes cardiac repair after myocardial infarction via histone acetylation. eLife. (2021) 23:10:e60311. doi: 10.10.7554/eLife.60311
91. Wang L, Du A, Lu Y, Zhao Y, Qiu M, Su Z, et al. Peptidase inhibitor 16 attenuates left ventricular injury and remodeling after myocardial infarction by inhibiting the HDAC1-Wnt3a-β-Catenin signaling sxis. J Am Heart Assoc. (2023) 12(10):e028866. doi: 10.1161/jaha.122.028866
92. Weng L, Ye J, Yang F, Jia S, Leng M, Jia B, et al. TGF-β1/SMAD3 regulates programmed cell death 5 that suppresses cardiac fibrosis post-myocardial infarction by inhibiting HDAC3. Circ Res. (2023) 133(3):237–51. doi: 10.1161/circresaha.123.322596
93. Jiang H, Jia D, Zhang B, Yang W, Dong Z, Sun X, et al. Exercise improves cardiac function and glucose metabolism in mice with experimental myocardial infarction through inhibiting HDAC4 and upregulating GLUT1 expression. Basic Res Cardiol. (2020) 115(3):28. doi: 10.1007/s00395-020-0787-1
94. Yang L, Liu N, Zhao W, Li X, Han L, Zhang Z, et al. Angiogenic function of astragaloside IV in rats with myocardial infarction occurs via the PKD1-HDAC5-VEGF pathway. Exp Ther Med. (2019) 17(4):2511–8. doi: 10.3892/etm.2019.7273
95. Nagata S, Marunouchi T, Tanonaka K. Histone deacetylase inhibitor SAHA treatment prevents the development of heart failure after myocardial infarction via an induction of heat-shock proteins in rats. Biol Pharm Bull. (2019) 42(3):453–61. doi: 10.1248/bpb.b18-00785
96. Liu F, Di Y, Ma W, Kang X, Li X, Ji Z. HDAC9 Exacerbates myocardial infarction via inactivating Nrf2 pathways. J Pharm Pharmacol. (2022) 74(4):565–72. doi: 10.1093/jpp/rgab065
97. Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. A survey of best practices for RNA-Seq data analysis. Genome Biol. (2016) 17:13. doi: 10.1186/s13059-016-0881-8
98. Jiang Z, Zhou X, Li R, Michal JJ, Zhang S, Dodson MV, et al. Whole transcriptome analysis with sequencing: methods, challenges and potential solutions. Cell Mol Life Sci. (2015) 72(18):3425–39. doi: 10.1007/s00018-015-1934-y
99. Sheng X, Fan T, Jin X. Identification of key genes involved in acute myocardial infarction by comparative transcriptome analysis. BioMed Res Int. (2020) 2020:1470867. doi: 10.1155/2020/1470867
100. You H, Dong M. Identification of immuno-inflammation-related biomarkers for acute myocardial infarction based on bioinformatics. J Inflamm Res. (2023) 16:3283–302. doi: 10.2147/jir.S421196
101. Zhuo LA, Wen YT, Wang Y, Liang ZF, Wu G, Nong MD, et al. LncRNA SNHG8 is identified as a key regulator of acute myocardial infarction by RNA-Seq analysis. Lipids Health Dis. (2019) 18(1):201. doi: 10.1186/s12944-019-1142-0
102. Deng B, Zhang X, Liang Y, Jiang H, Huang W, Wu Y, et al. Nonadherent culture method promotes MSC-mediated vascularization in myocardial infarction via miR-519d/VEGFA pathway. Stem Cell Res Ther. (2020) 11(1):266. doi: 10.1186/s13287-020-01780-x
103. Kaplan A, Abidi E, Diab R, Ghali R, Al-Awassi H, Booz GW, et al. Sex differences in cardiac remodeling post myocardial infarction with acute cigarette smoking. Biol Sex Differ. (2022) 13(1):36. doi: 10.1186/s13293-022-00446-y
104. Anderson DR, Poterucha JT, Mikuls TR, Duryee MJ, Garvin RP, Klassen LW, et al. IL-6 and its receptors in coronary artery disease and acute myocardial infarction. Cytokine. (2013) 62(3):395–400. doi: 10.1016/j.cyto.2013.03.020
105. Xiao S, Zhou Y, Wu Q, Liu Q, Chen M, Zhang T, et al. FCER1G And PTGS2 serve as potential diagnostic biomarkers of acute myocardial infarction based on integrated bioinformatics analyses. DNA Cell Biol. (2021) 40(8):1064–75. doi: 10.1089/dna.2020.6447
106. Zivotić I, Djurić T, Stanković A, Milasinovic D, Stankovic G, Dekleva M, et al. CDKN2B Gene expression is affected by 9p21.3 rs10757278 in CAD patients, six months after the MI. Clin Biochem. (2019) 73:70–6. doi: 10.1016/j.clinbiochem.2019.08.003
107. Lalem T, Zhang L, Scholz M, Burkhardt R, Saccheti V, Teren A, et al. Cyclin dependent kinase inhibitor 1 C is a female-specific marker of left ventricular function after acute myocardial infarction. Int J Cardiol. (2019) 274:319–25. doi: 10.1016/j.ijcard.2018.07.042
108. Zhang P, Shao L, Ma J. Toll-like receptors 2 and 4 predict new-onset atrial fibrillation in acute myocardial infarction patients. Int Heart J. (2018) 59(1):64–70. doi: 10.1536/ihj.17-084
109. Wang R, Zhang SF, Ma YT, Chen D, Ma J, Zhou YM, et al. Expression of tissue factor in monocytes and plasma during acute phase and recovery stage of patients with myocardial infarction. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue. (2011) 23(6):372–3.21672391
110. Li H, Jia X, Bai YQ, Wu P, Guo HL, Yun KM, et al. Gene expression profiles at different time points after acute myocardial infarction in mice. Fa Yi Xue Za Zhi. (2022) 38(3):343–9. doi: 10.12116/j.issn.1004-5619.2021.410505
111. Zhang Y, Zhong P, Xu Y, Wang B, Zhu T, Zhang W, et al. Differential expression of TXNIP isoforms in the peripheral leukocytes of patients with acute myocardial infarction. Dis Markers. (2018) 2018:9051481. doi: 10.1155/2018/9051481
112. Gobbi G, Carubbi C, Tagliazucchi GM, Masselli E, Mirandola P, Pigazzani F, et al. Sighting acute myocardial infarction through platelet gene expression. Sci Rep. (2019) 9(1):19574. doi: 10.1038/s41598-019-56047-0
113. Yu S, Qian H, Tian D, Yang M, Li D, Xu H, et al. Linggui zhugan decoction activates the SIRT1-AMPK-PGC1α signaling pathway to improve mitochondrial and oxidative damage in rats with chronic heart failure caused by myocardial infarction. Front Pharmacol. (2023) 14:1074837. doi: 10.3389/fphar.2023.1074837
114. Wang S, Cao N. Uncovering potential differentially expressed miRNAs and targeted mRNAs in myocardial infarction based on integrating analysis. Mol Med Rep. (2020) 22(5):4383–95. doi: 10.3892/mmr.2020.11517
115. Cederström S, Lundman P, Folkersen L, Paulsson-Berne G, Karadimou G, Eriksson P, et al. New candidate genes for ST-elevation myocardial infarction. J Intern Med. (2020) 287(1):66–77. doi: 10.1111/joim.12976
116. Zhang Y, Guo Z, Wu T, Liu J, Zhang B, Lai W, et al. SULT2B1b Inhibits reverse cholesterol transport and promotes cholesterol accumulation and inflammation in lymphocytes from AMI patients with low LDL-C levels. Clin Sci. (2020) 134(2):273–87. doi: 10.1042/cs20190459
117. Zhou L, Kou DQ. Correlation between acute myocardial infarction complicated with cerebral infarction and expression levels of MMP-2 and MMP-9. Eur Rev Med Pharmacol Sci. (2019) 23(1):297–302. doi: 10.26355/eurrev_201901_16776
118. Boileau A, Lalem T, Vausort M, Zhang L, Devaux Y. A 3-gene panel improves the prediction of left ventricular dysfunction after acute myocardial infarction. Int J Cardiol. (2018) 254:28–35. doi: 10.1016/j.ijcard.2017.10.109
119. Sharma AK, Bisht P, Gupta B, Sayeed Akhtar MD, Shaik Alavudeen S, Afzal O, et al. Investigating miRNA subfamilies: can they assist in the early diagnosis of acute myocardial infarction? Drug Discov Today. (2023) 28(10):103695. doi: 10.1016/j.drudis.2023.103695
120. Li J, Ma H, Wu X, Sun G, Yang P, Peng Y, et al. Promising roles of non-exosomal and exosomal non-coding RNAs in the regulatory mechanism and as diagnostic biomarkers in myocardial infarction. J Zhejiang Univ Sci B. (2023) 24(4):281–300. doi: 10.1631/jzus.B2200459
121. Li J, Han Y, Wang S, Wu X, Cao J, Sun T. Circular RNAs: biogenesis, biological functions, and roles in myocardial infarction. Int J Mol Sci. (2023) 24(4):4233. doi: 10.3390/ijms24044233
122. Almaghrbi H, Giordo R, Pintus G, Zayed H. Non-coding RNAs as biomarkers of myocardial infarction. Clin Chim Acta. (2023) 540:117222. doi: 10.1016/j.cca.2023.117222
123. Fadaei S, Zarepour F, Parvaresh M, Motamedzadeh A, Tamehri Zadeh SS, Sheida A, et al. Epigenetic regulation in myocardial infarction: non-coding RNAs and exosomal non-coding RNAs. Front Cardiovasc Med. (2022) 9:1014961. doi: 10.3389/fcvm.2022.1014961
124. Zhang L, Ding H, Zhang Y, Wang Y, Zhu W, Li P. Circulating microRNAs: biogenesis and clinical significance in acute myocardial infarction. Front Physiol. (2020) 11:1088. doi: 10.3389/fphys.2020.01088
125. Xie L, Zhang Q, Mao J, Zhang J, Li L. The roles of lncRNA in myocardial infarction: molecular mechanisms, diagnosis biomarkers, and therapeutic perspectives. Front Cell Dev Biol. (2021) 9:680713. doi: 10.3389/fcell.2021.680713
126. Mompeón A, Ortega-Paz L, Vidal-Gómez X, Costa TJ, Pérez-Cremades D, Garcia-Blas S, et al. Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: a systematic and paired comparative analysis. Sci Rep. (2020) 10(1):5373. doi: 10.1038/s41598-020-61507-z
127. Pinchi E, Frati P, Aromatario M, Cipolloni L, Fabbri M, La Russa R, et al. miR-1, miR-499 and miR-208 are sensitive markers to diagnose sudden death due to early acute myocardial infarction. J Cell Mol Med. (2019) 23(9):6005–16. doi: 10.1111/jcmm.14463
128. Zhao Y, Zhuang L, Tian P, Ma M, Wu G, Zhang Y. Rapid diagnosis of acute myocardial infarction based on reverse transcription-accelerated strand exchange amplification of miR-208a. Anal Methods. (2023) 15(35):4442–51. doi: 10.1039/d3ay01116j
129. Xu L, Tian L, Yan Z, Wang J, Xue T, Sun Q. Diagnostic and prognostic value of miR-486-5p, miR-451a, miR-21-5p and monocyte to high-density lipoprotein cholesterol ratio in patients with acute myocardial infarction. Heart Vessels. (2023) 38(3):318–31. doi: 10.1007/s00380-022-02172-2
130. Velle-Forbord T, Eidlaug M, Debik J, Sæther JC, Follestad T, Nauman J, et al. Circulating microRNAs as predictive biomarkers of myocardial infarction: evidence from the HUNT study. Atherosclerosis. (2019) 289:1–7. doi: 10.1016/j.atherosclerosis.2019.07.024
131. Zhu W, Luo L, Ye G, Ou J. Potential diagnostic value of N1LR and SNHG1 in acute myocardial infarction. BMC Med Genomics. (2023) 16(1):71. doi: 10.1186/s12920-023-01501-2
132. Tayae E, Amr E, Zaki A, Elkaffash D. LncRNA HIF1A-AS2: a potential biomarker for early diagnosis of acute myocardial infarction and predictor of left ventricular dysfunction. BMC Cardiovasc Disord. (2023) 23(1):135. doi: 10.1186/s12872-023-03164-4
133. Xie J, Liao W, Chen W, Lai D, Tang Q, Li Y. Circulating long non-coding RNA TTTY15 and HULC serve as potential novel biomarkers for predicting acute myocardial infarction. BMC Cardiovasc Disord. (2022) 22(1):86. doi: 10.1186/s12872-022-02529-5
134. Zhou Q, Boeckel JN, Yao J, Zhao J, Bai Y, Lv Y, et al. Diagnosis of acute myocardial infarction using a combination of circulating circular RNA cZNF292 and clinical information based on machine learning. MedComm. (2023) 4(3):e299. doi: 10.1002/mco2.299
135. Li Q, Wang Y, An Y, Wang J, Gao Y. The particular expression profiles of circular RNA in peripheral blood of myocardial infarction patients by RNA sequencing. Front Cardiovasc Med. (2022) 9:810257. doi: 10.3389/fcvm.2022.810257
136. Hromadka M, Motovska Z, Hlinomaz O, Kala P, Tousek F, Jarkovsky J, et al. MiR-126-3p and miR-223-3p as biomarkers for prediction of thrombotic risk in patients with acute myocardial infarction and primary angioplasty. J Pers Med. (2021) 11(6):508. doi: 10.3390/jpm11060508
137. Carvalho A, Ji Z, Zhang R, Zuo W, Qu Y, Chen X, et al. Inhibition of miR-195-3p protects against cardiac dysfunction and fibrosis after myocardial infarction. Int J Cardiol. (2023) 387:131128. doi: 10.1016/j.ijcard.2023.131128
138. Li B, Wu Y. LncRNA TUG1 overexpression promotes apoptosis of cardiomyocytes and predicts poor prognosis of myocardial infarction. J Clin Pharm Ther. (2020) 45(6):1452–6. doi: 10.1111/jcpt.13190
139. Yan L, Zhang Y, Zhang W, Deng SQ, Ge ZR. lncRNA-NRF is a potential biomarker of heart failure after acute myocardial infarction. J Cardiovasc Transl Res. (2020) 13(6):1008–15. doi: 10.1007/s12265-020-10029-0
140. Salgado-Somoza A, Zhang L, Vausort M, Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc. (2017) 17:33–6. doi: 10.1016/j.ijcha.2017.11.001
141. Feliciano RDS, Manchini MT, Atum ALB, da Silva GA, Antônio EL, Serra AJ, et al. Photobiomodulation therapy's effects on cardiac fibrosis activation after experimental myocardial infarction. Lasers Surg Med. (2022) 54(6):883–94. doi: 10.1002/lsm.23544
142. Feliciano RDS, Atum ALB, Ruiz ÉGDS, Serra AJ, Antônio EL, Manchini MT, et al. Photobiomodulation therapy on myocardial infarction in rats: transcriptional and posttranscriptional implications to cardiac remodeling. Lasers Surg Med. (2021) 53(9):1247–57. doi: 10.1002/lsm.23407
143. Su J, Hu Y, Cheng J, Li Z, Li J, Zheng N, et al. Comprehensive analysis of the RNA transcriptome expression profiles and construction of the ceRNA network in heart failure patients with sacubitril/valsartan therapeutic heterogeneity after acute myocardial infarction. Eur J Pharmacol. (2023) 944:175547. doi: 10.1016/j.ejphar.2023.175547
144. Li X, Yao Q, Cui H, Yang J, Wu N, Liu Y, et al. MiR-223 or miR-126 predicts resistance to dual antiplatelet therapy in patients with ST-elevation myocardial infarction. J Int Med Res. (2021) 49(6):3000605211016209. doi: 10.1177/03000605211016209
145. Sothivelr V, Hasan MY, Mohd Saffian S, Zainalabidin S, Ugusman A, Mahadi MK. Revisiting miRNA-21 as a therapeutic strategy for myocardial infarction: a systematic review. J Cardiovasc Pharmacol. (2022) 80(3):393–406. doi: 10.1097/fjc.0000000000001305
146. Liao Z, Chen Y, Duan C, Zhu K, Huang R, Zhao H, et al. Cardiac telocytes inhibit cardiac microvascular endothelial cell apoptosis through exosomal miRNA-21-5p-targeted cdip1 silencing to improve angiogenesis following myocardial infarction. Theranostics. (2021) 11(1):268–91. doi: 10.7150/thno.47021
147. Bejerano T, Etzion S, Elyagon S, Etzion Y, Cohen S. Nanoparticle delivery of miRNA-21 mimic to cardiac macrophages improves myocardial remodeling after myocardial infarction. Nano Lett. (2018) 18(9):5885–91. doi: 10.1021/acs.nanolett.8b02578
148. Park HJ, Hoffman JR, Brown ME, Bheri S, Brazhkina O, Son YH, et al. Knockdown of deleterious miRNA in progenitor cell-derived small extracellular vesicles enhances tissue repair in myocardial infarction. Sci Adv. (2023) 9(9):eabo4616. doi: 10.1126/sciadv.abo4616
149. Zhou R, Huang W, Fan X, Liu F, Luo L, Yuan H, et al. miR-499 released during myocardial infarction causes endothelial injury by targeting α7-nAchR. J Cell Mol Med. (2019) 23(9):6085–97. doi: 10.1111/jcmm.14474
150. Duan S, Wang C, Xu X, Zhang X, Su G, Li Y, et al. Peripheral serum exosomes isolated from patients with acute myocardial infarction promote endothelial cell angiogenesis via the miR-126-3p/TSC1/mTORC1/HIF-1α pathway. Int J Nanomed. (2022) 17:1577–92. doi: 10.2147/ijn.S338937
151. Li SN, Li P, Liu WH, Shang JJ, Qiu SL, Zhou MX, et al. Danhong injection enhances angiogenesis after myocardial infarction by activating MiR-126/ERK/VEGF pathway. Biomed Pharmacother. (2019) 120:109538. doi: 10.1016/j.biopha.2019.109538
152. Zhang XG, Wang LQ, Guan HL. Investigating the expression of miRNA-133 in animal models of myocardial infarction and its effect on cardiac function. Eur Rev Med Pharmacol Sci. (2019) 23(13):5934–40. doi: 10.26355/eurrev_201907_18338
153. Shyu KG, Wang BW, Cheng WP, Lo HM. MicroRNA-208a increases myocardial endoglin expression and myocardial fibrosis in acute myocardial infarction. Can J Cardiol. (2015) 31(5):679–90. doi: 10.1016/j.cjca.2014.12.026
154. Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH. Proteomics: technologies and their applications. J Chromatogr Sci. (2017) 55(2):182–96. doi: 10.1093/chromsci/bmw167
155. Cheng L, Wang X, Chou H, Liu T, Fu H, Li G. Proteomic sequencing of stellate ganglions in rabbits with myocardial infarction. Front Physiol. (2021) 12:687424. doi: 10.3389/fphys.2021.687424
156. Valdes-Marquez E, Clarke R, Hill M, Watkins H, Hopewell JC. Proteomic profiling identifies novel independent relationships between inflammatory proteins and myocardial infarction. Eur J Prev Cardiol. (2023) 30(7):583–91. doi: 10.1093/eurjpc/zwad020
157. Ma Y, Yuan Y, Aili Z, Maitusong M, Li H, Nijiati M. Proteomics analysis of coronary blood microparticles in patients with acute myocardial infarction. Cardiol J. (2023) 30(2):286–96. doi: 10.5603/CJ.a2022.0081
158. Ren YS, Li HH, Yao JC, Tan YJ, Pan LH, Peng T, et al. Application quantitative proteomics approach to identify differentially expressed proteins associated with cardiac protection mediated by cycloastragenol in acute myocardial infarction rats. J Proteomics. (2020) 222:103691. doi: 10.1016/j.jprot.2020.103691
159. Hage C, Savarese G. Proteomic profiling for investigating the pathophysiology of myocardial infarction. Eur J Prev Cardiol. (2023) 30(7):581–2. doi: 10.1093/eurjpc/zwad019
160. Das AA, Roy Choudhury K, Jagadeeshaprasad MG, Kulkarni MJ, Mondal PC, Bandyopadhyay A. Corrigendum to proteomic analysis detects deregulated reverse cholesterol transport in human subjects with ST-segment elevation myocardial infarction. J Proteomics. (2020) 224:103828. doi: 10.1016/j.jprot.2020.103828
161. Pan Y, Wang L, Xie Y, Tan Y, Chang C, Qiu X, et al. Characterization of differentially expressed plasma proteins in patients with acute myocardial infarction. J Proteomics. (2020) 227:103923. doi: 10.1016/j.jprot.2020.103923
162. Xie Y, Zhang H, Huang T. Quantitative proteomics reveal three potential biomarkers for risk assessment of acute myocardial infarction. Bioengineered. (2022) 13(3):4939–50. doi: 10.1080/21655979.2022.2037365
163. Kwon S, Park SH, Mun S, Lee J, Kang HG. Potential biomarkers to distinguish type 1 myocardial infarction in troponin-elevated diseases. Int J Mol Sci. (2023) 24(9):8097. doi: 10.3390/ijms24098097
164. Shi LY, Han YS, Chen J, Li ZB, Li JC, Jiang TT. Screening and identification of potential protein biomarkers for the early diagnosis of acute myocardial infarction. Ann Transl Med. (2021) 9(9):743. doi: 10.21037/atm-20-7891
165. Mohamed Bakrim N, Mohd Shah ANS, Talib NA, Ab Rahman J, Abdullah A. Identification of haptoglobin as a potential biomarker in young adults with acute myocardial infarction by proteomic analysis. Malays J Med Sci. (2020) 27(2):64–76. doi: 10.21315/mjms2020.27.2.8
166. Jacquet S, Yin X, Sicard P, Clark J, Kanaganayagam GS, Mayr M, et al. Identification of cardiac myosin-binding protein C as a candidate biomarker of myocardial infarction by proteomics analysis. Mol Cell Proteomics. (2009) 8(12):2687–99. doi: 10.1074/mcp.M900176-MCP200
167. Govindan S, Kuster DW, Lin B, Kahn DJ, Jeske WP, Walenga JM, et al. Increase in cardiac myosin binding protein-C plasma levels is a sensitive and cardiac-specific biomarker of myocardial infarction. Am J Cardiovasc Dis. (2013) 3(2):60–70.23785583
168. Liu Y, Huang D, Li Z, Zhou L, Cen T, Wei B, et al. A plasma proteomic approach in patients with heart failure after acute myocardial infarction: insights into the pathogenesis and progression of the disease. Front Cardiovasc Med. (2023) 10:1153625. doi: 10.3389/fcvm.2023.1153625
169. Wu X, You W, Wu Z, Ye F, Chen S. Serum biomarker analysis at the protein level on pulmonary hypertension secondary to old anterior myocardial infarction. Pulm Circ. (2020) 10(4):2045894020969079. doi: 10.1177/2045894020969079
170. Mao S, Liang Y, Chen P, Zhang Y, Yin X, Zhang M. In-depth proteomics approach reveals novel biomarkers of cardiac remodelling after myocardial infarction: an exploratory analysis. J Cell Mol Med. (2020) 24(17):10042–51. doi: 10.1111/jcmm.15611
171. Edfors R, Lindhagen L, Spaak J, Evans M, Andell P, Baron T, et al. Use of proteomics to identify biomarkers associated with chronic kidney disease and long-term outcomes in patients with myocardial infarction. J Intern Med. (2020) 288(5):581–92. doi: 10.1111/joim.13116
172. Heyse W, Vandewalle V, Marot G, Amouyel P, Bauters C, Pinet F. Identification of patient subtypes based on protein expression for prediction of heart failure after myocardial infarction. iScience. (2023) 26(3):106171. doi: 10.1016/j.isci.2023.106171
173. Chalise U, Becirovic-Agic M, Rodriguez-Paar JR, Konfrst SR, de Morais SDB, Johnson CS, et al. Harnessing the plasma proteome to mirror current and predict future cardiac remodeling after myocardial infarction. J Cardiovasc Transl Res. (2023) 16(1):3–16. doi: 10.1007/s12265-022-10326-w
174. Mansell DS, Bruno VD, Sammut E, Chiribiri A, Johnson T, Khaliulin I, et al. Acute regional changes in myocardial strain may predict ventricular remodelling after myocardial infarction in a large animal model. Sci Rep. (2021) 11(1):18322. doi: 10.1038/s41598-021-97834-y
175. Mohammad MA, Koul S, Egerstedt A, Smith JG, Noc M, Lang I, et al. Using proximity extension proteomics assay to identify biomarkers associated with infarct size and ejection fraction after ST-elevation myocardial infarction. Sci Rep. (2020) 10(1):18663. doi: 10.1038/s41598-020-75399-6
176. Foreback CC. Biochemical diagnosis of myocardial infarction. Henry Ford Hosp Med J. (1991) 39(3–4):159–64.1804820
177. Stătescu C, Anghel L, Tudurachi BS, Leonte A, Benchea LC, Sascău RA. From classic to modern prognostic biomarkers in patients with acute myocardial infarction. Int J Mol Sci. (2022) 23(16):9168. doi: 10.3390/ijms23169168
178. Panteghini M. Acute coronary syndrome: biochemical strategies in the troponin era. Chest. (2002) 122(4):1428–35. doi: 10.1378/chest.122.4.1428
179. Bormann J, Psyrakis DA, von Jeinsen B, Grün D, Elsner LK, Wolter JS, et al. Myeloid-related protein 8/14 and high-sensitivity cardiac troponin I to differentiate type 2 myocardial infarction. Int J Cardiol. (2020) 304:144–7. doi: 10.1016/j.ijcard.2020.01.043
180. Pandey AK, Duong T, Swiatkiewicz I, Daniels LB. A comparison of biomarker rise in type 1 and type 2 myocardial infarction. Am J Med. (2020) 133(10):1203–8. doi: 10.1016/j.amjmed.2020.02.024
181. Sheifer SE, Gersh BJ, Yanez ND 3rd, Ades PA, Burke GL, Manolio TA. Prevalence, predisposing factors, and prognosis of clinically unrecognized myocardial infarction in the elderly. J Am Coll Cardiol. (2000) 35(1):119–26. doi: 10.1016/s0735-1097(99)00524-0
182. Song N, Lu D, Wu G, Wang S, Zeng Y, Zhao J, et al. Serum proteomic analysis reveals the cardioprotective effects of Shexiang Baoxin pill and Suxiao Jiuxin pill in a rat model of acute myocardial infarction. J Ethnopharmacol. (2022) 293:115279. doi: 10.1016/j.jep.2022.115279
183. Yu F, Yu Y, Tian S, Zhou Y, Chen X, Ye J, et al. Quantitative proteomics reveals Shexiang Baoxin pill exerts cardioprotective effects by preserving energy metabolism in a rat model of myocardial infarction. J Ethnopharmacol. (2021) 266:113460. doi: 10.1016/j.jep.2020.113460
184. Wang Y, Liu L, Hu C, Cheng Y. Effects of Salviae mitiorrhizae and cortex moutan extract on the rat heart after myocardial infarction: a proteomic study. Biochem Pharmacol. (2007) 74(3):415–24. doi: 10.1016/j.bcp.2007.04.017
185. George MJ, Kleveland O, Garcia-Hernandez J, Palmen J, Lovering R, Wiseth R, et al. Novel insights into the effects of interleukin 6 antagonism in non-ST-segment-elevation myocardial infarction employing the SOMAscan proteomics platform. J Am Heart Assoc. (2020) 9(12):e015628. doi: 10.1161/jaha.119.015628
186. Bansal A, Dai Q, Chiao YA, Hakala KW, Zhang JQ, Weintraub ST, et al. Proteomic analysis reveals late exercise effects on cardiac remodeling following myocardial infarction. J Proteomics. (2010) 73(10):2041–9. doi: 10.1016/j.jprot.2010.06.009
187. Mao S, Zhang X, Chen M, Wang C, Chen Q, Guo L, et al. Beneficial effects of baduanjin exercise on left ventricular remodelling in patients after acute myocardial infarction: an exploratory clinical trial and proteomic analysis. Cardiovasc Drugs Ther. (2021) 35(1):21–32. doi: 10.1007/s10557-020-07047-0
188. López-Yerena A, Domínguez-López I, Vallverdú-Queralt A, Pérez M, Jáuregui O, Escribano-Ferrer E, et al. Metabolomics technologies for the identification and quantification of dietary phenolic compound metabolites: an overview. Antioxidants. (2021) 10(6):846. doi: 10.3390/antiox10060846
189. Xia JG, Li B, Zhang H, Li QX, Lam SM, Yin CL, et al. Precise metabolomics defines systemic metabolic dysregulation distinct to acute myocardial infarction associated with diabetes. Arterioscler Thromb Vasc Biol. (2023) 43(4):581–96. doi: 10.1161/atvbaha.122.318871
190. Zhang X, Cai Y, Su X, Jing Q, Liu H, Na K, et al. Untargeted metabolomics identified kynurenine as a predictive prognostic biomarker in acute myocardial infarction. Front Immunol. (2022) 13:950441. doi: 10.3389/fimmu.2022.950441
191. Wang X, Wang D, Wu J, Yu X, Lv J, Kong J, et al. Metabolic characterization of myocardial infarction using GC-MS-based tissue metabolomics. Int Heart J. (2017) 58(3):441–6. doi: 10.1536/ihj.16-432
192. Nogal A, Alkis T, Lee Y, Kifer D, Hu J, Murphy RA, et al. Predictive metabolites for incident myocardial infarction: a two-step meta-analysis of individual patient data from six cohorts comprising 7,897 individuals from the the COnsortium of METabolomic studies. Cardiovasc Res. (2023) cvad147. doi: 10.1093/cvr/cvad147. [Epub ahead of print]37706562
193. Liu W, Zhang L, Shi X, Shen G, Feng J. Cross-comparative metabolomics reveal sex-age specific metabolic fingerprints and metabolic interactions in acute myocardial infarction. Free Radical Biol Med. (2022) 183:25–34. doi: 10.1016/j.freeradbiomed.2022.03.008
194. Cao N, Wang Y, Bao B, Wang M, Li J, Dang W, et al. 12,13-diHOME And noradrenaline are associated with the occurrence of acute myocardial infarction in patients with type 2 diabetes mellitus. Diabetol Metab Syndr. (2023) 15(1):142. doi: 10.1186/s13098-023-01115-9
195. Khan A, Choi Y, Back JH, Lee S, Jee SH, Park YH. High-resolution metabolomics study revealing l-homocysteine sulfinic acid, cysteic acid, and carnitine as novel biomarkers for high acute myocardial infarction risk. Metabolisml. (2020) 104:154051. doi: 10.1016/j.metabol.2019.154051
196. Kharb S. Low blood glutathione levels in acute myocardial infarction. Indian J Med Sci. (2003) 57(8):335–7.12944689
197. Gundogdu G, Senol O, Demirkaya Miloglu F, Koza Y, Gundogdu F, Hacımüftüoğlu A, et al. Serum metabolite profiling of ST-segment elevation myocardial infarction using liquid chromatography quadrupole time-of-flight mass spectrometry. Biomed Chromatogr. (2020) 34(2):e4738. doi: 10.1002/bmc.4738
198. Usalp S, Altuntaş E, Bağırtan B, Karabay K. Comparison of serum lipoprotein(a) levels in young and middle-aged patients presenting for the first time with ST-elevation myocardial infarction: a single-centre study. Cardiovasc J Afr. (2023) 34:1–5. doi: 10.5830/cvja-2023-038
199. Jeong H, Han K, Yoo SJ, Kim MK. Low-density lipoprotein cholesterol level, statin use and myocardial infarction risk in young adults. J Lipid Atheroscler. (2022) 11(3):288–98. doi: 10.12997/jla.2022.11.3.288
200. Tian X, Zuo Y, Chen S, Li H, He Y, Zhang L, et al. Association of changes in lipids with risk of myocardial infarction among people without lipid-lowering therapy. Atherosclerosis. (2020) 301:69–78. doi: 10.1016/j.atherosclerosis.2020.03.026
201. Zafrir B, Khoury R, Saliba W. Remnant cholesterol and risk of myocardial infarction in patients with coronary artery disease undergoing revascularization. J Clin Lipidol. (2023) 17(3):332–41. doi: 10.1016/j.jacl.2023.03.009
202. Hou M, Ren YP, Wang R, Lu LX. Early cardiopulmonary resuscitation on serum levels of myeloperoxidase, soluble ST2, and hypersensitive C-reactive protein in acute myocardial infarction patients. World J Clin Cases. (2021) 9(34):10585–94. doi: 10.12998/wjcc.v9.i34.10585
203. Chiusolo S, Bork CS, Gentile F, Lundbye-Christensen S, Harris WS, Schmidt EB, et al. Adipose tissue n-3/n-6 fatty acids ratios versus n-3 fatty acids fractions as predictors of myocardial infarction. Am Heart J. (2023) 262:38–48. doi: 10.1016/j.ahj.2023.03.019
204. Hua T, Bao Q, He X, Cai W, He J. Lipidomics revealed alteration of sphingolipid metabolism during the reparative phase after myocardial infarction injury. Front Physiol. (2021) 12:663480. doi: 10.3389/fphys.2021.663480
205. Liu YT, Jia HM, Chang X, Ding G, Zhang HW, Zou ZM. The metabolic disturbances of isoproterenol induced myocardial infarction in rats based on a tissue targeted metabonomics. Mol BioSyst. (2013) 9(11):2823–34. doi: 10.1039/c3mb70222g
206. Delanghe J, De Buyzere M, De Scheerder I, Vogelaers D, Vandenbogaerde J, Van den Abeele AM, et al. Creatine determinations as an early marker for the diagnosis of acute myocardial infarction. Ann Clin Biochem. (1988) 25(Pt 4):383–8. doi: 10.1177/000456328802500410
207. Lee DY, Han K, Park S, Yu JH, Seo JA, Kim NH, et al. Glucose variability and the risks of stroke, myocardial infarction, and all-cause mortality in individuals with diabetes: retrospective cohort study. Cardiovasc Diabetol. (2020) 19(1):144. doi: 10.1186/s12933-020-01134-0
208. Park IH, Cho HK, Oh JH, Chun WJ, Park YH, Lee M, et al. Clinical significance of serum lactate in acute myocardial infarction: a cardiac magnetic resonance imaging study. J Clin Med. (2021) 10(22):5278. doi: 10.3390/jcm10225278
209. Gąsecka A, Fidali O, Kłębukowska A, Jasińska-Gniadzik K, Szwed P, Witkowska K, et al. Plasma concentration of TMAO is an independent predictor of adverse outcomes in patients after acute myocardial infarction. Postepy Kardiol Interwencyjnej. (2023) 19(1):31–9. doi: 10.5114/aic.2022.123884
210. McKirnan MD, Ichikawa Y, Zhang Z, Zemljic-Harpf AE, Fan S, Barupal DK, et al. Metabolomic analysis of serum and myocardium in compensated heart failure after myocardial infarction. Life Sci. (2019) 221:212–23. doi: 10.1016/j.lfs.2019.01.040
211. Vignoli A, Tenori L, Giusti B, Takis PG, Valente S, Carrabba N, et al. NMR-based metabolomics identifies patients at high risk of death within two years after acute myocardial infarction in the AMI-florence II cohort. BMC Med. (2019) 17(1):3. doi: 10.1186/s12916-018-1240-2
212. Zhao S, Tian Y, Wang S, Yang F, Xu J, Qin Z, et al. Prognostic value of gut microbiota-derived metabolites in patients with ST-segment elevation myocardial infarction. Am J Clin Nutr. (2023) 117(3):499–508. doi: 10.1016/j.ajcnut.2022.12.013
213. Li N, Wang Y, Zhou J, Chen R, Li J, Zhao X, et al. Association between the changes in trimethylamine N-oxide-related metabolites and prognosis of patients with acute myocardial infarction: a prospective study. J Cardiovasc Dev Dis. (2022) 9(11):380. doi: 10.3390/jcdd9110380
214. Gąsecka A, Szwed P, Jasińska K, Fidali O, Kłębukowska A, Eyileten C, et al. Symmetric dimethylarginine is altered in patients after myocardial infarction and predicts adverse outcomes. J Inflamm Res. (2021) 14:3797–808. doi: 10.2147/jir.S316078
215. Frangogiannis NG, Smith CW, Entman ML. The inflammatory response in myocardial infarction. Cardiovasc Res. (2002) 53(1):31–47. doi: 10.1016/s0008-6363(01)00434-5
216. Ward-Caviness CK, Xu T, Aspelund T, Thorand B, Montrone C, Meisinger C, et al. Improvement of myocardial infarction risk prediction via inflammation-associated metabolite biomarkers. Heart. (2017) 103(16):1278–85. doi: 10.1136/heartjnl-2016-310789
217. Mandurino-Mirizzi A, Cornara S, Somaschini A, Demarchi A, Galazzi M, Puccio S, et al. Elevated serum uric acid is associated with a greater inflammatory response and with short- and long-term mortality in patients with ST-segment elevation myocardial infarction undergoing primary percutaneous coronary intervention. NutrMetab Cardiovasc Dis. (2021) 31(2):608–14. doi: 10.1016/j.numecd.2020.10.020
218. Açıkgöz E, Açıkgöz SK, Yaman B, Kurtul A. Lower LDL-cholesterol levels associated with increased inflammatory burden in patients with acute ST-segment elevation myocardial infarction. Rev Assoc Med Bras (1992). (2021) 67(2):224–9. doi: 10.1590/1806-9282.67.02.20200548
219. Wahid M, Saqib F, Chicea L, Ahmedah HT, Sajer BH, Marc Vlaic RA, et al. Metabolomics analysis delineates the therapeutic effects of hydroethanolic extract of cucumis sativus L. Seeds on hypertension and isoproterenol-induced myocardial infarction. Biomed Pharmacother. (2022) 148:112704. doi: 10.1016/j.biopha.2022.112704
220. Wen W, Zhang Z, Jiang B, Hao Y. Orbitrap-MS-based untargeted metabolomics study on the therapeutic effect of colchicine on myocardial infarction. Biomed Chromatogr. (2021) 35(9):e5148. doi: 10.1002/bmc.5148
221. Wu G, Zhong J, Chen L, Gu Y, Hong Y, Ma J, et al. Effects of the Suxiao Jiuxin pill on acute myocardial infarction assessed by comprehensive metabolomics. Phytomedicine. (2020) 77:153291. doi: 10.1016/j.phymed.2020.153291
222. Wu G, Chen L, Gu Y, Hong Y, Ma J, Zheng N, et al. Exploring the mechanism underlying the cardioprotective effect of shexiang baoxin pill on acute myocardial infarction rats by comprehensive metabolomics. J Ethnopharmacol. (2020) 259:113001. doi: 10.1016/j.jep.2020.113001
223. Fang M, Meng Y, Du Z, Guo M, Jiang Y, Tu P, et al. The synergistic mechanism of total saponins and flavonoids in notoginseng-safflower against myocardial infarction using a comprehensive metabolomics strategy. Molecules. (2022) 27(24):8860. doi: 10.3390/molecules27248860
224. Yurista SR, Eder RA, Welsh A, Jiang W, Chen S, Foster AN, et al. Ketone ester supplementation suppresses cardiac inflammation and improves cardiac energetics in a swine model of acute myocardial infarction. Metabolism. (2023) 145:155608. doi: 10.1016/j.metabol.2023.155608
225. Zhao Z, Zhang Y, Liu L, Chen Y, Wang D, Jin X, et al. Metabolomics study of the effect of smoking and high-fat diet on metabolic responses and related mechanism following myocardial infarction in mice. Life Sci. (2020) 263:118570. doi: 10.1016/j.lfs.2020.118570
226. Misra BB, Langefeld CD, Olivier M, Cox LA. Integrated omics: tools, advances, and future approaches. J Mol Endocrinol. (2018) JME-18-0055. doi: 10.1530/jme-18-0055. [Epub ahead of print]30006342
227. Berg G, Rybakova D, Fischer D, Cernava T, Vergès MC, Charles T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome. (2020) 8(1):103. doi: 10.1186/s40168-020-00875-0
228. Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. (2018) 16(7):410–22. doi: 10.1038/s41579-018-0029-9
229. Chen J, Li Y, Tian Y, Huang C, Li D, Zhong Q, et al. Interaction between microbes and host intestinal health: modulation by dietary nutrients and gut-brain-endocrine-immune axis. Curr Protein Pept Sci. (2015) 16(7):592–603. doi: 10.2174/1389203716666150630135720
230. Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. (2012) 3:1245. doi: 10.1038/ncomms2266
231. Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. (2011) 108 Suppl 1(Suppl 1):4592–8. doi: 10.1073/pnas.1011383107
232. Organ CL, Otsuka H, Bhushan S, Wang Z, Bradley J, Trivedi R, et al. Choline diet and its gut microbe-derived metabolite, trimethylamine N-oxide, exacerbate pressure overload-induced heart failure. Circ Heart Fail. (2016) 9(1):e002314. doi: 10.1161/circheartfailure.115.002314
233. Beyazcicek E, Beyazcicek O. Protective effects of lacticaseibacillus rhamnosus on isoprenaline-induced myocardial infarction in rats. J Appl Microbiol. (2023) 134(1):lxac008. doi: 10.1093/jambio/lxac008
234. Mansuri NM, Mann NK, Rizwan S, Mohamed AE, Elshafey AE, Khadka A, et al. Role of gut microbiome in cardiovascular events: a systematic review. Cureus. (2022) 14(12):e32465. doi: 10.7759/cureus.32465
235. Anto L, Blesso CN. Interplay between diet, the gut microbiome, and atherosclerosis: role of dysbiosis and microbial metabolites on inflammation and disordered lipid metabolism. J Nutr Biochem. (2022) 105:108991. doi: 10.1016/j.jnutbio.2022.108991
236. Rekha K, Venkidasamy B, Samynathan R, Nagella P, Rebezov M, Khayrullin M, et al. Short-chain fatty acid: an updated review on signaling, metabolism, and therapeutic effects. Crit Rev Food Sci Nutr. (2022):1–29. doi: 10.1080/10408398.2022.2124231. [Epub ahead of print]36154353
237. Hu T, Wu Q, Yao Q, Jiang K, Yu J, Tang Q. Short-chain fatty acid metabolism and multiple effects on cardiovascular diseases. Ageing Res Rev. (2022) 81:101706. doi: 10.1016/j.arr.2022.101706
238. Han Y, Gong Z, Sun G, Xu J, Qi C, Sun W, et al. Dysbiosis of gut microbiota in patients with acute myocardial infarction. Front Microbiol. (2021) 12:680101. doi: 10.3389/fmicb.2021.680101
239. Wang Z, Zhao Y. Gut microbiota derived metabolites in cardiovascular health and disease. Protein Cell. (2018) 9(5):416–31. doi: 10.1007/s13238-018-0549-0
240. Hage RE, Al-Arawe N, Hinterseher I. The role of the gut microbiome and trimethylamine oxide in atherosclerosis and age-related disease. Int J Mol Sci. (2023) 24(3):2399. doi: 10.3390/ijms24032399
241. Zhang H, Jing L, Zhai C, Xiang Q, Tian H, Hu H. Intestinal flora metabolite trimethylamine oxide is inextricably linked to coronary heart disease. J Cardiovasc Pharmacol. (2023) 81(3):175–82. doi: 10.1097/fjc.0000000000001387
242. Khan I, Khan I, Kakakhel MA, Xiaowei Z, Ting M, Ali I, et al. Comparison of microbial populations in the blood of patients with myocardial infarction and healthy individuals. Front Microbiol. (2022) 13:845038. doi: 10.3389/fmicb.2022.845038
243. Zhou X, Li J, Guo J, Geng B, Ji W, Zhao Q, et al. Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome. (2018) 6(1):66. doi: 10.1186/s40168-018-0441-4
244. Jiang X, Huang X, Tong Y, Gao H. Butyrate improves cardiac function and sympathetic neural remodeling following myocardial infarction in rats. Can J Physiol Pharmacol. (2020) 98(6):391–9. doi: 10.1139/cjpp-2019-0531
245. Tan Y, Sheng Z, Zhou P, Liu C, Zhao H, Song L, et al. Plasma trimethylamine N-oxide as a novel biomarker for plaque rupture in patients with ST-segment-elevation myocardial infarction. Circ Cardiovasc Interv. (2019) 12(1):e007281. doi: 10.1161/circinterventions.118.007281
246. Aldujeli A, Patel R, Grabauskyte I, Hamadeh A, Lieponyte A, Tatarunas V, et al. The impact of trimethylamine N-oxide and coronary microcirculatory dysfunction on outcomes following ST-elevation myocardial infarction. J Cardiovasc Dev Dis. (2023) 10(5):197. doi: 10.3390/jcdd10050197
247. Li N, Zhou J, Wang Y, Chen R, Li J, Zhao X, et al. Association between trimethylamine N-oxide and prognosis of patients with acute myocardial infarction and heart failure. ESC Heart Fail. (2022) 9(6):3846–57. doi: 10.1002/ehf2.14009
248. Bin Waleed K, Lu Y, Liu Q, Zeng F, Tu H, Wei Y, et al. Association of trimethylamine N-oxide with coronary atherosclerotic burden in patients with non-ST-segment elevation myocardial infarction. Medicine. (2020) 99(27):e20794. doi: 10.1097/md.0000000000020794
249. Zununi Vahed S, Barzegari A, Zuluaga M, Letourneur D, Pavon-Djavid G. Myocardial infarction and gut microbiota: an incidental connection. Pharmacol Res. (2018) 129:308–17. doi: 10.1016/j.phrs.2017.11.008
250. Lam V, Su J, Koprowski S, Hsu A, Tweddell JS, Rafiee P, et al. Intestinal microbiota determine severity of myocardial infarction in rats. FASEB J. (2012) 26(4):1727–35. doi: 10.1096/fj.11-197921
251. Sadeghzadeh J, Vakili A, Sameni HR, Shadnoush M, Bandegi AR, Zahedi Khorasani M. The effect of oral consumption of probiotics in prevention of heart injury in a rat myocardial infarction model: a histopathological, hemodynamic and biochemical evaluation. Iran Biomed J. (2017) 21(3):174–81. doi: 10.18869/acadpub.ibj.21.3.174
252. Xu H, Yang F, Bao Z. Gut microbiota and myocardial fibrosis. Eur J Pharmacol. (2023) 940:175355. doi: 10.1016/j.ejphar.2022.175355
253. Zhong X, Zhao Y, Huang L, Liu J, Wang K, Gao X, et al. Remodeling of the gut microbiome by lactobacillus johnsonii alleviates the development of acute myocardial infarction. Front Microbiol. (2023) 14:1140498. doi: 10.3389/fmicb.2023.1140498
254. Li Z, Wang K, Ding Y, Ma W, Sun Y, Liu X, et al. Dapagliflozin modulates the faecal microbiota after myocardial infarction in non-diabetic mice. Clin Exp Pharmacol Physiol. (2023) 50(1):68–81. doi: 10.1111/1440-1681.13727
255. Hua F, Zhou P, Bao GH, Ling TJ. Flavonoids in Lu'an GuaPian tea as potential inhibitors of TMA-lyase in acute myocardial infarction. J Food Biochem. (2022) 46(7):e14110. doi: 10.1111/jfbc.14110
256. Zhou M, Li D, Xie K, Xu L, Kong B, Wang X, et al. The short-chain fatty acid propionate improved ventricular electrical remodeling in a rat model with myocardial infarction. Food Funct. (2021) 12(24):12580–93. doi: 10.1039/d1fo02040d
257. Zhou Q, Deng J, Pan X, Meng D, Zhu Y, Bai Y, et al. Gut microbiome mediates the protective effects of exercise after myocardial infarction. Microbiome. (2022) 10(1):82. doi: 10.1186/s40168-022-01271-6
258. Liu Z, Liu HY, Zhou H, Zhan Q, Lai W, Zeng Q, et al. Moderate-intensity exercise affects gut microbiome composition and influences cardiac function in myocardial infarction mice. Front Microbiol. (2017) 8:1687. doi: 10.3389/fmicb.2017.01687
259. Oliveira RA, Cabral V, Xavier KB. Microbiome-diet interactions drive antibiotic efficacy. Nat Microbiol. (2021) 6(7):824–5. doi: 10.1038/s41564-021-00926-8
260. He Z, Kong X, Shao T, Zhang Y, Wen C. Alterations of the gut microbiota associated with promoting efficacy of prednisone by bromofuranone in MRL/lpr mice. Front Microbiol. (2019) 10:978. doi: 10.3389/fmicb.2019.00978
261. Wang Z, Xiao H, Dong J, Li Y, Wang B, Chen Z, et al. Sexual dimorphism in gut microbiota dictates therapeutic efficacy of intravenous immunoglobulin on radiotherapy complications. J Adv Res. (2023) 46:123–33. doi: 10.1016/j.jare.2022.06.002
262. Chen W, Qian J, Fu J, Wu T, Lv M, Jiang S, et al. Changes in the gut microbiota may affect the clinical efficacy of oral anticoagulants. Front Pharmacol. (2022) 13:860237. doi: 10.3389/fphar.2022.860237
263. Pratapa A, Jalihal AP, Law JN, Bharadwaj A, Murali TM. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat Methods. (2020) 17(2):147–54. doi: 10.1038/s41592-019-0690-6
264. Chen L, Fan R, Tang F. Advanced single-cell omics technologies and informatics tools for genomics, proteomics, and bioinformatics analysis. Genomics Proteomics Bioinformatics. (2021) 19(3):343–5. doi: 10.1016/j.gpb.2021.12.001
265. Jin K, Gao S, Yang P, Guo R, Li D, Zhang Y, et al. Single-cell RNA sequencing reveals the temporal diversity and dynamics of cardiac immunity after myocardial infarction. Small Methods. (2022) 6(3):e2100752. doi: 10.1002/smtd.202100752
266. Tombor LS, John D, Glaser SF, Luxán G, Forte E, Furtado M, et al. Single cell sequencing reveals endothelial plasticity with transient mesenchymal activation after myocardial infarction. Nat Commun. (2021) 12(1):681. doi: 10.1038/s41467-021-20905-1
267. Ruiz-Villalba A, Romero JP, Hernández SC, Vilas-Zornoza A, Fortelny N, Castro-Labrador L, et al. Single-cell RNA sequencing analysis reveals a crucial role for CTHRC1 (collagen triple helix repeat containing 1) cardiac fibroblasts after myocardial infarction. Circulation. (2020) 142(19):1831–47. doi: 10.1161/circulationaha.119.044557
268. Wu X, Reboll MR, Korf-Klingebiel M, Wollert KC. Angiogenesis after acute myocardial infarction. Cardiovasc Res. (2021) 117(5):1257–73. doi: 10.1093/cvr/cvaa287
269. Qian J, Gao Y, Lai Y, Ye Z, Yao Y, Ding K, et al. Single-cell RNA sequencing of peripheral blood mononuclear cells from acute myocardial infarction. Front Immunol. (2022) 13:908815. doi: 10.3389/fimmu.2022.908815
270. Dick SA, Macklin JA, Nejat S, Momen A, Clemente-Casares X, Althagafi MG, et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol. (2019) 20(1):29–39. doi: 10.1038/s41590-018-0272-2
271. Li Z, Solomonidis EG, Meloni M, Taylor RS, Duffin R, Dobie R, et al. Single-cell transcriptome analyses reveal novel targets modulating cardiac neovascularization by resident endothelial cells following myocardial infarction. Eur Heart J. (2019) 40(30):2507–20. doi: 10.1093/eurheartj/ehz305
272. Vafadarnejad E, Rizzo G, Krampert L, Arampatzi P, Arias-Loza AP, Nazzal Y, et al. Dynamics of cardiac neutrophil diversity in murine myocardial infarction. Circ Res. (2020) 127(9):e232–e49. doi: 10.1161/circresaha.120.317200
273. Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife. (2019) 8:e43882. doi: 10.7554/eLife.43882
274. Forte E, Skelly DA, Chen M, Daigle S, Morelli KA, Hon O, et al. Dynamic interstitial cell response during myocardial infarction predicts resilience to rupture in genetically diverse mice. Cell Rep. (2020) 30(9):3149–63.e6. doi: 10.1016/j.celrep.2020.02.008
275. Xia N, Lu Y, Gu M, Li N, Liu M, Jiao J, et al. A unique population of regulatory T cells in heart potentiates cardiac protection from myocardial infarction. Circulation. (2020) 142(20):1956–73. doi: 10.1161/circulationaha.120.046789
276. Nguyen T, Wei Y, Nakada Y, Zhou Y, Zhang J. Cardiomyocyte cell-cycle regulation in neonatal large mammals: single nucleus RNA-Sequencing data analysis via an artificial-intelligence-based pipeline. Front Bioeng Biotechnol. (2022) 10:914450. doi: 10.3389/fbioe.2022.914450
277. Gladka MM. Single-cell RNA sequencing of the adult mammalian heart-state-of-the-art and future perspectives. Curr Heart Fail Rep. (2021) 18(2):64–70. doi: 10.1007/s11897-021-00504-3
278. Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. (2018) 22(3):600–10. doi: 10.1016/j.celrep.2017.12.072
279. Wu L, Li YF, Shen JW, Zhu Q, Jiang J, Ma SH, et al. Single-cell RNA sequencing of mouse left ventricle reveals cellular diversity and intercommunication. Physiol Genomics. (2022) 54(1):11–21. doi: 10.1152/physiolgenomics.00016.2021
280. Zhang Q, Guo Y, Zhang B, Liu H, Peng Y, Wang D, et al. Identification of hub biomarkers of myocardial infarction by single-cell sequencing, bioinformatics, and machine learning. Front Cardiovasc Med. (2022) 9:939972. doi: 10.3389/fcvm.2022.939972
281. Liu W, Li Y, Zhang Y, Li S, Chen Y, Han B, et al. Identification of biomarkers and immune infiltration in acute myocardial infarction and heart failure by integrated analysis. Biosci Rep. (2023) 43(7):BSR20222552. doi: 10.1042/bsr20222552
282. Chen Q, Yin Q, Song J, Liu C, Chen H, Li S. Identification of monocyte-associated genes as predictive biomarkers of heart failure after acute myocardial infarction. BMC Med Genomics. (2021) 14(1):44. doi: 10.1186/s12920-021-00890-6
283. Chen Q, Su L, Liu C, Gao F, Chen H, Yin Q, et al. PRKAR1A And SDCBP serve as potential predictors of heart failure following acute myocardial infarction. Front Immunol. (2022) 13:878876. doi: 10.3389/fimmu.2022.878876
284. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. (2017) 14(12):749–62. doi: 10.1038/nrclinonc.2017.141
285. Avard E, Shiri I, Hajianfar G, Abdollahi H, Kalantari KR, Houshmand G, et al. Non-contrast cine cardiac magnetic resonance image radiomics features and machine learning algorithms for myocardial infarction detection. Comput Biol Med. (2022) 141:105145. doi: 10.1016/j.compbiomed.2021.105145
286. Liu M, Xin A, Chen T, Chen F, Qian G, Zhang Y, et al. Non-contrast cine cardiac magnetic resonance derived-radiomics for the prediction of left ventricular adverse remodeling in patients with ST-segment elevation myocardial infarction. Korean J Radiol. (2023) 24(9):827–37. doi: 10.3348/kjr.2023.0061
287. Ma Q, Ma Y, Yu T, Sun Z, Hou Y. Radiomics of non-contrast-enhanced T1 mapping: diagnostic and predictive performance for myocardial injury in acute ST-segment-elevation myocardial infarction. Korean J Radiol. (2021) 22(4):535–46. doi: 10.3348/kjr.2019.0969
288. Ma Q, Ma Y, Wang X, Li S, Yu T, Duan W, et al. A radiomic nomogram for prediction of major adverse cardiac events in ST-segment elevation myocardial infarction. Eur Radiol. (2021) 31(2):1140–50. doi: 10.1007/s00330-020-07176-y
289. Chen BH, An DA, He J, Wu CW, Yue T, Wu R, et al. Myocardial extracellular volume fraction radiomics analysis for differentiation of reversible versus irreversible myocardial damage and prediction of left ventricular adverse remodeling after ST-elevation myocardial infarction. Eur Radiol. (2021) 31(1):504–14. doi: 10.1007/s00330-020-07117-9
290. Di Noto T, von Spiczak J, Mannil M, Gantert E, Soda P, Manka R, et al. Radiomics for distinguishing myocardial infarction from myocarditis at late gadolinium enhancement at MRI: comparison with subjective visual analysis. Radiol Cardiothorac Imaging. (2019) 1(5):e180026. doi: 10.1148/ryct.2019180026
291. Durmaz ES, Karabacak M, Ozkara BB, Kargın OA, Raimoglu U, Tokdil H, et al. Radiomics-based machine learning models in STEMI: a promising tool for the prediction of major adverse cardiac events. Eur Radiol. (2023) 33(7):4611–20. doi: 10.1007/s00330-023-09394-6
292. Liu J, Fan W, Liu Y, Bu H, Song J, Sun L. Association of epicardial and pericardial adipose tissue volumes with coronary artery calcification. Int Heart J. (2022) 63(6):1019–25. doi: 10.1536/ihj.22-006
293. Lin A, Kolossváry M, Yuvaraj J, Cadet S, McElhinney PA, Jiang C, et al. Myocardial infarction associates with a distinct pericoronary adipose tissue radiomic phenotype: a prospective case-control study. JACC Cardiovasc Imaging. (2020) 13(11):2371–83. doi: 10.1016/j.jcmg.2020.06.033
294. Si N, Shi K, Li N, Dong X, Zhu C, Guo Y, et al. Identification of patients with acute myocardial infarction based on coronary CT angiography: the value of pericoronary adipose tissue radiomics. Eur Radiol. (2022) 32(10):6868–77. doi: 10.1007/s00330-022-08812-5
295. Wang Z, Zhang J, Zhang A, Sun Y, Su M, You H, et al. The role of epicardial and pericoronary adipose tissue radiomics in identifying patients with non-ST-segment elevation myocardial infarction from unstable angina. Heliyon. (2023) 9(5):e15738. doi: 10.1016/j.heliyon.2023.e15738
296. Krieger G, Lupo O, Levy AA, Barkai N. Independent evolution of transcript abundance and gene regulatory dynamics. Genome Res. (2020) 30(7):1000–11. doi: 10.1101/gr.261537.120
297. Pischetsrieder M, Baeuerlein R. Proteome research in food science. Chem Soc Rev. (2009) 38(9):2600–8. doi: 10.1039/b817898b
298. Carlberg C, Molnár F. What is epigenomics? Human epigenomics. Singapore: Springer Singapore (2018). 3–18.
299. Yang J, Chen Y, Jing Y, Green MR, Han L. Advancing CAR T cell therapy through the use of multidimensional omics data. Nat Rev Clin Oncol. (2023) 20(4):211–28. doi: 10.1038/s41571-023-00729-2
300. Nevedomskaya E, Haendler B. From omics to multi-omics approaches for in-depth analysis of the molecular mechanisms of prostate cancer. Int J Mol Sci. (2022) 23(11):6281. doi: 10.3390/ijms23116281
301. Das T, Andrieux G, Ahmed M, Chakraborty S. Integration of online omics-data resources for cancer research. Front Genet. (2020) 11:578345. doi: 10.3389/fgene.2020.578345
302. Subramanian I, Verma S, Kumar S, Jere A, Anamika K. Multi-omics data integration, interpretation, and its application. Bioinform Biol Insights. (2020) 14:1177932219899051. doi: 10.1177/1177932219899051
303. Mu C, Zhao Q, Zhao Q, Yang L, Pang X, Liu T, et al. Multi-omics in Crohn's disease: new insights from inside. Comput Struct Biotechnol J. (2023) 21:3054–72. doi: 10.1016/j.csbj.2023.05.010
304. Yang Y, Shi X, Du Z, Zhou G, Zhang X. Associations between genetic variations in microRNA and myocardial infarction susceptibility: a meta-analysis and systematic review. Herz. (2022) 47(6):524–35. doi: 10.1007/s00059-021-05086-3
305. Luo X, Hu Y, Shen J, Liu X, Wang T, Li L, et al. Integrative analysis of DNA methylation and gene expression reveals key molecular signatures in acute myocardial infarction. Clin Epigenetics. (2022) 14(1):46. doi: 10.1186/s13148-022-01267-x
306. Han Y, Duan B, Wu J, Zheng Y, Gu Y, Cai X, et al. Analysis of time series gene expression and DNA methylation reveals the molecular features of myocardial infarction progression. Front Cardiovasc Med. (2022) 9:912454. doi: 10.3389/fcvm.2022.912454
307. Liu Y, Yan S, Zou L, Wen J, Fu W. Noise exposure and risk of myocardial infarction incidence and mortality: a dose-response meta-analysis. Environ Sci Pollut Res Int. (2022) 29(31):46458–70. doi: 10.1007/s11356-022-20377-w
308. Merlo AC, Troccolo A, Piredda E, Porto I, Gil Ad V. Myocardial infarction with non-obstructive coronary arteries: risk factors and associated comorbidities. Front Cardiovasc Med. (2022) 9:895053. doi: 10.3389/fcvm.2022.895053
309. Sagris M, Antonopoulos AS, Theofilis P, Oikonomou E, Siasos G, Tsalamandris S, et al. Risk factors profile of young and older patients with myocardial infarction. Cardiovasc Res. (2022) 118(10):2281–92. doi: 10.1093/cvr/cvab264
310. Wienbergen H, Boakye D, Günther K, Schmucker J, Mata Marín LA, Kerniss H, et al. Lifestyle and metabolic risk factors in patients with early-onset myocardial infarction: a case-control study. Eur J Prev Cardiol. (2022) 29(16):2076–87. doi: 10.1093/eurjpc/zwac132
311. Chen L, Wang H, Gao F, Zhang J, Zhang Y, Ma R, et al. Functional genetic variants in the SIRT5 gene promoter in acute myocardial infarction. Gene. (2018) 675:233–9. doi: 10.1016/j.gene.2018.07.010
312. Zhang P, Zhang J, Zhang Y, Wang S, Pang S, Yan B. Functional variants of the ATG7 gene promoter in acute myocardial infarction. Mol Genet Genomic Med. (2018) 6(6):1209–19. doi: 10.1002/mgg3.508
313. Sun Z, Pang S, Cui Y, Yan B. Genetic and functional variants analysis of the GATA6 gene promoter in acute myocardial infarction. Front Genet. (2019) 10:1100. doi: 10.3389/fgene.2019.01100
314. Wang H, Zhang S, Wang N, Zhang J, Chen M, He X, et al. Genetic variants of VEGFR-1 gene promoter in acute myocardial infarction. Hum Genomics. (2019) 13(1):56. doi: 10.1186/s40246-019-0243-1
315. Sedky NK, Abdel Rahman MF, Hassanein SI, Gad MZ. Genetic variants of CYP2R1 are key regulators of serum vitamin D levels and incidence of myocardial infarction in middle-aged Egyptians. Curr Pharm Biotechnol. (2018) 19(3):265–73. doi: 10.2174/1389201019666180528082737
316. Asif M, Bhat S, Nizamuddin S, Mustak MS. TG Haplotype in the LRP8 is associated with myocardial infarction in south Indian population. Gene. (2018) 642:225–9. doi: 10.1016/j.gene.2017.10.037
317. Semaev S, Shakhtshneider E, Orlov P, Ivanoshchuk D, Malyutina S, Gafarov V, et al. Association of RS708272 (CETP gene variant) with lipid profile parameters and the risk of myocardial infarction in the white population of Western Siberia. Biomolecules. (2019) 9(11):739. doi: 10.3390/biom9110739
318. Wang J, Lin B, Zhang Y, Ni L, Hu L, Yang J, et al. The regulatory role of histone modification on gene expression in the early stage of myocardial infarction. Front Cardiovasc Med. (2020) 7:594325. doi: 10.3389/fcvm.2020.594325
319. Corbin LJ, White SJ, Taylor AE, Williams CM, Taylor K, van den Bosch MT, et al. Epigenetic regulation of F2RL3 associates with myocardial infarction and platelet function. Circ Res. (2022) 130(3):384–400. doi: 10.1161/circresaha.121.318836
320. Liu Z, Huang S. Upregulation of SPI1 during myocardial infarction aggravates cardiac tissue injury and disease progression through activation of the TLR4/NFκB axis. Am J Transl Res. (2022) 14(4):2709–27.35559389
321. Wu J, Yan J, Hua Z, Jia J, Zhou Z, Zhang J, et al. Identification of molecular signatures in acute myocardial infarction based on integrative analysis of proteomics and transcriptomics. Genomics. (2023) 115(5):110701. doi: 10.1016/j.ygeno.2023.110701
322. Jia D, Zhang CZ, Qiu Y, Chen XF, Jia L, Chen AF, et al. Cardioprotective mechanisms of salvianic acid A sodium in rats with myocardial infarction based on proteome and transcriptome analysis. Acta Pharmacol Sin. (2019) 40(12):1513–22. doi: 10.1038/s41401-019-0265-1
323. Li Y, Wang C, Li T, Ma L, Fan F, Jin Y, et al. The whole transcriptome and proteome changes in the early stage of myocardial infarction. Cell Death Discov. (2019) 5:73. doi: 10.1038/s41420-019-0152-z
324. Liu W, Shen J, Li Y, Wu J, Luo X, Yu Y, et al. Pyroptosis inhibition improves the symptom of acute myocardial infarction. Cell Death Dis. (2021) 12(10):852. doi: 10.1038/s41419-021-04143-3
325. Contessotto P, Spelat R, Ferro F, Vysockas V, Krivickienė A, Jin C, et al. Reproducing extracellular matrix adverse remodelling of non-ST myocardial infarction in a large animal model. Nat Commun. (2023) 14(1):995. doi: 10.1038/s41467-023-36350-1
326. Jia D, Xiong L, Yu X, Chen X, Wang T, Chen AF, et al. Cardioprotective mechanism study of salvianic acid A sodium based on a proteome microarray approach and metabolomic profiling of rat serum after myocardial infarction. Molecular Omics. (2019) 15(4):271–9. doi: 10.1039/c9mo00005d
327. Yan T, Zhu X, Zhang X, Jia X, Liu J, Wang X, et al. The application of proteomics and metabolomics to reveal the molecular mechanism of Nutmeg-5 in ameliorating cardiac fibrosis following myocardial infarction. Phytomedicine. (2022) 105:154382. doi: 10.1016/j.phymed.2022.154382
328. Zhang P, Fang Z, Zhao M, Yi B, Huang Y, Yang H, et al. Ethanol extract of Pueraria lobata improve acute myocardial infarction in rats via regulating gut microbiota and bile acid metabolism. Phytother Res. (2023). doi: 10.1002/ptr.8005. [Epub ahead of print]
329. Chan MY, Efthymios M, Tan SH, Pickering JW, Troughton R, Pemberton C, et al. Prioritizing candidates of post-myocardial infarction heart failure using plasma proteomics and single-cell transcriptomics. Circulation. (2020) 142(15):1408–21. doi: 10.1161/circulationaha.119.045158
330. Li X, Lian Y, Wu Y, Ye Z, Feng J, Zhao Y, et al. Neonatal plasma exosomes contribute to endothelial cell-mediated angiogenesis and cardiac repair after acute myocardial infarction. Int J Mol Sci. (2023) 24(4):3196. doi: 10.3390/ijms24043196
331. Zhang X, Zhang X, Tong F, Cai Y, Zhang Y, Song H, et al. Gut microbiota induces high platelet response in patients with ST segment elevation myocardial infarction after ticagrelor treatment. eLife. (2022) 11:e70240. doi: 10.7554/eLife.70240
332. Dong C, Yang Y, Wang Y, Hu X, Wang Q, Gao F, et al. Gut microbiota combined with metabolites reveals unique features of acute myocardial infarction patients different from stable coronary artery disease. J Adv Res. (2023) 46:101–12. doi: 10.1016/j.jare.2022.06.008
333. Liao J, Zhang Y, Ma C, Wu G, Zhang W. Microbiome-metabolome reveals that the Suxiao Jiuxin pill attenuates acute myocardial infarction associated with fatty acid metabolism. J Ethnopharmacol. (2023) 312:116529. doi: 10.1016/j.jep.2023.116529
334. Kim SH, Lee KY, Chang K. The protective role of TREM2 in the heterogenous population of macrophages during post-myocardial infarction inflammation. Int J Mol Sci. (2023) 24(6):5556. doi: 10.3390/ijms24065556
335. Kuppe C, Ramirez Flores RO, Li Z, Hayat S, Levinson RT, Liao X, et al. Spatial multi-omic map of human myocardial infarction. Nature. (2022) 608(7924):766–77. doi: 10.1038/s41586-022-05060-x
336. Lavine K, Amrute J, Luo X, Penna V, Bredemeyer A, Yamawaki T, et al. Targeting immune-fibroblast crosstalk in myocardial infarction and cardiac fibrosis. Res Sq. (2023) rs.3.rs-2402606. doi: 10.21203/rs.3.rs-2402606/v1
337. Wang P, Shen C, Diao L, Yang Z, Fan F, Wang C, et al. Aberrant hypermethylation of aldehyde dehydrogenase 2 promoter upstream sequence in rats with experimental myocardial infarction. BioMed Res Int. (2015) 2015:503692. doi: 10.1155/2015/503692
338. Lan C, Chen C, Qu S, Cao N, Luo H, Yu C, et al. Inhibition of DYRK1A, via histone modification, promotes cardiomyocyte cell cycle activation and cardiac repair after myocardial infarction. EBioMedicine. (2022) 82:104139. doi: 10.1016/j.ebiom.2022.104139
339. Ward-Caviness CK, Agha G, Chen BH, Pfeiffer L, Wilson R, Wolf P, et al. Analysis of repeated leukocyte DNA methylation assessments reveals persistent epigenetic alterations after an incident myocardial infarction. Clin Epigenetics. (2018) 10(1):161. doi: 10.1186/s13148-018-0588-7
340. Hadas Y, Vincek AS, Youssef E, Żak MM, Chepurko E, Sultana N, et al. Altering sphingolipid metabolism attenuates cell death and inflammatory response after myocardial infarction. Circulation. (2020) 141(11):916–30. doi: 10.1161/circulationaha.119.041882
341. Lim SY, Lim FLS, Criado-Navarro I, Yeo XH, Dayal H, Vemulapalli SD, et al. Multi-omics investigation into acute myocardial infarction: an integrative method revealing interconnections amongst the metabolome, lipidome, glycome, and metallome. Metabolites. (2022) 12(11):1080. doi: 10.3390/metabo12111080
342. Guo AH, Baliira R, Skinner ME, Kumar S, Andren A, Zhang L, et al. Sirtuin 5 levels are limiting in preserving cardiac function and suppressing fibrosis in response to pressure overload. Sci Rep. (2022) 12(1):12258. doi: 10.1038/s41598-022-16506-7
343. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. (2002) 12(6):996–1006. doi: 10.1101/gr.229102
344. Zhernakova NI, Bunova SS, Agarkov NM, Lebedev DT, Aksenov VV. Vitamin D deficiency as an independent predictor of myocardial infarction in the elderly. Arch Razi Inst. (2021) 76(4):1069–76. doi: 10.22092/ari.2021.356047.1766
345. Liu X, Wu S, Song Q, Wang X. Visit-to-visit variability of lipid measurements and the risk of myocardial infarction and all-cause mortality: a prospective cohort study. Atherosclerosis. (2020) 312:110–6. doi: 10.1016/j.atherosclerosis.2020.09.003
346. Roos-Weil D, Decaudin C, Armand M, Della-Valle V, Diop MK, Ghamlouch H, et al. A recurrent activating missense mutation in waldenström macroglobulinemia affects the DNA binding of the ETS transcription factor SPI1 and enhances proliferation. Cancer Discov. (2019) 9(6):796–811. doi: 10.1158/2159-8290.Cd-18-0873
347. Niu X, Zhang J, Zhang L, Hou Y, Pu S, Chu A, et al. Weighted gene co-expression network analysis identifies critical genes in the development of heart failure after acute myocardial infarction. Front Genet. (2019) 10:1214. doi: 10.3389/fgene.2019.01214
348. Sun L, Ferreira JC, Mochly-Rosen D. ALDH2 Activator inhibits increased myocardial infarction injury by nitroglycerin tolerance. Sci Transl Med. (2011) 3(107):107ra11. doi: 10.1126/scitranslmed.3002067
349. Punwani D, Pelz B, Yu J, Arva NC, Schafernak K, Kondratowicz K, et al. Coronin-1A: immune deficiency in humans and mice. J Clin Immunol. (2015) 35(2):100–7. doi: 10.1007/s10875-015-0130-z
350. Katz MG, Hadas Y, Vincek A, Freage-Kahn L, Shtraizent N, Madjarov JM, et al. Acid ceramidase gene therapy ameliorates pulmonary arterial hypertension with right heart dysfunction. Respir Res. (2023) 24(1):197. doi: 10.1186/s12931-023-02487-2
351. Burtt N, Flannick J, Costanzo M, Duby M, Hoang Q, Jang DK, et al. Cardiovascular Disease Knowledge Portal (CVDKP) (2023). Available at: https://cvd.hugeamp.org/ (Accessed August 15, 2023).
352. Khomtchouk BB, Vand KA, Koehler WC, Tran DT, Middlebrook K, Sudhakaran S, et al. Heartbioportal. Circ Genom Precis Med. (2019) 12(4):e002426. doi: 10.1161/circgen.118.002426
353. Sollis E, Mosaku A, Abid A, Buniello A, Cerezo M, Gil L, et al. The NHGRI-EBI GWAS catalog: knowledgebase and deposition resource. Nucleic Acids Res. (2023) 51(D1):D977–D85. doi: 10.1093/nar/gkac1010
354. Athar A, Füllgrabe A, George N, Iqbal H, Huerta L, Ali A, et al. Arrayexpress update—from bulk to single-cell expression data. Nucleic Acids Res. (2019) 47(D1):D711–D5. doi: 10.1093/nar/gky964
355. GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet. (2013) 45(6):580–5. doi: 10.1038/ng.2653
356. Burgin J, Ahamed A, Cummins C, Devraj R, Gueye K, Gupta D, et al. The European nucleotide archive in 2022. Nucleic Acids Res. (2023) 51(D1):D121–D5. doi: 10.1093/nar/gkac1051
357. Deutsch EW, Bandeira N, Perez-Riverol Y, Sharma V, Carver JJ, Mendoza L, et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. (2023) 51(D1):D1539–D48. doi: 10.1093/nar/gkac1040
358. Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, et al. The PeptideAtlas project. Nucleic Acids Res. (2006) 34(Database issue):D655–8. doi: 10.1093/nar/gkj040
359. Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. (2013) 41(Database issue):D1063–9. doi: 10.1093/nar/gks1262
360. Burley SK, Bhikadiya C, Bi C, Bittrich S, Chao H, Chen L, et al. RCSB protein data bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. (2023) 51(D1):D488–D508. doi: 10.1093/nar/gkac1077
361. Lautenbacher L, Samaras P, Muller J, Grafberger A, Shraideh M, Rank J, et al. ProteomicsDB: toward a FAIR open-source resource for life-science research. Nucleic Acids Res. (2022) 50(D1):D1541–D52. doi: 10.1093/nar/gkab1026
362. Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based human protein atlas. Nat Biotechnol. (2010) 28(12):1248–50. doi: 10.1038/nbt1210-1248
363. Sud M, Fahy E, Cotter D, Azam K, Vadivelu I, Burant C, et al. Metabolomics workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. (2016) 44(D1):D463–70. doi: 10.1093/nar/gkv1042
364. Haug K, Cochrane K, Nainala VC, Williams M, Chang J, Jayaseelan KV, et al. Metabolights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. (2020) 48(D1):D440–D4. doi: 10.1093/nar/gkz1019
365. Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol. (2016) 34(8):828–37. doi: 10.1038/nbt.3597
366. Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The human cell atlas. eLife. (2017) 6:e27041. doi: 10.7554/eLife.27041
367. Tarhan L, Bistline J, Chang J, Galloway B, Hanna E, Weitz E. Single cell portal: an interactive home for single-cell genomics data. bioRxiv. (2023) 2023.07.13.548886. doi: 10.1101/2023.07.13.548886
368. Li M, Zhang X, Ang KS, Ling J, Sethi R, Lee NYS, et al. DISCO: a database of deeply integrated human single-cell omics data. Nucleic Acids Res. (2022) 50(D1):D596–D602. doi: 10.1093/nar/gkab1020
369. Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, et al. Database resources of the national center for biotechnology information in 2023. Nucleic Acids Res. (2023) 51(D1):D29–D38. doi: 10.1093/nar/gkac1032
370. Thakur M, Bateman A, Brooksbank C, Freeberg M, Harrison M, Hartley M, et al. EMBL's European bioinformatics institute (EMBL-EBI) in 2022. Nucleic Acids Res. (2023) 51(D1):D9–D17. doi: 10.1093/nar/gkac1098
371. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. (2023) 51(D1):D638–D46. doi: 10.1093/nar/gkac1000
372. Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. (2022) 50(D1):D687–D92. doi: 10.1093/nar/gkab1028
373. Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. (2023) 51(D1):D587–D92. doi: 10.1093/nar/gkac963
374. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. (2004) 5(10):R80. doi: 10.1186/gb-2004-5-10-r80
375. Rohart F, Gautier B, Singh A, Lê Cao KA. Mixomics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. (2017) 13(11):e1005752. doi: 10.1371/journal.pcbi.1005752
376. Mangiola S, Doyle MA, Papenfuss AT. Interfacing Seurat with the R tidy universe. Bioinformatics. (2021) 37(22):4100–7. doi: 10.1093/bioinformatics/btab404
377. Kramer O. Scikit-learn. Machine learning for evolution strategies. Cham: Springer International Publishing (2016). 45–53.
378. Liu C, Wang L, Liu Z. scGREAT: graph-based regulatory element analysis tool for single-cell multi-omics data. bioRxiv. (2023) 2023.01.27.525916. doi: 10.1101/2023.01.27.525916
379. Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. (2006) 34(Web Server issue):W729–32. doi: 10.1093/nar/gkl320
380. Fillbrunn A, Dietz C, Pfeuffer J, Rahn R, Landrum GA, Berthold MR. KNIME For reproducible cross-domain analysis of life science data. J Biotechnol. (2017) 261:149–56. doi: 10.1016/j.jbiotec.2017.07.028
381. Lin SH, Thakur R, Machiela MJ. LDexpress: an online tool for integrating population-specific linkage disequilibrium patterns with tissue-specific expression data. BMC Bioinformatics. (2021) 22(1):608. doi: 10.1186/s12859-021-04531-8
382. Liu T, Salguero P, Petek M, Martinez-Mira C, Balzano-Nogueira L, Ramšak Ž, et al. Paintomics 4: new tools for the integrative analysis of multi-omics datasets supported by multiple pathway databases. Nucleic Acids Res. (2022) 50(W1):W551–W9. doi: 10.1093/nar/gkac352
383. Gao B, Zhu J, Negi S, Zhang X, Gyoneva S, Casey F, et al. Quickomics: exploring omics data in an intuitive, interactive and informative manner. Bioinformatics. (2021) 37(20):3670–2. doi: 10.1093/bioinformatics/btab255
384. Gerstner N, Kehl T, Lenhof K, Müller A, Mayer C, Eckhart L, et al. Genetrail 3: advanced high-throughput enrichment analysis. Nucleic Acids Res. (2020) 48(W1):W515–W20. doi: 10.1093/nar/gkaa306
385. Conard AM, Goodman N, Hu Y, Perrimon N, Singh R, Lawrence C, et al. TIMEOR: a web-based tool to uncover temporal regulatory mechanisms from multi-omics data. Nucleic Acids Res. (2021) 49(W1):W641–W53. doi: 10.1093/nar/gkab384
386. Tuncbag N, Gosline SJ, Kedaigle A, Soltis AR, Gitter A, Fraenkel E. Network-based interpretation of diverse high-throughput datasets through the omics integrator software package. PLoS Comput Biol. (2016) 12(4):e1004879. doi: 10.1371/journal.pcbi.1004879
387. Ding J, Blencowe M, Nghiem T, Ha SM, Chen YW, Li G, et al. Mergeomics 2.0: a web server for multi-omics data integration to elucidate disease networks and predict therapeutics. Nucleic Acids Res. (2021) 49(W1):W375–W87. doi: 10.1093/nar/gkab405
388. Lavender CA, Shapiro AJ, Burkholder AB, Bennett BD, Adelman K, Fargo DC. ORIO (Online Resource for Integrative Omics): a web-based platform for rapid integration of next generation sequencing data. Nucleic Acids Res. (2017) 45(10):5678–90. doi: 10.1093/nar/gkx270
389. Wandy J, Daly R. Graphomics: an interactive platform to explore and integrate multi-omics data. BMC Bioinformatics. (2021) 22(1):603. doi: 10.1186/s12859-021-04500-1
390. Kuo TC, Tian TF, Tseng YJ. 3Omics: a web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst Biol. (2013) 7:64. doi: 10.1186/1752-0509-7-64
391. Anžel A, Heider D, Hattab G. MOVIS: a multi-omics software solution for multi-modal time-series clustering, embedding, and visualizing tasks. Comput Struct Biotechnol J. (2022) 20:1044–55. doi: 10.1016/j.csbj.2022.02.012
392. Uppal K, Ma C, Go YM, Jones DP, Wren J. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics. (2018) 34(4):701–2. doi: 10.1093/bioinformatics/btx656
393. Zhou G, Pang Z, Lu Y, Ewald J, Xia J. Omicsnet 2.0: a web-based platform for multi-omics integration and network visual analytics. Nucleic Acids Res. (2022) 50(W1):W527–W33. doi: 10.1093/nar/gkac376
394. Li J, Sang Y, Zeng S, Mo S, Zhang Z, He S, et al. Microbiosee: a web-based visualization toolkit for multi-omics of microbiology. Front Genet. (2022) 13:853612. doi: 10.3389/fgene.2022.853612
395. Li H, Shi M, Ren K, Zhang L, Ye W, Zhang W, et al. Visual omics: a web-based platform for omics data analysis and visualization with rich graph-tuning capabilities. Bioinformatics. (2023) 39(1):btac7. doi: 10.1093/bioinformatics/btac777
396. Harbig TA, Fratte J, Krone M, Nieselt K. OmicsTIDE: interactive exploration of trends in multi-omics data. Bioinform Adv. (2023) 3(1):vbac093. doi: 10.1093/bioadv/vbac093
397. Galaxy Community. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. (2022) 50(W1):W345–W51. doi: 10.1093/nar/gkac247
398. Shen B, Lin Y, Bi C, Zhou S, Bai Z, Zheng G, et al. Translational informatics for Parkinson's disease: from big biomedical data to amall actionable alterations. Genomics Proteomics Bioinformatics. (2019) 17(4):415–29. doi: 10.1016/j.gpb.2018.10.007
Keywords: myocardial infarction, multi-omics, personalized medicine, single-omics, integrative analysis
Citation: Zhan C, Tang T, Wu E, Zhang Y, He M, Wu R, Bi C, Wang J, Zhang Y and Shen B (2023) From multi-omics approaches to personalized medicine in myocardial infarction. Front. Cardiovasc. Med. 10:1250340. doi: 10.3389/fcvm.2023.1250340
Received: 30 June 2023; Accepted: 17 October 2023;
Published: 30 October 2023.
Edited by:
Emma Louise Robinson, University of Colorado, United StatesReviewed by:
Dafeng Yang, The Second Xiangya Hospital of Central South University, ChinaWilson Lek Wen Tan, Stanford University, United States
© 2023 Zhan, Tang, Wu, Zhang, He, Wu, Bi, Wang, Zhang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bairong Shen YmFpcm9uZy5zaGVuQHNjdS5lZHUuY24=
 Chaoying Zhan
Chaoying Zhan Tong Tang1
Tong Tang1 Rongrong Wu
Rongrong Wu Jiao Wang
Jiao Wang Bairong Shen
Bairong Shen