
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cardiovasc. Med. , 05 July 2023
Sec. Cardiovascular Genetics and Systems Medicine
Volume 10 - 2023 | https://doi.org/10.3389/fcvm.2023.1225014
This article is part of the Research Topic RNA-Chromatin Interactions: Biology, Mechanism, Disease and Therapeutics - Volume 2 View all 5 articles
The global leading cause of death is cardiovascular disease (CVD). Although advances in prevention and treatment have been made, the role of RNA epigenetics in CVD is not fully understood. Studies have found that RNA modifications regulate gene expression in mammalian cells, and m5C (5-methylcytosine) is a recently discovered RNA modification that plays a role in gene regulation. As a result of these developments, there has been renewed interest in elucidating the nature and function of RNA “epitranscriptomic” modifications. Recent studies on m5C RNA methylomes, their functions, and the proteins that initiate, translate and manipulate this modification are discussed in this review. This review improves the understanding of m5C modifications and their properties, functions, and implications in cardiac pathologies, including cardiomyopathy, heart failure, and atherosclerosis.
Cardiovascular disease (CVD) is the leading cause of death in many parts of the world, with an estimated 18 million deaths each year (1). By 2030, more than 23.3 million people will die from CVD, according to the World Health Organization (WHO) (2, 3). As the world’s leading cause of death, we must better understand the cellular, molecular, and genetic origins of CVD (4). There are currently a number of clinical trials investigating the safety and efficacy of RNA therapeutics in clinical conditions such as CVD (5). Over a hundred chemical modifications can be made to cellular RNA, and these modifications have been recently found to be important for posttranscriptional regulation (6–8). Most posttranscriptional modifications of RNA are conserved throughout evolution and can be found in all kingdoms of life (9). RNA methylation, which is the most prevalent epigenetic modification of RNA nucleotides, with over 170 different modifications reported thus far, primarily manifests as 7-methylguanosine (m7G), 5-methylcytosine (m5C) (Table 1), 5-hydroxymethylcytosine (5-hmC), N1-methyladenosine (m1A), N6-methyladenosine (m6A), N6, 2′-O-dimethyladenosine (m6Am), and 2′-O-methylation (2′-OMe) (10). M5C has been found to be highly abundant in eukaryotic mRNA molecules. Transcriptome analysis has identified more than 10,000 m5C sites (11). Archaea has also been reported to contain this modification, but it has not yet been identified in bacterial mRNA molecules (12). There is growing evidence that m5C RNA modifications play a broad role in RNA metabolism, including RNA export, the stability, efficiency, accuracy of translation, and long-distance RNA transport (13). The focus of this review article will be exploring the effect of m5C methyltransferases on CVD pathology since this modification, although there are a great number of other modifications, has been shown to be pivotal in CVD.
A methyltransferase (writer) and a demethylase (eraser) can reversibly regulate the methylation levels of m5C. The majority of m5C molecular functions are accomplished by binding proteins (readers). Recent studies have demonstrated that m5C plays multiple molecular roles in RNA processing, including mRNA export, RNA stability, translation and long-distance RNA transport (Figure 1) (13).
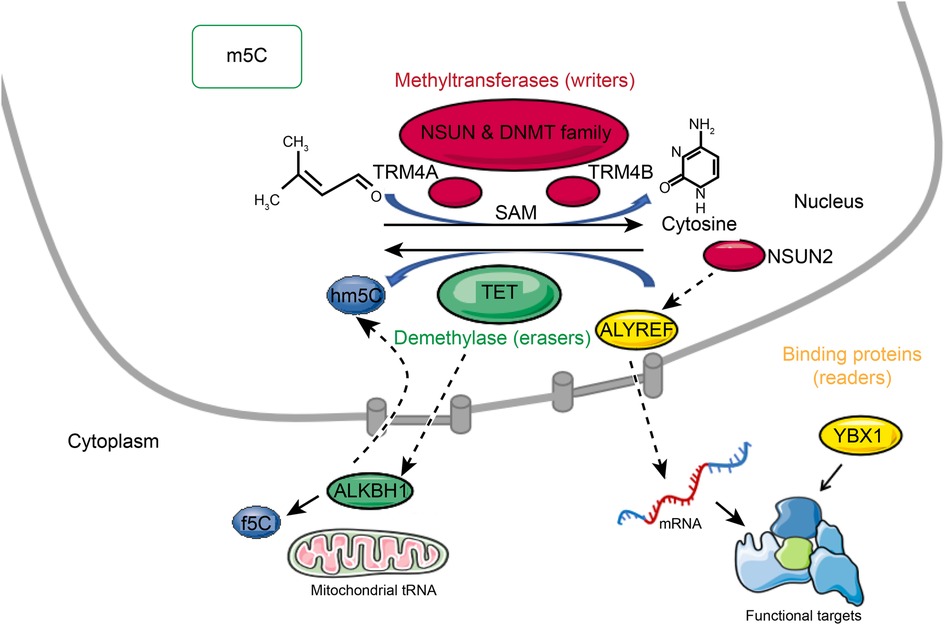
Figure 1. Reversible m5C mRNA modification. A methyltransferase (writer) and a demethylase (eraser) can reversibly regulate the methylation levels of m5C. The majority of m5C molecular functions are accomplished by binding proteins (readers). M5C is formed by methyltransferases that use S-adenosylmethionine (SAM) as the methyl donor and transfer the methyl group to cytosine. M5C methyltransferases include NSUN family members, DNMT, and TRDMT families. M5C can be oxidized by TET enzymes in mRNA to produce 5-hydroxymethylcytosine (hm5C), and f5C by alkb homolog 1 (ALKBH1) is formed at the mitochondrial tRNA wobble position. The binding partner ALYREF and the methyltransferase NSUN2 of m5C regulate mRNA export. YBX1 is a cytoplasmic mRNA m5C reader protein.
M5C is formed by methyltransferases that use S-adenosylmethionine (SAM) as the methyl donor and transfer the methyl group to cytosine (14). As many as 10 RNA m5C methyltransferases are known, including NOL1/NOP2/SUN domain (NSUN) family members, DNA methyltransferase (DNMT) homologs, and tRNA-specific methyltransferase (TRDMT) families. The NSUN family includes several variants (NSUN1 to NSUN7) and NSUN5a/b/c (15–17). In Arabidopsis thaliana, TRM4A and TRM4B are members of the TRDMT family (18, 19). Different cysteine residues are used in the catalysis of m5C RNA and m5C DNA methyltransferases. Enzymes of the NSUN and DNMT families contain amino acid motifs IV and VI. In the NSUN family, cysteine at position VI disrupts carbon 6 of the target cytosine by nucleophilic interaction (14, 20). As a result of hydrogen interactions with the proline and aspartate sidechains in motif IV, the nucleobase is oriented in an active position, and transient protonation is promoted. The methyl group from the donor SAM is then recognized by the activated base (14).
The role of m5C writers is now well-documented, but the role of m5C erasers is still controversial. It has been reported that m5C can be oxidized by enzymes of the ten-eleven translocator family (TET) in mRNA to produce 5-hydroxymethylcytosine (hm5C) (21, 22), and f5C by alkb homolog 1 (ALKBH1) is formed at the mitochondrial tRNA wobble position. Although f5C deposition in mitochondrial tRNAs has been established biologically, hm5C deposition in mRNAs remains unclear in terms of its biological relevance (23–25). In DNA, the TET (ten-eleven translocation) family proteins, including TET1, TET2, and TET3, catalyze 5mC to 5-hydroxymethylcytosine (5hmC). It has been shown that TETs are also RNA demethylases. Overexpression of TETs can significantly increase the RNA level of 5hmC (21). Moreover, TET1 mediates the oxidation of 5-formylcytosine (f5C) into 5-carboxycytosine (5caC) in RNA, and TET2 regulates the oxidation of m5C in mRNA, preventing 5-methylcytosine from forming double-stranded RNA. TET2-catalyzed RNA hm5C also involves RNA degradation, suggesting that it plays an important role in posttranscriptional regulation (26). ALKBH1 was identified as a mitochondrial DNA and RNA dioxygenase (27). On mt-tRNAMet and the anticodon of cytoplasmic tRNALeu, ALKBH1 catalyzes the conversion of m5C34 to hm5Cm34 (5-hydroxymethyl-2′-O-methylcytidine) and f5Cm34 (5-formyl-2′-O-methylcytidine). There was a significant reduction in mitochondrial translation and oxygen consumption upon deletion of ALKBH1, suggesting that ALKBH1-mediated RNA m5C metabolism might play an important role in mitochondrial function. It is also interesting to note that ALKBH1 is also involved in demethylating N1-methyladenosine (m1A) within cytoplasmic tRNAs (24, 28). ALKBH1 can also specifically act on histone H2A as a histone dioxygenase, in addition to different DNA or RNA chemical modifications (29, 30).
Most of the biological functions of RNA modifications are related to the proteins that bind to them. Recent studies have demonstrated the ability to pull down m5C-modified oligos by affinity chromatography-mass spectrometry in the mRNA of multiple species and have shown that 5-methylcytosine mediates the nuclear export of mRNAs. Knockdown of Aly/REF export factor (ALYREF), an mRNA transport adaptor, did not affect the nuclear export of nonm5C-bearing mRNA. Therefore, ALYREF seems to be a true m5C “reader” protein that regulates mRNA fate according to its m5C status (31). ALYREF has been reported to be involved in nuclear-cytoplasmic shuttling and is enriched in nuclear speckles containing pre-mRNA processing factors. ALYREF protein is retained in the nuclear compartment when NSUN2 levels are depleted, but its total cellular level remains the same. The binding partner ALYREF and the methyltransferase NSUN2 of m5C regulate mRNA export. In contrast to ALYREF, YBX1 (Y-box-binding protein 1) is a cytoplasmic mRNA m5C reader protein. YBX1 recognizes m5C through the indole ring of W65 in its cold-shock domain (32). Different tissues or species express different reader proteins, indicating that m5C modifications are functionally specific. In summary, it is necessary for the whole dynamic modification system mediated by m5C to be clarified along with the role of different components in m5C-mediated RNA metabolism, including the role of methyltransferases, demethyltransferases, and reader proteins, to help improve the understanding of RNA processing.
NSUN1, also known as p120, NOL1, and NOP2 (S. cerevisiae), directly binds to 60–80S preribosomal particles and catalyzes C4447 to m5C in human 28S rRNA and C2870 to m5C in yeast 25S rRNA (33). NSUN1 expression enhances the proliferation capacity of human cancer cells regardless of their histological origin. NSUN1 can participate in gene expression regulation through bromodomain-containing protein 4 (BRD4) and Pol II recruitment in 5-AZA-resistant leukemia cell lines (34–36). According to a recent study, this protein participates in the m5C methylation of the HIV TAR RNA (trans-activating response element) by and competing with the HIV-1 Tat protein for TAR binding (37).
There is another RNA m5C methyltransferase present in eukaryotes, NSUN2 (NOP2/SUN family, member 2) (14, 38–41). Researchers have found that NSUN2, which catalyzes 5mC methylation in tRNAs C48/49/50, contributes to the stability of tRNAs and protein synthesis by mediating methylation in the variable loop of tRNA (42). In contrast, m5C mediated by NSUN2 is widespread within the coding sequence (CDS) in mRNA. According to several studies, knockdown of NSUN2 significantly decreased the methylation levels of almost 40% (2,016 of 5,063) of m5C sites from 1,144 mRNAs. It was found that 92% (577 out of 629) of the m5C sites from 500 mRNAs were NSUN2-dependent in HeLa cells with NSUN2 knockout (43).
Over a decade ago, NSUN3 was found to belong to the NSUN (Nol1/Nop2/Sun domain) family of m5C RNA methyltransferases (44). In human cells, NSUN3 is localized to the mitochondrial matrix and is the last uncharacterized member of the m5C methyltransferase family. NSUN3 specifically interacts with mitochondrial tRNAMet, and is responsible for introducing a m5C modification at the swing position. NSUN family m5C methyltransferases replace the cysteine in motif IV of the TCT tripeptide with alanine (C265A). A conserved catalytic cysteine requirement and the efficient cross-linking of NSUN3 to 5-azacytidine (5-AzaC)-containing mitochondrial (mt-)tRNAMet strongly support NSUN3’s role as an active m5C RNA methyltransferase. NSUN3 uses the conserved mechanism of the NSUN family to mediate the m5C methylation of mitochondrial tRNAmet (24).
In previous studies, NSUN4 has been reported as a m5C-methyltransferase involved in methylating an unknown 16S rRNA residue (45–47). A subsequent study demonstrated that NSUN4 is capable of independently methylating C911 in human mitochondrial 12S rRNA (m5C911) (48). NSUN4 forms a stable complex with MTERF4 (methyltransferase-like 14) on the large subunit of the ribosome and is crucial to mitochondrial ribosome assembly and translation (45, 46).
It has been reported that NSUN5 catalyzes both m5C3782 and m5C3438 in human and mouse 28S rRNA, respectively (49). A nonmethylated C3782 position in 28S rRNA may inhibit global protein synthesis, promoting a translational stress response in gliomas (50).
PUA domains are RNA-binding domains found in many proteins, including YebU, YccW, RlmO, and m5C methyltransferases (51–55). PUA domains recognize the D-stem regions and CCA ends of tRNA, whereas methyltransferase domains recognize the base and ambient residues precisely. It has been shown that the CCA tail can be accurately identified by the PUA domain of NSUN6, then a precise recognition of the target site C72 by the NSUN6 catalytic core occurs when U73 binds to the NSUN6 RNA-recognition motif (RRM). It is important for tRNA recognition to have the second and third base pairs (2:71 and 3:70) of the acceptor stem. Lys248, Asp323, Cys326, and Cys373 are strictly conserved at the active site of NSUN6 in the RNA m5C methyltransferase (56). Additionally, NSUN6 interacts with two specific base pairs of the D-stem in the tRNA substrate (11:24 and 12:23) (57). Pyrococcus horikoshii OT3 (P. horikoshii) contains five genes that encode RNA m5C methyltransferases: PH1537, PH1374, PH1078, PH1991, and PH0851. It has been demonstrated that OT3 and PH1991 of P. horikoshii are homologs of human NSUN6 and are capable of catalyzing the modification of m5C72 along with tRNA. M5C72-modified tRNAs were slightly more thermally stable than nonmodified tRNAs (58).
Enhancer RNAs (eRNAs), which are transcriptionally regulated by enhancers in a tissue-specific manner, are a recent addition to the list of regulatory noncoding RNAS (ncRNAs) (59). Enhancer RNAs are activated by NSUN7 by promoting m5C deposition coupled to coactivator PGC-1α. PGC-1α is important for adaptive metabolic responses, suggesting that eRNA m5C modification by NSUN7 regulates metabolism (60).
Although Dnmt2 possesses motifs typical of DNA methyltransferases rather than RNA methyltransferases, it has been shown to methylate RNA. The enzyme also acts as a tRNA methyltransferase and is responsible for m5C38 in tRNAAsp (14, 61).
Atherosclerosis begins with vascular inflammation, which is a pivotal event (62). Endothelial and leukocytic adhesion molecules and their shedding are essential for mediating interactions between endothelial cells and blood components or extracellular matrix (63, 64). Intercellular adhesion molecule-1 (ICAM-1) plays a crucial role in inflammation and immune responses as an immunoglobulin-like protein found in leukocytes and endothelial cells (65). Adhesion molecules on the surface of circulating mononuclear cells mediate their attachment to vessel walls during atherosclerosis, including vascular cell adhesion molecule 1 (VCAM-1), platelet cell adhesion molecule 1 (PECAM-1), and ICAM-1. It was found that in apoE-deficient mice, gene deletion of ICAM-1 reduced monocyte recruitment, which resulted in protection from atherosclerotic lesions (66–68). Methylation of RNA is a critical modification that occurs extensively in ncRNA (10–12) and mRNAs (13, 14). The tRNA methyltransferase NSUN2 promotes cell proliferation in response to Myc activation. Recent studies have shown that homocysteine (Hcy)-induced NSUN2 methylates ICAM-1 mRNA and promotes ICAM-1 translation. NSUN2 plays a role in vascular inflammation and allograft avascular necrosis by increasing ICAM-1 levels (Table 2) (69). Atherogenesis induced by hyperhomocysteinemia (HHcy) is associated with the upregulation of IL-17A expression in T lymphocytes (70); however, the mechanism underlying this process remains unknown. Hcy is an intermediate product of the methionine cycle, which produces methyl donors (8). HHcy is a condition with elevated plasma Hcy levels, resulting in chronic inflammation (71, 72). It has been reported that IL-17A mRNA is methylated in the coding region (CR) by NSUN2. NSUN2 promotes the translational expression of IL-17A, which may be responsible for HHcy-induced activation of T lymphocytes. There is evidence that NSUN2-mediated RNA methylation may have a broader impact on aging-related declines and pathologies. The interest in understanding the impact of RNA modifications has increased, and studies have shown that RNA-modifying enzymes can regulate gene expression programs by influencing the levels of proliferative proteins through the RNA-methyltransferase NSUN2 (Table 2) (73). Additionally, Hcy-induced metabolic disorders have been shown to contribute to the development of CVDs through DNA methylation (12).
Altered mitochondrial function is currently recognized as an important factor in the initiation and progression of atherosclerosis. The barrier and metabolic functions of artery wall cells make them particularly vulnerable to mitochondrial dysfunction. Atherosclerosis occurs when mitochondria alter metabolic and respiratory processes and produce excessive quantities of reactive oxygen species (ROS), leading to oxidative stress. These processes contribute to vascular disease and chronic inflammation associated with atherosclerosis (74). Studies have shown that ALKBH1 is responsible for the biogenesis of 5-hydroxymethyl-2’-O-methylcytidine (hm5Cm)34 and f5Cm34 in cytoplasmic (ct)-tRNALeu (CAA) as well as f5C34 in mitochondrial (mt)-tRNAMet. In ALKBH1-knockout cells, mitochondrial translation and oxygen consumption were strongly reduced, indicating that ALKBH1 is required for mitochondrial function (Table 2) (28). Mitochondrial gene expression dysfunction caused by mutations in mitochondrial or nuclear genomes can also play a crucial role in human diseases, including atherosclerosis and CVD (75). Two pathogenic mutations (A4435G and C4437U) in mt-tRNAMet prevent NSUN3 from forming m5C34 (Table 2) (25). Mitochondrial diseases caused by these mutations include MIEH (maternally inherited essential hypertension) (76, 77). As a result of hypertension, atherosclerosis is more likely to develop (78). An increasing number of mutations have been identified in nuclear genes that shape mtRNA epitranscriptomes (23, 79–86). In the pathogenesis of atherosclerosis, genetic factors play a crucial role; however, the precise mechanisms are still not fully understood. Even so, family history analysis and gene variant screening are effective strategies to predict the development of atherosclerosis in individuals who are susceptible to hypertension (87).
Stress is a fundamental concept in biology that has been extensively applied in various fields, including psychology, physiology, social sciences, and environmental studies. As the concept of homeostasis becomes more defined, the concept of stress is also becoming more specific. For example, oxidative stress specifically refers to a disturbance in redox signaling and regulation.Endoplasmic reticulum stress is caused by the buildup of unfolded proteins in the endoplasmic reticulum, leading to cellular stress and dysfunction (88). The Integrated Stress Response (ISR) is a signaling network that is conserved throughout evolution and operates within cells to help them adapt to changing environmental conditions. Its role is to maintain the health of cells, tissues and organisms by enabling them to respond effectively to various stressors (89). Human diseases may be caused by aberrant posttranscriptional methylation of RNA. Radical reprogramming of protein translation is required by mammalian cells exposed to adverse environmental conditions (90). An integrated stress response (ISR) can be activated when eukaryotic cells are stimulated by diverse stimuli by phosphorylating eukaryotic initiation factor 2 alpha (eIF2a). Atherosclerosis in mice and humans shows persistent phosphorylation of eIF2a, a key factor in translation control (88). Angiopoietins (Angs) regulate angiogenesis. These proteins are known to promote the development of atherosclerosis (91). Angiogenin is a secreted ribonuclease, and angiogenin activates a stress-response program in mammalian cells by cleaving tRNA. The cleavage of tRNAs in eukaryotes is a conserved response to several stress stimuli (92–95). Researchers have found that angiopoietin and stress-induced tRNA cleavage are newly discovered components of mammalian stress responses. Moreover, activation of phospho-eIF2-independent translation was promoted by transfection of angiogenin-induced tRNA (94). A family of elF2α kinases triggered stress-induced translational arrest of mRNAs encoding “housekeeping” proteins by reducing elF2-GTP-tRNAiMet complexes required for translation (96). The researchers found that loss of RNA m5C methylation increased angiogenin-mediated endonucleolytic cleavage of tRNA, leading to small RNA fragment accumulation from 50 tRNA molecules. Protein translation rates were reduced, and stress pathways were activated when the tRNA fragments accumulated (42). There was an increase in small tRNA fragments in response to exogenous Ang treatment. Moreover, DNMT2 overexpression protected substrate tRNAs from angiogenin-mediated degradation. Overexpression of Dnmt2 did not affect tRNAMet-ATG, a nonsubstrate of Dnmt2. Several tRNAs are methylated by Dnmt2, specifically at C38, and this methylation protects tRNAs from cleavage by the angiogenin ribonuclease (Table 2) (97).
Approximately 1% to 2% of the global adult population suffer from heart failure, which is considered an epidemic in modern times. The pathophysiological causes of heart failure encompass myocardial damage, cardiac hypertrophy, cardiac overload, cardiac electrophysiological abnormalities, and other diseases. Heart failure occurs when the heart cannot supply the peripheral tissues with sufficient blood and oxygen to meet their metabolic needs. Many studies have focused on understanding the pathophysiological mechanisms of heart failure (98). It is characterized by a reduction in genes involved in fatty acid oxidation and mitochondrial ATP production, which may be caused by insufficient energy sources. The downregulation of PGC-1α target genes in failing hearts is attributed, in part, to a reduction in the occupancy and recruitment of polymerase II to promoter sites, which might be a novel mechanism of metabolic perturbations in the failing heart (99). Both Dnmt2-deficient mice and Dnmt2-deficient embryonic stem (ES) cells activate RNA polymerase II (pol II), one of the key players in cardiac hypertrophy. RNA pol II activation is tightly regulated by ncRNAs such as Rn7sk and B2 RNAs, in addition to proteins such as Cdk9. The transcriptional induction of Cdk9 following an injection of small noncoding RNAs (sncRNAs) in one-cell embryos results in cardiac hypertrophy (100). In summary, as a result of decreased dissociation of the negatively regulating ncRNA component Rn7sk, the positive transcription elongation factor b (P-TEFb) complex is activated, a critical step for cardiac growth. Dnmt2 reduces cardiac growth and ES cell differentiation by phosphorylating RNA pol II, which is controlled by the ncRNA Rn7sk (Table 2) (101).
Cardiomyopathy is a condition in which the heart structure and function are affected. The term idiopathic was used historically to denote an enigmatic etiology that specifically excluded cardiovascular or systemic diseases. There are morphological subtypes of cardiomyopathy, including hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), arrhythmogenic right ventricular cardiomyopathy (ARVC), and left ventricular noncompaction (LVNC) (102, 103). Cardiomyopathy and heart failure are prevalent presentations in mitochondrial disease resulting from inadequacies in the oxidative phosphorylation (OXPHOS) mechanism of mitochondria (104). Compared to other tissues, the muscle and heart tissue showed higher levels of m5C methylation in mice. A high methylation level and expression level of m5C-containing genes were associated with genes encoding mitochondrial and transport molecules in mouse muscle and heart tissue. In muscle and the heart tissue, the mouse VDAC1 gene contained three highly methylated sites. Generally, these data suggest that mitochondrial function may be affected by mRNA m5C modifications, particularly in organs that require the most energy, such as muscles and the heart (43). NSUN4 conditional inactivation in the heart causes respiratory chain abnormalities in germline knockouts as a result of impaired mitochondrial assembly and inhibition of mitochondrial translation. In 12S rRNA, NSUN4 increases the m5C modification at C911 residues, which regulates mitochondrial ribosome assembly and mitochondrial gene translation. Mitochondrial dysfunction occurs when Nsun4 is knocked out specifically in the heart. NSUN4 deletion in the heart caused cardiomyopathy, respiratory chain deficiency, and mitochondrial dysfunction, potentially as a result of suppressed mitoribosome assembly and mitochondrial translation (Table 2) (48). However, epigenetic modifications must be studied mechanistically to develop a more comprehensive CVD regulatory network. In the future, RNA m5C sites may prove to be an important target for assessing and treating clinical diseases. More research is needed on the molecular structures and functional pathways to fully understand the effect of RNA m5C modification and exploit it in clinical applications.
M5C sites are present in 34 different yeast tRNA species, but they are only found at the wobble position and position 48 of tRNALeuCAA. Upon cellular oxidative stress, there is a dynamic change in the distribution of m5C in tRNA, with an increased distribution at the wobble position and a decreased distribution at position 48, indicating a particular response to stress (105, 106). Under stress conditions, NSUN3-mediated m5C modifications in mitochondrial tRNAMet anticodon loops have been shown to decrease mitochondrial ROS production (107). Additionally, yeast, worms, and flies respond better to stress when NSUN5 is present (108). RCm1, the yeast homolog of NSUN5, catalyzes the formation of m5C2278 in 25S rRNA, which extends yeast lifespan. Specifically, Rcm1 loss alters ribosome conformation close to C2278 and translational stability, resulting in the recruitment of oxidative stress-responsive mRNAs into polysomes (109). Despite the fact that these stress responses are strongly associated with CVD, little is known about the interactions, and the detailed functions still need to be clarified.
The understanding of epigenetic modification in biological processes is increasing. In this review, we summarize the potential role of m5C methylation in CVDs and illustrate that m5C modification plays an important role in RNA epigenetics. We describe the “life cycle” of m5C methylation and its dominant function in CVDs (Figure 2). Moreover, we highlight the potential therapeutic role of interfering with m5C modifications, which could have a transformative effect on clinical medicine. Future research may uncover additional functions and underlying mechanisms of m5C methylation in CVDs, which would be important progress in the field of epigenetics.
YW, YL, YT, YZ, LC, and CZ performed the literature review. YW wrote most of the manuscript and revised it in its entirety. YT, YL, YZ, LC, and CZ wrote part of the manuscript and revised it in its entirety. CZ is responsible for ensuring that the descriptions are accurate and agreed upon by all authors. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81100106, 81670424), Hunan Provincial Natural Science Foundation of China (2021JJ30602), International Joint Laboratory for Arteriosclerotic Disease Research of Hunan Province (2018 WK4031), Outstanding Youth Project Supported by Scientific Research Fund of Hunan Provincial Education Department (19B478), Scientific Research Foundation for Doctor of University of South China (2012XQD37, 2017XQD04), and the Scientific Research Foundation for the Returned Overseas Scholars of University of South China (2017XQD29).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Maarman GJ, Chakafana G, Sliwa K. World heart day: a world heart federation communiqué on the future of basic sciences and translational medicine in global cardiovascular research. Am J Physiol Lung Cell Mol Physiol. (2020) 319(3):L545–6. doi: 10.1152/ajplung.00339.2020
2. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet. (2012) 380(9859):2095–128. doi: 10.1016/S0140-6736(12)61728-0
3. Lopez AD, Mathers CD. Measuring the global burden of disease and epidemiological transitions: 2002–2030. Ann Trop Med Parasitol. (2006) 100(5-6):481–99. doi: 10.1179/136485906X97417
4. Granger A, Emambokus N. Cell metabolism ♥ cardiovascular biology. Cell Metab. (2015) 21(2):151. doi: 10.1016/j.cmet.2015.01.018
5. Laina A, Gatsiou A, Georgiopoulos G, Stamatelopoulos K, Stellos K. RNA Therapeutics in cardiovascular precision medicine. Front Physiol. (2018) 9:953. doi: 10.3389/fphys.2018.00953
6. Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. (2014) 15:127–50. doi: 10.1146/annurev-genom-090413-025405
7. Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. (2016) 352(6292):1408–12. doi: 10.1126/science.aad8711
8. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. (2017) 169(7):1187–200. doi: 10.1016/j.cell.2017.05.045
9. Sarin LP, Leidel SA. Modify or die?–RNA modification defects in metazoans. RNA Biol. (2014) 11(12):1555–67. doi: 10.4161/15476286.2014.992279
10. Wu S, Zhang S, Wu X, Zhou X. Ma RNA methylation in cardiovascular diseases. Mol Ther. (2020) 28(10):2111–9. doi: 10.1016/j.ymthe.2020.08.010
11. Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. (2013) 9(6):e1003602. doi: 10.1371/journal.pgen.1003602
12. Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. (2016) 44(2):852–62. doi: 10.1093/nar/gkv1182
13. Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. (2021) 12(4):e1639. doi: 10.1002/wrna.1639
14. Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m⁵C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). (2019) 10(2):85–102. doi: 10.3390/genes10020102
15. Blanco S, Frye M. Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol. (2014) 31:1–7. doi: 10.1016/j.ceb.2014.06.006
16. Chi L, Delgado-Olguín P. Expression of NOL1/NOP2/sun domain (Nsun) RNA methyltransferase family genes in early mouse embryogenesis. Gene Expr Patterns. (2013) 13(8):319–27. doi: 10.1016/j.gep.2013.06.003
17. Reid R, Greene PJ, Santi DV. Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. (1999) 27(15):3138–45. doi: 10.1093/nar/27.15.3138
18. Cui X, Liang Z, Shen L, Zhang Q, Bao S, Geng Y, et al. 5-Methylcytosine RNA methylation in Arabidopsis Thaliana. Mol Plant. (2017) 10(11):1387–99. doi: 10.1016/j.molp.2017.09.013
19. David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, et al. Transcriptome-Wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell. (2017) 29(3):445–60. doi: 10.1105/tpc.16.00751
20. Liu Y, Santi DV. M5c RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc Natl Acad Sci USA. (2000) 97(15):8263–5. doi: 10.1073/pnas.97.15.8263
21. Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. (2014) 136(33):11582–5. doi: 10.1021/ja505305z
22. Shen Q, Zhang Q, Shi Y, Shi Q, Jiang Y, Gu Y, et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature. (2018) 554(7690):123–7. doi: 10.1038/nature25434
23. Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. (2016) 7:12039. doi: 10.1038/ncomms12039
24. Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, et al. NSUN3 And ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. (2016) 35(19):2104–19. doi: 10.15252/embj.201694885
25. Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 Methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. (2016) 12(7):546–51. doi: 10.1038/nchembio.2099
26. Guallar D, Bi X, Pardavila JA, Huang X, Saenz C, Shi X, et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat Gen. (2018) 50(3):443–51. doi: 10.1038/s41588-018-0060-9
27. Westbye MP, Feyzi E, Aas PA, Vågbø CB, Talstad VA, Kavli B, et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. (2008) 283(36):25046–56. doi: 10.1074/jbc.M803776200
28. Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, Suzuki T. ALKBH1 Is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. (2017) 45(12):7401–15. doi: 10.1093/nar/gkx354
29. Ougland R, Jonson I, Moen MN, Nesse G, Asker G, Klungland A, Larsen E. Role of ALKBH1 in the core transcriptional network of embryonic stem cells. Cell Physiol Biochem. (2016) 38(1):173–84. doi: 10.1159/000438619
30. Ougland R, Lando D, Jonson I, Dahl JA, Moen MN, Nordstrand LM, et al. ALKBH1 Is a histone H2A dioxygenase involved in neural differentiation. Stem Cells. (2012) 30(12):2672–82. doi: 10.1002/stem.1228
31. Dominissini D, Rechavi G. 5-methylcytosine Mediates nuclear export of mRNA. Cell Res. (2017) 27(6):717–9. doi: 10.1038/cr.2017.73
32. Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, et al. 5-methylcytosine Promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. (2019) 21(8):978–90. doi: 10.1038/s41556-019-0361-y
33. Bourgeois G, Ney M, Gaspar I, Aigueperse C, Schaefer M, Kellner S, et al. Eukaryotic rRNA modification by yeast 5-methylcytosine-methyltransferases and human proliferation-associated antigen p120. PloS One. (2015) 10(7):e0133321. doi: 10.1371/journal.pone.0133321
34. Trerè D, Migaldi M, Montanaro L, Pession A, Derenzini M. P120 expression provides a reliable indication of the rapidity of cell duplication in cancer cells independently of tumour origin. J Pathol. (2000) 192(2):216–20. doi: 10.1002/1096-9896(2000)9999:9999%3C::AID-PATH695%3E3.0.CO;2-L
35. Uchiyama B, Saijo Y, Kumano N, Abe T, Fujimura S, Ohkuda K, et al. Expression of nucleolar protein p120 in human lung cancer: difference in histological types as a marker for proliferation. Clin Cancer Res. (1997) 3(10):1873–7.9815576
36. Trixl L, Amort T, Wille A, Zinni M, Ebner S, Hechenberger C, et al. RNA Cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun. (2018) 9(1):1163. doi: 10.1038/s41467-018-03513-4
37. Kong W, Biswas A, Zhou D, Fiches G, Fujinaga K, Santoso N, Zhu J. Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with Tat for TAR binding and methylation. PLoS Pathog. (2020) 16(3):e1008430. doi: 10.1371/journal.ppat.1008430
38. Auxilien S, Guérineau V, Szweykowska-Kulińska Z, Golinelli-Pimpaneau B. The human tRNA m (5) C methyltransferase Misu is multisite-specific. RNA Biol. (2012) 9(11):1331–8. doi: 10.4161/rna.22180
39. Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA leu (CAA). Nucleic Acids Res. (2006) 34(20):6034–43. doi: 10.1093/nar/gkl765
40. Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, et al. 5-methylcytosine Promotes mRNA export—nSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. (2017) 27(5):606–25. doi: 10.1038/cr.2017.55
41. Sajini AA, Choudhury NR, Wagner RE, Bornelöv S, Selmi T, Spanos C, et al. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun. (2019) 10(1):2550. doi: 10.1038/s41467-019-10020-7
42. Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. (2014) 33(18):2020–39. doi: 10.15252/embj.201489282
43. Huang T, Chen W, Liu J, Gu N, Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Na Struct Mol Biol. (2019) 26(5):380–8. doi: 10.1038/s41594-019-0218-x
44. Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. (2004) 32(8):2453–63. doi: 10.1093/nar/gkh564
45. Cámara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, et al. MTERF4 Regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metabol. (2011) 13(5):527–39. doi: 10.1016/j.cmet.2011.04.002
46. Yakubovskaya E, Guja KE, Mejia E, Castano S, Hambardjieva E, Choi WS, Garcia-Diaz M. Structure of the essential MTERF4:nSUN4 protein complex reveals how an MTERF protein collaborates to facilitate rRNA modification. Structure. (2012) 20(11):1940–7. doi: 10.1016/j.str.2012.08.027
47. Spåhr H, Habermann B, Gustafsson CM, Larsson NG, Hallberg BM. Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc Natl Acad Sci USA. (2012) 109(38):15253–8. doi: 10.1073/pnas.1210688109
48. Metodiev MD, Spåhr H, Polosa PL, Meharg C, Becker C, Altmueller J, et al. NSUN4 Is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. (2014) 10(2):e1004110. doi: 10.1371/journal.pgen.1004110
49. Heissenberger C, Liendl L, Nagelreiter F, Gonskikh Y, Yang G, Stelzer EM, et al. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. (2019) 47(22):11807–25. doi: 10.1093/nar/gkz1043
50. Janin M, Ortiz-Barahona V, De Moura MC, Martínez-Cardús A, Llinàs-Arias P, Soler M, et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. (2019) 138(6):1053–74. doi: 10.1007/s00401-019-02062-4
51. Larsen LHG, Rasmussen A, Giessing AMB, Jogl G, Kirpekar F. Identification and characterization of the Thermus thermophilus 5-methylcytidine (m5C) methyltransferase modifying 23 S ribosomal RNA (rRNA) base C1942. J Biol Chem. (2012) 287(33):27593–600. doi: 10.1074/jbc.M112.376160
52. Andersen NM, Douthwaite S. Yebu is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J Mol Biol. (2006) 359(3):777–86. doi: 10.1016/j.jmb.2006.04.007
53. Pérez-Arellano I, Gallego J, Cervera J. The PUA domain—a structural and functional overview. FEBS J. (2007) 274(19):4972–84. doi: 10.1111/j.1742-4658.2007.06031.x
54. Purta E, O'Connor M, Bujnicki JM, Douthwaite S. Yccw is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J Mol Biol. (2008) 383(3):641–51. doi: 10.1016/j.jmb.2008.08.061
55. Seistrup KH, Rose S, Birkedal U, Nielsen H, Huber H, Douthwaite S. Bypassing rRNA methylation by RsmA/Dim1during ribosome maturation in the hyperthermophilic archaeon nanoarchaeum equitans. Nucleic Acids Res. (2017) 45(4):2007–15. doi: 10.1093/nar/gkw839
56. Liu RJ, Long T, Li J, Li H, Wang ED. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. (2017) 45(11):6684–97. doi: 10.1093/nar/gkx473
57. Long T, Li J, Li H, Zhou M, Zhou XL, Liu RJ, Wang ED. Sequence-specific and shape-selective RNA recognition by the human RNA 5-methylcytosine methyltransferase NSun6. J Biol Chem. (2016) 291(46):24293–303. doi: 10.1074/jbc.M116.742569
58. Li J, Li H, Long T, Dong H, Wang ED, Liu RJ. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. (2019) 47(4):2041–55. doi: 10.1093/nar/gky1236
59. Sartorelli V, Lauberth SM. Enhancer RNAs are an important regulatory layer of the epigenome. Nat Struct Mol Biol. (2020) 27(6):521–8. doi: 10.1038/s41594-020-0446-0
60. Aguilo F, Li S, Balasubramaniyan N, Sancho A, Benko S, Zhang F, et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep. (2016) 14(3):479–92. doi: 10.1016/j.celrep.2015.12.043
61. Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. (2006) 311(5759):395–8. doi: 10.1126/science.1120976
62. Libby P, Lichtman AH, Hansson GK. Immune effector mechanisms implicated in atherosclerosis: from mice to humans. Immunity. (2013) 38(6):1092–104. doi: 10.1016/j.immuni.2013.06.009
63. Mestas J, Ley K. Monocyte-endothelial cell interactions in the development of atherosclerosis. Trends Cardiovasc Med. (2008) 18(6):228–32. doi: 10.1016/j.tcm.2008.11.004
64. Fenyo IM, Gafencu AV. The involvement of the monocytes/macrophages in chronic inflammation associated with atherosclerosis. Immunobiol. (2013) 218(11):1376–84. doi: 10.1016/j.imbio.2013.06.005
65. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. (2013) 13(3):159–75. doi: 10.1038/nri3399
66. Glass CK, Witztum JL. Atherosclerosis. The road ahead. Cell. (2001) 104(4):503–16. doi: 10.1016/S0092-8674(01)00238-0
67. Collins RG, Velji R, Guevara NV, Hicks MJ, Chan L, Beaudet AL. P-Selectin or intercellular adhesion molecule (ICAM)-1 deficiency substantially protects against atherosclerosis in apolipoprotein E-deficient mice. J Experiment Med. (2000) 191(1):189–94. doi: 10.1084/jem.191.1.189
68. Huo Y, Ley K. Adhesion molecules and atherogenesis. Acta Physiol Scand. (2001) 173(1):35–43. doi: 10.1046/j.1365-201X.2001.00882.x
69. Luo Y, Feng J, Xu Q, Wang W, Wang X. NSun2 deficiency protects endothelium from inflammation via mRNA methylation of ICAM-1. Circ Res. (2016) 118(6):944–56. doi: 10.1161/CIRCRESAHA.115.307674
70. Feng J, Zhang Z, Kong W, Liu B, Xu Q, Wang X. Regulatory T cells ameliorate hyperhomocysteinaemia-accelerated atherosclerosis in apoE-/- mice. Cardiovasc Res. (2009) 84(1):155–63. doi: 10.1093/cvr/cvp182
71. Chen NC, Yang F, Capecci LM, Gu Z, Schafer AI, Durante W, et al. Relationship between serum homocysteine levels and depressive symptoms: the cooper center longitudinal study. J Clin Psych. (2012) 73(5):691–5. doi: 10.4088/JCP.11m07223
72. Gu P, DeFina LF, Leonard D, John S, Weiner MF, Brown ES. Regulation of homocysteine metabolism and methylation in human and mouse tissues. FASEB J. (2010) 24(8):2804–17. doi: 10.1096/fj.09-143651
73. Wang W. mRNA methylation by NSUN2 in cell proliferation. Wiley Interdiscip Rev RNA. (2016) 7(6):838–42. doi: 10.1002/wrna.1380
74. Salnikova D, Orekhova V, Grechko A, Starodubova A, Bezsonov E, Popkova T, Orekhov A. Mitochondrial dysfunction in vascular wall cells and its role in atherosclerosis. Int J Mol Sci. (2021) 22(16):8972–90. doi: 10.3390/ijms22168990
75. Wallace DC. Mitochondrial genetic medicine. Nat Gen. (2018) 50(12):1642–9. doi: 10.1038/s41588-018-0264-z
76. Lu Z, Chen H, Meng Y, Wang Y, Xue L, Zhi S, et al. The tRNAMet 4435A > G mutation in the mitochondrial haplogroup G2a1 is responsible for maternally inherited hypertension in a Chinese pedigree. Eur J Hum Genet. (2011) 19(11):1181–6. doi: 10.1038/ejhg.2011.111
77. Tang S, Wang J, Zhang VW, Li FY, Landsverk M, Cui H, et al. Transition to next generation analysis of the whole mitochondrial genome: a summary of molecular defects. Hum Mutat. (2013) 34(6):882–93. doi: 10.1002/humu.22307
78. Doyle AE. Hypertension and vascular disease. Am J Hypertens. (1991) 4(2 Pt 2):103S–6S. doi: 10.1093/ajh/4.2.103S
79. Garone C, D'Souza AR, Dallabona C, Lodi T, Rebelo-Guiomar P, Rorbach J, et al. Defective mitochondrial rRNA methyltransferase MRM2 causes MELAS-like clinical syndrome. Hum Mol Genet. (2017) 26(21):4257–66. doi: 10.1093/hmg/ddx314
80. Ghezzi D, Baruffini E, Haack TB, Invernizzi F, Melchionda L, Dallabona C, et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am J Hum Genet. (2012) 90(6):1079–87. doi: 10.1016/j.ajhg.2012.04.011
81. Kopajtich R, Nicholls TJ, Rorbach J, Metodiev MD, Freisinger P, Mandel H, et al. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am J Hum Genet. (2014) 95(6):708–20. doi: 10.1016/j.ajhg.2014.10.017
82. Nicholls TJ, Rorbach J, Minczuk M. Mitochondria: mitochondrial RNA metabolism and human disease. Int J Biochem Cell Biol. (2013) 45(4):845–9. doi: 10.1016/j.biocel.2013.01.005
83. Powell CA, Kopajtich R, D'Souza AR, Rorbach J, Kremer LS, Husain RA, et al. TRMT5 Mutations cause a defect in post-transcriptional modification of mitochondrial tRNA associated with multiple respiratory-chain deficiencies. Am J Hum Genet. (2015) 97(2):319–28. doi: 10.1016/j.ajhg.2015.06.011
84. Wedatilake Y, Niazi R, Fassone E, Powell CA, Pearce S, Plagnol V, et al. TRNT1 Deficiency: clinical, biochemical and molecular genetic features. Orphanet J Rare Dis. (2016) 11(1):90. doi: 10.1186/s13023-016-0477-0
85. Zeharia A, Shaag A, Pappo O, Mager-Heckel AM, Saada A, Beinat M, et al. Acute infantile liver failure due to mutations in the TRMU gene. Am J Hum Genet. (2009) 85(3):401–7. doi: 10.1016/j.ajhg.2009.08.004
86. Chakraborty PK, Schmitz-Abe K, Kennedy EK, Mamady H, Naas T, Durie D, et al. Mutations in TRNT1 cause congenital sideroblastic anemia with immunodeficiency, fevers, and developmental delay (SIFD). Blood. (2014) 124(18):2867–71. doi: 10.1182/blood-2014-08-591370
87. Erdmann J, Kessler T, Venegas LM, Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc Res. (2018) 114(9):1241–57. doi: 10.1093/cvr/cvy084
88. Onat UI, Yildirim AD, Tufanli Ö, Çimen I, Kocatürk B, Veli Z, et al. Intercepting the lipid-induced integrated stress response reduces atherosclerosis. J Am Coll Cardiol. (2019) 73(10):1149–69. doi: 10.1016/j.jacc.2018.12.055
89. Costa-Mattioli M, Walter P. The integrated stress response: from mechanism to disease. Science. (2020) 368(6489):eaat5314. doi: 10.1126/science.aat5314
90. Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Curr Opin Cell Biol. (2008) 20(2):222–6. doi: 10.1016/j.ceb.2008.01.013
91. Ou X, Gao JH, He LH, Yu XH, Wang G, Zou J, et al. Angiopoietin-1 aggravates atherosclerosis by inhibiting cholesterol efflux and promoting inflammatory response. Biochim Biophys Acta Mol Cell Biol Lipids. (2020) 1865(2):158535. doi: 10.1016/j.bbalip.2019.158535
92. Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. (2008) 14(10):2095–103. doi: 10.1261/rna.1232808
93. Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. (2009) 138(2):215–9. doi: 10.1016/j.cell.2009.07.001
94. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. (2009) 185(1):35–42. doi: 10.1083/jcb.200811106
95. Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. (2010) 285(14):10959–68. doi: 10.1074/jbc.M109.077560
96. Anderson P, Kedersha N. Stress granules: the tao of RNA triage. Trends Biochem Sci. (2008) 33(3):141–50. doi: 10.1016/j.tibs.2007.12.003
97. Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA Methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. (2010) 24(15):1590–5. doi: 10.1101/gad.586710
98. Tanai E, Frantz S. Pathophysiology of heart failure. Compr Physiol. (2015) 6(1):187–214. doi: 10.1002/cphy.c140055
99. Bhat S, Chin A, Shirakabe A, Ikeda Y, Ikeda S, Zhai P, et al. Recruitment of RNA polymerase II to metabolic gene promoters is inhibited in the failing heart possibly through PGC-1α (peroxisome proliferator-activated receptor-γ coactivator-1α) dysregulation. Circ Heart Fail. (2019) 12(3):e005529. doi: 10.1161/CIRCHEARTFAILURE.118.005529
100. Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. RNA Induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. (2008) 14(6):962–9. doi: 10.1016/j.devcel.2008.03.009
101. Ghanbarian H, Wagner N, Polo B, Baudouy D, Kiani J, Michiels JF, et al. Dnmt2/Trdmt1 as mediator of RNA polymerase II transcriptional activity in cardiac growth. PloS One. (2016) 11(6):e0156953. doi: 10.1371/journal.pone.0156953
102. Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, et al. Classification of the cardiomyopathies: a position statement from the European society of cardiology working group on myocardial and pericardial diseases. Eur Heart J. (2008) 29(2):270–6. doi: 10.1093/eurheartj/ehm342
103. Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, et al. Contemporary definitions and classification of the cardiomyopathies: an American heart association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation. (2006) 113(14):1807–16. doi: 10.1161/CIRCULATIONAHA.106.174287
104. Ahola S, Rivera Mejías P, Hermans S, Chandragiri S, Giavalisco P, Nolte H, Langer T. OMA1-mediated Integrated stress response protects against ferroptosis in mitochondrial cardiomyopathy. Cell Metab. (2022) 34(11):1875–91. doi: 10.1016/j.cmet.2022.08.017
105. Väre VYP, Eruysal ER, Narendran A, Sarachan KL, Agris PF. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules. (2017) 7(1):11–29. doi: 10.3390/biom7010029
106. Hou Y-M, Gamper H, Yang W. Post-transcriptional modifications to tRNA–a response to the genetic code degeneracy. RNA. (2015) 21(4):642–4. doi: 10.1261/rna.049825.115
107. Trixl L, Amort T, Wille A, Zinni M, Ebner S, Hechenberger C, et al. RNA Cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell Mol Life Sci. (2018) 75(8):1483–97. doi: 10.1007/s00018-017-2700-0
108. Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun. (2015) 6:6158. doi: 10.1038/ncomms7158
Keywords: RNA modification, m5C methyltransferases, cardiovascular diseases, atherosclerosis, mitochondrial dysfunction
Citation: Wang Y-Y, Tian Y, Li Y-Z, Liu Y-F, Zhao Y-Y, Chen L-H and Zhang C (2023) The role of m5C methyltransferases in cardiovascular diseases. Front. Cardiovasc. Med. 10:1225014. doi: 10.3389/fcvm.2023.1225014
Received: 18 May 2023; Accepted: 19 June 2023;
Published: 5 July 2023.
Edited by:
Shizuka Uchida, Aalborg University Copenhagen, DenmarkReviewed by:
Mirolyuba Ilieva, Aalborg University, Denmark© 2023 Wang, Tian, Li, Liu, Zhao, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chi Zhang emhhbmdjaGk5OTY2QGhvdG1haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.