
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 30 May 2023
Sec. Lipids in Cardiovascular Disease
Volume 10 - 2023 | https://doi.org/10.3389/fcvm.2023.1177054
 HariOm Singh1*
HariOm Singh1* Chandrashekhar Jori1
Chandrashekhar Jori1 Shyamveer1
Shyamveer1 Supriya D. Mahajan2
Supriya D. Mahajan2 Ravikumar Aalinkeel2
Ravikumar Aalinkeel2 Kathiravan Kaliyappan2
Kathiravan Kaliyappan2 Stanley A. Schwartz2
Stanley A. Schwartz2 Meenakshi Bhattacharya3
Meenakshi Bhattacharya3 Ruhi Shaikh4
Ruhi Shaikh4 Madhukar Salve4
Madhukar Salve4 Jyoti Deshmukh4
Jyoti Deshmukh4 Nemat Ali5
Nemat Ali5 Mohammad Khalid Parvez6
Mohammad Khalid Parvez6
HIV-associated lipodystrophy (HIVLD) is a metabolic condition with an irregularity in the production of lipoprotein particles, and its occurrence varies among HIV-infected patients. MTP and ABCG2 genes have a role in the transport of lipoproteins. The polymorphisms of MTP -493G/T and ABCG2 34G/A affect its expression and influence the secretion and transportation of lipoproteins. Hence, we investigated the MTP -493G/T and ABCG2 34G/A polymorphisms in 187 HIV-infected patients (64 with HIVLD and 123 without HIVLD) along with 139 healthy controls using polymerase chain reaction (PCR)-restriction fragment length polymorphism and expression analysis using real-time PCR. ABCG2 34A allele showed an insignificantly reduced risk of LDHIV severity [P = 0.07, odds ratio (OR) = 0.55]. MTP -493T allele exhibited a non-significantly reduced risk for the development of dyslipidemia (P = 0.08, OR = 0.71). In patients with HIVLD, the ABCG2 34GA genotype was linked with impaired low-density lipoprotein levels and showed a reduced risk for LDHIV severity (P = 0.04, OR = 0.17). In patients without HIVLD, the ABCG2 34GA genotype was associated with impaired triglyceride levels with marginal significance and showed an increased risk for the development of dyslipidemia (P = 0.07, OR = 2.76). The expression level of MTP gene was 1.22-fold decreased in patients without HIVLD compared with that in patients with HIVLD. ABCG2 gene was upregulated 2.16-fold in patients with HIVLD than in patients without HIVLD. In conclusion, MTP -493C/T polymorphism influences the expression level of MTP in patients without HIVLD. Individuals without HIVLD having ABCG2 34GA genotype with impaired triglyceride levels may facilitate dyslipidemia risk.
1. MTP -493C/T polymorphisms influence the expression level.
2. Individuals who have impaired triglyceride level with ABCG2 34GA genotype were likely to be associated with the risk of dyslipidemia.
3. Individuals who have impaired LDL, HDL, triglyceride, and cholesterol levels with MTP -493GT and MTP -493TT and ABCG2 234AA genotypes showed a trend of the risk for severity of LDHIV and dyslipidemia, respectively.
Though highly active antiretroviral therapy (HAART) has prominently decreased the rate of mortality in people living with HIV (PLWH), lipodystrophy in HIV-infected patients (LDHIV) and its metabolic complications remain a significant concern. HIV-associated lipodystrophy (HIVLD) usually develops into lipohypertrophy, lipoatrophy, or a combination of both in PLWH undergoing antiretroviral therapy (ART) and is associated with a higher risk for cardiovascular disease (CVD) (1). Dyslipidemia is a prominent metabolic condition that can occur due to irregularities in the production, processing, and degradation of lipoprotein particles. Literature reported that the occurrence of lipoatrophy varies from 13.3% to 52.9% (2–5). In India, the prevalence of lipodystrophy (LD) varies from 22% to 60.7% (6, 7). The effects of HIV and antiretroviral therapies on fat differ between individuals. However, studies reported that HIV-infected individuals who were not treated with ART had changes in their body fat composition (8–10). Variations in the effect of ART drugs among people have been probably linked to the genetic profiles of patients (11). Host genetic factors also play a role in producing these syndromes (12).
Microsomal triglyceride transfer protein (MTP) plays a role in the transportation of lipid molecules, primarily present in hepatocytes and enterocytes (13). It is necessary for the synthesis and secretion of ApoB-containing lipoproteins from the hepatocytes and enterocytes. The secretory pattern of ApoB-containing lipoproteins most probably affects the plasma low-density lipoprotein (LDL) and very-low-density lipoprotein (VLDL) levels (14). MTP plays an important role in transferring cholesterol esters (15) and triglycerides (TG) from the endoplasmic reticulum membranes to nascent ApoB lipoproteins, producing VLDL and removing lipids from the hepatocyte (16). The MTP gene is situated on chromosome 4q23. Genetic variations in the MTP gene affect the MTP expression and influence the secretion of lipoproteins in ER (17). Numerous single nucleotide polymorphisms (SNPs) in the MTP gene are known, and the promoter region -493G/T (rs1800591) polymorphism is one of the most commonly examined (18, 19). The G allele of MTP -493G/T polymorphism leads to decreased MTP transcription activity than the T allele, decreased hepatocyte triglyceride export, and increased intracellular TG accumulation (14, 20). Less manganese-superoxide dismutase (MnSOD) is transported to the mitochondria when the -493T allele is present in the mitochondrial targeting sequence (21). The MTP -493G/T polymorphism was linked to altered MTP gene expression and led to abnormal changes in export of lipids, causing deregulated hepatic lipid metabolism (22, 23). Non-alcoholic fatty liver disease risk was increased by the MTP -493G/T polymorphism (23).
ATP-binding cassette transporter G2 (ABCG2) protein belongs to the family of ABC transporters which is also called as breast cancer resistance protein (BCRP). It is involved in the transport of bile, lipoprotein, drug substrates, and other peptides (24). ABCG2 is widely expressed in healthy cells and organs, including the liver, kidney, and testes, as well as in the gastrointestinal tract, hematopoietic stem cells, and capillary endothelial cells. ABCG2 has a protective role in preventing the buildup of toxic xenobiotics in cells and organs (25). The overexpression of ABCG2 leads to an inter-individual differences in the bioavailability of drugs (26).
The ABCG2 gene is situated on chromosome 4q22; consisting of 16 exons encoding 655 amino acid residues which form a 72-kD membrane protein (27, 28). The ABCG2 gene is highly polymorphic. Numerous allelic variants in the ABCG2 gene are known; some of them may have an impact on the protein's function and/or expression which may also alter the transporter activity (29, 30). There are two significant functional variations among these ABCG2 34G/A (rs2231137, V12M), resulting in the substitution of Val12Met and ABCG2 421C/A (rs2231142, Q141K), causing a Glu141Lys substitution which were described and demonstrated to be associated with the negative effects of multiple drugs transported via ABCG2 (31, 32). These polymorphisms are linked to lower ABCG2 expression and protein production, which consequently results in decreased transporter activity (30, 32–34).
Until now, the report has not been published on the association of MTP -493G/T and ABCG2 34G/A polymorphisms with HIVLD worldwide. Hence, we evaluated the association of MTP -493G/T and ABCG2 34G/A polymorphisms with LDHIV along with its occurrence in patients without HIVLD and in healthy individuals.
This was a cross-sectional study. A total of 187 participants (HIV-infected individuals) were recruited from the OPD of Medicine Department, ART Plus Centre, Government Medical College (GMC) & Hospital, Aurangabad, Maharashtra, between 1 April 2021, and 1 May 2022. Out of 187 patients, a total 64 had HIVLD and show clinical evidence of lipodystrophy/lipoatrophy/dyslipidemia enrolled as cases. Clinical examinations of lipoatrophy/lipohypertrophy were done by experienced doctors (i.e., physicians) at the ART Plus Centre, GMC & Hospital, during routine clinic visits. The following requirements were taken care of while recruiting subjects for case studies: (a) HIV-1-infected patients who have been on ART for a prolonged period, for a minimum of 1 year; (b) HIV-1/AIDS patients suffering from metabolic disorder dyslipidemia, as determined by a study of lipid profile subsequent to a 14-h fast, and who had (i) triglyceride levels greater than 300 mg/dl, (ii) high-density lipoprotein (HDL) levels decreased below 35 mg/dl, (iii) LDL levels greater than 120 mg/dl, and (iv) cholesterol levels greater than 240 mg; and (c) HIV/AIDS patients with clinical signs of lipodystrophy and lipoatrophy. The following exclusion criteria were taken care of while recruiting subjects for case studies: (i) patients with HIV-1 infection who also have co-existing conditions such as tuberculosis (TB), diabetes mellitus, hepatitis B or C, autoimmune diseases, cancers, or cardiovascular disease history and (ii) patients with HIV-1 infection who regularly consume alcohol or recreational drug.
Out of 187 patients, a total of 123 were without HIVLD and enrolled as controls. The criteria for including participants in this group are as follows: (1) HIV-1-infected patients who were not related to dyslipidemia, lipoatrophy, or lipodystrophy syndrome, (2) gender and age were compatible, and (3) HIV-1-infected patients being treated with ART for at least 1 year. The exclusion criteria were the following: (1) patients with lipoatrophy or dyslipidemia; (2) HIV-1-infected patients who also have other illnesses such as cancer, diabetes mellitus, hepatitis B or C, TB, or any autoimmune disease; (3) patients who regularly consume alcohol; and (4) patients who indulged in recreational drug use.
The patients’ CD4 counts, blood glucose levels, and lipid profiles were measured routinely during clinic visits. A thorough clinical history after infection and before ART treatment, information related to current ART regimen given for treatment, and data on its duration were collected from each patient recruited for the study. A total of 139 individuals (those from the same family were excluded), free from HIV, hepatitis B and C and tuberculosis, age matched, and serum negative from HIV-ELISA test, were recruited as healthy controls. The institutional ethics committees (NARI) had approved the study, and a written informed consent from each participant was recorded.
Lipoatrophy was categorized as subcutaneous fat loss in one or more of the following regions: face (gaunt face and sunken eyes), buttocks, and limbs (skinny with obvious veins, muscle, or bones), whereas lipohypertrophy was characterized as vascular fat gain in at least one of the following areas: the trunk (wider waist circumference), neck or back base (buffalo hump), and the breasts. Dyslipidemia was diagnosed through the analysis of lipid profile, which includes triglycerides, LDL, cholesterol, and HDL-C.
Blood sample (2 ml) was withdrawn from recruited patients and kept at −8°C. Genomic DNA was extracted according to the given kit instructions from the pellets of peripheral blood samples using the QIAamp DNA Blood Mini Kit.
Polymerase chain reaction (PCR) and restriction fragment length polymorphism (PCR-RFLP) technique was utilized to genotype MTP -493G/T and ABCG2 34G/A polymorphisms in recruited subjects for each group. The MTP -493G/T and ABCG2 34G/A polymorphism primers were used as specified for particular gene amplification (14, 35). For 25μl PCR reaction, 10pmol FP and RP (primers), 10 mM dNTPs mix, 1 unit Taq DNA polymerase (Bangalore Genei, India), 100 mM Tris-HCl, and 1.5 mM MgCl2 containing PCR reaction buffer, 100–150 ng genomic DNA was taken as the standard for amplification of genes. The following PCR conditions were used for MTP -493G/T: initial denaturation temperature at 95°C for 5 min, 30 cycles of denaturation were at 95°C for 30 s, annealing temperature at 59°C for 30 s, extension at 72°C for 45 s, and a final extension at 72°C for 7 min. For ABCG2 34G/A, the following PCR conditions were used: initial denaturation temperature at 94°C for 5 min, 35 cycles of denaturation at 94°C for 45 s, annealing temperature at 60°C for 45 s, extension at 72°C for 1 min, and a final extension at 72°C for 7 min. The restriction enzymes HphI and BseMI (MBI Fermentas Inc., Glen Burnie, MD, United States) were used to digest the amplified products of MTP and ABCG2, respectively. Genotyping after restriction digestion of MTP and ABCG2 was performed on 15% of polyacrylamide gel using molecular weight markers and visualized after staining with ethidium bromide (EtBr). On the basis of sequence and location of SNP, genotypes of MTP -493G/T and ABCG2 34G/A were assigned as follows: for MTP: 109 bp for TT genotype; 109 bp, 89 bp, and 20 bp for GT genotype; and 109 bp and 89 bp for GG genotype; and for ABCG2: 291 bp for GG genotype, 291 bp, 261 bp, and 30 bp for GA genotype; and 261 bp and 30 bp for AA genotype. Veriti 96-well Thermal Cycler (Applied Biosystems, United States) was utilized to perform all the reaction. In a 2% agarose gel, PCR products were run with molecular weight markers and visualized the band with EtBr staining. Twenty percent of the samples were re-genotyped by other laboratory personnel to avoid differences in genotyping, and 10% of the samples underwent sequencing in order to prevent genotyping error.
For qPCR analysis, the oligonucleotide primers flanking a region of interest and DNA polymerase enzymes were used to amplify the sequence (Table 9 showed the sequence of the primer in the Supplementary material). The selective amplification and quantitative detection of MTP and ABCG2 genes were performed using a qPCR machine. The qPCR machine was connected to a computer that maintains a record of fluorimeter output and uses software to analyze the experiment's results employing user-defined data, such as control wells and standards. The qPCR assay was carried out in 96-well plates by using 7,500 fast real-time PCR equipment (Applied Biosystems, CA, United States). The qPCR reaction was conducted in a total of 24 μl reaction volume with 10 μl DNA (10–100 ng of genomic DNA) and 12 μl of master mix cyber green, with each primer containing 2 μl.
The mean ± standard deviation (SD) values were used to represent the age variable. Chi-square goodness-of-fit test was used to see if any healthy control subjects deviate from the Hardy–Weinberg equilibrium. To assess the genotype distribution in patients with and without HIVLD, as well as in patients without HIVLD compared to the healthy controls, χ2 statistics (Fisher's exact test for a cell size of <5) was utilized. Unconditional binary logistic regression was used to compute odds ratios (ORs) and 95% confidence interval (CI). The statistical analysis was conducted by using SPSS software version 17.0 (SPSS Inc., Released 2008; SPSS Statistics for Windows, Version 17.0, SPSS Inc., Chicago, IL, United States), and tests of statistical significance were two-sided and considered significant when a P-value was 0.05 (SPSS Inc., 2008). The qPCR data were analyzed on GraphPad prism directly using quantification Ct values, and the bar diagram was made in an Excel sheet.
The mean ages of the patients with HIVLD, the patients without HIVLD, and the healthy controls (years ± SD) were 38.37 ± 6.34 years, 37.87 ± 6.35 years, and 32.35 ± 7.45 years, respectively. Characteristics of the recruited patients with and without HIVLD along with the healthy controls are presented in Table 1.
The genotype and allele frequencies of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with HIVLD, patients without HIVLD, and healthy controls are shown in Table 2. MTP -493GG, MTP -493GT, and MTP -493TT and ABCG2 34GG, ABCG2 34GA, and ABCG2 34AA genotypes were distributed almost similarly between patients with and without HIVLD and patients with HIVLD and healthy controls (43.8% vs. 51.2%, 40.6% vs. 36.6%, 15.6% vs. 12.2%, 78.1% vs. 66.7%, 20.3% vs. 27.6%, and 1.6% vs. 5.7% and 43.8% vs. 43.9%, 40.6% vs. 36.0%, 15.6% vs. 20.1%, 78.1% vs. 65.5%, 65.5% vs. 31.7%, 1.6% vs. 2.9%). ABCG2 34A allele was dispersed dissimilarly between patients with and without HIVLD and showed a reduced risk for severity of LDHIV with borderline significance (11.71% vs. 19.51%; P = 0.07, OR = 0.55, 95% CI 0.28–1.06).
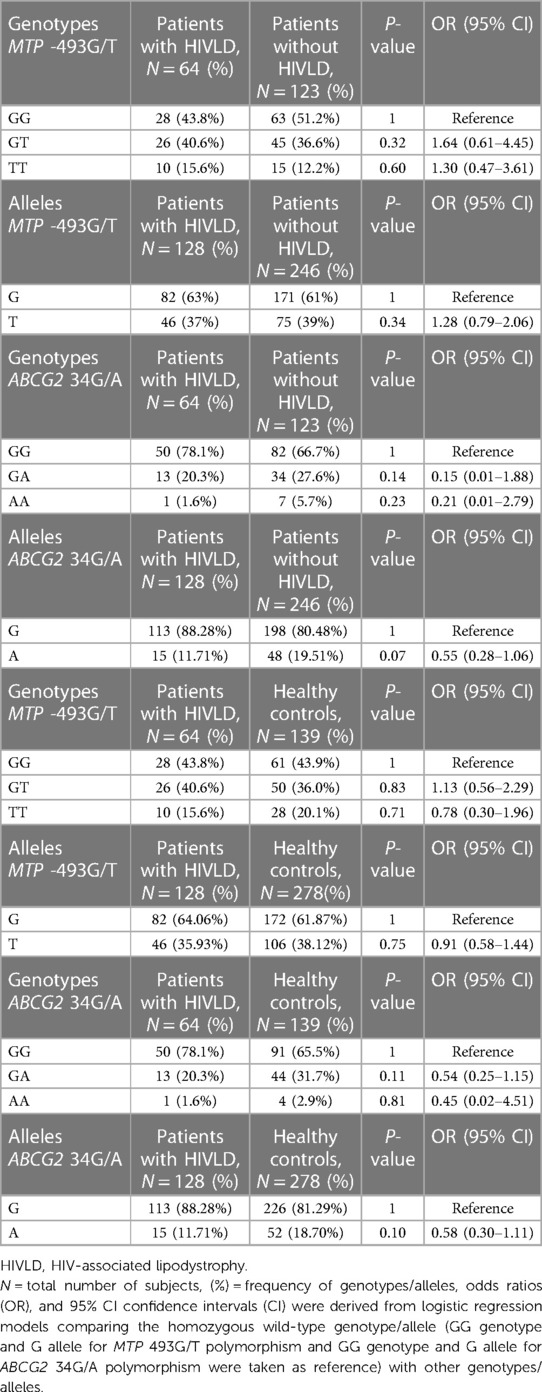
Table 2. Frequency distribution of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with HIVLD, patients without HIVLD, and healthy controls.
The genotype and allelic frequencies of MTP -493G/T and ABCG2 34G/A polymorphisms in patients without HIVLD and healthy controls are shown in Table 3. The genotype frequency of MTP -493G/T polymorphism did not follow the Hardy–Weinberg equilibrium (P = 0.005), while ABCG2 34G/A polymorphism in healthy controls were in the Hardy–Weinberg equilibrium (P = 0.63). The occurrences of MTP -493GG, MTP -493GT, and MTP -493TT and ABCG2 34GG, ABCG2 34GA, and ABCG2 34AA genotypes were almost alike between patients without HIVLD and healthy controls (51.2% vs. 43.9%, 36.6% vs. 36.0%, 12.2% vs. 20.1%, 66.7% vs. 65.5%, 27.6% vs. 31.7%, 5.7% vs. 2.9%). MTP -493T allele was distributed almost similarly between patients without HIVLD and healthy controls (37% vs. 38.12%).
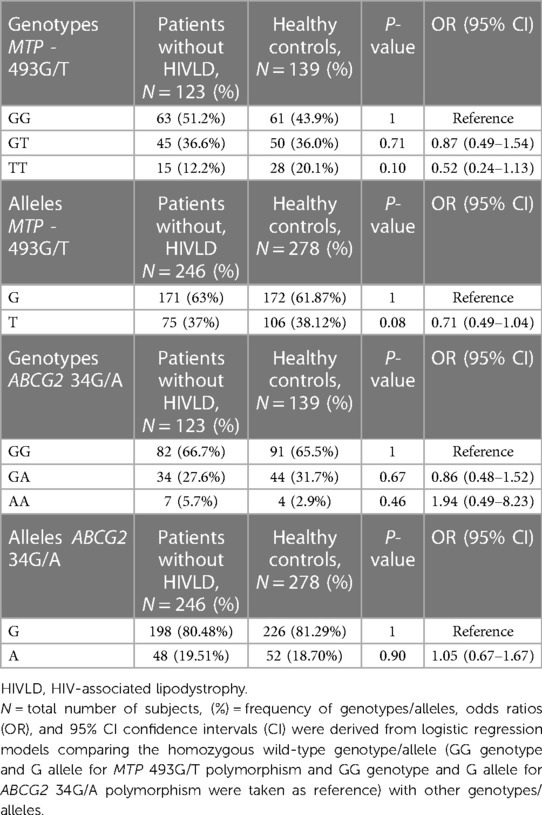
Table 3. Frequency distribution of MTP 493G/T and ABCG2 34G/A polymorphisms in between patients without HIVLD and healthy controls.
Haplotype frequency of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with and without HIVLD and healthy controls is shown in Table 4. In the patients with and without HIVLD, patients with HIVLD and healthy controls, and patients without HIVLD and healthy controls populations, there was no significant LD (D’) between both genes (P = 0.2247, P = 0.225, P = 0.2647). Comparing the LD values (Dij) between the patients with HIVLD and without HIVLD, patients with HIVLD and healthy controls, and patients without HIVLD and healthy controls subjects, no significant difference was found between both genes. It was expected that there might be additive or synergistic effect of these variations in modulation of pathogenesis of HIVLD.
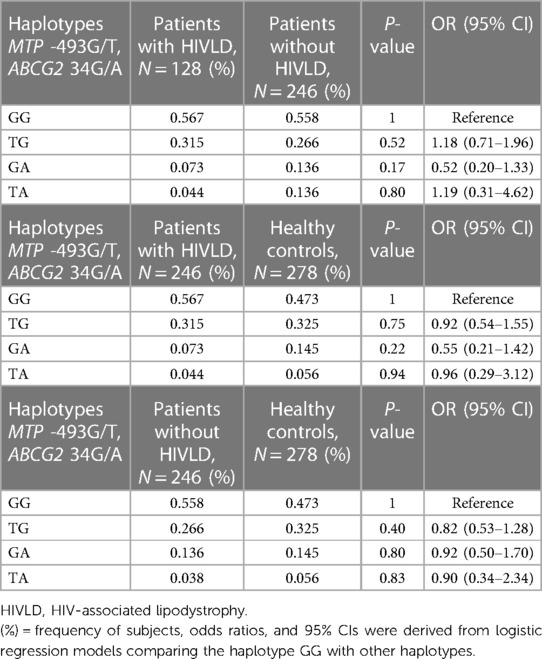
Table 4. Frequency distribution of haplotypes of MTP 493G/T and ABCG2 34G/A between patients with and without HIV-associated lipodystrophy and patients with HIV-associated lipodystrophy and healthy controls.
Haplotype GG (MTP *G/ABCG2*G) was taken as reference. The frequency of haplotype GG, TG, GA, and TA (MTP *G, *T, *G, *T/ABCG2*G, *G, *A, *A) did not differ significantly between patients with and without HIVLD, patients with HIVLD and healthy controls, and patients without HIVLD and healthy controls (56.7% vs. 55.8%, 31.5% vs. 26.6%, 7.3% vs. 13.6%, 4.4% vs. 13.6%, 56.7% vs. 47.3%, 31.5% vs. 32.5%, 7.3% vs. 14.5%, 4.4% vs. 5.6%, 55.8% vs. 47.3%, 26.6% vs. 32.5%, 13.6% vs. 14.5%, 3.8% vs. 5.6%).
The genotype frequencies of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with and without HIVLD who have impaired LDL level and normal LDL level are shown in Table 5. In patients with HIVLD, ABCG2 34GA genotype was distributed significantly lower in normal LDL level compared with impaired LDL level and showed a reduced risk for severity of LDHIV (7.1% vs. 30.6%; P = 0.04, OR = 0.17, 95% CI 0.02–0.94). ABCG2 34GG genotype was disseminated higher in impaired LDL level than in normal LDL level (92.9% vs. 66.7%). ABCG2 34GG, ABCG2 34GA, and ABCG2 34AA genotypes were dispersed almost alike in impaired LDL level and normal LDL level (64.7% vs. 68.1%, 31.4% vs. 25.0%, 3.9% vs. 6.9%) among patients with HIVLD. In patients with and without HIVLD, MTP -493GT and MTP -493TT genotypes were disseminated higher in impaired LDL level than in normal LDL level and showed an increased risk for severity of LDHIV and dyslipidemia, respectively (35.7% vs. 50.0%, P = 0.43, OR = 1.80, 95% CI 0.53–6.21 and 17.9% vs. 13.9%, P = 0.67, OR = 1.80, 95% CI 0.34–9.85 and 39.21% vs. 34.7%, P = 34.7%, OR = 1.30, 95% CI 0.56–3.04 and 13.72% vs. 11.1%, P = 0.75, OR = 1.42, 95% CI 0.40–5.07), while as MTP -493GG genotype was distributed lesser in impaired LDL level than in normal LDL level (47.05% vs. 54.2%).
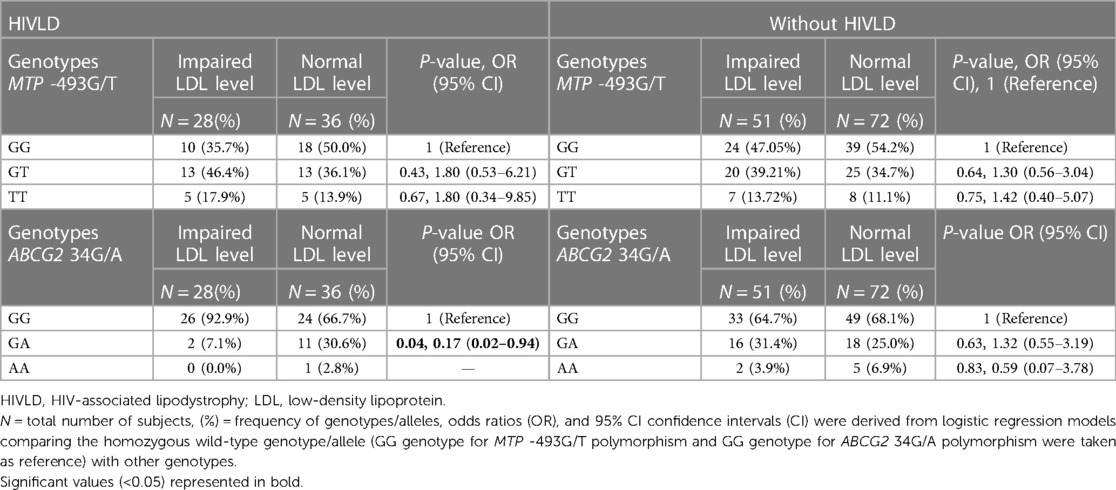
Table 5. Frequency distribution of MTP 493G/T and ABCG2 34G/A polymorphisms in patients with and without HIVLD who have impaired LDL level.
The genotype frequencies of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with and without HIVLD who have impaired HDL level and normal HDL level are shown in Table 6. In patients with and without HIVLD, MTP -493TT genotype displayed an increased risk for severity of LDHIV and dyslipidemia, respectively (37.5% vs. 12.5%, P = 0.19, OR = 5.57, 95% CI 0.58–62.28 and 20.0% vs. 11.1%, P = 0.62, OR = 2.00, 95% CI 0.35–10.61) when compared between impaired HDL level and normal HDL level, while the occurrences of MTP -493GG and MTP -493TT genotypes were disseminated lesser in impaired HDL level than in normal HDL level (25.0% vs. 46.4% and 37.5% vs. 41.1%; 46.66% vs. 51.9% and 33.33% vs. 37.0%). In patients with HIVLD, ABCG2 34GG genotype distributed lesser in impaired HDL level, while ABCG2 34GA genotype disseminated higher in impaired HDL level than in normal HDL level (75.0% vs. 78.6%, 25.0% vs. 19.6%). ABCG2 34GG genotype was distributed greater in impaired HDL level, while ABCG2 34GA genotype was disseminated lesser in impaired HDL level than in normal HDL level (86.7% vs. 63.9%, 13.3% vs. 29.6%) among patients without HIVLD.
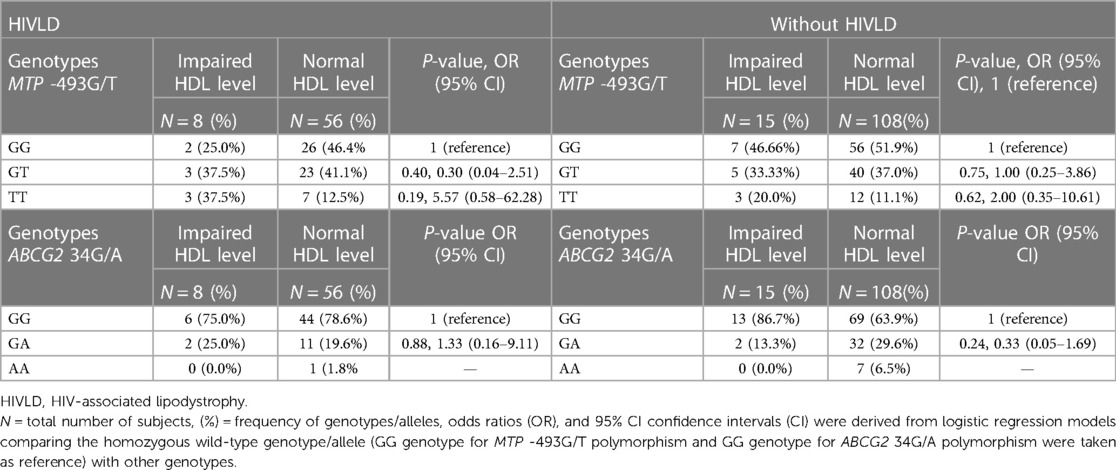
Table 6. Frequency distribution of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with HIVLD and patients without HIVLD who have impaired HDL level.
The genotype frequencies of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with and without HIVLD who have impaired triglyceride level and normal triglyceride level are shown in Table 7. MTP -493GT and MTP -493TT genotypes exhibited a risk for severity of LDHIV and dyslipidemia, respectively (50.0% vs. 39.3%, P = 0.59, OR = 2.36, 95% CI 0.32–20.8 and 25.0% vs. 14.3%, P = 0.59, OR = 3.25, 95% CI 0.270–40.83; 40.0% vs. 36.3%, P = 0.64, OR = 1.95, 95% CI 0.34–11.73 and 30.0% vs.10.6%, P = 0.14, OR = 5.00, 95% CI 0.69–36.83) when compared between impaired triglyceride level and normal triglyceride level among patients with and without HIVLD. In patients with and without HIVLD, ABCG2 34GG genotype distribution was dissimilar in impaired triglyceride level than in normal triglyceride level (87.5% vs. 76.8% and 70.0% vs. 66.4%). In patients without HIVLD, ABCG2 34GA genotype showed an increased risk for dyslipidemia with borderline significance when compared between impaired triglyceride level and normal triglyceride level (53.33% vs. 27.4%, P = 0.07, OR = 2.76, 95% CI 0.82–9.43). ABCG2 34GA genotype was distributed differently between impaired triglyceride level and normal triglyceride level (12.5% vs. 21.4%) among patients with HIVLD.
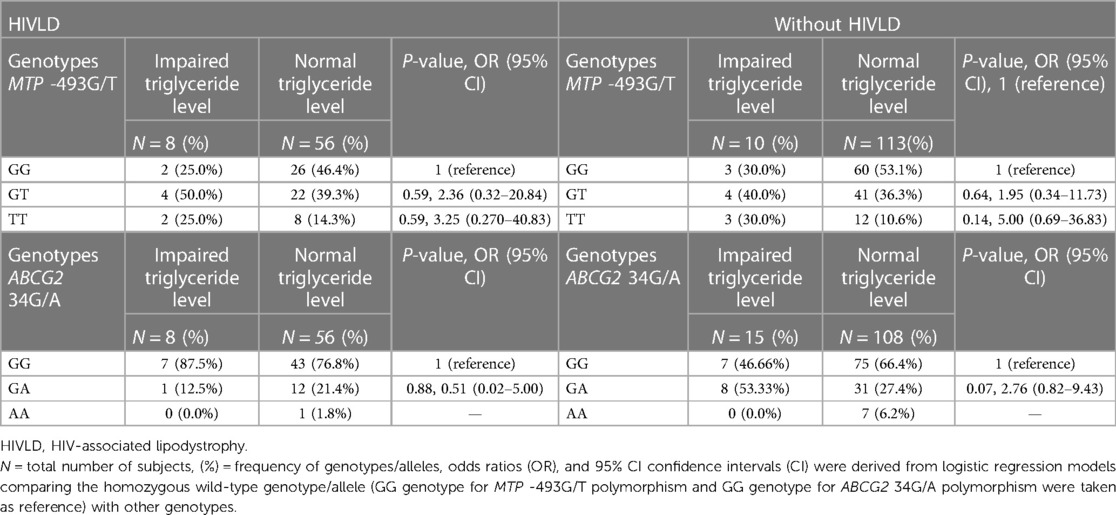
Table 7. Frequency distribution of MTP 493G/T and ABCG2 34G/A polymorphisms in patients with HIVLD and patients without HIVLD who have impaired triglyceride level.
The genotype frequencies of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with and without HIVLD who have impaired cholesterol level and normal cholesterol level are shown in Table 8. In patients with HIVLD, MTP -493GT and MTP -493TT genotypes showed a risk for severity of LDHIV (66.7% vs. 37.9%, P = 0.30, OR = 4.91, 95% CI 0.45–124.25 and 16.7% vs. 15.5%, P = 0.96, OR = 3.00, 95% CI 0.0–125.67) when compared between impaired cholesterol level and normal cholesterol level. MTP -493GT and MTP -493GG genotypes was distributed lesser in impaired cholesterol level than normal cholesterol level (16.7% vs. 46.6%). In patients without HIVLD, ABCG2 34AA genotype displayed a risk for dyslipidemia (13.3% vs. 13.3%, P = 0.44, OR = 3.24, 95% CI 0.37–23.98), and ABCG2 34GG and ABCG2 34GA genotypes were distributed almost similarly (60.0% vs. 67.6%; 26.7% vs. 27.8%) when compared between impaired cholesterol level and normal cholesterol level. MTP -493GG genotype was dispersed higher, and MTP -493GT genotype was dispersed lesser in impaired cholesterol level than in normal cholesterol level (73.3% vs. 48.1%; 26.7% vs. 38.0%).
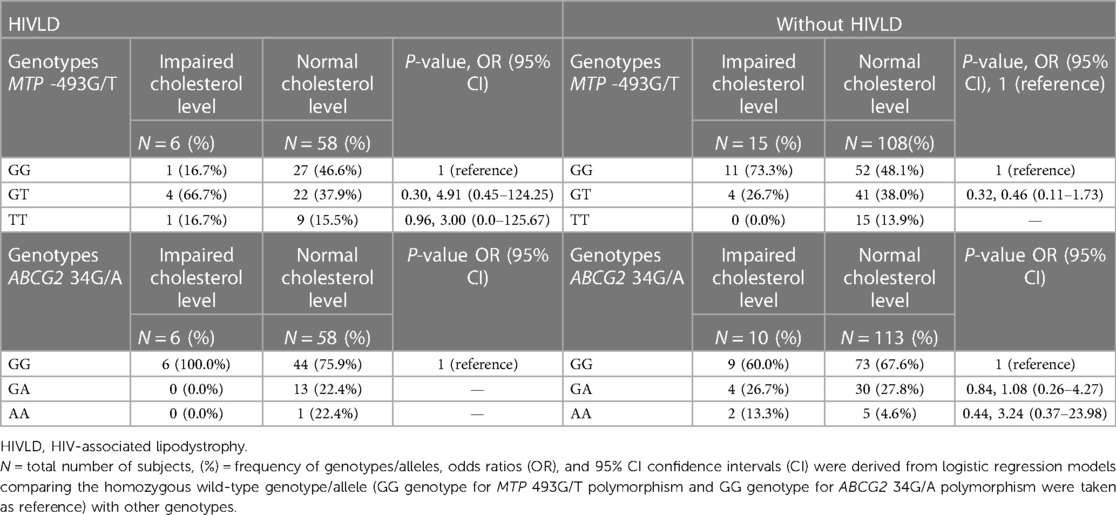
Table 8. Frequency distribution of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with HIVLD and patients without HIVLD who have impaired total cholesterol level.
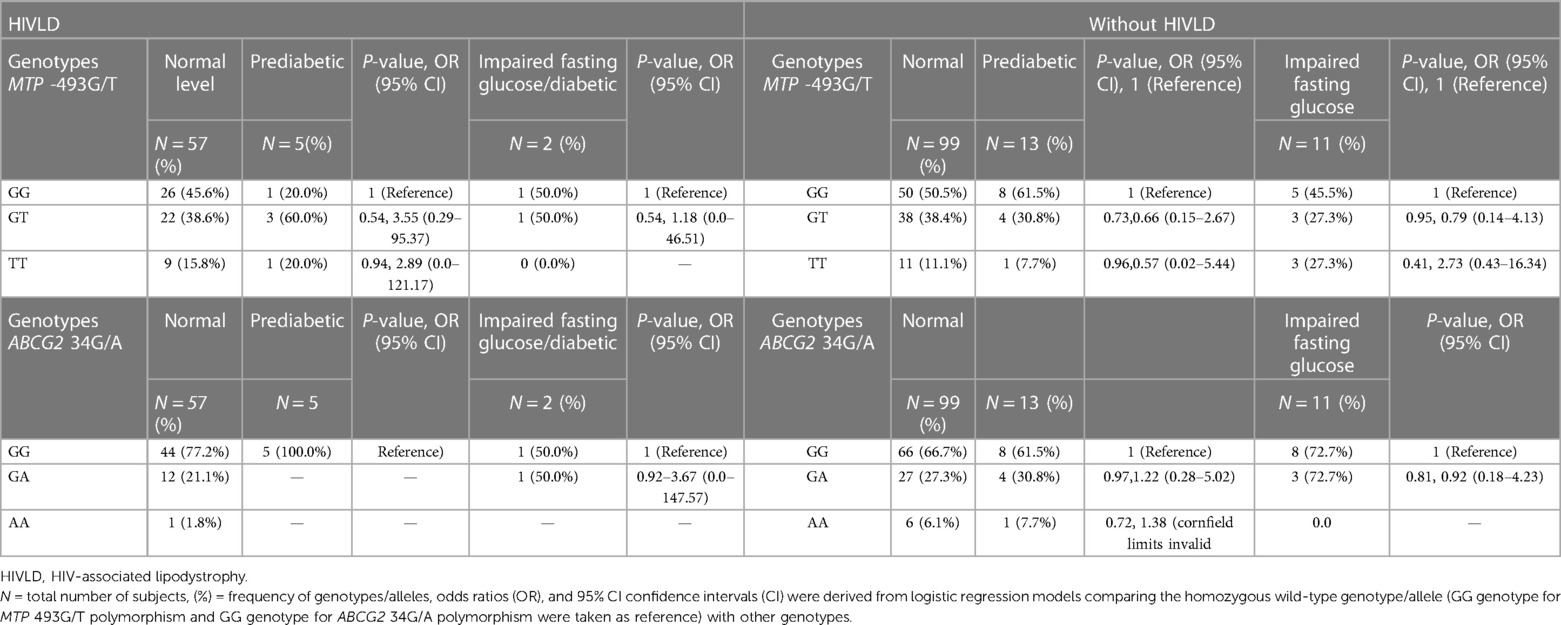
Table 9. Frequency distribution of MTP 493G/T and ABCG2 34G/A polymorphisms in patients with and without HIVLD who have impaired fasting glucose level.
The genotype frequencies of MTP -493G/T and ABCG2 34G/A polymorphisms in patients with and without HIVLD who have impaired fasting glucose level/diabetic, are prediabetic, and have normal glucose level are presented in Table 8. In patient with HIVLD, the occurrence of MTP 493GT genotypes was dissimilar between prediabetic and normal glucose level (60.0% vs. 38.6%, P = 0.54, OR = 3.55, 95% CI 0.29–95.37) and showed an insignificant risk for severity of HIVLD. In patient with HIVLD, the occurrence of ABCG2 34GA genotypes was different between diabetic and normal glucose level (50.0% vs. 21.1%, P = 0.92, OR = 3.67, 95% CI 0.0–147.57) and showed an insignificant risk for severity of HIVLD.
In patient without HIVLD, the occurrence of MTP 493GT genotypes was almost similar among diabetic, prediabetic, and normal glucose level (38.4% vs. 30.8% vs. 27.3%). In patients without HIVLD, the occurrence of MTP -493TT genotype was not significantly higher in diabetic glucose level than in normal glucose level and showed a risk for development of dyslipidemia (27.3 vs. 11.1%; P = 0.41, OR = 2.73, 95% CI 0.43–16.34). In patients without HIVLD, the occurrences of ABCG2 34GG and ABCG2 34GA genotypes were almost similar among diabetic, prediabetic, and normal glucose level (72.72% vs. 61.5% vs. 66.7% and 27.27% vs. 30.8% vs. 27.3%).
The expression analysis of MTP and ABCG2 genes in a total of 72 HIV-infected individuals (24 patients with HIVLD and 48 patients without HIVLD) is shown in Figure 1. The BAC2 gene was used as an internal control target. The MTP gene was downregulated 1.21-fold higher in patients without HIVLD than in patients with HIVLD (−1.11 vs. −2.33; 1.21-fold). ABCG2 gene was upregulated 2.16-fold higher in patients with HIVLD than in patients without HIVLD (+6.53 vs. +4.37; 2.16-fold).

Figure 1. Quantitative changes in gene expression levels in patients with HIV-associated lipodystrophy. Quantitative changes in gene expression levels of MTP and ABCG2 genes in patients with and without HIV-associated lipodystrophy.
This is the first study which independently evaluates the influence of the MTP -493G/T and ABCG2 34G/A polymorphisms and its haplotype with respect to the risk of dyslipidemia and severity of LDHIV from Western India. Metabolic complications of ART including LDHIV have grown to be a significant issue for the long-term, effective management of HIV infection. The effects of HIV and antiretroviral therapies on fat vary between individuals. Genetic constitution of any individual plays a significant role in inter-patient variability of fat transport, absorption, and elimination. The gene involved in lipid transport plays an important role to modulate plasma TG and HDL-C levels in HIV-infected patients (11). SNPs in genes having a role in lipid transport, absorption, and elimination may help understand individual differences in morphologic and metabolic abnormalities which may develop during HAART treatment (32).
MTP and ABCG2 play an important role in the transport of lipoproteins. MTP is necessary for the cellular synthesis and release of lipoproteins containing apolipoprotein B (ApoB) (15, 16). The crucial factor controlling lipoprotein secretion is the concentration of MTP in the ER (36). The plasma levels of VLDL and LDL are predicted to be influenced by the secretory pattern of ApoB-containing lipoproteins. Genetic variations can alter the amount or activity of MTP in the ER, as well as the lipoprotein secretory pattern and the function of MTP (17). ABCG2 is involved in the transport of bile and lipoprotein (25). The expression pattern and transporter activity of ABCG2 protein are influenced by the ABCG2 34G/A polymorphism.
In the present study, the prevalence of the MTP -493G/T polymorphism in healthy subjects was comparable to that in the studies of Gouda et al. (23) and incomparable to the studies of Oliveira et al. (37) and Ledmyr et al. (38). MTP -493T allele disclosed a non-significantly reduced dyslipidemia risk (P = 0.08, OR = 0.71) when compared between patients without HIVLD and healthy controls. MTP -493G/T polymorphism was anticipated to play a significant role in the development of dyslipidemia in type 2 diabetics (39). The occurrence of MTP -493GG genotype was greater in non-alcoholic steatohepatitis (NASH) patients compared with that in the controls (P = 0.002, 95% CI 2.9–392) (40). MTP -493G allele was slightly different between biopsy-proven NASH and simple steatosis (37).
In our study, the frequency of ABCG2 34G/A polymorphism in healthy individuals was different as compared with research done by Wang et al. (41), Wu et al. (35), and Sari et al. (42). ABCG2 34A allele showed a reduced risk for severity of LDHIV with borderline significance (P = 0.07, OR = 0.55). ABCG2 34AA genotype displayed a non-significant risk for development of dyslipidemia (P = 0.46, OR = 1.94). ABCG2 34GA and ABCG2 34AA genotypes were substantially linked to an increased risk of breast cancer (35). The ABCG2 34G/A polymorphism was frequently observed in a Korean population (43).
Our study has also sought to evaluate the significance of two important genes (MTP and ABCG2 genes) cluster both individually and collectively that are physically connected to each other in patients with and without HIVLD. Haplotype analysis, which evaluates interactions between two or more genes that are near to each other, has an advantage over a single-locus analysis in terms of disease susceptibility. In our study, none of haplotype was associated with the higher risk for severity of LDHIV and dyslipidemia.
In our study, ABCG2 34GA genotype was linked with the reduced risk for severity of LDHIV (P = 0.04, OR = 0.17), whereas MTP -493GT and MTP -493TT genotypes showed an insignificant risk for severity of LDHIV (P = 0.43, OR = 1.80; P = 0.67, OR = 1.80) when compared between impaired LDL level and normal LDL level. This suggests that patients with HIVLD who have impaired LDL level and MTP -493GT and MTP -493TT genotypes may have a risk for severity of LDHIV. It has been demonstrated that MTP -493G/T polymorphism affects transcriptional activity and is linked to low plasma levels of LDL cholesterol in healthy middle-aged males (14). In individuals with familial hypercholesterolemia (FH), the MTP-493T variant allele's LDL cholesterol-lowering function is switched to a triglyceride-lowering effect (44). Changes in VLDL and LDL were linked to MTP -493G/T polymorphism in individuals with type 2 diabetes (45).
In our study, MTP -493TT genotype displayed a non-significant risk for severity of LDHIV and dyslipidemia, respectively (P = 0.19, OR = 5.57; P = 0.62, OR = 2.00) when compared between impaired HDL level and normal HDL level among patients with and without HIVLD. This suggests that patients with and without HIVLD who have impaired HDL level and MTP-493TT genotype may have a risk for severity of LDHIV and dyslipidemia, respectively.
In the present study, MTP -493GT and MTP -493TT genotypes demonstrated an insignificant increased risk for severity of LDHIV and dyslipidemia, respectively (P = 0.59, OR = 2.36; P = 0.59, OR = 3.25 and P = 0.64, OR = 1.95; P = 0.14, OR = 5.00) when compared between impaired triglyceride level and normal triglyceride level among patients with and without HIVLD. ABCG2 34GA genotype was likely to be associated with impaired triglyceride level with marginal significance and have been shown an increased risk for dyslipidemia (P = 0.07, OR = 2.76) when compared between impaired triglyceride level and normal triglyceride level among patients without HIVLD. This suggests that patients with and without HIVLD who have impaired HDL level and MTP -493GT and MTP -493TT and ABCG2 34GA genotypes may have a risk for severity of LDHIV and dyslipidemia, respectively. The ABCG2 34G/A polymorphism affects the ABCG2 expression pattern and its transporter activity (31, 32). Fewer but more lipid-rich VLDL particles were found in the MTP -493TT variant genotype, corroborating the notion that MTP expression affects the hepatic secretion of triglyceride-rich, ApoB-containing lipoproteins (14).
MTP -493GT and MTP -493TT genotypes exhibited a non-significant risk for severity of LDHIV (P = 0.30, OR = 4.91; P = 0.96, OR = 3.00) when compared between impaired cholesterol level and normal cholesterol level among patients with HIVLD. ABCG2 234AA genotype exposed an insignificant risk for development of dyslipidemia (P = 0.44, OR = 3.24) when compared between impaired cholesterol level and normal cholesterol level among patients without HIVLD. This suggests that patients with and without HIVLD who have impaired cholesterol level and MTP -493GT and MTP -493TT and ABCG2 34AA genotypes may have a risk for severity of LDHIV and dyslipidemia, respectively.
In patients without HIVLD, individuals with MTP -493TT genotype have shown a risk for development of HIVLD (P = 0.41, OR = 2.73) when compared between diabetic and normal glucose level. However, risk could not reach statistical significance. This suggests that patients without HIVLD who have impaired fasting glucose level and MTP -493TT genotypes may have a risk for dyslipidemia. MTP -493G/T polymorphism was associated with gene expression and resulted in aberrant alterations of MTP synthesis and secretion, affecting the capacity for lipid export, which induces dysregulation of hepatic lipid metabolism (22, 23).
In the present study, MTP gene was downregulated 1.21-fold in patients without HIVLD because MTP -493T allele was expressed lesser in patients without HIVLD compared with that in patients with HIVLD and healthy subjects (P = 0.34, OR = 1.28; P = 0.08, OR = 0.71) and correlated with the decreased MTP expression. MTP -493G/T polymorphism was linked to altered gene expression and MTP secretion, which affected the ability to transport lipids (22, 23).
In our study, ABCG2 gene was upregulated 2.16-fold in patients with HIVLD. Reduced production of the ABCG2 protein and subsequently reduced transporter activity are linked to the ABCG2 34G/A polymorphism (30, 32–34). The overexpression of ABCG2 has been reported to lead to inter-individual difference in bioavailability of drugs (26).
This study has certain limitations. It could only evaluate the association and not decide causality. Initially, we allocated a ratio of 1:4 for case-controls. However, we were unable to manage matched enrollment in the controls. Despite this, our case-to-control ratio is approximately 1:2, which may be sufficient for this study. In addition, we had not assessed the plasma drug concentration in our subject participants. Hence, we were not able to address the correlation of MTP and ABCG2 polymorphisms with plasma drug levels in HIV patients.
The expression level of MTP gene was influenced by the MTP -493C/T polymorphism. Individuals who have impaired triglyceride level with ABCG2 34GA genotype may have a risk of dyslipidemia. Individuals with independently ABCG2 34A allele and ABCG2 34GA genotype with impaired LDL level may have a role to reduce the risk for severity of LDHIV. Similarly, MTP -493T allele may also reduce the risk for dyslipidemia. Further studies should be carried out in same and other populations with larger sample size for better understanding of the role of transporters among LDHIV.
The original contributions presented in the study are included in the article/Supplementary Materials, and further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the ICMR—National AIDS Research Institute, Pune. The patients/participants provided their written informed consent to participate in this study.
HS: Overall supervision and writing manuscript. CJ: Experimental work. SB: Data curation. SDM: Real time data sharing and analysis. RA: Real time standardization. KK: Real time experiment. SAS: Allow to work of Real time PCR and real time data analysis. MB: Clinical Support. RS, MS & JD: Recruitment of study subject. NA: Critical review. MKP: English language and critical scientific input. All authors contributed to the article and approved the submitted version.
This study was supported by ICMR (Grant No. HIV/50/206/09/2020/-ECD-II) and the Researchers Supporting Project (Grant no. RSP2023R379), King Saud University, Riyadh, Saudi Arabia for supporting this work.
We gratefully acknowledge the clinic staff Asefa Begum Khan, Shradha, and Sharad of ART Plus Centre, GMC & Hospital, Aurangabad, for the counseling of subject participants and collection of blood samples. We are also grateful to Sachin Dhaigude, ICMR—NARI, Pune, for the collection of blood samples. The authors are thankful to the Researchers Supporting Project (No. RSP2023R379), King Saud University, Riyadh, Saudi Arabia for supporting this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Loonam CR, Mullen A. Nutrition and the HIV-associated lipodystrophy syndrome. Nutr Res Rev. (2012) 25(2):267–87. doi: 10.1017/S0954422411000138
2. Miller J, Carr A, Emery S, Law M, Mallal S, Baker D, et al. HIV lipodystrophy: prevalence, severity and correlates of risk in Australia. HIV Med. (2003) 4(3):293–301. doi: 10.1046/j.1468-1293.2003.00159.x
3. Jacobson DL, Knox T, Spiegelman D, Skinner S, Gorbach S, Wanke C. Prevalence of, evolution of, and risk factors for fat atrophy and fat deposition in a cohort of HIV-infected men and women. Clin Infect Dis. (2005) 40(12):1837–45. doi: 10.1086/430379
4. Hansen AB, Lindegaard B, Obel N, Andersen O, Nielsen H, Gerstoft J. Pronounced lipoatrophy in HIV-infected men receiving HAART for more than 6 years compared with the background population. HIV Med. (2006) 7(1):38–45. doi: 10.1111/j.1468-1293.2005.00334.x
5. Mercier S, Gueye NF, Cournil A, Fontbonne A, Copin N, Ndiaye I, et al. Lipodystrophy and metabolic disorders in HIV-1-infected adults on 4- to 9-year antiretroviral therapy in Senegal: a case-control study. J Acquir Immune Defic Syndr. (2009) 51(2):224–30. doi: 10.1097/QAI.0b013e31819c16f4
6. Kalyanasundaram AP, Jacob SM, Hemalatha R, Sivakumar MR. Prevalence of lipodystrophy and dyslipidemia among patients with HIV infection on generic ART in rural south India. J Int Assoc Physicians AIDS Care. (2012) 11(5):329–34. doi: 10.1177/1545109711401750
7. Bhutia E, Hemal A, Yadav TP, Ramesh KL. Lipodystrophy syndrome among HIV infected children on highly active antiretroviral therapy in northern India. Afr Health Sci. (2014) 14(2):408–13. doi: 10.4314/ahs.v14i2.17
8. Diehl LA, Dias JR, Paes AC, Thomazini MC, Garcia LR, Cinagawa E, et al. Prevalência da lipodistrofia associada ao HIV em pacientes ambulatoriais brasileiros: relação com síndrome metabólica e fatores de risco cardiovascular [Prevalence of HIV-associated lipodystrophy in Brazilian outpatients: relation with metabolic syndrome and cardiovascular risk factors]. Arq Bras Endocrinol Metabol. (2008) 52(4):658–67 (in Portuguese). doi: 10.1590/S0004-27302008000400012
9. Lichtenstein KA, Ward DJ, Moorman AC, Delaney KM, Young B, Palella FJ Jr., et al. Clinical assessment of HIV-associated lipodystrophy in an ambulatory population. AIDS. (2001) 15(11):1389–98. doi: 10.1097/00002030-200107270-00008
10. Alencastro PR, Barcellos NT, Wolff FH, Ikeda ML, Schuelter-Trevisol F, Brandão AB, et al. People living with HIV on ART have accurate perception of lipodystrophy signs: a cross-sectional study. BMC Res Notes. (2017) 10(1):40. doi: 10.1186/s13104-017-2377-3
11. Tarr PE, Telenti A. Toxicogenetics of antiretroviral therapy: genetic factors that contribute to metabolic complications. Antivir Ther. (2007) 12(7):999–1013. doi: 10.1177/135965350701200714
12. Knox TA, Wanke C. 397 – gastrointestinal manifestations of HIV and AIDS. In: Goldman L, Schafer AI, editors. Goldman’s Cecil medicine (twenty-fourth edition). W.B. Saunders (2012). p. 2196–9. doi: 10.1016/B978-1-4377-1604-7.00397-3
13. Hussain MM, Rava P, Walsh M, Rana M, Iqbal J. Multiple functions of microsomal triglyceride transfer protein. Nutr Metab. (2012) 9:14. doi: 10.1186/1743-7075-9-14
14. Karpe F, Lundahl B, Ehrenborg E, Eriksson P, Hamsten A. A common functional polymorphism in the promoter region of the microsomal triglyceride transfer protein gene influences plasma LDL levels. Arterioscler Thromb Vasc Biol. (1998) 18(5):756–61. doi: 10.1161/01.ATV.18.5.756
15. Iqbal J, Rudel LL, Hussain MM. Microsomal triglyceride transfer protein enhances cellular cholesteryl esterification by relieving product inhibition. J Biol Chem. (2008) 283(29):19967–80. doi: 10.1074/jbc.M800398200
16. Wetterau JR, Combs KA, Spinner SN, Joiner BJ. Protein disulfide isomerase is a component of the microsomal triglyceride transfer protein complex. J Biol Chem. (1990) 265(17):9800–7. doi: 10.1016/S0021-9258(19)38742-3
17. Mirandola S, Bowman D, Hussain MM, Alberti A. Hepatic steatosis in hepatitis C is a storage disease due to HCV interaction with microsomal triglyceride transfer protein (MTP). Nutr Metab. (2010) 7:13. doi: 10.1186/1743-7075-7-13
18. Mirandola S, Osterreicher CH, Marcolongo M, Datz C, Aigner E, Schlabrakowski A, et al. Microsomal triglyceride transfer protein polymorphism (-493G/T) is associated with hepatic steatosis in patients with chronic hepatitis C. Liver Int. (2009) 29(4):557–65. doi: 10.1111/j.1478-3231.2008.01892.x
19. Siqueira ER, Oliveira CP, Correa-Giannella ML, Stefano JT, Cavaleiro AM, Fortes MA, et al. MTP -493G/T gene polymorphism is associated with steatosis in hepatitis C-infected patients. Braz J Med Biol Res. (2012) 45(1):72–7. doi: 10.1590/S0100-879X2011007500160
20. Bernard S, Touzet S, Personne I, Lapras V, Bondon PJ, Berthezène F, et al. Association between microsomal triglyceride transfer protein gene polymorphism and the biological features of liver steatosis in patients with type II diabetes. Diabetologia. (2000) 43(8):995–9. doi: 10.1007/s001250051481
21. Namikawa C, Shu-Ping Z, Vyselaar JR, Nozaki Y, Nemoto Y, Ono M, et al. Polymorphisms of microsomal triglyceride transfer protein gene and manganese superoxide dismutase gene in non-alcoholic steatohepatitis. J Hepatol. (2004) 40(5):781–6. doi: 10.1016/j.jhep.2004.01.028
22. Dowman JK, Tomlinson JW, Newsome PN. Pathogenesis of non-alcoholic fatty liver disease. QJM. (2010) 103(2):71–83. doi: 10.1093/qjmed/hcp158
23. Gouda W, Ashour E, Shaker Y, Ezzat W. MTP genetic variants associated with non-alcoholic fatty liver in metabolic syndrome patients. Genes Dis. (2017) 4(4):222–8. doi: 10.1016/j.gendis.2017.09.002
24. Ankathil R, Azlan H, Dzarr AA, Baba AA. Pharmacogenetics and the treatment of chronic myeloid leukemia: how relevant clinically? An update. Pharmacogenomics. (2018) 19(5):475–393. doi: 10.2217/pgs-2017-0193
25. Robey RW, To KK, Polgar O, Dohse M, Fetsch P, Dean M, et al. ABCG2: a perspective. Adv Drug Deliv Rev. (2009) 61(1):3–13. doi: 10.1016/j.addr.2008.11.003
26. Loscocco F, Visani G, Galimberti S, Curti A, Isidori A. BCR-ABL independent mechanisms of resistance in chronic myeloid leukemia. Front Oncol. (2019) 9:939. doi: 10.3389/fonc.2019.00939
27. Eyal S, Hsiao P, Unadkat JD. Drug interactions at the blood-brain barrier: fact or fantasy? Pharmacol Ther. (2009) 123(1):80–104. doi: 10.1016/j.pharmthera.2009.03.017
28. Vasiliou V, Vasiliou K, Nebert DW. Human ATP-binding cassette (ABC) transporter family. Hum Genomics. (2009) 3(3):281–90. doi: 10.1186/1479-7364-3-3-281
29. Bäckström G, Taipalensuu J, Melhus H, Brändström H, Svensson AC, Artursson P, et al. Genetic variation in the ATP-binding cassette transporter gene ABCG2 (BCRP) in a Swedish population. Eur J Pharm Sci. (2003) 18(5):359–64. doi: 10.1016/S0928-0987(03)00038-1
30. Zamber CP, Lamba JK, Yasuda K, Farnum J, Thummel K, Schuetz JD, et al. Natural allelic variants of breast cancer resistance protein (BCRP) and their relationship to BCRP expression in human intestine. Pharmacogenetics. (2003) 13(1):19–28. doi: 10.1097/00008571-200301000-00004
31. Kim HS, Sunwoo YE, Ryu JY, Kang HJ, Jung HE, Song IS, et al. The effect of ABCG2 V12M, Q141K and Q126X, known functional variants in vitro, on the disposition of lamivudine. Br J Clin Pharmacol. (2007) 64(5):645–54. doi: 10.1111/j.1365-2125.2007.02944.x
32. Imai Y, Nakane M, Kage K, Tsukahara S, Ishikawa E, Tsuruo T, et al. C421a polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low-level drug resistance. Mol Cancer Ther. (2002) 1(8):611–6.12479221
33. Kasza I, Várady G, Andrikovics H, Koszarska M, Tordai A, Scheffer GL, et al. Expression levels of the ABCG2 multidrug transporter in human erythrocytes correspond to pharmacologically relevant genetic variations. PLoS One. (2012) 7(11):e48423. doi: 10.1371/journal.pone.0048423
34. Kobayashi D, Ieiri I, Hirota T, Takane H, Maegawa S, Kigawa J, et al. Functional assessment of ABCG2 (BCRP) gene polymorphisms to protein expression in human placenta. Drug Metab Dispos. (2005) 33(1):94–101. doi: 10.1124/dmd.104.001628
35. Wu H, Liu Y, Kang H, Xiao Q, Yao W, Zhao H, et al. Genetic variations in ABCG2 gene predict breast carcinoma susceptibility and clinical outcomes after treatment with anthracycline-based chemotherapy. Biomed Res Int. (2015) 2015:279109. doi: 10.1155/2015/279109
36. Leung GK, Véniant MM, Kim SK, Zlot CH, Raabe M, Björkegren J, et al. A deficiency of microsomal triglyceride transfer protein reduces apolipoprotein B secretion. J Biol Chem. (2000) 275(11):7515–20. doi: 10.1074/jbc.275.11.7515
37. Oliveira CP, Stefano JT, Cavaleiro AM, Zanella Fortes MA, Vieira SM, Rodrigues Lima VM, et al. Association of polymorphisms of glutamate-cystein ligase and microsomal triglyceride transfer protein genes in non-alcoholic fatty liver disease. J Gastroenterol Hepatol. (2010) 25(2):357–61. doi: 10.1111/j.1440-1746.2009.06001.x
38. Ledmyr H, McMahon AD, Ehrenborg E, Nielsen LB, Neville M, Lithell H, et al. The microsomal triglyceride transfer protein gene-493T variant lowers cholesterol but increases the risk of coronary heart disease. Circulation. (2004) 109(19):2279–84. doi: 10.1161/01.CIR.0000130070.96758.7b
39. Chen L, Yoshino G, Maeda E, Zeng S. Effect of microsomal triglyceride transfer protein gene polymorphism in the promoter region on dyslipidemia in type 2 diabetic subjects. Chin Med J. (2003) 116(2):215–7.12775233
40. El-Koofy NM, El-Karaksy HM, Mandour IM, Anwar GM, El-Raziky MS, El-Hennawy AM. Genetic polymorphisms in non-alcoholic fatty liver disease in obese Egyptian children. Saudi J Gastroenterol. (2011) 17(4):265–70. doi: 10.4103/1319-3767.82582
41. Wang C, Xie L, Li H, Li Y, Mu D, Zhou R, et al. Associations between ABCG2 gene polymorphisms and isolated septal defects in a Han Chinese population. DNA Cell Biol. (2014) 33(10):689–98. doi: 10.1089/dna.2014.2398
42. Sari FM, Yanar HT, Ozhan G. Investigation of the functional single-nucleotide polymorphisms in the BCRP transporter and susceptibility to colorectal cancer. Biomed Rep. (2015) 3(1):105–9. doi: 10.3892/br.2014.383
43. Kim KA, Joo HJ, Park JY. ABCG2 polymorphisms, 34G> A and 421C> A in a Korean population: analysis and a comprehensive comparison with other populations. J Clin Pharm Ther. (2010) 35(6):705–12. doi: 10.1111/j.1365-2710.2009.01127.x
44. Lundahl B, Leren TP, Ose L, Hamsten A, Karpe F. A functional polymorphism in the promoter region of the microsomal triglyceride transfer protein (MTP -493G/T) influences lipoprotein phenotype in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. (2000) 20(7):1784–8. doi: 10.1161/01.ATV.20.7.1784
Keywords: genetic predisposition, MTP, ABCG2, HIVLD, association studies
Citation: Singh H, Jori C, Mahajan SD, Aalinkeel R, Kaliyappan K, Schwartz SA, Bhattacharya M, Shaikh R, Salve M, Deshmukh J, Ali N and Parvez MK (2023) Comparative analysis of MTP -493G/T and ABCG2 34G/A polymorphisms and theirs expression in HIV-associated lipodystrophy patients. Front. Cardiovasc. Med. 10:1177054. doi: 10.3389/fcvm.2023.1177054
Received: 1 March 2023; Accepted: 2 May 2023;
Published: 30 May 2023.
Edited by:
Kailash Gulshan, Cleveland State University, United StatesReviewed by:
Gregory Tchou, Cleveland Clinic, United States© 2023 Singh, Jori, Shyamveer, Mahajan, Aalinkeel, Kaliyappan, Schwartz, Bhattacharya, Shaikh, Salve, Deshmukh, Ali and Parvez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: HariOm Singh aHNpbmdoQG5hcmlpbmRpYS5vcmc=; aGFyaW9tc2dwZ2ltc0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.