- 1Division of Vascular Surgery, Department of Surgery, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, United States
- 2Department of Cellular and Regenerative Biology, University of Wisconsin-Madison School of Medicine and Public Health, Madison, WI, United States
There is gaining popularity in the use of single-cell technology and analysis in studying the pathogenesis of abdominal aortic aneurysm (AAA). As there are no current pharmacologic therapies for impeding aneurysm growth or preventing AAA rupture, identifying key pathways involved in AAA formation is critical for the development of future therapies. Single-cell RNA sequencing (scRNA-seq) technology provides an unbiased and global view of transcriptomic characteristics within each of the major cell types in aneurysmal tissues. In this brief review, we examine the current literature utilizing scRNA-seq for the analysis of AAA and discuss trends and future utility of this technology.
Introduction
Abdominal aortic aneurysm (AAA) is a complex multifactorial pathologic dilation of the abdominal aorta. AAAs are prevalent in roughly 1–2% of men over the age of 65 years, and nearly 0.5% of women over 70 years old, however, AAA rupture is the leading cause of death in the United States (1, 2). Aneurysms progressively dilate over time, and once reaching a threshold size, elective surgical repair with either open surgery or endovascular graft placement is recommended. Due to the lack of effective pharmacological therapies, surgical intervention remains the only available treatment for this disease. Without intervention, continued AAA sac expansion increases the risk of aneurysm rupture, which carries significant morbidity and mortality.
Due to the complex interplay of biochemical, hemodynamic, genetic, and environmental mechanisms, investigating the pathophysiology of AAA presents a challenge. Arterial wall damage related to advanced age, smoking, hypertension, male gender, and family predisposition results in an accumulation of chronic inflammation, oxidative stress and faulty repair mechanisms (2, 3). Irreversible changes in the vessel wall due to smooth muscle cell death, the activation of matrix metalloproteases, and altered extracellular matrix composition cause progressive dilation and weakening of the aortic wall (2). Disrupted arterial flow within the aneurysm sac places additional biomechanical stressors on the wall, further contributing to the pathologic process (4). Despite the identification of many mechanisms involved in AAA, there is limited understanding in the way these processes are interrelated and regulated at the cellular and molecular levels.
Single-cell RNA sequencing (scRNA-seq) is a technique used to analyze gene expression at the level of individual cells. Compared to conventional RNA sequencing, which analyzes an average gene expression in bulk tissue samples, scRNA-seq provides a more detailed and precise evaluation of cell heterogeneity within samples, as it can identify rare cell types and even predict cell-cell signaling pathways. Since the discovery of single-cell RNA sequencing (scRNA-seq) by Tang et al. in 2009, the use of this technology has become increasingly embraced in recent years by pathophysiological studies (5). Analyses of RNA transcripts have further demonstrated the vast complexity and heterogeneity of cells involved in cardiovascular disease. However, its use in the evaluation of AAA was unknown only until recently in 2018 (6). Today, scRNA-seq in AAA is gaining popularity, partially through the availability of public datasets. In this study, we review the current literature on scRNA-seq in AAA to demonstrate the potential of this technology in studying aneurysmal disease, and its challenges.
Methods
A literature search of publications regarding scRNA sequencing on AAA was performed using the PubMed search engine MEDLINE database. Search terms included (“single-cell RNA” OR “scRNA-seq”) AND “abdominal aortic aneurysm(s)” queried within all fields of text in order to capture the largest number of articles. Only original studies with full text published prior to 2023 were included. Studies were excluded if no new primary scRNA-seq data or in-depth analysis of existing datasets were performed. The model or tissue specimen to which scRNA-seq was performed, the number of cells and clusters, and the major scRNA-seq specific findings were then compared.
Results
The above search terms yielded 17 results, of which 2 studies were excluded. One study was excluded as the authors used a previously published scRNA-seq database to identify the presence of an RNA sequence of interest instead of analyzing the dataset (7). A review article was also excluded as it did not contain primary data (8). Of the remaining 15 manuscripts, 9 studies collected primary scRNA-seq data (Table 1), and 6 studies reanalyzed available scRNA-seq databases (Table 2).
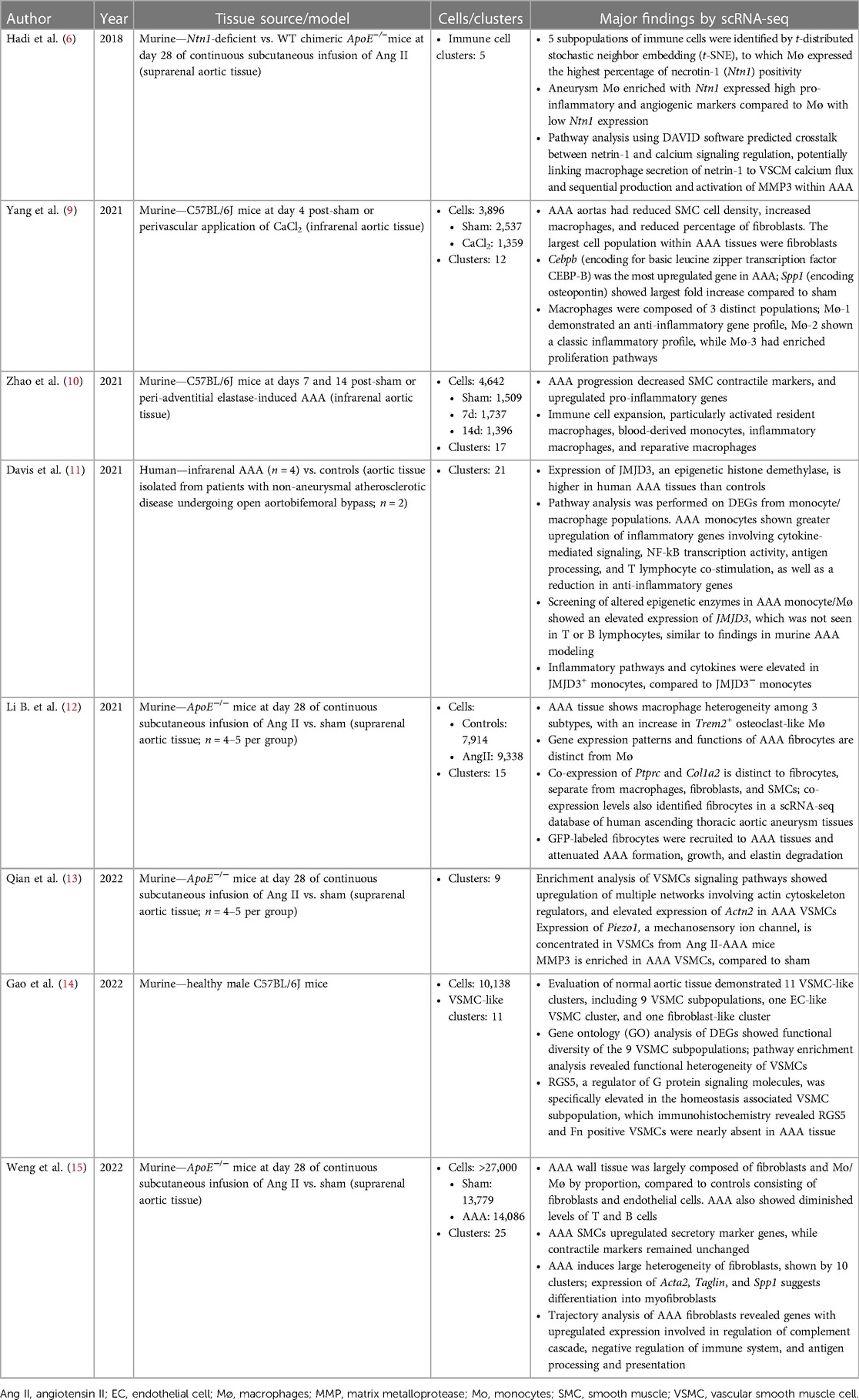
Table 1. Studies involving primary collection and analysis of single-cell RNA sequencing of AAA tissue.
The first publication utilizing scRNA-seq technology for the study of AAA was by Hadi et al. in 2018, using an angiotensin II (Ang II)-induced AAA murine model (6). A single article per year on the topic of scRNA-seq in AAA was published between 2018 and 2020. By 2021, popularity of scRNA-seq in AAA began to rise as 4 manuscripts were published, one by Davis et al. which is the first and only study to use this technology on human AAA tissues (11). In 2022, 9 manuscripts were published (excluding the 2 articles previously mentioned), resulting in a near exponential increase in new publications per year (Figure 1). The greater number of publicly accessible datasets allowed for more analytical research, demonstrated by 5 of the 6 database studies being conducted that year.
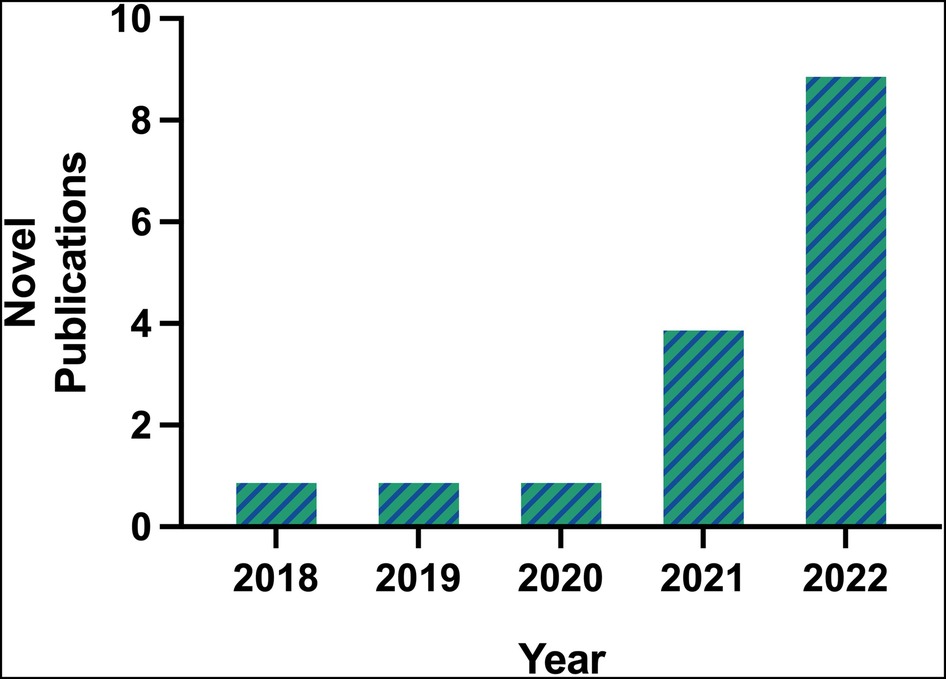
Figure 1. Number of novel publications on scRNA-seq in AAA available on PubMed prior to 2023, according to inclusion and exclusion criteria.
The currently available AAA scRNA-seq databases were derived from analyzing human and experimental aneurysmal aortic tissues. Three models of murine aneurysm that have been analyzed by scRNA-seq include the Ang II osmotic pump model, adventitial elastase model, and perivascular calcium chloride (CaCl2) models. All of these mouse studies used young male mice. In the human dataset, 2 of the 4 AAA tissue samples were from female patients, while both controls (n = 2) were acquired from male patients undergoing aortobifemoral bypass (11).
Of the database analysis studies, most compared the human dataset to murine AAA sets, or compared scRNA-seq data to other available genetic data, such as mRNA microarray expression datasets. Most studies reclustered available datasets and utilized differentially expressed genes (DEGs) to identify a specific sequence of interest. One study utilized CellChat analysis to determine intercellular communication pathways by identifying outgoing and incoming signals between cell clusters (17).
Consistent with the wide use of the Ang II model by other approaches, 4 of the original scRNA-seq studies were conducted with this model (Table 1). Each of these studies used ApoE−/− mice that were infused with Ang II (1,000 ng/kg/min) for 28 days, and primarily focused their analysis on a particular tissue cell type (6, 12, 13, 15). The majority of the published murine AAA scRNA-seq studies were performed on the suprarenal aortic tissue segments when the aneurysmal dilation was well established. While the study by Yang and colleagues examined the transcriptomic profiles of infrarenal aortic tissues acutely following the aneurysm induction by CaCl2 (9), Zhao and colleagues examined both 7- and 14-day tissues following aneurysm induction by elastase (10). Despite of using different AAA models, pathological stages, and aortic segments, all of the investigative groups of the published scRNA-seq studies discovered multiple cellular types and several commonly altered signaling pathways including those responsible inflammation, vascular smooth muscle cell phenotype changes and aortic wall remodeling (Tables 1,2).
Discussion
This brief systematic review serves as a resource for the current status of scRNA-seq in AAA. As identified above, scRNA-seq is gaining popularity and utility in AAA, as most studies have been conducted within the past year. Overall, this technology validates the existing understandings of the pathophysiologic mechanisms involved in AAA. Collectively, the scRNA-seq studies highlighted the importance of known pathological processes including inflammation, negative remodeling of extracellular matrix proteins, and phenotypic changes of vascular smooth muscle cells. However, with its capability of resolving transcriptome at a single cell resolution, scRNA-seq additionally provides a clear view on the heterogeneity of each of the major cell types found in aneurysmal tissues. Furthermore, the development of various bioinformatic analytical tools enables delineation of the specific interactions involved within complex pathways and predictions of cellular communications. For example, the scRNA-seq analysis conducted by Hadi et al. provides the precise mechanism infiltrated macrophages activate vascular smooth muscle cells to upregulate matrix metalloprotease-3 transcription and activation, which takes the known inflammatory response to a much deeper depth (6). Similarly, Davis et al.'s study recapitulates the critical involvement of inflammatory macrophages involved in AAA development, but also provides a molecular path through which macrophages transform (11).
Evaluating AAA using scRNA-seq has its limitations. In terms of scRNA-seq technology, evaluating changes in RNA at the transcriptome level does not necessarily reflect similar changes at the proteomic level. The high specificity of scRNA-seq comes at the cost of reduced sensitivity compared to bulk RNA sequencing, meaning low-abundance transcripts or rare cell phenotypes may be undetected or unrecognized using this method. Additionally, scRNA-seq typically generates low read coverage per cell and often uses short-read sequencing, which can make it difficult to identify alternative splicing events that occur at low levels. The manipulation of cells during ex vivo isolation may also lead to alterations in RNA prior to analysis. Using scRNA-seq for the evaluation of AAA is restricted by the limitations of current animal models and scRNA-seq databases. In mice, the available AAA models utilize only male mice in all the published scRNA-seq work. Although the majority of AAA occur in men, women, particularly postmenopausal women, are also affected by this highly lethal disease. The exclusion of female animals in the scRNA-seq study design diminishes the ability to evaluate sex differences in AAA. It is therefore a plausible progress made by Davis and colleagues to include AAA samples from female patients (11). However, it remains challenging to determine the optimal ratio between male and female samples, considering AAA disproportionally affects males.
In summary, scRNA-seq is a powerful research tool which enables unbiased investigation of AAA pathophysiology. Additional datasets from human AAA patients and controls are warranted. Further comparisons between human and murine datasets as well as between male and female datasets are useful in determining which of the complex molecular pathways are likely therapeutic targets.
Author contributions
JB and BL designed this study and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
This investigation was supported by the National Heart Lung and Blood Institute (NHLBI) of the National Institutes of Health (NIH) under 1R01HL149404-01A1 (BL), and the Ruth L. Kirschstein National Research Service Award T32 HL 007936 to the University of Wisconsin-Madison Cardiovascular Research Center (JB).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sampson UKA, Norman PE, Fowkes FGR, Aboyans V, Song Y, Harrell FE, et al. Estimation of global and regional incidence and prevalence of abdominal aortic aneurysms 1990 to 2010. Glob Heart. (2014) 9(1):159–70. doi: 10.1016/j.gheart.2013.12.009
2. Sakalihasan N, Michel JB, Katsargyris A, Kuivaniemi H, Defraigne JO, Nchimi A, et al. Abdominal aortic aneurysms. Nature Reviews Disease Primers 2018 4:1. (2018) 4(1):1–22. doi: 10.1038/s41572-018-0030-7
3. Shimizu K, Mitchell RN, Libby P. Inflammation and cellular immune responses in abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol. (2006) 26(5):987–94.. doi: 10.1161/01.ATV.0000214999.12921.4f
4. Inzoli F, Boschetti F, Zappa M, Longo T, Fumero R. Biomechanical factors in abdominal aortic aneurysm rupture. Eur J Vasc Surg. (1993) 7(6):667–74. doi: 10.1016/S0950-821X(05)80714-5
5. Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. (2009) 6(5):377–82. doi: 10.1038/nmeth.1315
6. Hadi T, Boytard L, Silvestro M, Alebrahim D, Jacob S, Feinstein J, et al. Macrophage-derived netrin-1 promotes abdominal aortic aneurysm formation by activating MMP3 in vascular smooth muscle cells. Nat Commun. (2018) 9(1):5022. doi: 10.1038/s41467-018-07495-1
7. Zhao G, Zhao Y, Lu H, Chang Z, Liu H, Wang H, et al. BAF60c Prevents abdominal aortic aneurysm formation through epigenetic control of vascular smooth muscle cell homeostasis. J Clin Invest. (2022) 132(21):e158309. doi: 10.1172/JCI158309
8. Deroo E, Stranz A, Yang H, Hsieh M, Se C, Zhou T. Endothelial dysfunction in the pathogenesis of abdominal aortic aneurysm. Biomolecules. (2022) 12(4):509. doi: 10.3390/biom12040509
9. Yang H, Zhou T, Stranz A, Deroo E, Liu B. Single-Cell RNA sequencing reveals heterogeneity of vascular cells in early stage murine abdominal aortic aneurysm—brief report. Arterioscler Thromb Vasc Biol. (2021) 41(3):1158–66. doi: 10.1161/ATVBAHA.120.315607
10. Zhao G, Lu H, Chang Z, Zhao Y, Zhu T, Chang L, et al. Single-cell RNA sequencing reveals the cellular heterogeneity of aneurysmal infrarenal abdominal aorta. Cardiovasc Res. (2021) 117(5):1402–16. doi: 10.1093/cvr/cvaa214
11. Davis FM, Tsoi LC, Melvin WJ, denDekker A, Wasikowski R, Joshi AD, et al. Inhibition of macrophage histone demethylase JMJD3 protects against abdominal aortic aneurysms. J Exp Med. (2021) 218(6):e20201839. doi: 10.1084/jem.20201839
12. Li B, Song X, Guo W, Hou Y, Hu H, Ge W, et al. Single-Cell transcriptome profiles reveal fibrocytes as potential targets of cell therapies for abdominal aortic aneurysm. Front Cardiovasc Med. (2021) 8:753711. doi: 10.3389/fcvm.2021.753711
13. Qian W, Hadi T, Silvestro M, Ma X, Rivera CF, Bajpai A, et al. Microskeletal stiffness promotes aortic aneurysm by sustaining pathological vascular smooth muscle cell mechanosensation via Piezo1. Nat Commun. (2022) 13(1):512. doi: 10.1038/s41467-021-27874-5
14. Gao YK, Guo RJ, Xu X, Huang XF, Song Y, Zhang DD, et al. A regulator of G protein signaling 5 marked subpopulation of vascular smooth muscle cells is lost during vascular disease. PLoS One. (2022) 17(3):e0265132. doi: 10.1371/journal.pone.0265132
15. Weng Y, Lou J, Bao Y, Cai C, Zhu K, Du C, et al. Single-Cell RNA sequencing technology revealed the pivotal role of fibroblast heterogeneity in angiotensin II-induced abdominal aortic aneurysms. DNA Cell Biol. (2022) 41(5):498–520. doi: 10.1089/dna.2021.0923
16. Liu Y, Wang X, Wang H, Hu T. Identification of key genes and pathways in abdominal aortic aneurysm by integrated bioinformatics analysis. J Int Med Res. (2019) 48(4):300060519894437. doi: 10.1177/0300060519894437
17. Yang H, DeRoo E, Zhou T, Liu B. Deciphering cell-cell communication in abdominal aortic aneurysm from single-cell RNA transcriptomic data. Front Cardiovasc Med. (2022) 9:831789. doi: 10.3389/fcvm.2022.831789
18. Li Y, Zhang Z, Liu D. Intracranial aneurysms induced by RUNX1 through regulation of NFKB1 in patients with hypertension-an integrated analysis based on multiple datasets and algorithms. Front Neurol. (2022) 13:877801. doi: 10.3389/fneur.2022.877801
19. Xiong T, Lv XS, Wu GJ, Guo YX, Liu C, Hou FX, et al. Single-Cell sequencing analysis and multiple machine learning methods identified G0S2 and HPSE as novel biomarkers for abdominal aortic aneurysm. Front Immunol. (2022) 13:907309. doi: 10.3389/fimmu.2022.907309
20. Davis FM, Tsoi LC, Ma F, Wasikowski R, Moore BB, Kunkel SL, et al. Single-cell transcriptomics reveals dynamic role of smooth muscle cells and enrichment of immune cell subsets in human abdominal aortic aneurysms. Ann Surg. (2022) 276(3):511–21. doi: 10.1097/SLA.0000000000005551
21. Cheng S, Liu Y, Jing Y, Jiang B, Wang D, Chu X, et al. Identification of key monocytes/macrophages related gene set of the early-stage abdominal aortic aneurysm by integrated bioinformatics analysis and experimental validation. Front Cardiovasc Med. (2022) 9:950961. doi: 10.3389/fcvm.2022.950961
Keywords: abdominal aortic aneursym, single-cell RNA sequencing, transcriptomic, inflammation, vascular smooth muscle cell (VSMC)
Citation: Bontekoe J and Liu B (2023) Single-cell RNA sequencing provides novel insights to pathologic pathways in abdominal aortic aneurysm. Front. Cardiovasc. Med. 10:1172080. doi: 10.3389/fcvm.2023.1172080
Received: 23 February 2023; Accepted: 9 May 2023;
Published: 23 May 2023.
Edited by:
Masanori Aikawa, Harvard Medical School, United StatesReviewed by:
Lin Chang, University of Michigan, United StatesA. Phillip Owens III, University of Cincinnati, United States
© 2023 Bontekoe and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Liu YmxpdTI0QHdpc2MuZWR1
 Jack Bontekoe
Jack Bontekoe Bo Liu
Bo Liu