
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 26 May 2023
Sec. Cardiovascular Genetics and Systems Medicine
Volume 10 - 2023 | https://doi.org/10.3389/fcvm.2023.1152851
This article is part of the Research Topic Cardiovascular Genomics, Stem Cells and Precision Medicine View all 5 articles
Objective: N6-Methyladenosine (m6A) modification is of great importance in both the pathological conditions and physiological process. The m6A single nucleotide polymorphisms (SNPs) are associated with cardiovascular diseases including coronary artery disease, heart failure. However, it is unclear whether m6A-SNPs are involved in atrial fibrillation (AF). Here, we aimed to explore the relationship between m6A-SNPs and AF.
Method: The relationship between m6A-SNPs and AF was evaluated by analyzing the AF genome-wide association study (GWAS) and m6A-SNPs annotated by the m6AVar database. Further, eQTL and gene differential expression analysis were performed to confirm the association between these identified m6A-SNPs and their target genes in the development of AF. Moreover, we did the GO enrichment analysis to figure out the potential functions of these m6A-SNPs affected genes.
Result: Totally, 105 m6A-SNPs were identified to be significantly associated with AF (FDR < 0.05), among which 7 showed significant eQTL signals on local genes in the atrial appendage. By using four public AF gene expression datasets, we identified genes SYNE2, USP36, and THAP9 containing SNPs rs35648226, rs900349, and rs1047564 were differentially expressed in AF population. Further, SNPs rs35648226 and rs1047564 are potentially associated with AF by affecting m6A modification and both of them might have an interaction with RNA-binding protein, PABPC1.
Conclusion: In summary, we identified m6A-SNPs associated with AF. Our study provided new insights into AF development as well as AF therapeutic target.
Atrial fibrillation (AF) is the most common arrhythmia, which can lead to severe stroke and heart failure and carries high risk of morbidity and mortality. AF affects millions of people worldwide with a prevalence of 2%–4% in the general population (1). The risk of developing AF is intimately associated with factors as diabetes, hypertension, heart failure, and smoking as well as alcohol et al. (2). Despite of over 100 years of research, the fundamental mechanisms of AF is not totally well known. AF results from interactions between triggers which are responsible for its initiation, and the substrate that is responsible for its perpetuation. Haïssaguerre et al. (3) first demonstrated in human patients that the atrial sleeves in the pulmonary veins harbor the vast majority of the ectopic electric triggers that initiate AF. His results have been confirmed by the following researchers (4). Since then, electrical isolation of the pulmonary veins became universal therapeutic means to prevent AF. Mechanisms underlying the perpetuation of AF are still debated. Multiple wavelets and localized (focal or reentrant) sources are largely accepted to drive AF. Besides, electrical remodeling, structural remodeling, genetic predisposition, and neuro-humoral contributors make the interactions between initiation and perpetuation more complex (5). AF appears to be highly heritable. Nearly one fifth of AF is familial, which suggests a genetic predisposition. To date, GWAS studies have identified a dozen of genetic loci associated with AF. Most of the gene mutation sites related to AF are from ion channel gene mutations, such as SCN5A, KCNQ1, ABCC9, etc., but non-ion channel gene mutations have also been found in patients with AF, including NPPA, TBX5, MYL4 genes (6). GWAS has also shown single-nucleotide polymorphism (SNP) is strongly associated with susceptibility to AF (7, 8).
N6-methyladenosine (m6A) modification, methylation of the sixth nitrogen atom on the RNA molecule adenosine, is the most abundant internal modification of messenger RNA (mRNA) in eukaryotic cells, which was also detected in transfer RNA (tRNA), ribosome RNA (rRNA) as well as non-coding RNA (ncRNA). m6A modification mainly occurs in the 3′-untranslated regions and nearby the stop codons of mRNAs (9). The process of m6A methylation is dynamic and reversible and is regulated by m6A methylation regulators including “writers”, “erasers” and “readers”. m6A writers are composed of RNA methyltransferase complex, such as methyltransferase like 3 (METTL3), methyltransferase like 14 (METTL14) as well as Wilm's tumor 1-associated protein (WTAP) (10). m6A erasers are demethylases including Fat mass and obesity-associated protein (FTO) and α-ketoglutarate-dependent dioxygenase alk B homolog 5 (ALKBH5) (11, 12). m6A readers are RNA-binding proteins (RBPs) that regulate the mRNA processing and metabolism containing the YT521-B homology (YTH) domain family (YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2), ELAV-like protein 1 (ELAVL1), insulin-like growth factors (IGF2BP1, IGF2BP2 and IGF2BP3), et al. (13). m6A mainly occurs within a highly-conserved consensus motif RRACH, in which R represents A or G, and H represents A, C or U (14). m6A methylation regulates the splicing, transport, storage, decay and translation of mRNAs, and plays crucial roles in various cellular pathways and processes such as cell differentiation, development and metabolism (14).
Evidences have shown m6A modification affects gene expression and plays an essential role in heart development and pathophysiological process of cardiovascular diseases including myocardial infraction (15), heart failure, myocardial hypertrophy (16), and heart regeneration (17). Wang et al. reported WTAP promoted myocardial ischemia-reperfusion injury through promoting endoplasmic reticulum stress and cell apoptosis by regulating m6A modification of the activating transcription factor 4 (ATF4) mRNA (15). Berulava et al. found m6A level is altered in heart hypertrophy and heart failure and m6A RNA methylation changes lead to changes in protein abundance, unconnected to mRNA levels, which contribute to heart failure progression (18). In another study, the FTO protein expression is reduced in failing heart, which increases m6A in RNA and decreases cardiomyocyte contractile function (19). Moreover, recent studies have confirmed that single-nucleotide polymorphisms (SNPs) can affect the m6A modification by changing the RNA sequence of the target site and key flanking nucleotides and these SNPs have been suggested to be associated with various diseases including coronary artery disease, ischemic stroke and cancer (20–22). However, it is unclear whether there is relationship between m6A-SNP and arrhythmia. Here, we investigated the potential roles of m6A-SNP and AF by using the public GWAS and m6Avar database, which may provide novel insights to AF development.
We first explored the effects of m6A-SNPs on AF using the published GWAS data (nielsen-thorolfsdottir-willer-NG2018-AFib-gwas-summary-statistics.tbl), which included 60,620 AF cases and 970,216 controls with 34,740,186 SNPs in European-American population. To identify the m6A-SNPs in these 34,740,186 SNPs, we obtained the m6A-SNPs from the m6AVar database (http://rmvar.renlab.org/download.html). This database contains three confidence levels: 217,241m6A-SNPs are in high confidence level, 506,235 m6A-SNPs are in medium confidence level, 678,338 m6A-SNPs are in low confidence level. We therefore tested the association between these m6A-SNPs and the risk of AF (Figure 1).
We performed the eQTL analysis to explore the possible functions for the AF associated m6A-SNPs in atrial appendage from European-American subjects by using the GTEx V8 database (https://gtexportal.org/home/), through downloaded significant variant gene pairs in GTEx, which contained 1,475,414 significant variant gene pairs. The nominal p-value threshold is for calling a variant gene pair signature for the gene. HaploReg browser (https://www.encodeproject.org/software/haploreg/), which aggregated eQTL data from 13 studies carried out in different human cells and tissues is used to validate whether the eQTL signal of the identified m6A-SNP is displayed. In addition, through HaploReg, potential effects on local gene expression and function annotation of the identified m6A-SNPs were analyzed to determine their possible roles in transcriptional regulation. This eQTL analysis was to find m6A-SNPs showed eQTL signals and get potential functional evidence of affecting AF for these SNPs.
GO database is a powerful system for gene annotation. Through GO analysis of identified AF-related genes, with which the m6A-SNPs showed eQTL signals, we can classify these genes according to different functional annotation and figure out which genes associated with AF are related to the functional changes. Gene list enrichments are identified in the following ontology categories: DisGeNET. The Metascape tools (https://metascape.org/gp/index.html#/main/step1) were used to do GO enrichment analysis and Quality Control and Association Analysis.
We further explored whether the expression of the AF-related m6A-SNPs regulated genes were related to AF. We therefore detected if the genes in which AF-associated m6A-SNPs located were expressed differently between AF population and healthy controls. GSE128188, GSE2240, GSE135445 and GSE79768 datasets were used for analyzing the different expressions between healthy controls and AF population. We used the online website network analyst (https://www.networkanalyst.ca/) to meta-analyze the differential genes of those datasets. The GEO dataset (http://www.ncbi.nlm.nih.gov/geo) collects microarray and sequencing information of gene expressions for various diseases. The DEmRNAs with combined p < 0.05 were considered to be significant in differential expression analysis.
To examine whether these AF-related m6A-SNPs play a role in m6A modification and predict its changes, a computational predictor of m6A modification site named sequence-based RNA adenosine methylation site predictor (SRAMP), was used to perform the analysis (23). After the reference sequence and changed sequence were input into SRAMP, the m6A modification sites near the m6A-SNP will be detected and the confidence of m6A modifications was displayed as well. Meanwhile, the potential functions of selected m6A-SNPs were analyzed by querying UCSC database.
We first used the AF GWAS and the m6Avar databases to find out the intersection and selected out these m6A-SNPs associated with AF. In this study, we analyzed 34,740,186 SNPs from the GWAS for atrial appendage and 1,401,814 m6A-SNPs from the m6Avar database, identifying a total of 42,711 m6A-SNPs and a Manhattan plot containing these m6A-SNPs was generated. Furthermore, we identified 105 m6A-SNPs that were significantly associated with AF risk with False Discovery Rate (FDR) was less than 0.05 (Figures 1, 2).
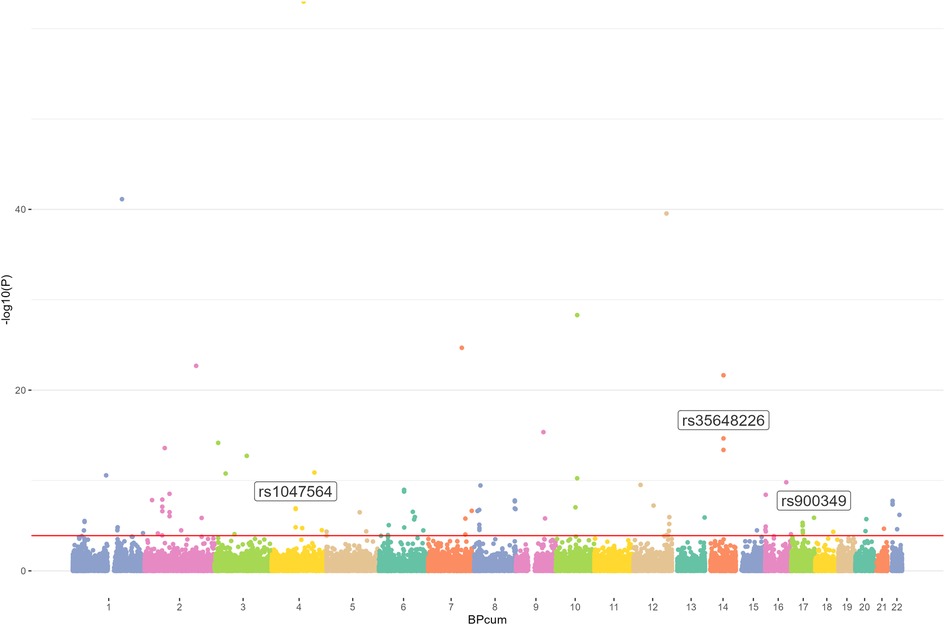
Figure 2. Genome-wide results for the relationship between m6A-SNPs and AF. Manhattan plot showed105 m6A-SNPs were associated with AF (FDR < 0.05). SNPs that may affect m6A modification of AF were annotated.
We then tried to figure out the potential roles underlying the 105 identified m6A-SNPs related to AF. Since m6A-SNPs may have effect on RNA modification as well as the risk of AF, they may act through changing the expression of local genes. We further investigated whether these AF-associated m6A-SNPs were related to the expression levels of local genes by using eQTL analysis. As a result, 7 of the 105 identified AF associated m6A-SNPs showed significant eQTL signals in atrial appendage using GTEx database with threshold: nominal p-values < the nominal p-value threshold (Table 1). Besides, all the 7 SNPs were loss of function variants.
To functionally annotate the identified SNPs, we carried out GO enrichment analysis. The GO enrichment analysis suggested that the biological functions of these genes of 105 m6A-SNPs include multicellular organismal process (GO:0032501), cellular process (GO:0009987), metabolic process (GO:0008152), regulation of biological process (GO:0050789), localization (GO:0051179), locomotion (GO:0040011), developmental process (GO:0032502), biological regulation (GO:0065007), response to stimulus (GO:0050896), negative regulation of biological process (GO:0048519), positive regulation of biological process (GO:0048518) (Figure 3A). As for quality control and association analysis of 7 genes regulated by 7 m6A-SNPs with eQTL signals, the top few enriched clusters (persistent atrial fibrillation:C2585653, familial atrial fibrillation:C3468561, paroxysmal atrial fibrillation:C0235480) are shown in the Figure 3B.
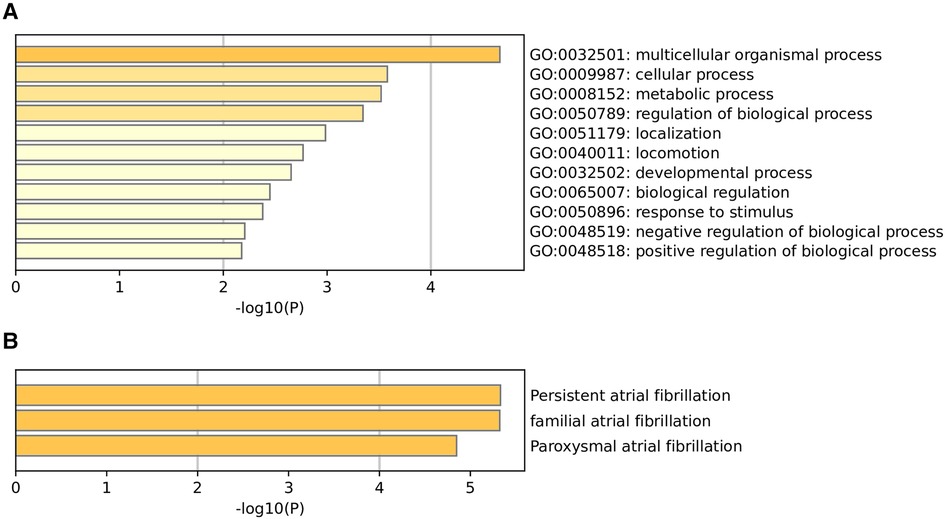
Figure 3. Go enrichment analysis (A) and quality control and association analysis (B) was used to elucidate the potential functions of genes corresponding to identified m6A-SNPs.
The above analysis found a total of 7 m6A-SNPs showing eQTL signals. In order to analyze the mRNA expression levels of the local genes containing these SNPs, we further compare the mRNA expression levels between AF cases and healthy ones in four RNA expression datasets (GSE128188, GSE2240, GSE135445 and GSE79768), consisting 40 patients with atrial fibrillation, 48 controls and 8,636 genes. The meta-analysis results showed all the 3 genes (SYNE2, USP36 and THAP9) corresponding to 3 m6A-SNPs (rs35648226, rs900349 as well as rs1047564) were differentially expressed (combined p < 0.05) (Table 2). This result suggested these m6A-SNPs might also have influence on AF risk by affecting the expression of local genes. The regulatory functions of the 3 m6A-SNPs, shown in Table 2, was performed to determine their possible roles in transcriptional regulation by HaploReg. SNPs with nominal p-values < the nominal p-value threshold were selected in European-American subjects and the GTEx V8 eQTL Calculator was used to further prioritize the eQTLs of SYNE2, USP36 and THAP9 in the atrial appendage (Figure 4).
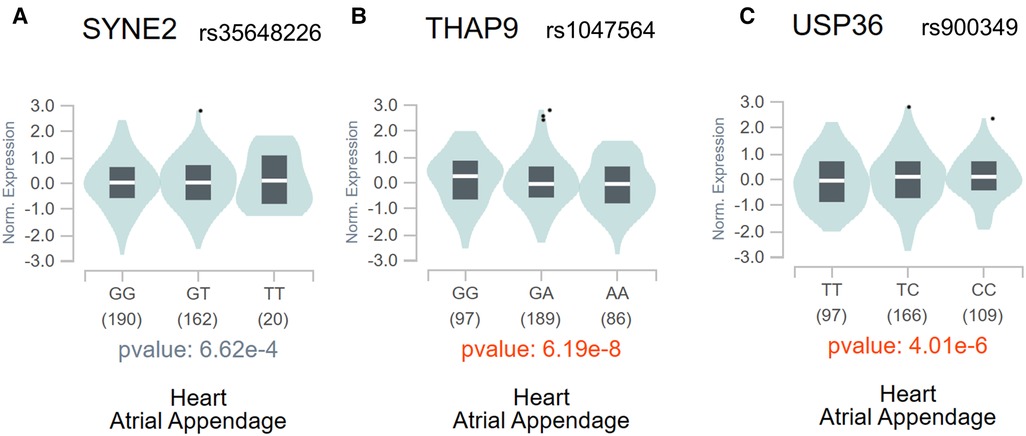
Figure 4. The violin plots of the eQTLs of SYNE2 (A), THAP9 (B), and USP36 (C) in the atrial appendage in the whole population through the GTEx V8 eQTL calculator.
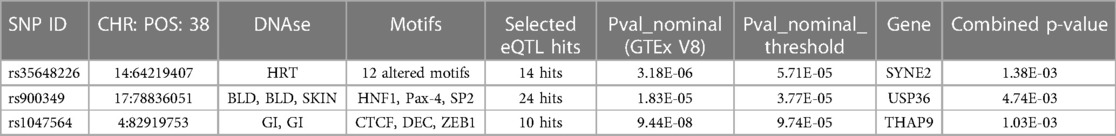
Table 2. 3 genes of m6A-SNPs were differentially expressed in European-American subjects and their annotations by HaploReg browser.
To investigate the possible m6A methylation sites of SYNE2 (rs35648226), USP36 (rs900349) and THAP9 (rs1047564), their transcript sequences or genomic sequences were predicted at the SRAMP website. According to the analysis results, the rs35648226 and rs1047564 that near to high convincible m6A-modified predicted peaks might have an influence on m6A modification. The rs35648226 is located in the CDS of SYNE2 and is a G > T change. It is located 3 bp from a predicted m6A site annotated by SRAMP databases (Figure 5A,B), and the G > T change was predicted to reduce the m6A level of SYNE2. The Figure 5C,D showed that this rs1047564 G > A polymorphism highly reduced the THAP9 m6A level. Previous studies (24–26) have suggested that alterations in the m6A level of RNA can impact the binding of RNA-binding proteins (RBPs), thereby influencing the gene expression level. Given that the SNPs identified in our study were found to affect the m6A level, we sought to further investigate which RBPs may be binding in the region where the SNPs are located. We next query the UCSC browser to analyze the potential function of the 2 m6A-SNPs. As is shown in Figure 6A, RIP-chip GeneST from ENCODE/SUNY Albany data suggested that SLBP and PABPC1 might have a potential interaction with rs35648226 (SYNE2). The SNP rs1047564, located on the 3′UTR of the THAP9 on chromosome 4, may interact with PABPC1 (Figure 6B). Our findings imply that these two SNPs may be related to AF susceptibility by lowering the degree of m6A modification that regulates gene expression. Besides, rs35648226 and rs1047564 may also have an impact on the local gene expression by binding with the RBPs SLBP and PABPC1.
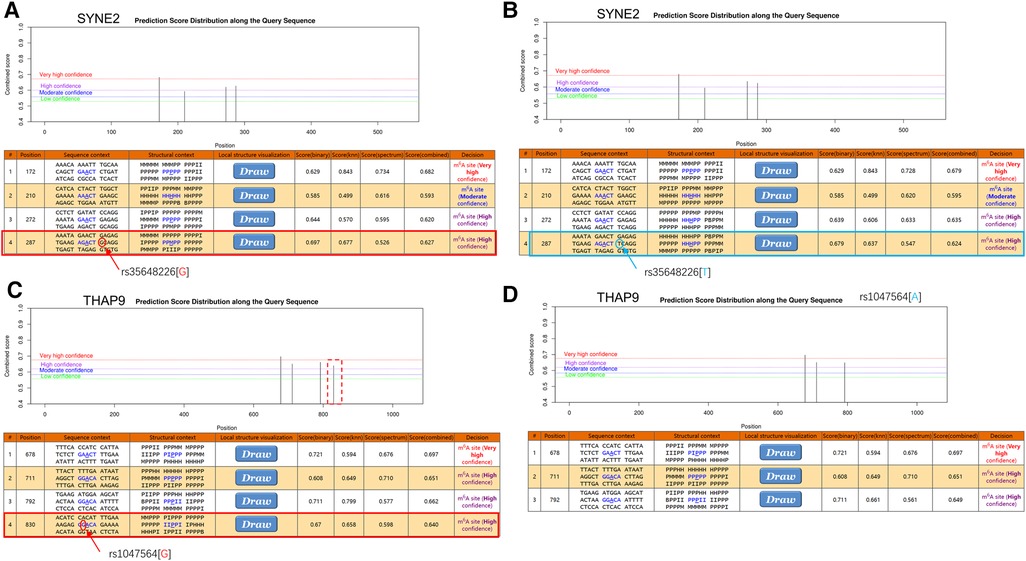
Figure 5. The m6A modification was predicted by using the genomic sequence of SYNE2 and THAP9 transcripts on website (http://www.cuilab.cn/scramp). (A,B) There were highly convincible m6A modification predicted peaks near SYNE2(rs35648226) and the G > T change was predicted to reduce the m6A level of SYNE2. (C,D) The red dashed box indicates the disappearance of the m6A modified peak near rs1047564 after inputting the altered sequence. The tracks sequentially show base position, FANTOM CAT, UCSC gene, transcription levels assayed by RNA-Seq on nine cell lines from ENCODE, DNase I hypersensitivity clusters, conservation and common SNPs.

Figure 6. Integrative analysis of the potential function of rs35648226 and rs1047564 by querying UCSC. (A) The regional association map of the rs35648226 locus. The SNP rs35648226 was located in the SYNE2 protein-coding region. RIP-chip GeneST from ENCODE/SUNY Albany data suggested that SLBP and PABPC1 might have a potential interaction with rs35648226. (B) The regional association map of the rs1047564 locus, which suggested that PABPC1 might have a potential interaction with rs1047564.
Studies have confirmed that m6A modification are important in cardiovascular diseases including myocardial infraction (15), heart failure (19), myocardial hypertrophy (16). But it's unclear whether m6A modification plays a role in AF risk and development. Here, our results reveal that m6A SNPs are associated with AF: First, we identify 105 m6A-SNPs that are associated with AF by analyzing data from AF GWAS and m6AVar database. Moreover, 7 m6A-SNPs show eQTL signals in atrial appendage. Besides, the results of our analysis reveal there are 3 m6A-SNPs corresponding to 3 genes are differently expressed in AF population. Further, 2 SNPs may influence AF risk by affecting m6A modification. These findings suggested that m6A-SNPs could be functional variants for AF.
As the most common post-transcription modification in eukaryotes, m6A modification can affect multiple physiological process and disease progression. m6A modification mediates cellular functions by regulating the mRNA metabolism process including nuclear export, stability, splicing, localization, and translation (27, 28). The m6A SNPs are usually adjacent to the methylation site. Therefore, the cellular functions involved in m6A modification will be influenced when m6A modification is altered by genetic variants (25). In addition, part of the m6A-SNPs are variants in UTR regions, which will affect transcriptional regulators and RNA binding protein. Besides, if the m6A-SNP is a missense variant, the amino acid composition as well as the secondary structure of the encoded protein would be changed. So, the m6A-SNPs have various functions which are worth being paid attention to. Thus, it's useful for exploring mechanism as well as the therapy strategy of AF to identify the m6A-SNPs associated with AF. Here, in our study, we find more than 100 m6A-SNPs are correlated with AF and our results suggest m6A-SNPs have vital roles in AF.
In the present study, we identified 105 m6A-SNPs associated with AF by analyzing the intersection of AF GWAS and m6Avar database, among which 7 showed eQTL signals in atrial appendage. Among these SNPs showing eQTL signals, rs2815301, rs1007654, rs900349 and rs28369993 were located in intron region. rs35648226 was located in CDS region. rs1047564 was located in 3′UTR region. rs1226590 was located in 5′UTR. We further prove the corresponding genes of the 3 m6A-SNPs (rs35648226, rs900349 as well as rs1047564) were differentially expressed by comparing the mRNA expression levels between AF patients and heathy controls, which suggests m6A-SNPs may be related to AF by influencing local gene expression. Moreover, SYNE2 (rs35648226) and THAP9 (rs1047564) were predicted to affect m6A modification of AF. When m6A modification is changed by the genetic variants, the biological process involved will be influenced as well. For example, if the variant is located in UTR, it will influence the transcriptional regulatory factors and RNA binding proteins. If the m6A-SNP is a missense mutation, not only will the m6A modification be affected, but the amino acid and secondary structure of the associated protein will be changed. Even if the variant is a synonymous one, it will make a difference in the RNA abundance transferred, which has an impact on the translation process (29). So, m6A-SNPs may affect the biological process by multiple ways. In our study, the AF-related m6A-SNP rs1047564 located in 3′UTR of the THAP9 on chromosome 4 was predicted to influence the m6A modification and protein binding, thus correlated with AF, which was not reported. rs35648226 located in CDS region of SYNE2 gene was also predicted to influence the m6A modification and protein binding. rs35648226 might have a potential interaction with SLBP and PABPC1, while rs1047564 might have a potential interaction with PABPC1, which suggest both of them are functional variants potentially by binding RBPs, which regulate the mRNA processing and metabolism. THAP9 was concerned in cancer (30). SYNE2 has previously reported to be related to AF (31, 32) and cardiac conduction (33). SNP rs1152591 located in an intron of SYNE2 was clearly associated with AF. Here, we report the m6A-SNPs located in different genes associated with AF, which is of great importance in explaining the role of m6A modification in AF.
Furthermore, GO enrichment analysis found the most significant GO processes included multicellular organismal process (GO:0032501), cellular process (GO:0009987), metabolic process (GO:0008152), regulation of biological process (GO:0050789). The mechanism of AF is complicated and unclear. The current consensus is that electrical remodeling, inflammation, fibrosis as well as genetics are the major substrate for AF (34, 35). In our analysis, these m6A-SNPs are involved in biological regulation and signaling, which may regulate the electrical remodeling, inflammation, fibrosis (36).
In summary, we are the first to explore the relationship between m6A-SNPs and AF. In this study, we reported over 100 m6A-SNPs associated with AF which may contribute to AF risk and development and proved the potential function. Though we found strong evidence to support the connection between m6A-SNPs and AF, further studies especially molecular experiments on animal models and cell lines are needed to elucidate the underlying mechanisms and functions of these m6A-SNPs. However, this study still has some limitations, such as the lack of large sample support of patients with atrial fibrillation and experimental data. Therefore, the potential marker genes and SNPs identified in this study need to be further verified to provide solid evidence for clinical diagnosis and prevention.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
YH, YT, YY and TS: conceptualized and designed the study. YH, YT, YY, LG and LH: prepared and analysed the data. YH, YT, YY and TS: drafted the manuscript. YH and YT: did most of the work during the revision. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (grant no. 82100331), Knowledge Innovation Program of Wuhan - Shuguang Project (grant no. 2022020801020484), Natural Science Foundation of Hubei Province (grant no. 2021CFB421), and Natural Science Research Project of Education Department of Anhui Province (grant no. KJ2021A1289, 2022AH040346).
We are grateful to Professor Jiang Chang at the department of Health Toxicology, Key Laboratory for Environment and Health, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, for their excellent technical assistance and helping edit the draft.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Richter S, Di Biase L, Hindricks G. Atrial fibrillation ablation in heart failure. Eur Heart J. (2019) 40(8):663–71. doi: 10.1093/eurheartj/ehy778
2. Sagris M, Vardas EP, Theofilis P, Antonopoulos AS, Oikonomou E, Tousoulis D. Atrial fibrillation: pathogenesis, predisposing factors, and genetics. Int J Mol Sci. (2021) 23(1):6. doi: 10.3390/ijms23010006
3. Haissaguerre M, Jais P, Shah DC, Takahashi A, Hocini M, Quiniou G, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. (1998) 339(10):659–66. doi: 10.1056/NEJM199809033391003
4. Enriquez A, Liang JJ, Santangeli P, Marchlinski FE, Riley MP. Focal atrial fibrillation from the superior vena Cava. J Atr Fibrillation. (2017) 9(6):1593. doi: 10.4022/jafib.1593
5. Cheniti G, Vlachos K, Pambrun T, Hooks D, Frontera A, Takigawa M, et al. Atrial fibrillation mechanisms and implications for catheter ablation. Front Physiol. (2018) 9:1458. doi: 10.3389/fphys.2018.01458
6. Roselli C, Rienstra M, Ellinor PT. Genetics of atrial fibrillation in 2020: GWAS, genome sequencing, polygenic risk, and beyond. Circ Res. (2020) 127(1):21–33. doi: 10.1161/CIRCRESAHA.120.316575
7. Wei Y, Wang L, Lin C, Xie Y, Bao Y, Luo Q, et al. Association between the rs2106261 polymorphism in the zinc finger homeobox 3 gene and risk of atrial fibrillation: evidence from a PRISMA-compliant meta-analysis. Medicine. (2021) 100(49):e27749. doi: 10.1097/MD.0000000000027749
8. Yang Y, Bartz TM, Brown MR, Guo X, Zilhao NR, Trompet S, et al. Identification of functional genetic determinants of cardiac troponin T and I in a multiethnic population and causal associations with atrial fibrillation. Circ Genom Precis Med. (2021) 14(6):e003460. doi: 10.1161/CIRCGEN.121.003460
9. Shi X, Cao Y, Zhang X, Gu C, Liang F, Xue J, et al. Comprehensive analysis of N6-methyladenosine RNA methylation regulators expression identify distinct molecular subtypes of myocardial infarction. Front Cell Dev Biol. (2021) 9:756483. doi: 10.3389/fcell.2021.756483
10. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
11. Yeo GS, O'Rahilly S. Uncovering the biology of FTO. Mol Metab. (2012) 1(1–2):32–6. doi: 10.1016/j.molmet.2012.06.001
12. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 Is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
13. Wu Y, Wang Z, Shen J, Yan W, Xiang S, Liu H, et al. The role of m6A methylation in osteosarcoma biological processes and its potential clinical value. Hum Genomics. (2022) 16(1):12. doi: 10.1186/s40246-022-00384-1
14. Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. (2020) 48(11):6251–64. doi: 10.1093/nar/gkaa347
15. Wang J, Zhang J, Ma Y, Zeng Y, Lu C, Yang F, et al. WTAP promotes myocardial ischemia/reperfusion injury by increasing endoplasmic reticulum stress via regulating m(6)A modification of ATF4 mRNA. Aging (Albany NY). (2021) 13(8):11135–49. doi: 10.18632/aging.202770
16. Dorn LE, Lasman L, Chen J, Xu X, Hund TJ, Medvedovic M, et al. The N(6)-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. (2019) 139(4):533–45. doi: 10.1161/CIRCULATIONAHA.118.036146
17. Gong R, Wang X, Li H, Liu S, Jiang Z, Zhao Y, et al. Loss of m(6)A methyltransferase METTL3 promotes heart regeneration and repair after myocardial injury. Pharmacol Res. (2021) 174:105845. doi: 10.1016/j.phrs.2021.105845
18. Berulava T, Buchholz E, Elerdashvili V, Pena T, Islam MR, Lbik D, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail. (2020) 22(1):54–66. doi: 10.1002/ejhf.1672
19. Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N, et al. FTO-dependent N(6)-methyladenosine regulates cardiac function during remodeling and repair. Circulation. (2019) 139(4):518–32. doi: 10.1161/CIRCULATIONAHA.118.033794
20. Mo XB, Lei SF, Zhang YH, Zhang H. Detection of m(6)A-associated SNPs as potential functional variants for coronary artery disease. Epigenomics. (2018) 10(10):1279–87. doi: 10.2217/epi-2018-0007
21. Liu H, Gu J, Jin Y, Yuan Q, Ma G, Du M, et al. Genetic variants in N6-methyladenosine are associated with bladder cancer risk in the Chinese population. Arch Toxicol. (2021) 95(1):299–309. doi: 10.1007/s00204-020-02911-2
22. Zhu R, Tian D, Zhao Y, Zhang C, Liu X. Genome-wide detection of m(6)A-associated genetic polymorphisms associated with ischemic stroke. J Mol Neurosci. (2021) 71(10):2107–15. doi: 10.1007/s12031-021-01805-x
23. Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. (2016) 44(10):e91. doi: 10.1093/nar/gkw104
24. Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, et al. Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell. (2018) 70(5):854–867 e859. doi: 10.1016/j.molcel.2018.05.001
25. Zheng Y, Nie P, Peng D, He Z, Liu M, Xie Y, et al. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res. (2018) 46(D1):D139–45. doi: 10.1093/nar/gkx895
26. Liu R, Liu L, Zhou Y. m6Adecom: analysis of m(6)A profile matrix based on graph regularized non-negative matrix factorization. Methods. (2022) 203:322–7. doi: 10.1016/j.ymeth.2022.01.007
27. Chao Y, Li HB, Zhou J. Multiple functions of RNA methylation in T cells: a review. Front Immunol. (2021) 12:627455. doi: 10.3389/fimmu.2021.627455
28. Willbanks A, Wood S, Cheng JX. RNA rpigenetics: fine-tuning chromatin plasticity and transcriptional regulation, and the implications in human diseases. Genes. (2021) 12(5):627. doi: 10.3390/genes12050627
29. Lin W, Xu H, Wu Y, Wang J, Yuan Q. In silico genome-wide identification of m6A-associated SNPs as potential functional variants for periodontitis. J Cell Physiol. (2020) 235(2):900–8. doi: 10.1002/jcp.29005
30. Su Y, Xie R, Xu Q. LncRNA THAP9-AS1 highly expressed in tissues of hepatocellular carcinoma and accelerates tumor cell proliferation. Clin Res Hepatol Gastroenterol. (2022) 46(10):102025. doi: 10.1016/j.clinre.2022.102025
31. Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. (2012) 44(6):670–5. doi: 10.1038/ng.2261
32. Hsu J, Gore-Panter S, Tchou G, Castel L, Lovano B, Moravec CS, et al. Genetic control of left atrial gene expression yields insights into the genetic susceptibility for atrial fibrillation. Circ Genom Precis Med. (2018) 11(3):e002107. doi: 10.1161/CIRCGEN.118.002107
33. Huang J, Luo R, Zheng C, Cao X, Zhu Y, He T, et al. Integrative analyses identify potential key genes and calcium-signaling pathway in familial atrioventricular nodal reentrant tachycardia using whole-exome sequencing. Front Cardiovasc Med. (2022) 9:910826. doi: 10.3389/fcvm.2022.910826
34. Dobrev D, Aguilar M, Heijman J, Guichard JB, Nattel S. Postoperative atrial fibrillation: mechanisms, manifestations and management. Nat Rev Cardiol. (2019) 16(7):417–36. doi: 10.1038/s41569-019-0166-5
35. Wijesurendra RS, Casadei B. Mechanisms of atrial fibrillation. Heart. (2019) 105(24):1860–7. doi: 10.1136/heartjnl-2018-314267
Keywords: N6-methyladenosine methylation, atrial fibrillation, single nucleotide polymorphism, genome-wide association study (GWAS), m6A modification
Citation: Huang Y, Tan Y, Yao Y, Gu L, Huang L and Song T (2023) Genome-wide detection of m6A-associated SNPs in atrial fibrillation pathogenesis. Front. Cardiovasc. Med. 10:1152851. doi: 10.3389/fcvm.2023.1152851
Received: 28 January 2023; Accepted: 10 May 2023;
Published: 26 May 2023.
Edited by:
Hua Zhong, University of Hawaii at Manoa, United StatesReviewed by:
Lu Ren, Stanford University, United States© 2023 Huang, Tan, Yao, Gu, Huang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Song c29uZ3Rhb193aHVlZHVAdG9tLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.