- 1Department of Cardiology and Vascular Medicine, Faculty of Medicine, Airlangga University—Dr. Soetomo General Hospital, Surabaya, Indonesia
- 2Faculty of Medicine, Duta Wacana Christian University, Yogyakarta, Indonesia
- 3Department of Pulmonology and Respiratory Medicine, Faculty of Medicine, Airlangga University—Dr. Soetomo General Hospital, Surabaya, Indonesia
Introduction: Peripartum cardiomyopathy (PPCM) is a potentially life-threatening pregnancy-related heart disease. Genetic roles such as gene polymorphisms may relate to the etiology of PPCM. This study analyzes the association between single nucleotide gene polymorphism (SNP) guanine nucleotide–binding protein beta-3 subunit (GNB3) C825T and insertion/deletion (I/D) of the angiotensin-converting enzyme (ACE) gene with the incidence of PPCM.
Methods: An analytic observational study with a case–control design was conducted at the Integrated Cardiac Service Center of Dr. Soetomo General Hospital, Surabaya, Indonesia. PPCM patients of the case and control groups were enrolled. Baseline characteristic data were collected and blood samples were analyzed for SNP in the GNB3 C825T gene and for I/D in the ACE gene by using the polymerase chain reaction, restriction fragment length polymorphism, and Sanger sequencing. We also assessed ACE levels among different ACE genotypes using a sandwich-ELISA test.
Results: A total of 100 patients were included in this study, with 34 PPCM cases and 66 controls. There were significant differences in GNB3 TT and TC genotypes in the case group compared with that in the control group (TT: 35.3% vs. 10.6%, p = 0.003; TC: 41.2% vs. 62.5%, p = 0.022). The TT genotype increased the risk of PPCM by 4.6-fold. There was also a significant difference in the ACE DD genotype in the case group compared with that in the control group (26.5% vs. 9.1%, p = 0.021). DD genotypes increased the risk of PPCM by 3.6-fold. ACE levels were significantly higher in the DD genotype group than in the ID and II genotype groups (4,356.88 ± 232.44 pg/mL vs. 3,980.91 ± 77.79 pg/mL vs. 3,679.94 ± 325.77 pg/mL, p < 0.001).
Conclusion: The TT genotype of GNB3 and the DD genotype of the ACE are likely to increase the risk of PPCM. Therefore, these polymorphisms may be predisposing risk factors for PPCM incidence. ACE levels were significantly higher in the DD genotype group, which certainly had clinical implications for the management of PPCM patients in the administration of ACE inhibitors as one of the therapy options.
1. Introduction
Maternal mortality ratio (MMR) is an indicator that describes national maternal health and welfare. Global MMR reached 214 per 100,000 live births in 2016 (1). In developing countries, MMR is 20 times higher than in developed countries (1). In 2012, Indonesia's MMR was 359 per 100,000 live births (2). An evaluation of the 2015 Millennium Development Goals revealed that 38 mothers in Indonesia died from diseases or complications related to pregnancy and childbirth every day (MMR: 305 per 100,000 live births). The causes of maternal death are mainly bleeding, infection, and cardiovascular disease, including hypertension during pregnancy and heart failure (3).
Peripartum cardiomyopathy (PPCM) is a potentially life-threatening pregnancy-related disease (4). PPCM is characterized by left ventricle (LV) dysfunction in the late peripartum period or in the first months of postpartum without a known history of heart disease (5). To date, there are many hypotheses about the etiology of PPCM, but none is considered as the primary explanation for all cases. PPCM is known to have a pathogenesis that involves many factors such as maternal autoimmune response, inflammation, oxidative stress, imbalance of cardiac proapoptotic factors and anti-angiogenic factors, micronutrient deficiencies, and genetic causes (6). Due to the complexity of the etiology, genetic factor, especially gene polymorphism, may play an essential role (7). Two major PPCM registries, Investigation of Pregnancy Associated Cardiomyopathy (IPAC) (8) and EURObservational Research Programme (EORP) (9), reported various incidence rates of PPCM among countries in different regions, which may be related to genetic predisposition in different races.
The guanine nucleotide–binding protein subunit β3 (GNB3) gene encodes the β3 subunit of G protein (Gβ3) located on chromosome 12p13 that consists of 11 exons and 10 introns. The single nucleotide polymorphism (SNP) of GNB3 at exon 10, C825T, is associated with an increased prevalence and poor outcome of PPCM in individuals of African progeny (10). T allele polymorphisms in the GNB3 gene are associated with increased intracellular signaling, increased risk of hypertension, low plasma renin, and cardiac remodeling (10). To date, there are no studies on GNB3 C825T gene polymorphism, especially in Asian populations.
The role of the insertion/deletion (I/D) 287-bp sequence inside intron 16 of the angiotensin-converting enzyme (ACE) gene and ACE activity in the etiology, pathogenesis, prognosis, and clinical implications of the cardiovascular system has been extensively studied. The deletion polymorphism of the ACE allele is associated with increased levels of ACE (11). In addition, the ACE DD genotype is positively correlated with specific cardiomyopathy such as ischemic cardiomyopathy (ICM), hypertrophic cardiomyopathy (HCM), alcoholic cardiomyopathy, and idiopathic dilated cardiomyopathy (IDCM) (12, 13). IDCM with low ejection fraction (EF) has a phenotype similar to PPCM, suggesting that there may be an association between the I/D of the ACE gene and PPCM. This study aims to determine the association between the SNP of the GNB3 C825T gene and the I/D of the ACE gene in women with PPCM.
2. Methods
2.1. Study design
An analytic observational study with a case–control study was conducted at the Integrated Cardiac Service Center, at Dr. Soetomo General Hospital and Institute of Tropical Diseases (ITD) Laboratory of Airlangga University, Surabaya, Indonesia from January 2021 to June 2022. The case group consisted of all women diagnosed with PPCM, while the control group comprised women without PPCM or a history of PPCM. The study was approved by the Dr. Soetomo General Hospital Surabaya Ethics Committee (0151/KEPK/II/2021). All procedures were approved by the relevant ethics committees and written informed consent was obtained from all study participants.
2.2. Patients and controls
All women who were 18–40 years’ old and who underwent examination and treatment at the Polyclinic Integrated Cardiac Service Center of Dr. Soetomo General Hospital Surabaya were included. PPCM was diagnosed according to the criteria of the European Society of Cardiology (ESC) Working Group on Peripartum Cardiology in 2010 (14). The criteria were: (1) Heart failure symptoms that appeared in the last 1 month of pregnancy to 5 months’ postpartum; (2) No history and other identifiable causes of heart failure; and (3) An left ventricular ejection fraction (LVEF) <45% based on echocardiography. All PPCM patients with previous history of heart failure, a history of coronavirus disease 2019 (COVID-19) infection complicated with any heart problems, and incomplete data were excluded. Controls were women with a history of pregnancy who had never been diagnosed with PPCM.
2.3. Detection of GNB3 C825T gene polymorphisms and ACE gene I/D
Patients selected on the basis of inclusion and exclusion criteria and signed a letter of informed consent to participate in the study. A 5 mL sample of cubital venous blood was collected in an ethylenediaminetetraacetic acid (EDTA) tube and rested for about 30 min. The tube was then centrifuged at 300 rpm for 10 min to separate the plasma. DNA extraction was carried out using the QiaAMP DNA Blood Mini Kit (Qiagen, Hilden, Germany) and stored at −20°C. DNA content was quantified by spectrophotometric absorption (Nanodrop Spectrophotometer, BioLab, Scoresby, VIC, Australia). All DNA samples were blind-tested.
The GNB3 C825T polymorphism was examined according to the procedure stipulated by Siffert et al. (15). We used 5′ TGACCCACTTGCCACCCGTGC 3′ as a sense primer and 5′ GCAGCAGCCAGGGCTGGC 3′ as an antisense primer. The polymerase chain reaction (PCR) was run using a Promega master mix reagent kit following the manufacturer's instructions (Promega, Madison, WI, United States). Amplifications were carried out in the T100TM Thermal Cycler (Bio-Rad, Hercules, CA, United States) as follows: 35 cycles with denaturation at 94°C for 1 min; annealing at 58°C for 45 s; extension at 72°C for 1 min; and a final extension at 72°C for 7 min. Amplification products were digested by using the restriction endonuclease enzyme, BseDI/BsaJI (MBI Fermentas, St. Leon-Rot, Germany), at 37°C in a water bath for 3 h and 80°C for 5 min. DNA fragments were obtained after the restriction enzyme was electrophoresed on a 2.5% agarose gel and stained with ethidium bromide and the BenchTop 1,000 bp DNA Ladder (Promega, Madison, WI, United States). The DNA fragments were imaged under ultraviolet (UV). The T allele was not digested by using the restriction endonuclease enzyme. It corresponded to the cDNA fragments of 256 bp (TT genotype), whereas the C allele corresponded to 152 bp and 104 bp (CC genotype). Thus, the CT genotype produced three bands, 256 bp, 152 bp, and 104 bp (Figure 1A). Three representative samples of each genotype (TT, TC, and CC) were confirmed with DNA sequencing using the Sanger method. DNA sequencing for the GNB3 C825T polymorphism was performed by using the ABI Prism 24-capillary 3,500xL Genetic Analyzer to confirm the PCR result. The sequence analysis of the DNA is shown in Figure 1B. The results were compared with the reference strains of the sequences that were published in GenBank using the Clone Manager Professional version 9.0.
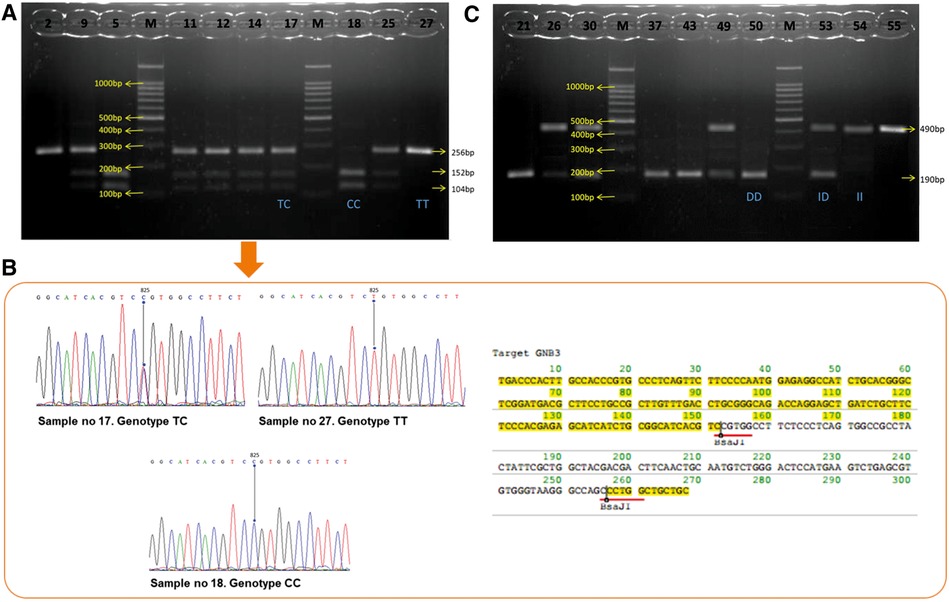
Figure 1. (A) Visualization of GNB3 PCR products. The results from 10 samples show TT genotypes: sample nos. 2 and 27; CC genotypes: sample nos. 5 and 18; TC genotypes: sample nos. 9, 11, 12, 14, 17, and 25. (B) DNA sequencing chromatogram and amino acid sequence restriction of GNB3. The lines point to nucleotide 825. Chromatogram of GNB3 shows TC genotypes at the upper left, TT genotypes at the upper right, and CC genotypes at the middle bottom. (C) Visualization of ACE gen PCR products. The results from 10 samples show DD genotypes: sample nos. 21, 37, 43, and 50; II genotypes: sample nos. 54 and 55; TC genotypes: sample nos. 26, 30, 49, and 53. GNB3, guanine nucleotide–binding protein beta-3 subunit; PCR, polymerase chain reaction; ACE, angiotensin-converting enzyme.
The I/D ACE polymorphism was examined as described by Rigat et al. (16). To amplify the ACE, a pair of primers 5′ CTGGAGACCACTCCCATCCTTTCT 3′ and an antisense primer 5′ GATGTGGCCATCTTCGTCAGA 3′ were used. The PCR amplification was processed as described for GNB3. The PCR product is a 490 bp fragment in the presence of the insertion (I) allele and a 190 bp fragment in the presence of the deletion (D) allele. Thus, each DNA sample revealed one of three possible patterns after electrophoresis: a 490 bp band (genotype II), a 190 bp band (genotype DD), or both 490 bp and 190 bp bands (genotype ID) (Figure 1C).
2.4. ACE ELISA test
Plasma from the PPCM group sample was separated after centrifugation and stored at −20°C until analysis. To determine the level of the ACE between different alleles and the genotype of ACE I/D, a sandwich-ELISA test was conducted using a Human Angiotensin-Converting Enzyme 1 ELISA (Elabscience, Hubei, China). The resulting optical density was read by using the BioRad ELISA Reader at 450 nm.
2.5. Statistical analysis
The data obtained were processed using SPSS (IBM Statistics 20.0) for Windows. The Hardy–Weinberg Equilibrium (HWE) was used to estimate the number of heterozygous and homozygous variant carriers in non-evolving populations on the basis of allele frequency. The χ2 test for the degree of freedom (dF) = 1 and a p-value = 0.05 were used to determine whether the observed genotypic distribution for GNB3 and ACE agreed with the HWE. The genotypes and alleles of GNB3 and ACE between the PPCM and the control groups were assessed using the χ2 test or Fisher's exact test according to the obtained data. Odds ratios (ORs) with a 95% confidence interval (95% CI) were determined to find the association of gene polymorphism intensity with disease. The normality of data was assessed using the Kolmogorov–Smirnov test. An independent Student’s t-test or a Mann–Whitney test was used for numerical data analysis of the two groups. For numerical data with >2 groups, analysis was performed using one-way ANOVA or the Kruskal–Wallis test as appropriate. Univariate and multivariate logistic regression analyses were done to determine whether gene polymorphism was the independent predictor of PPCM. Differences with p-values <0.05 were considered statistically significant.
3. Results
3.1. Characteristics of patients
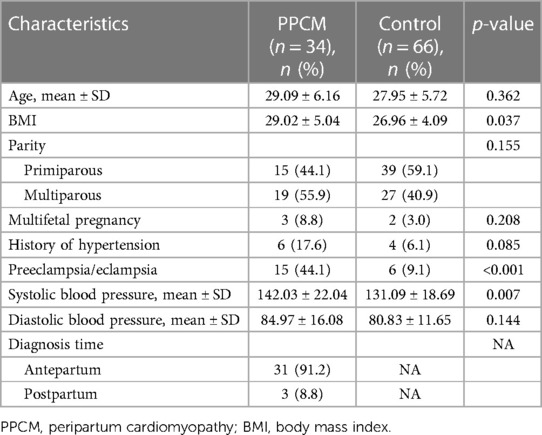
A total of 100 patients were included in the study, of which 34 were PPCM patients and 66 controls, and the characteristics of the case and control group patients are presented in Table 1. The mean BMI was higher in the PPCM group than in the control group (29.02 vs. 26.96, p = 0.037). The number of patients who had preeclampsia or eclampsia was significantly higher in the PPCM group than in the control group (44.1% vs. 9.1%, p < 0.001). The PPCM group had a higher mean systolic blood pressure than the control group (143.26 mmHg vs. 131.09 mmHg, p = 0.007). A total of 91.2% of patients with PPCM were diagnosed antepartum (Table 1). We did not find any deviations from the HWE in our population study.
3.2. GNB3 and ACE gene polymorphisms and the risk of PPCM
Of the total number of samples, the genotypes of GNB3 were mostly TC (n = 57, 57%), followed by CC (n = 24, 24%) and TT (n = 19, 19%). There were significant differences in the frequency of TT and TC genotypes of the GNB3 gene between the PPCM and the control groups (Table 2). Individuals with the TT genotype had a higher odds ratio of approximately 4.59 to have PPCM compared with those with the TC and CC genotypes (OR: 4.59; 95% CI: 1.60–13.17, p = 0.003) (Table 2). Although the frequency of T allele was higher in the PPCM group, the difference was not statistically significant compared with that in the control group (55.9% vs. 43.2%, p = 0.088).
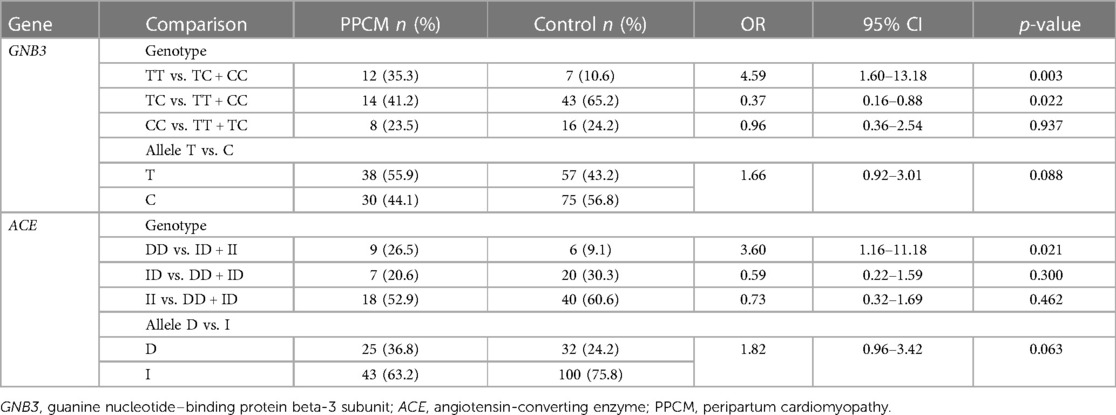
Table 2. Comparison of genotype and allele frequencies of the GNB3 and ACE genes in PPCM and control groups.
Our data indicated that 15 subjects had the DD genotype, 27 had the ID genotype, and 58 the II genotype of the ACE. The DD genotype was significantly higher in the PPCM group than in the control group (26.5% vs. 9.1%), and the presence of the DD genotype was associated with a higher risk of PPCM compared with individuals with the ID and II genotypes (OR: 3.60; 95% CI: 1.15–11.18, p = 0.021) (Table 2). The frequency of D allele was higher in the PPCM group, but the difference was not statistically significant compared with that in the control group (36.8% vs. 24.2%, p = 0.063) (Table 2). Univariate and multivariate logistic linear regression analyses were done on various variables, as presented in Table 3. The analysis from Table 3 showed that GNB3 TT and preeclampsia/eclampsia were independent predictors for PPCM.
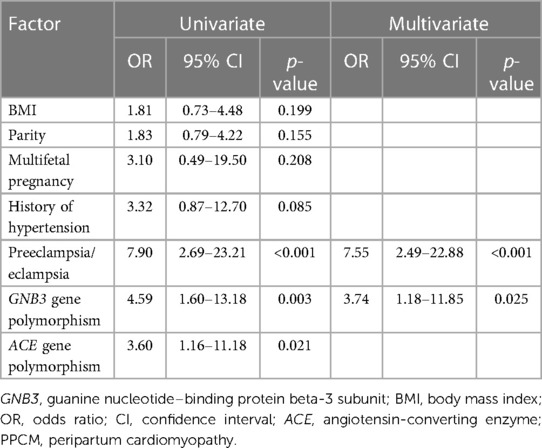
Table 3. Predictor of the PPCM determinate by univariate and multivariate logistic regression analyses.
3.3. Subanalysis of the GNB3 and ACE genotypes in the PPCM group
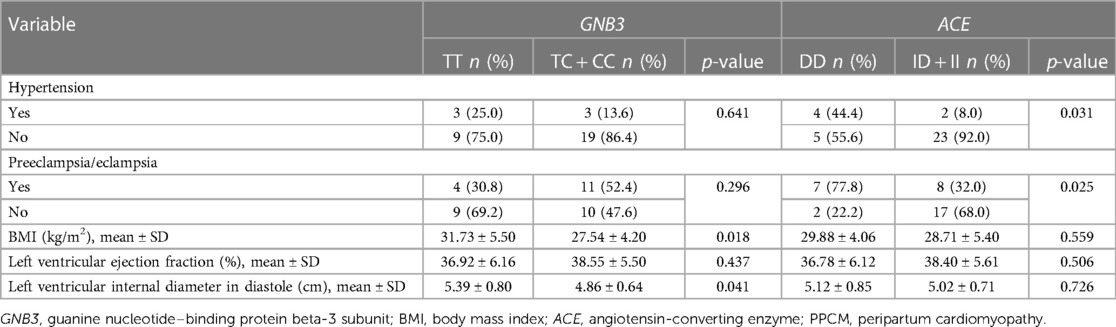
In PPCM patients, the frequency of the GNB3 genotype was significantly different and was based on BMI and left ventricular internal diameter in diastole (LVIDd) (Table 4). The BMI was higher in the TT genotype of GNB3 than in the TC and CC genotypes (31.73 vs. 27.54 kg/m2, p = 0.018). The mean of LVIDd was also higher in the TT genotype group than in the TC and CC groups (5.39 ± 0.80 vs. 4.86 cm ± 0.64 cm, p = 0.041). Hypertension and a history of preeclampsia/eclampsia were more frequent among those with the ACE DD genotype than among those with the ID and II genotypes; 44.4% vs. 8.0%, p = 0.031 and 77.8% vs. 32.0%, p = 0.025, respectively (Table 4).
3.4. Comparison of ACE levels based on ACE genotypes among PPCM patients
The ACE levels were measured among 30 of 34 PPCM patients because four subjects received ACE inhibitors that may cause bias. Our data revealed that the ACE levels in DD, ID, and II were 4,356.88, 3,980.91, and 3,679.94 pg/mL, respectively. The ACE levels were significantly higher in the DD genotype group than in the ID and II genotype groups, p < 0.001. The ACE levels in individuals with the ID genotype were also higher than in individuals with the II genotype (p = 0.020) (Figure 2).
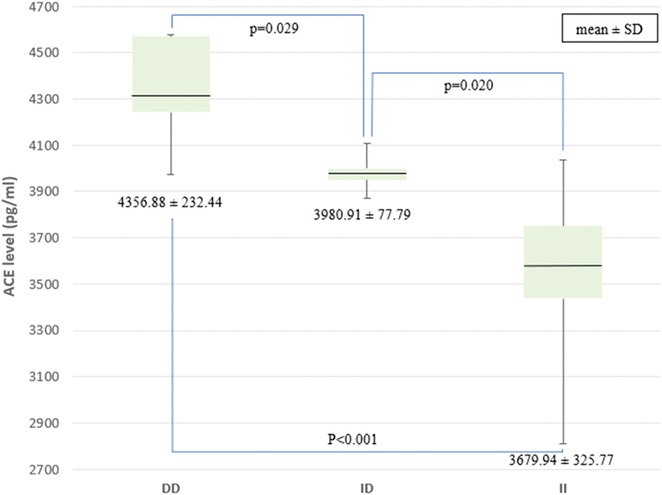
Figure 2. Comparison of ACE levels among the genotypes of the I/D ACE gene. ACE, angiotensin-converting enzyme; I/D, insertion/deletion.
4. Discussion
Despite the growing recognition of genetic predispositions as a risk factor for the development of PPCM, little is known about the impact of genomic background on racial differences. Our study was the first one to determine whether there was an association of the SNP GNB3 C825T gene and the incidence of PPCM in an Asian population. Although the frequency of the TT genotype was relatively rare, this genotype increased the risk of PPCM 4.6 times compared with the other CC and TC genotypes. Multivariate analysis also showed that GNB3 TT appeared to be an independent predictor for PPCM. A study conducted in North America with 97 subjects (30% were Blacks, 65% were Caucasian, and 5% were others), which assessed the relationship between different GNB3 genotypic backgrounds and their impact on improvement in LV remodeling in PPCM, found that the GNB3 TT genotype was more common in Blacks (10). GNB3 TT was also associated with a much higher incidence of PPCM and lower LVEF recovery (10).
Interestingly, our study found that the TC genotype (57%) was the most frequent genotype and may appear to afford protection from PPCM. However, there is limited evidence of the association between GNB3 TC polymorphism and PPCM incidence. The exact mechanism by which the GNB3 polymorphism contributes to the development of PPCM has not been fully understood, but it is thought to involve alterations in the G protein-coupled receptor (GPCR) signaling pathway (17). The GNB3 protein plays a key role in the signaling pathways through GPCR that control the contraction and relaxation of heart muscle cells (17, 18).
In addition, our study found that the most common genotypes of ACE were II (58%), followed by ID (27%) and DD (15%). The DD genotype increased the risk of PPCM 3.6 times compared with the other genotypes (II and ID). This result was similar to that of a study by Yaqoob et al., which found that the DD genotype was possibly a predisposing and independent risk factor for the pathophysiology of PPCM in the Kashmiri Indian population (13). The frequency of the DD genotype and D allele was also significantly higher in the PPCM population (13). The DD genotype was associated with poorer left ventricle systolic function in terms of ejection fraction, dimension, and left ventricle end-systolic and end-diastolic volumes (13).
Preeclampsia and eclampsia appear to be independent predictors for PPCM, as revealed by multivariate analysis. The pathophysiology of preeclampsia and eclampsia related to PPCM is still poorly understood, but several hypotheses suggest that hemodynamic stress caused by preeclampsia can contribute to the worsening of this condition (19). The EORP study states that the global preeclampsia incidence rate as a comorbid PPCM is 25%. A further investigation reveals that the rate of incidence in the Asia Pacific population reaches 46% (9). A meta-analysis of 22 observational studies with a total of 979 samples also reveals that 22% of PPCM patients develop preeclampsia/eclampsia (20).
No previous studies have reported an association of the GNB3 and ACE polymorphisms with hypertension and preeclampsia, specifically in the PPCM population. In our subanalysis, we found that the percentage of PPCM patients with hypertension (44.4% vs. 8%, p = 0.032) and a history of preeclampsia (77.8% vs. 32%, p = 0.025) was higher in the ACE DD genotype group than in the other genotype groups. A review study found that previous studies reported conflicting results, but the majority found that the DD genotype was associated with the incidence of hypertension and preeclampsia in pregnancy. A study of 121 pregnant women with a gestational age of 27–40 weeks reported a higher frequency of the DD genotype in the essential hypertension group than in the control group (21). A meta-analysis of 40 studies with a total of 3,977 cases and 7,065 controls concluded that the DD genotype increased the risk of preeclampsia compared with the DD and ID genotypes (52% vs. 17%), and D allele increased the risk of preeclampsia 1.29 times more than I allele (22).
Obesity is a risk factor for PPCM. Hemodynamic alterations, apoptosis, and inflammation are three potential causes of pathogenesis. Obesity causes excessive levels of circulating fat to alter blood volume, which increases stroke volume and stresses the LV wall, which, in turn, cause eccentric LV hypertrophy and, eventually, LV dysfunction (23). However, no previous studies have reported an association of the GNB3 and ACE gene polymorphisms with obesity in the PPCM population. A study of Caucasian, Chinese, and Black populations reported that the TT genotype had a higher mean BMI than other genotypes (TC and CC) (24). In our study, similar results were obtained, where the mean BMI in the TT genotype was significantly higher than that in the GNB3 TC and CC genotypes.
Although the mean LVEF in the GNB3 TT and ACE DD genotype groups has been reported to be lower in previous studies (10, 13), our data suggested no significant difference. Our results are in line with those of other studies, which showed no statistically significant difference in LVEF in the ACE DD genotype, although the mean LVEF was lower in the ACE DD genotype (13). However, the IPAC study reported that PPCM patients with the GNB3 TT genotype showed a lower LVEF at the initial stage of the study (10). After follow-up, LVEF was found to be significantly lower for GNB3 TT subjects at 6 months (p = 0.007) and 12 months (p < 0.001).
The geometry and thickness of the heart wall, especially the LV, are associated with cardiovascular risk. Our study found that GNB3 genotypes were associated with LVIDd, while the ACE was not. In contrast to our finding, a previous study by Poch et al., reported a lower mean of LVIDd in the GNB3 CC genotype group than in the TT and CT genotype groups in the essential hypertension population (25). Another study by Mahmood et al., also reported that the GNB3 TT genotype had a strong association with the incidence of LV hypertrophy (26). Similar to our study, a previous study found no difference in mean LVIDd among different genotypes of the ACE gene (13).
4.1. Comparison of ACE levels in the PPCM group
A study found that the I/D polymorphism of the ACE influenced the level of serum ACE in a healthy population (16). Observations of genetic polymorphisms may explain the interindividual variability in plasma ACE. In our study, the highest mean ACE levels were found in the DD group, followed by ID and II. This is in line with a study that found that the ACE levels increased twice as high in the DD genotype group as in the ID genotype group (16). Another study of pregnant women with hypertension in India reported significantly higher ACE levels in the DD genotype group than in the ID and DD genotype groups (27). The I/D of the ACE gene affects not only plasma ACE levels but also tissue ACE (28). Higher ACE levels would increase angiotensin II, which affects various systems, including the cardiovascular system (29). In the PPCM group, elevated ACE levels in the ACE DD genotype may be associated with the incidence of hypertension. An awareness on the part of clinicians about the existence of differences in ACE levels in each genotype will certainly have implications for the management of PPCM patients, one of which is the administration of ACE inhibitors as a therapy option in PPCM patients, especially those with the ACE DD genotype.
4.2. Limitations
The synergistic relationship between the GNB3 and the ACE gene could not be assessed in this study. In the PPCM group, only two patients had polymorphisms of both genes, while the control group had none. In this study, we did not analyze the levels of improvement in LVEF function in PPCM patients, which could have provided a better understanding of the issue. The reason for this lack of analysis was that we could not ask these patients to visit the hospital for an echocardiography examination because of the restrictions imposed by the COVID-19 pandemic, which has thrown many facets of the healthcare system out of gear.
5. Conclusion
This is a study determining the association of GNB3 C825T and ACE gene polymorphisms and the incidence of PPCM in an Asian population. The presence of the GNB3 TT genotype increases the risk of PPCM 4.6 times, while the ACE DD genotype potentially increases the risk of PPCM by 3.6 times. A subanalysis on PPCM patients found that those with TT had a higher BMI and LVIDd and also that those with the DD genotype were more prone to have hypertension and preeclampsia/eclampsia. ACE levels were significantly higher in the DD genotype group than in the ID and II genotype groups. These findings highlight the importance of gene polymorphisms in PPCM and, therefore, might be used as predictors and management strategies in the future.
Data availability statement
The data analyzed in this study are subject to the following licenses/restrictions: The datasets used are available from the corresponding author on reasonable request. Requests to access these datasets should be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Dr. Soetomo General Hospital Surabaya Ethics Committee. The patients/participants provided their written informed consent to participate in this study.
Author contributions
All authors listed have made equal substantial, direct, and intellectual contributions to the manuscript and approved the final manuscript for publication.
Acknowledgments
The authors would like to thank all the patients who were willing to participate in this study. The authors would also like to thank the staff at Dr. Soetomo Hospital for their cooperation in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Trends in maternal mortality 2000 to 2017: estimates by WHO, UNICEF, UNFPA, World Bank Group and the United Nations Population Division: executive summary. World Health Organization. (2019). p. 1–12.
2. Kementrian Kesehatan Republik Indonesia. Rencana Strategis Kementerian Kesehatan Tahun 2015–2019. (2015). p. 35–41.
3. Kotit S, Yacoub M. Cardiovascular adverse events in pregnancy: a global perspective. Glob Cardiol Sci Pract. (2021) 2021(1):e202105. doi: 10.21542/gcsp.2021.5
4. Arany Z, Elkayam U. Peripartum cardiomyopathy. Circulation. (2016) 133(14):1397–409. doi: 10.1161/CIRCULATIONAHA.115.020491
5. Stergiopoulos K, Lima FV. Peripartum cardiomyopathy—diagnosis, management, and long term implications. Trends Cardiovasc Med. (2019) 29(3):164–73. doi: 10.1016/j.tcm.2018.07.012
6. Ntusi NBA, Mayosi BM. Aetiology and risk factors of peripartum cardiomyopathy: a systematic review. Int J Cardiol. (2009) 131(2):168–79. doi: 10.1016/j.ijcard.2008.06.054
7. Dewi IP, Nugroho J. Genetic polymorphism in peripartum cardiomyopathy. Gynecol Obstet Reprod Med. (2021) 27(3):297–301. doi: 10.21613/GORM.2021.1072
8. McNamara DM, Elkayam U, Alharethi R, Damp J, Hsich E, Ewald G, et al. Clinical outcomes for peripartum cardiomyopathy in North America: results of the IPAC study (investigations of pregnancy-associated cardiomyopathy). J Am Coll Cardiol. (2015) 66(8):905–14. doi: 10.1016/j.jacc.2015.06.1309
9. Sliwa K, Petrie MC, van der Meer P, Mebazaa A, Hilfiker-Kleiner D, Jackson AM, et al. Clinical presentation, management, and 6-month outcomes in women with peripartum cardiomyopathy: an ESC EORP registry. Eur Heart J. (2020) 41(39):3787–97. doi: 10.1093/eurheartj/ehaa455
10. Sheppard R, Hsich E, Damp J, Elkayam U, Kealey A, Ramani G, et al. GNB3 C825T polymorphism and myocardial recovery in peripartum cardiomyopathy: results of the multicenter investigations of pregnancy-associated cardiomyopathy study. Circ Hear Fail. (2016) 9(3):1–6. doi: 10.1161/CIRCHEARTFAILURE.115.002683
11. Carluccio M, Soccio M, De Caterina R. Aspects of gene polymorphisms in cardiovascular disease: the renin-angiotensin system. Eur J Clin Invest. (2001) 31(6):476–88. doi: 10.1046/j.1365-2362.2001.00839.x
12. van Spaendonck-Zwarts KY, van Tintelen JP, van Veldhuisen DJ, van der Werf R, Jongbloed JDH, Paulus WJ, et al. Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation. (2010) 121(20):2169–75. doi: 10.1161/CIRCULATIONAHA.109.929646
13. Yaqoob I, Tramboo NA, Bhat IA, Pandith A, Beig JR, Hafeez I, et al. Insertion/deletion polymorphism of ACE gene in females with peripartum cardiomyopathy: a case-control study. Indian Heart J. (2018) 70(1):66–70. doi: 10.1016/j.ihj.2017.05.020
14. Sliwa K, Hilfiker-Kleiner D, Petrie MC, Mebazaa A, Pieske B, Buchmann E, et al. Current state of knowledge on aetiology, diagnosis, management, and therapy of peripartum cardiomyopathy: a position statement from the heart failure association of the European society of cardiology working group on peripartum cardiomyopathy. Eur J Heart Fail. (2010) 12(8):767–78. doi: 10.1093/eurjhf/hfq120
15. Siffert W, Rosskopf D, Siffert G, Busch S, Moritz A, Erbel R, et al. Association of a human G-protein β3 subunit variant with hypertension. Nat Genet. (1998) 18(1):45–8. doi: 10.1038/ng0198-45
16. Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. (1990) 86(4):1343–6. doi: 10.1172/JCI114844
17. Ricke-Hoch M, Pfeffer TJ, Hilfiker-Kleiner D. Peripartum cardiomyopathy: basic mechanisms and hope for new therapies. Cardiovasc Res. (2020) 116(3):520–31. doi: 10.1093/cvr/cvz252
18. Wang J, Gareri C, Rockman HA. G-protein-coupled receptors in heart disease. Circ Res. (2018) 123(6):716–35. doi: 10.1161/CIRCRESAHA.118.311403
19. Martinez BJT, Sison MCC, Acosta CS. Clinical profile and outcome of peripartum cardiomyopathy among teenager patients at the university of the Philippines—Philippine General Hospital. Acta Med Philipp. (2022) 56(7):5–11. doi: 10.47895/amp.vi0.3445
20. Bello N, Rendon ISH, Arany Z. The relationship between pre-eclampsia and peripartum cardiomyopathy: a systematic review and meta-analysis. J Am Coll Cardiol. (2013) 62(18):1715–23. doi: 10.1016/j.jacc.2013.08.717
21. Zotova TY, Lapaev NN, Azova MM, Blagonravov ML, Gigani OO, Ait Aissa A, et al. Distribution of polymorphisms of the renin–angiotensin system genes (ACE, AGT, and AGTR1), ITGB3, and FTO in pregnant patients with hypertensive disorders. Bull Exp Biol Med. (2019) 167(1):74–8. doi: 10.1007/s10517-019-04464-6
22. Wang C, Zhou X, Liu H, Huang S. Three polymorphisms of renin-angiotensin system and preeclampsia risk. J Assist Reprod Genet. (2020) 37(12):3121–42. doi: 10.1007/s10815-020-01971-8
23. Timoh T, Bloom ME, Siegel RR, Wagman G, Lanier GM, Vittorio TJ. A perspective on obesity cardiomyopathy. Obes Res Clin Pract. (2012) 6(3):e175–262. doi: 10.1016/j.orcp.2012.02.011
24. Siffert W, Forster P, Jöckel KH, Mvere DA, Brinkmann B, Naber C, et al. Worldwide ethnic distribution of the G protein beta3 subunit 825 T allele and its association with obesity in Caucasian, Chinese, and black African individuals. J Am Soc Nephrol. (1999) 10(9):1921–30. doi: 10.1681/ASN.V1091921
25. Poch E, González D, Gómez-Angelats E, Enjuto M, Paré JC, Rivera F, et al. G-protein β3 subunit gene variant and left ventricular hypertrophy in essential hypertension. Hypertension. (2000) 35(1):214–8. doi: 10.1161/01.HYP.35.1.214
26. Mahmood MS, Mian ZSU, Afzal A, Frossard PM. G-protein beta-3 subunit gene 825C > T dimorphism is associated with left ventricular hypertrophy but not essential hypertension. Med Sci Monit. (2005) 11(1):CR6–9.15614196
27. Aggarwal PK, Jain V, Jha V. Endothelial nitric oxide synthase, angiotensin-converting enzyme and angiotensinogen gene polymorphisms in hypertensive disorders of pregnancy. Hypertens Res. (2010) 33(5):473–7. doi: 10.1038/hr.2010.23
Keywords: single nucleotide polymorphism, peripartum cardiomyopathy, PPCM, guanine nucleotide–binding protein subunit β3, angiotensin-converting enzyme
Citation: Dewi IP, Wardhani LFK, Maghfirah I, Dewi KP, Subagjo A, Alsagaff MY and Nugroho J (2023) Association polymorphism of guanine nucleotide–binding protein β3 subunit (GNB3) C825T and insertion/deletion of the angiotensin-converting enzyme (ACE) gene with peripartum cardiomyopathy. Front. Cardiovasc. Med. 10:1096514. doi: 10.3389/fcvm.2023.1096514
Received: 14 November 2022; Accepted: 14 March 2023;
Published: 5 April 2023.
Edited by:
Amy Li, La Trobe University, AustraliaReviewed by:
Cristobal Dos Remedios, Victor Chang Cardiac Research Institute, AustraliaTimothy Spracklen, University of Cape Town, South Africa
© 2023 Dewi, Wardhani, Maghfirah, Dewi, Subagjo, Alsagaff and Nugroho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivana Purnama Dewi ZHJfaXZhbmFwZEBzdGFmZi51a2R3LmFjLmlk OTE2aXZhbmFAZ21haWwuY29t
Specialty Section: This article was submitted to Cardiovascular Genetics and Systems Medicine, a section of the journal Frontiers in Cardiovascular Medicine
 Ivana Purnama Dewi
Ivana Purnama Dewi Louisa Fadjri Kusuma Wardhani
Louisa Fadjri Kusuma Wardhani Irma Maghfirah
Irma Maghfirah Kristin Purnama Dewi
Kristin Purnama Dewi Agus Subagjo
Agus Subagjo Mochamad Yusuf Alsagaff
Mochamad Yusuf Alsagaff Johanes Nugroho
Johanes Nugroho