
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 02 March 2023
Sec. Cardiovascular Metabolism
Volume 10 - 2023 | https://doi.org/10.3389/fcvm.2023.1018422
 Renlingzi Zhang
Renlingzi Zhang Chong Di
Chong Di Hanlu Gao
Hanlu Gao Yunlou Zhu
Yunlou Zhu Congye Li
Congye Li Zhengfang Zhu
Zhengfang Zhu Qixing Wang
Qixing Wang Junjie Wang*
Junjie Wang* Feng Zhou*
Feng Zhou* Sheng Wang*
Sheng Wang*Background: Early diagnosis of septic cardiomyopathy is essential to reduce the mortality rate of sepsis. Previous studies indicated that iron metabolism plays a vital role in sepsis-induced cardiomyopathy. Here, we aimed to identify shared iron metabolism-related genes (IMRGs) in the myocardium and blood monocytes of patients with sepsis and to determine their prognostic signature.
Methods: First, an applied bioinformatics-based analysis was conducted to identify shared IMRGs differentially expressed in the myocardium and peripheral blood monocytes of patients with sepsis. Second, Cytoscape was used to construct a protein–protein interaction network, and immune infiltration of the septic myocardium was assessed using single-sample gene set enrichment analysis. In addition, a prognostic prediction model for IMRGs was established by Cox regression analysis. Finally, the expression of key mRNAs in the myocardium of mice with sepsis was verified using quantitative polymerase chain reaction analysis.
Results: We screened common differentially expressed genes in septic myocardium and blood monocytes and identified 14 that were related to iron metabolism. We found that HBB, SLC25A37, SLC11A1, and HMOX1 strongly correlated with monocytes and neutrophils, whereas HMOX1 and SLC11A1 strongly correlated with macrophages. We then established a prognostic model (HIF1A and SLC25A37) using the common differentially expressed IMRGs. The prognostic model we established was expected to better aid in diagnosing septic cardiomyopathy. Moreover, we verified these genes using datasets and experiments and found a significant difference between the sepsis and control groups.
Conclusion: Common differential expression of IMRGs was identified in blood monocytes and myocardium between sepsis and control groups, among which HIF1A and SLC25A37 might predict prognosis in septic cardiomyopathy. The study may help us deeply understand the molecular mechanisms of iron metabolism and aid in the diagnosis and treatment of septic cardiomyopathy.
Sepsis is a life-threatening clinical syndrome caused by a dysfunctional host response to an infection (1). It is the leading cause of death in patients who are critically ill, and its mortality significantly increases when combined with multiple dysfunctions, leading to functional failure of essential organs, such as the heart, liver, lungs, and kidneys (2). The fatality of sepsis is attributed to organ interdependence; that is, failure of one organ often leads to dysfunction or failure of other organs. The interdependence is particularly evident in cardiovascular failure, which reduces overall blood circulation, exacerbating mitochondrial dysfunction, tissue hypoxia, and metabolic imbalance (3). Immunosuppression in late sepsis contributes to progressive cardiovascular failure, which is often the last step before sepsis-induced death (4).
Iron metabolism plays a vital role in sepsis. High plasma ferritin levels can be identified as “macrophage activation-like syndrome” in patients with sepsis. Hyperferritinemia has been evaluated as a new inflammatory response biomarker that may provide a new strategy for the clinical study of anti-inflammatory effects (5). Lipid peroxidation is stimulated when large quantities of iron are released from necrotic tissue and damaged mitochondria, and hydroxyl radicals, the most effective reactive oxygen species, are produced (6). This chain reaction accelerates cell membrane damage, ultimately leading to cell death. Research has shown that iron metabolism imbalance is common in many cardiovascular diseases. Nutrient-related iron deficiency affects 75% of patients with heart failure (7). Both primary and secondary iron overload can induce heart disease through oxidative stress, but the specific mechanisms remain unknown (8–10). Excess iron in myocardial cells can also induce ferroptosis through the accumulation of peroxides in the cell membrane (9). Iron metabolism disorders play vital roles in the pathogenesis of septic cardiomyopathy (11, 12). Therefore, further understanding of iron disorders in septic cardiomyopathy would help discover effective targets for therapeutic intervention.
The pathological mechanism of septic cardiomyopathy has not been fully elucidated (13). Transcriptome sequencing in sepsis research has enabled us to gain an understanding of the differentially expressed genes (DEGs), how they are involved in sepsis, and the impact of signaling pathways. Previous studies have focused on transcriptomic differences in peripheral blood, but functions of the screened candidate genes could not be correlated with diseases involving multiple organ damage due to a large number of DEGs. Iron overload exists in septic cardiomyopathy; however, the regulatory genes of iron metabolism involved in the development of sepsis are unknown. Circulating immune cells are the link between local and systemic organ inflammation. To the best of our knowledge, this is the first integrated study to focus on iron metabolism-related genes (IMRGs) from transcriptomic data of septic cardiomyopathy and blood monocytes.
In this study, we performed applied bioinformatics analysis and screened for differentially expressed IMRGs in blood monocytes and heart samples of patients with sepsis. A prognostic model of IMRGs was established based on DEGs shared by septic myocardium and peripheral blood. Based on these candidate genes, we predicted targeted small-molecule drugs. Ultimately, this study may assist clinical diagnosis and treatment of septic cardiomyopathy.
We obtained clinical information from the Gene Expression Omnibus (GEO) database and datasets stored by Matkovich SJ (14) (GSE79962) and Biswas SK (15) (GSE46955) were downloaded. The cardiac tissue samples from 20 patients with septic cardiomyopathy and 11 patients without heart failure were included in the GSE79962 dataset. Blood monocyte samples in the GSE46955 dataset were collected from patients with gram-negative sepsis during infection and post-recovery, as well as from healthy donors. Raw data from GSE769962 and GSE65682 datasets were preprocessed using the R “affy” package (16) (version 3.8; R Foundation for Statistical Computing, Vienna, Austria), and raw data from the GSE46955 dataset were preprocessed using the R “limma” package (17) (version 3.42.2). A total of 507 IMRGs were obtained from the Molecular Signatures Database (MSigDB) version 7.4 (18).
The mRNA expression profiles of sepsis and control samples, including cardiac tissues and blood monocytes, were compared to identify DEGs with (|FC|) >1.5 and adjusted p < 0.05. Volcano plots and heatmaps of IMRGs were constructed using the “ggplot2” package (version 3.3.3) and R (version 3.6.3) (19). A t-test was used to determine p-values and adjusted p-values in the differential gene expression analysis. We identified the common IMRGs via an Online Venn diagram tool.1
The R “clusterProfiler” (version 3.14.3) package was used to visualize enrichment analysis and biological processes (20). To investigate potential biological functions between sepsis and controls, Gene ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted using R package “clusterProfiler” (21). Differences at p < 0.05 were considered statistically significant.
The STRING online database2 was used to assess functional relatedness and can be applied for protein–protein interactions (PPI) network analysis (22). Cytoscape (version 3.6.0) was used to visualize the network, with interaction scores set at >0.4. Genes and the relationship between genes are represented by nodes and edges, respectively. To identify key PPI network modules, molecular complex detection (MCODE) was used for gene network clustering analysis (23). CytoHubba in Cytoscape (version 0.3) was used to estimate essential nodes in the PPI network via different topological analysis methods (24).
To assess the immune response in sepsis, the single-sample gene set enrichment analysis (ssGSEA) algorithm was applied to estimate immune infiltration for each sample in the GSE79962 dataset (25). Additionally, a prognostic prediction model for IMRGs was constructed using a multivariate Cox regression analysis based on the univariate Cox regression analysis, and the patient’s risk scores were calculated for the GSE65682 dataset. Kaplan–Meier survival curve analyses were compared between high- and low-risk groups, and receiver operating characteristic (ROC) curves were used to evaluate the model (26).
The Drug Signatures Database is a collection of drug- and small-molecule-related gene sets. We searched for potential IMRGs in this database, and 10 predicted drugs are listed in Table 1 according to the combined score (27).
Twenty healthy C57BL/6 male mice between 8 and 10 weeks of age were provided by Shanghai Jisijie Laboratory Animal Co., Ltd. (Shanghai, China), and all experiments were performed at the Experimental Center of Tongji University (Shanghai, China) in a constant temperature and humidity environment. All mice in this study were raised at a regular circadian rhythm with free access to food and water. All animal procedures followed the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (Bethesda, MD, USA) in 1996, and all animal research was approved by the Animal Research Ethics Committee of Shanghai Tenth People’s Hospital (Shanghai, China).
Lipopolysaccharide (10 mg/kg) was administered to mice intraperitoneally. After 12 h, the mice were examined using transthoracic echocardiography and then euthanized with carbon dioxide anesthesia. Heart tissue was collected, fixed in 4% paraformaldehyde, and embedded in paraffin for histological analysis. According to standard technical protocols, the heart samples were sliced into sections of 4–5 mm and then stained with hematoxylin and eosin (H&E). All photomicrographs were acquired using a light microscope (TE2000-U; Nikon, Tokyo, Japan) at 200× magnification.
The left ventricular function of mice was examined using transthoracic echocardiography with an ultrasound system (Vevo 2,100; Vision Electronics, Toronto, ON, Canada) under isoflurane anesthesia. Accordingly, the left ventricular ejection fraction (LVEF) and fractional shortening (FS) were calculated. All measurements used for analysis were the average of three consecutive cardiac cycles.
Mice were euthanized after 12 h, and the total RNA in the myocardium was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. A Nanodrop Nd1000 analyzer (Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the quantity and purity of the total RNA. According to the manufacturer’s instructions, a Hieff 1st Strand cDNA Synthesis Kit (11121ES60; Yeasen, Shanghai, China) was used to reverse transcribe total RNA into cDNA. The Hieff SYBR Green Master Mix qPCR Kit (low Rox plus 11202es08; Yeasen) was used to perform quantitative polymerase chain reaction (qPCR) on a real-time PCR detector (QuanStudio; Applied Biosystems, Waltham, MA, USA). The primer pairs used are shown in Supplementary Table 1. Values were normalized to those of β-actin using the 2−ΔΔCt method.
We analyzed the relationship between IMRGs and cardiovascular and inflammatory diseases using the comparative toxicogenomics (CTD) database, and the inference scores (the degree to which iron metabolism genes are related to the disease) were determined.
Student’s t-test was used to compare continuous variables, and Chi-square or Fisher’s exact tests were used to compare categorical variables. The diagnostic accuracy of the two hub genes was analyzed using the survival curve analysis and expressed by the area under the ROC curve at a 95% confidence interval. Spearman’s rank test or Pearson correlation coefficient was used to analyze the correlation between potential genes and immune cells. All statistical analyses were performed using R software (version 3.6.3) and SPSS (version 19.0; SPSS Inc., Chicago, IL, USA). p-values less than 0.05 (bilateral) were considered statistically significant.
We downloaded the microarray RNA expression profiling datasets, GSE79962 and GSE46955, from the GEO database. In total, we identified 83 and 33 IMRGs differentially expressed in peripheral blood monocytes and myocardial tissues, respectively. Principal component analysis (PCA) was performed, which revealed that patients in GSE79962 were well separated in the two major dimensions (Figure 1A). Expression distribution across all genes between the non-heart failure and septic myocardium groups is depicted in a volcano plot (Figure 1B), and the common IMRGs are presented in a heatmap (Figure 1C). We identified 14 common DEGs upon comparing the iron metabolism-related DEGs between septic myocardial tissue and blood monocytes by overlapping the Venn diagrams (Figure 1D). PCA of the GSE46695 dataset indicated the relatedness of sample transcriptomes when comparing sepsis with non-sepsis groups (Figure 1E). Following analysis of the GSE46955 dataset, the DEGs between the control and sepsis groups were presented as a volcano plot (Figure 1F), and the common iron metabolism-related DEGs in a heatmap (Figure 1G).
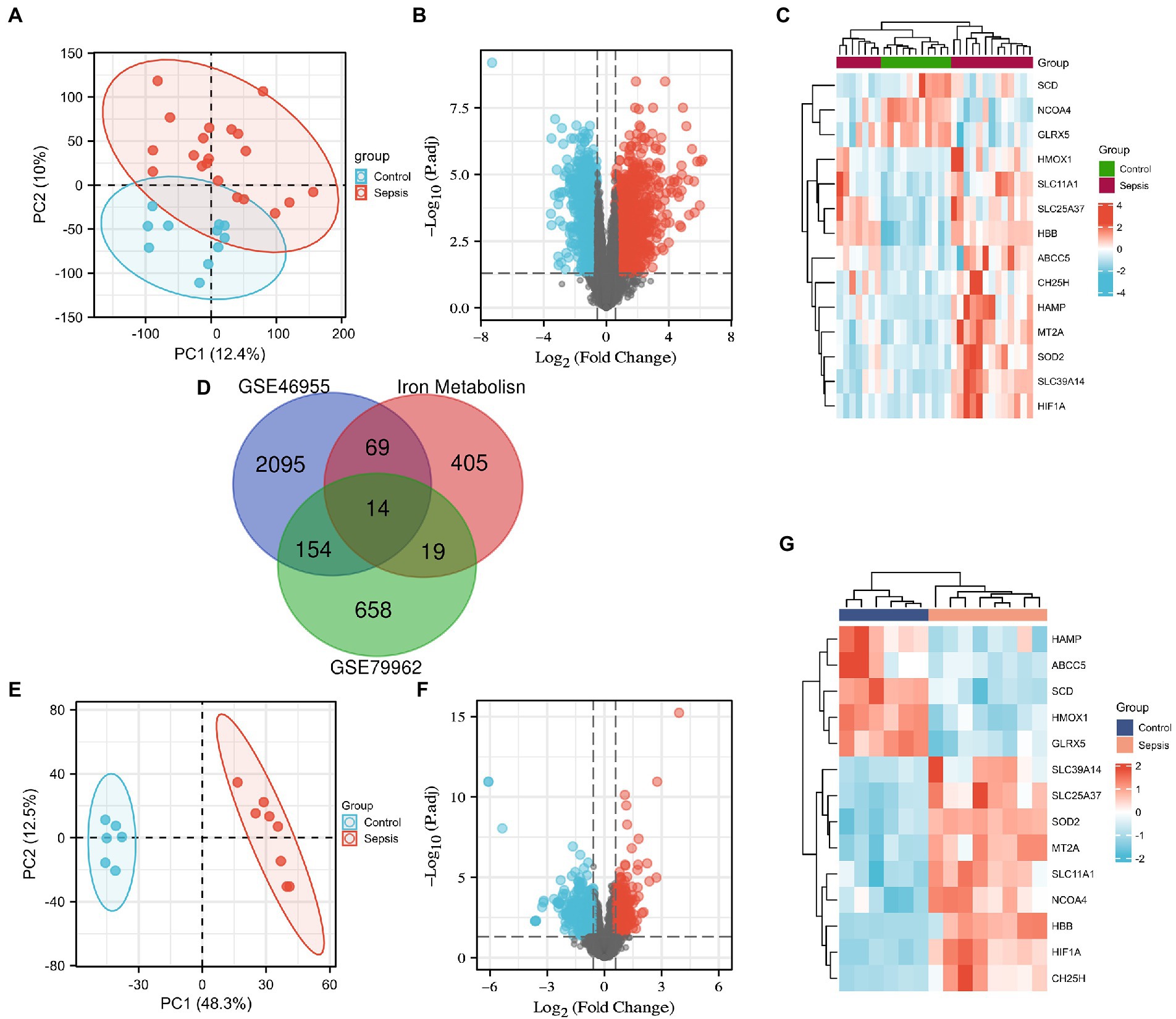
Figure 1. Screening of the differentially expressed iron metabolism-related genes (IMRGs). (A) Principal components analysis for GSE79962. (B) Volcano plot of GSE79962. (C) Heatmap of the common differentially expressed IMRGs in septic myocardiopathy and controls. (D) Venn diagram showing the overlap of genes among differentially expressed genes (DEGs) of GSE79962, GSE46955, and IMRGs. (E) Principal components analysis of GSE46955. (F) Volcano plot of GSE46955. (G) Heatmap of the common differentially expressed IMRGs in blood monocytes of sepsis and control groups.
Functional enrichment of the DEGs enabled the recognition of the main similarities and differences in the expression profiles of the IMRGs in myocardial samples and blood monocytes of patients with sepsis. GO and KEGG analyses were applied to identify the activated iron metabolic signaling pathways in the myocardium and blood monocytes of patients with sepsis. GSE79962 dataset analysis revealed that the most significantly enriched GO terms involved cellular transition metal ion homeostasis, blood microparticle, and iron ion binding (Figures 2A,B). Furthermore, GSE46955 dataset analysis revealed that the most significantly enriched GO terms involved mitotic nuclear division, spindle, and iron ion binding (Figures 2C,D). Most importantly, KEGG enrichment analysis revealed that the differentially expressed IMRGs from both datasets were mainly involved in the process of ferroptosis (Figures 2E,F).
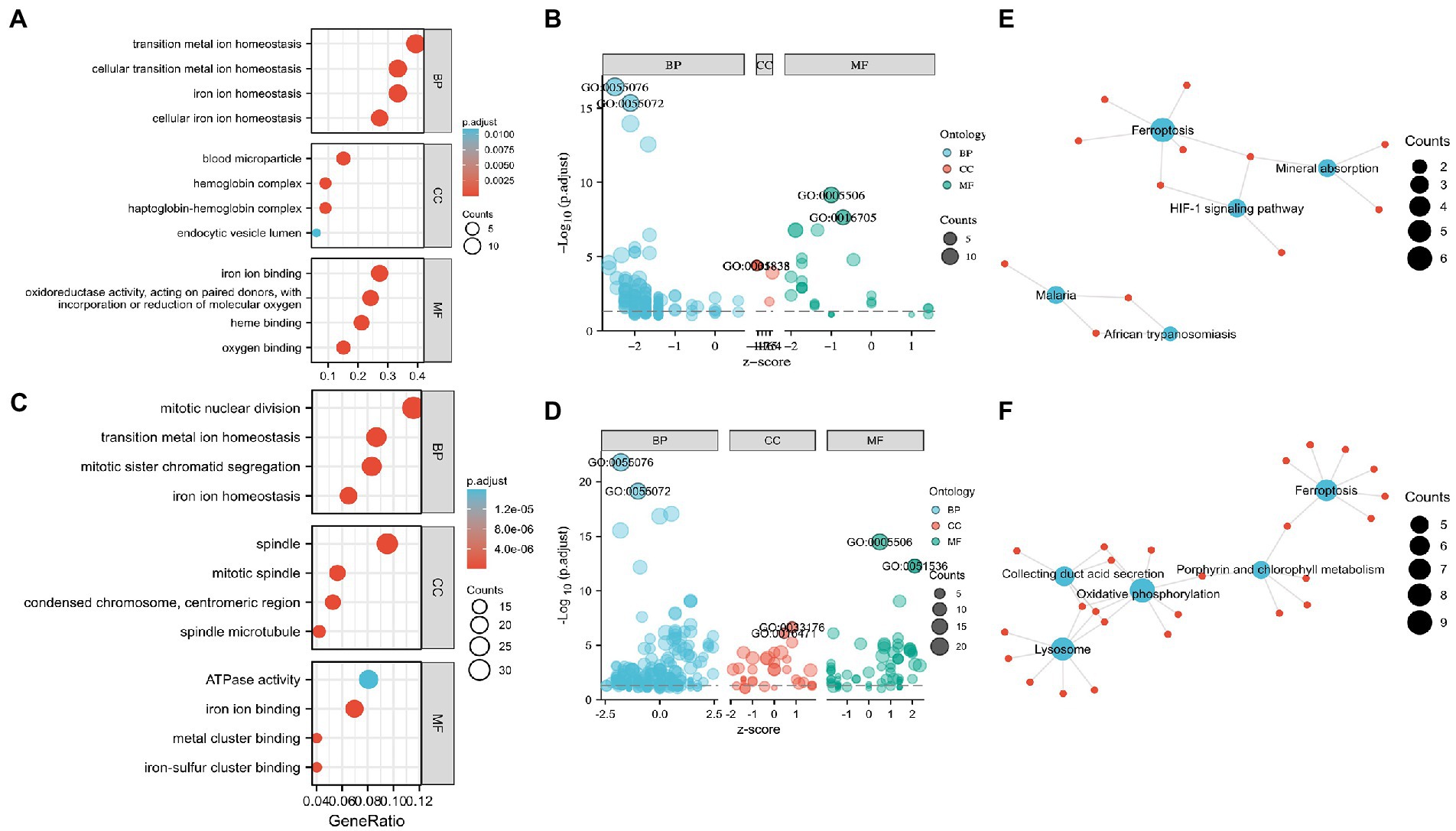
Figure 2. Enrichment analysis for the differentially expressed genes (DEGs) in circulation and the myocardium. (A) Gene Ontology (GO) enrichment analysis, (B) Bubble plot of enriched GO terms in GSE79962. (C) GO enrichment analysis, (D) Bubble plot of enriched GO terms in GSE46955. (E) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis in GSE79962. (F) KEGG pathway analysis in GSE46955. Abbreviations: BP, biological process; CC, cellular component; MF, molecular function.
The PPI network enabled the investigation of the molecular mechanisms of diseases and the identification of potential IMRGs. We performed a PPI analysis of 14 common IMRGs in blood monocytes and myocardium of patients with sepsis (Figure 3A). The MCODE plugin in Cytoscape revealed highly interconnected modules in the PPI network and a total of three functional modules were identified (Figures 3B–D). The top 10 hub genes were screened using the maximal clique centrality method with CytoHubba plugin (Figure 3E). These included HAMP, SLC25A37, SLC11A1, SLC39A14, HMOX1, SOD2, HIF1A, GLRX5, HBB, and NCOA4. The scores of hub genes identified by Cytohubba are shown in Supplementary Table 2. Using Metascape enrichment analysis, we established that these genes were related to iron overload, cardiomegaly, and transition metal ion homeostasis (Figures 3F–H).
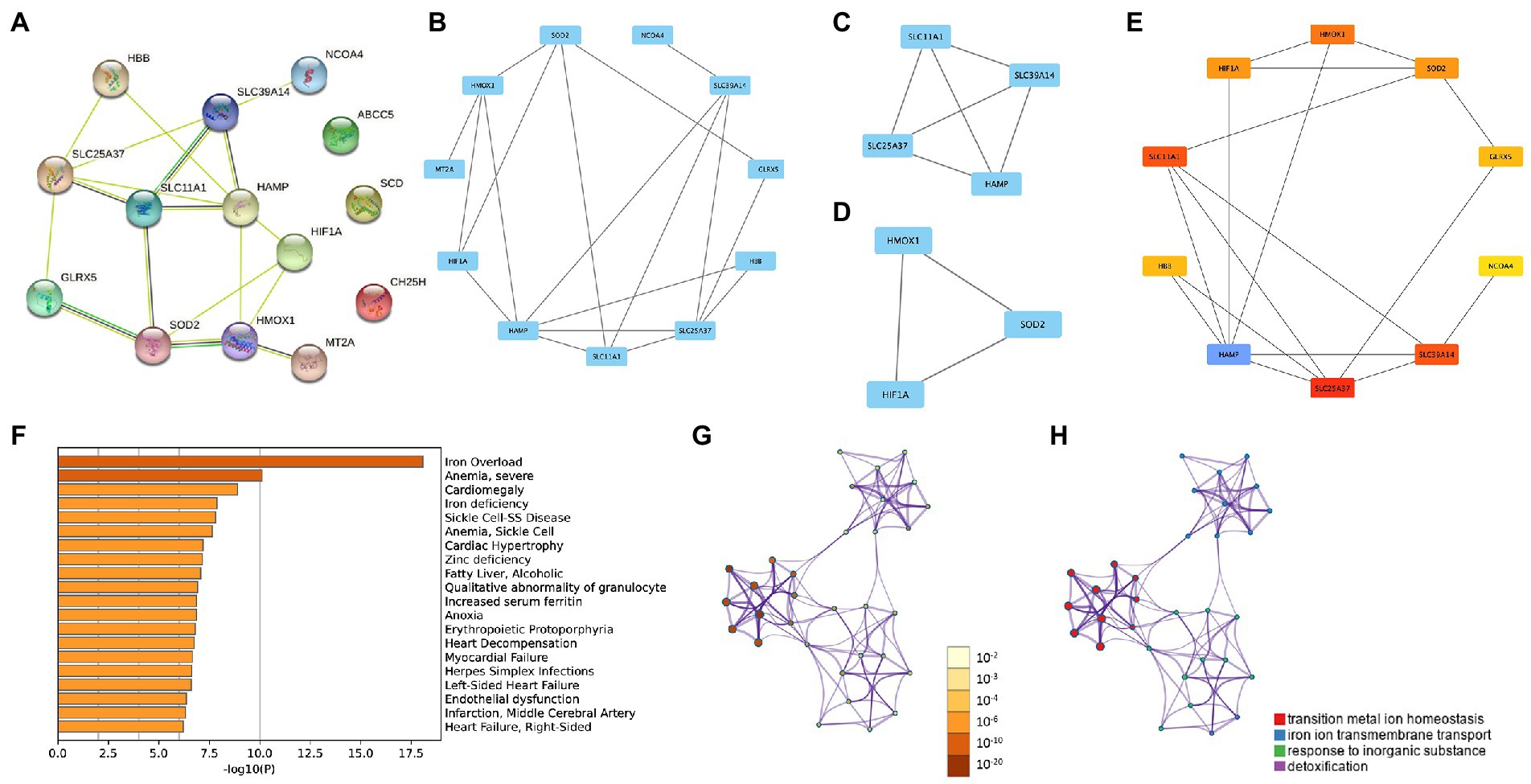
Figure 3. Protein–protein interaction (PPI) networks in common DEGs of iron metabolism. (A) PPI network formed by common differentially expressed IMRGs. (B–D) The top three MCODE complexes. (E) The top 10 hub genes identified by Cytohubba. (F–H) Enrichment analysis performed by Metascape.
To identify immune cell signatures in septic myocardium, the ssGSEA algorithm was used to analyze the proportion of immune cell infiltration. A heatmap depicting the proportions of immune cell types was constructed (Figure 4A). Box plots of 28 immune cells in sepsis samples (Figure 4B) and their correlations with immune cells were plotted (Figure 4C). For innate cells, neutrophils, monocytes, as well as activated dendritic, CD56 bright natural killer, immature dendritic, and plasmacytoid dendritic cells indicated significant differences between the septic myocardium and control groups. For adaptive immune cells, myeloid-derived suppressor, activated CD4 T, and type 2 T helper cells indicated significant differences. Correlation analysis between immune cells revealed that 15 immune cells were significantly correlated with monocytes. We evaluated the association between IMRGs and immune infiltration characterization in septic myocardium. Strong correlations were also observed between potential IMRGs (HMOX1, SLC11A1, SLC25A37, and HBB) and monocytes and neutrophils, whereas HMOX1 and SLC11A1 with macrophages. A negative correlation was observed between monocytes and GLRX5 and NCOA4 expression (Figure 4D). These results further support the notion that IMRGs may affect immune cell activity.
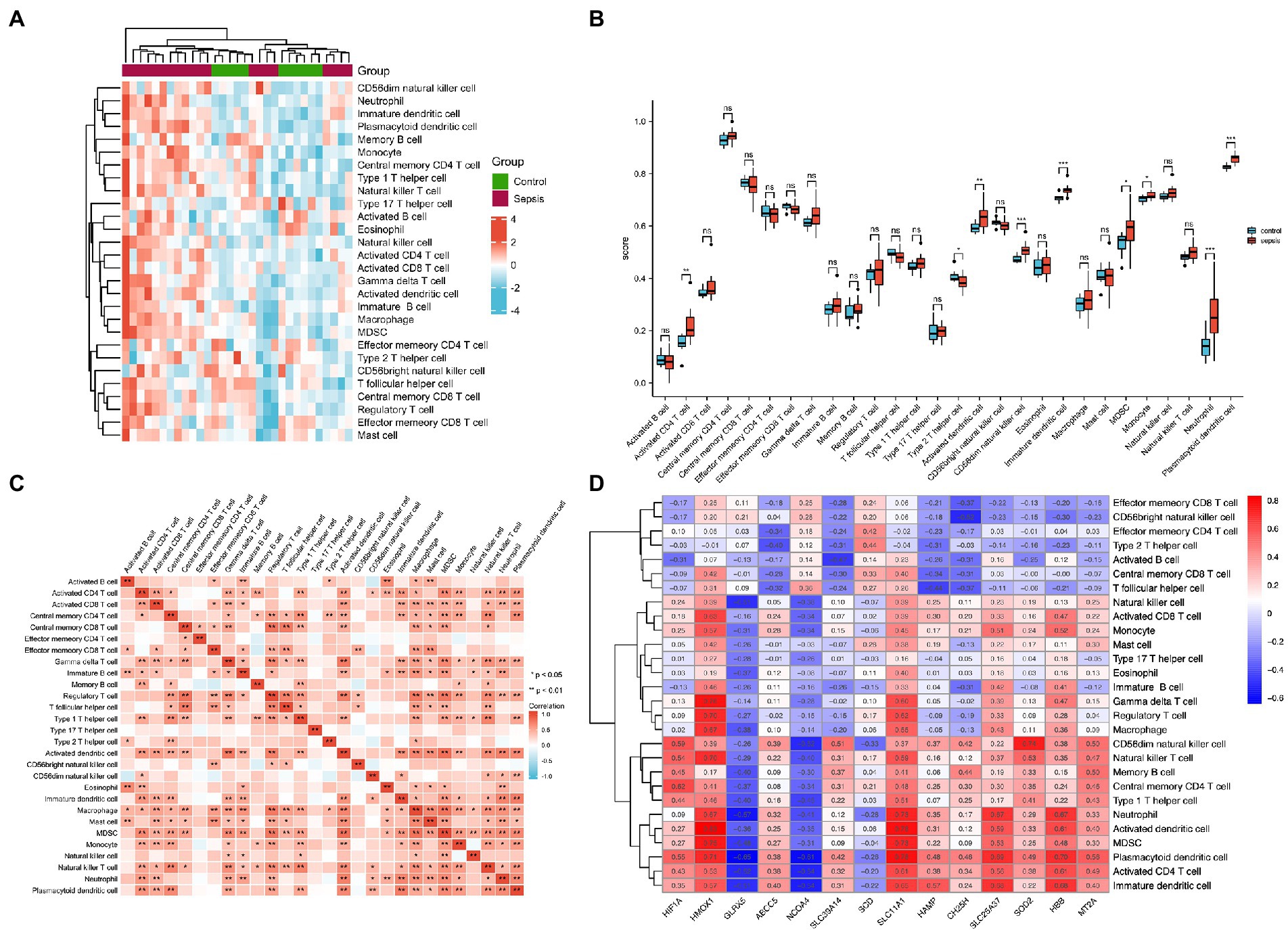
Figure 4. Landscape of immune infiltration in the myocardium of patients with sepsis and controls. (A) Heatmap of the proportions of the 28 immune cell types. (B) Box plots of immune cell proportions. (C) Correlation analysis of immune cells. (D) Correlation between common differentially expressed IMRGs and immune cells.
The potential IMRGs obtained by univariate Cox regression analysis were further incorporated into multivariate Cox regression analysis. This led to the identification of two iron metabolism genes related to sepsis prognosis (HIF1A and SLC25A37). The expression value and relative coefficients of two genes were used to calculate the survival risk score (Supplementary Table 3). The cutoff value of a patient’s risk score was set at 1.007. Based on the median risk score value, sepsis samples in GSE65682 were divided into high- and low-risk groups. After multivariate regression analysis, we found no statistically significant differences in age, gender, and diabetes except for risk score (Supplementary Figure 1). Using survival curve analysis, we observed a significant difference between the high- and low-risk group models (p = 2.052e-03; Figure 5A). Survival analysis revealed that two iron metabolism-related DEGs were associated with sepsis prognosis (Figure 5B). An ROC curve was plotted, and the area under the ROC curve for the experimental dataset GSE65682 was 0.708 (Figure 5C). We used the GSE65682 dataset to identify the expression of these two genes between sepsis and non-sepsis samples and found that their expressions changed significantly (Figure 5D).
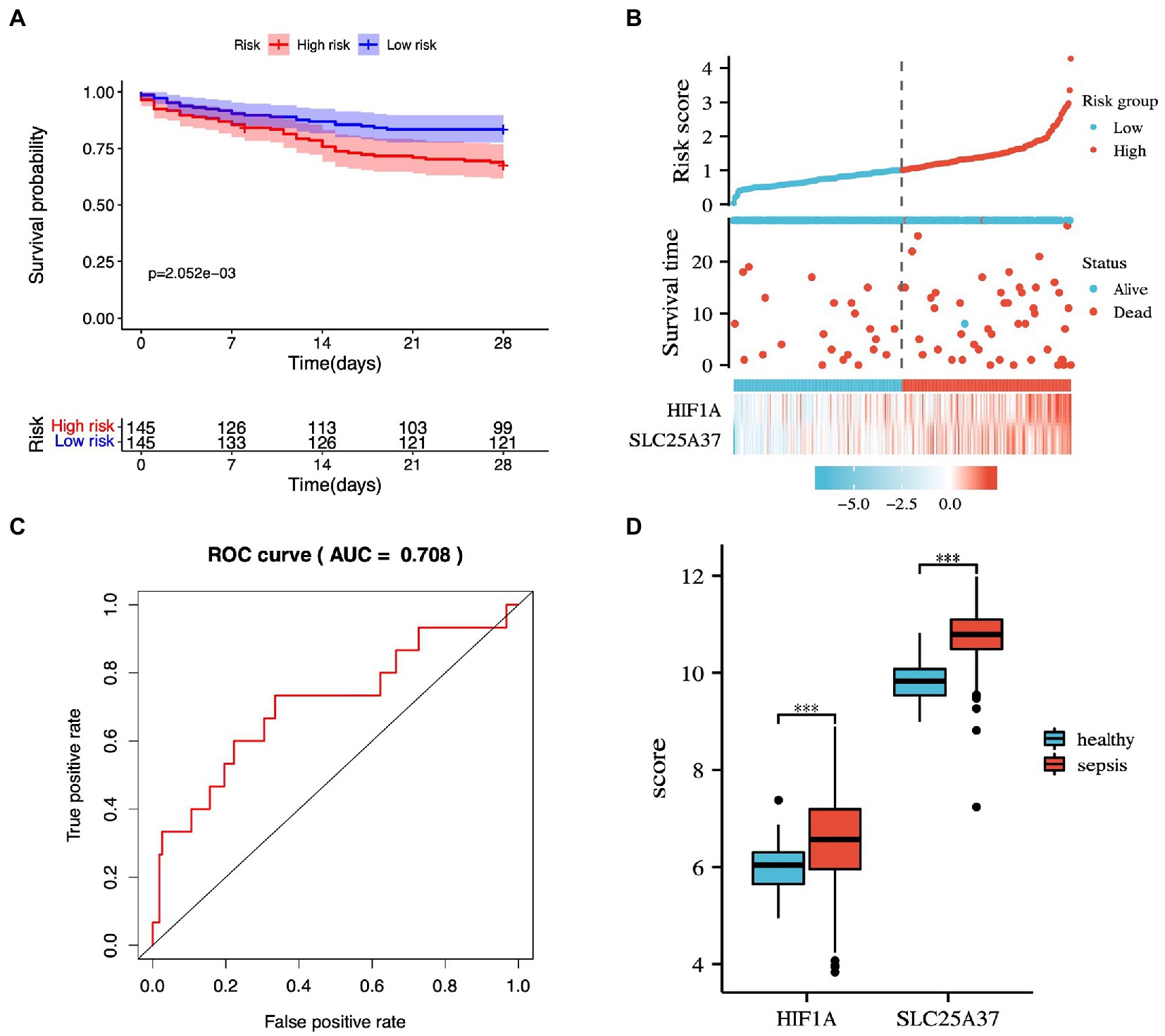
Figure 5. Prognostic prediction model based on common IMRGs. (A) Kaplan–Meier survival curves of 28-day mortality between high- and low-risk groups (p = 2.052e-03). (B) The risk score analysis between high-risk and low-risk groups and survival status analysis. (C) The receiver operating characteristic (ROC) curve of the two-gene model in patients with sepsis and controls (AUC:0.708). (D) The expression of the two genes between sepsis and healthy samples.
Echocardiography revealed that, compared with those in the control group, LPS treatment significantly reduced the LVEF (%) and FS (%) of the mice (Figure 6A). Furthermore, H&E staining revealed cardiomyocyte disarray in the LPS group compared with that of the control. (Figure 6B). To verify the reliability of the two IMRGs (HIF1A and SLC25A37), we evaluated the mRNA expression of key genes in the myocardium of mice. As shown in Figure 6C, the expressions of HIF1A and SLC25A37 were significantly higher in the sepsis group compared with that in the control group. The expression levels (logFC) of key genes with values from microarray and qPCR for comparison are listed in Supplementary Table 4.
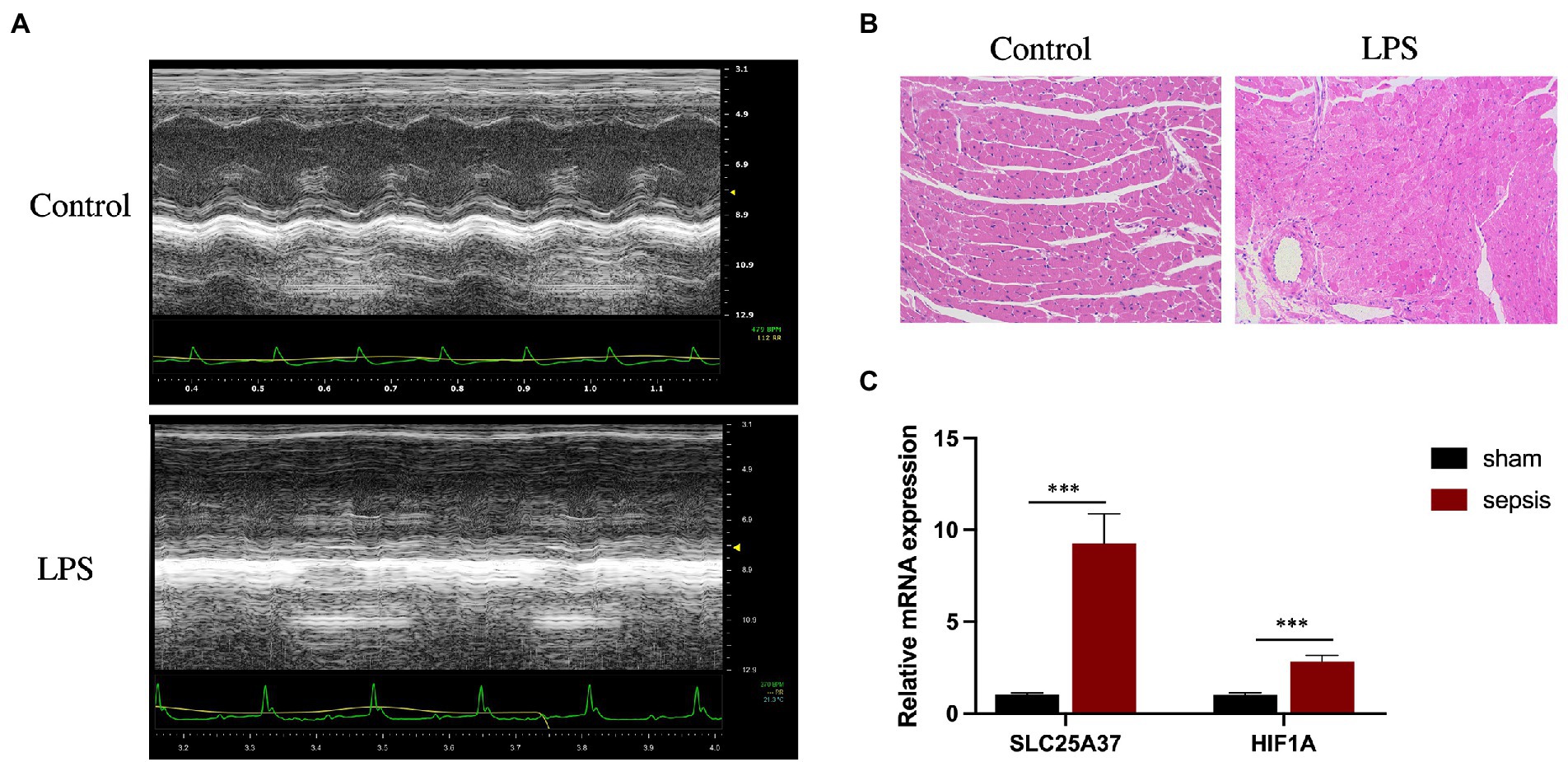
Figure 6. Validation of SLC25A37 and HIF1A in a mouse model of LPS-induced sepsis using quantitative PCR analysis. (A) Transthoracic echocardiography between sepsis and control groups. (B) Hematoxylin and eosin staining. (C) Relative expression of SLC25A37 and HIF1A between sepsis and control groups in mice, along with the expression of β-actin as an internal standard for normalization. Asterisks indicate a significant statistical p-value calculated using Student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001).
The CTD database was used to search for two common genes related to cardiovascular and inflammatory diseases. HIF1A was significantly associated with pneumonia (Inference Score: 161.07), inflammation (Inference Score: 467.34), and necrosis (Inference Score: 536.32), and was highly associated with certain cardiovascular diseases, including heart failure (Inference Score: 118.29), cardiomegaly (Inference Score: 190.62), and cardiomyopathies (Inference Score: 200.80) (Figure 7A). Similarly, SLC25A37 was associated with pneumonia (Inference Score: 56.82), inflammation (Inference Score: 119.76), and necrosis (Inference Score: 161.39), and was highly associated with cardiovascular diseases (Inference Score: 87.62), heart diseases (Inference Score: 82.00) and cardiomegaly (Inference Score: 48.33) (Figure 7B).
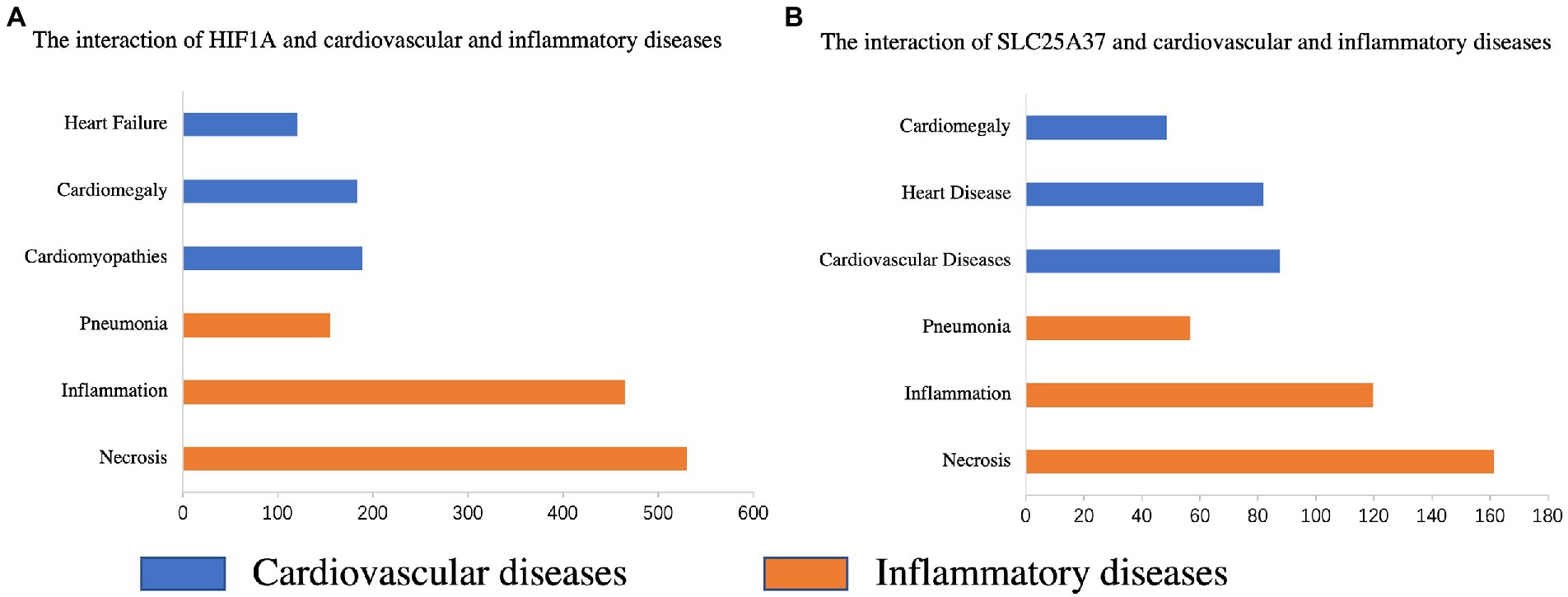
Figure 7. Relationship between the genes of the prognostic model and various diseases based on the comparative toxicogenomics database (CTD). Interactions between HIF1A (A) and SLC25A37 (B) and cardiovascular and inflammatory diseases, respectively.
Sepsis-induced cardiomyopathy or sepsis-induced myocardial dysfunction is increasingly recognized as a transient form of cardiac insufficiency during sepsis, with a notable impact on the prognosis of patients (28). Among the many pathogenic factors of septic cardiomyopathy, there is growing concern regarding the critical role that iron metabolism disorder plays in septic cardiomyopathy. Among the DEGs in septic cardiomyopathy tissue and peripheral blood monocytes, we found shared IMRGs and evaluated their correlation with the immune cells of septic cardiomyopathy. We established a prognostic model (HIF1A, SLC25A37) using the common differentially expressed IMRGs of septic myocardium and blood monocytes. The prognostic model we established might assist in the diagnosis of septic cardiomyopathy. We verified the potential genes in the myocardium of mice and found a significant difference between the sepsis and control groups.
Monocyte recruitment occurs after tissue damage caused by various infectious and aseptic inflammations. In cardiovascular disease, mobilization and infiltration of peripheral blood monocytes into diseased tissues are mainly adverse adaptation reactions (29). In a steady state, the heart of an adult mammal contains two macrophage populations: one from embryonic development and the other from blood monocytes (30). Under cardiac stress, monocyte-derived macrophages express high levels of pro-inflammatory genes, and their inappropriate activation in a sterile inflammatory environment may lead to pathological processes in the body (31). Recent advances in septic transcriptomics research have contributed to a better understanding of gene expression and its typical signaling pathways. Substantial evidence suggests significant changes in the leukocyte transcriptome in patients with sepsis; therefore, different patient phenotypes become apparent. Leukocytes of critically ill patients demonstrate profound changes in the transcriptome, in which 70–80% of RNA transcripts are differentially expressed compared with those in healthy individuals (32). The relationship between peripheral blood monocytes and cardiomyopathy in sepsis has not been clarified. Investigating potential IMRGs in peripheral blood monocytes and septic myocardium may provide a new approach to treating septic cardiomyopathy.
Sepsis can result in life-threatening organ dysfunction caused by the dysregulated host response to an underlying infection and is characterized by unbalanced inflammation and immunosuppression (32). The role of immunity in the pathogenesis of septic cardiomyopathy is self-evident. We used ssGSEA to analyze the immune infiltration of sequencing data in septic myocardium. Our research showed that innate immune cells, including neutrophils and monocytes, were significantly upregulated. Iron has multiple effects on the immune system, including regulating immune cell proliferation and differentiation, controlling various effector mechanisms of immune cells, and directly interfering with antimicrobial immune effector pathways. We analyzed the correlation between common IMRGs and immune cells of septic myocardium. Our study found that HBB, SLC25A37, SLC11A1, and HMOX1 have a strong correlation with monocytes and neutrophils, and HMOX1 and SLC11A1 have a strong correlation with macrophages. The incidence of sepsis-associated anemia is second only to iron deficiency anemia and is classified as a chronic anemic disease. When the expression of erythrocyte membrane proteins is altered, and erythrocyte deformability is reduced, hemolysis is more likely to occur during sepsis (33). Heme may increase host susceptibility to infection by inducing heme oxygenase 1 (HMOX-1) in immature neutrophils, thereby inhibiting the oxidative burst required to clear phagocytic bacteria (34). Hemolysis may also favor anti-inflammatory, immune cell polarization by inhibiting the dendritic cell maturation that is required for effector T cell responses. This may then induce monocyte differentiation into red plasma macrophages and drive regulatory T-cell expansion by regulating HMOX-1 expression in non-classical monocytes (35).
Inflammation causes changes in the genes related to iron metabolism. Ligands for Toll-like receptors 2, 4, and 6, BMP2, BMP6, α1 antitrypsin, activin B, IL-1, and IL-22 are examples of inflammation-driven hepcidin inducers (36–39). However, not all iron metabolism genes are equally regulated in monocytes and the myocardium. For example, hepcidin, known as hepcidin antimicrobial peptide (HAMP), is a 25-amino acid protein mainly produced by hepatocytes, but its expression and release are also detected in other organs, including the heart (40). When the iron levels in the body are high, hepcidin can promote iron-exporter ferroportin degradation and thus inhibit ferritin efflux. Hepcidin can also be released under conditions such as iron overload, red blood cell transfusion, iron therapy, or inflammation (36). We found that the mRNA expression level of HAMP is upregulated in the myocardium but downregulated in monocytes. When systemic hepcidin concentrations are significantly low, hepcidin produced by the myocardium acts as an autocrine or paracrine regulator of ferroportin and can protect tissues from ferroportin and extreme iron deficiency; however, this does not significantly affect systemic hepcidin (40).
We identified two IMRGs using the Cox regression method to establish a prognostic model (HIF1A and SLC25A37). The ROC curve shows that this model may have excellent diagnostic ability for sepsis. Juan José Martínez-García found that hypoxia-inducible factor (HIF)-1 α is activated, possibly through the influence of its P2X7 receptor on HIF-1 α, and that inhibition of NLRP3 leads to mitochondrial damage in the monocytes of patients with sepsis (41). Functional plasticity or reprogramming occurs in circulating monocytes during sepsis, and the expression and activity of HIF-1α are upregulated (15), which is consistent with our results. Studies have shown that SLC25A37 is related to mitochondrial iron accumulation in pancreatic cancer. SLC25A37 mediated by the PINK1-PARK2 pathway increases mitochondrial iron accumulation, which leads to the HIF1A-dependent Warburg effect and AIM2-dependent inflammasome activation in tumor cells (42).
There is a lack of effective treatment to regulate the complex pathophysiology of the host immune response. Traditional treatment of sepsis includes timely implementation of supportive treatment, such as fluid resuscitation, vasopressor and oxygen therapy, mechanical ventilation, timely source control, and medicines such as antibiotics (43). Previous clinical trials have partly focused on anti-inflammatory therapy, but were unsuccessful in combating the inflammatory storm of sepsis (43). Iron chelation has immunomodulatory effects and can improve the prognosis of sepsis models (44, 45). Therefore, iron chelators may become new targets for developing anti-sepsis medicine. In the present study, we predicted small-molecule drugs based on differential IMRGs, and studies have shown that some drugs have therapeutic effects on heart diseases in basic experiments (46–49).
Due to limited research data, this study presented some limitations. First, although we validated potential iron metabolism genes in a mouse model of septic cardiomyopathy, this was lacking in the peripheral blood of those with septic cardiomyopathy. Therefore, we require clinical randomized controlled studies on sepsis-induced cardiomyopathy for further validation. Second, the proportion of immune cells was inferred using the ssGSEA algorithm rather than a real test of sepsis cardiomyocytes, which may show differences from the actual results. Third, the specific regulatory mechanism of iron metabolism genes in septic cardiomyopathy found in this study has not been deeply elucidated and further exploration is required.
In conclusion, we identified the shared differential expression of IMRGs in myocardium tissue and blood monocytes and analyzed the relationship between shared IMRGs and immune cells of septic myocardium. We established a promising prognostic model that incorporates HIF1A and SLC25A37 for septic cardiomyopathy. These IMRGs may be potential therapeutic targets for the treatment of septic cardiomyopathy, and small-molecule drugs targeting them may help reduce the sepsis-related death rate.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by The ethics committee of Shanghai Tenth People’s Hospital, Tongji University.
RZ carried out the experiments and drafted the articles. RZ, CD, HG, YZ, CL, ZZ, and QW conducted the bioinformatics and statistical analyses. JW and FZ contributed to reviewing the article. SW edited and revised the article. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 81871540) and the Clinical Research Plan of Shanghai Hospital Development Center Foundation (Nos. SHDC2020CR6030-003, SHDC2020CR1028B, and SHDC12020122).
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2023.1018422/full#supplementary-material
1. ^https://online.visual-paradigm.com/diagrams/features/venn-diagram-tool/
2. ^http://string-db.org/, version 11.0.
1. Duran-Bedolla, J, Montes de Oca-Sandoval, MAM, Saldaña-Navor, V, Villalobos-Silva, JA, Rodriguez, MC, and Rivas-Arancibia, S. Sepsis, mitochondrial failure and multiple organ dysfunction. Clin Invest Med. (2014) 37:E58–69. doi: 10.25011/cim.v37i2.21087
2. Beesley, SJ, Weber, G, Sarge, T, Nikravan, S, Grissom, CK, Lanspa, MJ, et al. Septic cardiomyopathy. Crit Care Med. (2018) 46:625–34. doi: 10.1097/CCM.0000000000002851
3. Blanco, J, Muriel-Bombín, A, Sagredo, V, Taboada, F, Gandía, F, Tamayo, L, et al. Incidence, organ dysfunction and mortality in severe sepsis: a Spanish multicentre study. Crit Care. (2008) 12:R158. doi: 10.1186/cc7157
4. Karakike, E, and Giamarellos-Bourboulis, EJ. Macrophage activation-like syndrome: a distinct entity leading to early death in sepsis. Front Immunol. (2019) 10:55. doi: 10.3389/fimmu.2019.00055
5. Su, LJ, Zhang, JH, Gomez, H, Murugan, R, Hong, X, Xu, D, et al. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxidative Med Cell Longev. (2019) 2019:5080843. doi: 10.1155/2019/5080843
6. Jankowska, EA, Kasztura, M, Sokolski, M, Bronisz, M, Nawrocka, S, Oleśkowska-Florek, W, et al. Iron deficiency defined as depleted iron stores accompanied by unmet cellular iron requirements identifies patients at the highest risk of death after an episode of acute heart failure. Eur Heart J. (2014) 35:2468–76. doi: 10.1093/eurheartj/ehu235
7. Díez-López, C, Comín-Colet, J, and González-Costello, J. Iron overload cardiomyopathy: from diagnosis to management. Curr Opin Cardiol. (2018) 33:334–40. doi: 10.1097/HCO.0000000000000511
8. Fang, X, Ardehali, H, Min, J, and Wang, F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat Rev Cardiol. (2023) 20:7–23. doi: 10.1038/s41569-022-00735-4
9. Zhang, H, Zhabyeyev, P, Wang, S, and Oudit, GY. Role of iron metabolism in heart failure: from iron deficiency to iron overload. Biochim Biophys Acta Mol basis Dis. (2019) 1865:1925–37. doi: 10.1016/j.bbadis.2018.08.030
10. Li, N, Wang, W, Zhou, H, Wu, Q, Duan, M, Liu, C, et al. Ferritinophagy-mediated ferroptosis is involved in sepsis-induced cardiac injury. Free Radic Biol Med. (2020) 160:303–18. doi: 10.1016/j.freeradbiomed.2020.08.009
11. Xiao, Z, Kong, B, Fang, J, Qin, T, Dai, C, Shuai, W, et al. Ferrostatin-1 alleviates lipopolysaccharide-induced cardiac dysfunction. Bioengineered. (2021) 12:9367–76. doi: 10.1080/21655979.2021.2001913
12. Stevenson, EK, Rubenstein, AR, Radin, GT, Wiener, RS, and Walkey, AJ. Two decades of mortality trends among patients with severe sepsis: a comparative meta-analysis*. Crit Care Med. (2014) 42:625–31. doi: 10.1097/CCM.0000000000000026
13. Matkovich, SJ, Al Khiami, B, Efimov, IR, Evans, S, Vader, J, Jain, A, et al. Widespread down-regulation of cardiac mitochondrial and sarcomeric genes in patients with sepsis. Crit Care Med. (2017) 45:407–14. doi: 10.1097/ccm.0000000000002207
14. Shalova, IN, Lim, JY, Chittezhath, M, Zinkernagel, AS, Beasley, F, Hernández-Jiménez, E, et al. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity. (2015) 42:484–98. doi: 10.1016/j.immuni.2015.02.001
15. Gautier, L, Cope, L, Bolstad, BM, and Irizarry, RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. (2004) 20:307–15. doi: 10.1093/bioinformatics/btg405
16. Ritchie, ME, Phipson, B, Wu, D, Hu, Y, Law, CW, Shi, W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
17. Liberzon, A, Birger, C, Thorvaldsdóttir, H, Ghandi, M, Mesirov, JP, and Tamayo, P. The molecular signatures database (MsigDB) hallmark gene set collection. Cell Syst. (2015) 1:417–25. doi: 10.1016/j.cels.2015.12.004
18. Ito, K, and Murphy, D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol. (2013) 2:e79. doi: 10.1038/psp.2013.56
19. Yu, G, Wang, LG, Han, Y, and He, QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
20. Altermann, E, and Klaenhammer, TR. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics. (2005) 6:60. doi: 10.1186/1471-2164-6-60
21. Szklarczyk, D, Franceschini, A, Wyder, S, Forslund, K, Heller, D, Huerta-Cepas, J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. (2015) 43:D447–52. doi: 10.1093/nar/gku1003
22. Bader, GD, and Hogue, CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. (2003) 4:2. doi: 10.1186/1471-2105-4-2
23. Chin, CH, Chen, SH, Wu, HH, Ho, CW, Ko, MT, and Lin, CY. Cytohubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. (2014) 8:S11. doi: 10.1186/1752-0509-8-S4-S11
24. Sergushichev, A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv. (2016) 60012:1–9. doi: 10.1101/060012
25. Efron, B. Logistic regression, survival analysis, and the Kaplan-Meier curve. J Am Stat Assoc. (1988) 83:414–25. doi: 10.1080/01621459.1988.10478612
26. Yoo, M, Shin, J, Kim, J, Ryall, KA, Lee, K, Lee, S, et al. DSigDB: drug signatures database for gene set analysis. Bioinformatics. (2015) 31:3069–71. doi: 10.1093/bioinformatics/btv313
27. L’Heureux, M, Sternberg, M, Brath, L, Turlington, J, and Kashiouris, MG. Sepsis-induced cardiomyopathy: a comprehensive review. Curr Cardiol Rep. (2020) 22:35. doi: 10.1007/s11886-020-01277-2
28. Bajpai, G, Bredemeyer, A, Li, W, Zaitsev, K, Koenig, AL, Lokshina, I, et al. Tissue resident CCR2- and CCR2+ cardiac macrophages differentially orchestrate monocyte recruitment and fate specification following myocardial injury. Circ Res. (2019) 124:263–78. doi: 10.1161/CIRCRESAHA.118.314028
29. Epelman, S, Lavine, KJ, Beaudin, AE, Sojka, DK, Carrero, JA, Calderon, B, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. (2014) 40:91–104. doi: 10.1016/j.immuni.2013.11.019
30. Wynn, TA, and Vannella, KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. (2016) 44:450–62. doi: 10.1016/j.immuni.2016.02.015
31. van der Poll, T, van de Veerdonk, FL, Scicluna, BP, and Netea, MG. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol. (2017) 17:407–20. doi: 10.1038/nri.2017.36
32. Effenberger-Neidnicht, K, and Hartmann, M. Mechanisms of hemolysis during sepsis. Inflammation. (2018) 41:1569–81. doi: 10.1007/s10753-018-0810-y
33. Evans, C, Orf, K, Horvath, E, Levin, M, De La Fuente, J, Chakravorty, S, et al. Impairment of neutrophil oxidative burst in children with sickle cell disease is associated with heme oxygenase-1. Haematologica. (2015) 100:1508–16. doi: 10.3324/haematol.2015.128777
34. Zhong, H, and Yazdanbakhsh, K. Hemolysis and immune regulation. Curr Opin Hematol. (2018) 25:177–82. doi: 10.1097/MOH.0000000000000423
35. Armitage, AE, Eddowes, LA, Gileadi, U, Cole, S, Spottiswoode, N, Selvakumar, TA, et al. Hepcidin regulation by innate immune and infectious stimuli. Blood. (2011) 118:4129–39. doi: 10.1182/blood-2011-04-351957
36. Schaefer, B, Haschka, D, Finkenstedt, A, Petersen, BS, Theurl, I, Henninger, B, et al. Impaired hepcidin expression in alpha-1-antitrypsin deficiency associated with iron overload and progressive liver disease. Hum Mol Genet. (2015) 24:6254–63. doi: 10.1093/hmg/ddv348
37. Besson-Fournier, C, Latour, C, Kautz, L, Bertrand, J, Ganz, T, Roth, MP, et al. Induction of activin B by inflammatory stimuli up-regulates expression of the iron-regulatory peptide hepcidin through Smad1/5/8 signaling. Blood. (2012) 120:431–9. doi: 10.1182/blood-2012-02-411470
38. Lee, P, Peng, H, Gelbart, T, Wang, L, and Beutler, E. Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proc Natl Acad Sci U S A. (2005) 102:1906–10. doi: 10.1073/pnas.0409808102
39. Lakhal-Littleton, S, Wolna, M, Chung, YJ, Christian, HC, Heather, LC, Brescia, M, et al. An essential cell-autonomous role for hepcidin in cardiac iron homeostasis. elife. (2016) 5:e19804. doi: 10.7554/eLife.19804
40. Martínez-García, JJ, Martínez-Banaclocha, H, Angosto-Bazarra, D, de Torre-Minguela, C, Baroja-Mazo, A, Alarcón-Vila, C, et al. P2X7 receptor induces mitochondrial failure in monocytes and compromises NLRP3 inflammasome activation during sepsis. Nat Commun. (2019) 10:2711. doi: 10.1038/s41467-019-10626-x
41. Li, C, Zhang, Y, Cheng, X, Yuan, H, Zhu, S, Liu, J, et al. PINK1 and PARK2 suppress pancreatic tumorigenesis through control of mitochondrial iron-mediated immunometabolism. Dev Cell. (2018) 46:441–455.e8. doi: 10.1016/j.devcel.2018.07.012
42. Angus, DC, and van der Poll, T. Severe sepsis and septic shock. N Engl J Med. (2013) 369:840–51. doi: 10.1056/NEJMra1208623
43. Islam, S, Jarosch, S, Zhou, J, Parquet Mdel, C, Toguri, JT, Colp, P, et al. Anti-inflammatory and anti-bacterial effects of iron chelation in experimental sepsis. J Surg Res. (2016) 200:266–73. doi: 10.1016/j.jss.2015.07.001
44. Xia, Y, Farah, N, Maxan, A, Zhou, J, and Lehmann, C. Therapeutic iron restriction in sepsis. Med Hypotheses. (2016) 89:37–9. doi: 10.1016/j.mehy.2016.01.018
45. Goto, M, Suematsu, Y, Nunes, ACF, Jing, W, Khazaeli, M, Lau, WL, et al. Ferric citrate attenuates cardiac hypertrophy and fibrosis in a rat model of chronic kidney disease. Iran J Kidney Dis. (2019) 13:98–104.
46. Ito, K, Hirooka, Y, and Sunagawa, K. Cardiac sympathetic afferent stimulation induces salt-sensitive sympathoexcitation through hypothalamic epithelial Na+ channel activation. Am J Physiol Heart Circ Physiol. (2015) 308:H530–9. doi: 10.1152/ajpheart.00586.2014
47. Ichikawa, Y, Ghanefar, M, Bayeva, M, Wu, R, Khechaduri, A, Naga Prasad, SVN, et al. Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J Clin Invest. (2014) 124:617–30. doi: 10.1172/JCI72931
48. Tucker, A. Pulmonary vascular actions of the antihistamine oxatomide during hypoxia. Agents Actions. (1980) 10:207–12. doi: 10.1007/BF02025937
Keywords: sepsis, septic cardiomyopathy, iron metabolism, peripheral blood monocytes, diagnostic biomarkers
Citation: Zhang R, Di C, Gao H, Zhu Y, Li C, Zhu Z, Wang Q, Wang J, Zhou F and Wang S (2023) Identification of iron metabolism-related genes in the circulation and myocardium of patients with sepsis via applied bioinformatics analysis. Front. Cardiovasc. Med. 10:1018422. doi: 10.3389/fcvm.2023.1018422
Received: 13 August 2022; Accepted: 13 February 2023;
Published: 02 March 2023.
Edited by:
Francesca Innocenti, Careggi University Hospital, ItalyReviewed by:
Zhi Yang, General Hospital of Guangzhou Military Command, ChinaCopyright © 2023 Zhang, Di, Gao, Zhu, Li, Zhu, Wang, Wang, Zhou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junjie Wang, bWVnZW5laGFwcHlAMTI2LmNvbQ==; Feng Zhou, emZ5eXp6MDBAMTYzLmNvbQ==; Sheng Wang, c2hlbmd3YW5nMjZAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.