- 1Cardiovascular Division, Department of Medicine, University of Minnesota, Minneapolis, MN, United States
- 2Stem Cell Institute, University of Minnesota, Minneapolis, MN, United States
- 3Paul and Sheila Wellstone Muscular Dystrophy Center, University of Minnesota, Minneapolis, MN, United States
Cardiovascular disease (CVD) remains the number one cause of death worldwide. Ischemic heart disease contributes to heart failure and has considerable morbidity and mortality. Therefore, alternative therapeutic strategies are urgently needed. One class of epigenetic regulators known as pioneer factors has emerged as an important tool for the development of regenerative therapies for the treatment of CVD. Pioneer factors bind closed chromatin and remodel it to drive lineage specification. Here, we review pioneer factors within the cardiovascular lineage, particularly during development and reprogramming and highlight the implications this field of research has for the future development of cardiac specific regenerative therapies.
Introduction
Cardiovascular disease is the number one cause of death in the U.S and worldwide. Vascular and cardiac disease results in considerable morbidity and mortality (1, 2). The only curative disease for end-stage cardiovascular disease is orthotopic heart transplantation (3). While it is estimated that more than 100,000 Americans could benefit from cardiac transplantation, only 3,000 to 3,500 recipients receive such therapy due to limited donor organ availability (4). Therefore, new therapies are warranted.
Reprogramming of lineages has received intense interest and several outstanding reviews are available and provide a comprehensive overview of the field (5, 6). This field had its genesis, in part, based on the discovery of master regulators—those factors that promote lineage specific gene expression when overexpressed in somatic cells such as fibroblasts (6). These assays were referred to as conversion assays and the first master regulator to be described was Myod and its family members (7). Subsequently, more than 200 master regulators have been described and their functional roles have been explored using gene disruption technologies (8).
While the role(s) of master regulators focused primarily on their ability as transcription factors to govern lineage specific gene expression, pioneer factors function to bind nucleosomal DNA and relax the chromatin landscape upstream of lineage specific genes (6, 8, 9). In this fashion, pioneer factors reside at the very top of the hierarchical molecular cascade. Moreover, there are only a limited number of rigorously defined pioneer factors (9). Here, we highlight the criteria necessary for inclusion as a pioneer factor, we provide an overview of the field itself and highlight the role and mechanisms of pioneer factors that govern the cardiovascular field.
Coordinated role of networks and lineage specification during cardiovascular development
Cardiovascular development is a well-coordinated and complex process that requires the specification, proliferation, migration and differentiation of progenitor cells that become coupled to form a functional syncytium within the heart (10–14). Progenitor cells arising from the mesodermal germ layer form the early cardiac crescent and fuse to form the linear heart tube. Progenitor cell populations and their derivatives contribute to the first and second heart fields. These respective progenitor cell populations contribute to distinct structures within the mature heart, and are combinatorily regulated by distinct and overlapping transcriptional networks (15). Stage specific transcription factors and signaling pathways have been defined and function as key regulators of cardiovascular development. As previously outlined, master regulatory genes govern the transcriptional cascades and direct cellular lineages during differentiation and cellular reprogramming. However, within this group of master regulators, a small subset of transcription factors known as pioneer factors, have the unique capacity to bind and remodel silent and compacted regions of chromatin (nucleosomal DNA) to drive the expression of lineage specific genes that allow for development and reprogramming to occur (Figure 1). Due to their unique capacity to bind nucleosomal DNA and drive lineage development, pioneer factors have been shown to be critical factors for regenerative sciences and cancer biology.
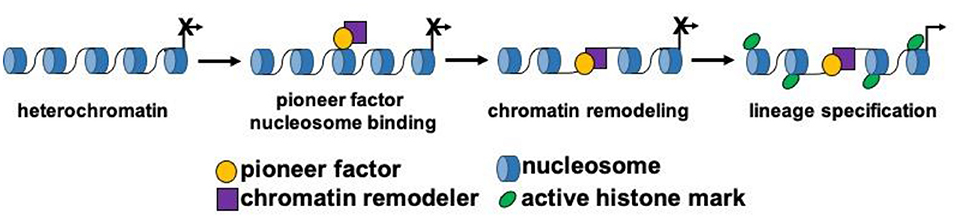
Figure 1. Pioneer factors drive lineage specification. Schematic model depicting the function of pioneer transcription factors during cellular lineage specification. Pioneer factors initially bind to nucleosomal DNA and then remodel chromatin by themselves or by recruiting a chromatin remodeling factor. These steps lead to the activation of gene expression and changes to the epigenetic landscape surrounding the DNA binding sites of the pioneer factor.
Role of master regulators during development
Essential transcription factors, known as master regulators, regulate cell fate and lineage commitment development. These master transcription factors regulate lineage commitment events, and can convert/reprogram cells (fibroblasts) to specific lineages (8, 16). In addition to the MYOD family, other prototypic master regulators include Pdx1, an important regulator of pancreas development (17). Global knockout of Pdx1 in the mouse results in the absence of the pancreas and similar to MYOD, ectopic overexpression of PDX1 converts somatic cells to pancreatic acinar cells (18, 19). Other master regulators have been identified for each lineage including (but not limited to): MESP1, MYF5, NEUROD, ASCL1/MASH1, GATA2, GATA4, PAX3, PAX7, FOXO, FOXA, SCL/TAL1, HIF1 and others (7, 17, 20–24). These lineage specific transcription factors or master regulators are essential for the different combinations of reprogramming factors used to develop organ specific cellular therapies (6). Among these master regulators, a subset of transcription factors known as pioneer factors, initiate lineage specific regulatory events to open up or relax compacted (heterochromatic) DNA to govern developmental and reprogramming processes (6, 25, 26) (Figure 1).
Role of pioneer factors during lineage specification
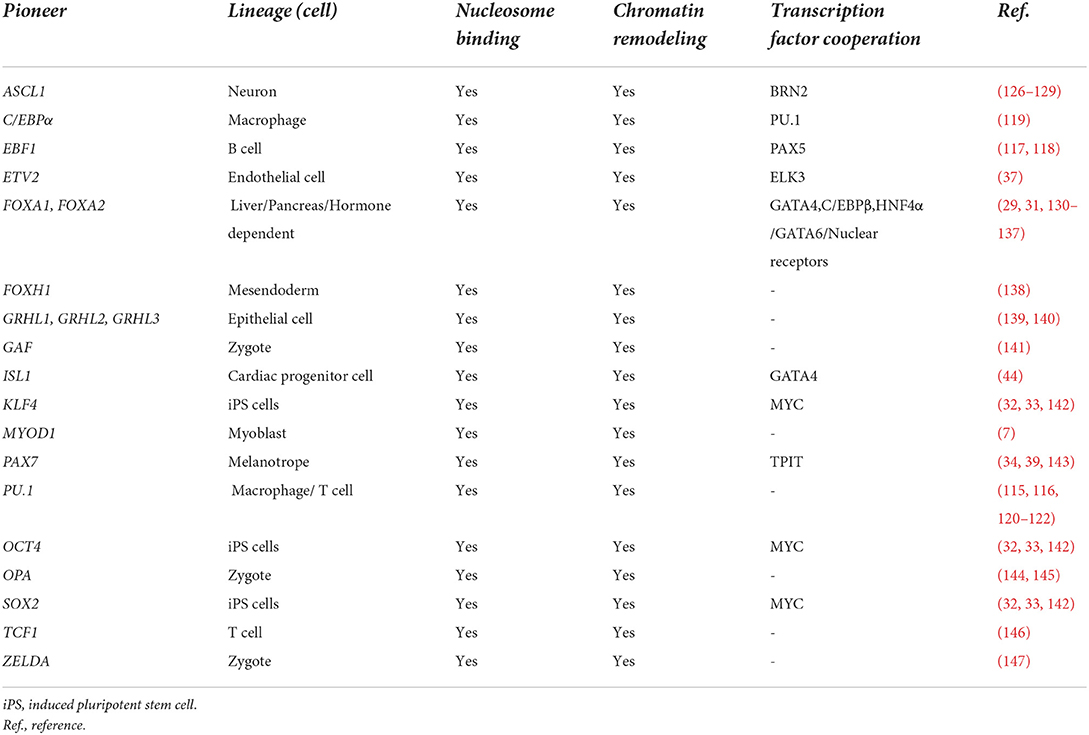
Pioneer factors are a specialized group of lineage-specific transcription factors that bind heterochromatic regions of DNA to promote chromatin relaxation and recruit non-pioneer transcription factors for lineage development or reprogramming to occur (Figure 1) (9, 25, 26). This important functional role is due to their unique capacity to scan heterochromatin, recognize partial (non-canonical) DNA motifs that are exposed on the surface on nucleosomes and bind to them. The complexes that are formed (chromatin remodeling and non-pioneer factors) following the binding of a pioneer factor are context dependent (i.e., cell type specific) and serve to dictate the diverse mechanisms whereby pioneer factors can regulate lineage specification and development. Pioneer factors were discovered with the dissection and definition of the mechanisms whereby liver specific regulatory complexes were bound to heterochromatin early during development (27, 28). These studies identified FOXA1 as the first prototypical pioneer factor that regulates hepatic lineage specification during early embryonic development (Table 1) (25, 29). A distinct feature regarding FOXA1 is that its DNA binding domain (forkhead/winged helix domain) shares a similar structure to that of linker histones, which enables this pioneer transcription factor to displace linker histones from nucleosomes to remodel chromatin and promote liver development (30, 31). The discovery of FOXA1 as a pioneer factor has led to the identification and characterization of other pioneer factors (Table 1). Perhaps, the most recognized examples of pioneer factors are OCT4, SOX2 and KLF4 (OSK), which promote the reprogramming of terminally differentiated fibroblasts to induced pluripotent stem cells (iPSCs) (32, 33). While c-MYC is also necessary for the reprogramming process of fibroblasts to iPSCs, unlike OSK, which can bind partial DNA motifs in nucleosomal DNA located in enhancers, c-MYC binds accessible regions in promoters and not nucleosomal DNA (25, 32, 33).
The molecular mechanisms whereby pioneer factors promote lineage specification and development remain ill-defined. One area of interest in the field is the effect of chromatin modifications (histones or DNA) on pioneer factor binding. A mechanism that has been observed is the binding of pioneer factors to nucleosomal DNA sites without any apparent effect. These regions are termed “pioneer factor resistant sites” and are being examined to define the co-regulatory mechanisms that are present in the chromatin environment to control cell fate by promoting or preventing chromatin remodeling and lineage specification by a pioneer factor following binding (34). For example, the pioneer factor PAX7 is able to bind facultative heterochromatin (H3K9me2) but not constitutive heterochromatin (H3M9me3) in order to regulate pituitary progenitor cell development (33–35). This action highlights the need to characterize the role of different histone and DNA modifying enzymes that regulate pioneer factor function by directly interacting with them or by indirectly modifying the chromatin landscape during lineage specification (33).
A majority of the pioneer factor studies have focused on the characterization of the activation of lineage transcriptional programs and the initial functions of pioneer factors. However, the mechanisms driving lineage repression and the later stages of lineage specification are not well understood and warrant further investigation. ASCL1 is one of the few pioneer factors whose repressive role has been explored and found to recruit cooperating factors such as the myelin transcription factor 1- like protein (MYT1L), which represses the myogenic lineage program during the reprogramming of fibroblasts to neuronal cells (36). Studies on these cooperating or settler factors have demonstrated recruitment of both non-pioneer transcription factors as well as chromatin remodeling enzymes (p300 and BRG1) to allow for lineage specification to occur by activating gene expression, modifying chromatin or stabilizing the binding of pioneer factors to their DNA binding sites (9, 37, 38). Gaining a deeper understanding on how pioneer factor function is regulated by these settler factors may enhance reprogramming strategies to more efficiently drive lineage development in vivo to treat cardiovascular disease (6).
Role of chromatin modifying factors for the function of pioneer factors
While pioneer factors are required for the initial binding to nucleosomal DNA, cooperation with other (non-pioneer factors) is necessary in order to drive lineage development and reprogramming (9, 25, 39). Two important events are required following the binding of a pioneer factor and these include: (1) chromatin relaxation and (2) recruitment/interaction with other transcription factors (Figure 1). These two events enable the effects of pioneer factors and lineage development to occur by amplifying the signal and providing context dependent mechanisms in different regions of the genome (9, 25). Chromatin relaxation is a crucial step during lineage development where pioneer factors have been shown to promote remodeling by themselves (FOXA1) or with the assistance of the SWI/SNF complex (9, 25). The SWI/SNF complex of proteins is one of the most studied chromatin remodeling complexes. This complex increases DNA accessibility to regulate the development or reprogramming of pluripotent, neuronal, cardiac and endothelial cells (37, 40). SMARCA4 (BRG1), the ATPase subunit of the SWI/SNF complex, is an important regulator of early embryonic development as Brg1 null embryos are lethal prior to implantation (41). Using in vitro differentiation and mouse studies, BRG1 has also been shown to be an important regulator of cardiovascular development and disease (42, 43). BRG1 also has been shown to be an important chromatin remodeler as it interacts with at least four different pioneer factors (OCT4, GATA3, ISL1 and ETV2) in a context dependent fashion to regulate chromatin remodeling and two of these pioneer factors are important regulators of cardiovascular development (Table 1) (37, 38, 44, 45).
Pioneer factors in the cardiovascular lineage
The cardiovascular lineage is composed of multiple lineages including: the muscle, vascular/endothelial and hematopoietic lineages (46–51). While many master regulators have been described and have important roles in the coordination of the development of the cardiovascular lineage, few pioneer factors have been identified within this lineage (8, 9). In part, this is due to the complexity associated with the different cellular lineages and structures within the cardiovascular system (13, 52). These pioneer factors are ISL1, GATA4 and ETV2, and in this section we will discuss the data supporting their pioneer role and function in the regulation of the cardiovascular lineage.
ISL1 is an important regulator for the development of the second heart field (SHF), and was recently identified as a pioneer factor (Table 1). Isl1 KO mice lack the right ventricle, outflow tract and portions of the atria because of its role as an important regulator of SHF cardiac progenitor cells (CPCs) (53–56). In recent studies by Gao et al., they described that ISL1, like other pioneer factors, regulated the development of SHF CPCs by binding nucleosomal DNA and relaxing chromatin by forming a complex with BRG1-BAF60C (44). They also identified GATA4 as a cooperating factor in selected sites bound to ISL1, suggesting a potential interaction for ISL1 in the regulation of cardiovascular development. Together, ISL1 and GATA4 were shown to bind regulatory DNA regions of important cardiac genes such as Hand2, Myocd, Ttn, Ryr2 and others. The exact mechanism whereby GATA4 promotes the pioneer function of ISL1 in these regulatory regions remains to be elucidated. These studies used both in vivo and in vitro assays to demonstrate that ISL1 binds nucleosomal DNA to regulate SHF development.
GATA4 is another important master regulator of cardiovascular development. Loss of Gata4 has been shown to lead to early cardiac defects and results in bifed (non-fused) heart fields and embryonic lethality (57–62). Additionally, GATA4 has the capacity (along with other master regulators) to reprogram fibroblasts to induced cardiomyocytes (iCMs) in vitro and in vivo (63–67). While GATA4 is a key regulator of cardiovascular development, its role as a pioneer factor has only been described in hepatic progenitors and reprogramming of fibroblasts to hepatic-like cells (29, 68). A recent study combined scRNAseq, ATACseq, ChIPseq and machine learning to define the molecular mechanisms governing iCM reprogramming using GATA4, MEF2C and TBX5 (GMT) and concluded that MEF2C and TBX5, but not GATA4 bind heterochromatin and promote chromatin remodeling during reprogramming (69). While these studies do not support the notion that GATA4 is a pioneer factor for the cardiac muscle lineage, further developmental studies are necessary to understand the heterochromatin binding and chromatin remodeling capabilities of GATA4 during cardiovascular development. The recent ISL1 studies suggest that GATA4 may bind to heterochromatin in the cardiovascular lineage independently but more mechanistic studies are needed (44).
More recently, ETV2 was identified as a pioneer factor for the cardiovascular lineage that regulates and reprograms endothelium (Table 1) (37). ETV2 is an essential transcription factor for the development of endothelial and hematopoietic lineages (51, 70–108). Its expression is observed in mesodermal progenitors and hemangioblasts that give rise to endocardial/endothelial and hematopoietic lineages, while repressing other lineages such as the cardiac and skeletal muscle lineages (49, 74, 79). Key regulatory genes and pathways for the cardiovascular lineage such as MESP1, NKX2-5, Wnt/Notch/BMP signaling (among others) have been shown to regulate ETV2 expression within the cardiovascular lineage (50, 74, 109, 110). Loss of Etv2 results in lethality by E8.5 in developing mouse embryos due to the lack of all vascular and blood lineages. Moreover, congenital heart defects in aborted developing human fetuses have been reported to harbor Etv2 mutations (50, 51, 111). Additionally, ETV2 overexpression (alone) reprograms terminally differentiated cells (fibroblasts) to endothelial cells both in vitro and in vivo (112). Our recent findings characterized the molecular mechanism whereby ETV2 regulates the endothelial lineage as a pioneer factor (37). ETV2 can scan the genome, bind nucleosomal DNA and remodel chromatin independent of its cellular context, whether it is related to fibroblast reprogramming or mouse embryonic stem cell (mESC) differentiation into endothelial progenitor cells (Figure 1). We characterized this functional role for ETV2 using scRNAseq, ATACseq, NOMEseq, ChIPseq and in vitro nucleosomal binding assays to unequivocally demonstrate that ETV2 binds nucleosomal DNA during endothelial lineage reprogramming/development. We identified canonical downstream targets for ETV2 such as Emcn, Lmo2, Rhoj and others that were bound by ETV2 during endothelial lineage development and reprogramming. Similar to ISL1, ETV2 recruits and directly interacts with BRG1. BRG1 is an essential co-factor for ETV2 to function as a pioneer factor as Brg1 knockdown and conditional knockout significantly affected the ability of ETV2 to remodel chromatin and drive endothelial lineage formation in both reprogrammed fibroblasts and differentiating mESCs, respectively (37). This ETV2-BRG1 interaction was verified using mass spectrometry, Co-IP assays and GST-pulldown assays. Additionally, we demonstrated that this interaction was important for enacting epigenetic changes during endothelial lineage development such as the deposition of histone 3 lysine 27 acetylation (H3K27Ac) in regions surrounding ETV2-BRG1 bound sites. Lastly, by screening co-factors we identified ELK3 as a transcription factor that was recruited to ETV2-BRG1 bound sites following chromatin remodeling and ELK3 has an important role in endothelial cell development (Figure 1 and Table 1). Understanding how other factors might regulate chromatin remodeling and the pioneer activity of ETV2, such as FOXC2 which is known to regulate Fox-Ets enhancer motifs during endothelial lineage development in combination of ETV2, will be important for further dissecting this molecular mechanism (113). Forkhead transcription factors are important in the field of pioneer factors and chromatin remodeling because of their unique protein structure that resembles linker histones and allows (some of the forkhead family members) to remodel chromatin (31). Furthermore, the expression of ETV2 is transient during development, understanding how other downstream co-factors (i.e., ELK3, FLI1, SCL/TAL1, etc.) direct the developmental machinery following the downregulation of ETV2 will be important for the development of therapeutic strategies using ETV2 to develop mature vasculature that can be used for ischemic diseases such as the transplantation of human vasculature (71, 114).
ETV2 possesses an Ets DNA binding domain (DBD) characterized by a winged helix-turn-helix structure which needs to be studied in terms of how it interacts with nucleosomal DNA to allow for chromatin binding and remodeling to occur (75). Previous studies on forkhead factors have demonstrated that the winged helix DBD of the pioneer factor FOXA resembles that of the structure of linker histones, while Ets factors have been shown to use their short α-helix structure to bind the major groove of DNA to target nucleosomes (25). We hypothesize that the winged helix of ETV2 will most likely behave like that of previously described Ets factors, but it remains to be explored.
Unlike ISL1 and GATA4, the pioneer function of ETV2 in the cardiovascular lineage is independent of its cellular context, whether it is cellular differentiation or reprogramming, it functions in a similar fashion in both model systems. Future studies will need to focus on further characterization of the molecular mechanisms driving endothelial cell development/reprogramming by ETV2 to enhance therapeutic approaches to develop mature vasculature for ischemic diseases. While ETV2 is an essential regulator of hematopoietic development, we did not define ETV2 as a pioneer factor for hematopoietic lineages and therefore we hypothesize that other co-factors and pioneer factors might facilitate this developmental process. For example, EBF1, PU.1 and C/EBPα regulate hematopoietic development and act as pioneer factors for the B cells, DN3 t cells and macrophages (115–122) (Table 1). Whether these factors are regulated by ETV2 early on or they act independently of ETV2 to regulate the development of hematopoietic lineages remains to be elucidated. Identifying this pioneer role for ETV2 has big implications for the development of regenerative therapies that aim to generate vasculature for ischemic diseases, particularly in the cardiovascular system (71, 112). Additionally, although not the focus of this review, the development of therapies that target pioneer factors in cancer will be very important. As ETV2 has been shown to have a role in cancer (82, 88, 98, 123), understanding whether its ability to remodel silent/compacted chromatin as a pioneer factor has important implications during angiogenesis, as therapeutic initiatives could target and inhibit ETV2 thereby impacting tumorigenesis (82, 98, 123). Additionally, it would be interesting to determine whether or not BRG1 or another chromatin remodeler also forms a complex with ETV2 in the context of cancer.
Conclusion(s)
More studies are emerging that claim to have characterized a novel pioneer transcription factor and this number will be expected to increase (8). This is in part due to the advances in molecular biology that facilitate the cellular characterization at the single cell level during embryogenesis. Further, these technologies will allow us to identify DNA binding sites for transcription factors (TF) and more importantly allow us to define the chromatin dynamics surrounding the DNA binding sites of such TFs. Importantly, the development of the Assay for Transposase-Accessible Chromatin followed by sequencing (ATAC-seq) allows for the definition of the chromatin landscape of differentiating or reprogramming cells using very few cells (50,000 cells or less) and support the claim that a TF is a pioneer factor (124, 125). While ATAC-seq characterization of cell populations can be insightful, we caution the reader that a more in-depth analysis is needed when assigning the role of pioneer factor. To designate a pioneer factor, three criteria need to be fulfilled: (1) pioneer factors need to bind nucleosomal DNA in vivo (sequencing) and in vitro (nucleosomal binding assay), (2) pioneer factors need to promote chromatin remodeling around DNA binding sites by themselves or by interacting with chromatin remodelers and (3) pioneer factors need to enact global epigenetic changes (i.e., demethylation) and recruit other co-factors that further promote the development or reprogramming of a cellular lineage (Figure 1; Table 1).
Further studies will be needed within the cardiovascular field to identify pioneer factors that regulate distinct cellular lineages and structures (i.e., first vs. second heart field) that comprise the four chambered organ. For example, while ETV2 sits at the top of the endothelial lineage developmental hierarchy, ISL1 and GATA4 are two of many regulators of the cardiac muscle lineage with very specific functions. We predict that multiple pioneer factors will be required to regulate cardiac muscle development and reprogramming. Other cellular lineages that were not discussed include: smooth muscle and cardiac fibroblasts as no pioneer factors have been identified for these lineages. Pioneer factors can be powerful tools for the development of regenerative therapies whose goal is to generate mature and functional cell lineages. Understanding the molecular mechanisms that drive lineage development by these and other pioneer factors within the cardiovascular lineage will be instrumental because coupling these pioneer factors along with chromatin remodelers and downstream targets genes can amplify the molecular effect needed to better develop regenerative therapies for cardiovascular disease.
Author contributions
JS-P and DG: conceived the review and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We are grateful for the support of the Department of Defense (Grant # 3002-11765-00089113).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Amini M, Zayeri F, Salehi M. Trend analysis of cardiovascular disease mortality, incidence, and mortality-to-incidence ratio: results from global burden of disease study 2017. BMC Public Health. (2021) 21:401. doi: 10.1186/s12889-021-10429-0
2. Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, et al. Global Burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J Am Coll Cardiol. (2020) 76:2982–3021.
3. Alraies MC, Eckman P. Adult heart transplant: indications and outcomes. J Thorac Dis. (2014) 6:1120–8. doi: 10.3978/j.issn.2072-1439.2014.06.44
4. Garry DJ, Masino AM, Naseem RH, Martin CM. Ponce de Leon's Fountain: stem cells and the regenerating heart. Am J Med Sci. (2005) 329:190–201. doi: 10.1097/00000441-200504000-00005
5. Aydin B, Mazzoni EO. Cell reprogramming: the many roads to success. Annu Rev Cell Dev Biol. (2019) 35:433–52. doi: 10.1146/annurev-cellbio-100818-125127
6. Wang H, Yang Y, Liu J, Qian L. Direct cell reprogramming: approaches, mechanisms and progress. Nat Rev Mol Cell Biol. (2021) 22:410–24. doi: 10.1038/s41580-021-00335-z
7. Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. (1988) 242:405–11. doi: 10.1126/science.3175662
8. Chan SS, Kyba M. What is a master regulator? J Stem Cell Res Ther. (2013) 3:114. doi: 10.4172/2157-7633.1000e114
9. Balsalobre A, Drouin J. Pioneer factors as master regulators of the epigenome and cell fate. Nat Rev Mol Cell Biol. (2022). doi: 10.1038/s41580-022-00464-z
10. Fishman MC, Olson EN. Parsing the heart: genetic modules for organ assembly. Cell. (1997) 91:153–6. doi: 10.1016/S0092-8674[00]80397-9
11. Olson EN, Srivastava D. Molecular pathways controlling heart development. Science. (1996) 272:671–6. doi: 10.1126/science.272.5262.671
12. Parmacek MS, Epstein JA. Pursuing cardiac progenitors: regeneration redux. Cell. (2005) 120:295–8. doi: 10.1016/j.cell.2005.01.025
13. Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. (2006) 126:1037–48. doi: 10.1016/j.cell.2006.09.003
14. Bruneau BG. The developmental genetics of congenital heart disease. Nature. (2008) 451:943–8. doi: 10.1038/nature06801
15. Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. (2010) 107:1428–44. doi: 10.1161/CIRCRESAHA.110.227405
16. Davis TL, Rebay I. Master regulators in development: views from the Drosophila retinal determination and mammalian pluripotency gene networks. Dev Biol. (2017) 421:93–107. doi: 10.1016/j.ydbio.2016.12.005
17. Vinogradova TV, Sverdlov ED. PDX1: a unique pancreatic master regulator constantly changes its functions during embryonic development and progression of pancreatic cancer. Biochemistry. (2017) 82:887–93. doi: 10.1134/S000629791708003X
18. Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, et al. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med. (2000) 6:568–72. doi: 10.1038/75050
19. Heller RS, Stoffers DA, Bock T, Svenstrup K, Jensen J, Horn T, et al. Improved glucose tolerance and acinar dysmorphogenesis by targeted expression of transcription factor PDX-1 to the exocrine pancreas. Diabetes. (2001) 50:1553–61. doi: 10.2337/diabetes.50.7.1553
20. Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. (1987) 51:987–1000. doi: 10.1016/0092-8674(87)90585-X
21. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. (2005) 122:947–56. doi: 10.1016/j.cell.2005.08.020
22. Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. (1996) 86:47–57. doi: 10.1016/S0092-8674(00)80076-8
23. Semenza GL. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev. (1998) 8:588–94. doi: 10.1016/S0959-437X(98)80016-6
24. Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. (2008) 3:69–84. doi: 10.1016/j.stem.2008.06.009
25. Zaret KS. Pioneer transcription factors initiating gene network changes. Annu Rev Genet. (2020) 54:367–85. doi: 10.1146/annurev-genet-030220-015007
26. Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. (2011) 25:2227–41. doi: 10.1101/gad.176826.111
27. Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. (1996) 10:1670–82. doi: 10.1101/gad.10.13.1670
28. McPherson CE, Shim EY, Friedman DS, Zaret KS. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. (1993) 75:387–98. doi: 10.1016/0092-8674(93)80079-T
29. Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. (2002) 9:279–89. doi: 10.1016/S1097-2765(02)00459-8
30. Iwafuchi-Doi M, Zaret KS. Cell fate control by pioneer transcription factors. Development. (2016) 143:1833–7. doi: 10.1242/dev.133900
31. Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, et al. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell. (2016) 62:79–91. doi: 10.1016/j.molcel.2016.03.001
32. Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. (2015) 161:555–68. doi: 10.1016/j.cell.2015.03.017
33. Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. (2012) 151:994–1004. doi: 10.1016/j.cell.2012.09.045
34. Mayran A, Khetchoumian K, Hariri F, Pastinen T, Gauthier Y, Balsalobre A, et al. Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat Genet. (2018) 50:259–69. doi: 10.1038/s41588-017-0035-2
35. Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L, et al. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science. (2019) 363:294–7. doi: 10.1126/science.aau0583
36. Lee QY, Mall M, Chanda S, Zhou B, Sharma KS, Schaukowitch K, et al. Pro-neuronal activity of Myod1 due to promiscuous binding to neuronal genes. Nat Cell Biol. (2020) 22:401–11. doi: 10.1038/s41556-020-0490-3
37. Gong W, Das S, Sierra-Pagan JE, Skie E, Dsouza N, Larson TA, et al. ETV2 functions as a pioneer factor to regulate and reprogram the endothelial lineage. Nat Cell Biol. (2022) 24:672–84. doi: 10.1038/s41556-022-00901-3
38. King HW, Klose RJ. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife. (2017) 6:e22631. doi: 10.7554/eLife.22631
39. Mayran A, Sochodolsky K, Khetchoumian K, Harris J, Gauthier Y, Bemmo A, et al. Pioneer and nonpioneer factor cooperation drives lineage specific chromatin opening. Nat Commun. (2019) 10:3807. doi: 10.1038/s41467-019-11791-9
40. Cenik BK, Shilatifard A. COMPASS and SWI/SNF complexes in development and disease. Nat Rev Genet. (2021) 22:38–58. doi: 10.1038/s41576-020-0278-0
41. Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. (2000) 6:1287–95. doi: 10.1016/S1097-2765(00)00127-1
42. Alexander JM, Hota SK, He D, Thomas S, Ho L, Pennacchio LA, et al. Brg1 modulates enhancer activation in mesoderm lineage commitment. Development. (2015) 142:1418–30. doi: 10.1242/dev.109496
43. Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. (2010) 466:62–7. doi: 10.1038/nature09130
44. Gao R, Liang X, Cheedipudi S, Cordero J, Jiang X, Zhang Q, et al. Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell Res. (2019) 29:486–501. doi: 10.1038/s41422-019-0168-1
45. Takaku M, Grimm SA, Shimbo T, Perera L, Menafra R, Stunnenberg HG, et al. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. (2016) 17:36. doi: 10.1186/s13059-016-0897-0
46. Ferretti E, Hadjantonakis AK. Mesoderm specification and diversification: from single cells to emergent tissues. Curr Opin Cell Biol. (2019) 61:110–6. doi: 10.1016/j.ceb.2019.07.012
47. Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. (2012) 74:41–68. doi: 10.1146/annurev-physiol-020911-153242
48. Tam PP, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev. (1997) 68:3–25. doi: 10.1016/S0925-4773(97)00123-8
49. Rasmussen TL, Kweon J, Diekmann MA, Belema-Bedada F, Song Q, Bowlin K, et al. ER71 directs mesodermal fate decisions during embryogenesis. Development. (2011) 138:4801–12. doi: 10.1242/dev.070912
50. Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc Natl Acad Sci. (2009) 106:814–9. doi: 10.1073/pnas.0807583106
51. Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. (2008) 2:497–507. doi: 10.1016/j.stem.2008.03.008
52. Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. (2000) 407:221–6. doi: 10.1038/35025190
53. Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D, et al. regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. (2009) 11:951–7. doi: 10.1038/ncb1906
54. Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. (2006) 127:1151–65. doi: 10.1016/j.cell.2006.10.029
55. Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. (2005) 433:647–53. doi: 10.1038/nature03215
56. Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. (2003) 5:877–89. doi: 10.1016/S1534-5807(03)00363-0
57. Oka T, Maillet M, Watt AJ, Schwartz RJ, Aronow BJ, Duncan SA, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. (2006) 98:837–45. doi: 10.1161/01.RES.0000215985.18538.c4
58. Bisping E, Ikeda S, Kong SW, Tarnavski O, Bodyak N, McMullen JR, et al. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci. (2006) 103:14471–6. doi: 10.1073/pnas.0602543103
59. Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, et al. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest. (2005) 115:1522–31. doi: 10.1172/JCI23769
60. Watt AJ, Battle MA Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci. (2004) 101:12573–8. doi: 10.1073/pnas.0400752101
61. Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. (1997) 11:1061–72. doi: 10.1101/gad.11.8.1061
62. Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. (1997) 11:1048–60. doi: 10.1101/gad.11.8.1048
63. Fu JD, Srivastava D. Direct reprogramming of fibroblasts into cardiomyocytes for cardiac regenerative medicine. Circ J. (2015) 79:245–54. doi: 10.1253/circj.CJ-14-1372
64. Nam YJ, Song K, Olson EN. Heart repair by cardiac reprogramming. Nat Med. (2013) 19:413–5. doi: 10.1038/nm.3147
65. Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. (2013) 1:235–47. doi: 10.1016/j.stemcr.2013.07.005
66. Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. (2012) 485:599–604. doi: 10.1038/nature11139
67. Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. (2010) 142:375–86. doi: 10.1016/j.cell.2010.07.002
68. Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. (2011) 475:386–9. doi: 10.1038/nature10116
69. Stone NR, Gifford CA, Thomas R, Pratt KJB, Samse-Knapp K, Mohamed TMA, et al. Context-specific transcription factor functions regulate epigenomic and transcriptional dynamics during cardiac reprogramming. Cell Stem Cell. (2019) 25:87–102 e9. doi: 10.1016/j.stem.2019.06.012
70. Singh BN, Sierra-Pagan JE, Gong W, Das S, Theisen JWM, Skie E, et al. ETV2 (Ets variant transcription factor 2)-Rhoj cascade regulates endothelial progenitor cell migration during embryogenesis. Arterioscler Thromb Vasc Biol. (2020) 40:2875–90. doi: 10.1161/ATVBAHA.120.314488
71. Das S, Koyano-Nakagawa N, Gafni O, Maeng G, Singh BN, Rasmussen T, et al. Generation of human endothelium in pig embryos deficient in ETV2. Nat Biotechnol. (2020) 38:297–302. doi: 10.1038/s41587-019-0373-y
72. Singh BN, Gong W, Das S, Theisen JWM, Sierra-Pagan JE, Yannopoulos D, et al. Etv2 transcriptionally regulates Yes1 and promotes cell proliferation during embryogenesis. Sci Rep. (2019) 9:9736. doi: 10.1038/s41598-019-45841-5
73. Singh BN, Tahara N, Kawakami Y, Das S, Koyano-Nakagawa N, Gong W, et al. Etv2-miR-130a-Jarid2 cascade regulates vascular patterning during embryogenesis. PLoS ONE. (2017) 12:e0189010. doi: 10.1371/journal.pone.0189010
74. Koyano-Nakagawa N, Garry DJ. Etv2 as an essential regulator of mesodermal lineage development. Cardiovasc Res. (2017) 113:1294–306. doi: 10.1093/cvr/cvx133
75. Garry DJ. Etv2 is a master regulator of hematoendothelial lineages. Trans Am Clin Climatol Assoc. (2016) 127:212–23.
76. Singh BN, Kawakami Y, Akiyama R, Rasmussen TL, Garry MG, Gong W, et al. The Etv2-miR-130a network regulates mesodermal specification. Cell Rep. (2015) 13:915–23. doi: 10.1016/j.celrep.2015.09.060
77. Koyano-Nakagawa N, Shi X, Rasmussen TL, Das S, Walter CA, Garry DJ. Feedback mechanisms regulate Ets variant 2 (Etv2) gene expression and hematoendothelial lineages. J Biol Chem. (2015) 290:28107–19. doi: 10.1074/jbc.M115.662197
78. Rasmussen TL, Martin CM, Walter CA, Shi X, Perlingeiro R, Koyano-Nakagawa N, et al. Etv2 rescues Flk1 mutant embryoid bodies. Genesis. (2013) 51:471–80. doi: 10.1002/dvg.22396
79. Koyano-Nakagawa N, Kweon J, Iacovino M, Shi X, Rasmussen TL, Borges L, et al. Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem Cells. (2012) 30:1611–23. doi: 10.1002/stem.1131
80. Choi K. ETS transcription factor ETV2/ER71/Etsrp in haematopoietic regeneration. Curr Opin Hematol. (2018) 25:253–8. doi: 10.1097/MOH.0000000000000430
81. Kim JY, Lee DH, Kim JK, Choi HS, Dwivedi B, Rupji M, et al. ETV2/ER71 regulates the generation of FLK1(+) cells from mouse embryonic stem cells through miR-126-MAPK signaling. Stem Cell Res Ther. (2019) 10:328. doi: 10.1186/s13287-019-1466-8
82. Lee TJ, Kang HK, Berry JC, Joo HG, Park C, Miller MJ, et al. ER71/ETV2 promotes hair regeneration from chemotherapeutic drug-induced hair loss by enhancing angiogenesis. Biomol Ther. (2021) 29:545–50. doi: 10.4062/biomolther.2021.022
83. Liu F, Li D, Yu YY, Kang I, Cha MJ, Kim JY, et al. Induction of hematopoietic and endothelial cell program orchestrated by ETS transcription factor ER71/ETV2. EMBO Rep. (2015) 16:654–69. doi: 10.15252/embr.201439939
84. Park C, Lee TJ, Bhang SH, Liu F, Nakamura R, Oladipupo SS, et al. Injury-Mediated Vascular Regeneration Requires Endothelial ER71/ETV2. Arterioscler Thromb Vasc Biol. (2016) 36:86–96. doi: 10.1161/ATVBAHA.115.306430
85. Xu CX, Lee TJ, Sakurai N, Krchma K, Liu F, Li D, et al. ETV2/ER71 regulates hematopoietic regeneration by promoting hematopoietic stem cell proliferation. J Exp Med. (2017) 214:1643–53. doi: 10.1084/jem.20160923
86. Zhao H, Choi K. A CRISPR screen identifies genes controlling Etv2 threshold expression in murine hemangiogenic fate commitment. Nat Commun. (2017) 8:541. doi: 10.1038/s41467-017-00667-5
87. Zhao H, Xu C, Lee TJ, Liu F, Choi K. ETS transcription factor ETV2/ER71/Etsrp in hematopoietic and vascular development, injury, and regeneration. Dev Dyn. (2017) 246:318–27. doi: 10.1002/dvdy.24483
88. Baltrunaite K, Craig MP, Palencia Desai S, Chaturvedi P, Pandey RN, Hegde RS, et al. ETS transcription factors Etv2 and Fli1b are required for tumor angiogenesis. Angiogenesis. (2017) 20:307–23. doi: 10.1007/s10456-017-9539-8
89. Casie Chetty S, Sumanas S. Ets1 functions partially redundantly with Etv2 to promote embryonic vasculogenesis and angiogenesis in zebrafish. Dev Biol. (2020) 465:11–22. doi: 10.1016/j.ydbio.2020.06.007
90. Craig MP, Grajevskaja V, Liao HK, Balciuniene J, Ekker SC, Park JS, et al. Etv2 and fli1b function together as key regulators of vasculogenesis and angiogenesis. Arterioscler Thromb Vasc Biol. (2015) 35:865–76. doi: 10.1161/ATVBAHA.114.304768
91. Palencia-Desai S, Kohli V, Kang J, Chi NC, Black BL, Sumanas S. Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development. (2011) 138:4721–32. doi: 10.1242/dev.064998
92. Sumanas S, Choi K. ETS Transcription Factor ETV2/ER71/Etsrp in hematopoietic and vascular development. Curr Top Dev Biol. (2016) 118:77–111. doi: 10.1016/bs.ctdb.2016.01.005
93. Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood. (2008) 111:4500–10. doi: 10.1182/blood-2007-09-110569
94. Wong KS, Proulx K, Rost MS, Sumanas S. Identification of vasculature-specific genes by microarray analysis of Etsrp/Etv2 overexpressing zebrafish embryos. Dev Dyn. (2009) 238:1836–50. doi: 10.1002/dvdy.21990
95. Chen PC, Hsueh YW, Lee YH, Tsai HW, Tsai KJ, Chiang PM, et al. primes angioblast formation by inducing ETV2 and LMO2 via FGFR1/BRAF/MEK/ERK. Cell Mol Life Sci. (2021) 78:2199–212. doi: 10.1007/s00018-020-03630-8
96. Chestnut B, Casie Chetty S, Koenig AL, Sumanas S. Single-cell transcriptomic analysis identifies the conversion of zebrafish Etv2-deficient vascular progenitors into skeletal muscle. Nat Commun. (2020) 11:2796. doi: 10.1038/s41467-020-16515-y
97. DiTacchio L, Bowles J, Shin S, Lim DS, Koopman P, Janknecht R. Transcription factors ER71/ETV2 and SOX9 participate in a positive feedback loop in fetal and adult mouse testis. J Biol Chem. (2012) 287:23657–66. doi: 10.1074/jbc.M111.320101
98. Kabir AU, Lee TJ, Pan H, Berry JC, Krchma K, Wu J, et al. Requisite endothelial reactivation and effective siRNA nanoparticle targeting of Etv2/Er71 in tumor angiogenesis. JCI Insight. (2018) 3:e97349. doi: 10.1172/jci.insight.97349
99. Kataoka H, Hayashi M, Kobayashi K, Ding G, Tanaka Y, Nishikawa S. Region-specific Etv2 ablation revealed the critical origin of hemogenic capacity from Hox6-positive caudal-lateral primitive mesoderm. Exp Hematol. (2013) 41:567–81 e9. doi: 10.1016/j.exphem.2013.02.009
100. Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, et al. Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRalpha+ primitive mesoderm. Blood. (2011) 118:6975–86. doi: 10.1182/blood-2011-05-352658
101. Kobayashi K, Ding G, Nishikawa S, Kataoka H. Role of Etv2-positive cells in the remodeling morphogenesis during vascular development. Genes Cells. (2013) 18:704–21. doi: 10.1111/gtc.12070
102. Lammerts van Bueren K, Black BL. Regulation of endothelial and hematopoietic development by the ETS transcription factor Etv2. Curr Opin Hematol. (2012) 19:199–205. doi: 10.1097/MOH.0b013e3283523e07
103. Moore JC, Sheppard-Tindell S, Shestopalov IA, Yamazoe S, Chen JK, Lawson ND. Post-transcriptional mechanisms contribute to Etv2 repression during vascular development. Dev Biol. (2013) 384:128–40. doi: 10.1016/j.ydbio.2013.08.028
104. Salanga MC, Meadows SM, Myers CT, Krieg PA. ETS family protein ETV2 is required for initiation of the endothelial lineage but not the hematopoietic lineage in the Xenopus embryo. Dev Dyn. (2010) 239:1178–87. doi: 10.1002/dvdy.22277
105. Veldman MB, Lin S. Etsrp/Etv2 is directly regulated by Foxc1a/b in the zebrafish angioblast. Circ Res. (2012) 110:220–9. doi: 10.1161/CIRCRESAHA.111.251298
106. Veldman MB, Zhao C, Gomez GA, Lindgren AG, Huang H, Yang H, et al. Transdifferentiation of fast skeletal muscle into functional endothelium in vivo by transcription factor Etv2. PLoS Biol. (2013) 11:e1001590. doi: 10.1371/journal.pbio.1001590
107. Wareing S, Eliades A, Lacaud G, Kouskoff V. ETV2 expression marks blood and endothelium precursors, including hemogenic endothelium, at the onset of blood development. Dev Dyn. (2012) 241:1454–64. doi: 10.1002/dvdy.23825
108. Yamamizu K, Matsunaga T, Katayama S, Kataoka H, Takayama N, Eto K, et al. PKA/CREB signaling triggers initiation of endothelial and hematopoietic cell differentiation via Etv2 induction. Stem Cells. (2012) 30:687–96. doi: 10.1002/stem.1041
109. Chan SS, Shi X, Toyama A, Arpke RW, Dandapat A, Iacovino M, et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell. (2013) 12:587–601. doi: 10.1016/j.stem.2013.03.004
110. Shi X, Zirbes KM, Rasmussen TL, Ferdous A, Garry MG, Koyano-Nakagawa N, et al. The transcription factor Mesp1 interacts with cAMP-responsive element binding protein 1 (Creb1) and coactivates Ets variant 2 (Etv2) gene expression. J Biol Chem. (2015) 290:9614–25. doi: 10.1074/jbc.M114.614628
111. Basel-Salmon L, Ruhrman-Shahar N, Barel O, Hagari O, Marek-Yagel D, Azulai N, et al. Biallelic variants in ETV2 in a family with congenital heart defects, vertebral abnormalities and preaxial polydactyly. Eur J Med Genet. (2021) 64:104124. doi: 10.1016/j.ejmg.2020.104124
112. Lee S, Park C, Han JW, Kim JY, Cho K, Kim EJ, et al. Direct reprogramming of human dermal fibroblasts into endothelial cells using ER71/ETV2. Circ Res. (2017) 120:848–61. doi: 10.1161/CIRCRESAHA.116.309833
113. De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. (2008) 135:1053–64. doi: 10.1016/j.cell.2008.10.049
114. Garry DJ, Sierra-Pagan JE. Mechanisms that Govern Endothelial Lineage Development and Vasculogenesis. Advanced Technologies in Cardiovascular Bioengineering. Cham: Springer (2022). p. 31–48. doi: 10.1007/978-3-030-86140-7_3
115. Minderjahn J, Schmidt A, Fuchs A, Schill R, Raithel J, Babina M, et al. Mechanisms governing the pioneering and redistribution capabilities of the non-classical pioneer PU. 1 Nat Commun. (2020) 11:402. doi: 10.1038/s41467-019-13960-2
116. Ungerback J, Hosokawa H, Wang X, Strid T, Williams BA, Sigvardsson M, et al. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU. 1). Genome Res. (2018) 28:1508–19. doi: 10.1101/gr.231423.117
117. Li R, Cauchy P, Ramamoorthy S, Boller S, Chavez L, Grosschedl R. Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev. (2018) 32:96–111. doi: 10.1101/gad.309583.117
118. Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, et al. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity. (2016) 44:527–41. doi: 10.1016/j.immuni.2016.02.021
119. van Oevelen C, Collombet S, Vicent G, Hoogenkamp M, Lepoivre C, Badeaux A, et al. C/EBPalpha activates pre-existing and De Novo macrophage enhancers during induced pre-B cell transdifferentiation and myelopoiesis. Stem Cell Reports. (2015) 5:232–47. doi: 10.1016/j.stemcr.2015.06.007
120. Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, et al. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell. (2014) 54:844–57. doi: 10.1016/j.molcel.2014.04.006
121. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. (2010) 38:576–89. doi: 10.1016/j.molcel.2010.05.004
122. Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, et al. PU. 1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. (2008) 105:6057–62. doi: 10.1073/pnas.0711961105
123. Zhao C, Gomez GA, Zhao Y, Yang Y, Cao D, Lu J, et al. ETV2 mediates endothelial transdifferentiation of glioblastoma. Signal Transduct Target Ther. (2018) 3:1–11. doi: 10.1038/s41392-018-0007-8
124. Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. (2015) 523:486–90. doi: 10.1038/nature14590
125. Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. (2015) 109:21. doi: 10.1002/0471142727.mb2129s109
126. Luo C, Lee QY, Wapinski O, Castanon R, Nery JR, Mall M, et al. Global DNA methylation remodeling during direct reprogramming of fibroblasts to neurons. Elife. (2019) 8:e40197. doi: 10.7554/eLife.40197
127. Raposo A, Vasconcelos FF, Drechsel D, Marie C, Johnston C, Dolle D, et al. Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. (2015) 10:1544–56. doi: 10.1016/j.celrep.2015.02.025
128. Wapinski OL, Lee QY, Chen AC Li R, Corces MR, Ang CE, et al. Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. (2017) 20:3236–47. doi: 10.1016/j.celrep.2017.09.011
129. Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. (2013) 155:621–35. doi: 10.1016/j.cell.2013.09.028
130. Carroll JS, Liu XS, Brodsky AS Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. (2005) 122:33–43. doi: 10.1016/j.cell.2005.05.008
131. Cernilogar FM, Hasenoder S, Wang Z, Scheibner K, Burtscher I, Sterr M, et al. Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. (2019) 47:9069–86. doi: 10.1093/nar/gkz627
132. Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the cover: location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. (2005) 102:11651–6. doi: 10.1073/pnas.0505575102
133. Lee K, Cho H, Rickert RW, Li QV, Pulecio J, Leslie CS, et al. FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep. (2019) 28:382–93 e7. doi: 10.1016/j.celrep.2019.06.034
134. Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. (2008) 132:958–70. doi: 10.1016/j.cell.2008.01.018
135. Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. (2011) 475:390–3. doi: 10.1038/nature10263
136. Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. (2011) 21:555–65. doi: 10.1101/gr.111534.110
137. Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. (2009) 138:245–56. doi: 10.1016/j.cell.2009.04.056
138. Charney RM, Forouzmand E, Cho JS, Cheung J, Paraiso KD, Yasuoka Y, et al. Foxh1 occupies cis-regulatory modules prior to dynamic transcription factor interactions controlling the mesendoderm gene program. Dev Cell. (2017) 40:595–607 e4. doi: 10.1016/j.devcel.2017.02.017
139. Jacobs J, Atkins M, Davie K, Imrichova H, Romanelli L, Christiaens V, et al. The transcription factor grainy head primes epithelial enhancers for spatiotemporal activation by displacing nucleosomes. Nat Genet. (2018) 50:1011–20. doi: 10.1038/s41588-018-0140-x
140. Nevil M, Gibson TJ, Bartolutti C, Iyengar A, Harrison MM. Establishment of chromatin accessibility by the conserved transcription factor Grainy head is developmentally regulated. Development. (2020) 147:dev185009. doi: 10.1242/dev.185009
141. Gaskill MM, Gibson TJ, Larson ED, Harrison MM. GAF is essential for zygotic genome activation and chromatin accessibility in the early Drosophila embryo. Elife. (2021) 10:e66668. doi: 10.7554/eLife.66668.sa2
142. Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, et al. Cooperative binding of transcription factors orchestrates reprogramming. Cell. (2017) 168:442–59 e20. doi: 10.1016/j.cell.2016.12.016
143. Budry L, Balsalobre A, Gauthier Y, Khetchoumian K, L'Honore A, Vallette S, et al. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. (2012) 26:2299–310. doi: 10.1101/gad.200436.112
144. Koromila T, Gao F, Iwasaki Y, He P, Pachter L, Gergen JP, et al. Odd-paired is a pioneer-like factor that coordinates with Zelda to control gene expression in embryos. Elife. (2020) 9:e59610. doi: 10.7554/eLife.59610.sa2
145. Soluri IV, Zumerling LM, Payan Parra OA, Clark EG, Blythe SA. Zygotic pioneer factor activity of odd-paired/Zic is necessary for late function of the Drosophila segmentation network. Elife. (2020) 9:e53916. doi: 10.7554/eLife.53916.sa2
146. Johnson JL, Georgakilas G, Petrovic J, Kurachi M, Cai S, Harly C, et al. Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T cells. Immunity. (2018) 48:243–57 e10. doi: 10.1016/j.immuni.2018.01.012
Keywords: pioneer factors, ETV2/ER71, ETS factors, cardiac regeneration, chromatin remodeling, reprogramming, epigenetics
Citation: Sierra-Pagan JE and Garry DJ (2022) The regulatory role of pioneer factors during cardiovascular lineage specification – A mini review. Front. Cardiovasc. Med. 9:972591. doi: 10.3389/fcvm.2022.972591
Received: 18 June 2022; Accepted: 03 August 2022;
Published: 23 August 2022.
Edited by:
Ajit Magadum, Temple University, United StatesReviewed by:
Robert Kelly, UMR7288 Institut de Biologie du Développement de Marseille (IBDM), FranceCopyright © 2022 Sierra-Pagan and Garry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel J. Garry, Z2FycnlAdW1uLmVkdQ==
 Javier E. Sierra-Pagan1
Javier E. Sierra-Pagan1 Daniel J. Garry
Daniel J. Garry