
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Cardiovasc. Med. , 14 September 2022
Sec. Cardiovascular Genetics and Systems Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fcvm.2022.959028
Background and aims: Macrophage migration inhibitory factor (MIF) gene rs755622 G/C polymorphism was suggested to be associated with CAD risk. However, due to the different results among the individual studies, no agreement has been reached till now. Therefore, the meta-analysis on the association of MIF gene rs755622 G/C polymorphism with CAD was performed.
Methods and results: The association between them was evaluated by calculating the pooled odds ratios (ORs) and the corresponding 95% confidence intervals (CIs). The random-effects models were used because of the significant heterogeneity among them. In this meta-analysis, 8,488 subjects from 9 studies were included. The MIF gene rs755622 G/C polymorphism was significantly associated with CAD under the allelic (OR: 1.213, 95% CI: 1.039–1.417, P = 0.014), recessive (OR: 1.945, 95% CI: 1.214–3.115, P = 0.006), dominant (OR: 0.781, 95% CI: 0.617–0.989, P = 0.041), homozygous (OR: 2.057, 95% CI: 1.289–3.284, P = 0.003), and additive (OR: 1.327, 95% CI: 1.081–1.630, P = 0.007) genetic models.
Conclusion: MIF gene rs755622 G/C polymorphism was significantly related to CAD, especially in the Chinese population. Persons with the C allele of the MIF gene rs755622 G/C polymorphism might be susceptible to CAD.
Coronary artery disease (CAD) is a serious disease that endangers human health and life. In developed Western countries, such as Europe and the United States, CAD has decreased incidence compared with 30 years ago but remains the main cause of death in the population (1). In China, with the improvement of people’s living standards, changes in diet structure and eating habits, and increased social pressure, the incidence and mortality of CAD increase. At present, the number of cardiovascular diseases in China has reached 330 million. In 2018, the crude mortality rate of CAD was 120.18/100,000 in urban areas and 128.24/100,000 in rural areas. The CAD has become a heavy burden on Chinese individuals and society (2).
The pathogenesis of CAD is the atherosclerosis of the coronary artery. Risk factors for CAD include smoking, hyperlipidemia, hypertension, and diabetes. Studies found that CAD is also a chronic inflammatory disease, and a variety of inflammatory factors play important roles in the occurrence and development of CAD. The macrophage migration inhibitor (MIF) is a homologous trimer protein that can locally inhibit macrophage migration and promote the inflammatory response. MIF can be expressed in many cells and tissues, including mononuclear phagocytes, vascular smooth muscle cells, and cardiomyocytes (3). MIF is an autocrine factor of macrophages, which plays an important role in inflammation and is a key mediator of arteriosclerosis (4).
The human MIF gene, located in 21q11.2, spans about 2,119 bp, contains 3 exons and 2 introns, and encodes 115 amino acids. The MIF gene –173G/C polymorphism (rs755622) is located in the promoter, which is located at the binding site of the response element of transcription factor activator protein 4 (AP4). This rs755622 polymorphism may affect the affinity of the response element of AP4, thus affecting the transcription level of the MIF gene (5). The MIF gene polymorphism and increased MIF protein concentration are associated with the incidence of ulcerative colitis, psoriasis, and tuberculosis. These diseases are often accompanied by inflammatory reactions, which illustrate the key role of MIF in promoting the occurrence and development of these diseases (3).
Many studies suggested that the MIF gene –173 G/C polymorphism is associated with the risk of CAD, but some studies confirmed no association between MIF gene –173 G/C polymorphism and CAD. Ji et al. found that patients with CAD have higher frequencies of C and –173CC genotypes than the control. The MIF gene rs755622 polymorphism is suggested to be related to CAD in a Chinese population, and the plasma MIF level is a predictor of plaque stability (6). Similarly, Fahriye et al. found that the low frequency of the GG genotype and G allele of the MIF gene –173 G/C polymorphism may be associated with the etiopathogenesis of patients with CAD in a Turkish population (7). Nevertheless, Tereshchenko et al. found no significant difference in the distribution of MIF gene –173G/C genotypes between CAD and control groups in Czech or Russian populations (8). Additionally, Wu et al. reported that the –173G/C polymorphism of the MIF gene does not contribute to CAD risk in another Chinese population (9).
Recently, similar meta-analyses on the association of MIF gene –173G/C polymorphism and CAD have been published. Li et al. carried out a meta-analysis to evaluate the association of MIF gene –173 G/C polymorphism and CAD susceptibility. Evidence of associations between them in Asian and Caucasian populations is also found. However, only six individual studies are included in their meta-analysis. Therefore, Li et al.’s meta-analysis results may not be as comprehensive as those obtained from the current meta-analysis (10). Lian et al. published a meta-analysis to investigate the relationship between them and also found that the MIF gene promoter –173 locus is significantly correlated with CAD risk and that the C allele is a risk factor for CAD (11). However, two studies published by a similar author group are included in their meta-analysis (12, 13). The adopted data may overlap, thus affecting the credibility of the results. Hence, Lian’s results may not be as objective as those from the current meta-analysis.
In this study, case–control studies on the correlation between MIF promoter region –173C/G polymorphism and CAD risk are collected, and systematic analysis is conducted to understand the association between CAD risk and MIF promoter region –173C/G polymorphism (Supplementary Table 1).
The relevant studies in the literature were searched from electronic databases such as the China National Knowledge Infrastructure, WanFang database, Weipu database, Web of Science, and PubMed. In the search process, keywords as “CAD,” “coronary heart disease,” “macrophage migration inhibitory factor,” “MIF,” “rs755622,” “gene,” “mutation,” and “polymorphism” were used. The final search was updated on 31 May 2022. The qualified studies were published between 2006 and 2018.
The included studies should conform to the subsequent criteria. The included studies should (a) evaluate the association of MIF gene rs755622 G/C polymorphism with CAD, and (b) the diagnosis of CAD was defined as the stenosis degree of the left main trunk, left anterior descending branch, left circumflex branch, right coronary artery, and any one of the main branches ≥ 50% by coronary angiography examination. (c) The research should be case-control or cohort studies. If multiple studies were retrieved by one author group, the data would be analyzed. If there was overlapped data, the data could be merely adopted for one time. The data with more sample size would be adopted. The studies that did not provide enough data should not be analyzed. The selected studies’ quality would be evaluated by using the Newcastle-Ottawa Scale (NOS) score.
The data extraction process was carried out by three researchers following the standard procedures. Two investigators completed literature retrieval, quality evaluation, and data extraction independently. If the two investigators have differences, they will discuss them with the third investigator. However, this did not often occur and had less effect on the overall NOS score. The obtained literature was then scored according to the NOS score evaluation system. According to the scoring rules, the score of the literature was between 0 and 9 points. Two independent researchers graded strictly according to the grading guidelines. A literature score greater than or equal to 6 is considered high quality. Data that met the inclusion criteria were extracted from the literature, and data extraction tables were developed. The table includes the first author, publication year, study region, race, number of genotypes in the CAD group and control group, genotyping method, the sample size in each group, and P-value of Hardy-Weinberg equilibrium (HWE) in the control group. The population in the case group might be affected by disease pressure (a type of selection pressure), which would make its genotype distribution deviate from HWE. However, the genotype distribution of the control group without disease should be consistent with HWE. Therefore, in the meta-analysis of genetic association studies, it is necessary to test whether the genotype distribution of the control group in the case-control study is consistent with HWE.
In this study, there were six genetic models, namely, allelic (C allele distribution frequency), recessive (CC vs. GG + GC), dominant (GG vs. GC + CC), heterozygous (GC vs.GG), homozygous (CC vs. GG), and additive (C allele vs. G allele). The pooled odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were adopted to assess the association between MIF gene rs755622 G/C polymorphism and CAD.
The Pheterogeneity and I2 were calculated to assess the heterogeneity among the included studies (14). On the condition of Pheterogeneity < 0.05, the random-effects models would be utilized to analyze the data. Conversely, the data analysis would be performed by using the fixed effects models (15). Z-test was used to calculate the pooled ORs under the six genetic models. The open-source software R was used to perform Fisher’s exact test and evaluate the HWE (16). The publication bias was evaluated by using Begg’s test (17, 18). P < 0.05 was considered statistically significant difference in the above tests. All data were analyzed using Stata12.0 software.
There were nine studies in the meta-analysis. The NOS scores were greater than 6 in all the included studies, which suggested the included literature qualities were high. In total, 3,843 CAD cases and 4,645 controls were identified according to the evaluation indexes (Table 1) (3, 6–9, 12, 19–21). Of the nine studies, seven were concentrated on the Chinese population (3, 6, 9, 12, 19–21), while the other two studies were concentrated on Caucasians (7, 8).
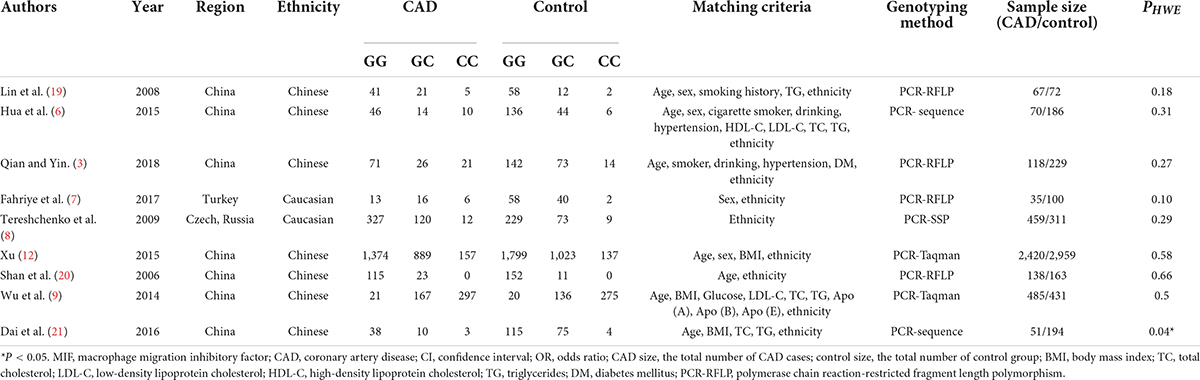
Table 1. Characteristics of the studies of the association between the MIF gene rs755622 polymorphism and CAD included in the meta-analysis.
A total of 20 studies in the literature were screened after a preliminary literature search. Among them, nine were eligible for the current meta-analysis. One paper deviating from the HWE was also included because the sensitivity analysis would be performed subsequently (21). Three excluded papers were published by the same author group to the study published by Luo et al. (13), Luo et al. (22), and Du et al. (23). Considering some data was overlapped among these studies, only Xu’s study was included because the sample size in this study was the largest of all of the four studies by the same author group. Five review literature studies were rejected, and three literature studies were excluded because they were not associated with either the MIF gene rs755622 G/C polymorphism or CAD (Supplementary Table 2).
A total population of 9 articles and 8,488 participants contributed to those results. The Chinese subgroup reflected 7 articles and 7,583 participants. The Caucasian subgroup reflected 2 articles and 905 participants. In the Caucasian subgroup analysis, Russian, Czech, and Turkish populations were included. Moreover, the Turkish study is a new addition to the previous study by Li et al. (10), and it appeared to change the results significantly in comparison to the findings of Li.
In the total population, the MIF gene rs755622 G/C polymorphism was significantly associated with CAD under the allelic (OR: 1.213, 95% CI: 1.039–1.417, P = 0.014), recessive (OR: 1.945, 95% CI: 1.214–3.115, P = 0.006), dominant (OR: 0.781, 95% CI: 0.617–0.989, P = 0.041), homozygous (OR: 2.057, 95% CI: 1.289–3.284, P = 0.003), and additive (OR: 1.327, 95% CI: 1.081–1.630, P = 0.007) genetic models. No significant relation between them was found under the heterozygous (OR: 1.146, 95% CI: 0.818–1.496, P = 0.316) genetic model (Table 2 and Figures 1–6).
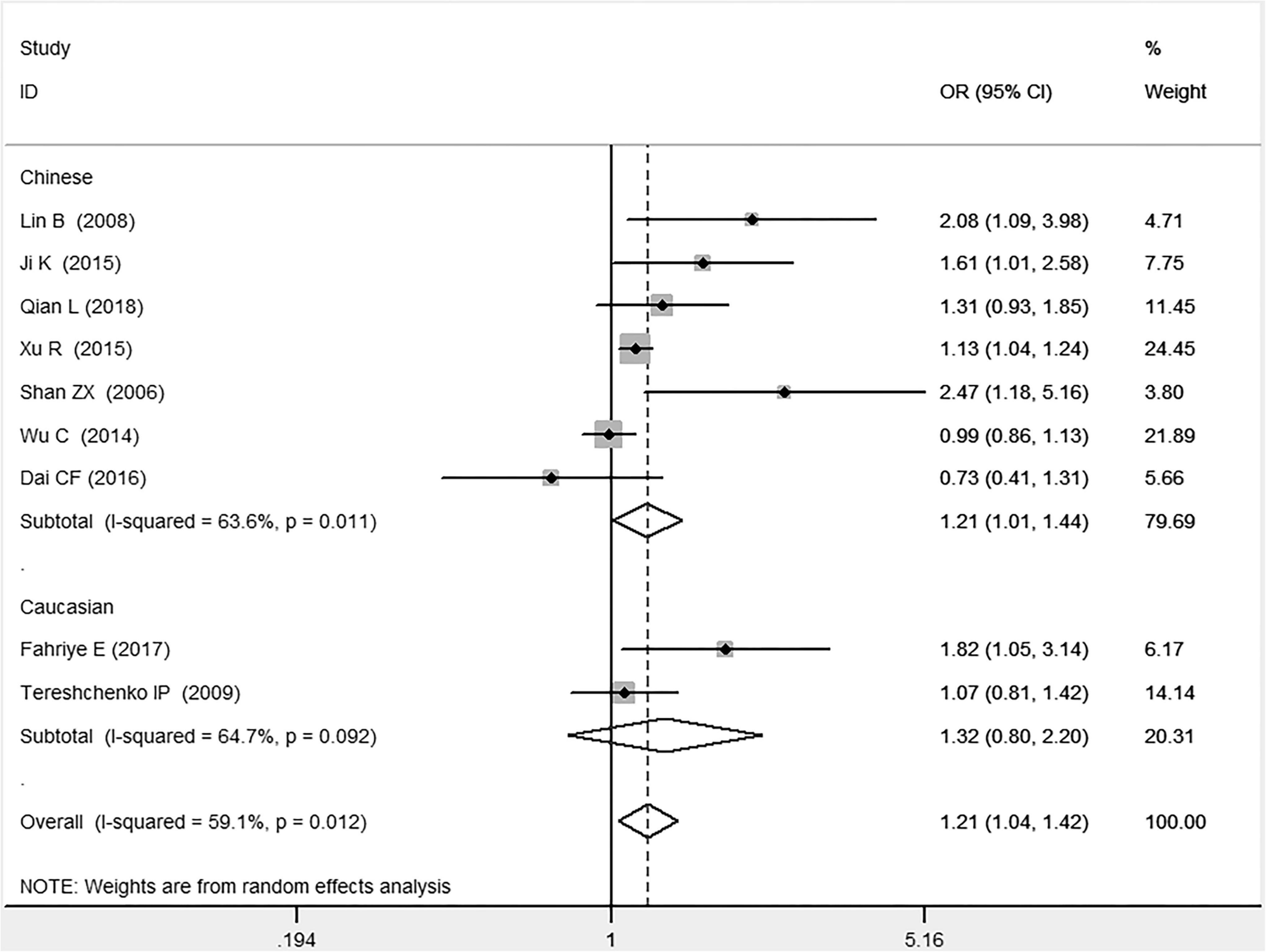
Figure 1. Forest plot of CAD associated with MIF gene rs755622 G/C polymorphism under an allelic genetic model (i.e., C allele vs. G allele + C allele of MIF gene rs755622 G/C polymorphism).
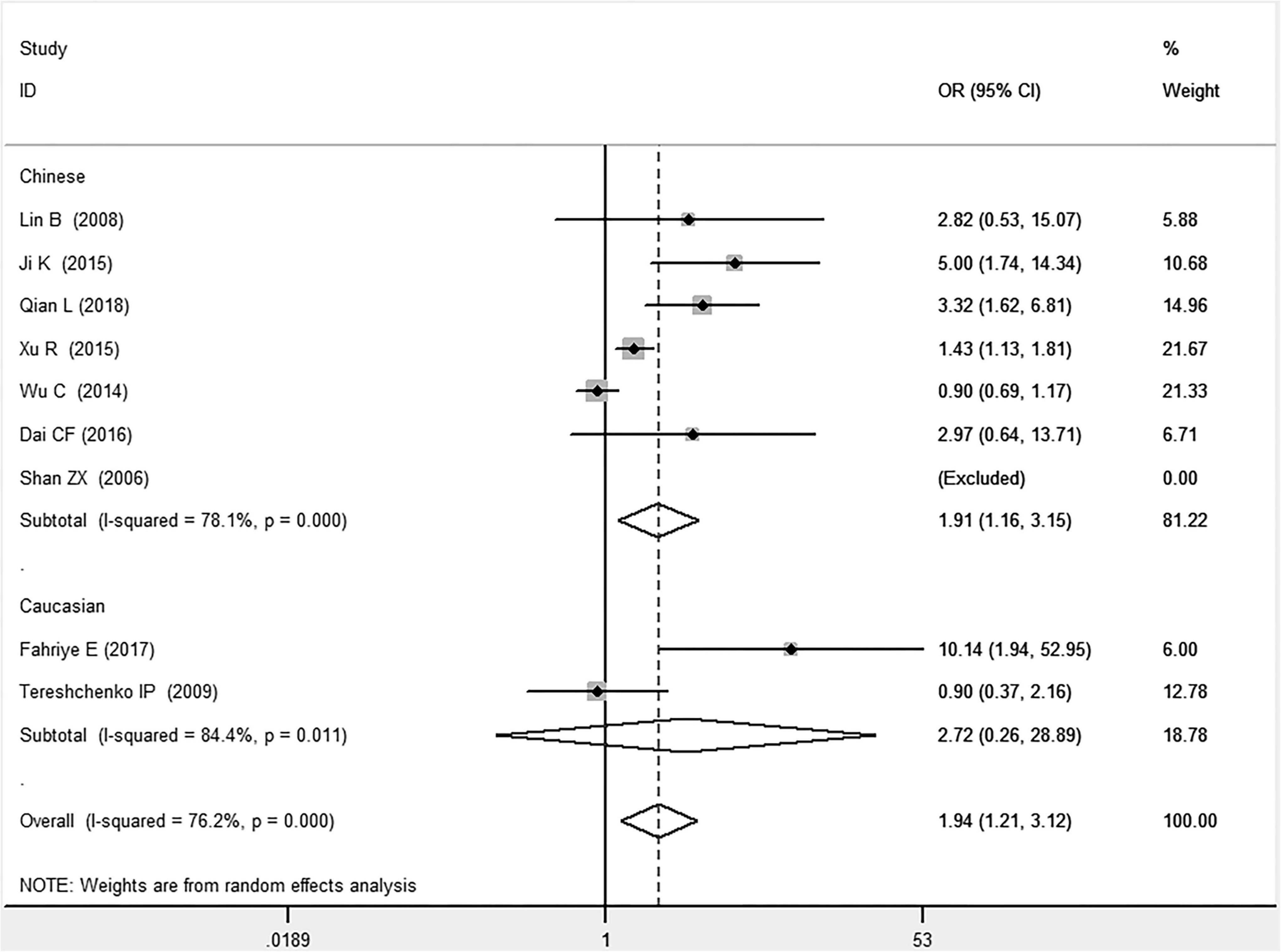
Figure 2. Forest plot of CAD associated with MIF gene rs755622 G/C polymorphism under a recessive genetic model (i.e., CC vs. GG + GC of MIF gene rs755622 G/C polymorphism).
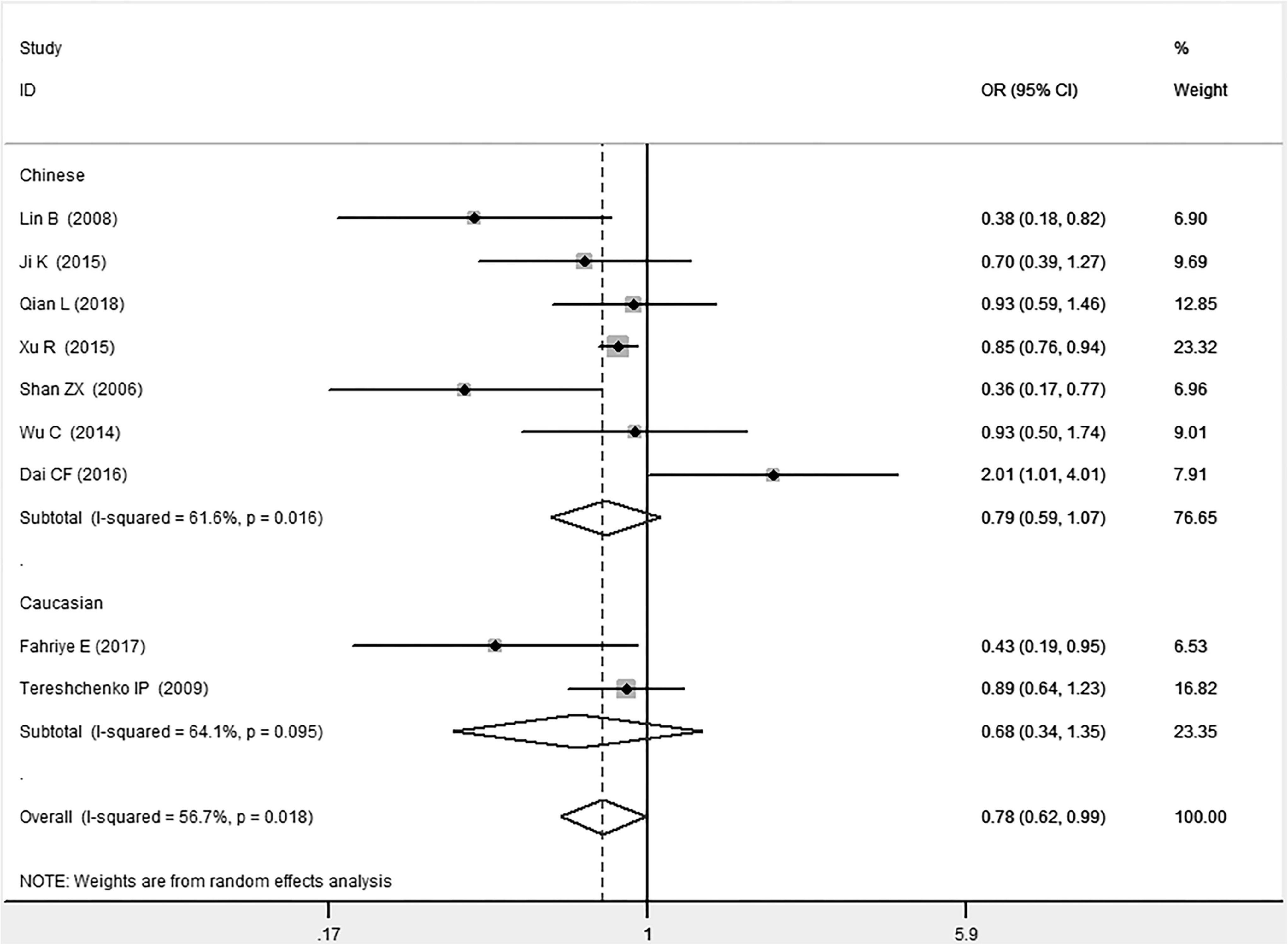
Figure 3. Forest plot of CAD associated with MIF gene rs755622 G/C polymorphism under a dominant genetic model (i.e., GG vs. GC + CC of MIF gene rs755622 G/C polymorphism).
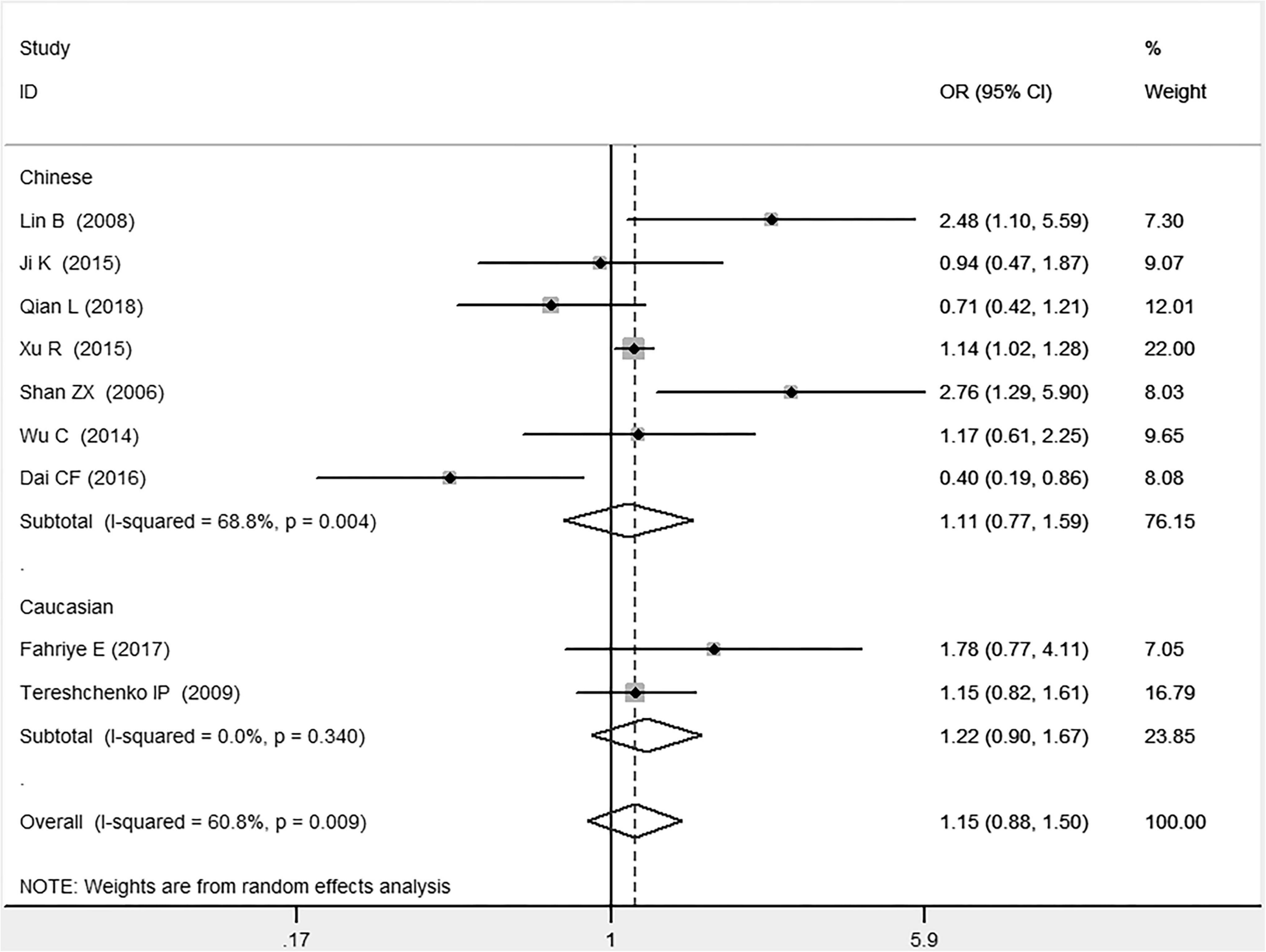
Figure 4. Forest plot of CAD associated with MIF gene rs755622 G/C polymorphism under a heterozygous genetic model (i.e., GC vs. GG of MIF gene rs755622 G/C polymorphism).
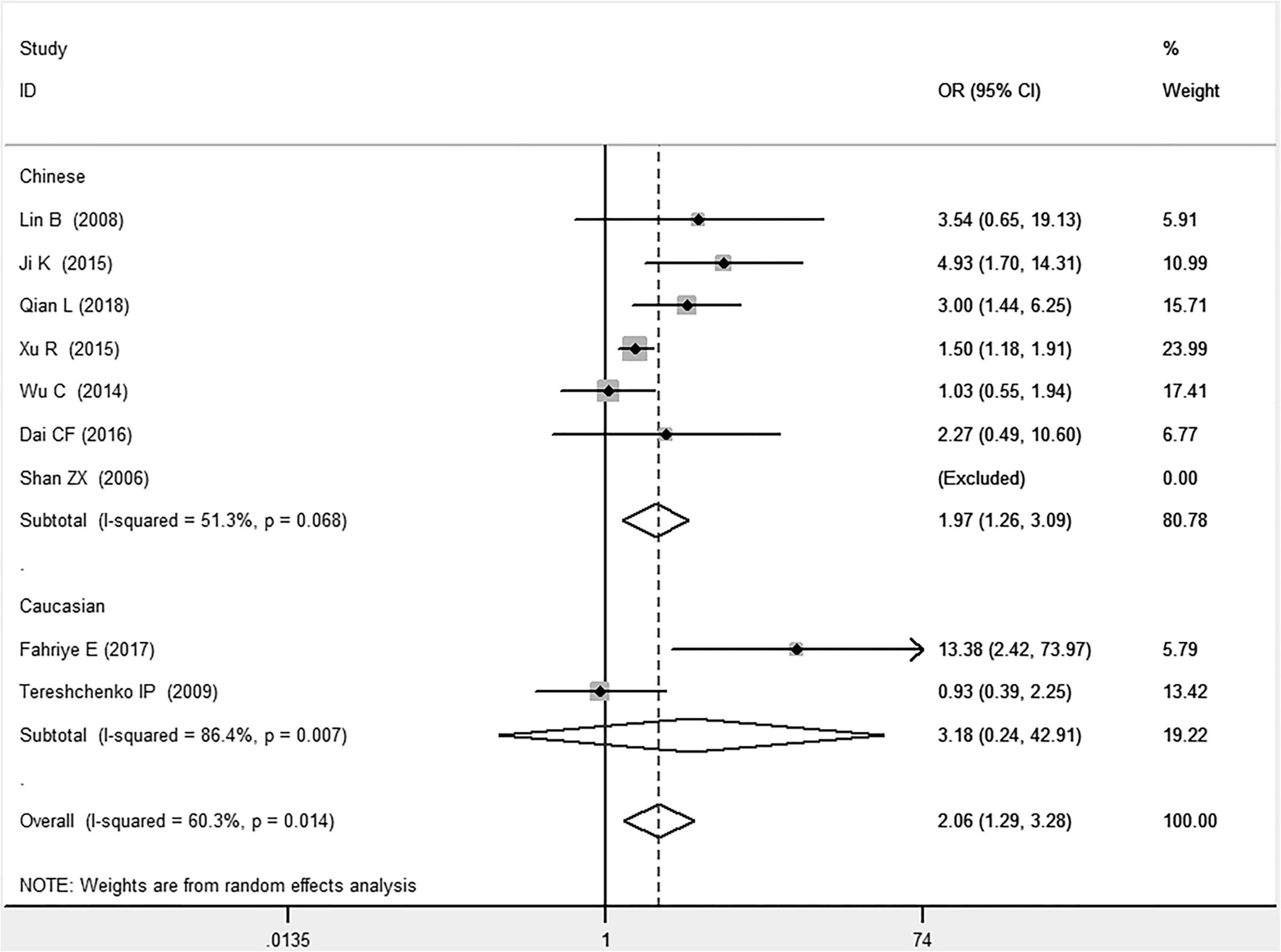
Figure 5. Forest plot of CAD associated with MIF gene rs755622 G/C polymorphism under a homozygous genetic model (i.e., CC vs. GG of MIF gene rs755622 G/C polymorphism).

Figure 6. Forest plot of CAD associated with MIF gene rs755622 G/C polymorphism under an additive genetic model (i.e., C allele vs. G allele of MIF gene rs755622 G/C polymorphism).
In the Chinese subgroup, a significant association between MIF gene rs755622 G/C polymorphism and CAD was observed under the allelic (OR: 1.207, 95% CI: 1.009–1.443, P = 0.039), recessive (OR: 1.912, 95% CI: 1.160–3.152, P = 0.011), homozygous (OR: 1.971, 95% CI: 1.259–3.086, P = 0.003), and additive (OR: 1.306, 95% CI: 1.030–1.657, P = 0.028) genetic models. There was no significant association between them under the dominant (OR: 0.793, 95% CI: 0.586–1.073, P = 0.132) or heterozygous (OR: 1.106, 95% CI: 0.768–1.592, P = 0.588) genetic models.
In the Caucasian subgroup, no significant association between them was observed under the allelic (OR: 1.323, 95% CI: 0.797–2.195, P = 0.279), recessive (OR: 2.719, 95% CI: 0.256–28.891, P = 0.407), dominant (OR: 0.676, 95% CI: 0.339–1.349, P = 0.267), heterozygous (OR: 1.224, 95% CI: 0.896–1.672, P = 0.205), homozygous (OR: 3.183, 95% CI: 0.236–42.908, P = 0.383), or additive (OR: 1.534, 95% CI: 0.719–3.272, P = 0.269) genetic models.
As there was significant heterogeneity under all of the genetic models (Pheterogeneity < 0.05), the random-effects models were utilized. After the population was stratified according to ethnicity, the heterogeneity was reduced in the subgroups under the heterozygous and the homozygous genetic models. Under the heterozygous genetic model, the I2 was reduced from 60.8% in the total population to 0 in the Caucasian subgroup. Under the homozygous genetic model, the I2 was reduced from 60.3% in the total population to 51.3% in the Chinese subgroup. It was indicated that the different ethnicity was associated with the heterogeneity source. In general, if Pheterogeneity is large, I2 must be small. The smaller Pheterogeneity is, the greater the heterogeneity is, and I2 represents how big it is. In Table 2, under the allelic, recessive, dominant, and additive genetic models, it was found that the Pheterogeneity value has the same change trend with the I2 in the Caucasian subgroup. In the heterogeneity test, the results of the Q-test are inconsistent with those of the H and I2-test, which is due to the small number of studies in the Caucasian subgroup, and the Q-test and interval range estimation are often inaccurate (24). Q-value changes with the change in the number of studies. H and I2-test statistics are often used to correct the influence of the study’s number on the Q-value. Their value would not change with the study’s number, and the heterogeneity test results are more robust and reliable.
No significant publication bias was observed through the Begg’s test under the allelic (T = 2.18, P = 0.066), dominant (T = –0.04, P = 0.973), heterozygous (T = 0.87, P = 0.414), homozygous (T = 1.89, P = 0.107), or additive genetic models (T = 1.93, P = 0.095) (Figure 7). It was suggested that there was no significant publication bias in the current meta-analysis under these genetic models. A significant publication bias was only observed under the recessive genetic models (T = 2.59, P = 0.041).
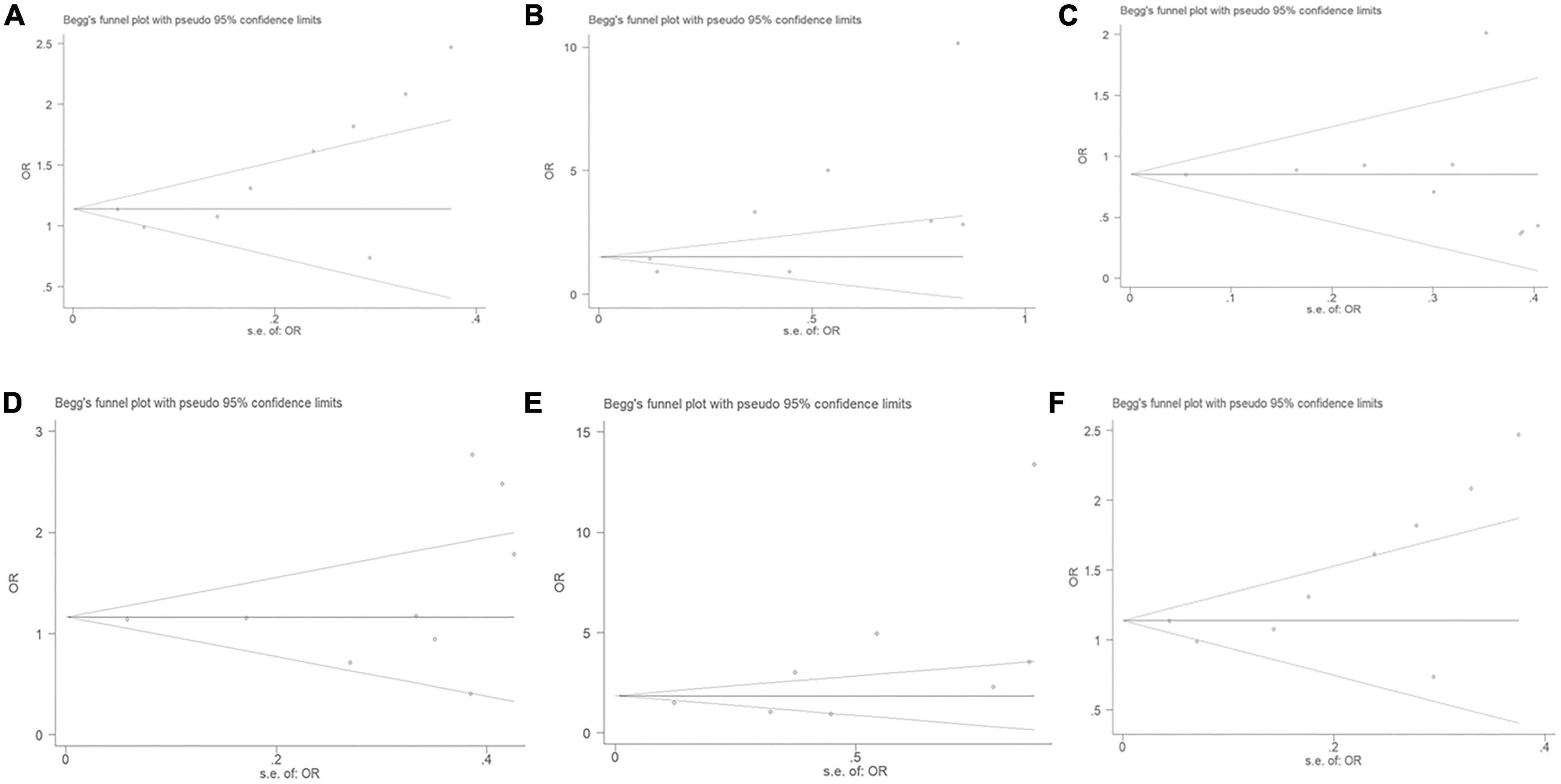
Figure 7. The Begg’s funnel plot for studies of the association of CAD and MIF gene rs755622 G/C polymorphism. (A) The Begg’s funnel plot for studies of the association under an allelic genetic model (i.e., C allele vs. G allele + C allele of MIF gene rs755622 G/C polymorphism). (B) The Begg’s funnel plot for studies of the association under a recessive genetic model (i.e., CC vs. GG + GC of MIF gene rs755622 G/C polymorphism). (C) The Begg’s funnel plot for studies of the association under a dominant genetic model (i.e., GG vs. GC + CC of MIF gene rs755622 G/C polymorphism). (D) The Begg’s funnel plot for studies of the association under a heterozygous genetic model (i.e., GC vs. GG of MIF gene rs755622 G/C polymorphism). (E) The Begg’s funnel plot for studies of the association under a homozygous genetic model (i.e., CC vs. GG of MIF gene rs755622 G/C polymorphism). (F) The Begg’s funnel plot for studies of the association under an additive genetic model (i.e., C allele vs. G allele of MIF gene rs755622 G/C polymorphism). The horizontal and vertical axes correspond to the OR on log scale and SE(logOR), respectively. OR, odds ratio; SE, standard error.
The sensitivity analysis has been performed under the six genetic models to analyze the results’ stability. It was observed that even if any of the individual studies were eliminated from this meta-analysis, the primary outcome was not influenced. It was suggested that the present meta-analysis’s results were considerably stable (Figure 8).
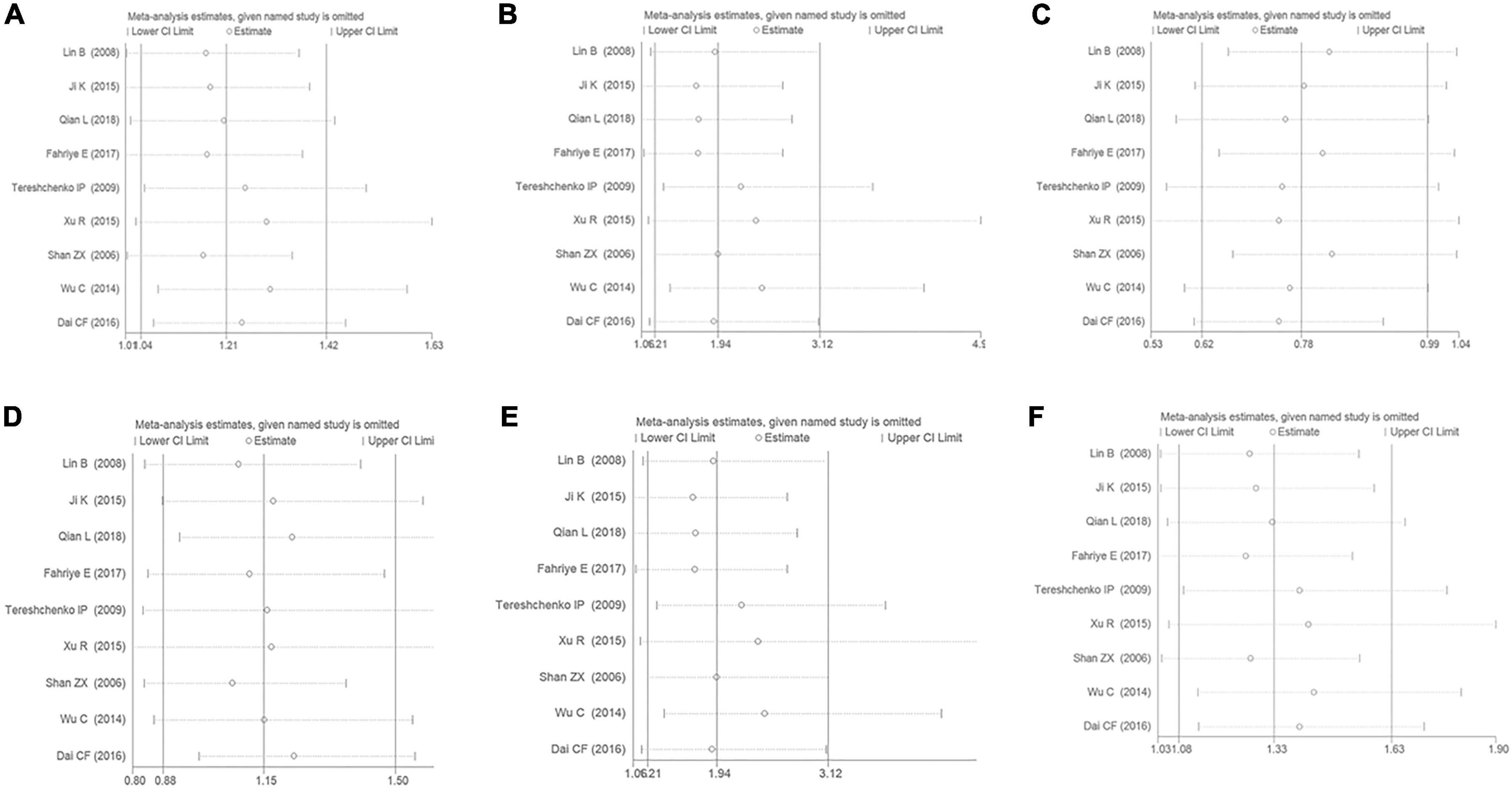
Figure 8. The sensitivity analysis on the association of CAD and MIF gene rs755622 G/C polymorphism. (A) The sensitivity analysis on the association under an allelic genetic model (i.e., C allele vs. G allele + C allele of MIF gene rs755622 G/C polymorphism). (B) The sensitivity analysis on the association under a recessive genetic model (i.e., CC vs. GG + GC of MIF gene rs755622 G/C polymorphism). (C) The sensitivity analysis on the association under a dominant genetic model (i.e., GG vs. GC + CC of MIF gene rs755622 G/C polymorphism). (D) The sensitivity analysis on the association under a heterozygous genetic model (i.e., GC vs. GG of MIF gene rs755622 G/C polymorphism). (E) The sensitivity analysis on the association under a homozygous genetic model (i.e., CC vs. GG of MIF gene rs755622 G/C polymorphism). (F) The sensitivity analysis on the association under an additive genetic model (i.e., C allele vs. G allele of MIF gene rs755622 G/C polymorphism).
In the current meta-analysis, the MIF gene rs755622 G/C polymorphism is found to be remarkably associated with the development of CAD under allelic (OR: 1.213), recessive (OR: 1.945), dominant (OR: 0.781), homozygous (OR: 2.057), and additive (OR: 1.327) genetic models. The C allele of the MIF gene –173G/C polymorphism may increase the CAD risk. The significant association between MIF gene rs755622 G/C polymorphism and CAD is found in the Chinese subgroup (P < 0.05) but not in the Caucasian subgroup (P > 0.05). Given that only two individual studies are included in the Caucasian subgroup, the result in this subgroup needs to be confirmed in more studies in the future. Only the random-effects models are used because of the significant heterogeneity among individual studies. Heterogeneity is distinctly reduced in the subgroup analysis under heterozygous and homozygous genetic models. The heterogeneity source is suggested to be related to different ethnicities.
CAD is a complex multifactorial disease resulting from the interaction of genetic and environmental factors. CAD is common in the elderly, and the formation of atherosclerotic plaque in the coronary artery is the main cause of CAD. Atherosclerosis is a disease of fat deposition, and inflammation plays a key role in the formation and development of atherosclerotic thrombosis. Many inflammatory factors, chemokines, and enzymes are involved in the pathogenesis of atherosclerosis (25).
MIF was discovered by Bloom and Bennett in a hypersensitivity experiment on guinea pigs in 1966 and was named for its ability to inhibit macrophage mobility (26). Later studies found that MIF can participate in a series of stress responses of the host to microbial infection; activate macrophages; inhibit migration; and enhance adhesion, phagocytosis, and tumor-killing activity (27–29). MIF is widely expressed in T cells, mononuclear macrophages, various hematopoietic cells, central and peripheral nerve cells, renal tubular epithelial cells, fat cells, vascular endothelial cells, liver cells, lung, prostate, and testis. MIF may be involved in various physiological and pathological processes of the body, including inflammatory response, immune response, tumor genesis, and tissue damage and repair (30).
When atherosclerotic plaques occur, serum MIF levels increase rapidly, suggesting that MIF may be involved in the occurrence of atherosclerosis. MIF can be rapidly released from macrophages, cardiomyocytes, and vascular endothelial cells under the stimulation of harmful factors (31). The released MIF can promote the transformation of macrophages into foam cells and accelerate the progression of atherosclerosis (32). As an important proinflammatory factor and chemokine, MIF can recruit macrophages, T lymphocytes, and other inflammatory immune cells to participate in the inflammatory response in atherosclerotic plaque (33, 34) and stimulate macrophages and lymphocytes to secrete inflammatory factors, including interleukin-6, interleukin-8, tumor necrosis factor-α, and intracellular adhesion factor. MIF can remarkably improve the level of inflammatory response in atherosclerotic plaques (35). Hence, considering the current research results, MIF may be a biomarker for the early diagnosis of CAD and a new target for the treatment of CAD, especially in the Chinese population.
MIF gene has multiple polymorphic loci, including –173G/C, 254T/C, and 656C/G. Among these polymorphic loci, –173G/C polymorphism is closely related to CAD. Given that the MIF gene –173 C allele has an AP4 response element-binding site, the difference in the mononucleotide may result in different affinities with the AP4 response element, thus affecting the MIF expression level. Donn et al. found that healthy volunteers with the –173C genotype (GC + CC) have significantly higher serum MIF levels than those with the –173GG genotype (36). Hence, the MIF gene –173G/C mutation can lead to a high MIF level and promote CAD development, which is in line with the outcome of the present meta-analysis.
In the meta-analysis by Li et al. (10), OR values are larger under the C/G, CC/GG, and CC/CG + GG genetic models than the results obtained from the current meta-analysis. Under the CG/CC and GG/CG + CC genetic models, the OR values in their meta-analysis are smaller than the current result. Although similar methodological techniques are used in Li’s paper, the current work has added value. A total of 8,488 participants analyzed herein compared with the 2,736 participants evaluated in Li mitigate that concern of value-added. In addition, five of six individual studies in Li’s meta-analysis are also included herein. The study of Luo et al. (13) is excluded due to replication of the study by Xu (12), which is included in the current research. In another meta-analysis by Lian et al. (11), two individual studies published by a similar author group are included in their meta-analysis (12, 13). Despite the similar method used and similar effect size obtained in their meta-analysis and the current research, overlapping data may affect the results.
Limitations are present in this study. The sample size is relatively small. The data on the Caucasian population are especially limited. More studies from a large sample size of Caucasian populations are needed to cover the shortage. In addition, the environmental factors of hyperlipidemia, hypertension, and diabetes also have significant effects on CAD susceptibility. Other gene polymorphisms, e.g., PCSK9 gene E670G polymorphism (37), ALDH2 gene G487A polymorphism (38), CDKN2B-AS1 gene rs4977574 A/G polymorphism (39), and FVII gene R353Q polymorphism (40), may also be involved with CAD susceptibility.
The MIF gene –173C/G polymorphism is associated with CAD risk. The C allele of the MIF gene –173C/G polymorphism may be the risk factor for CAD, especially in the Chinese population. More studies on the relationship between MIF gene –173C/G polymorphism and CAD need to be conducted to further verify this conclusion. MIF gene expression studies are necessary for confirmation.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Y-YL and HW researched the data. Y-YL wrote the manuscript, researched the data, contributed to the discussion, and reviewed/edited the manuscript. Y-YL and Y-YZ reviewed/edited the manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the National Natural Science Foundation of China (NSFC 81100073 to Y-YL), the Excellent Young and Middle-Aged Teachers Assistance Program of Nanjing Medical University for Y-YL (2013–2015, JX2161015034), the Jiangsu Overseas Research and Training Program for University Prominent Young and Middle-aged Teachers and Presidents, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). This study was also funded by the Natural Science Foundation of Jiangsu Province (BK 2012648 to HW) and “Six Talent Peaks” project in Jiangsu Province (2015-WSN-033).
We thank all our colleagues working in The First Affiliated Hospital of Nanjing Medical University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.959028/full#supplementary-material
Supplementary Table 1 | PRISMA checklist.
Supplementary Table 2 | PRISMA flow diagram.
1. Sheridan SL, Draeger LB, Pignone MP, Sloane PD, Samuel-Hodge C, Finkelstein EA, et al. Designing and implementing a comparative effectiveness study of two strategies for delivering high quality CHD prevention: methods and participant characteristics for the Heart to Health study. Contemp Clin Trials. (2013) 36:394–405. doi: 10.1016/j.cct.2013.07.013
2. The Writing Committee of the Report on Cardiovascular Health and Diseases in China. Report on cardiovascular health and diseases Burden in China: an updated summary of 2020. Chine Circu J. (2021) 36: 521–45.
3. Qian L, Yin RP. Macrophage migration inhibitory factor promoter polymorphisms (–173G/C) : relationship with coronary atherosclerotic disease subjects. J Med Res. (2018) 47:32–5.
4. Lin B. Association of MIF-173 Single Nucleotide Polymorphism with Coronary Heart Disease in Chinese Population. Master’s thesis. Zhejiang: Zhejiang University (2008). In Chinese.
5. Fei BY, Lv HX, Yang JM, Ye ZY. Association of MIF-173 gene polymorphism with inflammatory bowel disease in Chinese Han population. Cytokine. (2008) 41:44–7. doi: 10.1016/j.cyto.2007.10.010
6. Ji K, Wang X, Li J, Lu Q, Wang G, Xue Y, et al. Macrophage migration inhibitory factor polymorphism is associated with susceptibility to inflammatory coronary heart disease. Biomed Res Int. (2015) 2015:315174. doi: 10.1155/2015/315174
7. Fahriye E, Sacide P, Mustafa BE, Ayşen B, Sibel OB, Birol Y, et al. MBL2 and MIF gene polymorphisms in cardiovascular patients with atherosclerotic lesions undergoing heart valve replacement. Biotechnol Biotechnol Equip. (2017) 31:1173–7. doi: 10.1080/13102818.2017.1375864
8. Tereshchenko IP, Petrkova J, Mrazek F, Lukl J, Maksimov VN, Romaschenko AG, et al. The macrophage migration inhibitory factor (MIF) gene polymorphism in Czech and Russian patients with myocardial infarction. Clin Chim Acta. (2009) 402:199–202. doi: 10.1016/j.cca.2008.12.034
9. Wu C, Gong Y, Zhu X, Sun A, Yuan J, Zou Y, et al. The polymorphisms rs2516839 of USF1 and –173G/C of MIF were not associated with coronary artery disease but dyslipidemia in a Chinese population. J Cardiovasc Dis Diagn. (2014) 2:177. doi: 10.4172/2329-9517.1000177
10. Li DY, Zhang JY, Chen QJ, Liu F, Zhao Q, Gao XM, et al. MIF –173G/C (rs755622) polymorphism modulates coronary artery disease risk: evidence from a systematic meta-analysis. BMC Cardiovasc Disord. (2020) 20:300. doi: 10.1186/s12872-020-01564-4
11. Lian Z, Yu SR, Cui YX, Li SF, Song JX, Li ZY, et al. Association between polymorphisms in MIF promoter –173 locus and risk of coronary artery disease: a meta-analysis. Chongqing Med. (2021) 50:1012–7.
12. Xu, R. Association. of the MIF Gene Polymorphism with Coronary Artery Disease in Han, Uygur and Kazakh in Chinese Xinjiang. Xinjiang: Xinjiang Medical University (2016) 35 p. In Chinese.
13. Luo JY, Xu R, Li XM, Zhou Y, Zhao Q, Liu F, et al. MIF Gene Polymorphism rs755622 is associated with coronary artery disease and severity of coronary lesions in a Chinese Kazakh population: a case-control study. Medicine (Baltimore). (2016) 95:e2617. doi: 10.1097/MD.0000000000002617
14. Umans DS, Hallensleben ND, Verdonk RC, Bouwense SAW, Fockens P, vanSantvoort HC, et al. Recurrence of idiopathic acute pancreatitis after cholecystectomy: systematic review and meta-analysis. Br J Surg. (2020) 107:191–9. doi: 10.1002/bjs.11429
15. Delgado-Rodríguez M, Sillero-Arenas M. Systematic review and meta-analysis. Med Intensiva. (2018) 42:444–53. doi: 10.1016/j.medin.2017.10.003
16. Hua S, Ma C, Zhang J, Li J, Wu W, Xu N, et al. Influence of APOA5 locus on the treatment efficacy of three statins: evidence from a randomized pilot study in chinese subjects. Front Pharmacol. (2018) 9:352. doi: 10.3389/fphar.2018.00352
17. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. (1994) 50:1088–101.
18. Kassardjian V, Varma S, Andiappan M, Creugers NHJ, Bartlett D. A systematic review and meta analysis of the longevity of anterior and posterior all-ceramic crowns. J Dent. (2016) 55:1–6. doi: 10.1016/j.jdent.2016.08.009
19. Lin B, Fu GS, Li Cl, Huang H, Mao HH, Zhang P, et al. Association of MIF –173 gene polymorphism with coronary hear t disease in Chinese cigar ette smoking Population. Clin Educ Gen Pract. (2008) 6:31–4. In Chinese,
20. Shan ZX, Fu YH, Yu XY, Deng CY, Tan HH, Lin QX, et al. Association of the polymorphism of macrophage migration inhibitory factor gene with coronary heart disease in Chinese population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2006) 23:548–50. In Chinese,
21. Dai CF, Liu XD, Luo HL, Liu FF, Na MJ, Mou ZY. Correlation between the plasma level of MIF and its related gene –173G/C polymorphism and risk factors of atherosclerosis in pilots. Med J Chin PLA. (2016) 41:153–7. In Chinese,
22. Luo JY, Fang BB, Du GL, Liu F, Li YH, Tian T, et al. Association between MIF gene promoter rs755622 and susceptibility to coronary artery disease and inflammatory cytokines in the Chinese Han population. Sci Rep. (2021) 11:8050.
23. Du GL, Luo JY, Wang D, Li YH, Fang BB, Li XM, et al. MIF gene rs755622 polymorphism positively associated with acute coronary syndrome in Chinese Han population: case-control study. Sci Rep. (2020) 10:140. doi: 10.1038/s41598-019-56949-z
24. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. (2002) 21:1539–58. doi: 10.1002/sim.1186
25. Noels H, Bernhagen J, Weber C. Macrophage migration inhibitory factor: a noncanonical chemokine important in atherosclerosis. Trends Cardiovasc Med. (2009) 19:76–86. doi: 10.1016/j.tcm.2009.05.002
26. Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. (1966) 153:80–2. doi: 10.1126/science.153.3731.80
27. Mehta JL, Romeo F. Inflammation, infection and atherosclerosis: do antibacterials have a role in the therapy of coronary artery disease? Drugs. (2000) 59:159–70. doi: 10.2165/00003495-200059020-00001
28. Donn RP, Shelley E, Ollier WE, Thomson W, British Paediatric Rheumatology Study Group. A novel 5′-flanking region polymorphism of macrophage migration inhibitory factor is associated with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. (2001) 44:1782–5. doi: 10.1002/1529-0131(200108)44:8<1782::AID-ART314>3.0.CO;2-#
29. Dambacher J, Staudinger T, Seiderer J, Sisic Z, Schnitzler F, Pfennig S, et al. Macrophage migration inhibitory factor (MIF) –173G/C promoter polymorphism influences upper gastrointestinal tract involvement and disease activity in patients with Crohn’s disease. Inflamm Bowel Dis. (2007) 13:71–82. doi: 10.1002/ibd.20008
30. Grieb G, Merk M, Bernhagen J, Bucala R. Macrophage migration inhibitory factor (MIF): a promising biomarker. Drug News Perspect. (2010) 23:257–64. doi: 10.1358/dnp.2010.23.4.1453629
31. Lin SG, Yu XY, Chen YX, Huang XR, Metz C, Bucala R, et al. De novo expression of macrophage migration inhibitory factor in atherogenesis in rabbits. Circ Res. (2000) 87:1202–8. doi: 10.1161/01.res.87.12.1202
32. Burger-Kentischer A, Goebel H, Seiler R, Fraedrich G, Schaefer HE, Dimmeler S, et al. Expression of macrophage migration inhibitory factor in different stages of human atherosclerosis. Circulation. (2002) 105:1561–6. doi: 10.1161/01.cir.0000012942.49244.82
33. Schober A, Bernhagen J, Weber C. Chemokine-like functions of MIF in atherosclerosis. J Mol Med (Berl). (2008) 86:761–70. doi: 10.1007/s00109-008-0334-2
34. Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. (2007) 13:587–96. doi: 10.1038/nm1567
35. Burger-Kentischer A, Göbel H, Kleemann R, Zernecke A, Bucala R, Leng L, et al. Reduction of the aortic inflammatory response in spontaneous atherosclerosis by blockade of macrophage migration inhibitory factor (MIF). Atherosclerosis. (2006) 184:28–38. doi: 10.1016/j.atherosclerosis.2005.03.028
36. Donn R, Alourfi Z, De Benedetti F, Meazza C, Zeggini E, Lunt M, et al. Mutation screening of the macrophage migration inhibitory factor gene: positive association of a functional polymorphism of macrophage migration inhibitory factor with juvenile idiopathic arthritis. Arthritis Rheum. (2002) 46:2402–9. doi: 10.1002/art.10492
37. Li YY, Wang H, Yang XX, Geng HY, Gong G, Lu XZ. PCSK9 Gene E670G polymorphism and coronary artery disease: an updated meta-analysis of 5,484subjects. Front Cardiovasc Med. (2020) 7:582865. doi: 10.3389/fcvm.2020.582865
38. Li YY, Wang H, Wu JJ, Kim HJ, Yang XX, Geng HY, et al. ALDH2 gene G487A polymorphism and coronary artery disease: a meta-analysis including 5644 participants. J Cell Mol Med. (2018) 22:1666–74. doi: 10.1111/jcmm.13443
39. Li YY, Wang H, Zhang YY. CDKN2B-AS1 gene rs4977574 A/G polymorphism and coronary heart disease: a meta-analysis of 40,979 subjects. J Cell Mol Med. (2021) 25:8877–89. doi: 10.1111/jcmm.16849
Keywords: MIF, rs755622, polymorphism, coronary artery disease, genetic, macrophage migration inhibitory factor
Citation: Li Y-y, Wang H and Zhang Y-y (2022) Macrophage migration inhibitory factor gene rs755622 G/C polymorphism and coronary artery disease: A meta-analysis of 8,488 participants. Front. Cardiovasc. Med. 9:959028. doi: 10.3389/fcvm.2022.959028
Received: 21 June 2022; Accepted: 22 August 2022;
Published: 14 September 2022.
Edited by:
Joanne E. Curran, The University of Texas Rio Grande Valley, United StatesReviewed by:
Sacide Pehlivan, Istanbul University, TurkeyCopyright © 2022 Li, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-yan Li, bHl5bmptdTEyM0AxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.