
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 27 July 2022
Sec. Cardiovascular Genetics and Systems Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fcvm.2022.921803
 Cheng Wang1,2
Cheng Wang1,2 Puliang Yu3
Puliang Yu3 Lizhi Hu2
Lizhi Hu2 Minglu Liang2
Minglu Liang2 Yi Mao1
Yi Mao1 Qiutang Zeng1
Qiutang Zeng1 Xiang Wang1
Xiang Wang1 Kai Huang1,2
Kai Huang1,2 Jin Yan4
Jin Yan4 Li Xie5
Li Xie5 Fengxiao Zhang1,2*
Fengxiao Zhang1,2* Feng Zhu1,2*
Feng Zhu1,2*Background: Familial hypercholesterolemia (FH) can elevate serum low-density lipoprotein cholesterol (LDL-C) levels, which can promote the progression of acute coronary syndrome (ACS). However, the effect of FH on the prognosis of ACS remains unclear.
Methods: In this prospective cohort study, 223 patients with ACS having LDL-C ≥ 135.3 mg/dL (3.5 mmol/L) were enrolled and screened for FH using a multiple-gene FH panel. The diagnosis of FH was defined according to the ACMG/AMP criteria as carrying pathogenic or likely pathogenic variants. The clinical features of FH and the relationship of FH to the average 16.6-month risk of cardiovascular events (CVEs) were assessed.
Results: The prevalence of molecularly defined FH in enrolled patients was 26.9%, and coronary artery lesions were more severe in patients with FH than in those without (Gensini score 66.0 vs. 28.0, respectively; P < 0.001). After lipid lowering, patients with FH still had significantly higher LDL-C levels at their last visit (73.5 ± 25.9 mg/dL vs. 84.7 ± 37.1 mg/dL; P = 0.013) compared with those without. FH increased the incidence of CVEs in patients with ACS [hazard ratio (HR): 3.058; 95% confidence interval (CI): 1.585–5.900; log-rank P < 0.001].
Conclusion: FH is associated with an increased risk of CVEs in ACS and is an independent risk factor for ACS. This study highlights the importance of genetic testing of FH-related gene mutations in patients with ACS.
Familial hypercholesterolemia (FH) is a common genetic disorder that causes a remarkable increase in serum low-density lipoprotein cholesterol (LDL-C) levels, and results in xanthoma in the skin, subcutis, or tendon sheaths (1). FH can be divided into heterozygote FH, homozygote FH, compound heterozygote, and double heterozygote. Among these variants, heterozygote FH is more common. The prevalence of homozygote FH is estimated to be 1–3 in 1,000,000 in the general population, whereas that of heterozygote FH is higher, reaching 1 in 500 to 1 in 200 (1, 2). In China, adults with persistent serum LDL-C ≥ 135.3 mg/dL (3.5 mmol/L) without an apparent secondary cause of hypercholesterolemia should be screened for FH. The cutoff points are much lower than that followed by Western countries (3).
Elevated LDL-C level is a well-established factor that increases the risk of acute coronary syndrome (ACS) (4). The incidence of ACS in patients with untreated FH is 15–20 times higher than that in patients without FH (5, 6). Early identification of patients with FH can be helpful in adjusting their treatment regimen; moreover, the incidence of ACS can be reduced if appropriate treatment is administered before ACS onset. Therefore, it is important to accurately diagnose and screen FH in patients with ACS. However, the incidence of FH varies considerably in different regions owing to dietary and genetic factors. Among patients with ACS, the prevalence of FH was more than 5% in Europe (7, 8) and 2.7% in Japan (9).
Genetic testing is the gold standard for FH that identifies the presence of heterozygous pathogenic mutations in the genes associated with LDL-C metabolism. When combined with clinical features, next-generation sequencing can show high specificity and sensitivity in the identification of FH (10). However, genetic testing is rarely used to detect FH in patients with ACS owing to high economic costs among other reasons (1, 11, 12).
In this study, we evaluated the prevalence of molecularly defined FH in Chinese patients with ACS having LDL-C ≥ 135.3 mg/dL (3.5 mmol/L) and the effects of mutation on the incidence of cardiovascular events (CVEs) in a prospective observational cohort.
This study was registered in the Chinese Clinical Trial Registry (identifier number: ChiCTR-EOC-17011463). Patients with ACS [ST-segment elevation myocardial infarction (STEMI), non-ST segment elevation myocardial infarction (NSTEMI), and unstable angina] having LDL-C levels ≥ 135.3 mg/dL (95th percentile of LDL-C among Chinese), and those > 18 years were consecutively recruited from March 1, 2017, to February 19, 2019, in the coronary care unit (CCU) of Union Hospital, Wuhan, China. The study was reviewed and approved by the ethical review board (Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, China). Written informed consent was obtained from all participants enrolled in this study.
Blood samples from patients were collected for DNA analysis. Genomic DNA samples were extracted, amplified, and sequenced following a previously reported procedure (13). The inactivating variants in genes with mutations known to cause FH (LDLR, APOB, PCSK9, and LDLRAP1), myopathy-associated variant in the SLCO1B1 gene (rs4149056, rs4363657, and rs2306283), and genes functioning within the cholesterol-metabolism pathways (APOE, CETP, LIPA, LPL, and STAP1) were identified using a commercial FH Multigene Panel (Lipopro-1, Aegicare, China) (14). An average of 9.85 M (± 1.13 M) reads with 150 base paired end-sequencing strategy was obtained for the designed FH panel over all samples. The mean depth over targeted regions was 11628X (± 1215X), offering a high coverage of the regions of our interest. The mean coverage of selected target gene regions is shown in Supplementary Figure 1.
To identify FH-associated single nucleotide variants (SNVs) and Indels (short insertions and/or deletions of nucleotides < 50 bp), sequencing data were analyzed using a previously described in-house pipeline (13). Briefly, raw FASTQ files were processed with FASTP1 to detect and trim adapters and exclude low-quality reads. Clean reads were mapped to the GRCh37/hg19 reference genome using the Burrows-Wheeler Aligner (15). SNPs/Indels were identified using the Haplotype Caller tool of GATK and annotated using ANNOVAR (16). Annotation included genomic coordinates, functional role on protein coding, minor allele frequency, dbSNP identifiers, ClinVar identifiers, and deleteriousness-prediction scores of several prediction tools for pathogenicity of the variant (17). On average, 182 variants (± 15) were called for each sample, consisting of 154 SNPs (± 13) and 28 Indels (± 2).
A CNV kit was used to determine CNV in the LDLR gene (18). Normal references used for CNV identification were constructed using sequencing data from healthy subjects over the same DNA-sequencing protocol. During reference creation, the circular binary segmentation algorithm was chosen for CNV event segmentation, and the threshold parameter for copy-number calling was set to “−1.6, −0.8, 0.5, 1.” A previously published procedure was followed for process strategies that included normal reference creation, CNV event segmentation, and copy number reverting (13), except for the “target amplicon sequencing” mode, which was chosen to exclude off-target regions from calculation and disable edge correction. The detected CNVs were annotated using AnnotSV (19).
The identified variants were classified as pathogenic (P), likely pathogenic (LP), variants of uncertain significance (VUS), likely benign, or benign, following the ACMG criteria 8. Molecularly defined FH was defined as carrying the P or LP variants.
The deadline for follow-up in this study was December 31, 2019. Follow-up details are described in Supplementary Material. CVEs were defined as cardiovascular death, non-fatal myocardial infarction, unstable angina pectoris requiring admission, unplanned coronary revascularization (unplanned percutaneous coronary intervention or coronary artery bypass graft driven by coronary ischemia), or heart failure requiring admission after discharge from the CCU.
The exclusion criteria, clinical measurements, and follow-up procedures are detailed in Supplementary Material.
Baseline characteristic variables of continuous data are presented as mean ± standard deviation (SD), or median and interquartile range (IQR), as appropriate. Categorical variables are presented as absolute numbers (N) and percentages. Student’s t-test, one-way analysis of variance (ANOVA), or non-parametric tests were used to compare differences between groups of continuous parameters, as appropriate. Categorical variables were compared using the Chi-square test or Fisher’s exact test. A Kaplan-Meier curve with log-rank test was used to assess the percent of free time from CVEs. The prevalence of molecularly defined FH among patients with ACS and its corresponding 95% confidence interval (CI) was determined using normal approximation methods. Missing data in the lipid value at the last visit were replaced using the “last observation carried forward” method. Cox proportional hazards model was used to explore the relationship between FH and CVEs. Adjusted hazard ratio (HR) with 95% CI was used to measure the strength of association between CVEs and potential predictors. Statistical analyses were performed using R statistics version 4.0.0 (Arbor Day) and IBM SPSS Statistics version 22.0 (IBM SPSS Statistics, IBM Corporation). Statistical significance was defined using two-sided P < 0.05.
A total of 261 patients hospitalized with ACS with elevated LDL-C levels of ≥ 135.3 mg/dL (Figure 1) were identified. Of these, 39 patients were excluded for the following reasons: rejection to participate (n = 15), in-hospital death before enrollment (n = 5), cancer (n = 3), untreated hypothyroidism (n = 7), nephrotic syndrome (n = 3), severe liver insufficiency (n = 2), and alcohol abuse (n = 3). Lastly, 223 patients were subjected to genetic testing and data analysis. CVE onset led to 51 patients ending the follow-up before December 31, 2019. There were 167 patients who ended follow-up on December 31, 2019. Five patients were lost to follow-up before the last scheduled visit of the study, and the last follow-up time was considered the end of the follow-up time.
Baseline demographics and clinical characteristics of enrolled patients with or without molecularly defined FH are shown in Table 1. All patients were of Asian (Chinese) descent. A total of 56.5% of study participants had a history of hypertension, 28.3% had a history of diabetes mellitus, 2.2% had a history of stroke, 38.1% were current smokers, and 45.7% had peripheral arteriosclerosis. In the sample, 24.7% of patients reported having previous coronary artery disease (CAD), and 13 patients (5.8%) were found to have xanthoma on physical examination. A total of 72.2% of study participants had angina pectoris and the remaining had acute myocardial infarction (AMI).
Of the 223 patients in the study, FH was molecularly diagnosed in 60 (26.9%) patients, who carried 61 P and LP variants (Supplementary Table 1). Fifty-nine patients (26.5%) were monoallelic FH variant carriers (57 heterozygous LDLR variant carriers, 2 heterozygous APOB variant carriers) and one patient was a biallelic FH variant carrier (compound heterozygous for LDLR variants). The 61 P/LP variants included 34 missenses, 9 non-senses, 3 frameshift indels, 12 splicing variants, and 3 CNVs (Supplementary Table 1). Twenty-two VUS were detected in 22 patients (Supplementary Table 2).
In general, there were no significant differences between the two groups with respect to age, gender, body mass index (BMI), prevalence of smoking; history of hypertension, diabetes mellitus, and stroke; and the incidence of peripheral arteriosclerosis or xanthoma. Notably, the prevalence of a prior history of CAD was much higher in patients with FH than in those without (45.0% vs. 17.2%; P < 0.001) (Table 1). The proportion of AMI was significantly higher in patients with FH compared with those without (41.5% vs. 22.3%; P = 0.001). The Gensini score revealed that coronary artery lesions in patients with FH were more severe than in those without FH (Gensini score 66.0 vs. 28.0, respectively; P < 0.001). Moreover, there were no significant differences in revascularization treatment among patients with and without FH. All patients received a statin alone and 106 (47.5%) received a statin/ezetimibe combination on discharge from the hospital. There were no significant differences in the rate of statin or ezetimibe treatment between the two groups (statins: 100% vs.100%, P = 1.000; ezetimibe: 55% vs. 44.8%, P = 0.226) (Table 2).
The baseline lipid profiles of all patients were obtained before commencing lipid-lowering therapy (Table 1). The presence of pathogenic FH in patients resulted in significantly higher levels of both total cholesterol (TC) and LDL-C compared with those without FH (TC: 281.1 mg/dL vs. 251.4 mg/dL, P < 0.001; LDL: 212.7 mg/dL vs. 179.0 mg/dL, P < 0.001), but a decrease in HDL-C levels (39.1 mg/dL vs. 45.2 mg/dL; P = 0.002). There were no significant differences in triglyceride levels (134.6 mg/dL vs. 148.8 mg/dL; P = 0.484) between the groups.
Changes in lipid profiles in the laboratory results and the statin drug treatment during the out-of-hospital follow-up are reported in Table 2. During follow-up, the missing data from analyzed patients was < 10%. LDL-C levels were available at follow-up for all 223 patients. The mean TC and LDL-C levels at the last visit were 138 and 76.6 mg/dL, respectively. At the last visit, 34 patients (15.2%) had LDL-C levels ≥ 100 mg/dL, 83 patients (37.2%) had LDL-C levels between 70 and 100 mg/dL, and 104 patients (46.6%) had LDL-C levels < 100 mg/dL (Table 2).
Although there were no significant differences between discharged patients with and without FH among those receiving statin treatment, the FH pathogenic variants led to an increase in median LDL-C at the last visit (84.7 mg/dL vs. 73.5 mg/dL; P = 0.013) and an absolute reduction in LDL-C (134.6 mg/dL vs. 111.8 mg/dL; P < 0.001). There was no significant difference in the proportion of decline (60.4% vs. 61.3%; P = 0.677) between the two groups (Table 2). Moreover, the percentage of patients with FH having LDL-C levels < 70 mg/dL was slightly lower than those without FH; however, there were no differences between the two groups in the proportion of patients with LDL-C levels between 70 and 100 mg/dL or LDL-C levels ≥ 100 mg/dL at the last visit.
During the follow-up period (16.6 ± 7.1 months), 51 (22.9%) patients developed CVEs. Seven patients (3.1%) died (four patients with FH), four (1.8%) had a non-fatal myocardial infarction, 30 (13.5%) experienced unstable angina pectoris requiring admission, two (0.9%) received unplanned revascularization, and five (2.2%) experienced heart failure. Moreover, there was no difference in the proportion of patients receiving statin drug treatment during the follow-up period between those with and without FH.
Next, the correlation between pathogenic variants and risk factors with CVEs was analyzed. Among the 51 CVEs, 24 and 27 CVEs were observed in patients with and without FH, respectively. Kaplan-Meier curves revealed that patients with the FH P/LP variants had a much higher incidence of CVEs (HR: 3.058; 95% CI: 1.585–5.900 log-rank P < 0.001) (Figure 2). As the carrier with VUS was classified as the non-FH group, which could affect the relationship between FH P/LP variants and the risk of CVEs, Kaplan-Meier analysis was performed when patients with VUS were classified in the FH group or excluded (Supplementary Figure 2). The incidence of CVEs was significantly higher in patients with the FH P/LP variants than those with the FH likely benign/benign variants (HR: 2.717; 95% CI: 1.437–5.138, log-rank P < 0.001) (Supplementary Figure 2A), and the incidence of CVEs was also much higher in patients with the FH P/LP/VUS variants than in those with the FH likely benign/benign variants (HR: 1.917; 95% CI: 1.071–3.431, log-rank P = 0.016) (Supplementary Figure 2B). These findings confirmed the robustness of the results with VUS in patients with FH. The risk of CVD is associated with gender, age, BMI, Gensini score, history of smoking, hypertension, and diabetes. Multivariate Cox regression analysis was used to estimate these risk factors. The forest map shows that the incidence of CVEs is not significantly different between patients of different genders, ages, and BMI. Moreover, a history of hypertension and diabetes did not affect the rate of CVEs in this study population (Figure 3), whereas a history of smoking promoted the rate of CVEs in patients with ACS (HR: 2.11; 95% CI: 1.006–4.426, P = 0.048) (Figure 3). Moreover, the results showed that a high Gensini score was associated with higher CVE rates (HR: 1.01; 95% CI: 1.001–1.014, P = 0.030) (Figure 3). Moreover, patients with FH P/LP variants had an increased risk of CVEs (HR: 2.72; 95% CI: 1.447–5.126, P = 0.002) (Figure 3), independent of these factors.
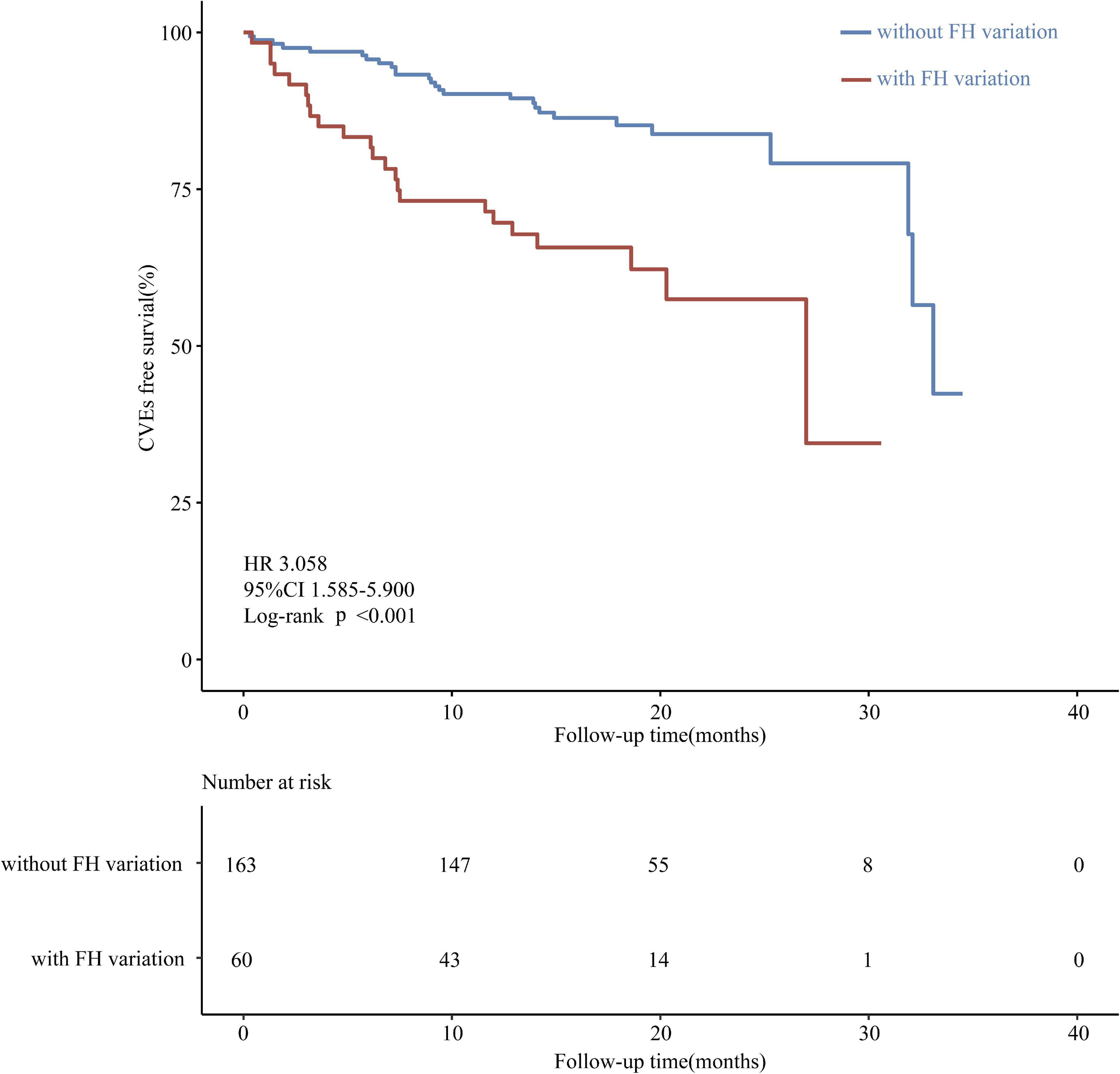
Figure 2. Kaplan-Meier curve of the cumulative event-free survival analyzed according to variant classification based on the ACMG/AMP criteria.
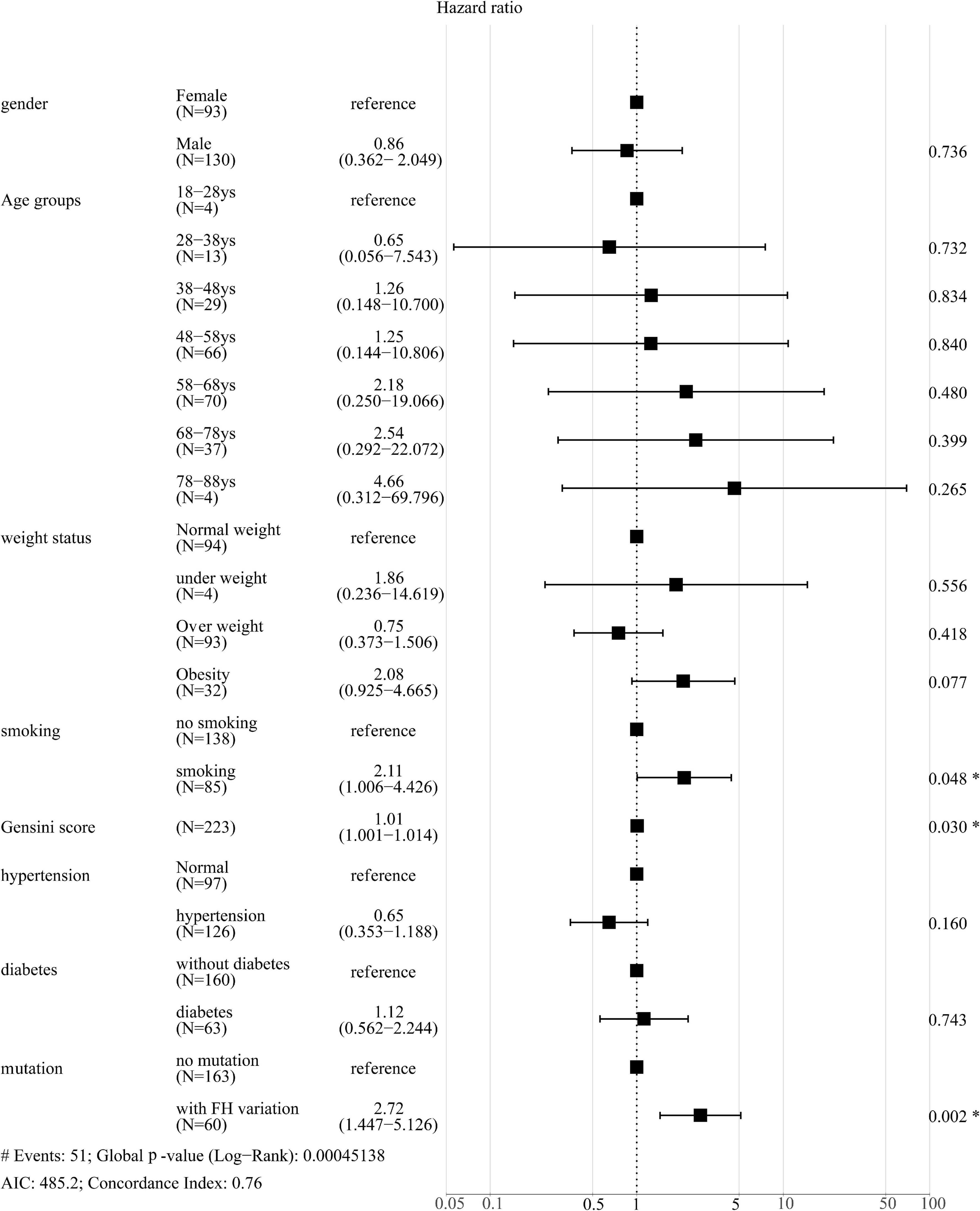
Figure 3. Forest plot of hazard ratios using multivariate Cox regression analysis of the effect of different parameters on CVEs. *P < 0.05.
In this study, we have described a genetic analysis of FH in suspected ACS patients with LDL-C levels ≥ 135.3 mg/dL. The prevalence of molecularly defined FH in these patients was 26.9%. Coronary artery lesions in patients with FH were more severe than in those without FH. After a follow-up of 16.6 months, on average, we found that the FH pathogenic variants caused an increase in the incidence of CVEs.
A previous study has reported a considerably low prevalence of FH in China (20). This low detection rate of FH diagnosis is attributable to the lack of emphasis on genetic screening in the general population, especially in many developing countries. We observed that 26.9% of patients with ACS having LDL-C levels ≥ 135.3 mg/dL carried FH-related mutations, which was also much higher than that reported previously in Western countries and Japan (5, 21, 22). It is well known that adults hospitalized for ACS have a higher prevalence of FH than the general population (7). Besides, three CNV carriers were found in our study, which also improved the detection rate of FH. The above differences may be attributed to different nationalities and the lower average level of cholesterol in the Chinese population.
With changes in the nutrient composition and an increased intake of high-calorie foods, the number of patients with hypercholesterolemia identified based on physical examination has rapidly increased in China. The discovery of a large number of patients with ACS with hypercholesterolemia suggests that FH is a common disease and that its diagnosis and treatment in China are not adequate. Thus, it represents a global public health burden as it is a major cause of early CAD (1). Early detection of FH can lead to targeted dietary control and more effective treatment program with pharmacological agents. Expanded genetic screening and cascade testing have also been recommended.
As the prevalence of FH is significantly higher in patients with ACS than in the general population, FH gene testing is necessary for patients with ACS. Previous clinical studies on lipid profiles have mainly assessed the prevalence and cardiovascular prognosis of patients with FH as suggested by the Simon Broome (SB) algorithms in the United Kingdom and the Dutch Lipid Clinic (DLC) algorithm, and not by genetic screening. However, these clinical algorithms do not apply to children for the diagnosis of FH, as the clinical DLC and SB algorithms may be not valid in these populations and in subjects with normal LDL-C levels in the absence of molecular genetic testing, which may lead to the inaccurate diagnosis of FH (23–26). Genetic tests can provide a more accurate molecular diagnosis for patients with FH; thus, these tests should be considered the standard diagnostic procedure for patients with possible FH, which could help reduce the incidence of ACS (27).
The findings from our study suggest that FH increases the risk of CVEs after multivariate adjustment for common risk factors. Moreover, we found that the pathogenic variants of FH led to an increase in LDL-C levels even after treatment with lipid-lowering drugs (statins combined with ezetimibe). These findings indicate that knowledge on the FH pathogenic variation status can provide information on the additional risks, apart from the clinical data, in identifying individuals with ACS who are at the highest risk. Accordingly, these patients should be instructed to initiate early lipid-lowering therapy to improve the clinical outcomes of ACS. In patients with ACS with pathogenic FH, more intensive lipid-lowering therapy (PCSK9 inhibitors, higher-intensity statins) may be necessary to further reduce LDL-C levels.
An increasing number of lipid-lowering treatments, such as PCSK9 inhibitors and lipid adsorption therapy are available to treat FH. Moreover, the novel gene-editing CRISPR technology could actually change these gene-driven diseases (28, 29). The intake of dietary supplements and functional foods and performance of physical activities are also considered beneficial in maintaining a normal lipid profile (30, 31).
We found that FH was present in 26.9% of patients in China with ACS having LDL-C ≥ 135.3 mg/dL, which may be a more beneficial approach compared with earlier genetic screening. FH-associated pathogenic variants can provide the prognostic evidence for ACS and risk-stratification information that is independent of LDL-C levels. Early recognition of FH by genetic testing can help change the treatment strategy for patients with ACS and accordingly alter their prognosis.
Our study has several limitations. First, this is a single-center, prospective cohort study with a small sample size, where only a few CVEs were observed. Further in-depth studies including multicenter clinical studies are, therefore, needed to confirm our study findings. Second, the sequencing panel used in our study only contained eight most common genes and may have been missing the polygenic FH gene and other genes related to lipid metabolism. The majority of mutations were found in the LDLR gene. Third, as the clinical data in this study were obtained before PCSK9 inhibitors were marketed in China, patients in this study were not treated with PCSK9 inhibitors. Fourth, the original planned follow-up time of this study was 3 years. Due to the onset of the COVID-19 pandemic in Wuhan, the follow-up study could not be conducted. As positive results were observed in this study, it was terminated early. Lastly, unknown confounding factors may exist, although multiple confounding factors were adjusted for in the Cox regression analysis.
The raw data reported in this article are deposited in https://doi.org/10.6084/m9.figshare.20304711.v1.
The studies involving human participants were reviewed and approved by the Ethics Committee of Wuhan Union Hospital in China. The patients/participants provided their written informed consent to participate in this study.
FXZ and FZ conceived and designed the study. CW, FXZ, and FZ wrote the manuscript. All authors performed the experiments, analyzed the data, contributed to the article, and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (No. 81570348).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Yongchu Liu, Ph.D., for manuscript editing and technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.921803/full#supplementary-material
FH, Familial hypercholesterolemia; ACS, Acute coronary syndrome; CVE, Cardiovascular event; STEMI, ST-segment elevation myocardial infarction; NSTEMI, Non-ST segment elevation myocardial infarction; CCU, Coronary care unit; AMI, Acute myocardial infarction; CNV, Copy number variation; TC, Total cholesterol; LDL-C, Low-density lipoprotein cholesterol; PCSK9, Proprotein convertase subtilisin/kexin type 9; CAD, Coronary artery disease; VUS, Variant of uncertain significance; LP, Likely pathogenic; ACMG, American College of Medical Genetics and Genomics; AMP, The Association for Molecular Pathology.
1. Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European atherosclerosis society. Eur Heart J. (2013) 34:3478–90a.
2. Nohara A, Tada H, Ogura M, Okazaki S, Ono K, Shimano H, et al. Homozygous familial hypercholesterolemia. J Atheroscler Thromb. (2021) 28:665–78. doi: 10.5551/jat.RV17050
3. Atherosclerosis and Coronary Heart Disease Group of the Chinese Society of Cardiology of Chinese Medical Association, Editorial Board of Chinese Journal of Cardiology. [Chinese expert consensus on screening, diagnosis and treatment of familial hypercholesterolemia]. Zhonghua Xin Xue Guan Bing Za Zhi. (2018) 46:99–103.
4. Cholesterol Treatment Trialists’ [CTT] Collaboration, Baigent C, Blackwell L, Emberson J, Holland LE, Reith C, et al. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. (2010) 376:1670–81. doi: 10.1016/S0140-6736(10)61350-5
5. Singh A, Gupta A, Collins BL, Qamar A, Monda KL, Biery D, et al. Familial hypercholesterolemia among young adults with myocardial infarction. J Am Coll Cardiol. (2019) 73:2439–50. doi: 10.1016/j.jacc.2019.02.059
6. Benedek P, Eriksson M, Duvefelt K, Freyschuss A, Frick M, Lundman P, et al. Genetic testing for familial hypercholesterolemia among survivors of acute coronary syndrome. J Intern Med. (2018) 284:674–84. doi: 10.1111/joim.12812
7. Nanchen D, Gencer B, Auer R, Raber L, Stefanini GG, Klingenberg R, et al. Prevalence and management of familial hypercholesterolaemia in patients with acute coronary syndromes. Eur Heart J. (2015) 36:2438–45. doi: 10.1093/eurheartj/ehv289
8. Dyrbus K, Gasior M, Desperak P, Osadnik T, Nowak J, Banach M. The prevalence and management of familial hypercholesterolemia in patients with acute coronary syndrome in the polish tertiary centre: results from the TERCET registry with 19,781 individuals. Atherosclerosis. (2019) 288:33–41. doi: 10.1016/j.atherosclerosis.2019.06.899
9. Harada-Shiba M, Ako J, Arai H, Hirayama A, Murakami Y, Nohara A, et al. Prevalence of familial hypercholesterolemia in patients with acute coronary syndrome in Japan: results of the EXPLORE-J study. Atherosclerosis. (2018) 277:362–8. doi: 10.1016/j.atherosclerosis.2018.06.856
10. Youngblom E, Pariani M, Knowles JW. Familial hypercholesterolemia. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, et al. editors. GeneReviews®. (Seattle, WA: University of Washington) (1993).
11. Brænne I, Kleinecke M, Reiz B, Graf E, Strom T, Wieland T, et al. Systematic analysis of variants related to familial hypercholesterolemia in families with premature myocardial infarction. Eur J Hum Genet. (2015) 24:191. doi: 10.1038/ejhg.2015.100
12. Nordestgaard BG, Benn M. Genetic testing for familial hypercholesterolaemia is essential in individuals with high LDL cholesterol: who does it in the world? Eur Heart J. (2017) 38:1580–3. doi: 10.1093/eurheartj/ehx136
13. Sun D, Liu Y, Cai W, Ma J, Ni K, Chen M, et al. Detection of disease-causing SNVs/Indels and CNVs in single test based on whole exome sequencing: a retrospective case study in epileptic encephalopathies. Front Pediatr. (2021) 9:635703. doi: 10.3389/fped.2021.635703
14. Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, et al. Whole-genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation. (2019) 139:1593–602. doi: 10.1161/CIRCULATIONAHA.118.035658
15. Fb UB, Cau L, Tafazzoli A, Mechin MC, Wolf S, Romano MT, et al. Mutations in three genes encoding proteins involved in hair shaft formation cause uncombable hair syndrome. Am J Hum Genet. (2016) 99:1292–304. doi: 10.1016/j.ajhg.2016.10.004
16. Odumpatta R, Mohanapriya A. Next generation sequencing exome data analysis aids in the discovery of SNP and INDEL patterns in Parkinson’s disease. Genomics. (2020) 112:3722–8. doi: 10.1016/j.ygeno.2020.04.025
17. Wei CH, Phan L, Feltz J, Maiti R, Hefferon T, Lu Z. tmVar 2.0: integrating genomic variant information from literature with dbSNP and ClinVar for precision medicine. Bioinformatics. (2018) 34:80–7. doi: 10.1093/bioinformatics/btx541
18. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. (2016) 12:e1004873. doi: 10.1371/journal.pcbi.1004873
19. Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. (2018) 34:3572–4. doi: 10.1093/bioinformatics/bty304
20. Shi Z, Yuan B, Zhao D, Taylor AW, Lin J, Watts GF. Familial hypercholesterolemia in China: prevalence and evidence of underdetection and undertreatment in a community population. Int J Cardiol. (2014) 174:834–6. doi: 10.1016/j.ijcard.2014.04.165
21. Takasaki A, Kurita T, Masuda J, Hoshino K, Seko T, Tanigawa T, et al. Prevalence and prognosis of familial hypercholesterolemia in patients with acute coronary syndrome in Mie prefecture, Japan- report from Mie ACS registry. Circ J. (2020) 85:9–18. doi: 10.1253/circj.CJ-20-0112
22. Nanchen D, Gencer B, Muller O, Auer R, Aghlmandi S, Heg D, et al. Prognosis of patients with familial hypercholesterolemia after acute coronary syndromes. Circulation. (2016) 134:698–709. doi: 10.1161/CIRCULATIONAHA.116.023007
23. Amor-Salamanca A, Castillo S, Gonzalez-Vioque E, Dominguez F, Quintana L, Lluis-Ganella C, et al. Genetically confirmed familial hypercholesterolemia in patients with acute coronary syndrome. J Am Coll Cardiol. (2017) 70:1732–40. doi: 10.1016/j.jacc.2017.08.009
24. Cao YX, Wu NQ, Sun D, Liu HH, Jin JL, Li S, et al. Application of expanded genetic analysis in the diagnosis of familial hypercholesterolemia in patients with very early-onset coronary artery disease. J Transl Med. (2018) 16:345. doi: 10.1186/s12967-018-1737-7
25. Simon Broome Register Group. Risk of fatal coronary heart disease in familial hypercholesterolaemia: scientific steering committee on behalf of the simon broome register group. BMJ. (1991) 303:893–6. doi: 10.1136/bmj.303.6807.893
26. Wiegman A, Gidding SS, Watts GF, Chapman MJ, Ginsberg HN, Cuchel M, et al. Familial hypercholesterolaemia in children and adolescents: gaining decades of life by optimizing detection and treatment. Eur Heart J. (2015) 36:2425–37. doi: 10.1093/eurheartj/ehv157
27. Versmissen J, Oosterveer DM, Yazdanpanah M, Defesche JC, Basart DC, Liem AH, et al. Efficacy of statins in familial hypercholesterolaemia: a long term cohort study. BMJ. (2008) 337:a2423. doi: 10.1136/bmj.a2423
28. Cesaro A, Fimiani F, Gragnano F, Moscarella E, Schiavo A, Vergara A, et al. New frontiers in the treatment of homozygous familial hypercholesterolemia. Heart Fail Clin. (2022) 18:177–88. doi: 10.1016/j.hfc.2021.07.008
29. Tarpley AJ. New frontier in lipids: PCSK9 inhibitors and implications for the life insurance industry. J Insur Med. (2018) 47:230–5. doi: 10.17849/insm-47-4-1-6.1
30. Wang Y, Xu D. Effects of aerobic exercise on lipids and lipoproteins. Lipids Health Dis. (2017) 16:132. doi: 10.1186/s12944-017-0515-5
Keywords: acute coronary syndrome, cardiovascular events, familial hypercholesterolemia, low-density lipoprotein cholesterol, lipid
Citation: Wang C, Yu P, Hu L, Liang M, Mao Y, Zeng Q, Wang X, Huang K, Yan J, Xie L, Zhang F and Zhu F (2022) Prevalence and prognosis of molecularly defined familial hypercholesterolemia in patients with acute coronary syndrome. Front. Cardiovasc. Med. 9:921803. doi: 10.3389/fcvm.2022.921803
Received: 16 April 2022; Accepted: 06 July 2022;
Published: 27 July 2022.
Edited by:
Marat V. Ezhov, Ministry of Health of the Russian Federation, RussiaReviewed by:
Andrea Vergara, University of Campania Luigi Vanvitelli, ItalyCopyright © 2022 Wang, Yu, Hu, Liang, Mao, Zeng, Wang, Huang, Yan, Xie, Zhang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengxiao Zhang, eGlhb3lvdTMyMUBmb3htYWlsLmNvbQ==; Feng Zhu, emh1ZmVuZ0BodXN0LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.