- 1Department of Cardiology, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan, China
- 2Department of Endocrinology, China Resources and WISCO General Hospital, Wuhan, China
- 3Department of Geratology, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan, China
- 4Institute of Myocardial Injury and Repair, Wuhan University, Wuhan, China
Background: Recent studies have shown that the 4G/5G insertion/deletion variant of SERPINE1 (rs1799889) is closely linked to coronary artery disease (CAD). This study aims to clarify the effects of the rs1799889 variant on lipid levels and to insight into the mechanisms underlying the rs1799889 variant and CAD.
Methods and Results: By searching PubMed and the Cochrane databases for studies published before 31 October 2021, 40 studies conducted on a total of 13,117 subjects were included for the analysis. The consistent findings for the effects of the 5G allele of rs1799889 variant on lipid metabolism were the significantly decreased triglycerides (TG) [standardized mean difference (SMD) = –0.12, 95% CI = –0.21 to 0.03, P = 0.01], total cholesterol (TC) (SMD = –0.12, 95% CI = –0.17 to 0.06, P < 0.001), and low-density lipoprotein cholesterol (LDL-C) (SMD = –0.13, 95% CI = –0.23 to 0.03, P = 0.01) levels. Intriguingly, the significant effects of the rs1799889 variant on LDL-C (SMD = –0.15, 95% CI = –0.26 to 0.05, P < 0.01) and TC (SMD = –0.17, 95% CI = –0.27 to 0.07, P < 0.01) levels were primarily observed in the Asian population. However, the significant effect of the rs1799889 variant on high-density lipoprotein cholesterol (HDL-C) (SMD = 0.26, 95% CI = 0.03–0.48, P = 0.03) levels was detected only in female subjects.
Conclusion: The rs1799889 variant of SERPINE1 is a protective genetic factor against CAD, the Asian population with the 5G allele of the rs1799889 variant may have a reduced CAD risk.
Introduction
The plasminogen activator inhibitor-1 (PAI-1), also known as SERPINE1, is a member of the serine protease inhibitor superfamily and plays a major role in regulating fibrinolysis by inhibiting plasminogen activity.
The SERPINE1 gene is located on the long arm of human chromosome 7 (7q21.3-22). The most-studied rs1799889 variant, also known as 4G/5G insertion/deletion sequence, is located in the SERPINE1 promoter region and formed by the 5th guanine (G base) insertion or deletion in the 4G sequence at position-675th base. The 5G allele has lower transcriptional activity than the 4G allele (1) and subjects with homozygous 5G allele have about 25% lower circulating PAI-1 concentrations than subject’s possession of homozygous 4G allele (2).
Notably, a set of elegant experiments showed that SERPINE1 may affect lipid metabolism. For instance, SERPINE1 knockout (SERPINE1–/–) inhibits the expression of adipogenic genes (3) and ameliorates dyslipidemia (4) in mice. Consistently, transplanting white adipose tissue from SERPINE1–/– mice into obese mice reduces plasma triglycerides (TG) and total cholesterol (TC) levels (5). Moreover, inhibition of SERPINE1 expression decreases proprotein convertase subtilisin/kexin type 9 (PCSK9) and increases low-density lipoprotein receptor (LDLR) levels (6). Taken together, these indicate that the expression levels of SERPINE1 are closely linked to lipid metabolism. Therefore, the 5G allele of the rs1799889 variant may affect lipid levels due to its profound impact on SERPINE1 expression levels (1, 2).
Currently, a series of meta-analyses (7–10) showed that the 5G allele of the rs1799889 variant largely decreased coronary artery disease (CAD) risk; however, the specific mechanism is unknown. Therefore, this study is conducted to investigate the effects of the rs1799889 variant on lipid levels and to clarify the mechanistic basis for the correlations between the rs1799889 variant and CAD.
Materials and Methods
The study design followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (11).
Literature Search
The literature search was carried out by using PubMed, Medline, Embase, Cochrane Library, Web of Science, Google Scholar, Foreign Medical Journal Service, and Excerpta Medica for manuscripts published before 31 October 2021, by entering the following keywords: (“SERPINE1,” “PAI-1,” or “plasminogen activator inhibitor-1”), (“4G/5G,” “insertion/deletion,” or “rs1799889”), (“variant,” “mutant,” or “polymorphism”), and (“lipid,” “lipids,” “lipid metabolism,” “lipoprotein,” “cholesterol,” “blood lipid,” “serum lipid,” or “circulating lipid”).
Inclusion Criteria and Exclusion Criteria
The inclusion criteria of this meta-analysis were as follows. (1) The studies investigated the effects of the rs1799889 variant on lipid levels. (2) The studies at least provided one of the four lipid parameters (TG, TC, LDL-C, and HDL-C). (3) The studies provided the genotype frequencies of the rs1799889 variant. (4) The rs1799889 variant offered the mean lipid levels with standard deviation (SD) or standard error (SE) by genotypes. (5) The interventional studies provided pre-intervention data. (6) The language of eligible studies was restricted to English or Chinese. The exclusion criteria of this meta-analysis include (1) studies that were not related to the rs1799889 variant, (2) studies that were not related to lipid levels, (3) studies not presenting genotype or allele counts, (4) studies having invalid data, (5) studies having incomplete data, (6) pedigree studies (7), overlapping studies, and (8) abstract, review, case report, meta-analysis, and animal studies.
Subgroup Analysis
Subgroup analysis was carried out by ethnicity, gender, and health status. The ethnicity was divided into Caucasian, Asian, Turkish, and other ethnics. The healthy status was divided into patients with CAD, patients with type 2 diabetes mellitus (T2DM), and healthy subjects.
Other Items
Refer to the previous publication (12) for more details about data extraction, data analysis, heterogeneity processing, and publication bias tests.
Results
Study Selection
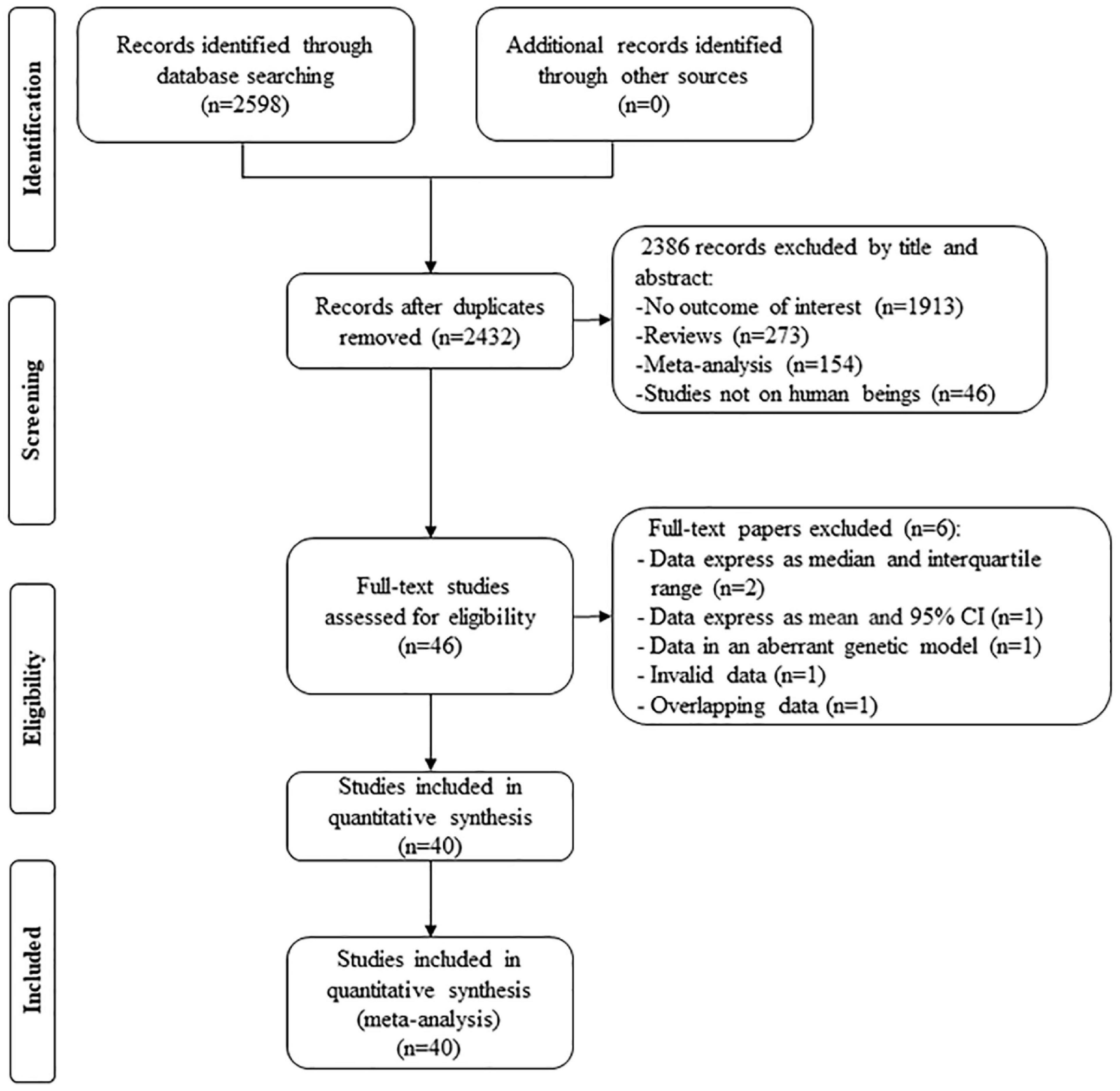
By searching the PubMed and Cochrane database, 2,598 studies were screened; after excluding duplicates, 2,386 studies were removed by their title and abstract. Then, 46 full-text studies were included for assessing eligibility, in which two studies (13, 14) provided lipid data by the genotype of rs1799889 but expressed as a median and interquartile range, 1 study (15) provided lipid data by the genotype of rs1799889 but expressed as mean and 95% CI, 1 study (16) provided lipid levels by the genotype of rs1799889 but in an aberrant genetic model [(4G5G + 4G4G) vs. 5G5G], 1 study (17) provided invalid data, and 1 study (18) had subjects overlapping with other publications (19). Therefore, six studies were further excluded. Finally, 40 studies (13,117 subjects) were eligible for the analysis (Figure 1).
The characteristics of the eligible studies were summarized in Supplementary Table 1. The circulating lipid levels by the genotype of the SERPINE1 rs1799889 variant were presented in Supplementary Table 2.
Effects of the rs1799889 Variant on Circulating Lipid Levels
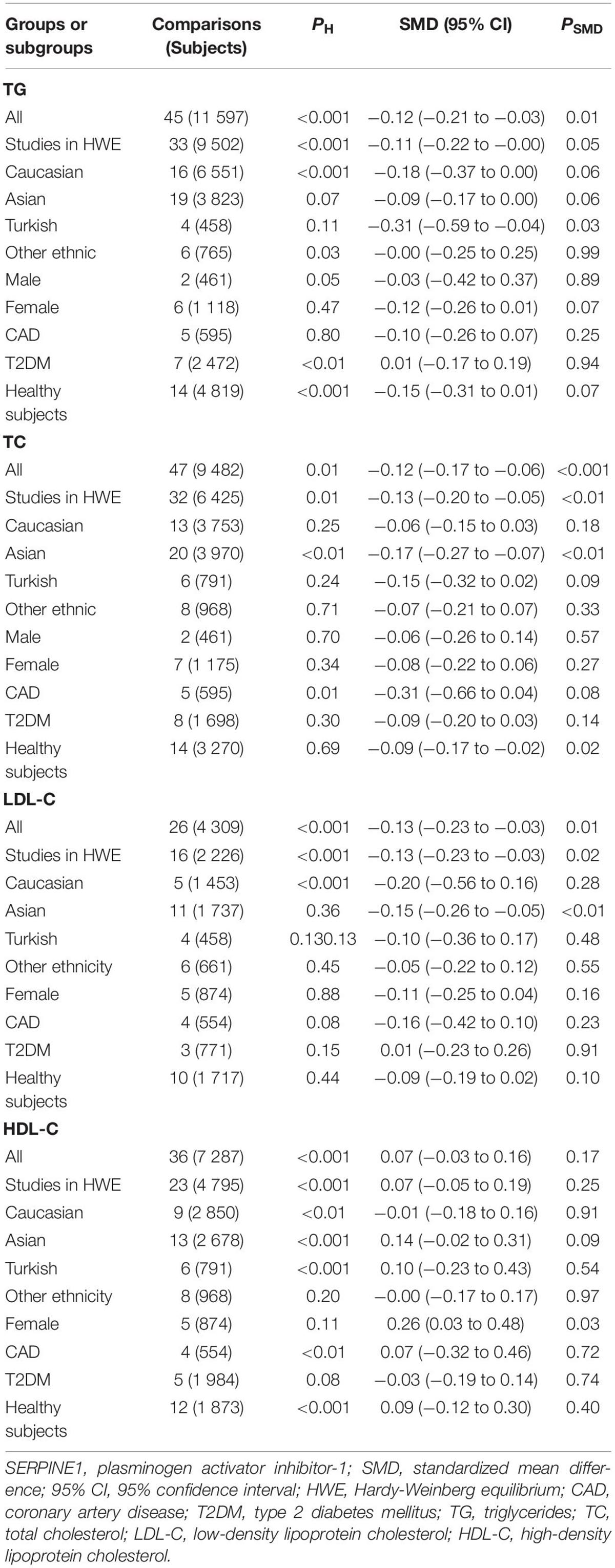
The consistent findings for the effects of the rs1799889 variant on lipid metabolism were the significantly decreased LDL-C [standardized mean difference (SMD) = –0.13, 95% CI = –0.23 to 0.03, P = 0.01], TC [SMD = –0.12, 95% CI = –0.17 to 0.06, P < 0.001], and TG [SMD = –0.12, 95% CI = –0.21 to 0.03, P = 0.01] levels (Table 1 and Figures 2–4). After excluding the population that deviated from Hardy–Weinberg equilibrium (HWE), the significant effects of the rs1799889 variant on LDL-C, TC, and TG levels were also detected (Table 1).
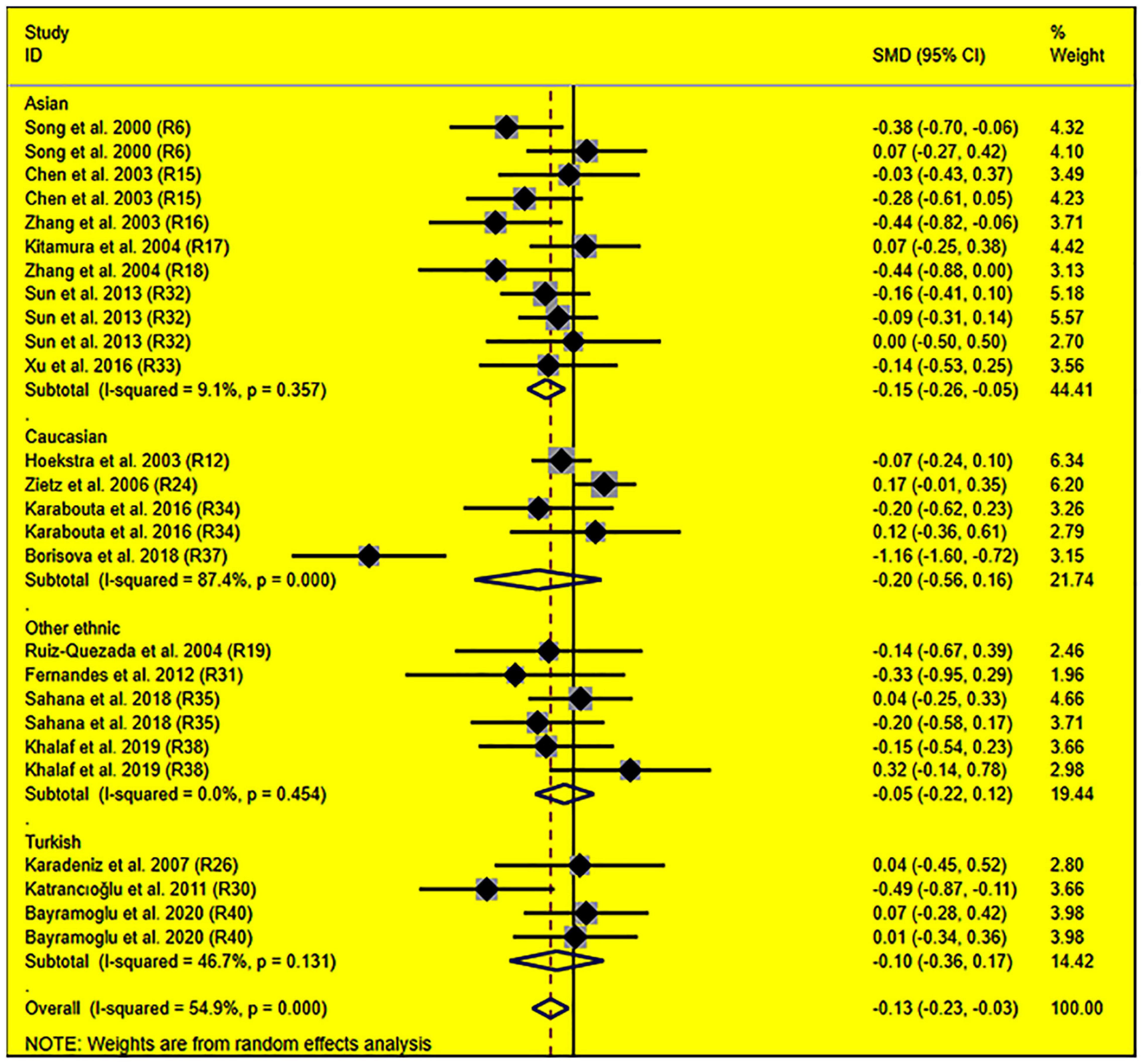
Figure 2. Forest plot of the meta-analysis between SERPINE1 rs1799889 polymorphism and circulating LDL-C levels.
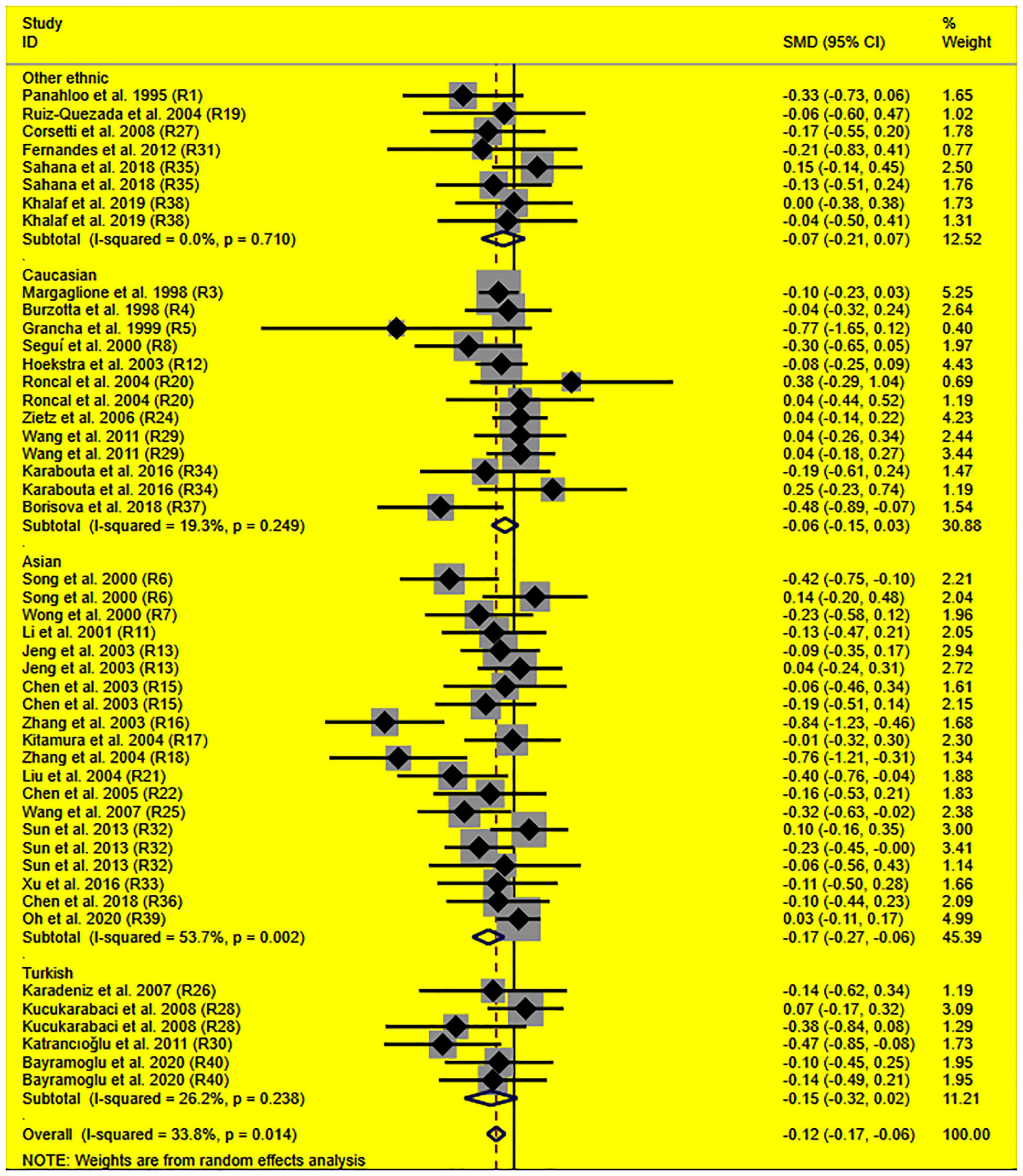
Figure 3. Forest plot of the meta-analysis between SERPINE1 rs1799889 polymorphism and circulating TC levels.
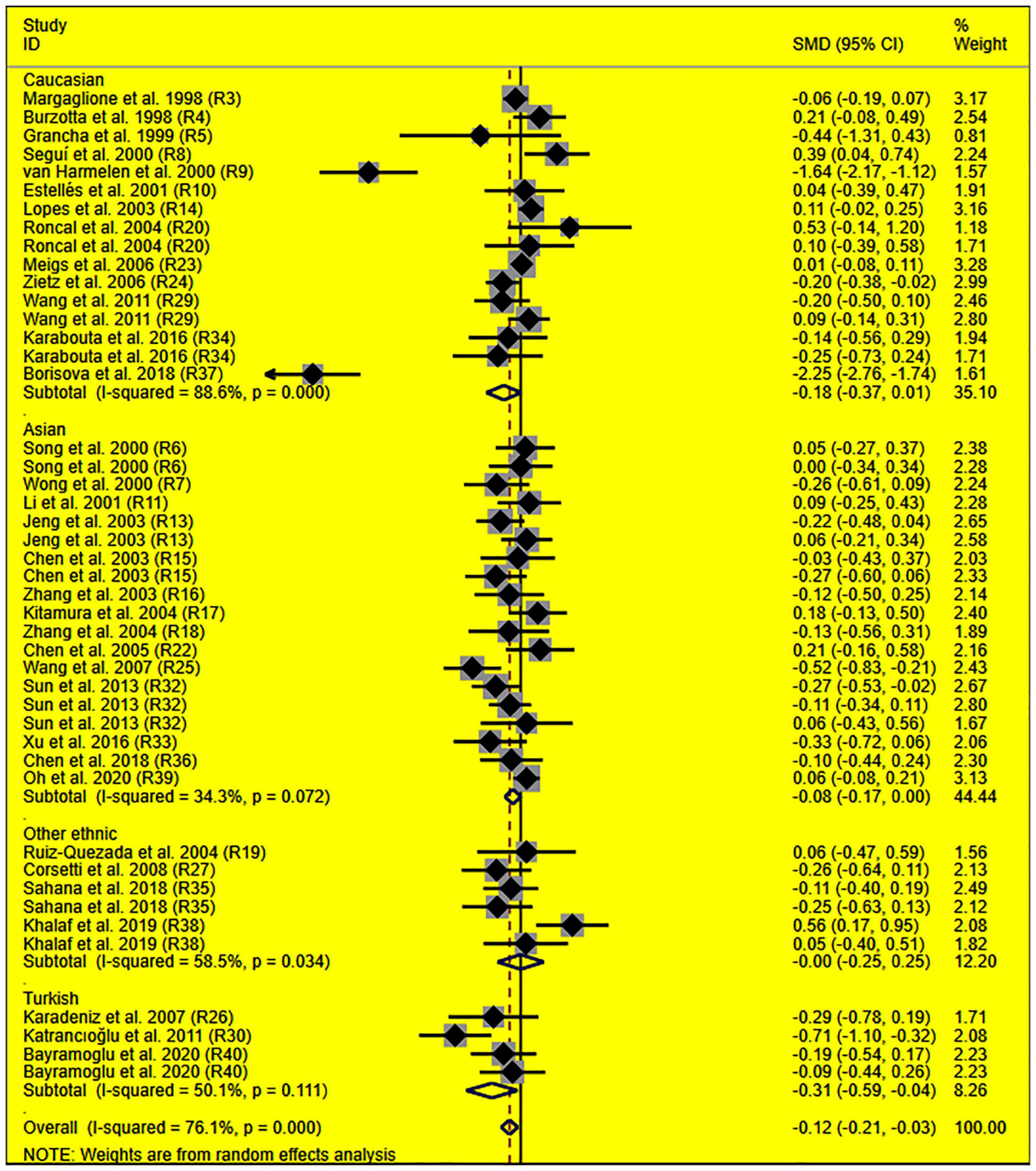
Figure 4. Forest plot of the meta-analysis between SERPINE1 rs1799889 polymorphism and circulating TG levels.
Subgroup analysis by ethnicity showed that the significant effects of the rs1799889 variant on LDL-C (SMD = –0.15, 95% CI = –0.26 to 0.05, P < 0.01) and TC (SMD = –0.17, 95% CI = –0.27 to 0.07, P < 0.01) levels were only observed in the Asian population. Meanwhile, the significant effect of the rs1799889 variant on TG (SMD = –0.31, 95% CI = –0.59 to 0.04, P = 0.03) level was only observed in the Turkish population. Moreover, a marginally significant effect of the rs1799889 variant on TG (SMD = –0.09, 95% CI = –0.17 to 0.00, P = 0.06) and HDL-C [SMD = 0.14, 95% CI = –0.02 to 0.31, P = 0.09; Figure 5] levels was observed in the Asian population (Table 1). Subgroup analysis by gender showed that the significant effect of the rs1799889 variant on HDL-C level was only detected in female subjects (Table 1).
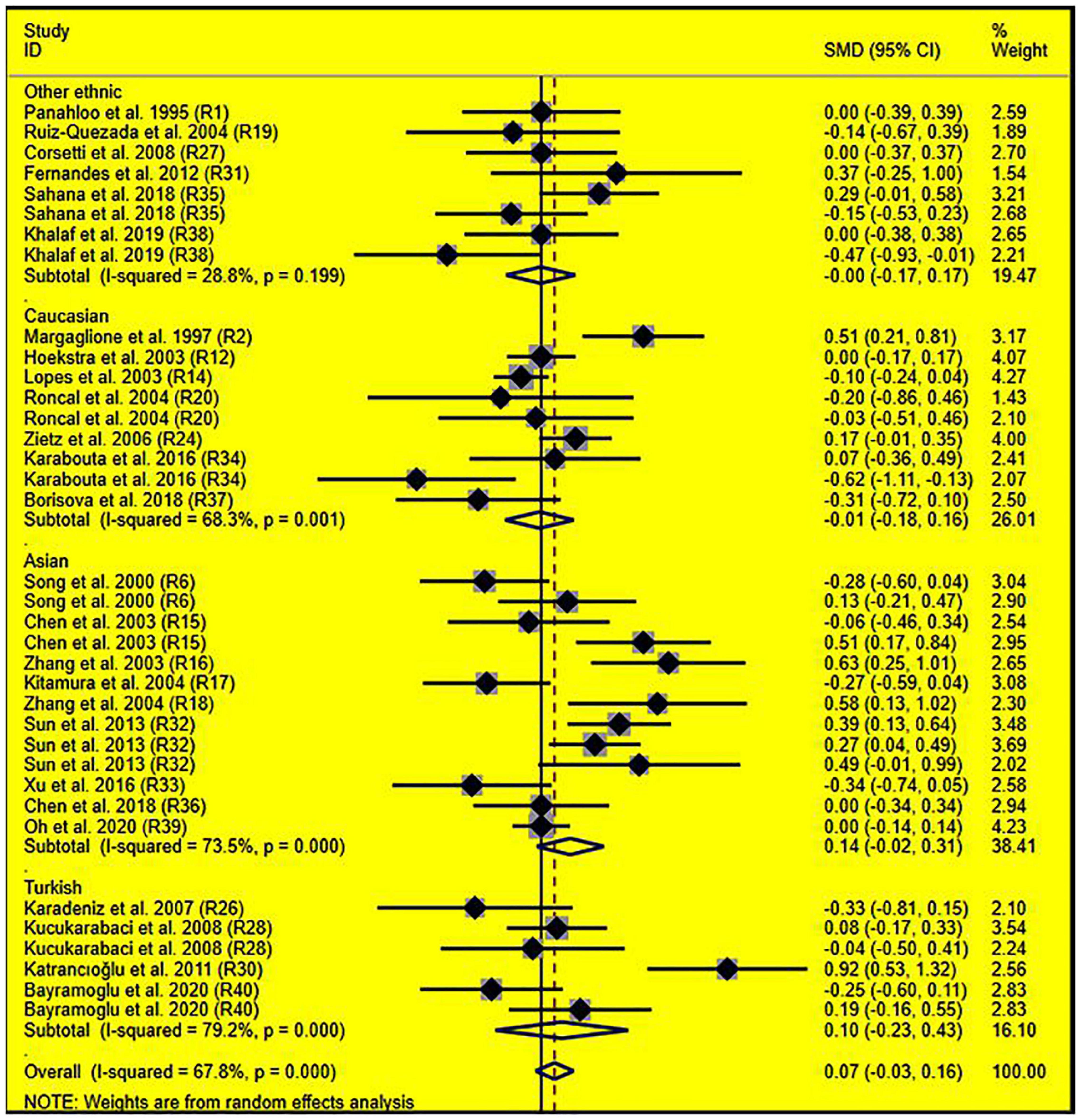
Figure 5. Forest plot of the meta-analysis between SERPINE1 rs1799889 polymorphism and circulating HDL-C levels.
The analyses were re-performed after excluding the studies with heterogeneity (Table 2). Notably, the effects of the rs1799889 variant on lipid levels did not change substantially (Table 2), indicating the synthetic results (the analysis data reflect the effect of the rs1799889 variant on lipid profile at total or subgroup level) were robust (refer to Table 2 for more details).
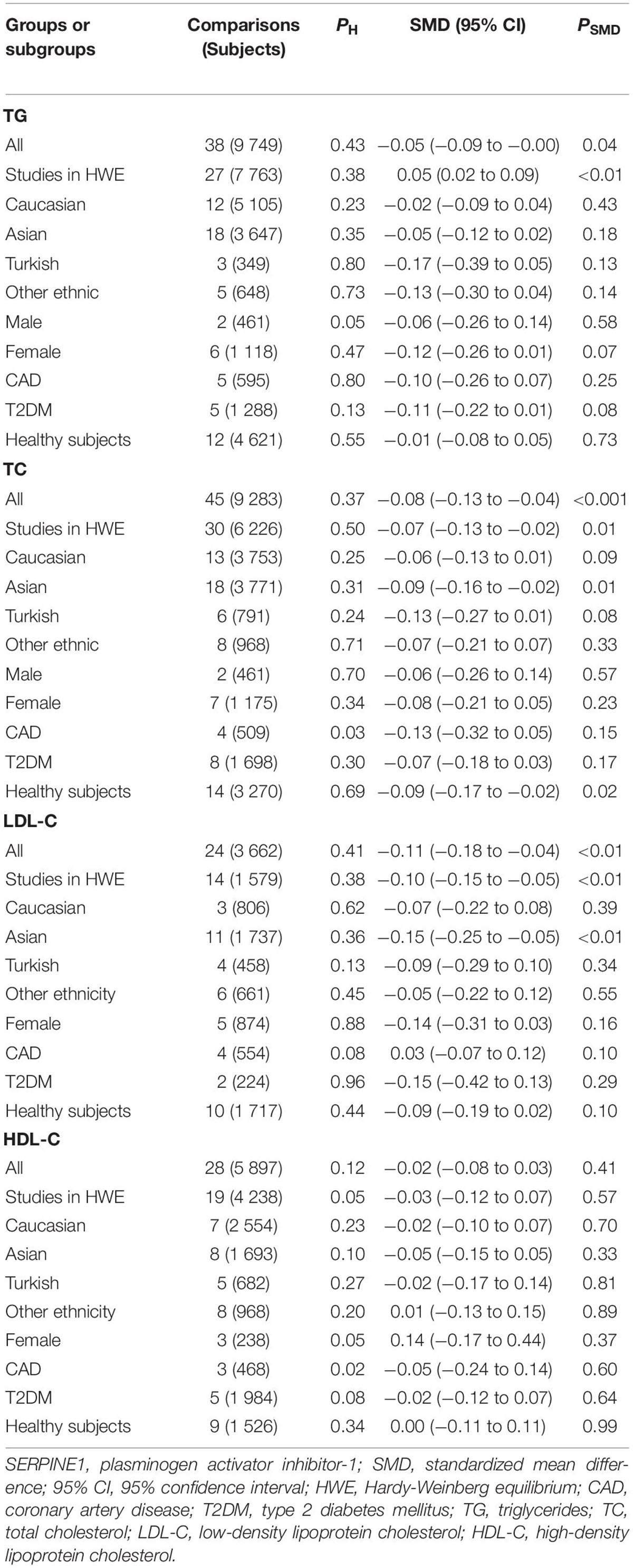
Table 2. Meta-analysis of SERPINE1 rs1799889 polymorphism with lipid levels (after eliminating the studies with heterogeneity).
Evaluation of Heterogeneity
In analyzing the effects of the rs1799889 variant on lipid profile, significant heterogeneity was observed (Table 1). Seven (Seguí, 2000; van Harmelen, 2000; Lopes, 2003; Wang, 2007; Katrancıoğlu, 2011; Borisova, 2018; Khalaf, 2019), two (Zhang, 2003, 2004), two (Zietz, 2006; Borisova, 2018), and eight (Margaglione, 1997; Chen, 2003; Zhang, 2003, 2004; Katrancıoğlu, 2011; Sun, SK1 2013, Sun SK2 2013, Karabouta, 2016) comparisons (refer to Supplementary Material for more details about reference citations) were identified as the potential sources of heterogeneity to TG, TC, LDL-C, and HDL-C, respectively. However, the recalculated results did not change significantly after excluding these comparisons (refer to Table 2 for more details).
Publication Bias Test
There was no publication bias in the synthetic results (refer to Supplementary Figures 1–4 for more details), indicating the synthetic results were reliable.
Discussion
Our data showed that the rs1799889 variant had significant effects on circulating TG, TC, and LDL-C levels. Subgroup analysis further indicated the significant effects of the rs1799889 variant on TC and LDL-C levels primarily in the Asian population.
Several underlying mechanisms could be proposed to explain the effects of the rs1799889 variant on lipids levels. (1) By repressing peroxisome proliferator-activated receptor γ (PPARγ) expression: PPARγ, a known adipogenic gene (20), plays a major role in TG synthesis (21), the inhibition of PAI-1 activity caused by rs1799889 (1, 2) may repress the expression of PPARγ (3). As a result, the synthesis of TG was reduced; accordingly, the plasma levels of TG were decreased (Tables 1, 2). (2) By disturbing the expression of PCSK9: PCSK9 plays a central role in the degradation of LDLR; the decreased PAI-1 activity caused by rs1799889 (1, 2) may reduce PCSK9 expression (6), therefore, upregulating LDLR; therefore, circulating LDL-C levels was decreased (Tables 1, 2). (3) By modulating the expression of low-density lipoprotein receptor-related protein (LRP). SERPINE1 contains a high-affinity binding site for LRP (22, 23), and the fulfilling of some important physiological functions depends on the cooperation between SERPINE1 and LRP (24, 25). Therefore, the largely decreased PAI-1 activity caused by rs1799889 (1, 2) may affect the expression of LRP.
This analysis indicated the 5G allele of the rs1799889 variant significantly decreased TG, TC, and LDL-C levels (Tables 1, 2), when combined with previous meta-analyses (7–10), indicating the 5G allele of the rs1799889 variant was a causal genetic marker for decreased CAD risk.
According to the 2018 ACC/AHA (26), the 2019 ESC/EAS (27), and the adult treatment panel III (ATP III) cholesterol guidelines (28), LDL-C was regarded as the major initiating factor for CAD and used as the main intervention target, while TC, HDL-C, and TG were used as the secondary targets. In this study, significantly reduced LDL-C levels were observed in subjects with the rs1799889 variant (Tables 1, 2), indicating the 4G/5G insertion/deletion sequence of SERPINE1 potentially be the target for CAD therapy. Moreover, subgroup analysis by ethnicity showed that the significant effects of the rs1799889 variant on LDL-C and TC levels were primarily in Asians (Tables 1, 2), indicating the Asians with the 5G allele of the rs1799889 variant may have a reduced CAD risk. Intriguingly, this speculation was verified by Li et al. (29).
Strengths and Limitations
To the best of our knowledge, this is the first meta-analysis to investigate the effects of the SERPINE1 4G/5G insertion/deletion sequence on circulating lipid levels. Several strengths of the meta-analysis should be noted. For instance, all data were recalculated after excluding the studies with heterogeneity, which no doubt advances the preciseness of conclusions drawn in this manuscript. In addition, the analysis results benefit insight into the mechanism underlying the rs1799889 variant and CAD. More importantly, our data indicate that the 4G/5G insertion/deletion sequence of the SERPINE1 gene potentially be the target for CAD therapy. However, this meta-analysis did not investigate the interaction between the rs1799889 variant and environmental factors on lipid levels due to the lack of available data from the included studies.
Conclusion
The rs1799889 variant of SERPINE1 is a protective genetic factor against CAD, and the Asian population with the 5G allele of the rs1799889 variant may have a reduced CAD risk.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
ZL, BW, JW, and YL conceived and designed this study as well as drafted the manuscript and performed the statistical analyses. HL, YZ, YP, XL, and YF carried out the searches and collected the data. All authors reviewed and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.859979/full#supplementary-material
References
1. Dawson S, Hamsten A, Wiman B, Henney A, Humphries S. Genetic variation at the plasminogen activator inhibitor-1 locus is associated with altered levels of plasma plasminogen activator inhibitor-1 activity. Arterioscler Thromb. (1991) 11:183–90. doi: 10.1161/01.atv.11.1.183
2. Eriksson P, Kallin B, van’t Hooft FM, Båvenholm P, Hamsten A. Allele-specific increase in basal transcription of the plasminogen-activator inhibitor 1 gene is associated with myocardial infarction. Proc Natl Acad Sci USA. (1995) 92:1851–5. doi: 10.1073/pnas.92.6.1851
3. Tamura Y, Kawao N, Okada K, Yano M, Okumoto K, Matsuo O, et al. Plasminogen activator inhibitor-1 is involved in streptozotocin-induced bone loss in female mice. Diabetes. (2013) 62:3170–9. doi: 10.2337/db12-1552
4. Tamura Y, Kawao N, Yano M, Okada K, Matsuo O, Kaji H. Plasminogen activator inhibitor-1 deficiency ameliorates insulin resistance and hyperlipidemia but not bone loss in obese female mice. Endocrinology. (2014) 155:1708–17. doi: 10.1210/en.2013-1888
5. Liu S, Li Y, Fan X, Li K, Xu C, Zhang L, et al. Transplantation of adipose tissue lacking PAI-1 improves glucose tolerance and attenuates cardiac metabolic abnormalities in high-fat diet-induced obesity. Adipocyte. (2020) 9:170–8. doi: 10.1080/21623945.2020.1748961
6. Levine JA, Oleaga C, Eren M, Amaral AP, Shang M, Lux E, et al. Role of PAI-1 in hepatic steatosis and dyslipidemia. Sci Rep. (2021) 11:430. doi: 10.1038/s41598-020-79948-x
7. Nikolopoulos GK, Bagos PG, Tsangaris I, Tsiara CG, Kopterides P, Vaiopoulos A, et al. The association between plasminogen activator inhibitor type 1 (PAI-1) levels, PAI-1 4G/5G polymorphism, and myocardial infarction: a Mendelian randomization meta-analysis. Clin Chem Lab Med. (2014) 52:937–50. doi: 10.1515/cclm-2013-1124
8. Gong LL, Peng JH, Han FF, Zhu J, Fang LH, Wang YH, et al. Association of tissue plasminogen activator and plasminogen activator inhibitor polymorphism with myocardial infarction: a meta-analysis. Thromb Res. (2012) 130:e43–51. doi: 10.1016/j.thromres.2012.06.015
9. Liang Z, Jiang W, Ouyang M, Yang K. PAI-1 4G/5G polymorphism and coronary artery disease risk: a meta-analysis. Int J Clin Exp Med. (2015) 8:2097–107.
10. Xu K, Liu X, Yang F, Cui D, Shi Y, Shen C, et al. PAI-1 -675 4G/5G polymorphism in association with diabetes and diabetic complications susceptibility: a meta-analysis study. PLoS One. (2013) 8:e79150. doi: 10.1371/journal.pone.0079150
11. Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. (2009) 62:e1–34.
12. Liu F, Wang S, Luo Z. Associations of the miRNA-146a rs2910164 and the miRNA-499a rs3746444 polymorphisms with plasma lipid levels: a meta-analysis. Front Genet. (2021) 12:746686. doi: 10.3389/fgene.2021.746686
13. Funk M, Endler G, Exner M, Marculescu R, Endler L, Abrahamian H, et al. PAI-1 4G/5G insertion/deletion promoter polymorphism and microvascular complications in type 2 diabetes mellitus. Wien Klin Wochenschr. (2005) 117:707–10. doi: 10.1007/s00508-005-0425-9
14. Mahmutbegovic N, Mehicevic A, Adler G, Omerhodzic I, Mahmutbegovic E, Valjevac A, et al. Bosnian study on markers of ischaemic stroke in adults 20-50 years old (SMISAO): preliminary report. Folia Biol (Praha). (2020) 66:169–78.
15. Al-Hamodi ZH, Saif-Ali R, Ismail IS, Ahmed KA, Muniandy S. Plasminogen activator inhibitor-1 4G/5G polymorphism is associated with metabolic syndrome parameters in Malaysian subjects. J Clin Biochem Nutr. (2012) 50:184–9. doi: 10.3164/jcbn.11-48
16. Rallidis LS, Gialeraki A, Merkouri E, Liakos G, Dagres N, Sionis D, et al. Reduced carriership of 4G allele of plasminogen activator inhibitor-1 4G/5G polymorphism in very young survivors of myocardial infarction. J Thromb Thrombolysis. (2010) 29:497–502. doi: 10.1007/s11239-009-0398-z
17. de la Cruz-Mosso U, Elena Ramos-Arellano L, Francisco Muñoz-Valle J, Berenice Salgado-Bernabé A, Salgado-Goytia L, Castro-Alarcón N, et al. PAI-1 haplogenotype confers genetic susceptibility for obesity and hypertriglyceridemia in Mexican children. Invest Clin. (2016) 57:246–58.
18. Zietz B, Buechler C, Drobnik W, Herfarth H, Schölmerich J, Schäffler A. Allelic frequency of the PAI-1 4G/5G promoter polymorphism in patients with type 2 diabetes mellitus and lack of association with PAI-1 plasma levels. Endocr Res. (2004) 30:443–53. doi: 10.1081/erc-200035728
19. Zietz B, Leonhardt K, Schäffler A. Kandidatengene des diabetes mellitus typ 2 Gibt es einen Gen-Dosiseffekt für Risikofaktoren sowie mikro-und makrovaskuläre Folgeerkrankungen? Med Klin (Munich). (2006) 101:605–16.
20. Yoda E, Hachisu K, Kuwata H, Nakatani Y, Hara S. Gene Deletion of calcium-independent phospholipase A(2)gamma (iPLA(2)gamma) suppresses adipogenic differentiation of mouse embryonic fibroblasts. Biol Pharm Bull. (2020) 43:1375–81. doi: 10.1248/bpb.b20-00321
21. Botta M, Audano M, Sahebkar A, Sirtori CR, Mitro N, Ruscica M. PPAR agonists and metabolic syndrome: an established role? Int J Mol Sci. (2018) 19:1197. doi: 10.3390/ijms19041197
22. Stefansson S, Muhammad S, Cheng XF, Battey FD, Strickland DK, Lawrence DA. Plasminogen activator inhibitor-1 contains a cryptic high affinity binding site for the low density lipoprotein receptor-related protein. J Biol Chem. (1998) 273:6358–66. doi: 10.1074/jbc.273.11.6358
23. Vash B, Phung N, Zein S, DeCamp D. Three complement-type repeats of the low-density lipoprotein receptor-related protein define a common binding site for RAP, PAI-1, and lactoferrin. Blood. (1998) 92:3277–85.
24. Cao C, Lawrence DA, Li Y, Von Arnim CA, Herz J, Su EJ, et al. Endocytic receptor LRP together with tPA and PAI-1 coordinates Mac-1-dependent macrophage migration. EMBO J. (2006) 25:1860–70. doi: 10.1038/sj.emboj.7601082
25. Teesalu T, Blasi F, Talarico D. Embryo implantation in mouse: fetomaternal coordination in the pattern of expression of uPA, uPAR, PAI-1 and alpha 2MR/LRP genes. Mech Dev. (1996) 56:103–16. doi: 10.1016/0925-4773(96)00515-1
26. Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: a report of the American college of cardiology/American heart association task force on clinical practice guidelines. Circulation. (2019) 139:e1082–143.
27. Mach F, Baigent C, Catapano AL, Koskinas KC, Casula M, Badimon L, et al. 2019 ESC/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J. (2020) 41:111–88.
28. National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III). Third report of the national cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III) final report. Circulation. (2002) 106:3143–421.
Keywords: rs1799889, polymorphism, lipid, coronary artery disease, Asian
Citation: Luo Z, Liu Y, Li H, Zhou Y, Peng Y, Lin X, Fang Y, Wan J and Wei B (2022) Systematic Review and Meta-Analysis of SERPINE1 4G/5G Insertion/Deletion Variant With Circulating Lipid Levels. Front. Cardiovasc. Med. 9:859979. doi: 10.3389/fcvm.2022.859979
Received: 17 February 2022; Accepted: 30 May 2022;
Published: 23 June 2022.
Edited by:
Neil Morgan, University of Birmingham, United KingdomReviewed by:
Guangyu Zhang, City of Hope National Medical Center, United StatesYingfeng Wan, University of Michigan, United States
Copyright © 2022 Luo, Liu, Li, Zhou, Peng, Lin, Fang, Wan and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Wan, wanjing2017@163.com; Baozhu Wei, wbz9810@163.com
†These authors have contributed equally to this work and share first authorship
 Zhi Luo
Zhi Luo Yang Liu2†
Yang Liu2† Yawen Zhou
Yawen Zhou Baozhu Wei
Baozhu Wei