
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med., 14 December 2022
Sec. Cardiovascular Genetics and Systems Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fcvm.2022.1061771
Abdominal aortic aneurysm (AAA) is a severe life-threatening disease that is generally asymptomatic and is diagnosed at a very late stage. The genetic component underpinning AAA is considerable, with an estimated heritability of up to 70%. Therefore, identifying genetic biomarkers for AAA is valuable for predicting high-risk populations. We used integrative bioinformatics and cellular AAA model-based validation to reveal that the gene encoding protein tyrosine phosphatase non-receptor type 22 (PTPN22) may be a potentially useful diagnostic biomarker for AAA. Integrative bioinformatics analyses of clinical specimens showed that PTPN22 expression was consistently upregulated in aortic tissues and peripheral blood mononuclear cells (PBMCs) derived from patients with AAA. Moreover, transcriptomics data revealed that PTPN22 is a potential biomarker for AAA with limited diagnostic value in patients with thoracic aortic aneurysm/dissection. Single-cell RNA sequencing-based findings further highlight PTPN22 expression in aortic immune cells and vascular smooth muscle cells (VSMCs) is consistently upregulated in patients with AAA. A cellular AAA model was eventually employed to verify the increase in PTPN22 expression. Collectively, the results indicate that PTPN22 could be a potentially useful diagnostic biomarker for AAA.
Abdominal aortic aneurysm (AAA) is a severe life-threatening disease with an overall incidence of 6% in men and 1.6% in women (1). The hallmark of AAA is localized enlargement of the infrarenal aorta with a diameter of > 3.0 cm (2). Unless there is a rapid increase in size or rupture, AAA is generally asymptomatic and is diagnosed at a very late stage (1). The occurrence of AAA is highly associated with an unhealthy lifestyle, such as a history of smoking or hypertension (3). However, the genetic component underpinning AAA is substantial, with an estimated heritability of up to 70% (4). Difficulties remain in terms of the implementation of screening programs, and current knowledge on the genetic component of AAA is insufficient to guide early screening in the clinic. Therefore, identifying biomarkers for AAA is valuable for predicting high-risk populations (5).
High-throughput platform-based biomarker identification has been highlighted as a promising approach for the diagnosis and prevention of AAA and other diseases (6–8). The discovery of therapeutic targets is generally based on a consistent increase or decrease in the expression of target genes in patients and/or experimental models, followed by genetic manipulation and/or targeted drug-based screening, which allow confirmation of the therapeutic targets and subsequent identification of feasible treatment options (9–12). Integrative bioinformatics and experimental validation are highly efficient tools for identifying biomarkers to accurately predict the occurrence of AAA (6, 13). As proof-of-concept, several publications have employed these research strategies to identify biomarkers with diagnostic and prognostic value in several diseases (7, 8, 14–16).
In the current study, we used a comprehensive bioinformatics-based analysis and experimental validation to verify the gene encoding protein tyrosine phosphatase non-receptor type 22 (PTPN22) as a potentially specific biomarker for patients with AAA.
To profile the genetic alterations in aortic tissues collected from patients with AAA, two datasets with similar grouping characteristics were retrieved (Figure 1A). As shown in Figure 1B, differentially expressed genes (DEGs) were identified in patients with AAA based on the GSE47472 dataset (Supplementary Figure 1). Among these DEGs, 169 were significantly upregulated, while 155 were significantly downregulated. The upregulated DEGs were highly enriched in biological processes, such as the perinuclear region of the cytoplasm and regulation of transcription (Figure 1C). Based on the GSE7084 dataset (Supplementary Figure 2), 1,653 AAA-specific DEGs were identified (Figure 1D), which consisted of 698 significantly upregulated and 955 significantly downregulated genes. Different from the GSE47472 dataset, these upregulated DEGs were highly enriched in the inflammatory response and the immune response (Figure 1E). To obtain more reliable candidates associated with AAA, we cross-compared the upregulated genes from both datasets. Four genes, including PTPN22, CPVL, ARHGDIB, and ANGPTL6, were concurrently upregulated (Figure 1F). The receiver operating characteristic (ROC) curve results based on the GSE47472 (Figure 1G) and GSE7084 (Figure 1H) datasets further revealed that these four candidates were highly predictive of the AAA diagnosis. These results suggest that PTPN22, CPVL, ARHGDIB, and ANGPTL6 are potential diagnostic biomarkers for patients with AAA.
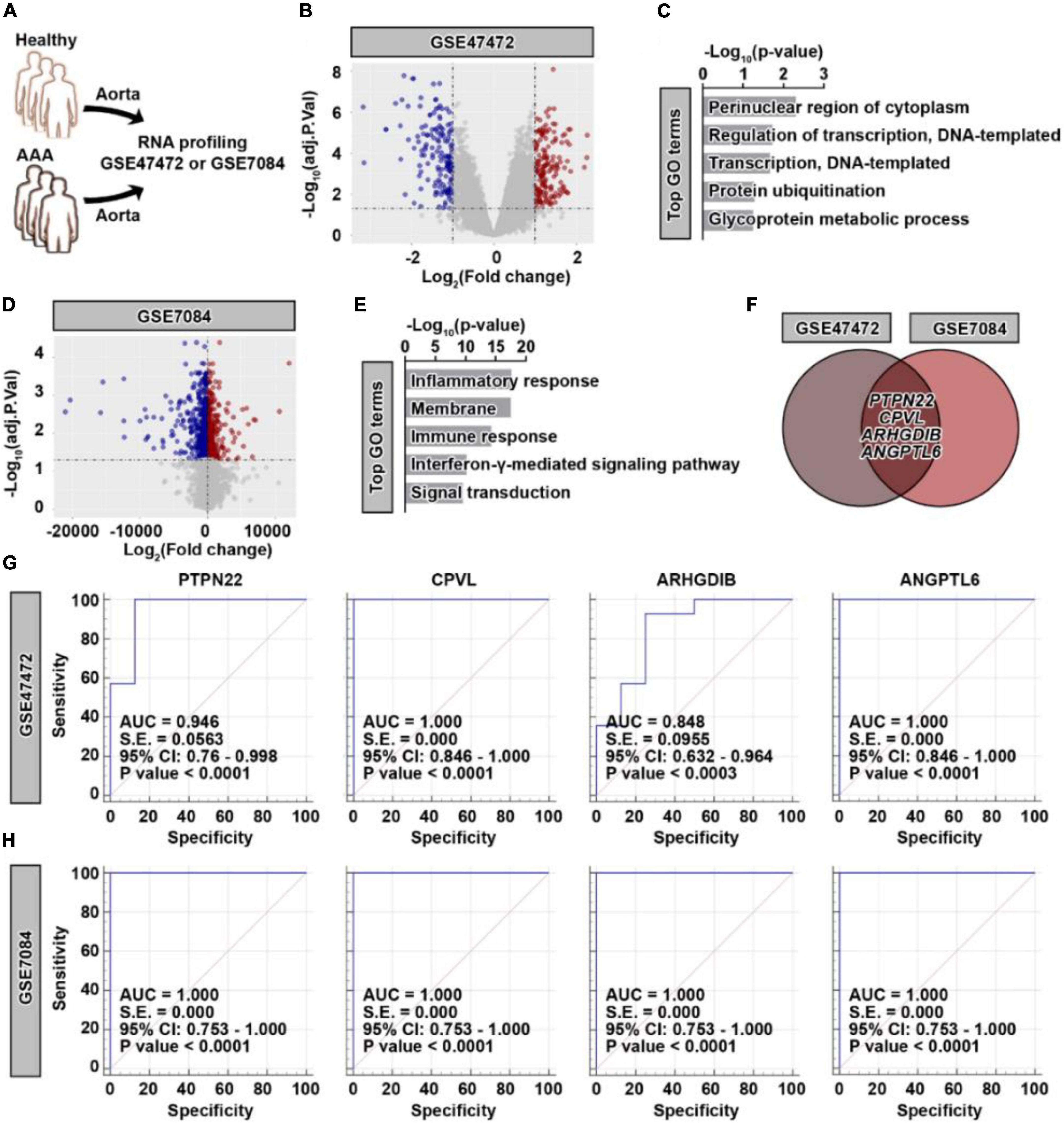
Figure 1. Identification of four potential genetic biomarkers for abdominal aortic aneurysm (AAA). (A) Schematic of the GSE47472 and GSE7084 datasets. (B) Volcano plot of differentially expressed genes (DEGs) in AAA samples based on the GSE47472 dataset. Red, significantly upregulated genes; blue, significantly downregulated genes; gray, no significant difference. A fold change of ≥ 2 and a p-value of < 0.05 were considered statistically significant. (C) The top enriched Gene Ontology (GO) terms of significantly upregulated DEGs in AAA samples. (D) Volcano plot of DEGs in AAA samples based on the GSE7084 dataset. Red, significantly upregulated genes; blue, significantly downregulated genes; gray, no significant difference. A fold change of ≥ 2 and a p-value of < 0.05 were considered statistically significant. (E) The top enriched GO terms of significantly upregulated DEGs in AAA samples. (F) Venn diagram of significantly upregulated DEGs between the GSE47472 and GSE7084 datasets. Receiver operating characteristic curve analysis of PTPN22, CPVL, ARHGDIB, and ANGPTL6 based on the GSE47472 (G) and GSE7084 (H) datasets.
Considering that the aortic diameter in AAA is highly associated with the expression of genetic biomarkers, the GSE57691 dataset was analyzed to precisely identify reliable biomarkers (Figure 2A and Supplementary Figure 3). Among the four identified genes (PTPN22, CPVL, ARHGDIB, and ANGPTL6), we found that PTPN22 expression was consistently upregulated in both small and large AAA samples (Figure 2B). To explore whether the increase in PTPN22 expression is conserved among species, the GSE109039 dataset was retrieved (Supplementary Figures 4A,B). An exclusive increase in the expression of PTPN22 was observed in the experimental AAA model (Supplementary Figure 4C). To further validate the expression and diagnostic value of PTPN22, an independent cohort of peripheral blood mononuclear cells (PBMCs) from patients with AAA was derived (17). As expected, PTPN22 expression was significantly increased in PBMCs from AAA patients (Figure 2C). The diagnostic value (p = 0.0067) of PTPN22 in patients with AAA was also confirmed by the ROC curve analysis (Figure 2D). Taken together, these results reveal that PTPN22 is a genetic biomarker for patients with AAA.
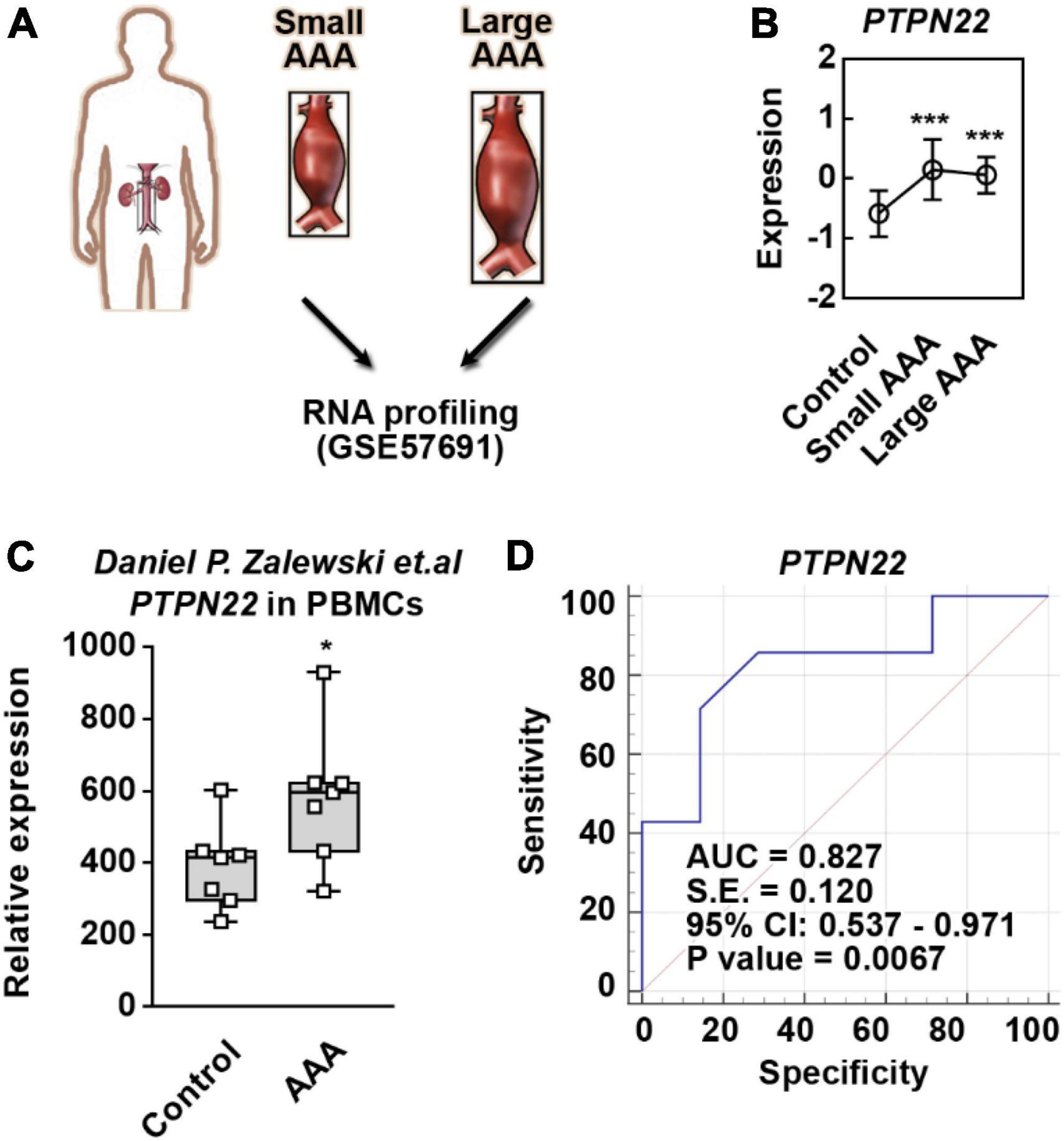
Figure 2. PTPN22 is a genetic biomarker for abdominal aortic aneurysm (AAA). (A) Schematic of the GSE57691 dataset. Abdominal aortic tissues collected from patients with small and large AAA were profiled by RNA sequencing. Small AAA, mean maximum aortic diameter of 54.3 ± 2.3 mm; large AAA, mean maximum aortic diameter of 68.4 ± 14.3 mm. (B) PTPN22 expression based on the GSE57691 dataset. The data are shown as the mean ± standard deviation (SD) of at least 10 patients. * Significantly different from control; ***p < 0.001. The expression (C) and receiver operating characteristic curve (D) analyses of PTPN22 in peripheral blood mononuclear cell (PBMC) samples collected from control participants and patients with AAA based on an independent cohort. The data are shown as the mean ± SD of seven patients. *Significantly different from control; ***p < 0.001.
Single-cell RNA sequencing is a powerful tool that can be used to dissect cellular heterogeneity within tissues (18). To explore the potential cellular origin of upregulated PTPN22 within aortic tissues from patients with AAA, the GSE166676 dataset, which is consisted of two healthy volunteers and four patients with AAA (18), was retrieved (Figure 3A). First, the clustering analysis revealed 10 distinct cell populations within aortic tissues (Figure 3B), which is consistent with a previous report (18). As shown in Figure 3C, PTPN22 expression was consistently upregulated in aortic tissues from patients with AAA. To determine the potential origin, aortic cells were re-clustered using lineage-specific biomarkers, including EPCAM (epithelial cells), CLDN5 (endothelial cells), COL1A2 (mesenchymal cells), and PTPRC (immune cells) (Figure 3D). As shown in Figure 3E, the distribution of PTPN22-expressing cells was highly co-localized with clusters assigned as immune cells, suggesting that AAA-upregulated PTPN22 may be derived from aortic immune cells. Collectively, these findings suggest that PTPN22 expression in aortic immune cells is remarkably upregulated in patients with AAA.
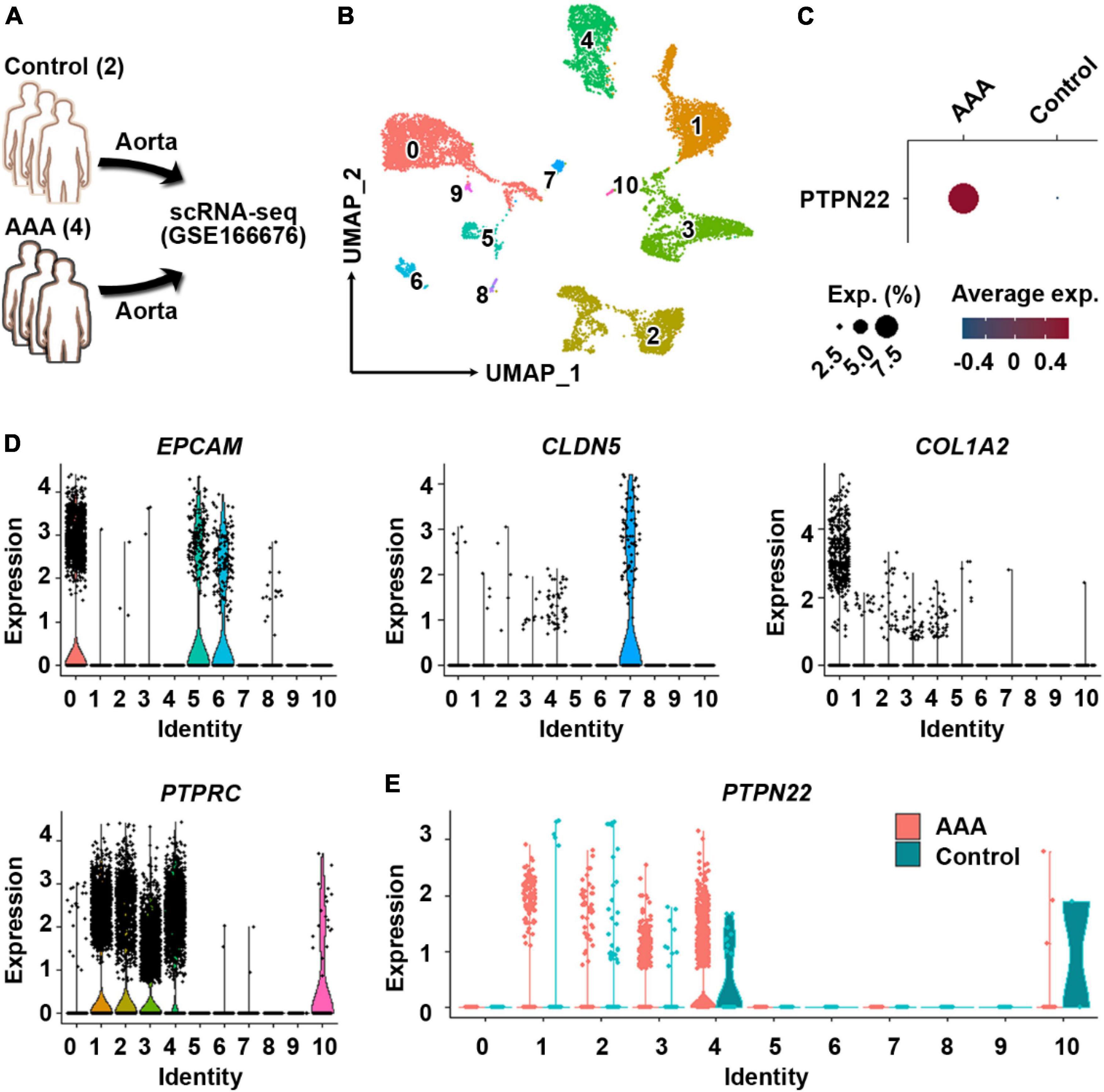
Figure 3. PTPN22 upregulation in aortic tissues from patients with abdominal aortic aneurysm (AAA) is largely distributed in resident immune cells. (A) Schematic of the GSE166676 dataset. scRNA-seq, single-cell RNA sequencing. (B) Uniform manifold approximation and projection (UMAP) plot showing that aortic tissues were divided into 11 distinct clusters. (C) Dot plot showing that PTPN22 expression was highly increased in aortic tissues from patients with AAA. (D) Violin plots showing the expression of EPCAM, CLDN5, COL1A2, and PTPRC among the 11 distinct clusters. (E) Violin plot showing the expression of PTPN22 between control participants and patients with AAA among the 11 distinct clusters.
To precisely determine the cellular origin of AAA-upregulated PTPN22, immune cells were extracted (Figure 4A) and re-clustered with their identities annotated using reported marker genes (Figure 4B). As expected, PTPN22 expression in immune cells was also upregulated in AAA samples (Figure 4C) when compared with control samples. As shown in Figure 4D, PTPN22 expression was uniquely distributed in aortic immune cell subpopulations, including T cells, natural killer (NK) cells, B cells, neutrophils, and dendritic cells (DCs), but not in basophils and monocytes. Together, these results reveal that upregulated PTPN22 expression in immune cells from AAA patients is mainly distributed in the subpopulations, including T cells, NK cells, and B cells.
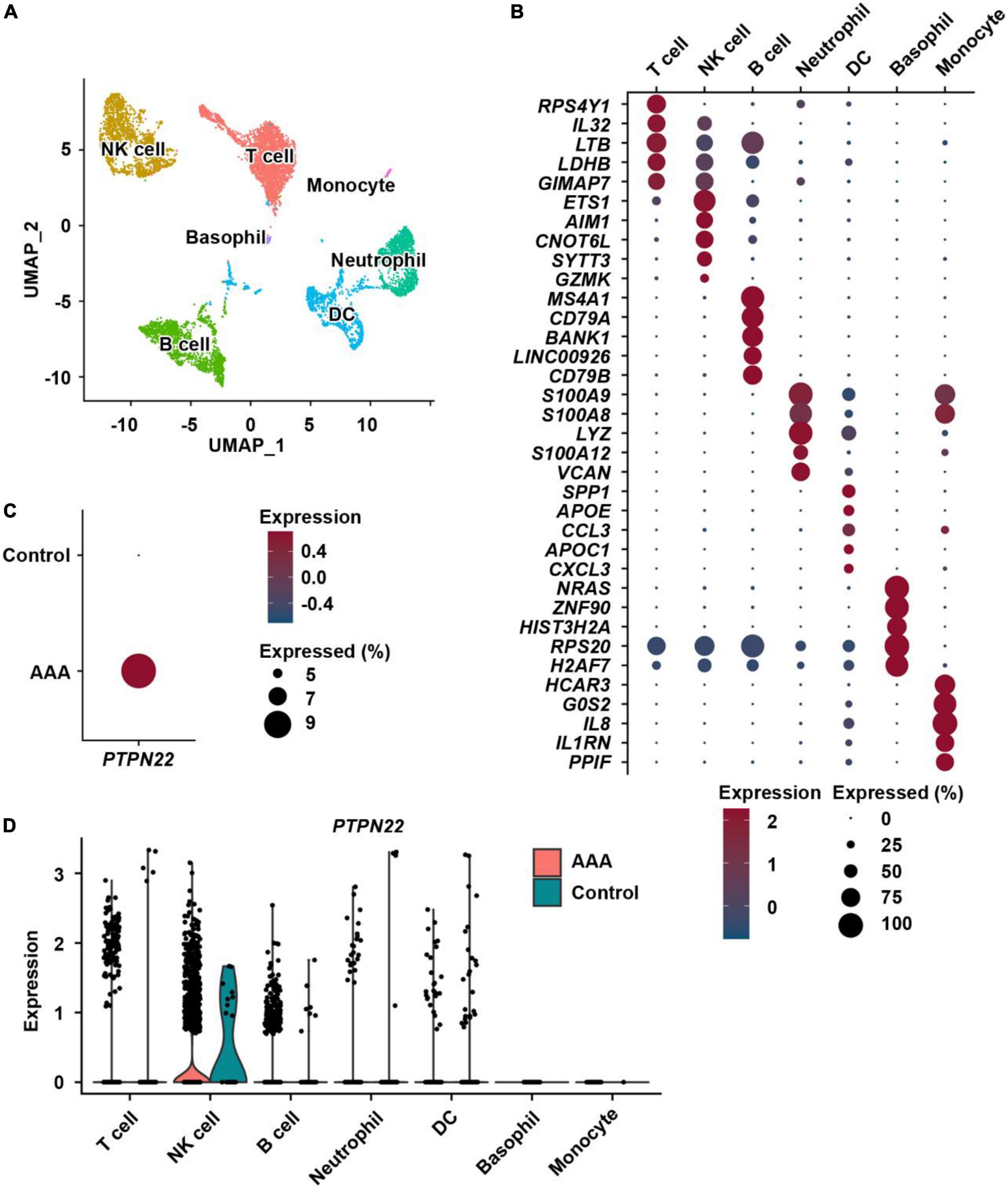
Figure 4. Distribution analysis of PTPN22 within resident immune cells. (A) UMAP plot showing immune cell subpopulations within aortic tissues. NK cell, natural killer cell; DC, dendritic cell. (B) Dot plot of the top five marker genes for each immune cell subpopulation. (C) Dot plot showing that the expression of PTPN22 was highly increased in immune cells from patients with AAA. (D) Violin plot showing the expression of PTPN22 between control participants and patients with AAA among the distinct immune cell subpopulations.
To evaluate the expression and diagnostic value of PTPN22 in patients with TAAD, thoracic aortic tissues from 11 patients with TAAD and 8 control participants were profiled by RNA sequencing (Figure 5A). We identified 909 DEGs that were significantly upregulated in TAAD samples, and the expression patterns of these genes among various samples are shown using heatmap (Figure 5B). The significantly upregulated DEGs were highly enriched in certain Gene Ontology (GO) terms, including programmed cell death and metabolic processes (Figure 5C), revealing the prominent role of cell death within aortic tissues during TAAD progression. However, unexpectedly, the expression of PTPN22 in TAAD samples was unaltered when compared with control samples (Figure 5D). As shown in Figure 5E, the ROC curve showed that the expression of PTPN22 lacked diagnostic value for TAAD in our aortic tissue-based analysis (p = 0.9388). Although TAAD and AAA are anatomically inextricable, they share several diversities (19, 20). The GSE9106 dataset, which included peripheral blood cell-based RNA profiling of TAA, was retrieved to evaluate the association of PTPN22 with TAA (Supplementary Figures 5A,B). However, PTPN22 expression was unaltered between controls and patients with TAA (Supplementary Figure 5C). The ROC curve analysis further showed that the area under the curve (AUC) for PTPN22 (p = 0.6081) was 0.531 (Supplementary Figure 5D), indicating the limited diagnostic value of PTPN22 for TAA. Collectively, these clinical TAAD sample-based findings confirm that the expression and diagnostic value of PTPN22 is restricted to AAA, rather than TAAD.
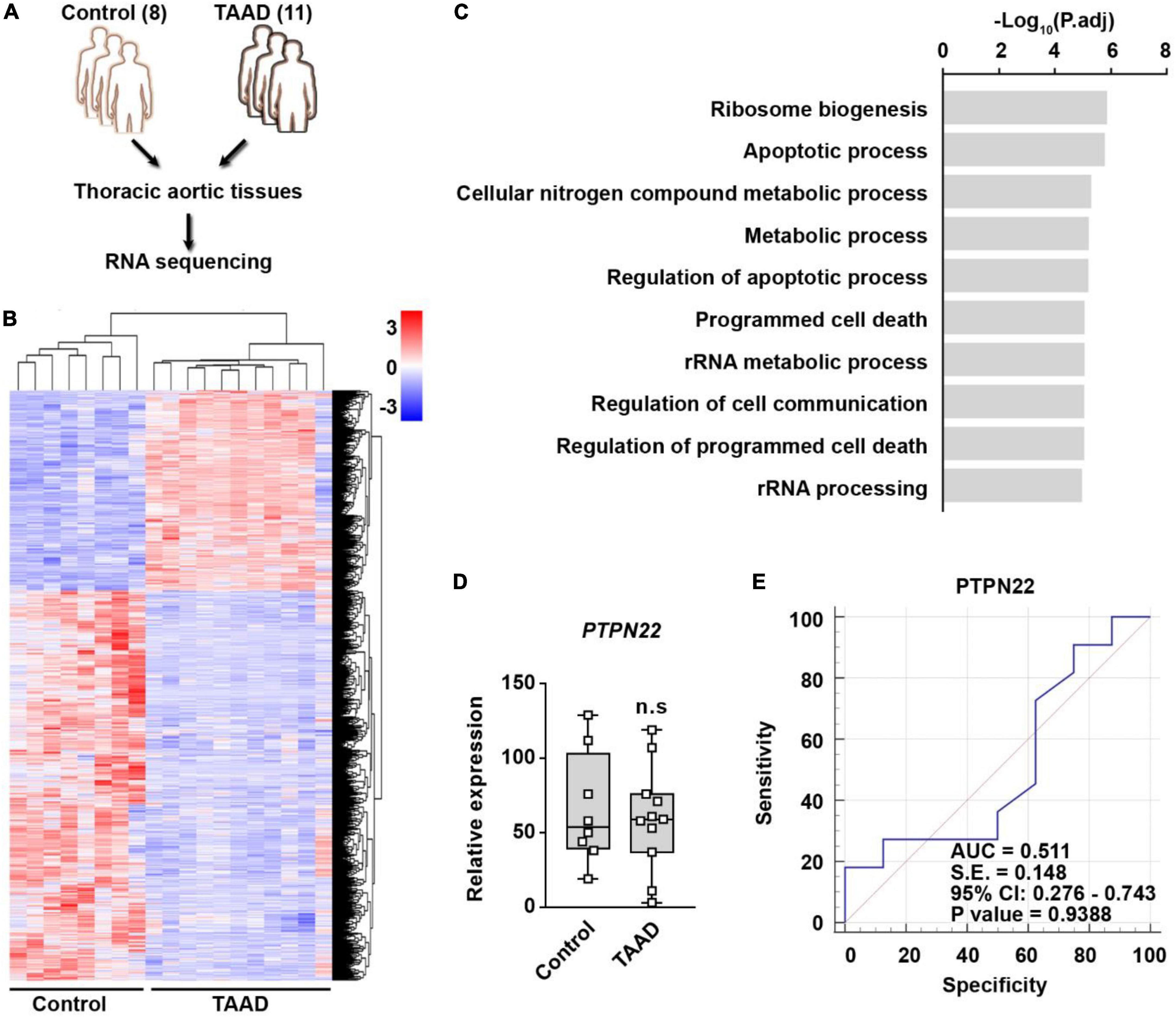
Figure 5. PTPN22 lacks diagnostic value for TAAD. (A) Schematic of the experimental design. Thoracic aortic tissues collected from 11 patients with TAAD and 8 control participants were subjected to RNA sequencing. (B) Heatmap showing differentially expressed genes (DEGs) between control and TAAD samples. (C) The top enriched Gene Ontology terms of the significantly upregulated DEGs in TAAD samples. Expression (D) and receiver operating characteristic (ROC) curve (E) analyses of PTPN22 in aortic tissues collected from control participants and patients with TAA. n.s., not significant.
VSMCs are the only cell type within the aortic media, and loss of VSMCs resulting from apoptotic cell death contributes etiologically to both human and experimental AAA (21, 22). To explore the potential role of PTPN22 in aortic VSMCs, cells expressing ACTA2, a marker gene for VSMC, were further extracted from scRNA-seq data analyzed above (Supplementary Figure 6). As expected, the expression of PTPN22 was consistently increased in VSMCs from AAA patients (Figure 6A). Although a murine AAA model induced by calcium chloride (CaCl2) has been well-established (23, 24), there is still a lack of ideal cellular AAA models for in vitro studies (25). To confirm that PTPN22 expression was increased in the in vitro AAA model, a CaCl2-based apoptotic VSMC model was adopted (Figure 6B) (24, 26). As shown in Figure 6C, VSMCs were morphologically changed in response to CaCl2 treatment in a concentration-dependent manner. Consistent with a previous report (26), VSMCs treated with CaCl2 at a concentration of 100 mmol/L showed remarkably diminished α-smooth muscle actin (α-SMA) expression that did not extend entirely across the cell body (Figures 6D,E), suggesting that VSMCs treated with CaCl2 (100 mmol/L) can reproduce the aortic aneurysmal phenotype in vitro. Moreover, PTPN22 expression was significantly increased in VSMCs treated with CaCl2, as assessed by quantitative real-time polymerase chain reaction (qRT-PCR) (Figure 6F). Taken together, these results further highlight the important role of PTPN22 in AAA.
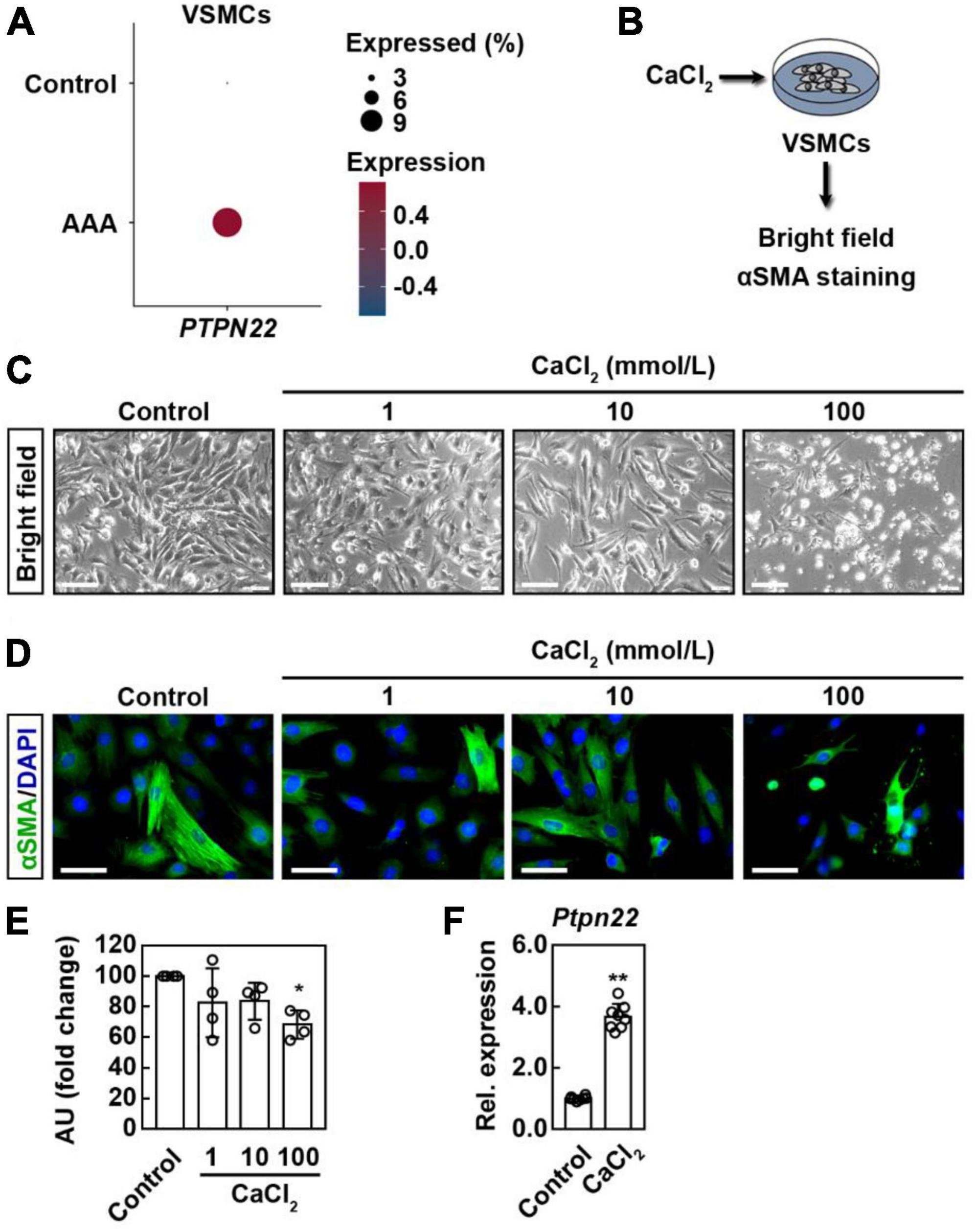
Figure 6. PTPN22 expression is consistently upregulated in VSMCs from AAA patients and cellular model. (A) Dot plot showing that the expression of PTPN22 was highly increased in vascular smooth muscle cells (VSMCs) from patients with AAA based on scRNA-seq data. (B) Schematic of the experimental design. Representative bright field (C) and immunofluorescence (D) images of VSMCs treated with different concentrations of calcium chloride (CaCl2). For (C), representative images of four biological replicates from two independent experiments are shown. Scale bar, 100 μm. For (D), the cells were stained with α-smooth muscle actin (α-SMA) (green), and the nuclei were labeled with DAPI (blue). Representative images of four biological replicates from two independent experiments are shown. Scale bar, 50 μm. For (E), the data are shown as the mean ± standard deviation of four biological replicates. *Significantly different from control; *p < 0.05. AU, Arbitrary Unit. (F) VSMCs were treated with CaCl2 (100 mmol/L), and relative (Rel.) expression of cellular PTPN22 was assessed by quantitative real-time polymerase chain reaction. The data are shown as the mean ± standard deviation of four biological replicates. *Significantly different from control; **p < 0.01.
AAA is generally asymptomatic before rupture; thus, diagnosing AAA at an early stage and elucidating the underlying mechanisms leading to progressive dilatation are two major challenges in the clinic. Identification of genetic biomarkers is useful to develop novel diagnostic and therapeutic approaches (2). Previous studies have identified several potential biomarkers for AAA through studying the levels of different molecules related to the pathological mechanisms of AAA (27, 28). In this study, we demonstrated that PTPN22 may be a valuable AAA-specific diagnostic biomarker. PTPN22 was first associated with type 1 diabetes mellitus, and it has since demonstrated involvement in the pathogenesis of various autoimmune diseases, such as rheumatoid arthritis (29). Lymphoid tyrosine phosphatase, which is encoded by PTPN22, is expressed in all leukocyte linages. Although the increased expression and diagnostic value of PTPN22 were verified using a peripheral blood sample-based cohort in this study, more PBMC-based examinations of PTPN22 between healthy controls and patients with AAA need be conducted to confirm its clinical value as a biomarker for AAA. Moreover, PTPN22 in circulating extracellular vesicles in peripheral blood also deserves to be investigated (30, 31).
People with high cholesterol and a history of smoking are at a high risk of AAA, and biomarker-based monitoring of patients diagnosed with AAA at an early stage also has long-term benefits. Thus, genetic screening among these populations is valuable (32). Two independent cohort-based genome-wide association studies for AAA have identified nine risk loci, and several genes have been reported to have vital functions in the pathogenesis of AAA (13, 33). Previous studies have also highlighted that targeting these crucial molecules could prevent AAA development (34, 35). For instance, imatinib can prevent aneurysm progression by inhibiting the expression and activation of matrix metallopeptidase 9 in experimental AAA models (35). Here, we identified that PTPN22 may be a potential therapeutic target for AAA, which is supported by the observation that PTPN22 expression is well replicated in experimental AAA models induced by CaCl2. Identification of therapeutic targets is generally based on a consistent upregulation or downregulation in the expression of target genes. Then, genetic manipulation and/or targeted drug-based screening allow the therapeutic targets to be confirmed, which could in turn help to identify feasible treatment options (9–12). Therefore, genetic deletion or small molecule-based inhibition of PTPN22, such as with PTPN22-IN-1, would be helpful for the development of therapeutic agents to prevent AAA progression (36).
AAA results from changes in the aortic wall structure, including thinning of the media and adventitia due to loss of VSMCs and extracellular matrix degradation (2). However, as a protein tyrosine phosphatase, the intracellular distribution of the protein encoded by PTPN22 is largely restricted to the nucleus and cytoplasm. Here we provide the first evidence at single-cell level that a remarkable increase in the expression of PTPN22 in both aortic immune cells and VSMCs from patients with AAA. However, functionally, the protein encoded by PTPN22 serves as a intracellularly negative regulator of T cell receptor signaling. Therefore, it seems unlikely that this protein would be secreted from immune cells and VSMCs as its upregulated (37). To the best of our knowledge, no previous studies have determined whether the protein encoded by PTPN22 can be secreted by aortic immune cells and/or VSMCs, as well as its intermediate role between these two type of cells within aortic tissues during the pathogenesis of AAA. We thus speculate that the steady state within local aortic tissues would be disrupted as a result of a remarkable increase in PTPN22 expression in aortic immune cells and/or VSMCs. We believe that a cascade amplification reaction resulting from immune cells and/or VSMCs mediated by PTPN22 upregulation may play a critical role in the occurrence and progression of AAA.
Although AAA and TAAD share multiple similarities, they also demonstrate several differences, including the population prevalence, mode of inheritance, and predisposing genes (19). Genetic disorders and congenital factors are common causes of TAAD. For example, mutations in the gene encoding fibrillin-1 can cause progressive ascending aortic dilatation. The aortic wall of the ascending aorta also has a higher blood pressure than the abdominal aorta (38). Atherosclerosis is the most important risk factor for AAA (19), and atherosclerotic tissues generally demonstrate excessive resident immune cell activation and aberrant activation of VSMCs (39, 40). This microenvironment may lead to a cascade amplification of PTPN22 production by immune cells and VSMCs, resulting in a significant increase in expression in local arterial immune cells and VSMCs in patients with AAA. The difference in the expression of PTPN22 between patients with TAAD and patients with AAA may be largely due to differences in the mechanisms leading to these diseases. We thus believe that more in-depth basic and clinical research is needed to understand the difference in the expression of PTPN22 between patients with AAA and those with TAAD.
In conclusion, our integrative bioinformatics analyses have highlighted the potential diagnostic and therapeutic value of PTPN22 as a biomarker for AAA. However, more experimental and clinical studies should be conducted to further elucidate the importance of PTPN22 in AAA.
The datasets were retrieved from the Gene Expression Omnibus database1 and were analyzed using RStudio. An adjusted p-value of < 0.05 and a | Log2FC| of ≥ 1 were considered statistically significant. The GSE47472 dataset contained 8 control aortic tissues and 14 AAA neck tissues (41), and the GSE7084 dataset contained 8 control abdominal aortic tissues and 7 AAA tissues. The GPL2507 platform was adopted (42). The GSE57691 dataset consisted of 10 control aortic tissues from organ donors, 20 aortic tissues from patients with small AAA (mean maximum aortic diameter of 54.3 ± 2.3 mm), and 29 aortic tissues from patients with large AAA (mean maximum aortic diameter of 68.4 ± 14.3 mm) (43). The GSE109639 dataset contained tissues from 6 sham mice and 6 mice with experimental AAA induced by CaCl2 (44). The GSE16676 dataset contained tissues from two healthy volunteers and four patients with AAA (18). A count matrix consisting of transcriptomics data of PBMCs from patients with AAA and healthy volunteers was deposited previously (17).
To explore the potential biological functions of the identified DEGs, the Database for Annotation, Visualization and Integrated Discovery2 was employed to conduct the GO analysis (45).
The ROC curve was generated using MedCalc software (version 19.0.7), and the AUC value with 95% confidence interval was calculated.
VSMCs (American Type Culture Collection, Cat#PCS-100-012) were kindly provided by Dr. Tao Zhuang (Shanghai East Hospital, Tongji University School of Medicine) (46). VSMCs were maintained in Dulbecco’s Modified Eagle Medium (Gibco, US) supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin (Gibco), and 100 μg/mL streptomycin (Gibco). VSMCs were incubated at 37°C in a humidified atmosphere supplemented with 5% carbon dioxide. VSMCs at passages 12–15 were used in the present study. To build a reliable experimental AAA model in vitro, CaCl2 at different concentrations (0, 1, 10, and 100 mmol/L) was used to treat VSMCs for 24 h, as described previously (24, 26). Treatment with 100 mmol/L CaCl2 for 24 h was ultimately selected to induce AAA in vitro.
For α-SMA immunofluorescence staining, the cells were fixed with 4% paraformaldehyde and blocked with phosphate-buffered saline containing 10% normal goat serum, as previously described (47). The cells were then incubated with primary antibody against α-SMA (Cat#ab7817, Abcam) followed by the corresponding secondary antibody. After, the cells were incubated with DAPI and images were captured using a microscope.
Total RNA was purified using TRIzol (Invitrogen, US), and cDNA libraries were synthesized with the Reverse Transcription Reagent kit (Takara, Japan). cDNA was submitted for qRT-PCR using SYBR Green mix (Takara) with GAPDH normalization. Primers (Supplementary Table 1) were purchased from Sangon Biological Engineering (Shanghai, China). The qRT-PCR data were analyzed using the comparative Ct (Δ Δ Ct) method (48).
The study was performed in accordance with the Declaration of Helsinki and was approved by the ethics committee of our institution, and all patients gave written informed consent before operation (approval number: 2021-KY-079). Thoracic aortic tissues were collected from 11 patients with TAAD and eight control participants. Written informed consent was obtained from all of the study participants.
The data are shown as the mean ± standard deviation. Paired groups were compared using the two-tailed Student’s t-test, while multiple groups were compared using the one-way analysis of variance with Tukey’s multiple comparisons test. A p-value of < 0.05 was considered statistically significant. Statistical analyses were performed using GraphPad Prism 7.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found in the article/Supplementary material.
The studies involving human participants were reviewed and approved by The First Affiliated Hospital of USTC (approval number: 2021-KY-079). The patients/participants provided their written informed consent to participate in this study.
XK and JG: conceptualization, supervision, project administration, and funding acquisition. PR and LG: methodology, software, and formal analysis. PR: validation, resources, and writing—original draft preparation. XK and PR: investigation. TC and HJ: data curation. XK: writing—review and editing. TC: visualization. All authors have read and agreed to the final version of the manuscript.
This study was funded by Anhui Institute of Cardiovascular Diseases (KF2018010).
We would like to thank the Gene Expression Omnibus program for providing the high-quality data, and Tao Zhuang for providing the VSMC cell line.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.1061771/full#supplementary-material
1. Kent K. Clinical practice. Abdominal aortic aneurysms. N Engl J Med. (2014) 371:2101–8. doi: 10.1056/NEJMcp1401430
2. Sakalihasan N, Michel J, Katsargyris A, Kuivaniemi H, Defraigne J, Nchimi A, et al. Abdominal aortic aneurysms. Nat Rev Dis Primers. (2018) 4:34. doi: 10.1038/s41572-018-0030-7
3. Stackelberg O, Wolk A, Eliasson K, Hellberg A, Bersztel A, Larsson S, et al. Lifestyle and risk of screening-detected abdominal aortic aneurysm in men. J Am Heart Assoc. (2017) 6:e004725. doi: 10.1161/JAHA.116.004725
4. Wahlgren C, Larsson E, Magnusson P, Hultgren R, Swedenborg J. Genetic and environmental contributions to abdominal aortic aneurysm development in a twin population. J Vasc Surg. (2010) 51:3–7; discussion7. doi: 10.1016/j.jvs.2009.08.036
5. Yokoyama U, Arakawa N, Ishiwata R, Yasuda S, Minami T, Goda M, et al. Proteomic analysis of aortic smooth muscle cell secretions reveals an association of myosin heavy chain 11 with abdominal aortic aneurysm. Am J Physiol Heart Circ Physiol. (2018) 315:H1012–8. doi: 10.1152/ajpheart.00329.2018
6. Li J, Pan C, Zhang S, Spin J, Deng A, Leung L, et al. Decoding the genomics of abdominal aortic aneurysm. Cell. (2018) 174: 1361–72.e10. doi: 10.1016/j.cell.2018.07.021
7. You S, Gao L. Identification of NMU as a potential gene conferring alectinib resistance in non-small cell lung cancer based on bioinformatics analyses. Gene. (2018) 678:137–42. doi: 10.1016/j.gene.2018.08.032
8. Gao L, Wu X, Zhang L, Dai Y, Zhu Z, Zhi Y, et al. REG4 is a potential biomarker for radiochemotherapy sensitivity in colorectal cancer. Onco Targets Ther. (2021) 14:1605–11. doi: 10.2147/OTT.S296031
9. Carss K, Deaton A, Del Rio-Espinola A, Diogo D, Fielden M, Kulkarni D, et al. Using human genetics to improve safety assessment of therapeutics. Nat Rev Drug Discov. (2022) [Epub ahead of print]
10. Petroni G, Buqué A, Coussens L, Galluzzi L. Targeting oncogene and non-oncogene addiction to inflame the tumour microenvironment. Nat Rev Drug Discov. (2022) 21:440–62. doi: 10.1038/s41573-022-00415-5
11. Yamamoto J, Ito T, Yamaguchi Y, Handa H. Discovery of CRBN as a target of thalidomide: a breakthrough for progress in the development of protein degraders. Chem Soc Rev. (2022) 51:6234–50. doi: 10.1039/D2CS00116K
12. Cohen P, Cross D, Jänne P. Kinase drug discovery 20 years after imatinib: progress and future directions. Nat Rev Drug Discov. (2021) 20:551–69. doi: 10.1038/s41573-021-00195-4
13. Jones G, Tromp G, Kuivaniemi H, Gretarsdottir S, Baas A, Giusti B, et al. Meta-analysis of genome-wide association studies for abdominal aortic aneurysm identifies four new disease-specific risk loci. Circ Res. (2017) 120:341–53.
14. Huang R, Wang K, Gao L, Gao W. TIMP1 is A potential key gene associated with the pathogenesis and prognosis of ulcerative colitis-associated colorectal cancer. Onco Targets Ther. (2019) 12:8895–904. doi: 10.2147/OTT.S222608
15. Huang R, Gu W, Sun B, Gao L. Identification of COL4A1 as a potential gene conferring trastuzumab resistance in gastric cancer based on bioinformatics analysis. Mol Med Rep. (2018) 17:6387–96. doi: 10.3892/mmr.2018.8664
16. Huang R, Gao L. Identification of potential diagnostic and prognostic biomarkers in non-small cell lung cancer based on microarray data. Oncol Lett. (2018) 15:6436–42. doi: 10.3892/ol.2018.8153
17. Zalewski D, Ruszel K, Stêpniewski A, Gałkowski D, Bogucki J, Kołodziej P, et al. Identification of transcriptomic differences between lower extremities arterial disease, abdominal aortic aneurysm and chronic venous disease in peripheral blood mononuclear cells specimens. Int J Mol Sci. (2021) 22:3200. doi: 10.3390/ijms22063200
18. Davis F, Tsoi L, Melvin W, denDekker A, Wasikowski R, Joshi A, et al. Inhibition of macrophage histone demethylase JMJD3 protects against abdominal aortic aneurysms. J Exp Med. (2021) 218:e20201839. doi: 10.1084/jem.20201839
19. Pinard A, Jones G, Milewicz D. Genetics of thoracic and abdominal aortic diseases. Circ Res. (2019) 124:588–606. doi: 10.1161/CIRCRESAHA.118.312436
20. Alegret J, Ligero C, Vernis J, Beltrán-Debón R, Aragonés G, Duran I, et al. Factors related to the need for surgery after the diagnosis of bicuspid aortic valve: one center’s experience under a conservative approach. Int J Med Sci. (2013) 10:176–82. doi: 10.7150/ijms.5399
21. Henderson E, Geng Y, Sukhova G, Whittemore A, Knox J, Libby P. Death of smooth muscle cells and expression of mediators of apoptosis by T lymphocytes in human abdominal aortic aneurysms. Circulation. (1999) 99:96–104. doi: 10.1161/01.CIR.99.1.96
22. Morgan S, Yamanouchi D, Harberg C, Wang Q, Keller M, Si Y, et al. Elevated protein kinase C-δ contributes to aneurysm pathogenesis through stimulation of apoptosis and inflammatory signaling. Arterioscler Thromb Vasc Biol. (2012) 32:2493–502. doi: 10.1161/ATVBAHA.112.255661
23. Longo G, Xiong W, Greiner T, Zhao Y, Fiotti N, Baxter B. Matrix metalloproteinases 2 and 9 work in concert to produce aortic aneurysms. J Clin Invest. (2002) 110:625–32. doi: 10.1172/JCI0215334
24. Liu Z, Fitzgerald M, Meisinger T, Batra R, Suh M, Greene H, et al. CD95-ligand contributes to abdominal aortic aneurysm progression by modulating inflammation. Cardiovasc Res. (2019) 115:807–18. doi: 10.1093/cvr/cvy264
25. Paraskevas K, Mikhailidis D, Perrea D. Experimental models of abdominal aortic aneurysms: an overview. Curr Pharm Des. (2008) 14:325–37. doi: 10.2174/138161208783497697
26. Guo D, Pannu H, Tran-Fadulu V, Papke C, Yu R, Avidan N, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. (2007) 39:1488–93. doi: 10.1038/ng.2007.6
27. Martinez-Pinna R, Ramos-Mozo P, Madrigal-Matute J, Blanco-Colio L, Lopez J, Calvo E, et al. Identification of peroxiredoxin-1 as a novel biomarker of abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. (2011) 31:935–43. doi: 10.1161/ATVBAHA.110.214429
28. Golledge J, Tsao P, Dalman R, Norman P. Circulating markers of abdominal aortic aneurysm presence and progression. Circulation. (2008) 118:2382–92. doi: 10.1161/CIRCULATIONAHA.108.802074
29. Mustelin T, Bottini N, Stanford S. The contribution of PTPN22 to rheumatic disease. Arthritis Rheumatol. (2019) 71:486–95. doi: 10.1002/art.40790
30. Hoshino A, Kim H, Bojmar L, Gyan K, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. (2020) 182: 1044–1061.e18.
31. Cocozza F, Grisard E, Martin-Jaular L, Mathieu M, Théry C. SnapShot: extracellular vesicles. Cell. (2020) 182: 262–262.e1. doi: 10.1016/j.cell.2020.04.054
32. Sidloff D, Stather P, Dattani N, Bown M, Thompson J, Sayers R, et al. Aneurysm global epidemiology study: public health measures can further reduce abdominal aortic aneurysm mortality. Circulation. (2014) 129:747–53. doi: 10.1161/CIRCULATIONAHA.113.005457
33. Tang W, Saratzis A, Pattee J, Smith J, Pankratz N, Leavy O, et al. Replication of Newly identified genetic associations between abdominal aortic aneurysm and SMYD2, LINC00540, PCIF1/MMP9/ZNF335, and ERG. Eur J Vasc Endovasc Surg. (2020) 59:92–7. doi: 10.1016/j.ejvs.2019.02.017
34. Harrison S, Smith A, Jones G, Swerdlow D, Rampuri R, Bown M, et al. Interleukin-6 receptor pathways in abdominal aortic aneurysm. Eur Heart J. (2013) 34:3707–16. doi: 10.1093/eurheartj/ehs354
35. Yao F, Yao Z, Zhong T, Zhang J, Wang T, Zhang B, et al. Imatinib prevents elastase-induced abdominal aortic aneurysm progression by regulating macrophage-derived MMP9. Eur J Pharmacol. (2019) 860:172559. doi: 10.1016/j.ejphar.2019.172559
36. Wang X, Wei G, Ding Y, Gui X, Tong H, Xu X, et al. Protein tyrosine phosphatase PTPN22 negatively modulates platelet function and thrombus formation. Blood. (2022) 140:1038–51. doi: 10.1182/blood.2022015554
37. Bottini N, Peterson E. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu Rev Immunol. (2014) 32:83–119. doi: 10.1146/annurev-immunol-032713-120249
38. Lu H, Daugherty A. Aortic aneurysms. Arterioscler Thromb Vasc Biol. (2017) 37:e59–65. doi: 10.1161/ATVBAHA.117.309578
39. Björkegren J, Lusis A. Atherosclerosis: recent developments. Cell. (2022) 185:1630–45. doi: 10.1016/j.cell.2022.04.004
40. Roy P, Orecchioni M, Ley K. How the immune system shapes atherosclerosis: roles of innate and adaptive immunity. Nat Rev Immunol. (2022) 22:251–65. doi: 10.1038/s41577-021-00584-1
41. Biros E, Moran C, Rush C, Gäbel G, Schreurs C, Lindeman J, et al. Differential gene expression in the proximal neck of human abdominal aortic aneurysm. Atherosclerosis. (2014) 233:211–8.
42. Lenk G, Tromp G, Weinsheimer S, Gatalica Z, Berguer R, Kuivaniemi H. Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms. BMC Genomics. (2007) 8:237. doi: 10.1186/1471-2164-8-237
43. Biros E, Gäbel G, Moran C, Schreurs C, Lindeman J, Walker P, et al. Differential gene expression in human abdominal aortic aneurysm and aortic occlusive disease. Oncotarget. (2015) 6:12984–96.
44. Furusho A, Aoki H, Ohno-Urabe S, Nishihara M, Hirakata S, Nishida N, et al. Involvement of B cells, immunoglobulins, and syk in the pathogenesis of abdominal aortic aneurysm. J Am Heart Assoc. (2018) 7:e007750.
45. Huang da W, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. (2009) 4:44–57.
46. Zhuang T, Liu J, Chen X, Pi J, Kuang Y, Wang Y, et al. Cell-specific effects of GATA (GATA Zinc finger transcription factor family)-6 in vascular smooth muscle and endothelial cells on vascular injury neointimal formation. Arterioscler Thromb Vasc Biol. (2019) 39:888–901.
47. Pi J, Tao T, Zhuang T, Sun H, Chen X, Liu J, et al. A MicroRNA302-367-Erk1/2-Klf2-S1pr1 pathway prevents tumor growth via restricting angiogenesis and improving vascular stability. Circ Res. (2017) 120:85–98.
Keywords: abdominal aortic aneurysm, diagnosis, biomarker, PTPN22, cellular AAA model
Citation: Ruan P, Gao L, Jiang H, Chu T, Ge J and Kong X (2022) Identification of PTPN22 as a potential genetic biomarker for abdominal aortic aneurysm. Front. Cardiovasc. Med. 9:1061771. doi: 10.3389/fcvm.2022.1061771
Received: 05 October 2022; Accepted: 29 November 2022;
Published: 14 December 2022.
Edited by:
Lu Gan, Sichuan University, ChinaReviewed by:
Lin Deng, Southern University of Science and Technology, ChinaCopyright © 2022 Ruan, Gao, Jiang, Chu, Ge and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianjun Ge, emtkZ2pqQHVzdGMuZWR1LmNu; Xiang Kong, ZHJrb25neGlhbmdAdXN0Yy5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.