- 1State Key Laboratory of Component-Based Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin, China
- 2Biomedical Engineering, Imperial College London, London, United Kingdom
- 3Tianjin Jinghai District Hospital, Tianjin, China
- 4Department of Cardiovascular-Thoracic Surgery, Northwestern University Feinberg School of Medicine, Chicago, IL, United States
- 5Department of Pediatrics, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL, United States
Introduction: Machine learning (ML) has gained intensive popularity in various fields, such as disease diagnosis in healthcare. However, it has limitation for single algorithm to explore the diagnosing value of dilated cardiomyopathy (DCM). We aim to develop a novel overall normalized sum weight of multiple-model MLs to assess the diagnosing value in DCM.
Methods: Gene expression data were selected from previously published databases (six sets of eligible microarrays, 386 samples) with eligible criteria. Two sets of microarrays were used as training; the others were studied in the testing sets (ratio 5:1). Totally, we identified 20 differently expressed genes (DEGs) between DCM and control individuals (7 upregulated and 13 down-regulated).
Results: We developed six classification ML methods to identify potential candidate genes based on their overall weights. Three genes, serine proteinase inhibitor A3 (SERPINA3), frizzled-related proteins (FRPs) 3 (FRZB), and ficolin 3 (FCN3) were finally identified as the receiver operating characteristic (ROC). Interestingly, we found all three genes correlated considerably with plasma cells. Importantly, not only in training sets but also testing sets, the areas under the curve (AUCs) for SERPINA3, FRZB, and FCN3 were greater than 0.88. The ROC of SERPINA3 was significantly high (0.940 in training and 0.918 in testing sets), indicating it is a potentially functional gene in DCM. Especially, the plasma levels in DCM patients of SERPINA3, FCN, and FRZB were significant compared with healthy control.
Discussion: SERPINA3, FRZB, and FCN3 might be potential diagnosis targets for DCM, Further verification work could be implemented.
1. Introduction
Machine learning (ML), composed of various intricate algorithms, is recently commonly applied to explore potential biomarkers (e.g., lipidome, metabolome, and transcriptome) and prognosis (1, 2), especially in variable filtration (3–5). For example, MLs can recognize patterns better representing the individual risk compared to classical surgical risk scores (6). ML includes various types, such as support vector machine (SVM) (7, 8), random forest (RF) (9), decision tree (DT) (10–12), and so on. Different ML has its specialty and shortcoming. For example, least absolute shrinkage and selection operator (LASSO) processed a precision matrix of Gaussian variables using an ℓ1-penalty (13) until small values to zero but eliminated too many variables. For SVM, separated hyperplanes allow for correct partitioning and maximize geometric spacing but may be worse in a small sample size (14) compared with other MLs (15). Different ML algorithms possess both characteristics and limitations which cannot be ignored, especially in the choice of variables. Many researchers (16–18) only focus on single or two MLs which might ignore their potential shortcomings. In our previous research (19), five MLs show different weights even with the same genes. So just intersecting the top N genes may unconsciously delete some dominant genes (20–23). And ignoring the weights of genes may result in an imbalance of filtration (19, 24).
Dilated cardiomyopathy (DCM), not only the primary myocardial disease but also the dominant trigger in chronic heart failure (HF) (25), manifests clinically in systolic heart insufficiency and dilatation of the left ventricle (26, 27). Although there are already clinical diagnosis criteria for DCM, by the time the clinical diagnosis is clear, most of the patient’s underlying condition is poor (27). Though drugs (e.g., ivabradine) for HF are used to treat DCM and improve the prognosis in the short term (28), the long-term prognosis remains poor (29). Therefore, early diagnosis with identifying markers of DCM is necessary. Previous studies had indicated the diagnosis value of genes (30, 31) (e.g., TBX20 or Gab1) in DCM but with few microarrays (32), which means a small sample size and non-universality. Thus, developing a predictive model for DCM genetic diagnosis with multiple microarrays is necessary.
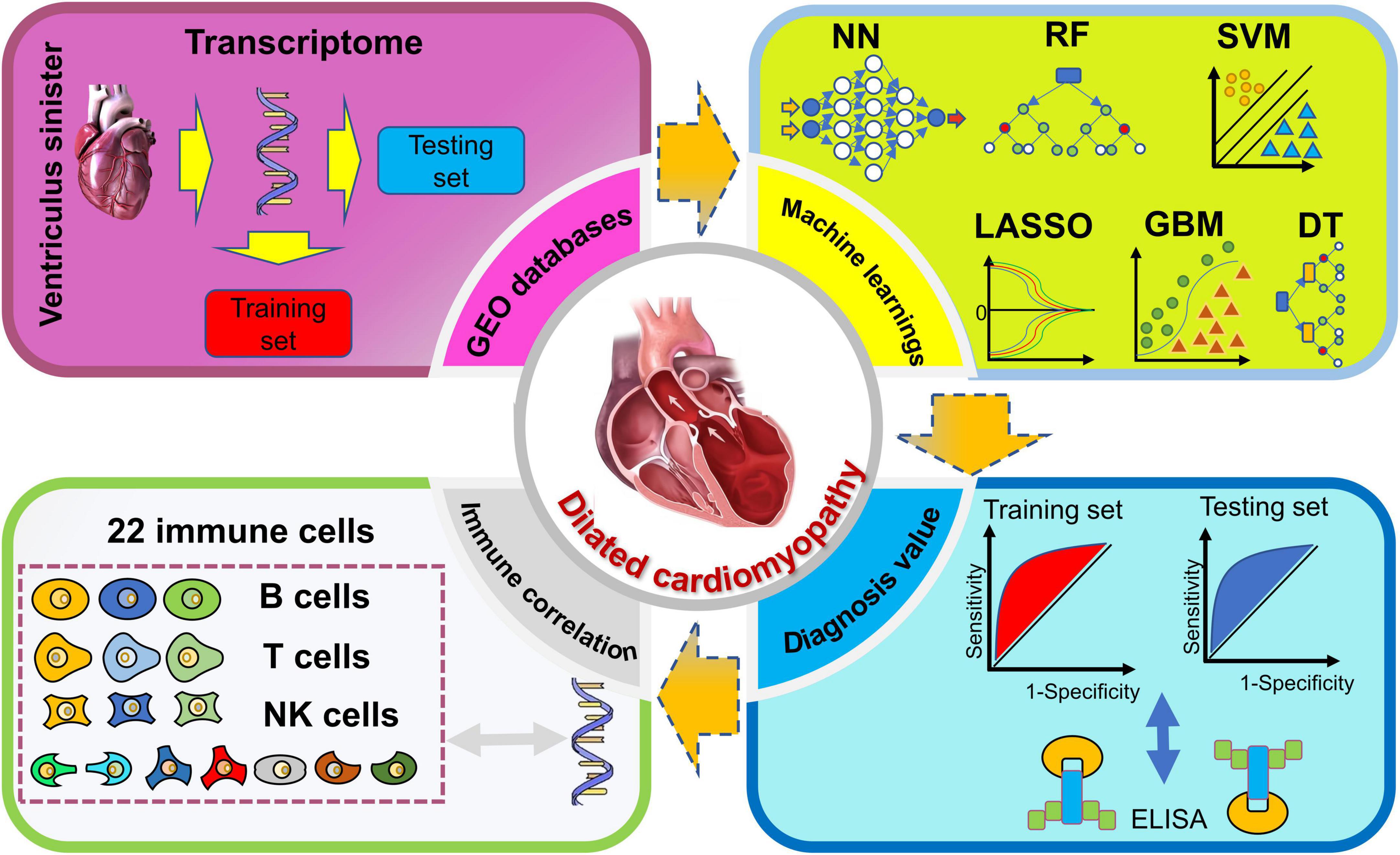
In this study, we identified potential transcriptomic information regarding DCM diagnosis with the overall weights in MLs of multiple microarrays. Furthermore, we further developed an immune correlation analysis between diagnosis genes and immune cells. Finally, DCM patients and healthy control were recruited for validation of related proteins of genes. The process of the following analysis (Figure 1) was shown in the flow chart.
2. Materials and methods
2.1. Data acquisition
We derived the transcriptome information of DCM from Gene Expression Omnibus (GEO). According to the following criteria, the primary data were derived with the keyword of “DCM”: (1) inclusion criteria (i) sample of the left ventricle with a diagnosis of DCM patients; (ii) transcriptome; (iii) primary data was free and accessible. (2) exclusion criteria (i) suspected carcinoma, ischemic cardiomyopathy, heart valve disease, and other diseases; (ii) intervention(s) in DCM patients.
2.2. Data processing
Firstly, the sva R package (version 3.36) was applied to eliminate branch effects and quantile normalization with the specific function of ComBat. Secondly, we divide all microarrays into training or testing sets with a ratio of 5:1 (33). Briefly, the training set for developing the potential diagnosis value, and the testing for verifying the results. Thirdly, we identify the differentially expressed genes (DEGs). The functional analysis of DEGs was applied through the Kyoto Encyclopedia of Genes and Genomes Gene Set Enrichment Analysis (KEGG-GSEA), Gene Ontology (GO), and Disease Ontology (DO) enrichment based on three packages, DOSE (version 3.22.1), clusterProfiler (version 4.4.4), and enrichplot (version 1.16.2). The GO consist of three parts, molecular function (MF), biological process (BP), and cellular components (CCs). Moreover, six MLs algorithms were applied to the classification model and filtered the candidate diagnosis genes. As for the testing group, we identify the diagnosis value of potential candidate genes. Lastly, the immune correlation between the above genes was developed.
2.3. Searching for DEGs
The R package, limma (version 3.52.4), was adopted to average the same gene expression with the function of avereps and then identify the DEGs. After quantile normalization, primary data sets were transformed into log2. P-value was adjusted to the false discovery rate based on Benjamini and Hochberg method. Two thresholds were set, the absolute value of fold change (|logFC|) > 1, and the false discovery rate < 0.001. With the DEGs, the heatmap and volcano plot were applied with the pheatmap (version 1.0.12) and ggplot2 (version 3.3.6).
2.4. Classification models with six MLs
Based on the above DEGs, we further developed classification models with six MLs algorithms, SVM, LASSO, RF, gradient boosting machine (GBM), DT, and neural network (NN) to assess the classification value. Briefly, we constructed the six MLs classification models with optimized parameters in the training sets, and the testing was adopted for the validation of the six MLs. All ML models are cross-validated 10-fold to ensure stability. The accuracy value was adopted to estimate the value for six MLs and greater accuracy indicates the better classification value of the model.
The first ML (LASSO) was developed with the glmnet (version 4.1-4) R package. The function cv.glmnet was applied to optimize the value of lambda. For basic parameters, the following settings were the scale of lambda between 0 and 2,000 with one step size, the family of “binomial,” and the type measure of “class.” With the min lambda, the function glmnet was applied to the LASSO model in training sets with alpha (equal to 1) and a family of “binomial.”
The second ML (SVM) was adopted with e1071 R package (version 1.7-11). The function tune.svm was utilized to optimize the settings parameter. For basic parameters, the following settings were the kernel of “linear,” and the cost between 1 and 20. With the best number of support vectors, the classification model was built.
The third ML (DT) was finished with two R packages, rpart (version 4.1.16) and rpart.plot (version 3.1.1). The rpart function was applied to the model with the method of “class,” cp value of 0.00001.
The fourth ML (RF) was adopted with randomForest (version 4.7-1.1). In randomForest, the tuneRF was served to optimize 500 trees and 1 step size. With the optimal trees for min error rate, the classification model of training sets was accomplished.
The fifth ML (NN) was developed with neuralnet R package (version 1.44.2). In neuralnet, the neuralnet was served with five layers (containing an input layer, an output layer, and three hidden layers), the err.fct of “sse,” and the output of linear.
The last ML, GBM, was different from the above five algorithms with more steps and prone to making. The GMB was accomplished with h2o (version 3.38.0.1). Only JAVA operating environment that the h2o can process the classification model. Thereby, we had to timely download and installed java development kit (JDK). Necessary for running memory with h2o.init in GBM and we adjusted the model memory of GBM to 16G. Due to the h2o data type being indispensable for GBM, we transform the data format with as.h2o in both the training set and testing set. Finally, h2o.gbm was applied to tune the parameters and model (we set the distribution of “bernoulli,” 200 trees, 0.001 for a learning rate, 0.9 for a sample rate).
Importantly, based on the above weights of six MLs for DEGs, we calculated the normalized six MLs weights of DEGs as the function in R: . For example, if the weight of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in six MLs was 15, −11, 10, −1, 160, and −4. And the max weights of absolute value in the above model were 30, 44, 40, 4, 320, and 8, respectively. The overall weight of GAPDH was |15| /30 + |−11| /44 + |10| /40 + |−1| /4 + |160| /320 + |−4| /8 = 2.25. Then, we filter the candidate genes for ROC (pROC, version 1.18.0) and immune correlation (CIBERSORT function) with overall weights > 1. Area under the curve (AUC) was calculated to judge the diagnosis value between control and DCM individuals.
2.5. Access to clinical samples
The trial complied with the Declaration of Helsinki and was approved by the Ethics Committees of the participating hospitals. All DCM patients and healthy volunteers provided written informed consent from September 20, 2022 to October 31, 2022. Ethics Committee/Institutional Review Board: Ethics Review Committee Jinghai District Hospital, Plan 11. Diary number: JHYYLL-2022-0307.
Briefly, according to the Chinese guidance (27), the inclusion criteria of DCM contain three parts, (1) left ventricular end-diastolic diameter > 5.0 cm (women) or > 5.5 cm (men); (2) left ventricular ejection fraction < 45%, left ventricular fractional shortening < 25%; (3) no other heart-related diseases and >20 years old. Blood samples were collected in ethylene diamine tetraacetic acid (EDTA)-containing tubes after a 10-h overnight fast and centrifuged at 4°C, 3,000 g for 10 min, then plasma was stored at −80°C. All the plasma levels of SERPINA3, FCN3, and FRZB were measured by ELISA kits (SERPINA3 Human ELISA Kit, Abcam, Cambridge, UK; Hycult Biotechnology, Uden, The Netherlands; R&D Systems, Minneapolis, MN, USA, respectively).
2.6. Statistical analysis
All the statistical analyses were processed by R software (version 4.1.1). CIBERSORT was adopted for immune correlation analysis. We estimate the immune correlates of 22 immune cells and visualization in the corrplot R package (version 0.92). For continuous variables, the independent Student’s t-test was adopted if the variables met Gaussian distribution, if not, the Wilcoxon test was used. A two-sided p-value < 0.05 was considered to be significant.
3. Results
3.1. Incorporation of microarrays
Among six microarrays (Table 1) (386 sample sizes) were finally obtained, including GSE5406, GSE57338, GSE1145, GSE1869, GSE3585, and GSE42955. According to the random ratio of 5:1, the training set was integrated with two microarrays (168 DCM and 152 healthy control), including GSE5406 and GSE57338. At the same time, the testing set was integrated with four (39 DCM and 27 control), composed of GSE1145, GSE1869, GSE3585, and GSE42955.
3.2. Searching for DEGs
Among 20 DEGs with biological significance (Supplementary Table 1) from 12,937 RNAs were identified in the training sets. Compared to the healthy control, 13 genes down-regulated (SERPINA3, PLA2G2A, IL1RL1, CD163, SERPINE1, FCN3, CYP4B1, LYVE1, S100A8, SLCO4A1, MYOT, ANKRD2, and VSIG4) and 7 genes up-regulated (MXRA5, FRZB, HBB, LUM, SFRP4, NPPA, and ASPN) in the DCM individuals (Figure 2).
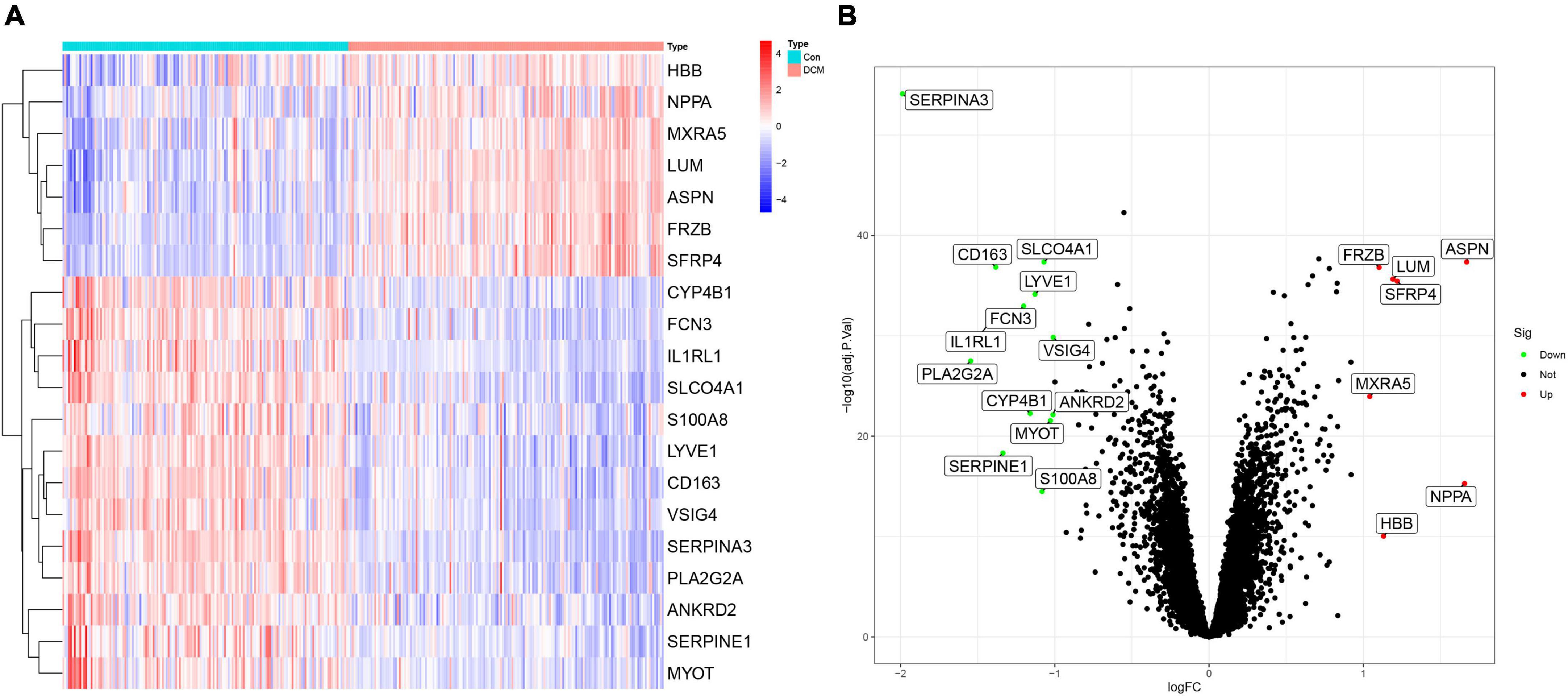
Figure 2. The heatmap and volcano plot of 20 differently expressed genes (DEGs) in dilated cardiomyopathy (DCM) and healthy. (A) The heatmap of 20 DEGs; (B) the volcano plot of 20 DEGs.
3.3. Functional enrichment analysis
Based on the above DEGs, we identified 21 GSEA terms (Supplementary Table 2) and show the top 5 (Figures 3A, B), 102 GO terms (Supplementary Table 3) and show the top 4 (Figure 3C), 68 DO terms (Supplementary Table 4) and show the top 10 (Figure 3D). Among GSEA-KEGG enrichments, the top 3 presented significance in Type I diabetes mellitus, graft versus host disease, and allograft rejection. Regarding the GO terms in BP, the top 3 presented significant enrichments in the cellular zinc ion homeostasis, positive regulation of inflammatory response, and zinc ion homeostasis. In terms of DO, the top 3 diseases presented were atherosclerosis, arteriosclerotic cardiovascular disease, and arteriosclerosis.
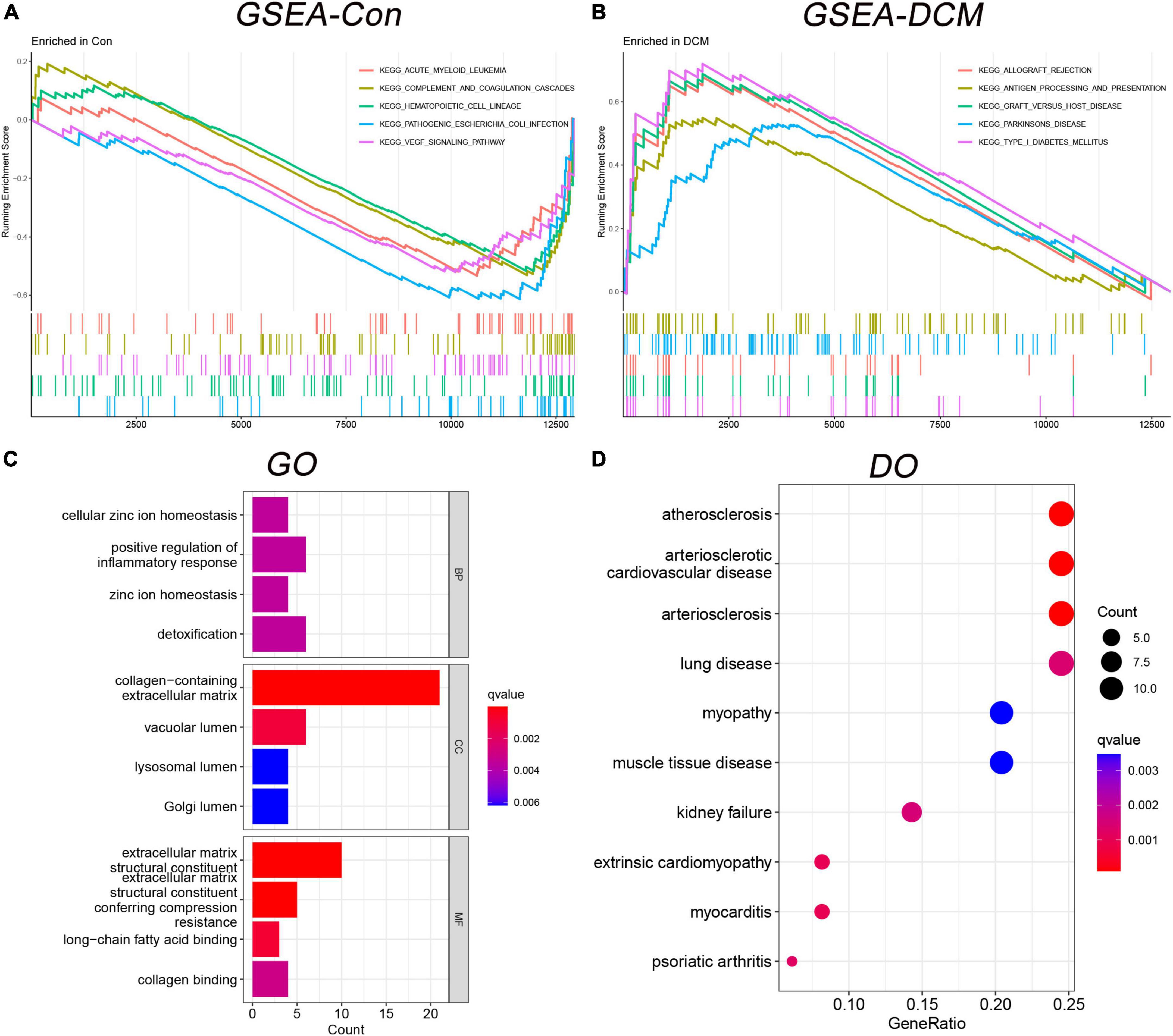
Figure 3. Functional enrichment analysis in gene set enrichment analysis-kyoto encyclopedia of genes and genomes (GSEA-KEGG), gene ontology (GO), and disease ontology (DO). (A) The GSEA-KEGG enrichment in control; (B) the GSEA-KEGG enrichment in DCM; (C) the GO enrichment term; and (D) the DO enrichment term.
3.4. Six MLs algorithms for classification model and candidate genes
Six classification models of MLs were successfully established (Figure 4), and we calculated the accuracy (Table 2) of both training sets and testing sets. In LASSO (Figure 4A), we filtered nine candidate genes. Disappointed, LASSO’s accuracy of the two sets were only 52.5 and 59.09%. In SVM, 19 genes were identified (Figure 4B), and the accuracies of the two sets were unstable, 90.94 and 51.52%. In RF (Figure 4C), the error rate of the classification model decreases as the number of trees increases, until 234 trees the error rate is minimized and smoothed. Surprisingly, the accuracy of the two sets was 100%. In DT (Figure 4D), thresholds of 7.2 in SERPINA3 can discriminate the health and DCM, but the accuracies of the two sets were also unstable like SVM, 93.75 and 53.03%. In GBM (Figure 4E), we developed six folds models to explore the candidate genes, but the accuracies of the two sets were also unstable, 96.03 and 53.03%. In NN (Figure 4F), enough in three hidden layers to discriminate the health and DCM, and the accuracies of both sets were 100%. Among all those models, the most important genes with the primary weights were identified (Supplementary Table 5). In the six MLs, both the RF and NN show the optimal and stable classification value. The accuracy of both MLs was 100%. Furthermore, the summation (Table 3) of normalized weights (dividing the absolute value by max weights) was calculated to screen the diagnosis genes. And nine genes (SERPINA3, CD163, FCN3, LYVE1, SLCO4A1, LUM, FRZB, PLA2G2A, and SFRP4) talent showing themselves with overall weights > 1 (Table 3).
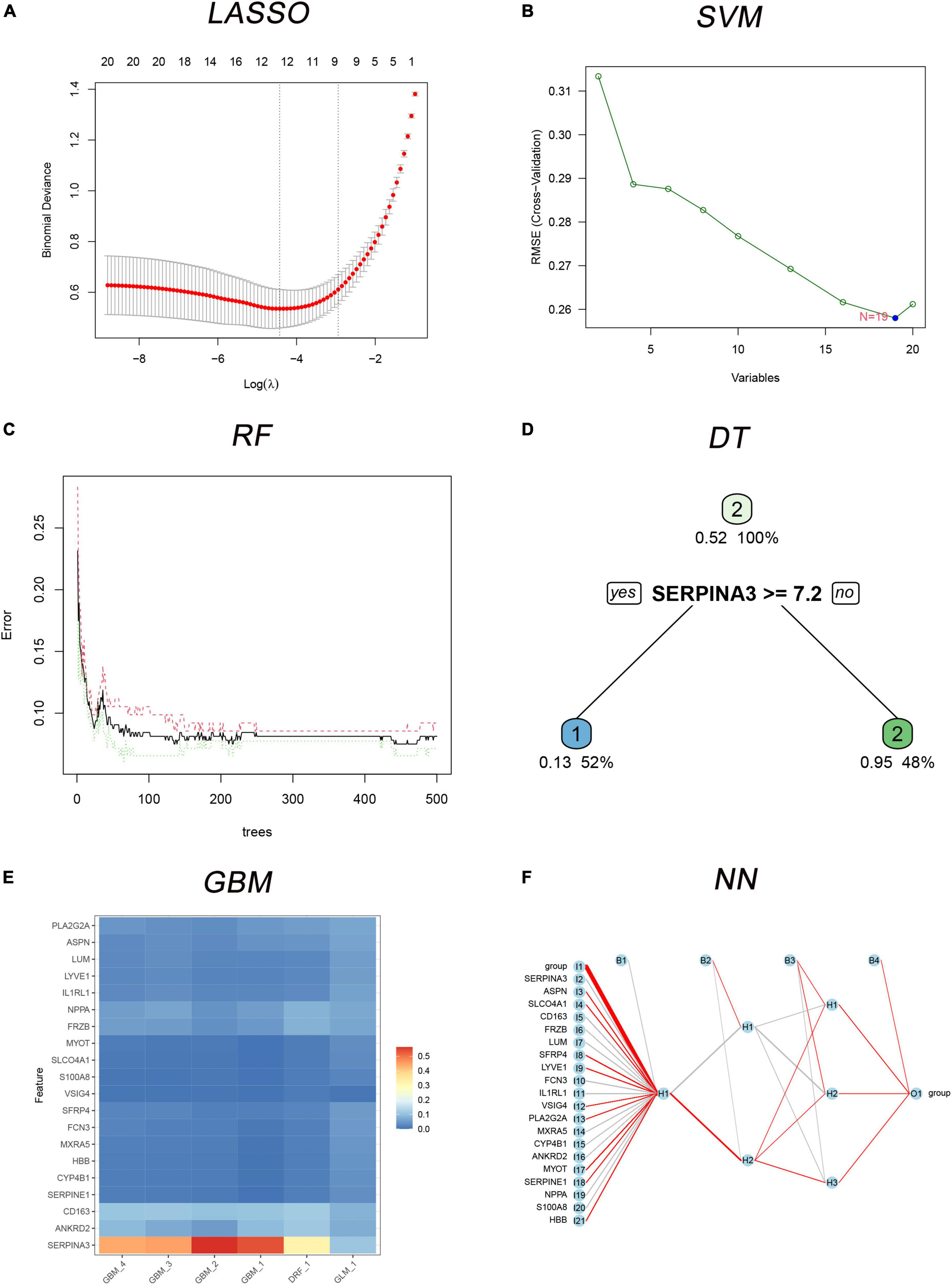
Figure 4. The six MLs classification models built with 20 differently expressed genes (DEGs). (A) Least absolute shrinkage and selection operator (LASSO) for 9 candidate genes; (B) support vector machine (SVM) for 19 candidates DEGs; (C) the error rate of the random forest (RF) classification model with increasing trees; (D) the decision tree (DT) for classification of control and dilated cardiomyopathy (DCM) individuals; (E) multiple gradient boosting machine (GBM) classification models of control and DCM individuals; (F) neural network (NN) for classification of control and DCM individuals.
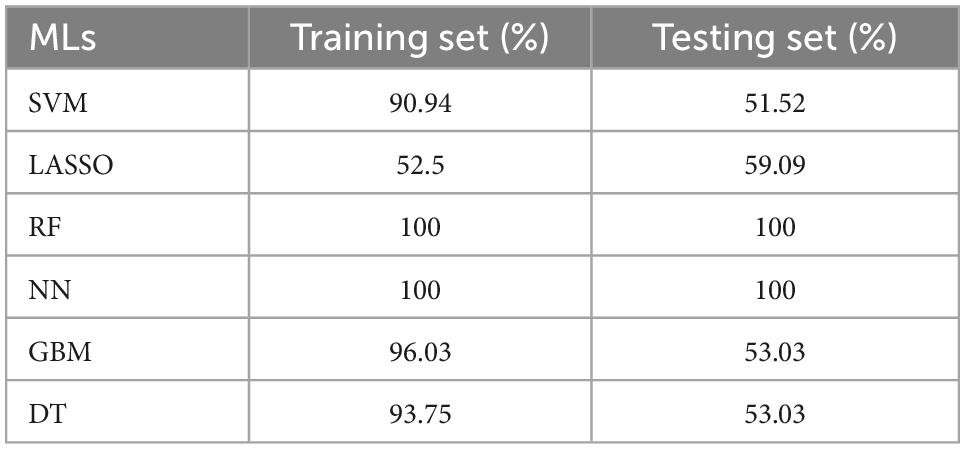
Table 2. The accuracy of six classification machine learnings (MLs) in the training and testing sets.
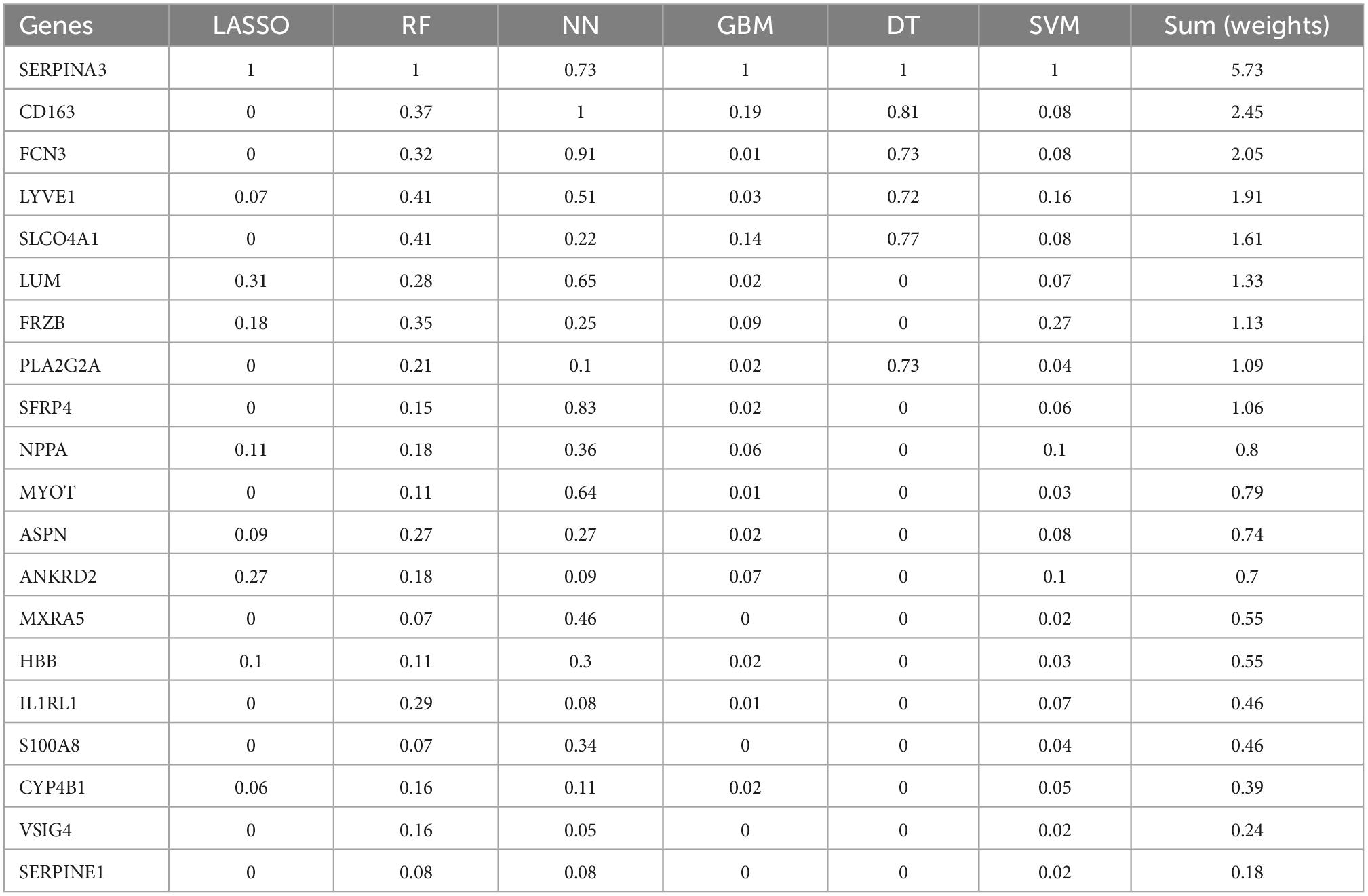
Table 3. The summed normalized weights of 20 differently expressed genes (DEGs) in six classifications machine learnings (MLs).
Based on the summed normalized weights > 1, nine candidates genes were chosen for diagnosis in DCM and healthy individuals. Next, we validate the nine candidate genes in the testing set, and except for SLCO4A1, the other eight show significance (Figure 5).
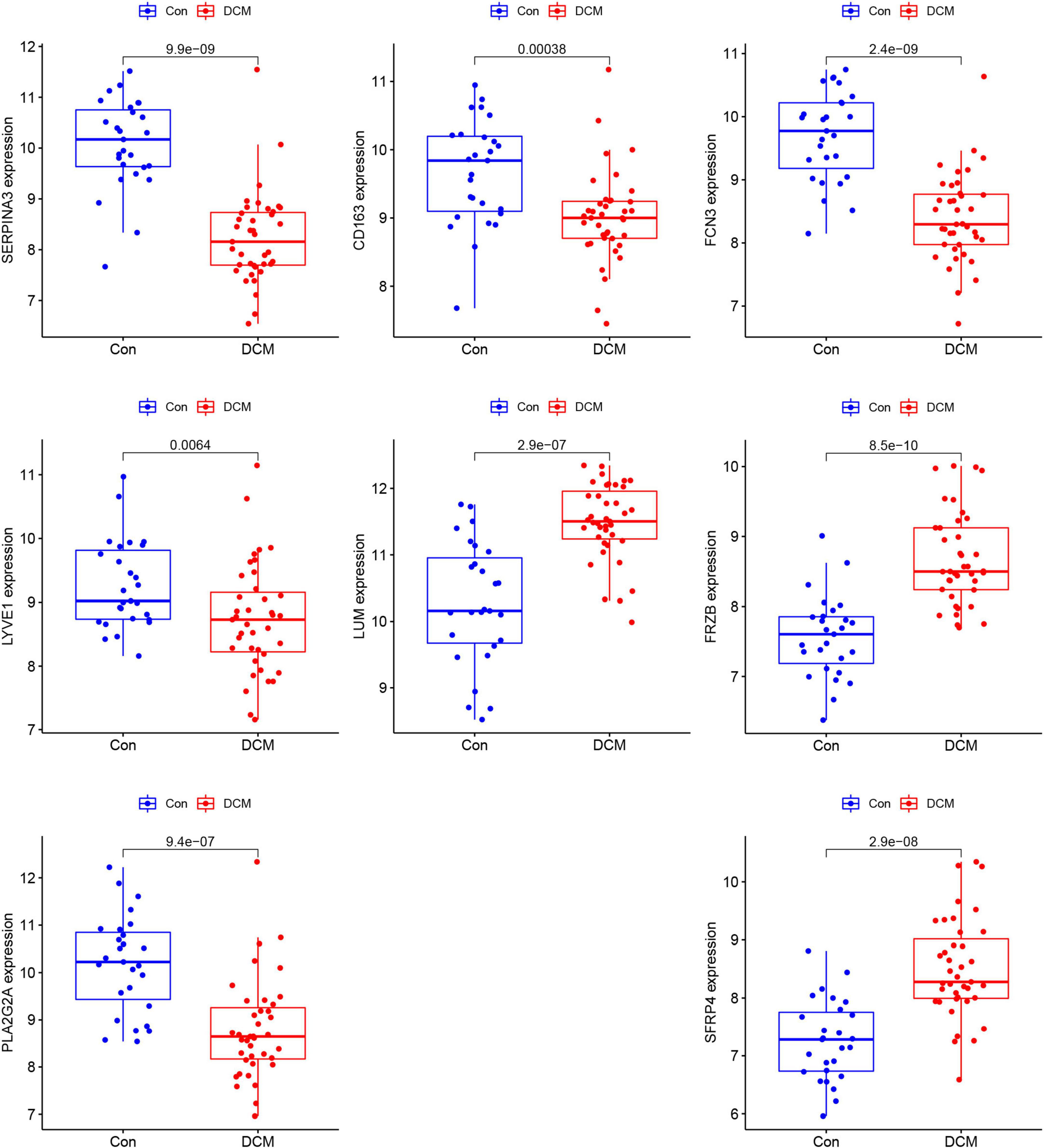
Figure 5. The comparison of the 8 genes between dilated cardiomyopathy (DCM) and healthy in testing sets.
3.5. Evaluation of the diagnosis value
Eight genes (just mentioned above) were taken into the ROC curve (Supplementary Figures 1, 2). AUC values of SERPINA3, FCN3, LUM, FRZB, PLA2G2A, and SFRP4 were higher than 0.8 in both two sets. Moreover, three genes SERPINA3, FCN3, and FRZB were higher than 0.88 (Figure 6) in the training sets and even >0.9 in the testing sets. Especially, SERPINA3 was higher than 0.9 in both sets. In a word, three genes, SERPINA3, FCN3, and FRZB may be the potential diagnosis genes compared with DCM and healthy control.
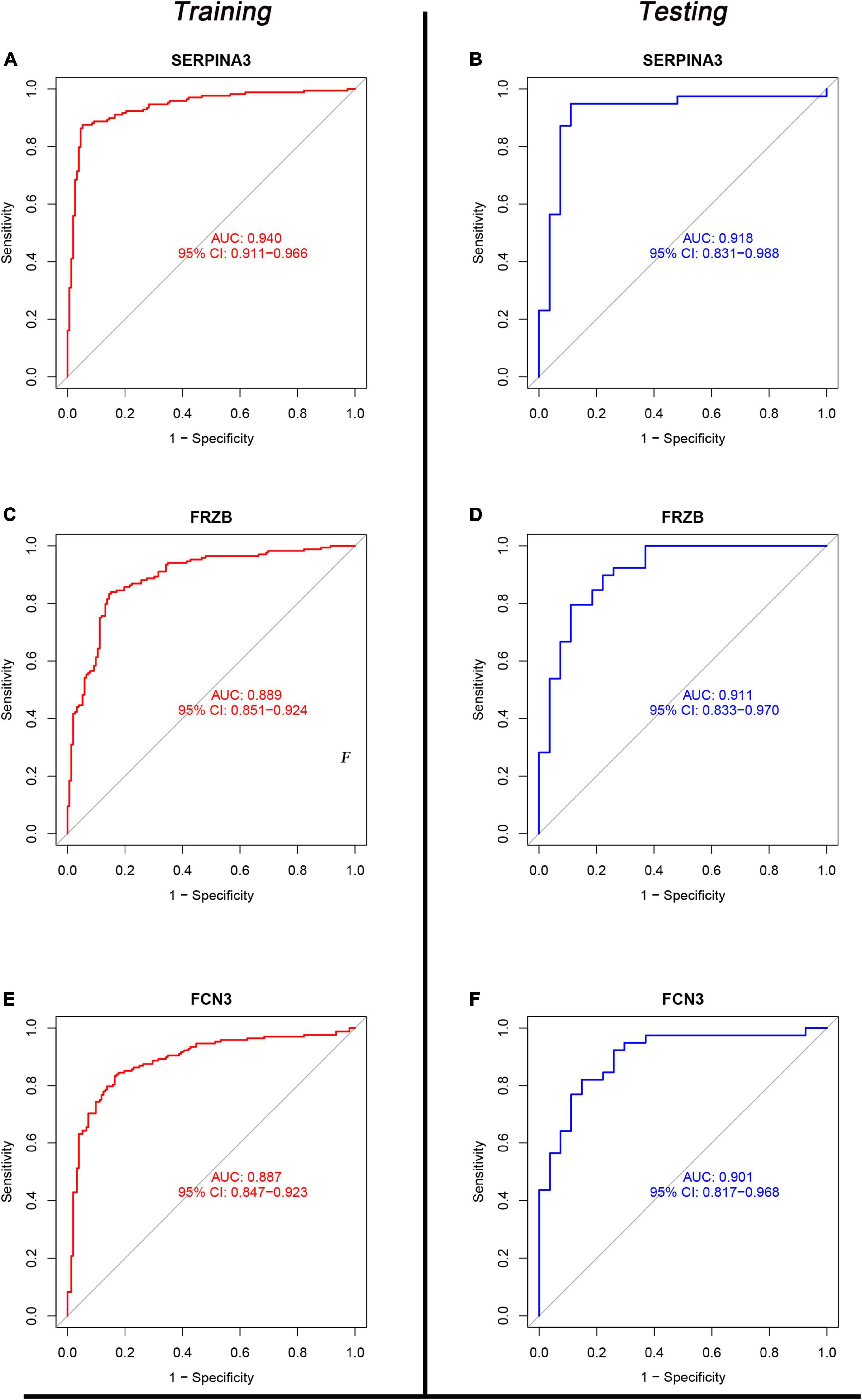
Figure 6. The receiver operating characteristic (ROC) of SERPINA3, FRZB, and FCN3 between the control and dilated cardiomyopathy (DCM) groups. (A,C,E) The ROC of SERPINA3, FRZB, and FCN3 in the Training set; (B,D,F) the ROC of SERPINA3, FRZB, and FCN3 in the testing set.
3.6. Immune correlation
The immune correlation between signal genes and 22 immune cells was applied to all 386 samples of six microarrays (Supplementary Figure 3). SERPINA3 (Figure 7A) shows significant correlations in Monocytes, T cells CD8, and Plasma cells. Regarding FRZB (Figure 7B), the T cells CD4 memory resting, plasma cells, monocytes, and T cells regulatory (Tregs) show significant correlations. In FCN3 (Figure 7C), the mast cells activated, macrophages M0, and plasma cells show significant correlations. These three genes show a typical significant immune cell, plasma cells. All three genes were correlated with plasma cells.
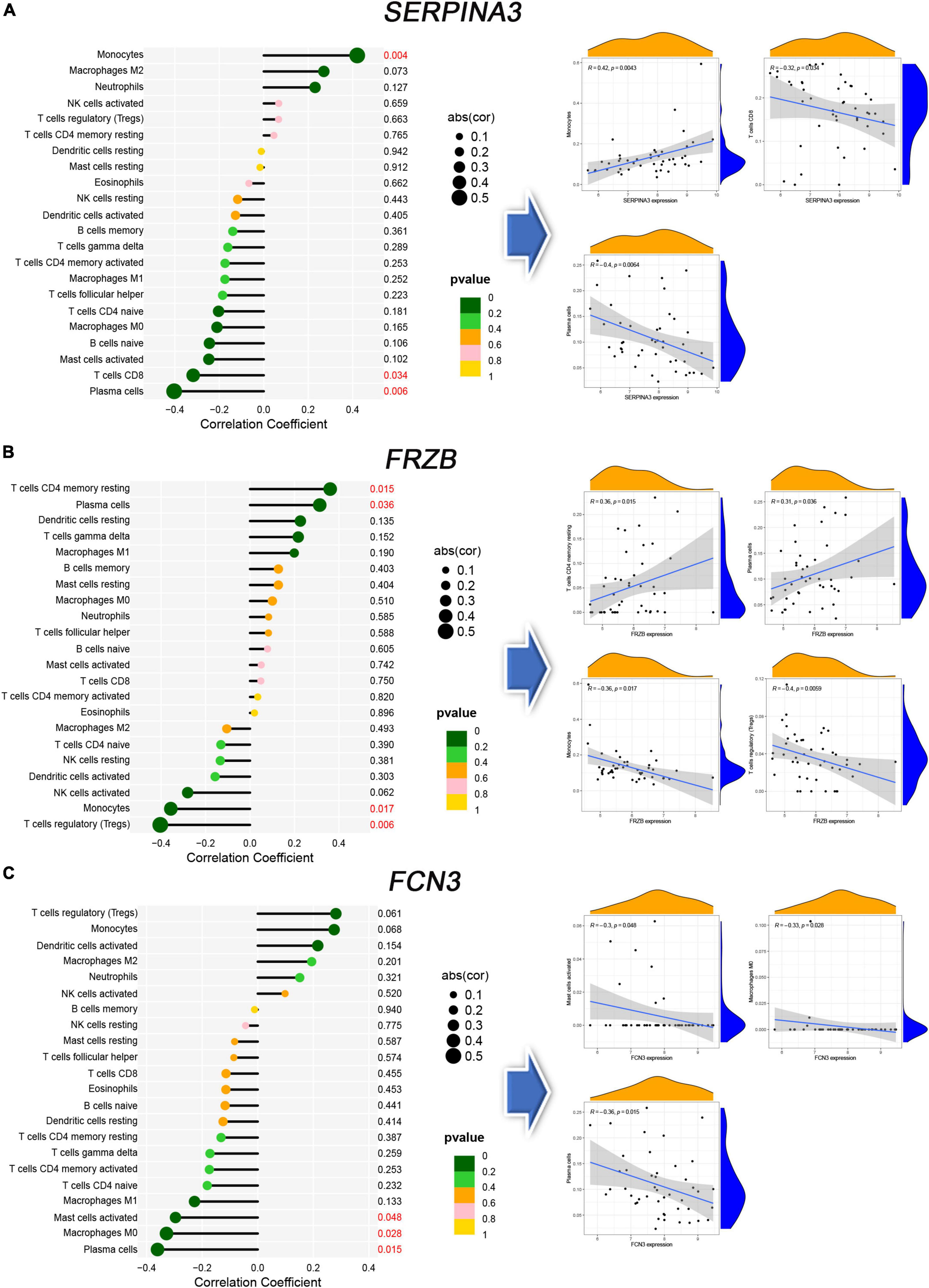
Figure 7. The immune correlation between diagnosis genes and immune cells. (A–C) The lollipop map and linear regression map in SERPINA3, FRAB, and FCN3.
3.7. Differences in plasma proteins
Finally, 24 individuals (12 healthy controls and 12 DCM patients) were recruited. We measured the plasma levels (Figure 8) of SERPINA3, FRZB, and FCN3. The plasma levels of SERPINA3 in DCM patients (397.17 ± 49.22 μg/ml) were higher (P < 0.001) than in healthy individuals (221.25 ± 14.15 μg/ml). Similarly, the plasma levels of FRZB in DCM patients (2,042.75 ± 292.62 pg/ml) were higher (P < 0.001) than in healthy individuals (784.58 ± 55.85 pg/ml). In FCN3, the plasma levels in DCM (13.67 ± 2.69 μg/ml) were lower than in the healthy control (20.92 ± 1.38 μg/ml). More importantly, all of the protein levels of these three genes were significant in DCMs compared with healthy controls.
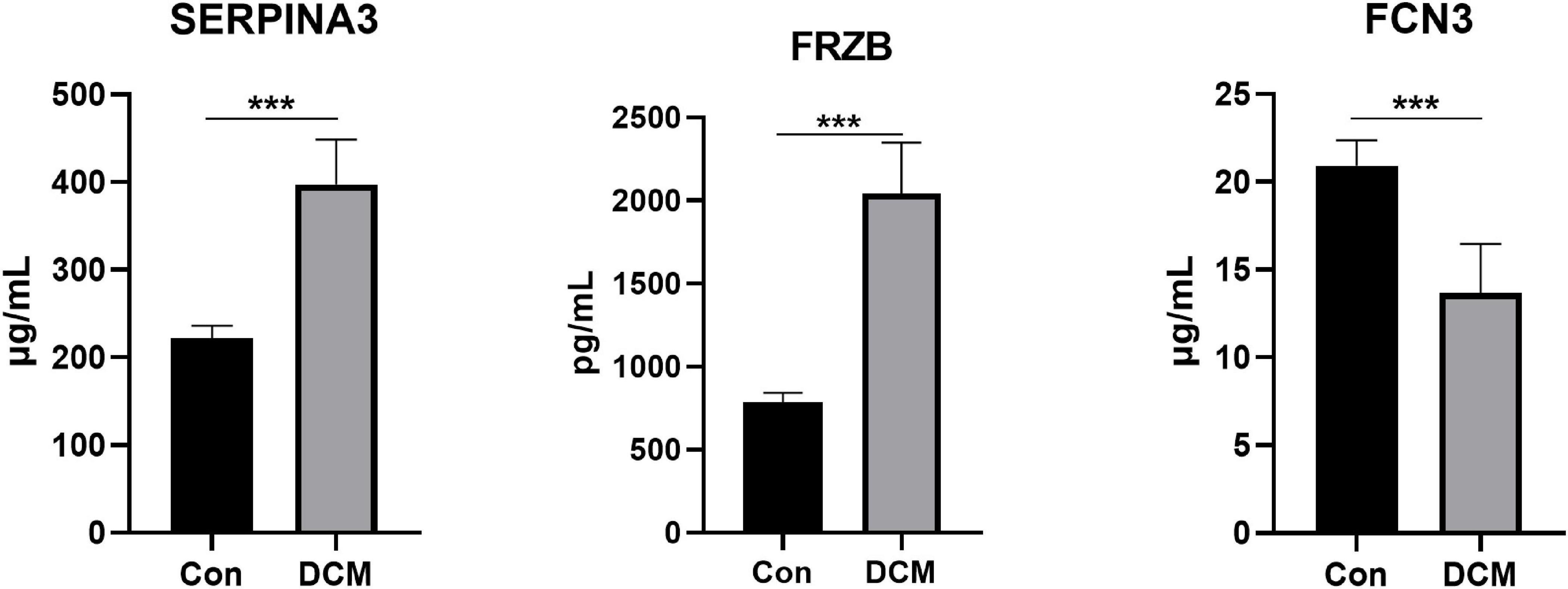
Figure 8. The plasma levels of SERPINA3, FRZB, and FCN3 in dilated cardiomyopathy (DCM) patients and healthy controls. ***Mean P < 0.001.
4. Discussion
To our knowledge, this is the first work with normalized overall weights to filter candidate genes in DCM. Three genes, SERPINA3, FRZB, and FCN3 show the AUC values in the training set (0.940, 0.889, and 0.887, respectively) and testing set (0.918, 0.911, and 0.901, respectively). In plasma protein, SERPINA3, FRZB, and FCN3 in DCM were significant compared with the control.
MLs have been extensively performed in four types of analysis, filtration of variables, classification, congression, and cluster. In bioinformatics, many studies take only one or two MLs, such as WGCNA (34), LASSO, and SVM. Nevertheless, a single ML might ignore the dominant variables. In our work (Table 1), the FCN3 will be missed if just take the intersection of LASSO and SVM like the previous study (35). Various MLs showed their advantages. For instance, SVM and NN show their talents in the diagnosis of pigmented skin lesions (36). And in the pre-operative prediction of postsurgical mortality (37), GBM was the most MLs compared with DT, RF, and SVM. In our work, both RF and NN show their talent discrimination value in both training and testing sets with an accuracy of 100%. The normalized weights may be different even in the same variable (Table 1) in various MLs. So our work takes the sum of the normalized weights of different MLs into the following diagnosis value. Three tRNA, SERPINA3, FRZB, and FCN3, were filtered with a potential diagnosis of DCM. Furthermore, our method finds two potential diagnosis genes (FRZB and FCN3) in DCM that have never been reported before. Compared with previous studies, SERPIAN3 presented the diagnosis value (38) in HF, and this work expanded its scale into DCM with the same point as Asakura and Kitakaze (39). Furthermore, Yang et al. (40) emphasizes the therapeutic value of FRZB, and our study expands its treatment potential to diagnosis value. Regarding FCN3, though studies pay attention to the diagnosis value for HF (41), no study reports the diagnosis value for DCM to our knowledge.
Serine proteinase inhibitor A3 (SERPINA3), also known as alpha-1 antichymotrypsin, has been shown to promote the development of cancer (42) and cardiac remodeling in patients with HF. In HF, though Delrue et al. (43) had confirmed that SERPINA3 is still an independent predictor of all-cause mortality, studies have paid little attention to the effect of pharmacological treatment of DCM. Spironolactone (44–47) dominates an important treated role in DCM. The previous study identified that spironolactone and lisinopril can downregulate SERPINA3 and treat mice with Duchenne muscular dystrophy, which suggests that SERPINA3 may be related to the salt corticosteroid receptor (48). Another study (49) came to a similar conclusion, SERPINA3 was both upregulated in vivo (mice of mineralocorticoid receptor cardiac upregulation) and in vitro (H9C2 cells with aldosterone 24 h). The above studies indicated that the up-regulated of SERPINA3 might be correlated with the mineralocorticoid receptor. However, few studies pay attention to DCM to our knowledge. And this work emphasizes the important role of SERPINA3 in DCM.
FRZB, sFRP3 also named, is one of a frizzled-related proteins (FRPs) family (the other three were sFRP-1, sFRP-2, and sFRP-4). The sFRP-3 and 4, can modulate apoptosis susceptibility in ventricular myocytes (50). However, though a previous study indicated that FRP contributed to the pathogenesis of DCM by down-regulated Wnt/β-catenin signaling pathway (51), no description of which of the four subtypes is associated. In DCM children (52), the serum circulating sFRP1 will trigger ventricular remodeling and cardiomyocyte fibrosis. And sFRP-1 knockout mice (53) indicated an abnormal cardiac structure present with increasing age. And the sFRP2 can prevent the conversion of inflammatory precursor components and the transformation of cardiomyocytes to pathogenic myofibroblasts (54) in DCM. However, no studies emphasized the function of FRZB in DCM, especially in plasma circulation. And our work first reported the diagnosis value of RZB in DCM. Furthermore, this work identified the significant upregulation of the circulation of FRZB protein in DCM.
FCN3, ficolin 3, was the most effective activator of the lectin pathway of complement (55) and more focus in rheumatic heart disease (56, 57). The FCN3 is inversely associated with the severity of HF (58). Furthermore, lower FCN3 is associated with the severity and outcome of HF (59). In congenital heart disease (60), the protein of FCN3 may prolong bleeding time and increase susceptibility to lung infection in the Fallot. However, few studies contribute to DCM. And our work first reported the diagnosis value of FCN3 in DCM. Furthermore, this work identified the significant downregulation of the circulation FCN3 protein in DCM.
Some limitations exist in our work. At first, inadequate validation is a common limitation in bioinformatics research. To decrease inadequate validation, three methods were taken, increase the sample size, developed the testing sets, and add little sample size clinical validation. However, additional studies should be conducted to validate, including but not limited to large sample size clinical trials or animal experiments for reliable verification of our predicted results. Secondly, MLs models exists some inevitable limitations, such as black box phenomenon (61), especially in NN (62) which contains various layers (e.g., an input layer, an output layer, and several hidden layers). Finally, few clinical features can be obtained, such as the age (63) or race (64) of the patient, which might trigger the bias of the result. In summary, further subgroup analyses are expected to assess more valuable conclusions in future works.
5. Conclusion
The overall weights methods for the filtration of genes in six MLs were developed, and we successfully found validation of three diagnosis genes, SERPINA3, FRZB, and FCN3. Further verification work could be implemented.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5406; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57338; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1145; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1869; https://www.ncbi. nlm.nih.gov/geo/query/acc.cgi?acc=GSE3585; and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42955.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Review Committee Jinghai District Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
LZ and YL: conceptualization, investigation, and data curation. LZ and KW: methodology. LZ: software and writing—original draft preparation. YL, KW, LZ, LH, HY, XF, XG, and JZ: validation. KW: writing—review and editing. ZL, HY, and XF: supervision. HZ and LH: project administration. HY: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
We are grateful for the foundation of the Science and Technology Program of Tianjin (No. 22ZYJDSS00100).
Acknowledgments
We thank for the support from the Tianjin University of Traditional Chinese Medicine.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.1044443/full#supplementary-material
References
1. Elmarakeby H, Hwang J, Arafeh R, Crowdis J, Gang S, Liu D, et al. Biologically informed deep neural network for prostate cancer discovery. Nature. (2021) 598:348–52. doi: 10.1038/s41586-021-03922-4
2. Frazer J, Notin P, Dias M, Gomez A, Min J, Brock K, et al. Disease variant prediction with deep generative models of evolutionary data. Nature. (2021) 599:91–5. doi: 10.1038/s41586-021-04043-8
3. Cawley G, Talbot N. Gene selection in cancer classification using sparse logistic regression with bayesian regularization. Bioinformatics. (2006) 22:2348–55. doi: 10.1093/bioinformatics/btl386
4. Han L, van Hemert J, Baldock R. Automatically identifying and annotating mouse embryo gene expression patterns. Bioinformatics. (2011) 27:1101–7. doi: 10.1093/bioinformatics/btr105
5. Kouyos R, von Wyl V, Hinkley T, Petropoulos C, Haddad M, Whitcomb J, et al. Assessing predicted Hiv-1 replicative capacity in a clinical setting. PLoS Pathog. (2011) 7:e1002321. doi: 10.1371/journal.ppat.1002321
6. Montisci A, Palmieri V, Vietri M, Sala S, Maiello C, Donatelli F, et al. Big data in cardiac surgery: real world and perspectives. J Cardiothorac Surg. (2022) 17:277. doi: 10.1186/s13019-022-02025-z
7. Vo Ngoc L, Huang C, Cassidy C, Medrano C, Kadonaga J. Identification of the human Dpr core promoter element using machine learning. Nature. (2020) 585:459–63. doi: 10.1038/s41586-020-2689-7
8. Yan J, Qiu Y, Ribeiro Dos Santos A, Yin Y, Li Y, Vinckier N, et al. Systematic analysis of binding of transcription factors to noncoding variants. Nature. (2021) 591:147–51. doi: 10.1038/s41586-021-03211-0
9. Dutta A, Goldman T, Keating J, Burke E, Williamson N, Dirmeier R, et al. Machine learning predicts biogeochemistry from microbial community structure in a complex model system. Microbiol Spectr. (2022) 10:e0190921. doi: 10.1128/spectrum.01909-21
10. Bouraffa T, Yan L, Feng Z, Xiao B, Wu Q, Xia Y. Context-aware correlation filter learning toward peak strength for visual tracking. IEEE Trans Cybern. (2021) 51:5105–15. doi: 10.1109/tcyb.2019.2935347
11. Wen J, Wang G, Xie X, Lin G, Yang H, Luo K, et al. Prognostic value of a four-mirna signature in patients with lymph node positive locoregional esophageal squamous cell carcinoma undergoing complete surgical resection. Ann Surg. (2021) 273:523–31. doi: 10.1097/sla.0000000000003369
12. Koga S, Zhou X, Dickson D. Machine learning-based decision tree classifier for the diagnosis of progressive supranuclear palsy and corticobasal degeneration. Neuropathol Appl Neurobiol. (2021) 47:931–41. doi: 10.1111/nan.12710
13. Wysocki A, Rhemtulla M. On penalty parameter selection for estimating network models. Multivariate Behav Res. (2021) 56:288–302. doi: 10.1080/00273171.2019.1672516
14. Crabtree N, Moore J, Bowyer J, George N. Multi-class computational evolution: development, Benchmark evaluation and application to Rna-Seq biomarker discovery. Biodata Min. (2017) 10:13. doi: 10.1186/s13040-017-0134-8
15. Li C, Wang J, Ge L, Zhou Y, Zhou S. Optimization of sample construction based on Ndvi for cultivated land quality prediction. Int J Environ Res Public Health. (2022) 19:7781. doi: 10.3390/ijerph19137781
16. Zhao S, Dong X, Shen W, Ye Z, Xiang R. Machine learning-based classification of diffuse large B-Cell lymphoma patients by eight gene expression profiles. Cancer Med. (2016) 5:837–52. doi: 10.1002/cam4.650
17. He Y, Ma J, Ye XA. Support vector machine classifier for the prediction of osteosarcoma metastasis with high accuracy. Int J Mol Med. (2017) 40:1357–64. doi: 10.3892/ijmm.2017.3126
18. Li C, Zeng X, Yu H, Gu Y, Zhang W. Identification of hub genes with diagnostic values in pancreatic cancer by bioinformatics analyses and supervised learning methods. World J Surg Oncol. (2018) 16:223. doi: 10.1186/s12957-018-1519-y
19. Zhang L, Mao R, Lau C, Chung W, Chan J, Liang F, et al. Identification of useful genes from multiple microarrays for ulcerative colitis diagnosis based on machine learning methods. Sci Rep. (2022) 12:9962. doi: 10.1038/s41598-022-14048-6
20. Liu Z, Li H, Pan S. Discovery and validation of key biomarkers based on immune infiltrates in Alzheimer’s disease. Front Genet. (2021) 12:658323. doi: 10.3389/fgene.2021.658323
21. Lu M, Qiu S, Jiang X, Wen D, Zhang R, Liu Z. Development and validation of epigenetic modification-related signals for the diagnosis and prognosis of hepatocellular carcinoma. Front Oncol. (2021) 11:649093. doi: 10.3389/fonc.2021.649093
22. Yao Y, Zhao J, Zhou X, Hu J, Wang Y. Potential role of a three-gene signature in predicting diagnosis in patients with myocardial infarction. Bioengineered. (2021) 12:2734–49. doi: 10.1080/21655979.2021.1938498
23. Yu J, Zhu M, Lv M, Wu X, Zhang X, Zhang Y, et al. Characterization of a five-microrna signature as a prognostic biomarker for esophageal squamous cell carcinoma. Sci Rep. (2019) 9:19847. doi: 10.1038/s41598-019-56367-1
24. Wang K, Zhang L, Li L, Wang Y, Zhong X, Hou C, et al. Identification of drug-induced liver injury biomarkers from multiple microarrays based on machine learning and bioinformatics analysis. Int J Mol Sci. (2022) 23:11945. doi: 10.3390/ijms231911945
25. McDonagh T, Metra M, Adamo M, Gardner R, Baumbach A, Böhm M, et al. 2021 Esc guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. (2021) 42:3599–726. doi: 10.1093/eurheartj/ehab368
26. Heidenreich P, Bozkurt B, Aguilar D, Allen L, Byun J, Colvin M, et al. 2022 Aha/Acc/Hfsa guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association joint committee on clinical practice guidelines. Circulation. (2022) 145:e895–1032. doi: 10.1161/cir.0000000000001063
27. Writing Group For Practice Guidelines For Diagnosis and Treatment Of Genetic Diseases Medical Genetics Branch Of Chinese Medical Association, Sun J, Han S, Hu J, Jiang C, Wang Q, et al. [Clinical practice guidelines for hereditary cardiomyopathy]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2020) 37:300–7. doi: 10.3760/cma.j.issn.1003-9406.2020.03.013
28. Abdel-Salam Z, Rayan M, Saleh A, Abdel-Barr M, Hussain M, Nammas W. I(F) current inhibitor ivabradine in patients with idiopathic dilated cardiomyopathy: impact on the exercise tolerance and quality of life. Cardiol J. (2015) 22:227–32. doi: 10.5603/CJ.a2014.0057
29. Nakano S, Miyamoto S, Movsesian M, Nelson P, Stauffer B, Sucharov C. Age-related differences in phosphodiesterase activity and effects of chronic phosphodiesterase inhibition in idiopathic dilated cardiomyopathy. Circ Heart Fail. (2015) 8:57–63. doi: 10.1161/circheartfailure.114.001218
30. Zhao C, Bing S, Song H, Wang J, Xu W, Jiang J, et al. Tbx20 loss-of-function mutation associated with familial dilated cardiomyopathy. Clin Chem Lab Med. (2016) 54:325–32. doi: 10.1515/cclm-2015-0328
31. Zhao J, Yin M, Deng H, Jin F, Xu S, Lu Y, et al. Cardiac Gab1 deletion leads to dilated cardiomyopathy associated with mitochondrial damage and cardiomyocyte apoptosis. Cell Death Differ. (2016) 23:695–706. doi: 10.1038/cdd.2015.143
32. Zhou L, Liu C, Zou Y, Chen Z. Development and verification of the nomogram for dilated cardiomyopathy gene diagnosis. Sci Rep. (2022) 12:8908. doi: 10.1038/s41598-022-13135-y
33. Wang Y, Guan Q, Lao I, Wang L, Wu Y, Li D, et al. Using deep convolutional neural networks for multi-classification of thyroid tumor by histopathology: a large-scale pilot study. Ann Transl Med. (2019) 7:468. doi: 10.21037/atm.2019.08.54
34. Radulescu E, Jaffe A, Straub R, Chen Q, Shin J, Hyde T, et al. Identification and prioritization of gene sets associated with schizophrenia risk by co-expression network analysis in human brain. Mol Psychiatry. (2020) 25:791–804. doi: 10.1038/s41380-018-0304-1
35. Choi B, Bair E, Lee J. Nearest shrunken centroids via alternative genewise shrinkages. PLoS One. (2017) 12:e0171068. doi: 10.1371/journal.pone.0171068
36. Dreiseitl S, Ohno-Machado L, Kittler H, Vinterbo S, Billhardt H, Binder MA. Comparison of machine learning methods for the diagnosis of pigmented skin lesions. J Biomed Inform. (2001) 34:28–36. doi: 10.1006/jbin.2001.1004
37. Chiew C, Liu N, Wong T, Sim Y, Abdullah H. Utilizing machine learning methods for preoperative prediction of postsurgical mortality and intensive care unit admission. Ann Surg. (2020) 272:1133–9. doi: 10.1097/sla.0000000000003297
38. Dang H, Ye Y, Zhao X, Zeng Y. Identification of candidate genes in ischemic cardiomyopathy by gene expression omnibus database. BMC Cardiovasc Disord. (2020) 20:320. doi: 10.1186/s12872-020-01596-w
39. Asakura M, Kitakaze M. Global gene expression profiling in the failing myocardium. Circ J. (2009) 73:1568–76. doi: 10.1253/circj.cj-09-0465
40. Yang Y, Liu P, Teng R, Liu F, Zhang C, Lu X, et al. Integrative bioinformatics analysis of potential therapeutic targets and immune infiltration characteristics in dilated cardiomyopathy. Ann Transl Med. (2022) 10:348. doi: 10.21037/atm-22-732
41. Li D, Lin H, Li L. Multiple feature selection strategies identified novel cardiac gene expression signature for heart failure. Front Physiol. (2020) 11:604241. doi: 10.3389/fphys.2020.604241
42. Meijers W, Maglione M, Bakker S, Oberhuber R, Kieneker L, de Jong S, et al. Heart failure stimulates tumor growth by circulating factors. Circulation. (2018) 138:678–91. doi: 10.1161/circulationaha.117.030816
43. Delrue L, Vanderheyden M, Beles M, Paolisso P, Di Gioia G, Dierckx R, et al. Circulating Serpina3 improves prognostic stratification in patients with a de novo or worsened heart failure. ESC Heart Fail. (2021) 8:4780–90. doi: 10.1002/ehf2.13659
44. Bell S, Adkisson D, Lawson M, Wang L, Ooi H, Sawyer D, et al. Antifailure therapy including spironolactone improves left ventricular energy supply-demand relations in nonischemic dilated cardiomyopathy. J Am Heart Assoc. (2014) 3:e000883. doi: 10.1161/jaha.114.000883
45. Nakagawa H, Oberwinkler H, Nikolaev V, Gaßner B, Umbenhauer S, Wagner H, et al. Atrial natriuretic peptide locally counteracts the deleterious effects of cardiomyocyte mineralocorticoid receptor activation. Circ Heart Fail. (2014) 7:814–21. doi: 10.1161/circheartfailure.113.000885
46. Verma A, Wulffhart Z, Lakkireddy D, Khaykin Y, Kaplan A, Sarak B, et al. Incidence of left ventricular function improvement after primary prevention Icd implantation for non-ischaemic dilated cardiomyopathy: a multicentre experience. Heart. (2010) 96:510–5. doi: 10.1136/hrt.2009.178061
47. Wang Y, Xu Y, Zou R, Wu L, Liu P, Yang H, et al. Effect of levocarnitine on the therapeutic efficacy of conventional therapy in children with dilated cardiomyopathy: results of a randomized trial in 29 children. Paediatr Drugs. (2018) 20:285–90. doi: 10.1007/s40272-018-0284-2
48. Chadwick J, Hauck J, Lowe J, Shaw J, Guttridge D, Gomez-Sanchez C, et al. Mineralocorticoid receptors are present in skeletal muscle and represent a potential therapeutic target. FASEB J. (2015) 29:4544–54. doi: 10.1096/fj.15-276782
49. Latouche C, Sainte-Marie Y, Steenman M, Castro Chaves P, Naray-Fejes-Toth A, Fejes-Toth G, et al. Molecular signature of mineralocorticoid receptor signaling in cardiomyocytes: from cultured cells to mouse heart. Endocrinology. (2010) 151:4467–76. doi: 10.1210/en.2010-0237
50. Schumann H, Holtz J, Zerkowski H, Hatzfeld M. Expression of secreted frizzled related proteins 3 and 4 in human ventricular myocardium correlates with apoptosis related gene expression. Cardiovasc Res. (2000) 45:720–8. doi: 10.1016/s0008-6363(99)00376-4
51. Le Dour C, Macquart C, Sera F, Homma S, Bonne G, Morrow J, et al. Decreased Wnt/B -catenin signalling contributes to the pathogenesis of dilated cardiomyopathy caused by mutations in the Lamin a/C gene. Hum Mol Genet. (2017) 26:333–43. doi: 10.1093/hmg/ddw389
52. Jeffrey D, Pires Da Silva J, Garcia A, Jiang X, Karimpour-Fard A, Toni L, et al. Serum circulating proteins from pediatric patients with dilated cardiomyopathy cause pathologic remodeling and cardiomyocyte stiffness. JCI Insight. (2021) 6:e148637. doi: 10.1172/jci.insight.148637
53. Sklepkiewicz P, Shiomi T, Kaur R, Sun J, Kwon S, Mercer B, et al. Loss of secreted frizzled-related protein-1 leads to deterioration of cardiac function in mice and plays a role in human cardiomyopathy. Circ Heart Fail. (2015) 8:362–72. doi: 10.1161/circheartfailure.114.001274
54. Blyszczuk P, Müller-Edenborn B, Valenta T, Osto E, Stellato M, Behnke S, et al. Transforming growth factor-B -dependent Wnt secretion controls myofibroblast formation and myocardial fibrosis progression in experimental autoimmune myocarditis. Eur Heart J. (2017) 38:1413–25. doi: 10.1093/eurheartj/ehw116
55. Michalski M, Świerzko A, Pa̧gowska-Klimek I, Niemir Z, Mazerant K, Domżalska-Popadiuk I, et al. Primary ficolin-3 deficiency–is it associated with increased susceptibility to infections? Immunobiology. (2015) 220:711–3. doi: 10.1016/j.imbio.2015.01.003
56. Beltrame M, Catarino S, Goeldner I, Boldt A, de Messias-Reason I. The lectin pathway of complement and rheumatic heart disease. Front Pediatr. (2014) 2:148. doi: 10.3389/fped.2014.00148
57. Elshamaa M, Hamza H, El Rahman N, Emam S, Elghoroury E, Farid T, et al. Association of Ficolin-2 (Fcn2) functional polymorphisms and protein levels with rheumatic fever and rheumatic heart disease: relationship with cardiac function. Arch Med Sci Atheroscler Dis. (2018) 3:e142–55. doi: 10.5114/amsad.2018.80999
58. Li H, Zhang F, Zhang D, Tian X. Changes of serum ficolin-3 and C5b-9 in patients with heart failure. Pak J Med Sci. (2021) 37:1860–4. doi: 10.12669/pjms.37.7.4151
59. Prohászka Z, Munthe-Fog L, Ueland T, Gombos T, Yndestad A, Förhécz Z, et al. Association of Ficolin-3 with severity and outcome of chronic heart failure. PLoS One. (2013) 8:e60976. doi: 10.1371/journal.pone.0060976
60. Xuan C, Gao G, Yang Q, Wang X, Liu Z, Liu X, et al. Proteomic study reveals plasma protein changes in congenital heart diseases. Ann Thorac Surg. (2014) 97:1414–9. doi: 10.1016/j.athoracsur.2013.11.069
61. Regazzoni F, Chapelle D, Moireau P. Combining data assimilation and machine learning to build data-driven models for unknown long time dynamics-applications in cardiovascular modeling. Int J Numer Method Biomed Eng. (2021) 37:e3471. doi: 10.1002/cnm.3471
62. Peng J, Ran Z, Shen J. Seasonal variation in onset and relapse of Ibd and a model to predict the frequency of onset, relapse, and severity of Ibd based on artificial neural network. Int J Colorectal Dis. (2015) 30:1267–73. doi: 10.1007/s00384-015-2250-6
63. Kalkan I, Dağli U, Oztaş E, Tunç B, Ulker A. Comparison of demographic and clinical characteristics of patients with early Vs. adult Vs. late onset ulcerative colitis. Eur J Intern Med. (2013) 24:273–7. doi: 10.1016/j.ejim.2012.12.014
Keywords: diagnosis value, dilated cardiomyopathy, machine learning, SERPINA3, FRZB, FCN3
Citation: Zhang L, Lin Y, Wang K, Han L, Zhang X, Gao X, Li Z, Zhang H, Zhou J, Yu H and Fu X (2023) Multiple-model machine learning identifies potential functional genes in dilated cardiomyopathy. Front. Cardiovasc. Med. 9:1044443. doi: 10.3389/fcvm.2022.1044443
Received: 14 September 2022; Accepted: 22 December 2022;
Published: 11 January 2023.
Edited by:
Vittorio Palmieri, Azienda Ospedaliera dei Colli, ItalyReviewed by:
Maria Teresa Vietri, Seconda Università degli Studi di Napoli, ItalyLingfang Zhuang, Shanghai Jiao Tong University, China
Xiang Ma, First Affiliated Hospital of Xinjiang Medical University, China
Copyright © 2023 Zhang, Lin, Wang, Han, Zhang, Gao, Li, Zhang, Zhou, Yu and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heshui Yu,  aHNfeXUwOEAxNjMuY29t; Xuebin Fu,
aHNfeXUwOEAxNjMuY29t; Xuebin Fu,  eGZ1QGx1cmllY2hpbGRyZW5zLm9yZw==
eGZ1QGx1cmllY2hpbGRyZW5zLm9yZw==
†These authors have contributed equally to this work
 Lin Zhang
Lin Zhang Yexiang Lin2†
Yexiang Lin2† Lifeng Han
Lifeng Han Xiumei Gao
Xiumei Gao Xuebin Fu
Xuebin Fu