
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 30 November 2021
Sec. Cardiovascular Genetics and Systems Medicine
Volume 8 - 2021 | https://doi.org/10.3389/fcvm.2021.757362
Cardiac injury is a common complication of coronavirus disease 2019 (COVID-19), but the exact mechanisms have not been completely elucidated. The virus receptors on subsets of cells are key determinants of susceptibility to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Due to its high sequence similarity to SARS-CoV, SARS-CoV-2 also utilizes ACE2 as the cell entry receptor. A growing number of studies have indicated that other receptors apart from ACE2 are involved in SARS-CoV-2 infection. This study aimed to elucidate the expression characteristics of SARS-CoV-2 cellular receptors in the heart. We first investigated ACE2 expression in a comprehensive transcriptional landscape of the human heart comprising single-nucleus RNA-seq (snRNA-seq) data for >280,000 cells. Then, the expression distributions of novel SARS-CoV-2 receptors were analyzed at the single-cell level to clarify the cardiovascular complications in COVID-19. We observed a higher percentage of ACE2-positive cells in pericytes (8.3%), fibroblasts (5.1%), and adipocytes (4.4%) in the human heart, compared to other cell types. The frequency of ACE2-positive cells in each cell type from the ventricles was significantly higher than that in the atria, suggesting that the ventricular cells are more susceptible to SARS-CoV-2 infection. The distribution patterns of other receptors (BSG, HSPA5, KREMEN1, NRP1, ANPEP, AXL) were significantly different from those of ACE2, demonstrating higher expression levels in ventricular cardiomyocytes. Moreover, our results suggest that fibroblasts and adipocytes, aside from pericytes, may be vulnerable targets for SARS-CoV-2 infection in the human heart. Our study presents potential targets for future clinical studies and interventions for cardiac injury in patients with COVID-19.
The coronavirus disease 2019 (COVID-19), caused by a pathogenic coronavirus known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1), has become a serious global public health issue (2). Currently, there are no specific therapeutics available for the treatment of COVID-19 (3). Respiratory system symptoms were the predominant clinical manifestations in patients with COVID-19, similar to severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (4). However, patients with COVID-19 seem to have a diverse range of extrapulmonary manifestations (5–8). Cardiac injury is a common complication of COVID-19 and is observed in 7–20% of patients infected with SARS-CoV-2 (9). Moreover, SARS-CoV-2 infection in the myocardium was verified by reverse transcription polymerase chain reaction (RT-qPCR) and histopathology (8, 10). The presence of myocardial injury is an independent risk factor for mortality in patients with COVID-19 (11). However, the exact mechanisms underlying cardiac injury in patients with COVID-19 have not been completely elucidated.
As essential elements of virus transmission, the expression and distribution of virus receptors on subsets of cells are key determinants of susceptibility to SARS-CoV-2 infection (12). SARS-CoV-2 has ~79.6% genome sequence homology with SARS-CoV (13, 14). Thus, angiotensin-converting enzyme 2 (ACE2) is thought to play an important role in the attachment and entry of SARS-CoV-2 and the pathogenesis of COVID-19 (4, 12, 15). Cells expressing ACE2 may be considered as a therapeutic target for COVID-19 (16). Some studies have demonstrated that ACE2 is highly expressed in pericytes in the human heart (17), but it has not been well-studied because of the lack of a comprehensive human single-cell atlas. It is of great significance to gain deeper insights into the expression characteristics of SARS-CoV-2 cellular receptors in the heart.
It is worth noting that certain organs with little ACE2 expression was also detected for virus invasion (18). The different clinical features of COVID-19 and SARS (19) also demonstrated that there may be other receptors and/or co-receptors of SARS-CoV-2, which are involved in the entry of the virus into cells (20). High-throughput receptor profiling for SARS-CoV-2 revealed that the S protein could specifically bind to the following cellular receptors with high affinity: ASGR1, KREMEN1 (21), ADAM17, GRP78, and CD147 (22). Recent studies have also suggested the possible involvement of NRP1 (23), ANPEP (20, 24), and AXL (25) in SARS-CoV-2 attachment, subsequent entry into the cell, and infection. Therefore, the expression features of the recently discovered SARS-CoV-2 receptors in human cardiac cells should be explored.
In this study, we first investigated the ACE2 expression in a comprehensive transcriptional landscape of the human heart comprising single-nucleus RNA sequencing (snRNA-seq) data for >280,000 cells (26). Then, the expression distributions of the novel SARS-CoV-2 receptors were also analyzed at the single-cell level to advance our understanding of cardiovascular complications in COVID-19.
We acquired the available snRNA-seq data derived from seven normal human heart samples from the Broad Institute's Single Cell Portal1 under the Study ID SCP498 online. In which, “healthy_human_4chamber_map_unnormalized_V3.h5ad” was used in the downstream analyses. We used the gene expression matrix and cell type annotation provided by the original article (26).
Since the low-quality cells were excluded from the original publication, we used all the data available. The downloaded unique molecular identifier count matrix was converted to Seurat object using the R package Seurat v.3.2.2 (27). We then normalized the raw gene expression matrix using the NormalizeData function and visualize the expression level using the Violin plot function in Seurat. Uniform manifold approximation and projection (UMAP)2 was used to visualize the results.
Downstream analyses, including highly variable gene detection (FindVariableFeatures, method= “vst,” nfeatures = 2,000), data feature scaling (ScaleData), PCA (Principal Component Analysis, RunPCA, from highly variable genes), an integration procedure (RunHarmony, npcs = 23), neighbor construction (FindNeighbors, using the output of RunHarmony), and clustering (FindClusters, resolution = 0.4), were performed using the R package Seurat (v.3.2.2) and harmony (v1.0). Cell type labels were assigned manually using the annotation provided in the original article (26). We further investigated clusters for which the genes indicated additional diversity and specificity.
Unless otherwise stated, tests for differentially expressed genes (DEGs) were performed using the FindAllMarker or FindMarker function in the Seurat package (with the default test method: Wilcoxon rank-sum test). To compute for the DEGs, all genes were included and expressed in at least 25% of cells in either of the two populations compared.
Cell–cell interaction weights for each ligand-receptor pair were computed as the product of the fold change of ligands in sender-cell types and the fold change of the corresponding receptors in receiver cell types. We used the “layout_nicely” function in the R package “igraph” to visualize the cell-cell interaction.
The R and Python scripts used to analyze the data in this study are available from the corresponding authors upon reasonable request (Table 1).
All statistical tests were performed using the R statistical programming language (V.3.6.2, R Foundation for Statistical Computing, Vienna, Austria). For analysis of snRNA-seq data, we used Wilcoxon rank-sum tests (between two groups) or Kruskal–Wallis test (for comparisons of multiple groups) to detect differential genes, with P-values adjusted based on Bonferroni correction. Statistical significance was set at P < 0.05.
We obtained single-nucleus transcriptome data of seven human donor hearts containing 287,269 nuclei sequenced by 10X Genomics from the previous study (26). The basic characteristics of the human donors are shown in Supplementary Table 1. As shown in Figure 1A, unsupervised graph-based clustering revealed 11 cell types in the human heart, of which multiple cell types expressed the ACE2 gene (Figure 1B). Clear differences in the proportion of ACE2-positive cells were found among the different cell types (Figure 1C). Furthermore, high percentages of ACE2-positive cells were observed in the pericytes (8.3%), fibroblasts (5.1%), and adipocytes (4.4%) in the heart (Table 2). These results are slightly different from those reported in the literature (17). In addition to pericytes, we predicted that fibroblasts and adipocytes could be vulnerable targets for SARS-CoV-2 infection. Similar to previous reports, the pericyte showed a significant intercellular interaction with the endothelium (Figure 1D). In addition, fibroblast demonstrated significant reciprocal interactions with vascular smooth muscle cells (Figure 1D). These results suggest that vascular injury plays an important role in the occurrence and development of cardiac injury in COVID-19. After binding to the cell surface, SARS-CoV-2 requires S protein cleavage proteases, such as TMPRSS2, FURIN, CTSL, and CTSB, to facilitate cell entry by inducing the fusion of cellular and viral membranes. TMPRSS2/4/11A, the well-known co-receptor for SARS-CoV-2 in other organs, demonstrated almost no co-expression (<1%) with ACE2 in various cell types of the human heart (Figure 1E; Supplementary Table 2). However, FURIN and CTSL/B seemed to play significant roles as co-receptors for SARS-CoV-2 entry into the human heart (Figure 1F). Further studies are required to elucidate the exact mechanism.
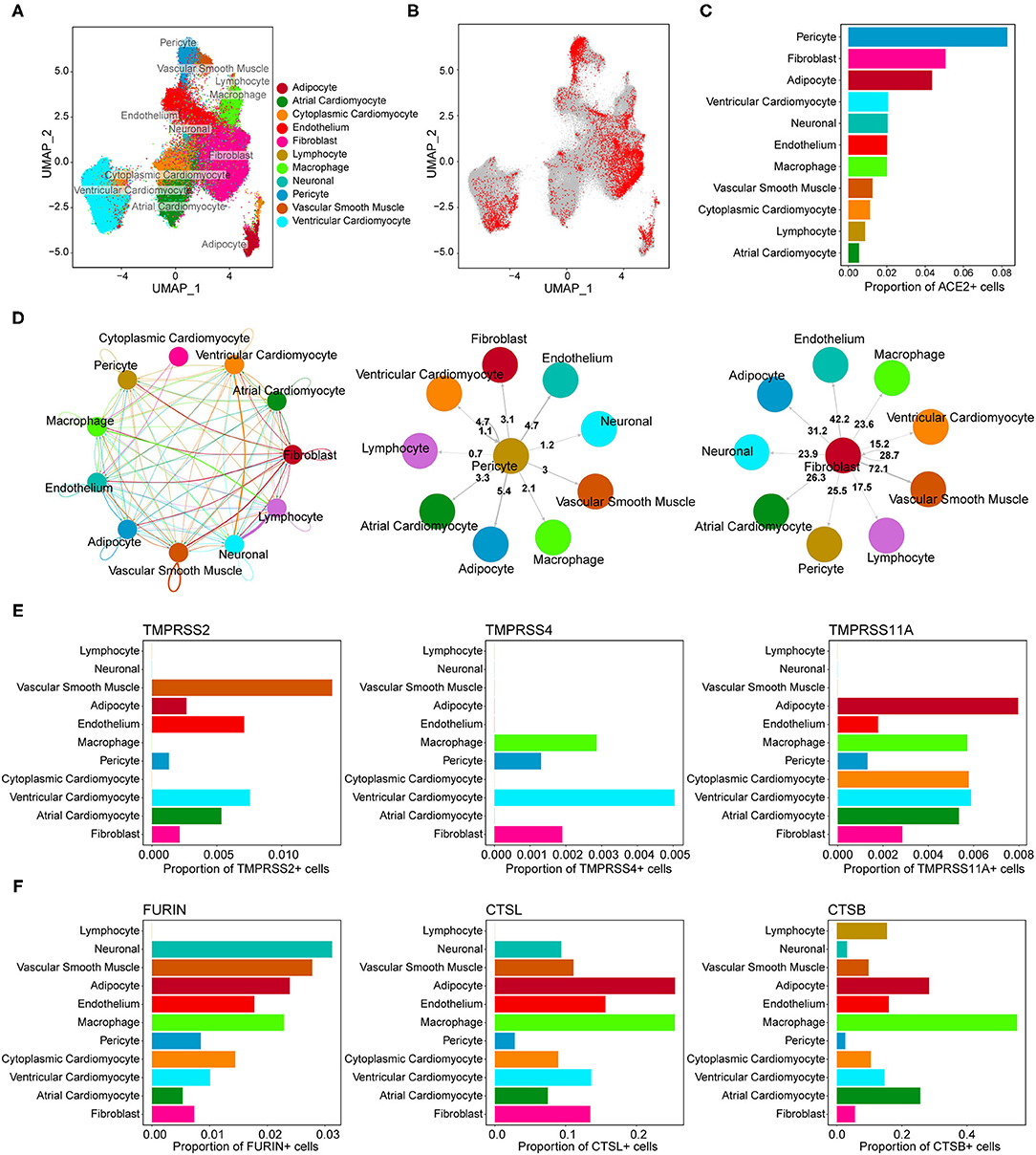
Figure 1. Gene expression characterizations of angiotensin-converting enzyme 2 (ACE2) and its co-receptors in the human heart. (A) Uniform manifold approximation and projection (UMAP) plot of cell clusters in the human heart. (B) Feature plot of ACE2 expression across all cell clusters. (C) The proportion of ACE2-positive cells in each cell population. (D) Reciprocal interaction network between different cell types. (Middle panel) Cell-cell interaction between pericyte (ligand) and other cell types (receptors). (Right panel) Cross-talks from fibroblast to other cell types. (E,F) The co-expression percentage of ACE2 and accessory proteases, such as TMPRSS2, FURIN, CTSL, and CTSB.
It has been proven that the male sex is a risk factor for disease severity in COVID-19 (28). The heart is composed of different chambers, which have distinct structural and functional characteristics. Hence, gaining a deeper understanding of the expression characteristics of SARS-CoV-2 cellular receptors in the human heart at spatial and sex-specific levels is of great importance. No significant differences in the overall distribution of cardiac cells between the sexes were observed (Figure 2A). The proportion of ACE2-positive cells was further compared between different genders in each cell type. As shown in Figure 2B, the ratio of ACE2-positive cells had a gender-differentiated effect in pericytes, fibroblasts, and the endothelium. However, it is interesting to note that even with a lower ACE2 proportion, higher expression levels of ACE2 were observed in pericytes, fibroblasts, and endothelium in male hearts (Figure 2C). This might be a reason why there is no significant difference observed in adverse cardiac events between patients with COVID-19 of different genders.
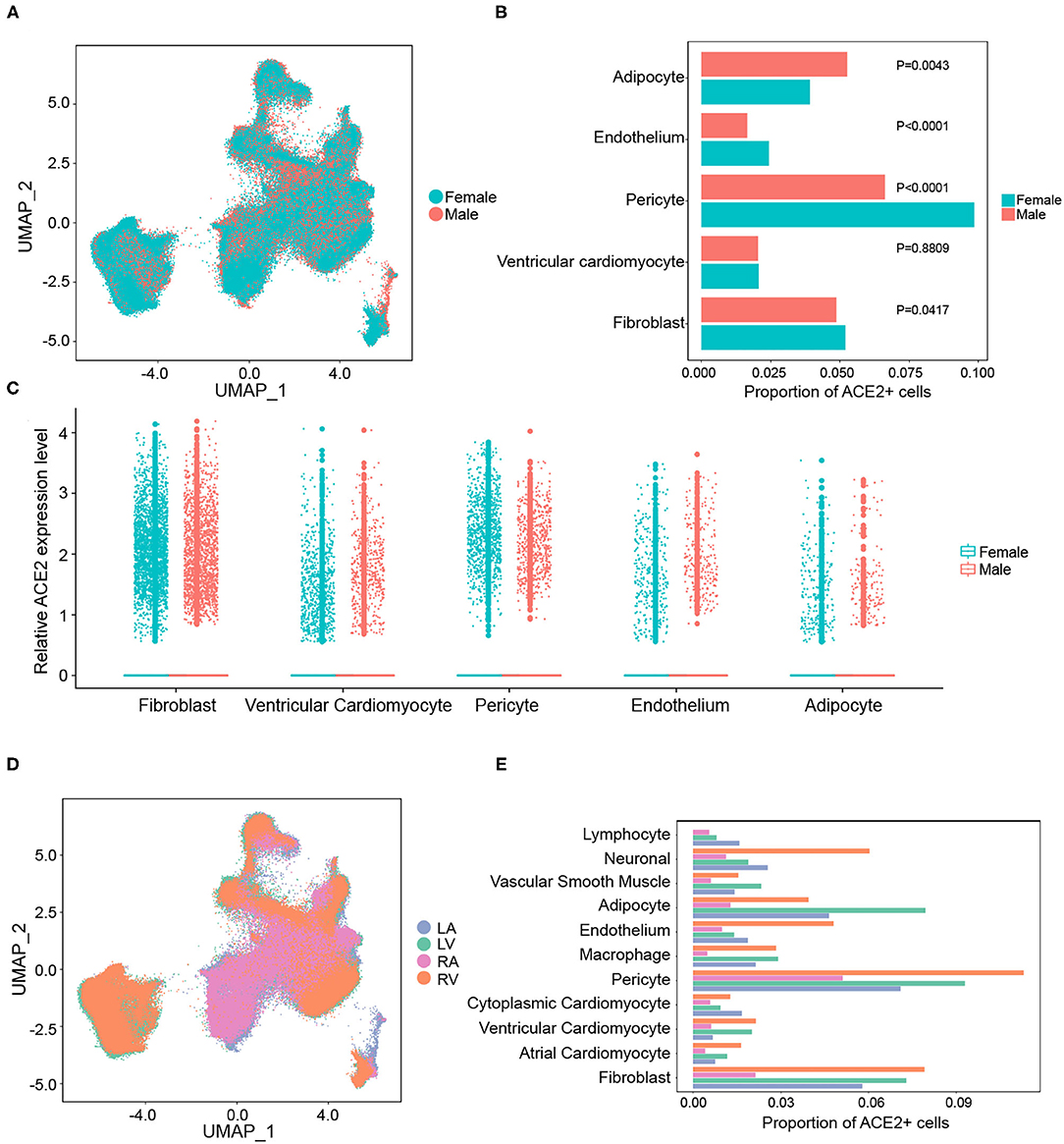
Figure 2. Gender and regional differences in the expression of ACE2 in the human heart. (A) The overall distribution of cardiac cells between sexes. (B) The gender-differentiated effect on the percentage of ACE2-positive cells in main cell clusters. (C) The expression levels of ACE2 between sexes in different cell types. (D) The distributions of gene expression in cells from different chambers of the heart. (E) The proportion of ACE2-positive cells in each cell cluster from different cardiac chambers.
However, there were remarkable differences in the gene expression between the atria and ventricles of the human heart (Figure 2D). The frequency of ACE2-positive cells in each cell type from the ventricles was significantly higher than that in the atria (Figure 2E). For instance, the proportion of ACE2-positive cells in ventricular cardiomyocytes (2.1%) were significantly higher than that in the atrial cardiomyocytes (0.5%). This suggests that human ventricular cells are more susceptible to SARS-CoV-2 infection.
Previous studies have reported potential receptors involved in SARS-CoV-2 binding and infection other than the ACE2 (20–25). Each potential receptor exhibited a distinct expression pattern (Figure 3A). The human cardiac cells showed nearly no expression of ASGR1, while the other receptors (BSG, HSPA5, KREMEN1, NRP1, ANPEP, and AXL) showed scattered expression in each cell type (Figure 3A; Supplementary Tables 3–5). The expression distributions and correlations of the potential receptors and ACE2 were analyzed using feature plots. There was no significant association between the distribution of ACE2 and the potential receptors (Figures 3B,C). Dissimilarities among the expression characteristics of the potential receptors, compared with ACE2, might be the reason for the different clinical features of COVID-19 and SARS. Significantly different from the distribution pattern of ACE2, positive expression of BSG, KREMEN1, and NRP1 were highest in the ventricular cardiomyocytes (Figures 3D,E). These results may shed light on the susceptibility of human ventricular cardiomyocytes to SARS-CoV-2.
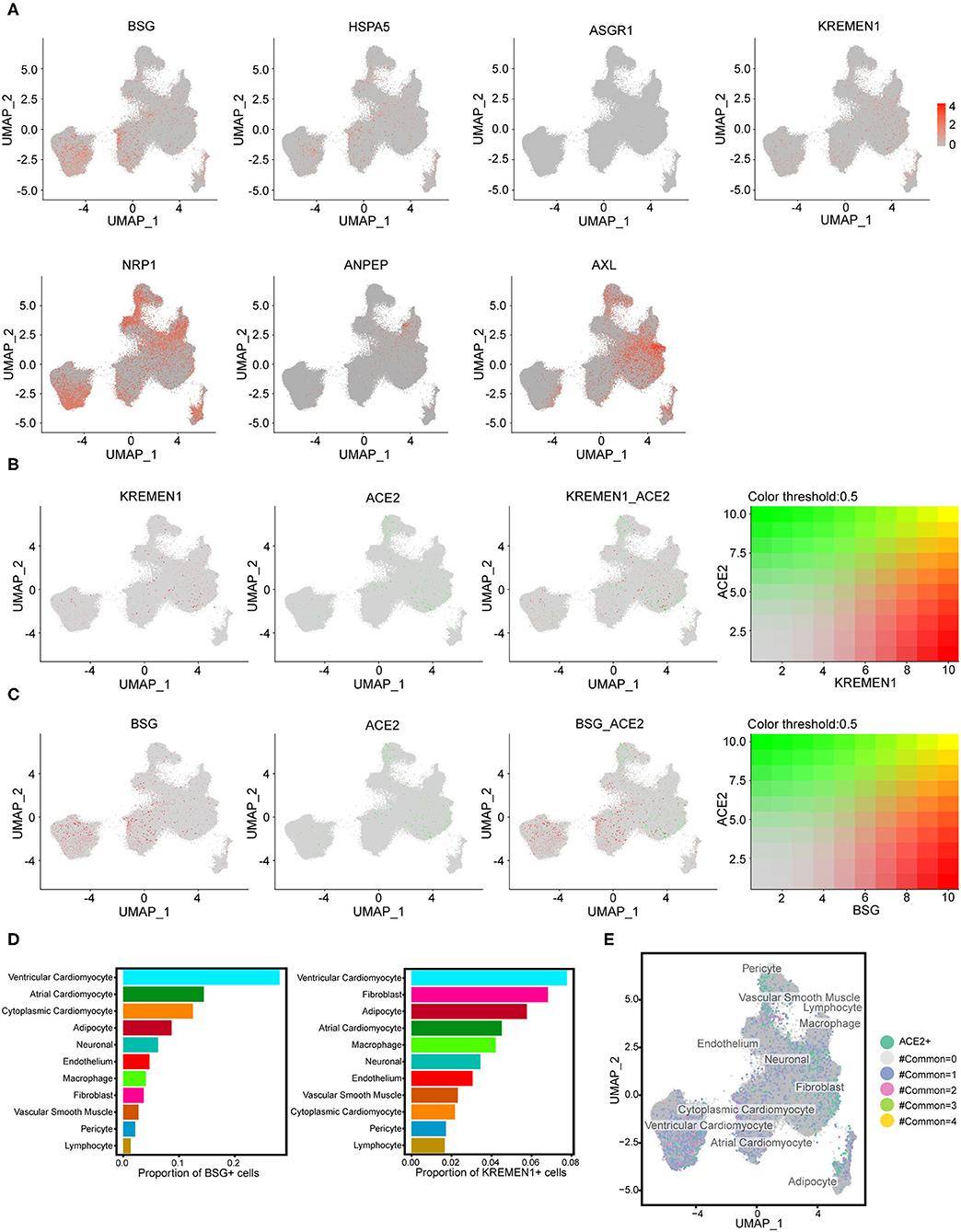
Figure 3. The expression signatures of potential receptors in the human heart. (A) Expression features of potential SARS-CoV-2 receptors (BSG, HSPA5, ASGR1, KREMEN1, NRP1, ANPEP, and AXL) across all cell clusters. (B) The expression distributions of KREMEN1 and its correlation with ACE2. (C) The expression distributions of BSG and its correlations with ACE2. For (B,C) right panels: green represents BSG/KREMEN1 positive cells, red represents ACE2 positive, gray indicates double-negative cells, and yellow indicates double-positive cells. (D) The proportion of BSG-positive or KREMEN1-positive cells in each cell population. (E) The expression characterizations of the four known receptors (ACE2, BSG, HSPA5, and KREMEN1). The numbers represent the receptors with a coexistence state.
The heart is one of the involved organs in COVID-19 (29). Using the results from the detailed expression patterns of ACE2 and the other potential receptors of SARS-CoV-2 in the human heart, we suggested that fibroblasts and adipocytes could serve as vulnerable targets for SARS-CoV-2 other than pericytes (17, 30). Moreover, human ventricular cells are more susceptible to SARS-CoV-2. Our study provides more information on the infectious mechanism of SARS-CoV-2 in the human heart.
Cardiac fibroblasts play important roles in normal physiology and pathological states, such as fibrosis (31). Fibroblasts, a major cell type in the heart, support the structural framework of tissue and maintain tissue homeostasis (32). Impaired consecutive fibroblasts can cause progressive cardiac dysfunction and increase the risk of sudden death (33, 34). Meanwhile, an imbalance or dysfunction of adipocytes could lead to inflammatory cascade reactions by secreting proinflammatory mediators (adipokines, IL-1, TNF-α, etc.) (35, 36), and these inflammatory markers could further play a role in the activation of immune cells (37). Epidemiological investigations have also demonstrated that obesity is a risk factor for a poor prognosis of COVID-19 (38). These observations suggest that fibroblasts and adipocytes are additional vulnerable targets for SARS-CoV-2. In addition, the mammalian atria and ventricles display striking differences in their structural features and gene expression (39). More than 2,000 genes appear to be differentially expressed in human ventricles and atria (26). The results of our study suggest that ventricular cells, especially cardiomyocytes, fibroblasts, adipocytes, and pericytes, are more likely to suffer from SARS-CoV-2 infection.
Recently, various investigations have suggested the existence of other receptors involved in SARS-CoV-2 infection, in addition to ACE2 (20–25). In this study, these receptors had relatively scattered distributions than ACE2 in the human heart. It is noteworthy that the expression levels of these receptors were significantly higher in ventricular cardiomyocytes, suggesting higher susceptibility of ventricular cardiomyocytes to SARS-CoV-2 infection. These potential SARS-CoV-2 receptors may play an important role in promoting viral infection of the human cardiac system and may be potential targets for future clinical cardiac intervention strategies.
First, our results and conclusions are data-driven, which needed further laboratory verifications. Additionally, the original sample size was relatively small, which restricted its further implications. Finally, the snRNA-seq data we included was from normal human hearts, which could, therefore, limit further analysis of the correlation between gene expression features and clinical characteristics in patients with COVID-19.
Publicly available datasets were analyzed in this study. This data can be found here: Broad Institute's Single Cell Portal (https://singlecell.broadinstitute.org/single_cell/study/SCP498/transcriptional-and-cellulardiversity-of-the-human-heart) under study ID SCP498.
XS contributed to the study's conception and design. Material preparation and data collection were performed by JR, YZ, and XL. SL and XL analyzed the data. The first draft of the manuscript was written by JR. All the authors commented on previous versions of the manuscript. All the authors read and approved the final manuscript.
This study was supported by the Beijing Municipal Science and Technology Commission (Z181100001718197).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the authors who uploaded their single-cell sequence data for others to perform further research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2021.757362/full#supplementary-material
1. ^https://singlecell.broadinstitute.org/single_cell/study/SCP498/transcriptional-and-cellulardiversity-of-the-human-heart.
1. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. (2020) 579:265–9. doi: 10.1038/s41586-020-2008-3
2. Nishiga M, Wang DW, Han Y, Lewis DB, Wu JC. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat Rev Cardiol. (2020) 17:543–58. doi: 10.1038/s41569-020-0413-9
3. Jeyanathan M, Afkhami S, Smaill F, Miller MS, Lichty BD, Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. (2020) 20:615–32. doi: 10.1038/s41577-020-00434-6
4. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. (2019) 17:181–92. doi: 10.1038/s41579-018-0118-9
5. Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. (2020) 20:363–74. doi: 10.1038/s41577-020-0311-8
6. Wadman M, Couzin-Frankel J, Kaiser J, Matacic C. A rampage through the body. Science. (2020) 368:356–60. doi: 10.1126/science.368.6489.356
7. Puelles VG, Lutgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. (2020) 383:590–2. doi: 10.1056/NEJMc2011400
8. Hu H, Ma F, Wei X, Fang Y. Coronavirus fulminant myocarditis saved with glucocorticoid and human immunoglobulin. Eur Heart J. (2020) 42:206. doi: 10.1093/eurheartj/ehaa190
9. Clerkin KJ, Fried JA, Raikhelkar J, Sayer G, Griffin JM, Masoumi A, et al. COVID-19 and cardiovascular disease. Circulation. (2020) 141:1648–55. doi: 10.1161/CIRCULATIONAHA.120.046941
10. Tavazzi G, Pellegrini C, Maurelli M, Belliato M, Sciutti F, Bottazzi A, et al. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur J Heart Fail. (2020) 22:911–5. doi: 10.1002/ejhf.1828
11. Shi S, Qin M, Shen B, Cai Y, Liu T, Yang F, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. (2020) 5:802–10. doi: 10.1001/jamacardio.2020.0950
12. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271-80.e8. doi: 10.1016/j.cell.2020.02.052
13. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. (2020) 26:450–2. doi: 10.1038/s41591-020-0820-9
14. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
15. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol. (2009) 7:226–36. doi: 10.1038/nrmicro2090
16. Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. (2020) 588:682–7. doi: 10.1038/s41586-020-2852-1
17. Chen L, Li X, Chen M, Feng Y, Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc Res. (2020) 116:1097–100. doi: 10.1093/cvr/cvaa078
18. Hikmet F, Mear L, Edvinsson A, Micke P, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. (2020) 16:e9610. doi: 10.15252/msb.20209610
19. Chu H, Chan JF, Yuen TT, Shuai H, Yuan S, Wang Y, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. (2020) 1:e14–23. doi: 10.1016/S2666-5247(20)30004-5
20. Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. (2020) 26:681–7. doi: 10.1038/s41591-020-0868-6
21. Lu Q, Liu J, Zhao S, Gomez Castro MF, Laurent-Rolle M, Dong J, et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. (2021) 54:1304-19.e9. doi: 10.1016/j.immuni.2021.05.006
22. Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A, et al. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J. (2020) 56:2001123. doi: 10.1183/13993003.01123-2020
23. Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. (2020) 370:856–60. doi: 10.1126/science.abd2985
24. Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. (2020) 526:135–40. doi: 10.1016/j.bbrc.2020.03.044
25. Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. (2021) 31:126–40. doi: 10.1038/s41422-020-00460-y
26. Tucker NR, Chaffin M, Fleming SJ, Hall AW, Parsons VA, Bedi K Jr, et al. Transcriptional and Cellular Diversity of the Human Heart Circulation. (2020) 142:466–82. doi: 10.1161/CIRCULATIONAHA.119.045401
27. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell. (2019) 177:1888-902.e21. doi: 10.1016/j.cell.2019.05.031
28. Gebhard C, Regitz-Zagrosek V, Neuhauser HK, Morgan R, Klein SL. Impact of sex and gender on COVID-19 outcomes in Europe. Biol Sex Differ. (2020) 11:29. doi: 10.1186/s13293-020-00304-9
29. Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med. (2020) 382:1708–20. doi: 10.1056/NEJMoa2002032
30. Liu H, Gai S, Wang X, Zeng J, Sun C, Zhao Y, et al. Single-cell analysis of SARS-CoV-2 receptor ACE2 and spike protein priming expression of proteases in the human heart. Cardiovasc Res. (2020) 116:1733–41. doi: 10.1093/cvr/cvaa191
31. Baudino TA, Carver W, Giles W, Borg TK. Cardiac fibroblasts: friend or foe? Am J Physiol Heart Circ Physiol. (2006) 291:H1015–26. doi: 10.1152/ajpheart.00023.2006
32. Gaudesius G, Miragoli M, Thomas SP, Rohr S. Coupling of cardiac electrical activity over extended distances by fibroblasts of cardiac origin. Circ Res. (2003) 93:421–8. doi: 10.1161/01.RES.0000089258.40661.0C
33. Banerjee I, Yekkala K, Borg TK, Baudino TA. Dynamic interactions between myocytes, fibroblasts, and extracellular matrix. Ann N Y Acad Sci. (2006) 1080:76–84. doi: 10.1196/annals.1380.007
34. Bowers SL, Baudino TA. Cardiac myocyte-fibroblast interactions and the coronary vasculature. J Cardiovasc Transl Res. (2012) 5:783–93. doi: 10.1007/s12265-012-9407-2
35. Meftahi GH, Jangravi Z, Sahraei H, Bahari Z. The possible pathophysiology mechanism of cytokine storm in elderly adults with COVID-19 infection: the contribution of “inflame-aging.” Inflamm Res. (2020) 69:825–39. doi: 10.1007/s00011-020-01372-8
36. Patel VB, Basu R, Oudit GY. ACE2/Ang 1-7 axis: a critical regulator of epicardial adipose tissue inflammation and cardiac dysfunction in obesity. Adipocyte. (2016) 5:306–11. doi: 10.1080/21623945.2015.1131881
37. Mery G, Epaulard O, Borel AL, Toussaint B, Le Gouellec A. COVID-19: underlying adipokine storm and angiotensin 1-7 umbrella. Front Immunol. (2020) 11:1714. doi: 10.3389/fimmu.2020.01714
38. Kruglikov IL, Shah M, Scherer PE. Obesity and diabetes as comorbidities for COVID-19: underlying mechanisms and the role of viral-bacterial interactions. Elife. (2020) 9:e61330. doi: 10.7554/eLife.61330
Keywords: SARS-CoV-2, cardiac injury, cellular receptors, ACE2, expression features
Citation: Ren J, Zhang Y, Liu S, Li X and Sun X (2021) Detailed Analyses of the Expression Patterns of Potential Severe Acute Respiratory Syndrome Coronavirus 2 Receptors in the Human Heart Using Single-Nucleus RNA Sequencing. Front. Cardiovasc. Med. 8:757362. doi: 10.3389/fcvm.2021.757362
Received: 12 August 2021; Accepted: 11 October 2021;
Published: 30 November 2021.
Edited by:
George W. Booz, University of Mississippi Medical Center School of Dentistry, United StatesReviewed by:
Theresa Li-Yun Chang, Rutgers, The State University of New Jersey, United StatesCopyright © 2021 Ren, Zhang, Liu, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaogang Sun, eGlhb2dhbmdzdW5sQDE2My5jb20=; Xiangjie Li, ZWxlNzE3QDE2My5jb20=
†These authors have contributed equally to this work
‡These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.