- 1Department of Cardiology, Xiamen Key Laboratory of Cardiac Electrophysiology, Xiamen Institute of Cardiovascular Diseases, The First Affiliated Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, China
- 2Department of Internal Medicine, Cardiovascular Center and Division of Cardiology, National Taiwan University Hospital and National Taiwan University College of Medicine, Taipei, China
- 3Cardio-X Center, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology and Center for Human Genome Research, Huazhong University of Science and Technology, Wuhan, China
Objective: To investigate the genetic characteristics and transcriptional regulation of the SCN5A gene of Brugada syndrome (BrS) patients in China.
Methods: Using PubMed, Medline, China National Knowledge Internet (CNKI), and Wanfang Database, Chinese patients with BrS who underwent SCN5A gene testing were studied.
Results: A total of 27 suitable studies involving Chinese BrS patients who underwent the SCN5A gene test were included. A total of 55 SCN5A gene mutations/variations were reported in Chinese BrS patients, including 10 from southern China and 45 from northern China. Mutations/variations of BrS patients from southern China mostly occurred in the regions of the α-subunit of Nav1.5, including DIII (Domain III), DIV, DIII-DIV, C-terminus regions, and the 3'UTR region. Furthermore, we analyzed the post-transcriptional modifications (PTMs) throughout the Nav1.5 protein encoded by SCN5A and found that the PTM changes happened in 72.7% of BrS patients from southern China and 26.7% from northern China.
Conclusions: SCN5A mutations/variations of BrS patients in southern China mostly occurred in the DIII-DIV to C-terminus region and the 3'-UTR region of the SCN5A gene, different from northern China. PTM changes were consistent with the mutation/variation distribution of SCN5A, which might be involved in the regulation of the pathogenesis of BrS patients.
Introduction
Brugada syndrome (BrS) is an inheritable arrhythmogenic disease. The typical electrocardiographic manifestations include ST segment elevation ≥2 mm and T wave inversion on the right thoracic lead (V1–V3) of ECG. BrS is prone to polymorphic ventricular tachycardia, ventricular fibrillation, and sudden cardiac death while the heart structure is normal (1, 2). It is more prevalent in Asian population with an onset age of 30–40 years old, and the ratio of male to female is 8–10:1 (3, 4). The mortality accounted for 4–12% of sudden death each year and even for 20% sudden death without organic heart disease in Southeast Asia (5).
Up to date, 23 genes have been confirmed to be related to BrS (5–7), including gene mutations/variations that lead to ion channel dysfunction such as sodium, calcium, and potassium ions. About 30–35% of BrS patients were identified with pathogenic mutant genes, while SCN5A mutations/variations encoding the Nav1.5 α-subunit of the cardiac sodium channel accounted for 20 to 30% (5, 8–10). Currently, the recommended treatment for BrS is ICD implantation and medication (quinidine, isoproterenol, etc.); however, therapeutic effect is unsatisfactory.
The mutations/variations from BrS patients can significantly reduce the inward Na+ current by delaying activation, accelerating inactivation, delaying reactivation, or reducing the membrane expression of ion channels (11, 12). Furthermore, the decrease in the inward Na+ current has influence on the depolarization and repolarization of the cardiac action potential, thus causing the generation of typical ECG of BrS (2). Genetic distribution characteristics of BrS on the SCN5A mutations/variations from the world (9) and Japan (13) had been reported. However, there are no genetic distribution analyses on the SCN5A mutation/variation location sites in Chinese BrS patients until now. In the present study, we aim to analyze the reported SCN5A mutation/variation location sites of Chinese BrS patients and predict the PTMs affected by the mutations/variations.
Materials and Methods
Information Retrieval and Inclusion Criteria
Two investigators searched Medline, PubMed, CNKI, and Wanfang Database. The query terms were “Brugada syndrome” “China” and “SCN5A.” Articles published in Chinese or English in peer-reviewed journals that met the following criteria were included in our study.
A. Inclusion of subjects with BrS were as previously defined (14).
B. Chinese patients.
C. Who underwent SCN5A gene DNA sequencing.
In addition, we also contacted the corresponding author of several studies in order to obtain more specific experimental data that were not included in the article. In order to resolve any difference or uncertainty between the two investigators, a third investigator was responsible for reexamining the source data and consultation.
Predictive Analysis the PTMs of SCN5A
First, we retrieved the amino acid sequence and analyzed the domain details of the α-subunit of SCN5A protein in the following website: https://www.uniprot.org/uniprot/Q14524, then predicted the PTM sites with the software on the amino acid sites and mapped on the mutation/variation sites reported from the literature in China. Furthermore, we analyzed the change of PTMs in the natural variant.
Statistical Analysis
Categorical variables were expressed as percentage and analyzed using the chi-square test. All analyses were conducted using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). Statistical significance was set at p < 0.05.
Results
Studies Retrieved and Information Extraction, and Distribution of the SCN5A Mutations/Variations on Nav1.5 Protein
A flow chart of the data research is shown in Figure 1. We excluded 306 unqualified studies that did not match the inclusion criteria or were duplicates. A total of 27 suitable studies were included, and details are shown in Table 1. Duplicated loci were removed, and a total of 55 mutations/variations including 45 sites from northern China and 10 sites from southern China (Guangdong, Guangxi, Hong Kong, Hainan, Jiangxi, Fujian, Taiwan) were included. Further analysis of these loci was performed as detailed below.
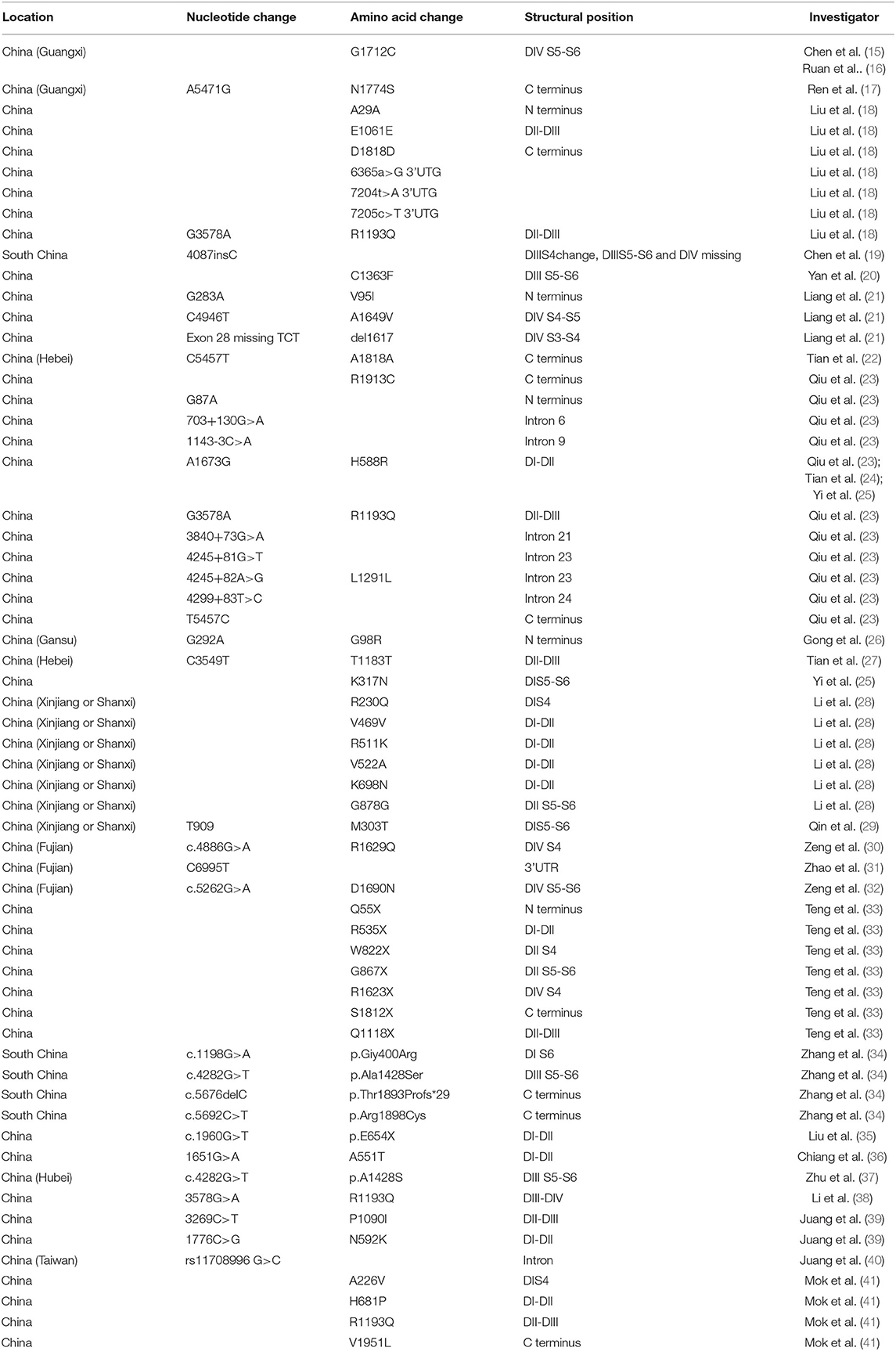
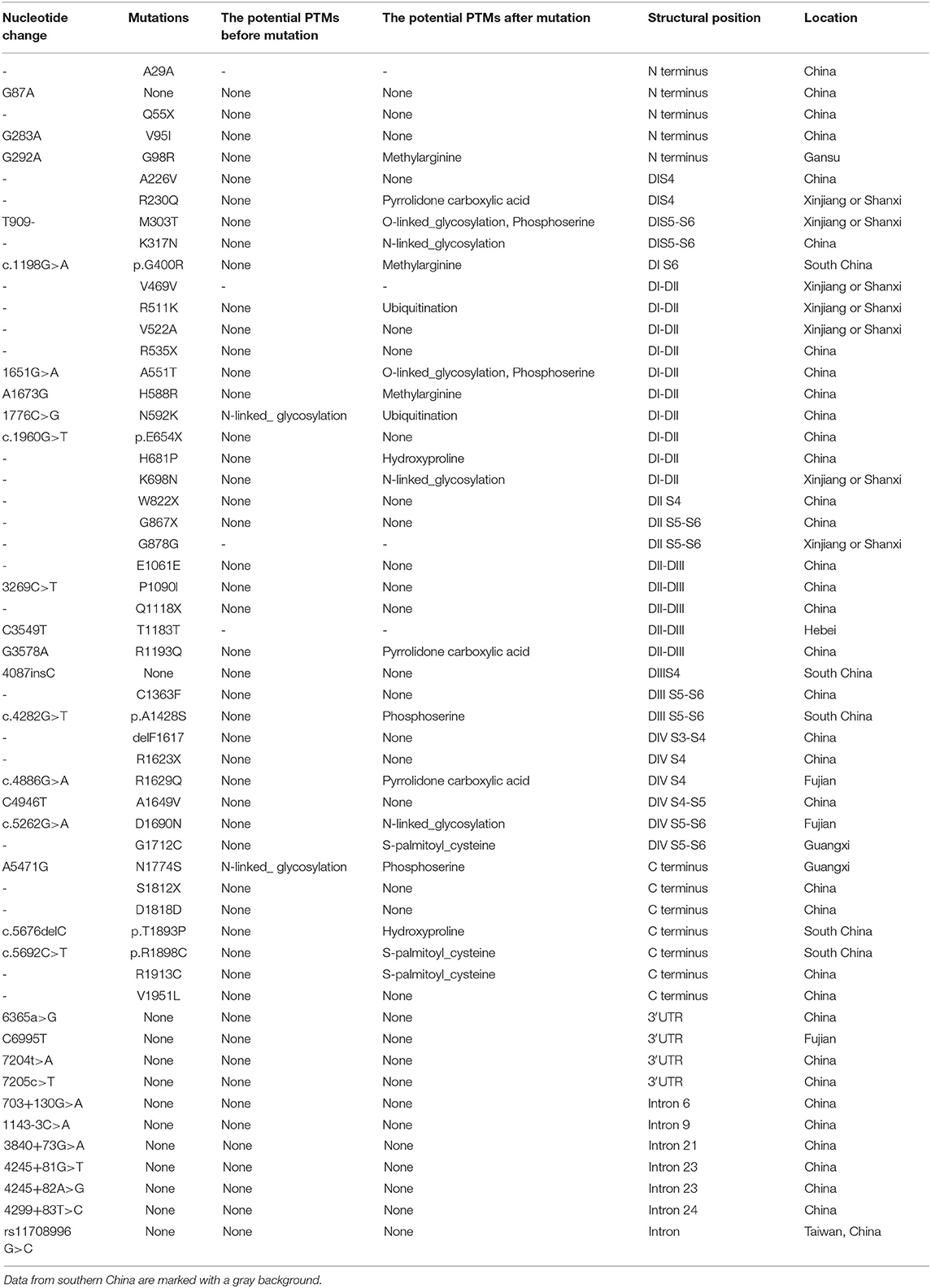
Table 1. Distribution characteristics of SCN5A gene mutation/variation sites in Chinese BrS patients.
The Nav1.5 channel has four highly conserved homologous transmembrane-spanning domains (DI–DIV) that are connected by an interdomain linker (IDL), and each domain consists of six transmembrane α spins (S1–S6). In order to visualize the distribution of these loci, we marked the mutation/variation sites on Nav1.5 protein (excluding introns and 3'UTR, which were not translated into amino acids), as shown in Figure 2. The blue dots represented the SCN5A mutations/variations from northern China, and the red dots represented the SCN5A mutations/variations from southern China.
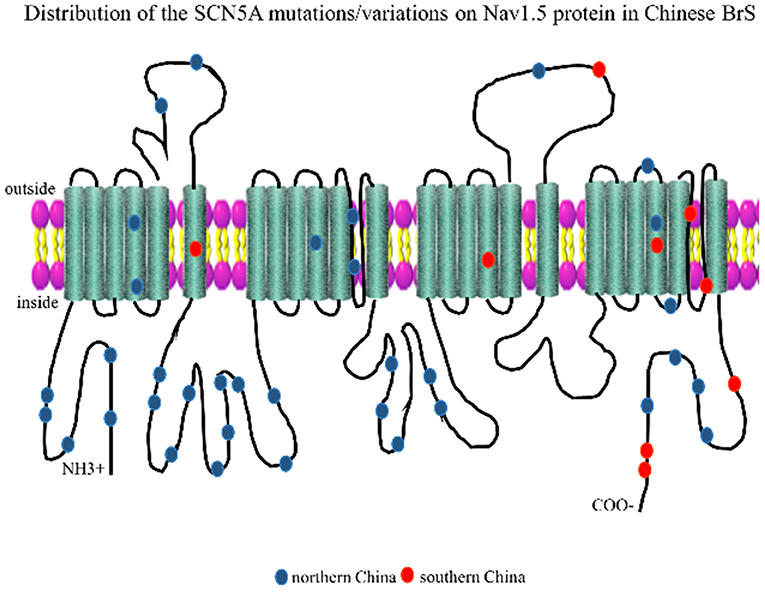
Figure 2. Representation of SCN5A gene mutations/variations was labeled on the corresponding location of the Nav1.5 protein structure. The blue dots represented mutations/variations from southern China. The red dots represented mutations/variations from northern China.
Locational Distribution of SCN5A Gene Mutations/Variations in China
The whole SCN5A gene was divided into 5′-UTR, N-Term (N-terminus), D I, IDL I-II, DII, IDL II-III, DIII, IDL III-IV, DIV, C-Term (C-terminus), 3′-UTR, and other regions. Mutation/variation loci distribution is shown in Figure 3. Distinguished by those 12 parts, the distribution in southern China and northern China were 5′-UTR (0 vs. 0), N-Term (0 vs. 11.1%), DI (10 vs. 8.9%), IDL I-II (0 vs. 22.2%), DII (0 vs. 6.7%), IDL II-III (0 vs. 11.1%), DIII (10 vs. 2.2%), IDL III-IV (0 vs. 0), DIV (30 vs. 6.7%), C-Term (30 vs. 8.9%), 3′-UTR (10 vs. 2.2%), and other (10 vs. 20%), respectively. It was found that SCN5A mutations/variations in northern China were mainly concentrated (60%) in 5′-UTR, N-Term, DI, IDL I-II, DII, and IDLII-III while mostly distributed (80%) in DIII, IDL III-IV, DIV, C-Term, and 3′-UTR in southern China.
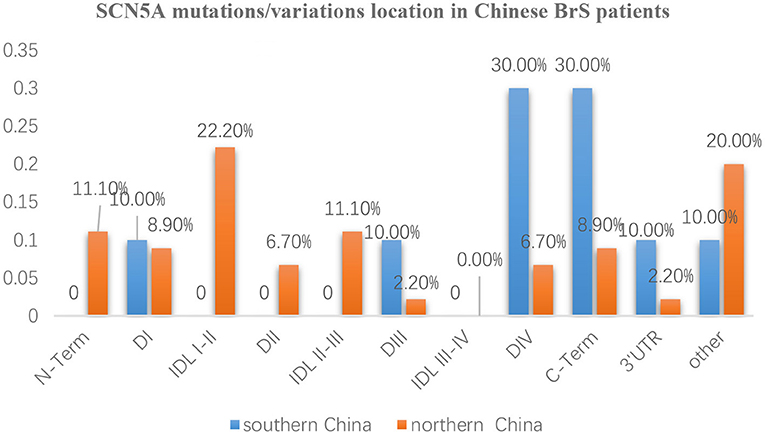
Figure 3. The distribution characteristics of mutation/variation sites in different parts of the SCN5A gene in Chinese BrS patients The blue columns represent the distribution of gene mutations/variations from southern China. The orange columns represent the distribution of gene mutations/variations from northern China.
Furthermore, the locations of SCN5A gene mutations/variants were divided into five parts as follows: N-Term, Transmembrane regions, IDL, C-Term, and others as shown in Figure 4A. The mutation/variation site distributions of SCN5A in southern China and northern China were 0, 50, 0, 30, and 20%, and 11.1, 24.5, 33.3, 8.9, and 22.2%, respectively (p = 0.000).
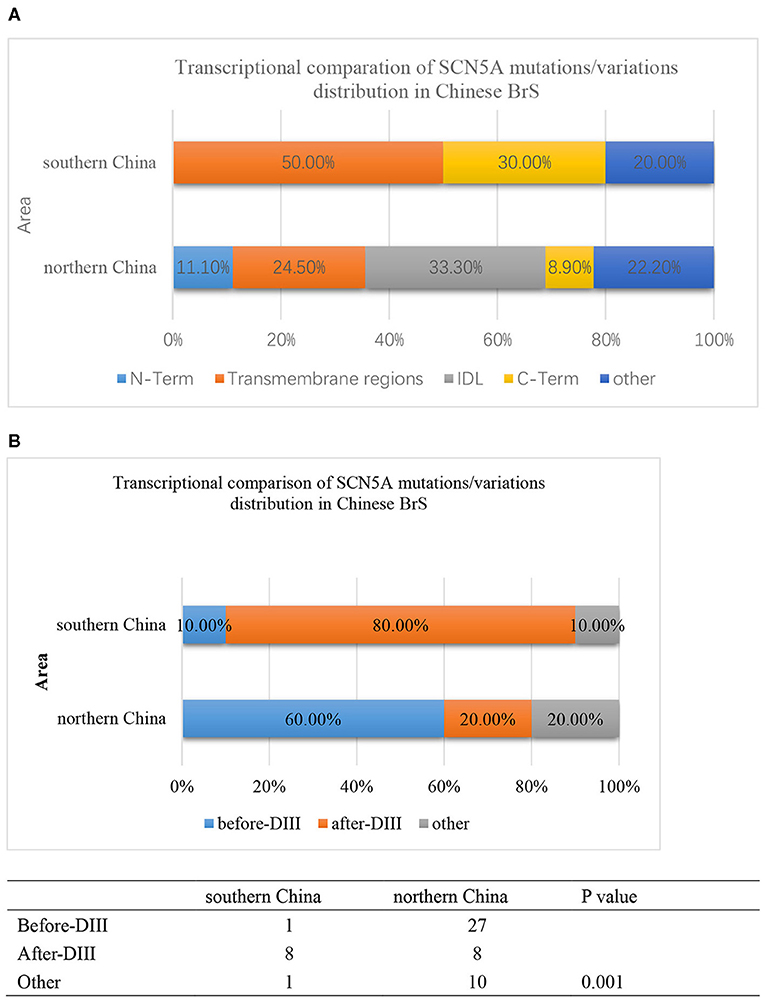
Figure 4. (A) The transcriptional comparison of the SCN5A mutation/variation distribution between southern China and northern China, which was distinguished by N-term, Transmembrane, IDL, C-term, and other parts of the SCN5A gene. The light blue bar represents N-terminus. The orange bar represents transmembrane regions. The gray bar represents the IDL region. The yellow bar represents the C-terminus. The deep blue bar represents other regions. (B) The transcriptional comparison of the SCN5A mutation/variation distribution between southern China and northern China, which was distinguished by before-DIII, after-DIII, and other parts of the SCN5A gene. The blue bar represents before-DIII parts. The orange bar represents after-DIII parts. The gray bar represents other parts.
On the other hand, the locations of SCN5A gene mutations/variants were divided into three parts as follows: before-DIII, after-DIII, and others as shown in Figure 4B. The mutation/variation site distributions of SCN5A in southern China and northern China were 10, 80, and 10% and 60, 20, and 20%, respectively (p = 0.001).
Then, we compared the distributions of mutations/variations among China (southern China and northern China), Japan, and the world, as shown in Figure 5A. Japanese data refer to a Japanese multicenter register (42), and the global data are from the website http://triad.fsm.it/cardmoc/. We distinguished the SCN5A mutation sites on the protein Nav1.5 structure by N-term, Transmembrane regions, IDL, and C-term and found 0, 62.5, 0, and 37.5% for each part in southern China; 14.3, 31.4, 42.9, and 11.4% in northern China; 4.4, 66.7, 20, and 8.9% in Japan; and 4.8, 71, 18.4, and 5.8% in the world, respectively (p = 0.000).
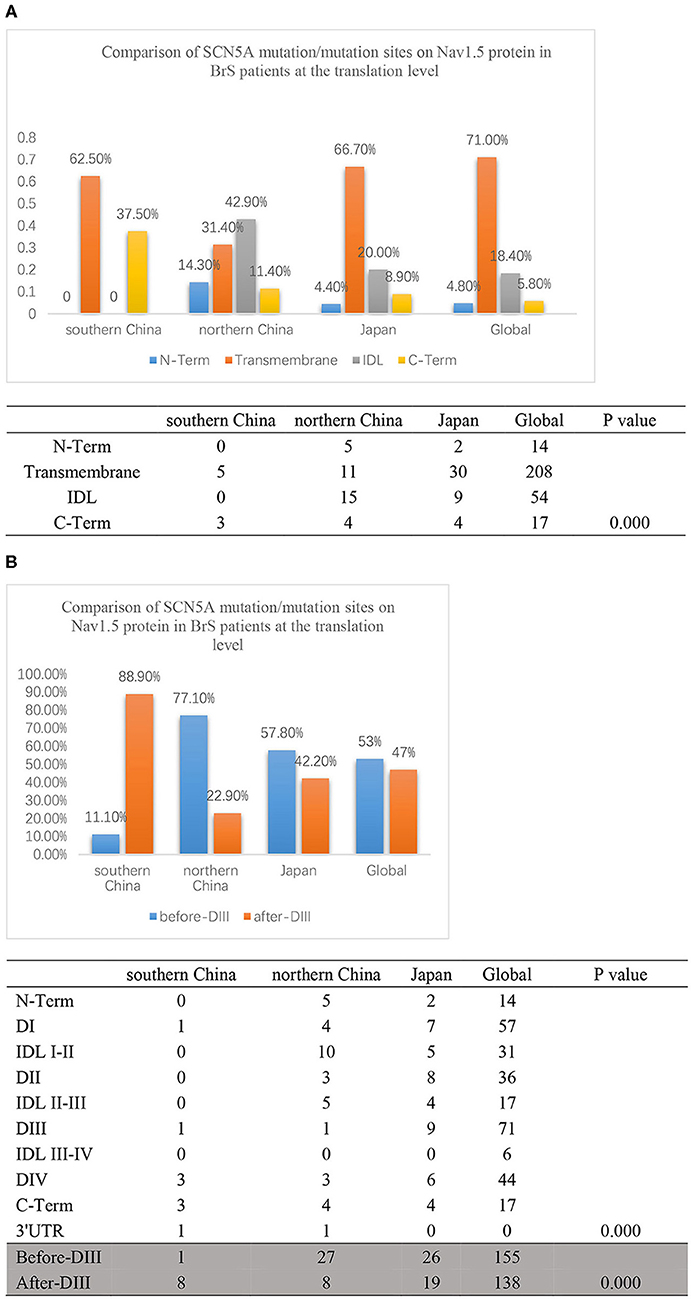
Figure 5. (A) The translation comparison of the SCN5A mutation/variation distribution on the Nav1.5 protein structure between southern China and northern China. Nav1.5 protein was distinguished by N-term, Transmembrane regions, IDL, C-term, and other parts. The light blue columns represent the N-terminus region. The orange columns represent the transmembrane region. The gray columns represent the IDL region. The yellow columns represent the C-terminus region. (B) The translation comparison of the SCN5A mutation/variation distribution on the Nav1.5 protein structure between southern China and northern China. Nav1.5 protein was distinguished by before-DIII and after-DIII. The blue columns represent before-DIII parts. The orange columns represent after-DIII parts.
In addition, the structure of Nav1.5 protein was divided into two parts: DI, IDL I-II, DII, and IDL II-III were set as the first half part (before-DIII), and DIII, IDL III-IV, DIV, and C-Term were set as the second half part (post-DIII) as shown in Figure 5B. The results indicated that 88.9% of the mutation sites were located in the post-DIII region in southern China, while only 22.9% in northern China, 42.2% in Japan, and 47% in the world (p = 0.000).
PTMs Prediction for Nav1.5 Protein Change
As mutations that cause changes in amino acids may have influences on protein modification, we predictively analyzed the PTMs of SCN5A with software and mapped on the mutations/variations in China (Table 2), and found a tendency for amino acids to acquire more modification sites after mutation. PTM change was likely to occur in 72.7% of BrS patients from southern China and 26.7% from northern China (p = 0.000).
Discussion
Our main findings in the study of the genetic characteristics of SCN5A in Chinese BrS patients were as follows: (1) More SCN5A gene mutations/variations were found in northern China than in southern China. (2) SCN5A mutations/variations of BrS patients in southern China mostly occurred in the DIII–DIV to C-terminus region and the 3′-UTR region of the SCN5A gene. (3) PTM changes were consistent with the mutation/variation distribution of SCN5A, which might be involved in the regulation of the pathogenesis of BrS patients.
BrS can be found all over the world, and the prevalence of BrS can reach 0.5%0 in high-prevalence areas. BrS is the leading cause of death for men less than 40 years old, only second to the death rate of traffic accidents in Southeast Asian countries (42, 43). In southern China, BrS patients were anticipated to have a relatively high incidence rate. However, our study revealed that SCN5A gene mutations published were found to be more in northern China than southern China. The possible reasons may be that BrS is a rare disease, and the total number of cases reported at present was not large and patients in many studies did not undergo DNA sequencing, which results in data bias. We will continue to pay attention to relevant reports and continue to collect cases to further confirm the data.
Further analysis showed that the locations of mutation sites had their own characteristics in southern China. Most mutation sites were clustered in the transmembrane regions in southern China statistically different from northern China. Mutation sites were mostly located in the second half part of the protein structure (post-DIII) in southern China, while in the first half part positions (before-DIII) in northern China, Japan, and the world.
The SCN5A gene, located on chromosome 3p21, contains 28 exons with a total length of about 80 kb and encodes the α-subunit protein Nav1.5. Some mutations lead to a decrease in current density, others do not lead to a decrease in INa, while some location-specific SCN5A mutations resulted in poorer outcomes during follow-up (44). As different mutation locations lead to different pathological changes, we try to analyze whether protein functional modifications are involved in the mechanism.
PTM is a crucial modification method for protein transcription, such as phosphorylation, acetylation, ubiquitination, and glycosylation, which may bring a broad range of effects, such as protein stability, enzymatic activity, subcellular localization, and interactions. Multiple kinases including cyclic AMP-dependent protein kinase (PKA), protein kinase C (PKC), and calcium/calmodulin-dependent kinase II (CaMK II) phosphorylate regulate Nav1.5 channel physiology and pathology (45–48) including SUMOylation (49), ubiquitination (50), acetylation (51), etc. In our previous study, we revealed that miR-192-5p bound to the 3′-UTR of human SCN5A to negatively regulate the expression of Nav1.5 and reduce INa density. Our study demonstrated an important post-transcriptional role of miR-192-5p in post-transcriptional regulation of Nav1.5 (31).
Hence, we predicted the PTM sites with software and mapped on the mutations/variations in China. PTM change was likely to occur in 72.7% of BrS patients in southern China and 26.7% in northern China, suggesting that PTMs might be involved in the regulation of the pathogenesis of BrS, which provided new ideas and directions to further study the role of Nav1.5 in the pathogenesis of BrS.
Conclusion
The mutation sites of BrS patients from southern China mostly distributed in the DIII–DIV to C-terminus region and the 3′-UTR region of the SCN5A gene, which was different from northern China, Asia, and other countries around the world. PTM change might be involved in the regulation of the pathogenesis of BrS.
Contribution to the Field Statement
While BrS is a rare disease, it is especially young-male predominant and accounts for 20% sudden death without organic heart disease in Southeast Asia, which causes more and more concerns. We analyzed the genetic characteristics of SCN5A mutations/variations and found that SCN5A mutations/variations of BrS patients from southern China mostly occurred in the DIII–DIV to C-terminus region and the 3′-UTR region of the SCN5A gene, which was different from northern China, Japan, and the world. PTM changes predicted by the mutations/variations may be involved in the regulation of the pathogenesis of BrS. Our findings provide new ideas and directions to further study the role of Nav1.5 in the pathogenesis of BrS.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
All authors participated in the design of the study, analysis and interpretation of the data, and review of the manuscript and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81870183).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. (1992) 20:1391–6. doi: 10.1016/0735-1097(92)90253-J
2. Berne P, Brugada J. Brugada syndrome 2012. Circ J. (2012) 76:1563–71. doi: 10.1253/circj.CJ-12-0717
3. Mizusawa Y, Wilde AA. Brugada syndrome. Circ Arrhythm Electrophysiol. (2012) 5:606–16. doi: 10.1161/CIRCEP.111.964577
4. Brugada P, Brugada J, Roy D. Brugada syndrome 1992-2012: 20 years of scientific excitement, and more. Eur Heart J. (2013) 34:3610–5. doi: 10.1093/eurheartj/eht113
5. Antzelevitch C, Brugada P, Borggrefe M, Brugada J, Brugada R, Corrado D. Brugada syndrome: report of the second consensus conference: endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation. (2005) 111:659–70. doi: 10.1161/01.CIR.0000152479.54298.51
6. Juang JJ, Horie M. Genetics of Brugada syndrome. J Arrhythmia. (2016) 32:418–25. doi: 10.1016/j.joa.2016.07.012
7. Cerrone M. Controversies in Brugada syndrome. Trends Cardiovasc Med. (2018) 28:284–92. doi: 10.1016/j.tcm.2017.11.003
8. Wang QI, Ohno S, DING WG, Fukuyama M, Miyamoto A, Itoh H, et al. Gain-of-function KCNH2 mutations in patients with Brugada syndrome. J Cardiovasc Electrophysiol. (2014) 25:522–30. doi: 10.1111/jce.12361
9. Kapplinger JD, Tester DJ, Alders M, Benito B, Berthet M, Brugada J. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. (2010) 7:33–46. doi: 10.1016/j.hrthm.2009.09.069
10. Le Scouarnec S, Karakachoff M, Gourraud JB, Lindenbaum P, Bonnaud S, Portero V. Testing the burden of rare variation in arrhythmia-susceptibility genes provides new insights into molecular diagnosis for Brugada syndrome. Human molecular genetics. (2015) 24:2757–63. doi: 10.1093/hmg/ddv036
11. Makiyama T, Akao M, Haruna Y, Tsuji K, Doi T, Ohno S. Mutation analysis of the glycerol-3 phosphate dehydrogenase-1 like (GPD1L) gene in Japanese patients with Brugada syndrome. Circ J. (2008) 72:1705–6. doi: 10.1253/circj.CJ-08-0508
12. Kattygnarath D, Maugenre S, Neyroud N, Balse E, Ichai C, Denjoy I. MOG1: a new susceptibility gene for Brugada syndrome. Circ Cardiovasc Genet. (2011) 4:261–8. doi: 10.1161/CIRCGENETICS.110.959130
13. Yamagata K, Horie M, Aiba T, Ogawa S, Aizawa Y, Ohe T. Genotype-phenotype correlation of SCN5A mutation for the clinical and electrocardiographic characteristics of probands with brugada syndrome: a japanese multicenter registry. Circulation. (2017) 135:2255–70. doi: 10.1161/CIRCULATIONAHA.117.030845
14. Brugada J, Campuzano O, Arbelo E, Sarquella-Brugada GBrugada R. Present status of brugada syndrome: JACC state-of-the-art review. J Am Coll Cardiol. (2018) 72:1046–59. doi: 10.1016/j.jacc.2018.06.037
15. Chen YY, Liu SR, Xie LZ, Zhu TY, Chen YZ, Deng XJ, et al. Functional analysis of a novel SCN5A mutation G1712C identified in Brugada syndrome. J South Med Univ. (2016) 37:256–60. doi: 10.3969/j.issn.1673-4254.2017.02.19
16. Ruan FH, Meng SR, Chen ZM, Peng J, Wang W, Yang RH. Detection of a novel SCN5A gene mutation (G1712C) in patients w ith asymptomatic Burgada syndrome. Chin H eart J. (2007) 19:218–21. doi: 10.13191/j.chj.2007.02.100.ruanfh.032
17. Faxin Ren JY, Wei L, Jun H, CaiLian K, Tao X. The novel SCN5A mutation of Brugada syndrome in a Chinese male patient. Chin J Cardiac Arrhyth. (2003) 7:336–9.
18. Fuqiang Liu YL, Yong X, Donghua Z, Zhongbo X, Wenyan L, Jian P. Detection of SCN5A gene mutation in patients with Brugada syndrome. Chinese J Pract Med. (2012) 28:266–7.
19. Junzhu Chen LZ, Jingxiang L, Juan P, Yunpeng S, Qianmin T, Furong Z, et al. The frameshift mutation of gene SCN5A leads to Brugada syndrome. Chin J Cardiol. (2002) 30:538–41.
20. Aizhen Yan XL, Yanhong L, Zhihong W, Fenghua L. Identification and analysis of a new SCN5A gene mutation of in a Brugada syndrome family. (2015) p. 238–9.
21. Liang P, Liu WL, Hu DY Li CL, Tao WH Li L. Novel SCN5A gene mutations associated with Brugada syndrome: V95I, A1649V and delF1617. Zhonghua xin xue guan bing za zhi. (2006) 34:616–9.
22. Tian L, Zhu JF, Du R, Ke Q. Mutation detection of SCN5A gene in two members of a Chinese family with Brugada syndrome. Shandong Med J. (2013) 53:11–5.
23. Qiu X, Hu D, Lu Y, Li L, Sun Y, Li C. The gene screening of SCN5A variation in Chinese patients with diagnostic and suspected Brugada syndrome. Chin J Cardiac Pacification Electrophysiol. (2009) 23:204–8. doi: 10.13333/j.cnki.cjcpe.2009.03.005
24. Li T, Zhang L, Zhu Y, Zhu J. Detection of SCN5A gene mutations in sporadic Chinese patients with Brugada syndrome. Shandong Med J. (2013) 53:9–10.
25. Meng SYDXJQS. Functional expression of a novel SCN5A mutation K317N identified in a Chinese Brugada syndrome family. J Clin Exp Med. (2011) 10:1655–8.
26. Gong H. Genetic epidemiology investigation and SCN5A gene mutations screening in familial Brugada Syndrome. Directed Research. (2011).
27. Tian L, Zhu JF, Yang JG, Qinmei K. Mutation detection of SCN5A gene in two members of a Chinese family with Brugada syndrome. Shandong Med J. (2013) 53:11–2.
28. Li F. Two Brugada symdrome family SCN5A genetic screening resuelts, and relevant clinical features. Directed Research. (2013).
29. Qin H. SCN5A gene analysis and a novel mutation detection in two Brugada symdrome nuclear pedigree. Directed Research. (2013).
30. Zeng Z, Zhou J, Hou Y, Liang X, Zhang Z, Xu X. Electrophysiological characteristics of a SCN5A voltage sensors mutation R1629Q associated with Brugada syndrome. PLoS ONE. (2013) 8:e78382. doi: 10.1371/journal.pone.0078382
31. Zhao Y, Huang Y, Li W, Wang Z, Zhan S, Zhou M. Post-transcriptional regulation of cardiac sodium channel gene SCN5A expression and function by miR-192-5p. Biochim Biophys. (2015) 1852:2024–34 doi: 10.1016/j.bbadis.2015.07.016
32. Zeng Z, Xie Q, Huang Y, Zhao Y, Li W, Huang Z. pD1690N sodium voltage-gated channel α subunit 5 mutation reduced sodium current density and is associated with Brugada syndrome. Molec Med Rep. (2016) 13:5216–22 doi: 10.3892/mmr.2016.5162
33. Teng S, Gao L, Paajanen V, Pu JFan Z. Readthrough of nonsense mutation W822X in the SCN5A gene can effectively restore expression of cardiac Na+ channels. Cardiovasc Res. (2009) 83:473–80. doi: 10.1093/cvr/cvp116
34. Zhang L, Tester DJ, Lang D, Chen Y, Zheng J, Gao R. Does sudden unexplained nocturnal death syndrome remain the autopsy-negative disorder: a gross, microscopic, and molecular autopsy investigation in southern China. Mayo Clinic Proc. (2016) 91:1503–14. doi: 10.1016/j.mayocp.2016.06.031
35. Liu X, Zheng J, Fan ZRao L. Case report: an unusual case of Brugada syndrome combined with a ventricular septal defect: a case report. Medicine. (2017) 96:e8695. doi: 10.1097/MD.0000000000008695
36. Chiang KC, Lai LPShieh RC. Characterization of a novel Nav15 channel mutation, A551T, associated with Brugada syndrome. J Biomed Sci. (2009) 16:76. doi: 10.1186/1423-0127-16-76
37. Zhu JF, Du LL, Tian Y, Du YM, Zhang L, Zhou T. Novel heterozygous mutation c4282G>T in the SCN5A gene in a family with Brugada syndrome. Exp Therapeutic Med. (2015) 9:1639–45. doi: 10.3892/etm.2015.2361
38. Li L, Ruan Y, Liu N, Zhao Q, Zhang M, Li X. “Pill-in-the-Pocket” treatment of propafenone unmasks ECG brugada pattern in an atrial fibrillation patient with a common SCN5A R1193Q polymorphism. Front Physiol. (2019) 10:353. doi: 10.3389/fphys.2019.00353
39. Juang JM, Lu TP, Lai LC, Hsueh CH, Liu YB, Tsai CT. Utilizing multiple in silico analyses to identify putative causal SCN5A variants in Brugada syndrome. Sci Rep. (2014) 4:3850. doi: 10.1038/srep03850
40. Jimmy Juang JM, Liu YB, Julius Chen CY Yu QY, Chattopadhyay A, Lin LY. Validation and disease risk assessment of previously reported genome-wide genetic variants associated with brugada syndrome: SADS-TW BrS registry. Circ Genomic Prec Med. (2020) 13:e002797. doi: 10.1161/CIRCGEN.119.002797
41. Bloise R, Chan HW, Lam C, Tsang HH. Clinical profile and genetic basis of Brugada syndrome in the Chinese population. Hong Kong Med J. (2004) 10:32–7.
42. Fowler SJ, Priori SG. Clinical spectrum of patients with a Brugada ECG. Curr Opin Cardiol. (2009) 24:74–81. doi: 10.1097/HCO.0b013e32831cb920
43. Priori SG, Wilde AA, Horie M, Cho Y, Behr ER, Berul C. Executive summary: HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes. Europace. (2013) 15:1389–406. doi: 10.1093/europace/eut272
44. Hedley PL, Jørgensen P, Schlamowitz S, Moolman-Smook J, Kanters JK, Corfield VA. The genetic basis of Brugada syndrome: a mutation update. Human Mutation. (2009) 30:1256–66. doi: 10.1002/humu.21066
45. Aye TT, Scholten A, Taouatas N, Varro A, Van Veen TA, Vos MA. Proteome-wide protein concentrations in the human heart. Molecular bioSyst. (2010) 6:1917–27. doi: 10.1039/c004495d
46. Liu X, Chen Z, Han Z, Liu Y, Wu X, Peng Y. AMPK-mediated degradation of Nav15 through autophagy. FASEB J. (2019) 33:5366–76. doi: 10.1096/fj.201801583RR
47. Lorenzini M, Burel S, Lesage A, Wagner E, Charrière C, Chevillard PM. Proteomic and functional mapping of cardiac NaV15 channel phosphorylation sites. J Gen Physiol. (2021) 153:e202012646 doi: 10.1085/jgp.202012646
48. Burel S, Coyan FC, Lorenzini M, Meyer MR, Lichti CF, Brown JH. C-terminal phosphorylation of Na(V)15 impairs FGF13-dependent regulation of channel inactivation. J Biol Chem. (2017) 292:17431–48 doi: 10.1074/jbc.M117.787788
49. Plant LD, Xiong D, Romero J, Dai HGoldstein SAN. Hypoxia produces pro-arrhythmic late sodium current in cardiac myocytes by SUMOylation of Na(V)15 channels. Cell Rep. (2020) 30:2225–36e4 doi: 10.1016/j.celrep.2020.01.025
50. Hu Y, Bai X, Zhang C, Chakrabarti S, Tang B, Xiong H. Ubiquitination-activating enzymes UBE1 and UBA6 regulate ubiquitination and expression of cardiac sodium channel Nav15. Biochem J. (2020) 477:1683–700. doi: 10.1042/BCJ20200138
Keywords: brugada, SCN5A, China, genetic characteristics, post-transcriptional modifications
Citation: Zhang Z, Chen H, Chen W, Zhang Z, Li R, Xu J, Yang C, Chen M, Liu S, Li Y, Wang T, Tu X and Huang Z (2021) Genetic Characteristics and Transcriptional Regulation of Sodium Channel Related Genes in Chinese Patients With Brugada Syndrome. Front. Cardiovasc. Med. 8:714844. doi: 10.3389/fcvm.2021.714844
Received: 26 May 2021; Accepted: 05 July 2021;
Published: 05 August 2021.
Edited by:
Ding-Sheng Jiang, Huazhong University of Science and Technology, ChinaReviewed by:
CHENG HU, Shanghai Jiao Tong University, ChinaShan Lu, University of California, Davis, United States
Wen-lin Cheng, Wuhan University, China
Copyright © 2021 Zhang, Chen, Chen, Zhang, Li, Xu, Yang, Chen, Liu, Li, Wang, Tu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengrong Huang, aHVhbmd6aGVuZ3JvbmdAeG11LmVkdS5jbg==; TzungDau Wang, dGR3YW5nQG50dS5lZHUudHc=; Xin Tu, eHR1QGh1c3QuZWR1LmNu
 Ziguan Zhang
Ziguan Zhang Hongwei Chen1
Hongwei Chen1 Runjing Li
Runjing Li Cui Yang
Cui Yang Minwei Chen
Minwei Chen TzungDau Wang
TzungDau Wang Xin Tu
Xin Tu Zhengrong Huang
Zhengrong Huang