- 1Department of Cardiology, Institute of Cardiovascular Diseases, The First Affiliated Hospital, Guangxi Medical University, Nanning, China
- 2Department of Neurology, The First Affiliated Hospital, Guangxi Medical University, Nanning, China
Background: The current study aimed to investigate the effects of synaptotagmin-like 3 (SYTL3) and solute carrier family 22 member 3 (SLC22A3) single nucleotide polymorphisms (SNPs) and gene-environment (G × E) interactions on blood lipid levels as well as the risk of coronary artery disease (CAD) and ischaemic stroke (IS) in the Southern Chinese Han population.
Methods: The genetic makeup of 6 SYTL3-SLC22A3 SNPs in 2269 unrelated participants (controls, 755; CAD, 758 and IS, 756) of Chinese Han ethnicity was detected by the next-generation sequencing techniques.
Results: The allele and genotype frequencies of the SYTL3 rs2129209 and SLC22A3 rs539298 SNPs were significantly different between the case and control groups. The SLC22A3 rs539298 SNP was correlated with total cholesterol (TC) levels in controls, the rs539298G allele carriers maintained lower TC levels than the rs539298G allele non-carriers. At the same time, the SLC22A3 rs539298 SNP interacted with alcohol consumption reduced the risk of CAD and IS. The SYTL3-SLC22A3 A-C-A-A-A-A, G-T-C-G-C-A and A-T-A-A-C-A haplotypes increased and the A-C-A-A-C-G haplotype reduced the risk of CAD, whereas the SYTL3-SLC22A3 A-C-A-A-A-A, G-T-C-G-A-G and A-T-A-A-C-A haplotypes increased and the A-C-A-A-A-G and A-C-A-A-C-G haplotypes reduced the risk of IS. In addition, several SNPs interacted with alcohol consumption, body mass index ≥ 24 kg/m2 and cigarette smoking to affect serum lipid parameters such as triglyceride, high-density lipoprotein cholesterol, TC, and apolipoprotein A1 levels.
Conclusions: Several SYTL3-SLC22A3 variants, especially the rs539298 SNP, several haplotypes, and G × E interactions, were related to blood lipid parameters and the risk of CAD and IS in the Southern Chinese Han population.
Introduction
Ischaemic cardiovascular and cerebrovascular diseases, including coronary artery disease (CAD) and ischaemic stroke (IS), are the primary contributors to global disability and death despite the vast therapeutic and diagnostic methods innovated in the last 10 years. Several recent studies have shown that as a multifactorial and complex disorder, CAD or IS occurs due to numerous factors, such as unhealthy lifestyles, alterations of serum lipid levels, genomic background and environmental factors and interactions among these factors (1–3). As the common pathological basis of CAD (4) and IS (5), atherosclerosis is the result of chronic inflammation (6) and abnormal lipid metabolism, such as increased levels of triglyceride (TG) (7), apolipoprotein (Apo) B (8), low-density lipoprotein cholesterol (LDL-C) (9), and total cholesterol (TC) (10), along with reduced levels of ApoA1 (8) and high-density lipoprotein cholesterol (HDL-C) (11) in serum.
Many genes and genetic loci associated with CAD (12) or IS (13) have been identified and reported in previous genome-wide association studies (GWASes). Similarly, a large number of genes or loci associated with lipid metabolism have also been associated with CAD and/or IS (1–3). Currently, several compelling genes closely associated with blood lipid parameters, including synaptotagmin-like 3 (SYTL3) and solute carrier family 22-member 3 (SLC22A3), have been identified in the European population by GWASes (14). The SYTL3 (also named SLP3, gene ID: 94120, OMIM: 608441, HGNC:15587) is located on chromosome 6q25.3 (exon count: 22), and its encoded protein plays a key role in vesicular trafficking. Several compelling studies have suggested that the transport of lipids and proteins between eukaryotic cells via secretion and endocytosis is primarily facilitated by vesicular trafficking (15, 16). Muse et al. (17) reported that the SYTL3 was associated with the incidence of acute myocardial infarction (AMI). However, its mechanism remains unclear.
The SLC22A3 (also known as OCT3; EMTH; EMT, gene ID: 6581, OMIM: 604842, HGNC:10967) is located on chromosome 6q25.3 (exon count: 15), and it encodes an organic cation transporter 3 protein, which is a member of the peripheral membrane protein family and plays a key role in biogenic histamine deactivation and synthesis (18, 19). As an effective proinflammatory mediator, histamine can enhance the deposition of LDL-C in vascular endothelial cells by inducing inflammatory mediators such as adhesion molecules, cytokines, chemokines and others. Inflammatory mediators further increase the permeability of vascular endothelial cells, so histamine can create a favorable environment for the formation of atherosclerotic plaques (20, 21). Previous studies showed that the SLC22A3 silencing could effectively inhibit the synthesis of histamine and proinflammatory mediators (MCP-1, IL-8 and IL-6) and reduce the infiltration of monocytes and the adhesion between leukocytes and the endothelium (22). The SLC22A3 expression is widespread, and it can be found in skeletal muscle, heart, brain, and placenta. The SLC22A3 is highly expressed in the human heart, with the strongest SLC22A3 immunoreactivity found in vascular endothelial cells (23, 24). A series of studies have confirmed that the SLC22A3 rs1810126, rs2048327 and rs3088442 SNPs reduce the risk of CAD by downregulating SLC22A3 transcription and protein levels (22, 25). Chen et al. (26) found that the SLC22A3 could be regarded as a gene vector for the association between lipid metabolism and CAD. Nevertheless, the association between the SYTL3 rs9364496, rs6455600, rs2129209, rs9456350, SLC22A3 rs446809 and rs539298 SNPs and the risk of CAD and IS is still unclear and not reported in the Chinese Han population. Thus, this research aimed to understand the relationship between the six selected SNPs and the risk of CAD and IS in the Southern Chinese Han population.
Methods
Subjects
A case-control study was conducted among the Chinese Han participants in Guangxi Zhuang Autonomous Region, a province in Southern China. A total of 2,269 samples were collected from the First Affiliated Hospital of Guangxi Medical University, including controls, 755; CAD patients, 758; and IS cases, 756. CAD was defined as significant coronary artery stenosis (≥ 50%) in at least one of the three major coronary arteries or their major branches (branch diameter ≥ 2 mm) (27). All patients suffering from IS received detailed and rigorous neurological examination as well as brain magnetic resonance imaging (MRI) scans. The diagnostic criteria for IS were based on the International Classification of Diseases (9th Revision). Subjects with a history of neoplastic, autoimmune, type 1 diabetes mellitus, haematologic, thyroid, renal or liver diseases were excluded. Patients suffering from IS had no history of CAD, and patients suffering from CAD also had no history of IS.
After adjusting for sex and age, all of the healthy control participants were randomly recruited from the Physical Examination Center of the First Affiliated Hospital of Guangxi Medical University during the same period. All participants were healthy, and none of them had a history of myocardial infarction (MI), type 2 diabetes mellitus (T2DM), CAD or IS as judged by a critical clinical examination and medical history collection. All subjects in the current research signed written informed consent forms. The research proposal was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University (No: Lunshen-2014-KY-Guoji-001; Mar. 7, 2014).
Biochemical Index Detection
After fasting for at least 12 h, a sample of 5 ml of peripheral venous blood was drawn from each participant. A portion of the blood sample (2 ml) was used to measure serum lipid levels, and another portion of the blood sample (3 ml) was used to extract genomic DNA. The methods for measuring serum ApoB, LDL-C, ApoA1, TC, HDL-C, and TG were described in detail in our previous study (28). All determinations were performed using an autoanalyser (Type 7170A; Hitachi Ltd., Tokyo, Japan) in the Clinical Science Experiment Center of the First Affiliated Hospital, Guangxi Medical University (29).
Diagnostic Criteria
The definition for normal values of serum ApoB (0.80–1.05 g/L), TG (0.56–1.70 mmol/L), ApoA1 (1.20–1.60 g/L), LDL-C (2.70–3.10 mmol/L), TC (3.10–5.17 mmol/L), HDL-C (1.16–1.42 mmol/L), and the ApoA1/ApoB ratio (1.00–2.50) was based on our previous study (30). The diagnostic criteria for hyperlipidaemia (31), hypertension (32, 33), obesity, overweight, and normal weight (34, 35) were described in detail in previous studies (31–35). Participants with a previous diagnosis of diabetes or a fasting plasma glucose ≥ 7.0 mmol/L or 2 h postprandial plasma glucose ≥ 11.1 mmol/L were defined as diabetic patients (36).
SNP Selection and Genotyping
Six SNPs located on SYTL3 and SLC22A3 were chosen based on predetermined criteria: (1) The SYTL3-SLC22A3 cluster was selected from previous GWASes associated with blood lipid levels. (2) Tagging SNPs were identified via Haploview (Broad Institute of MIT and Harvard, USA, version 4.2), and potential lipid metabolism-associated functional SNPs were predicted using the latest version of the 1000 Genome Project Database. (3) More complete details of the selected SNPs were collected from NCBI dbSNP Build 132. (4) Regarding SNP selection, we also referenced a previous study by Ober et al. (14), and the minor allele frequency (MAF) of all SNPs was more than 1% and associated with blood lipid profiles in a previous study. (5) Six SNPs of SYTL3 (rs9364496, rs6455600, rs2129209 and rs9456350) and SLC22A3 (rs446809 and rs539298) were selected using the block-based method, which involves marking the association of linkage disequilibrium (LD) amongst the chosen SNPs (r2 > 0.8). White blood cells were used for genomic DNA extraction with phenol-chloroform (37). All obtained DNA samples were numbered and maintained at −20°C until further studies were carried out. Next-generation sequencing technology (NGS) was used to analyse the genotypes of the 6 selected SNPs at the Center for Human Genetics Research, Shanghai Genesky Bio-Tech Co. Ltd., China (38). Supplementary Table 1 depicts the relevant primer sequences. Detailed steps for multiplex PCR and high throughput sequencing are also shown in Supplementary Material.
Statistical Analyses
SPSS (Version 22.0) software was used to analyse all data collected from the current research. An independent sample t-test was used to evaluate the continuous data (mean ± SD) that were normally distributed between control and patient groups. TG levels that were not normally distributed were expressed using median and quartile ranges and were evaluated using the Wilcoxon-Mann-Whitney test. Data such as the genotype distribution, sex ratio, and the number of smokers and drinkers were analyzed by the chi-square test. The standard goodness-of-fit test was used to test the Hardy-Weinberg equilibrium (HWE). The correlation between genotypes and blood lipid levels was tested by the covariance analysis (ANCOVA). Any variants related to the blood lipid parameter at a value of P ≤ 0.008 were considered statistically significant after Bonferroni correction. Age, alcohol consumption, sex, cigarette smoking and body mass index (BMI) were adjusted for the statistical analysis. Unconditional logistic regression analysis was used to calculate the 95% confidence interval (CI) and the odds ratio (OR). The interactions of the six selected SNPs with cigarette smoking, sex, age, alcohol consumption and BMI ≥ 24 kg/m2 on blood lipid levels and the risk of CAD and IS were performed by using a factorial regression analysis after controlling for potential confounders. A P ≤ 0.00125 was considered statistically significant after Bonferroni correction. Haploview (Broad Institute of MIT and Harvard, USA, version 4.2) software was used to calculate the haplotype frequencies and pair wise LD among the six detected SNPs. The heatmap of the inter-locus models was measured by using R software (version 4.1.0).
Results
Common and Biochemical Characteristics
As presented in Table 1, there were no differences in the proportion of smokers, diastolic blood pressure, age, the sex ratio, or serum ApoB and LDL-C levels between the controls and cases. The glucose, pulse pressure, systolic blood pressure, BMI, serum TG levels were significantly lower, and the serum TC, ApoA1, and HDL-C levels; the proportion of drinkers; and the ApoA1/ApoB ratio were significantly higher in controls than in CAD and IS patients.
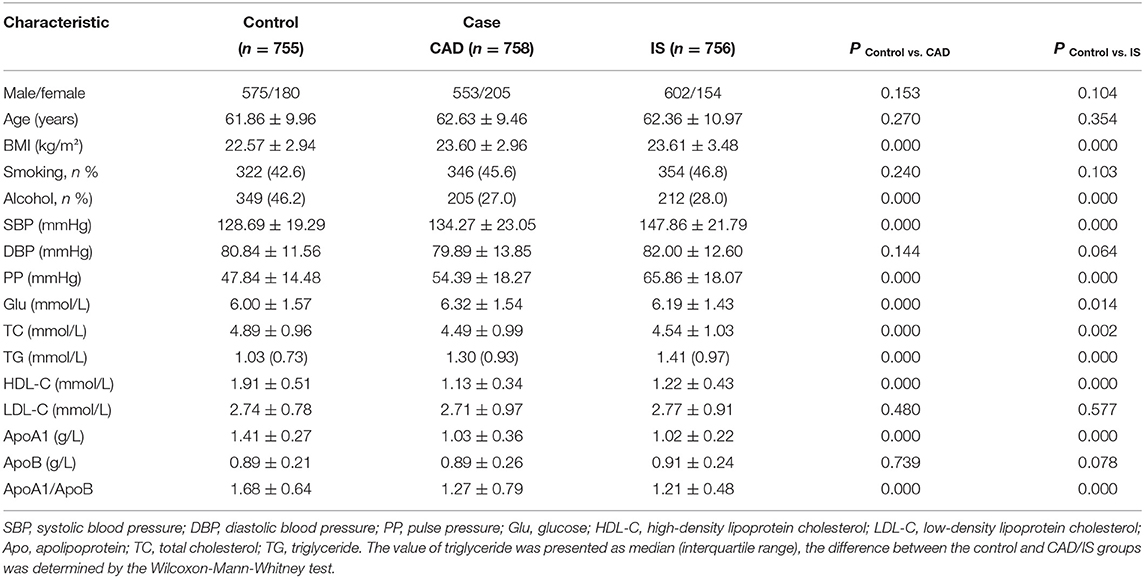
Table 1. Comparison of demographic, lifestyle characteristics, and serum lipid levels of the participants.
Genotypic and Allelic Frequencies
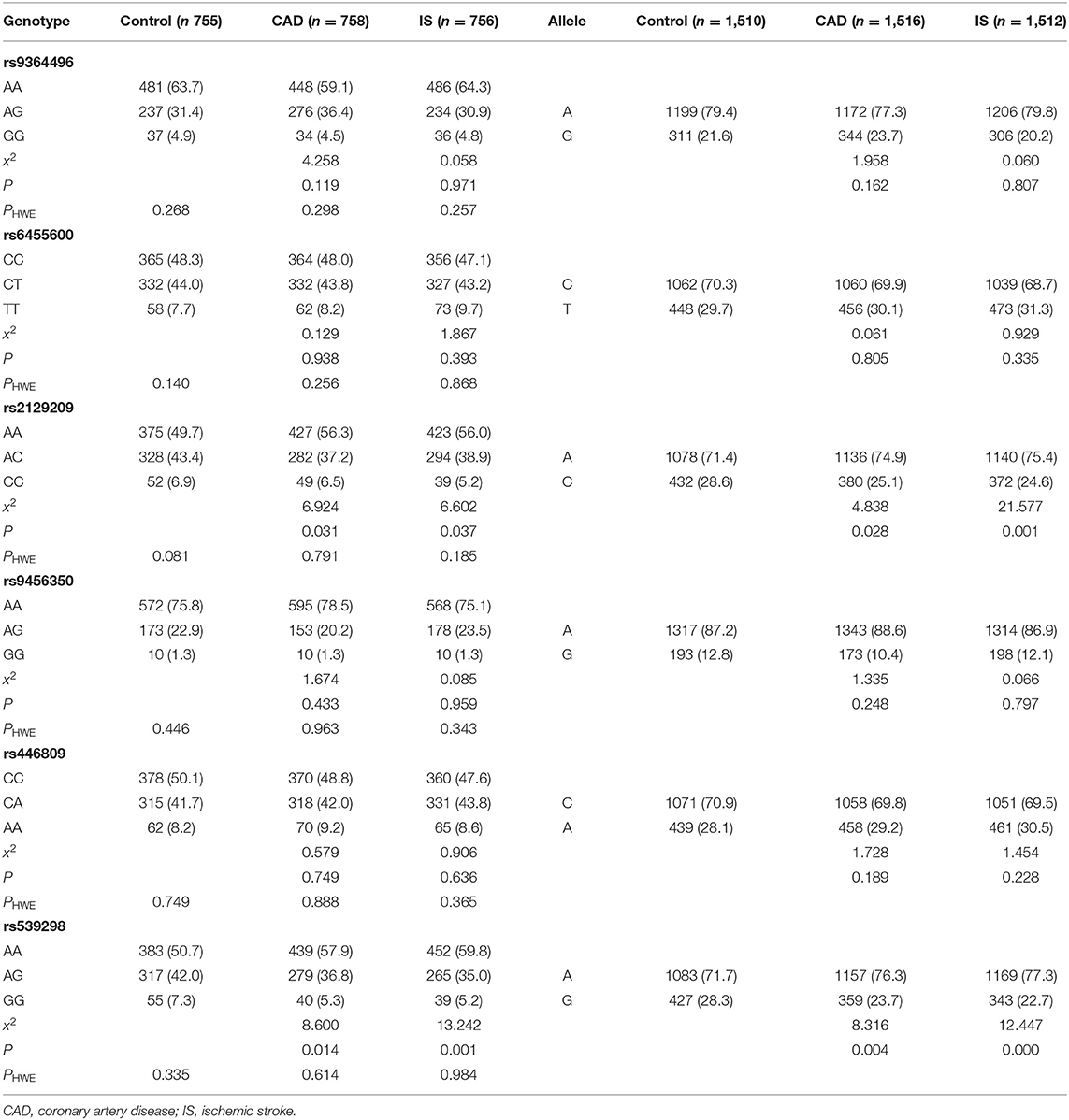
As shown in Table 2, all detected SNPs in the CAD, IS and control groups conformed to HWE (P > 0.05). The frequencies of the rs2129209AC/CC genotypes and the rs2129209C allele were lower in CAD (AC/CC, 43.7%; C, 25.1%) and IS (AC/CC, 44.1%; C, 24.6%) patients than in control participants (AC/CC, 50.3%; C, 28.6%, P < 0.05-0.01 for all). The frequencies of the rs539298AG/GG genotypes and rs539298G allele were also lower in CAD (AG/GG, 42.1%; G, 23.7%) and IS (AG/GG, 40.2%; G, 22.7%) patients than in control subjects (AG/GG, 49.3%; G, 28.3%, P < 0.05-0.001 for all). The specific and detailed genotype distribution of the six selected SNPs in the control, CAD, and IS groups is shown in Supplementary Material.
Genotypes and the Risk of Diseases
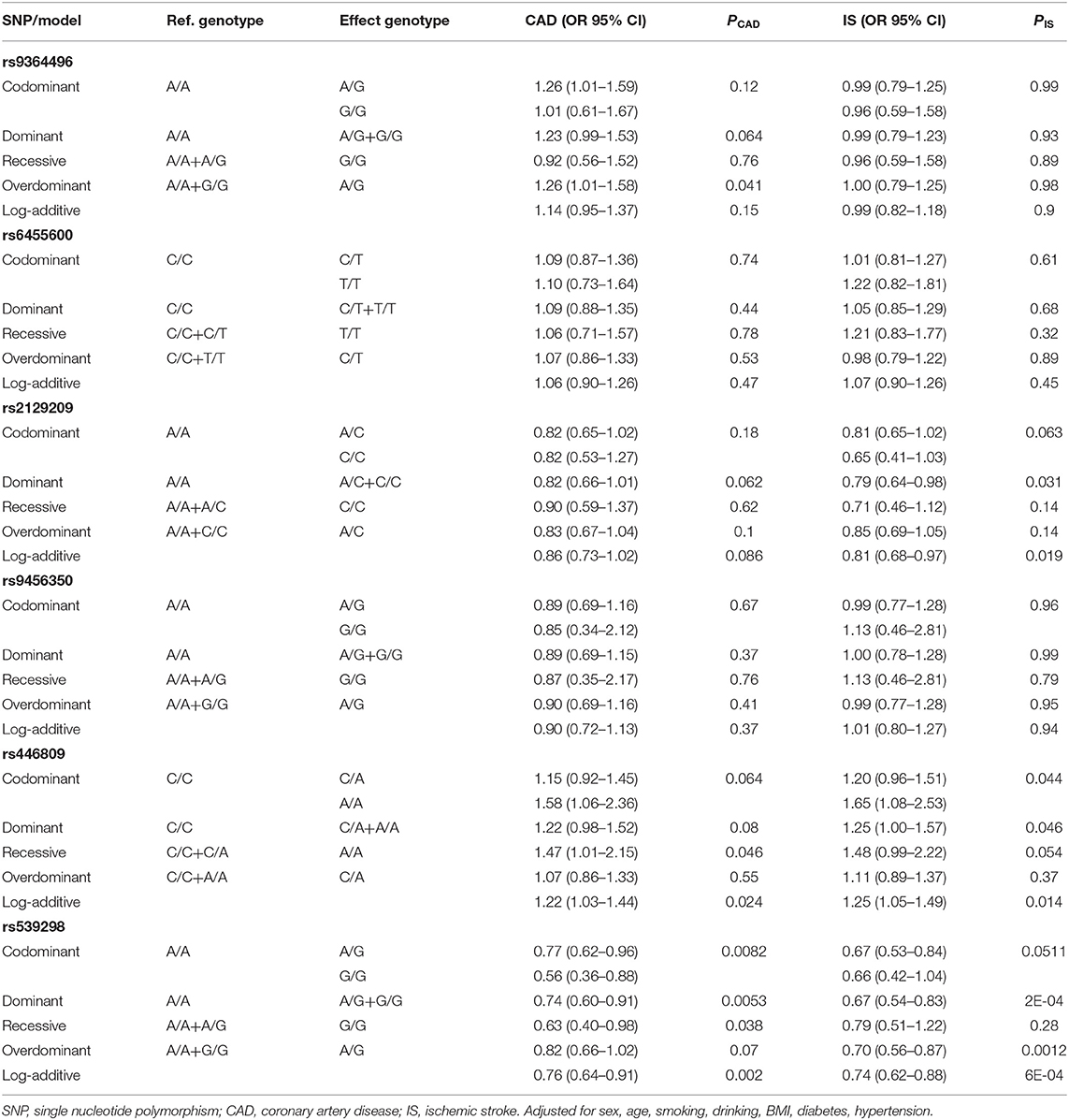
As described in Table 3, only the rs539298 SNP decreased the risk of CAD and IS (P < 0.008 was considered statistically significant after Bonferroni correction). The dominant model (AG/GG vs. AA) for CAD group showed OR = 0.74, 95% CI = 0.60–0.91, P = 0.0053; and the log-additive model (A vs. G) illustrated OR = 0.76, 95% CI = 0.64–0.91, P = 0.002. The dominant model (AG/GG vs. AA) for IS group displayed OR = 0.67, 95% CI = 0.54–0.83, P = 2E-04; and the log-additive model (A vs. G) revealed OR = 0.74, 95% CI = 0.62–0.88, P = 6E-04.
Interaction Between the rs539298 SNP and Environmental Factors on the Risk of Diseases
As shown in Figure 1, the interaction between the rs539298AG/GG genotypes and alcohol consumption reduced the risk of CAD (OR = 0.53, 95% CI = 0.37–0.77, P < 0.01) and IS (OR = 0.44, 95% CI = 0.30–0.65, P < 0.01).
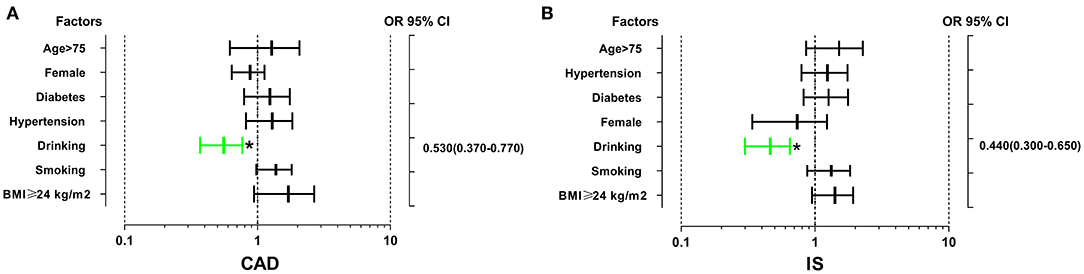
Figure 1. The interactions of the SLC22A3 rs539298 SNP and diabetes, drinking, hypertension, smoking, age, sex, and BMI on the risk of CAD and IS. The rs539298AG/GG genotypes interacted with alcohol consumption to decrease the risk of CAD (A) (OR = 0.53, 95% CI = 0.37–0.77) and IS (B) (OR = 0.44, 95% CI = 0.30–0.65). *P < 0.01.
Haplotypes and the Risk of Diseases
As presented in Figure 2, strong LD was found among the SYTL3 rs9364496, rs6455600, rs2129209 and rs9456350 SNPs as well as the SLC22A3 rs446809 and rs539298 SNPs in the CAD (Figure 2A) and IS (Figure 2B) groups (D' = 0.64–0.95).
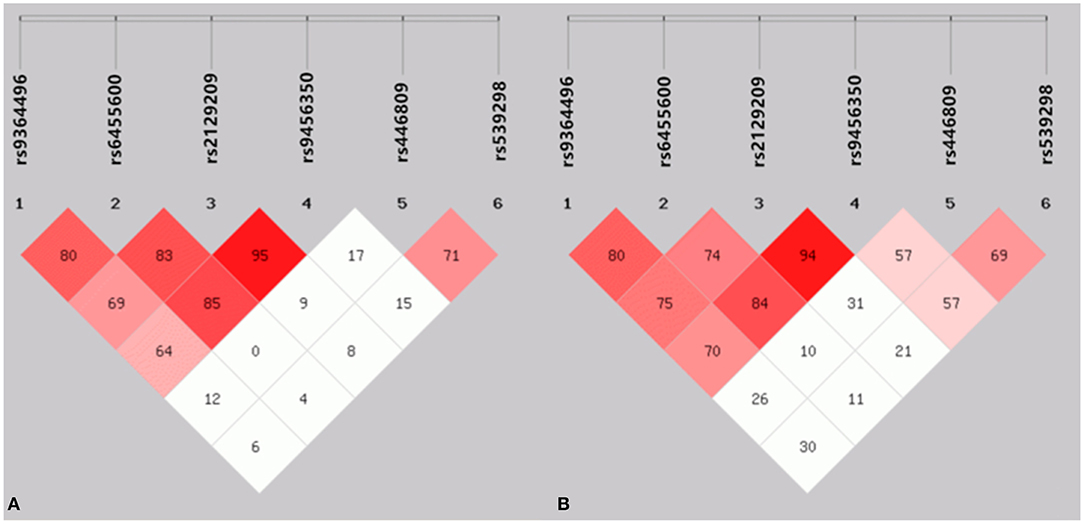
Figure 2. The linkage disequilibrium (LD) represents pair wise D' × 100 in the CAD (A) and IS (B) groups.
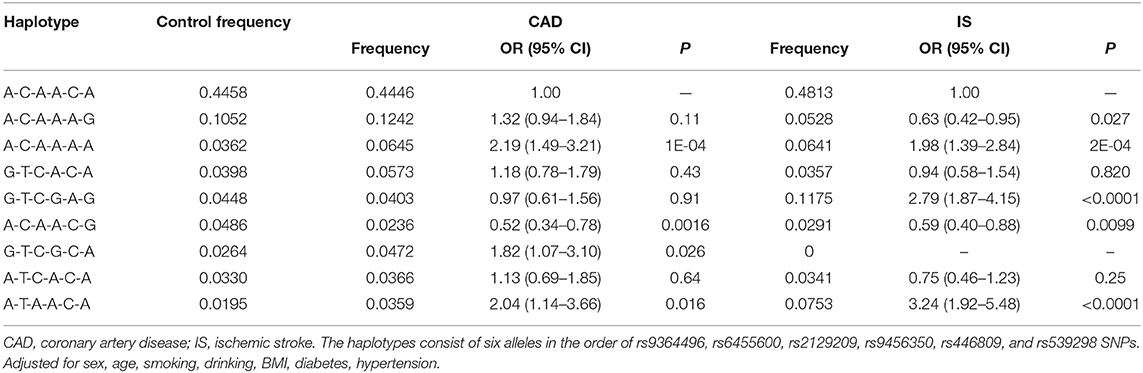
Haplotype analysis among the six selected SNPs showed that the main haplotype was the SYTL3-SLC22A3 A-C-A-A-C-A (Table 4). The SYTL3-SLC22A3 A-C-A-A-A-A (adjusted OR = 2.19, 95% CI = 1.49–3.21, P = 1E-04), G-T-C-G-C-A (adjusted OR = 1.82, 95% CI = 1.07–3.10, P = 0.026) and A-T-A-A-C-A (adjusted OR = 2.04, 95% CI = 1.14–3.66, P = 0.016) haplotypes increased the risk of CAD, while the SYTL3-SLC22A3 A-C-A-A-C-G (adjusted OR = 0.52, 95% CI = 0.34–0.78, P = 0.0016) haplotype reduced the risk of CAD. Meanwhile, the SYTL3-SLC22A3 A-C-A-A-A-A (adjusted OR = 1.98, 95% CI = 1.39–2.84, P = 2E-04), G-T-C-G-A-G (adjusted OR = 2.79, 95% CI = 1.87–4.15, P < 0.0001) and A-T-A-A-C-A (adjusted OR = 3.24, 95% CI = 1.92–5.48, P < 0.0001) haplotypes increased the risk of IS, whereas the SYTL3-SLC22A3 A-C-A-A-A-G (adjusted OR = 0.63, 95% CI = 0.42–0.95, P = 0.027) and A-C-A-A-C-G (adjusted OR = 0.59, 95% CI = 0.40–0.88, P = 0.0099) haplotypes reduced the risk of IS.
Interactions Between the Haplotypes and Environmental Factors on the Risk of Diseases
The interactions between the SYTL3-SLC22A3 haplotypes and several environmental factors on the risk of CAD and IS are shown in Figure 3. The interactions of the SYTL3-SLC22A3 A-C-A-A-A-A-smoking (adjusted OR = 3.70, 95% CI = 2.29–5.96, P < 0.01), SYTL3-SLC22A3 A-C-A-A-A-A-age > 75 (adjusted OR = 1.91, 95% CI = 1.28–2.84, P < 0.05), SYTL3-SLC22A3 G-T-C-G-C-A-smoking (adjusted OR = 2.25, 95% CI = 1.43–3.54, P < 0.01), SYTL3-SLC22A3 G-T-C-G-C-A-BMI ≥ 24 kg/m2 (adjusted OR = 2.80, 95% CI = 1.78–4.40, P < 0.01), SYTL3-SLC22A3 A-T-A-A-C-A-age > 75 (adjusted OR = 2.72, 95% CI = 1.62–4.56, P < 0.01), and SYTL3-SLC22A3 A-T-A-A-C-A-smoking (adjusted OR = 2.38, 95% CI = 1.27–4.43, P < 0.01) increased the risk of CAD, whereas the interaction of the SYTL3-SLC22A3 A-C-A-A-C-G-drinking (adjusted OR = 0.31, 95% CI = 0.14–0.67, P < 0.01) decreased the risk of CAD.
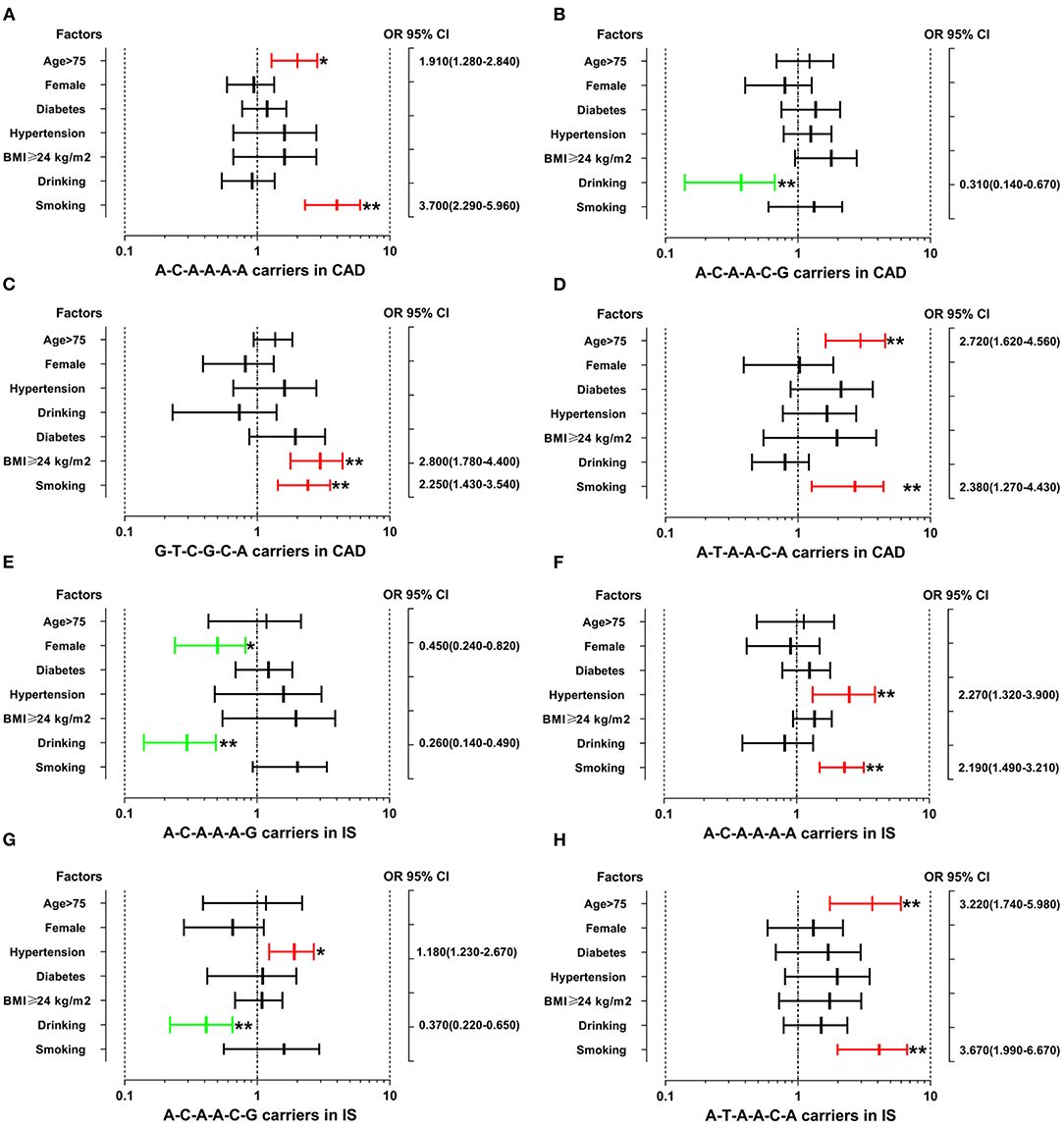
Figure 3. The interactions between the SYTL3-SLC22A3 haplotypes and the risk of CAD and IS. CAD, coronary artery disease; IS, ischaemic stroke. (A) The SYTL3-SLC22A3 A-C-A-A-A-A haplotype interacted with age > 75 and smoking to increase the risk of CAD. (B) The interaction between the SYTL3-SLC22A3 A-C-A-A-C-G haplotype and alcohol consumption decreased the risk of CAD. (C) The interactions between SYTL3-SLC22A3 G-T-C-G-C-A and cigarette smoking and BMI ≥ 24 kg/m2 increased the risk of CAD. (D) The SYTL3-SLC22A3 A-T-A-A-C-A haplotype interacted with age > 75 and smoking to increase the risk of CAD. (E) The interactions between the SYTL3-SLC22A3 A-C-A-A-A-G haplotype and female and alcohol consumption decreased the risk of IS. (F) The SYTL3-SLC22A3 A-C-A-A-A-A haplotype interacted with hypertension and smoking to increase the risk of IS. (G) The interactions between the SYTL3-SLC22A3 A-C-A-A-C-G haplotype and hypertension increased the risk of IS, and alcohol consumption decreased the risk of IS. (H) The interactions between the SYTL3-SLC22A3 A-T-A-A-C-A haplotype and age > 75 and smoking increased the risk of IS. *P < 0.05 and **P < 0.01.
The interactions of the SYTL3-SLC22A3 A-C-A-A-A-A-hypertension (adjusted OR = 2.27, 95% CI = 1.32–3.90, P < 0.01), SYTL3-SLC22A3 A-C-A-A-A-A-smoking (adjusted OR = 2.19, 95% CI = 1.49–3.21, P < 0.01), SYTL3-SLC22A3 A-C-A-A-C-G-hypertension (adjusted OR = 1.18, 95% CI = 1.23–2.67, P < 0.05), SYTL3-SLC22A3 A-T-A-A-C-A-age > 75 (adjusted OR = 3.22, 95% CI = 1.74–5.98, P < 0.01), SYTL3-SLC22A3 A-T-A-A-C-A-smoking (adjusted OR = 3.67, 95% CI = 1.99–6.67, P < 0.01) increased the risk of IS, whereas the interactions of the SYTL3-SLC22A3 A-C-A-A-A-G-female (adjusted OR = 0.45, 95% CI = 0.24–0.82, P < 0.05), SYTL3-SLC22A3 A-C-A-A-A-G-drinking (adjusted OR = 0.26, 95% CI = 0.14–0.49, P < 0.01), and SYTL3-SLC22A3 A-C-A-A-C-G-drinking (adjusted OR = 0.37, 95% CI = 0.22–0.65, P < 0.01) decreased the risk of IS. However, there was no significant interaction between the SYTL3-SLC22A3 G-T-C-G-A-G haplotype and several environmental factors including gender, age, diabetes, hypertension, BMI, smoking and drinking.
Relationship Between Genotypes and Serum Lipid Parameters
The correlation between the SYTL3-SLC22A3 SNPs and serum lipid parameters in the control group is shown in Figure 4. There were significant differences in TC levels between the rs539298AA and rs539298AG/GG genotypes (P < 0.008 was considered statistically significant after Bonferroni correction), the subjects with rs539298AG/GG genotypes had lower serum TC levels than those with rs539298AA genotype. However, there was no significant correlation between the remaining 5 SNPs and serum lipid levels in the control group (P ≥ 0.008 for all).
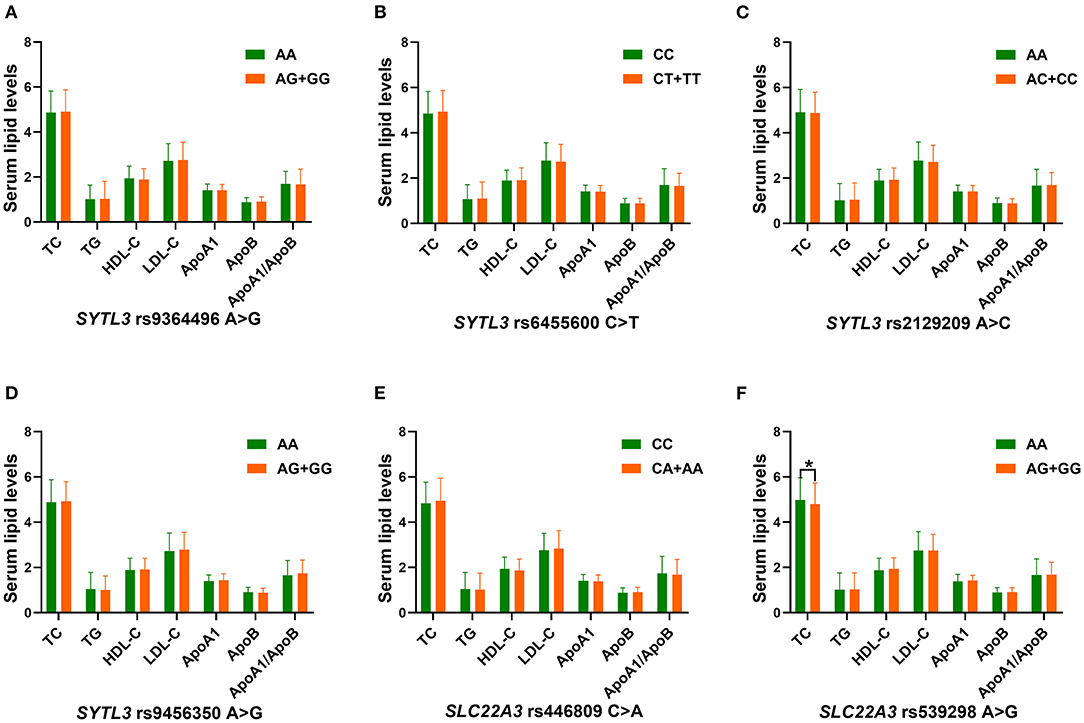
Figure 4. The association between the genotypes of six SYTL3-SLC22A3 SNPs and blood lipid levels in the control group. HDL-C, high-density lipoprotein cholesterol; TC, total cholesterol; ApoA1, apolipoprotein A1; TG, triglyceride; ApoB, apolipoprotein B; LDL-C, low-density lipoprotein cholesterol; ApoA1/ApoB, the ratio of apolipoprotein A1 to apolipoprotein B. A *P < 0.008 (after adjusting for 6 independent tests by the Bonferroni correction) was considered statistically significant. (A), rs9364496; (B), rs6455600; (C), rs2129209; (D), rs9456350; (E), rs446809; and (F), rs539298 SNPs.
Interactions Between the SYTL3-SLC22A3 SNPs and Several Environmental Factors on Serum Lipid Parameters and the Risk of CAD/IS
As listed in Table 5, several interactions between the SYTL3-SLC22A3 SNPs and some environmental factors on serum lipid parameters were detected in the control group. The interactions of the SYTL3 rs6455600-smoking, SYTL3 rs6455600-BMI, and SYTL3 rs9456350-BMI affected TG levels. The interaction of the SYTL3 rs2129209- and SLC22A3 rs539298-BMI influenced TC levels; and the interaction of the SYTL3 rs9364496-smoking or SYTL3 rs9364496-BMI influenced TG or TC levels. The interaction of the SYTL3 rs2129209-, SYTL3 rs9456350- and SLC22A3 rs446809-alcohol consumption influenced HDL-C levels; and the interaction of the SLC22A3 rs539298-alcohol consumption affected HDL-C and ApoA1 levels.
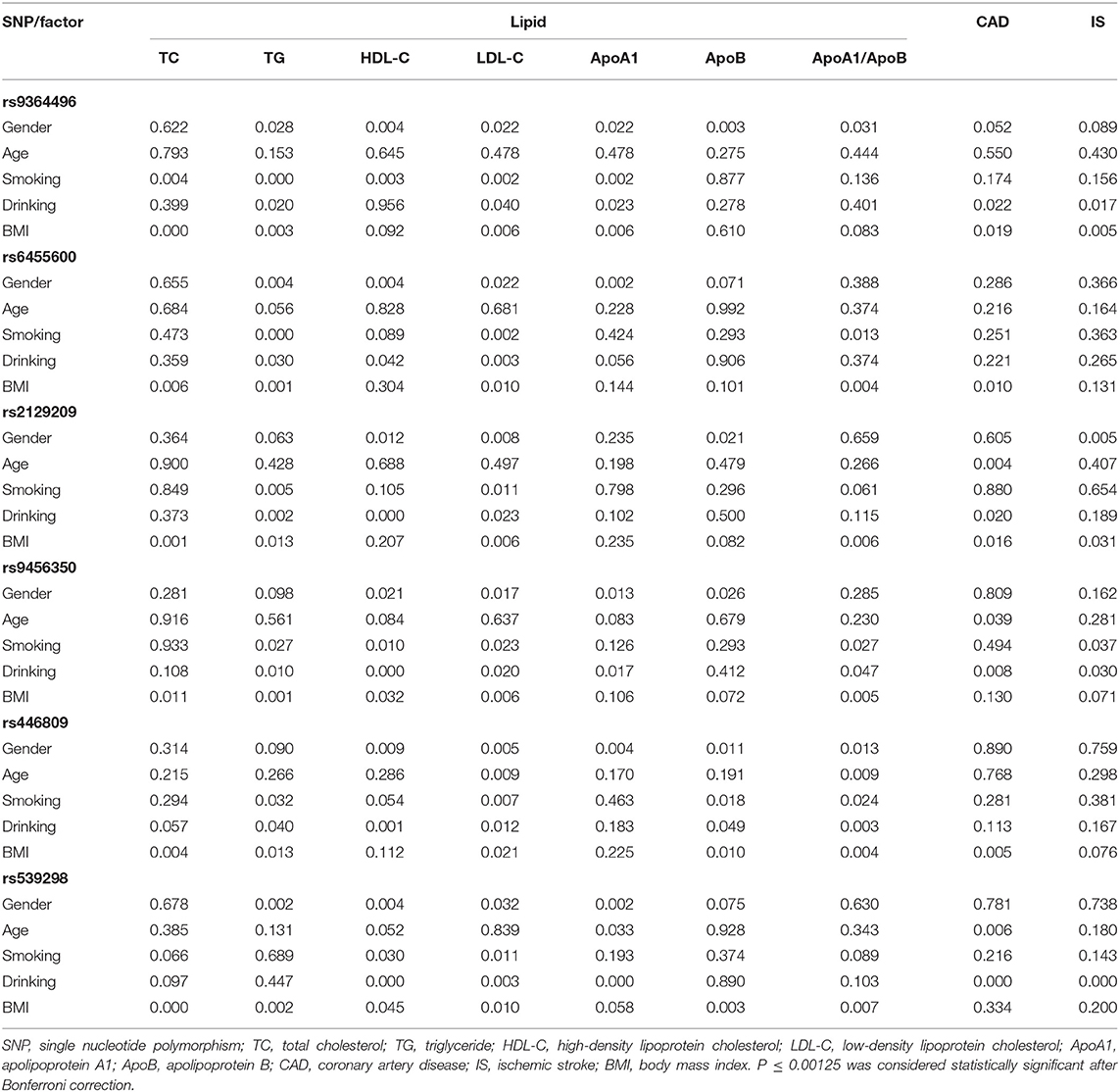
Table 5. The P-values for the interactions of genotypes and gender, age, drinking, smoking, and BMI on serum lipid levels and the risk of CAD and IS.
As presented in Figure 5, the interactions of the SYTL3 rs6455600CT/TT-BMI ≥ 24 kg/m2 or smoking, and SYTL3 rs9456350AG/GG-BMI ≥ 24 kg/m2 increased TG levels. The interactions of the SLC22A3 rs539298AG/GG-BMI <24 kg/m2, and SYTL3 rs2129209AC/CC-BMI <24 kg/m2 reduced TC levels; and the interaction of SYTL3 rs9364496AG/GG-BMI ≥ 24 kg/m2 or smoking increased TC or TG levels, respectively. The interactions of the SYTL3 rs9456350AG/GG-, SLC22A3 rs446809CA/AA-, and SYTL3 rs2129209AC/CC-alcohol consumption increased HDL-C levels; and the interaction of the SLC22A3 rs539298AG/GG-alcohol consumption increased HDL-C and ApoA1 levels.
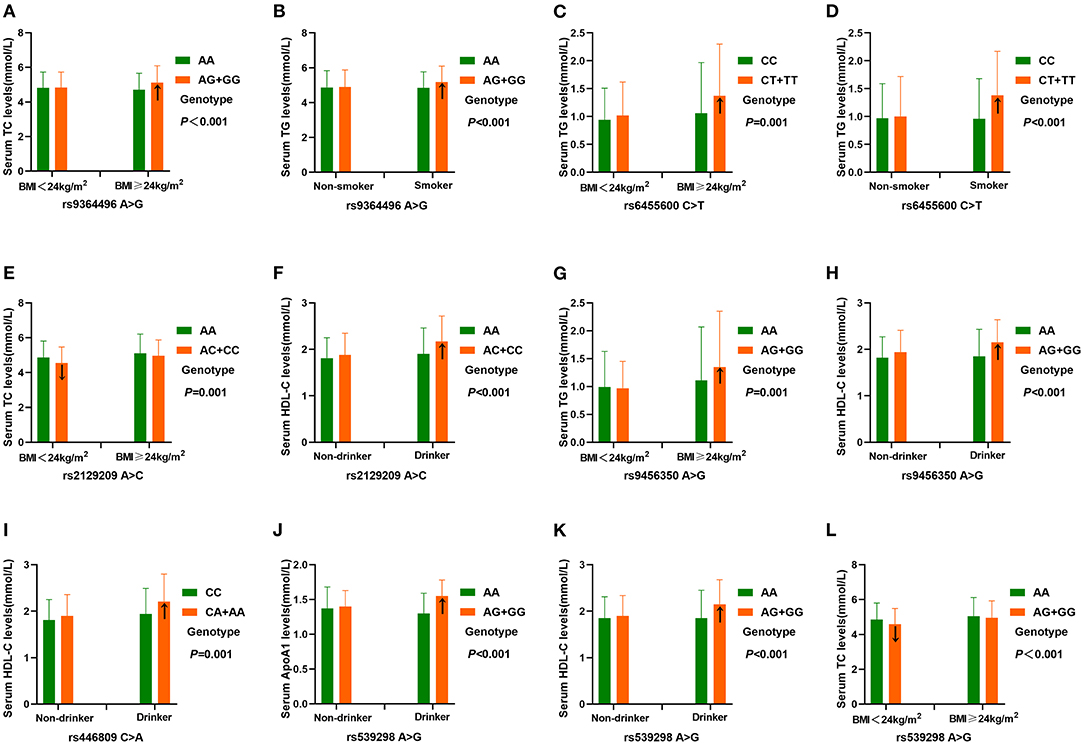
Figure 5. The interactions of the SYTL3-SLC22A3 SNPs and drinking, BMI and smoking on serum lipid levels. TC, total cholesterol; TG, triglyceride; BMI, body mass index; ApoA1, apolipoprotein A1; HDL-C, high-density lipoprotein cholesterol. A P ≤ 0.00125 (corresponding to P < 0.05 after adjusting for five environmental exposures multiplied by eight outcomes by the Bonferroni correction) was considered statistically significant. (A), rs9364496-BMI on TC; (B), rs9364496-smoking on TG; (C), rs6455600-BMI on TG; (D), rs6455600-smoking on TG; (E), rs2129209-BMI on TC; (F), rs2129209-drinking on HDL-C; (G), rs9456350-BMI on TG; (H), rs9456350-drinking on HDL-C; (I), rs446809-drinking on HDL-C; (J), rs539298-drinking on ApoA1; (K), rs539298-drinking on HDL-C; (L), rs539298-BMI on TC.
Relative Factors for Serum Lipid Parameters
As shown in Figure 6, Pearson correlation analysis revealed that there were significant correlations between the 6 SNPs and several environmental factors, including alcohol consumption, BMI, cigarette smoking, blood glucose, age, blood pressure, sex and serum lipid profiles, in the CAD (Figure 6A) and IS groups (Figure 6B).
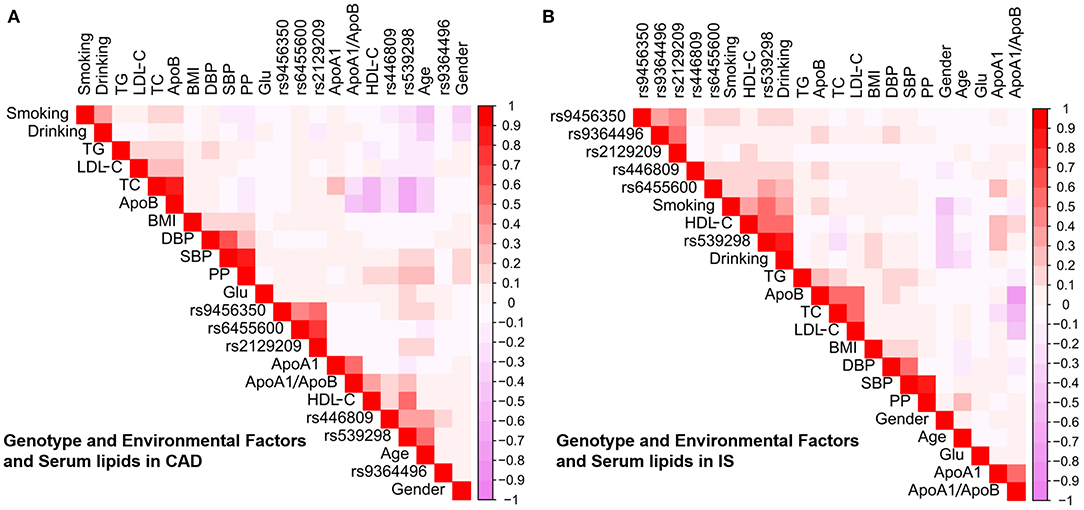
Figure 6. Association between environmental exposures as well as the candidate loci and serum lipid parameters in CAD (A) or IS (B). CAD, coronary artery disease; IS, ischaemic stroke. SBP, systolic blood pressure; TC, total cholesterol; BMI, body mass index; TG, triglyceride; ApoA1/B, the ratio of apolipoprotein A1 to apolipoprotein B; HDL-C, high-density lipoprotein cholesterol; DBP, diastolic blood pressure; LDL-C, low-density lipoprotein cholesterol; ApoA1, apolipoprotein A1; PP, pulse pressure; ApoB, apolipoprotein B; Glu, glucose.
Discussion
The main findings of the current research included the following aspects: (1) The allelic and genotypic frequencies of the SYTL3 rs2129209 and SLC22A3 rs539298 SNPs were different between controls and CAD/IS patients. The rs2129209AC/CC genotype and rs2129209C allele frequencies as well as the rs539298AG/GG genotype and rs539298G allele frequencies were lower in CAD/IS patients than in controls, respectively. (2) The SLC22A3 rs539298 SNP was correlated with TC levels in the control group, the rs539298G allele carriers had lower TC levels than the rs539298G allele non-carriers. At the same time, the SLC22A3 rs539298 SNP interacted with alcohol consumption would reduce the risk of CAD and IS. (3) Several haplotypes were associated with an increased or decreased risk of CAD and IS, and the haplotypes could explain more changes in the risk of CAD/IS than any single SNP alone. In addition, the interactions between several haplotypes and different environmental factors on the onset of CAD and IS were also observed. (4) The interactions between the SYTL3-SLC22A3 SNPs and several environmental factors, including smoking, alcohol consumption and BMI, could affect serum lipid parameters such as TC, TG, HDL-C and ApoA1 levels.
The genotypic and allelic frequencies of the SLC22A3 rs539298 SNP in different racial/ethnic groups are not well-known. Based on the data derived from the International 1000 Genomes database (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/), the frequencies of the rs539298G allele and the AG, GG genotypes were, respectively, 52.21, 49.56, and 27.43% in Europeans; 30.23, 41.86 and 9.30% in Japanese; 59.82, 53.57, and 33.04% in Sub-Saharan Africans; 52.04, 59.18, and 22.45% in Americans of African Ancestry in the Southwestern USA (ASW); 39.20, 48.86, and 14.77% in Gujarati Indians in Houston (GIH); 51.67, 45.56, and 28.89% in Luhya in Webuye, Kenya (LWK); 49.00, 46.00, and 25.00% in those with Mexican ancestry in Los Angeles, California (MEX); 58.10, 50.00, and 33.10% in Maasai in Kinyawa, Kenya (MKK); and 26.83, 48.84, and 4.65% in Han Chinese in Beijing (CHB). In the current study, we noticed that the allele and genotype frequencies of the SLC22A3 rs539298 SNP were significantly different between the controls and cases. The frequencies of the G allele and AG and GG genotypes in our study populations were 28.28, 41.99, and 7.28% in controls; 23.68, 36.81, and 5.28% in CAD patients; and 22.69, 35.05, and 5.16% in IS cases (P < 0.05–0.001, respectively). These findings suggest that the SLC22A3 rs539298 SNP may have racial/ethnic and population specificity. However, these findings need to be confirmed in other populations or ethnic groups with larger sample size.
Hyperlipidaemia is closely related to the occurrence and development of atherosclerotic cardiovascular and cerebrovascular diseases (39, 40). Previous studies showed that the SLC22A3 rs2048327 SNP was significantly associated with cardiovascular disease events in familial hypercholesterolemia subjects (41). A genome-wide haplotype association (GWHA) research revealed that the SLC22A3-LPAL2-LPA cluster was a strong susceptibility locus for CAD (42). Furthermore, Wang, et al. suggested that the rs3088442G allele might inhibit miR-147a binding to the 3' UTR region of SLC22A3, resulting in increased the expression levels of SLC22A3, and ultimately lead to increased risk of CAD (43). In addition, another case control study noticed that four SNPs (rs2048327, rs3127599, rs7767084 and rs10755578) in the SLC22A3-LPAL2-LPA cluster were not significantly associated with the risk of CAD (44). However, no study has yet well-elucidated the potential association between the six selected SNPs in the SYTL3-SLC22A3 cluster and serum lipid levels and the risk of CAD and IS. To meet this need, the potential correlation between the six selected SNPs in the SYTL3-SLC22A3 cluster and serum lipid levels in humans has been well-elaborated in our previous research (45). We noticed that the rs6455600, rs2129209 and rs539298 SNPs were associated with TC levels; and the rs446809 SNP was associated with TG and LDL-C levels in the Chinese Han population. Furthermore, we also observed that the rs539298G allele carriers had lower serum TC levels than the rs539298G allele non-carriers, and the dominant model of the rs539298 SNP reduced the morbidity of hyperlipidaemia in the Chinese Han population. In the current research, we obtained the similar findings to our previous study. We noticed that serum TC levels in the control group were significantly different between the rs539298AA and rs539298AG/GG genotypes, and participants with the rs539298AG/GG genotypes had lower serum TC levels than those with the rs539298AA genotype. In addition, we also found that the rs539298G allele carriers had lower risk of CAD (adjusted OR = 0.74, 95% CI: 0.60–0.91) and IS (adjusted OR = 0.67, 95% CI: 0.54–0.83) than the rs539298G allele non-carriers. These findings suggested that the SLC22A3 rs539298A allele may be a genetic risk factor for ischaemic cardiovascular and cerebrovascular diseases, and the SLC22A3 rs539298G allele may reduce the risk of CAD and IS by affecting serum TC levels. However, the correlation between the SLC22A3 rs539298 SNP and serum TC levels needs to be confirmed by proteomics studies in future research.
Haplotype analysis showed that the SYTL3-SLC22A3 A-C-A-A-A-A, G-T-C-G-C-A and A-T-A-A-C-A haplotypes increased the risk of CAD, while the SYTL3-SLC22A3 A-C-A-A-C-G haplotype reduced the risk of CAD. Meanwhile, the SYTL3-SLC22A3 A-C-A-A-A-A, G-T-C-G-A-G and A-T-A-A-C-A haplotypes increased the risk of IS, whereas the SYTL3-SLC22A3 A-C-A-A-A-G and A-C-A-A-C-G haplotypes reduced the risk of IS. These results suggest that haplotypes could explain more changes in the risk of CAD/IS than any single SNP alone.
A large number of studies have suggested that as a multifactorial and complex disorder, CAD or IS occurs due to numerous pathogenic factors, including genetic background, environmental exposures and their interactions (46–48). However, the potential mutual effect between the SLC22A3 rs539298 SNP and environmental factors on blood lipid levels and the risk of CAD and IS is still unknown. In the current research, we firstly noticed that the interaction of the SLC22A3 rs539298-alcohol consumption increased serum ApoA1 as well as HDL-C levels; and SLC22A3 rs539298-BMI <24 kg/m2 reduced serum TC levels. These findings could partly account for a decreased risk of CAD and IS in rs539298G allele carriers. Several other potential interactions between haplotypes and environmental factors on the risk of CAD and IS were also observed in this study. The interactions between the SYTL3-SLC22A3 A-C-A-A-A-A and A-T-A-A-C-A haplotypes and age > 75 and smoking increased the risk of CAD. The interactions between the SYTL3-SLC22A3 G-T-C-G-C-A haplotype and smoking and BMI ≥ 24 kg/m2 increased the risk of CAD. The interaction between the SYTL3-SLC22A3 A-C-A-A-C-G haplotype and alcohol consumption reduced the risk of CAD. In addition, the interaction between the SYTL3-SLC22A3 A-C-A-A-A-G haplotype and female sex and alcohol consumption reduced the risk of IS. The interactions between the SYTL3-SLC22A3 A-C-A-A-A-A and smoking and hypertension increased the risk of IS. The interactions between the SYTL3-SLC22A3 A-T-A-A-C-A and smoking and age > 75 increased the risk of IS. The interactions between the SYTL3-SLC22A3 A-C-A-A-C-G haplotype and hypertension increased the risk of IS, while its interaction with alcohol consumption reduced the risk of IS. However, these findings have yet to be confirmed by additional research.
The current study may have several limitations. First, compared to other studies, the number of controls and patients was relatively small. Larger samples are necessary to validate our results in future research. Second, most of the patients with CAD or IS received some secondary prevention drugs. All of these drugs may have a certain effect on blood lipid levels. Third, to improve the accuracy of the statistical analysis, we adjusted for several environmental factors, such as BMI, age, smoking, sex and alcohol consumption, but the potential effects of the above factors on blood lipid levels and the risk of CAD and IS could not be completely eliminated. Finally, in vitro and in vivo studies are necessary to strengthen the significance of our results.
Conclusions
In conclusion, the current research showed that the allelic and genotypic frequencies of the SLC22A3 rs539298 SNP were significantly different between controls and CAD/IS patients. The SLC22A3 rs539298G carriers in the control group had lower serum TC levels than the G allele non-carriers. The interaction between the SLC22A3 rs539298 SNP and alcohol consumption reduced the risk of CAD and IS. In addition, haplotypes could explain more changes in the risk of CAD/IS than any single SNP alone. Several potential interactions between the haplotypes and some environmental factors affected the risk of CAD and IS. These findings revealed that the SLC22A3 rs539298A allele may be a new genetic marker for CAD and IS in our study populations. The correlation between the SLC22A3 rs539298 SNP and CAD/IS may be partly explained by its association with decreased serum TC levels in this study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of the First Affiliated hospital, Guangxi Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
P-FZ conceived the study, participated in the design, performed the statistical analyses, and drafted the manuscript. R-XY conceived the study, participated in the design, carried out the epidemiological survey, collected the samples, and helped to draft the manuscript. X-LC, W-XC, J-ZW, and FH carried out the epidemiological survey and collected the samples. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81460169) and the Youthful Science Foundation of Guangxi Province (No. 2017GXNSFBA198067). There was no role of the funding body in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2021.713068/full#supplementary-material
Abbreviations
ANCOVA, covariance analysis; Apo, apolipoprotein; BMI, body mass index; CAD, coronary artery disease; CI, confidence interval; DNA, deoxyribonucleic acid; GWAS, genome-wide association study; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; OR, odds ratio; SYTL3, synaptotagmin like 3; SLC22A3, solute carrier family 22 member 3; SNP, single nucleotide polymorphisms; T2DM, type 2 diabetes mellitus; TC, total cholesterol; TG, triglyceride; AMI, acute myocardial infarction; MRI, magnetic resonance imaging; MI, myocardial infarction; LD, linkage disequilibrium; NGS, next-generation sequencing; ASW, Americans of African Ancestry in the Southwestern USA; GIH, Gujarati Indians in Houston; LWK, Luhya in Webuye, Kenya; MEX, Mexican ancestry in Los Angeles, California; MEX, Maasai in Kinyawa, Kenya; CHB, Han Chinese in Beijing.
References
1. Zhang QH, Yin RX, Chen WX, Cao XL, Wu JZ. TRIB1 and TRPS1 variants, G × G and G × E interactions on serum lipid levels, the risk of coronary heart disease and ischemic stroke. Sci Rep. (2019) 9:2376. doi: 10.1038/s41598-019-38765-7
2. Zheng PF, Yin RX, Deng GX, Guan YZ, Wei BL, Liu CX. Association between the XKR6 rs7819412 SNP and serum lipid levels and the risk of coronary artery disease and ischemic stroke. BMC Cardiovasc Disord. (2019) 19:202–2. doi: 10.1186/s12872-019-1179-z
3. Li KG, Yin RX, Huang F, Chen WX, Wu JZ, Cao XL. XKR6 rs7014968 SNP increases serum total cholesterol levels and the risk of coronary heart disease and ischemic stroke. Clin Appl Thromb-Hem. (2020) 26:1076029620902844. doi: 10.1177/1076029620902844
4. Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation. (2005) 111 3481–8. doi: 10.1161/CIRCULATIONAHA.105.537878
5. Holmstedt CA, Turan TN, Chimowitz MI. Atherosclerotic intracranial arterial stenosis: risk factors, diagnosis, and treatment. Lancet Neurol. (2013) 12:1106–14. doi: 10.1016/S1474-4422(13)70195-9
6. Li B, Li W, Li X, Zhou H. Inflammation: a novel therapeutic target/direction in atherosclerosis. Curr Pharm Des. (2017) 23:1216–27. doi: 10.2174/1381612822666161230142931
7. Criqui MH, Heiss G, Cohn R, Cowan LD, Suchindran CM, Bangdiwala S, et al. Plasma triglyceride level and mortality from coronary heart disease. N Engl J Med. (1993) 328:1220–5. doi: 10.1056/NEJM199304293281702
8. Kwiterovich PO Jr, Coresh J, Smith HH, Bachorik PS, Derby CA, Pearson TA. Comparison of the plasma levels of apolipoproteins B and A-1, and other risk factors in men and women with premature coronary artery disease. Am J Cardiol. (1992) 69:1015–21. doi: 10.1016/0002-9149(92)90856-T
9. Crouse JR, Parks JS, Schey HM, Kahl FR. Studies of low density lipoprotein molecular weight in human beings with coronary artery disease. J Lipid Res. (1985) 26:566–74. doi: 10.1016/S0022-2275(20)34343-1
10. Shekelle RB, Shryock AM, Paul O, Lepper M, Stamler J, Liu S, et al. Diet, serum cholesterol, and death from coronary heart disease. The Western Electric study. N Engl J Med. (1981) 304:65–70. doi: 10.1056/NEJM198101083040201
11. Silbernagel G, Schottker B, Appelbaum S, Scharnagl H, Kleber ME, Grammer TB, et al. High-density lipoprotein cholesterol, coronary artery disease, and cardiovascular mortality. Eur Heart J. (2013) 34:3563–71. doi: 10.1093/eurheartj/eht343
12. Sabater-Lleal M, Huang J, Chasman D, Naitza S, Dehghan A, Johnson AD, et al. Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. (2013) 128:1310–24. doi: 10.1161/CIRCULATIONAHA.113.002251
13. Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NL, Jackson C, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. (2012) 43:3161–7. doi: 10.1161/STROKEAHA.112.665760
14. Ober C, Nord AS, Thompson EE, Pan L, Tan Z, Cusanovich D, et al. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. J Lipid Res. (2009) 50:798–806. doi: 10.1194/jlr.M800515-JLR200
15. Agmon E, Stockwell BR. Lipid homeostasis and regulated cell death. Curr Opin Chem Biol. (2017) 39:83–9. doi: 10.1016/j.cbpa.2017.06.002
16. Stefan CJ, Trimble WS, Grinstein S, Drin G, Reinisch K, De Camilli P, et al. Membrane dynamics and organelle biogenesis-lipid pipelines and vesicular carriers. BMC Biol. (2017) 15:102. doi: 10.1186/s12915-017-0432-0
17. Muse ED, Kramer ER, Wang H, Barrett P, Parviz F, Novotny MA, et al. A whole blood molecular signature for acute myocardial infarction. Sci Rep. (2017) 7:12268. doi: 10.1038/s41598-017-12166-0
18. Verhaagh S, Schweifer N, Barlow DP, Zwart R. Cloning of the mouse and human solute carrier 22a3 (Slc22a3/SLC22A3) identifies a conserved cluster of three organic cation transporters on mouse chromosome 17 and human 6q26-q27. Genomics. (1999) 55:209–18. doi: 10.1006/geno.1998.5639
19. Schneider E, Machavoine F, Pléau JM, Bertron AF, Thurmond RL, Ohtsu H, et al. Organic cation transporter 3 modulates murine basophil functions by controlling intracellular histamine levels. J Exp Med. (2005) 202:387–93. doi: 10.1084/jem.20050195
20. Rozenberg I, Sluka SH, Rohrer L, Hofmann J, Becher B, Akhmedov A, et al. Histamine H1 receptor promotes atherosclerotic lesion formation by increasing vascular permeability for low-density lipoproteins. Arterioscler Thromb Vasc Biol. (2010) 30:923–30. doi: 10.1161/ATVBAHA.109.201079
21. Kimura S, Wang KY, Tanimoto A, Murata Y, Nakashima Y, Sasaguri Y. Acute inflammatory reactions caused by histamine via monocytes/macrophages chronically participate in the initiation and progression of atherosclerosis. Pathol Int. (2004) 54:465–74. doi: 10.1111/j.1440-1827.2004.01653.x
22. Li L, He M, Zhou L, Miao X, Wu F, Huang S, et al. A solute carrier family 22 member 3 variant rs3088442 G → A associated with coronary heart disease inhibits lipopolysaccharide-induced inflammatory response. J Biol Chem. (2015) 290:5328–40. doi: 10.1074/jbc.M114.584953
23. Chen L, Pawlikowski B, Schlessinger A, More SS, Stryke D, Johns SJ, et al. Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet Genomics. (2010) 20:687–99. doi: 10.1097/FPC.0b013e32833fe789
24. Solbach TF, Grube M, Fromm MF, Zolk O. Organic cation transporter 3: expression in failing and nonfailing human heart and functional characterization. J Cardiovasc Pharmacol. (2011) 58:409–17. doi: 10.1097/FJC.0b013e3182270783
25. Zhao Q, Wei H, Liu D, Shi B, Li L, Yan M, et al. PHACTR1 and SLC22A3 gene polymorphisms are associated with reduced coronary artery disease risk in the male Chinese Han population. Oncotarget. (2017) 8:658–63. doi: 10.18632/oncotarget.13506
26. Chen L, Yao Y, Jin C, Wu S, Liu Q, Li J, et al. Integrative genomic analysis identified common regulatory networks underlying the correlation between coronary artery disease and plasma lipid levels. BMC Cardiovasc Disord. (2019) 19:310. doi: 10.1186/s12872-019-01271-9
27. Franceschini N, Carty C, Buzkova P, Reiner AP, Garrett T, Lin Y, et al. Association of genetic variants and incident coronary heart disease in multiethnic cohorts: the PAGE study. Circ Cardiovasc Genet. (2011) 4:661–72. doi: 10.1161/CIRCGENETICS.111.960096
28. Sun JQ, Yin RX, Shi GY, Shen SW, Chen X, Bin Y, et al. Association of the ARL15 rs6450176 SNP and serum lipid levels in the Jing and Han populations. Int J Clin Exp Pathol. (2015) 8:12977–94.
29. Guo T, Yin RX, Nie RJ, Chen X, Bin Y, Lin WX. Suppressor of cytokine signaling 3 A+930–>G (rs4969168) polymorphism is associated with apolipoprotein A1 and low-density lipoprotein cholesterol. Int J Clin Exp Pathol. (2015) 8:7305–17.
30. Li WJ, Yin RX, Huang JH, Bin Y, Chen WX, Cao XL. Association between the PPP1R3B polymorphisms and serum lipid traits, the risk of coronary artery disease and ischemic stroke in a southern Chinese Han population. Nutr Metab. (2018) 15:27. doi: 10.1186/s12986-018-0266-y
31. Zheng PF, Yin RX, Liu CX, Deng GX, Guan YZ, Wei BL. SYNE1-QK1 SNPs, G × G and G × E interactions on the risk of hyperlipidaemia. J Cell Mol Med. (2020) 24:5772–85. doi: 10.1111/jcmm.15239
32. Yin RX, Aung LH, Long XJ, Yan TT, Cao XL, Huang F, et al. Interactions of several genetic polymorphisms and alcohol consumption on blood pressure levels. Biofactors. (2015) 41:339–51. doi: 10.1002/biof.1234
33. Yin RX, Wu DF, Wu JZ, Cao XL, Aung LHH, Miao L, et al. Interactions of several lipid-related gene polymorphisms and cigarette smoking on blood pressure levels. Int J Biol Sci. (2012) 8:685–96. doi: 10.7150/ijbs.4401
34. Zhou BF, Cooperative Meta-Analysis Group of the Working Group on Obesity in China. Predictive values of body mass index and waist circumference for risk factors of certain related diseases in Chinese adults—study on optimal cut-off points of body mass index and waist circumference in Chinese adults. Biomed Environ Sci. (2002) 15:83–96.
35. Wildman RP, Gu D, Reynolds K, Duan X, He J. Appropriate body mass index and waist circumference cutoffs for categorization of overweight and central adiposity among Chinese adults. Am J Clin Nutr. (2004) 80:1129–36. doi: 10.1093/ajcn/80.5.1129
36. Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. (1998) 15: 539–53.
37. Yin RX, Wang Y, Chen GQ, Lin WX, Yang DZ, Pan SL. Lipoprotein lipase gene polymorphism at the PvuII locus and serum lipid levels in Guangxi Hei Yi Zhuang and Han populations. Clin Chem Lab Med. (2006) 44:1416–21. doi: 10.1515/CCLM.2006.273
38. Onda Y, Takahagi K, Shimizu M, Inoue K, Mochida K. Multiplex PCR targeted amplicon sequencing (MTA-Seq): simple, flexible, and versatile SNP genotyping by highly multiplexed PCR amplicon sequencing. Front Plant Sci. (2018) 9:201. doi: 10.3389/fpls.2018.00201
39. Yeramaneni S, Kleindorfer DO, Sucharew H, Alwell K, Moomaw CJ, Flaherty ML, et al. Hyperlipidemia is associated with lower risk of poststroke mortality independent of statin use: a population-based study. Int J Stroke. (2017) 12:152–60. doi: 10.1177/1747493016670175
40. Zhang X, Patel A, Horibe H, Wu Z, Barzi F, Rodgers A, et al. Cholesterol, coronary heart disease, and stroke in the Asia Pacific region. Int J Epidemiol. (2003) 32:563–72. doi: 10.1093/ije/dyg106
41. Paquette M, Bernard S, Baass A. SLC22A3 is associated with lipoprotein (a) concentration and cardiovascular disease in familial hypercholesterolemia. Clin Biochem. (2019) 66:44–8. doi: 10.1016/j.clinbiochem.2019.02.008
42. Trégouët DA, König IR, Erdmann J, Munteanu A, Braund PS, Hall AS, et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL; 2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. (2009) 41:283–5. doi: 10.1038/ng.314
43. Wang L, Chen J, Zeng Y, Wei J, Jing J, Li G, et al. Functional variant in the SLC22A3-LPAL2-LPA gene cluster contributes to the severity of coronary artery disease. Arterioscler Thromb Vasc Biol. (2016) 36:1989–96. doi: 10.1161/ATVBAHA.116.307311
44. Lv X, Zhang Y, Rao S, Liu F, Zuo X, Su D, et al. Lack of association between four SNPs in the SLC22A3-LPAL2-LPA gene cluster and coronary artery disease in a Chinese Han population: a case control study. Lipids Health Dis. (2012) 11:128. doi: 10.1186/1476-511X-11-128
45. Zheng PF, Yin RX, Guan YZ, Wei BL, Liu CX, Deng GX. SYTL3-SLC22A3 SNPs, G × G and G × E interactions on the risk of hyperlipidaemia. Front Genet. (2021) 12:679027. doi: 10.3389/fgene.2021.679027
46. Yang Q, Yin RX, Cao XL, Huang F, Zhou YJ, Chen WX. ANGPTL4 variants and their haplotypes are associated with serum lipid levels, the risk of coronary artery disease and ischemic stroke and atorvastatin cholesterol-lowering responses. Nutr Metab (Lond). (2018) 15:70. doi: 10.1186/s12986-018-0308-5
47. Li WJ, Yin RX, Cao XL, Chen WX, Huang F, Wu JZ. DOCK7-ANGPTL3 SNPs and their haplotypes with serum lipid levels and the risk of coronary artery disease and ischemic stroke. Lipids Health Dis. (2018) 17:30. doi: 10.1186/s12944-018-0677-9
Keywords: coronary artery disease, ischaemic stroke, SYTL3, SLC22A3, single nucleotide polymorphism, haplotype, lipids
Citation: Zheng P-F, Yin R-X, Cao X-L, Chen W-X, Wu J-Z and Huang F (2021) Effect of SYTL3-SLC22A3 Variants, Their Haplotypes, and G × E Interactions on Serum Lipid Levels and the Risk of Coronary Artery Disease and Ischaemic Stroke. Front. Cardiovasc. Med. 8:713068. doi: 10.3389/fcvm.2021.713068
Received: 21 May 2021; Accepted: 21 July 2021;
Published: 12 August 2021.
Edited by:
Seitaro Nomura, The University of Tokyo, JapanReviewed by:
Liu Miao, Liuzhou People's Hospital, ChinaRaj Sewduth, VIB KU Leuven Center for Cancer Biology, Belgium
Copyright © 2021 Zheng, Yin, Cao, Chen, Wu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui-Xing Yin, eWlucnVpeGluZ0AxNjMuY29t; orcid.org/0000-0001-7883-4310
 Peng-Fei Zheng
Peng-Fei Zheng Rui-Xing Yin
Rui-Xing Yin Xiao-Li Cao2
Xiao-Li Cao2 Wu-Xian Chen
Wu-Xian Chen Feng Huang
Feng Huang