
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cardiovasc. Med. , 07 July 2021
Sec. General Cardiovascular Medicine
Volume 8 - 2021 | https://doi.org/10.3389/fcvm.2021.682357
This article is part of the Research Topic Non-Coding RNA and Coronary Artery Disease View all 12 articles
Circular RNAs (circRNAs) represent a novel class of widespread and diverse endogenous RNA molecules. This unusual class of RNA species is generated by a back-splicing event of exons or introns, resulting in a covalently closed circRNA molecule. Accumulating evidence indicates that circRNA plays an important role in the biological functions of a network of competing endogenous RNA (ceRNA). CircRNAs can competitively bind to miRNAs and abolish the suppressive effect of miRNAs on target RNAs, thus regulating gene expression at the posttranscriptional level. The role of circRNAs as ceRNAs in the pathogenesis of cardiovascular and cerebrovascular diseases (CVDs) has been recently reported and highlighted. Understanding the underlying molecular mechanism could aid the discovery of therapeutic targets or strategies against CVDs. Here, we review the progress in studying the role of circRNAs as ceRNAs in CVDs, with emphasis on the molecular mechanism, and discuss future directions and possible clinical implications.
Cardiovascular and cerebrovascular diseases (CVDs) are general terms used to refer to all cardiac and cerebral diseases related to vasculopathy. Despite improvements in pharmacotherapy and surgical interventions, as well as lifestyle modifications, morbidity and mortality in patients with CVDs remain high in recent years (1). CVDs are still the leading cause of death worldwide (1). Current treatments primarily alleviate symptoms or slow down disease progression. To develop novel preventive and therapeutic strategies, we should elucidate and understand the underlying molecular mechanisms of these diseases.
Circular RNAs (circRNAs) were first discovered in plant viroids more than 40 years ago (2). A few years later, circRNAs were observed by electron microscopy in the cytoplasmic fractions of eukaryotic cells (3). However, they were mainly considered to be errors of the normal splicing process and did not receive much attention (4). In recent years, advances in the high-throughput sequencing technology and circRNA-specific bioinformatics algorithms have resulted in the discovery and identification of thousands of circRNAs (5–9). Indeed, circRNAs are diverse, abundant, and expressed in a tissue- and developmental stage-specific manner (5, 8). Although the functions of most circRNAs remain elusive, a select number of circRNAs are known to function as competitive endogenous RNAs (ceRNAs) by decoying miRNAs from other target transcripts, thereby controlling gene expression at the posttranscriptional level (10).
Recent studies have identified circRNAs as ceRNAs in many diseases including CVDs (11–13). Here, we reviewed the role of circRNAs as ceRNAs in atherosclerosis, myocardial infarction, cardiac fibrosis, heart failure, aneurysm, and stroke, with a special focus on the molecular mechanism, which provides a new direction for further research on the pathogenesis and treatment of CVDs.
Several years before the discovery of ceRNAs, or natural miRNA sponges, many studies had found that artificial miRNA sponges were able to specifically and effectively inhibit miRNA activity (14–16). These synthetic miRNA sponges are usually expressed from strong promoters, engineered to carry multiple binding sites for a miRNA or miRNA family of interest and have been shown to derepress miRNA targets both in vitro and in vivo (14). The sponging constructs are not only invaluable tools for miRNA loss-of-function studies in vitro and in vivo, but also critical for the development of RNA-based therapeutic applications (15, 16).
The theoretical and practical development of the artificial miRNA sponge laid an important foundation for the ceRNA hypothesis. In 2011, Salmena et al. postulated that all endogenous RNA transcripts sharing common miRNA response elements (MREs) can communicate with and regulate each other through competition for a limited pool of miRNAs (17). This hypothesis not only attributes new functions for non-coding RNA transcripts, but also implies coding-independent functions of protein-coding messenger RNAs (17). Given that any transcripts containing MREs can theoretically serve as ceRNAs, they may represent a widespread form of posttranscriptional gene regulation in a variety of physiological and pathophysiological processes.
In 2013, two independent groups firstly discovered that circRNAs can serve as ceRNAs (18, 19). Hansen et al. showed that ciRS-7 (also known as CDR1as) contains 73 conserved binding sites for miR-7 (19). ciRS-7 strongly binds to miR-7 as well as the miRNA effector Argonaute protein 2 (AGO2) and efficiently suppresses miR-7 activity, thereby attenuating the availability of miR-7 to bind to its target mRNAs. In situ hybridization showed that miR-7 and ciRS-7 displayed clear co-expression in mouse brain sections and in primary cells isolated from mouse brain, suggesting a high level of endogenous interactivity between them. Another research group demonstrated that ectopic expression of human ciRS-7 in zebrafish impairs midbrain development, consistent with the effect of miR-7 inhibition induced by morpholinos injection (18). The midbrain reduction induced by ciRS-7 expression in zebrafish could be partially rescued by expressing miR-7, indicating the interaction between ciRS-7 and miR-7. These two studies also serve as the first functional analysis of naturally expressed circRNAs. Acting as ceRNAs, circRNAs can bind to miRNAs and inhibit the function of miRNAs, thereby upregulating miRNA target gene expression (Figure 1). Because of the lack of free ends, circRNAs are more resistant to exonucleases and more stable than linear RNAs (18, 19). These natures of circRNAs are thought to facilitate their effectiveness as ceRNAs. Indeed, so far many circRNAs have been found to act as ceRNAs both under normal physiological conditions and diseased conditions (20).
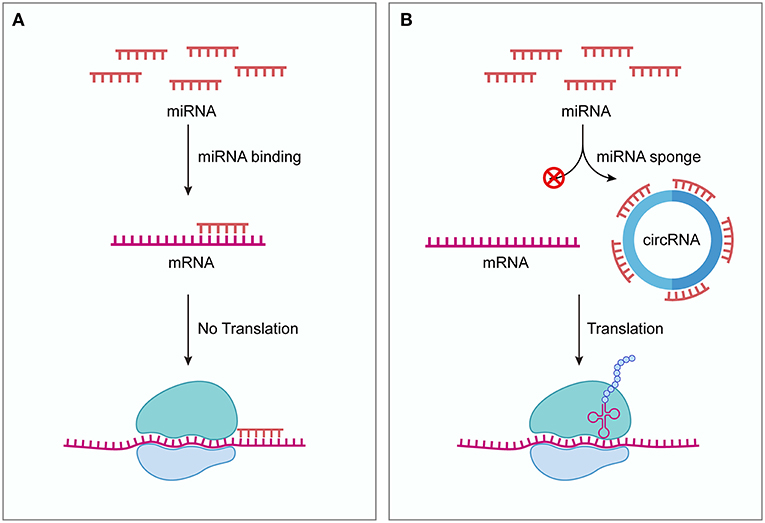
Figure 1. circRNAs function as ceRNAs. (A) Without competing transcripts circRNAs, miRNAs bind to target mRNAs through sequence complementarity, blocking the translation of target mRNAs. (B) circRNAs can competitively sequestrate miRNAs and derepress their target mRNAs, thus promoting the expression of target genes. ceRNA, competing endogenous RNA; circRNA, circular RNA.
Atherosclerosis (AS) is one group of the most common vasculopathies and a common pathological basis for many cardiovascular diseases, characterized by abnormal lipid metabolism and inflammatory reactions. A number of cells are involved in the pathology and pathophysiology of AS, including vascular endothelial cells (VECs) and vascular smooth muscle cells (VSMCs). Recent studies have found that circRNAs can effectively regulate the proliferation and migration of VECs and VSMCs by acting as ceRNAs, thus modulating the progression of AS (21, 22).
VECs comprise the inner layer of blood vessels and play crucial roles in vascular homeostasis. Endothelial cell injury and dysfunction are critical events that trigger the development of atherosclerotic lesions. Importantly, ox-LDL has been identified to contribute to the progression of AS by inducing VEC injury and apoptosis (23, 24). Zhang et al. found that the circRNA circ_0003204, which is upregulated in ox-LDL-induced human aortic endothelial cells (HAECs), can promote the expression of TGFβR2 and its downstream phospho-SMAD3 by sponging miR-370, leading to inhibition of the proliferation, migration, and capillary-like formation of HAECs exposed to ox-LDL (21). Another study revealed that circ_0124644 exacerbates ox-LDL-induced HUVECs injury through the miR-149-5p/PAPP-A axis (25). In addition, circ-USP36 can bind miR-98-5p, which results in increased levels of VCAM1, thus accelerating ox-LDL-induced apoptosis, inflammatory and viability inhibition of HUVEC cells (26).
VSMCs reside in the tunica media and maintain vascular tension. Abnormal proliferation of VSMCs accelerates the progression of atherosclerosis. A previous study reported that circACTA2 competitively binds miR-548f-5p and upregulates smooth muscle α-actin (α-SMA) expression, thereby facilitating stress fiber formation and cell contraction in human arterial smooth muscle cells (HASMCs) (27). α-SMA participates in the formation of filaments that are major components of the cytoskeleton which is essential for VSMC proliferation and migration (27). Another study reported that circ_Lrp6 competes endogenously by binding to miR-145, thus regulating VSMC migration, proliferation, and differentiation (22). More recently, it has been reported that CircMap3k5 functions as a miRNA sponge to decrease miR-22-3p repression of TET2 (13). As a consequence, CircMap3k5 inhibits the proliferation of smooth muscle cells (SMCs) and attenuates intimal hyperplasia (13). ox-LDL can promote the growth, migration, and differentiation of VSMCs, leading to VSMC dysfunction (28). Studies have shown that hsa_circ_0029589 and circ_0010283 are up-regulated in ox-LDL-induced VSMCs (29–32). Knockdown of these circRNAs ameliorates the dysfunction of VSMCs induced by ox-LDL and they exert these biological functions by serving as ceRNAs. VSMCs may preserve phenotype alterations from a highly quiescent and differentiated phenotype (contractile phenotype) to a proliferative and dedifferentiated phenotype (secretory phenotype) (33). The VSMC phenotypic switching is an early event in AS. circ_RUSC2, circ-SATB2, circDHCR24 and circ-Sirt1 have been found to regulate the phenotypic transformation of VSMCs by functioning as ceRNAs (11, 34–36). These circRNAs may be targets for regulating VSMC phenotypic modulation and serve as potential diagnostic biomarkers and therapeutic targets for AS.
Myocardial infarction (MI), also called acute MI, is characterized by cardiac injury and remodeling caused by a sudden loss of oxygen supply due to acute coronary artery occlusion. Apoptosis has shown great contribution to cardiomyocyte loss during acute MI. Several studies have shown that circRNAs are involved in the regulation of cardiomyocyte apoptosis (37–41). The mitochondrial fission and apoptosis-related circRNA (MFACR) can mediate cardiomyocyte apoptosis and MI by exerting an endogenous competitive effect and regulating the expression of MTP18 (37). circNCX1 is increased in response to ROS and promotes the expression of the pro-apoptosis factor CDIP1 by targeting miR-133a-3p, eventually leading to the death of cardiomyocytes (38). Cdr1as is upregulated in MI mice, and promotes myocardial cell apoptosis and increases cardiac infarct size by targeting the Cdr1as/miR-7a axis (39). Conversely, circMACF1 can attenuate cardiomyocyte apoptosis through the miR-500b-5p-EMP1 axis (40). circ-Ttc3, as a miR-15b-5p sponge, promotes the expression of Arl2 and plays a cardioprotective role in MI (41).
The stimulation of endogenous cardiac regeneration is a promising approach to replace the lost myocardium after MI. Cardiomyocyte proliferation and angiogenesis are essential for structural and functional repair during cardiac regeneration. Gao et al. demonstrated that circFASTKD1 binds directly to miR-106a and relieve its inhibition of LATS1 and LATS2, thereby suppressing the YAP signaling pathway (42). The downregulation of circFASTKD1 ameliorates MI by promoting angiogenesis through the miR-106a-LATS1/2-YAP axis (42). Another study showed that circHipk3 acts as a sponge for miR-133a to promote CTGF expression (43). Upregulation of circHipk3 induces cardiac regeneration and angiogenesis, and decreases the infarct size after MI (43). Huang et al. clarified that circNfix can sponge miR-214 to promote Gsk3β expression and that the knockdown of circNfix promotes cardiomyocyte proliferation and angiogenesis after MI, attenuating cardiac dysfunction and improving prognosis (44).
Cardiac fibrosis is defined as the replacement of normal myocardial tissues with non-beating fibrous tissues caused by the hyperfunction of fibroblasts and differentiation into myofibroblasts. It is a common pathological feature of the most adverse ventricular remodeling, such as MI and diabetic cardiomyopathy. Some circRNAs have been reported to exert pro-fibrosis effect in cardiac fibroblasts (CFs). circRNA_010567 and circRNA_000203 were upregulated in diabetic mouse myocardium and Ang-II-induced CFs (45, 46). circRNA_010567 can competitively bind to miR-141 and remove the inhibition of TGF-β1 by miR-141, thus increasing TGF-β1 levels (45). circRNA_000203 can competitively bind to anti-fibrotic miR-26b-5p, enhancing the fibrotic phenotype in CFs (46). Similarly, circPAN3 exhibits pro-fibrotic effects via the miR-221/FoxO3/ATG7 axis (47). Another study showed that circHIPK3 is markedly upregulated in CFs and heart tissues treated with Ang-II (48). Knockdown of circHIPK3 prevents Ang-II-induced cardiac fibrosis and improves diastolic function by sponging miR-29b-3p (48). Several circRNAs have been shown to exert anti-fibrotic effect in CFs. circNFIB, which is decreased in post-MI heart samples of mice and TGF-β-treated CFs, can directly sponge miR-433 to inhibit fibroblast proliferation (49). circ_LAS1L can inhibit the activation, proliferation, and migration of CFs by targeting the miR-125b-SFRP5 signaling axis (50). Herein, a better understanding of the molecular mechanisms that regulate cardiac fibrosis could aid the discovery of novel therapeutic approaches for the development of cardiac remodeling.
Cardiac hypertrophy is characterized by the enlargement of cardiomyocytes and thickening of the ventricular walls. It initially develops as an adaptive response to diverse patho-physiological stimuli; however, chronic sustained cardiac hypertrophy may eventually progress to heart failure (HF). Several reports have revealed that circRNAs function as ceRNAs in the development and progression of HF. Wang et al. found that overexpression of heart-related circRNA (HRCR) increases the expression of ARC by inhibiting miR-223 activity, which attenuates the development of cardiac hypertrophy and HF (51). Lim et al. found that circSlc8a1 inhibition could attenuate cardiac hypertrophy and HF by sponging miR-133a (52). It has been reported that circRNA_000203 exacerbates cardiac hypertrophy by competitively sponging miR-26b-5p and miR-140-3p to derepress Gata4, a pro-hypertrophic transcription factor (53). A recent study showed that circHIPK3 is upregulated in pressure-overload induced cardiomyocytes (54). Knockdown of circHIPK3 plays a protective role against cardiac hypertrophy by sponging miR-185-3p (54). These circRNAs hold promise as prospective therapeutic targets for HF.
Aneurysm is defined as a pathological condition featured by permanent localized dilation of the vessel wall, involving all layers. Growth impediment and apoptosis of VSMCs can contribute to aneurysm development. Recently, interactions between circRNAs and miRNAs have also been demonstrated in aneurysms. For example, hsa_circRNA_101238 can sequester miR-320a, increasing the expression of MMP9 and leading to thoracic aortic dissection (55). circ_0020397 plays an important role in the phenotypic modulation of intracranial aneurysms by promoting VSMC viability via the miR-502-5p/GREM1 axis (56). Additionally, CDR1as serves as a sponge for miR-7, which increases the expression of CKAP4 to facilitate the proliferation and suppress the apoptosis of VSMCs, leading to VSMC remodeling and progression of abdominal aortic aneurysm (AAA) (57). circCBFB facilitates VSMC proliferation and inhibits VSMC apoptosis via the circCBFB/miR-28-5p/GRIA4/LYPD3 axis (58). circCCDC66 sponges miR-342-3p to upregulate its host gene CCDC66 in VSMCs and regulates VSMC apoptosis and proliferation (59). These newly identified mechanisms may provide novel options for the treatment of aneurysm.
Stroke is a common cardiovascular and cerebrovascular disease and one of the major causes of death and disability worldwide. During the hours after an ischemic stroke, neurons become permanently damaged and undergo cell death. Therefore, novel therapeutic approaches to rescue damaged neurons are urgently needed. The latest study conducted by Dai et al. revealed that knockdown of circHECTD1 attenuates neuronal injury caused by cerebral ischemia via the miR-133b/ TRAF3 axis (60). Chen et al. revealed that circUCK2 functions as a miR-125b-5p sponge to inhibit miR-125b-5p activity, resulting in an increase in GDF11 expression and a subsequent amelioration of neuronal injury (61). Wu et al. demonstrated that circTLK1 acts as an endogenous miR-335-3p sponge to increase the expression of TIPARP, resulting in the exacerbation of neuronal injury (62). It has been shown that circSHOC2 in ischemic-preconditioned astrocyte-derived exosomes attenuates ischemia-induced neuronal apoptosis and ameliorates neuronal damage by acting on the miR-7670-3p/SIRT1 axis (63). The above circRNAs may serve as potential targets for inducing neuroprotection against ischemic stroke.
Astrocytes are the most abundant cells in the brain and play important roles in maintaining normal brain function. Studies have demonstrated that treatments capable of decreasing infarct size are often accompanied by attenuated astrocyte responses (64, 65). Han et al. found that circHECTD1 acts as an endogenous miR-142 sponge to increase the expression of TIPARP, resulting in astrocyte activation, and thus contributes to cerebral infarction (66). Specific blockage of circHECTD1 may be a potential therapeutic target for the inhibition of astrocyte activation in stroke patients.
In addition, Bai et al. firstly reported that endothelial-mesenchymal transition (EndoMT) contributes to blood-brain barrier (BBB) damage during stroke pathogenesis (67). circDLGAP4 competitively binds miR-143 and remove the inhibition of HECTD1 by miR-143, thus increasing HECTD1 levels (67). Overexpression of circDLGAP4 significantly attenuates neuronal deficits, decreases infarct areas, and ameliorates BBB damage through the inhibition of EndoMT (67).
To investigate the interaction between circRNAs and miRNAs, several bioinformatic tools can be used. For instance, different miRNA-target prediction algorithms, including TargetScan (68), miRanda (69), and PITA (70) have greatly facilitated the discovery of circRNA-miRNA interactions. In addition, some tools incorporate gene expression data including AGO crosslinking-immunprecipitation (CLIP) data, which improve the capability to predict physiologically relevant circRNA-miRNA interactions. starBase 2.0 (http://starbase.sysu.edu.cn/) is the first database containing circRNA-miRNA interactions based on Ago and RNA binding protein (RBP) binding sites (71). It provides comprehensive interaction networks of ncRNAs, mRNA and RBP from 108 CLIP-Seq datasets. CircInteractome (http://circinteractome.nia.nih.gov/) combines many features from other websites, such as circBase, TargetScan, StareBase (72). Similarly, it can be used to explore the interacting miRNAs of circRNAs. Since every database uses distinct rules of targeting and produces different prediction outcomes, a combination of various databases may provide information in a reliable way.
In addition to the above mentioned prediction methods, experimental validation is required to prove that a circRNA has a bona fide miRNA sponging function. First, dual-luciferase reporter assays can be utilized to screen the potential interacting miRNAs. The circRNA sequences containing wild-type (WT) or mutated miRNA binding sites are inserted into luciferase reporter vectors. Transfection of a miRNA mimic can reduce the luciferase activities in the WT group, but not in the mutated group. Second, RNA fluorescence in situ hybridization (RNA-FISH) can be used to assess the colocalization of circRNAs with miRNAs in cells and tissues. RNA-FISH probe should target circRNA backspliced junctions in order to avoid recognition of their cognate linear RNAs. It is important to note that RNA-FISH signals for circRNAs and miRNAs appear as distinct puncta in the cell, not as the diffuse signals typically seen when detecting protein. Third, AGO2 RNA immunoprecipitation (RIP) can be employed to confirm a physical interaction between the circRNA, miRNA and AGO2. It is a more direct evidence supporting the miRNA sponging role of a circRNA. Fourth, RNA pull-down assay can be performed using biotinylated probes designed specifically for the circRNA/miRNA of interest. After confirming that the circRNA/miRNA is successfully enriched in the pull-down material, the interacting miRNA/circRNA molecules can be studied by RT-qPCR analysis.
As discussed in this review, many circRNAs serve as ceRNAs to mediate miRNA functions in CVDs (Table 1). These circRNA-miRNA-mRNA competitive endogenous RNA regulatory networks represent a novel section in the pathogenesis and development of CVDs. As lots of circRNAs are deregulated or show a disease-specific profile in CVDs and might play a role in the progression of CVDs, they might serve as promising biomarkers for early diagnosis or therapeutic targets in the future. However, how to effectively regulate circRNA levels in target cells is now still unresolved. Extracellular vesicles are naturally occurring RNA carriers that can induce physiological changes in target cells through the transfer of numerous molecules. These vesicles can be adapted for the delivery of circRNAs or siRNAs targeting the specific backspliced sequence of circRNA. Therefore, exogenous upregulation or downregulation of circRNAs to regulate miRNAs could become a novel therapy for CVD treatment. In addition, a recent study found that artificial circRNA sponges can competitively inhibit pro-hypertrophic miR-132 and−212, thereby attenuating pressure overload-induced cardiac hypertrophy (73). Thus, introducing synthetic circRNAs sponges into target cells might also serve as new efficient approaches for future CVD therapy. Indeed, the superior ability of circRNA in sponging miRNAs and its unique cellular stability confer them great potential for use as therapeutic drugs. Nevertheless, the real clinical application of circRNAs as molecular drugs or targets is challenging. How to ensure therapeutic efficacy and safety, and avoid off-target adverse effects awaits additional and extensive investigation.
Despite growing evidence for the phenotypic effects of circRNA deregulation in CVDs, the current ceRNA hypothesis is still controversial regarding whether they are really active in the endogenous cellular context. Importantly, optimal ceRNA-mediated regulation is observed when miRNA and ceRNA levels are near equimolarity. The levels of circRNA need to be high enough to relieve the miRNA repression on mRNA. Thus, methods to quantitatively determine absolute expression levels of circRNA and miRNA will allow a more precise determination of ceRNA effectiveness of circRNAs in diverse physiological and pathological conditions. Another important factor is the number of miRNA-binding sites. CircRNAs that harbor multiple binding sites for the same miRNAs may serve as better ceRNA candidates than those with fewer sites. Among the circRNAs we discussed in this review, with the exception of MFACR (37), circSlc8a1 (52), circNCX1 (38), circLrp6 (22), HRCR (51), which contains 15, 10, 8, 7, 6 binding sites for miR-652-3p, miR-133a, miR-133a-3p, miR-145, miR-223, respectively, the majority of circRNAs harbor only a single or very few binding sites for the same miRNAs (Table 1). To function as efficient miRNA sponges, the copy numbers of these circRNAs that contain minimal numbers of miRNA-binding sites should attain levels comparable to the miRNAs. However, most of the researches are lack of assessments of absolute levels of circRNAs and miRNAs. Therefore, assessing the stoichiometry of a circRNA with the interacting miRNA is clearly needed in future research. In addition, factors such as miRNA/circRNA binding affinity and RBPs that could occupy MREs can also affect the ceRNA activity of circRNAs. The binding affinity of miRNA and its RNA target, to a great extent, is influenced by matching between MREs and seed sequences. In general, 8-mer seed match has higher affinity than those with 7-mer, and 6-mer has weakest miRNA target-site efficacy (74). As a competitor, circRNA should have a better affinity to miRNA than target mRNAs. RBP can interfere with circRNA-miRNA interactions by directly occupying RNA target sites, or by altering the circRNA secondary structure to influence its affinity for miRNAs. Therefore, technologies such as HITS-CLIP/iCLIP/PAR-CLIP, the mass-spectrometry based quantitative proteomic approaches for capturing RNA-interactome and measuring circRNA-binding protein abundance will help to assess the effect of competitive binding of RBP to MREs.
In summary, circRNAs acting as ceRNAs play important roles in CVDs. Research on the circRNA-mediated ceRNA regulatory network will open up new avenues for basic CVD research, as well as stimulate the development of novel diagnostic and therapeutic strategies for CVDs.
XM and D-lL: wrote the manuscript. X-dX: critically reviewed the manuscript. All authors contributed to the article and approved the submitted version.
The study was supported by grants from the National Natural Science Foundation of China (81871120 and 82071576), the Natural Science Foundation of Guangdong Province (2019A1515010334 and 2019KZDXM059), the discipline construction project of Guangdong Medical University (4SG21008G) and the program for Training High-level Talents of Dongguan (201901019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, et al. Heart disease and stroke statistics-2018 update: a report from the American heart association. Circulation. (2018) 137:e67–492. doi: 10.1161/CIR.0000000000000558
2. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AKJPotNAoS. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
3. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. (1979) 280:339–40. doi: 10.1038/280339a0
4. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. (1993) 7:155–60. doi: 10.1096/fasebj.7.1.7678559
5. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. doi: 10.1261/rna.035667.112
6. Wang PL, Bao Y, Yee M-C, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. (2014) 9:e90859. doi: 10.1371/journal.pone.0090859
7. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. (2014) 9:1966–80. doi: 10.1016/j.celrep.2014.10.062
8. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. (2013) 9:e1003777. doi: 10.1371/journal.pgen.1003777
9. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. (2015) 10:170–7. doi: 10.1016/j.celrep.2014.12.019
10. Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. (2017) 8:73271–81. doi: 10.18632/oncotarget.19154
11. Kong P, Yu Y, Wang L, Dou YQ, Zhang XH, Cui Y, et al. circ-Sirt1 controls NF-kappaB activation via sequence-specific interaction and enhancement of SIRT1 expression by binding to miR-132/212 in vascular smooth muscle cells. Nucleic Acids Res. (2019) 47:3580–93. doi: 10.1093/nar/gkz141
12. Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. (2020) 19:65. doi: 10.1186/s12943-020-01152-2
13. Zeng Z, Xia L, Fan S, Zheng J, Qin J, Fan X, et al. Circular RNA CircMAP3K5 acts as a MicroRNA-22-3p sponge to promote resolution of intimal hyperplasia via TET2-mediated smooth muscle cell differentiation. Circulation. (2021) 143:354–71. doi: 10.1161/CIRCULATIONAHA.120.049715
14. Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. (2007) 4:721–6. doi: 10.1038/nmeth1079
15. Brown BD, Cantore A, Annoni A, Sergi LS, Lombardo A, Della Valle P, et al. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. (2007) 110:4144–52. doi: 10.1182/blood-2007-03-078493
16. Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. (2009) 6:63–6. doi: 10.1038/nmeth.1277
17. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
18. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
19. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
20. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. (2013) 3:1113–21. doi: 10.1158/2159-8290.CD-13-0202
21. Zhang S, Song G, Yuan J, Qiao S, Xu S, Si Z, et al. Circular RNA circ_0003204 inhibits proliferation, migration and tube formation of endothelial cell in atherosclerosis via miR-370-3p/TGFbetaR2/phosph-SMAD3 axis. J Biomed Sci. (2020) 27:11. doi: 10.1186/s12929-019-0595-9
22. Hall IF, Climent M, Quintavalle M, Farina FM, Schorn T, Zani S, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res. (2019) 124:498–510. doi: 10.1161/CIRCRESAHA.118.314240
23. Chen M, Masaki T, Sawamura T. LOX-1, the receptor for oxidized low-density lipoprotein identified from endothelial cells: implications in endothelial dysfunction and atherosclerosis. Pharmacol Ther. (2002) 95:89–100. doi: 10.1016/s0163-7258(02)00236-x
24. Rueckschloss U, Galle J, Holtz J, Zerkowski HR, Morawietz H. Induction of NAD(P)H oxidase by oxidized low-density lipoprotein in human endothelial cells: antioxidative potential of hydroxymethylglutaryl coenzyme a reductase inhibitor therapy. Circulation. (2001) 104:1767–72. doi: 10.1161/hc4001.097056
25. Wang G, Li Y, Liu Z, Ma X, Li M, Lu Q, et al. Circular RNA circ_0124644 exacerbates the ox-LDL-induced endothelial injury in human vascular endothelial cells through regulating PAPP-A by acting as a sponge of miR-149-5p. Mol Cell Biochem. (2020) 471:51–61. doi: 10.1007/s11010-020-03764-0
26. Peng K, Jiang P, Du Y, Zeng D, Zhao J, Li M, et al. Oxidized low-density lipoprotein accelerates the injury of endothelial cells via circ-USP36/miR-98-5p/VCAM1 axis. IUBMB Life. (2021) 73:177–87. doi: 10.1002/iub.2419
27. Sun Y, Yang Z, Zheng B, Zhang XH, Zhang ML, Zhao XS, et al. A novel regulatory mechanism of smooth muscle alpha-actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circ Res. (2017) 121:628–35. doi: 10.1161/CIRCRESAHA.117.311441
28. Pirillo A, Norata GD, Catapano AL. LOX-1, OxLDL, and atherosclerosis. Mediators Inflamm. (2013) 2013:152786. doi: 10.1155/2013/152786
29. Huang Z, Li P, Wu L, Zhang D, Du B, Liang C, et al. Hsa_circ_0029589 knockdown inhibits the proliferation, migration and invasion of vascular smooth muscle cells via regulating miR-214-3p and STIM1. Life Sci. (2020) 259:118251. doi: 10.1016/j.lfs.2020.118251
30. Yang L, Yang F, Zhao H, Wang M, Zhang Y. Circular RNA circCHFR facilitates the proliferation and migration of vascular smooth muscle via miR-370/FOXO1/Cyclin D1 pathway. Mol Ther Nucleic Acids. (2019) 16:434–41. doi: 10.1016/j.omtn.2019.02.028
31. Zhuang JB, Li T, Hu XM, Ning M, Gao WQ, Lang YH, et al. Circ_CHFR expedites cell growth, migration and inflammation in ox-LDL-treated human vascular smooth muscle cells via the miR-214-3p/Wnt3/beta-catenin pathway. Eur Rev Med Pharmacol Sci. (2020) 24:3282–92. doi: 10.26355/eurrev_202003_20696
32. Feng Z, Zhu Y, Zhang J, Yang W, Chen Z, Li B. Hsa-circ_0010283 regulates oxidized low-density lipoprotein-induced proliferation and migration of vascular smooth muscle cells by targeting the miR-133a-3p/pregnancy-associated plasma protein A axis. Circ J. (2020) 84:2259–69. doi: 10.1253/circj.CJ-20-0345
33. Owens GKKM, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. (2004) 84:767–801. doi: 10.1152/physrev.00041.2003
34. Sun J, Zhang Z, Yang S. Circ_RUSC2 upregulates the expression of miR-661 target gene SYK and regulates the function of vascular smooth muscle cells. Biochem Cell Biol. (2019) 97:709–14. doi: 10.1139/bcb-2019-0031
35. Mao YY, Wang JQ, Guo XX, Bi Y, Wang CX. Circ-SATB2 upregulates STIM1 expression and regulates vascular smooth muscle cell proliferation and differentiation through miR-939. Biochem Biophys Res Commun. (2018) 505:119–25. doi: 10.1016/j.bbrc.2018.09.069
36. Peng W, Li T, Pi S, Huang L, Liu Y. Suppression of circular RNA circDHCR24 alleviates aortic smooth muscle cell proliferation and migration by targeting miR-149-5p/MMP9 axis. Biochem Biophys Res Commun. (2020) 529:753–9. doi: 10.1016/j.bbrc.2020.06.067
37. Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. (2017) 24:1111–20. doi: 10.1038/cdd.2017.61
38. Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, et al. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. (2018) 8:5855–69. doi: 10.7150/thno.27285
39. Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on Its target genes expression. PLoS ONE. (2016) 11:e0151753. doi: 10.1371/journal.pone.0151753
40. Zhao B, Li G, Peng J, Ren L, Lei L, Ye H, et al. CircMACF1 attenuates acute myocardial infarction through miR-500b-5p-EMP1 axis. J Cardiovasc Transl Res. (2021) 14:161–72. doi: 10.1007/s12265-020-09976-5
41. Cai L, Qi B, Wu X, Peng S, Zhou G, Wei Y, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol. (2019) 130:10–22. doi: 10.1016/j.yjmcc.2019.03.007
42. Gao WQ, Hu XM, Zhang Q, Yang L, Lv XZ, Chen S, et al. Downregulation of circFASTKD1 ameliorates myocardial infarction by promoting angiogenesis. Aging. (2020) 13:3588–604. doi: 10.18632/aging.202305
43. Si X, Zheng H, Wei G, Li M, Li W, Wang H, et al. circRNA Hipk3 induces cardiac regeneration after myocardial infarction in mice by binding to notch1 and miR-133a. Mol Ther Nucleic Acids. (2020) 21:636–55. doi: 10.1016/j.omtn.2020.06.024
44. Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. (2019) 139:2857–76. doi: 10.1161/CIRCULATIONAHA.118.038361
45. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun. (2017) 487:769–75. doi: 10.1016/j.bbrc.2017.04.044
46. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. (2017) 7:40342. doi: 10.1038/srep40342
47. Li F, Long TY, Bi SS, Sheikh SA, Zhang CL. circPAN3 exerts a profibrotic role via sponging miR-221 through FoxO3/ATG7-activated autophagy in a rat model of myocardial infarction. Life Sci. (2020) 257:118015. doi: 10.1016/j.lfs.2020.118015
48. Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol. (2019) 292:188–96. doi: 10.1016/j.ijcard.2019.04.006
49. Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao L, et al. Upregulation of circular RNA CircNFIB attenuates cardiac fibrosis by sponging miR-433. Front Genet. (2019) 10:564. doi: 10.3389/fgene.2019.00564
50. Sun LY, Zhao JC, Ge XM, Zhang H, Wang CM, Bie ZD. Circ_LAS1L regulates cardiac fibroblast activation, growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem Funct. (2020) 38:443–50. doi: 10.1002/cbf.3486
51. Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. (2016) 37:2602–11. doi: 10.1093/eurheartj/ehv713
52. Lim TB, Aliwarga E, Luu TDA, Li YP, Ng SL, Annadoray L, et al. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res. (2019) 115:1998–2007. doi: 10.1093/cvr/cvz130
53. Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan R, et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res. (2020) 116:1323–34. doi: 10.1093/cvr/cvz215
54. Xu X, Wang J, Wang X. Silencing of circHIPK3 inhibits pressure overload-induced cardiac hypertrophy and dysfunction by sponging miR-185-3p. Drug Des Devel Ther. (2020) 14:5699–710. doi: 10.2147/DDDT.S245199
55. Zou M, Huang C, Li X, He X, Chen Y, Liao W, et al. Circular RNA expression profile and potential function of hsa_circRNA_101238 in human thoracic aortic dissection. Oncotarget. (2017) 8:81825–37. doi: 10.18632/oncotarget.18998
56. Yin K, Liu X. Circ_0020397 regulates the viability of vascular smooth muscle cells by up-regulating GREM1 expression via miR-502-5p in intracranial aneurysm. Life Sci. (2021) 265:118800. doi: 10.1016/j.lfs.2020.118800
57. Zhao F, Chen T, Jiang N. CDR1as/miR-7/CKAP4 axis contributes to the pathogenesis of abdominal aortic aneurysm by regulating the proliferation and apoptosis of primary vascular smooth muscle cells. Exp Ther Med. (2020) 19:3760–6. doi: 10.3892/etm.2020.8622
58. Yue J, Zhu T, Yang J, Si Y, Xu X, Fang Y, et al. CircCBFB-mediated miR-28-5p facilitates abdominal aortic aneurysm via LYPD3 and GRIA4. Life Sci. (2020) 253:117533. doi: 10.1016/j.lfs.2020.117533
59. Yang R, Wang Z, Meng G, Hua L. Circular RNA CCDC66 facilitates abdominal aortic aneurysm through the overexpression of CCDC66. Cell Biochem Funct. (2020) 38:830–8. doi: 10.1002/cbf.3494
60. Dai Q, Ma Y, Xu Z, Zhang L, Yang H, Liu Q, et al. Downregulation of circular RNA HECTD1 induces neuroprotection against ischemic stroke through the microRNA-133b/TRAF3 pathway. Life Sci. (2021) 264:118626. doi: 10.1016/j.lfs.2020.118626
61. Chen W, Wang H, Feng J, Chen L. Overexpression of circRNA circUCK2 attenuates cell apoptosis in cerebral ischemia-reperfusion injury via miR-125b-5p/GDF11 signaling. Mol Ther Nucleic Acids. (2020) 22:673–83. doi: 10.1016/j.omtn.2020.09.032
62. Wu F, Han B, Wu S, Yang L, Leng S, Li M, et al. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J Neurosci. (2019) 39:7369–93. doi: 10.1523/JNEUROSCI.0299-19.2019
63. Chen W, Wang H, Zhu Z, Feng J, Chen L. Exosome-shuttled circSHOC2 from IPASs regulates neuronal autophagy and ameliorates ischemic brain injury via the miR-7670-3p/SIRT1 axis. Mol Ther Nucleic Acids. (2020) 22:657–72. doi: 10.1016/j.omtn.2020.09.027
64. Wang W, Redecker C, Yu ZY, Xie MJ, Tian DS, Zhang L, et al. Rat focal cerebral ischemia induced astrocyte proliferation and delayed neuronal death are attenuated by cyclin-dependent kinase inhibition. J Clin Neurosci. (2008) 15:278–85. doi: 10.1016/j.jocn.2007.02.004
65. Fang SH, Wei EQ, Zhou Y, Wang ML, Zhang WP, Yu GL, et al. Increased expression of cysteinyl leukotriene receptor-1 in the brain mediates neuronal damage and astrogliosis after focal cerebral ischemia in rats. Neuroscience. (2006) 140:969–79. doi: 10.1016/j.neuroscience.2006.02.051
66. Han B, Zhang Y, Zhang Y, Bai Y, Chen X, Huang R, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. (2018) 14:1164–84. doi: 10.1080/15548627.2018.1458173
67. Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. (2018) 38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017
68. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. (2009) 19:92–105. doi: 10.1101/gr.082701.108
69. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. (2004) 2:e363. doi: 10.1371/journal.pbio.0020363
70. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. (2007) 39:1278–84. doi: 10.1038/ng2135
71. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. (2014) 42:D92–7. doi: 10.1093/nar/gkt1248
72. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. (2016) 13:34–42. doi: 10.1080/15476286.2015.1128065
73. Lavenniah A, Luu TDA, Li YP, Lim TB, Jiang J, Ackers-Johnson M, et al. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol Ther. (2020) 28:1506–17. doi: 10.1016/j.ymthe.2020.04.006
Keywords: circRNA, ceRNA, cardiovascular and cerebrovascular diseases, molecular mechanism, clinical implication
Citation: Min X, Liu D-l and Xiong X-d (2021) Circular RNAs as Competing Endogenous RNAs in Cardiovascular and Cerebrovascular Diseases: Molecular Mechanisms and Clinical Implications. Front. Cardiovasc. Med. 8:682357. doi: 10.3389/fcvm.2021.682357
Received: 18 March 2021; Accepted: 15 June 2021;
Published: 07 July 2021.
Edited by:
En-Zhi Jia, Nanjing Medical University, ChinaReviewed by:
Annadoray Lavenniah, National University of Singapore, SingaporeCopyright © 2021 Min, Liu and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xing-dong Xiong, eGlvbmd4aW5nZG9uZ0AxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.