- 1Laboratory of Rescue Center of Severe Wound and Trauma Chinese People's Liberation Army, Department of Cardiovascular Surgery, General Hospital of Northern Theater Command of China Medical University, Shenyang, China
- 2Laboratory of Rescue Center of Severe Wound and Trauma Chinese People's Liberation Army, Emergency Medicine Department of General Hospital of Northern Theater Command, Shenyang, China
- 3The Second Affiliated Hospital of Shenyang Medical College, The Veterans General Hospital of Liaoning Province, Shenyang, China
- 4Shenyang Medical College, Shenyang, China
Circular RNA (circRNA) is a subclass of non-coding RNAs that enables the circular transcripts resistant to the exonuclease digestion. Iron homeostasis is essential for the body to maintain normal physiological functions. At present, the relationship among circRNA, iron metabolism and heart failure remains largely unknown. This study aimed to explore the regulatory mechanism of circRNA and iron metabolism in heart failure. We obtained circRNA, miRNA and mRNA data from public databases and built a ceRNA network. The prediction results were verified in the myocardial tissues of pressure overload-induced heart failure mice through the use of histopathological staining methods, iron and malondialdehyde (MDA) measurement tests, quantitative real-time PCR (qRT-PCR), Western blot analysis and luciferase reporter assay. A total of 4 genes related to iron metabolism and oxidative stress were identified, and a ceRNA network involving 7 circRNAs, 7 miRNAs, and 4 mRNAs was constructed using bioinformatics tools. The results of qRT-PCR and Western blot analyses indicated that the expression level of FTH1 was similar with that predicted by bioinformatics analysis. Echocardiographic measurement showed that heart failure mice have lower fractional shortening and ejection fraction. Moreover, the myocardium of heart failure mice displayed obvious fibrosis as well as increased levels of iron and MDA compared to control mice. Besides, circSnx12 could act as an endogenous sponge to bind with miR-224-5p, and the 3'UTR region of FTH1 also had miRNA binding sites. A circRNA-miRNA-mRNA regulatory network was successfully constructed by identifying differentially expressed genes related to iron metabolism. This new approach reveals potential circRNA targets for the treatment of heart failure.
Introduction
Heart failure (HF) has become a major healthcare burden worldwide, as it is one of the leading causes of morbidity and mortality globally. HF can be caused by myocardial infarction, myocarditis and hypertension. Prior research has made great progress in understanding the pathophysiological mechanisms of HF, but little is known about its gene-level alterations. Recent findings suggest that non-coding RNA can play a vital role in mediating the pathological process of HF. Thus, in-depth exploration of the molecular mechanisms underlying HF pathogenesis can provide new ideas for optimal HF therapy and gain new insights into HF biomarkers and drug development (1). At present, the common animal model of HF is transverse aortic constriction (TAC). By increasing the cardiac afterload, the myocardium of the mice becomes damaged and scarred, ultimately resulting in fibrotic. Hence, in this study, the expression profiling datasets of TAC-induced HF animal model were used for subsequent analysis. This model is the same as the modeling method in the datasets.
Circular RNA (circRNA) is a new type of non-coding RNA that has been discovered in plant viruses (2), and is generated by direct back-splicing of exons via covalent bonding. The lack of 5′ cap and 3′ poly A makes circRNA more resistant to exonuclease digestion, suggesting its better stability compared to linear RNA. Previous studies shown that circRNAs were involved in the pathogenesis of cardiovascular diseases, including vascular disease (3), atherosclerosis (4), cardiomyopathy (5, 6) and HF (7, 8). CircRNAs not only exhibit dynamic expression patterns in a series of physiological and pathological conditions, but also play significant roles in regulating gene expression and transcription (9–11). Recent decades have witnessed the great progress of miRNAs as one of the most significant molecular targets for studying the mechanisms of cardiovascular disease. David Port et al. (12) found that a number of miRNAs are differentially regulated in the failing heart, including miR-224. Annalisa Angelini et al. (13) obtained more than 2257 mature miRNA from endomyocardial biopsies. They found that miRNA profiling on formalin-fixed paraffin-embedded endomyocardial biopsies differentiates form different types of heart transplant rejection. Most endogenous circRNAs have conserved miRNA target sites that competitively bind miRNAs to mRNAs, thereby preventing miRNA-mediated post-translational inhibition and regulating the expression levels of target genes (14). Recent circRNA findings have provided new insights into the mechanisms of miRNA regulation in a broader cellular context. The circRNA CDR1 was the first circRNA identified to function as a miRNA sponge, which has 74 binding sites for miR-7 (15).
Iron is an essential element for various enzymatic reactions such as DNA replication and repair, cellular respiration and energy metabolism (16, 17). In the heart, systemic iron homeostasis is essential for maintaining normal cardiac function. However, excess iron in the cardiomyocytes can result in an uncontrolled condition called iron overload, which ultimately lead to adverse effects. At the cellular level, iron overload can increase the levels of reactive oxygen species (ROS), thus generating oxidative damage to large molecules such as DNA, membrane lipids and proteins (18). In addition, iron overload disrupts the dynamic balance between mitochondrial division and fusion in cells (19). Therefore, assessing the changes in these RNA molecules is critical to understanding the pathophysiological mechanisms of iron-induced HF. We constructed a circRNA-miRNA-mRNA network based on competitive endogenous RNA (ceRNA), which may be associated with iron overload during HF, by means of bioinformatics and experimental approaches.
Materials and Methods
Data Acquisition
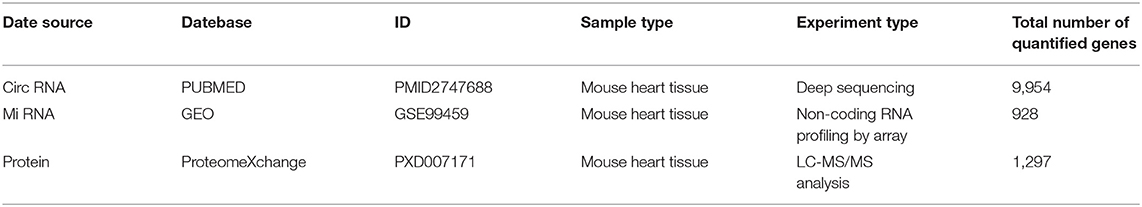
Gene Expression Omnibus (GEO) is a public functional genomic database. We searched HF-related expression profiling datasets through GEO database and selected the miRNA array data of HF mice induced by pressure overload. MiRNA dataset (GSE99459) was downloaded from GEO, and the corresponding platform annotation information were also retrieved to convert the probe information into the corresponding gene name. Rudebusch et al. (20) obtained the protein expression profiles of HF through mass spectrometry (MS), and the raw date are available via ProteomeXchange with identifier PXD007171. Werfel et al. (21) obtained the circRNA expression data of HF through deep sequencing techniques and the raw date were published on PUBMED with identifier PMID27476877 (Table 1).
Identification of Differential Expression Profiles of circRNAs, miRNAs, and mRNAs
Raw data were downloaded from GEO and PUBMED database. “Limma” package in R software was used to identify the differentially expressed miRNAs with thresholds of |logFC|>1 and P < 0.05. Both mRNA and circRNA data were extracted from the research results of Rudebusch et al. (20) and Werfel et al. (21), respectively, which had been uploaded to PUBMED.
GO Functional Enrichment Analysis
UNIPROT (https://uniprot.org/) is publicly accessible database that consists of various biological resources and analysis tools. It also contains gene annotation and protein sequence data. To gain insights into the function of differentially expressed mRNAs in the network, UNIPROT database was employed for GO enrichment analysis. The significance level was set at P < 0.05. The R software packages ggplot2 and GOplot were used to visualize the results of the enrichment analysis.
Prediction of circRNA-miRNA and miRNA-mRNA Binding Sites
Subsequently, TargetScan and RegRNA 2.0 database was utilized for assessing the regulatory relationships between miRNAs and mRNAs. Given that the miRNAs belonging to the same family may have similar seed sequences, only one miRNA ID was retained for each family group. To construct a ceRNA-based network, we screened miRNA-mRNAs with reverse expression relationships, as miRNAs often negatively regulate mRNA expression (22). Next, starBase database was used to predict the regulatory relationship between circRNAs and miRNAs. It is worth noting that the circRNA primarily functions as a miRNA sponge and its expression level may not affect miRNA expression. Finally, a circRNA-miRNA-mRNA regulatory network was established using Cytoscape 3.7.2 software based on the interactions of both miRNA-mRNA and circRNA-miRNA groups.
Animal Models
The animal experiment protocol was approved by the ethics committee of the General Hospital of Shenyang Military Region. Six-week-old male C57/BL6J mice (weighting ~18–20 g) were supplied by the Shenyang Military Region General Hospital Experimental Animal Center. The mice were fed a standard diet and had unlimited access to drinking water. After 2 weeks of feeding, the mice were randomly assigned to TAC group (n = 15) and SHAM group (n = 16), and then anesthetized by intraperitoneal injection of 50 mg/kg pentobarbital sodium. To allow direct access to the transverse aorta, the mice a horizontal incision (5 mm in length) was made at the suprasternal notch. TAC operation was then performed by ligating the aorta between the right innominate and left carotid arteries using a 27G needle tied with 7-0 silk suture. The needle was promptly withdrawn, leaving the aortic constriction in place. The surgical procedure of mice in SHAM group was similar to that of mice in TAC group, except that the 7-0 silk suture was only crossed through the aortic arch without ligation.
Echocardiographic Assessment
An echocardiogram was conducted on mice at 6 weeks post-TAC following anesthesia with 2.0% isoflurane and 1 L/min oxygen. Transthoracic echocardiographic images (two-dimensional and M-mode) were acquired using an ultrasound system (Vevo 2100, VisualSonics) equipped with a high-frequency (30 MHz) transducer. To evaluate left ventricular fractional shortening and ejection fraction, M-mode measurements were carried out in the short-axis view.
Histopathological Analysis
Cardiac tissues were collected from 6 mice in each group, fixed in 10% neural-buffered formalin, embedded in paraffin, and then sectioned at 4-μm thickness. After hematoxylin and eosin (HE) staining, the tissue sections were examined using an optical microscope (OLYMPUS) at × 200 magnification. To evaluate myocardial fibrosis, the tissue sections were stained with a Masson's trichrome staining kit (BASO, BA-4079). Myocardium was stained red, nucleus were stained black and collagen fibers were stained blue. The collagen volume fraction was determined as the percentage of collagen (blue-stained area) to the total heart tissue area. Wheat germ agglutinin (WGA; GeneTex, GTX01502) staining was carried out to assess the size of the cardiomyocytes. All data were processed and analyzed by ImageJ software.
Immunohistochemistry Analysis
Immunohistochemical staining was performed using an immunohistochemistry kit (MXB, KIT-9710). To inhibit endogenous peroxidase, the tissue sections were first exposed to 1% H2O2 and then subjected to heat-induced antigen retrieval at 97°C for 15 min. After incubation with GPX4, ACSL4, MYH6, and MYH7 primary antibodies for 1 h, the sections were rinsed with phosphate-buffered solution and incubated again with the corresponding secondary antibodies. Finally, the sections were stained with 3,3-diaminobenzidine (DAB; MXB, DAB-0031) and counterstained with hematoxylin, followed by microscopic examination. The cells with brownish-yellow particles in their nuclei and/or cytoplasm were classified as immunoreactive cells.
Evaluation of Malondialdehyde (MDA) and Iron Concentrations
The levels of MDA and iron in mouse heart tissues were measured using the Lipid Peroxidation (MDA) Assay Kit (ab118970, Abcam) and Iron Assay Kit (MAK025, Sigma-Aldrich), respectively, according to the manufacturer's instructions.
qRT-PCR Analysis
The myocardial tissues of 5 mice in each TAC group and SHAM group were used for qRT-PCR experiments. Total RNA was extracted from the myocardial tissues using Trizol reagent (Invitrogen), and cDNA was then synthesized using a First Strand cDNA synthesis kit (Geneseed). Quantitative RT-PCR analysis was conducted on a Real-Time PCR System (Applied Biosystems) using SYBR Green Master Mix (Geneseed), cDNA template and specific primers (Table 2). β-actin was used to normalize circRNA and mRNA expression, while U6 (also known as RNU6) was used to normalize miRNA expression. In the majority of miRNA expression studies, miRNAs or other small RNAs were used as reference genes (23). It is questionable as to whether mRNAs can be used reference genes in miRNA expression studies, because longer RNAs may have different isolation efficiencies compared to miRNAs, they may be degraded faster due to their length (24). So U6, as a snRNA, was used to mormalize miRNA expression. All reactions were performed in triplicate, and the relative expression levels of the target genes were determined by 2−ΔΔCt method.
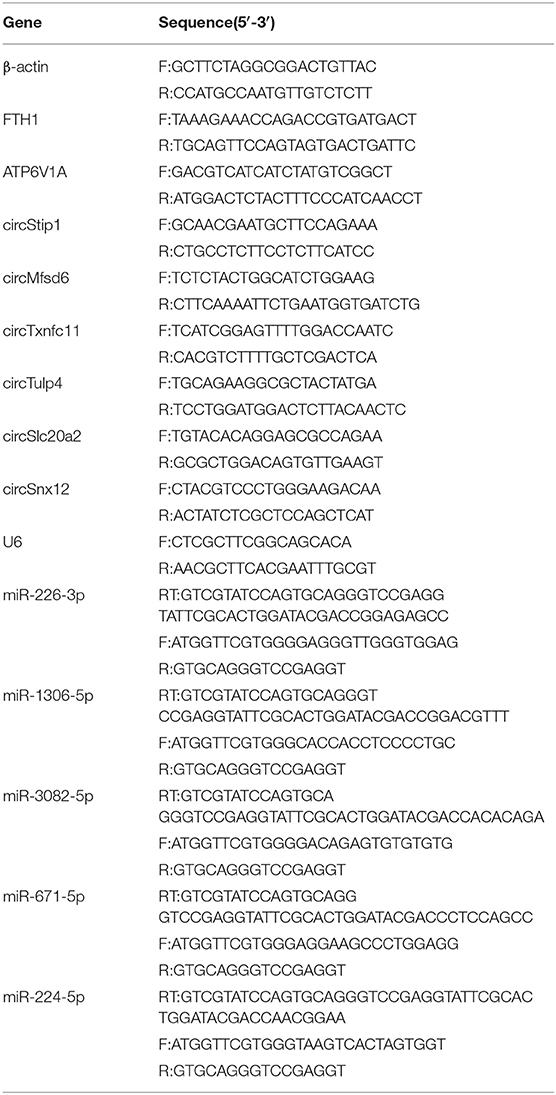
Table 2. Primer sequences of circRNA, miRNA, mRNA, and internal reference used for validation by qRT-PCR.
Western Blotting
The myocardial tissues were lysed with lysis buffer containing using protease and phosphatase inhibitors. Total protein content was assessed using BCA reagent. Equivalent amounts of protein extract were separated on SDS-PAGE, and subsequently transferred onto PVDF membranes. After blocking with 5% skim milk for 1 h, the membranes were incubated with primary antibodies such as ANP (Abcam, ab180649), GPX4 (Abcam, ab125066), NOX1 (Abcam, ab131088), ACSL4 (Abcam, ab155282), FTH1 (Abcam, ab183781), ATP6V1A (Abcam, ab199326), and GAPDH (Cell Signaling, 2118) overnight at 4°C. After washing, the membranes were incubated with HRP-conjugated secondary antibodies and visualized using an enhanced chemiluminescence substrate kit (Bio-Rad Laboratories).
Dual-Luciferase Reporter Assay
To construct luciferase reporter plasmids, the fragments of FTH1 3'UTR and circSnx12 as well their mutant sequences were synthesized, and then introduced into a psiCHECK-2.0 vector. HL-1 cell line was cultured in 24-well plates and transfected with miR-224-5p mimic/luciferase reporter construct or negative control miRNA construct. After transfection for 48 h, the cells were quenched, and the luciferase reporter activity was determined using a dual-luciferase reporter assay system. Renilla luciferase was used as an internal normalization standard.
Statistical Analysis
All results were presented as mean ± SD. Statistical differences between two groups were compared using the Student's t-test. A p < 0.05 was deemed statistically significant. All statistical tests were carried out by GraphPad Prism V. 6.0 and SPSS V. 26.0.
Results
Differential Expression Profiles of circRNAs, miRNAs, and mRNAs
According to Julia Rudebusch's research (20), after TAC at day 28, the expression levels of 94 transcripts were altered in the left ventricle. Of them, 63 were up-regulated and 31 were down-regulated. In addition, Stanislas Werfel found that there were 96 up-regulated and 10 down-regulated circRNAs in the heart tissues of TAC mice as compared to SHAM mice. In this study, we downloaded miRNA expression datasets from GEO (GSE99459), and identified that 280 miRNAs were differentially expressed, including 183 up-regulated and 97 down-regulated miRNAs (Figure 1A, Supplementary Table 1).
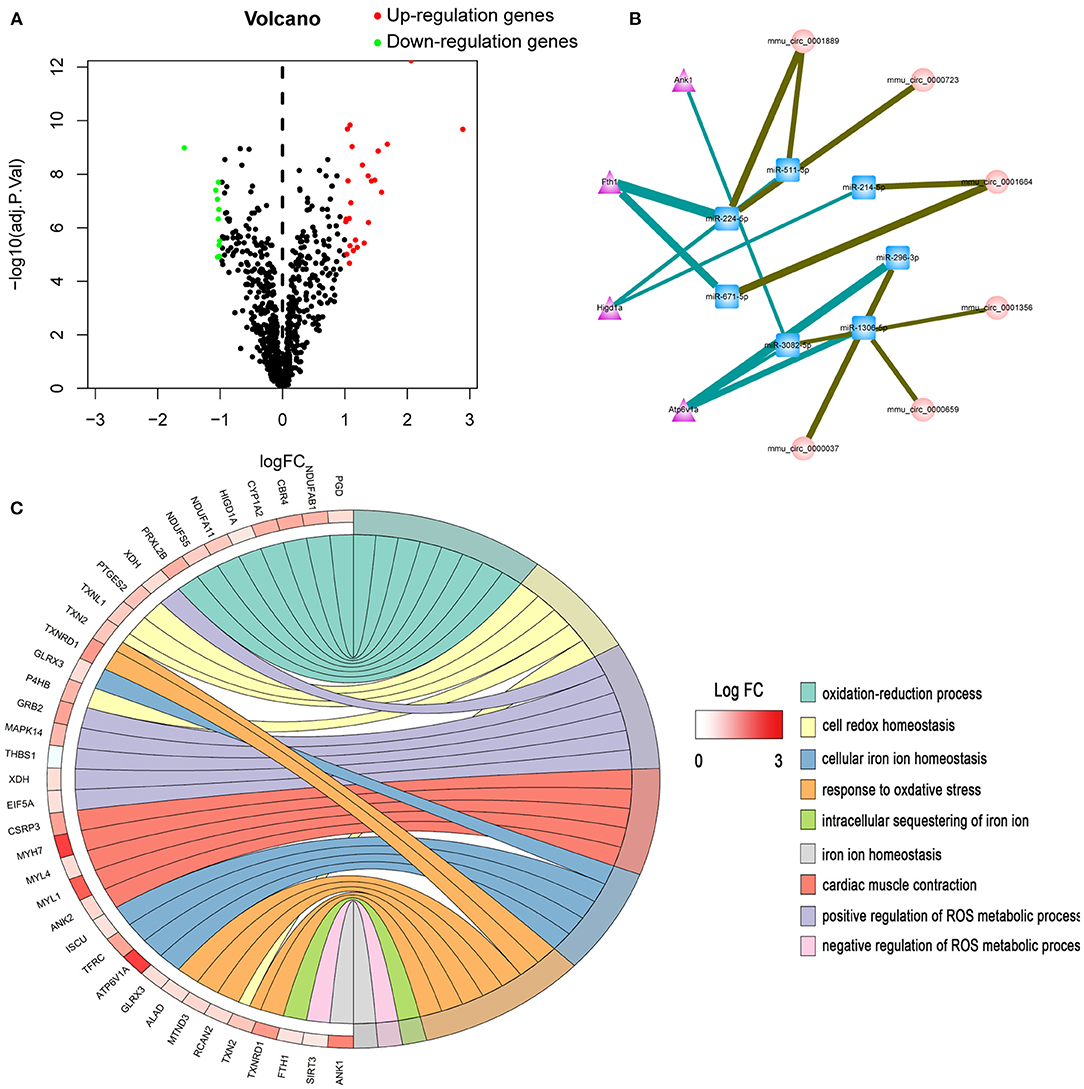
Figure 1. Bioinformatics analysis of differentially expressed genes. (A) Volcano plot of differentially expressed miRNAs. (B) The circRNA-miRNA-mRNA competing endogenous RNA network. The triangle nodes represent mRNAs in purple, circular nodes indicate circRNAs in pink, and square nodes represent miRNAs in blue. The thicker the edge, the higher the weight score of the interaction. (C) GO enrichment analysis of differentially expressed genes.
Screening of ceRNAs Associated With Iron Metabolism
Ferroptosis is an iron-dependent form of regulated cell death. Dysregulation of iron metabolism can lead to iron deficiencies and, eventually, human diseases. First, the mRNAs related to iron metabolism and oxidative stress were screened. GO enrichment analysis was performed on all differentially expressed mRNAs. In total, 37 mRNAs related to iron metabolism and oxidative stress were screened out (Figure 1C, Supplementary Table 2), among which 11 proteins showed significant changes within 28 days after TAC operation. Next, the most significantly differentially expressed genes (ATP6V1A, FTH1, HIGD1A, and ANK1) were chosen for target gene prediction. Based on the results of miRNA-mRNA interactions predicted by TargetScan and RegRNA 2.0, 7 differential expressed miRNAs and 4 differential expressed mRNAs were significantly interacted with each other. The results of circRNA-miRNA network prediction by starBase demonstrated that 7 circRNAs were interacting with the above miRNAs. Based on these findings, we constructed a circRNA-miRNA-mRNA network by using Cytoscape 3.8.0. A total of 7 circRNAs, 7 miRNAs and 4 mRNAs were included in the network (Figure 1B).
TAC Mice Display Myocardial Injury and Fibrosis
In this experiment, we randomly divided C57/BL6J mice into two groups, namely TAC and SHAM. Echocardiographic measurement showed that TAC remarkably decreased both fractional shortening and ejection fraction in the left ventricle of HF mice (Figures 2A,B). The heart of the TAC group was larger than the heart of the SHAM group (Figure 2C). Meanwhile, chronic TAC significantly increased the left ventricular mass of HF mice (Figure 2E). Western blotting and immunohistochemical staining were carried out to identify the myocardial hypertrophy factors in mouse heart tissues. It was observed that the expression levels of ANP and MYH7 were markedly higher (P < 0.05) in TAC group than in SHAM group (Figures 3A–E). HE staining showed that cell necrosis and inflammatory cell infiltration were observed in the heart tissues of HF mice induced by pressure overload. Masson staining and WGA staining showed that the mice in TAC group had obvious myocardial fibrosis and myocyte hypertrophy (Figures 2D,F,G).
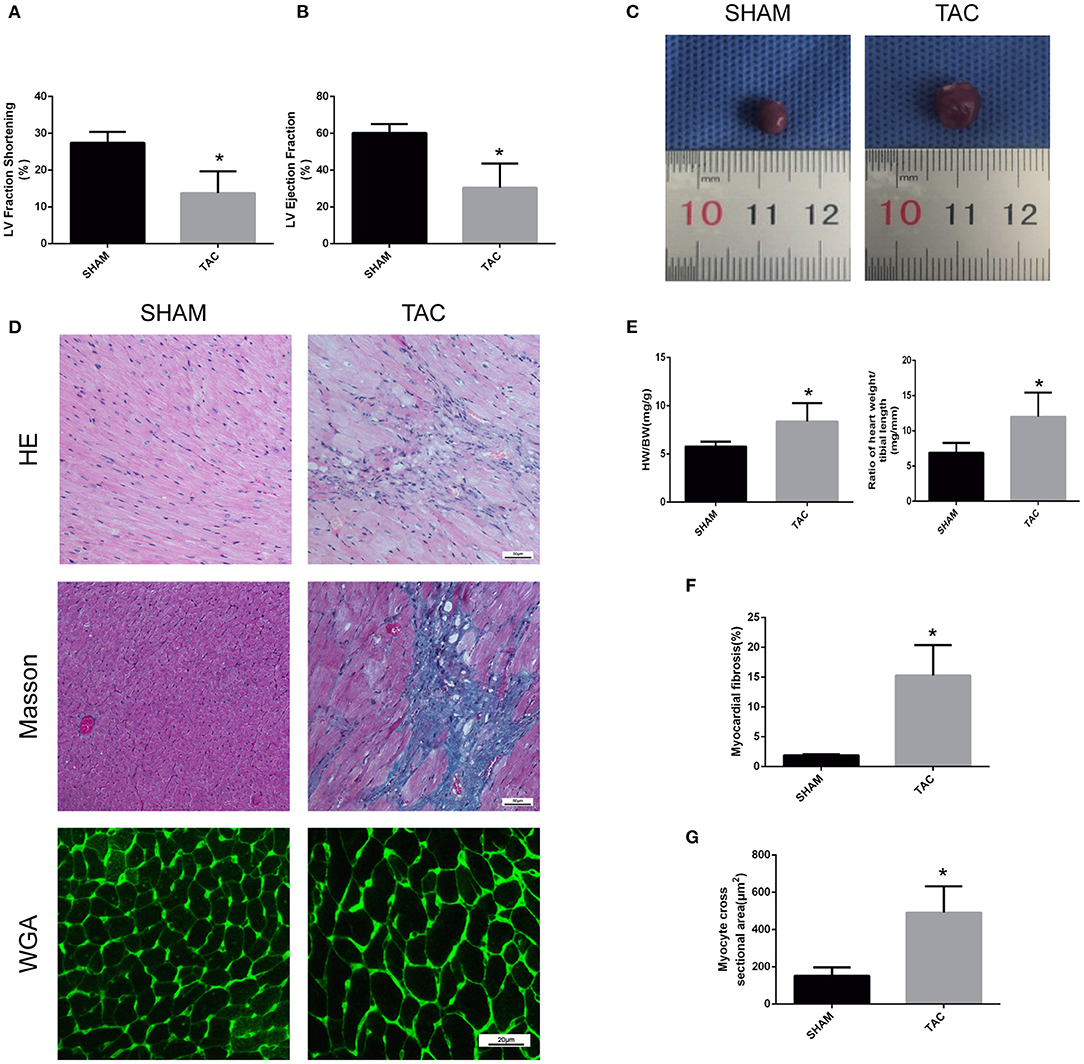
Figure 2. Functional and pathological analysis of heart tissues in TAC mice. (A,B) Left ventricular fraction shorting and ejection fraction. (C) Heart size. (D) Representative HE- and Masson-stained myocardial sections. WGA-stained myocardial sections were used to measure myocyte cross sectional area. (E) The heart weight/body weight and heart weight/tibia length ratios of TAC mice. (F) Quantitative data of myocardial fibrosis (%). (G) Quantitative data of myocyte cross sectional areas. *P < 0.05.
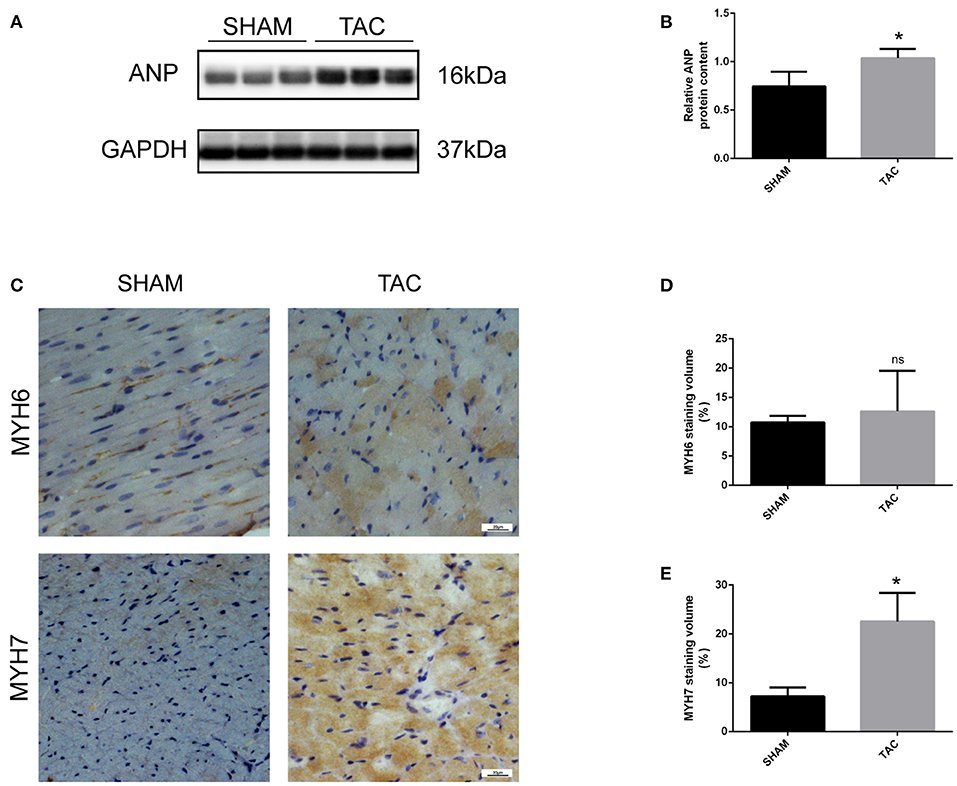
Figure 3. Western blotting and immunohistochemical analysis of TAC mice. (A,B) Representative Western blot image and quantitative data of ANP in myocardial tissues. (C–E) Representative immunohistochemical staining images and quantitative data of MYH6 and MYH7 in myocardial tissues. *P < 0.05; ns, non-significant.
Ferroptosis of Heart Tissue Is Induced by TAC
Western blot and immunohistochemistry analyses were performed to identify the regulatory factors of ferroptosis in mouse heart tissues and to determine the occurrence of ferroptotic-like death in TAC-induced HF mice. As shown in Figures 4A–D, the expression of GPX4 was decreased and the levels of ACSL4 and NOX1 were increased in TAC group when compared to SHAM group (P < 0.05). Furthermore, colorimetric assays were used to detect the content of ferrous ion and MDA in the mouse cardiomyocytes. Notably, the levels of ferrous and MDA were significantly higher in TAC group than in SHAM group (Figures 4E,F). This indicates that the myocardial tissues of mice in TAC surgery group are not only overloaded with iron, but also have increased lipid peroxidation.
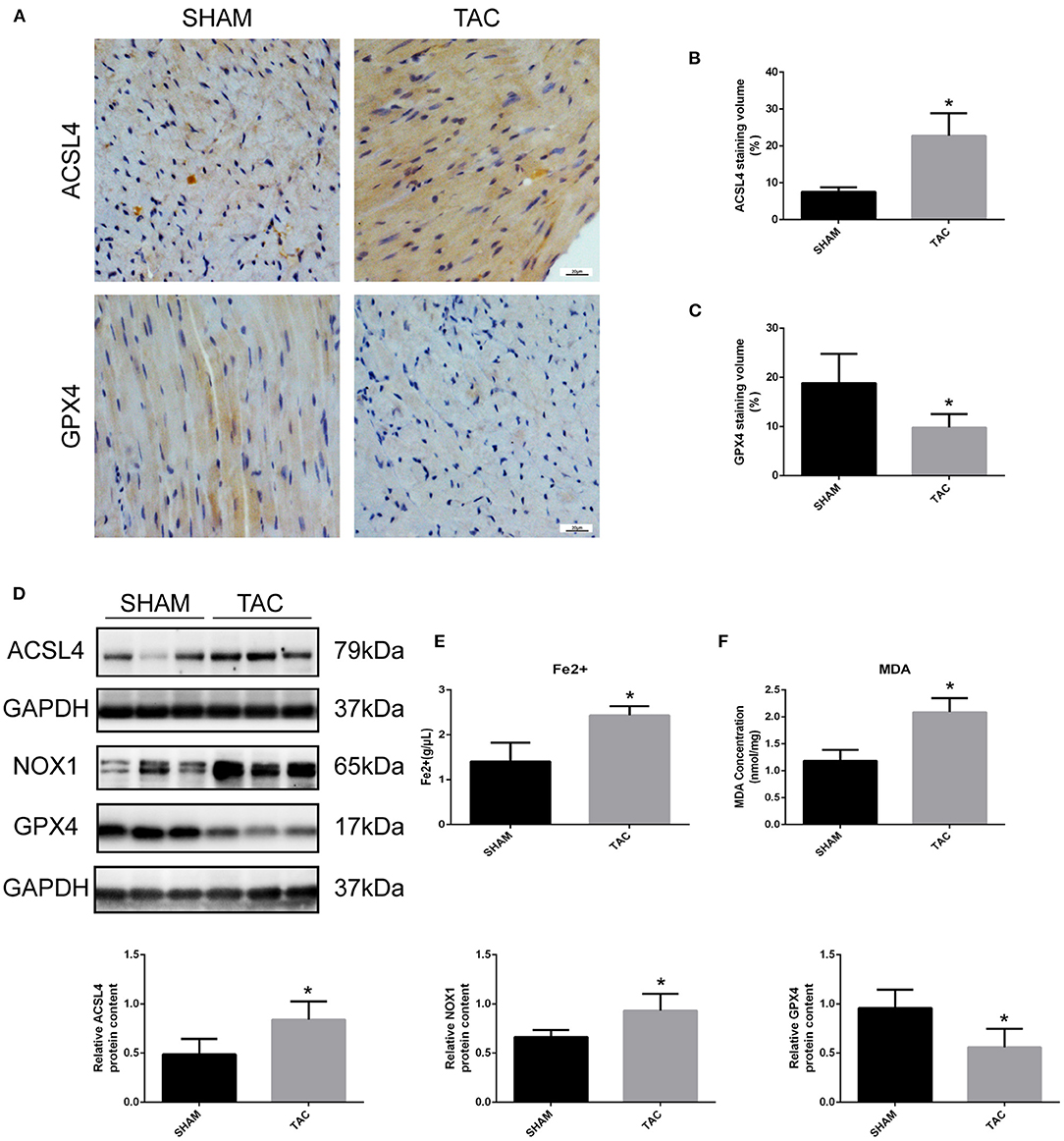
Figure 4. Detection of ferroptosis-associated genes in TAC mice. (A–C) Representative immunohistochemical staining images and quantitative data of ACSL4 and GPX4 in myocardial tissues. (D) Representative Western blot image and quantitative data of ACSL4, NOX1 and GPX4 in myocardial tissues. The concentrations of ferrous ion (E) and MDA (F) in TAC mouse heart tissues. *P < 0.05.
Analysis of the Candidate Genes Through qRT-PCR and Western Blot
Among the four candidate genes, we selected two ferroptosis-related genes (ATP6V1A and FTH1) for qRT-PCR and Western blot detection. In addition, 5 miRNAs and 7 circRNAs that may interact with ATP6V1A and FTH1 in the ceRNA network were selected for qRT-PCR verification. We found that FTH1 was down-regulated, as consistent with the target prediction results (Figure 5A). miR-3082-5p was down-regulate in the TAC group (Figure 6A), but miR-1306-5p and miR-224-5p was up-regulate (Figures 6B,E). There was no significant difference in the expression of miR-296-3p between the two groups (Figure 6D). In addition, circSLC20A2, circTULP4 and circSNX12, which may interact with FTH1, were also significantly down-regulated (Figures 6H,J,K). However, the expression pattern of miR-671-5p was contrary to the prediction results (Figure 6C). Moreover, the result of ATP6V1A expression was also contrary to the prediction results (Figure 5B), and the circRNAs (circSTIP1, circMFSD6, circTXNDC11 and circFRYL) that may interact with ATP6V1A were also significantly down-regulated (Figures 6F,G,I,L). Western blot analysis was performed to further confirm the protein levels of FTH1 and ATP6V1A. We found that the expression levels of FTH1 and ATP6V1A were lower in TAC group than in SHAM group (Figures 5C–F), which were consistent with the qRT-PCR data.
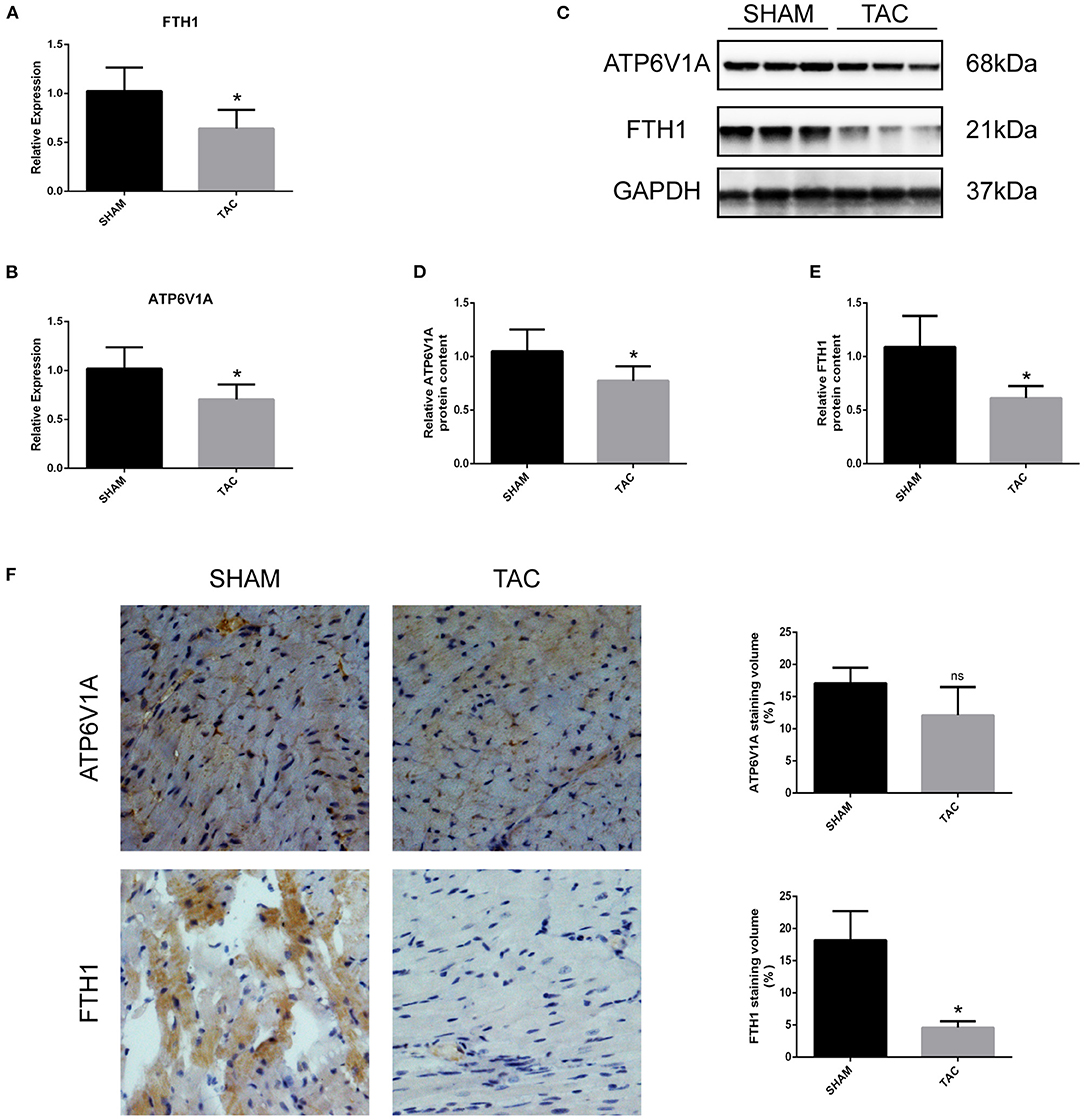
Figure 5. QRT-PCR and Western blot analyses of ATH6V1A and FTH1 in myocardial tissues. (A,B) Representative qRT-PCR results of FTH1 and ATP6V1A in myocardial tissues. (C–F) Representative Western blot images, immunohistochemistry, and quantitative data of ATP6V1A and FTH1 in myocardial tissues. *P < 0.05; ns, non-significant.
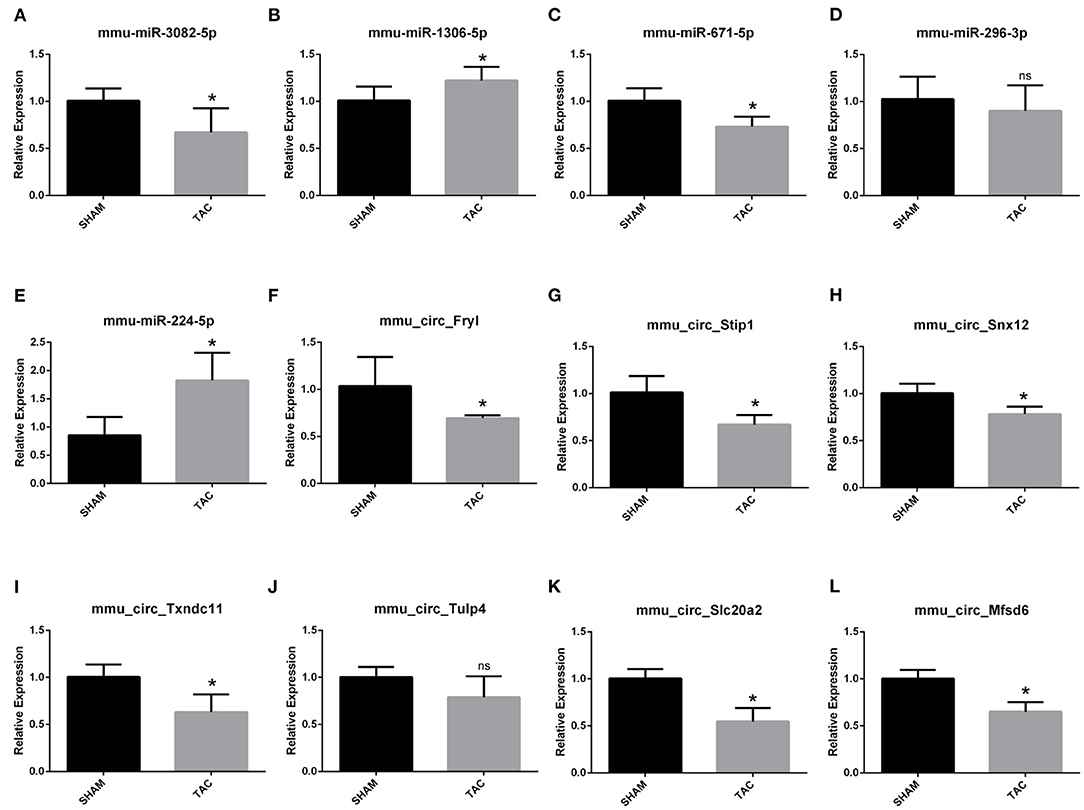
Figure 6. QRT-PCR validation of the differentially expressed circRNAs and miRNAs. The expression results of miRNAs (A–E) and circRNAs (F–L). *P < 0.05; ns, non-significant.
miR-224-5p Directly Targets FTH1 and Interacts With circSnx12
Furthermore, we assessed the binding relationship between the ferroptosis-associated gene FTH1 and its target miR-224-5p. The results of luciferase reporter assay showed that miR-224-5p mimics could reduce the luciferase activity of FTH1 3′-UTR, but did not significantly affected that of FTH1 3′-UTR mutant (Figure 7A). These findings imply that FTH1 is a direct target of miR-224-5p. Additionally, we demonstrated the regulatory relationship between miR-224-5p and circSnx12. Sequence analysis revealed that there was a potential binding site for miR-224-5p in circSnx12 sequences. Consequently, wild-type and mutant circSnx12 were cloned into the psiCHECK2.0 vector and then co-transfected into HL-1 cells with miR-224-5p mimic or negative controls. The luciferase activity levels was remarkably lower in HL-1 cells co-transfected with miR-224-5p mimic and circSnx12 than in control group (Figure 7B). These findings suggest that miR-224-5p can directly interact with circSnx12.
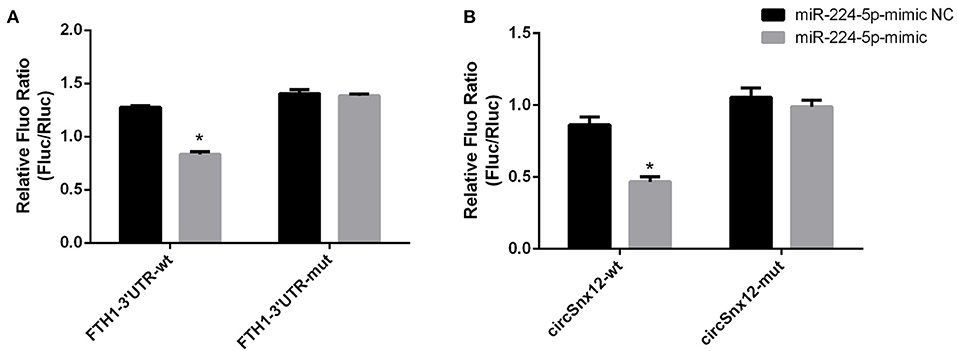
Figure 7. Dual-luciferase reporter activities. The interrelationships of miR-224-5p with FTH1 (A) and circSnx12 (B) were confirmed using dual-luciferase reporter assays. *P < 0.05.
Discussion
The iron content in the mitochondria of cardiomyocytes is higher than that of other cells. Therefore, it is important to maintain the iron homeostasis of the mitochondria of cardiomyocytes (25). Iron overload can impair heart function by affecting cardiac mitochondrial dynamics and promoting cardiac mitochondrial deterioration (26). Chen et al. (27) indicated that cardiac cell death during HF can be reduced by inhibiting autophagy and ferroptosis. Through the use of HF mouse model induced by pressure overload, we found that the iron content of myocardial cells was noticeably increased compared to SHAM group. Moreover, the iron metabolism- and ferroptosis-related genes, such as FTH1, GPX4, NOX1, and ACSL4, were also differentially expressed. GPX4 is considered to be the primary enzyme that prevents ferroptosis (28). The decreased expression of GPX4 and increased expression of NOX1 and ACSL4 indicate that lipid peroxidation occurs in cardiomyocytes during HF, and the increase in MDA content also proves this phenomenon. Excessive iron accumulation in cardiomyocytes can generate ROS through Fenton reaction, thereby sensitizing cardiomyocytes to ferroptosis. In this study, we found that the expression levels of iron metabolism-associated genes were associated with HF, and these genes might interact with circRNAs and miRNAs. Moreover, we observed that ATP6V1A expression was down-regulated in the myocardium of TAC mice compared to SHAM mice, which appeared to be contrary to the bioinformatics prediction results. ATP6V1A is a catalytic subunit of the peripheral V1 complex of vacuolar ATPase that participates in systemic pH regulation in both lysosome and endosome (29). Lowering the pH of the endosome by ATP6V1A can release iron ions into the cytoplasm, thus disrupting systemic iron homeostasis in the body (30). Hence, downregulated ATP6V1A expression may be a sign of lysosomal acidification disorder. A recent study (31) showed that a decrease in V-ATPase-mediated lysosomal acidification could lead to the deposition of enlarged non-functional lysosomes and complete blockage of autophagic flux, which in turns causes damage to heart muscle cells. Another study (32) found that the abnormal expression of APT6V1A could dysregulate the function of V-ATPase in myocardial ischemia/reperfusion injury model, which in turn leads to myocardial injury and cell death. ROS could affect the function of lysosomes, thus leading to the accumulation of dysfunctional mitochondria and ultimately cell death (32). Nevertheless, the relationship between ATP6V1A and HF needs to be further verified by experiments. More importantly, we found that FTH1 was markedly down-regulated in TAC mice. FTH1 is an important component of ferritin. As a selective ferritin autophagic degradation process, ferritinophagy is also responsible for the regulation of systemic iron homeostasis and other iron-dependent pathophysiological processes (33, 34). Ferritin heavy chain (FHC) is involved in the first step of iron storage by catalyzing the ferroxidation of Fe2+; while ferritin light chain (FLC) enables the storage of Fe3+ by promoting the nucleation of ferrihydrite (35). Sun et al. (34) reported that the suppression of ferritinophagy by NCOA4 or Atg5/Atg7 siRNA could prevent pUL38-deficient human cytomegalovirus infection-induced cell apoptosis. Accumulating data have indicated that ferritinophagy is required for the induction of ferroptosis (36, 37). In this study, we found that the expression of FTH1 is repressed during HF, which in turn releases a large amount of ferrous ions into the cytoplasm. It has also been reported that excessive ferrous ions can increase the levels of ROS through the Fenton reaction, and radically attack cardiomyocyte mitochondria and cell membrane. Lipid peroxidation, as evidenced by the increased content of MDA, can contribute to cardiomyocyte dysfunction and ultimately cell death.
Growing evidence have suggested that circRNAs are important molecular regulators for HF pathogenesis. CircRNAs can act as miRNA sponges to counteract miRNA-mediated repression of mRNA, and thus interfering with its mRNA-targeting activity. However, it is unclear why linear RNA does not have a similar sponge function. CircRNAs can bind to several miRNAs or contain multiple binding sites for a single miRNA (38). In this study, we aimed to establish a putative regulatory network involving circRNAs, miRNAs and mRNAs in an animal model of HF induced by TAC. GO functional enrichment analysis revealed the potential roles of non-coding RNAs and coding RNAs in advanced HF. We first identified a protein associated with iron metabolism (FTH1), predicted that miR-224-5p might bind to FTH1 3'UTR by TargetScan and RegRNA 2.0 database, and further determined that miR-224-5p could be a potential binding target for circSnx12 through starBase database. The results of qRT-PCR verification indicated that circSnx12, miR-224-5p and FTH1 were correlated with each other, and double luciferase assay revealed that miR-224-5p was indeed the downstream target of circSnx12, and miR-224-5p could bind to the 3'UTR region of FTH1 and regulate its expression level. Therefore, it is speculated that low circSnx12 expression and high miR-224-5p expression can downregulate FTH1 expression, directly regulate iron overload in myocardial cells, and ultimately leads to cardiac cell death.
The changes in gene/protein expression can affect cell phenotypes. In this study, we determined the expression levels of proteins related to iron metabolism and oxidative stress, predicted the non-coding RNAs that interact with the target gene/proteins, constructed a ceRNA-based network, and performed experimental verification. A significant interrelationship among circRNA, miRNA and mRNA was identified in HF mice. However, the main limitation of our study was that we did not detect the expression level of circSnx12 that may affect miR-224-5p expression. In the near future, we will verify the effects of circSnx12 and miR-224-5p overexpression or knockdown on iron overload-induced mitochondrial abnormalities and myocardial cell death. In addition, we will prove the interaction between CircRNA and AGO2 by RNA pull-down and RNA IP to support that the revealed circRNAs can act as sponges of miRNAs.
Conclusions
In summary, we successfully constructed a circRNA-miRNA-mRNA regulatory network through identification of iron metabolism-associated genes and further conducted experimental verification using TAC mouse model. This new approach reveals potential circRNA targets for the treatment of HF.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at: PUBMED(PMID2747688); GEO(GSE99459); ProteomeXchange(PXD007171).
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of the General Hospital of Northern Theater Command.
Author Contributions
HZ collected the tissue sample, analyzed the data, and wrote the manuscript. LS and CT helped with histopathological staining. YL made the revision of the manuscript. MH made the revision of the manuscript and designed the research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by PLA foundation of China (No. BWS16J010; No. AWS14L008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors grateful for having free access to the GEO database. We would like to express our gratitude to EditSprings (https://www.editsprings.com/) for the expert linguistic services provided.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2021.656093/full#supplementary-material
References
1. Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan R, et al. Circular rna circrna_000203 aggravates cardiac hypertrophy via suppressing mir-26b-5p and mir-140-3p binding to gata4. Cardiovasc Res. (2019) 116:1323–34. doi: 10.1093/cvr/cvz215
2. Kolakofsky D. Isolation and characterization of sendai virus di-RNAS. Cell. (1976) 8:547–55. doi: 10.1016/0092-8674(76)90223-3
3. Boeckel JN, Jaé, AW Heumüller, Chen W, Dimmeler S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circulat Res. (2015) 117:884–90. doi: 10.1161/CIRCRESAHA.115.306319
4. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding rna anril modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. (2016) 7:12429. doi: 10.1038/ncomms12429
5. Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. (2017) 136:1629–42. doi: 10.1161/CIRCULATIONAHA.117.029004
6. Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by depressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. (2017) 7:40342. doi: 10.1038/srep40342
7. Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. (2017) 24:1111–20. doi: 10.1038/cdd.2017.61
8. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. (2017) 7:3842–55. doi: 10.7150/thno.19764
9. Han J, Gao L, Dong J, Bai J, Zhang M, Zheng J. The expression profile of developmental stage-dependent circular RNA in the immature rat retina. Mol Vis. (2017) 23:457–69.
10. Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. (2018) 52:743–54. doi: 10.3892/ijo.2018.4265
11. Wang M, Su P, Liu Y, Zhang X, Yan J, An X, et al. Abnormal expression of circRNA_089763 in the plasma exosomes of patients with post-operative cognitive dysfunction after coronary artery bypass grafting. Mol Med Rep. (2019) 20:2549–62. doi: 10.3892/mmr.2019.10521
12. Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: functional correlates. J Mol Cell Cardiol. (2008) 45:185–92. doi: 10.1016/j.yjmcc.2008.04.014
13. Di Francesco A, Fedrigo M, Santovito D, Natarelli L, Castellani C, De Pascale F, et al. MicroRNA signatures in cardiac biopsies and detection of allograft rejection. J Heart Lung Transplant. (2018) 37:1329–40. doi: 10.1016/j.healun.2018.06.010
14. Hall IF, Climent M, Quintavalle M, Farina FM, Schorn T, Zani S, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res. (2019) 124:498–510. doi: 10.1161/CIRCRESAHA.118.314240
15. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
16. Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry. (2012) 51:5705–24. doi: 10.1021/bi300752r
17. Loréal O, Cavey T, Bardou-Jacquet E, Guggenbuhl P, Ropert M, Brissot P. Iron, hepcidin, and the metal connection. Front Pharmacol. (2014) 5:128. doi: 10.3389/fphar.2014.00128
18. Anderson ER, Shah YM. Iron homeostasis in the liver. Compr Physiol. (2013) 3:315–30. doi: 10.1002/cphy.c120016
19. Khamseekaew J, Kumfu S, Wongjaikam S, Kerdphoo S, Jaiwongkam T, Srichairatanakool S, et al. Effects of iron overload, an iron chelator and a T-type calcium channel blocker on cardiac mitochondrial biogenesis and mitochondrial dynamics in thalassemic mice. Eur J Pharmacol. (2017) 799:118–27. doi: 10.1016/j.ejphar.2017.02.015
20. Rüdebusch J, Benkner A, Poesch A, Dörr M, Völker U, Grube K, et al. Dynamic adaptation of myocardial proteome during heart failure development. PLoS ONE. (2017) 12:e0185915. doi: 10.1371/journal.pone.0185915
21. Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. (2016) 98:103–7. doi: 10.1016/j.yjmcc.2016.07.007
22. Lin X, Chen Y. Identification of potentially functional CircRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit Basic Res. (2018) 24:70–8. doi: 10.12659/MSMBR.909737
23. Schaefer A, Jung M, Miller K, Lein M, Kristiansen G, Erbersdobler A, et al. Suitable reference genes for relative quantification of miRNA expression in prostate cancer. Exp Mol Med. (2010) 42:749–58. doi: 10.3858/emm.2010.42.11.076
24. Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. (2008) 14:844–52. doi: 10.1261/rna.939908
25. Wofford JD, Chakrabarti M, Lindahl PA. Mössbauer spectra of mouse hearts reveal age-dependent changes in mitochondrial and ferritin iron levels. J Biol Chem. (2017) 292:5546–54. doi: 10.1074/jbc.M117.777201
26. Sumneang N, Siri-Angkul N, Kumfu S, Chattipakorn SC, Chattipakorn N. The effects of iron overload on mitochondrial function, mitochondrial dynamics, and ferroptosis in cardiomyocytes. Arch Biochem Biophys. (2020) 680:108241. doi: 10.1016/j.abb.2019.108241
27. Chen X, Xu S, Zhao C, Liu B. Role of TLR4/NADPH oxidase 4 pathway in promoting cell death through autophagy and ferroptosis during heart failure. Biochem Biophys Res Commun. (2019) 516:37–43. doi: 10.1016/j.bbrc.2019.06.015
28. Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. (2017) 171:273–85. doi: 10.1016/j.cell.2017.09.021
29. Chen R, Jäättelä M, Liu B. Lysosome as a central hub for rewiring PH homeostasis in tumors. Cancers. (2020) 12:2437. doi: 10.3390/cancers12092437
30. Miles AL, Burr SP, Grice GL, Nathan JA. The vacuolar-ATPase complex and assembly factors, TMEM199 and CCDC115, control HIF1α prolyl hydroxylation by regulating cellular iron levels. Elife. (2017) 6:e22693. doi: 10.7554/eLife.22693
31. Mauvezin C, Nagy P, Juhász G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. (2015) 6:7007. doi: 10.1038/ncomms8007
32. Ma X, Liu H, Murphy JT, Foyil SR, Godar RJ, Abuirqeba H, et al. Regulation of the transcription factor EB-PGC1α axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress. Mol Cell Biol. (2015) 35:956–76. doi: 10.1128/MCB.01091-14
33. Tang M, Chen Z, Wu D, Chen L. Ferritinophagy/ferroptosis: Iron-related newcomers in human diseases. J Cell Physiol. (2018) 233:9179–90. doi: 10.1002/jcp.26954
34. Sun Y, Bao Q, Xuan B, Xu W, Pan D, Li Q, et al. Human cytomegalovirus protein pUL38 prevents premature cell death by binding to ubiquitin-specific protease 24 and regulating iron metabolism. J Virol. (2018) 92:e00191–18. doi: 10.1128/JVI.00191-18
35. Li JY, Paragas N, Ned RM, Qiu A, Viltard M, Leete T, et al. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev Cell. (2009) 16:35–46. doi: 10.1016/j.devcel.2008.12.002
36. Masaldan S, Clatworthy SAS, Gamell C, Meggyesy PM, Rigopoulos AT, Haupt S, et al. Iron accumulation in senescent cells is coupled with impaired ferritinophagy and inhibition of ferroptosis. Redox Biol. (2018) 14:100–15. doi: 10.1016/j.redox.2017.08.015
37. Doll S, Conrad M. Iron and ferroptosis: a still ill-defined liaison. IUBMB Life. (2017) 69:423–34. doi: 10.1002/iub.1616
Keywords: heart failure, RNA, untranslated, computational biology, ferroptosis
Citation: Zheng H, Shi L, Tong C, Liu Y and Hou M (2021) circSnx12 Is Involved in Ferroptosis During Heart Failure by Targeting miR-224-5p. Front. Cardiovasc. Med. 8:656093. doi: 10.3389/fcvm.2021.656093
Received: 21 January 2021; Accepted: 25 March 2021;
Published: 21 April 2021.
Edited by:
Christian Schulte, University Heart and Vascular Center Hamburg (UHZ), GermanyReviewed by:
Konstantinos Theofilatos, King's College London, United KingdomDonato Santovito, Hospital of the University of Munich, Germany
Copyright © 2021 Zheng, Shi, Tong, Liu and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunen Liu, bHllOTkwMUAxNjMuY29t; Mingxiao Hou, aG91bWluZ3hpYW8xODhAMTYzLmNvbQ==
 Haoyuan Zheng
Haoyuan Zheng Lin Shi2
Lin Shi2 Changci Tong
Changci Tong Yunen Liu
Yunen Liu