
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cardiovasc. Med. , 04 December 2020
Sec. Atherosclerosis and Vascular Medicine
Volume 7 - 2020 | https://doi.org/10.3389/fcvm.2020.584987
This article is part of the Research Topic What do we know about COVID-19 implications for cardiovascular disease? View all 109 articles
 Jia Teng Sun1,2†
Jia Teng Sun1,2† Zhongli Chen3†
Zhongli Chen3† Peng Nie1,2†
Peng Nie1,2† Heng Ge1,2
Heng Ge1,2 Long Shen1,2
Long Shen1,2 Fan Yang1,2
Fan Yang1,2 Xiao Long Qu1,2
Xiao Long Qu1,2 Xiao Ying Ying1,2
Xiao Ying Ying1,2 Yong Zhou1,2
Yong Zhou1,2 Wei Wang1,2
Wei Wang1,2 Min Zhang1,2
Min Zhang1,2 Jun Pu1*
Jun Pu1*Background: Emerging studies have described and analyzed epidemiological, clinical, laboratory, and radiological features of COVID-19 patients. Yet, scarce information is available regarding the association of lipid profile features and disease severity and mortality.
Methods: We conducted a prospective observational cohort study to investigate lipid profile features in patients with COVID-19. From 9 February to 4 April 2020, a total of 99 patients (31 critically ill and 20 severely ill) with confirmed COVID-19 were included in the study. Dynamic alterations in lipid profiles were recorded and tracked. Outcomes were followed up until 4 April 2020.
Results: We found that high-density lipoprotein-cholesterol (HDL-C) and apolipoprotein A-1 (apoA-1) levels were significantly lower in the severe disease group, with mortality cases showing the lowest levels (p < 0.0001). Furthermore, HDL-C and apoA-1 levels were independently associated with disease severity (apoA-1: odds ratio (OR): 0.651, 95% confidence interval (CI): 0.456–0.929, p = 0.018; HDL-C: OR: 0.643, 95% CI: 0.456–0.906, p = 0.012). For predicting disease severity, the areas under the receiver operating characteristic curves (AUCs) of HDL-C and apoA-1 levels at admission were 0.78 (95% CI, 0.70–0.85) and 0.85 (95% CI, 0.76–0.91), respectively. For in-hospital deaths, HDL-C and apoA-1 levels demonstrated similar discrimination ability, with AUCs of 0.75 (95% CI, 0.61–0.88) and 0.74 (95% CI, 0.61–0.88), respectively. Moreover, patients with lower serum concentrations of apoA-1 (<0.95 g/L) or HDL-C (<0.84 mmol/l) had higher mortality rates during hospitalization (log-rank p < 0.001). Notably, levels of apoA-1 and HDL-C were inversely proportional to disease severity. The survivors of severe cases showed significant recovery of apoA-1 levels at the end of hospitalization (vs. midterm apoA-1 levels, p = 0.02), whereas the mortality cases demonstrated continuously lower apoA-1 levels throughout hospitalization. Correlation analysis revealed that apoA-1 and HDL-C levels were negatively correlated with both admission levels and highest concentrations of C-reactive protein and interleukin-6.
Conclusions: Severely ill COVID-19 patients featured low HDL-C and apoA-1 levels, which were strongly correlated with inflammatory states. Thus, low apoA-1 and HDL-C levels may be promising predictors for severe disease and in-hospital mortality in patients suffering from COVID-19.
As Coronavirus Disease 2019 (COVID-19) continues to spread worldwide, millions of people across hundreds of countries have been impacted. Epidemiological data show that although most cases are mild, severely ill patients rapidly progress to acute respiratory disease, multi-organ failure, and septic shock, with a remarkably increased mortality rate. Therefore, early identification of risk factors for COVID-19 severity and progression is of great importance.
Mounting evidence suggests that an impaired immune function and hyper-inflammatory response are characteristics of COVID-19 severity and mortality (1–3). Systemic inflammation and sepsis are prevalent metabolic disorders accompanying severe COVID-19 (4). Furthermore, proteome analysis suggests that patients with severe COVID-19 display dysregulated lipid metabolism (5). Dyslipidemia is associated with damage to the immune, respiratory, and cardiovascular systems, along with high levels of proinflammatory cytokines. Furthermore, dyslipidemia is casually associated with increased risk of thrombotic complications, endothelial dysfunction, and higher platelet activity (6). Thus, lipid dysregulation may contribute to morbidity and mortality from COVID-19 infection. However, the characteristics and dynamic changes in lipid profiles in COVID-19 patients, as well as their predictive value in disease severity and mortality, remain largely unknown.
Here, we performed an observational cohort study to investigate the lipid profile features of patients with COVID-19 and illuminate the associations between lipid features and disease severity/mortality.
This observational cohort study prospectively included 99 COVID-19-confirmed inpatients treated from 9 February to 4 April 2020 in Leishenshan Hospital, an urgently constructed hospital designated for COVID-19 patients located in Wuhan, China. All patients were diagnosed with COVID-19 according to interim guidance provided by the World Health Organization (WHO) (7). COVID-19 severity was classified according to the Guidelines on the Diagnosis and Treatment of COVID-19 released by the National Health Commission of China (version 7). Criteria for severe cases included any of the following: (1) respiratory rate ≥ 30 per min; (2) blood oxygen saturation (SPO2) ≤ 93% at rest; (3) partial pressure of arterial oxygen to fraction of inspired oxygen ratio <300; (4) more than 50% of lung infiltrates within 24–48 h; or (5) patients needing mechanical respiratory support or presenting with septic shock or multi-organ dysfunction or failure. All patients had a definite outcome (discharged, continued treatment, deceased) before data analysis.
Time from symptom onset to hospitalization and length of hospital stay were recorded. All epidemiological, clinical, laboratory, and outcome data were collected with standardized data collection forms from the electronic medical records system at Leishenshan Hospital. Personal history, including comorbidities, was confirmed with patients or family members. For information not available from the electronic medical records, researchers also communicated directly with patients or their families to obtain additional epidemiological and symptom data. Lipid profiles, including total cholesterol (TC), triglycerides (TG), low-density lipoprotein-cholesterol (LDL-C), high-density lipoprotein-cholesterol (HDL-C), apolipoprotein A-1 (apoA-1), and apolipoprotein B (apoB), were first determined within 24 h of admission. A subset of patients had multiple lipid and cytokine metrics (i.e., collected more than once); therefore, these data were included for longitudinal analysis. Dynamic alterations in the above indicators were recorded. The Sequential Organ Failure Assessment (SOFA) score (https://www.mdcalc.com/sequential-organ-failure-assessmment-sofa-score) were calculated for each participant on admission. Two researchers independently reviewed the forms to double-check the data collected.
Outcomes were followed up until 4 April 2020. The primary outcome in the study was defined as in-hospital death.
No preliminary sample size calculation was evaluated, considering the observational nature of our study about this emerging infectious disease. Continuous variables were expressed as medians with interquartile ranges (IQR) and compared using unpaired Student's t-test or Mann-Whitney U test. Categorical data were expressed as absolute values and percentages and were compared using chi-square or Fisher's exact tests. Univariate and multivariable analyses were conducted to examine the associations between lipids and disease severity. To assess the discrimination ability of each lipid marker for outcome, receiver operating characteristic (ROC) curves were calculated, and the optimal cutoff values were determined by maximizing the Youden index. Spearman tests were used to analyze the correlations between lipids and inflammatory factors. Survival differences among groups with different lipid concentrations were compared by Kaplan-Meier analysis using the log-rank test. Significance levels were set based on two-sided α < 0.05. Data analyses were performed in statistical packages R (The R Foundation; http://www.r-project.org; version 3.6.1) and SPSS 22.0. Diagrams were plotted by GraphPad Prism 8.0 (GraphPad Software, USA).
A total of 99 laboratory-confirmed COVID-19 patients were prospectively enrolled in this study. As shown in Table 1, the median time from symptom onset to admission was comparable between mild and severe cases [20.00 (IQR: 14.00–26.00) days vs. 19.00 (IQR, 10.25–30.00) days, p = 0.841] as well as between severe-surviving and severe-non-surviving groups [20.00 (IQR: 10.50–30.00) days vs. 17.00 (IQR, 10.00–30.00) days, p = 0.663]. Compared with mild cases, severely ill patients were older (severe: median 70.5 years: IQR, 61.3–81.8 vs. mild: 52 years: IQR, 42.0–62.0) and more likely to have comorbidities (severe: 84% vs. mild: 59.2%) and higher SOFA scores (severe: median, 5, IQR, 2–7 vs. mild: median, 0, IQR, 0–1). No sex differences were found between the mild and severe groups. Fourteen patients received mechanical ventilation in the severe group, whereas no mechanical ventilation was used in the mild cases. A total of 15 severe group patients died in hospital. Mechanical ventilation was more frequently applied among non-survivors. Severe-non-surviving cases presented significantly higher SOFA scores (median, 8.00, IQR, 7.50–10.00) than severe-surviving cases (median, 3.00, IQR, 1.25–5.00). Statin and antiviral treatment were similar among the groups. However, corticosteroid and antibiotic use differed significantly between severe and mild patients. Of note, more deceased patients received corticosteroid therapy compared with severe-surviving patients. The time from symptom onset to admission was comparable between the mild and severe groups [20 IQR (14–26) days vs. 19 IQR (10.25–30) days, p = 0.841] as well as between the severe-surviving and severe-non-surviving groups [20 IQR (10.5–30) days vs. 17 IQR (10–30) days, p = 0.663]. Mild patients experienced a longer hospitalization stay compared to severe patients [20 IQR (15–25) days vs. 15 IQR (9–20.5) days, p = 0.012]. Length of hospitalization was similar between the severe-surviving and severe-non-surviving groups [15 IQR (9–22.5) days vs. 15 IQR (10–18.5) days, p = 0.706].
For major laboratory characteristics, mild and severe COVID-19 cases demonstrated significant deviation in terms of blood cell proportions, coagulation functions, cardiac and renal functions, inflammatory indicators, and lipid profiles. Hierarchical clustering was performed to visualize the differences in laboratory parameters between mild and severe COVID-19 patients. The resulting heatmap illustrated different enrichment in blood indicators between mild and severe cases (Figure 1, Supplementary Figure 1). Notably, inflammatory cytokines, which are organ injury-associated indicators, were found at higher concentrations in the severe cases, whereas certain blood indicators, including lymphocytes, erythrocytes, hemoglobin, and albumin, were higher in the mild group.
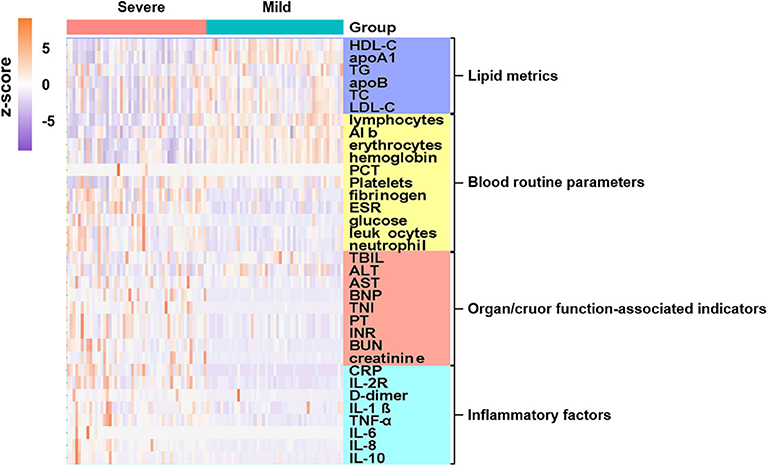
Figure 1. Admission characteristics of laboratory parameters between mild and severe COVID-19 patients. Hierarchical clustering was applied based on laboratory parameters. Heatmap indicates enriched concentration of laboratory indicators in mild and severe cases. Levels of laboratory metrics were scaled by calculating z-scores (subtracting mean, then dividing by standard deviation of each row). Laboratory metrics were categorized into four major groups, i.e., lipid metrics, routine blood parameters, organ/cruor function-associated indicators, and inflammatory factors, with color bars on right side of plot indicating each analyte category. Y-axis represents laboratory values after z-scoring by row; x-axis represents individual cases. Annotations show severe cases in pink and mild cases in cyan.
In terms of lipid profiles, we detected lower concentrations of HDL-C, apoA-1, LDL-C, and TC in the severe group compared with the mild group (Figures 2A–D). The TG level was significantly increased in the severe-non-surviving cases compared with the severe-surviving cases (Figure 2E), while HDL-C, apoA-1, LDL-C, TC and apoB concentrations were comparable between these two groups (Figures 2A–D,F).
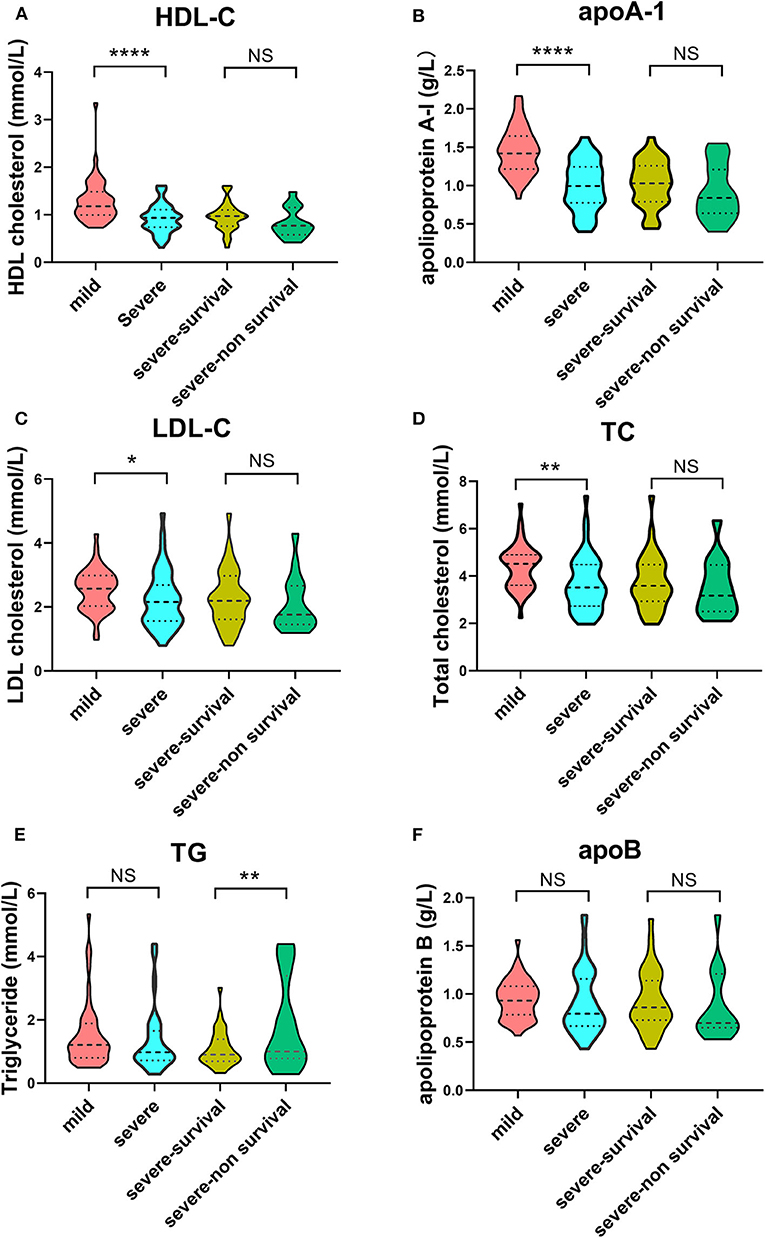
Figure 2. Violin plots of lipid features of mild vs. severe and severe survivors vs. severe non-survivors. Plots demonstrate lipid concentration within each group. Horizontal dotted lines represent first and third quartiles; horizontal dashed lines within plot indicate median of lipid levels. Dunnett's test was applied to assess significance of differences with mild cases serving as the control. (****p < 0.0001, **p < 0.01, *p < 0.05).
Based on the distinct lipid profile features between the severe and mild cases, we performed univariate and multivariate logistic regression analyses to explore the associations between lipid concentrations and disease severity. According to univariate analysis, TC, HDL-C, and apoA-1 levels were associated with severe disease as both continuous and categorical variables (divided by tertiles), whereas LDL-C and TG did not reach statistical significance. Remarkably, based on multivariate analysis, we found that apoA-1 (OR: 0.651 95% CI: 0.456–0.929, p = 0.018) and HDL-C (OR: 0.643 95% CI: 0.456–0.906, p = 0.012) were still independently associated with severity after adjusting for well-recognized risk factors: i.e., age and albumin, D-dimer, C-reactive protein (CRP), and interleukin-6 (IL-6) levels (Table 2). Moreover, patients with the highest tertile of HDL-C and apoA-1 displayed the lowest risk for severe COVID-19. Even after considering comorbidities and SOFA scores for further adjustment, apoA-1 and HDL-C levels remained independently associated with severe status of the disease (Supplementary Table 1). The ROC curves confirmed the significant predictive value of HDL-C and apoA-1 for the presence of severe cases. As shown in Table 3, apoA-1 ≤ 1.16 g/L predicted severity with a specificity of 0.86, sensitivity of 0.66, and area under ROC curve (AUC) of 0.85 (95% CI: 0.76–0.91; p < 0.001). An optimal serum HDL-C cut-off of 1.00 mmol/L provided diagnostic specificity and sensitivity of 75.5 and 68.2%, respectively, for severe cases. TC also displayed prognostic capability, but LDL-C, apoB, and TG showed weak discrimination of the severe condition.
We further detected the predictive performance of lipid profiles for in-hospital death. Notably, ROC analysis revealed that HDL-C and apoA-1 remained valuable for predicting in-hospital death. At a threshold of 0.95 g/L, the AUC of the ROC curve of apoA-1 for death was 0.74 (95% CI 0.61–0.88, p = 0.002). With a cut-off of 0.84 mmol/L, the AUC of HDL-C for death was 0.75 (95% CI: 0.61–0.88, p = 0.002) (Table 4). Moreover, the Kaplan-Meier survival curves and log-rank tests demonstrated that patients with lower apoA-1 or HDL-C levels had a higher rate of in-hospital mortality (divided according to the best threshold) (Figure 3).
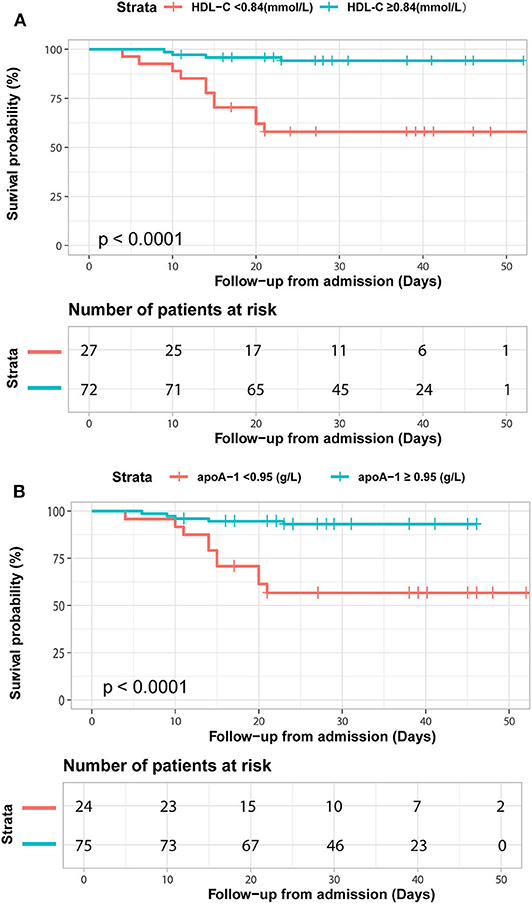
Figure 3. Kaplan-Meier survival curves for in-hospital deaths based on dichotomized HDL-C and apoA-1 concentrations. COVID-19 patients with apoA-1 (A) and HDL-C (B) levels above and below the optimal cutoff value (calculated by ROC analysis) showed obvious disparity in survival time (p < 0.0001).
Figure 4 shows the changes in inflammatory factors and lipid profiles in the mild, severe-surviving, and severe-non-surviving groups from hospital admission, mid-term hospitalization, and end of hospitalization. As illustrated in Figures 4A,B, throughout hospitalization, CRP and IL-6 levels were significantly and continuously high in the severe-surviving and mortality cases but showed low levels among mild cases. Notably, compared with that in the severe-surviving group, both CRP and IL-6 levels in mortality cases were significantly higher at the end of hospitalization (p < 0.05).
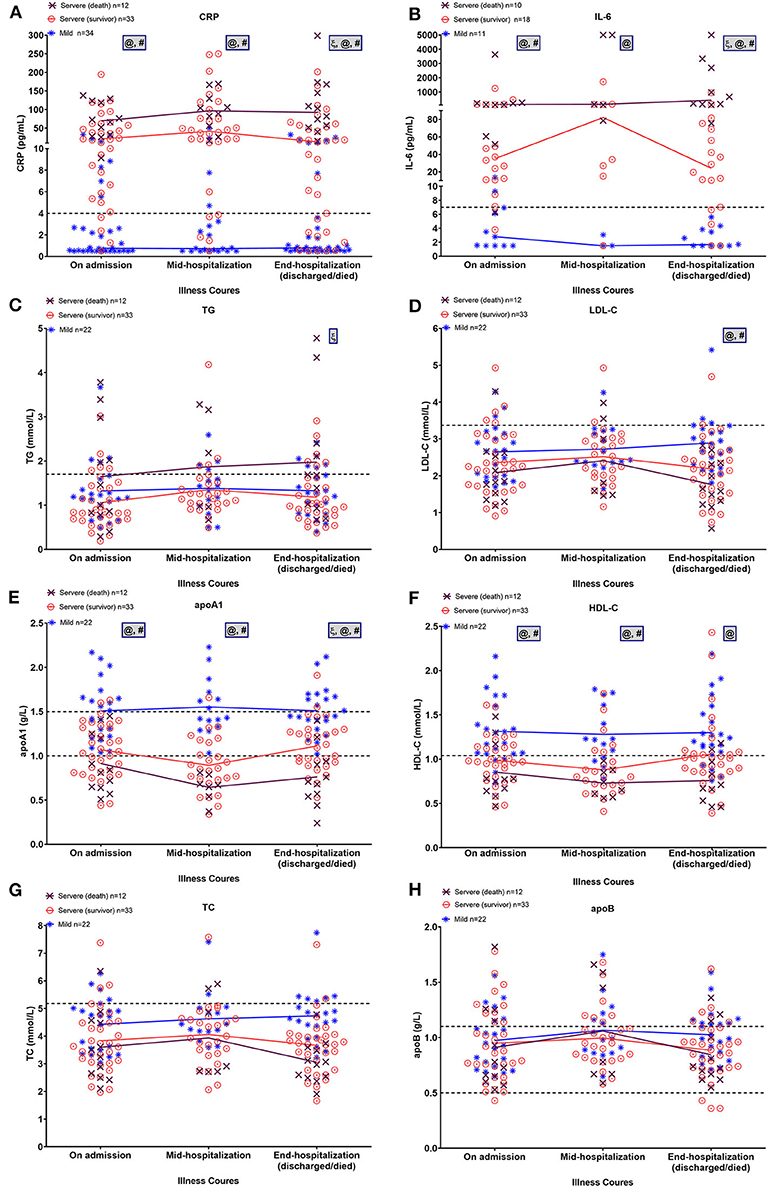
Figure 4. Dynamic alterations in lipid and major laboratory markers from admission in COVID-19 patients. Temporal changes in CRP (A), IL-6 (B), TG (C), LDL-C (D), apoA-1 (E), HDL-C (F), TC (G), and apoB (H) in a subset of COVID-19 patients with ≥2 longitudinal data across three time periods, including on admission, mid-hospitalization, and end of hospitalization. Horizontal dashed lines indicate normal reference range of factors. Mean values of normally distributed parameters (lipid metrics) and median values of non-normally distributed factors (CRP and IL-6) in each group at three time periods are linked by lines. Significant differences among three groups at each time point were compared using one-way ANOVA with Tukey's multiple comparisons test or Kruskal-Wallis test as appropriate. Statistical significance (p < 0.05) is indicated by ξ between severe (death) and severe (survivor) cases, @ between severe (death) and mild cases, and # between severe (survivor) and mild cases. IL-6, interleukin-6; CRP, C-reactive protein; TG, triglycerides; TC, total cholesterol; LDL-C, low-density lipoprotein-cholesterol; HDL-C, high-density lipoprotein-cholesterol, apoA-1, apolipoprotein A-1; apoB, apolipoprotein B.
On admission, regardless of severity or outcome, most patients presented comparable TG and LDL-C levels (Figures 4C,D). By the end of hospitalization, however, TG levels displayed a slight upward trend in the mortality cases and were significantly higher than that in the severe survivors (p = 0.013); in addition, LDL-C levels were significantly lower in severe survivors and non-survivors compared to that in the mild cases (both p < 0.01). Levels of apoA-1 and HDL-C were inversely proportional to disease severity, with mortality cases showing continuously lower levels across hospitalization (Figures 4E,F). Of note, after a slight downward trend in mid-term apoA-1 levels, severe survivors showed a significant recovery in apoA-1 levels at the end of hospitalization (vs. mid-term apoA-1 levels, p = 0.02). By the end of hospitalization, the lowest apoA-1 levels were found in severe cases with a fatal outcome (p < 0.01). For TC and apoB, no significant differences were observed among the three groups across the three time points (Figures 4G,H).
Correlation analysis was performed to detect potential factors related to lipid characteristics. As shown in Figure 5A, admission lipid profiles, especially apoA-1 and HDL-C, were negatively correlated with inflammatory factors, such as CRP and IL-6. Admission apoA-1 and HDL-C levels were inversely correlated with peak CRP and IL-6 concentrations during the clinical course of the disease (Figure 5B).
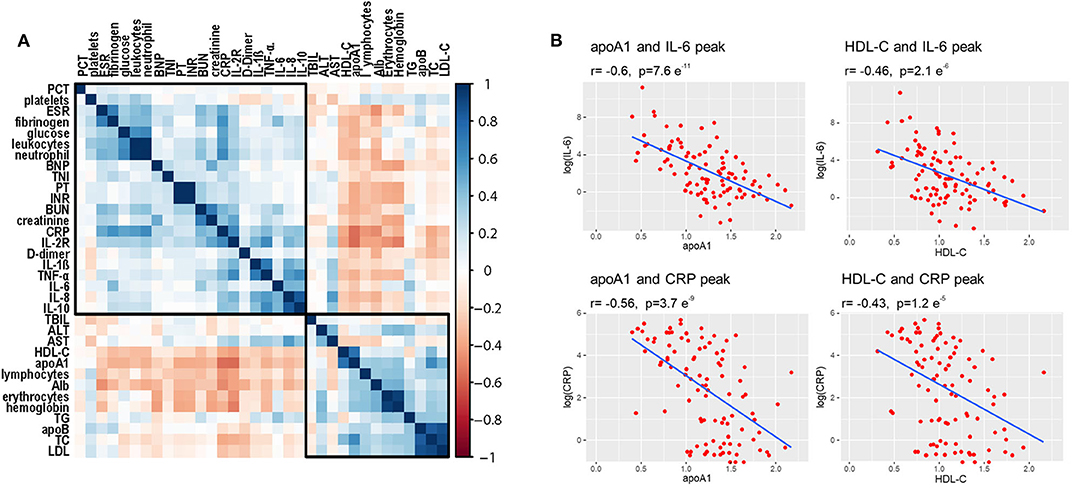
Figure 5. Correlations among lipid profiles and laboratory parameters. (A) Heatmap values represent pairwise Spearman rank correlation coefficients. Blue indicates positive correlation, red indicates negative correlation. (B) Spearman correlation coefficient analysis shows that initial HDL-C and apoA-1 levels were significantly inversely correlated with peak values of CRP and IL-6 during disease course.
Our study highlighted an important association between lipid profiles and fatal clinical outcomes in COVID patients. The main findings are as follows: (1) COVID-19 patients in severe disease were characterized by decreased apoA-1 and HDL-C levels; (2) low apoA-1 and HDL-C levels on admission were able to predict COVID-19 severity and mortality during hospitalization; and (3) apoA-1 and HDL-C levels were strongly correlated with inflammatory indicators, and deviated markedly from the normal reference range in severe cases throughout the course of the disease.
Previous studies have shown that infection and sepsis are accompanied by a metabolic change in the lipid profile, featuring hypertriglyceridemia and reduced HDL-C levels in serum (4, 8). Lipid metabolism dysregulation has also been confirmed in septic patients secondary to both community and hospital-acquired pneumonia (9, 10). In the context of COVID-19, excessive cytokine activation in response to SARS-CoV-2 infection appears to contribute to multiple organ dysfunction. As a result, sepsis and septic shock are frequently observed complications in severe COVID-19 patients (11, 12). Therefore, it is not surprising that serum apoA-1 and HDL-C levels were lower in severely ill patients, especially non-survivors, compared to mild cases.
Both apoA-1 (r = −0.55; p < 0.001) and HDL-C (r = −0.45; p < 0.001) levels were negatively related to SOFA scores, a common diagnostic tool for identifying sepsis severity (13). Based on multivariate analyses, decreased apoA-1 and HDL-C levels were independently associated with COVID-19 severity after adjusting for established indicators of severity, such as age, low albumin, and increased D-dimer, CRP, and IL-6 levels (14, 15). These covariates were included in the multivariate analysis due to their close association with sepsis development reported in previous studies (16, 17). In addition, ROC analysis illustrated that decreased apoA-1 and HDL-C levels were strong predictors of COVID-19 severity. In line with our findings, Groin et al. found that low serum HDL-C concentration on admission is a risk factor for the development of severe sepsis (18).
Our results also highlighted the predictive value of decreased HDL/apoA-1 levels on admission to in-hospital death in COVID-19 patients. Almost half of our research population developed into severe cases, with a relatively high mortality rate of 15.1%. This may be because Leishenshan Hospital was a designated hospital for treating complicated patients transferred from other local hospitals. Our study, for the first time, illustrated that in-hospital death increased significantly in patients with low serum apoA-1 (<0.95 g/L) or HDL-C (<0.84 mmol/L). In addition, ROC analysis verified the predictive value of HDL-C and apoA-1 levels for in-hospital death among COVID-19 patients. This is in agreement with previous study, which found that low apoA-1 concentration is independently associated with the 30-day mortality rate in septic patients (19). Interestingly, here, the temporal recording of lipid profiles showed that the initial decrease in apoA-1/HDL-C levels in survivors began to recover at the end of hospitalization. A similar tendency in HDL-C change has also been observed in patients recovering from sepsis (20). Here, however, apoA-1 rapidly deteriorated in non-survivors throughout the clinical course of the disease.
The underlying mechanisms of HDL-C reduction in severe COVID-19 patients and its association with increased mortality are not fully understood. HDL-C and its major structural protein (apoA-1) directly exert anti-inflammatory effects by neutralizing lipopolysaccharides (LPS), thus playing an important role in host resistance to bacterial, viral, and parasitic infection (21). The protective role of apoA-1 is also evidenced in acute lung injury and acute respiratory distress syndrome. Specifically, apoA-1-deficient mice exhibit enhanced recruitment of neutrophils and monocytes to airspace under LPS inhalation (22). However, both HDL-C and its beneficial effects can be disturbed by inflammation (23, 24). For example, pro-inflammatory cytokines like IL-6 and CRP directly inhibit apolipoprotein synthesis enzyme activity, resulting in reduced apoA-1 and HDL-C production (25). In our study, IL-6 and CRP concentrations were significantly higher in the severe group, and were negatively correlated with lipid indicators apoA-1 and HDL-C. We also found that serum amyloid A (SAA), an acute phase protein, was markedly increased in severe patients. SAA-enriched HDL is reported to clear more rapidly from circulation than normal HDL (26). Hence, the inflammatory-induced humoral innate response to scavenge lipoprotein from circulation may be another potential mechanism leading to low-HDL-C levels. As a result, a vicious cycle occurs in severely ill COVID-19 patients, with a deficiency in HDL-C resulting in cytokine overproduction and a further depletion of HDL-C.
In our study, TC and LDL-C levels in severe patients tended to follow a pattern similar to that of HDL-C. Low TC and LDL-C levels are considered as markers of malnutrition, as nutrition provides the basic substrate for cholesterol synthesis (27). Furthermore, early enteral nutrition is reported to accelerate the recovery of TC levels (20). Consistently, the nutrition states of patients deteriorated in our study, as reflected by continuously decreased levels of albumin in the severe group. Like HDL-C, inflammatory mediators also participate in impaired LDL-C synthesis. Thus, hypocholesterolemia may reflect both malnutrition and an overactive inflammatory status in severe COVID-19 patients.
Although admission TG levels were comparable between mild and severe cases, TG levels were remarkably elevated in non-survivors. Serum TG frequently increases under a septic environment due to reduced TG hydrolysis. Inflammatory cytokines also contribute to inhibit LPL activity, overproduction of free fatty acid, and TG synthesis (26). Besides, after comparing the survival rates between four groups of patients stratified by TG and apoA-1 levels, we found that patients with lower apoA-1 levels and elevated TG levels displayed the unfavorable prognosis with the lowest survival rate (Supplementary Figure 2). Thus, we considered that elevated TG levels, together with persistently low lipoprotein cholesterol concentrations, might be a marker of uncontrolled inflammation and increased risk of death in COVID-19 patients. And further assessment in larger cohorts are required for validation.
There are several limitations in our study. First, given the small sample size, to avoid overfitting, we only calculated the Kaplan-Meier survival curve to evaluate the prognostic values of apoA-1 and HDL-C but did not conduct multivariate cox regression to assess the independent prognostic values of these lipid metrics. Thus, further larger cohorts are warranted to verify our conclusions. Second, some patients were already in poor condition when transferred from the local hospital to Leishenshan Hospital, resulting in a higher rate of severe cases in our study. Further studies on outpatients and other mobile hospitals are required to provide a more complete picture of the relationship between lipid profiles and disease progression. Third, our study only focused on lipid concentrations rather than their quality. Therefore, whether lipid particle composition and functional alteration can affect COVID-19 outcomes deserves further investigation.
Lipid metabolism disorders, characterized by low HDL-C and apoA-1 levels, were found in severely ill COVID-19 patients. The altered HDL-C and apoA-1 levels were negatively correlated with inflammatory indicators. Low apoA-1 and HDL-C levels on admission exhibited predictive value in discriminating disease severity and mortality during hospitalization. Our study examined COVID-19 in regard to lipid metabolism, and thus provides new insights into the disease.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by The Ethics Commission of Renji Hospital. The patients/participants provided their written informed consent to participate in this study.
JS and JP: conceived and designed the experiments. ZC, PN, HG, LS, FY, XQ, WW, MZ, XY, and YZ: collected and analyzed the data. JS, ZC, and JP: wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Science Fund for Distinguished Young Scholars (81625002), National Natural Science Foundation of China (81800223, 81930007, 81770238), Shanghai Sailing Program (18YF1413500), and Shanghai Outstanding Academic Leaders Program (18XD1402400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2020.584987/full#supplementary-material
1. Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. (2020) 20:669–77. doi: 10.1016/S1473-3099(20)30243-7
2. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
3. Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. (2020) 146:110–18. doi: 10.1016/j.jaci.2020.04.006
4. Golucci A, Marson FAL, Ribeiro AF, Nogueira RJN. Lipid profile associated with the systemic inflammatory response syndrome and sepsis in critically ill patients. Nutrition. (2018) 55–6:7–14. doi: 10.1016/j.nut.2018.04.007
5. Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell (2020) 182:59–72.e15. doi: 10.1016/j.cell.2020.05.032
6. Sorokin AV, Karathanasis SK, Yang ZH, Freeman L, Kotani K, Remaley AT. Covid-19-associated dyslipidemia: implications for mechanism of impaired resolution and novel therapeutic approaches. Faseb J. (2020). doi: 10.1096/fj.202001451. [Epub ahead of print].
7. World Health Organization. Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (2019-nCoV) Infection is Suspected: Interim Guidance. World Health Organization (2020). Available online at: https://apps.who.int/iris/handle/10665/330893
8. Tanaka S, Diallo D, Delbosc S, Genève C, Zappella N, Yong-Sang J, et al. High-density lipoprotein (HDL) particle size and concentration changes in septic shock patients. Ann Intensive Care. (2019) 9:68. doi: 10.1186/s13613-019-0541-8
9. Sharma NK, Ferreira BL, Tashima AK, Brunialti MKC, Torquato RJS, Bafi A, et al. Lipid metabolism impairment in patients with sepsis secondary to hospital acquired pneumonia, a proteomic analysis. Clin Proteomics. (2019) 16:29. doi: 10.1186/s12014-019-9252-2
10. Sharma NK, Tashima AK, Brunialti MKC, Ferreira ER, Torquato RJS, Mortara RA, et al. Proteomic study revealed cellular assembly and lipid metabolism dysregulation in sepsis secondary to community-acquired pneumonia. Sci Rep. (2017) 7:15606. doi: 10.1038/s41598-017-15755-1
11. Alhazzani W, Møller MH, Arabi YM, Loeb M, Gong MN, Fan E, et al. Surviving sepsis campaign: guidelines on the management of critically Ill adults with coronavirus disease 2019 (COVID-19). Crit Care Med. (2020) 48:e440–69. doi: 10.1097/CCM.0000000000004363
12. Li H, Liu L, Zhang D, Xu J, Dai H, Tang N, et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. (2020) 395:1517–20. doi: 10.1016/S0140-6736(20)30920-X
13. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. (2016) 315:801–10. doi: 10.1001/jama.2016.0287
14. Feng Y, Ling Y, Bai T, Xie Y, Huang J, Li J, et al. COVID-19 with different severity: a multi-center study of clinical features. Am J Respir Crit Care Med. (2020) 201:1380–88. doi: 10.1164/rccm.202002-0445OC
15. Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. (2020) 368:m1295. doi: 10.1136/bmj.m1091
16. Rodelo JR, De la Rosa G, Valencia ML, Ospina S, Arango CM, Gómez CI, et al. D-dimer is a significant prognostic factor in patients with suspected infection and sepsis. Am J Emerg Med. (2012) 30:1991–9. doi: 10.1016/j.ajem.2012.04.033
17. Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit Care. (2010) 14:R15. doi: 10.1186/cc8872
18. Grion CM, Cardoso LT, Perazolo TF, Garcia AS, Barbosa DS, Morimoto HK, et al. Lipoproteins and CETP levels as risk factors for severe sepsis in hospitalized patients. Eur J Clin Invest. (2010) 40:330–8. doi: 10.1111/j.1365-2362.2010.02269.x
19. Chien JY, Jerng JS, Yu CJ, Yang PC. Low serum level of high-density lipoprotein cholesterol is a poor prognostic factor for severe sepsis. Crit Care Med. (2005) 33:1688–93. doi: 10.1097/01.CCM.0000171183.79525.6B
20. Marik PE. Dyslipidemia in the critically ill. Crit Care Clin. (2006) 22:151–9, viii. doi: 10.1016/j.ccc.2005.08.008
21. Tanaka S, Couret D, Tran-Dinh A, Duranteau J, Montravers P, Schwendeman A, et al. High-density lipoproteins during sepsis: from bench to bedside. Crit Care. (2020) 24:134. doi: 10.1186/s13054-020-02860-3
22. Gordon EM, Figueroa DM, Barochia AV, Yao X, Levine SJ. High-density lipoproteins and apolipoprotein a-i. Potential new players in the prevention and treatment of lung disease. Front Pharmacol. (2016) 7:323. doi: 10.3389/fphar.2016.00323
23. Jahangiri A, de Beer MC, Noffsinger V, Tannock LR, Ramaiah C, Webb NR, et al. HDL remodeling during the acute phase response. Arterioscler Thromb Vasc Biol. (2009) 29:261–7. doi: 10.1161/ATVBAHA.108.178681
24. Sun JT, Liu Y, Lu L, Liu HJ, Shen WF, Yang K, et al. Diabetes-invoked high-density lipoprotein and its association with coronary artery disease in patients with type 2 diabetes mellitus. Am J Cardiol. (2016) 118:1674–9. doi: 10.1016/j.amjcard.2016.08.044
25. Pirillo A, Catapano AL, Norata GD. HDL in infectious diseases and sepsis. Handb Exp Pharmacol. (2015) 224:483–508. doi: 10.1007/978-3-319-09665-0_15
26. Wendel M, Paul R, Heller AR. Lipoproteins in inflammation and sepsis. II. Clinical aspects. Intensive Care Med. (2007) 33:25–35. doi: 10.1007/s00134-006-0433-x
Keywords: HDL-C, apoA-1, inflammation, lipid, COVID-19
Citation: Sun JT, Chen Z, Nie P, Ge H, Shen L, Yang F, Qu XL, Ying XY, Zhou Y, Wang W, Zhang M and Pu J (2020) Lipid Profile Features and Their Associations With Disease Severity and Mortality in Patients With COVID-19. Front. Cardiovasc. Med. 7:584987. doi: 10.3389/fcvm.2020.584987
Received: 19 July 2020; Accepted: 02 November 2020;
Published: 04 December 2020.
Edited by:
Shuyang Zhang, Peking Union Medical College Hospital, ChinaReviewed by:
Sasha A. Singh, Harvard Medical School, United StatesCopyright © 2020 Sun, Chen, Nie, Ge, Shen, Yang, Qu, Ying, Zhou, Wang, Zhang and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Pu, pujun310@hotmail.com
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.