
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cancer Control Soc., 29 July 2024
Sec. Multifaceted Approaches in Cancer Support
Volume 2 - 2024 | https://doi.org/10.3389/fcacs.2024.1414881
This article is part of the Research TopicMechanistic Roles of Exercise on Cancer Progression, Recurrence and Survival OutcomesView all articles
Background: Consumption of tobacco and its associated products has been linked with a high incidence of oral cancer in the Indian subcontinent. The present study aims to assess the differential effects of areca nut, tobacco, and slaked lime on the pathogenesis and progression of oral cancer.
Methodology: Extracts of areca nut (ANE), tobacco leaf (TLE), and lime water (LWE) were prepared and characterized. Oral keratinocyte cells (BICR10) were treated with the prepared extracts alone and in combinations. Cell apoptosis, necrosis, nuclear aberrations, and transcript expression profiles were assessed to study the cancer pathogenesis.
Results: An increase in apoptosis and necrosis was observed in the cells treated with ANE and ANE+LWE, while a decrease in apoptosis was observed in cells treated with TLE. We also found a variation in the size of nuclei and an increase in the number of multinuclei in the cells treated with TLE and TLE+LWE. Out of 24 head and neck cancer-related genes, we found significant upregulation in seven genes in ANE, six in ANE+LWE, 13 in TLE, 20 in LWE, and 14 in TLE+LWE treated cells. On pathway enrichment analysis, there were alterations in the “pathways in cancer,” “focal adhesion,” and “amoebiasis.”
Conclusion: Although areca nut has been regarded as a carcinogen, its carcinogenic potential is lesser than tobacco and lime. Moreover, adjunctive use of tobacco and lime along with areca nut may be an essential factor in the onset and progression of oral cancer.
Habitual and recreational use of betel quid, along with some other components like areca nut and slaked lime, is a common practice in the Indian subcontinent (1). It has been traditionally used as a mouth freshener for centuries. However, it is harmful; some consumers prefer to add chewing tobacco to it, making it prone to addiction (2). These being common everywhere, other ingredients and flavoring agents are also added based on local practices and preferences. In the past few decades, consumption of its processed form, namely 'paan masala,' has increased due to its ease of usage and shelf life (3). Moreover, with its usage, the prevalence of several potentially malignant oral disorders and oral cancer has also increased (4).
Areca nut and tobacco have been classified as carcinogenic to humans (Group 1) by the International Agency for Research on Cancer (IARC) (5). The constituents of areca nut include arecoline, arecaidine, guvacoline, and guvacine (6). Arecoline, being the abundant ingredient, is grouped as possibly carcinogenic to humans (Group 2B) (7). On the other hand, nicotine, being the abundant constituent of tobacco, itself is not considered to be a carcinogen by IARC (8); however, other ingredients, i.e., N'-nitrosonornicotine (NNN) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) have been regarded as Group 1 carcinogens (9). Nicotine can undergo conversion to NNN and NNK during the process of curing (10). Slaked lime, which predominantly comprises calcium hydroxide, is also added, which increases the adsorption of the constituents of areca nut and tobacco into the bloodstream, increasing their bioavailability and making them more pleasurable to the chewer (11). Lime also hydrolyzes almost all arecoline to arecaidine (12). Hence, it possibly increases the carcinogenicity of both areca nut and tobacco.
Areca nut is known to cause oral submucous fibrosis (OSMF), a potentially malignant oral disorder (13). It mainly affects fibroblasts in the connective tissue but also affects keratinocytes in the oral epithelium (14). Its malignant transformation to Oral squamous cell carcinoma (OSCC) is found in as many as 13% of OSMF patients (15). OSCC is a lesion of epithelial origin and comprises around 90% of head and neck cancers (16). However, whether areca nuts alone or the additional components that lead to this malignant transformation to oral cancer is still unanswered.
Hence, the current paper aims to study the differential effects of areca nut, tobacco, and slaked lime and their combinations on the progression of oral squamous cell carcinoma. Owing to the role of these adjunctive components in cancer pathogenesis, we considered preparing and using extract instead of purified compounds. We studied cell viability, apoptosis, necrosis, and genomic instability to assess the effect of these components on epithelial keratinocytes. We also identified top altered genes from previous reports and performed gene expression analysis to understand their impact on cancer pathogenesis and progression.
The active ingredients of ANE and TLE were confirmed by LCMS. The prevalent peak at m/z=156.1 shows the presence of arecoline (MW −155.19 g/mol) in the ANE (Supplementary Figure S1A). A peak at m/z=142.1 indicates the presence of arecaidine in ANE.
The prevalent peak at m/z = 163.12 shows the presence of nicotine (MW −162.23 g/mol) in TLE (Supplementary Figure S1A). The presence of trace amounts of NNN (MW −177.2 g/mol) and NNK (207.23 g/mol) was confirmed by a peak at m/z = 178.14 and 208.04, respectively.
MTT assay shows cell proliferation at lower concentrations of ANE (up to 12.5 μg/ml); however, cell inhibition at higher concentrations. The IC50 of ANE is 98.71 μg/ml, and LWE is 32.32% (Supplementary Figures S2A, C). A sub-cytotoxic dose of ANE (10 μg/ml) and LWE (5%) was used in further experiments. However, the IC50 of the cells treated with TLE could not be determined as the cells were vital, even at a concentration as high as 1,000 μg/ml (Supplementary Figure S2B). Hence, TLE was used at a 100 μg/ml concentration to treat the cells.
EB-AO staining reveals a significantly higher number of cells treated with ANE and ANE+LWE showing apoptosis and necrosis. In contrast, the cells treated with TLE showed a reduction in the number of cells with necrosis (Figures 1A, B). Western blot shows a significant increase in the expression of RIPK1 in cells treated with ANE and ANE+LWE (Figures 1C, D). Moreover, we also performed the transcript expression profile of apoptotic genes (fadd, apaf1, bid, noxa, bim and bax) as seen in Figure 1E. We found a significant increase in all these genes in the ANE-treated cells. There was a significant increase in fadd, bid, noxa, and bax expression in ANE+LWE-treated cells. LWE and TLE+LWE also show significant increases in the expression of apaf1, noxa, bim, and bax. As expected, the expression of these genes was downregulated in TLE-treated cells.
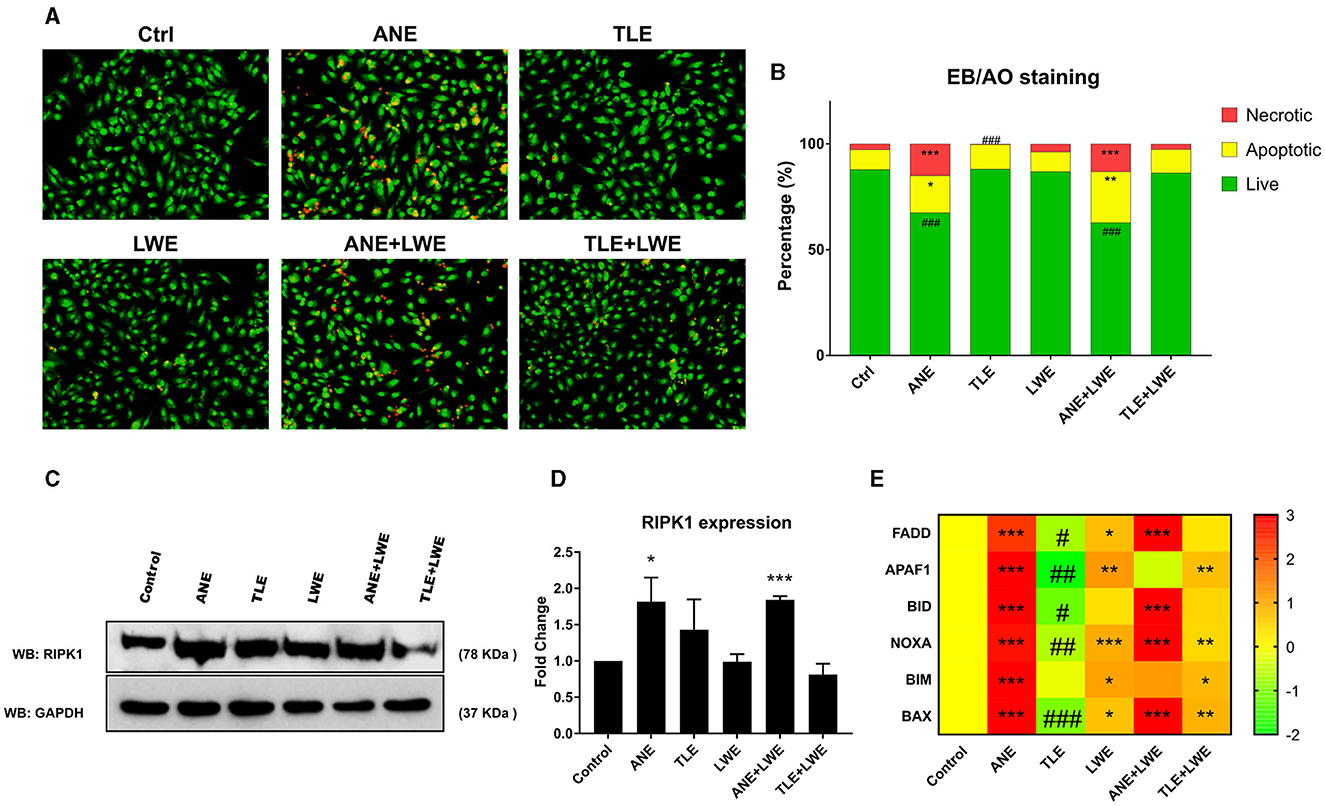
Figure 1. Status of apoptosis and necrosis in cells treated with ANE, TLE and LWE. Representative images showing green colored cells as live, yellow to orange colored cells as apoptotic and dark orange to red colored cells as necrotic by EB-AO dual staining (A), quantification of live apoptotic and necrotic cells by cell counting (B) Western blot of RIPK1 protein (C) and its quantification (D) showing increased necrosis in ANE and ANE+LWE treated cells. Heat map showing expression of apoptotic genes fadd, apaf1, bid, noxa, bim, and bax (E), indicating decreased expression in cells treated with TLE but increased expression with ANE, LWE, ANE+LWE and TLE+LWE treated cells. All the experiments were performed in three biological replicates with two technical replicates in each set. The data is presented as mean values ± SEM. Intergroup comparison was performed using one-way ANOVA followed by Tukey's Post hoc analysis (#/*p < 0.05, ##/**p < 0.01, ###/***p < 0.001).
The presence of multinuclei and micronuclei was assessed by staining the treated cells with DAPI (Figure 2A). It was found that there is a significant increase (p < 0.05) in the count of multinuclei in cells treated with TLE and TLE+LWE (Figure 2B). Micronlcei were increased in all the groups except for cells treated with LWE (Figure 2C). There was a significant variation in nuclei size in the cells treated with TLE and TLE+LWE (Figure 2D).
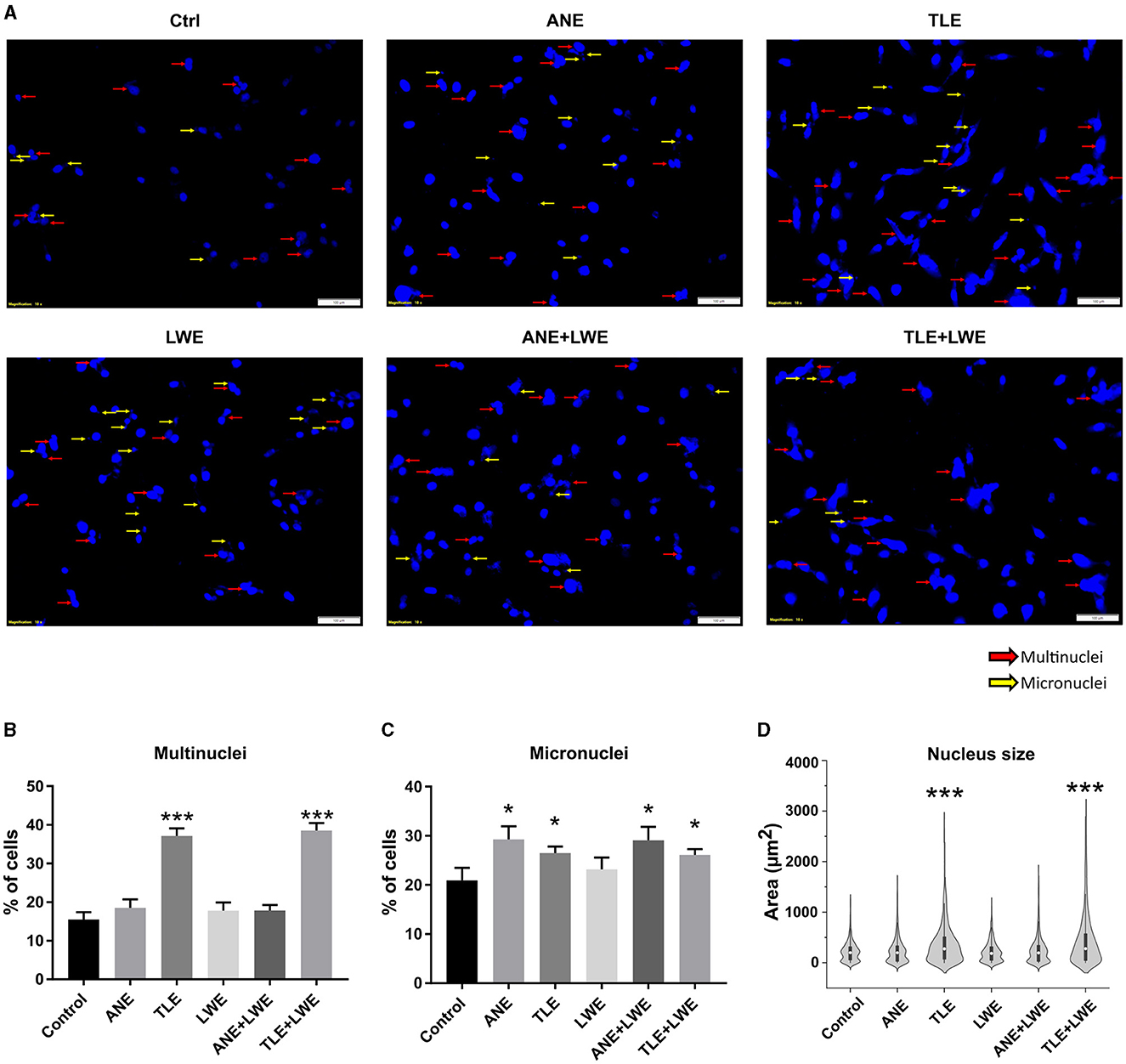
Figure 2. Genomic instability in cells treated with ANE, TLE, and LWE. Representative images showing nucleus stained with DAPI with red arrows showing multinuclei and yellow arrows showing micronuclei (A), higher number of cells with mutinuclei were observed in cells treated with TLE and TLE+LWE (B), and higher number of cells with micronuclei were observed in cells treated with ANE, LWE, ANE+LWE and TLE+LWE (C) A significant variation in the size of nuclei was observed in cells treated with TLE and TLE+LWE (D). All the experiments were performed in three biological replicates with two technical replicates in each set. The data is presented as mean values ± SEM. Intergroup comparison was performed using one-way ANOVA followed by Tukey's Post hoc analysis (*p < 0.05, **p < 0.01, ***p < 0.001).
Figure 3A shows a correlation heat map of the genes studied in the treated cells (also see Supplementary Table S2). It was observed that genes were clustered into two groups where isg15, ca9, hoxd11, mmp11, adam12 and inhba were in one group while col5a1, col5a2, col1a1, if16, fn1, spp1, tgfb1, lamc2, aim2, postn, hoxb7, flna, anxa4, nell2, mmp1, mmp3, pcsk9 and tm4sf19 in another group. There was a positive correlation of the genes within each set and a negative correlation between both sets. A heat map with the gene expression profile is shown in Figure 3B (also see Supplementary Table S3, supporting information). Out of a total of 24 genes, there was significant upregulation (p < 0.05) in seven genes in ANE, six in ANE+LWE, 13 in TLE, 20 in LWE, and 14 genes in TLE+LWE treated cells. There was a significant downregulation (p < 0.05) of 14 genes in ANE, three genes in TLE, none in LWE, nine genes in ANE+LWE, and one gene in TLE+LWE treated cells. The slaked lime itself leads to a significant upregulation of most of these genes but also enhances the expression of ca9, inhba, adam12, nell2, and pcsk9 in addition to areca nut and tobacco. The expression profile has also been summarized in Figure 5.
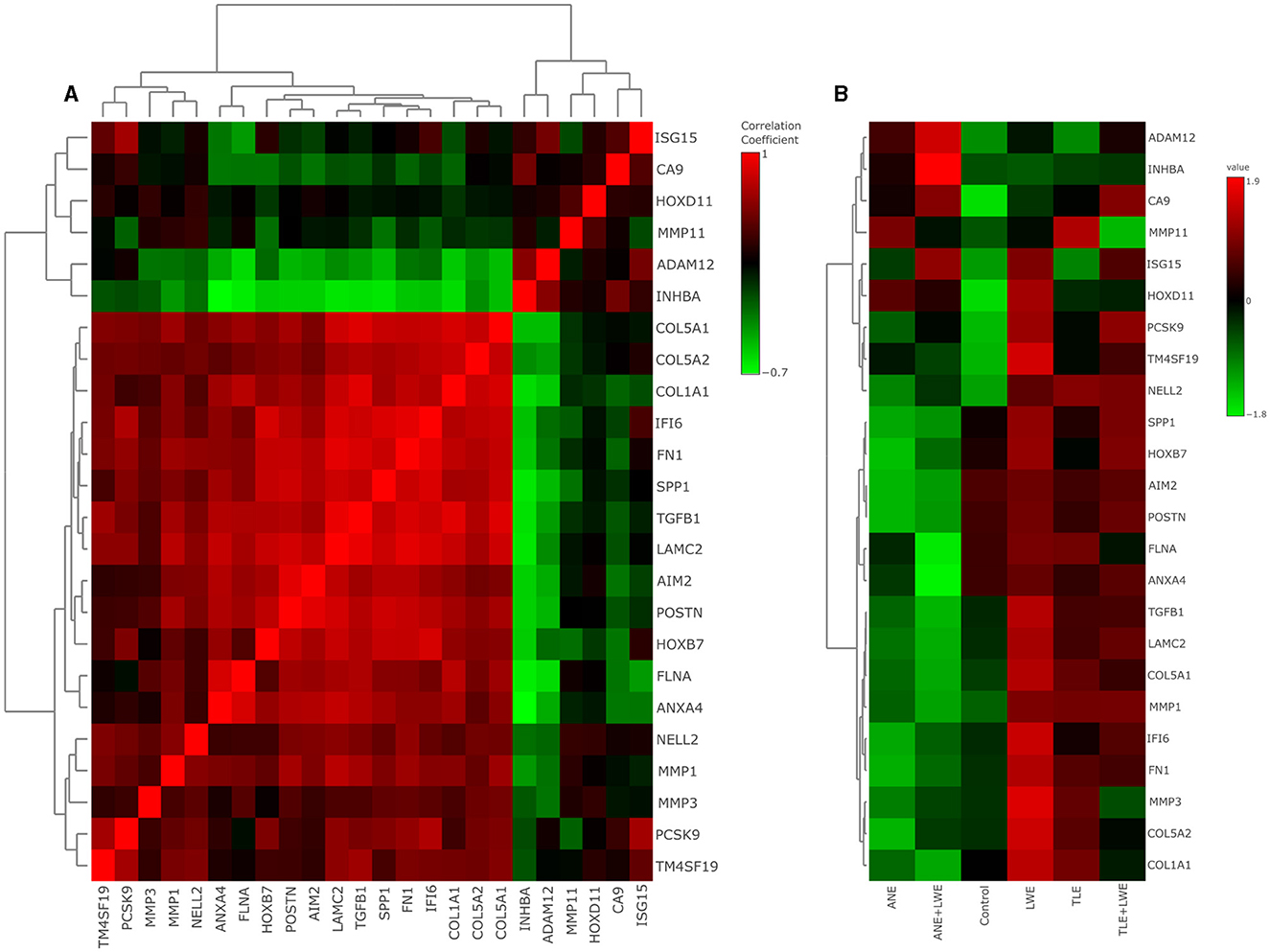
Figure 3. Gene expression profile by qRTPCR in cells treated with ANE, TLE and LWE. Correlation heat map (A) shows gene clustering into two sets with isg15, ca9, hoxd11, mmp11, adam12, and inhba in one group while the remaining genes are in another group. There was a positive correlation between the genes within each group but a negative correlation between the groups. Gene expression heat map (B) shows the upregulation of seven genes in ANE, six in ANE+LWE, thirteen in TLE, twenty in LWE, and fourteen genes in TLE+LWE treated cells. There was a significant downregulation of fourteen genes in ANE, three genes in TLE, none in LWE, nine genes in ANE+LWE, and one in TLE+LWE treated cells. All the experiments were performed in three biological replicates with two technical replicates in each set. Intergroup comparison was performed using one-way ANOVA followed by Tukey's post hoc analysis (α = 5%).
On treatment with ANE, TLE, and LWE, alteration in the gene expression was found to be associated with several metabolic pathways, as shown in Figure 4A (also see Supplementary Figure S4, Supplementary Tables S4–S7). Supplementary Figures S3A, S3B show box plots with normalized data (log2 transformed) of the qRT-PCR data in all the groups and PCA plots, respectively. The three most altered pathways were “focal adhesion,” “amoebiasis,” and “pathways in cancer.” Other altered pathways were the “PI3K-Akt signaling pathway”, “ECM-receptor interaction,” “AGE-RAGE signaling pathways in diabetic complications,” “proteoglycans in cancer,” and “MAPK signaling pathway”. Figure 4B shows a Venn diagram showing the number of differentially expressed pathways in all the groups (see Supplementary Table S6). It was observed that twelve pathways were altered commonly in all the groups. Three pathways, namely the PPAR signaling pathway, Bladder cancer, and IL-17 signaling pathway, were altered between ANE vs. TLE-treated cells and ANE+LWE vs. TLE+LWE-treated cells. We found that adding LWE to ANE and TLE enhances the effect of cancer by affecting focal adhesion the most.
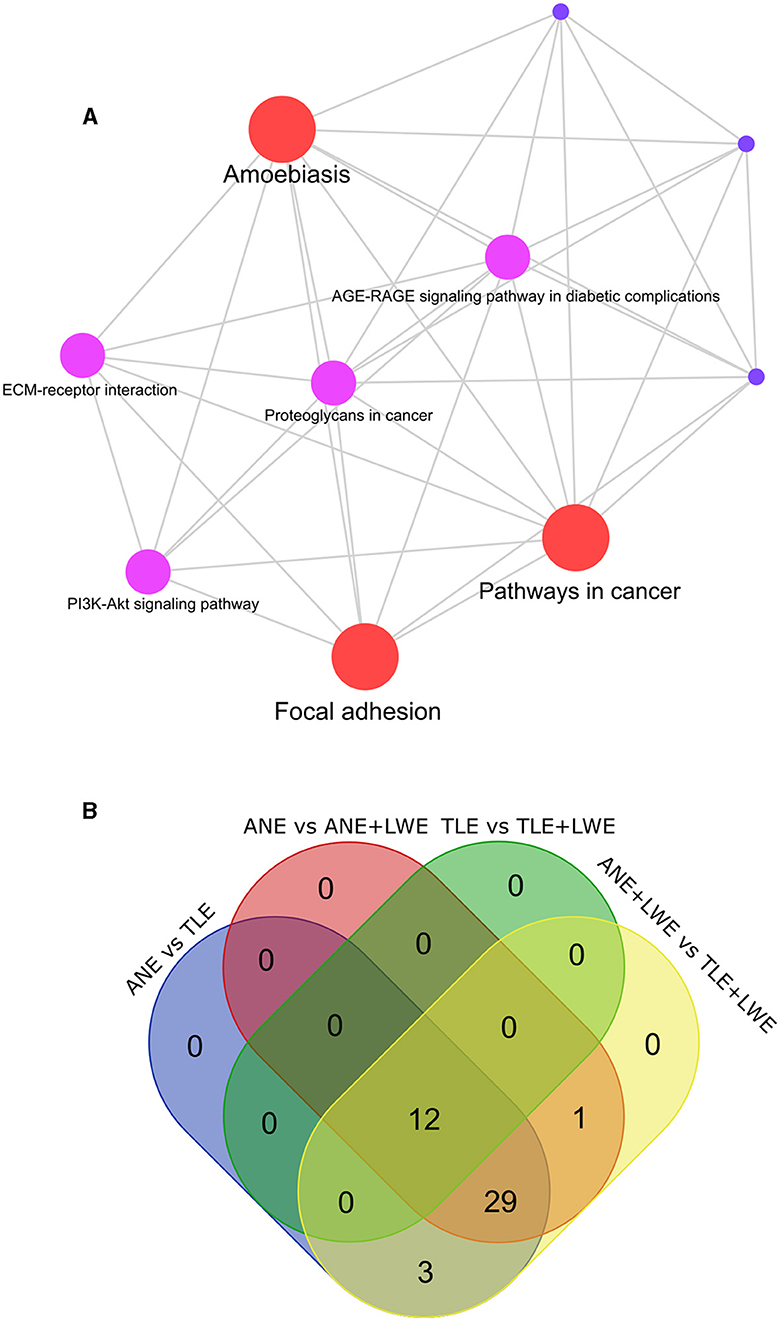
Figure 4. Pathway analysis. The gene expression profile in all the groups was subjected to pathway enrichment (A). The top altered pathways identified in all the treatment groups were “focal adhesion,” “amoebiasis,” and “pathways in cancer”. The Venn diagram (B) shows pathways common between the different groups. Twelve pathways were altered commonly in all the groups. Compared to areca nut treatment, tobacco leads to alteration in three pathways, namely the PPAR signaling pathway, Bladder cancer, and IL-17 signaling pathway.
Oral cancer is a rising concern in India, and consumption of tobacco and its associated products, like betel quid, is considered to be the major culprit in its etiopathogenesis (17). It is interesting to note that not all consumers develop cancerous lesions. Besides the genetic and environmental factors, several other risk factors can also play an important role in causing cancer (18). A person is more likely to develop cancer based on habits such as the number of years of consumption, frequency per day, location of placement in mouth, and duration of placement (19). A synergistic effect due to the simultaneous use of more than one form of tobacco or its associated products can also affect its etiopathogenesis (20). It is intriguing to know which of the known components in the betel quid plays a role in cancer pathogenesis the most. Hence, the current study was designed to explore and distinguish the effect of three major carcinogenic substances of betel quid (areca nut, tobacco, and slaked lime) on cancer progression. We treated the oral epithelial keratinocyte cell line (BICR10) with extracts prepared from areca nut, tobacco, and lime. We then assessed the active ingredients of tobacco and lime, followed by the status of apoptosis, necrosis, necroptosis, genomic instability, and OSCC gene expression analysis in the cells. The cells were treated with these extracts named ANE, TLE, and LWE alone and in combinations (ANE+LWE and TLE+LWE). During the initial experiments, a group with a combination of all three (ANE+TLE+LWE) was also included; however, even with multiple permutations and combinations of treatment doses, the vitality of cells was compromised to a great extent (data not shown), ultimately affecting the outcome of our experiments. Hence, we decided not to include this group in the study. This was unfortunate because many consumers use these components together in the betel quid. However, this did not affect the purpose of our investigation, which was to ascertain which of these components is more important in the onset of cancer or even the advancement of an existing disease.
The working dose of ANE, TLE, and LWE was decided based on the results of MTT, and a subcyotoxic dose was used for further experiments. We found cell proliferation at lower concentrations while cell inhibition at higher concentrations on treatment with ANE. Similar effects of areca nut on epithelial keratinocytes have also been reported in previous studies (21, 22). We also found cytotoxic effects of LWE on epithelial keratinocytes. The proliferative/cytotoxic effects of slaked lime on mucosal epithelium have not been studied much. However, its proliferative effect on fibroblasts have been reported (11). So it can play an important role in pathogenesis of potentially malignant lesions like oral submucous fibrosis. Interestingly, TLE did not induce cell death, even at high concentrations used in the study (1,000 μg/ml). This supports the understanding of evasion of death in cancer cells. Probably, even a higher concentration of TLE was required to induce cell death; however, the study aimed not to assess the cytotoxicity but to evaluate the carcinogenicity of tobacco and compare it with other components. Hence, for further experiments, the working dose of TLE was decided by studying other factors like nuclear aberrations and gene expression profile, while a sub-cytotoxic dose of ANE and LWE that showed cancer gene expression was used.
Evasion of apoptosis is one of the hallmarks of cancer that promotes tumor pathogenesis and resistance to treatment. We studied the apoptosis and necrosis in the cells treated with ANE, TLE, and LWE by EB-AO staining, as well as mRNA expression profiles (23). Necrosis of cancer cells in the form of necroptosis has also been reported to show both tumor-promoting and tumor-suppressing effects (24). RIPK1, which is a marker of necroptosis, has been reported in various cancers (25, 26). Its role in oral cancer has also been reported in several studies (27–29). Hence, we also studied necroptosis by assessing the expression of RIPK1 on western blot. However, we used a cell proliferative dose of ANE; it was observed that there was an increase in apoptosis and necrosis in the cells treated with ANE and ANE+LWE. This suggests a simultaneous proliferative and inhibitory effect of areca nut. Moreover, it explains the heterogeneity in the cell behavior where the dominant behavior shows up, which ultimately affects the outcome of the tumor (30). In fact, a study suggests that areca nut can serve as an anticancer agent through its capability to induce caspase-dependent apoptosis (31). However, this can be dose-dependent and should be cautiously examined before appreciation. Another study suggests that the apoptosis in epithelial cells is induced by arecoline in the nuts by upregulating tropomyosin- through the transforming growth factor-β/Smad pathway (32). Further, the increased apoptosis due to ANE may also be compared to its effect on the oral mucosa, leading to epithelial atrophy, which, histologically, is seen as a decrease in the number of cell rows and loss of rete pegs. This atrophy is also mediated by the presence of arecoline and copper in the ANE (33, 34). The role of apoptosis in OSMF-related epithelial atrophy has also been reported previously (35, 36). As expected, there was a significant decrease in apoptosis in the TLE-treated cells. This decreased apoptosis corroborates the increased cell viability in TLE-treated cells, indicating its role in cancer progression. Contrary to our findings, studies reports that smokeless tobacco causes apoptosis in fibroblasts and keratinocytes (37, 38). This apoptosis has been considered a result of free radical activity and oxidative tissue damage (39).
We also report genomic instability by assessing the multnuclei and micronuclei in the cells treated with ANE, TLE, and LWE. Genomic instability is an evident feature of cancer, which is caused by defects in certain processes that control the way cells divide (40). The presence of micronuclei and multinuclei is one of the key features of cancer cells (41, 42). Large multinucleated cells in cancer occur due to acytokinetic cell division or cell fusion (43). In comparison, micronuclei are small structures created by chromosomal fragments surrounded by nuclear membranes that fail to reorganize in the primary nucleus after cell division (44). In our study, we also found a significant increase in micronuclei in ANE and ANE+LWE; however, multinucleation and variation in size were not observed in cells treated with ANE, LWE, and ANE+LWE-treated cells. Several previous studies have reported an increase in the number of micronuclei and multinuclei in areca nut chewers (45, 46). The increased nuclear aberrations due to areca nut have been associated with increased cellular reactive oxygen species and deregulation of actin filaments (47). As expected, we found cells treated with TLE and TLE+LWE showed a higher number of cells with multinuclei and micronuclei. Moreover, there was a significant variation in the size and morphology of the nucleus. Histologically, such variation in size is seen in advanced cancers and is named as anisonucleosis or nuclear pleomorphism (48). Similar nuclear aberrations have also been shown in several clinical reports from the oral epithelial cells of OSCC patients (49).
Next, we selected OSCC-related genes by identifying the top upregulating genes reported in head and neck cancer from the UALCAN portal and performed gene expression analysis in ANE, TLE, and LWE-treated cells. We observed that 13 genes were significantly upregulated on treatment with TLE while only six genes (isg15, ca9, hoxd11, mmp11, adam12, and inhba) were upregulated both in ANE and ANE+LWE treated cells. These genes have been reported previously to be involved in cancer progression (50, 51). The isg15 has been previously reported to show both tumor inhibition by apoptosis (51) and tumor progression (52). While ca9, a cell surface protein, has been linked to cancer and is induced by tissue hypoxia (50), which is one of the important features in areca nut mediated lesion—OSMF and plays an important role in its malignant transformation (53). Another gene, hoxd11, is associated with potentially malignant disorders and their progression to oral cancer (54). Adam 12 has also been shown to regulate cancer cell proliferation and survival (55). We also found that the addition of LWE to ANE and TLE further increased the expression of OSCC genes. Slaked lime has also been reported to favor the carcinogenic effect of betel quid components by altering the environmental conditions (11). The possible mechanism reported includes an increase in inflammation and ROS generation, which in turn affects nucleic acids and other macromolecules (56). Interestingly, several OSCC genes that were upregulated on treatment with TLE, were downregulated on treatment with ANE. However, areca nut is regarded as a carcinogen, a previous study suggest that arecoline in areca nut can inhibit mitochondrial acetyl-CoA acetyltransferase 1 (ACAT1), which in turn can inhibit the cancer pathogenesis (57). The downregulated genes in the ANE and ANE+LWE-treated cells indicate the limited role of areca nuts in cancer onset and progression or it could be inferred that areca nut leads to OSCC through mechanisms different from tobacco. Hence, we also studied the possible molecular pathways that can be involved differently in areca nut, tobacco and lime mediated oral cancer progression.
The pathways that were altered most in all the groups were pathways in cancer, amoebiasis, focal adhesion, ECM-receptor interaction, and PI3K-AKT signaling pathway, compared to untreated cells. These pathways have also previously been reported to be altered in oral cancer (58) and other extraoral cancers like gastric cancer (59, 60) and cervical cancer (61). We found that compared to areca nut, tobacco treatment also leads to the most alteration in focal adhesion. Moreover, the addition of lime to areca nut or tobacco leads to the most alteration in the focal adhesion pathway. The focal adhesion signaling molecules critically regulate cell behavior and impact tumor cell survival (62). The oncogenic focal adhesion proteins contribute to multiple facets of cancer, including increased cell proliferation, resistance to apoptosis, elevated cell motility and invasion, and promotion of angiogenesis (63). The pathways in cancer through several mechanisms have also been reported to cause tissue invasion and metastasis, cell proliferation, evasion of apoptosis, sustained angiogenesis, genomic damage, genomic instability, insensitivity to antigrowth signals, resistance to chemotherapy, immortality, and block of differentiation (64). We also found that compared to areca nut, both tobacco, and lime alter the ECM receptor interaction and PI3K-Akt pathways. The ECM receptor interaction pathway plays an important role in tumor shedding, adhesion, degradation, movement, and hyperplasia (65). PI3K-Akt pathway is an intracellular signal transduction pathway that promotes metabolism, proliferation, cell survival, growth, and angiogenesis in response to extracellular signals (66).
The summary of the results of the current study is shown in Figure 5. The results of the present study indicate that tobacco consumption increases the malignant properties in the cells significantly compared to areca nut. Adjunctive use of tobacco and lime may be an important factor in the onset and progression of OSCC. Moreover, the addition of slaked lime to areca nut and tobacco further upregulates several cancer-related genes, of which pcsk9 and inhba were upregulated the most. Studying these transcripts and proteins can further help understand oral cancer's pathogenesis. Moreover, they may serve as a biomarker for diagnosis, disease severity assessment, or prognosis of OSCC. The results of the current study are limited to the in vitro experimental setup and need to be validated in further clinical observational studies.
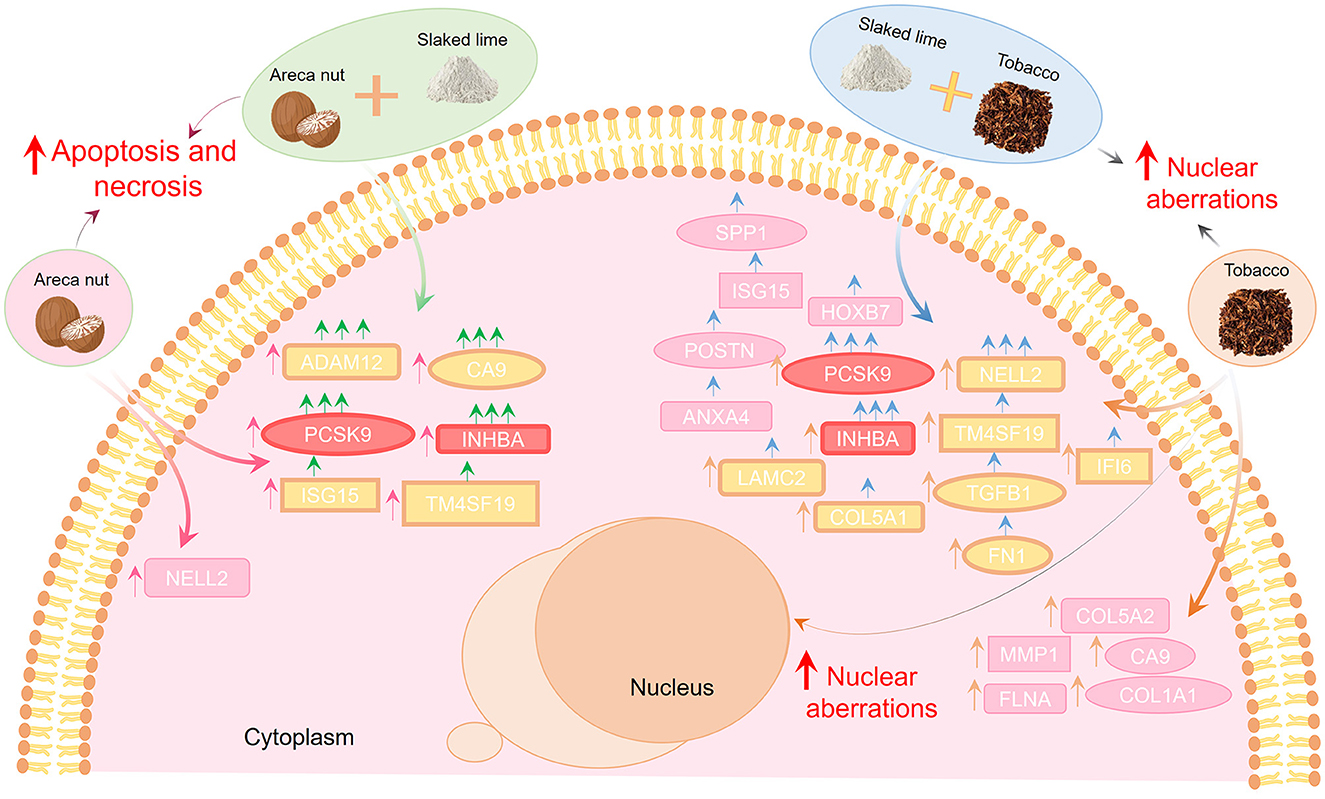
Figure 5. Areca nut, lime, and tobacco modulate the oral cancer progression differentially. Areca nut leads to an increase in apoptosis, while tobacco leads to an increase in nuclear aberrations, indicating that tobacco consumption increases the malignant properties in the cells significantly compared to areca nut. Adding lime to areca nut and tobacco can enhance the expression of OSCC genes (pcsk9, inhba, nell2, adam12, and ca9), where pcsk9 and inhba were common in both groups. Hence, adjunctive use of tobacco and lime may be an important factor in the onset and progression of OSCC. Pink arrows indicate treatment with areca nut, orange arrows treatment with tobacco, green arrows indicate areca nut with limes, and blue arrows indicate tobacco with lime.
Areca nuts (from Areca catechu), dried tobacco leaves (from Nicotiana tabacum), and slaked lime were procured from an authorized dealer in Indore, Madhya Pradesh, India. The authenticity of areca nut and tobacco was confirmed by Dr. Sanjay Vyas, Professor, Plant Taxonomy, Govt. Holkar Science College, Indore. The extracts for areca nut and tobacco leaves were prepared as described previously (67). Briefly, they were ground into a fine powder, and 10 g of each was added to 100 ml of ice-cold deionized distilled water. The mixture was incubated at 4°C for 4 h, followed by centrifugation, and the supernatant was filtered with a 0.22 μm syringe filter. The filtrate was then lyophilized (Alpha 1-2 LDplus, Martin Christ) and stored at −80°C. For all the experiments, a fresh solution of areca nut extract (ANE) and tobacco leaf extract (TLE) was prepared in a concentration of 10 mg/ml each, where the lyophilized powder was suspended in sterile distilled water followed by filtration with 0.22 μm syringe filters. The presence of active ingredients (Arecoline in ANE and Nicotine in TLE) was confirmed by Liquid chromatography- Mass spectrometry (LCMS, Bruker MicrOTOF QII HRMS) as reported previously (67). 10 μl samples were injected, and a flow rate of 0.25 ml/min was maintained. The MS data were collected in centroid mode between m/z 80 and 700 in ESI-positive mode.
A saturated aqueous solution of slaked lime was prepared by adding 20 g lime to 100 ml DMEM and kept on a magnetic stirrer at 4°C for 12 h. The suspension was then centrifuged, and the clear supernatant was filtered with a 0.22 μm syringe filter and then stored at 4°C. The lime water extract (LWE) was directly added to the cell culture media in the required concentration for all the experiments.
We are thankful to Prof. Sanjeev Shukla from the Indian Institute of Science Education and Research—Bhopal, India, who kindly provided keratinocyte cells (BICR10). The cells were cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco—11995065) supplemented with 10% fetal bovine serum (FBS, Gibco—10270106) Hydrocortisone (Sigma-Aldrich—H0888) and antibiotics (Penicillin 10000 U/mL and Streptomycin-10,000 μg/mL, 15140122, Gibco) at 37°C and 5% CO2. The seeding density of cells for all the experiments was 2 × 104 cells/cm2. All the experiments were performed in DMEM (Himedia labs—AT063A) with 3.7 g/L Sodium bicarbonate, without FBS. Before treatment with ANE, TLE, and LWE, the cells were acclimatized in DMEM supplemented with low FBS (2.5%) without antibiotics for 12 h.
To assess the cell viability, an MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazolium bromide) assay was performed as reported previously (68). Briefly, the cells were cultured in a 96-well plate and treated with ANE, TLE, and LWE. The concentration for ANE was 0–100 μg/ml, TLE 0–1,000 μg/ml, and LWE 0–20%. After 24 h incubation, the media was discarded, and the 50 μl of MTT (0.5 mg/ml) was added to each well and incubated for 3 h at 37°C in the dark. Further, the MTT was discarded, 100 μl dimethyl sulphoxide (DMSO) was added, and the plate was incubated at room temperature for 2 h on an orbital shaker. The absorbance was recorded at 570 nm and normalized with the absorbance of DMSO. Percentage viability and IC50 were calculated, and the graph was plotted on GraphPad Prism.
For all further experiments, the cells were treated with a sub-cytotoxic dose of ANE (10 μg/ml), TLE (100 μg/ml), and LWE (5%) for 24 h. The treatment strategy included six groups where the cells were treated with ANE, TLE, and LWE alone and a combination of doses (ANE+LWE and TLE+LWE). Untreated cells were considered as controls.
Ethidium bromide and acridine orange dual staining were performed to assess the status of apoptosis and necrosis in cells treated with ANE, TLE, and LWE for 24 h, as described previously (69). The media was discarded, and the cells were washed with phosphate-buffered saline (PBS). A mixture of acridine orange and ethidium Bromide (Sigma-Aldrich, St. Louis, MO, USA) (100 μg/ml each) was added to the cells and incubated in the dark for 5 min at 37°C followed by washing with PBS. The cells were visualized for fluorescence, and images were acquired (Olympus IX83, Olympus, Tokyo, Japan). The nuclei of cells that appeared green were considered to be alive, yellow to orange were considered to be apoptotic, and red was considered to be necrotic (70). The analysis was performed by cell counting after setting a color threshold (green—live, yellow to orange—apoptotic, and red—necrotic) on the cellSens Dimension imaging software (Olympus, Tokyo, Japan). The cells lying in either of the color ranges were counted automatically using the software.
The treated cells were washed and fixed using 4% paraformaldehyde (PFA) for 30 min at room temperature. The cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 1 h at room temperature and washed with PBS. The cells were visualized for fluorescence, and images were acquired (Olympus IX83, Olympus, Tokyo, Japan). The percentage of cells with micronuclei and multinuclei were counted in at least five fields of view in each of the three biological replicates. The area of the nucleus in the two-dimensional images was also evaluated on ImageJ software to assess the variation in the size of the nucleus after treatment (71).
The relative mRNA expression profile of apoptotic genes and OSCC genes was performed as described previously (72). Total RNA was isolated using Ribozol reagent (VWR™ Cat No. N580) as per the manufacturer's instruction. The cDNA synthesis was performed using the PrimeScript 1st strand cDNA Synthesis Kit (Takara, Cat No. RR820Q). The top twenty-five upregulating genes related to head and neck squamous cell carcinoma were identified from the TCGA gene database on the UALCAN portal (The University of Alabama at Birmingham Cancer Data Analysis Portal, https://ualcan.path.uab.edu). The gene identified as OSCC genes were ca9, mmp11, inhba, mmp9, hoxd11, mmp12, adam12, lamc2, hoxb7, isg15, postn, aim2, cdkn2a, mmp1, pthlh, tgfbi, col5a2, tm4sf19, col5a1, col1a1, nell2, mmp3, fn1, ifi6, and pcsk9. Three other genes (anxa4, flna, and spp1) reported altered in OSCC were also included (73–75). The apoptotic genes included were fadd, apaf1, bid, noxa, bim and bax. The list of primers is shown in Supplementary Table S1. The genes were amplified on AriaMx Real-Time PCR System (Agilent Technologies 5301 Stevens Creek Blvd Santa Clara, CA 95051 USA) using 18sRNA as the housekeeping gene. The relative expression was calculated using a previously reported method (76).
Expression of RIPK1 was assessed using a western blot, as reported previously (77). Briefly, total protein was isolated using Radio-Immunoprecipitation Assay (RIPA) buffer. SDS PAGE was performed to separate the proteins, then transferred to a nitrocellulose membrane. Bovine serum albumin (BSA) and antibodies were diluted in TBST (Tris 0.05 M, pH 7.4 NaCl 0.15 M, 0.05% Tween 20). The blocking was performed using 4.5% BSA for 1 h. The membranes were incubated at 4°C with the primary antibody—RIPK1 (Catalog number 3493, Cell Signaling Technology, Danvers, USA) overnight. After washing with TBST, a secondary antibody tagged with horseradish peroxidase was added and incubated for 40 min. The membranes were developed using Enhanced Chemiluminescence (ECL) substrates (Clarity Western ECL Substrate, Bio-Rad). The visualization and imaging were performed on Gel Doc XR + Gel documentation system (Bio-Rad). The protein level was normalized against the Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression level.
All the data was represented as mean and standard error of the mean. The data analysis was performed using GraphPad Prism (Version 7.04, GraphPad Software, Inc.). The Student's unpaired t-test (two-tailed) was performed to determine the difference between the two groups. Mann-Whitney U test was used as an alternative to Student's unpaired t-test when the data was not found to be normally distributed. One-way ANOVA was used to determine differences between all six groups. Kruskal Wallis test was used as a non-parametric alternative to ANOVA when the data was not found to be normally distributed. The analysis of the relative gene expression was performed, and the heat maps were plotted using MetaboAnalyst 5.0 (www.metaboanalyst.ca) (78). Correlation analysis was performed on the clustering of genes to study the magnitude of the relation between them. The expression profile was also subjected to pathway enrichment on ExpressAnalyst (www.expressanalyst.ca) (79). The level of significance was considered to be 5% for all the analyses.
The data presented in the manuscript are available from the corresponding author upon reasonable request. The source data has been submitted and can be accessed at https://www.ebi.ac.uk/biostudies/studies/S-BSST1495.
TV: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft. SA: Conceptualization, Investigation, Methodology, Software, Writing – review & editing. MK: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. HJ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors received financial support from the Prime Minister's Research Fellowship, the Ministry of Education (PMRF ID: 2100556) and the Indian Council of Medical Research, India (BMI/12(82)/2021).
We acknowledge the Department of Science and Technology (DST), Government of India, under the FIST scheme (project No. SR/FST/LS-I/2020/621) for providing us with various instruments (HPLC, LC-2030C 3D plus, Shimadzu; LC-HRMS, Dionex Ultimate 3000 HPLC coupled with Bruker MicrOTOF QII; MP-AES 4210, Agilent Technologies, and Olympus IX83 microscope). We thank Mr. Ghanashyam Bhavsar for helping in instrument handling and data acquisition on LCMS instruments. We thank Ms. Annu Rani for helping us to prepare the figures for the current manuscript. We appreciate lab members of Infection Bioengineering Group for their insightful discussions and advice. We gratefully acknowledge the Sophisticated Instrumentation Centre, Indian Institute of Technology Indore, for providing facilities and support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcacs.2024.1414881/full#supplementary-material
1. Humans IWG on the E of CR to. Betel-quid and areca-nut chewing. In: Betel-quid and Areca-nut Chewing and Some Areca-nut-derived Nitrosamines. Lyon: International Agency for Research on Cancer (2004). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK316574/ (accessed February 28, 2024).
2. Papke RL, Hatsukami DK, Herzog TA. Betel quid, health, and addiction. Subst Use Misuse. (2020) 55:1528–32. doi: 10.1080/10826084.2019.1666147
3. Kumar S, Gawde N, Pednekar MS. Social determinants and the prevalence of Paan masala use among adults in India: results from Global Adult Tobacco Survey, 2016-17. Asian Pac J Cancer Prev. (2023) 24:3773–81. doi: 10.31557/APJCP.2023.24.11.3773
4. Nair U, Bartsch H, Nair J. Alert for an epidemic of oral cancer due to use of the betel quid substitutes Gutkha and pan masala: a review of agents and causative mechanisms. Mutagenesis. (2004) 19:251–62. doi: 10.1093/mutage/geh036
5. IARC. Acrolein, Crotonaldehyde, and Arecoline. Available online at: https://publications.iarc.fr/Book-And-Report-Series/Iarc-Monographs-On-The-Identification-Of-Carcinogenic-Hazards-To-Humans/Acrolein-Crotonaldehyde-And-Arecoline-2021 (accessed February 28, 2024).
6. Myers AL. Metabolism of the areca alkaloids – toxic and psychoactive constituents of the areca (betel) nut. Drug Metab Rev. (2022) 54:343–60. doi: 10.1080/03602532.2022.2075010
7. Marques MM, Beland FA, Lachenmeier DW, Phillips DH, Chung F-L, Dorman DC, et al. Carcinogenicity of acrolein, crotonaldehyde, and arecoline. Lancet Oncol. (2021) 22:19–20. doi: 10.1016/S1470-2045(20)30727-0
8. Murphy SE. Biochemistry of nicotine metabolism and its relevance to lung cancer. J Biol Chem. (2021) 296:100722. doi: 10.1016/j.jbc.2021.100722
9. Humans IWG on the E of CR to. N′-Nitrosonornicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Personal Habits and Indoor Combustions. International Agency for Research on Cancer (2012) Available online at: https://www.ncbi.nlm.nih.gov/books/NBK304392/ (accessed February 26, 2024).
10. Xue J, Yang S, Seng S. Mechanisms of cancer induction by tobacco-specific NNK and NNN. Cancers (Basel). (2014) 6:1138–56. doi: 10.3390/cancers6021138
11. Zhang P, Chua NQE, Dang S, Davis A, Chong KW, Prime SS, et al. Molecular mechanisms of malignant transformation of oral submucous fibrosis by different betel quid constituents—does fibroblast senescence play a role? Int J Mol Sci. (2022) 23:1637. doi: 10.3390/ijms23031637
12. Zhang P, Sari EF, McCullough MJ, Cirillo N. Metabolomic profile of Indonesian betel quids. Biomolecules. (2022) 12:1469. doi: 10.3390/biom12101469
13. Arakeri G, Patil SG, Aljabab AS, Lin K-C, Merkx MAW, Gao S, et al. Oral submucous fibrosis: an update on pathophysiology of malignant transformation. J Oral Pathol Med. (2017) 46:413–7. doi: 10.1111/jop.12582
14. Ekanayaka RP, Tilakaratne WM. Oral submucous fibrosis: review on mechanisms of malignant transformation. Oral Surg Oral Med Oral Pathol Oral Radiol. (2016) 122:192–9. doi: 10.1016/j.oooo.2015.12.018
15. Kumar V, Kumari N, Ealla KKR, Gour S, Srivastava H, Rallabhandi S. Comparative analysis of trace elements in the saliva and serum of patients with oral submucous fibrosis and squamous cell carcinoma. Mol Clini Oncol. (2024) 20:1–8. doi: 10.3892/mco.2024.2716
16. Tan Y, Wang Z, Xu M, Li B, Huang Z, Qin S, et al. Oral squamous cell carcinomas: state of the field and emerging directions. Int J Oral Sci. (2023) 15:1–23. doi: 10.1038/s41368-023-00249-w
17. Ghalige HS, Sultania M, Muduly DK, Kar M. 21th FHNO Conference (Virtual), 2021. J Head Neck Phys Surg. (2022) 10:S1.
18. Institute Institute of Medicine (US) Roundtable on Environmental Health Sciences R, Wilson S, Jones L, Couseens C, Hanna K. The links between environmental factors, genetics, and the development of cancer. In: Cancer and the Environment: Gene-Enviroment Interaction. Washington, D.C.: National Academies Press (US) (2002). Available online at: https://www.ncbi.nlm.nih.gov/books/NBK220897/ (accessed May 24, 2024).
19. Me S, Shakeel S. Presence of oral mucosal lesions associated with tobacco use – among the patients visiting OPD: a prevalence study, at the oxford dental college Bangalore. Int J Appl Dent Sci. (2022) 8:370–6. doi: 10.22271/oral.2022.v8.i2f.1530
20. Singh AG, Chaturvedi P. Areca nut and oral cancer. Oral Dis. doi: 10.1111/odi.14943. [Epub ahead of print].
21. Chen Q, Jiao J, Wang Y, Mai Z, Ren J, He S, et al. Egr-1 mediates low-dose arecoline induced human oral mucosa fibroblast proliferation via transactivation of Wnt5a expression. BMC Mol Cell Biol. (2020) 21:80. doi: 10.1186/s12860-020-00325-7
22. Dey N, Chattopadhyay DJ, Chatterjee IB. Molecular mechanisms of cigarette smoke-induced proliferation of lung cells and prevention by vitamin C. J Oncol. (2011) 2011:e561862. doi: 10.1155/2011/561862
23. Fulda S. Evasion of apoptosis as a cellular stress response in cancer. Int J Cell Biol. (2010) 2010:370835. doi: 10.1155/2010/370835
24. Liu Z, Jiao D. Necroptosis, tumor necrosis and tumorigenesis. Cell Stress 4:1–8. doi: 10.15698/cst2020.01.208
25. Ermine K, Yu J, Zhang L. Role of receptor interacting protein (RIP) kinases in cancer. Genes Dis. (2021) 9:1579–93. doi: 10.1016/j.gendis.2021.10.007
26. Li F, Sun H, Yu Y, Che N, Han J, Cheng R, et al. RIPK1-dependent necroptosis promotes vasculogenic mimicry formation via eIF4E in triple-negative breast cancer. Cell Death Dis. (2023) 14:1–16. doi: 10.1038/s41419-023-05841-w
27. Huang K, Gu X, Xu H, Li H, Shi M, Wei D, et al. Prognostic value of necroptosis-related genes signature in oral squamous cell carcinoma. Cancers. (2023) 15:4539. doi: 10.3390/cancers15184539
28. Krishnan RP, Pandiar D, Ramani P, Jayaraman S. Necroptosis in human cancers with special emphasis on oral squamous cell carcinoma. J Stomatol Oral Maxillofacial Surg. (2023) 124:101565. doi: 10.1016/j.jormas.2023.101565
29. Li J, Huang S, Zeng L, Li K, Yang L, Gao S, et al. Necroptosis in head and neck squamous cell carcinoma: characterization of clinicopathological relevance and in vitro cell model. Cell Death Dis. (2020) 11:391. doi: 10.1038/s41419-020-2538-5
30. Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. (2010) 1805:105. doi: 10.1016/j.bbcan.2009.11.002
31. Sari LM, Subita GP, Auerkari EI. Areca nut extract demonstrated apoptosis-inducing mechanism by increased caspase-3 activities on oral squamous cell carcinoma. F1000Res. (2018) 7:723. doi: 10.12688/f1000research.14856.5
32. Li L, Gu L, Yao Z, Wang Y, Tang Z, Wu X. Arecoline suppresses epithelial cell viability by upregulating tropomyosin-1 through the transforming growth factor-β/Smad pathway. Pharm Biol. (2020) 58:1253–1260. doi: 10.1080/13880209.2020.1851729
33. Kapoor R, Sansare K, Tamgadge S, Karjodkar F, Mehra A, Mishra I, et al. Epithelial atrophy, fibrosis and vascularity correlation with epithelial dysplasia in oral submucous fibrosis, a prospective study. J Microsc Ultrastruct. (2020) 10:1–6. doi: 10.4103/JMAU.JMAU_36_20
34. Khan I, Pant I, Narra S, Radhesh R, Ranganathan K, Rao SG, et al. Epithelial atrophy in oral submucous fibrosis is mediated by copper (II) and arecoline of areca nut. J Cell Mol Med. (2015) 19:2397–412. doi: 10.1111/jcmm.12622
35. Li M, Gao F, Zhou Z-S, Zhang H-M, Zhang R, Wu Y-F, et al. Arecoline inhibits epithelial cell viability by upregulating the apoptosis pathway: implication for oral submucous fibrosis. Oncol Rep. (2014) 31:2422–8. doi: 10.3892/or.2014.3091
36. Zhu B, Jiang Q, Que G, Dai Z, Wu Y. Role of autophagy and apoptosis in atrophic epithelium in oral submucous fibrosis. J Oral Sci. (2020) 62:184–8. doi: 10.2334/josnusd.19-0170
37. Bagchi M, Balmoori J, Bagchi D, Ray SD, Kuszynski C, Stohs SJ. Smokeless tobacco, oxidative stress, apoptosis, and antioxidants in human oral keratinocytes. Free Radical Biol Med. (1999) 26:992–1000. doi: 10.1016/S0891-5849(98)00286-X
38. Li L, Zhou X, Wang Y. Smokeless tobacco extract inhibits proliferation and promotes apoptosis in oral mucous fibroblasts. Oncol Lett. (2018) 16:5066–74. doi: 10.3892/ol.2018.9252
39. Ramage L, Jones AC, Whelan CJ. Induction of apoptosis with tobacco smoke and related products in A549 lung epithelial cells in vitro. J Inflamm (Lond). (2006) 3:3. doi: 10.1186/1476-9255-3-3
40. Yao Y, Dai W. Genomic instability and cancer. J Carcinog Mutagen. (2014) 5:1000165. doi: 10.4172/2157-2518.1000165
41. Ye CJ, Sharpe Z, Alemara S, Mackenzie S, Liu G, Abdallah B, et al. Micronuclei and genome chaos: changing the system inheritance. Genes (Basel). (2019) 10:366. doi: 10.3390/genes10050366
42. Weihua Z, Lin Q, Ramoth AJ, Fan D, Fidler IJ. Formation of solid tumors by a single multinucleated cancer cel. Cancer. (2011) 117:4092–9. doi: 10.1002/cncr.26021
43. Ariizumi T, Ogose A, Kawashima H, Hotta T, Umezu H, Endo N. Multinucleation followed by an acytokinetic cell division in myxofibrosarcoma with giant cell proliferation. J Exp Clini Cancer Res. (2009) 28:44. doi: 10.1186/1756-9966-28-44
44. Krupina K, Goginashvili A, Cleveland DW. Causes and consequences of micronuclei. Curr Opin Cell Biol. (2021) 70:91–9. doi: 10.1016/j.ceb.2021.01.004
45. Sanjeeta N, Banerjee S, Mukherjee S, Devi TP, Nandini DB, Aparnadevi P. Correlation of the activities of antioxidant enzymes superoxide dismutase and glutathione reductase with micronuclei counts among areca nut chewers of Manipuri population using exfoliative cytology: a preliminary study. J Cancer Res Ther. (2022) 18:984–9. doi: 10.4103/jcrt.JCRT_1227_20
46. Lu S-Y, Chang K-W, Liu C-J, Tseng Y-H, Lu H-H, Lee S-Y, et al. Ripe areca nut extract induces G1 phase arrests and senescence-associated phenotypes in normal human oral keratinocyte. Carcinogenesis. (2006) 27:1273–84. doi: 10.1093/carcin/bgi357
47. Lin C-C, Chang M-C, Chang H-H, Wang T-M, Tseng W-Y, Tai T-F, et al. Areca nut-induced micronuclei and cytokinesis failure in Chinese hamster ovary cells is related to reactive oxygen species production and actin filament deregulation. Environ Mol Mutagen. (2009) 50:367–74. doi: 10.1002/em.20463
48. Wenig BM. Squamous cell carcinoma of the upper aerodigestive tract: dysplasia and select variants. Mod Pathol. (2017) 30:S112–8. doi: 10.1038/modpathol.2016.207
49. Sánchez-Romero C, Carlos R, Dantas Soares C, Paes de Almeida O. Unusual multinucleated giant cell reaction in a tongue squamous cell carcinoma: histopathological and immunohistochemical features. Head Neck Pathol. (2018) 12:580–6. doi: 10.1007/s12105-018-0892-8
50. Pastorekova S, Gillies RJ. The role of carbonic anhydrase IX in cancer development: links to hypoxia, acidosis, and beyond. Cancer Metastasis Rev. (2019) 38:65–77. doi: 10.1007/s10555-019-09799-0
51. Zhou M-J, Chen F-Z, Chen H-C, Wan X-X, Zhou X, Fang Q, et al. ISG15 inhibits cancer cell growth and promotes apoptosis. Int J Mol Med. (2017) 39:446–52. doi: 10.3892/ijmm.2016.2845
52. Desai SD. ISG15: a double edged sword in cancer. Oncoimmunology. (2015) 4:e1052935. doi: 10.1080/2162402X.2015.1052935
53. Chaudhary M, Bajaj S, Bohra S, Swastika N, Hande A. The domino effect: Role of hypoxia in malignant transformation of oral submucous fibrosis. J Oral Maxillofac Pathol. (2015) 19:122–7. doi: 10.4103/0973-029X.164519
54. Padam KSR, Morgan R, Hunter K, Chakrabarty S, Kumar NAN, Radhakrishnan R. Identification of HOX signatures contributing to oral cancer phenotype. Sci Rep. (2022) 12:10123. doi: 10.1038/s41598-022-14412-6
55. Piotrowski KB, Blasco LP, Samsøe-Petersen J, Eefsen RL, Illemann M, Oria VO, et al. ADAM12 expression is upregulated in cancer cells upon radiation and constitutes a prognostic factor in rectal cancer patients following radiotherapy. Cancer Gene Ther. (2023) 30:1369–81. doi: 10.1038/s41417-023-00643-w
56. Islam S, Muthumala M, Matsuoka H, Uehara O, Kuramitsu Y, Chiba I, et al. How each component of betel quid is involved in oral carcinogenesis: mutual interactions and synergistic effects with other carcinogens-a review article. Curr Oncol Rep. (2019) 21:53. doi: 10.1007/s11912-019-0800-8
57. Fan J, Lin R, Xia S, Chen D, Elf SE, Liu S, et al. Tetrameric acetyl-CoA acetyltransferase 1 is important for tumor growth. Mol Cell. (2016) 64:859–74. doi: 10.1016/j.molcel.2016.10.014
58. Bajrai LH, Sohrab SS, Mobashir M, Kamal MA, Rizvi MA, Azhar EI. Understanding the role of potential pathways and its components including hypoxia and immune system in case of oral cancer. Sci Rep. (2021) 11:19576. doi: 10.1038/s41598-021-98031-7
59. Darang E, Pezeshkian Z, Mirhoseini SZ, Ghovvati S. Bioinformatics and pathway enrichment analysis identified hub genes and potential biomarker for gastric cancer prognosis. Front Oncol. (2023) 13:1187521. doi: 10.3389/fonc.2023.1187521
60. Dey L, Mukhopadhyay A. A systems biology approach for identifying key genes and pathways of gastric cancer using microarray data. Gene Reports. (2021) 22:101011. doi: 10.1016/j.genrep.2020.101011
61. Wu K, Yi Y, Liu F, Wu W, Chen Y, Zhang W. Identification of key pathways and genes in the progression of cervical cancer using bioinformatics analysis. Oncol Lett. (2018) 16:1003–9. doi: 10.3892/ol.2018.8768
62. Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. (2015) 31:65–75. doi: 10.1016/j.semcancer.2014.07.009
64. KEGG PATHWAY: Pathways in Cancer - Homo sapiens (human). Available online at: https://www.genome.jp/pathway/hsa05200 (accessed March 1, 2024).
65. Bao Y, Wang L, Shi L, Yun F, Liu X, Chen Y, et al. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol Biol Lett. (2019) 24:38. doi: 10.1186/s11658-019-0162-0
66. Shi X, Wang J, Lei Y, Cong C, Tan D, Zhou X. Research progress on the PI3K/AKT signaling pathway in gynecological cancer (Review). Mol Med Rep. (2019) 19:4529–35. doi: 10.3892/mmr.2019.10121
67. Verma TP, Adhikari S, Singh S, Rani C, Kumar R, Jha HC. A comprehensive analysis of metabolomics and lipidomics in areca nut mediated oral submucous fibrosis progression through LCMS and Raman spectroscopy. J Raman Spectrosc. 55:797–808. doi: 10.1002/jrs.6679
68. Tiwari D, Singh VK, Baral B, Pathak DK, Jayabalan J, Kumar R, et al. Indication of neurodegenerative cascade initiation by amyloid-like aggregate-forming EBV proteins and peptide in Alzheimer's disease. ACS Chem Neurosci. (2021) 12:3957–67. doi: 10.1021/acschemneuro.1c00584
69. Jakhmola S, Jha HC. Glial cell response to Epstein-Barr Virus infection: A plausible contribution to virus-associated inflammatory reactions in the brain. Virology. (2021) 559:182–95. doi: 10.1016/j.virol.2021.04.005
70. Ribble D, Goldstein NB, Norris DA, Shellman YG. A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol. (2005) 5:12. doi: 10.1186/1472-6750-5-12
71. Schneider CA, Rasband WS, Eliceiri KW, NIH. Image to ImageJ: 25 years of image analysis. Nat Methods. (2012) 9:671–5. doi: 10.1038/nmeth.2089
72. Sonkar C, Verma T, Chatterji D, Jain AK, Jha HC. Status of kinases in Epstein-Barr virus and Helicobacter pylori Coinfection in gastric Cancer cells. BMC Cancer. (2020) 20:925. doi: 10.1186/s12885-020-07377-0
73. Wei B, Guo C, Liu S, Sun M-Z. Annexin A4 and cancer. Clinica Chimica Acta. (2015) 447:72–8. doi: 10.1016/j.cca.2015.05.016
74. Zhang N, Gao L, Ren W, Li S, Zhang D, Song X, et al. Fucoidan affects oral squamous cell carcinoma cell functions in vitro by regulating FLNA-derived circular RNA. Ann N Y Acad Sci. (2020) 1462:65–78. doi: 10.1111/nyas.14190
75. Subramani VN, Narasimhan M, Thiyagarajan M, Munuswamy BD, Jayamani L. Expression of osteopontin in oral squamous cell carcinoma and its surgical margins-an immunohistochemical study. J Clin Diagn Res. (2015) 9:ZC66–9. doi: 10.7860/JCDR/2015/12777.6836
76. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–δδct method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
77. Baral B, Saini V, Tandon A, Singh S, Rele S, Dixit AK, et al. SARS-CoV-2 envelope protein induces necroptosis and mediates inflammatory response in lung and colon cells through receptor interacting protein kinase 1. Apoptosis. (2023) 28:1596–617. doi: 10.1007/s10495-023-01883-9
78. Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 50: narrowing the gap between raw spectra and functional insights. Nucl Acids Res. (2021) 49:W388–96. doi: 10.1093/nar/gkab382
Keywords: oral cancer, cancer pathogenesis, areca nut, tobacco, slaked lime
Citation: Verma TP, Adhikari S, Kandpal M and Jha HC (2024) Differential effects of areca nut, tobacco, and lime on oral squamous cell carcinoma progression. Front. Cancer Control Soc. 2:1414881. doi: 10.3389/fcacs.2024.1414881
Received: 09 April 2024; Accepted: 08 July 2024;
Published: 29 July 2024.
Edited by:
Vesile Yildiz Kabak, Hacettepe University, TürkiyeReviewed by:
Jose Alberto Garcia Gomez, Hospital General Universitario de Elche, SpainCopyright © 2024 Verma, Adhikari, Kandpal and Jha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hem Chandra Jha, aGVtY2poYUBpaXRpLmFjLmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.