- 1Biology Department, Institute of Science Education and Research (IISER), Pune, India
- 2Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan
- 3Department of Biomedical Sciences, National Taiwan University, Taipei, Taiwan
- 4International Institute for Sustainability with Knotted Chiral Meta Matter (SKCM2), Hiroshima University, Higashi-Hiroshima, Japan
Ubiquitin C-terminal hydrolases (UCHs) are crucial enzymes within the ubiquitin-proteasome system, characterized by a characteristic Gordian knotted topology. Another important structural feature of the UCH family is a hydrophobic β-sheet core containing a conserved catalytic triad of cysteine, histidine, and aspartate wrapped by several α-helices and a crossover loop. The catalytic triad cleaves the (iso) peptide bond at the C-terminus of ubiquitin via a nucleophilic attack. The highly dynamic crossover loop is involved in substrate binding and selectivity. UCHs play vital roles in various cellular processes, such as cell signaling, DNA repair, neuroprotection, and tumor suppression. Point mutations in catalytic and non-catalytic residues of UCHs are linked to various diseases, including cancers and neurodegeneration. Additionally, post-translational modifications (PTMs), such as oxidation, impact the deubiquitinase activity of UCHs and increase aggregation propensity. This review focuses on how disease-associated point mutations, PTMs, and interactions with different binding partners modulate the structural and functional dynamics of UCHs and how perturbations of these functional dynamics are characterized using a battery of biophysical techniques to gain insights into the molecular mechanisms underlying UCH dysfunction and diseases.
Introduction
Deubiquitinating enzymes (DUBs) are a family of proteases crucial for regulating the ubiquitin-proteosome system (UPS). They act as erasers, counteracting the effects of the E1/E2/E3 ubiquitination machinery as well as modifying the ubiquitin code, which is essential for maintaining protein health, cellular signaling, transcription regulation, and other biological functions (Komander et al., 2009; Reyes-Turcu et al., 2009; Wilkinson, 2009; Clague et al., 2013; Mevissen and Komander, 2017). DUBs are classified into two major classes: cysteine proteases and metalloproteases. Among the cysteine proteases, DUBs are further classified into six families based on their structural features and catalytic mechanisms: ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCH), ovarian tumor proteases (OTUs), Machado-Josephine domain (MJD), motif interacting with Ub-containing novel DUB (MINDY), and zinc finger with UFM1-specific peptidase domain protein (zUFSP) (Clague et al., 2013; Abdul Rehman et al., 2016).
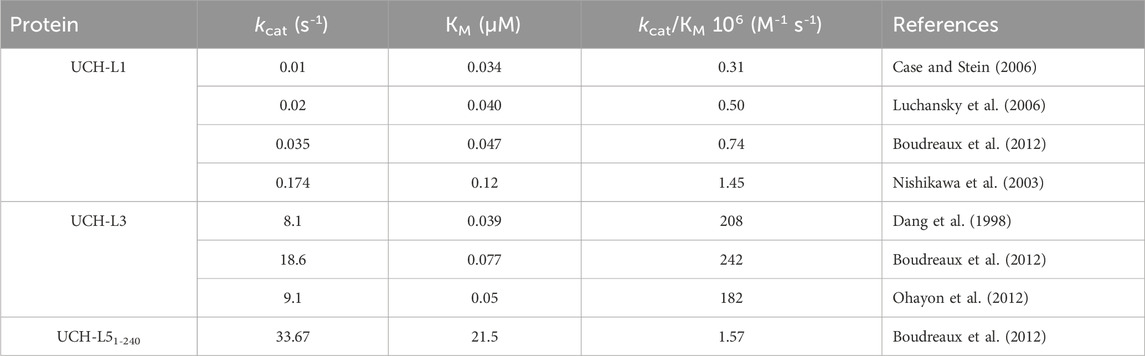
This review focuses on the human UCHs: UCH-L1, UCH-L3, UCH-L5 (also known as UCH37), and BRCA1-associated protein-1 (BAP1). UCH-L1 and UCH-L3 are single-domain proteins, sharing 52% sequence identity. UCH-L5 and BAP1 are multidomain proteins with the UCH domains located at their N-termini (Johnston et al., 1997; Fang and Shen, 2017) (Figure 1A). The UCH domains of all human paralogs are approximately 230 amino acids in length (Figure 1B). Structurally, UCHs fold into a 52 (Gordian) knotted topology with a central β-sheet surrounded by several α-helices (Johnston et al., 1997; Jarmolinska et al., 2019; Bishop et al., 2016). Their knotted topology is essential for DUB activity (Ferreira et al., 2024). They share a conserved catalytic triad consisting of cysteine, histidine, and aspartate residues responsible for cleaving ubiquitin from substrates (Figure 1C) (Das et al., 2006; Johnston et al., 1997; Misaghi et al., 2005; Nishio et al., 2009; Boudreaux et al., 2010). Being knotted also provides exceptional mechanostability to withstand pulling/unfolding-coupled proteasomal degradation, which is functionally relevant for UCH-L5, whose DUB activity is activated upon binding to the proteasomal subunit ADRM1/RPN13 (Sahtoe et al., 2015; Vanderlinden et al., 2015; Sriramoju et al., 2018; Sriramoju et al., 2020) Additionally, these proteins are also important model systems for studying the folding pathways of complex Gordian knotted proteins (Andersson et al., 2009; Zhang and Jackson, 2016; Lou et al., 2016; Ziegler et al., 2016; Lee and Hsu, 2018; Puri S. and Hsu S. T. D., 2022).
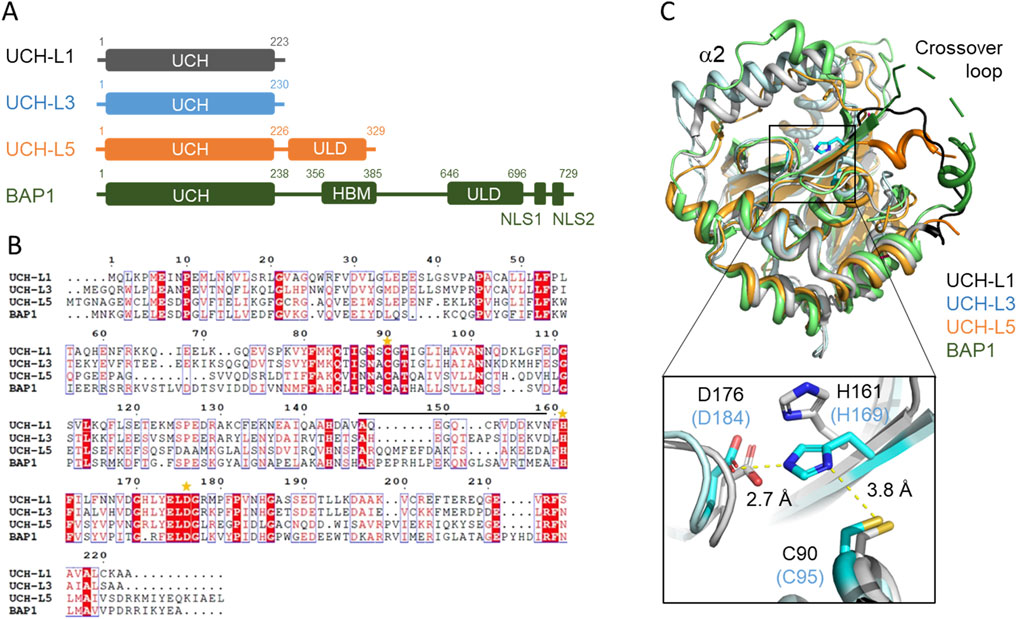
Figure 1. Multiple sequences and structural alignments of UCH-L1, UCH-L3, UCH-L5, and BAP1. (A) Domain organization of human UCHs; ULD: UCH37-liked domain; HBM: HCF binding domain; NLS: nuclear localization sequence. (B) Multiple sequence alignment of the human UCH domains. Asterisks indicate the conserved catalytic cysteine, histidine, and aspartate. The crossover loop residues are highlighted by a black line above the sequences. (C) Structural alignment of UCH-L1 (white; PDB ID: 2ETL), UCH-L3 (cyan; PDB ID: 1UCH), UCH-L5 UCH domain (gold; PDB ID: 3RII), and BAP1 UCH domain (green; PDB ID: 8H1T). The crossover loops are shown in darker colors. The crossover loops of UCH-L3 and BAP1 are disordered in the crystal structures. Inset: The catalytic triad of UCH-L1 and UCH-L3 with identities of the individual residues indicated in black and cyan for UCH-L1 and UCH-L3, respectively. The distances between the catalytic side-chain atoms are indicated.
UCHs are cysteine proteases that hydrolyze the (iso) peptide bond between ubiquitin and modified substrates through a nucleophilic attack by the catalytic cysteine to form a thioester intermediate between the catalytic cysteine and C-terminal glycine of ubiquitin. The final release of ubiquitin is mediated by a second nucleophilic attack by a water molecule (Boudreaux et al., 2010; Hanpude et al., 2017). The lengths of crossover loops, through which the ubiquitinated substrate accesses the catalytic sites of UCH proteins, vary among the four human paralogs, leading to different substrate specificities and catalytic efficiencies (Das et al., 2006; Misaghi et al., 2005; Nishio et al., 2009; Zhou et al., 2012; Mevissen and Komander, 2017). Having the shorter crossover loops, UCH-L1 and UCH-L3 cannot hydrolyze the iso-peptide bonds of K48-linked di-ubiquitin (Zhou et al., 2012), such that they are thought to be responsible for removing short peptide fragments on ubiquitin for ubiquitin recycling. In some cases, UCHs also process linear polyubiquitin chains (pro-ubiquitin) into mono ubiquitin for their proper functions (Ciechanover, 1998).
In addition to their role in the UPS, UCHs are also implicated in regulating different signaling pathways, DNA repair, neuroprotection, protection against oxidative stress, cell cycle regulation, mitochondrial function, tumor suppression, and oncogenesis (Komander et al., 2009; Clague et al., 2013; Bishop et al., 2016). Overall, these functions of the UCH family highlight their importance in a myriad of cellular processes and their potential significance in human health and diseases, including neurodegeneration and cancers (Lee and Hsu, 2017; Fang and Shen, 2017). UCHs are susceptible to post-translational modifications (PTMs, particularly redox-dependent oxidation of cysteines and methionines, which can be particularly detrimental to the DUB activities of UCHs (Lee et al., 2013). Therefore, the functional implications of oxidative stresses on the UCHs’ physiological roles are of biomedical importance.
UCHs exhibit abundant conformational plasticity, resulting in distinct open and closed conformations corresponding to the inactive (protonated catalytic cysteine) and active state (deprotonated catalytic cysteine) to allow substrate binding, catalysis, and interaction with regulatory molecules (Reyes-Turcu et al., 2009; Hsu, 2016). UCH-L3 undergoes a substantial folding-upon-binding process that sees the crossover loop wrap around ubiquitin to fold into a well-ordered conformation while being highly disordered without ubiquitin (Misaghi et al., 2005). Conformational changes upon ubiquitin binding are very subtle in the case of UCH-L1. Ubiquitin binding triggers a cascade of side-chain rearrangements to align the imidazole ring of histidine 161 (H161) towards the catalytic cysteine 90 (C90) in an appropriate geometry for efficient enzyme reactions (Das et al., 2006; Boudreaux et al., 2010). In the case of UCH-L5, much more subtle side-chain rearrangements are associated with ubiquitin binding. The ubiquitin-binding-induced conformational changes manifest in very different enzyme kinetics characteristics that show very different KM and kcat values in the Michaelis-Menten analyses of UCHs based on the fluorogenic ubiquitin 7-amido-4-methylcoumarin (Ub-AMC) hydrolysis assay (Table 1) (Hsu, 2016). While UCH-L1 and UCH-L3 exhibit comparable substrate binding affinities (KM) in the high nM range, the kcat values differ by two orders of magnitudes. This may be attributed to the misalignment of the catalytic triad of UCH-L1 in its apo form (Figure 1C). By contrast, the catalytic domain of UCH-L5 (residues 1–240, UCH-L51-240) exhibits poor substrate binding but a high turnover rate, resulting in a similar kcat/KM as that of UCH-L1, making them both poor enzymes in the context of Ub-AMC hydrolysis. The poor substrate binding of UCH-L51-240 may be attributed to the long crossover loop that could interfere with substrate binding while the catalytic triad is well-aligned to hydrolyze the substrate.
A large number of familial mutations, truncations, and PTMs, such as phosphorylation, ubiquitination, acetylation, and oxidation, have been documented in the literature for the human UCHs, contributing to altered enzymatic activities, functional dynamics, folding stability, subcellular localization, and protein-protein interactions (Wilkinson et al., 1989; Ventii et al., 2008; Eletr and Wilkinson, 2011; Lee and Hsu, 2017; Affar and Carbone, 2018; Wang and Wang, 2021). Small molecule inhibitors against human UCHs with high selectivity and affinities have actively been pursued (Panyain et al., 2020; Hewitt et al., 2022; Grethe et al., 2022). Considering their important biomedical implications and the underlying dynamics that cover a broad spectrum of timescales, it is vital to obtain a comprehensive understanding regarding how changes in structures and dynamics of UCHs upon protein-protein interaction, PTMs, and alternations of other environmental factors contribute to their biological functions.
To achieve this, one must go beyond the static snapshots of conformational states by protein crystallography and cryo-electron microscopy (cryo-EM) single particle analysis. On the one hand, solution-state nuclear magnetic resonance (NMR) spectroscopy is exceptionally versatile in gleaning atomic insights into protein dynamics across a broad spectrum of timescales (Kleckner and Foster, 2011; Tzeng and Kalodimos, 2011; Sekhar and Kay, 2019; Palmer and Koss, 2019; Alderson and Kay, 2021; Dyson and Wright, 2021). On the other hand, small-angle X-ray scattering (SAXS) provides a global view of protein structures and dynamics in solution (Hammel, 2012). Recently, hydrogen-deuterium exchange mass spectroscopy (HDX-MS) has become a robust tool to characterize protein dynamics and map binding interfaces without the need for elaborated stable isotope labeling needed for advanced NMR spectroscopy (Masson et al., 2017; Masson et al., 2019; Hodge, et al., 2019; Trabjerg et al., 2018). These advanced experimental tools are complemented by long-term and multiscale molecular dynamics (MD) simulations to investigate protein dynamics with increased spatial and temporal resolutions (Zheng et al., 2019; Paissoni et al., 2020; Peacock and Komives, 2021; Kenny et al., 2024; Paissoni et al., 2024). In the review, we shall discuss our attempt to integrate experimental biophysical tools to understand the intrinsic dynamics and effects of pathogenic mutations and PTMs on the structures and functions of human UCHs.
Main text
Impacts of disease-associated mutations on the structures and dynamics of UCH-L1
UCH-L1 is one of the most abundant proteins in human neurons, constituting 1–2% of total soluble protein in neuronal cells. Like other UCH members, UCH-L1 is a multifunctional enzyme and has been implicated in different biological processes, such as neuronal development, synaptic transmission, axonal transport, protection from oxidative stress, and ubiquitin homeostasis (Lee and Hsu, 2017). Specifically, UCH-L1 has been suggested to be an important biomarker for brain injury (Liu et al., 2019; Mi and Graham, 2023). At a molecular level, UCH-L1 is proposed to be a potentiator of cyclin-dependent kinase to enhance cell proliferation independent of its DUB activity (Kabuta et al., 2013). UCH-L1 variants harboring R63A and H185A mutations have been shown to bind to LAMP-2A associated with chaperone-mediated autophagy (Kabuta et al., 2008). UCH-L1 has even been shown to exhibit ubiquitin ligase activity toward α-synuclein (Liu et al., 2002). Nonetheless, the substrate specificity of UCH-L1 remains elusive.
UCH-L1 harbors a large number of clinically reported mutations, many of which autosomal dominant in the context Parkinson’s disease (PD) (https://www.uniprot.org/uniprotkb/P09936/entry) (Matuszczak et al., 2020). Specifically, the I93M mutation is an autosomal dominant risk factor found in PD patients Leroy et al., 1998) while the S18Y polymorphism is found be prevalent in Caucasian PD population but unlikely to be pathogenic (Lincoln et al., 1999; Healy et al., 2006). The E7A mutation is autosomal recessive transmission associated with childhood onset blindness (Bilguvar et al., 2013). Recombinant E7A variant shows >100-fold reduction in the DUB activity compared to the wild type. The R178Q and A216D mutations are compound heterozygous mutations found in a Norwegian family with early onset optic atrophy (Rydning et al., 2016). The expression of the A216D variant cannot be detected in patient cells while that of R178Q is 25% compared to control cells. Furthermore, bacteria-expressed recombinant A216D variant is insoluble, leading to loss of function, while recombinant R178Q variant exhibits an increased DUB activity based on the Ub-AMC assay (Rydning et al., 2016).
In this section, we shall focus on how two well-known mutations–I93M (UCH-L1I93M) and R178Q (UCH-L1R178Q) – affect the function of UCH-L1 through dynamic alterations in the catalytic triad and other regions without affecting the overall 3D structure according to X-ray crystallography (Boudreaux et al., 2010; Andersson et al., 2011; Sriramoju et al., 2015; Kenny et al., 2024). UCH-L1I93M is a mutant identified in a German family with PD (Lincoln et al., 1999). Recombinant UCH-L1I93M exhibits reduced DUB activity towards the model substrate, Ub-AMC (Healy et al., 2004). An animal study showed that over-expression of α-synuclein resulted in a significant loss of dopaminergic cells in UCH-L1I93M transgenic mice as compared to the wild-type (UCH-L1WT) mice, suggesting that the I93M mutation is linked to the progression of PD (Liu et al., 2002). Whole-exome sequencing of a Norwegian twin affected with early-onset neurodegenerative symptoms revealed heterozygous variants in UCH-L1 with R178Q (UCH-L1R178Q) and A216D (UCH-L1A216D) mutations (Nyberg-Hansen and Refsum, 1972; Rydning et al., 2016; Kenny et al., 2024). UCH-L1R178Q exhibits enhanced DUB activity and is proposed to play a neuroprotective role in maintaining cognition in these patients.
Nonetheless, the crystal structures of UCH-L1WT and UCH-L1I93M are virtually identical with a root mean square deviation (RMSD) of <0.3 Å for the backbone Cα atoms; the catalytic side-chains also align very well (Boudreaux et al., 2010; Kenny et al., 2024). In other words, the static snapshots of protein structures are insufficient to explain the loss or gain in the DUB activity (Figure 2A). To address this issue, we applied multidimensional and heteronuclear NMR spectroscopy to characterize the structures and dynamics of the UCH-L1 variants in solution states. In the case of UCH-L1, we showed that the backbone amide and side-chain methyl groups of UCH-L1I93M display large chemical shift perturbations far beyond the mutation site, indicating significant structural perturbations that cannot be probed by X-ray crystallography (Andersson et al., 2011; Sriramoju et al., 2015). Furthermore, NMR HDX analysis showed that the hydrophobic core residues in UCH-L1I93M exhibit increased fluctuations in their hydrogen bonding network, leading to an overall increase of the HDX rate by about 10-fold compared to that of UCH-L1WT, which is consistent with the decreased folding stability and increase aggregation propensity due to the I93M mutation. NMT HDX analysis of UCH-L1 probes very slow dynamics: The amide protons of the core residues in UCH-L1WT are fully exchanged after about 1 month whereas those of UCH-L1I9M become fully exchange with bulk solvent within days (Andersson et al., 2011). Equilibrium unfolding of UCH-L1 variants by urea showed a three-state unfolding process. The I93M mutation results in a reduced free energy of unfolding from the native state (N) to the intermediate state (I), ΔΔGI-N, by 1.36 ± 0.16 kcal mol-1 and a reduced free energy of unfolding from the intermediate state (I) to the denatured state (D), ΔΔGD-I, by 2.57 ± 0.27 kcal mol-1. The results were derived from global fitting to the changes in intrinsic fluorescence and far-UV circular dichroism (CD) signals as a function of urea concentration (Andersson et al., 2011) (Figure 2B).
Figure 2. Effects of disease-associated mutations on the structures and dynamics of UCHL1. (A) Structural alignment of the crystal structures of UCH-L1WT (white; PDB ID: 2ETL), UCH-L1I93M (magenta; PDB ID: 3IRT), and UCH-L1R178Q (green; PDB ID: 8DY8). Inset: structural alignment of the catalytic residues C90, H161, and D176 shown in sticks. The carbon atoms of UCH-L1WT, UCH-L1I93M, and UCH-L1R178Q are shown in white, magenta, and green, respectively. The nitrogen, oxygen, and sulfur atoms are shown in blue, red, and gold, respectively. (B) Structural mapping of chemical shift perturbations (CSPs) induced by the I93M mutation. The backbone amide nitrogen and side methyl carbon atoms that exhibit significant CSPs are shown in cyan and light pink spheres, respectively. The sizes of the backbone amide nitrogen atoms are shown in small, medium, and large sizes corresponding to composite (15N+1H) CSPs greater than 0.2, 0.5, and 1.0 ppm, respectively. The mutation site is shown in dark blue spheres. (C) Structural mapping of CSPs induced by the R178Q mutation. The backbone amide nitrogen atoms that exhibit significant CSPs–defined as the composite (15N+1H) CSPs greater than two standard deviations, 0.08 ppm–are shown in cyan spheres; those that are broadened beyond detection are shown in yellow spheres. The severe line-broadening effects are distributed globally.
UCH-L1 is metastable with a highly populated partially unfolding form (PUF) under native conditions (Lou et al., 2016). The PUF of UCH-L1 shares a highly stable core structure with a urea-induced folding intermediate, which can form a well-defined dimeric assembly despite being partially unfolded according to SAXS analysis (Lee and Hsu, 2018). Such a dimeric intermediate can be formed reversibly upon the removal and addition of urea under equilibrium conditions. However, the destabilization due to the I93M mutation results in rapid and irreversible aggregation of UCH-L1I93M in the presence of 3–4 M urea (Lee and Hsu, 2018). Note that the intrinsic disorder of UCH-L1 intermediate is inferred from the Kratky plot analysis that reports on the time-averaged X-ray scattering contributions from the conformational ensemble so there is no timescale information.
Contrary to UCH-L1I93M, which showed a well-resolved fingerprint two-dimensional backbone amide 15N-1H correlation spectrum and the three-dimensional triple resonance (1H, 13C, and 15N) spectra that enable complete backbone resonance assignments, more than 40 backbone amide 15N-1H correlations could not be traced in UCH-L1R178Q, which was attributed to altered protein dynamics on the µs-ms timescale that lead to unfavorable line broadening beyond detection. Structural mapping of these disappeared residues showed that the alterations in protein dynamics affect residues more than 20 Å away from the R178Q mutation site (Kenny et al., 2024) (Figure 2C). Despite the widespread changes in protein dynamics, likely on the µs-ms timescale, comparison of the SAXS/WAXS data of UCH-L1WT and UCH-L1R178Q suggested no changes in the global dimension, i.e., no unfolding, according to the Guinier plot analysis, and a minimal increase in the global dynamics according to the Kratky plot analysis (Kenny et al., 2024). Collectively, the NMR and SAXS/WAXS data indicated subtle dynamic changes within UCH-L1R178Q that are sufficient to generate significant functional changes (a 3-fold increase in the DUB activity). X-ray crystallography under crystalline states cannot probe such a dynamic change.
Mapping of oxidation-mediated altered dynamics in UCH-L1
UCH-L1 undergoes various PTMs, including phosphorylation, ubiquitination, and oxidation. Recently, it has been established that many DUBs, including UCH-L1, protect cells from oxidative stress by absorbing reactive oxygen species (ROS) through oxidation of their catalytic cysteine (Lee et al., 2013). Previous studies on PD patient samples show that C90 and C152 of UCH-L1 are irreversibly oxidized (C-SO2H and C-SO3H) and aggregate into Lewy bodies, a hallmark of PD (Choi et al., 2004). We set out to investigate how UCH-L1 changes its structure, dynamics, and function during the oxidation-reduction cycle. To mimic the cellular oxidation, we treated recombinant UCH-L1 with H2O2 and mapped the oxidized residues using mass spectrometry. We find that residues M1, M6, M12, C90, and C152 are highly sensitive to oxidation (Puri and Hsu, 2021). Furthermore, we dissected the effects of oxidization of individual residues on the structure, dynamics, and function of UCH-L1. We showed that oxidation of catalytic C90 is highly deleterious to both structure and function due to an increase in polarity of the hydrophobic core of UCH-L1, which contributes to partial unfolding, reduced stability, soluble oligomerization, and functional loss (Puri and Hsu, 2021).
In contrast, the oxidation of M1, M6, M12, and C152 under mild H2O2 stress (treatment with 30-fold H2O2 for 60–120 min) did not affect protein structure as the in-solution structural features captured by far-UV CD, intrinsic and ANS-based extrinsic fluorescence show no structural differences between the native and oxidized forms. Nonetheless, we found a reduction in the DUB activity that reflects a two-fold decrease in substrate binding, i.e., KM. This finding prompts us to investigate the effect of oxidation on the dynamics of UCH-L1. We find that oxidation of M1, M6, M12, and C152 impacts the dynamics of α-helix α1 and crossover loop, contributing to its reduced substrate affinity and hence DUB activity (Figure 3) (Puri and Hsu, 2021). Moreover, PTMs by bulky groups like cyclopentenone prostaglandin and 4-hydroxy-2-nonenal (HNE) lead to significant unfolding and loss of DUB activity in UCH-L1 protein due to disruption of hydrophobic interaction network in the folded UCH-L1 protein which ultimately leads to aggregation and functional loss (Koharudin et al., 2010; Jackson et al., 2012). Note that the half-life of UCH-L1 in sera is relatively long (<12 h) and the long half-life has been exploited to serve as a biomarker to monitor brain trauma together with glial fibrillary acidic protein (GFAP) (Diaz-Arrastia et al., 2014; Korley et al., 2022). The high abundance and long half-life of UCH-L1 could potentially contribute to a physiology role in absorbing ROS. Indeed, upregulation of UCH-L1 expression is induced by oxidative stress and UCH-L1 is proposed to be part of an anti-oxidative stress response in podocytes (Reichelt et al., 2023).
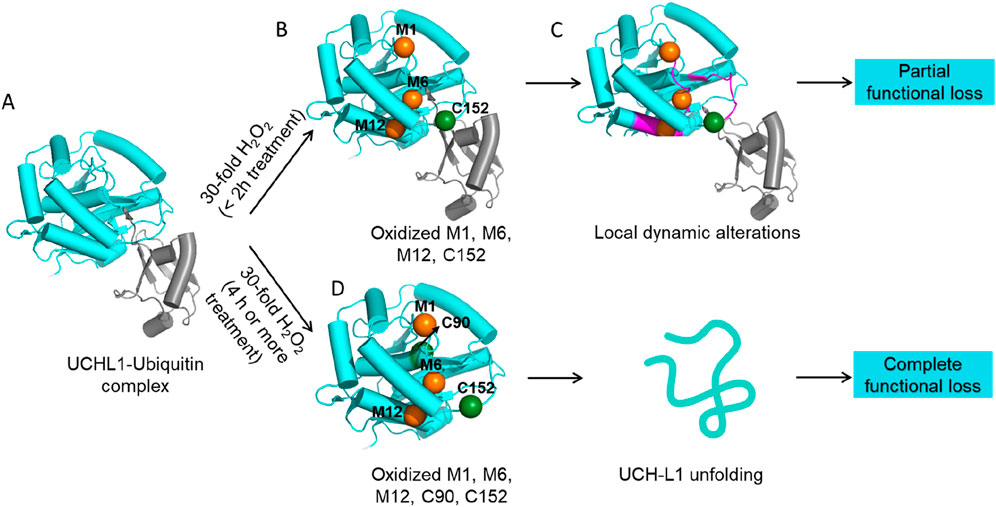
Figure 3. Dynamic changes in UCH-L1 upon oxidation. (A) Native UCH-L1 interacts with ubiquitin through α-helix α1 and crossover loop. (B) Mild oxidation of UCH-L1 results in oxidation of M1, M6, M12 (orange spheres), and C152 (green sphere) residues. (C) Oxidized M1, M6, M12, and C152 cause local dynamic alterations in UCH-L1 in α-helix α1, and a crossover loop is shown in magenta, which results in poor affinity for ubiquitin and hence partial loss in function. (D) Moderate oxidation of UCH-L1 (treatment with 30-fold H2O2 for over 4 h) results in the oxidation of catalytic C90 in addition to M1, M6, M12, and C152. Oxidation of C90 and the change in polarity in the hydrophobic core collectively contribute to the unfolding, loss of ubiquitin binding, and complete functional loss in UCH-L1.
Structures and dynamics of UCH-L3
Unlike UCH-L1, UCH-L3 is ubiquitously expressed in all tissue types. It is a main player in apoptosis and tumorigenesis, including breast, non-small lung, prostate, and ovarian cancers. Its interaction with ubiquitin and Nedd8 is critical in germ-cell apoptosis (Wada et al., 1998). However, its exact contribution to the progression or inhibition of cancer remains controversial. UCH-L3 shares a 52% sequence similarity with UCH-L1. The apo form crystal structures of UCH-L1 and UCH-L3 are also very similar: the overall pairwise positional RMSD of the resolved Cα atoms is only 0.8 Å. Nonetheless, the α-helix α2 exhibits a clear translational displacement between UCH-L1 and UCH-L3, and the crossover loop of UCH-L3 is highly disordered in the apo form (Figure 4A). A more subtle but functionally critical structural difference is the side-chain arrangements of the catalytic triad between UCH-L1 and UCH-L3. The imidazole side-chain of H161 of UCH-L1 is pointing away from the thiol side-chain of the catalytic C90, which the general base cannot stabilize until ubiquitin binding to UCH-L1 that triggers a cascade of side-chain conformational changes to align the catalytic side-chains to a productive conformation (Boudreaux et al., 2010) (Figure 1C). This misalignment may contribute to their significantly different DUB activities, as UCH-L3 is nearly 200 times more efficient as a DUB compared to UCH-L1 (Table 1) (Johnston et al., 1997; Misaghi et al., 2005; Hafez et al., 2022). Upon binding to ubiquitin, UCH-L3 undergoes substantial folding upon binding for the crossover loop and the following α-helix (Misaghi et al., 2005) (Figure 4B). The presence of another ubiquitin that covalently modifies that proximal ubiquitin through the K27 iso-peptide bond linkage does not significantly perturb the overall structure of the ubiquitin-bound UCH-L3 (van Tilburg et al., 2021) (Figure 4C).
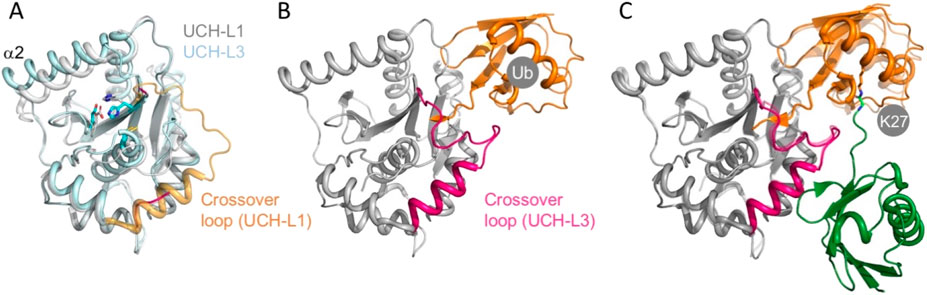
Figure 4. Structural plasticity of UCH-L3. (A) Structural alignment of UCH-L1 (white, PDB ID: 2ETL) and apo UCH-L3 (light cyan, PDB ID: 1XD3). The crossover loops and the following α-helices of UCH-L1 and UCH-L3 are colored gold and magenta, respectively; the latter is highly disordered. A significant displacement between the two structures can be seen for α-helix 2. (B) The crystal structure of UCH-L3 in complex with ubiquitin (Ub; PDB ID: 1XD3). Ubiquitin (Ub) is colored orange. The crossover loop and the following helix become ordered upon binding to Ub. (C) The crystal structure of UCH-L3 in complex with a K27-linked di-ubiquitin (PDB ID: 6QML). The proximal and distal Ubs are colored orange and green, respectively. The side-chain of K27 of the proximal Ub and that of the C-terminal G76 of the distal Ub are shown in sticks with matching colors. The structure is superimposed with the highly transparent structure as in (B), which shows minimal structural changes in UCH-L3 and a minor rotation of the proximal Ub.
Structures and dynamics of UCH-L5
UCH-L5 is also known as UCH37; it is closely related to BAP1 with >40% sequence identity. UCH-L5 contains a coiled-coil C-terminal UCH37-like domain (ULD), which inhibits the DUB activity of UCH-L5 in cis by covering the ubiquitin-binding site, which can be relieved by binding to the helical bundle DEUBAD domain of the proteasomal subunit, RPN13 (Figure 5A). In contrast, the nuclear factor related to kappa-B-binding protein (NFRKB) – also known as INO80 – binds to the ULD of UCH-L5 to inhibit the DUB activity by covering the ubiquitin-binding interface of UCH-L5 (Figure 5B). The Hill and Sixma groups independently solved the two structures with virtually identical results (Vanderlinden et al., 2015; Sahtoe et al., 2015). RPN13 and INO80/NFRKB bind to the same ULD domain with opposing effects by changing the conformation of the long α-helix of the ULD connecting the UCH domain. In the activated form, RPN13 DEUBAD keeps the long α-helix straight, while in the inactivated form, NFRKB/INO80 bends the long α-helix of the ULD and uses its long helix to cover the ubiquitin entrance.
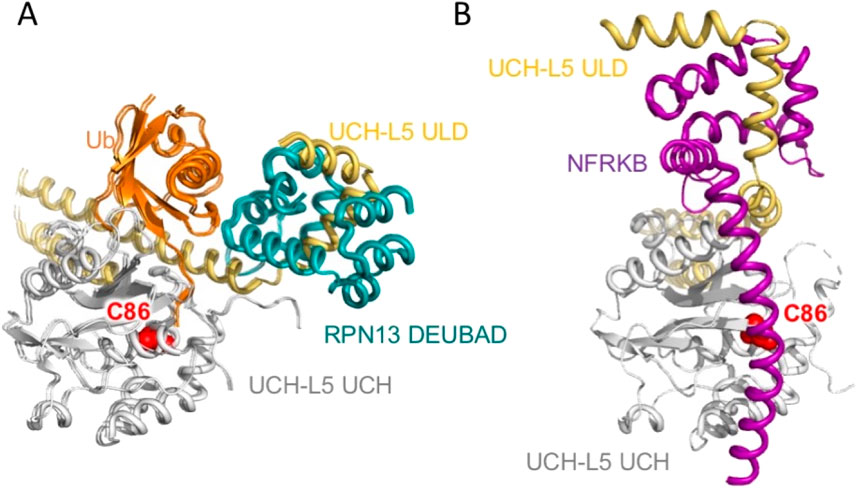
Figure 5. Structure of UCH-L5 in complex with ubiquitin and the DEUBAD domain of RPN13. (A) Cartoon representation of the ternary complex of the UCH domain and ULD of UCH-L5, Ub, and the DEUBAD domain of RPN13, which are colored white, yellow, orange, and teal, respectively (PDB ID: 4WLR). (B) Cartoon representation of the binary complex of the UCH domain and ULD of UCH-L5, and the nuclear factor related to NFRKB, also known as INO80, which are colored white, yellow, orange, and purple, respectively (PDB ID: 4WLP). Both structures are aligned with respect to the UCH domain of UCH-L5. The catalytic C86 of UCH-L5 is shown in red spheres in both panels and indicated with its residue name.
In addition to the plasticity of the ULD of UCH-L5, the long crossover loop of UCH-L5 has been demonstrated to be pivotal for its ability to hydrolyze the K48-linked di-ubiquitin whereas UCH-L1 and UCH-L3 cannot. Grafting the crossover loop of UCH-L5 onto UCH-L1 and UCH-L3 renders the chimeras capable of processing the K48-linked di-ubiquitin (Zhou et al., 2012). Nonetheless, the atomic structure of UCH-L5 in complex with K48-linked di-ubiquitin is currently unknown. It is unclear how the long crossover loop of UCH-L5 would flap up or down to accommodate the K48-linked di-ubiquitin, which requires a lot more space than the insertion of the C-terminus of a mono-ubiquitin (Lee et al., 2017).
Mapping of mutation-mediated dynamics in BAP1
BAP1 is a master regulator of tumor suppression (Carbone et al., 2013). It is 729 amino acids long with the N-terminal UCH domain, followed by the host cell factor binding motif (HBM), non-organizing region (NORS), UCH37-like domain (ULD), and two nuclear localization signals (NLS1 and NLS2) at the C-terminus (Figure 1A) (Jensen et al., 1998; Ventii et al., 2008; Masclef et al., 2021). These functional domains interact with many cellular proteins, including host cell factor 1 (HCF-1), INO80, transcription factor Yin Yang 1 (YY1), BRCA1-associated Ring domain protein 1 (BRAD1), and sex comb-like transcription regulators 1/2 (ASXL1/2). BAP1 contributes to various cellular processes, ranging from chromatin remodeling, cell cycle progression, regulation of transcription, cell proliferation, apoptosis, and regulation of chromosome stability (Pan et al., 2015; Carbone et al., 2013; Sime et al., 2018). Many germline mutations in the BAP1 gene are linked to the high incidence of aggressive cancers, such as uveal melanoma, renal cell carcinoma, and cutaneous melanoma. In contrast, low-frequency somatic mutations contribute to lung, breast, and colon cancers. According to the Catalog of Somatic Mutations in Cancer (COSMIC) database, 60% of cancer-associated mutations in BAP1 are located within the UCH domain (BAP1-UCH) (Bhattacharya et al., 2015). Additionally, substitution, deletion, and truncation in the UCH domain of BAP1 all contribute to the onset and progression of different cancers. We and other groups have characterized many catalytic and non-catalytic mutations within BAP1-UCH, including S10N, G45R, I47F, S63C, N78S, F81V, C91G, C91S, C91W, A95D, G128R, H169Q, Y173D, G178V and W196G, to understand their impacts on structure, dynamics, stability, and DUB activity (Bhattacharya et al., 2015; Puri et al., 2022).
We have shown by differential scanning calorimetry (DSC) that all BAP1-UCH variants (residues 1–238) exhibit two distinct thermal unfolding transitions (Puri et al., 2022). The two melting temperatures of wild type BAP1-UCH are 46.9 and 50.0°C. Most of the cancer-associated variants exhibit reduced melting temperatures by as much as 7°C compared to wild type. In contrast, the catalytically inactive C91G variant shows significantly increased melting temperatures by 5.6 and 4.6°C compared to those of wild type. In addition, significant loss of the enthalpy of unfolding is observed that correlate with the reduced melting temperatures. Furthermore, the compactness of the BAP1-UCH variants defined by their radii of gyration (Rg) derived from small angle X-ray scattering (SAXS) also correlates well with the enthalpy of unfolding. The most destabilized variants, F81V and G128R have an Rg value of 26.6 ±0.4 and 26.9 ± 0.3 Å compared to 21.4 ± 0.1 Å for wild type. These two highly destabilized variants exhibited increased aggregation propensity evidenced by their fast aggregation kinetics monitored by 8-Anilinonathalene-1-sulfonic acid (ANS) and thioflavin T (ThT) fluorescence. They also exhibit significantly reduce the DUB activity despite their distant location (more than 20 Å away) from the catalytic triad.
To examine how these cancer-associated mutations impact on the folding stability of BAP1-UCH, we used HDX-MS to probe the folding dynamics on the minute-to-hour timescale manifested in deuterium uptakes of individual peptides. The deuterium uptake of peptide backbone amide groups reflect solvent exposure and hydrogen bond stability. An increased deuterium uptake for a given peptide segment indicates increased fluctuations of the corresponding local structure. Pairwise comparison of the deuterium uptakes of F81V and F128R with respect to WT shows significantly higher deuterium uptake in the β-sheet-rich hydrophobic core of the mutants, which may contribute to the mutants’ increased aggregation propensity and reduced DUB activity.
Another interesting mutation is N78S, which does not affect structural features and stability (similar mid-point of denaturation to WT BAP1 in a thermal unfolding experiment) but shows a two-fold increase in the DUB activity of BAP1. HDX-MS data showed that N78S mutation results in enhanced dynamics in the catalytic triad, which could contribute to the increased DUB activity. For other high-frequency cancer-associated mutations, we also observed long-range perturbation of the deuterium uptakes far away from the mutation sites (Figure 6) (Puri et al., 2022).
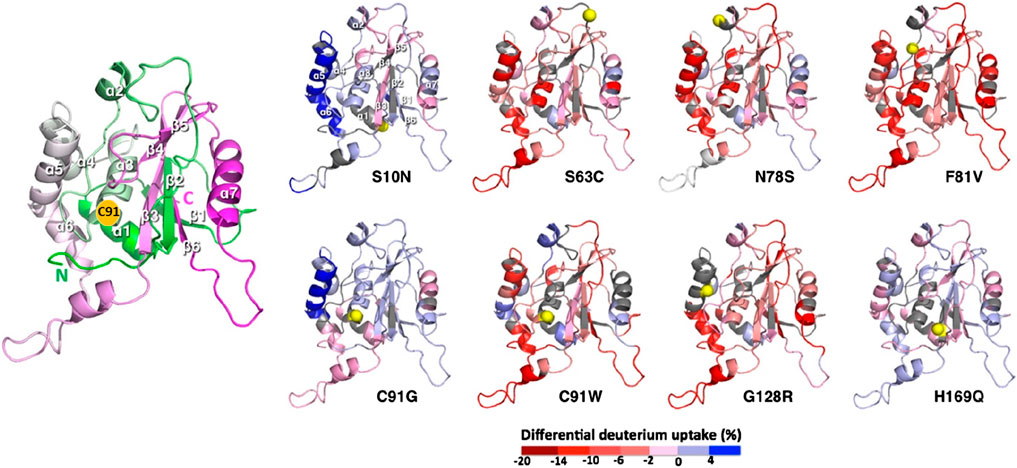
Figure 6. HDX-MS analysis of BAP1 variants harboring cancer-associated mutations. Structural mapping of differential deuterium uptakes of cancer-associated variants to WT. The cartoon representations of BAP1-UCH variants are color-coded blue-to-dark red, corresponding to reduced-to-increased deuterium uptakes of individual peptides relative to WT. The mutation sites are indicated in yellow spheres. The uncovered regions are colored in gray. An enlarged reference cartoon representation of BAP1-UCH is shown on the left, which is color-ramped from green to white to magenta for the residues from the N terminus to the C terminus. The individual secondary structural elements are labeled accordingly (Figure adapted from Puri et al. (2022), with permission from Elsevier).
Mapping of oxidation-mediated altered dynamics in BAP1-UCH
Similar to other DUBs, BAP1 undergoes various PTMs, including phosphorylation, ubiquitination, and oxidation (Pan et al., 2015; Puri and Hsu, 2022b). Unlike UCH-L1, however, oxidation of cysteine residues is highly deleterious for the structure and function of the UCH domain of BAP1 (BAP1-UCH) (Puri and Hsu, 2022a). BAP1-UCH contains three cysteines hidden inside the hydrophobic core. In such a scenario, catalytic cysteine (C91) undergoes oxidation first due to its lower pKa followed by the other two cysteines (C39 and C102) (Puri and Hsu, 2022b). Oxidation of catalytic cysteine increases the polarity in the hydrophobic core and results in partial unfolding and irreversible aggregation of BAP1-UCH, which was reversible in the case of UCH-L1 due to a surface-exposed cysteine C152. This work highlighted that the overall organization of cysteine residues in 3D structures plays a critical role in making UCH-L1 a ROS scavenger (Puri and Hsu, 2022b; Puri and Hsu, 2021). However, oxidation of BAP1 might be pathogenic to the cells due to its complete loss of function and aggregation, which needs to be verified with future cell-based or model organism-based experiments.
Mapping of binding partners induced dynamic alterations in BAP1
The DUB activity of BAP1 is allosterically modulated by its binding partners. One of the most explored mechanisms for this is the enhancement of the DUB activity of BAP1 due to the binding of polycomb group-like protein ASXL1/2 proteins with the ULD domain of BAP1. The ternary complex of the BAP1-UCH domain, BAP1-ULD domain, and ASXL2-AB domain has a 1:1 stoichiometry with a dissociation constant, Kd, of 4.26 µM. The modest Kd is appropriate for maintaining reversible binding. Biochemical investigations on the binding sites showed that ASXL2-AB binds to residues R666-H669 of the BAP1-ULD domain. The binding stabilizes the crossover loop without affecting the conformation of the catalytic residues. Stabilization of the crossover loop contributes to the higher binding affinity of the UCH domain for ubiquitin and, therefore, enhanced DUB activity. The enhanced activity of BAP1 is responsible for the deubiquitylation of histone H2A on chromatin to regulate the activity of transcription factors (Peng et al., 2018; Peng et al., 2021; De et al., 2019).
Despite the biomedical importance of BAP1, it has been refractory to crystallization for high-resolution crystallographic structure determination. We have been able to use one-dimensional methyl NMR spectra to assess the conformational differences between different cancer-associated variants of BAP1, but the structural details are limited. Recently, the cryo-EM structure of BAP1 in complex with ubiquitinated H2A as part of the Polycom repressive deubiquitinase complex (PR-DUB) has been independently reported, representing a breakthrough in our structural understanding of BAP1 (Thomas et al., 2023; Ge et al., 2023). Nonetheless, the local resolutions of BAP1-UCH are limited due to the intrinsic dynamics of the UCH domain outside the core histone complex (Figure 7). Subsequent local refinement has improved the cryo-EM map of the BAP1-ASXL1 (1–378)-Ub sub-complex has been improved to 3.9 Å to enable unambiguous definition of the backbone structure (Ge et al., 2023).
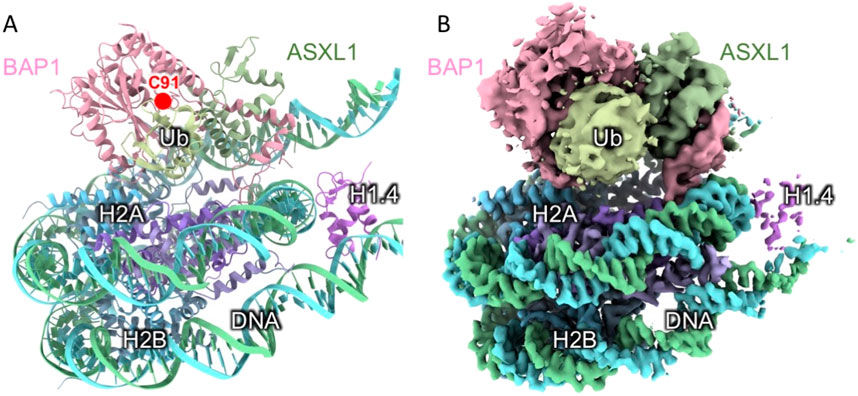
Figure 7. Cryo-EM structure of BAP1 as part of the PR-DUB complex. The atomic model (PDB ID: 8H1T) (A) and the associated cryo-EM map (EMD-34431) (B) of the PR-DUB complex. The individual subunits are differentially colored and indicated accordingly. The local resolution of BAP1 is insufficient to resolve the secondary structures as opposed to the well-resolved base pairing of the DNA and the α-helix of H2A. The cryo-EM map is displayed at the recommended contour level of 4.6 σ according to EMDataResource (https://www.emdataresource.org/EMD-34431). The position of the catalytic C91 of BAP1 is indicated in (A).
Mechanostability in human UCHs
All UCHs share an intricately knotted backbone topology with five projected non-reducible crossings to form a 52 Gordian knot (Hsu, 2023). The folding mechanisms of UCHs have been experimentally characterized in detail (Andersson et al., 2009; Andersson et al., 2011; Lee et al., 2017; Lou et al., 2016). Similar to the much larger SPOUT RNA methyltransferases superfamily, the knotted structural elements are required to bind to substrates or co-factors, including the Ub binding motif of UCHs (Hsu, 2023). Untying the topological knots of 31 trefoil-knotted YbeA and YibK by circular permutation abolished their co-factor binding pertinent to the RNA methylation function while maintaining the native contacts and three-dimensional structures (Ko et al., 2019; Chuang et al., 2019). Progressive truncation of the N-terminus of UCH-L1 also abolishes its DUB activity, as predicted by molecular dynamics simulations (Ferreira et al., 2024).
The intricate entanglements of topologically knotted proteins can be tightened into tight knots when subject to pulling-induced unfolding from one or both ends. Indeed, mechanical pulling experiments have been applied to several topologically knotted proteins, including UCH-L1 (Bornschlögl et al., 2009; He et al., 2014; Ziegler et al., 2016). When pulled from different positions other than the N- and C-termini, UCH-L1 can be transformed into other knot types as a result of partial disentanglement (Ziegler et al., 2016). In addition to the single-molecule force microscopy analysis of knotted proteins, their mechanostability has been examined biochemically with the aid of the bacterial proteasome ClpXP (Cordova et al., 2014). By appending a short ssrA tag to the C-termini of target proteins, ClpX would bind to the ssrA tag and exert mechanical unfolding force fueled by ATP hydrolysis to pull the unfolded substrate into the downstream ClpP proteolytic chamber for degradation. Compared to the small 31 knotted MJ0366 (San Martin et al., 2017), human UCHs are much more resistant to ClpXP-mediated force unfolding-coupled proteolysis (Sriramoju et al., 2018; Sriramoju et al., 2020). Specifically, UCH-L5 exhibits the slowest ClpXP proteolysis rate, which is four orders of magnitude slower than that of the green fluorescent protein, GFP, despite the much lower thermal and chemical stability of UCH-L5 compared to that of GFP (Sriramoju et al., 2018). The truncation of the first 11 residues, UCH-L1∆11, resulted in accelerated ClpXP proteolysis; the truncation also results in the loss of the 52 knot as UCHs have a shallow knot at the N-termini, which could be unknotted by removing the first 6-8 residues. As UCH-L5 functions on the 26S proteasome after being activated upon binding to the RPN13 subunit (Figure 5A), being mechanically stable may have functional advantages when working on the proteasome that constantly unfolds substrates for subsequent proteolysis similar to ClpXP.
Conclusion
This review highlights how mutations, PTMs, and protein-protein interactions alter the functional dynamics of human UCHs. The molecular understanding of the underlying mechanisms regulating the DUB activity of individual UCHs is essential for defining the molecular basis of substrate specificity and their implications in different biological pathways. While the physiological substrates of BAP1 are well-documented and under detailed scrutiny, little is known about the bona fide substrates of UCH-L1, UCH-L3, and UCH-L5. This review also underscores the need for an integrative structural biology and biophysics approach as the DUB activities of UCHs are tightly coupled with their dynamics under different physiological conditions that include the effects of PTMs, mutations, and substrate binding. These dynamic features may not be accessible to crystallography and cryo-EM analyses that afford high-resolution structural information as snapshots of functional states. Long-term molecular dynamics simulations, single molecule FRET, and quantitative NMR analyses about the conformational exchanges across different timescales could potentially be employed to glean a comprehensive view of the functional dynamics of UCHs, which are critical for a better understanding of UCH biology.
Author contributions
SP: Visualization, Writing–original draft. S-TD: Funding acquisition, Resources, Visualization, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Academia Sinica intramural fund, an Academia Sinica Career Development Award, Academia Sinica to STDH (AS-CDA-109-L08), and the National Science and Technology Council (NSTC), Taiwan (110-2113-M-001-050-MY3 and NSTC113-2123-M-001-010-) to STDH. SP acknowledges the Department of Science and Technology for Inspire faculty funding DST/INSPIRE/04/2023/000261, IISER Pune, India, and Fondazione Veronesi, Italy for postdoctoral work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdul Rehman, S., Kristariyanto, Y. A., Choi, S.-Y., Nkosi, P. J., Weidlich, S., Labib, K., et al. (2016). MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 63, 146–155. doi:10.1016/j.molcel.2016.05.009
Affar, E. B., and Carbone, M. (2018). BAP1 regulates different mechanisms of cell death. Cell Death and Dis. 9, 1151. doi:10.1038/s41419-018-1206-5
Alderson, T. R., and Kay, L. E. (2021). NMR spectroscopy captures the essential role of dynamics in regulating biomolecular function. Cell 184 (3), 577–595. doi:10.1016/j.cell.2020.12.034
Andersson, F. I., Pina, D. G., Mallam, A. L., Blaser, G., and Jackson, S. E. (2009). Untangling the folding mechanism of the 5 2 -knotted protein UCH-L3. FEBS J. 276, 2625–2635. doi:10.1111/j.1742-4658.2009.06990.x
Andersson, F. I., Werrell, E. F., McMorran, L., Crone, W. J. K., Das, C., Hsu, S.-T. D., et al. (2011). The effect of Parkinson’s-Disease-associated mutations on the deubiquitinating enzyme UCH-L1. J. Mol. Biol. 407, 261–272. doi:10.1016/j.jmb.2010.12.029
Bhattacharya, S., Hanpude, P., and Maiti, T. K. (2015). Cancer-associated missense mutations in BAP1 catalytic domain induce amyloidogenic aggregation: a new insight in enzymatic inactivation. Sci. Rep. 5, 18462. doi:10.1038/srep18462
Bilguvar, K., Tyagi, N. K., Ozkara, C., Tuysuz, B., Bakircioglu, M., Choi, M., et al. (2013). Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 110 (9), 3489–3494. doi:10.1073/pnas.1222732110
Bishop, P., Rocca, D., and Henley, J. M. (2016). Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution, and roles in brain function and dysfunction. Biochem. J. 473, 2453–2462. doi:10.1042/BCJ20160082
Bornschlögl, T., Anstrom, D. M., Mey, E., Dzubiella, J., Rief, M., and Forest, K. T. (2009). Tightening the knot in phytochrome by single-molecule atomic force microscopy. Biophysical J. 96, 1508–1514. doi:10.1016/j.bpj.2008.11.012
Boudreaux, D. A., Chaney, J., Maiti, T. K., and Das, C. (2012). Contribution of active site glutamine to rate enhancement in ubiquitin C-terminal hydrolases. FEBS J. 279, 1106–1118. doi:10.1111/j.1742-4658.2012.08507.x
Boudreaux, D. A., Maiti, T. K., Davies, C. W., and Das, C. (2010). Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCH-L1 into productive conformation. Proc. Natl. Acad. Sci. U.S.A. 107, 9117–9122. doi:10.1073/pnas.0910870107
Carbone, M., Yang, H., Pass, H. I., Krausz, T., Testa, J. R., and Gaudino, G. (2013). BAP1 and cancer. Nat. Rev. Cancer 13, 153–159. doi:10.1038/nrc3459
Case, A., and Stein, R. L. (2006). Mechanistic studies of ubiquitin C-terminal hydrolase L1. Biochemistry 45, 2443–2452. doi:10.1021/bi052135t
Choi, J., Levey, A. I., Weintraub, S. T., Rees, H. D., Gearing, M., Chin, L.-S., et al. (2004). Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J. Biol. Chem. 279, 13256–13264. doi:10.1074/jbc.M314124200
Chuang, Y.-C., Hu, I.-C., Lyu, P.-C., and Hsu, S.-T. D. (2019). Untying a protein knot by circular permutation. J. Mol. Biol. 431, 857–863. doi:10.1016/j.jmb.2019.01.005
Ciechanover, A. (1998). The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17, 7151–7160. doi:10.1093/emboj/17.24.7151
Clague, M. J., Barsukov, I., Coulson, J. M., Liu, H., Rigden, D. J., and Urbé, S. (2013). Deubiquitylases from genes to organism. Physiol. Rev. 93, 1289–1315. doi:10.1152/physrev.00002.2013
Cordova, J. C., Olivares, A. O., Shin, Y., Stinson, B. M., Calmat, S., Schmitz, K., et al. (2014). Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell 158, 647–658. doi:10.1016/j.cell.2014.05.043
Dang, L. C., Melandri, F. D., and Stein, R. L. (1998). Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry 37, 1868–1879. doi:10.1021/bi9723360
Das, C., Hoang, Q. Q., Kreinbring, C. A., Luchansky, S. J., Meray, R. K., Ray, S. S., et al. (2006). Structural basis for conformational plasticity of the Parkinson’s disease-associated ubiquitin hydrolase UCH-L1. Proc. Natl. Acad. Sci. U.S.A. 103, 4675–4680. doi:10.1073/pnas.0510403103
De, I., Chittock, E. C., Grötsch, H., Miller, T. C. R., McCarthy, A. A., and Müller, C. W. (2019). Structural basis for the activation of the deubiquitinase calypso by the polycomb protein ASX. Structure 27, 528–536.e4. doi:10.1016/j.str.2018.11.013
Diaz-Arrastia, R., Wang, K. K. W., Papa, L., Sorani, M. D., Yue, J. K., Puccio, A. M., et al. (2014). Acute biomarkers of traumatic brain injury: relationship between plasma levels of ubiquitin C-terminal hydrolase-L1 and glial fibrillary acidic protein. J. Neurotrauma 31 (1), 19–25. doi:10.1089/neu.2013.3040
Dyson, H. J., and Wright, P. E. (2021). NMR illuminates intrinsic disorder. Curr. Opin. Struct. Biol. 70, 44–52. doi:10.1016/j.sbi.2021.03.015
Eletr, Z. M., and Wilkinson, K. D. (2011). An emerging model for BAP1’s role in regulating cell cycle progression. Cell Biochem. Biophys. 60, 3–11. doi:10.1007/s12013-011-9184-6
Fang, Y., and Shen, X. (2017). Ubiquitin carboxyl-terminal hydrolases: involvement in cancer progression and clinical implications. Cancer Metastasis Rev. 36, 669–682. doi:10.1007/s10555-017-9702-0
Ferreira, S. G. F., Sriramoju, M. K., Hsu, S.-T. D., Faísca, P. F. N., and Machuqueiro, M. (2024). Is there a functional role for the knotted topology in protein UCH-L1? J. Chem. Inf. Model. 64, 6827–6837. doi:10.1021/acs.jcim.4c00880
Ge, W., Yu, C., Li, J., Yu, Z., Li, X., Zhang, Y., et al. (2023). Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase. Nature 616, 176–182. doi:10.1038/s41586-023-05841-y
Grethe, C., Schmidt, M., Kipka, G.-M., O’Dea, R., Gallant, K., Janning, P., et al. (2022). Structural basis for specific inhibition of the deubiquitinase UCH-L1. Nat. Commun. 13, 5950. doi:10.1038/s41467-022-33559-4
Hafez, N., Modather El-Awadly, Z., and Arafa, R. K. (2022). UCH-L3 structure and function: insights about a promising drug target. Eur. J. Med. Chem. 227, 113970. doi:10.1016/j.ejmech.2021.113970
Hammel, M. (2012). Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS). Eur. Biophysics J. 41, 789–799. doi:10.1007/s00249-012-0820-x
Hanpude, P., Bhattacharya, S., Kumar Singh, A., and Kanti Maiti, T. (2017). Ubiquitin recognition of BAP1: understanding its enzymatic function. Biosci. Rep. 37, BSR20171099. doi:10.1042/BSR20171099
He, C., Lamour, G., Xiao, A., Gsponer, J., and Li, H. (2014). Mechanically tightening a protein slipknot into a trefoil knot. J. Am. Chem. Soc. 136, 11946–11955. doi:10.1021/ja503997h
Healy, D. G., Abou-Sleiman, P. M., Casas, J. P., Ahmadi, K. R., Lynch, T., Gandhi, S., et al. (2006). UCHL-1 is not a Parkinson's disease susceptibility gene. Ann. Neurology 59 (4), 627–633. doi:10.1002/ana.20757
Healy, D. G., Abou-Sleiman, P. M., and Wood, N. W. (2004). Genetic causes of Parkinson's disease: UCH-L-1. Cell Tissue Res. 318, 189–194. doi:10.1007/s00441-004-0917-3
Hewitt, C. S., Das, C., and Flaherty, D. P. (2022). Rational development and characterization of a ubiquitin variant with selectivity for ubiquitin C-terminal hydrolase L3. Biomolecules 12, 62. doi:10.3390/biom12010062
Hodge, E. A., Benhaim, M. A., and Lee, K. K. (2019). Bridging protein structure, dynamics, and function using hydrogen/deuterium exchange mass spectrometry. Protein Sci. 29, 843–855. doi:10.1002/pro.3790
Hsu, S.-T. D. (2016). “Folding dynamics and structural basis of the enzyme mechanism of Ubiquitin C-Terminal Hydroylases,” in Understanding enzymes (Singapore: Jenny Stanford Publishing), 191–226. doi:10.1201/b19951-8
Hsu, S.-T. D. (2023). Folding and functions of knotted proteins. Curr. Opin. Struct. Biol. 83, 102709. doi:10.1016/j.sbi.2023.102709
Jackson, S., Werrell, E. F., Hsu, S.-T. D., Murphy, B., Crone, W., and Wetzel et al, S. (2012). The effect of HNE modification on the structure and function of the neuronal protein UCH-L: links to neurodegenerative disease. Free Radic. Biol. Med. 53 (Suppl. 1), S241–S242. doi:10.1016/j.freeradbiomed.2012.08.025
Jarmolinska, A. I., Perlinska, A. P., Runkel, R., Trefz, B., Ginn, H. M., Virnau, P., et al. (2019). Proteins’ knotty problems. J. Mol. Biol. 431, 244–257. doi:10.1016/j.jmb.2018.10.012
Jensen, D. E., Proctor, M., Marquis, S. T., Gardner, H. P., Ha, S. I., Chodosh, L. A., et al. (1998). BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16, 1097–1112. doi:10.1038/sj.onc.1201861
Johnston, S. C., Larsen, C. N., Cook, W. J., Wilkinson, K. D., and Hill, C. P. (1997). Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 Å resolution. EMBO J. 16, 3787–3796. doi:10.1093/emboj/16.13.3787
Kabuta, T., Furuta, A., Aoki, S., Furuta, K., and Wada, K. (2008). Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J. Biol. Chem. 283 (35), 23731–23738. doi:10.1074/jbc.M801918200
Kabuta, T., Mitsui, T., Takahashi, M., Fujiwara, Y., Kabuta, C., Konya, C., et al. (2013). Ubiquitin C-terminal hydrolase L1 (UCH-L1) acts as a novel potentiator of cyclin-dependent kinases to enhance cell proliferation independently of its hydrolase activity. J. Biol. Chem. 288 (18), 12615–12626. doi:10.1074/jbc.M112.435701
Kenny, S., Lai, C.-H., Chiang, T.-S., Brown, K., Hewitt, C. S., Krabill, A. D., et al. (2024). Altered protein dynamics and a more reactive catalytic cysteine in a neurodegeneration-associated UCH-L1 mutant. J. Mol. Biol. 436, 168438. doi:10.1016/j.jmb.2024.168438
Kleckner, I. R., and Foster, M. P. (2011). An introduction to NMR-based approaches for measuring protein dynamics. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1814, 942–968. doi:10.1016/j.bbapap.2010.10.012
Ko, K.-T., Hu, I.-C., Huang, K.-F., Lyu, P.-C., and Hsu, S.-T. D. (2019). Untying a knotted SPOUT RNA methyltransferase by circular permutation results in a domain-swapped dimer. Structure 27, 1224–1233.e4. doi:10.1016/j.str.2019.04.004
Koharudin, L. M. I., Liu, H., Di Maio, R., Kodali, R. B., Graham, S. H., and Gronenborn, A. M. (2010). Cyclopentenone prostaglandin-induced unfolding and aggregation of the Parkinson disease-associated UCH-L1. Proc. Natl. Acad. Sci. U.S.A. 107, 6835–6840. doi:10.1073/pnas.1002295107
Komander, D., Clague, M. J., and Urbé, S. (2009). Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563. doi:10.1038/nrm2731
Korley, F. K., Jain, S., Sun, X., Puccio, A. M., Yue, J. K., Gardner, R. C., et al. (2022). Prognostic value of day-of-injury plasma GFAP and UCH-L1 concentrations for predicting functional recovery after traumatic brain injury in patients from the US TRACK-TBI cohort: an observational cohort study. Lancet Neurology 21 (9), 803–813. doi:10.1016/S1474-4422(22)00256-3
Lee, J.-G., Baek, K., Soetandyo, N., and Ye, Y. (2013). Reversible inactivation of deubiquitinases by reactive oxygen species in vitro and in cells. Nat. Commun. 4, 1568. doi:10.1038/ncomms2532
Lee, Y.-T. C., Chang, C.-Y., Chen, S.-Y., Pan, Y.-R., Ho, M.-R., and Hsu, S.-T. D. (2017). Entropic stabilization of a deubiquitinase provides conformational plasticity and slow unfolding kinetics beneficial for functioning on the proteasome. Sci. Rep. 7, 45174. doi:10.1038/srep45174
Lee, Y.-T. C., and Hsu, S.-T. D. (2017). Familial mutations and post-translational modifications of UCH-L1 in Parkinson’s Disease and neurodegenerative disorders. CPPS 18, 733–745. doi:10.2174/1389203717666160217143721
Lee, Y.-T. C., and Hsu, S.-T. D. (2018). A natively monomeric deubiquitinase UCH-L1 forms highly dynamic but defined metastable oligomeric folding intermediates. J. Phys. Chem. Lett. 9, 2433–2437. doi:10.1021/acs.jpclett.8b00815
Leroy, E., Boyer, R., Auburger, G., Leube, B., Ulm, G., Mezey, E., et al. (1998). The ubiquitin pathway in Parkinson’s disease. Nature 395, 451–452. doi:10.1038/26652
Lincoln, S., Vaughan, J., Wood, N., Baker, M., Adamson, J., Gwinn-Hardy, K., et al. (1999). Low frequency of pathogenic mutations in the ubiquitin carboxy-terminal hydrolase gene in familial Parkinson’s disease. Neuroreport 10 (2), 427–429. doi:10.1097/00001756-199902050-00040
Liu, H., Povysheva, N., Rose, M. E., Mi, Z., Banton, J. S., Li, W., et al. (2019). Role of UCH-L1 in axonal injury and functional recovery after cerebral ischemia. Proc. Natl. Acad. Sci. U.S.A. 116, 4643–4650. doi:10.1073/pnas.1821282116
Liu, Y., Fallon, L., Lashuel, H. A., Liu, Z., and Lansbury, P. T. (2002). The UCH-L1 gene encodes two opposing enzymatic activities that affect α-synuclein degradation and Parkinson’s disease Susceptibility. Cell 111, 209–218. doi:10.1016/S0092-8674(02)01012-7
Lou, S.-C., Wetzel, S., Zhang, H., Crone, E. W., Lee, Y.-T., Jackson, S. E., et al. (2016). The knotted protein uch-l1 exhibits partially unfolded forms under native conditions that share common structural features with its kinetic folding intermediates. J. Mol. Biol. 428, 2507–2520. doi:10.1016/j.jmb.2016.04.002
Luchansky, S. J., Lansbury, P. T., and Stein, R. L. (2006). Substrate recognition and catalysis by UCH-L1. Biochemistry 45, 14717–14725. doi:10.1021/bi061406c
Masclef, L., Ahmed, O., Estavoyer, B., Larrivée, B., Labrecque, N., Nijnik, A., et al. (2021). Roles and mechanisms of BAP1 deubiquitinase in tumor suppression. Cell Death Differ. 28, 606–625. doi:10.1038/s41418-020-00709-4
Masson, G. R., Burke, J. E., Ahn, N. G., Anand, G. S., Borchers, C., Brier, S., et al. (2019). Recommendations for performing, interpreting, and reporting hydrogen-deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 16, 595–602. doi:10.1038/s41592-019-0459-y
Masson, G. R., Jenkins, M. L., and Burke, J. E. (2017). An overview of hydrogen-deuterium exchange mass spectrometry (HDX-MS) in drug discovery. Expert Opin. Drug Discov. 12, 981–994. doi:10.1080/17460441.2017.1363734
Matuszczak, E., Tylicka, M., Komarowska, M. D., Debek, W., and Hermanowicz, A. (2020). Ubiquitin carboxy-terminal hydrolase L1 – physiology and pathology. Cell Biochem. and Funct. 38, 533–540. doi:10.1002/cbf.3527
Mevissen, T. E. T., and Komander, D. (2017). Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192. doi:10.1146/annurev-biochem-061516-044916
Mi, Z., and Graham, S. H. (2023). Role of UCH-L1 in the pathogenesis of neurodegenerative diseases and brain injury. Ageing Res. Rev. 86, 101856. doi:10.1016/j.arr.2023.101856
Misaghi, S., Galardy, P. J., Meester, W. J. N., Ovaa, H., Ploegh, H. L., and Gaudet, R. (2005). Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 280, 1512–1520. doi:10.1074/jbc.M410770200
Nishikawa, K., Li, H., Kawamura, R., Osaka, H., Wang, Y. L., Hara, Y., et al. (2003). Alterations of structure and hydrolase activity of parkinsonism-associated human ubiquitin carboxyl-terminal hydrolase L1 variants. Biochem. Biophysical Res. Commun. 304, 176–183. doi:10.1016/s0006-291x(03)00555-2
Nishio, K., Kim, S.-W., Kawai, K., Mizushima, T., Yamane, T., Hamazaki, J., et al. (2009). Crystal structure of the deubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain. Biochem. Biophysical Res. Commun. 390, 855–860. doi:10.1016/j.bbrc.2009.10.062
Nyberg-Hansen, R., and Refsum, S. (1972). Spastic paraparesis associated with optic atrophy in monozygotic twins. Acta neurol. Scand. Suppl. 51, 261–263.
Ohayon, S., Spasser, L., Aharoni, A., and Brik, A. (2012). Targeting deubiquitinases enabled by chemical synthesis of proteins. J. Am. Chem. Soc. 134, 3281–3289. doi:10.1021/ja2116712
Paissoni, C., Jussupow, A., and Camilloni, C. (2020). Determination of protein structural ensembles by hybrid-resolution saxs restrained molecular dynamics. J. Chem. Theory Comput. 16, 2825–2834. doi:10.1021/acs.jctc.9b01181
Paissoni, C., Puri, S., Broggini, L., Sriramoju, M. K., Maritan, M., Russo, R., et al. (2024). A conformational fingerprint for amyloidogenic light chains. doi:10.1101/2024.07.12.603200
Palmer, A. G., and Koss, H. (2019). Chemical exchange. Methods Enzym. 615, 177–236. doi:10.1016/bs.mie.2018.09.028
Pan, H., Jia, R., Zhang, L., Xu, S., Wu, Q., Song, X., et al. (2015). BAP1 regulates cell cycle progression through E2F1 target genes and mediates transcriptional silencing via H2A monoubiquitination in uveal melanoma cells. Int. J. Biochem. and Cell Biol. 60, 176–184. doi:10.1016/j.biocel.2015.01.001
Panyain, N., Godinat, A., Lanyon-Hogg, T., Lachiondo-Ortega, S., Will, E. J., Soudy, C., et al. (2020). Discovery of a potent and selective covalent inhibitor and activity-based probe for the deubiquitylating enzyme UCH-L1, with antifibrotic activity. J. Am. Chem. Soc. 142, 12020–12026. doi:10.1021/jacs.0c04527
Peacock, R. B., and Komives, E. A. (2021). Hydrogen/deuterium exchange and nuclear magnetic resonance spectroscopy reveal dynamic allostery on multiple time scales in the serine protease thrombin. Biochemistry 60, 3441–3448. doi:10.1021/acs.biochem.1c00277
Peng, H., Cassel, J., McCracken, D. S., Prokop, J. W., Sementino, E., Cheung, M., et al. (2021). Kinetic characterization of asxl1/2-mediated allosteric regulation of the BAP1 deubiquitinase. Mol. Cancer Res. 19, 1099–1112. doi:10.1158/1541-7786.MCR-20-0080
Peng, H., Prokop, J., Karar, J., Park, K., Cao, L., Harbour, J. W., et al. (2018). Familial and somatic bap1 mutations inactivate ASXL1/2-mediated allosteric regulation of BAP1 deubiquitinase by targeting multiple independent domains. Cancer Res. 78, 1200–1213. doi:10.1158/0008-5472.CAN-17-2876
Puri, S., Chen, S.-N., Chiu, Y.-H., Draczkowski, P., Ko, K.-T., Yang, T.-J., et al. (2022). Impacts of cancer-associated mutations on the structure-activity relationship of BAP1. J. Mol. Biol. 434, 167553. doi:10.1016/j.jmb.2022.167553
Puri, S., and Hsu, S.-T. D. (2021). Crossover loop cysteine C152 acts as an antioxidant to maintain the folding stability and deubiquitinase activity of UCH-L1 under oxidative stress. J. Mol. Biol. 433, 166879. doi:10.1016/j.jmb.2021.166879
Puri, S., and Hsu, S. T. D. (2022a). “Elucidation of folding pathways of knotted proteins,” in Methods in enzymology (Elsevier), 275–297. doi:10.1016/bs.mie.2022.07.012
Puri, S., and Hsu, S.-T. D. (2022b). Oxidation of catalytic cysteine of human deubiquitinase BAP1 triggers misfolding and aggregation in addition to functional loss. Biochem. Biophysical Res. Commun. 599, 57–62. doi:10.1016/j.bbrc.2022.02.029
Reichelt, J., Sachs, W., Frömbling, S., Fehlert, J., Studencka-Turski, M., Betz, A., et al. (2023). Non-functional ubiquitin C-terminal hydrolase L1 drives podocyte injury through impairing proteasomes in autoimmune glomerulonephritis. Nat. Commun. 14, 2114. doi:10.1038/s41467-023-37836-8
Reyes-Turcu, F. E., Ventii, K. H., and Wilkinson, K. D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397. doi:10.1146/annurev.biochem.78.082307.091526
Rydning, S. L., Backe, P. H., Sousa, M. M., Iqbal, Z., Øye, A. M., Sheng, Y., et al. (2017). Novel UCHL1 mutations reveal new insights into ubiquitin processing. Hum. Mol. Genet. 26, 1031–1040. doi:10.1093/hmg/ddw391
Sahtoe, D. D., van Dijk, W., El Oualid, F., Ekkebus, R., Ovaa, H., and Sixma, T. K. (2015). Mechanism of UCH-L5 activation and inhibition by deubad domains in RPN13 and INO80G. Mol. Cell 57, 887–900. doi:10.1016/j.molcel.2014.12.039
San Martín, Á., Rodriguez-Aliaga, P., Molina, J. A., Martin, A., Bustamante, C., and Baez, M. (2017). Knots can impair protein degradation by ATP-dependent proteases. Proc. Natl. Acad. Sci. U.S.A. 114, 9864–9869. doi:10.1073/pnas.1705916114
Sekhar, A., and Kay, L. E. (2019). An NMR view of protein dynamics in health and disease. Annu. Rev. Biophysics 48, 297–319. doi:10.1146/annurev-biophys-052118-115647
Sime, W., Niu, Q., Abassi, Y., Masoumi, K. C., Zarrizi, R., Køhler, J. B., et al. (2018). BAP1 induces cell death via interaction with 14-3-3 in neuroblastoma. Cell Death Dis. 9, 458. doi:10.1038/s41419-018-0500-6
Sriramoju, M. K., Chen, Y., and Hsu, S.-T. D. (2020). Protein knots provide mechano-resilience to an AAA+ protease-mediated proteolysis with profound ATP energy expenses. Biochimica Biophysica Acta (BBA) - Proteins Proteomics 1868, 140330. doi:10.1016/j.bbapap.2019.140330
Sriramoju, M. K., Chen, Y., Lee, Y.-T. C., and Hsu, S.-T. D. (2018). Topologically knotted deubiquitinases exhibit unprecedented mechanostability to withstand the proteolysis by an AAA+ protease. Sci. Rep. 8, 7076. doi:10.1038/s41598-018-25470-0
Sriramoju, M. K., Lyu, P. C., and Hsu, S.-T. D. (2015). Structural perturbation of the Parkinson’s disease-associated I93M mutation in human UCH-L1 revealed by solution-state NMR spectroscopy. Chin. J. Magnetic Reson. 32, 329–340. doi:10.11938/cjmr20150215
Thomas, J. F., Valencia-Sánchez, M. I., Tamburri, S., Gloor, S. L., Rustichelli, S., Godínez-López, V., et al. (2023). Structural basis of histone H2A lysine 119 deubiquitination by polycomb repressive deubiquitinase BAP1/ASXL1. Sci. Adv. 9, 32. doi:10.1126/sciadv.adg9832
Trabjerg, E., Nazari, Z. E., and Rand, K. D. (2018). Conformational analysis of complex protein states by hydrogen/deuterium exchange mass spectrometry (HDX-MS): challenges and emerging solutions. TrAC Trends Anal. Chem. 106, 125–138. doi:10.1016/j.trac.2018.06.008
Tzeng, S. R., and Kalodimos, C. G. (2011). Protein dynamics and allostery: an NMR view. Curr. Opin. Struct. Biol. 21 (1), 62–67. doi:10.1016/j.sbi.2010.10.007
VanderLinden, R., Hemmis, C. W., Schmitt, B., Ndoja, A., Whitby, F. G., Robinson, H., et al. (2015). Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol. Cell 57, 901–911. doi:10.1016/j.molcel.2015.01.016
van Tilburg, G. B. A., Murachelli, A. G., Fish, A., van der Heden van Noort, G. J., Ovaa, H., and Sixma, T. K. (2021). K27-Linked diubiquitin inhibits UCHL3 via an unusual kinetic trap. Cell Chem. Biol. 28, 191–201.e8. doi:10.1016/j.chembiol.2020.11.005
Ventii, K. H., Devi, N. S., Friedrich, K. L., Chernova, T. A., Tighiouart, M., Van Meir, E. G., et al. (2008). BRCA1-Associated Protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 68, 6953–6962. doi:10.1158/0008-5472.CAN-08-0365
Wada, H., Kito, K., Caskey, L. S., Yeh, E. T. H., and Kamitani, T. (1998). Cleavage of the c-terminus of NEDD8 by UCH-L3. Biochem. Biophysical Res. Commun. 251, 688–692. doi:10.1006/bbrc.1998.9532
Wang, Y., and Wang, F. (2021). Post-translational modifications of deubiquitinating enzymes: expanding the ubiquitin code. Front. Pharmacol. 12, 685011. doi:10.3389/fphar.2021.685011
Wilkinson, K. D., Lee, K., Deshpande, S., Duerksen-Hughes, P., Boss, J. M., and Pohl, J. (1989). The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science 246, 670–673. doi:10.1126/science.2530630
Zhang, H., and Jackson, S. E. (2016). Characterization of the folding of a 5 2 -knotted protein using engineered single-tryptophan variants. Biophysical J. 111, 2587–2599. doi:10.1016/j.bpj.2016.10.029
Zheng, J., Strutzenberg, T., Pascal, B. D., and Griffin, P. R. (2019). Protein dynamics and conformational changes explored by hydrogen/deuterium exchange mass spectrometry. Curr. Opin. Struct. Biol. 58, 305–313. doi:10.1016/j.sbi.2019.06.007
Zhou, Z.-R., Zhang, Y.-H., Liu, S., Song, A.-X., and Hu, H.-Y. (2012). Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem. J. 441, 143–149. doi:10.1042/BJ20110699
Keywords: Deubiquitinase enzymes (DUBs), ubiquitin C-terminal hydrolases (UCHs), DUB activity, post-translational modifications, protein dynamics, NMR spectroscopy, mass spectrometry, hydrogen-deuterium exchange
Citation: Puri S and Hsu S-TD (2024) Functional dynamics of human ubiquitin C-terminal hydrolases. Front. Biophys. 2:1479898. doi: 10.3389/frbis.2024.1479898
Received: 13 August 2024; Accepted: 28 October 2024;
Published: 06 November 2024.
Edited by:
Albert Guskov, University of Groningen, NetherlandsReviewed by:
Ulrich Weininger, Martin Luther University of Halle-Wittenberg, GermanyDerek P. Claxton, Vanderbilt University, United States
Copyright © 2024 Puri and Hsu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shang-Te Danny Hsu, c3Roc3VAZ2F0ZS5zaW5pY2EuZWR1LnR3
 Sarita Puri
Sarita Puri Shang-Te Danny Hsu
Shang-Te Danny Hsu