
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Biophys. , 08 January 2024
Sec. Membrane Pores, Channels, and Transporters
Volume 1 - 2023 | https://doi.org/10.3389/frbis.2023.1334804
Mitochondrial Carrier Family proteins (MCFs) are located in the mitochondrial inner membrane and play essential roles in various cellular processes. Due to the relatively low abundance of many members of the family, in vitro structure and function determination of most MCFs require over-expression and purification of recombinant versions of these proteins. In this study, we report on a new method for overexpression of MCFs in Escherichia coli (E. coli) membranes, efficient purification of native-like proteins, and their reconstitution in mitochondrial inner membrane lipid mimics. cDNAs of Uncoupling Protein 4 (UCP4), Adenine Nucleotide Translocase (ANT) and Phosphate Translocase (PiT) were subcloned into the pET26b (+) expression vector such that fusion proteins with a short N-terminal pelB leader sequence and a six-histidine tag were produced to target the proteins toward the inner membrane of E. coli and facilitate affinity purification, respectively. Utilizing a modified autoinduction method, these proteins were overexpressed and extracted from the membrane of E. coli BL21 (DE3) and two modified strains, E. coli BL21 C43 (DE3) and E. coli BL21 Lobstr (DE3), in high yields. The proteins were then purified by immobilized metal affinity chromatography as monomers. Purity, identity, and concentration of the eluted monomers were determined by semi-native SDS-PAGE, Western blotting and mass spectrometry, and a modified Lowry assay, respectively. Cleavage of the pelB leader sequence from proteins was verified by mass spectrometric analysis. The purified proteins, surrounded by a shell of bacterial membrane lipids, were then reconstituted from the mild non-denaturing octyl glucoside (OG) detergent into phospholipid liposomes. Monomeric UCP4 spontaneously self-associated to form stable tetramers in lipid membranes, which is consistent with our previous studies. However, PiT and ANT remained dominantly monomeric in both detergent and liposome milieus, as detected by a combination of spectroscopic and electrophoretic methods. Native-like helical conformations of proteins were then confirmed by circular dichroism spectroscopy. Overall, this study demonstrates that targeting mitochondrial carrier family proteins to E. coli membranes provides an effective expression system for producing this family of proteins for biophysical studies.
In contrast to the mitochondrial outer membrane, which contains pores that allow the passage of ions and molecules up to the size of small proteins, the mitochondrial inner membrane (MIM) is more selectively permeable. This low permeability is partly due to the presence of a superfamily of membrane transporters, known as the mitochondrial carrier family (MCF) proteins or the solute carrier family (SLC25 protein family), that facilitate the transport of specific substrates between the mitochondrial intermembrane space and matrix (Palmieri and Pierri, 2010a; Palmieri and Monné, 2016). Interestingly, there are some reports of transporters belonging to this family that are found in organelles other than mitochondria, such as chloroplasts, peroxisomes, and mitosomes (Palmieri, 2013). MCF proteins, which are encoded by nuclear DNA (Ferramosca and Zara, 2013; Horten et al., 2020), play essential roles in cellular metabolism as they provide a link between metabolic reactions taking place in the mitochondrial matrix and cytosol by catalyzing the translocation of various solutes across the MIM. While most of these transporters have established functions, one-third remain ‘orphan’ with no experimentally determined substrate or transport activity (Nicholls, 2021). MCFs have different spatial distributions; some are present in almost all tissues, whereas others are tissue-specific reflecting their specialized functions (Palmieri and Pierri, 2010a).
The most abundant protein in the MIM is Adenine Nucleotide translocase (ANT), also known as the ADP/ATP translocase. Humans possess four ANT isoforms (ANT1-ANT4) that are expressed in different proportions in various tissues (Bround et al., 2020). ANTs exchange mitochondrial ATP4- for cytosolic ADP3-, resulting in a transfer of negative charge. ATP is synthesized from ADP and inorganic phosphate (Pi) by oxidative phosphorylation, and the supply of Pi in the mitochondrial matrix is maintained by phosphate translocase (PiT). PiT mediates the uptake of hydrophilic Pi into the matrix, either by proton co-transport across the MIM or in exchange for hydroxyl ions; either way the uptake of Pi is electroneutral (Runswick et al., 1987). There are two tissue-specific isoforms of PiT found in human tissues, PiT-A and PiT-B, that share 70% sequence identity, and differ primarily in their sequence near the N-terminus. PiT-A is abundantly expressed only in heart and muscle, whereas PiT-B is expressed at lower levels in all tissues (Palmieri, 2013). Dependence on cardiolipin has been reported for PiT function whereby the addition of cardiolipin almost doubled the phosphate transport rate for purified protein reconstituted into liposomes. Moreover, the activity of recombinant PiT could be increased by more than 5 times by adding purified cardiolipin to liposomes (Kadenbach et al., 1982; Mende et al., 2005; Seifert et al., 2015).
Another family of related MIM carrier proteins are the uncoupling proteins (UCPs). A general feature of UCPs is their ability to transport protons in accordance with their concentration gradient from the intermembrane space to the matrix, therefore dissipating the proton-motive force and uncoupling ATP production from the respiratory chain processes (Diehl and Hoek, 1999; Ardalan et al., 2022). Anion transporter UCPs have been reported in fungi, plants, and other eukaryotes, including all mammals (Jarmuszkiewicz et al., 2000; Sluse and Jarmuszkiewicz, 2002; Jarmuszkiewicz et al., 2010; Busiello et al., 2015; Demine et al., 2019). Among five human UCPs (UCP1-UCP5), UCP4 and UCP5 are predominantly expressed in brain tissues. The amount of UCP4 varies in different brain regions, with the highest levels found in the mitochondria of the cortex (Smorodchenko et al., 2009). UCPs 2, 4 and 5 are considered as neuronal UCPs, as they are mainly found and play neuroprotective roles in the central nervous system (CNS). UCP4 and UCP5 share the lowest amino acid sequence identity to other members in family (30% and 34% sequence identity to UCP1, respectively). Relatively few conducted in vitro studies on these proteins indicate that UCP4 and UCP5 can catalyse proton (and chloride) transport across lipid bilayers (Hoang et al., 2015). Therefore, a regulatory role in ROS mitigation by uncoupling has been proposed (Zhao et al., 2019). However, further experiments are required to determine the physiological roles of these proteins in their native environment (Hoang et al., 2012).
Considering the essential roles of mitochondrial carrier proteins in cell metabolism and their great potential as drug targets (Armstrong, 2007; Rask-Andersen et al., 2013; Wu et al., 2019), a more detailed molecular understanding of their native structure, binding properties, and functional dynamics in physiologically relevant environments is essential for deciphering their mutually interconnected action in mitochondria. These membrane proteins should be isolated and purified in conformations close to their native state, which is an arduous task. So far, mammalian cell lines, yeast, and bacteria have been the recombinant expression systems used most often to produce these proteins in quantities (Andréll and Tate, 2013; Goehring et al., 2014; He et al., 2014). Due to the low quantities of the majority of proteins in their host species, the heterologous expression systems of 1) the yeast Saccharomyces cerevisiae, that contains mitochondria to which active recombinant proteins can be targeted (Winkler et al., 2001); and 2) the bacterium E. coli (Escherichia coli), from which purified recombinant proteins can be obtained in soluble mixed micelles or in inclusion bodies (Echtay et al., 2018), have been most extensively used. Among UCPs, UCP1 is the only member that has been isolated from mammalian tissues, while the other UCPs have been expressed in E. coli and yeast (Klingenberg, 2001). There are also several reports of functional and structural studies of ANT (Knirsch et al., 1989; Haferkamp et al., 2002; Bertholet et al., 2022) and PiT (Dolce et al., 1996; Fiermonte et al., 1998; Schroers et al., 1998) in all three expression systems. So far, the most commonly used methods for recombinant expression of these proteins in E. coli have relied on purifying and refolding them from inclusion bodies. In this study, we report on a new cost-effective method for overexpression of MCFs (with focus on ANT, PiT and UCPs) in E. coli membranes, the extraction and purification of native-like forms in high-yield, and their reconstitution in mitochondrial inner membrane lipid models for structural and functional analysis.
Egg yolk extract containing at least 60% (w/w) phosphatidylcholine (egg yolk PC or L-α-PC) was obtained from Sigma-Aldrich (St. Louis, MO; Cat. No P5394); the remaining ∼40% of the egg yolk lipid extract was mostly phosphatidylethanolamine (PE) and small amounts of other lipids. octyl glucoside (OG) and octytetraoxyethylene (C8E4) were obtained from BioShop Canada Inc. (Burlington, ON) and Bachem (Bubendorf, Switzerland), respectively. The fluorescent probe dye, 6-methoxy-N-(3-sulfopropyl) quinolinum; (SPQ) (99%) was from Biotium Inc. (Fremont, California). HPLC grade water, acetonitrile (ACN) and formic acid were obtained from Merck (Darmstadt, Germany). Trypsin was purchased from Worthington Biochemical Corp (Lakewood, NJ)). All other chemicals were purchased from Sigma (St. Louis, MO).
Codon-optimized cDNA of the human PiT (GenBank accession no. CAB56612.1 Q00325-2) and bovine ANT (Genbank accession no. AAI02995.2) were synthesized and subcloned into pET26b (+) (gene synthesis and cloning services provided by Biobasic, Markham, ON). UCP4 cDNA (GenBank accession no. AAQ89951.1) was synthesized by GenScript Corp. and processed to subclone into pET26b (+) as discussed in Hoang et al. (2015). The target recombinant proteins comprised an N-terminus pelB leader sequence, followed by a His6-tag, and the protein sequence. The pelB leader sequence is designed to direct proteins to the cell membrane and is cleaved by a periplasmic peptidase, which has been confirmed with our previous study on UCP1 (Hoang et al., 2013) and was further verified by mass spectrometry in the current study (Supplementary Figure S1). The pET26b (+) vectors encoding recombinant UCP4, ANT, and PiT were transformed into E. coli BL21 Codon Plus-RIPL (BL21 CD+) [in case of constructs that were not codon-optimized], BL21 (DE3), BL21 Lobstr (DE3), and BL21 C43 (DE3) strains, by heat shock (42°C, 90 s).
Recombinant versions of PiT, ANT, and neuronal UCP4 were overexpressed in E. coli BL21 (DE3) [with the exception of UCP4, which was introduced into E. coli BL21 CodonPlus (DE3)-RIPL] cells as well as in its two modified expression strains, E. coli BL21 Lobstr (DE3) and E. coli BL21 C43 (DE3), using a modified autoinduction method (Studier, 2005). Specifically, the cells were grown overnight in 500 mL or 1 L autoinduction culture media (1% w/v Tryptone, 0.5% w/v yeast extract, 1 mM MgSO4, 0.5% w/v glycerol, 0.05% w/v glucose, 0.2% w/v lactose, 25 mM (NH4)2SO4, 50 mM KH2PO4, 50 mM Na2HPO4) at three different temperatures (room temperature, 30°C, and 16°C) for 22 h before collection. Bacterial cells were collected by centrifugation at 4,000 × g for 1 h at 4°C using Beckman Coulter Avanti JXN-26 Centrifuge and JLA 10.500 rotor. Cell pellets were resuspended in 20 mL extraction buffer [500 mM NaCl, 5 mM MgCl2, 20 mM Tris-HCl pH 8.0, half (for 1 L culture) or one-quarter (for 500 mL culture) of a tablet of enediaminetetraacetic acid (EDTA)-free protease inhibitor (Roche Applied Science), one small scoop of DNase, and lysozyme (BioShop Canada Inc.) at a final concentration of 0.1–0.2 mg/mL]. Bacterial cell lysis was achieved using a high-pressure cell disruptor (Constant Systems Limited, Daventry, United Kingdom), operating at 20 kPsi. The cell lysate was cleared of cell debris and inclusion bodies by centrifugation (Beckman Coulter Avanti JXN-26, 20,000 × g, 20 min, 4°C) using a JA 25.5 rotor (Beckman Coulter). Subsequently, the supernatant was ultracentrifuged (Beckman Coulter Optima™ MAX Ultracentrifuge) at 50,000 × g (MLA 80 rotor, Beckman Coulter, Brea, CA) for 1 h to collect the bacterial membranes in the pellet fraction (Figure 1).
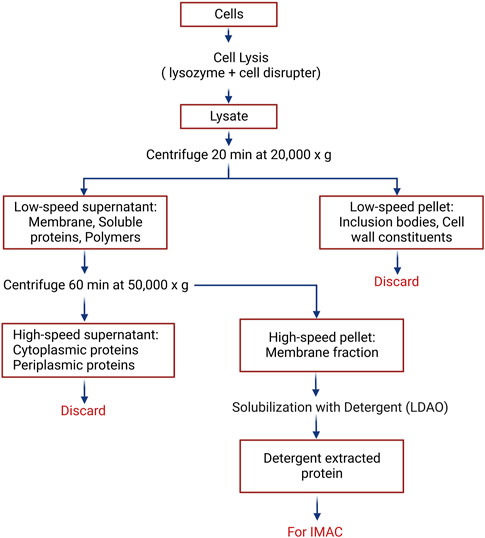
FIGURE 1. Procedure for the extraction of membrane-embedded proteins from Escherichia coli. Detailed protocol is provided in the Materials and Methods Section, 3.2.
Pellets containing UCP4 in bacterial membranes were solubilized using a 1–3 mL syringe and G21-gauge needle to resuspend the membrane pellet in binding buffer [10 mM imidazole, 1% w/v lauryldimethylamine oxide (LDAO detergent), 500 mM NaCl, 1 mM Tris (hydroxypropyl) phosphine (THP), and 20 mM Tris-HCl pH 8.0], and then incubated overnight in the cold room. In some instances, the membrane fraction was then further cleared of any remaining debris by extra centrifugation (Beckman Coulter Avanti JXN-26, 8,000 × g, 10 min, 4°C, JA 25.5 rotor). The solution was then rotated with 1 mL of equilibrated nickel-nitrilotriacetic acid (Ni-NTA) resin (Bio-rad, Waltham, Massachusetts) suspended in binding buffer in a 1 cm diameter glass column for 1 h. The resin was allowed to settle and the flow-through was collected. The resin was washed with binding buffer (8 mL), and again with wash buffer [30 mM imidazole, 1% w/v octyl glucoside (OG), 500 mM NaCl, 1 mM Tris (hydroxypropyl) phosphine (THP), and 20 mM Tris-HCl pH 8.0] (5 mL), and eluted stepwise with wash buffer containing increasing concentrations of imidazole [100 mM (2 mL), 250 mM (2 mL), and 400 mM (6 mL, collected as six 1 mL fractions) imidazole]. The same protocol was used for UCP4, PiT and ANT. Imidazole was removed from the purified proteins (collected upon elution with 400 mM imidazole in 1% OG) using Econo-Pac 10DG desalting spin columns (Bio-Rad, Hercules, California). Fully denaturing and semi-native sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and modified Lowry assay (Peterson, 1977) were used to assess the purity and concentration of the proteins, respectively.
The stepwise elution described above permitted the separation of monomers, which eluted alone from the IMAC column at the highest imidazole concentrations (i.e. 400 mM), from tetramers, which tended to elute from the column at lower imidazole concentrations (i.e., 100 and 250 mM). The purification conditions could therefore be modified to isolate a mixture of monomeric and oligomeric forms by omitting the stepwise elution. The specific wash and elution conditions used to isolate a mixture of monomers and tetramers were the sequential application of binding buffer (10 mL), wash buffer (6.5 mL) and elution buffer containing 400 mM imidazole (5 mL).
To analyze proteins using semi-native PAGE (Voulhoux et al., 2003), sodium dodecyl sulfate (SDS) was excluded from the sample buffer, and its concentration was reduced in the gel and running buffer as compared to that of fully denaturing conditions in SDS-PAGE (0.1% w/v in semi-native vs 1% w/v in denaturing conditions). Moreover, the samples were not heated before loading on the gel. The gels were run at 110 V, stained with 0.2% w/v solution of Coomassie Brilliant Blue R-250 in methanol: acetic acid: water (45:10:45 by volume) for 30–60 min and destained for 2 h.
The identity of overexpressed His6-tagged UCP4, PiT and ANT was confirmed by both Western blot and MS analyses. For immunoblotting, 5–10 µg of purified recombinant proteins were run on a 12% w/v polyacrylamide semi-native PAGE gel and transferred to nitrocellulose membrane using the semi-dry method (20 V for 90 min). Membranes were stained with 0.1% w/v Amido Black (in 45:10:45 MeOH: acetic acid: H2O) to verify the efficiency of transfer, and then were blocked in TBS-T (50 mM Tris-HCl, pH 7.5, 104 mM NaCl, 0.1% v/v Tween 20) containing 5% w/v skim milk on a slow rocker at room temperature for 1 h. Membranes were incubated overnight at 4°C with the primary antibodies (Mendez-Romero et al., 2019). A mouse monoclonal IgM κ UCP4 antibody (Santa Cruz Biotechnology Inc. Dallas, Texas), a mouse monoclonal IgM κ PiT antibody (Santa Cruz Biotechnology Inc. Dallas, Texas), and a rabbit polyclonal ANT antibody (Novus Biologicals), each diluted in TBS 1:1,000 (v/v), were used for the detection of UCP4, PiT and ANT, respectively. Membranes were then washed with TBS-T (2×10 mL) and rinsed with TBS, and then incubated at room temperature for 1 h with secondary antibodies. A horseradish peroxidase (HRP)-conjugated pre-adsorbed antibody (1:2,500 dilution), raised in rabbit against Mouse IgG (Rockland, Limerick, Pennsylvania), was used as the secondary antibody for UCP4 and PiT. And a HRP-conjugated antibody (1:2,500 dilution), raised in goat against rabbit IgG (Rockland, Limerick, Pennsylvania), was utilized as the secondary antibody for ANT. Membranes were washed again as described above, and the detection of the secondary antibody was achieved using the chemiluminescent reagent Luminata Crescenda Western HRP substrate (Millipore Sigma). The specificity of the mouse monoclonal antibody for UCP4 and PiT was assessed using an excess amount of blocking peptide (Santa Cruz Biotechnology Inc. Dallas, Texas). The image was captured using a Bio-Rad VersaDoc imaging system (exposure time: 320 s).
To further confirm the identity of purified monomeric proteins in OG by mass spectrometry analysis, filter-aided sample preparation (FASP) was modified for protein digestion (Liebler and Ham, 2009). The mass spectrometric analysis was performed by Proteomics Resource Centre at the University of Ottawa. Briefly, samples were loaded on a 10 K filter (Amicon® Ultra-0.5, MilliPore). 8 M urea was used to displace the original buffer. Protein reduction [with 10 mM dithiothreitol (DTT) in 50 mM ammonium bicarbonate (ABC)] and alkylation (with 10 mM IAA in 50 mM ABC) were done sequentially in the filter. For digestion, a mass ratio of 1:50 between trypsin and protein was used with continuous shaking overnight at 37°C. Digested peptides were then desalted on 200 μL filter-tip columns packed in-house with 10 μm ReproSil-Pur C18 beads (200 Å; Dr. Maisch GmbH, Germany) and dried down using a SpeedVac (ThermoFisher Scientific, San Jose, CA). Dried samples were reconstituted in 30 μL 0.5% v/v formic acid, and 2 µL was loaded for MS analysis. For LC-MS analysis, Dionex ultimate RS3000 was hooked up with Exploris 480 mass spectrometer (ThermoFisher Scientific, San Jose, CA), operated with a nano-electrospray interface operated in positive ion mode. The solvent system consists of buffer A of 0.1% v/v formic acid in water, and buffer B of 0.1% v/v formic acid in 80% v/v acetonitrile. Reconstituted peptides were loaded on a 75 μm I.D. × 150 mm fused silica analytical column packed in-house with 3 μm ReproSil-Pur C18 beads (100 Å; Dr. Maisch GmbH, Ammerbuch, Germany). The flow rate was set to 200 nL/min, and the gradient was set as 5–35% buffer B in 45 min, followed by 5 min from 35% to 80%, 5 min of 80%, and 5 min of re-equilibration. The spray voltage was set to 2.2 kV and the temperature of the heated capillary was 300°C. One full MS scan from 350 to 1,200 m/z followed by a data-dependent MS/MS scan of the 15 most intense ions with a dynamic exclusion repeat count of 1 in 20 s. The mass resolution is 60,000 for ms1 and 15,000 for ms2. A real-time internal calibration by the lock mass of background ion 445.120025 was used. All data were recorded with Xcalibur software (ThermoFisher Scientific, San Jose, CA). For data analysis, the peak lists of the raw files were processed and analyzed with MaxQuant software (Version 1.6.3.4) (Liebler and Ham, 2009) against the three target proteins along with the built-in contaminant sequences including commonly observed contaminants. Cysteine carbamidomethylation was selected as a fixed modification; the methionine oxidation, protein N-terminal acetylation ubiquitination were set for variable modification. Enzyme specificity was set to trypsin, not allowing for cleavage N-terminal to proline. Other parameters were used as default.
Reconstitution of UCP4, ANT, and PiT into PC vesicles was achieved using a previously described detergent-mediated reconstitution method (Echtay et al., 2000; Jaburek and Garlid, 2003; Ardalan et al., 2021a), with minor modifications (Supplementary Figure S2). Briefly, a thin layer of lipid was formed in a round-bottomed flask by drying egg yolk PC in methanol/chloroform (1:3) under vacuum overnight. The lipid was rehydrated in reconstitution buffer containing 20 mM Tris-HCl, pH 8.0, 50 mM NaCl, and 1% w/v Glycerol. The phospholipids were then solubilized with C8E4 to a final detergent: phospholipid ratio of 2.5 (w/w) and incubated on ice for 40 min. Purified, desalted proteins (∼10 µM) were then added to the transparent solution containing mixed lipid/detergent micelles and incubated at 4°C for 1.5 h. Spontaneous formation of liposomes was achieved by removing the C8E4 detergent using SM-2 Biobeads (Bio-Rad). The samples were transferred into 1 cm diameter columns containing 1 cm of packed biobeads, which had been equilibrated with reconstitution buffer, incubated for 2 h, and then the Biobeads were collected by centrifugation (1,000 ×g for 1 min). The supernatant containing the proteoliposomes was transferred to a column containing fresh, equilibrated biobeads and incubated for 1 h to remove residual detergent. The Biobeads were collected by centrifugation and the supernatant containing the proteoliposomes was collected. Protein-free liposomes were prepared as a control. For circular dichroism (CD) conformational studies, the final protein: phospholipid molar ratio was about 1:1,000.
The size and homogeneity of liposomes and proteoliposomes were determined by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, United Kingdom). The results were the average of 3–5 measurements.
Far-UV CD measurements were performed at 1 nm resolution at 25°C, using an AVIV 215 spectropolarimeter (Aviv Biomedical, Lakewood, New Jersey). All far UV-CD measurements (in OG and liposomes) were taken in quartz cells with 0.1 cm path length (25°C). Ellipticities were converted to mean residue ellipticity [θ]. All individual CD spectra were an average of at least two independent measurements. The concentrations of proteins were 5–8 µM for samples in 1% OG and ∼1 µM in proteoliposomes.
Due to its rapid growth and ease of genetic manipulation and handling, E. coli is a broadly used host for heterologous expression of a wide variety of membrane proteins. To examine the expression level, yield, and purity of the proteins of interest, the E. coli BL21 (DE3), or E. coli BL21 Codon Plus-RIPL and two modified expression strains, E. coli BL21 Lobstr (DE3) and E. coli BL21 C43 (DE3), were selected as expression hosts for the current study. Escherichia coli BL21 C43 (DE3) is designed to reduce expression-induced toxicity (Andersen et al., 2013) and E. coli BL21 Lobstr (DE3) [low background strain] is ideal for proteins with poor expression level and to reduce contamination from endogenous E. coli proteins SlyD and ArnA whose Ni-binding characteristics mimic that of His-tagged proteins, thus resulting in fewer purification steps (Andersen et al., 2013). To produce native-like folded proteins, the cDNAs of UCP4, ANT and PiT were subcloned into the pET26b (+) expression vector. The resulting recombinant proteins include the pelB signal peptide fused to the N-terminus, which targets the proteins to the inner membrane of E. coli and is cleaved by a periplasmic signal peptidase (Hoang et al., 2013) (Supplementary Figure S1). The proteins are retained in the bacterial membrane, rather than reaching the periplasm, due to the hydrophobic nature of the six-TM domains. The pelB leader is immediately followed by a six-histidine (His6) affinity tag which is revealed upon cleavage of the leader and facilitates affinity purification. The His6-tagged proteins were purified on nickel containing columns after isolation and solubilization of the bacterial membrane. Expression of proteins in bacterial membranes allows the protein and any strongly associated membrane lipids to be extracted together with mild detergents and copurified. Membrane lipids that copurify with proteins can protect the protein from potential denaturing interactions with the solubilizing detergent and, therefore, stabilize proteins during the extraction and purification process to maintain proper folding.
To elucidate the effects of culture conditions on the production of UCP4, ANT, and PiT proteins in the membrane of E. coli BL21 (DE3) and/or E. coli BL21 Codon Plus-RIPL, E. coli BL21 Lobstr (DE3) and E. coli BL21 C43 (DE3), cell growth was monitored at three different expression temperatures (room temperature, 30°C, and 16°C) and two different culture media volumes (500 and 1,000 mL) using a modified autoinduction method (see Section 2.3) (Studier, 2005). Interestingly, no significant protein expression was observed in the 1 L cultures at any of temperatures (data not shown). However, 500 mL cultures grown at room temperature resulted in the highest expression levels and resulted in the highest yield of purified protein in all strains. Under the optimal growth conditions, all three strains demonstrated comparable protein expression (Figure 2, Supplementary Figure S3 and Table 1). While the yield of each protein was somewhat less than can be achieved when expressing in inclusion bodies (Galluccio, et al., 2022), the current method provides proteins in ample yields and has the advantage of producing native-like folded proteins ready for biophysical studies. Neuronal UCP4, overexpressed with this method, was purified in monomeric and tetrameric forms observed as bands of approximately 36 and 144 kDa on semi-native SDS-PAGE gels (Figure 2, lane 1 and Supplementary Figure S3 lanes 1–2). The identity of these bands was confirmed by Western blot analysis probed with a mouse monoclonal UCP4 antibody which reacted with both bands suggesting that they represent monomeric and tetrameric forms of UCP4, respectively, as has been observed previously (Hoang et al., 2015) (Figure 2, lane 2). Further purification was performed to purify monomeric UCP4 (Figure 2, lane 3). Under similar purification conditions, only monomeric forms were detected for PiT and ANT (Figure 2, lanes 6 and 9, respectively) as monitored by mouse monoclonal anti-PiT and rabbit polyclonal anti-ANT antibodies (Figure 2, lanes 7 and 10, respectively). The specificity of both antibodies was confirmed by blocking their immunoreactivity using excess amounts of a competing peptide (data not shown). On fully denaturing SDS-PAGE gels the monomeric UCP4, ANT and PiT proteins migrated differently than each other (Supplementary Figure S4). Moreover, the mass spectrometric analysis of the purified recombinant monomeric forms of UCP4, ANT, and PiT verified the identities of the proteins as well as the cleavage of the pelB peptide and presence of the His-tag. (Supplementary Figure S1).
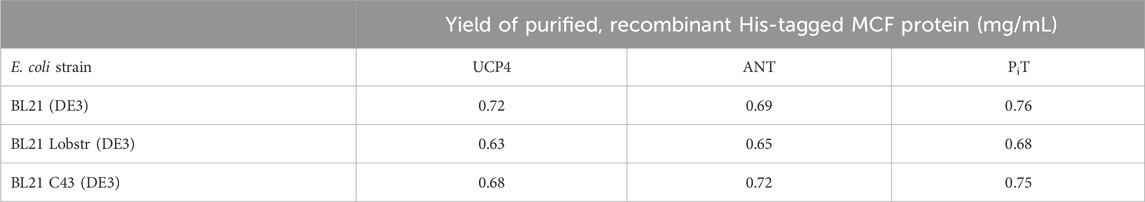
TABLE 1. Yields of purified, recombinant MCF proteins UCP4, ANT and PiT expressed in three different strains of Escherichia coli (BL21 (DE3), BL21 Lobstr (DE3), and BL21 C43 (DE3)) using a modified autoinduction expression protocol. Data are expressed as mg protein per mL of bacterial culture.
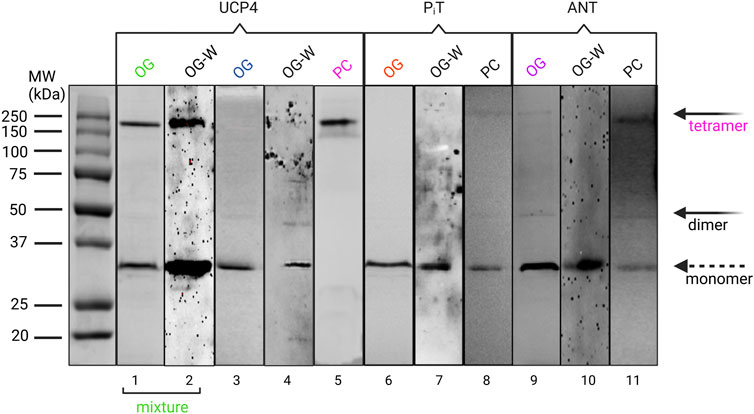
FIGURE 2. Semi-native PAGE analysis of purified UCP4, PiT, and ANT, extracted from membranes of Escherichia coli BL21 (DE3) in OG detergent and reconstituted in liposomes made with egg yolk extract containing 60% phosphatidylcholine (PC) detected by Coomassie blue staining (lanes 1, 2, 4, 5, 7, 8, 10) and Western blot (W) (lane 3, 6, 9) methods. Mixture and monomer terms at the bottom refer to the mixture of monomeric and tetrameric forms of UCP4 in OG. The solid arrows indicate the locations of oligomers, and the open arrow shows the monomers. Western blots detecting UCP4, PiT, and ANT in OG detergent were probed with mouse monoclonal anti-UCP4, anti-PiT, and a rabbit polyclonal anti-ANT, respectively. Collectively, the data confirm the spontaneous formation of tetrameric form in UCP4 in lipid vesicles, but not for PiT, and ANT.
Knowing the self-association behaviour of UCPs in lipid vesicles (Hoang et al., 2013; Hoang et al., 2015; Ardalan et al., 2021a; Ardalan et al., 2021b), monomeric forms of UCP4 were isolated from the E. coli membranes in the presence of mild OG detergent (Figure 2, lane 3) to assess its ability to spontaneously oligomerize when reconstituted in lipid membranes. The selection of OG as a mild detergent was based on our previous study of the conformational stability of UCP1 in different detergents, which showed that the overall structural integrity of the protein was most stable when purified and stored in the presence of 1% OG micelles (Hoang et al., 2013). The monomeric forms of UCP4 were reconstituted in egg yolk PC, as a model for mitochondrial membranes for structural analysis. Egg yolk PC (about 60% PC) was chosen as it contains other forms of phospholipids (such as PE) that are also found in the MIM. DLS measurements confirmed the presence of a monodisperse population with radii in the range of large unilamellar vesicles (∼80 nm). Interestingly, semi-native electrophoresis showed that when UCP4 was reconstituted, all monomers self-associated into tetramers; monomers were no longer detected on semi-native gels after reconstitution (Figure 2, lane 5). This has also been shown in the case of other UCPs (Hoang et al., 2013; Hoang et al., 2015; Ardalan et al., 2021b). To investigate whether conformationally comparable PiT and ANT proteins also self-associated into oligomers in lipid membranes, purified monomeric proteins were reconstituted under the same experimental conditions as UCP4. In contrast to UCP4, neither ANT nor PiT proteins showed evidence for forming only tetramers in liposomes; instead, they maintained their original dominantly monomeric forms as in OG detergent (Figure 2, lanes 8 and 11, respectively).
Conformations of purified UCP4, ANT, and PiT from all three E. coli strains either solubilized in OG or reconstituted into liposomes were compared using CD spectroscopy, which can be used to estimate secondary structure content and overall conformation of proteins in solution and lipid vesicle dispersions (Johnson, 1988). As shown by the far-UV CD spectra (Figure 3A, and S5), all three proteins in 1% OG displayed the distinctive spectral features of α-helical backbone secondary structures with double negative maximum ellipticities at ∼ 222 and ∼ 208 nm and a strong positive maximum at ∼193 nm (not shown), corresponding to π → π* (193 and 208 nm) and n → π* (222 nm) transitions of the peptide bond (Greenfield and Fasman, 1969). The θ208/θ222 ratios of the CD spectra in OG were 1.11, 1.28, 1.32 for monomeric ANT, PiT, and UCP4, respectively, confirming the predominant α-helical structures in the OG detergent (Figure 3A) (Toniolo et al., 1996; Hoang et al., 2013). The ellipticity ratio was 1.13 for the mixture of monomeric and tetrameric UCP4 (decreased in comparison to the ratio of the monomer, Figure 3B), which implies the influence of the associated tetrameric form. As expected, CD spectra of the monomeric forms of UCP4 and PiT in OG detergent exhibited comparable helical conformations (Figure 3A). These dominantly helical conformations were also predicted by AlphaFold (Jumper et al., 2021; Ruff and Pappu, 2021; David et al., 2022) (Supplementary Figures S6A, C). Overlaying the AlphaFold structures of UCP4 and PiT with the crystal structure of ANT reveals an overall structural similarity of the three proteins (Supplementary Figures S6B, D). It is worth mentioning that the overlay of the X-ray crystal structure of bovine ANT (PDB ID: 1OKC) and the AlphaFold predicted structure of the human protein revealed a reliable match of helical conformations (Supplementary Figure S7). The slight difference in the CD spectrum of monomeric ANT in OG (as compared to those of UCP4 and PiT) may be related to the presence of trace oligomeric forms, which are weakly detectable in semi-native SDS-PAGE (Figure 2). Moreover, the superimpositions of the X-ray crystal structure of ANT with AlphaFold-predicted structures of UCP4 and PiT show that the transmembrane helical bundle of ANT is in a looser configuration compared to that of UCP4 and PiT (Figure 4); this is consistent with the differences observed in the CD spectra (Figure 3A). In UCP4, the lower θ208/θ222 ellipticity ratios for the mixture of oligomeric states (that includes monomers and tetramers) (1.13) compared to the exclusively monomeric form in OG (1.32) provides further evidence for the presence of multimers (Figure 3B and S3) (Johnson, 1988). Moreover, the far-UV CD signal of UCP4 was notably different after reconstitution into liposomes as compared to that of the protein in OG detergent (Figure 3B). Specifically, in lipid vesicles, a shoulder-like π → π* parallel transition band at ∼ 208 nm replaced the negative maximum band in OG, while the intense negative maximum band was observed for the n → π* transition band at ∼ 222 nm. This conformational transformation resulted in a θ208/θ222 ellipticity ratio of 0.6 (lower than one) for UCP4 after reconstitution into liposomes (Figure 3B). The marked conformational changes underlines the important role of the lipid environment on the structural stability and folding of UCP4, which is in good agreement with our previously reported studies on other UCPs (Hoang et al., 2012; Hoang et al., 2015; Ardalan et al., 2021a; Ardalan et al., 2021b). The θ208/θ222 ellipticity ratios lower than one have been formerly reported for human UCPs’ helical bundle motifs and their self-associated multimers (Hoang et al., 2015).
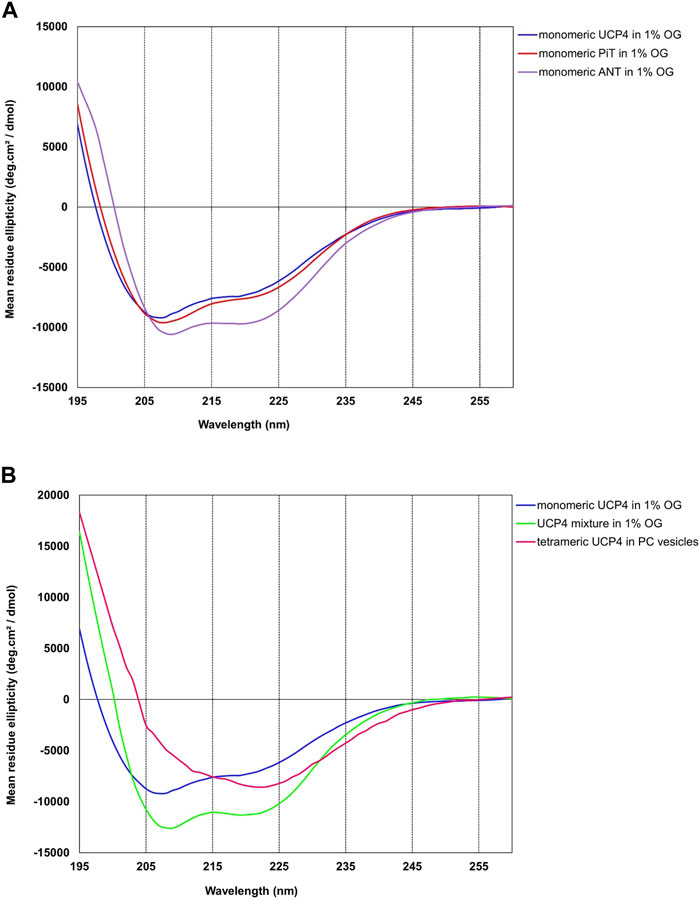
FIGURE 3. Far UV-CD spectra of neuronal UCP4, ANT, and PiT purified from the membrane of Escherichia coli BL21 (DE3), at 25°C. (A) All three proteins showed an overall α-helical structure in 1% OG detergent. (B) Tetramer formation of reconstituted UCP4 in liposomes resulted in a strong change in the CD spectrum. Each reported CD spectrum is an average of a minimum of six scans. The concentrations of proteins were 5–9 µM in OG and 1 µM in lipid vesicles.
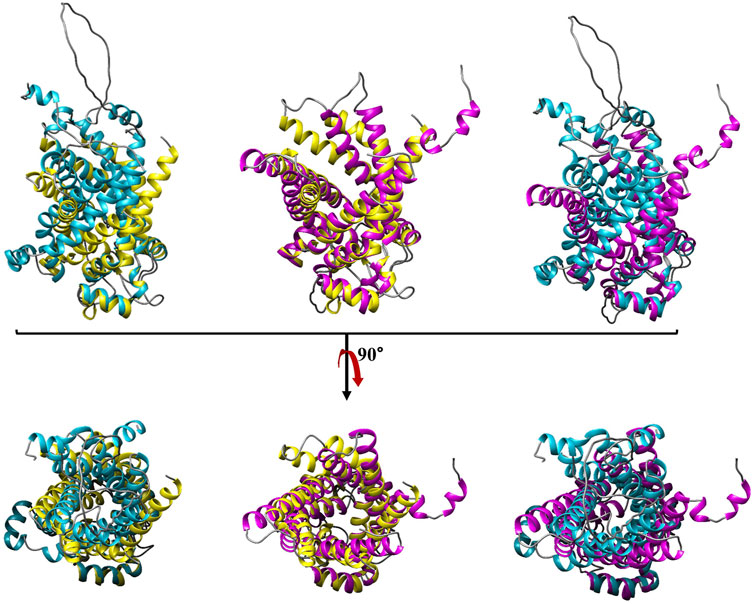
FIGURE 4. Structural comparison of bovine ANT with human UCP4 and PiT. The secondary structures of ANT, UCP4 and PiT are shown in gold, magenta, and blue, respectively. Root mean square deviation values (RMSDs) for the corresponding pairs of ANT-PiT, UCP4-PiT, ANT-UCP4 are reported in Supplementary Table S1 as 1.84, 5.09, and 5.39 (A°), respectively. The images in the top row have been rotated 90° from the top toward the reader in the bottom row.
ANT was the first member of the MCF whose primary structure became known by amino acid sequencing (Aquila et al., 1985). ANT is also the only member of the family for which high resolution X-ray structures are available. The structure of this carrier protein (bovine AAC1 or ANT1) in complex with the inhibitor carboxyatractyloside, was originally reported in 2003 (PDB ID:1OKC) (Pebay-Peyroula et al., 2003). ANT, like all MCF family members, contains a tripartite structure composed of three tandem repeats (Figure 5A). Each repeat comprises ∼100 amino acids folded into two transmembrane (TM) α-helices connected by a short amphipathic helix parallel to the membrane plane at the matrix end (Pebay-Peyroula et al., 2003). A proline residue within the conserved PX [D/E]XX [K/R] motif creates a kink in each odd-numbered helix, and the six TM helices surround a large open pocket oriented toward the cytosol. The six charged residues from the three PX [D/E]XX [K/R] motifs form a cyclic inter-helical salt bridge network near the matrix side of the MIM known as the matrix-gate or m-gate, and has been proposed to be an essential factor in stabilizing the cytosolic conformation of ANT. Topologically, ANT has both of its N- and C- termini, as well as two cytosolic loops, in the intermembrane space. The matrix-facing side of ANT is composed of three hydrophilic loops that are available for interaction of the protein with other molecules (Runswick et al., 1987; Palmieri and Pierri, 2010b). The structure of ANT most likely represents the common structural features of all MCF members of known function and has therefore been used as a template for building homology models of various MIM carriers (Palmieri, 2013). The only known exceptions are the Ca2+- binding aspartate/glutamate and ATP-Mg/Pi carriers that include an additional extensive N-terminal domain that fulfills a regulatory (non-transport) function (Palmieri and Pierri, 2010a).
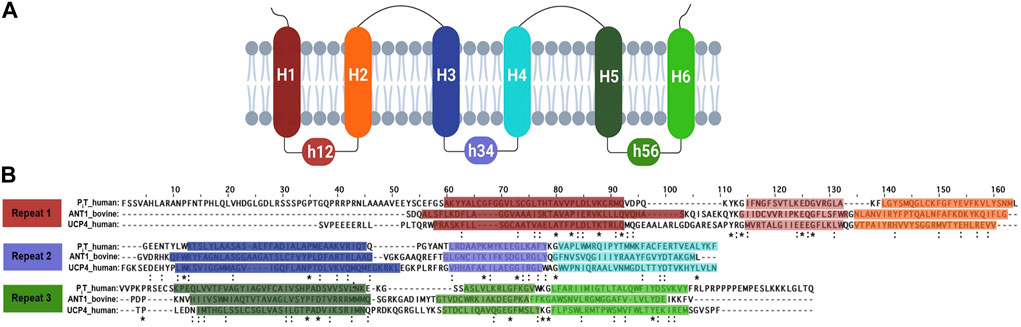
FIGURE 5. (A) A schematic diagram of the carrier protein secondary structure and membrane topology. Transmembrane helices and their connector helix are labelled as H and h, respectively. (B) Sequence alignment of carrier proteins involved in energy transfer. Alignment of the amino acid sequence of human PiT, human UCP4, and bovine ANT1, based on the EBI Clustal Omega Multiple Sequence Alignment Tool. Each line displays a repeat structure for the protein, consisting of two transmembrane helices linked by a short helix, represented within horizontal boxes that transition from darker to lighter shades. To indicate the identical residues and the conservation between residues with strongly similar properties, the asterisk (*) and colon (:) were used, respectively in every repeat across all proteins. Methionine, the initiator amino acid for protein biosynthesis, is not included in any sequences.
UCP4 and PiT share the typical tripartite structure of MCFs consisting of three tandem repeats (Figure 5A) and display overall homology with ANT; UCP4 and PiT are 23% and 21% identical to ANT1, respectively. Similarities were also evident when the predicted structures were aligned (Figure 4). The degree of similarity was assessed using root-mean-square deviations (RMSD) of the backbone α-carbon atoms and template modelling scores (TM-scores) (Supplementary Table S1). The ANT-PiT pair exhibits RMSD and TM-align values of 1.84 and 0.87, respectively, suggesting a higher likelihood of structural similarity compared to the other two pairs, which had RMSD values above 5.0 (Figure 4 and Supplementary Table S1). The higher RMSD values for the UCP4-ANT and UCP4-PiT pairs explain why the superposition of all three proteins (Supplementary Figure S6) is not stronger. Moreover, the amino acid sequences for all three proteins (bovine ANT (P02722), human phosphate translocase (Q00325), and human uncoupling protein 4 (O95847) obtained from the UniProtKB database) were submitted to Clustal Omega for multiple sequence alignment (MSA) using the default parameters. The MSA was visualized in the Jalview software environment. Identical and similar amino acids, as well as the conserved tandem repeats, are highlighted in the aligned sequences shown in Figure 5B. The striking presence of conserved critical residues, specifically glycine, proline, charged, and aromatic residues throughout all nine repeats (Figure 5B) is consistent with the overall similarities among the three proteins, and the conserved tandem repeats in particular.
Some of the characteristics of amino acid composition for three carriers, size ranging from 298 to 361 residues, are compared in Table 2. For these membrane proteins a relatively high percentage of polar amino acids, with a significant prevalence of basic residues over acidic ones, resulted a large positive net charge which is about twice as high in ANT and PiT as in UCP4. In summary, Table 2 reveals similarities in amino acid composition and properties among the three proteins, but also highlights differences in the number of cysteines, prolines, aromatic and charged residues and the degree of hydrophobicity, all of which contribute to the observed conformational and physicochemical properties of the proteins.
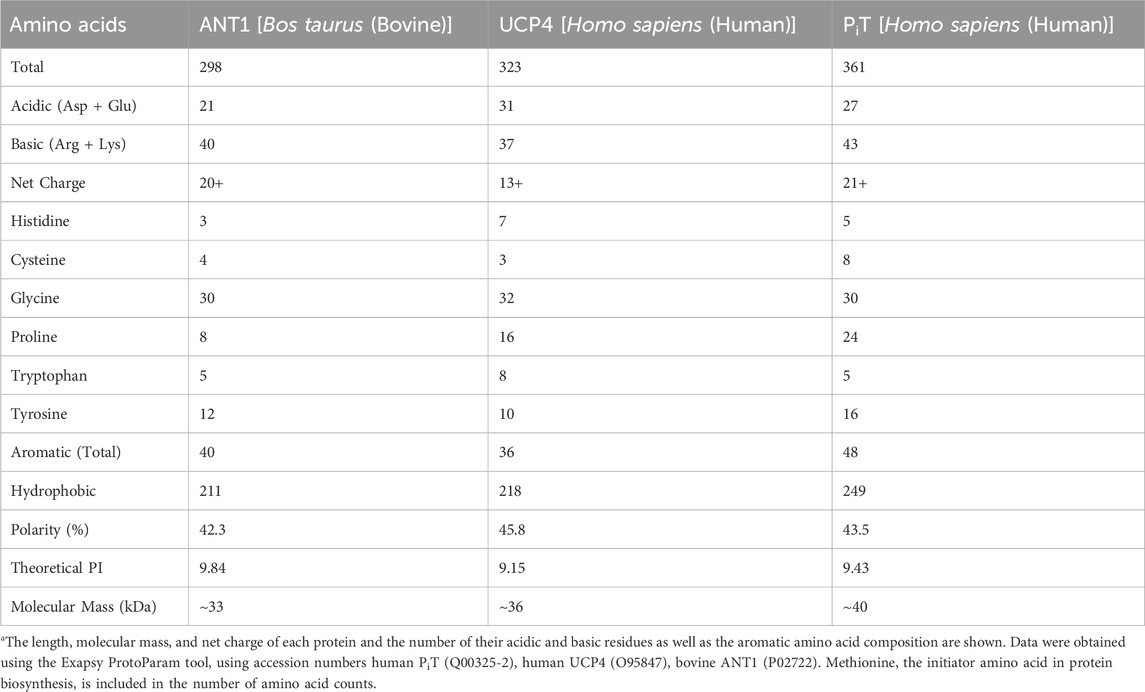
TABLE 2. Comparison of amino acid characteristicsa of the carrier proteins involved in energy transfer.
The production of recombinant membrane proteins for fundamental biophysical structural and functional research, as well as pharmaceutical and biotechnological applications, has been growing in recent years (Assenberg et al., 2013). Concomitantly, the interest in advanced novel approaches for the expression, purification and characterization of recombinant membrane proteins has also increased. In the current study we describe a method for obtaining native-like recombinant eukaryotic MCF proteins (using UCP4, ANT, and PiT as examples) in high yields from bacterial expression systems. We employed the combination of a short pelB leader sequence to target the proteins to the bacterial membrane, a modified autoinduction expression method, use of mild OG and comparable detergents to extract the proteins from membranes, and optimized IMAC purification conditions for efficient isolation and purification of properly folded monomeric proteins for molecular biophysical studies. A particularly interesting finding of this study, which is consistent with our previous studies on other UCP homologues, is the spontaneous self-association of monomeric UCP4 in lipid membranes to form stable tetrameric forms, whereas the other conformationally related proteins (ANT and PiT) remained primarily monomeric in both detergent and liposome milieus. Research on interaction and functional interrelation between these three proteins will be undertaken as a next step of this study. These hitherto less understood connections are essential for understanding the principles of the structural and functional activity of MCFs in the highly complex inner mitochondrial membrane.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
MT: Conceptualization, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. MS: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Supervision, Writing–review and editing. MJ-N: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research project was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery and the Canada Foundation for Innovation (CFI) grants to MJ-N. (05900 and 6786) and MS. (05437 and 11292). MT has been a recipient of the Ontario Graduate Scholarship for the year 2022/2023.
We thank Dr. W. A. Houry (Department of Biochemistry, University of Toronto, Canada) for generously providing us with the BL21-Lobstr (DE3) cells. K. Amani (Neurosnap Inc.) and M. Pottier (Faculty of Science, Wilfrid Laurier University) are gratefully acknowledged for bioinformatics and technical assistance, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frbis.2023.1334804/full#supplementary-material
Andersen, K. R., Leksa, N. C., and Schwartz, T. U. (2013). Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins 81, 1857–1861. doi:10.1002/prot.24364
Andréll, J., and Tate, C. G. (2013). Overexpression of membrane proteins in mammalian cells for structural studies. Mol. Membr. Biol. 30, 52–63. doi:10.3109/09687688.2012.703703
Aquila, H., Link, T. A., and Klingenberg, M. (1985). The uncoupling protein from brown fat mitochondria is related to the mitochondrial ADP/ATP carrier. Analysis of sequence homologies and of folding of the protein in the membrane. EMBO J. 4, 2369–2376. doi:10.1002/j.1460-2075.1985.tb03941.x
Ardalan, A., Smith, M. D., and Jelokhani-Niaraki, M. (2022). Uncoupling proteins and regulated proton leak in mitochondria. Int. J. Mol. Sci. 23 (3), 1528–1623. doi:10.3390/ijms23031528
Ardalan, A., Sowlati-Hashjin, S., Oduwoye, H., Uwumarenogie, S. O., Karttunen, M., Smith, M. D., et al. (2021a). Biphasic proton transport mechanism for uncoupling proteins. J. Phys. Chem. B 125, 9130–9144. doi:10.1021/acs.jpcb.1c04766
Ardalan, A., Sowlati-Hashjin, S., Uwumarenogie, S. O., Fish, M., Mitchell, J., Karttunen, M., et al. (2021b). Functional oligomeric forms of uncoupling protein 2: strong evidence for asymmetry in protein and lipid bilayer systems. J. Phys. Chem. B 125, 169–183. doi:10.1021/acs.jpcb.0c09422
Armstrong, J. S. (2007). Mitochondrial medicine: pharmacological targeting of mitochondria in disease. Br. J. Pharmacol. 151, 1154–1165. doi:10.1038/sj.bjp.0707288
Assenberg, R., Wan, P. T., Geisse, S., and Mayr, L. M. (2013). Advances in recombinant protein expression for use in pharmaceutical research. Curr. Opin. Struct. Biol. 23, 393–402. doi:10.1016/j.sbi.2013.03.008
Bertholet, A. M., Natale, A. M., Bisignano, P., Suzuki, J., Fedorenko, A., Hamilton, J., et al. (2022). Mitochondrial uncouplers induce proton leak by activating AAC and UCP1. Nature 606, 180–187. doi:10.1038/s41586-022-04747-5
Bround, M. J., Bers, D. M., and Molkentin, J. D. (2020). A 20/20 view of ANT function in mitochondrial biology and necrotic cell death. J. Mol. Cell. Cardiol. 144, A3–A13. doi:10.1016/j.yjmcc.2020.05.012
Busiello, R. A., Savarese, S., and Lombardi, A. (2015). Mitochondrial uncoupling proteins and energy metabolism. Front. Physiol. 6, 36–37. doi:10.3389/fphys.2015.00036
David, A., Islam, S., Tankhilevich, E., and Sternberg, M. J. (2022). The AlphaFold database of protein structures: a biologist’s guide. J. Mol. Biol. 434, 167336. doi:10.1016/j.jmb.2021.167336
Demine, S., Renard, P., and Arnould, T. (2019). Mitochondrial uncoupling: a key controller of biological processes in physiology and diseases. Cells 8, 795. doi:10.3390/cells8080795
Diehl, A. M., and Hoek, J. B. (1999). Mitochondrial uncoupling: role of uncoupling protein anion carriers and relationship to thermogenesis and weight control “The benefits of losing control”. J. Bioenergetics Biomembr. 31, 493–506. doi:10.1023/A:1005452624640
Dolce, V., Fiermonte, G., and Palmieri, F. (1996). Tissue-specific expression of the two isoforms of the mitochondrial phosphate carrier in bovine tissues. FEBS Lett. 399, 95–98. doi:10.1016/S0014-5793(96)01294-X
Echtay, K. S., Bienengraeber, M., Mayinger, P., Heimpel, S., Winkler, E., Druhmann, D., et al. (2018). Uncoupling proteins: martin Klingenberg’s contributions for 40 years. Archives Biochem. Biophysics 657, 41–55. doi:10.1016/j.abb.2018.09.006
Echtay, K. S., Winkler, E., Frischmuth, K., and Klingenberg, M. (2000). Uncoupling proteins 2 and 3 are highly active H+ transporters and highly nucleotide sensitive when activated by coenzyme Q (ubiquinone). Proc. Natl. Acad. Sci. 98, 1416–1421. doi:10.1073/pnas.98.4.1416
Ferramosca, A., and Zara, V. (2013). Biogenesis of mitochondrial carrier proteins: molecular mechanisms of import into mitochondria. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1833, 494–502. doi:10.1016/j.bbamcr.2012.11.014
Fiermonte, G., Dolce, V., and Palmieri, F. (1998). Expression in Escherichia coli, functional characterization, and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J. Biol. Chem. 273, 22782–22787. doi:10.1074/jbc.273.35.22782
Galluccio, M., Console, L., Pochini, L., Scalise, M., Giangregorio, N., and Indiveri, C. (2022). Strategies for successful over-expression of human membrane transport systems using bacterial hosts: future perspectives. Int. J. Mol. Sci. 23, 3823–3850. doi:10.3390/ijms23073823
Goehring, A., Lee, C. H., Wang, K. H., Michel, J. C., Claxton, D. P., Baconguis, I., et al. (2014). Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585. doi:10.1038/nprot.2014.173
Greenfield, N., and Fasman, G. D. (1969). Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 8, 4108–4116. doi:10.1021/bi00838a031
Haferkamp, I., Hackstein, J. H. P., Voncken, F. G. J., Schmit, G., and Tjaden, J. (2002). Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur. J. Biochem. 269, 3172–3181. doi:10.1046/j.1432-1033.2002.02991.x
He, Y., Wang, K., and Yan, N. (2014). The recombinant expression systems for structure determination of eukaryotic membrane proteins. Protein Cell 5, 658–672. doi:10.1007/s13238-014-0086-4
Hoang, T., Kuljanin, M., Smith, M. D., and Jelokhani-Niaraki, M. (2015). A biophysical study on molecular physiology of the uncoupling proteins of the central nervous system. Biosci. Rep. 35, 002266–e315. doi:10.1042/BSR20150130
Hoang, T., Smith, M. D., and Jelokhani-Niaraki, M. (2012). Toward understanding the mechanism of ion transport activity of neuronal uncoupling proteins UCP2, UCP4, and UCP5. Biochemistry 51, 4004–4014. doi:10.1021/bi3003378
Hoang, T., Smith, M. D., and Jelokhani-Niaraki, M. (2013). Expression, folding, and proton transport activity of human uncoupling protein-1 (ucp1) in lipid membranes. J. Biol. Chem. 288, 36244–36258. doi:10.1074/jbc.M113.509935
Horten, P., Colina-Tenorio, L., and Rampelt, H. (2020). Biogenesis of mitochondrial metabolite carriers. Biomolecules 10, 1008–1013. doi:10.3390/biom10071008
Jaburek, M., and Garlid, K. D. (2003). Reconstitution of recombinant uncoupling proteins. J. Biol. Chem. 278, 25825–25831. doi:10.1074/jbc.M302126200
Jarmuszkiewicz, W., Woyda-Ploszczyca, A., Antos-Krzeminska, N., and Sluse, F. E. (2010). Mitochondrial uncoupling proteins in unicellular eukaryotes. Biochimica Biophysica Acta (BBA) - Bioenergetics 1797, 792–799. doi:10.1016/j.bbabio.2009.12.005
Jarmuszkiewicz, W. X., Milani, G., Fortes, F., Schreiber, A. Z., Sluse, F. E., and Vercesi, A. E. (2000). First evidence and characterization of an uncoupling protein in fungi kingdom: CpUCP of Candida parapsilosis. FEBS Lett. 467, 145–149. doi:10.1016/S0014-5793(00)01138-8
Johnson, W. C. (1988). Secondary structure of proteins through circular dichroism spectroscopy. Annu. Rev. Biophysics Biophysical Chem. 17, 145–166. doi:10.1146/annurev.bb.17.060188.001045
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kadenbach, B., Mende, P., Kolbe, H., Stipani, I., and Palmieri, F. (1982). The mitochondrial phosphate carrier has an essential requirement for cardiolipin. FEBS Lett. 139, 109–112. doi:10.1016/0014-5793(82)80498-5
Klingenberg, M. (2001). Uncoupling proteins - how do they work and how are they regulated. IUBMB Life 52, 175–179. doi:10.1080/15216540152845975
Knirsch, M., Gawaz, M. P., and Klingenberg, M. (1989). The isolation and reconstitution of the ADP/ATP carrier from wild-type Saccharomyces cerevisiae Identification of primarily one type (AAC-2). FEBS Lett. 244, 427–432. doi:10.1016/0014-5793(89)80577-0
Liebler, D. C., and Ham, A. J. L. (2009). Spin filter-based sample preparation for shotgun proteomics. Nat. Methods. 6, 785. doi:10.1038/nmeth1109-785a
Mende, P., Kolbe, H. V. J., Kadenbach, B., Stipani, I., and Palmieri, F. (2005). Reconstitution of the isolated phosphate-transport system of pig-heart mitochondria. Eur. J. Biochem. 128, 91–95. doi:10.1111/j.1432-1033.1982.tb06937.x
Mendez-Romero, O., Uribe-Carvajal, S., Chiquete-Felix, N., and Muhlia-Almazan, A. (2019). Mitochondrial uncoupling proteins UCP4 and UCP5 from the Pacific white shrimp Litopenaeus vannamei. J. Bioenerg. Biomembr. 51, 103–119. doi:10.1007/s10863-019-09789-5
Nicholls, D. G. (2021). Mitochondrial proton leaks and uncoupling proteins. Biochimica Biophysica Acta (BBA) - Bioenergetics 1862, 148428. doi:10.1016/j.bbabio.2021.148428
Palmieri, F. (2013). The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol. Aspects Med. 34, 465–484. doi:10.1016/j.mam.2012.05.005
Palmieri, F., and Monné, M. (2016). Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1863, 2362–2378. doi:10.1016/j.bbamcr.2016.03.007
Palmieri, F., and Pierri, C. L. (2010a). Mitochondrial metabolite transport. Essays Biochem. 47, 37–52. doi:10.1042/BSE0470037
Palmieri, F., and Pierri, C. L. (2010b). Structure and function of mitochondrial carriers - role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 584, 1931–1939. doi:10.1016/j.febslet.2009.10.063
Pebay-Peyroula, E., Dahout-Gonzalez, C., Kahn, R., Trézéguet, V., Lauquin, G. J. M., and Brandolin, G. (2003). Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44. doi:10.1038/nature02056
Peterson, G. L. (1977). A simplification of the protein assay method of Lowry etal. Which is more generally applicable. Anal. Biochem. 83, 346–356. doi:10.1016/00032697(77)90043-4
Rask-Andersen, M., Masuram, S., Fredriksson, R., and Schiöth, H. B. (2013). Solute carriers as drug targets: current use, clinical trials and prospective. Mol. Aspects Med. 34, 702–710. doi:10.1016/j.mam.2012.07.015
Ruff, K. M., and Pappu, R. V. (2021). AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 433, 167208. doi:10.1016/j.jmb.2021.167208
Runswick, M. J., Powell, S. J., Nyren, P., and Walker, J. E. (1987). Sequence of the bovine mitochondrial phosphate carrier protein: structural relationship to ADP/ATP translocase and the brown fat mitochondria uncoupling protein. EMBO J. 6, 1367–1373. doi:10.1002/j.1460-2075.1987.tb02377.x
Schroers, A., Burkovski, A., Wohlrab, H., and Krämer, R. (1998). The phosphate carrier from yeast mitochondria: dimerization is a prerequisite for function. J. Biol. Chem. 273, 14269–14276. doi:10.1074/jbc.273.23.14269
Seifert, E. L., Ligeti, E., Mayr, J. A., Sondheimer, N., and Hajnóczky, G. (2015). The mitochondrial phosphate carrier: role in oxidative metabolism, calcium handling and mitochondrial disease. Biochem. Biophysical Res. Commun. 464, 369–375. doi:10.1016/j.bbrc.2015.06.031
Sluse, F. E., and Jarmuszkiewicz, W. (2002). Uncoupling proteins outside the animal and plant kingdoms: functional and evolutionary aspects. FEBS Lett. 510, 117–120. doi:10.1016/S0014-5793(01)03229-X
Smorodchenko, A., Rupprecht, A., Sarilova, I., Ninnemann, O., Bräuer, A. U., Franke, K., et al. (2009). Comparative analysis of uncoupling protein 4 distribution in various tissues under physiological conditions and during development. Biochimica Biophysica Acta (BBA) - Biomembr. 1788, 2309–2319. doi:10.1016/j.bbamem.2009.07.018
Studier, F. W. (2005). Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234. doi:10.1016/j.pep.2005.01.016
Toniolo, C., Polese, A., Formaggio, F., Crisma, M., and Kamphuis, J. (1996). Circular dichroism spectrum of a peptide 310-helix. J. Am. Chem. Soc. 118, 2744–2745. doi:10.1021/ja9537383
Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M., and Tommassen, J. (2003). Role of a highly conserved bacterial protein in outer membrane protein assembly Science. Science 299, 262–265. doi:10.1126/science.1078973
Winkler, E., Heidkaemper, D., Klingenberg, M., Liu, Q., and Caskey, T. (2001). UCP3 expressed in yeast is primarily localized in extramitochondrial particles. Biochem. Biophysical Res. Commun. 282, 334–340. doi:10.1006/bbrc.2001.4563
Wu, Y., Chen, M., and Jiang, J. (2019). Mitochondrial dysfunction in neurodegenerative diseases and drug targets via apoptotic signaling. Mitochondrion 49, 35–45. doi:10.1016/j.mito.2019.07.003
Keywords: mitochondrial carrier proteins, adenine nucleotide translocase (ANT), phosphate translocase (PiT), uncoupling proteins (UCPs), autoinduction, expression and purification, native like folding, reconstitution
Citation: Tabefam M, Smith MD and Jelokhani-Niaraki M (2024) Expression, purification and folding of native like mitochondrial carrier proteins in lipid membranes. Front. Biophys. 1:1334804. doi: 10.3389/frbis.2023.1334804
Received: 07 November 2023; Accepted: 22 December 2023;
Published: 08 January 2024.
Edited by:
Olga Vinogradova, University of Connecticut, United StatesReviewed by:
Jose A. Brito, Universidade Nova de Lisboa, PortugalCopyright © 2024 Tabefam, Smith and Jelokhani-Niaraki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masoud Jelokhani-Niaraki, bWplbG9raGFuaUB3bHUuY2E=; Matthew D. Smith, bXNtaXRoQHdsdS5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.