- Department of Biomedical Engineering, The University of Texas at Austin, Austin, TX, United States
Cardiac tissue engineering (CTE) holds promise in addressing the clinical challenges posed by cardiovascular disease, the leading global cause of mortality. Human induced pluripotent stem cells (hiPSCs) are pivotal for cardiac regeneration therapy, offering an immunocompatible, high density cell source. However, hiPSC-derived cardiomyocytes (hiPSC-CMs) exhibit vital functional deficiencies that are not yet well understood, hindering their clinical deployment. We argue that machine learning (ML) can overcome these challenges, by improving the phenotyping and functionality of these cells via robust mathematical models and predictions. This review paper explores the transformative role of ML in advancing CTE, presenting a primer on relevant ML algorithms. We focus on how ML has recently addressed six key address six key challenges in CTE: cell differentiation, morphology, calcium handling and cell-cell coupling, contraction, and tissue assembly. The paper surveys common ML models, from tree-based and probabilistic to neural networks and deep learning, illustrating their applications to better understand hiPSC-CM behavior. While acknowledging the challenges associated with integrating ML, such as limited biomedical datasets, computational costs of learning data, and model interpretability and reliability, we examine suggestions for improvement, emphasizing the necessity for more extensive and diverse datasets that incorporate temporal and imaging data, augmented by synthetic generative models. By integrating ML with mathematical models and existing expert knowledge, we foresee a fruitful collaboration that unites innovative data-driven models with biophysics-informed models, effectively closing the gaps within CTE.
1 Introduction
Cardiac tissue engineering (CTE) is poised to improve clinical outcomes of cardiovascular disease (CVD), the leading cause of death worldwide. Affecting over 80 million Americans every year (Benjamin et al., 2018), the impact of cardiovascular disease is magnified by the heart’s inability to repair and self-regenerate. Cardiomyocytes (CMs) have a limited ability to proliferate and generate new tissue after birth (Laflamme and Murry, 2011). Thus, infarction events that cause the death of cardiac tissue commonly result in permanent cardiac output deficiencies (Anderson and Morrow, 2017). Current treatment strategies for CVD seek to resolve infarction triggers, like coronary artery atherosclerosis, and prevent future events (Reddy et al., 2015). While effective, these strategies do not repair or regenerate already-damaged cardiac tissue.
Human induced pluripotent stem cells (hiPSCs) are pivotal in realizing the potential of cardiac regeneration therapy. They address immunogenicity concerns associated with allogeneic cell sources and, more importantly, facilitate the achievement of high cell densities essential for restoring the over one billion cardiac cells damaged during myocardial infarction. hiPSCs are derived from patient tissue, minimizing the immunogenicity of implanted engineered tissues. Additionally, the self-renewal and proliferative behavior of hiPSCs ensure a theoretically limitless supply of CMs, enabling the engineering of patient-specific cardiac macrotissues at physiological cell densities. Yet, hiPSC differentiated cardiomyocytes (hiPSC-CM) exhibit abnormal morphology and decreased functionality compared to adult CMs with regards to contraction force and electrochemical coupling (Feric and Radisic, 2016). These deficiencies compound to significantly increase the potential for arrhythmias when hiPSC-CMs are implanted in vivo (Chong et al., 2014). Numerous groups have demonstrated the role of electrical (Nunes et al., 2013; Ruan et al., 2016), mechanical (McCain and Parker, 2011; Ruan et al., 2016), and biochemical stimuli (Horikoshi et al., 2019) on hiPSC-CM development and functionality, working to close the gap between hiPSC-CMs and in vivo cardiac tissues (Ronaldson-Bouchard et al., 2018). Even so, the discovery and optimization of these stimuli requires complex experimental methods and extended timelines, greatly slowing the rate of progress. Artificial intelligence, and more specially machine learning, has recently emerged to be a powerful tool, poised to accelerate experimental methods and parallel, even predict, in vitro work.
Herein we provide a primer on relevant machine learning algorithms, review current work applying machine learning to CTE and trends of focus, and discuss key ideas for further development. Specifically, we identify the following six challenges of CTE: cell differentiation, morphology, calcium handling and cell-cell coupling, cell contraction, and tissue assembly. Historically, biomedical applications of ML have primarily focused on processing large quantitative sets like RNA sequencing data or MRI imaging datasets (Wang et al., 2018; Cascianelli et al., 2020; Petegrosso et al., 2020). Here we discuss how this work have been translated to the field of tissue engineering to resolve the challenges related to hiPSC-CM maturity. It is worth noting others have sought to review the applications of machine learning in stem cell therapies (Coronnello and Francipane, 2022), cardiovascular pathology and treatment (Bizopoulos and Koutsouris, 2019; Glass et al., 2022; Kresoja et al., 2023), 3D bioprinting (An et al., 2021), organ-on-a-chip methodologies (Koyilot et al., 2022), and biomedical engineering as a whole (Shajun Nisha and Nagoor Meeral, 2021), yet few focus on the specific challenges of CTE and provide an in-depth review of how machine learning may be leveraged to improve the maturation of hiPSC-CMs.
It is important to note that ML remains a highly dynamic field of research, particularly within tissue engineering. In our literature review, we specifically targeted research articles that employ ML models to enhance the comprehension of hiPSC-CM behavior. We deliberately excluded studies that integrate ML into the use of hiPSC-CMs as models, like drug cardiotoxicity or disease progression (Heylman et al., 2015; Lee et al., 2017; Maddah et al., 2020; Grafton et al., 2021; Juhola et al., 2021; Kowalczewski et al., 2022; Yang et al., 2023). To assemble the cohort of research papers, a comprehensive literature search was conducted using the PubMed-NCBI database with the following search terms:
1) cardiac tissue engineering[MeSH:noexp] AND artificial intelligence[MeSH:noexp];
2) cardiac tissue engineering[MeSH:noexp] AND machine learning[MeSH:noexp];
3) cardiac maturation[MeSH:noexp] AND machine learning[MeSH:noexp]; and
4) scaffold optimization hiPSC-CM machine learning.
Other searches with relevant key terms yielded no results; therefore, they were omitted from this list. By selecting papers from the last decade that align with our specific focus, we curated a cohort of 23 research papers spanning 2013–2023.
In review, we observe that efforts within each challenge of CTE work to 1) improve the functionality of hiPSC-CMs and 2) improve physiological phenotyping and characterization of tissues to accelerate innovation. This distinction is made in our discussion of each implementation of ML in hiPSC-CM immaturity.
2 Artificial intelligence and machine learning
Artificial intelligence (AI) has become a transformative force across numerous disciplines, promising groundbreaking advancements and technological revolutions. Within the expansive landscape of AI, machine learning (ML) emerges as a pivotal subset, fundamentally reshaping problem-solving and decision-making processes. In our exploration of recent ML methods in the realm of CTE, it is imperative to navigate through the layers of AI and ML, grasping their core principles and understanding their relevance in the broader field of biomedical engineering (BME).
At its essence, AI strives to emulate intelligent behavior and decision-making in computers, pushing the boundaries of what machines can achieve. AI systems are designed to undertake tasks requiring human-like intelligence, including learning, reasoning, problem-solving, perception, and language understanding. Within this multifaceted field, ML stands out as an integral component as it falls within the vast data science umbrella of AI, empowering systems to learn patterns and make decisions from data without explicit programming. The hallmark of ML lies in its capacity to enhance performance iteratively through learning from experience and statistical inferences. This computationally expensive learning process is a crucial feature, especially in addressing the complex challenges posed by CTE, where the interplay of biological components demands robust and well-guided solutions. The intersection of ML and BME has exposed unprecedented possibilities in healthcare, diagnostics, and therapeutic interventions (Jovel and Greiner, 2021; Strzelecki and Badura, 2022). In the context of CTE, ML algorithms offer the potential to decipher intricate relationships within biological systems. Mathematical or physics-based models traditionally attempt to decipher systems like cellular interactions and tissue behavior, yet many times fall short (Park et al., 2018). To supplement these efforts, researchers have begun to harness ML in the recent decade to further analyze and explore novel avenues in CTE. Within the broader ML landscape lies deep learning (DL) with its neural network architectures, showing remarkable success in 2D and 3D image and signal processing (Litjens et al., 2017; Zhou et al., 2017). DL models with multiple layers (usually more than one hidden layer between the input and output layers) excel in capturing intricate hierarchical representations and features from complex data, when enough data is provided (Cybenko, 1989; Hornik et al., 1989; Yarotsky, 2017). Within CTE, this translates to enhanced capabilities in biomedical image analysis, signal processing, and predictive modeling, offering a deeper understanding of cardiac dynamics and responses to various perturbations.
The integration of AI and its subfields is not without challenges. Biomedical data, either in signals or images, is often complex, heterogeneous, and characterized by high dimensionality (Dinov, 2016; Hulsen et al., 2019). Ensuring the robustness and generalizability of ML models across diverse datasets is a persistent challenge. Additionally, ML models are often viewed as “black boxes,” which naturally creates distrust regarding the transparency and interpretability of decision-making processes in critical biomedical applications and healthcare, specifically in clinical settings with diagnoses and treatment plans. The challenges of ML in CTE are further amplified by the intricate nature of cardiac tissue. Modeling the dynamic and multifaceted aspects of cardiac function and response requires sophisticated ML approaches. Furthermore, DL models are known for their demand for substantial amounts of labeled data, posing challenges in scenarios where obtaining sufficiently large training sets may be difficult or impractical. Nevertheless, as this review explores, ML methods, including DL architectures, prove to be successful in CTE, especially in cases where mathematical models are limited. It is noteworthy that these physics-based models demand extensive time and computational resources for numerically solving complex systems of equations spanning multiple scales, all the while data-driven methods often fall short in achieving high accuracy and generalization capabilities (Peek et al., 2014; Rueckert and Schnabel, 2020; Regazzoni et al., 2022). We expect the balancing of these trade-offs will expedite the evaluation and accuracy of mathematical models and tackle issues associated with many-query problems in CTE applications. By understanding these challenges, researchers and practitioners can harness the potential of ML methodologies and advance the frontiers of cardiac tissue research and engineering.
3 Common machine learning models
In the last decade, ML has become integral to BME, particularly in CTE, by providing powerful tools to extract meaningful insights from complex data. We first discuss common ML models and then delve into how these are used to tackle the CTE challenges we have identified in this review.
3.1 Linear models
Two of the most intuitive and straightforward supervised ML models are linear and logistic regression (James et al., 2023). Linear regression learns the relationship between the dependent variable and one or more independent variables by fitting a linear equation to observed data. It is widely used in CTE for tasks, such as predicting cell culture properties based on a set of initial parameters. Despite its simplicity, linear regression provides interpretable insights into the impact of individual features on the outcome. Applying a threshold to the output classifies data into multiple categories, such as different types of tissue structures or cell types. Similarly, logistic regression is for binary classification tasks, making use of the logistic or sigmoid function to map the linear combination of input features into a probability distribution between 0 and 1 (Fukunaga and Hostetler, 1975). Logistic regression is valuable in scenarios like cell-type classification, where it is essential understanding the likelihood of a cell culture having a specific content of successfully differentiated cells or having specific types of cell phenotypes, as we will discuss in forthcoming sections.
3.2 Tree-based models
More involved, but well-established supervised ML models are decision trees and random forests, both of which serve for both regression and classification tasks. Decision trees partition the feature space into regions based on feature thresholds, making decisions by traversing the tree from the root to the leaves (Quinlan, 1986). They are susceptible to overfitting, but standard techniques like pruning and ensemble methods mitigate this. Decision trees are intuitive and interpretable, while capturing complex decision boundaries, making them valuable for feature selection and identifying critical factors. Random forest models build an ensemble of decision trees, each trained on a subset of the data and features (Breiman, 2001). They leverage the diversity among trees to improve generalization and robustness, favored for its ability to handle high-dimensional data and nonlinear relationships. The aggregation of multiple trees enhances predictive performance and provides insights into feature importance.
3.3 Probabilistic models
Naïve Bayes is a probabilistic classification algorithm based on Bayes’ theorem, which describes the probability of an event based on prior knowledge of conditions that might be related to the event (Manning, 2008; Witten et al., 2011). The “naïve” assumption is that features are conditionally independent given the class label, which simplifies the computation of probabilities, but may not hold in all cases. Thanks to its framework that is computationally efficient, simple to implement, and often requires smaller amounts of training data, naïve Bayes performs well in tasks with high-dimensional data, like predicting diseases or drugs based on cellular features. Gaussian Processes (GPs) are one of the most flexible models for regression tasks, providing a probabilistic framework for learning relationships in data by considering entire distributions of functions (Rasmussen and Williams, 2006). Unlike traditional regression models, GPs offer a non-parametric approach, making them suitable for capturing complex patterns and uncertainties in diverse datasets. The strength of GPs lies in their ability to provide not only predictions, but also estimates of prediction uncertainty, valuable in CTE scenarios where uncertainty quantification is crucial for cell maturation. Linear discriminant analysis (LDA) and its variants, like quadratic discriminant analysis (QDA), are dimensionality reduction and classification techniques (Izenman, 2008; Hastie et al., 2009). These models find combinations of features that maximize the distance between class means and minimize the spread within each class, making them valuable for task in biomedical signal processing and imaging, like feature extraction and cell-type classification.
3.4 Other models
Support vector machines (SVMs) aim to find the hyperplane that maximally separates data points of different classes (Cortes and Vapnik, 1995). They operate effectively in high-dimensional spaces and are applicable for tasks such as classifying cell cultures based on complex profiles. An SVM uses kernels to compute the similarity between pairs of data points, enabling it to handle nonlinear relationships and making it versatile in capturing intricate patterns in biomedical data. It is mostly used in classification tasks, but it can be extended to regression. As it implies, k-nearest neighbors (kNN) classifies data points based on the majority class among their k-nearest neighbors (Cover and Hart, 1967). kNN is valued for its simplicity and effectiveness, especially in scenarios where data points form well-defined clusters. kNN is sensitive to the choice of distance metric and the number of neighbors, making careful parameter selection crucial in achieving optimal results.
3.5 Neural networks and deep learning
Artificial neural networks (ANN) vary significantly from the mentioned models, consisting of interconnected nodes organized in layers that are trained using backpropagation (Haykin, 1998, 2008; Bishop and Nasrabadi, 2006). The input layer receives the initial data, usually preprocessed, and computations take place within intermediate “hidden” layers, helping the network learn complex patterns and representations. The output layer produces the final output or prediction. Each connection between neurons or “nodes” in different layers is associated with a weight. The network learns by adjusting these weights during training, where the activation of neurons in the hidden and output layers is determined by applying certain activation functions, like sigmoidal, hyperbolic tangent, or leaky rectified linear unit (ReLu). Fully-connected or “dense” layers are those that have each neuron connected to every neuron in the adjacent layer, creating a dense network of connections and, hence, more unknowns to solve for. A common ANN is a feedforward neural network, where information flows from the input layer through one or more hidden layers to the output layer without forming cycles or loops (Rumelhart et al., 1986; Haykin, 2008). Feedforward neural networks are foundational and used in various machine learning tasks like cell-type classification or bioprinting parameter regression based on multimodal data.
While feedforward networks are powerful and can be considered DL with several hidden layers, more complex architectures, such as convolutional neural networks (CNNs) and recurrent neural networks (RNNs), have been developed to address specific challenges in different types of data through either supervised or unsupervised learning (Goodfellow et al., 2016). CNNs are designed for grid-like data, leveraging convolutional layers for spatial feature extraction (Szegedy et al., 2015; Venkatesan and Li, 2017; Shanmugamani, 2018). CNNs excel in medical image analysis, detecting patterns and structures, by applying filters to learn hierarchical representations that identify localized features critical in tasks, such as cell detection and quality assessment of maturing hiPSC-CMs. RNNs maintain hidden states to capture temporal dependencies in sequential data with feedback loops and are commonly applied to time-series data, allowing tasks like physiological signal analysis and predicting disease progression (Rumelhart et al., 1986; Pascanu et al., 2013). However, traditional RNNs face challenges in capturing long-term dependencies, leading to the development of more advanced architectures like Long Short-Term Memory (LSTM). LSTM networks are a type of RNN designed to address the vanishing gradient problem, modeling long-range dependencies in sequential data (Gers et al., 2000). These networks are crucial for tasks requiring an understanding of temporal relationships across even irregular intervals, such as subtleties of cellular interactions and tissue development and analyzing dynamic physiological signals. Their ability to capture context over extended sequences makes them well-suited for time-sensitive medical data.
These handful of models collectively form a diverse toolkit for addressing various challenges in BME, offering solutions tailored to different data types, complexities, and desired outcomes. Other models found in the literature that are more tailored to specific CTE challenges will be discussed in the following sections. Although the field of ML can be overwhelming, numerous resources exist to support researchers from diverse backgrounds in acquiring the skills to program, implement, and utilize ML models in Python, R, and MATLAB (Conway and White, 2012; Murphy, 2012; Kelleher et al., 2015; Theodoridis, 2015; Müller and Guido, 2016; Raschka and Mirjalili, 2017; Géron, 2019).
4 ML in cardiac cell differentiation
Numerous protocols for CM differentiation exist that result in varying degrees of purity and functionality. Post differentiation purification schemes leveraging metabolic selection have become a standard part of cardiac differentiation to ensure high percentages of CMs and to prime lipid metabolism seen in adult CMs (Sharma et al., 2015). These strategies are inherently reactive and cannot proactively ensure efficient differentiation. Given that cardiac differentiation is a multifactor process with complex interactions, experimental optimization can be expensive both in time and resources. ML techniques have been leveraged to identify important factors in driving efficient cardiac differentiation and predict the purity of differentiating hiPSC-CMs at early timepoints.
Williams et al. were among the first to leverage ML techniques to identify the most influential experimental variables that may be controlled during cardiac differentiation and sought to establish a robust classification and prediction of CM purity during differentiation in advanced stirred tank bioreactors (STBRs) (Williams et al., 2020). STBRs facilitate scale-up of differentiation protocols and continuous monitoring of environmental conditions (such as media dissolved oxygen and pH level) and have been used extensively for tissue culture over the last decade (Kehoe et al., 2010; Kempf et al., 2016; Halloin et al., 2019). Manual sampling can also enable evaluation of cell viability and proliferation. During the differentiation process, spanning up to 5 days after initiation, 39 variables were measured relating to dissolved oxygen, pH, cell density, aggregate size, nutrient concentration, and preculture conditions. Post differentiation, CM purity was analyzed via flow cytometry using canonical CM markers cardiac troponin T and myosin heavy chain. Cultures yielding >90% CMs were coded to be successful or insufficient otherwise. Data from 58 unique cardiac differentiations were split into training (n = 42) and testing (n = 16) datasets and applied to random forest, GP regression, and multivariate adaptive regression splines (MARS) algorithms. The latter is a non-parametric regression technique for modeling complex relationships between input features and output variables, well-suited for data with underlying nonlinear interactions and discontinuities (Friedman, 1991). Random forest and GP regression models yielded 90% accuracy and 90% precision in identifying experiments that would yield sufficient CM purity, while random forest models yielded 84% accuracy at identifying if an experiment would have an insufficient final CM content. Importantly, this work details how ML may be used to predict CM purity after only 5 days of differentiation, using less than 16% of collected data.
Similarly, Mohammadi et al. studied random forest, GP classification, and SVM methods to identify important differentiation factors and conduct quality checks on hiPSC-CM embedded PEG-fibrinogen microspheres (Mohammadi et al., 2022). While extruded cardiac microspheres enhance 3D culture and differentiation efficiency, potentially facilitating scalable hiPSC-CM production, they may affect CM purity due to variations in physical properties. By tuning variables such as CHIR concentration, fibrinogen concentration, differentiation media, size and shape of extruded microspheres, and cells seeding density, this group aimed to develop predictive models for resulting CM purity. Principal component analysis (PCA) was first employed to determine features that correlate strongly with efficient CM differentiation (65% CM or higher). PCA is a method commonly used to transform high-dimensional data into a lower-dimensional space by identifying the principal components that capture the most variance (Pearson, 1901; Hotelling, 1936). This uncovers latent patterns in datasets with numerous correlated features while retaining as much information as possible. In this case, random forest, GP classification, and SVMs were trained on select input features including postfreeze passage number, PGE-fibrinogen concentration, and the ratio between the CHIR concentration and microsphere surface to volume ratio with CM purity as output. GP classification models yielded 70% accuracy with 56% precision at classifying high purity cultures. It should be noted that the principal components used in the best-performing model explained only 18% of the input data variance. This suggests that the variance of the input set did not entirely account for the variance in the output data, indicating the need for more features and indicators within a larger dataset.
One year later, the same supervised classification task was tested, this time employing CNNs (Mohammadi et al., 2023). Two models were created to predict the percentage of CM content on Day 10 of differentiation, using pre-differentiation experimental features and nondestructive images from Days 3 or 5. One CNN utilized only phase-contrast image, while another incorporated a combination of both images and the features identified in earlier work (Mohammadi et al., 2022). With an accuracy of 85% and precision of 92%, the best-performing model was the combination CNN using images from Day 5, outperforming the other ML models, including GP with features only, SVM with images only, and SVM with a combination of images and features. Although successful on only about 300 images, the model developed a dependence on the specific cell line used. Evaluation on another cell line yielded a significantly lower accuracy of 46%, emphasizing the substantial challenges in generalizing predictions across diverse cell lines.
5 ML in morphology
CMs exhibit a strong structure-function relationship, causing morphological differences to have significant impact on cardiac function. Healthy, adult CMs exhibit elongated, rod-shaped, and anisotropic morphologies with high aspects ratios (7:1 to 9.5:1) (Gerdes et al., 1992). hiPSC-CMs instead exhibit round, polygonal morphologies with significantly smaller aspect ratios, trending more to isotropic orientations (Feric and Radisic, 2016). Morphological assessment has historically required extensive time and resources to execute and is prone to user bias. ML methods trained on images of CMs have accelerated and standardized morphological assessment of hiPSC-CMs.
Pasquilini et al. were among the first to bring ML to analyze hiPSC-CM morphology and intracellular structure (Pasqualini et al., 2015). After gathering fluorescent images of rat primary ventricular monocytes (rpCMs) and hiPSC-CMs, stained for sarcomeric α-actinin and fibronectin, 11 metrics were assessed. Among others, metrics included sarcomere length, sarcomeric packing density, and Z-disk relative coherence. Naïve Bayes, a feedforward neural network, and a bootstrap aggregation of decision trees were trained to classify rat primary cardiomyocytes (rpCMs) as mature or immature using the 11 metrics as prediction elements and a user-defined maturity label as output elements. All models, trained with the same dataset, yielded similar degrees of hiPSC-CMs immaturity accuracy, with the naïve Bayes classifier yielding 70%, the neural network yielding 71%, and the tree bagging classifier yielding 77%.
Orita et al. addressed the variability in the viability of cultured hiPSC-CMs through a CNN, emphasizing the need for accurate and high-throughput screening methods due to limitations in manual inspection (Orita et al., 2019). A VGG16 architecture, pretrained with the ImageNet dataset for transfer learning, was further trained on experimental images labeled as ‘normal’ or ‘abnormal’ by an expert, however, the authors did not elaborate on what parameters warranted normal or abnormal classification. The VGG16 architecture used contained a total of 16 layers: 13 convolutional layers and 3 fully connected layers. The former are used for extracting image features, such as edges and colors, while the latter are used for image classification through nonlinear combination of the extracted features. All convolutional layers of VGG16 were frozen and used as fixed feature extractors, except for the eighth fully connected layer, which was modified from 1000 nodes to 2 nodes for binary classification. After implementing data augmentation methods to expand the dataset to 18,000 images, the model successfully reached 90% accuracy, showcasing its capability for automating quality control. Nevertheless, the study acknowledges the necessity for a more extensive dataset, especially given the trade-off between calculation speed and performance. The findings suggest that full retraining of VGG16 or exploring more advanced pretrained models could further enhance performance and robustness in classifying hiPSC-CM cultures.
Khadangi et al. introduced CardioVinci, a deep-learning framework combining a U-net and GAN for unsupervised segmentation and synthesis of 3D electron microscopy (EM) datasets of cardiac cells (Khadangi et al., 2022). 3D Electron microscopy datasets excel in providing high resolution images of intracellular organelles and extracellular matrix organization. But their large data size cause processing and analysis steps to be time and energy intensive. Generative adversarial networks (GANs) excel in biomedical imaging by generating realistic synthetic data through adversarial training of a generator and discriminator network, fostering advancements in image synthesis, data augmentation, and disease simulation for enhanced machine learning model training (Goodfellow et al., 2014). U-net, a versatile convolutional neural network, proves indispensable in biomedical image segmentation, effectively delineating and identifying structures with its distinctive U-shaped architecture, thereby contributing to precise and efficient medical image analysis and diagnosis (Ronneberger et al., 2015). The U-net segments mitochondria, myofibrils, and Z-discs, achieving notable accuracy as confirmed by niche evaluation metrics, like compactness, flatness, sphericity, elongation, and surface area to volume ratio (SA:V). Next in the CardioVinci pipeline, a GAN is encoded as a probabilistic model of CM architecture to extract morphological metrics and spatial distributions of mitochondria, myofibrils, and Z-discs in semantic segmentations. StyleGAN is popular type of GAN architecture with densely connected convolutions motivated by style transfer problems (Karras et al., 2019). Here, StyleGAN was used to minimize the Fréchet inception distance and was trained on experimentally collected 3D EM datasets of left ventricular cardiomyocyte extracted from a type 1 diabetic rat tissue sample to generate a probability distribution of 2D CM structures found within the image slices of the 3D EM segmented volume datasets, offering a novel approach to generative modeling of CM ultrastructure. Despite its ability to generate statistically similar geometries, CardioVinci requires an initial segmentation step and accurate stacking of 2D images into 3D volumes, posing challenges for large tissue samples. Nevertheless, it represents a notable advancement in automating the analysis of cardiac ultrastructure.
6 ML in calcium handling and cell-cell coupling
Functional adult CMs exhibit mature intracellular and efficient intercellular calcium handing. Effective intracellular calcium handling is dictated by high expression of voltage-gated Ca2+, Na+, and K+ ion channels in adult CMs (Bodi et al., 2005; Sartiani et al., 2007). Responding to varying membrane potentials, these channels trigger intracellular calcium release and take-up, allowing for efficient contraction cycles. Relative expressions of these ion channels are significantly lower in hiPSC-CMs vs. adult CMs, resulting in deficient function action potential and contraction cycling (Feric and Radisic, 2016). Similarly, hiPSC-CMs exhibit lower levels of gap junction proteins, preventing efficient intercellular transduction of cardiac action potentials between neighboring CMs. Significant work has been conducted to accurately classify calcium transient signals into normal or abnormal to study the contributions of calcium handling to cardiac disorders and overall disfunction. Application of these methods in characterizing calcium transients of hiPSC-CMs may help identify the root causes of deficient calcium cycling and improve the electrophysiology of hiPSC-CMs to match adult CMs.
Through spectral clustering, Gorospe et al. (2014) automatically classified a population of hESC-CMs into different phenotypes based on electrophysiology signals. Spectral clustering is a ML technique that utilizes the eigenvalues and eigenvectors of the data matrix to project the data into a lower-dimensional space for the purpose of clustering data points (Ng et al., 2001). Heterogeneity of differentiated CMs may be characterized by electrophysiology as action potential (AP) waveforms differ significantly across pacemaker, atrial, and ventricular phenotypes. The classification task is based on the hypothesis that APs of the same phenotype exhibit more similar shapes than those of different phenotypes. This distinction is captured via spectral clustering in an experimentally collected dataset comprising 6940 APs from nine cardiac clusters, analyzed using Euclidean distance to quantify the similarity between APs. Then, these similarities were fed as inputs to a simple spectral grouping algorithm with the aim to objectively distinguish populations of cardiac APs with distinct phenotypes. The best grouping fitness was achieved when identifying 2 phenotypes, as indicated by normalized cut cost and Davies-Bouldin Index metrics. However, due to the simplicity of the clustering algorithm and diversity in waveforms, the strength of this method diminished when attempting to identify three and four phenotypes per cell cluster. Notably, grouping the population based on standard AP features, like duration at 30% (APD30) and 80% (APD80) repolarization, and their differential, was effective for two groups, while using the entire AP waveform was effective for two, three, or four groups.
Researchers often use microelectrode arrays (MEAs) to measure APs in cardiac cells. This technique enables simultaneous recordings from multiple locations, allowing researchers to capture the spatiotemporal dynamics of electrical activity across the entire cell population. However, simultaneous recordings from multiple locations generate a large amount of data, and the interpretation of such complex datasets may pose challenges. To tackle this, Jurkiewicz et al. (2021) implemented an SVM to analyze signals recorded from hiPSC-CMs on MEAs. Within MEA readings, intracellular APs can be directly measured via electroporation or indirectly via extracellular field potentials (FPs). The SVM was used to filter MEA recorded signals into APs and FPs, classify signals as usable or unusable, and extract AP features from the signals. Collected signals were first split into APs and FPs based on time of electroporation. Signals collected prior to electroporation were identified as FPs, while signal collected after electroporation were classified as action potentials. APs and FPs were then sorted into usable and unusable signals based on deterministic noise and signal fidelity rules. The SVM was then trained on AP and FPs signal as predictive inputs to identify usable and unusable signals. The classifier achieved 100% accuracy on the training set due to linear separability. Finally, once AP signals were identified, 3 classical features were computed - basic cycle length (BCL), APD30 and APD80. This analysis facilitated an in-depth investigation of cardiac restitution, exploring the relationship between APD for a given beat to the diastolic interval (DI) of the preceding beat, DI = BCL − APD. The application of an SVM with MEA readings accelerates the location and classification of APs and allows for quick extraction of AP features within MEA datasets, but is limited in its translatability to patch clamp experimental data.
Juhola et al. (2015) extended the analysis to calcium cycling signals, using ML techniques to characterize and classify hiPSC-CMs based on calcium cycling signals. Calcium cycling, downstream of cardiac action potentials, provides insight into cardiac calcium handing and contraction. In this study, calcium-reporting fluorescent images of hiPSCs-CMs derived from patients with catecholaminergic polymorphic ventricular tachycardia (CVPT) were collected. After pre-processing, calcium waveforms were generated by plotting the average intensity in regions of interest within calcium images, resulting in 280 unique signals. Peaks within each waveform were identified and classified as normal or abnormal via deterministic rules associated with changes in absolute fluorescence, peak amplitude, and symmetry. Next, seven peak parameters, including left/right side amplitude, duration, and time interval between peaks were calculated for each identified peak. These seven predictive elements were reduced via PCA, which yielded a first principal component explaining 95% of sample variance and a second principal component explaining 5%. After testing a variety of ML models, including kNN and SVMs, QDA was found to perform the best with an 80% accuracy compared to human expert assessment. It should be noted that the deterministic rules guiding classification were rooted in identifying at least one abnormal peak in waveforms, i.e., abnormal waveforms were classified as such if they contain at least one abnormal peak. In a follow-up study, Juhola et al. (2018) then applied this method to discriminate specific genetic diseases. Applying previous methods to hiPSC-CMs derived from patients with catecholaminergic polymorphic ventricular tachycardia (CPVT), long QT syndrome (LQT), and hypertrophic cardiomyopathy (HCM), yielded classification accuracies of 79%–88%.
Building on this work, Hwang et al. (2020) incorporated expert determined ground truths, leveraging shared characteristics of normal and abnormal Ca2+ transient peaks and signals across all samples. The peaks in 200 training calcium transients were identified and characterized according to 14 peak characteristics similar to Juhola et al. (2015). Notably, the peak detection algorithm was improved by using derivatives of calcium transient over raw amplitudes and additional peak characteristics were calculated, such as nearby peak distance varying peak amplitude, and peak asymmetry. A peak-level SVM classifier was then trained using the 14 peak variables as predictive features and expert normal/abnormal assessment as outcome variables. Taking the outcomes of peak-level classification, overall calcium transient signals were classified as normal/abnormal using a cell-level SVM. For training, peak normality assessments along with additional cell variables, like proportion of abnormal peaks per signal, variance of peak amplitude per signal, variance of peak distances per signal, and variance of peak areas per signal, were taken as predictive features, while human-expert assessments were taken as outcomes. This two-tiered signal analysis pipeline results in an 88% training accuracy and 87% test accuracy, but these models were trained and tested on similar signals potentially overfitting or biasing the model. In general, training and testing on more varied data sets is recommended.
Yang et al. (2022) introduced an alternative ML method to identify drug treatment of hiPSC-CMs, of 63 unique compounds, using calcium waveforms. The custom CardioWave pipeline extracts 38 parameters from waveforms, including metrics on rise/decay time, number of peaks, peak frequency, amplitude, and shoulder characteristics. With 303 samples across 63 compounds, PCA was used to project all samples parameters into a 2D space. Next, a random forest model was trained with calcium waveforms as predictive elements and the identity of the dosed compound as ground truth elements. Employing leave-one-compound-out cross-validation to evaluate the random forest model yielded an overall accuracy rate of 86%. Gaps in the experimental data set, notably varying drug concentrations and forced hyper-parameterization of training inputs, may be reducing the accuracy of this model.
Aghasafari et al. (2021) introduced a multitask network that simultaneously classifies iPSC-CM AP traces into categories representing drugged CMs and drug-free CMs and translates them into adult-CM AP waveforms. Action potentials of drug-free iPSC-CM and adult human CMs were generated via pacing in silico models using the Kernik iPSC-CMs in silico model (Kernik et al., 2020) and the O’Hara–Rudy human adult-CMs in silico model (O’Hara et al., 2011). Drugged action potentials were collected via simulation of the above models modified with a simple drug-induced IKr block model of hERG channel conduction, reducing conductance by 1%–50%, and complex Markov model of conformation-state dependent IKr block in the presence of hERG blocking drug dofetilide. A deep learning algorithm was built on an RNN architecture with LSTM layers, where the layers remember the most important iPSC-CM AP features needed for the synchronized classification and translation tasks. The model used preprocessed iPSC-CM APs as the network input and adult-CM APs along with corresponding drug-free and drugged labels as network outputs. The supervised tasks of classification and translation were achieved independently of each other through individual fully-connected layers. Addressing the challenge of immature hiPSC-CM models, the algorithm navigates the translation task by learning from synthetic data, exhibiting proficiency in predicting drug-free and drugged states with approximately 90% accuracy. Although the model also successfully translates hiPSC-CM APs into adult-CM APs with less than 0.003 mean-squared error, it requires multiple high-quality datasets for effective training, with potential improvements including addressing data sparsity and exploring diverse approaches beyond supervised learning. The LSTM-based multitask network showcases the capability to discern and translate cardiac APs, offering promise for applications in drug testing and disease modeling.
7 ML in cell contraction
Contraction is intricately tied to intracellular calcium handling in CMs. Thus, the abnormal calcium cycling seen in hiPSC-CMs further diminishes contractile force generation and contributes to a large functionality gap between hiPSC-CMs and adult, and even neonatal, CMs. The active stress generated by freshly isolated contracting human ventricular CMs was measured to be >50 mN/mm2 (van der Velden et al., 1998) and neonatal (<2 weeks) ventricular CMs generated of 0.8–1.7 mN/mm2 (Wiegerinck et al., 2009). hiPSC-CMs, by comparison are measured to produce 0.15–0.30 mN/mm2 (Kita-Matsuo et al., 2009; Hazeltine et al., 2012). Numerous mechanical and electrical stimuli such as static and cyclic mechanical stretch (Kensah et al., 2011; Turnbull et al., 2014; Ribeiro et al., 2019), electrical stimulation (Nunes et al., 2013; Ruan et al., 2016), and supporting cell co-cultures (Richards et al., 2017; Beauchamp et al., 2020), have been noted to improve contractile properties of hiPSCs. Given the multitude of factors influencing cardiac contraction development, machine learning emerges as a powerful tool for accelerating the characterization and optimization of hiPSC-CMs’ contraction.
Lee et al. (2015) provided one of the first applications of ML to evaluate the contraction of CMs. Leveraging an SVM, brightfield image sequences of beating CMs were classified as normal or abnormal. Brightfield images were first processed with optimal flow algorithms to generate matrices of x- and y-directional vectors that describe CM motion. PCA summarized vector information into a singular variable that described bulk cardiac contraction over time. Using user-identified contraction and relaxation peaks, 12 contraction parameters were calculated including peak duration, amplitude, rise time, area under curve, and frequency. An SVM was then trained with over 200 unique samples to classify samples as normal or abnormal using the contraction parameters as predictive variables and known experimental conditions as the outcome variable. The trained model achieved an accuracy rate of 83%–99% in identifying cells treated with known contraction modulators, E-4031, verapamil, and blebbistatin, at higher concentrations. Notably, the model faced challenges in predicting outcomes for drug dosing concentrations lower than 10 nM.
Orita et al. (2020) automated the process of contraction identification and employed a trained SVM to classify hiPSC-CM cultures as normal (experimentally usable) or abnormal (experimentally unusable). 556 brightfield image sequences of beating hiPSC-CMs were captured and classified by experts as abnormal (n = 190) or normal (n = 366). The contraction properties of the imaged CMs were calculated to be the 200-frame simple moving average (SMA) around the frame of interest. Data augmentation, via the sliding window method, enabled each contraction profile to be split into individual contraction waves, increasing the data set six-fold. Then, a fast Fourier transform converted contraction waves (n = 3,336) to the frequency domain and UMAP was employed to reduce the dimensionality of contraction waves to two dimensions. Uniform manifold approximation and projection (UMAP) is a nonlinear dimensionality reduction technique that is particularly effective for preserving both local and global structures in high-dimensional data (McInnes et al., 2020). Finally, an SVM was trained, using UMAP reduced dimensions as inputs and expert-assessment as ground truths, yielding an 89% accuracy and 92% precision rate. Notably, this pipeline was trained on global features within image sequences, potentially overlooking important local features that could impact accuracy negatively.
Similarly, Teles et al. (2021) employed ML to differentiate healthy CMs from CMs derived from patients with Timothy syndrome (TS). Cardiomyocytes were differentiated from healthy iPSC lines (WTC-11 and BS2) and from iPSCs derived from patients with TS. Brightfield videos of contracting hiPSC-CMs were collected and processed using custom MATLAB software to assess contractility parameters like beat frequency, peak-to-peak time, interbeat variability, and rise time. Contractility parameters and experimentally defined ground truths were trained on kNN, decision trees, naive Bayes classifier, QDA, and SVM. QDA and decision trees yielded a 92% accuracy when discriminating TS and healthy WTC-11s, while decision trees and SVM were best preforming at differentiating TS and healthy BS2 cells with 88% and 87% accuracy respectively. Finally, WTC-11 and BS2 derived cardiomyocytes were differentiated from each other with accuracies above 90% using decision trees, naïve Bayes with normal kernel, and SVM with quadratic and cubic kernel. Given the strongest predictive parameter was unable to be identified due to dimension reduction pre-processing, the authors argue for the generation of more diverse datasets of contractility, varying race, ethnicity, sex, and disease of donor cells.
Another automated approach for assessing drug-induced effects on CM electrophysiology used a feed-forward neural network that processes mechanical beating signals from hiPSC-CMs (Ouyang et al., 2022). A custom feature extraction program was applied to partition CM beating signals into distinct beating patterns and extract specific features in the time domain. These features were then manually analyzed to identify various drugs and predict their cardiotoxic concentrations. A multi-labeled neural network (MLNN) was constructed to identify different drugs and drug concentrations based on the specialized features of individual CM beating patterns. Nodes with dual labels in the output layers were designed to represent both binary drug types and numeric drug concentrations. This approach aimed to reduce the number of output classes in the classifier, preventing overfitting, especially when training with a limited dataset. By combining automatic feature extraction and MLNN, the model accurately classified six different drugs as either cardiotoxic or not and predicted their cardiotoxic concentrations, achieving 98% and 96% for training and testing accuracies, respectively. Despite the introduction of noise, the MLNN demonstrated robust performance, maintaining accuracy, precision, and recall at approximately 96%. Although the MLNN outperforms other DL methods like CNN and RNN frameworks at low concentrations, there is a need for more extensive datasets to enable a comprehensive comparison with other deep-learning methods.
8 ML in tissue assembly
The assembly of in vitro differentiated CMs into functional tissue is hindered by the complex nature of healthy myocardium, characterized by the high cell density, multicellularity, ordered structural composition. While the self-assembly of hiPSC-CMs into engineered heart tissues has produced largely functional 3D myocardium, its scalability and lack of vasculature pose limitations (Ronaldson-Bouchard et al., 2018). Various approaches, including scaffold or scaffold free systems (Feinberg et al., 2013; Nunes et al., 2013; Kobayashi et al., 2019; Qasim et al., 2019) such as bioprinting have been explored for 3D cardiac tissue assembly (Ong et al., 2018; Esser et al., 2023; Finkel et al., 2023). ML offers a potential avenue to expedite the optimization of scaffold and bioink parameters for supporting multicellular culture and facilitating functional tissue assembly.
Rafieyan et al. (2023) provide one of the few direct applications of ML to hiPSC-CM scaffold optimization. Their main objective was to overcome limitations created by small experimental datasets. To address this, they first developed a comprehensive, multi-cell line, muti-fabrication method dataset through an extensive literature review that connected material properties to CM function. Subsequently, they identified the most accurate algorithm to predict cell behavior on CTE scaffolds. Briefly, scaffold materials in this dataset included synthetic and natural materials like polycaprolactone and Fibroblasts-derived ECM. Fabrication methods in the dataset were comprised of electrospinning, hydrogel encapsulation, and 3D printing among others. Finally, cell lines ranged from cardiac progenitor cells to hiPSCs. Cellular response with these materials was categorized into 4 classes: (0) no evaluation of cellular behavior preformed, (1) poor cell viability, growth, and proliferations, (2) cell viability, growth, and proliferation seen up to 3 days, and (3) cell viability, growth, and proliferation seen past 3 days. The dataset, encompassing 33 different materials, 16 different cell lines, and 6 different fabrication methods, was used to train 23 algorithms using material parameters as predictive inputs and the 4 classifications of cell response as outcomes. XGBoost, a gradient boosting machine for regression and classification problems, yielded the highest classification accuracy (87%), while quadratic discriminant analysis yielded the lowest (33%). XGBoost also indicated bioactive ECM as a defining feature driving CM function. This study demonstrates the importance of comprehensive datasets in achieving high accuracy in ML methods and illustrates an example wherein ML may help identify important contributors of CM behavior, leveraging a diverse and disparate dataset.
Among various cardiac tissue assembly methods, bioprinting stands out as particularly promising. Not surprisingly, there is a notable emphasis on the application of hierarchical machine learning (HML) to optimizing bioprinting parameters specifically addressing the complexities of bioprinting for cardiac tissues, where achieving precise and accurate spatial arrangements of cells is crucial for functional outcomes. HML, a hybrid approach integrating physical and statistical methodologies, is particularly designed for small experimental datasets (Menon et al., 2017). In this approach, predictors are linked to the system response through an intermediary layer with variables parameterized by established physiochemical relationships. Regression techniques are then employed to establish connections between the middle layer and the system response, enabling effective prediction and optimization. Bone et al. (2020) used HML to optimize ink-related variables and printer settings for improved print fidelity in the Freeform Reversible Embedding of Suspended Hydrogels (FRESH) bioprinting modality. FRESH printing, while successfully employed for assembling 3D cardiac tissues, is plagued by print fidelity issues difficult to optimize (Finkel et al., 2023). With 48 prints generated and assessed for print fidelity by varying normalized flow rate, nozzle speed, alginate concentration, and nozzle diameter, an HML model was constructed. The model included a bottom layer composed of predictor system variables, a middle layer leveraging known physical relationships of the flow-gelation process of alginate, and a top layer representing print fidelity rating. Despite the low sample size, the model revealed that faster nozzle speed and higher alginate concentrations were the primary factors contributing to print fidelity. The model was then used to predict print scores based on synthetic parameters yielding R2 = 0.5 for the fidelity of lines and corners. Although exhibiting low accuracy, this model primary leverages physical knowledge to enhance predictive power and achieve interpretability of the results for downstream analysis, even with limited training data.
Another innovative ML approach to optimize bioprinting fidelity was employed by Conev et al. (2020), taking one step further by training a regression ML model to estimate the printing quality metric based on given material and printing parameters. Utilizing a previously collected data set that varied weight percent of poly(propylenefumarate) (PPF) composition, fiber spacing, printing speed, and printing pressure, two ML models were explored for their accuracy in predicting print fidelity. The first approach involved a classification-based model, using random forests with user defined high and low print fidelity ground truths. The second approach tested a regression-based model to circumvent the need for user-defined thresholds for print fidelity. A random forest model was trained to predict machine precision and material accuracy, followed by applying a threshold to characterize the printing as either “low” or “high” quality. These models were compared to a standard linear regression model that predicts material accuracy based on printing speed and printing pressure. The random forest classifier and regressor models yielded accuracies of 74% and 75%, respectively. The high performance of this model should be weighted by the limited scope of this study. Only one polymer, PPF, made up the entire training and testing data set, albeit at varying weight percentages.
Lee et al. (2020) focused on optimizing bioink composition, particularly for CTE, with a keen consideration of properties like elastic modulus and yield stress that are crucial for efficient 3D printing. For the optimization process, they employed inductive logic programming (ILP), a ML technique that learns general rules or patterns from specific instances by combining logic programming with inductive reasoning (Muggleton and de Raedt, 1994). Concentrations of type 1 collagen, fibrin, and hyaluronic acid as components of the bioink were varied to create 19 unique bioink formulations. By training a model using the fidelity of these formulations, the authors can predict rheological parameters and printing results. The ML analysis revealed that the main contributing factor determining shape fidelity is the bioink’s elastic modulus, accounting for 85% accuracy, while the dominant factor determining extrusion efficiency was the bioink’s yield stress, accounting for 90% accuracy. To validate the model’s findings, the study successfully produced 3D structures of fibroblast-laden hydrogels with high shape fidelity, maintaining the shape for 28 days without collapsing or contracting. Confocal microscopy observations showed an increase in the number of living cells over time, emphasizing the bioink’s potential for sustained cell culture. While the predictive model demonstrated effectiveness, the study underscores the need for a more comprehensive understanding of the complex relationships among bioink variables and parameters in the context of 3D bioprinting for CTE applications.
Similarly, Ruberu et al. (2021) utilized Bayesian optimization (BO) to identify the optimal printing parameters of gelatin methacryloyl (GelMA) and hyaluronic acid methacrylate (HAMA) bioinks with minimal experimentation. Notably, this pipeline leveraged ML to adaptively inform sampling during the experimentation process, surpassing classical Design of Experiment (DoE) methods that determine a sampling pattern prior to taking observations. This enables development of a database to predict printability and to make recommendations to the experimenter. A data set of score prints were generated varying GelMA and GelMA/HAMA composition, ink reservoir temperature, print pressure, print speed, and platform temperature. This seed data set was used to initialize the Bayesian optimizer. Within the optimizer, a probabilistic model of the system was built and used to recommend the next batch of experiments (printer settings) to be conducted. After executing the recommended experiments, prints were scored and fed again into optimizer in a continuous loop until optimal print conditions were reached. Optimal prints were achieved in 4-47 experiments, depending on the concentration of GelMA, drastically under the 6,000 to 10,000 possible combinations in the Bayesian algorithm. Here, BO efficiently navigates the complex parameter space of 3D bioprinting via theoretical convergence guarantees and collaborative adaptability. However, there is still a significant reliance on trial-and-error experimentation, and larger batch sizes potentially encompassing experiments with lower confidence.
9 Discussion
The lack of effective curative treatments for CVD generates a large need that hiPSC-CMs may fill if their functionality can be better understood and improved. Traditional brute-force methods at improving cardiac maturity have yielded some results, but there still exists a large gap between engineered cardiac tissue and the tissue they aim to repair or replace. The high experimental costs of cardiac optimization create opportunities for the utilization of ML.
Several groups have employed mathematical and numerical modeling of cardiac systems to help guide future areas of focus. Sobie and Wehrens (2009) reviewed computational and experimental models that may be used to explain the mechanisms of Ca2+-dependent arrythmias, like CPVT. Montero-Calle et al. (2022) have utilized computational modeling to guide the fabrication of biomaterials to support the assembly of cardiac microtissues. Furthermore, other researchers have generated in silico mathematical models of hiPSC-CMs to estimate adult-CM behavior (Koivumäki et al., 2018; Tveito et al., 2018). These methods, while effective, are inherently deterministic as they require extensive model parameter definition and incorporate assumptions on relationships and system operation. The computational challenges of traditional models are summarized in long response times to solve the equations at hand, high monetary costs for computational resources for every evaluation with a new set of parameters, and a high environmental footprint precisely due to the repeated long response times. On the other hand, training a ML model is the most arduous computational step that is done once, while response times are typically 1 s or less for a new input or set of parameters.
Table 1 summarizes the ML methods reviewed here in connection to the functional deficiencies of hiPSC-CM they aim to address. Methods are implemented primarily in Python using experimental data (Figures 1A, C). The top three common models are SVMs, random forests, and kNNs, successful at learning from limited datasets with relatively low training times. However, these supervised models require the selection of specialized features that may make-or-break the task at hand. Meanwhile, deep convolutional networks benefit from the ability to process images and signals without such careful selection of features, since they learn the features themselves, but they require significantly more data than the supervised models. This yields higher computation costs with the longer training times, leading to their lower utilization (Figure 1D). The frequency of model usage within each CTE challenge show the utility of SVMs within various problem cases and the specialization of neural networks for the analysis of image data sets within CM morphology (Figure 2). A handful of studies emphasize the potential of DL for automating quality control, while acknowledging the trade-off between calculation speed and performance compared to the more common ML methods cited in this review. Two of the more critical items that should be measured for all ML models are the computational resources and the amount of time required for successful implementation and training. However, many authors do not provide this. Finally, there exists a distinct focus on physiological phenotyping in the field, and that is reflected in this review. These studies are critical to inform the necessary interventions that may improve hiPSC-CM functionality. This need places further emphasis on the importance of training data bias, for if bias exists in phenotyping methods, it may skew the results of interventions designed to these parameters.

Figure 1. Of the 23 papers reviewed, we describe the frequency of programming language used (A), the specific python package used (B), the source of training data (C), and if deep learning methods were used (D).
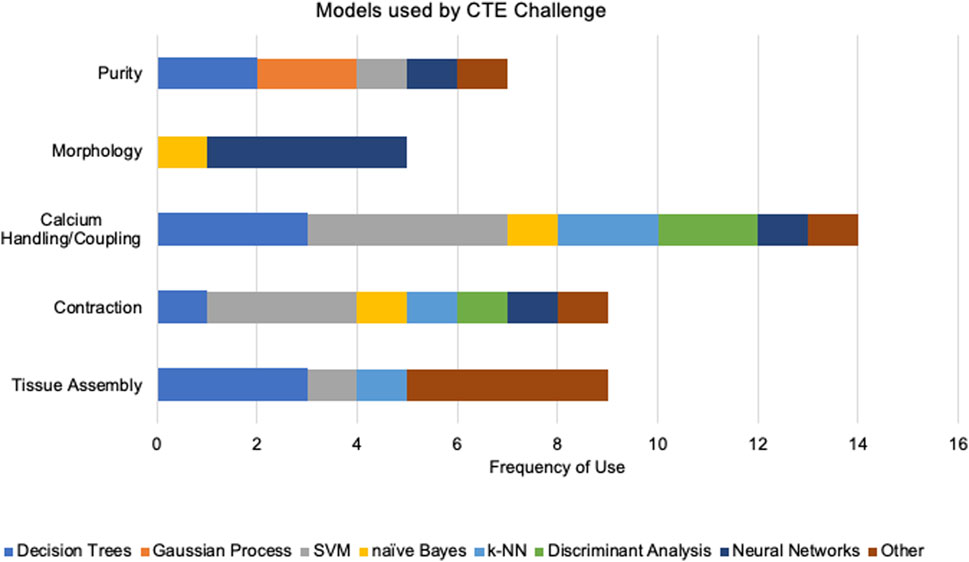
Figure 2. We segment the frequency of models used within each cardiac tissue engineering challenge. We categorized models as “decision trees” (RF, tree bagging, classification tree, XGBoost), “Gaussian processes” (GPR, GP), “SVM”, “naïve Bayes”, “kNN”, “discriminant analysis” (LDA, QDA), “neural networks” (CNN, NN, U-net, StyleGAN, RNN, feed forward NN, hierarchical machine learning), and “other” (MARS, spectral grouping, UMAP, hierarchical ML, inductive logic, Bayesian optimization, logistic regression).
In addition, the training size itself is a crucial factor for almost every AI approach. Notably, the studies reviewed here rely on experimental data, particularly from cardiac differentiation experiments, as a primary data source. The experimental data consists of variables related to cell density, bioprinting parameters, drug concentrations, and preculture conditions. While these datasets capture various aspects of the differentiation process, they remain low in sample sizes. Researchers often choose to focus on different variables, even when addressing the same challenges, such that this variability poses a barrier when attempting to join datasets across studies (Rafieyan et al., 2023). This review highlights the need for more features and larger datasets to comprehensively explain the variance in output data. Closing this gap is vital for understanding the complete set of factors influencing cardiac differentiation.
The lack of proper training data sets could be addressed by incorporating more temporal data, integrating diverse types of data from imaging and variable cell sources, and implementation of generative models to synthesize larger datasets. Firstly, long-term dependencies and variations in cardiac differentiation need to be considered, especially when aiming for clinically relevant outcomes. Although binary classification tasks are proactive ways to quantify if a particular environment will be successful or not, how the environment succeeds or fails over time can lead to new insights for early correction approaches. These insights can also aid in more accurate mathematical modeling, emphasizing the symbiotic relationship between data-driven and physics-based techniques. Secondly, collecting data from diverse cell lines and experimental conditions, like 3D bioprinting settings, is crucial to improving the robustness and generalizability of ML models. Consistent data collection across the field is needed to compare and combine disparate datasets. Integrating various features and modalities, including imaging data, can enhance the predictive power of nowadays pragmatic ML models. However, prioritizing or even measuring the vast possible number of features is overwhelming. There is a gradual shift toward unsupervised or semi-supervised learning to capture nuanced information, especially now that these approaches are more practical. The ultimate goal is to generate digital twins (An et al., 2021; Zimmermann et al., 2021) of hiPSC-CMs, producing robust and diverse datasets spanning various configurations. Although this goal is still out of reach, we anticipate that ML coupled with mathematical modeling will play a crucial role in achieving it. In the meantime, generative models can close the gap, augmenting existing data with tailored synthetic data. Some emerging approaches rely on DL frameworks, like GANs and autoencoders (Goodfellow et al., 2014; Goodfellow et al., 2016), but with sufficient modeling and understanding of the task at hand, non-DL approaches many times are sufficient, particularly hidden Markov models and Gaussian mixture models (Bishop and Nasrabadi, 2006).
10 Conclusion
The application of ML in CTE is progressing toward more sophisticated frameworks capable of handling diverse data types. These observations underscore the multidimensional nature of CTE challenges, the innovative integration of ML with computational models, and the ongoing need for refining ML frameworks to balance accuracy, interpretability, and computational efficiency. Through combining ML with mathematical models and expert knowledge, we envision a rich collaboration between novel data-driven and biophysics-informed models to bridge knowledge and computational gaps.
Author contributions
NK: Conceptualization, Formal Analysis, Methodology, Project administration, Writing–original draft. JC: Conceptualization, Formal Analysis, Methodology, Writing–original draft. EC: Funding acquisition, Supervision, Writing–review and editing. JZ: Funding acquisition, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We gratefully acknowledge the support of the National Science Foundation (2002652, awarded to JZ) and National Institutes of Health (R01CA236857, awarded to JC and EC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aghasafari, P., Yang, P. C., Kernik, D. C., Sakamoto, K., Kanda, Y., Kurokawa, J., et al. (2021). A deep learning algorithm to translate and classify cardiac electrophysiology. eLife 10, e68335. doi:10.7554/eLife.68335
An, J., Chua, C. K., and Mironov, V. (2021). Application of machine learning in 3D bioprinting: focus on development of big data and digital twin. Int. J. Bioprinting 7 (1), 342. doi:10.18063/ijb.v7i1.342
Anderson, J. L., and Morrow, D. A. (2017). Acute myocardial infarction. N. Engl. J. Med. 376 (21), 2053–2064. doi:10.1056/NEJMra1606915
Beauchamp, P., Jackson, C. B., Ozhathil, L. C., Agarkova, I., Galindo, C. L., Sawyer, D. B., et al. (2020). 3D Co-culture of hiPSC-derived cardiomyocytes with cardiac fibroblasts improves tissue-like features of cardiac spheroids. Front. Mol. Biosci. 7, 14. doi:10.3389/fmolb.2020.00014
Benjamin, E. J., Virani, S. S., Callaway, C. W., Chamberlain, A. M., Chang, A. R., Cheng, S., et al. (2018). Heart disease and stroke statistics—2018 update: a report from the American heart association. Circulation 137 (12), e67–e492. doi:10.1161/CIR.0000000000000558
Bizopoulos, P., and Koutsouris, D. (2019). Deep learning in cardiology. IEEE Rev. Biomed. Eng. 12, 168–193. doi:10.1109/RBME.2018.2885714
Bodi, I., Mikala, G., Koch, S. E., Akhter, S. A., and Schwartz, A. (2005). The L-type calcium channel in the heart: the beat goes on. J. Clin. Investigation 115 (12), 3306–3317. doi:10.1172/JCI27167
Bone, J. M., Childs, C. M., Menon, A., Póczos, B., Feinberg, A. W., LeDuc, P. R., et al. (2020). Hierarchical machine learning for high-fidelity 3D printed biopolymers. ACS Biomaterials Sci. Eng. 6 (12), 7021–7031. doi:10.1021/acsbiomaterials.0c00755
Cascianelli, S., Molineris, I., Isella, C., Masseroli, M., and Medico, E. (2020). Machine learning for RNA sequencing-based intrinsic subtyping of breast cancer. Sci. Rep. 10 (1), 14071. doi:10.1038/s41598-020-70832-2
Chong, J. J. H., Yang, X., Don, C. W., Minami, E., Liu, Y. W., Weyers, J. J., et al. (2014). Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510 (7504), 273–277. doi:10.1038/nature13233
Conev, A., Litsa, E. E., Perez, M. R., Diba, M., Mikos, A. G., and Kavraki, L. E. (2020). Machine learning-guided three-dimensional printing of tissue engineering scaffolds. Part A 26 (23–24), 1359–1368. doi:10.1089/ten.tea.2020.0191
Conway, D., and White, J. (2012). Machine learning for hackers: case studies and algorithms to get you started. First Edition. Sebastopol, CA: O’Reilly.
Coronnello, C., and Francipane, M. G. (2022). Moving towards induced pluripotent stem cell-based therapies with artificial intelligence and machine learning. Stem Cell Rev. Rep. 18 (2), 559–569. doi:10.1007/s12015-021-10302-y
Cortes, C., and Vapnik, V. (1995). Support-vector networks. Mach. Learn. 20, 273–297. doi:10.1007/bf00994018
Cover, T., and Hart, P. (1967). Nearest neighbor pattern classification. IEEE Trans. Inf. theory 13 (1), 21–27. doi:10.1109/tit.1967.1053964
Cybenko, G. (1989). Approximation by superpositions of a sigmoidal function. Math. Control, Signals Syst. 2 (4), 303–314. doi:10.1007/BF02551274
Dinov, I. D. (2016). Volume and value of big healthcare data. J. Med. Statistics Inf. 4, 3. doi:10.7243/2053-7662-4-3
Esser, T. U., Anspach, A., Muenzebrock, K. A., Kah, D., Schrüfer, S., Schenk, J., et al. (2023). Direct 3D-bioprinting of hiPSC-derived cardiomyocytes to generate functional cardiac tissues. Adv. Mater. 35, 2305911. doi:10.1002/adma.202305911
Feinberg, A. W., Ripplinger, C., van der Meer, P., Sheehy, S., Domian, I., Chien, K., et al. (2013). Functional differences in engineered myocardium from embryonic stem cell-derived versus neonatal cardiomyocytes. Stem Cell Rep. 1 (5), 387–396. doi:10.1016/j.stemcr.2013.10.004
Feric, N. T., and Radisic, M. (2016). Maturing human pluripotent stem cell-derived cardiomyocytes in human engineered cardiac tissues. Adv. Drug Deliv. Rev. 96, 110–134. doi:10.1016/j.addr.2015.04.019
Finkel, S., Sweet, S., Locke, T., Smith, S., Wang, Z., Sandini, C., et al. (2023). FRESH™ 3D bioprinted cardiac tissue, a bioengineered platform for in vitro pharmacology. Apl. Bioeng. 7 (4), 046113. doi:10.1063/5.0163363
Friedman, J. H. (1991). Multivariate adaptive regression splines. Ann. Statistics 19 (1), 1–67. doi:10.1214/aos/1176347963
Fukunaga, K., and Hostetler, L. (1975). The estimation of the gradient of a density function, with applications in pattern recognition. IEEE Trans. Inf. Theory 21 (1), 32–40. doi:10.1109/TIT.1975.1055330
Gerdes, A. M., Kellerman, S. E., Moore, J. A., Muffly, K. E., Clark, L. C., Reaves, P. Y., et al. (1992). Structural remodeling of cardiac myocytes in patients with ischemic cardiomyopathy. ’, Circ. 86 (2), 426–430. doi:10.1161/01.CIR.86.2.426
Géron, A. (2019). Hands-on machine learning with scikit-learn, keras, and TensorFlow: concepts, tools, and techniques to build intelligent systems. 2nd edition. Beijing China ; Sebastopol, CA: O’Reilly Media.
Gers, F. A., Schmidhuber, J., and Cummins, F. (2000). Learning to forget: continual prediction with LSTM. Neural Comput. 12 (10), 2451–2471. doi:10.1162/089976600300015015
Glass, C., Lafata, K. J., Jeck, W., Horstmeyer, R., Cooke, C., Everitt, J., et al. (2022). The role of machine learning in cardiovascular pathology. Can. J. Cardiol. 38 (2), 234–245. doi:10.1016/j.cjca.2021.11.008
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep learning. Illustrated edition. Cambridge, Mass: The MIT Press.
Goodfellow, I. J., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2014). Generative adversarial networks. arXiv. doi:10.48550/arXiv.1406.2661
Gorospe, G., Zhu, R., Millrod, M. A., Zambidis, E. T., Tung, L., and Vidal, R. (2014). Automated grouping of action potentials of human embryonic stem cell-derived cardiomyocytes. IEEE Trans. Biomed. Eng. 61 (9), 2389–2395. doi:10.1109/TBME.2014.2311387
Grafton, F., Ho, J., Ranjbarvaziri, S., Farshidfar, F., Budan, A., Steltzer, S., et al. (2021). Deep learning detects cardiotoxicity in a high-content screen with induced pluripotent stem cell-derived cardiomyocytes. eLife 10, e68714. doi:10.7554/eLife.68714
Halloin, C., Coffee, M., Manstein, F., and Zweigerdt, R. (2019). Production of cardiomyocytes from human pluripotent stem cells by bioreactor technologies. Methods Mol. Biol. 1994, 55–70. doi:10.1007/978-1-4939-9477-9_5
Hastie, T., Tibshirani, R., and Friedman, J. (2009). The elements of statistical learning. New York, NY: Springer (Springer Series in Statistics. doi:10.1007/978-0-387-84858-7
Haykin, S. (1998). Neural networks: a comprehensive foundation. Subsequent edition. Upper Saddle River, N.J: Prentice Hall.
Hazeltine, L. B., Simmons, C. S., Salick, M. R., Lian, X., Badur, M. G., Han, W., et al. (2012). Effects of substrate mechanics on contractility of cardiomyocytes generated from human pluripotent stem cells. Int. J. Cell Biol. 2012, 1–13. doi:10.1155/2012/508294
Heylman, C., Datta, R., Sobrino, A., George, S., and Gratton, E. (2015). Supervised machine learning for classification of the electrophysiological effects of chronotropic drugs on human induced pluripotent stem cell-derived cardiomyocytes. PLOS ONE 10 (12), e0144572. doi:10.1371/journal.pone.0144572
Horikoshi, Y., Yan, Y., Terashvili, M., Wells, C., Horikoshi, H., Fujita, S., et al. (2019). Fatty acid-treated induced pluripotent stem cell-derived human cardiomyocytes exhibit adult cardiomyocyte-like energy metabolism phenotypes. Cells 8 (9), 1095. doi:10.3390/cells8091095
Hornik, K., Stinchcombe, M., and White, H. (1989). Multilayer feedforward networks are universal approximators. Neural Netw. 2 (5), 359–366. doi:10.1016/0893-6080(89)90020-8
Hotelling, H. (1936). Simplified calculation of principal components. Psychometrika 1 (1), 27–35. doi:10.1007/bf02287921
Hulsen, T., Jamuar, S. S., Moody, A. R., Karnes, J. H., Varga, O., Hedensted, S., et al. (2019). From big data to precision medicine. Front. Med. 6, 34. doi:10.3389/fmed.2019.00034
Hwang, H., Liu, R., Maxwell, J. T., Yang, J., and Xu, C. (2020). Machine learning identifies abnormal Ca 2+ transients in human induced pluripotent stem cell-derived cardiomyocytes. Sci. Rep. 10 (1), 16977. doi:10.1038/s41598-020-73801-x
Izenman, A. J. (2008). Modern multivariate statistical techniques. New York, NY: Springer (Springer Texts in Statistics). doi:10.1007/978-0-387-78189-1
James, G., Witten, D., Hastie, T., Tibshirani, R., and Taylor, J. (2023). An introduction to statistical learning: with applications in pyth. Cham: Springer International Publishing (Springer Texts in Statistics). doi:10.1007/978-3-031-38747-0
Jovel, J., and Greiner, R. (2021). An introduction to machine learning approaches for biomedical research. Front. Med. 8, 771607. doi:10.3389/fmed.2021.771607
Juhola, M., Joutsijoki, H., Penttinen, K., and Aalto-Setälä, K. (2018). Detection of genetic cardiac diseases by Ca 2+ transient profiles using machine learning methods. Sci. Rep. 8 (1), 9355–9410. doi:10.1038/s41598-018-27695-5
Juhola, M., Penttinen, K., Joutsijoki, H., and Aalto-Setälä, K. (2021). Analysis of drug effects on iPSC cardiomyocytes with machine learning. Ann. Biomed. Eng. 49 (1), 129–138. doi:10.1007/s10439-020-02521-0
Juhola, M., Penttinen, K., Joutsijoki, H., Varpa, K., Saarikoski, J., Rasku, J., et al. (2015). Signal analysis and classification methods for the calcium transient data of stem cell-derived cardiomyocytes. Comput. Biol. Med. 61, 1–7. doi:10.1016/j.compbiomed.2015.03.016
Jurkiewicz, J., Kroboth, S., Zlochiver, V., and Hinow, P. (2021). Automated feature extraction from large cardiac electrophysiological data sets. J. Electrocardiol. 65, 157–162. doi:10.1016/j.jelectrocard.2021.02.003
Karras, T., Laine, S., and Aila, T. (2019). A style-based generator architecture for generative adversarial networks. arXiv. doi:10.48550/arXiv.1812.04948
Kehoe, D. E., Jing, D., Lock, L. T., and Tzanakakis, E. S. (2010). Scalable stirred-suspension bioreactor culture of human pluripotent stem cells. Tissue Eng. Part A 16 (2), 405–421. doi:10.1089/ten.tea.2009.0454
Kelleher, J. D., Namee, B. M., and D’Arcy, A. (2015). Fundamentals of machine learning for predictive data analytics: algorithms, worked examples, and case studies. 1st edition. Cambridge, Massachusetts: The MIT Press.
Kempf, H., Andree, B., and Zweigerdt, R. (2016). Large-scale production of human pluripotent stem cell derived cardiomyocytes. Adv. Drug Deliv. Rev. 96, 18–30. doi:10.1016/j.addr.2015.11.016
Kensah, G., Gruh, I., Viering, J., Schumann, H., Dahlmann, J., Meyer, H., et al. (2011). A novel miniaturized multimodal bioreactor for continuous in situ assessment of bioartificial cardiac tissue during stimulation and maturation. Methods 17 (4), 463–473. doi:10.1089/ten.tec.2010.0405
Kernik, D. C., Yang, P. C., Kurokawa, J., Wu, J. C., and Clancy, C. E. (2020). A computational model of induced pluripotent stem-cell derived cardiomyocytes for high throughput risk stratification of KCNQ1 genetic variants. PLOS Comput. Biol. 16 (8), e1008109. doi:10.1371/journal.pcbi.1008109
Khadangi, A., Boudier, T., Hanssen, E., and Rajagopal, V. (2022). CardioVinci: building blocks for virtual cardiac cells using deep learning. Philosophical Trans. R. Soc. B Biol. Sci. 377 (1864), 20210469. doi:10.1098/rstb.2021.0469
Kita-Matsuo, H., Barcova, M., Prigozhina, N., Salomonis, N., Wei, K., Jacot, J. G., et al. (2009). Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLOS ONE 4 (4), e5046. doi:10.1371/journal.pone.0005046
Kobayashi, J., Kikuchi, A., Aoyagi, T., and Okano, T. (2019). Cell sheet tissue engineering: cell sheet preparation, harvesting/manipulation, and transplantation. J. Biomed. Mater. Res. Part A 107 (5), 955–967. doi:10.1002/jbm.a.36627
Koivumäki, J. T., Naumenko, N., Tuomainen, T., Takalo, J., Oksanen, M., Puttonen, K. A., et al. (2018). Structural immaturity of human iPSC-derived cardiomyocytes: in silico investigation of effects on function and disease modeling. Front. Physiology 9, 80. doi:10.3389/fphys.2018.00080
Kowalczewski, A., Sakolish, C., Hoang, P., Liu, X., Jacquir, S., Rusyn, I., et al. (2022). Integrating nonlinear analysis and machine learning for human induced pluripotent stem cell-based drug cardiotoxicity testing. J. Tissue Eng. Regen. Med. 16 (8), 732–743. doi:10.1002/term.3325
Koyilot, M. C., Natarajan, P., Hunt, C. R., Sivarajkumar, S., Roy, R., Joglekar, S., et al. (2022). Breakthroughs and applications of organ-on-a-chip technology. Cells 11 (11), 1828. doi:10.3390/cells11111828
Kresoja, K.-P., Unterhuber, M., Wachter, R., Thiele, H., and Lurz, P. (2023). A cardiologist’s guide to machine learning in cardiovascular disease prognosis prediction. Basic Res. Cardiol. 118 (1), 10. doi:10.1007/s00395-023-00982-7
Laflamme, M. A., and Murry, C. E. (2011). Heart regeneration. Nature 473 (7347), 326–335. doi:10.1038/nature10147
Lee, E. K., Kurokawa, Y. K., Tu, R., George, S. C., and Khine, M. (2015). Machine learning plus optical flow: a simple and sensitive method to detect cardioactive drugs. Sci. Rep. 5 (1), 11817. doi:10.1038/srep11817
Lee, E. K., Tran, D. D., Keung, W., Chan, P., Wong, G., Chan, C. W., et al. (2017). Machine learning of human pluripotent stem cell-derived engineered cardiac tissue contractility for automated drug classification. Stem Cell Rep. 9 (5), 1560–1572. doi:10.1016/j.stemcr.2017.09.008
Lee, J., Oh, S. J., Kim, W. D., and Kim, S. H. (2020). Machine learning-based design strategy for 3D printable bioink: elastic modulus and yield stress determine printability. Biofabrication 12 (3), 035018. doi:10.1088/1758-5090/ab8707
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., et al. (2017). A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88. doi:10.1016/j.media.2017.07.005
Maddah, M., Mandegar, M. A., Dame, K., Grafton, F., Loewke, K., and Ribeiro, A. J. (2020). Quantifying drug-induced structural toxicity in hepatocytes and cardiomyocytes derived from hiPSCs using a deep learning method. J. Pharmacol. Toxicol. Methods 105, 106895. doi:10.1016/j.vascn.2020.106895
Manning, C. D. (2008). Intro to information retrieval. Cambridge, UNITED KINGDOM: Cambridge University Press Textbooks. Available at: http://ebookcentral.proquest.com/lib/utxa/detail.action?docID=5119897 (Accessed December 15, 2023).
McCain, M. L., and Parker, K. K. (2011). Mechanotransduction: the role of mechanical stress, myocyte shape, and cytoskeletal architecture on cardiac function. Pflügers Archiv - Eur. J. Physiology 462 (1), 89–104. doi:10.1007/s00424-011-0951-4
McInnes, L., Healy, J., and Melville, J. (2020). UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv. doi:10.48550/arXiv.1802.03426
Menon, A., Gupta, C., Perkins, K. M., DeCost, B. L., Budwal, N., Rios, R. T., et al. (2017). Elucidating multi-physics interactions in suspensions for the design of polymeric dispersants: a hierarchical machine learning approach. Mol. Syst. Des. Eng. 2 (3), 263–273. doi:10.1039/c7me00027h
Mohammadi, S., Hashemi, M., Finklea, F., Williams, B., Lipke, E., and Cremaschi, S. (2022). Classification of cardiac differentiation outcome, percentage of cardiomyocytes on day 10 of differentiation, for hydrogel-encapsulated hiPSCs. J. Adv. Manuf. Process. 5 (2), e10148. doi:10.1002/amp2.10148
Mohammadi, S., Hashemi, M., Finklea, F. B., Lipke, E. A., and Cremaschi, S. (2023). Differentiating engineered tissue images and experimental factors to classify cardiomyocyte content. Tissue Eng. Part A 29 (1–2), 58–66. doi:10.1089/ten.tea.2022.0122
Montero-Calle, P., Flandes-Iparraguirre, M., Mountris, K., S de la Nava, A., Laita, N., Rosales, R. M., et al. (2022). Fabrication of human myocardium using multidimensional modelling of engineered tissues. Biofabrication 14 (4), 045017. doi:10.1088/1758-5090/ac8cb3
Muggleton, S., and de Raedt, L. (1994). Inductive logic programming: theory and methods. J. Log. Program. 19–20, 629–679. doi:10.1016/0743-1066(94)90035-3
Müller, A. C., and Guido, S. (2016). Introduction to machine learning with Python: a guide for data scientists. Sebastopol, CA: O’Reilly Media, Inc.
Ng, A., Jordan, M., and Weiss, Y. (2001). “On Spectral Clustering: Analysis and an algorithm,” in (Editors) D. Dietterich, S. Becker, and Z. Ghahramani Advances in Neural Information Processing Systems. MIT Press. Available at: https://proceedings.neurips.cc/paper_files/paper/2001/file/801272ee79cfde7fa5960571fee36b9b-Paper.pdf.
Nunes, S. S., Miklas, J. W., Liu, J., Aschar-Sobbi, R., Xiao, Y., Zhang, B., et al. (2013). Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes. Nat. Methods 10 (8), 781–787. doi:10.1038/nmeth.2524
O’Hara, T., Virág, L., Varró, A., and Rudy, Y. (2011). Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLOS Comput. Biol. 7 (5), e1002061. doi:10.1371/journal.pcbi.1002061
Ong, C. S., Nam, L., Ong, K., Krishnan, A., Huang, C. Y., Fukunishi, T., et al. (2018). 3D and 4D bioprinting of the myocardium: current approaches, challenges, and future prospects. BioMed Res. Int. Hindawi 2018, 1–11. doi:10.1155/2018/6497242
Orita, K., Sawada, K., Koyama, R., and Ikegaya, Y. (2019). Deep learning-based quality control of cultured human-induced pluripotent stem cell-derived cardiomyocytes. J. Pharmacol. Sci. 140 (4), 313–316. doi:10.1016/j.jphs.2019.04.008
Orita, K., Sawada, K., Matsumoto, N., and Ikegaya, Y. (2020). Machine-learning-based quality control of contractility of cultured human-induced pluripotent stem-cell-derived cardiomyocytes. Biochem. Biophysical Res. Commun. 526 (3), 751–755. doi:10.1016/j.bbrc.2020.03.141
Ouyang, Q., Yang, W., Wu, Y., Xu, Z., Hu, Y., Hu, N., et al. (2022). Multi-labeled neural network model for automatically processing cardiomyocyte mechanical beating signals in drug assessment. Biosens. Bioelectron. 209, 114261. doi:10.1016/j.bios.2022.114261
Park, K. M., Shin, Y. M., Kim, K., and Shin, H. (2018). Tissue engineering and regenerative medicine 2017: a year in review. Tissue Eng. Part B Rev. 24 (5), 327–344. doi:10.1089/ten.teb.2018.0027
Pascanu, R., Mikolov, T., and Bengio, Y. (2013). “On the difficulty of training recurrent neural networks,” in International conference on machine learning (Atlanta, Georgia, USA: Pmlr), 1310–1318.
Pasqualini, F. S., Sheehy, S., Agarwal, A., Aratyn-Schaus, Y., and Parker, K. (2015). Structural phenotyping of stem cell-derived cardiomyocytes. Stem Cell Rep. 4 (3), 340–347. doi:10.1016/j.stemcr.2015.01.020
Pearson, K. (1901). LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin philosophical Mag. J. Sci. 2 (11), 559–572. doi:10.1080/14786440109462720
Peek, N., Holmes, J. H., and Sun, J. (2014). Technical challenges for big data in biomedicine and Health: data sources, infrastructure, and analytics. Yearb. Med. Inf. 23 (1), 42–47. doi:10.15265/IY-2014-0018
Petegrosso, R., Li, Z., and Kuang, R. (2020). Machine learning and statistical methods for clustering single-cell RNA-sequencing data. Briefings Bioinforma. 21 (4), 1209–1223. doi:10.1093/bib/bbz063
Qasim, M., Arunkumar, P., Powell, H. M., and Khan, M. (2019). Current research trends and challenges in tissue engineering for mending broken hearts. Life Sci. 229, 233–250. doi:10.1016/j.lfs.2019.05.012
Quinlan, J. R. (1986). Induction of decision trees. Mach. Learn. 1 (1), 81–106. doi:10.1007/BF00116251
Rafieyan, S., Vasheghani-Farahani, E., Baheiraei, N., and Keshavarz, H. (2023). MLATE: machine learning for predicting cell behavior on cardiac tissue engineering scaffolds. Comput. Biol. Med. 158, 106804. doi:10.1016/j.compbiomed.2023.106804
Raschka, S., and Mirjalili, V. (2017). Machine learning and deep learning with Python, scikit-learn, and TensorFlow. Python Machine Learning, Second Edition. Birmingham Mumbai: Packt Publishing.
Reddy, K., Khaliq, A., and Henning, R. J. (2015). Recent advances in the diagnosis and treatment of acute myocardial infarction. World J. Cardiol. 7 (5), 243–276. doi:10.4330/wjc.v7.i5.243
Regazzoni, F., Salvador, M., Dede’, L., and Quarteroni, A. (2022). A machine learning method for real-time numerical simulations of cardiac electromechanics. Comput. Methods Appl. Mech. Eng. 393, 114825. doi:10.1016/j.cma.2022.114825
Ribeiro, A. J. S., Guth, B. D., Engwall, M., Eldridge, S., Foley, C. M., Guo, L., et al. (2019). Considerations for an in vitro, cell-based testing platform for detection of drug-induced inotropic effects in early drug development. Part 2: designing and fabricating microsystems for assaying cardiac contractility with physiological relevance using human iPSC-cardiomyocytes. Front. Pharmacol. 10, 934. doi:10.3389/fphar.2019.00934
Richards, D. J., Coyle, R. C., Tan, Y., Jia, J., Wong, K., Toomer, K., et al. (2017). Inspiration from heart development: biomimetic development of functional human cardiac organoids. Biomaterials 142, 112–123. doi:10.1016/j.biomaterials.2017.07.021
Ronaldson-Bouchard, K., Ma, S. P., Yeager, K., Chen, T., Song, L., Sirabella, D., et al. (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556 (7700), 239–243. doi:10.1038/s41586-018-0016-3
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net: convolutional networks for biomedical image segmentation. arXiv. doi:10.48550/arXiv.1505.04597
Ruan, J.-L., Tulloch, N. L., Razumova, M. V., Saiget, M., Muskheli, V., Pabon, L., et al. (2016). Mechanical stress conditioning and electrical stimulation promote contractility and force maturation of induced pluripotent stem cell-derived human cardiac tissue. Circulation 134 (20), 1557–1567. doi:10.1161/CIRCULATIONAHA.114.014998
Ruberu, K., Senadeera, M., Rana, S., Gupta, S., Chung, J., Yue, Z., et al. (2021). Coupling machine learning with 3D bioprinting to fast track optimisation of extrusion printing. Appl. Mater. Today 22, 100914. doi:10.1016/j.apmt.2020.100914
Rueckert, D., and Schnabel, J. A. (2020). Model-based and data-driven strategies in medical image computing. Proc. IEEE 108 (1), 110–124. doi:10.1109/JPROC.2019.2943836
Rumelhart, D. E., Hinton, G. E., and Williams, R. J. (1986). Learning representations by back-propagating errors. Nature 323 (6088), 533–536. doi:10.1038/323533a0
Sartiani, L., Bettiol, E., Stillitano, F., Mugelli, A., Cerbai, E., and Jaconi, M. E. (2007). Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells Dayt. 25 (5), 1136–1144. doi:10.1634/stemcells.2006-0466
Shajun Nisha, S., and Nagoor Meeral, M. (2021). “9 - applications of deep learning in biomedical engineering,” in Handbook of deep learning in biomedical engineering. Editors V. E. Balas, B. K. Mishra, and R. Kumar (Academic Press), 245–270. doi:10.1016/B978-0-12-823014-5.00008-9
Shanmugamani, R. (2018). Deep learning for computer vision: expert techniques to train advanced neural networks using TensorFlow and keras. Birmingham Mumbai: Packt Publishing.
Sharma, A., Li, G., Rajarajan, K., Hamaguchi, R., Burridge, P. W., and Wu, S. M. (2015). Derivation of highly purified cardiomyocytes from human induced pluripotent stem cells using small molecule-modulated differentiation and subsequent glucose starvation. J. Vis. Exp. JoVE Prepr. (97), 52628. doi:10.3791/52628
Sobie, E. A., and Wehrens, X. H. T. (2009). Computational and experimental models of Ca2+-dependent arrhythmias. Drug Discov. Today Dis. Models 6 (3), 57–61. doi:10.1016/j.ddmod.2010.04.002
Strzelecki, M., and Badura, P. (2022). Machine learning for biomedical application. Appl. Sci. 12 (4), 2022. doi:10.3390/app12042022
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., et al. (2015). “Going deeper with convolutions,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 1–9.
Teles, D., Kim, Y., Ronaldson-Bouchard, K., and Vunjak-Novakovic, G. (2021). Machine learning techniques to classify healthy and diseased cardiomyocytes by contractility profile. ACS biomaterials Sci. Eng. 7 (7), 3043–3052. doi:10.1021/acsbiomaterials.1c00418
Turnbull, I. C., Karakikes, I., Serrao, G. W., Backeris, P., Lee, J., Xie, C., et al. (2014). Advancing functional engineered cardiac tissues toward a preclinical model of human myocardium. FASEB J. 28 (2), 644–654. doi:10.1096/fj.13-228007
Tveito, A., Jæger, K. H., Huebsch, N., Charrez, B., Edwards, A. G., Wall, S., et al. (2018). Inversion and computational maturation of drug response using human stem cell derived cardiomyocytes in microphysiological systems. Sci. Rep. 8 (1), 17626. doi:10.1038/s41598-018-35858-7
van der Velden, J., Klein, L., van der Bijl, M., Huybregts, M., Stooker, W., Witkop, J., et al. (1998). Force production in mechanically isolated cardiac myocytes from human ventricular muscle tissue. Cardiovasc. Res. 38 (2), 414–423. doi:10.1016/s0008-6363(98)00019-4
Venkatesan, R., and Li, B. (2017). Convolutional neural networks in visual computing: a concise guide. Boca Raton: CRC Press.
Wang, L., Xi, Y., Sung, S., and Qiao, H. (2018). RNA-seq assistant: machine learning based methods to identify more transcriptional regulated genes. BMC Genomics 19 (1), 546. doi:10.1186/s12864-018-4932-2
Wiegerinck, R. F., Cojoc, A., Zeidenweber, C. M., Ding, G., Shen, M., Joyner, R. W., et al. (2009). Force frequency relationship of the human ventricle increases during early postnatal development. Pediatr. Res. 65 (4), 414–419. doi:10.1203/PDR.0b013e318199093c
Williams, B., Löbel, W., Finklea, F., Halloin, C., Ritzenhoff, K., Manstein, F., et al. (2020). Prediction of human induced pluripotent stem cell cardiac differentiation outcome by multifactorial process modeling. Front. Bioeng. Biotechnol. 8, 851. doi:10.3389/fbioe.2020.00851
I. H. Witten, E. Frank, and M. A. Hall (2011). “Front matter,” Data mining: practical machine learning tools and techniques. Third Edition (Boston: Morgan Kaufmann (The Morgan Kaufmann Series in Data Management Systems), i–iii. doi:10.1016/B978-0-12-374856-0.00018-3
Yang, H., Obrezanova, O., Pointon, A., Stebbeds, W., Francis, J., Beattie, K. A., et al. (2023). Prediction of inotropic effect based on calcium transients in human iPSC-derived cardiomyocytes and machine learning. Toxicol. Appl. Pharmacol. 459, 116342. doi:10.1016/j.taap.2022.116342
Yang, H., Stebbeds, W., Francis, J., Pointon, A., Obrezanova, O., Beattie, K. A., et al. (2022). Deriving waveform parameters from calcium transients in human iPSC-derived cardiomyocytes to predict cardiac activity with machine learning. Stem Cell Rep. 17 (3), 556–568. doi:10.1016/j.stemcr.2022.01.009
Yarotsky, D. (2017). Error bounds for approximations with deep ReLU networks. Neural Netw. 94, 103–114. doi:10.1016/j.neunet.2017.07.002
Zhou, S. K., Greenspan, H., and Shen, D. (2017). “Front matter,” Deep learning for medical image analysis (Academic Press), i–iii. doi:10.1016/B978-0-12-810408-8.00026-2
Keywords: cardiac tissue engineering, machine learning, hiPSC-CM, cardiac maturation, deep learning
Citation: Kalkunte N, Cisneros J, Castillo E and Zoldan J (2024) A review on machine learning approaches in cardiac tissue engineering. Front. Front. Biomater. Sci. 3:1358508. doi: 10.3389/fbiom.2024.1358508
Received: 19 December 2023; Accepted: 21 March 2024;
Published: 03 April 2024.
Edited by:
Silviya Petrova Zustiak, Saint Louis University, United StatesReviewed by:
Arul Prakash Francis, Saveetha Dental College And Hospitals, IndiaNathaniel Huebsch, Washington University in St. Louis, United States
Dilip Thomas, Stanford University, United States
Copyright © 2024 Kalkunte, Cisneros, Castillo and Zoldan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janet Zoldan, emphbmV0QHV0ZXhhcy5lZHU=
†These authors have contributed equally to this work
 Nikhith Kalkunte
Nikhith Kalkunte Jorge Cisneros
Jorge Cisneros Edward Castillo
Edward Castillo Janet Zoldan
Janet Zoldan