
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol., 01 April 2025
Sec. Organoids and Organ-On-A-Chip
Volume 13 - 2025 | https://doi.org/10.3389/fbioe.2025.1568389
This article is part of the Research TopicAdvancements in Image Processing and Analysis Techniques for Microphysiological SystemsView all articles
 Nina Truong1†
Nina Truong1† Abir Zahra1†
Abir Zahra1† Ryan C. V. Lintao2,3
Ryan C. V. Lintao2,3 Rahul Chauhan1
Rahul Chauhan1 Giovana Fernanda Bento4
Giovana Fernanda Bento4 Manuel Vidal Jr.3,5,6
Manuel Vidal Jr.3,5,6 Sungjin Kim7
Sungjin Kim7 Po Yi Lam7
Po Yi Lam7 Thomas Conrads8,9
Thomas Conrads8,9 Kelly Conrads8,9
Kelly Conrads8,9 Arum Han7
Arum Han7 Ramkumar Menon3
Ramkumar Menon3 Lauren S. Richardson3*
Lauren S. Richardson3*Over the past decade, organ-on-chip technology (microphysiological systems or tissue chips) has reshaped in-vitro physiological and pathological modeling and pharmaceutical drug assessment. FDA Modernization Act 2.0 allows for alternatives to animal testing or the use of appropriate non-animal models/new approach methods (NAMs), such as Organ-on-chips (OC) platforms or in silico simulation models, to generate pre-clinical in-vitro drug trial data for regulatory purposes primes the microfluidic field to have exponential growth in the coming years. The changes in the approaches of regulatory agencies could significantly impact the development of therapeutics for use during pregnancy. However, limitations of the devices and molecular and biochemical assay shortfalls hinder the progress of the OOC field. This review describes available reproductive and pregnancy-related OOC platforms, and the current methodologies utilized to generate endpoint datasets (e.g., microscopic imaging, immunocytochemistry, real-time polymerase chain reaction, cytokine multiplex analysis). Microfluidic platform limitations, such as fewer number of cells or low supernatant volumes and restrictions regarding fabrication materials, are described. Novel approaches (e.g., spatial transcriptomics, imaging cytometry by time of flight, exosomes analysis using Exoview) to overcome these challenges are described. OOC platforms are primed to provide biologically relevant and clinically translational data that can revolutionize in-vitro physiological modeling, drug discovery, and toxicologic risk assessment. However, engineering adaptations to increase the throughput of devices (i.e., device arrays) and biological advancements to improve data throughput are both needed for these platforms to reach their full potential.
Organ-on-chips (OOC) or tissue chips are microphysiological systems that mimic human organ architecture and physiology. Functionality OOCs can be utilized to better understand the development of a disease and disease progression and analyze adverse drug effects in organs of interest. Various OOCs have been constructed to model single human organs such as the brain (van der Helm et al., 2016a; Sances et al., 2018; Jeong et al., 2018), liver (Materne et al., 2015; Maschmeyer et al., 2015; Gori et al., 2016; Deng et al., 2019), kidney (An et al., 2015; Paoli and Samitier, 2016; Wilmer et al., 2016; Ashammakhi et al., 2018), lung (Huh, 2015; Konar et al., 2016; Shrestha et al., 2020), and heart (Zhang et al., 2015; Jastrzebska et al., 2016; Ribas et al., 2016), as well as multi-organ systems (Materne et al., 2015; Maschmeyer et al., 2015; Abaci and Shuler, 2015; Jodat et al., 2018; Baert et al., 2020) such as the reproductive systems (Xiao et al., 2017). These 3D systems allow for environmental manipulations such as cell patterning, flow rate, concentration gradients, culture conditions, and organ-organ interactions allowing investigators to control experimental parameters to perform reliable, and reproducible humanized research (Ma et al., 2021). For decades, animal models have been used as the gold standard for in-vivo experiments, but due to species differences (anatomical and functional differences between animals and humans) (Ruberte et al., 2023; Dolenšek et al., 2015; Sheng et al., 2010; Soncin et al., 2015; Soncin et al., 2018), financial costs, and ethical implications, animal models remain far from the ideal (Han, 2023). Ex-vivo tissue explant models retain human context and biological complexity though they have a short half-life (∼48 h), are difficult to dissect without inducing damage, and must be collected fast to ensure adequate viability (Hughes et al., 2021; Grivel and Margolis, 2009). While in-vitro 2-D cell cultures can provide informative data, they lack the cellular context and ability to simulate human organs’ physiological complexity and interactions with their surroundings (Wu et al., 2020). However, the advanced technology of microfluidic OOC systems can help to overcome these limitations and bridge the gap between cell cultures, animal models, and human trials to advance research (Wu et al., 2020).
Within the vast arena of OOC devices, a small portion of platforms exist in the field of women’s health and reproduction. Reproduction is a fundamental life process crucial to the continuation and diversification of all species. The specialized tissues, organs, and hormones of the reproductive system work in concert to enable fertilization and support the ensuing process of embryonic and fetal development. Many aspects of human reproduction are being explored and imitated by OOC devices, such as the female and male reproductive tract (Xiao et al., 2017; Tantengco et al., 2022a), vaginal microbiome (Mahajan et al., 2022), cervix (Yu et al., 2022; Tantengco et al., 2021; Lee M. et al., 2021; Takeuchi et al., 2019; Tantengco et al., 2022b), ovary (Nagashima et al., 2018; Aziz et al., 2017; Saha et al., 2021; Aziz et al., 2020; Lee JH. et al., 2021), testis (Baert et al., 2020), and endometrium (Park et al., 2020; Gnecco et al., 2019; Gnecco et al., 2017; De Bem et al., 2021; Ahn et al., 2021; Bruner-Tran et al., 2017; Govindasamy et al., 2023). Like many other OOC devices, alteration and combinations of these organ structures allow for a wide variety of microenvironments and complex systems to be created. Under different experimental conditions/treatments, numerous properties of these organs/organ systems may be explored, including implantation (Park et al., 2022), 28-day menstrual cycle (Xiao et al., 2017), spermatogenesis, sperm mobility (Yu et al., 2022; Lee M. et al., 2021), and pathologic processes such as cancer development and metastasis (Saha et al., 2021). Furthermore, these devices have been shown to have utility in assessing reproductive organs’ responses to therapeutics (Saha et al., 2021; Shojaei et al., 2021) or toxicant exposure (Aziz et al., 2020).
A subset of these devices has been reported to model pregnancy-related interfaces by recapitulating several tissue layers, organs, and anatomical systems. A few of these OOC models include the placenta (Shojaei et al., 2021; Lee et al., 2016; Mosavati et al., 2020; Pemathilaka et al., 2019; Yin et al., 2019; Blundell et al., 2018; Abostait et al., 2022a; Mandt et al., 2018; Zhu et al., 2018; Kreuder et al., 2020; Boos et al., 2021; Jeong et al., 2024; Vidal et al., 2024; Abostait et al., 2022b; Richardson et al., 2022; Miura et al., 2015; Rabussier et al., 2023a; Mosavati et al., 2022; Ghorbanpour et al., 2023; Blundell et al., 2016), amnion (Zhu et al., 2020; Richardson et al., 2019a), cervix (Yu et al., 2022; Tantengco et al., 2021; Lee M. et al., 2021; Takeuchi et al., 2019; Tantengco et al., 2022b; Dadkhah et al., 2023), endometrium (Park et al., 2020; Gnecco et al., 2019; Gnecco et al., 2017; De Bem et al., 2021; Ahn et al., 2021; Bruner-Tran et al., 2017), decidua (Rogers et al., 2018; Richardson et al., 2023a; Lintao et al., 2024), vagina (Mahajan et al., 2022; Czechowicz et al., 2022; Tantengco et al., 2022c; Tantengco et al., 2022d), fetal membrane (Park et al., 2022; Richardson L. et al., 2020; Richardson et al., 2020b; Kim et al., 2022), and a combination of these to form organ systems (i.e., vagina-cervix-decidua (Tantengco et al., 2022c; Tantengco OR. et al., 2022), uterine-endometrium-ovary (Park et al., 2020), placenta-decidua-fetal membrane (Kammala et al., 2023; Safarzadeh et al., 2024)). Besides some differences in the process of fabrication and design of the placental-environment OOCs, multiple studies have aimed to investigate the aspects that are related to this organ during pregnancy in different conditions, such as diseases and tests of drug transportation (Mosavati et al., 2022; Ghorbanpour et al., 2023; Pemathilaka et al., 2022; Ko et al., 2022; Cherubini et al., 2023). These microphysiological systems provide countless avenues for innovative research by providing platforms to study 1) the development, characterization, and remodeling of tissues during pregnancy (e.g., sperm locomotion (Yu et al., 2022), trophoblast invasion during implantation (Park et al., 2022), amnion membrane remodeling during gestation (Zhu et al., 2020; Richardson et al., 2019b)); 2) contribute to a better understanding of the physiologic crosstalk between different pregnancy-related feto-maternal organs by providing cellular level resolution of interactions (Park et al., 2020; Zhu et al., 2020; Richardson L. et al., 2020); 3) providing human-based, ethical experimentation models for drug transport across the feto-maternal interfaces (Pemathilaka et al., 2019; Blundell et al., 2018; Richardson et al., 2022; Kammala et al., 2023; Safarzadeh et al., 2024) (i.e., placenta and fetal membrane) and investigate the effects of therapeutics prior to human trials (Saha et al., 2021; Shojaei et al., 2021); 4) creating models to evaluate fetal risk assessment of after maternal toxicant exposures (Kim et al., 2022; Richardson et al., 2019c); 5) modeling inflammatory responses to bacterial infection [i.e., chorioamnionitis, ascending urea plasma parvum infection (Tantengco et al., 2022b; Zhu et al., 2018; Richardson et al., 2020b; Bento et al., 2023)] and adverse pregnancy outcomes (Tantengco et al., 2021; Lee et al., 2016; Mosavati et al., 2020; Zhu et al., 2018).
Progress has been made in recent decades regarding women’s health and basic and translational research in obstetrics. However, much work utilizing these new approach methods is still needed as dictated by the alarmingly high rate of pregnancy complications. Pregnancy is a complex system due to the interaction of two unique physiologies, the mother and the fetus, and much is still unknown regarding the inner workings of this multifaceted reproductive process. The challenges posed by the many limitations of animal models, current in-vivo and ex-vivo study designs, and ethical concerns associated with the direct investigation of pregnancy in human subjects hinder the depth of understanding one can obtain through research. OOC devices can accelerate research in this area and advance the understanding of pregnancy health and adverse pregnancy events by more accurately emulating the complexity of human reproduction in experimental settings. In this review article, we discuss the current methodologies and limitations of reproductive and obstetrics-based OOC platforms and highlight the advances in high throughput data acquisition techniques that will enhance the biological and translational potential of data collected from microfluidic experiments.
Though OOC platforms more accurately represent human anatomy and biological function than current in-vitro systems, they do have limitations hindering their utility in different scientific fields (i.e., bench research, toxicology, and pharmacology). These restrictions include fabrication materials, modeling vascular flow, and the lack of biologically relevant multi-organ systems. The sections below will elaborate on this list of limitations and the challenges in overcoming them with current technological approaches.
Polydimethylsiloxane (PDMS) is one of the most used materials in microfluidics as it allows easy prototyping through simple soft lithography techniques (Gokaltun et al., 2017). By casting PDMS prepolymer mixed with the curing agent onto a master mold, replicas with designed structures can be fabricated after incubation in an oven. The PDMS-enclosed microenvironment is supportive of cell growth due to its biocompatibility and gas permeability (Shakeri et al., 2021). Most importantly, PDMS is transparent to visible light (Dizon et al., 2019) and its low autofluorescence (Gokaltun et al., 2017) allows bright-field and fluorescent microscopy to monitor cell morphology and various marker expressions. The major disadvantage of PDMS is the adsorption of lipophilic proteins and small molecules (e.g., estrogen) because of the hydrophobic surface (Dizon et al., 2019; Ren et al., 2010). Recent studies related to reproductive OOCs have shown that PDMS devices do not adsorb hydrophilic compounds such as cytokines (Richardson et al., 2020b) (i.e., Interluken-6), biological signalers (Radnaa et al., 2021) (i.e., exosomes), endocrine factors (progesterone), chemical compounds (Richardson LS. et al., 2023) (i.e., nicotine), nor pharmacological compounds (Richardson et al., 2022) (i.e., Pravastatin, Rosuvastatin, Aspirin, Heparin), and environmental toxicants [i.e., cadmium (10µM ∼ 20% adsorption) (Kim et al., 2022)] (Figure 1). It is important to identify OOC material’s constraints, and pilot studies should be set up to determine the adsorption properties of analytes under investigation. For example, though PDMS is suspected of adsorbing endocrine factors, our models have shown that cells grown on-chip still produce functionally active naïve progesterone and progesterone synthesis can be inhibited by pathologic agents (i.e., lipopolysaccharide) (Richardson et al., 2023c) (Figure 1E). We were also able to test the functional effect of progesterone on cells grown on chips by disturbing its receptors (Lintao et al., 2024). These results highlight the detectability of high concentrations of biologically relevant progesterone even in the wake of slight adsorption by PDMS.
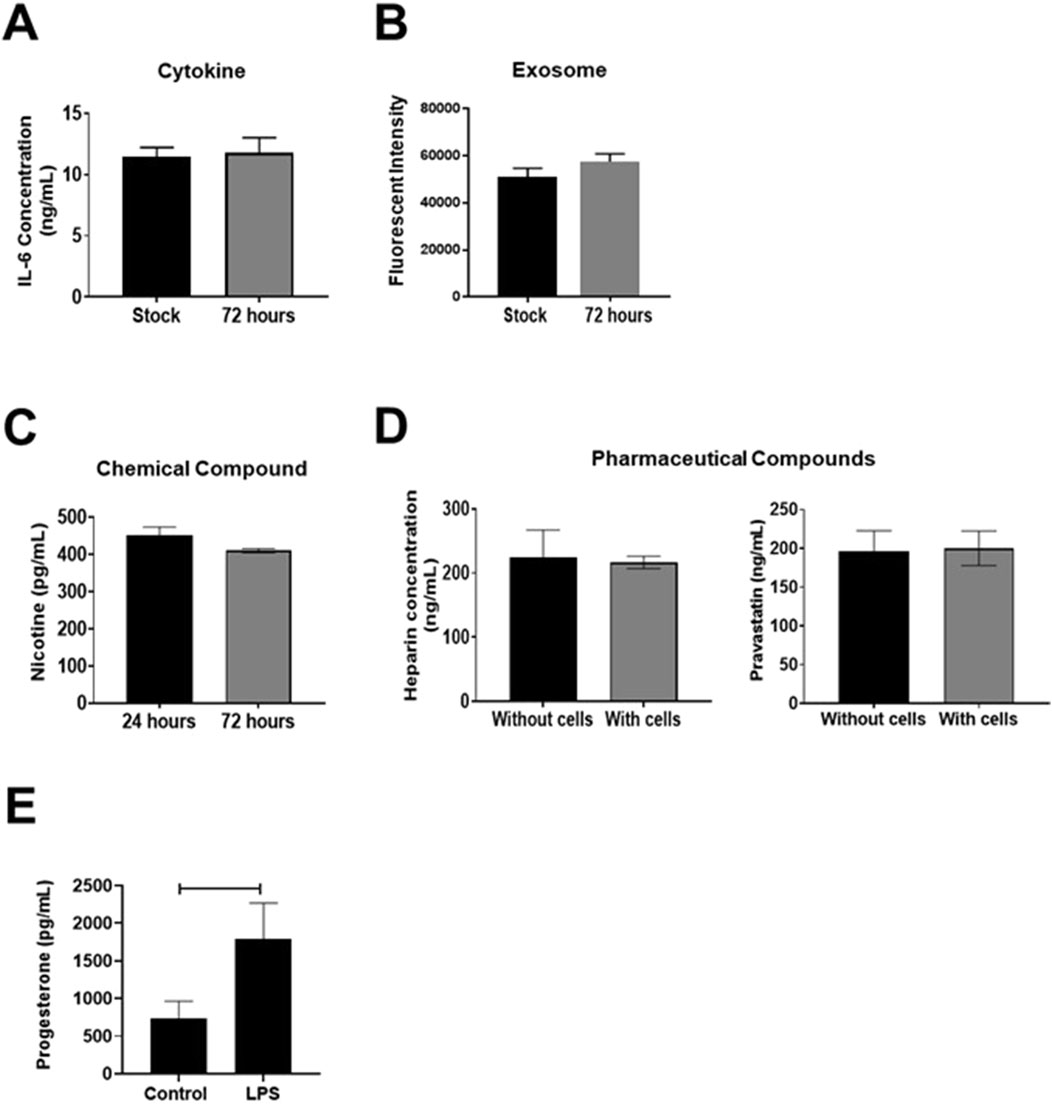
Figure 1. PDMS adsorption of compounds. (A, B) ELISA measured pro-inflammatory cytokine Interleukin (IL)-6 concentrations or CSFE stained exosomes within an OOC device, showing no difference over a 72-h period, indicating no or limited adsorption to the PDMS device or reservoir system (n = 3). Values are expressed as mean ± SEM. This figure is adapted from Richardson et al. (2020b) and Radnaa et al. (2021). (C, D) Targeted mass spectrometry analysis measured nicotine, heparin, or pravastatin concentration within an OOC device. Nicotine levels did not change over 72-h, and heparin, nor pravastatin concentrations differed after 8-h in devices with or without placental cells (n = 3). Values are expressed as mean ± SEM. Figures are adapted from Richardson et al. (2022), Richardson LS. et al. (2023). (E) ELISA-based evaluation of progesterone adsorption within OOC devices. Within a choriodecidua OOC, maternal decidua LPS treatment induced significant progesterone production within the neighboring chorion cells (p = 0.04). Values are expressed as mean ± SEM. The figure is adapted from Richardson et al. (2023a).
Many approaches have been suggested and studied to increase the hydrophilicity of PDMS surface to reduce PDMS-to-compound binding. First, oxygen plasma treatment can increase the surface energy on PDMS quickly without affecting the transparency for imaging, but the plasma effect declines rapidly and is lost within hours (Miranda et al., 2021). To achieve long-term treatment, additional coating with a zwitterionic polymethacrylate copolymer has been proposed, in which the hydrophilic surfaces could last for at least 200 h (Dizon et al., 2019; Miranda et al., 2021). Investigation on using 5% Pluronic F127 coating on PDMS devices to block non-selective adsorption of hydrophobic small molecules has been done and shows that the Pluronic F127 layer could effectively separate the interaction between PDMS surface and culture media and reduce the adsorption of Nile Red without affecting cell viability (Hawkins et al., 2022). Instead of treating the surfaces of PDMS after curing, Gökaltun et al. suggested mixing liquid PDMS with a smart copolymer additive containing PDMS as the base elastomer and 60%–70% hydrophilic polyethylene glycol (PEG) before curing (Gokaltun et al., 2017). This copolymer additive self-organizes on the surface when exposed to water and has the PEG segment in contact with the aqueous environment to prevent adsorptions of small molecules without additional steps, while the hydrophilic property could last for 20 months (Gokaltun et al., 2019). Thermoplastics such as polycarbonate, polymethyl methacrylate, and polyvinyl chloride are favorable in manufacturing for scale-up production (Shakeri et al., 2021; Miranda et al., 2021) using injection molding, carbon dioxide laser cutting, and the adsorption of small molecules could be avoided (Gokaltun et al., 2017; Busek et al., 2021). However, it requires more complicated equipment and expensive molds for developing new devices (Sonmez et al., 2020), which might not be a suitable approach for prototyping. Additionally, many of these methods restrict the number of cell culture chambers that can be fabricated within an OOC platform, limiting their ability to recreate anatomically correct organ structures. Further research is needed to validate and assess a variety of fabrication materials and their utility in housing multiple cellular and collagen layers to create physiological OOC devices.
In OOC platforms, media is perfused into a cell culture chamber by either a syringe pump and peristaltic pump perfusion (Novak et al., 2020a; Wang et al., 2018; Hsieh et al., 2019; Novak et al., 2020b) or by gravity feed media reservoirs (Richardson et al., 2019a; Richardson et al., 2020b; Richardson LS. et al., 2023; Phan et al., 2017; Wang and Shuler, 2018; Esch et al., 2016). Media perfusion is most common as it serves to mimic the circulatory system that transports sufficient nutrients for long-term cell culture and recapitulates a better physiologically relevant micro-environment compared to a static culture system for organs that contain physiological blood flow (Gijzen et al., 2021; de Graaf et al., 2022). Unlike conventional blood flow in-vivo, modeling in-utero fluid dynamics (i.e., placental blood flow or amniotic fluid movement) is challenging. Maternal to fetal blood flow at the placental interface is specifically challenging to model within pregnancy-related OOCs due to the numerous lobules of the placenta and maternal spiral arteries that are inter-branched, allowing for non-unform slow blood flow. As it is difficult to calculate the exact amount of fluid flow and shear stress experienced by the placental cells in-utero, researchers have utilized arbitrary fluid flow values (i.e., 50–100 μl/h) (Pemathilaka et al., 2019; Blundell et al., 2018; Blundell et al., 2016) to create low levels of fluid perfusion. Though fluid flow across placental trophoblast cells has been shown to upregulated microvilli formation (Miura et al., 2015; Blundell et al., 2016), it is unknown if any negative or non-physiological effects are present due to these conditions. In addition, the amniotic fluid that surrounds the fetus slowly moves back and forth due to maternal and fetal movement, resulting in low levels of shear stress on the amnion epithelial cell (Morandi et al., 2023; Richardson et al., 2020c). However, studying this motion has been limited due to the lack of proper in-vitro platforms. Kim et al. recently studied the effect of amniotic fluid on amnion membrane disruption using an existing OOC by slow shaking (Richardson et al., 2019a; Kim et al., 2024). Programmable syringe pumps with the desired flow rate were connected to the device to control the flow direction and time interval. However, the experimental setup has not fully mimicked the in-vivo situation as it only created back-and-forth fluidic movement, while the physiological situation is much more complicated, including vibration and random influx and outflux of fluids. Integrating electrical or mechanical engineering modules to improve the system should be considered to recreate a higher degree of physiological properties and functions of targeted tissues or organs. Real-world validation techniques, such as using patient-derived tissues or microvascular modeling, could be helpful to try and tease out proper flow rates that should be incorporated into these key interfaces.
Traditionally, to establish biologically relevant physiologic or pathologic states on-chip, cellular components are assessed by a variety of standard molecular assays to determine functional relevance to the human organ of interest. Cellular components are either kept on-chip and analyzed for morphologic, cytotoxic, or migratory changes (bright-field microscopy or live imaging), protein expression (Immunocytochemistry [ICC]), or barrier function (transendothelial electrical resistance [TEER]). Collection of cells, or cellular components, from devices through trypsinization, cell lysis, or TRIzol allows protein analysis by Western blot, gene expression by RT-PCR, or cell fate and marker analysis using flow cytometry.
The ability of human primary, immortalized cells or iPSCs grown in OOC platforms to biologically recreate human physiology is one of the most significant advancements of this methodology in the field of science. However, when it comes to recreating reproductive or pregnancy-related organs, there are many hurdles in acquiring primary tissue or cell lines for use within said platforms, especially since iPSC lines are not available for many organs of interest nor iPSCs will not undergo the natural aging of fetal organs as expected in-utero. Few labs obtain local access to reproductive or pregnancy-related tissue to derive primary cells, and fewer have the resources to establish validated cell lines from said organs. Thus, restricting the use of physiologically accurate cells to be cultured within OOCs. Furthermore, commercially available cell lines do not always mimic the in-vivo cell type they are derived from, or they are collected from carcinomas or diseased tissue that could affect their functional responses on-chip. Often these cellular restrictions force scientists to use improper cell lines (e.g., BeWO, JEG, WISH cells) or to exclude cell types altogether due to the current state of the field’s resources. This can often be seen with commercial platforms that only allow for two cell culture chambers and are trying to recreate complex multi-layered tissue. This cellular challenge is most detrimental, as it can lead to biologically inaccurate results and cast doubt on OOCs within different fields.
Bright-field and fluorescence microscopy are basic optical microscopy technique that uses transmitted light and filter cubes to produce images of a sample, commonly used to visualize the structure of cells and tissues and observe morphological changes (Tantengco et al., 2021; Gnecco et al., 2019; Gnecco et al., 2017; Kreuder et al., 2020; Rogers et al., 2018; Richardson L. et al., 2020; Richardson et al., 2020b; Kim et al., 2022). It is simple, inexpensive, and can be combined with other techniques. Bright-field microscopy can be used in OOC studies to visualize live cell and their structure, assess cell health and function, and monitor morphological changes in response to stimuli. This technique can provide important information to optimize conditions and improve understanding of organ responses in a controlled, reproducible manner. However, microscopy-related results only provide a “snapshot” of one timepoint and one region within a device leading to unwanted bias when assessing cell morphology endpoints. To overcome this limitation, many groups have turned to live cell imaging to evaluate cell responses and movement in real-time. This technique is a valuable method for visualizing cells and their adaptations to stimuli in real-time within a microscale environment such as OOC devices (Yu et al., 2022; Nagashima et al., 2018; Park et al., 2022; Boos et al., 2021; Richardson et al., 2020b). Boos et al. used live imaging to monitor placental proliferation and growth in the development of their microfluidic device that models the embryo-maternal interface (Boos et al., 2021). This provides valuable information on cellular responses to various stimuli and enables the development of more accurate organ models.
ICC is a laboratory technique to visualize and analyze the distribution and localization of specific proteins or other cellular components in fixed cells or tissues. It uses antibodies that bind specifically to target antigens and visualizes the binding with a detectable label, such as a fluorescent dye, to obtain information about the presence and location of the target within the cell or tissue. ICC can be used in OOC studies to analyze the distribution and localization of specific cellular components (Richardson et al., 2019a) within the cells and tissues on the chip (Xiao et al., 2017; Ahn et al., 2021; Park et al., 2022; Mosavati et al., 2020; Pemathilaka et al., 2019; Yin et al., 2019; Blundell et al., 2018; Abostait et al., 2022a; Mandt et al., 2018; Ghorbanpour et al., 2023; Pemathilaka et al., 2022; Saha et al., 2020), providing insight into cellular and molecular functions (Baert et al., 2020; Tantengco et al., 2022a; Tantengco et al., 2021; Tantengco et al., 2022b; Saha et al., 2021; Park et al., 2020; Gnecco et al., 2019; Gnecco et al., 2017; Kreuder et al., 2020; Boos et al., 2021; Blundell et al., 2016; Rogers et al., 2018; Richardson et al., 2020b; Kim et al., 2022) and advancing the development of these devices for various applications such as disease modeling and drug screening. Mahajan et al. used immunostaining on their vagina-on-a-chip to visualize the presence of cytoskeletal proteins (i.e., Cytokeratin [CK]14, CK15, CK5, CK13, F-actin), junction markers (i.e., Involucrin, Zonula occludin-1, E-cadherin), and desmosomes (i.e., Desmoglein [DSG]-3, DSG-1) in the squamous stratified vaginal epithelium (Mahajan et al., 2022). ICC is limited, however, in the number of fluorophores (i.e., markers) that can be evaluated at one time. Traditional fluorescent microscopy allows four excitation/emission cubes to simultaneously evaluate four tagged antibodies. This still restricts the throughput of analysis conducted within cell culture chambers.
The on-chip measurement of cell barrier function using TEER measurement is very commonly used approach. Blundell et al. confirmed the integrity of OOC in their placental drug transport studies using TEER, which assessed the structural integrity of the placental barrier (i.e., trophoblast-endothelial layers) and its ability to mimic drug transport across the maternal-fetal interface in the human placenta (Blundell et al., 2018). This method provides a quantitative determination of the barrier function of cell cultures for OOC experiments to assess system functionality, drug toxicity, and efficacy (Gnecco et al., 2017; Yin et al., 2019; Blundell et al., 2018; Boos et al., 2021). However, current TEER machines provide “chopstick” style electrodes that are only compatible with transwell-orientated OOC platforms. This greatly restricts the usability of this methodology across OOC systems. Advanced engineering of TEER electrodes etched on the glass surfaces of the OOC can record TEER in real-time, overcoming the challenges of the current TEER platforms.
To analyze target RNA in a cell sample, RT qPCR is commonly used by combining the principles of PCR with real-time fluorescence detection. It is a rapid, sensitive, and specific tool used in molecular biology and diagnostics. RT qPCR can be used to assess gene expression in cells grown on OOC in microscales cultures. Data can be used validate functional and molecular similarities to in-vivo situations, providing insights into gene regulation and cellular responses (Baert et al., 2020; Mahajan et al., 2022; Tantengco et al., 2022b; Nagashima et al., 2018; Saha et al., 2021; Park et al., 2020; Ahn et al., 2021; Aziz et al., 2020; Yin et al., 2019; Saha et al., 2020). It can also be used for quality control. Yin et al. used qPCR to evaluate the expression of Iinterluken-6 (an inflammatory cytokine) secretion from the system after nanoparticle exposure to investigate the in-vitro experimental placental response in a pathological condition (Yin et al., 2019). Unfortunately, due to the low number of cells seeded within OOC chambers, collection of high abundance pure RNA is very challenging and limits the throughput of transcript analysis.
Western blotting, which is a semi-quantitative laboratory technique for analyzing specific protein markers in a sample by separating the proteins, can be used in OOC studies to analyze the protein expression profiles of cells collected from microfluidic devices. This provides insight into the health and function of the cells and the overall system (Tantengco et al., 2022a; Saha et al., 2021; Park et al., 2020). Park et al. used Western blotting to analyze the enhanced mRNA and protein levels of plasminogen activator inhibitor-2, a reliable reproductive toxicity marker, in endometrial and ovarian follicular cells cultured in their dual reproductive OOC device following toxic exposure (Park et al., 2020). Conversely, semiquantitative protein marker analysis can be conducted through flow cytometry, that analyzes the physical and chemical characteristics of individual cells in a fluid stream by using lasers and fluorescence to detect and profile the cells based on their size, shape, contents and other parameters (Saha et al., 2020). It is commonly used in OOC experiments to allow real-time analysis within the system to quantify cells in the system, assess cell viability and phenotype, and measure the secretion of specific cytokines or other biomarkers that may indicate proper organ function (Park et al., 2020; Park et al., 2022; Boos et al., 2021; Kim et al., 2022). This technique was used by Kim et al. to analyze the cell cycle of maternal decidua cells and determine the effect of cadmium exposure on cell proliferation inside their feto-maternal interface organ-on-chip (FMi-OOC) (Kim et al., 2022). However, a general limitation of microfluidic platforms is the use of minimal amounts of cells (i.e., 50,000–100,000 per chamber) to establish the organ system. While this can increase the throughput of devices that can be run in a single experiment and minimize reagent costs, it also restricts the type of endpoint assay that can be conducted without pooling time points or devices (e.g., flow cytometry and traditional Western blotting).
ELISA is a type of assay that uses an enzyme-labeled antibody to detect and quantify the presence of a specific protein in a sample (Xiao et al., 2017). It is commonly used for detecting and quantifying hormones, cytokines (Mahajan et al., 2022; Richardson et al., 2019a; Saha et al., 2020), and antigens (Baert et al., 2020; Xiao et al., 2017; Tantengco et al., 2022a; Mahajan et al., 2022; Tantengco et al., 2022b; Aziz et al., 2017; Gnecco et al., 2019; Gnecco et al., 2017; Park et al., 2022; Aziz et al., 2020; Abostait et al., 2022a; Blundell et al., 2016; Rogers et al., 2018; Richardson et al., 2020b; Kim et al., 2022). Xiao et al. used ELISA to detect ovarian hormone production, Interluken-8, vascular endothelial growth factor, and albumin between microfluidic and static cultures (Xiao et al., 2017). The limitation of this methodology lies in the required supernatant volume needed for analysis (i.e., ∼100 μl per analyte), which utilizes a majority of one cell chamber’s media volume. To overcome this, researchers have turned to Luminex multiplexing, which is a multiplexed immunoassay system used to simultaneously detect the presence of many specific proteins or other analytes by capturing specific targets onto spherical beads in suspension (Tantengco et al., 2022a; Tantengco et al., 2021; Tantengco et al., 2022b; Saha et al., 2021; Richardson et al., 2020b; Kim et al., 2022). Luminex multiplexing technology can be used in OOC studies to simultaneously measure multiple biological markers (i.e., up to 20–40 analytes depending on their compatibility within kits) in small fluid volumes (i.e., ∼25 µL) collected from chips (Baert et al., 2020; Xiao et al., 2017; Tantengco et al., 2022a; Tantengco et al., 2021; Tantengco et al., 2022b; Aziz et al., 2017; Saha et al., 2021; Gnecco et al., 2019; Gnecco et al., 2017; Park et al., 2022; Abostait et al., 2022a; Rogers et al., 2018; Richardson et al., 2020b; Kim et al., 2022). This provides valuable information about the health and function of the cells and tissues, which can be used to optimize the performance of these devices.
Targeted mass spectrometry is an analytical tool that measures the mass-to-charge ratio of charged particles, allowing for an in-depth identification of a specific subset of compounds within a complex sample. It allows for highly sensitive and selective quantification of targeted analytes, providing more specific information than untargeted mass spectrometry approaches. Targeted mass spectrometry can be useful in OOC studies as it can analyze biological markers in small fluid volumes collected from the chip, allowing researchers to understand cellular responses and biomolecular interactions (De Bem et al., 2021; Kim et al., 2022). For example, Park et al. used mass spectrometry to quantify proteins from conditioned media on an implantation-on-a-chip to explore the established functions of the proteins found in relation to implantation and placentation and to discover new proteins that may have an unknown role in these processes (Park et al., 2022). Pemathilaka et al. (2022) used liquid chromatography/mass spectrometry (LC-MS) to measure Naltrexone concentrations, a drug commonly used for opioid treatment to determine if it crosses the placenta.
The limitations described above have started to be addressed with novel molecular and biological methodologies that specialize in using low numbers of cells and supernatants to generate libraries of data. However, these novel techniques are only utilized by a minority of OOC platforms, and therefore, to provide examples of each technique’s utility, the below section will describe results generated from pregnancy-related OOCs. These sections will be divided by RNA and protein analysis of cells and supernatants (Figure 2).
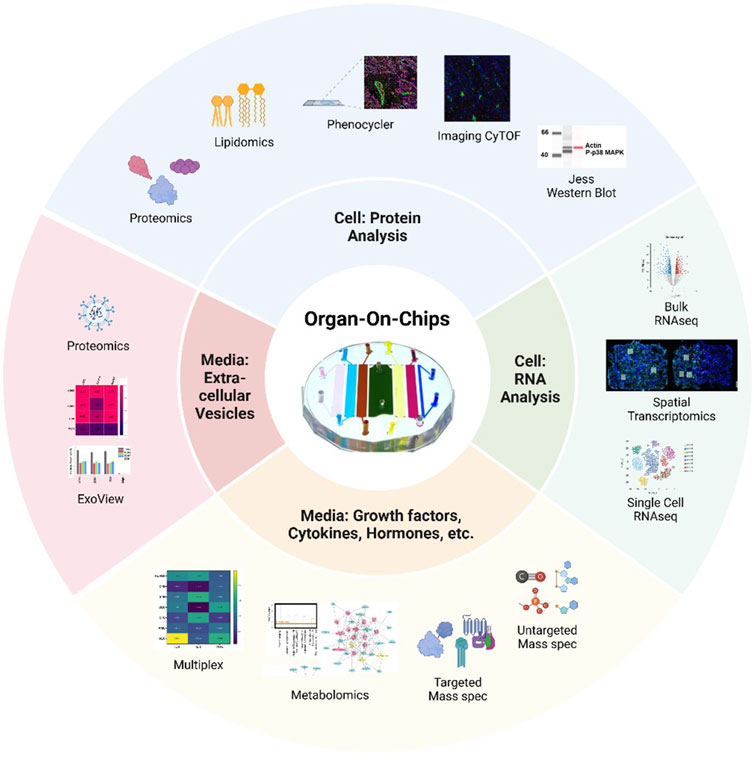
Figure 2. Summary of advanced methodologies to study cells and media from organ-on-chips A variety of protein analysis strategies can be used to assess cell phenotypes on-chip, including proteomics, lipidomic, PhenoCycler or imaging CyTOF multiplexing of antibody staining, or Jess Western blotting. RNA collected from OOCs can be analyzed by bulk or single-cell RNAseq, while spatial transcriptomics allows for regions of interest (white boxes) to be selected from cell on-chip. Media collected from individual chambers can be analyzed for a variety of secreted factors by multiplexing for specific targets or by conducting mass spectrometry for metabolites, targeted, or untargeted molecules. Extracellular vesicles, like exosomes, can be evaluated from small volumes of OOC media by methods such as ExoView or proteomics. The device in the center is the multi-organ fetal membrane and placenta feto-maternal interface OOC (Kammala et al., 2023; Safarzadeh et al., 2024).
As mentioned above, current OOC users face multiple challenges when trying to generate transcriptome-based cellular data. Specifically, difficulties in isolating high quantity, pure RNA and problems collecting cells out of cell culture chambers are quite common. Two high throughput technologies, RNAseq and spatial transcriptomics (Figure 2), can help to alleviate these challenges. RNAseq is a technique that uses next-generation sequencing to produce transcriptome data (e.g., pooled RNA snapshot of the cells’ status) from cellular samples (Wang et al., 2009). In general, RNA isolated from chips are converted into cDNA library fragments with adaptors, amplified, and sequenced in a high throughput manner to generate reads through multiple platforms (i.e., Illumina IG, Applied Biosystems SOLiD). This allows for high-throughput and complex analysis even with low RNA yield arising from generally low cell numbers in OOC platforms. This technology has recently been utilized with chorion and decidua-derived RNA from the chorio-decidua immune interface OOC (Lintao et al., 2024). This library of transcriptome data generated a heatmap of inflammation (e.g., Toll-like receptors, interferons, chemokines) and immunity genes (e.g., clusters of differentiation, human leukocyte antigens) that were differentially expressed in chorion trophoblasts and decidual stromal cells (Figure 3). It provides a large RNA library that can now be investigated for future biological implications highlighting its application within OOC platforms. Sometimes, cells are extremely difficult to remove from cell chambers. Planar OOC devices can be adapted to use spatial transcriptomics methodologies that enable researchers to spatially localize and quantify gene expression on-chip. This technology was recently used to select targeted regions of in-vivo tissue or in-vitro cell culture, compare the transcriptomic profile of human decidua to decidual cells grown within an OOC device surrounded by other pregnancy-related cellular layers. This type of analysis allows researchers to specifically select regions of interest within OOC platforms or tissues and conduct full transcriptome analysis without removal of the cells from their biological context.
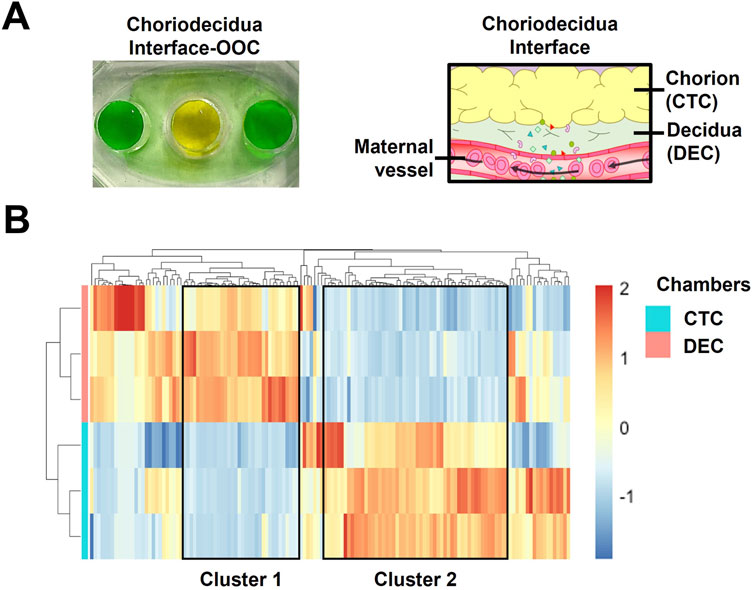
Figure 3. Utilizing targeted RNAseq to characterize the chorion and decidua cells on-chip (A) Color dye image of a two-chamber chorio-decidua interface OOC and the corresponding anatomical structure. (B) Heatmaps of transcripts involved in inflammation and immunity with >1,000 reads identified in chorion trophoblasts (n = 3) and decidual stromal cells (n = 3) derived from the chorio-decidual interface-on-chip. Transcript intensities were shown as z-scores and were displayed as colors ranging from red (highest) to blue (lowest). The rows and columns are clustered using correlation distance and average linkage. The figure is adapted from Lintao et al. (2024).
In parallel with RNA analysis, protein-expression data is often necessary to determine disease states or decipher the biological implications of differential expression of transcripts. Protein data derived from cells or media from OOC experiments face limitations as described above. However, several novel technologies can overcome these challenges to derive the protein signature. These technologies include both on-chip (i.e., imaging CyTOF or PhenoCycler) and off-chip (i.e., proteomics and Jess Simple Western blotting) platforms (Figure 2). Multiplex protein analysis of cells on-chip is only accomplished by imaging CyTOF which is an application of mass cytometry used to quantify metal-labeled targets (i.e., up to 30) on the surface and interior of single cells. Cells are immunostained with pre-conjugated metal antibodies that are then laser ablated, collecting protein data for downstream analysis. Recent experiments utilizing the chorio-decidua immune interface OOC assess the status of migratory primary immune cells into the chorion chamber using this approach (Richardson et al., 2023c). Figure 4 shows a subset of these data highlighting imaging CyTOFs ability to identify and characterize unique immune cell sub-types on-chip (i.e., NK cells, dendritic cells, and neutrophils) after control or lipopolysaccharide (LPS) treatment (Figure 4B). These results emphasize the unique ability of this technology to document multiplexing of protein-level data without removing cells from their culture chambers.
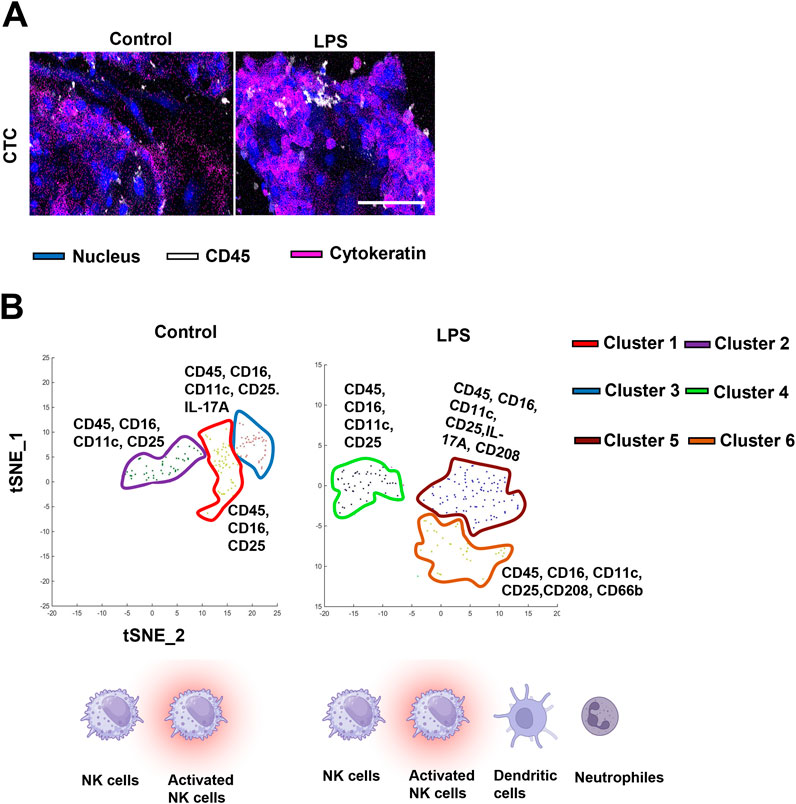
Figure 4. Adapting OOC devices for imaging CyTOF analysis of immune cell activation at the choriodecidua interface (A) CyTOF images showing migratory leukocytes identified by their CD45 staining (white) next to chorion cells (blue nuclear stain and pink cytokeratin stain) in the center chorion chamber. Scale bars, 100 µm. (B) Phenographs overlaid on t-SNE plots showing the specific immune cell clusters identified in each group. Different colors represent different histoCAT-identified cell clusters which were dominant in differentially expressed immune cell markers. Background clusters that did not meet the inclusion criteria were removed from these graphs. The figure is adapted from Richardson et al. (2023a).
If collection of cellular components is possible from chip experiments, two sensitive methodologies can be implemented to screen for overall protein profiles using mass spectrometry and eukaryotic libraries (i.e., proteomics), followed by Jess Simple Western blotting to confirm protein expression in a multiplex scale using microliters of sample. Proteomic analysis is possible from single-cell chambers of most OOC platforms that contain around 100,000–200,000 cells (i.e., ∼9–15 μg of total protein), though recovery rates differ across devices. Isolation methods vary greatly depending on local expertise or sup-types of downstream mass spectrometry analysis; however, different styles of isolation (i.e., RIPA lysis then buffer transfer into TEAB or isolation with TEAB) greatly affect differential protein expression and should be standardized across experiments. To further confirm these protein results, automated Jess Simple Western blotting can evaluate protein expression using minimal sample volume (2–5 μl; i.e., typically 0.2–0.4 mg/mL of protein) (Bento et al., 2023) (Figure 5). This analysis can be extended to incorporate antibody multiplexing with chemiluminescence and fluorescent-based antibodies. Thus, a novel method for protein-level Western blot analysis within single-cell chambers in a single OOC device is established.

Figure 5. Quantifying protein expression from individual devices and various cells within OOC platforms (A) Color dye image of a four-chamber fetal membrane feto-maternal interface OOC and the corresponding anatomical structure. (B) Protein analysis using Jess Simple Western blotting platform. Optimization of protein quantity per lain (i.e., 0.2–0.4 mg/mL) and p38 MAPK antibody titration. The figure is adapted from Bento et al. (2023).
Depending on the OOC platform, supernatant collection and volumes (i.e., small volume size or over-diluted) can limit the number of endpoint assays accomplished at a single time point within one device. While multiplexing of cytokine and hormone analysis is quite common to overcome these limitations (i.e., requiring 25 μl of sample to assess 5–40 analyte, instead of 100ul per analyte), other methodologies are available to generate additional high throughput data from small volumes. One such approach is utilizing mass spectrometry to assess proteins, secreted metabolites, or lipid profiles in an untargeted manner and then comparing these results to documented proteomics, metabolomic, or lipidomic libraries (Figure 2). By utilizing “multi-omics” approaches, a biological profile can be generated from each cell layer within OOC devices, contributing to the establishment and validation of these systems. Ongoing studies suggest this type of assessment can be conducted on a single-cell chamber to investigate a particular cell layer’s function or metabolism of a drug/toxicant or utilized to investigate the overall organ’s response. Though these results document normal metabolic pathways and processes found within these cellular layers, targeted metabolic approaches can be used to document drug or toxicant propagation and metabolism to generate translationally impactful data from low sample volumes.
Over the past 10 years, exosome signaling at the organ and cellular level has been documented to play critical roles in endocrine or paracrine signaling and the development of disease states. Exosome analysis is typically restricted to media samples greater than 500ul in volume; within those samples, the quality and quantity of exosomes are minimal. ExoView™ R100 platform is an immuno-affinity-based technology that allows specific exosome populations (i.e., CD63, CD81, and CD9) to bind in a multiplexed manner to a functional ExoView™ chip (Figure 2). The instrument provides multi-level and comprehensive exosome measurements for particle size analysis, and phenotyping (i.e., CD markers). Ongoing studies document that 100 μl of samples collected straight from OOC cell chambers (i.e., “unisolated” and cultured in fetal bovine serum-free media) can be analyzed using ExoView™ chip to determine exosome quantity and characterize CD marker expression. Immunostaining for proteins of interest within the lumen or membrane of exosomes is also possible with this technology (Kammala et al., 2022). Still, there are challenges with exosome research on OOCs. Downstream characterization of exosome cargo using proteomics and other OMICS or additional functional studies can be challenging as the chips may not yield a very high quantity of exosomes.
OOC technology offers improved cell culture capability to create physiologically more relevant human tissue environments, leading to better predictive in-vitro models. However, creating in-vitro human physiology is often made at the expense of throughput, whether low throughput regarding the number of devices used in an experiment or low throughput regarding low data collection. Therefore, creating next-generation OOCs that allow for sufficient data collection requires novel methodologies (described above) merged with advanced microfluidic engineering architecture together with the corresponding biology. Scaling down OOC size (i.e., Mimetas placenta chip) (Rabussier et al., 2023b) or interconnecting arrays of devices, as shown in Figure 6, can be two alternative design strategies to increase the experimental throughput of OOC platforms. In arrayed models, multiple compartments are interconnected through microfluidic channels to operate multiple assays simultaneously, which enables semi-independent drug treatment (Figures 6A,B). A recent study has introduced a high throughput platform for drug development (Azizgolshani et al., 2021) that adapts this strategy. This platform supports multiple human tissue models, including liver, vascular, gastrointestinal, and kidney. In this system, 96 individual devices are integrated with programmable flow control systems for high throughput data collection. Electrical components, such as electrodes, are also integrated for electrochemical sensing within the design to produce higher readouts (Zhang et al., 2017; Bonk et al., 2015; Jin et al., 2020; Dornhof et al., 2022). Electrical sensors can also be embedded to quantify the barrier function through a real-time TEER (Bossink et al., 2021; Henry et al., 2017; Maoz et al., 2017; van der Helm et al., 2016b; Renous et al., 2021). Therefore, this system enables high throughput drug screening capability, increasing predictive accuracy.
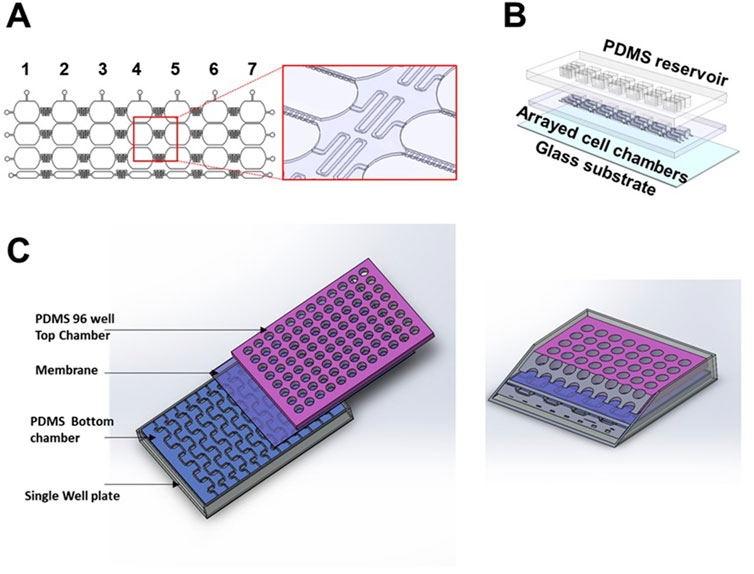
Figure 6. Development of array OOC platforms to model the fetal membrane feto-maternal interface (A) 2D and 3D schematics of an arrayed OOC device used to model the feto-maternal interface. The cropped region highlights the delay channels connecting each of the seven devices and microchannels connecting each cellular chamber within said device. (B) 3D drawing of the individual components of this array platform. (C) Highlights the concept of a vascularized feto-maternal interface model with cross-section.
Numerous biosensors have been developed and employed for a variety of sensing applications in microphysiological systems. The capability to obtain real-time measurements has enabled researchers to conduct experiments noninvasively and collect quantitative analytical data more efficiently. For example, impedance-based techniques have allowed for the study of single cells, providing insights into the cell cytoplasm through the membrane and enabling the investigation of cellular senescence (Xu et al., 2016).Additionally, oxygen sensors have been instrumental in detecting cellular oxygen deprivation and oxidative stress (Moya et al., 2018; Azimzadeh et al., 2021; Bossink et al., 2020). Various methods, such as aptamer-based tracking (Ferguson et al., 2013; Liu et al., 2015; Hao et al., 2018), electrochemical detection (Russell et al., 2019; Lu et al., 2021), and photonic biosensing (Cognetti et al., 2023), have been explored for real-time detection of inflammatory markers. These biosensors can also be integrated into the reproductive field to monitor the feto-maternal interface and predict preterm birth. Impedance-based sensors can assess the integrity of placental cells at the maternal-fetal interface. In contrast, oxygen sensors can monitor oxygen levels and identify potential hypoxia, a known risk factor for preterm birth. Moreover, various sensors capable of detecting inflammatory markers and hormonal levels (estrogen and progesterone) in real-time could be crucial for identifying intrauterine inflammation, which is a significant contributor to preterm labor. Yet, the integration of sensing technology to pregnancy-related applications has not been fully utilized as it requires complex fabrication processes with expensive core facility usage, significant effort on optimizing the sensors for desired sensitivity, thorough training of AI-driven systems to generate automated readouts, and consideration of biocompatibility. However, the field is making progress to integrate these systems into devices to enable continuous, noninvasive monitoring (Izadifar et al., 2024). Thereby making OOCs an effective tool for developing pathophysiological models and facilitating drug discovery.
Aside from biosensors, OOCs have started to reflect more complex physiology that is seen in-vivo. Among the advances in modeling the vasculature include the assembly of vascular cells into networks and their remodeling to form vascular beds underflow (Mandrycky et al., 2021). Optimizing the vasculature dynamics remains unresolved when creating a vascularized model of the placenta. This is particularly important in tissues and disease states where immune cells play a significant role in tissue homeostasis and pathogenesis (Van Os et al., 2023). In the context of pregnancy, given that labor is an inflammatory process, the inclusion of immune cells in pregnancy-related OOCs would better provide the immunological context of such a process, as seen in the chorio-decidual immune interface-on-chip experiments we have conducted (Richardson et al., 2023a). Current research is ongoing to develop a vertical, stacked, arrayed microfluidic device with vasculature on the bottom and a 96-well style array on the top for feto-maternal interface cell culture (Figure 6C). Overall, integrating vascular and immune cells into OOCs could potentially provide a mechanistic picture of physiologic and pathophysiologic processes seen in-vivo.
Utilizing high throughput methodologies and arrayed platforms is the future of the OOC field to meet the mandate stated by the FDA Modernization Act 2.0, which is expected to revolutionize the current state of pre-clinical drug testing. The FDA Modernization Act 2.0 allows for alternatives to animal testing (NAMs), such as OOC platforms or in silico simulation models, to generate drug-related data for regulatory purposes (Han, 2023; Haderspeck et al., 2019). This is especially relevant when it comes to developmental and reproductive toxicity (DART) studies and drug development for pregnant women, as pre-clinical studies are needed to provide safe, reliable, and reproducible data on drugs and to accelerate phase clinical trials for reproductive and pregnancy-related adverse outcomes. These experiments are essential as pregnant women are currently therapeutic orphans and are not included routinely in clinical trials (Illamola et al., 2018). Issues arising from DART testing are responsible for 10% of preclinical toxicology-related drug attrition, which in turn accounts for 20% of overall attrition during preclinical drug development (Guengerich and MacDonald, 2007). Currently, developmental and reproductive toxicity in pharmaceutical risk assessment is being done preclinically on two animal species, routinely rodents and rabbits (Stokes, 2015). Alternative models for reproductive and developmental toxicity, such as rodent whole embryo cultures, mouse embryonic stem cells, and zebrafish, have gained traction over the past decades (Brannen et al., 2016). These models are insufficient in generating data required by regulatory authorities for consideration of certain drugs, specifically therapeutics to be given during pregnancy (McCormack and Best, 2014; Adam et al., 2011; Sheffield et al., 2014). With OOC technology, researchers have been able to recreate reproductive tissues in-vitro that reflect in-vivo physiological processes. A microfluidic culture of murine ovarian follicles developed by Xiao et al., (2017) integrated various endocrine loops of the female reproductive tract to reflect a 28-day menstrual cycle hormone profile seen in human, which has potential implications on how potential drug candidates can disrupt hormonal profiles in females. Recently, a human testis-on-a-chip was able to demonstrate the reciprocal crosstalk between Sertoli and Leydig cells that are important in germ cell development and spermatogenesis (Park et al., 2024). Using this platform, researchers were able to identify SERPINB2 as a marker of male reproductive toxicity, which has implications for toxicity screening of potential drug candidates.
In addition to DART testing, OOC technology can be a solution to the lack of drug data in the pregnant cohort due to ethical concerns with including pregnant women in clinical trials (Young and Huh, 2021). Specifically, reproductive and pregnancy-related OOCs can be utilized at every stage of the drug discovery pipeline, including early drug discovery phases that target the identification of novel compounds along with physiological or disease modeling. While intestine, liver, and kidney OOCs can provide information on drug absorption, metabolism, and excretion profiles, pregnancy-related OOCs can provide insights into how the drug is distributed in both maternal and fetal compartments. Such information provides preliminary data on whether the drug would reach therapeutic dosage for the mother and the fetus, depending on who is being treated, and whether it reaches toxic levels in the fetal compartment (Kammala et al., 2023; Menon et al., 2024). Overall, the field is poised to make substantial changes in the area of pharmacology and provide stringent testing for therapeutics before phase trials. Time will only tell if existing OOC platforms will advance and overcome the current limitations of these systems to achieve their ultimate utility and purpose in the field of translational research.
As the reproductive OOC field moves forward devices will need to 1) be scaled down and “miniaturized” to create high throughput platforms, 2) integrate automatic cell/media loading to increase reproducibility and scalability, 3) incorporate patient-specific samples to create personalized medical approaches, 4) increase their biological complexity (incorporating immune cells, microbiomes, and linking multi-organ systems), 5) utilize physiologically relevant cutting edge endpoints, and 6) develop combinatorial novel approach methods such as artificial intelligence and machine learning to extrapolate and predict clinically relevant results from in-vitro experiments. Regardless of the methodology, OOC experiments should be detailed in a manner that allows for easy replication of results, thereby improving the reproducibility of findings and strengthening researchers, clinicians, and federal regulators’ trust in these platforms. STAR methods (Menke et al., 2020; Tonzani and Fiorani, 2021) should be incorporated into manuscripts utilizing OOCs and data generated from these devices should be deposited into Eva Analytics [MPS database (Gough et al., 2016)] to promote the wide-scale use of these devices as key tools to move the scientific field forward.
NT: Investigation, Writing – original draft. AZ: Investigation, Writing – original draft. RL: Investigation, Writing – original draft. RC: Investigation, Writing – original draft. GB: Investigation, Writing – original draft. MV: Investigation, Writing – original draft. SK: Investigation, Writing – original draft. PL: Investigation, Writing – original draft. TC: Investigation, Writing – original draft. KC: Investigation, Writing – original draft. AH: Investigation, Writing – original draft. RM: Investigation, Writing – original draft. LR: Investigation, Writing – original draft.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by UG3 TR003283 (NIH/NCATs/NICHD) to AH and RM and R01HD100729-01S1 (NIH/NICHD) to RM, and R01HD110400 (NIH/NICHD) to LR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The content is solely the authors’ responsibility and does not necessarily represent the official views of the National Institutes of Health.
Abaci, H. E., and Shuler, M. L. (2015). Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integr. Biol. (Camb). 7 (4), 383–391. doi:10.1039/c4ib00292j
Abostait, A., Tyrrell, J., Abdelkarim, M., Shojaei, S., Tse, W. H., El-Sherbiny, I. M., et al. (2022a). Placental nanoparticle uptake-on-a-chip: the impact of trophoblast syncytialization and shear stress. Mol. Pharm. 19, 3757–3769. doi:10.1021/acs.molpharmaceut.2c00216
Abostait, A., Tyrrell, J., Abdelkarim, M., Shojaei, S., Tse, W. H., El-Sherbiny, I. M., et al. (2022b). Placental nanoparticle uptake-on-a-chip: the impact of trophoblast syncytialization and shear stress. Mol. Pharm. 19 (11), 3757–3769. doi:10.1021/acs.molpharmaceut.2c00216
Adam, M. P., Polifka, J. E., and Friedman, J. M. (2011). Evolving knowledge of the teratogenicity of medications in human pregnancy. Am. J. Med. Genet. C Semin. Med. Genet. 157c (3), 175–182. doi:10.1002/ajmg.c.30313
Ahn, J., Yoon, M. J., Hong, S. H., Cha, H., Lee, D., Koo, H. S., et al. (2021). Three-dimensional microengineered vascularised endometrium-on-a-chip. Hum. Reprod. 36 (10), 2720–2731. doi:10.1093/humrep/deab186
An, F., Qu, Y., Liu, X., Zhong, R., and Luo, Y. (2015). Organ-on-a-Chip: new platform for biological analysis. Anal. Chem. Insights 10, ACI.S28905–45. doi:10.4137/aci.s28905
Ashammakhi, N., Wesseling-Perry, K., Hasan, A., Elkhammas, E., and Zhang, Y. S. (2018). Kidney-on-a-chip: untapped opportunities. Kidney Int. 94 (6), 1073–1086. doi:10.1016/j.kint.2018.06.034
Azimzadeh, M., Khashayar, P., Amereh, M., Tasnim, N., Hoorfar, M., and Akbari, M. (2021). Microfluidic-based oxygen (O(2)) sensors for on-chip monitoring of cell, tissue and organ metabolism. Biosens. (Basel) 12 (1), 6. doi:10.3390/bios12010006
Aziz, A. R., Yu, X., Jiang, Q., Zhao, Y., Deng, S., Qin, K., et al. (2020). Doxorubicin-induced toxicity to 3D-cultured rat ovarian follicles on a microfluidic chip. Toxicol. Vitro 62, 104677. doi:10.1016/j.tiv.2019.104677
Aziz, A. U. R., Fu, M., Deng, J., Geng, C., Luo, Y., Lin, B., et al. (2017). A microfluidic device for culturing an encapsulated ovarian follicle. Micromachines (Basel) 8 (11), 335. doi:10.3390/mi8110335
Azizgolshani, H., Coppeta, J. R., Vedula, E. M., Marr, E. E., Cain, B. P., Luu, R. J., et al. (2021). High-throughput organ-on-chip platform with integrated programmable fluid flow and real-time sensing for complex tissue models in drug development workflows. Lab. Chip 21 (8), 1454–1474. doi:10.1039/d1lc00067e
Baert, Y., Ruetschle, I., Cools, W., Oehme, A., Lorenz, A., Marx, U., et al. (2020). A multi-organ-chip co-culture of liver and testis equivalents: a first step toward a systemic male reprotoxicity model. Hum. Reprod. 35 (5), 1029–1044. doi:10.1093/humrep/deaa057
Bento, G. F. C., Richardson, L. S., da Silva, M. G., Tantengco, O. A. G., and Menon, R. (2023). Modeling an ascending infection by Ureaplasma parvum and its cell signaling and inflammatory response at the feto-maternal interface. Am. J. Reprod. Immunol. 90 (4), e13770. doi:10.1111/aji.13770
Blundell, C., Tess, E. R., Schanzer, A. S., Coutifaris, C., Su, E. J., Parry, S., et al. (2016). A microphysiological model of the human placental barrier. Lab. Chip 16 (16), 3065–3073. doi:10.1039/c6lc00259e
Blundell, C., Yi, Y. S., Ma, L., Tess, E. R., Farrell, M. J., Georgescu, A., et al. (2018). Placental drug transport-on-a-chip: a microengineered in vitro model of transporter-mediated drug efflux in the human placental barrier. Adv. Healthc. Mater 7 (2). doi:10.1002/adhm.201700786
Bonk, S. M., Stubbe, M., Buehler, S. M., Tautorat, C., Baumann, W., Klinkenberg, E. D., et al. (2015). Design and characterization of a sensorized microfluidic cell-culture system with electro-thermal micro-pumps and sensors for cell adhesion, oxygen, and pH on a glass chip. Biosens. (Basel). 5 (3), 513–536. doi:10.3390/bios5030513
Boos, J. A., Misun, P. M., Brunoldi, G., Furer, L. A., Aengenheister, L., Modena, M., et al. (2021). Microfluidic Co-culture platform to recapitulate the maternal-placental-embryonic Axis. Adv. Biol. (Weinh) 5 (8), e2100609. doi:10.1002/adbi.202100609
Bossink, E., Zakharova, M., de Bruijn, D. S., Odijk, M., and Segerink, L. I. (2021). Measuring barrier function in organ-on-chips with cleanroom-free integration of multiplexable electrodes. Lab. Chip 21 (10), 2040–2049. doi:10.1039/d0lc01289k
Bossink, E. G. B. M., Slob, J. V. M., Wasserberg, D., Segerink, L. I., and Odijk, M. (2020). Versatile fabrication and integration method of optical oxygen sensors in organ-on-chips, 1–4. doi:10.1109/SENSORS47125.2020.9278735
Brannen, K. C., Chapin, R. E., Jacobs, A. C., and Green, M. L. (2016). Alternative models of developmental and reproductive toxicity in pharmaceutical risk assessment and the 3Rs. Ilar J. 57 (2), 144–156. doi:10.1093/ilar/ilw026
Bruner-Tran, K. L., Gnecco, J., Ding, T., Glore, D. R., Pensabene, V., and Osteen, K. G. (2017). Exposure to the environmental endocrine disruptor TCDD and human reproductive dysfunction: translating lessons from murine models. Reprod. Toxicol. 68, 59–71. doi:10.1016/j.reprotox.2016.07.007
Busek, M., Novik, S., Aizenshtadt, A., Amirola-Martinez, M., Combriat, T., Grünzner, S., et al. (2021). Thermoplastic elastomer (TPE)-Poly(Methyl methacrylate) (PMMA) hybrid devices for active pumping PDMS-free organ-on-a-chip systems. Biosens. (Basel) 11 (5), 162. doi:10.3390/bios11050162
Cherubini, M., Erickson, S., Padmanaban, P., Haberkant, P., Stein, F., Beltran-Sastre, V., et al. (2023). Flow in fetoplacental-like microvessels in vitro enhances perfusion, barrier function, and matrix stability. Sci. Adv. 9 (51), eadj8540. doi:10.1126/sciadv.adj8540
Cognetti, J. S., Moen, M. T., Brewer, M. G., Bryan, M. R., Tice, J. D., McGrath, J. L., et al. (2023). A photonic biosensor-integrated tissue chip platform for real-time sensing of lung epithelial inflammatory markers. Lab. Chip 23 (2), 239–250. doi:10.1039/d2lc00864e
Czechowicz, P., Nowicka, J., Neubauer, D., Chodaczek, G., Krzyżek, P., and Gościniak, G. (2022). Activity of novel ultrashort cyclic lipopeptides against biofilm of Candida albicans isolated from VVC in the ex vivo animal vaginal model and BioFlux biofilm model-A pilot study. Int. J. Mol. Sci. 23 (22), 14453. doi:10.3390/ijms232214453
Dadkhah, E., Hajari, M. A., Abdorahimzadeh, S., Shahverdi, A., Esfandiari, F., Ziarati, N., et al. (2023). Development of a novel cervix-inspired tortuous microfluidic system for efficient, high-quality sperm selection. Lab. Chip 23 (13), 3080–3091. doi:10.1039/d3lc00037k
De Bem, T. H. C., Tinning, H., Vasconcelos, E. J. R., Wang, D., and Forde, N. (2021). Endometrium on-a-chip reveals insulin- and glucose-induced alterations in the transcriptome and proteomic secretome. Endocrinology 162 (6), bqab054. doi:10.1210/endocr/bqab054
de Graaf, M. N. S., Vivas, A., van der Meer, A. D., Mummery, C. L., and Orlova, V. V. (2022). Pressure-driven perfusion system to control, multiplex and recirculate cell culture medium for organs-on-chips. Micromachines (Basel) 13 (8), 1359. doi:10.3390/mi13081359
Deng, J., Wei, W., Chen, Z., Lin, B., Zhao, W., Luo, Y., et al. (2019). Engineered liver-on-a-chip platform to mimic liver functions and its biomedical applications: a review. Micromachines (Basel) 10 (10), 676. doi:10.3390/mi10100676
Dizon, G. V., Clarin, M. T. R., Venault, A., Tayo, L., Chiang, H. C., Zheng, J., et al. (2019). A nondestructive surface zwitterionization of polydimethylsiloxane for the improved human blood-inert properties. ACS Appl. Bio Mater 2 (1), 39–48. doi:10.1021/acsabm.8b00212
Dolenšek, J., Rupnik, M. S., and Stožer, A. (2015). Structural similarities and differences between the human and the mouse pancreas. Islets 7 (1), e1024405. doi:10.1080/19382014.2015.1024405
Dornhof, J., Kieninger, J., Muralidharan, H., Maurer, J., Urban, G. A., and Weltin, A. (2022). Microfluidic organ-on-chip system for multi-analyte monitoring of metabolites in 3D cell cultures. Lab. Chip 22 (2), 225–239. doi:10.1039/d1lc00689d
Esch, M. B., Ueno, H., Applegate, D. R., and Shuler, M. L. (2016). Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab. Chip 16 (14), 2719–2729. doi:10.1039/c6lc00461j
Ferguson, B. S., Hoggarth, D. A., Maliniak, D., Ploense, K., White, R. J., Woodward, N., et al. (2013). Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci. Transl. Med. 5 (213), 213ra165. doi:10.1126/scitranslmed.3007095
Ghorbanpour, S. M., Richards, C., Pienaar, D., Sesperez, K., Aboulkheyr Es., H., Nikolic, V. N., et al. (2023). A placenta-on-a-chip model to determine the regulation of FKBPL and galectin-3 in preeclampsia. Cell Mol. Life Sci. 80 (2), 44. doi:10.1007/s00018-022-04648-w
Gijzen, L., Yousef Yengej, F. A., Schutgens, F., Vormann, M. K., Ammerlaan, C. M. E., Nicolas, A., et al. (2021). Culture and analysis of kidney tubuloids and perfused tubuloid cells-on-a-chip. Nat. Protoc. 16 (4), 2023–2050. doi:10.1038/s41596-020-00479-w
Gnecco, J. S., Ding, T., Smith, C., Lu, J., Bruner-Tran, K. L., and Osteen, K. G. (2019). Hemodynamic forces enhance decidualization via endothelial-derived prostaglandin E2 and prostacyclin in a microfluidic model of the human endometrium. Hum. Reprod. 34 (4), 702–714. doi:10.1093/humrep/dez003
Gnecco, J. S., Pensabene, V., Li, D. J., Ding, T., Hui, E. E., Bruner-Tran, K. L., et al. (2017). Compartmentalized culture of perivascular stroma and endothelial cells in a microfluidic model of the human endometrium. Ann. Biomed. Eng. 45 (7), 1758–1769. doi:10.1007/s10439-017-1797-5
Gokaltun, A., Kang, Y. B. A., Yarmush, M. L., Usta, O. B., and Asatekin, A. (2019). Simple surface modification of poly(dimethylsiloxane) via surface segregating smart polymers for biomicrofluidics. Sci. Rep. 9 (1), 7377. doi:10.1038/s41598-019-43625-5
Gokaltun, A., Yarmush, M. L., Asatekin, A., and Usta, O. B. (2017). Recent advances in nonbiofouling PDMS surface modification strategies applicable to microfluidic technology. Technol. (Singap World Sci.) 5 (1), 1–12. doi:10.1142/S2339547817300013
Gori, M., Simonelli, M. C., Giannitelli, S. M., Businaro, L., Trombetta, M., and Rainer, A. (2016). Investigating nonalcoholic fatty liver disease in a liver-on-a-chip microfluidic device. PLoS One 11 (7), e0159729. doi:10.1371/journal.pone.0159729
Gough, A., Vernetti, L., Bergenthal, L., Shun, T. Y., and Taylor, D. L. (2016). The microphysiology systems database for analyzing and modeling compound interactions with human and animal organ models. Appl Vitro Toxicol 2 (2), 103–117. doi:10.1089/aivt.2016.0011
Govindasamy, N., Long, H., Ranga, A., Trappmann, B., and Bedzhov, I. (2023). 3D biomimetic environment enabling ex utero trophoblast invasion and co-culture of embryos and somatic cells. Star. Protoc. 4 (3), 102456. doi:10.1016/j.xpro.2023.102456
Grivel, J. C., and Margolis, L. (2009). Use of human tissue explants to study human infectious agents. Nat. Protoc. 4 (2), 256–269. doi:10.1038/nprot.2008.245
Guengerich, F. P., and MacDonald, J. S. (2007). Applying mechanisms of chemical toxicity to predict drug safety. Chem. Res. Toxicol. 20 (3), 344–369. doi:10.1021/tx600260a
Haderspeck, J. C., Chuchuy, J., Kustermann, S., Liebau, S., and Loskill, P. (2019). Organ-on-a-chip technologies that can transform ophthalmic drug discovery and disease modeling. Expert Opin. Drug Discov. 14 (1), 47–57. doi:10.1080/17460441.2019.1551873
Han, J. J. (2023). FDA Modernization Act 2.0 allows for alternatives to animal testing. Artif. Organs 47 (3), 449–450. doi:10.1111/aor.14503
Hao, Z., Wang, Z., Li, Y., Zhu, Y., Wang, X., De Moraes, C. G., et al. (2018). Measurement of cytokine biomarkers using an aptamer-based affinity graphene nanosensor on a flexible substrate toward wearable applications. Nanoscale 10 (46), 21681–21688. doi:10.1039/c8nr04315a
Hawkins, J., Miao, X., Cui, W., and Sun, Y. (2022). Surface functionalization of poly(dimethylsiloxane) substrates facilitates culture of pre-implantation mouse embryos by blocking non-selective adsorption. J. R. Soc. Interface 19 (189), 20210929. doi:10.1098/rsif.2021.0929
Henry, O. Y. F., Villenave, R., Cronce, M. J., Leineweber, W. D., Benz, M. A., and Ingber, D. E. (2017). Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab. Chip 17 (13), 2264–2271. doi:10.1039/c7lc00155j
Hsieh, H. L., Nath, P., and Huang, J. H. (2019). Multistep fluidic control network toward the automated generation of organ-on-a-chip. ACS Biomater. Sci. Eng. 5 (9), 4852–4860. doi:10.1021/acsbiomaterials.9b00912
Hughes, D. L., Hughes, A., Soonawalla, Z., Mukherjee, S., and O'Neill, E. (2021). Dynamic physiological culture of ex vivo human tissue: a systematic review. Cancers (Basel) 13 (12), 2870. doi:10.3390/cancers13122870
Huh, D. D. (2015). A human breathing lung-on-a-chip. Ann. Am. Thorac. Soc. 12 (Suppl. 1), S42–S44. doi:10.1513/AnnalsATS.201410-442MG
Illamola, S. M., Bucci-Rechtweg, C., Costantine, M. M., Tsilou, E., Sherwin, C. M., and Zajicek, A. (2018). Inclusion of pregnant and breastfeeding women in research - efforts and initiatives. Br. J. Clin. Pharmacol. 84 (2), 215–222. doi:10.1111/bcp.13438
Izadifar, Z., Charrez, B., Almeida, M., Robben, S., Pilobello, K., van der Graaf-Mas, J., et al. (2024). Organ chips with integrated multifunctional sensors enable continuous metabolic monitoring at controlled oxygen levels. Biosens. Bioelectron. 265, 116683. doi:10.1016/j.bios.2024.116683
Jastrzebska, E., Tomecka, E., and Jesion, I. (2016). Heart-on-a-chip based on stem cell biology. Biosens. Bioelectron. 75, 67–81. doi:10.1016/j.bios.2015.08.012
Jeong, S., Fuwad, A., Yoon, S., Jeon, T. J., and Kim, S. M. (2024). A microphysiological model to mimic the placental remodeling during early stage of pregnancy under hypoxia-induced trophoblast invasion. Biomimetics (Basel). 9 (5), 289. doi:10.3390/biomimetics9050289
Jeong, S., Kim, S., Buonocore, J., Park, J., Welsh, C. J., Li, J., et al. (2018). A three-dimensional arrayed microfluidic blood-brain barrier model with integrated electrical sensor array. IEEE Trans. Biomed. Eng. 65 (2), 431–439. doi:10.1109/TBME.2017.2773463
Jin, Z. H., Liu, Y. L., Fan, W. T., and Huang, W. H. (2020). Integrating flexible electrochemical sensor into microfluidic chip for simulating and monitoring vascular mechanotransduction. Small 16 (9), e1903204. doi:10.1002/smll.201903204
Jodat, Y. A., Kang, M. G., Kiaee, K., Kim, G. J., Martinez, A. F., Rosenkranz, A., et al. (2018). Human-derived organ-on-a-chip for personalized drug development. Curr. Pharm. Des. 24 (45), 5471–5486. doi:10.2174/1381612825666190308150055
Kammala, A., Benson, M., Ganguly, E., Radnaa, E., Kechichian, T., Richardson, L., et al. (2022). Fetal membranes contribute to drug transport across the feto-maternal interface utilizing the breast cancer resistance protein (BCRP). Life (Basel) 12 (2), 166. doi:10.3390/life12020166
Kammala, A. K., Richardson, L. S., Radnaa, E., Han, A., and Menon, R. (2023). Microfluidic technology and simulation models in studying pharmacokinetics during pregnancy. Front. Pharmacol. 14, 1241815. doi:10.3389/fphar.2023.1241815
Kim, S., Lam, P. Y., Richardson, L., Menon, R., and Han, A. (2024). A dynamic flow fetal membrane organ-on-a-chip system for modeling the effects of amniotic fluid motion. Res. Sq. doi:10.21203/rs.3.rs-4372328/v1
Kim, S., Richardson, L., Radnaa, E., Chen, Z., Rusyn, I., Menon, R., et al. (2022). Molecular mechanisms of environmental toxin cadmium at the feto-maternal interface investigated using an organ-on-chip (FMi-OOC) model. J. Hazard. Mater. 422, 126759. doi:10.1016/j.jhazmat.2021.126759
Ko, G., Jeon, T. J., and Kim, S. M. (2022). Trophoblast migration with different oxygen levels in a gel-patterned microfluidic system. Micromachines (Basel) 13 (12), 2216. doi:10.3390/mi13122216
Konar, D., Devarasetty, M., Yildiz, D. V., Atala, A., and Murphy, S. V. (2016). Lung-on-A-chip technologies for disease modeling and drug development. Biomed. Eng. Comput. Biol. 7 (Suppl. 1), BECB.S34252–27. doi:10.4137/becb.s34252
Kreuder, A. E., Bolaños-Rosales, A., Palmer, C., Thomas, A., Geiger, M. A., Lam, T., et al. (2020). Inspired by the human placenta: a novel 3D bioprinted membrane system to create barrier models. Sci. Rep. 10 (1), 15606. doi:10.1038/s41598-020-72559-6
Lee, J. H., Linden, C., Diaz, F. J., and Wong, P. K. (2021b). A reconfigurable microfluidic building block platform for high-throughput nonhormonal contraceptive screening, 72–75.
Lee, J. S., Romero, R., Han, Y. M., Kim, H. C., Kim, C. J., Hong, J. S., et al. (2016). Placenta-on-a-chip: a novel platform to study the biology of the human placenta. J. Matern. Fetal Neonatal Med. 29 (7), 1046–1054. doi:10.3109/14767058.2015.1038518
Lee, M., Park, J. W., Kim, D., Kwon, H., Cho, M. J., Lee, E. J., et al. (2021a). Viscous cervical environment-on-a-chip for selecting high-quality sperm from human semen. Biomedicines 9 (10), 1439. doi:10.3390/biomedicines9101439
Lintao, R. C. V., Richardson, L. S., Kammala, A. K., Chapa, J., Yunque-Yap, D. A., Khanipov, K., et al. (2024). PGRMC2 and HLA-G regulate immune homeostasis in a microphysiological model of human maternal-fetal membrane interface. Commun. Biol. 7 (1), 1041. doi:10.1038/s42003-024-06740-2
Liu, Y., Matharu, Z., Rahimian, A., and Revzin, A. (2015). Detecting multiple cell-secreted cytokines from the same aptamer-functionalized electrode. Biosens. Bioelectron. 64, 43–50. doi:10.1016/j.bios.2014.08.034
Lu, Y., Zhou, Q., and Xu, L. (2021). Non-invasive electrochemical biosensors for TNF-α cytokines detection in body fluids. Front. Bioeng. Biotechnol. 9, 701045. doi:10.3389/fbioe.2021.701045
Ma, C., Peng, Y., Li, H., and Chen, W. (2021). Organ-on-a-Chip: a new paradigm for drug development. Trends Pharmacol. Sci. 42 (2), 119–133. doi:10.1016/j.tips.2020.11.009
Mahajan, G., Doherty, E., To, T., Sutherland, A., Grant, J., Junaid, A., et al. (2022). Vaginal microbiome-host interactions modeled in a human vagina-on-a-chip. Microbiome 10 (1), 201. doi:10.1186/s40168-022-01400-1
Mandrycky, C. J., Howard, C. C., Rayner, S. G., Shin, Y. J., and Zheng, Y. (2021). Organ-on-a-chip systems for vascular biology. J. Mol. Cell Cardiol. 159, 1–13. doi:10.1016/j.yjmcc.2021.06.002
Mandt, D., Gruber, P., Markovic, M., Tromayer, M., Rothbauer, M., Kratz, S. R. A., et al. (2018). Fabrication of biomimetic placental barrier structures within a microfluidic device utilizing two-photon polymerization. Int. J. Bioprint 4 (2), 144. doi:10.18063/IJB.v4i2.144
Maoz, B. M., Herland, A., Henry, O. Y. F., Leineweber, W. D., Yadid, M., Doyle, J., et al. (2017). Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab. Chip 17 (13), 2294–2302. doi:10.1039/c7lc00412e
Maschmeyer, I., Lorenz, A. K., Schimek, K., Hasenberg, T., Ramme, A. P., Hübner, J., et al. (2015). A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab. Chip 15 (12), 2688–2699. doi:10.1039/c5lc00392j
Materne, E. M., Maschmeyer, I., Lorenz, A. K., Horland, R., Schimek, K. M. S., Busek, M., et al. (2015). The multi-organ chip-a microfluidic platform for long-term multi-tissue coculture. J. Vis. Exp. (98), e52526. doi:10.3791/52526
McCormack, S. A., and Best, B. M. (2014). Obstetric pharmacokinetic dosing studies are urgently needed. Front. Pediatr. 2, 9. doi:10.3389/fped.2014.00009
Menke, J., Roelandse, M., Ozyurt, B., Martone, M., and Bandrowski, A. (2020). The rigor and transparency index quality metric for assessing biological and medical science methods. iScience 23 (11), 101698. doi:10.1016/j.isci.2020.101698
Menon, R., Richardson, L., and Kammala, A. K. (2024). New approach methods on the bench side to accelerate clinical trials during pregnancy. Expert Opin. Drug Metab. Toxicol. 20 (7), 555–560. doi:10.1080/17425255.2024.2353944
Miranda, I., Souza, A., Sousa, P., Ribeiro, J., Castanheira, E. M. S., Lima, R., et al. (2021). Properties and applications of PDMS for biomedical engineering: a review. J. Funct. Biomater. 13 (1), 2. doi:10.3390/jfb13010002
Miura, S., Sato, K., Kato-Negishi, M., Teshima, T., and Takeuchi, S. (2015). Fluid shear triggers microvilli formation via mechanosensitive activation of TRPV6. Nat. Commun. 6, 8871. doi:10.1038/ncomms9871
Morandi, F., Airoldi, I., Faini, A., Horenstein, A., Malavasi, F., Matysiak, N., et al. (2023). Immune-regulatory properties carried by human amnion epithelial cells: focus on the role of HLA-G and adenosinergic ectoenzymes. Hum. Immunol. 84, 359–365. doi:10.1016/j.humimm.2023.04.006
Mosavati, B., Oleinikov, A., and Du, E. (2022). 3D microfluidics-assisted modeling of glucose transport in placental malaria. Sci. Rep. 12 (1), 15278. doi:10.1038/s41598-022-19422-y
Mosavati, B., Oleinikov, A. V., and Du, E. (2020). Development of an organ-on-a-chip-device for study of placental pathologies. Int. J. Mol. Sci. 21 (22), 8755. doi:10.3390/ijms21228755
Moya, A., Ortega-Ribera, M., Guimerà, X., Sowade, E., Zea, M., Illa, X., et al. (2018). Online oxygen monitoring using integrated inkjet-printed sensors in a liver-on-a-chip system. Lab. Chip 18 (14), 2023–2035. doi:10.1039/c8lc00456k
Nagashima, J. B., El Assal, R., Songsasen, N., and Demirci, U. (2018). Evaluation of an ovary-on-a-chip in large mammalian models: species specificity and influence of follicle isolation status. J. Tissue Eng. Regen. Med. 12 (4), e1926–e1935. doi:10.1002/term.2623
Novak, R., Ingram, M., Marquez, S., Das, D., Delahanty, A., Herland, A., et al. (2020a). Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 4 (4), 407–420. doi:10.1038/s41551-019-0497-x
Novak, R., Ingram, M., Marquez, S., Das, D., Delahanty, A., Herland, A., et al. (2020b). Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 4 (4), 407–420. doi:10.1038/s41551-019-0497-x
Paoli, R., and Samitier, J. (2016). Mimicking the kidney: a key role in organ-on-chip development. Micromachines (Basel) 7 (7), 126. doi:10.3390/mi7070126
Park, J. Y., Mani, S., Clair, G., Olson, H. M., Paurus, V. L., Ansong, C. K., et al. (2022). A microphysiological model of human trophoblast invasion during implantation. Nat. Commun. 13 (1), 1252. doi:10.1038/s41467-022-28663-4
Park, S. R., Kim, S. R., Lee, J. W., Park, C. H., Yu, W. J., Lee, S. J., et al. (2020). Development of a novel dual reproductive organ on a chip: recapitulating bidirectional endocrine crosstalk between the uterine endometrium and the ovary. Biofabrication. 10 (1), 015001. doi:10.1088/1758-5090/abbd29
Park, S. R., Kook, M. G., Kim, S. R., Lee, C. M., Lee, J. W., Park, J. K., et al. (2024). Development of a novel testis-on-a-chip that demonstrates reciprocal crosstalk between Sertoli and Leydig cells in testicular tissue. Exp. Mol. Med. 56 (7), 1591–1605. doi:10.1038/s12276-024-01258-3
Pemathilaka, R. L., Alimoradi, N., Reynolds, D. E., and Hashemi, N. N. (2022). Transport of maternally administered pharmaceutical agents across the placental barrier in vitro. ACS Appl. Bio Mater 5 (5), 2273–2284. doi:10.1021/acsabm.2c00121
Pemathilaka, R. L., Caplin, J. D., Aykar, S. S., Montazami, R., and Hashemi, N. N. (2019). Placenta-on-a-Chip: in vitro study of caffeine transport across placental barrier using liquid chromatography mass spectrometry. Glob. Chall. 3 (3), 1800112. doi:10.1002/gch2.201800112
Phan, D. T. T., Wang, X., Craver, B. M., Sobrino, A., Zhao, D., Chen, J. C., et al. (2017). A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab. Chip 17 (3), 511–520. doi:10.1039/c6lc01422d
Rabussier, G., Bunter, I., Bouwhuis, J., Soragni, C., van Zijp, T., Ng, C. P., et al. (2023a). Healthy and diseased placental barrier on-a-chip models suitable for standardized studies. Acta Biomater. 164, 363–376. doi:10.1016/j.actbio.2023.04.033
Rabussier, G., Bünter, I., Bouwhuis, J., Soragni, C., van Zijp, T., Ng, C. P., et al. (2023b). Healthy and diseased placental barrier on-a-chip models suitable for standardized studies. Acta Biomater. 164, 363–376. doi:10.1016/j.actbio.2023.04.033
Radnaa, E., Richardson, L. S., Sheller-Miller, S., Baljinnyam, T., de Castro Silva, M., Kumar Kammala, A., et al. (2021). Extracellular vesicle mediated feto-maternal HMGB1 signaling induces preterm birth. Lab. Chip 21 (10), 1956–1973. doi:10.1039/d0lc01323d
Ren, K., Zhao, Y., Su, J., Ryan, D., and Wu, H. (2010). Convenient method for modifying poly(dimethylsiloxane) to be airtight and resistive against absorption of small molecules. Anal. Chem. 82 (14), 5965–5971. doi:10.1021/ac100830t
Renous, N., Kiri, M. D., Barnea, R. A., Rauti, R., Leichtmann-Bardoogo, Y., and Maoz, B. M. (2021). Spatial trans-epithelial electrical resistance (S-TEER) integrated in organs-on-chips. Lab. Chip 22 (1), 71–79. doi:10.1039/d1lc00789k
Ribas, J., Sadeghi, H., Manbachi, A., Leijten, J., Brinegar, K., Zhang, Y. S., et al. (2016). Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl Vitro Toxicol 2 (2), 82–96. doi:10.1089/aivt.2016.0002
Richardson, L., Gnecco, J., Ding, T., Osteen, K., Rogers, L. M., Aronoff, D. M., et al. (2019c). Fetal membrane organ-on-chip: an innovative approach to study cellular interactions. Reprod. Sci. 27, 1562–1569. doi:10.1007/s43032-020-00184-9
Richardson, L., Gnecco, J., Ding, T., Osteen, K., Rogers, L. M., Aronoff, D. M., et al. (2020a). Fetal membrane organ-on-chip: an innovative approach to study cellular interactions. Reprod. Sci. 27 (8), 1562–1569. doi:10.1007/s43032-020-00184-9
Richardson, L., Jeong, S., Kim, S., Han, A., and Menon, R. (2019a). Amnion membrane organ-on-chip: an innovative approach to study cellular interactions. FASEB J. 33 (8), 8945–8960. doi:10.1096/fj.201900020RR
Richardson, L., Jeong, S., Kim, S., Han, A., and Menon, R. (2019b). Amnion membrane organ-on-chip: an innovative approach to study cellular interactions. Faseb J. 33, 8945–8960. doi:10.1096/fj.201900020RR
Richardson, L., Radnaa, E., Lintao, R. C. V., Urrabaz-Garza, R., Maredia, R., Han, A., et al. (2023a). A microphysiological device to model the choriodecidual interface immune status during pregnancy. J. Immunol. 210 (9), 1437–1446. doi:10.4049/jimmunol.2200821
Richardson, L., Radnaa, E., Lintao, R. C. V., Urrabaz-Garza, R., Maredia, R., Han, A., et al. (2023c). A microphysiological device to model the choriodecidual interface immune status during pregnancy. J. Immunol. 210, 1437–1446. doi:10.4049/jimmunol.2200821
Richardson, L. S., Kammala, A. K., Kim, S., Lam, P. Y., Truong, N., Radnaa, E., et al. (2023b). Development of oxidative stress-associated disease models using feto-maternal interface organ-on-a-chip. Faseb J. 37 (7), e23000. doi:10.1096/fj.202300531R
Richardson, L. S., Kim, S., Han, A., and Menon, R. (2020b). Modeling ascending infection with a feto-maternal interface organ-on-chip. Lab. Chip 20, 4486–4501. doi:10.1039/d0lc00875c
Richardson, L. S., Radnaa, E., Urrabaz-Garza, R., Lavu, N., and Menon, R. (2020c). Stretch, scratch, and stress: suppressors and supporters of senescence in human fetal membranes. Placenta 99, 27–34. doi:10.1016/j.placenta.2020.07.013
Richardson, L. S., A, K. K., Costantine, M. M., Fortunato, S. J., Radnaa, E., Kim, S., et al. (2022). Testing of drugs using human feto-maternal interface organ-on-chips provide insights into pharmacokinetics and efficacy. Lab. Chip 22 (23), 4574–4592. doi:10.1039/d2lc00691j
Rogers, L. M., Anders, A. P., Doster, R. S., Gill, E. A., Gnecco, J. S., Holley, J. M., et al. (2018). Decidual stromal cell-derived PGE2 regulates macrophage responses to microbial threat. Am. J. Reprod. Immunol. 80 (4), e13032. doi:10.1111/aji.13032
Ruberte, J., Schofield, P. N., Sundberg, J. P., Rodriguez-Baeza, A., Carretero, A., and McKerlie, C. (2023). Bridging mouse and human anatomies; a knowledge-based approach to comparative anatomy for disease model phenotyping. Mamm. Genome 34 (3), 389–407. doi:10.1007/s00335-023-10005-4
Russell, C., Ward, A. C., Vezza, V., Hoskisson, P., Alcorn, D., Steenson, D. P., et al. (2019). Development of a needle shaped microelectrode for electrochemical detection of the sepsis biomarker interleukin-6 (IL-6) in real time. Biosens. Bioelectron. 126, 806–814. doi:10.1016/j.bios.2018.11.053
Safarzadeh, M., Richardson, L. S., Kammala, A. K., Mosebarger, A., Bettayeb, M., Kim, S., et al. (2024). A multi-organ, feto-maternal interface organ-on-chip, models pregnancy pathology and is a useful preclinical extracellular vesicle drug trial platform. Extracell. Vesicle 3doi, 100035. doi:10.1016/j.vesic.2024.100035
Saha, B., Mathur, T., Handley, K. F., Hu, W., Afshar-Kharghan, V., Sood, A. K., et al. (2020). OvCa-Chip microsystem recreates vascular endothelium-mediated platelet extravasation in ovarian cancer. Blood Adv. 4 (14), 3329–3342. doi:10.1182/bloodadvances.2020001632
Saha, B., Mathur, T., Tronolone, J. J., Chokshi, M., Lokhande, G. K., Selahi, A., et al. (2021). Human tumor microenvironment chip evaluates the consequences of platelet extravasation and combinatorial antitumor-antiplatelet therapy in ovarian cancer. Sci. Adv. 7 (30), eabg5283. doi:10.1126/sciadv.abg5283
Sances, S., Ho, R., Vatine, G., West, D., Laperle, A., Meyer, A., et al. (2018). Human iPSC-derived endothelial cells and microengineered organ-chip enhance neuronal development. Stem Cell Rep. 10 (4), 1222–1236. doi:10.1016/j.stemcr.2018.02.012
Shakeri, A., Khan, S., and Didar, T. F. (2021). Conventional and emerging strategies for the fabrication and functionalization of PDMS-based microfluidic devices. Lab a Chip 21 (16), 3053–3075. doi:10.1039/d1lc00288k
Sheffield, J. S., Siegel, D., Mirochnick, M., Heine, R. P., Nguyen, C., Bergman, K. L., et al. (2014). Designing drug trials: considerations for pregnant women. Clin. Infect. Dis. 59 (Suppl. 7), S437–S444. doi:10.1093/cid/ciu709
Sheng, S. R., Wang, X. Y., Xu, H. Z., Zhu, G. Q., and Zhou, Y. F. (2010). Anatomy of large animal spines and its comparison to the human spine: a systematic review. Eur. Spine J. 19 (1), 46–56. doi:10.1007/s00586-009-1192-5
Shojaei, S., Ali, M. S., Suresh, M., Upreti, T., Mogourian, V., Helewa, M., et al. (2021). Dynamic placenta-on-a-chip model for fetal risk assessment of nanoparticles intended to treat pregnancy-associated diseases. Biochim. Biophys. Acta Mol. Basis Dis. 1867 (7), 166131. doi:10.1016/j.bbadis.2021.166131
Shrestha, J., Razavi Bazaz, S., Aboulkheyr Es, H., Yaghobian Azari, D., Thierry, B., Ebrahimi Warkiani, M., et al. (2020). Lung-on-a-chip: the future of respiratory disease models and pharmacological studies. Crit. Rev. Biotechnol. 40 (2), 213–230. doi:10.1080/07388551.2019.1710458
Soncin, F., Khater, M., To, C., Pizzo, D., Farah, O., Wakeland, A., et al. (2018). Comparative analysis of mouse and human placentae across gestation reveals species-specific regulators of placental development. Development 145 (2), dev156273. doi:10.1242/dev.156273
Soncin, F., Natale, D., and Parast, M. M. (2015). Signaling pathways in mouse and human trophoblast differentiation: a comparative review. Cell Mol. Life Sci. 72 (7), 1291–1302. doi:10.1007/s00018-014-1794-x
Sonmez, U. M., Coyle, S., Taylor, R. E., and LeDuc, P. R. (2020). Polycarbonate heat molding for soft lithography. Small 16 (16), e2000241. doi:10.1002/smll.202000241
Stokes, W. S. (2015). Animals and the 3Rs in toxicology research and testing: the way forward. Hum. Exp. Toxicol. 34 (12), 1297–1303. doi:10.1177/0960327115598410
Takeuchi, M., Nagasaka, K., Yoshida, M., Kawata, Y., Miyagawa, Y., Tago, S., et al. (2019). On-chip immunofluorescence analysis of single cervical cells using an electroactive microwell array with barrier for cervical screening. Biomicrofluidics 13 (4), 044107. doi:10.1063/1.5089796
Tantengco, O. A. G., Richardson, L. S., Medina, P. M. B., Han, A., and Menon, R. (2021). Organ-on-chip of the cervical epithelial layer: a platform to study normal and pathological cellular remodeling of the cervix. FASEB J. 35 (4), e21463. doi:10.1096/fj.202002590RRR
Tantengco, O. A. G., Richardson, L. S., Radnaa, E., Kammala, A. K., Kim, S., Medina, P. M. B., et al. (2022a). Modeling ascending Ureaplasma parvum infection through the female reproductive tract using vagina-cervix-decidua-organ-on-a-chip and feto-maternal interface-organ-on-a-chip. FASEB J. 36 (10), e22551. doi:10.1096/fj.202200872R
Tantengco, O. A. G., Richardson, L. S., Radnaa, E., Kammala, A. K., Kim, S., Medina, P. M. B., et al. (2022b). Exosomes from Ureaplasma parvum-infected ectocervical epithelial cells promote feto-maternal interface inflammation but are insufficient to cause preterm delivery. Front. Cell Dev. Biol. 10, 931609. doi:10.3389/fcell.2022.931609
Tantengco, O. A. G., Richardson, L. S., Radnaa, E., Kammala, A. K., Kim, S., Medina, P. M. B., et al. (2022c). Exosomes from Ureaplasma parvum-infected ectocervical epithelial cells promote feto-maternal interface inflammation but are insufficient to cause preterm delivery. Front. Cell Dev. Biol. 10, 931609. doi:10.3389/fcell.2022.931609
Tantengco, O. A. G., Richardson, L. S., Radnaa, E., Kammala, A. K., Kim, S., Medina, P. M. B., et al. (2022d). Exosomes from Ureaplasma parvum-infected ectocervical epithelial cells promote feto-maternal interface inflammation but are insufficient to cause preterm delivery. Front. Cell Dev. Biol. 10, 931609. doi:10.3389/fcell.2022.931609
Tantengco, O., Richardson, L., Radnaa, E., Han, A., Medina, P., Menon, R., et al. (2022e). Cervix-on-a-chip for investigating ascending Ureaplasma parvum infection in pregnancy. Am. J. Obstetrics Gynecol. 226 (1), S12, S13. doi:10.1016/j.ajog.2021.11.068
Tonzani, S., and Fiorani, S. (2021). The STAR Methods way towards reproducibility and open science. iScience 4, 102137. doi:10.1016/j.isci.2021.102137
van der Helm, M. W., Odijk, M., Frimat, J. P., van der Meer, A. D., Eijkel, J. C., van den Berg, A., et al. (2016b). Direct quantification of transendothelial electrical resistance in organs-on-chips. Biosens. Bioelectron. 85, 924–929. doi:10.1016/j.bios.2016.06.014
van der Helm, M. W., van der Meer, A. D., Eijkel, J. C., van den Berg, A., and Segerink, L. I. (2016a). Microfluidic organ-on-chip technology for blood-brain barrier research. Tissue Barriers 4 (1), e1142493. doi:10.1080/21688370.2016.1142493
Van Os, L., Engelhardt, B., and Guenat, O. T. (2023). Integration of immune cells in organs-on-chips: a tutorial. Front. Bioeng. Biotechnol. 11, 1191104. doi:10.3389/fbioe.2023.1191104
Vidal, M. S., Richardson, L. S., Kumar Kammala, A., Kim, S., Lam, P. Y., Cherukuri, R., et al. (2024). Endocrine-disrupting compounds and their impact on human placental function: evidence from placenta organ-on-chip studies. Lab. Chip 24 (6), 1727–1749. doi:10.1039/d3lc00998j
Wang, Y., Wang, L., Guo, Y., Zhu, Y., and Qin, J. (2018). Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system. RSC Adv. 8 (3), 1677–1685. doi:10.1039/c7ra11714k
Wang, Y. I., and Shuler, M. L. (2018). UniChip enables long-term recirculating unidirectional perfusion with gravity-driven flow for microphysiological systems. Lab. Chip 18 (17), 2563–2574. doi:10.1039/c8lc00394g
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10 (1), 57–63. doi:10.1038/nrg2484
Wilmer, M. J., Ng, C. P., Lanz, H. L., Vulto, P., Suter-Dick, L., and Masereeuw, R. (2016). Kidney-on-a-Chip technology for drug-induced nephrotoxicity screening. Trends Biotechnol. 34 (2), 156–170. doi:10.1016/j.tibtech.2015.11.001
Wu, Q., Liu, J., Wang, X., Feng, L., Wu, J., Zhu, X., et al. (2020). Organ-on-a-chip: recent breakthroughs and future prospects. Biomed. Eng. Online 19 (1), 9. doi:10.1186/s12938-020-0752-0
Xiao, S., Coppeta, J. R., Rogers, H. B., Isenberg, B. C., Zhu, J., Olalekan, S. A., et al. (2017). A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat. Commun. 8, 14584. doi:10.1038/ncomms14584
Xu, Y., Xie, X., Duan, Y., Wang, L., Cheng, Z., and Cheng, J. (2016). A review of impedance measurements of whole cells. Biosens. Bioelectron. 77, 824–836. doi:10.1016/j.bios.2015.10.027
Yin, F., Zhu, Y., Zhang, M., Yu, H., Chen, W., and Qin, J. (2019). A 3D human placenta-on-a-chip model to probe nanoparticle exposure at the placental barrier. Toxicol Vitro 54, 105–113. doi:10.1016/j.tiv.2018.08.014
Young, R. E., and Huh, D. D. (2021). Organ-on-a-chip technology for the study of the female reproductive system. Adv. Drug Deliv. Rev. 173, 461–478. doi:10.1016/j.addr.2021.03.010
Yu, S. X., Liu, Y., Wu, Y., Luo, H., Huang, R., Wang, Y. J., et al. (2022). Cervix chip mimicking cervical microenvironment for quantifying sperm locomotion. Biosens. Bioelectron. 204, 114040. doi:10.1016/j.bios.2022.114040
Zhang, Y. S., Aleman, J., Arneri, A., Bersini, S., Piraino, F., Shin, S. R., et al. (2015). From cardiac tissue engineering to heart-on-a-chip: beating challenges. Biomed. Mater 10 (3), 034006. doi:10.1088/1748-6041/10/3/034006
Zhang, Y. S., Aleman, J., Shin, S. R., Kilic, T., Kim, D., Mousavi Shaegh, S. A., et al. (2017). Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. U. S. A. 114 (12), E2293–E2302. doi:10.1073/pnas.1612906114
Zhu, Y., Wang, H., Yin, F., Guo, Y., Li, F., Gao, D., et al. (2020). Amnion-on-a-chip: modeling human amniotic development in mid-gestation from pluripotent stem cells. Lab. Chip 20 (17), 3258–3268. doi:10.1039/d0lc00268b
Keywords: organ-on-chip, pregnancy, reproduction, in-vitro tools, cell culture
Citation: Truong N, Zahra A, Lintao RCV, Chauhan R, Bento GF, Vidal Jr. M, Kim S, Lam PY, Conrads T, Conrads K, Han A, Menon R and Richardson LS (2025) Modeling reproductive and pregnancy-associated tissues using organ-on-chip platforms: challenges, limitations, and the high throughput data frontier. Front. Bioeng. Biotechnol. 13:1568389. doi: 10.3389/fbioe.2025.1568389
Received: 29 January 2025; Accepted: 14 March 2025;
Published: 01 April 2025.
Edited by:
Stephanie J. Hachey, University of California, Irvine, United StatesReviewed by:
Christopher Long, Hesperos Inc., United StatesCopyright © 2025 Truong, Zahra, Lintao, Chauhan, Bento, Vidal Jr., Kim, Lam, Conrads, Conrads, Han, Menon and Richardson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lauren S. Richardson, bGVzdGFmZm9AdXRtYi5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.