
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 10 March 2025
Sec. Industrial Biotechnology
Volume 13 - 2025 | https://doi.org/10.3389/fbioe.2025.1564764
The degradation of hemicellulose, including xylan, is an important industrial process as it provides cheap and sustainable source of economically valuable monosaccharides. β-xylosidases are key enzymes required for complete degradation of xylan and are used in the production of monosaccharides, such as xylose. In this study, we characterized a novel, xylose-tolerant β-xylosidase isolated from Limosilactobacillus fermentum SK152. Sequence analysis and protein structure prediction revealed that the putative β-xylosidase belongs to the glycoside hydrolase (GH) family 43 subfamily 11 and exhibits high homology with other characterised GH43 β-xylosidases from fungal and bacterial sources. The putative β-xylosidase was named LfXyl43. The catalytic residues of LfXyl43, which are highly conserved among GH 43 β-xylosidases, were predicted. To fully characterise LfXyl43, the gene encoding it was heterologously expressed in Escherichia coli. Biochemical characterisation revealed that the recombinant LfXyl43 (rLfXyl43) was active against artificial and natural substrates containing β-1,4-xylanopyranosyl residues, such as p-nitrophenyl-β-D-xylopyranoside (pNPX) and oNPX. Moreover, it demonstrated weak α-L-arabinofuranosidase activity. The optimal activity of rLfXyl43 was obtained at pH 7.0 at 35°C. rLfXyl43 could degrade xylo-oligosaccharides, such as xylobiose, xylotriose, and xylotetraose, and showed hydrolysing activity towards beechwood xylan. Moreover, rLfXyl43 demonstrated synergy with a commercial xylanase in degrading rye and wheat arabinoxylan. The activity of rLfXyl43 was not affected by the addition of metal ions, chemical reagents, or high concentrations of NaCl. Notably, rLfXyl43 exhibited tolerance to high xylose concentrations, with a Ki value of 100.1, comparable to that of other xylose-tolerant GH 43 β-xylosidases. To our knowledge, this is the first β-xylosidase identified from a lactic acid bacterium with high tolerance to salt and xylose. Overall, rLfXyl43 exhibits great potential as a novel β-xylosidase for use in the degradation of lignocellulosic material, especially xylan hemicellulose. Its high activity against xylo-oligosaccharides, mild catalytic conditions, and tolerance to high xylose concentrations makes it a suitable enzyme for industrial applications.
Xylans are hemicelluloses with a β-D-xylanopyranosyl backbone, linked either via a β-1,4- or β-1,3-glycosidic bond (Li et al., 2023). Xylans with β-1,3-glycosidic bonds are found in some chlorophyte and red algae, while β-1,4-xylanopyranosyl backbone is the most predominant hemicelluloses in vascular plant cell walls (Moreira and Filho, 2016; Qaseem et al., 2025). Arabinose, glucuronic acid, ferulic acid, and p-coumaric acid are among the main side chains attached to the xylan backbone (Saha, 2003a; Moreira and Filho, 2016; Naidu et al., 2018; Li et al., 2023). Different plant sources contain different xylans, with varying types and degrees of side-chain substitutions (Saha, 2003a; Moreira and Filho, 2016; Naidu et al., 2018). Xylans are cheap and eco-friendly feedstock that are processed into valuable industrial and pharmacological products. Owing to the complexity of side-chain substitutions, complete degradation of xylans requires concerted activities of different enzymes including endo-β-1,4-xylanases, β-D-xylosidases, as well as debranching enzymes. Endo-xylanases hydrolyse the β-1,4 glycosidic bonds, freeing up xylo-oligosaccharides (XOS) of various lengths, whereas β-D-xylosidases attack these XOS to produce xylose as the end product (Rohman et al., 2019; Li et al., 2023). The activity β-D-xylosidases relieves the inhibitory effect of XOS on the endo-xylanase, resulting in more efficient breakdown of the xylan substrate. Xylose is an important product of xylan degradation, which can be converted to other valuable products, such as xylitol, ethanol, and lactate (Saha, 2003a). In addition, the degradation of xylan and XOS in the gastrointestinal tract via these enzymes has been reported to contribute to the intestinal health by promoting the production of beneficial short-chain fatty acids, such as butyrate, propionate, and acetate, by the commensal microbiome (Singh et al., 2015; Lin et al., 2016). This underscores the importance of xylan degradation, not only for industrial applications but also in promoting health.
β-Xylosidases (E.C. 3.2.1.37) are diverse enzymes varying in structures, hydrolytic activities, biochemical properties, and substrate preferences (Rohman et al., 2019; Li et al., 2023). According to the Carbohydrate-active Enzyme (CAZy) database, β-xylosidases are currently classified into 11 glycoside hydrolase (GH) families based on their structural properties: GH 1, 3, 5, 30, 39, 43, 51, 52, 54, 116, and 120. GH family 43 (GH 43) is the second largest GH family in the CAZy database, comprising not only of β-xylosidases but also other GH enzymes, such α-L-arabinofuranosidases, arabinases, and xylanases. The GH 43 family is further divided into 39 sub-families based on the structure of the C-terminal auxiliary domain (Rohman et al., 2019; Li et al., 2023). Unlike other β-xylosidases belonging to the other GH families, GH 43 β-xylosidases demonstrate an inverting mechanism (Rohman et al., 2019; Li et al., 2023). A number of GH 43 β-xylosidases and bi-functional β-xylosidases/α-L-arabinofuranosidases have been characterised from different bacterial and fungal sources. Aside from pH and thermal stability, robust nature, and high expression yield, which are valuable characteristics for industrial applications, some of these β-xylosidases have shown tolerance to high xylose concentrations. β-Xylosidases isolated from Bacteroides ovatus (Jordan et al., 2017), Thermobifida halotolerans YIM 90462T (Yin et al., 2019), Humicola insolens Y1 (Yang et al., 2014), and Enterobacter sp. (Ontañon et al., 2018) are some of the reported GH 43 β-xylosidases with exceptionally high tolerance to xylose.
Lactic acid bacteria (LAB) form a diverse group of bacteria, generally used in food production, and are beneficial members of the intestinal microbiota in animal and human hosts (Axelsson and Ahrné, 2000; Bintsis, 2018). LAB include various species from Lactobacillus, Pediococcus, Leuconostoc, Weisella, Oenococcus, and Streptococcus among other genera (Axelsson and Ahrné, 2000; König and Fröhlich, 2017). Owing to their ecological niches, LAB are valuable sources of GH enzymes used for the degradation of plant biomass, including xylans and XOS (Vasquez et al., 2024). In fact, LAB can carry single or multiple copies of GH enzymes, including GH 43 β-xylosidase. Previous studies have demonstrated the capability of LAB to utilize XOS as carbon source indicating the presence and importance of β-xylosidases in their metabolic activities (Ohara et al., 2006; Moura et al., 2007; Jang and Kim, 2011; Maria et al., 2014; Iliev et al., 2020). However, β-xylosidases from LAB have been isolated and characterised in only a few studies. For instance, two β-xylosidases of the GH 43 family were identified, heterologously expressed, and characterised from Levilactobacillus brevis DSM 20054, and these exhibited activity towards XOS (Michlmayr et al., 2013). Falck et al. (2016) also reported a GH 43 β-xylosidase from Weisella sp. 92 that can hydrolyse short XOS. Similarly, a GH 43 β-xylosidase was identified from Lactobacillus rossiae, which could liberate xylose from XOS (Pontonio et al., 2016). Nevertheless, reports of xylose tolerant GH 43 β-xylosidases from LAB with xylan-degrading ability are scarce.
This study was aimed at investigating a putative GH 43 β-xylosidase, LfXyl43, identified from the genome of Limosilatobacillus fermentum SK152. We compared the molecular structure of LfXyl43 with those of other known GH 43 enzymes to elucidate its mechanism of action. Moreover, we heterologously expressed and characterised β-xylosidase, and determined its tolerance to xylose and salt. Finally, we explored the ability of this newly characterised β-xylosidase to degrade XOS and xylan.
Multiple sequence analysis of amino acid sequences of LfXyl43 and closely related GH 43 β-xylosidases/α-L-arabinofuranosidases was performed using UniProt Align (https://www.uniprot.org/align) after retrieving their sequences from the NCBI database (https://www.ncbi.nlm.nih.gov/). The phylogenetic tree was constructed using neighbour-joining method with Poisson correction in MEGA 11 (Tamura et al., 2021). Structural homology analysis and function prediction were performed using the Phyre 2.2 web server (Kelley et al., 2015). CAZy (https://www.cazy.org/) annotation and signal peptide identification was done via the dbCAN2 server (Zhang et al., 2018). The three-dimensional model of LfXyl43 was determined using AlphaFold2 (Jumper et al., 2021) via ColabFold (Mirdita et al., 2022). Protein–ligand docking was performed using CB-Dock2 (Liu et al., 2022), with p-nitrophenyl-β-D-xylopyranoside (pNPX; ChEBI ID 90148), xylobiose (ChEBI ID 28309), and xylotetraose (ChEBI ID 62972) used as ligands. Visualisation was performed using PyMol (ver. 3.0) (Schrödinger and DeLano, 2020).
All bacterial strains, plasmids, and primer sequences used in this study are summarised in Table 1. Lm. fermentum SK152 was cultured in Man Rogosa Sharpe (MRS) broth (BD Difco, United States) at 37°C, without aeration. For cloning and protein expression, E. coli DH5α and Escherichia coli BL21 (DE3) competent cells were used, respectively. E. coli strains were grown in Luria-Bertani (LB) broth (BD Difco, United States) supplemented with ampicillin (100 μg/mL) at 37°C, with aeration.
The genomic DNA of Lm. fermentum SK152 was extracted following the protocol described by Yoo et al. (2017) and used as the template for amplifying the β-xylosidase/α-L-arabinofuranosidase gene (Supplementary Figure S1), Lfxyl43, using the primers listed in Table 1. PCR was performed following the method described by Vasquez et al. (2022). For constructing the expression vector, the amplified Lfxyl43 gene was digested using the restriction enzymes NdeI and XhoI (TaKaRa, Tokyo, Japan), and then cloned into the pET21b (+) (Novagen) vector using T4 ligase (TaKaRa, Tokyo, Japan). The resulting expression vector with the Lfxyl43 gene was transformed into E. coli DH5α to verify the sequence, and into E. coli BL21 (DE3) for protein overexpression.
For expressing the 6× histidine-tagged recombinant LfXyl43 (rLfXyl43), transformed E. coli BL21 (DE3) was grown overnight in LB broth (with ampicillin, 100 μg/mL final concentration) at 37°C, with shaking. The culture was then diluted at 1:100 in LB broth with ampicillin and allowed to grow to an optical density at 600 nm (O.D.600) of 0.5 before induction with 0.1 mM isopropyl- β-D-thiogalactopyranoside (IPTG). After IPTG addition, the culture was incubated at 20°C for 12 h, with aeration. The cells were harvested by centrifugation (10,000 × g for 10 min) and the pellet, thus obtained, was washed twice with 1× phosphate-buffered saline (PBS). The cells were resuspended in 50 mM sodium phosphate buffer (pH 7.0) and sonicated for 6–7 cycles (10 s sonication, 15 s pause) on ice. Cell-free extract was collected by centrifugation (15,000 × g for 20 min at 4°C) and then filtered through a 0.22 µm filter to remove cell debris. For purifying the recombinant protein, the cell-free extract was applied to an equilibrated nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen, Hilden, Germany) for 1 h at 4°C. The protein was eluted with 50 mM sodium phosphate buffer containing 250 mM imidazole. The purified protein was dialysed against 50 mM sodium phosphate buffer with 20% glycerol (pH 7.0) using a dialysis membrane (30,000 MWCO). The concentration of purified protein was determined using the Bradford assay, with bovine serum albumin used as a standard. The purified proteins were stored at −20°C until further use. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE, 12%) was performed according to Vasquez et al. (2022) to verify expression and purification. The protein marker (MW: 10–245 kDa) used in this study was purchased from LPS Solution (Daejon, Republic of Korea).
For measuring the enzymatic activity of rLfXyl43, the method by Falck et al. (2016) was followed, with some modifications. Briefly, the standard enzyme assay was performed at 35°C for 5 min in 50 mM sodium phosphate buffer (pH 7.0) with 10 µg purified rLfXyl43 and 2 mM pNPX (Sigma, MO, United States). The reaction was stopped by adding an equal volume of 1 M Na2CO3. The release of p-nitrophenol was measured spectrophotometrically at 405 nm (SpectraMax, Molecular Diagnostics, United States). One unit of enzyme activity was defined as the amount of enzyme needed to release 1 μmol of p-nitrophenol per minute under the assay conditions. All assays were performed in triplicates.
The optimum pH for the β-xylosidase and α-L-arabinofuranosidase activity of rLfXyl43 was determined by performing the standard enzyme assay at different pH (2.0–10.0) (Supplementary Table S1). While the optimal temperature was determined by performing the standard enzyme assay at different temperatures (20°C–80°C). The substrates used were pNPX or p-nitrophenyl-α-L-arabinofuranoside (pNPAf; Sigma, MO, United States). The effect of pH on the activity of rLfXyl43 was assessed by pre-incubating the enzyme at different pH (2.0–10.0) in the absence of the substrate at 20°C for 1 h. The substrate was added after the pre-incubation, and then the standard assay was performed. The thermal stability of rLfXyl43 was measured by pre-incubating the enzyme (in 50 mM sodium phosphate buffer, pH 7.0) without the substrate at different temperatures (25, 30, 35, 40, and 45°C) for 0, 30, 60, 90, and 120 min. After the pre-incubation, the mixtures were cooled before adding the substrate. The standard enzyme assay was performed as described above.
The substrate specificity of rLfXyl43 was determined by performing the standard assay with different p-nitrophenyl or o-nitrophenyl substrates (2 mM), including pNPX, pNPAf, and o-nitrophenyl-β-D-xylopyranoside (oNPX; Sigma, MO, United States). All assays were performed in triplicates.
The activity of rLfXyl43 towards XOS, xylans, and arabinoxylan was measured by performing the standard assay using 2 mM xylobiose, xylotriose, or xylotetraose (Megazyme, Ireland) or 1% (w/v) beechwood xylan (Serva, Germany), rye or wheat arabinoxylan (Megazyme, Ireland) in 50 mM sodium phosphate buffer (pH 7.0). The reaction mixture was incubated at 35°C for 30 min. The release of reducing sugar was measured by adding an equal volume of 3, 5-dinitrosalycylic acid (DNSA), followed by boiling for 5 min (Miller, 1959). The reaction tubes were cooled down and the absorbance was measured at 540 nm. One unit of enzyme activity was defined as the amount of enzyme needed to release 1 µmol of reducing sugar equivalent to xylose per minute under the assay conditions. All assays were performed in triplicates.
The kinetic parameters (Km, Vmax, kcat, and kcat/Km) for rLfXyl43 were determined by performing the standard enzyme assay using different concentrations of pNPX, pNPAf, or XOS (1–10 mM). The kinetic parameters were calculated by fitting the non-linear regression to the Michaelis–Menten equation in GraphPad Prism for Windows (ver. 10.0). All assays were performed in triplicates.
The effect of metal ions and other chemical reagents on the activity of rLfXyl43 was determined by individually adding various metal ion solutions (K+, Ca2+, Fe3+, Mg2+, Mn2+, and Ni2+) or chemical reagents (EDTA, SDS, and DTT) to the reaction mixture at a final concentration of 1 or 10 mM. Standard assays were performed as previously described and the values were compared with those for the control without any addition.
The tolerance of rLfXyl43 to xylose and NaCl was evaluated according to Yang et al. (2014), with some modifications. Briefly, the enzyme was pre-incubated in increasing concentrations of xylose or NaCl (0–1,500 mM) at 35°C for 30 min before adding the substrate (2 mM pNPX). The standard enzyme assay was then performed to measure the residual activity. The competitive inhibitor constant, Ki, for xylose was then calculated and defined as the amount of xylose required to inhibit half of the β-xylosidase activity under the assay conditions.
The degree of synergy between rLfXyl43 and a commercial xylanase from Trichoderma viridae (Sigma, MO, United States) was determined by reacting 1 U/mL of rLfXyl43 alone or xylanase alone, or in combination with 1% (w/v) beechwood xylan, rye, or wheat arabinoxylan in 50 mM sodium phosphate buffer (pH 7.0). Each reaction mixture was incubated at 35°C for 30 min. The activity was measured as described above using the DNSA method. The degree of synergy was defined as the ratio of released xylose when the assay was performed with the two enzymes simultaneously to the sum of released xylose when the assay was performed with each enzyme alone. Thin layer chromatography (TLC) was performed by double ascending the samples on a silica gel F254 plates (Merck, MA, United States) using 1-butanol:acetic acid:water (2:1:1, v/v/v) as the mobile phase. The migration of hydrolysis products was detected by spraying diphenylamine in aniline:acetone:phosphoric acid solution (2:2:100:15, w/v/v/v) onto the plates, followed by heating until the bands appeared.
All data in this study were reported as mean ± standard deviation (SD) of triplicate experiments. The SD was less than 5%.
The Lfxyl43 gene, which contains 1,656 base pairs and encodes a 551-amino acid protein (Supplementary Figure S2), was identified from the genome of Lm. fermentum SK152 (GenBank Accession No. PQ818275). The gene does not contain any signal peptides, suggesting that the encoded protein is cytoplasmic. Annotation of the amino acid sequence using the CAZy database via dbCAN2 predicted that the encoded protein, LfXyl43, belongs to the GH 43 subfamily 11. Phylogenetic analysis (Figure 1A) with other GH 43 subfamily 11 members revealed that LfXyl43 had high sequence similarity (67.6%) with XynB1 from L. brevis ATCC 14869 compared to other enzymes belonging to the same subfamily. Preliminary analysis of the tertiary structure and function of the protein using the Phyre 2.2 server revealed high structural homology of LfXyl43 with GH 43 enzymes, including β-xylosidase from Bacillus halodurans C-125 (PDB ID: 1YRZ), B. subtilis (PDB ID: 1YIF), and Geobacillus stearothermophilus T-6 (PDB ID: 2EXH), among others. The GH 43 β-xylosidase from B. halodurans C-125 (PDB ID: 1YRZ) is reported to have three highly conserved catalytic residues: Asp16, Asp129, Glu188. MSA analysis showed that LfXyl43, together with other characterised β-xylosidases, contains these highly conserved residues (Figure 1B). The amino acid residues Pro249, Asp250, Ala251, Gly252, Arg299, Gly517, Glu518, Ile519, and Phe522 were predicted to have key roles in substrate binding.
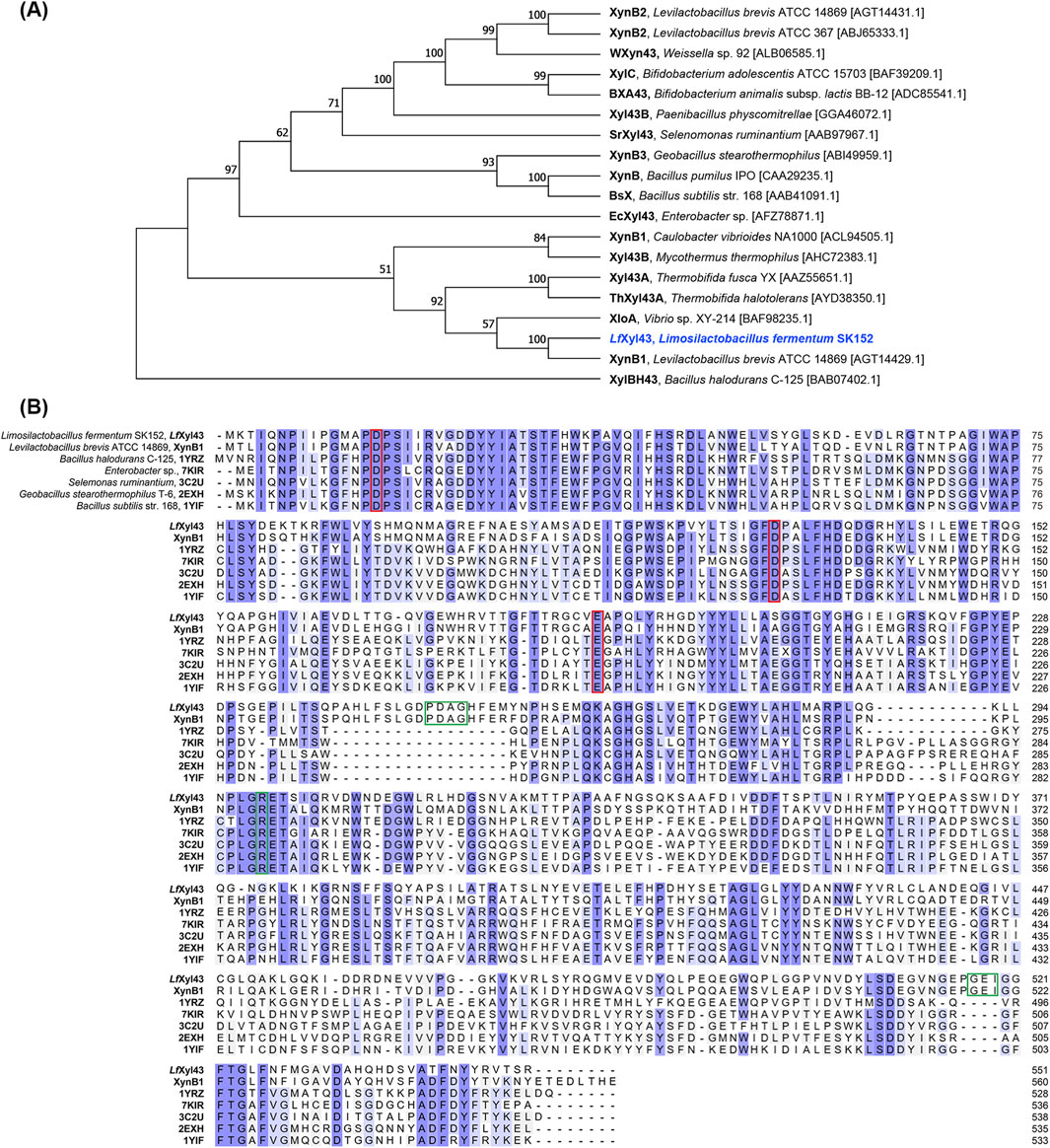
Figure 1. Sequence analysis of the β-xylosidase/α-L-arabinofuranosidase, LfXyl43, from Limosilactobacillus fermentum SK152. Phylogenetic tree showing the relationship of the amino acid sequence of LfXyl43 with those of other glycoside hydrolase (GH) 43 subfamily 11 enzymes (A). Reference sequences were retrieved from NCBI GenBank. The phylogenetic tree was built using neighbour-joining method with bootstrap analysis of 1,000 and Poisson correction method in MEGA 11. Bootstrap values (in %) are indicated at each node. Multiple sequence alignment of the amino acid sequence of LfXyl43 against those of other GH 43 β-xylosidases and α-L-arabinofuranosidases (B). Conserved sequences are shaded in blue. The putative catalytic residues are indicated with the red box, while the predicted contact residues are indicated in green box.
For further studying the molecular structure of the putative β-xylosidase LfXyl43, its three-dimensional structure was modelled using AlphFold2 via ColabFold. Alignment of the structure of LfXyl43 with that of XylBH43 of B. halodurans C-125 (PDB ID: 1YRZ) revealed 94% homology (Figure 2A). In addition, the tertiary structure of LfXyl43 showed an N-terminal five-bladed β-propeller domain (Ile4 to Trp313), a C-terminal β-sandwich domain (Asp346 to Val548), and a loop (Leu314 to Val345) connecting the two structural domains (Figure 2A). Moreover, the catalytic residues of 1YRZ aligned with Asp15, Asp129, and Glu188 (Figure 2B), suggesting that these residues are responsible for the catalytic activity of LfXyl43. The putative catalytic pocket was predicted using CB-Dock2 to be positioned within the N-terminal β-propeller domain, with a predicted volume of 1,090 Å3 (Figure 2C). The catalytic pocket was complexed with xylobiose to show subsites −1 and +1. Protein–ligand docking was also performed to predict the interaction of LfXyl43 with its potential ligands. Figures 2D–F show the predicted position of pNPX (Vina score: −8.2), xylobiose (Vina score: −6.9), and xylotetraose (Vina score: −7.9) relative to the putative catalytic residues and other contact residues.

Figure 2. Three-dimensional model of β-xylosidase/α-L-arabinofuranosidase, LfXyl43, from Limosilactobacillus fermentum SK152 and its interactions with its ligands. Alignment of the predicted tertiary structure of LfXyl43 (blue) with that of XylBH43 (gray) from Bacillus halodurans C-125 (PDB ID: 1YRZ) showing the N-terminal 5-bladed β-propeller and the auxiliary β-sandwich domain connected via the loop (A). Alignment of the catalytic residues of LfXyl43 (blue) and XylBH43 (gray) (B). Surface representation of LfXyl43 and the location of the catalytic pocket complexed with a ligand at sub-sites −1 and +1 (C). The putative catalytic residues Asp15, Asp129, and Asp188 and their predicted poses relative to the ligands: p-nitrophenyl-β-D-xylopyranoside (D), xylobiose (E), and xylotetraose (F). See the main text for the Vina score for each protein–ligand complex.
For further studying the activity of LfXyl43, the Lfxyl43 gene was successfully cloned into the pET21b expression vector using the restriction enzymes NdeI and XhoI to produce a recombinant protein with a 6× histidine tag at its C-terminus. After overexpression, the enzyme was expressed as soluble protein and found mostly in the cell-free extract of E. coli BL21 (DE3) (Figure 3). The crude protein from the cell-free extract of E. coli was then purified using Ni-NTA agarose resin and dialysed against a protein storage buffer. rLfXyl43 was highly expressed, with a total protein yield of 121.2 mg/L of the original culture (1,138 U). To confirm its size, the purified protein was loaded on a 12% SDS-PAGE gel. Based on the amino acid sequence, the protein with the 6× histidine tag was found to have an expected size of 62.5 kDa, which was consistent with the SDS-PAGE result showing a single band at around the expected size (Figure 3A). To determine the oligomeric state of rLfXyl43, the purified protein was loaded on a native PAGE gel (Figure 3B), which after electrophoresis showed multiple bands with sizes exceeding 62 kDa, indicating that rLfXyl43 may exist in dimeric, trimeric, or tetrameric form.
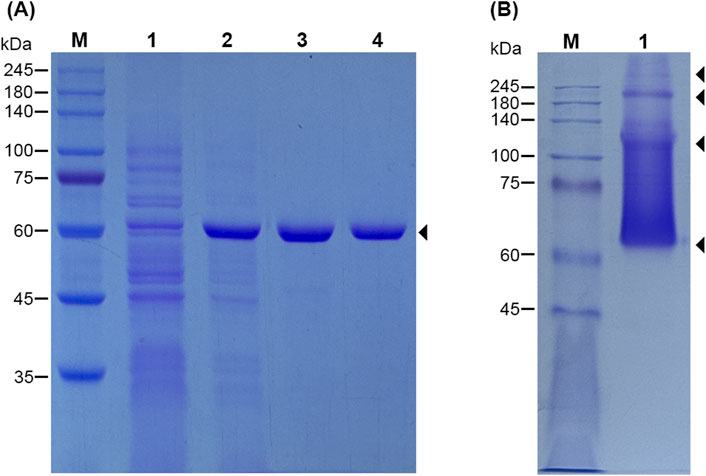
Figure 3. SDS-PAGE analysis of the recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152 (A). Lane M, protein marker (range 10–245 kDa); lane 1, cell-free extract of uninduced Escherichia coli BL21 (DE3); lane 2, cell-free extract of IPTG-induced E. coli BL21 (DE3) after overexpression for 12 h at 20°C; lane 3, Ni-NTA-purified rLfXyl43; lane 4, rLfXyl43 after dialysis. The protein size is 62.5 kDa (including the 6× histidine tag). Native-PAGE analysis rLfXyl43 showing possible oligomeric states of the protein (B). Lane M, protein marker; lane 1, purified rLfXyl43 in native PAGE loading buffer.
We performed enzyme assays for biochemical characterisation of rLfXyl43 (Figure 4). rLfXyl43 showed β-xylosidase and α-L-arabinofuranosidase activities over a pH range from 5.0 to 9.0, with the highest β-xylosidase activity observed between pH 7.0 and 8.0, and the highest α-L-arabinofuranosidase activity noted at pH 7.0 (Figure 4A). At the optimal pH (7.0), rLfXyl43 showed β-xylosidase activity from 20°C to 45°C, with maximum activity at 35°C (Figure 4B). Moreover, the α-L-arabinofuranosidase activity of rLfXyl43 was observed from 25°C to 45°C, with an optimum at 35°C (Figure 4B). The recombinant LfXyl43 showed no β-xylosidase or α-L-arabinofuranosidase activity at temperatures higher than 50°C. It showed residual activity after pre-incubation at pH 5.0 to 8.0 but was most stable at pH 7.0 (Figure 4C). The thermal stability of rLfXyl43 was also determined (Figure 4D). The enzyme retained approximately 90% of its activity after 120 min at 25, 30, and 35°C but gradually lost its activity to 40% after 120 min at 40°C. Finally, rLfXyl43 showed loss in residual activity after 30 min at 45°C.
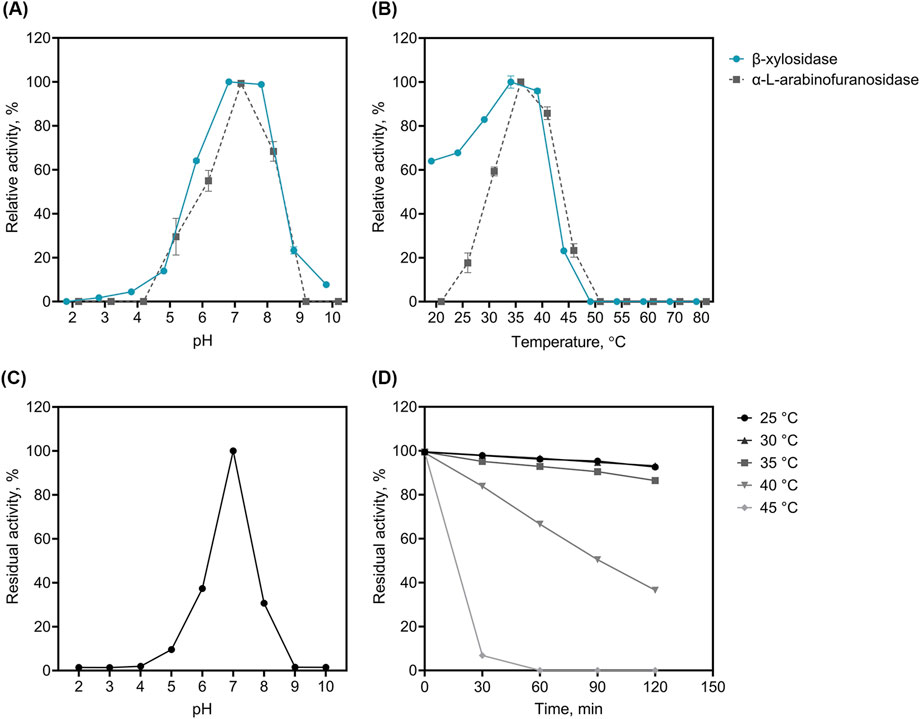
Figure 4. Biochemical characterization of the recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152. The optimal pH (A) and temperature (B) of rLfXyl43 determined using either p-nitrophenyl-β-D-xylopyranoside or p-nitrophenyl-α-L-arabinofuranoside. The stability of rLfXyl43 at different pH (C) was measured after 1 h incubation of the enzyme without the substrate in buffers with different pH. The thermal stability (D) of rLfXyl43 was determined by incubating the enzyme at different temperatures (25, 30, 35, 40, and 45°C) and the residual activity was measured at specific time points. Data are reported as the mean ± standard deviation of triplicate experiments. Some error bars smaller than the symbol are not shown.
The substrate specificities of rLfXyl43 were determined using different artificial substrates, XOS, xylans, and arabinoxylans (Table 2). rLfXyl43 showed high activity towards pNPX and oNPX, with specific activities of 9.4 and 3.7 U/mg, respectively. It also showed weak activity (0.7 U/mg) towards pNPAf, indicating that β-xylosidase is the dominant activity of rLfXyl43. By contrast, rLfXyl43 showed no activity against other artificial substrates. rLfXyl43 showed high hydrolytic activities towards xylobiose (35.8 U/mg), xylotriose (25.3 U/mg) and xylotetraose (30.2 U/mg). Conversely, the enzyme demonstrated weak activity towards xylans (0.7 U/mg for beechwood xylan) and no detectable activity towards rye or wheat arabinoxylans.
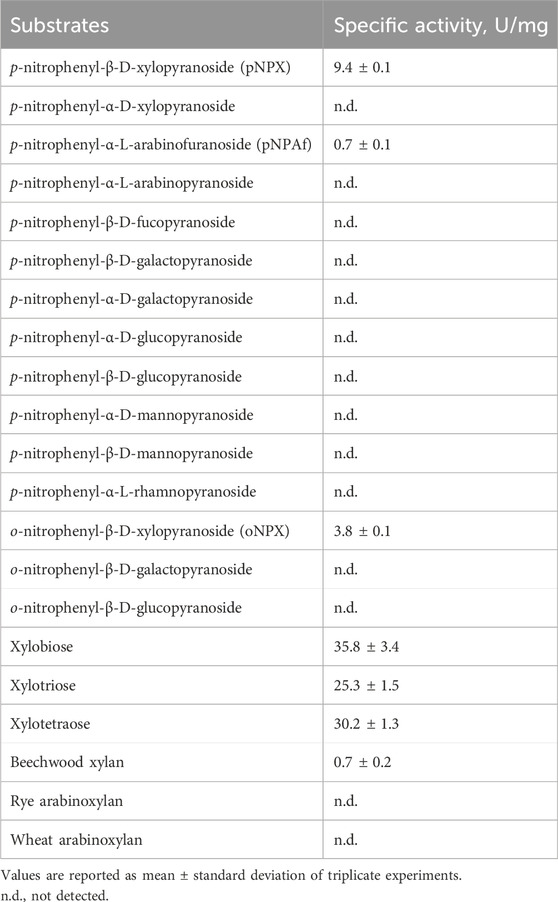
Table 2. Substrate specificities of recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152.
The kinetic constants for the hydrolysis of pNPX, pNPAf, and XOS were determined and are summarized in Table 3. The apparent Km for the artificial substrates pNPX and pNPAf was 2.0 and 1.5 mM and Vmax was 19.7 and 0.4 μmol min−1 mg−1, respectively. The turnover rate (kcat) for pNPX and pNPAf was 141.8 and 2.8 s−1 and the catalytic efficiency (kcat/Km) was 7.2 and 1.9 s−1 mM−1, respectively, demonstrating higher catalytic efficiency with pNPX than with pNPAf. rLfXyl43 demonstrated increasing Km values towards longer XOS (xylobiose < xylotriose < xylotetraose), indicating higher affinity for XOS with higher degree of polymerization. Similarly, the enzyme also demonstrated higher turnover and catalytic efficiency towards xylotetraose (3,526 s−1 and 41.9 s−1 mM−1) than towards xylobiose (3.2-fold) or xylotriose (1.6-fold).

Table 3. Kinetic parameters of recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152.
The effect of ions and other reagents on the activity of rLfXyl43 was also investigated (Table 4). The addition of 1 mM ions to the reaction mixture had little effect on the β-xylosidase activity of rLfXyl43. The addition of Ca2+, Mn2+, Mg2+, and K+ increased the residual activity of rLfXyl43 slightly. At 10 mM, the presence of Ca2+, Fe3+, Mn2+, Mg2+, and K+ reduced the activity of rLfXyl43, whereas Ni2+ dramatically weakened the hydrolysis of pNPX to 68.6%. The presence of 1 or 10 mM EDTA and SDS did not drastically affect the activity of rLfXyl43 towards pNPX. However, DTT reduced its activity to 85.4% at 1 mM and to 72.9% at 10 mM.
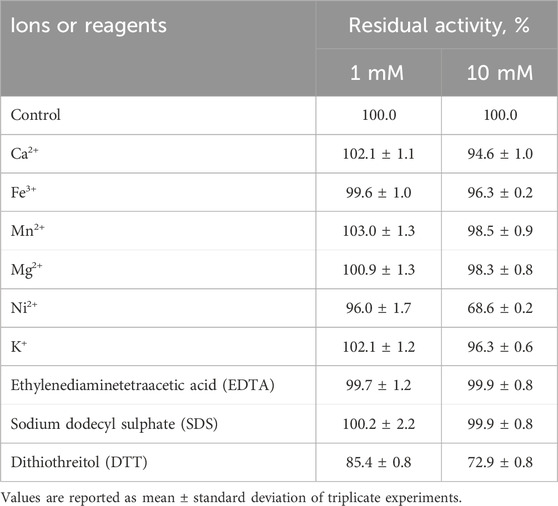
Table 4. Effect of metal ions and other reagents on the activity of recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152.
We determined the tolerance of rLfXyl43 to xylose (Figure 5A). At 100 mM xylose, rLfXyl43 retained 60% of its activity towards pNPX. At 500 mM xylose, rLfXyl43 still exhibited 20% activity relative to the control and eventually decreased to 6% at 1,500 mM xylose. The calculated Ki for xylose as a competitive inhibitor was 100.1 mM. Table 5 shows the reported Ki values for other GH 43 enzymes from bacterial or fungal sources. rLfXyl43 showed higher tolerance to xylose compared to the β-xylosidases/α-L-arabinofuranosidases from Lv. brevis ATCC 367, Fusarium proliferatum, B. ovatus, Thermobifida halotorelans, Thermomyces lanuginosus, H. insolens (Xyl43A), and Enterobacter sp. Conversely, β-xylosidases from Paecilomyces thermophila and H. insolens (Xyl43B) demonstrated better tolerance to xylose than did rLfXyl43. The tolerance of rLfXyl43 to NaCl was also determined (Figure 5B). Increasing concentration of NaCl did not affect the activity of rLfXyl43. The protein exhibited 100% activity at 100–1,000 mM NaCl, and 94% residual activity at 1,500 mM NaCl.
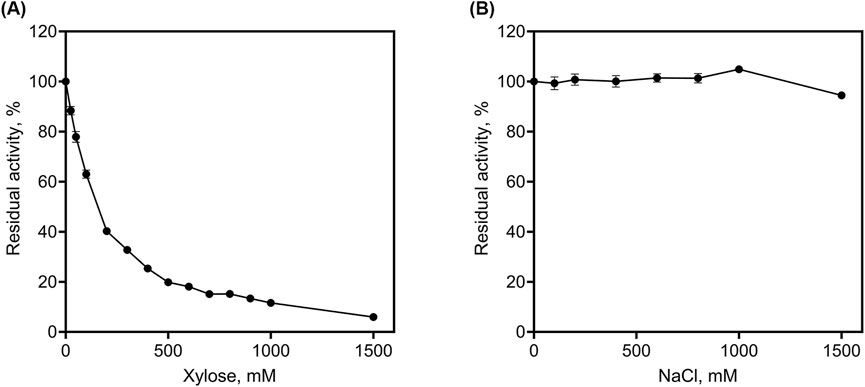
Figure 5. Determination of tolerance of the recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152 to xylose (A) and NaCl (B).
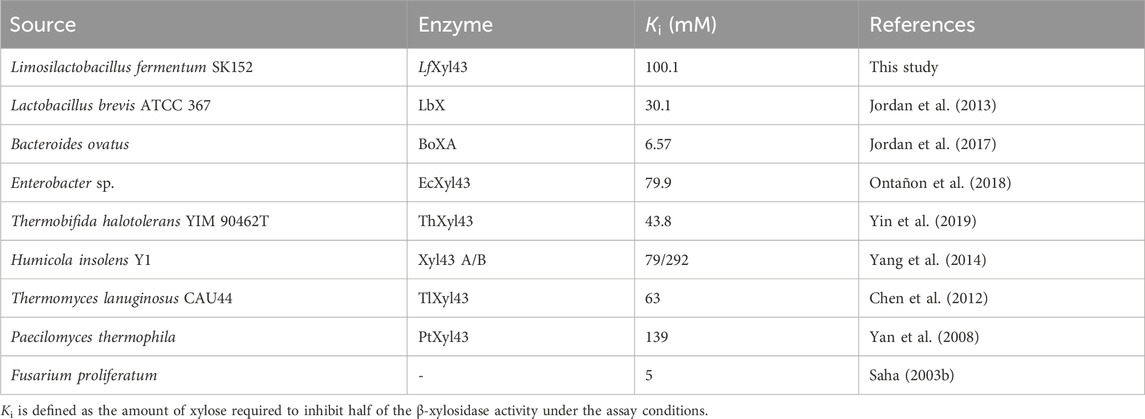
Table 5. Comparison of the xylose tolerance of recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152 with other characterized GH 43 enzymes with known Ki values.
The synergistic activity of rLfXyl43 with xylanases was investigated using a commercial endo-xylanase (Table 6). Equimolar amounts (1 U/mL each) of both rLfXyl43 and xylanase (Xyn) in simultaneous hydrolysis resulted in a modest degree of synergy towards beechwood xylan (1.33), rye arabinoxylan (1.2), and wheat arabinoxylan (1.2). TLC analysis (Figure 6) indicated the actions of individual enzymes against the substrates. For the three substrates, xylanase facilitated the endo-hydrolysis the xylan backbone producing shorter XOS, which were eventually hydrolysed by rLfXyl43 to produce xylose.
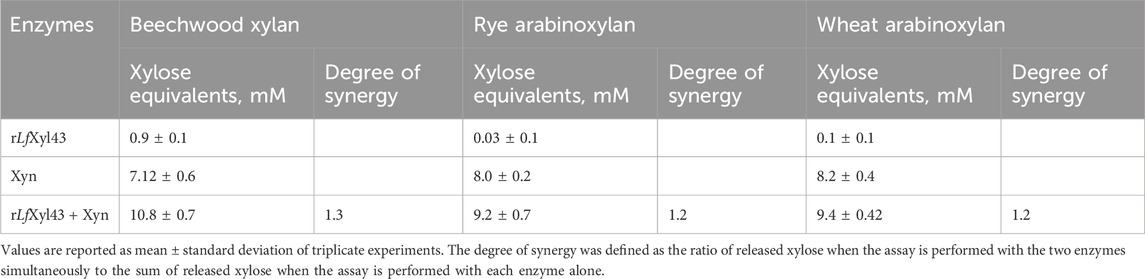
Table 6. Synergistic degradation of xylan and arabinoxylans by recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152, and a commercial xylanase (Xyn).
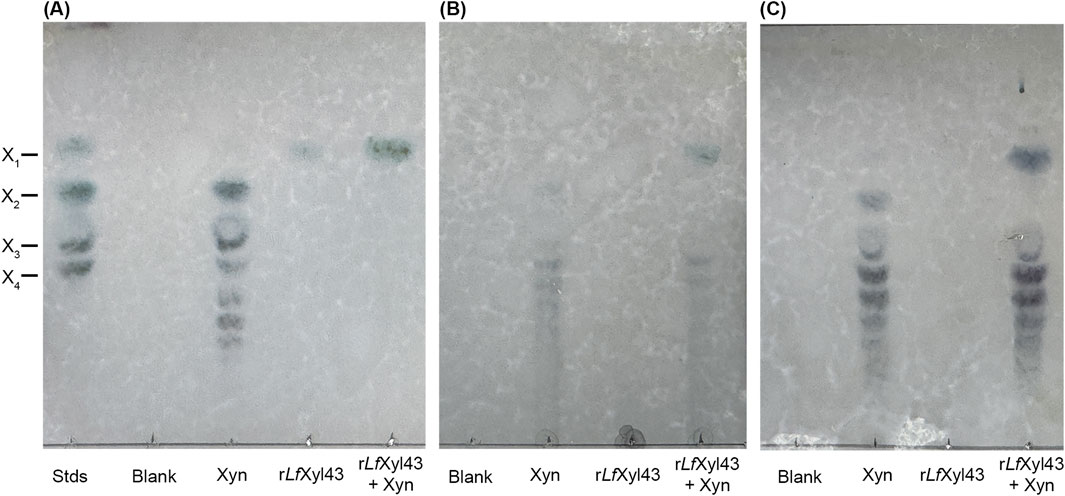
Figure 6. Synergism of the recombinant β-xylosidase/α-L-arabinofuranosidase, rLfXyl43, from Limosilactobacillus fermentum SK152 and a commercial xylanase. Thin layer chromatography analysis of the degradation of beechwood xylan (A), rye arabinoxylan (B), and wheat arabinoxylan (C) using 1 U/mL of rLfXyl43, commercial xylanase (Xyn), or both (simultaneous reaction). X1, xylose; X2, xylobiose; X3, xylotriose; and X4, xylotetraose.
Lm. fermentum is an obligate hetero-fermentative LAB, typically associated with plant, dairy, and meat fermentation (Naghmouchi et al., 2020). Many of the Lm. fermentum strains are probiotic LAB, with anti-microbial and immuno-modulatory properties (Mikelsaar and Zilmer, 2009; Zhao et al., 2019; Yoo et al., 2023). Together with other LAB genera, Lm. fermentum has been reported to utilise XOS potentially due to the presence of multiple enzymes, including β-xylosidase (Ohara et al., 2006; Moura et al., 2007; Jang and Kim, 2011; Madhukumar and Muralikrishna, 2012; Michlmayr et al., 2012). However, as of date, β-xylosidase from Lm. fermentum has not been reported or characterised. Herein, we have characterised a putative β-xylosidase identified from the genome of Lm. fermentum SK152 isolated from kimchi. Preliminary analysis showed that Lm. fermentum SK152 can degrade xylan (Supplementary Figure S3). Genome annotation of Lm. fermentum SK152 revealed the presence of several GH enzymes, potentially used for the degradation of plant materials during fermentation or in the gut (Yoo et al., 2017). Analysis of the protein sequence of the putative β-xylosidase revealed that it belongs to the GH 43 subfamily 11. Furthermore, it showed sequence homology with other GH 43 subfamily 11 β-xylosidases, most notably with XynB1 of Lv. brevis ATCC 14869. β-Xylosidases in the GH 43 subfamily 11 are characterised by an N-terminal five-bladed β-propeller domain, where the catalytic domain can be found, and an auxiliary domain of β-sandwich domain at the C-terminus. Ontañon et al. (2018) demonstrated that, although the function of the auxiliary C-terminal domain is unclear, it is critical for the activity of the enzyme as some residues participate in its catalytic activity. Indeed, some residues found at the C-terminus of the LfXyl43 (Gly517, Glu518, Ile519, and Phe522) were predicted to be critical contact residues between LfXyl43 and its ligand. Similarly, amino acids Pro249, Asp250, Ala251, and Gly252 may also act as binding residues. Notably, these residues were only found between LfXyl43 and XynB1 of Lv. brevis ATCC 14869. These additional residues may offer advantages in terms of substrate specificity or stability but should be empirically determined in future investigations. Meanwhile, GH 43 enzymes have three highly conserved residues critical for their inverting function: an Asp, which acts as a general nucleophile/base, a Glu residue, which acts as a proton donor/acid, and another Asp residue, which acts as a pKa modulator of the proton donor. When compared with other characterised GH 43 β-xylosidase, LfXyl43 exhibited high structural homology with the β-xylosidase of B. halodurans C-125 (1YRZ), confirming that the putative enzyme belongs to the GH 43 family of enzymes. Furthermore, the catalytic residues of GH 43 enzymes are well conserved among GH 43 enzymes, including LfXyl43. Thus, we propose that the Asp15 and Glu188 residues of LfXyl43 act as general base and acid, respectively, whereas the Asp129 residue is a pKa modulator. The results of protein–ligand studies support this hypothesis, as ligands, such as pNPX, xylobiose, and xylotetraose, interacted with Asp15 and Glu188 residues at the −1 subsite.
The oligomeric state of β-xylosidase is also critical for its activity. Most characterised GH 43 β-xylosidases are homo-dimeric, homo-trimeric, or homo-tetrameric. The auxiliary C-terminal domain is believed to have a role in the formation of multimers in this enzyme family (Falck et al., 2016; Ontañon et al., 2018). Due to its high structural homology with other GH 43 β-xylosidases, we predicted that LfXyl43 might also have a multimeric state. Its closest homologue, 1YRZ, is characterised as homo-dimeric, whereas most of the GH 43 β-xylosidases, such as 8UWS, 3C2U, 2EXH, and 1YIF, are characterised as homo-tetrameric. Native PAGE analysis revealed that the recombinant LfXyl43 exists largely as a monomer, but can also exist as dimer, trimer, or tetramer. This is consistent with other data showing that β-xylosidase can exists as monomer or multimers in solution (Falck et al., 2016). The oligomeric state of β-xylosidases affects its functionality (Corradini et al., 2021); however, in the case of LfXyl43, it remains to be investigated.
For examining the biochemical characteristics and substrate specificities of rLfXyl43, the Lfxyl43 gene was cloned and expressed in E. coli. The recombinant expression of LfXyl43 in the current study provided high enzyme yield and purity which is necessary for the characterization of the enzyme. The recombinant β-xylosidase showed high activity towards pNPX and oNPX, confirming its β-xylosidase activity. Activity towards oNPX among the GH 43 enzymes, especially those from LAB, has rarely been reported. rLfXyl43 also showed weak activity towards pNPAf, which indicates that it is a bi-functional enzyme. Other reported GH 43 enzymes have also demonstrated bi-functionality with higher activity as a β-xylosidase than as an α-L-arabinofuranosidase. For instance, XylBH43 from B. halodurans C-125, EcXyl43 from Enterobacter sp., BX43A from Bifidobacterium animalis BB-12, and XynB2 from Lv. brevis DSM 20054 exhibited higher catalytic efficiency towards pNPX than towards pNPAf (Liang et al., 2009; Michlmayr et al., 2013; Viborg et al., 2013; Ontañon et al., 2018). The bi-functionality of these enzymes may be attributable to the similarity in the spatial configuration of the −OH groups of D-xylose and L-arabinose (Liang et al., 2009). Regardless of the bi-functionality of rLfXyl43, its higher activity towards pNPX suggests that its major activity is as a β-xylosidase, rather than as a α-L-arabinofuranosidase. rLfXyl43 also exhibited high activity towards natural substrates, such as XOS and xylan. In fact, the steady-state kinetics of rLfXyl43 showed higher catalytic efficiency for XOS substrates than pNPX, with kcat/Km values increasing in the order xylobiose < xylotriose < xylotetraose. Although belonging to the same GH family, not all GH 43 β-xylosidases exhibit similar catalytic efficiency towards XOS. For example, WXyn43 from Weissella sp. 92 exhibits higher catalytic efficiency towards xylobiose and xylotriose than towards xylotetraose (Falck et al., 2016). Conversely, increasing catalytic efficiency towards longer XOS was observed for BoXA (from B. ovatus), PcAxy43B (from Paenibacillus curdanolyticus B-6), EcXyl43A, and BXA43, similar to that for rLfXyl43 (Viborg et al., 2013; Jordan et al., 2017; Ontañon et al., 2018; Limsakul et al., 2021). Although it remains unclear as to why such phenomenon exists among enzymes of the same GH family, the presence of additional contact residues not present in other GH 43 enzymes may provide additional catalytic subsites that can accommodate longer substrates (Ontañon et al., 2018).
The effects of pH and temperature on the β-xylosidase/α-L-arabinofuranosidase activity of rLfXyl43 were also investigated in the present study. The activity of rLfXyl43 was highest at pH 7.0. This is consistent with reports for other GH 43 β-xylosidases from fungal and bacterial sources, for which the optimal pH ranges from 6 to 8. Recombinant β-xylosidase from P. thermophila (PtXyl43), H. insolens Y1 (Xyl43B), T. halotolerans YIM 90462T (ThXyl43A), and Bacillus pumilus TCCC 11350 (Xyl) showed pH optima within this range (Yan et al., 2008; Yang et al., 2014; Liu et al., 2019; Yin et al., 2019). The optimal temperature for the activity of rLfXyl43 was found to be 35°C. rLfXyl43 also showed narrower pH and thermal stability compared with other GH 43 enzymes. The thermostability of rLfXyl43 was found to be much lower than that of other β-xylosidases (40°C–60°C) but is not uncommon among GH 43 β-xylosidases (Li et al., 2023). For example, β-xylosidases isolated from B. ovatus (BoXA) and B. pumilus TCCC 11350 (Xyl) exhibit optimal activity at 25°C and 30°C, respectively (Jordan et al., 2017; Liu et al., 2019). Cold-adapted enzymes are believed to form more flexible tertiary structures than those active at high temperatures (Liu et al., 2019; Li et al., 2023). The optimal activity of rLfXyl43 could potentially be related to niche adaptation to fermentation and gut environments, wherein the conditions are typically milder. This makes rLfXyl43 desirable for industrial degradation of lignocellulosic materials as it does not need high temperatures to retain its catalytic activity and would be suitable for a more sustainable and cost-effective process.
The effect of metal ions and other reagents on the activity of rLfXyl43 was also investigated. The activity of rLfXyl43 towards pNPX was slightly, but not dramatically, improved by the addition of Ca2+, Mn2+, and K+, and only at lower concentrations. At 10 mM, the addition of these ions, most notably by Ni2+, hindered the activity of rLfXyl43. Some GH 43 β-xylosidases are activated by the addition of divalent metal ions, such as Ca2+, Mg2+, and Mn2+. For instance, the β-xylosidases RS223-BX and RUM630-BX, isolated from cow rumen and wastewater metagenomic library, respectively, demonstrated enhanced catalytic activities in the presence of metal ions (Lee et al., 2013; Jordan et al., 2015). Moreover, a β-xylosidase from compost metagenome, CoXyl43, is also activated by Ca2+ ions (Matsuzawa et al., 2017). The β-xylosidase BoXA is also reported to be activated by Ca2+ (Jordan et al., 2017). These metal ion-activated β-xylosidases have metal-binding sites located at their catalytic N-termini. Whether LfXyl43 also has a metal-binding site similar to these β-xylosidases would need to be elucidated using crystallographic analysis.
During the hydrolysis of xylans by β-xylosidases, the concentration of monosaccharides, including xylose, increases which, in turn, inhibit the enzyme by blocking its catalytic pockets (Li et al., 2023). Despite this, some β-xylosidases showed exceptional tolerance to xylose. Bacterial β-xylosidases EcXyl43 from Enterobacter sp. and ThXyl43 from T. halotolerans are highly tolerant to xylose inhibition (Ki = 79.9 and 43.8, respectively) (Ontañon et al., 2018; Yin et al., 2019). β-xylosidases from fungal sources, such as Xyl43 A/B (from H. insolens), TlXyl43 (from T. lanuginosus), and PtXyl43 (from P. thermophila), also show high tolerance to xylose inhibition (Ki = 79/292, 63, and 139, respectively) (Yan et al., 2008; Chen et al., 2012; Yang et al., 2014). Notably, rLfXyl43 also demonstrated exceptional tolerance to xylose (Ki = 100.1). The Ki value of rLfXyl43 for xylose was found to be higher than that for previously reported β-xylosidases (except for Xyl43B and PtXyl43). Currently, there is limited data on xylose-tolerant β-xylosidases from LAB. The β-xylosidase from Lv. brevis ATCC 367, LbX, was found to have a Ki value of 30.1 for xylose (Jordan et al., 2013). In another study, Michlmayr et al. (2013) reported that XynB1 and XynB2 from Lv. brevis ATCC 14869 retained 80% and 33.8% of their respective activities at 100 mM xylose; however they did not report the Ki values for these enzymes. It has been shown that substrate affinity is directly linked to substrate inhibition. Previous studies have demonstrated that mutations in highly conserved residues Trp147 (in XylBH43 of B. halodurans C-125) and Trp145 (in SXA of S. ruminantium) to Gly147 and Gly145, respectively, resulted to lower substrate affinity and lower substrate inhibition (Fan et al., 2010; Wagschal et al., 2012). Although, the Trp147 residue of LfXyl43 is unaltered, neighbouring residues Glu146 and Glu148 (which are also present in XynB1, a potential xylose-tolerant β-xylosidase) could alter the interaction of the substrate at +1 subsite, decreasing its affinity. Moreover, to the best of our knowledge, no salt-tolerant β-xylosidase from LAB has been described as of date. The salt tolerance observed for rLfXyl43 may be due to the presence of highly negatively charged surface at the catalytic pocket (Supplementary Figure S4), which stabilizes binding of water molecules during protein-substrate interaction (Zhang et al., 2019; Wu et al., 2023; Cao et al., 2024). However, the tolerance mechanism of LfXyl43 towards salt and xylose must be validated through mutagenesis experiments. The tolerance of rLfXyl43 to high xylose and NaCl concentration offers promise for uses in industrial applications, mostly requiring enzymes from food-grade LAB.
Finally, in this study, we demonstrated that rLfXyl43 can hydrolyse xylans, specifically beechwood xylan, but not rye or wheat arabinoxylan. Although these xylans have similar 1,4-β-D-xylopyranoside backbones, rye and wheat arabinoxylan possess α-L-arabinofuranoside residues at either O-2 or O-3 positions unlike the 4-O-methylglucuronic acid substitution in beechwood xylan (Piro et al., 2023). Among the three xylans, rye is the most substituted, followed by wheat then beechwood–the differences in the degree of side-chain substitutions could influence the hydrolytic activity of xylanolytic enzymes (Rudjito et al., 2023). Despite exhibiting α-L-arabinofuranosidase activity, rLfXyl43 alone was not able to release xylose either from rye or wheat arabinoxylan indicating that rLfXyl43 cannot hydrolyse these linkages, which prohibits its access to the 1,4-β-D-xylopyranoside backbone. However, through synergistic activity with an endo-β-1,4-xylanase, rLfXyl43 contributed to the higher hydrolysis of these arabinoxylans to produce xylose (1.2-fold higher for both rye and wheat arabinoxylan). Several GH 43 β-xylosidases demonstrate synergy with xylanase. β-Xylosidase from B. pumilus TCCC 11350 exhibited a high degree of synergy with a xylanase during simultaneous degradation of beechwood xylan (Liu et al., 2019). Additionally, EcXyl43 also demonstrated cooperative activity with a xylanase in degrading xylan (Ontañon et al., 2018), comparable to the activity of rLfXyl43. This further underscores the potential applicability of rLfXyl43 in the degradation of hemicellulose for industrial purposes.
LfXyl43 is a β-xylosidase belonging to the GH 43 subfamily 11, isolated from Lm. fermentum SK152. It showed high homology with other GH 43 β-xylosidases. We found that rLfXyl43 exhibited high activity towards pNPX, XOS, and xylan, and weak α-L-arabinofuranosidase activity towards pNPAf. The optimal pH and temperature for the activity of rLfXyl43 were determined to be 7.0°C and 35°C, respectively. rLfXyl43 showed synergism with an endo-xylanase in degrading xylans and arabinoxylans. Finally, rLfXyl43 was found to be tolerant to high concentrations of NaCl and xylose, similar to other xylose-tolerant β-xylosidase reported previously. To our knowledge, LfXyl43 is the first salt and xylose-tolerant β-xylosidase characterised from this LAB genus. Overall, this new β-xylosidase is a potential candidate enzyme for the degradation of xylan hemicelluloses that could be exploited for industrial and biomedical applications.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
RV: Conceptualization, Formal Analysis, Investigation, Methodology, Software, Visualization, Writing–original draft. JS: Investigation, Writing–review and editing. JL: Investigation, Writing–review and editing. SK: Data Curation, Investigation, Writing–original draft. D-KK: Conceptualization, Funding acquisition, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agricultural Microbiome R&D Program for Advancing innovative technology Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (RS-2024-0040347740982119420101). It was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-RS-2023-00275307).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2025.1564764/full#supplementary-material
Axelsson, L., and Ahrné, S. (2000). “Lactic acid bacteria,” in Applied microbial systematics (Dordrecht: Springer Netherlands), 367–388. doi:10.1007/978-94-011-4020-1_13
Bintsis, T. (2018). Lactic acid bacteria: their applications in foods. J. Bacteriol. Mycol. Open Access 6. doi:10.15406/jbmoa.2018.06.00182
Cao, L., Lin, M., Ning, J., Meng, X., Pu, X., Zhang, R., et al. (2024). Critical roles of acidic residues in loop regions of the structural surface for the salt tolerance of a GH39 β-d-Xylosidase. J. Agric. Food Chem. 72, 5805–5815. doi:10.1021/acs.jafc.3c07957
Chen, Z., Jia, H., Yang, Y., Yan, Q., Jiang, Z., and Teng, C. (2012). Secretory expression of a β-xylosidase gene fromThermomyces lanuginosusinEscherichia coliand characterization of its recombinant enzyme: β-Xylosidase fromThermomyces lanuginosus. Lett. Appl. Microbiol. 55, 330–337. doi:10.1111/j.1472-765X.2012.03299.x
Corradini, F. A. S., Milessi, T. S., Gonçalves, V. M., Ruller, R., Sargo, C. R., Lopes, L. A., et al. (2021). High stabilization and hyperactivation of a recombinant β-xylosidase through immobilization strategies. Enzyme Microb. Technol. 145, 109725. doi:10.1016/j.enzmictec.2020.109725
Falck, P., Linares-Pastén, J. A., Adlercreutz, P., and Karlsson, E. N. (2016). Characterization of a family 43 β-xylosidase from the xylooligosaccharide utilizing putative probioticWeissellasp. strain 92. Glycobiology 26, 193–202. doi:10.1093/glycob/cwv092
Fan, Z., Yuan, L., Jordan, D. B., Wagschal, K., Heng, C., and Braker, J. D. (2010). Engineering lower inhibitor affinities in β-d-xylosidase. Appl. Microbiol. Biotechnol. 86, 1099–1113. doi:10.1007/s00253-009-2335-7
Iliev, I., Vasileva, T., Bivolarski, V., Momchilova, A., and Ivanova, I. (2020). Metabolic profiling of xylooligosaccharides by lactobacilli. Polym. (Basel) 12, 2387–2418. doi:10.3390/polym12102387
Jang, M. H., and Kim, M. D. (2011). β-1,4-Xylosidase activity of Leuconostoc lactic acid bacteria isolated from kimchi. Korean J. Food Sci. Technol. 43, 169–175. doi:10.9721/KJFST.2011.43.2.169
Jordan, D. B., Braker, J. D., Wagschal, K., Lee, C. C., Chan, V. J., Dubrovska, I., et al. (2015). X-Ray crystal structure of divalent metal-activated β-xylosidase, RS223BX. Appl. Biochem. Biotechnol. 177, 637–648. doi:10.1007/s12010-015-1767-z
Jordan, D. B., Stoller, J. R., Lee, C. C., Chan, V. J., and Wagschal, K. (2017). Biochemical characterization of a GH43 β-xylosidase from Bacteroides ovatus. Appl. Biochem. Biotechnol. 182, 250–260. doi:10.1007/s12010-016-2324-0
Jordan, D. B., Wagschal, K., Grigorescu, A. A., and Braker, J. D. (2013). Highly active β-xylosidases of glycoside hydrolase family 43 operating on natural and artificial substrates. Appl. Microbiol. Biotechnol. 97, 4415–4428. doi:10.1007/s00253-012-4475-4
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi:10.1038/nprot.2015.053
König, H., and Fröhlich, J. (2017). “Lactic acid bacteria,” in Biology of Microorganisms on grapes, in Must and in wine. Cham: Springer International Publishing, 3–41. doi:10.1007/978-3-319-60021-5_1
Lee, C. C., Braker, J. D., Grigorescu, A. A., Wagschal, K., and Jordan, D. B. (2013). Divalent metal activation of a GH43 β-xylosidase. Enzyme Microb. Technol. 52, 84–90. doi:10.1016/j.enzmictec.2012.10.010
Li, N., Zhang, R., Zhou, J., and Huang, Z. (2023). Structures, biochemical characteristics, and functions of β-xylosidases. J. Agric. Food Chem. 71, 7961–7976. doi:10.1021/acs.jafc.3c01425
Liang, Y., Li, X., Shin, H., Rachel, C. R., and Mao, Z. (2009). Expression and characterization of a xylosidase (Bxyl) from Bacillus halodurans C-125. Shengwu Gongcheng Xuebao/Chinese J. Biotechnol. 25, 1386–1393.
Limsakul, P., Phitsuwan, P., Waeonukul, R., Pason, P., Tachaapaikoon, C., Poomputsa, K., et al. (2021). A novel multifunctional arabinofuranosidase/endoxylanase/β-xylosidase GH43 enzyme from Paenibacillus curdlanolyticus B-6 and its synergistic action to produce arabinose and xylose from cereal arabinoxylan. Appl. Environ. Microbiol. 87, e0173021. doi:10.1128/AEM.01730-21
Lin, S.-H., Chou, L.-M., Chien, Y.-W., Chang, J.-S., and Lin, C.-I. (2016). Prebiotic effects of xylooligosaccharides on the improvement of microbiota balance in human subjects. Gastroenterol. Res. Pract. 2016, 1–6. doi:10.1155/2016/5789232
Liu, Y., Huang, L., Zheng, D., Xu, Z., Li, Y., Shao, S., et al. (2019). Biochemical characterization of a novel GH43 family β-xylosidase from Bacillus pumilus. Food Chem. 295, 653–661. doi:10.1016/j.foodchem.2019.05.163
Liu, Y., Yang, X., Gan, J., Chen, S., Xiao, Z.-X., and Cao, Y. (2022). CB-Dock2: improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 50, W159–W164. doi:10.1093/nar/gkac394
Madhukumar, M. S., and Muralikrishna, G. (2012). Fermentation of xylo-oligosaccharides obtained from wheat bran and Bengal gram husk by lactic acid bacteria and bifidobacteria. J. Food Sci. Technol. 49, 745–752. doi:10.1007/s13197-010-0226-7
Maria, A., Margarita, T., Iilia, I., and Iskra, I. (2014). Gene expression of enzymes involved in utilization of xylooligosaccharides by Lactobacillus strains. Biotechnol. Biotechnol. Equip. 28, 941–948. doi:10.1080/13102818.2014.948257
Matsuzawa, T., Kaneko, S., Kishine, N., Fujimoto, Z., and Yaoi, K. (2017). Crystal structure of metagenomic β-xylosidase/α-l-arabinofuranosidase activated by calcium. J. Biochem. 162, 173–181. doi:10.1093/jb/mvx012
Michlmayr, H., Eder, R., Kulbe, K. D., and Del Hierro, A. (2012). β-Glycosidase activities of Oenococcus oeni: current state of research and future challenges. Mittl. Klosterneubg 62, 87–96.
Michlmayr, H., Hell, J., Lorenz, C., Böhmdorfer, S., Rosenau, T., and Kneifel, W. (2013). Arabinoxylan oligosaccharide hydrolysis by family 43 and 51 glycosidases from Lactobacillus brevis DSM 20054. Appl. Environ. Microbiol. 79, 6747–6754. doi:10.1128/AEM.02130-13
Mikelsaar, M., and Zilmer, M. (2009). Lactobacillus fermentum ME-3 – an antimicrobial and antioxidative probiotic. Microb. Ecol. Health Dis. 21, 1–27. doi:10.1080/08910600902815561
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. doi:10.1021/ac60147a030
Mirdita, M., Schütze, K., Moriwaki, Y., Heo, L., Ovchinnikov, S., and Steinegger, M. (2022). ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682. doi:10.1038/s41592-022-01488-1
Moreira, L. R. S., and Filho, E. X. F. (2016). Insights into the mechanism of enzymatic hydrolysis of xylan. Appl. Microbiol. Biotechnol. 100, 5205–5214. doi:10.1007/s00253-016-7555-z
Moura, P., Barata, R., Carvalheiro, F., Gírio, F., Loureiro-Dias, M. C., and Esteves, M. P. (2007). In vitro fermentation of xylo-oligosaccharides from corn cobs autohydrolysis by Bifidobacterium and Lactobacillus strains. Lwt 40, 963–972. doi:10.1016/j.lwt.2006.07.013
Naghmouchi, K., Belguesmia, Y., Bendali, F., Spano, G., Seal, B. S., and Drider, D. (2020). Lactobacillus fermentum: a bacterial species with potential for food preservation and biomedical applications. Crit. Rev. Food Sci. Nutr. 60, 3387–3399. doi:10.1080/10408398.2019.1688250
Naidu, D. S., Hlangothi, S. P., and John, M. J. (2018). Bio-based products from xylan: a review. Carbohydr. Polym. 179, 28–41. doi:10.1016/j.carbpol.2017.09.064
Ohara, H., Owaki, M., and Sonomoto, K. (2006). Xylooligosaccharide fermentation with Leuconostoc lactis. J. Biosci. Bioeng. 101, 415–420. doi:10.1263/jbb.101.415
Ontañon, O. M., Ghio, S., Marrero Díaz de Villegas, R., Piccinni, F. E., Talia, P. M., Cerutti, M. L., et al. (2018). EcXyl43 β-xylosidase: molecular modeling, activity on natural and artificial substrates, and synergism with endoxylanases for lignocellulose deconstruction. Appl. Microbiol. Biotechnol. 102, 6959–6971. doi:10.1007/s00253-018-9138-7
Piro, M. C., Muylle, H., and Haesaert, G. (2023). Exploiting rye in wheat quality breeding: the case of arabinoxylan content. Plants 12, 737. doi:10.3390/plants12040737
Pontonio, E., Mahony, J., Di Cagno, R., O’Connell Motherway, M., Lugli, G. A., O’Callaghan, A., et al. (2016). Cloning, expression and characterization of a β-d-xylosidase from Lactobacillus rossiae DSM 15814T. Microb. Cell Fact. 15, 72. doi:10.1186/s12934-016-0473-z
Qaseem, M. F., Zhang, W., Dupree, P., and Wu, A.-M. (2025). Xylan structural diversity, biosynthesis, and functional regulation in plants. Int. J. Biol. Macromol. 291, 138866. doi:10.1016/j.ijbiomac.2024.138866
Rohman, A., Dijkstra, B. W., and Puspaningsih, N. N. T. (2019). β-Xylosidases: structural diversity, catalytic mechanism, and inhibition by monosaccharides. Int. J. Mol. Sci. 20, 5524. doi:10.3390/ijms20225524
Rudjito, R. C., Jiménez-Quero, A., Muñoz, M. D. C. C., Kuil, T., Olsson, L., Stringer, M. A., et al. (2023). Arabinoxylan source and xylanase specificity influence the production of oligosaccharides with prebiotic potential. Carbohydr. Polym. 320, 121233. doi:10.1016/j.carbpol.2023.121233
Saha, B. C. (2003a). Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 30, 279–291. doi:10.1007/s10295-003-0049-x
Saha, B. C. (2003b). Purification and properties of an extracellular β-xylosidase from a newly isolated Fusarium proliferatum. Bioresour. Technol. 90, 33–38. doi:10.1016/S0960-8524(03)00098-1
Schrödinger, L., and DeLano, W. (2020). The PyMOL molecular graphics system. Available at: http://www.pymol.org/pymol.
Singh, R. D., Banerjee, J., and Arora, A. (2015). Prebiotic potential of oligosaccharides: a focus on xylan derived oligosaccharides. Bioact. Carbohydrates Diet. Fibre 5, 19–30. doi:10.1016/j.bcdf.2014.11.003
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi:10.1093/molbev/msab120
Vasquez, R., Bagon, B. B., Song, J. H., Han, N. S., and Kang, D. K. (2022). A novel, non-GMO surface display in Limosilactobacillus fermentum mediated by cell surface hydrolase without anchor motif. BMC Microbiol. 22, 190. doi:10.1186/s12866-022-02608-9
Vasquez, R., Song, J. H., Park, Y. S., Paik, H. D., and Kang, D. K. (2024). Application of probiotic bacteria in ginsenoside bioconversion and enhancing its health-promoting benefits: a review. Food Sci. Biotechnol. doi:10.1007/s10068-024-01734-6
Viborg, A. H., Sørensen, K. I., Gilad, O., Steen-Jensen, D. B., Dilokpimol, A., Jacobsen, S., et al. (2013). Biochemical and kinetic characterisation of a novel xylooligosaccharide-upregulated GH43 β-d-xylosidase/α-l-arabinofuranosidase (BXA43) from the probiotic Bifidobacterium animalis subsp. lactis BB-12. Amb. Express 3, 56. doi:10.1186/2191-0855-3-56
Wagschal, K., Jordan, D. B., and Braker, J. D. (2012). Catalytic properties of β-d-xylosidase XylBH43 from Bacillus halodurans C-125 and mutant XylBH43-W147G. Process Biochem. 47, 366–372. doi:10.1016/j.procbio.2011.07.009
Wu, Y., Hu, J., Du, Y., Lu, G., Li, Y., Feng, Y., et al. (2023). Mechanistic insights into the halophilic xylosidase xylo-1 and its role in xylose production. J. Agric. Food Chem. 71, 15375–15387. doi:10.1021/acs.jafc.3c05045
Yan, Q. J., Wang, L., Jiang, Z. Q., Yang, S. Q., Zhu, H. F., and Li, L. T. (2008). A xylose-tolerant β-xylosidase from Paecilomyces thermophila: characterization and its co-action with the endogenous xylanase. Bioresour. Technol. 99, 5402–5410. doi:10.1016/j.biortech.2007.11.033
Yang, X., Shi, P., Huang, H., Luo, H., Wang, Y., Zhang, W., et al. (2014). Two xylose-tolerant GH43 bifunctional β-xylosidase/α-arabinosidases and one GH11 xylanase from Humicola insolens and their synergy in the degradation of xylan. Food Chem. 148, 381–387. doi:10.1016/j.foodchem.2013.10.062
Yin, Y.-R., Xian, W.-D., Han, M.-X., Zhou, E.-M., Liu, L., Alkhalifah, D. H. M., et al. (2019). Expression and characterisation of a pH and salt tolerant, thermostable and xylose tolerant recombinant GH43 β-xylosidase from Thermobifida halotolerans YIM 90462T for promoting hemicellulose degradation. Ant. Van Leeuwenhoek 112, 339–350. doi:10.1007/s10482-018-1161-2
Yoo, D., Bagon, B. B., Valeriano, V. D. V., Oh, J. K., Kim, H., Cho, S., et al. (2017). Complete genome analysis of Lactobacillus fermentum SK152 from kimchi reveals genes associated with its antimicrobial activity. FEMS Microbiol. Lett. 364. doi:10.1093/femsle/fnx185
Yoo, J. M., Song, J. H., Vasquez, R., Hwang, I.-C., Lee, J. S., and Kang, D.-K. (2023). Characterization of novel amylase-sensitive, anti-listerial class IId bacteriocin, agilicin C7 produced by ligilactobacillus agilis C7. Food Sci. Anim. Resour. 43, 625–638. doi:10.5851/kosfa.2023.e24
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi:10.1093/nar/gky418
Zhang, R., Li, N., Liu, Y., Han, X., Tu, T., Shen, J., et al. (2019). Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 β-xylosidase: activity and instability at high neutral salt concentrations. Food Chem. 301, 125266. doi:10.1016/j.foodchem.2019.125266
Keywords: beta-xylosidase, GH family 43, lactic acid bacteria, xylan degradation, xylose tolerant, xylo-oligosaccharide
Citation: Vasquez R, Song JH, Lee JS, Kim S and Kang D-K (2025) Heterologous expression and characterization of xylose-tolerant GH 43 family β-xylosidase/α-L-arabinofuranosidase from Limosilactobacillus fermentum and its application in xylan degradation. Front. Bioeng. Biotechnol. 13:1564764. doi: 10.3389/fbioe.2025.1564764
Received: 22 January 2025; Accepted: 24 February 2025;
Published: 10 March 2025.
Edited by:
Noha M. Mesbah, Suez Canal University, EgyptReviewed by:
Eleonora Campos, National Scientific and Technical Research Council (CONICET), ArgentinaCopyright © 2025 Vasquez, Song, Lee, Kim and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dae-Kyung Kang, ZGtrYW5nQGRhbmtvb2suYWMua3I=
†Present address: Sanghoon Kim, College of Pharmacy and Pharmaceutical Sciences, Florida A&M University, Tallahassee, FL, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.