
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 31 March 2025
Sec. Organoids and Organ-On-A-Chip
Volume 13 - 2025 | https://doi.org/10.3389/fbioe.2025.1505963
This article is part of the Research Topic Tissue Chip Platforms to Mimic Oral-Systemic Disease Crosstalk View all 3 articles
 Mauricio G. C. Sousa1,2,3
Mauricio G. C. Sousa1,2,3 Danielle S. K. Brasino4
Danielle S. K. Brasino4 Madeline Krieger2
Madeline Krieger2 Duygu A. Dindar2
Duygu A. Dindar2 Rebekka Duhen1,2
Rebekka Duhen1,2 Zhenzhen Zhang5
Zhenzhen Zhang5 Cristiane Miranda Franca1,2,3
Cristiane Miranda Franca1,2,3 Luiz E. Bertassoni1,2,3,5,6*
Luiz E. Bertassoni1,2,3,5,6*The tumor microbiota has emerged as a pivotal contributor to a variety of cancers, impacting disease development, progression, and therapeutic resistance. Due to the complexity of the tumor microenvironment, reproducing the interactions between the microbes, tumor cells, and the immune system remains a great challenge for both in vitro and in vivo studies. To this end, significant progress has been made toward leveraging tumor-on-a-chip model systems to replicate critical hallmarks of the native disease in vitro. These microfluidic platforms offer the ability to mimic essential components of the tumor microenvironment, including controllable fluid flow conditions, manipulable extracellular matrix dynamics, and intricate 3D multi-cellular communication. The primary objective of this review is to discuss recent challenges and advances in engineering host-microbiota and tumor interactions on-a-chip. Ultimately, overcoming these obstacles will help us gain deeper insights into tumor-microbe interactions and enhance avenues for developing more effective cancer therapies.
Although the contribution of the microbiome to the development of cancer has long been proposed, recent evidence has demonstrated that the microbiota plays multifaceted roles in cancer development, progression, and treatment (Fu et al., 2023; Zhao et al., 2023; Battaglia et al., 2024). These recent discoveries have sparked significant interest in understanding the type, localization, mechanisms of action, and interactions of different types of microorganisms with the tumor microenvironment (TME) and its components (Lythgoe et al., 2022; Li et al., 2024). Many tumor-resident microbes (TRMi) are opportunistic, facultative anaerobes and intracellular pathogens that induce DNA damage (Espinoza and Minami, 2018; Gu X. et al., 2020), produce genotoxins (Cao et al., 2022; Joo et al., 2024), and cause inflammation (Lu et al., 2024; Zhang et al., 2024). TRMi can inactivate anti-cancer drugs and modulate the immune system, leading to poorer prognoses for therapies (Jiang et al., 2023; Sfanos, 2023; Kang et al., 2024). The recent reports of the involvement of certain microorganisms with some tumors such as colorectal cancer (Zepeda-Rivera et al., 2024) has attracted the attention of health authorities and clinicians, urging elucidation of their roles in the induction of malignant processes, dissecting intra- and extracellular effects, and clinical outcomes.
A substantial body of work has demonstrated, through comprehensive 16S rRNA analysis on several solid tumor types (Nejman et al., 2020) and spatial profiling combined with RNA sequencing (Galeano Nino et al., 2022), that TRMi interact with cancer cells, immune cells, and the extracellular matrix. This leads to the recruitment of immune cells and transcriptional changes in cancer cells and cancer-associated fibroblasts (Galeano Nino et al., 2022; Narunsky-Haziza et al., 2022; Wong-Rolle et al., 2022; Zepeda-Rivera et al., 2024). However, elucidating the exact mechanistic events linking the presence of microorganisms and activity with cancer is challenging due to the need for reliable pre-clinical models that emulate patient conditions (Rodrigues et al., 2021;Vitale et al., 2024). In vivo models of host-microbiota interactions remain the most reliable tools for understanding cancer biology (Ladaycia et al., 2021; Rodrigues et al., 2021; Li G. et al., 2023), and are well-established in the literature. Nevertheless, these models cannot accurately capture the extensive heterogeneity of native microorganisms observed in the human microbiome, which is shaped by unique genetic and immune composition (Aizpurua et al., 2023; Cao et al., 2024). Furthermore, traditional cell culture and co-cultures of mammalian cells and microorganisms often inadequately represent the spatial organization and ratios inherent in the tumor microenvironment (Asghar et al., 2015; Kapalczynska et al., 2018).
Alternative models such as organotypic cultures that combine engineering and biology provide a real-time simulation of critical elements of the interactions between TRMi and the TME in vitro (Radhakrishnan et al., 2020; Neufeld et al., 2022). These systems include 3D extracellular matrices (Yang et al., 2024), vasculature (Zervantonakis et al., 2012), and the interplay among stromal, immune, and cancer cells with microorganisms (Jouybar et al., 2024; Shin et al., 2024). Tumors-on-a-chip, for instance, emerged as attractive in vitro models that can tackle several challenges associated with accurately replicating the unique properties of the tumor microenvironment (Sontheimer-Phelps et al., 2019; Maulana et al., 2024). These microdevices incorporate microfluidic channels and manipulable fluid flow to model multiple hallmarks of the disease, including shear stress (Ortega Quesada et al., 2024), diverse cell types, an extracellular matrix, control of hypoxic gradients, and metabolic exchange between microorganisms and mammalian cells (Shah et al., 2016; Thacker et al., 2020; Shin et al., 2024), among other features. These platforms may enable scientists to decipher the interfaces between TRMi and different phases of cancer development by modulating patient-derived tissues in chips and capturing critical biomechanical elements of the TME.
In this review, we describe opportunities and challenges associated with using tumor-on-a-chip models to investigate interactions between the TME and TRMi. We discuss the requirements for designing chips that represent the specificities of the TME and exemplify some applications of these platforms in tumors directly known to harbor microorganisms.
Despite using microfluidic devices to study tumor-microbe interactions represents a relatively new development, the relationship between microorganisms and healthy tissues has been modeled for at least 20 years (Liu and Zhu, 2005; Kim and Sung, 2024). In the organs on-a-chip field, the gut was one of the first targets for chip modeling (Huh et al., 2013), and gut-on-a-chip devices have become very popular among researchers (Kim and Sung, 2024). These models have been improving in incorporating characteristics of native tissues and their interfaces, facilitating the growth of native-like microbial species (Ballerini et al., 2025). As a result, they have significantly enhanced the diversity and heterogeneity of microbial models, enabling the inclusion of bacteria, fungi (Srinivasan et al., 2011), viruses (Bein et al., 2021), and parasites (Nikolaev et al., 2020). Such advancements are critical for studying microbial species that may directly or indirectly contribute to dysbiosis, inflammation, and tumor establishment (Figure 1).
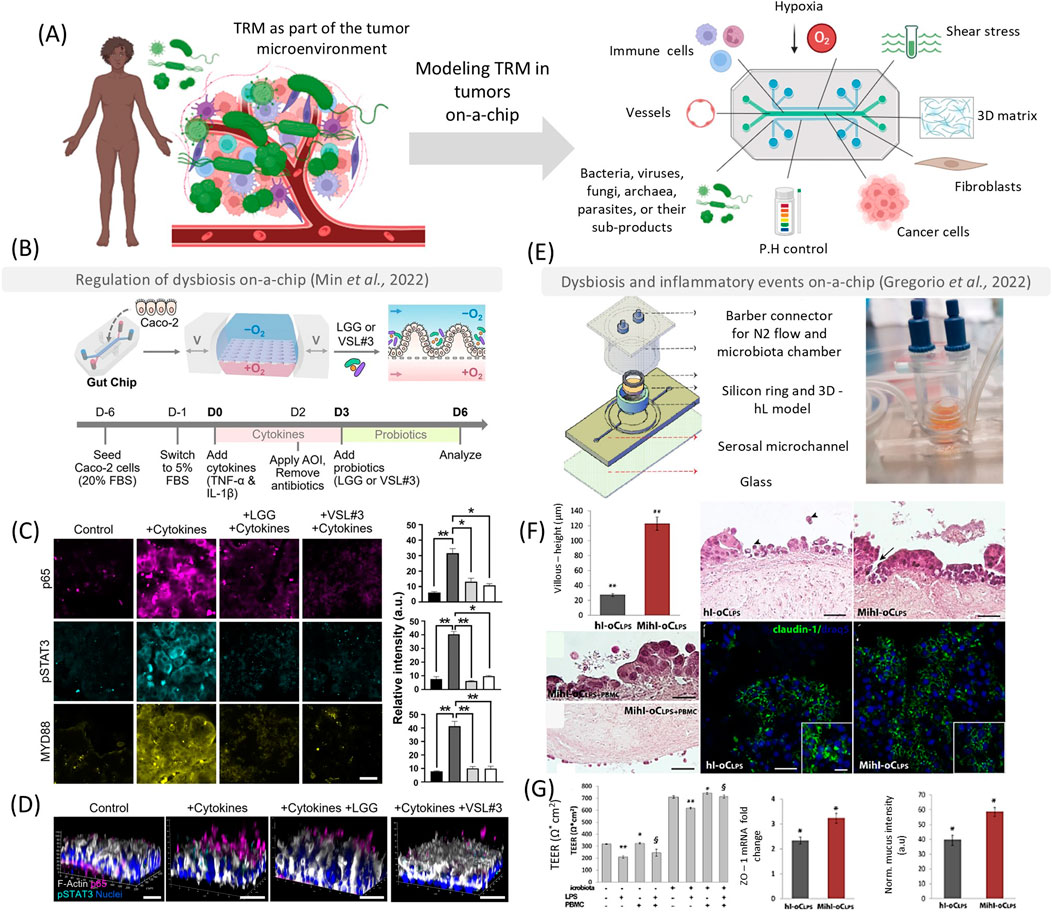
Figure 1. Tumor on-a-chip as platforms to emulate the host-microbe interactions in early oncogenesis and inflammation. These models can be used to emulate essential hallmarks of the tumor microenvironment, such as shear stress, hypoxia, a 3D matrix, different cell types, and interactions with several bacteria, fungi, viruses, archaea, parasites, and their sub-products (A). In (B) reproduced from Min et al. (2022) a polydimethylsiloxane (PDMS) chip was used to understand the role of probiotics in reestablishing gut homeostasis. Chips that were treated with Lactobacillus rhamnosus (LLG) or a complex mixture VSL#3 could reduce inflammatory and carcinogenic signaling pathways (p65 – magenta, pSTAT3 - cyan, and MYD88) after 3 days [(C, D) scale bar – 50 µm]. In (E, F), reproduced from de Gregorio et al. (2022), the authors developed a device that features an open central chamber accommodating a transwell membrane separating the lumen and serosal sides to emulate a human intestine on-a-chip (hi-oC) (E). The authors observed that lipopolysaccharide (hl-oCLPS) would stimulate a higher inflammatory response compared to the hi-oC (F), and the probiotics (Mihl-oCLPS) improved tight junction formation and mucus production (G). Scales bars in (F, G) represent 100 µm. Parts of this figure were created with biorender.com. This figure was licensed under a common creative attribution [CC by 4.0 Spring Nature–(B–D) and Elsevier (E–G)].
In the context of tumor initiation, microfluidic technologies hold significant potential to elucidate the causative mechanisms underlying the association between dysbiosis on epithelial barriers and cancer development (Poceviciute and Ismagilov, 2019). However, reproducing multispecies cultures in vitro presents challenges due to the complex interactions of these microorganisms on their native tissue, which occur under varying oxygen conditions (e.g., skin, colorectal, and oral cavities) and dynamic metabolic interactions (Moossavi et al., 2022). Static systems can promote microorganism overgrowth, which may overwhelm the rate of mammalian/microorganism cell interactions (Jalili-Firoozinezhad et al., 2019; Shin et al., 2024). Microfluidic devices can emulate tight junction formation, epithelial layers, mucus, and peristaltic movements related to shear stress to adopt microbial diversity and interactions with epithelial cells during early cancer development (Sibilio et al., 2024). These devices also serve as excellent platforms for studying bacterial-carcinogenic signaling pathways (Valiei et al., 2023). For instance, a PDMS chip was designed to mimic the gut, featuring multiple rows of microgut culture chambers (Lee et al., 2024). The microgut was generated using Caco cell layers in collagen gels, creating a villus structure subjected to shear stress, hypoxia (1% oxygen to facilitate bacterial growth for 1 day), and flow to support epithelial tissue formation. After 6 days, the monolayer exhibited a well-differentiated brush border, various intestinal cells (including goblet cells, enterocytes, and Paneth cells), defined tight junctions, and mucin production. The system supported the growth of both enterotoxigenic (ETBF8 and ETBF9) and non-toxigenic (NTBF) strains of Bacteroides fragilis. In this model, only the ETBF9 strain significantly stimulated pre-oncogenic signaling pathways, such as WNT-β-catenin and p-STAT3, compared to the microorganism-free group. Introducing Lactobacillus rhamnosus into the microgut significantly inhibited ETBF9 growth. These findings were previously observed only in vivo and demonstrate that microfluidic devices can replicate substantial biomimetic physiological and pre-carcinogenic events (Owens et al., 2021).
Another important area of exploration in recent years is the crosstalk between microbiota and the immune system in cancer (Jayakrishnan et al., 2024). Dysbiosis can lead to pathobiont proliferation and inflammation (Biragyn and Ferrucci, 2018). Opportunistic microorganisms, such as F. nucleatum and Candida albicans, proliferate under dysbiosis conditions (Gu P. et al., 2020; Li et al., 2022). Their interaction with epithelial and cancer cells triggers inflammatory cytokines (e.g., interleukins IL-6, IL-17, IL-8) and pathways (e.g., NF-kB, STAT3), promoting epithelial-to-mesenchymal transition, immune chemoattractants, and cancer cell proliferation (Narunsky-Haziza et al., 2022; Wang and Fang, 2023; Wang et al., 2023b). Additionally, microorganisms interact with the immune system in cancer through toll-like receptors (TLR2 and TLR4) and myeloid-derived suppressor cells, triggering an immunosuppressive environment (Yang et al., 2017; Wong and Yu, 2023). Some of these interactions have therapeutic consequences, especially in immunotherapies. Recent evidence has shown that F. nucleatum can produce succinic acid which neutralizes the effect of anti-PD1 antibodies (Jiang et al., 2023).
Combining all elements of the epithelium, the microbiome, and cancer cells together with immune cells in vitro is challenging. However, microfluidic devices have recapitulated some of these complexities by providing an architecture that supports immune cell infiltration, differentiation, and communication with microorganisms, which can be directly applied to the TME (Figures 1A–D). Researchers have developed a microfluidic device to understand the immune-responsive human microbiota-intestine axis (De Gregorio et al., 2022) (Figures 1E–G). This device features an open central chamber to accommodate a trans-well membrane separating the lumen and serosal sides. This chamber is housed within a poly(methyl methacrylate)-derived cap to provide N2 flow for an anaerobic luminal environment to support the growth of L. rhamnosus and Bifidobacterium longum. The microorganisms studied exhibited a protective immunoregulatory effect in the presence of lipopolysaccharide (LPS, derived from E. coli). Peripheral blood mononuclear cells (PBMCs) introduced to the chip produced higher levels of anti-inflammatory cytokines (such as IL-10) and lower levels of inflammatory cytokines (such as IL-2), demonstrating that the immunomodulatory role of probiotics can be recapitulated in complex models (De Gregorio et al., 2022) (Figures 1E–G).
Considering the immune system’s complexity, which requires antigen presentation and interaction with other systems, such as the circulatory and lymphatic systems, leaves several opportunities remain to explore host-microbe interactions in cancer. However, as blood and lymphatic vessels can be engineered in organs-on-a-chip, integrating multiple aspects of the microbiome, epithelial tissues, and immune cells with the vasculature, will provide robust tools to predict cancer development and progression.
Due to the diversity and heterogeneity of microbes across various tissues, there is a compelling need to simulate their interactions within specific physiological environments in the human body (Shin et al., 2024). For instance, the gastrointestinal tract and oral cavity can be inhabited by anaerobic pathobionts (Zhao et al., 2023; Liao et al., 2024), while the mammary glands can harbor aerobic species (Tzeng et al., 2021). Furthermore, numerous uncertainties persist regarding how local and tissue-specific microbes might influence the initiation, progression, and resistance of distant tissue tumors (Fu et al., 2023). To address many of these biological questions, organ-on-a-chip technologies enable the investigation of microbe interactions with specific tissues (such as the tumor microenvironment) and their spread to other tissues (metastasis). In this section, we elucidate the applications of these advanced models in studying various cancer types, such as colorectal, breast, and head and neck cancers (Figure 2).
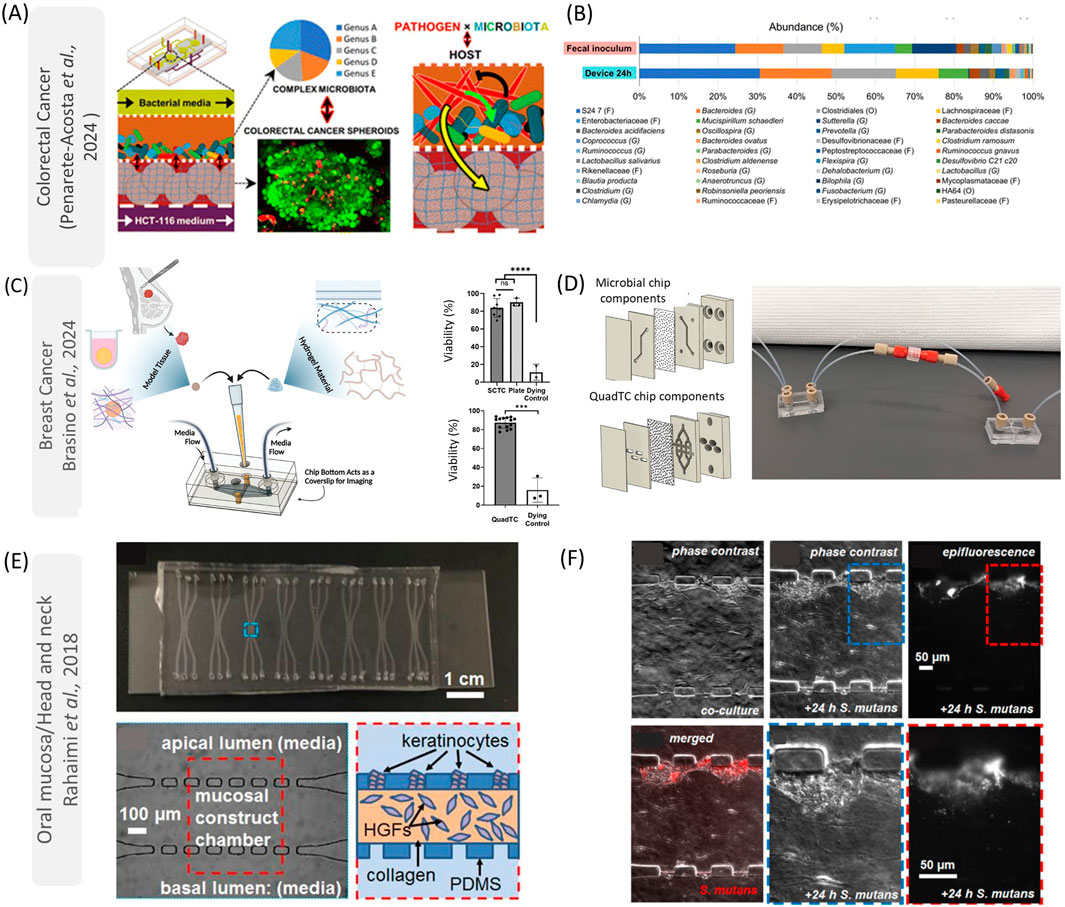
Figure 2. Applications of on-chip models to understand host-microbe interactions in cancer for colorectal cancer (A, B), breast cancer (C, D), and oral mucosa/head and neck (E, F). In (A), Penarete-Acosta et al. (2024) developed a colorectal tumor on-a-chip to evaluate the effect of F. nucleatum in modulating the microbiome. They found that this microorganism is responsible for increasing the abundance of pro-inflammatory microorganisms (B). In (C), reproduced from Brasino et al., developed microfluidic devices that can keep patient-derived organoid viability (C) while communicating with the gut microbiome (D). These polycarbonate chips allow researchers to study hypoxic environments (gut) while connecting those systems with other distal tissues (D). In (E, F), reproduced from Rahimi et al. (2018), a PDMS device was designed to mimic the oral mucosa interaction with microorganisms. These devices have a central chamber containing gingival fibroblasts (HGFs) in collagen and a layer of keratinocytes (E). Although the authors just evaluated the species S. mutans (F), this platform can be used to study how oral pathobiont microorganisms can interact with head and neck cancers. This figure was licensed under a common creative attribution (CC by 4.0 Royal Chemical Society, Wiley and Elsevier). (E, F) was reproduced from [Christopher Rahimi, Benjamin Rahimi, Dominic Padova, Seyed A. Rooholghodos, Diane R. Bienek, Xiaolong Luo, Gili Kaufman, Christopher B. Raub; Oral mucosa-on-a-chip to assess layer-specific responses to bacteria and dental materials. Biomicrofluidics 1 September 2018; 12 (5): 054106. https://doi.org/10.1063/1.5048938], with the permission of AIP Publishing.
According to the World Health Organization, colorectal cancers (CRCs) are the second leading cause of cancer-related deaths worldwide and were responsible for causing almost one million deaths in 2023 (World Health Organization, 2023a). Mounting evidence shows that our microbiota is involved in several aspects of CRC development (Lynch and Pedersen, 2016; Cui et al., 2024). Several bacterial species may play a role in the initiation or progression of GI cancer (Lotstedt et al., 2023). Gut dysbiosis can cause variations in the normal pH balance, generate genotoxins, trigger adhesion receptor interactions, induce inflammation, trigger epithelial-mesenchymal transition, chemoattract immunosuppressive cells, and produce a hypoxic environment that favors the growth of anaerobic microorganisms (Rath and Haller, 2022; Simpson et al., 2022; White and Sears, 2023). All these factors are critical to establish the tumor microenvironment bioarchitecture. Organ-on-a-chip models to study host-microbe interactions in CRC have been shown to mimic dysbiosis in vitro, to elucidate in real-time key signals, proteins, metabolites, and cytokines involved with CRC initiation (Udayasuryan et al., 2020).
One of the benefits of using microfluidic devices to study gut microbiota and colonocytes is that these interactions can be manipulated to provide an understanding of contact-independent mechanisms and metabolic exchanges between specific microorganisms and cancer cells in real-time (Mitrofanova et al., 2024). For example, researchers have developed a microfluidic device to mimic the non-physical interactions between F. nucleatum, colonocytes, and commensal microbiota (Penarete-Acosta et al., 2024). The polydimethylsiloxane (PDMS) microfluidic device used in this study had four microfluidic layers: two in the middle for mammalian cells and bacteria, and one top and bottom layer for media. These layers are each separated by porous polyester membranes to prevent physical interaction between the bacteria and colonocytes (HCT116 cell line). The colonocytes were cultivated in Matrigel, and the chip was then transferred to an anaerobic chamber to mimic the hypoxic colorectal environment. In this model, F. nucleatum stimulated genes related to inflammation/cell signaling (such as IL-8 and VEGF), and epithelial-mesenchymal transition (SNAI2). Additionally, microorganisms from the murine fecal microbiota were seeded directly into the chip model and used to study the impact of F. nucleatum on microbial composition. Although a decrease in mammalian cellular viability was observed over 24 h due to the challenges in cultivating mammalian cells in an acidic and anaerobic environment enriched with microorganism metabolites, the presence of F. nucleatum appeared to modulate the microbiome, upregulating the presence of some genera such as Suterella and Clostridium (Penarete-Acosta et al., 2024) (Figures 2A, B).
The potential use of microfluidic devices to explore the gut microbiome and colonocytes is promising; however, maintaining the viability of different microbial species and mammalian cells in the same device is still a major challenge. Furthermore, incorporating integral aspects of the innate and adaptive immune system such as neutrophils, macrophages, and T cells will aid in the understanding of how dysbiosis impacts tumor microenvironment initiation and establishment. A recent study demonstrated the development of human intestinal immune-organoids to recapitulate and investigate drug-induced intestinal inflammation (Recaldin et al., 2024). Thus, combining patient-derived colorectal organoids, immune cells, fecal microbiota, we can envision to establish complex microphysiological systems that could serve as personalized diagnostics, leveraging live biopsies that can be cultivated, manipulated and treated in real-time.
Breast cancers (BC) are heterogeneous diseases derived from the ductal epithelium of mammary tissue (Danenberg et al., 2022) and are the leading type of cancer in women (Siegel et al., 2024). BC are composed of a microenvironment rich in myoepithelial cells and stromal cells, including immune cells, fibroblasts, and adipocytes condensed in an extracellular matrix (Chung et al., 2017; World Health Organization, 2023b). Recent studies have identified several breast bacterial taxa that appear to be associated with BC, such as the genera Staphylococcus, Enterococcus, and Streptococcus (Banerjee et al., 2021; Tzeng et al., 2021). Healthy versus cancer tissues have demonstrated differences in microbiome composition, with malignant environments presenting a higher composition of facultative anaerobic microorganisms (Fu et al., 2022). The presence of intratumoral bacteria is also thought to be critical to metastasis, regulating the cellular resistance to fluid stress by reorganizing the cytoskeleton (Fu et al., 2022).
The metabolic and hypoxic environment in breast cancer dictates a challenging task to be reproduced in vitro. Furthermore, although several pieces of evidence illustrate the role of gut microorganisms affecting estrogen metabolism and the immune system in breast cancer (Urbaniak et al., 2016; Plaza-Diaz et al., 2019; Heath et al., 2024), the communication between these distant tissues is not recapitulated in current tissue culture models. Microfluidic models may enrich our understanding of the dynamics linked with intratumoral or gut microbiota and breast cancer. For instance, Brasino et al. (2024) fabricated chips with gas-impermeable polycarbonate without tapes or gaskets. The authors cultivated colon epithelial cells (Caco2) for 6 days to form a villus-like structure followed by introducing Lactobacillus plantarum or a breast cancer-associated bacterium, Blautia coccoides. It was demonstrated that bacteria could be found on villus surfaces following 2 days of co-culture while maintaining epithelial viability, showing the functionality of interactions between facultative or obligate anaerobes and the colon-like layer structure. The authors also demonstrated that cultivating breast tumor spheroids (MCF7 cells cultivated in collagen) in microfluidics could provide a platform to elucidate the interactions between the gut microbiota and breast tumors (Figures 2C, D). By directing the flow within the chip from the microbial compartment (gut) to the breast tumor spheroid, the researchers facilitated metabolite exchange between gut microbes and the tumor. Interestingly, spheroids downstream of gut chips cultured with Estradiol-Glow-supplemented media exhibited significantly elevated expression of GREB1, an estrogen-responsive gene transcript, compared to those treated with the vehicle control (Brasino et al., 2024). These findings provide novel insights into the bidirectional relationship between gut microbes and estrogen signaling, highlighting potential mechanisms of microbial influence on tumor biology.
The possibility of creating microfluidic devices replicating hallmarks of ductal epithelium and its connection with vasculature might direct the knowledge of metastatic niches and metabolic interactions of microorganisms with distant tissues, such as the gut-breast axis. More studies are needed however to evaluate the role of specific extracellular microorganisms and the immune system in breast cancer progression and metastasis.
Head and Neck Cancers (HNCs) are the sixth most common cancer and are known for their high heterogeneity and disparity in therapeutic conduct (World Health Organization, 2024). Despite some progress in understanding the mechanism of this disease over time, some hallmarks of its complex microenvironment still need to be explored, mainly due to the need for more advanced in vitro models (Almela et al., 2021). The oral cavity is a unique environment hosting more than 700 species of microorganisms with a spectrum of oxygen requirements, interfaces with soft and hard tissues, and connection with the gastrointestinal tract (Baker et al., 2023). Within this diverse milieu, key microorganisms such as F. nucleatum, Prevotella intermedia, Porphiromonas gingivalis, C. albicans, and human papillomavirus (HPV), among others correlate with HNC initiation, progression, and treatment resistance (Yu et al., 2017; Galeano Nino et al., 2022; Wang et al., 2023a; Wang et al., 2023c). With the increasing number of HPV + HNCs worldwide (Califano et al., 2023; Ramos da Silva et al., 2023), developing biological systems to reproduce HNCs in vitro, including host-microbe interactions accurately, remains a substantial barrier to further research, especially in early cancers. This challenge partly arises because some of the TRMis are opportunistic, and their pre-cancer mechanisms are still poorly understood. Furthermore, a critical barrier persists in elucidating how slight alterations in dysbiosis, and the associated metabolites may influence oncogenesis in cancer development. These phenomena raise numerous unanswered epigenetic questions that require further investigation.
The oral mucosa is the main barrier in the oral cavity and is constantly in contact with oral saliva and several microorganisms in the oral cavity (Baker et al., 2024). The host-microbe interactions happen mainly in the epithelial layer, where epithelial cells participate directly in the inflammatory response to dysbiosis and can undergo epithelial-to-mesenchymal transition which progresses the disease (Cai et al., 2024). It has been reported that some microorganisms involved with periodontal plaques such as F. nucleatum and P. gingivalis, participate in the tumor microenvironment, especially in immunosuppressive areas (Galeano Nino et al., 2022). Although the mechanisms involved with oral microorganisms and oncogenesis are still to be fully understood, microfluidic devices are essential tools to recapitulate those phenomena, especially by providing organotypic functions and interactions with microbes and their subproducts, while being visualized under the microscope for long-term cultures (da Costa Sousa et al., 2024). In this direction, several models of gingiva and oral mucosa have been developed in the literature, especially to evaluate biomaterials-soft tissue interactions, and response to radiation (Tabatabaei et al., 2020; Ly et al., 2021; Ly et al., 2022; Muniraj et al., 2023) (Figures 2E, F). For example, Muniraj et al. (2023) developed a microphysiological model of gingiva on-a-chip. This microfluidic device was prepared by thermal bonding of polymethyl methacrylate sheets with different thicknesses. Both lower (metabolic waste) and upper chambers were connected to independent inlets and outlets. The main chamber was used to cultivate gingival equivalents, separated from the lower chamber by a polycarbonate porous membrane. The authors observed that after 3 weeks of culture, the samples did not contract and maintained the epithelial structure presenting multi-layered, ortho-keratinized, squamous epithelium over a cellular lamina propria. Furthermore, the engineered tissue presented a high expression of filaggrin, loricrin (epithelial barrier), collagen-IV (basement membrane), and laminin-V (epithelial junction). Also, it was observed that the gingiva-on-a-chip could respond to mucosal irritation (alcohol mouthwash), providing outputs of cell metabolic activity similar to those obtained using gingiva inserts in MTT and LDH tests (Muniraj et al., 2023). In another investigation, Ly et al. (2022) developed mucositis on-a-chip using PDMS with 3 channels. The central channel was used to create three-layer gingival keratinocytes (GIE), human gingival fibroblasts (HGF), and human dermal microvascular endothelial cells (HMEC) in collagen that generated a mucosa-like tissue that could be cultivated for 21 days. The model was used to emulate the mucositis induced by cisplatin and radiation, which could be observed especially on day 12 of treatment, by the higher release of LDH and cellular viability (Ly et al., 2022). Although the authors did not evaluate the effect of the microbiota in mucositis development, this platform opens several opportunities to study host-microbe interactions and oral cancer on-a-chip.
Considering the complexity and diversity of HNCs, future models should integrate aspects of the saliva, oral epithelium, and other microorganisms that have been associated with oral cancers, such as the fungal genus Candida (Wang et al., 2023a), and HPV (Sathish et al., 2014). Additionally, these devices could be used to simulate the interactions between alcohol and smoking, both of which are known to modulate oral dysbiosis and cancer progression (Benam et al., 2016). By including immune components such as tertiary lymphoid structures (Li H. et al., 2023) these systems might also provide an advanced understanding of the dynamics of inflammatory stages, adaptive responses, and their effect on immunotherapies.
The microbiome is relevant to most, if not all, cancers, beyond those outlined here, including malignancies of the skin, lung, urogenital system, and bone marrow. Therefore, there is a need to develop models that can mimic the physiology of these tissues and their connections with the microbiome (Ramirez-Labrada et al., 2020; Martin et al., 2022; Voigt et al., 2022; Woerner et al., 2022). Despite documentation of interactions between the microbiome and cancer development, many opportunities remain to further explore the potential of organ on-a-chip modeling within the tumor microenvironment, such as their mechanistic interactions with the immune system in the TME, immunosuppression, metastasis, and immunotherapies. Furthermore, the connection of tissue-specific microbiota with distant cancers and metastasis should be explored, primarily because the gut microbiome has been correlated with metastasis to several tissues (Fu et al., 2023).
While the outlook for host-microbes on-a-chip models in cancer studies is promising, several challenges are still to be overcome. Current models often struggle to capture the full complexity of the microbiome, including the diversity and density of microorganisms that accurately reflect local and systemic niches. Variability in environmental factors such as temperature, oxygen levels, growth media, and shear stress further complicates the standardization of experimental conditions, reducing reproducibility and comparability across studies. Furthermore, achieving the precise organization of tissues, including accurate cell-cell interactions and extracellular matrix composition, remains a significant technical challenge. Finally, the integration of advanced technologies, such as artificial intelligence and high-throughput systems, is still in its early stages, limiting the predictive power and scalability of these platforms.
Overcoming these challenges will require a multi-disciplinary approach that integrates expertise in tissue engineering, microbiology, and computational sciences. Addressing the need for microbiome diversity, optimizing growth conditions for heterogeneous microorganisms, and refining tissue engineering techniques are essential steps toward enhancing the fidelity and translational of these models. Additionally, incorporating cutting-edge technologies will not only improve data analysis but also enable the development of more robust and scalable systems. Once these limitations are addressed, organ-on-a-chip models have the potential to transform our understanding of microorganisms in cancer and develop specific immune and microbiological therapies. These platforms could serve as “live biopsies” or personalized tissue avatars, allowing clinicians to evaluate patient-specific responses to treatments in real-time or to assess microbiota profiles and correlate them with disease progression. Also, these platforms can be used to test engineered microorganisms by synthetic biology to compete with TRMis and attract, polarize, and stimulate tumor-killing immune cells. Such applications will generate several opportunities for more precise therapeutic interventions, ultimately improving treatment outcomes and enhancing the quality of life for cancer patients.
In conclusion, while the path to fully realizing the potential of organ-on-a-chip models in microbes-cancer research is linked with technical and biological challenges, their promise opens multiple doors to answer unique biological questions. These innovative platforms represent a significant step forward in understanding and treating cancer through a microbiome-informed perspective, offering a future where precision medicine can address the complexities of cancer biology accurately.
MS: Writing–original draft, Writing–review and editing. DB: Writing–original draft, Writing–review and editing. MK: Writing–original draft, Writing–review and editing. DD: Writing–original draft. RD: Writing–original draft, Writing–review and editing. ZZ: Writing–original draft, Writing–review and editing. CF: Writing–original draft, Writing–review and editing. LB: Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research and/or publication of this article. We acknowledge funding from the Cancer Early Detection Advanced Research (CEDAR) Center at Oregon Health & Science University’s Knight Cancer Institute. LB acknowledges NIH/NCI/NIDCR funding: R01DE026170, 3R01DE026170-03S1, R01DE029553, CA263860, 5R21CA263860-02, and T90DE030859. LB thanks the Friends of Doernbecher Grant Program at OHSU. MS acknowledges funding from NIDCR R90DE031533-01. CM acknowledges OHSU for the Collins Medical Trust (102327) and NIDCR for K01030484.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aizpurua, O., Blijleven, K., Trivedi, U., Gilbert, M. T. P., and Alberdi, A. (2023). Unravelling animal-microbiota evolution on a chip. Trends Microbiol. 31 (10), 995–1002. doi:10.1016/j.tim.2023.04.010
Almela, T., Tayebi, L., and Moharamzadeh, K. (2021). 3D bioprinting for in vitro models of oral cancer: toward development and validation. Bioprinting 22, e00132. doi:10.1016/j.bprint.2021.e00132
Asghar, W., El Assal, R., Shafiee, H., Pitteri, S., Paulmurugan, R., and Demirci, U. (2015). Engineering cancer microenvironments for in vitro 3-D tumor models. Mater TodayKidlingt. 18 (10), 539–553. doi:10.1016/j.mattod.2015.05.002
Baker, J. L., Mark Welch, J. L., Kauffman, K. M., McLean, J. S., and He, X. (2023). The oral microbiome: diversity, biogeography and human health. Nat. Rev. Microbiol. 22, 89–104. doi:10.1038/s41579-023-00963-6
Baker, J. L., Mark Welch, J. L., Kauffman, K. M., McLean, J. S., and He, X. (2024). The oral microbiome: diversity, biogeography and human health. Nat. Rev. Microbiol. 22 (2), 89–104. doi:10.1038/s41579-023-00963-6
Ballerini, M., Galie, S., Tyagi, P., Catozzi, C., Raji, H., Nabinejad, A., et al. (2025). A gut-on-a-chip incorporating human faecal samples and peristalsis predicts responses to immune checkpoint inhibitors for melanoma. Nat. Biomed. Eng. doi:10.1038/s41551-024-01318-z
Banerjee, S., Wei, Z., Tian, T., Bose, D., Shih, N. N. C., Feldman, M. D., et al. (2021). Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 12 (9), 831. doi:10.1038/s41419-021-04092-x
Battaglia, T. W., Mimpen, I. L., Traets, J. J. H., van Hoeck, A., Zeverijn, L. J., Geurts, B. S., et al. (2024). A pan-cancer analysis of the microbiome in metastatic cancer. Cell 187 (9), 2324–2335.e19. doi:10.1016/j.cell.2024.03.021
Bein, A., Kim, S., Goyal, G., Cao, W., Fadel, C., Naziripour, A., et al. (2021). Enteric coronavirus infection and treatment modeled with an immunocompetent human intestine-on-A-chip. Front. Pharmacol. 12, 718484. doi:10.3389/fphar.2021.718484
Benam, K. H., Novak, R., Nawroth, J., Hirano-Kobayashi, M., Ferrante, T. C., Choe, Y., et al. (2016). Matched-comparative modeling of normal and diseased human airway responses using a microengineered breathing lung chip. Cell Syst. 3 (5), 456–466.e4. doi:10.1016/j.cels.2016.10.003
Biragyn, A., and Ferrucci, L. (2018). Gut dysbiosis: a potential link between increased cancer risk in ageing and inflammaging. Lancet Oncol. 19 (6), e295–e304. doi:10.1016/s1470-2045(18)30095-0
Brasino, D. S. K., Speese, S. D., Schilling, K., Schutt, C. E., and Barton, M. C. (2024). A linkable, polycarbonate gut microbiome-distal tumor chip platform for interrogating cancer promoting mechanisms. Adv. Sci. (Weinh) 11, e2309220. doi:10.1002/advs.202309220
Cai, L., Zhu, H., Mou, Q., Wong, P. Y., Lan, L., Ng, C. W. K., et al. (2024). Integrative analysis reveals associations between oral microbiota dysbiosis and host genetic and epigenetic aberrations in oral cavity squamous cell carcinoma. NPJ Biofilms Microbiomes 10 (1), 39. doi:10.1038/s41522-024-00511-x
Califano, J., Yousef, A., Mostafa, H., Valsamakis, A., Zhang, X., Batis, N., et al. (2023). Lead time to recurrence after posttreatment plasma and saliva HPV DNA testing in patients with low-risk HPV oropharynx cancer. JAMA Otolaryngol. Head. Neck Surg. 149 (9), 812–819. doi:10.1001/jamaoto.2023.1730
Cao, Y., Oh, J., Xue, M., Huh, W. J., Wang, J., Gonzalez-Hernandez, J. A., et al. (2022). Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science 378 (6618), eabm3233. doi:10.1126/science.abm3233
Cao, Y., Xia, H., Tan, X., Shi, C., Ma, Y., Meng, D., et al. (2024). Intratumoural microbiota: a new frontier in cancer development and therapy. Signal Transduct. Target Ther. 9 (1), 15. doi:10.1038/s41392-023-01693-0
Chung, W., Eum, H. H., Lee, H. O., Lee, K. M., Lee, H. B., Kim, K. T., et al. (2017). Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 8, 15081. doi:10.1038/ncomms15081
Cui, W., Guo, M., Liu, D., Xiao, P., Yang, C., Huang, H., et al. (2024). Gut microbial metabolite facilitates colorectal cancer development via ferroptosis inhibition. Nat. Cell Biol. 26 (1), 124–137. doi:10.1038/s41556-023-01314-6
da Costa Sousa, M. G., Vignolo, S. M., Franca, C. M., Mereness, J., Alves Fraga, M. A., Silva-Sousa, A. C., et al. (2024). Engineering models of head and neck and oral cancers on-a-chip. Biomicrofluidics 18 (2), 021502. doi:10.1063/5.0186722
Danenberg, E., Bardwell, H., Zanotelli, V. R. T., Provenzano, E., Chin, S. F., Rueda, O. M., et al. (2022). Breast tumor microenvironment structures are associated with genomic features and clinical outcome. Nat. Genet. 54 (5), 660–669. doi:10.1038/s41588-022-01041-y
De Gregorio, V., Sgambato, C., Urciuolo, F., Vecchione, R., Netti, P. A., and Imparato, G. (2022). Immunoresponsive microbiota-gut-on-chip reproduces barrier dysfunction, stromal reshaping and probiotics translocation under inflammation. Biomaterials 286, 121573. doi:10.1016/j.biomaterials.2022.121573
Espinoza, J. L., and Minami, M. (2018). Sensing bacterial-induced DNA damaging effects via natural killer group 2 member D immune receptor: from dysbiosis to autoimmunity and carcinogenesis. Front. Immunol. 9, 52. doi:10.3389/fimmu.2018.00052
Fu, A., Yao, B., Dong, T., and Cai, S. (2023). Emerging roles of intratumor microbiota in cancer metastasis. Trends Cell Biol. 33 (7), 583–593. doi:10.1016/j.tcb.2022.11.007
Fu, A., Yao, B., Dong, T., Chen, Y., Yao, J., Liu, Y., et al. (2022). Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 185 (8), 1356–1372.e26. doi:10.1016/j.cell.2022.02.027
Galeano Nino, J. L., Wu, H., LaCourse, K. D., Kempchinsky, A. G., Baryiames, A., Barber, B., et al. (2022). Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611 (7937), 810–817. doi:10.1038/s41586-022-05435-0
Guo P., P., Tian, Z., Kong, X., Yang, L., Shan, X., Dong, B., et al. (2020). FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J. Exp. Clin. Cancer Res. 39 (1), 202. doi:10.1186/s13046-020-01677-w
Gu X., X., Song, L. J., Li, L. X., Liu, T., Zhang, M. M., Li, Z., et al. (2020). Fusobacterium nucleatum causes microbial dysbiosis and exacerbates visceral hypersensitivity in a colonization-independent manner. Front. Microbiol. 11, 1281. doi:10.3389/fmicb.2020.01281
Heath, H., Mogol, A. N., Santaliz Casiano, A., Zuo, Q., and Madak-Erdogan, Z. (2024). Targeting systemic and gut microbial metabolism in ER(+) breast cancer. Trends Endocrinol. Metab. 35 (4), 321–330. doi:10.1016/j.tem.2023.12.008
Huh, D., Kim, H. J., Fraser, J. P., Shea, D. E., Khan, M., Bahinski, A., et al. (2013). Microfabrication of human organs-on-chips. Nat. Protoc. 8 (11), 2135–2157. doi:10.1038/nprot.2013.137
Jalili-Firoozinezhad, S., Gazzaniga, F. S., Calamari, E. L., Camacho, D. M., Fadel, C. W., Bein, A., et al. (2019). A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3 (7), 520–531. doi:10.1038/s41551-019-0397-0
Jayakrishnan, T. T., Sangwan, N., Barot, S. V., Farha, N., Mariam, A., Xiang, S., et al. (2024). Multi-omics machine learning to study host-microbiome interactions in early-onset colorectal cancer. NPJ Precis. Oncol. 8 (1), 146. doi:10.1038/s41698-024-00647-1
Jiang, S. S., Xie, Y. L., Xiao, X. Y., Kang, Z. R., Lin, X. L., Zhang, L., et al. (2023). Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe 31 (5), 781–797.e9. doi:10.1016/j.chom.2023.04.010
Joo, J. E., Chu, Y. L., Georgeson, P., Walker, R., Mahmood, K., Clendenning, M., et al. (2024). Intratumoral presence of the genotoxic gut bacteria pks(+) E. coli, Enterotoxigenic Bacteroides fragilis, and Fusobacterium nucleatum and their association with clinicopathological and molecular features of colorectal cancer. Br. J. Cancer 130 (5), 728–740. doi:10.1038/s41416-023-02554-x
Jouybar, M., de Winde, C. M., Wolf, K., Friedl, P., Mebius, R. E., and den Toonder, J. M. J. (2024). Cancer-on-chip models for metastasis: importance of the tumor microenvironment. Trends Biotechnol. 42 (4), 431–448. doi:10.1016/j.tibtech.2023.10.001
Kang, X., Lau, H. C., and Yu, J. (2024). Modulating gut microbiome in cancer immunotherapy: harnessing microbes to enhance treatment efficacy. Cell Rep. Med. 5 (4), 101478. doi:10.1016/j.xcrm.2024.101478
Kapalczynska, M., Kolenda, T., Przybyla, W., Zajaczkowska, M., Teresiak, A., Filas, V., et al. (2018). 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch. Med. Sci. 14 (4), 910–919. doi:10.5114/aoms.2016.63743
Kim, R., and Sung, J. H. (2024). Recent advances in gut- and gut-organ-axis-on-a-chip models. Adv. Healthc. Mater 13, e2302777. doi:10.1002/adhm.202302777
Ladaycia, A., Loretz, B., Passirani, C., Lehr, C. M., and Lepeltier, E. (2021). Microbiota and cancer: in vitro and in vivo models to evaluate nanomedicines. Adv. Drug Deliv. Rev. 170, 44–70. doi:10.1016/j.addr.2020.12.015
Lee, J., Menon, N., and Lim, C. T. (2024). Dissecting gut-microbial community interactions using a gut microbiome-on-a-chip. Adv. Sci. (Weinh) 11 (20), e2302113. doi:10.1002/advs.202302113
Li, G., Sun, Y., Huang, Y., Lian, J., Wu, S., Luo, D., et al. (2023). Fusobacterium nucleatum-derived small extracellular vesicles facilitate tumor growth and metastasis via TLR4 in breast cancer. BMC Cancer 23 (1), 473. doi:10.1186/s12885-023-10844-z
Li, H., Miao, M. X., Jia, C. L., Cao, Y. B., Yan, T. H., Jiang, Y. Y., et al. (2022). Interactions between Candida albicans and the resident microbiota. Front. Microbiol. 13, 930495. doi:10.3389/fmicb.2022.930495
Li, H., Zhu, S. W., Zhou, J. J., Chen, D. R., Liu, J., Wu, Z. Z., et al. (2023). Tertiary lymphoid structure raises survival and immunotherapy in HPV(-) HNSCC. J. Dent. Res. 102 (6), 678–688. doi:10.1177/00220345231151685
Li, L., Chandra, V., and McAllister, F. (2024). Tumor-resident microbes: the new kids on the microenvironment block. Trends Cancer 10 (4), 347–355. doi:10.1016/j.trecan.2023.12.002
Liao, C., Rolling, T., Djukovic, A., Fei, T., Mishra, V., Liu, H., et al. (2024). Oral bacteria relative abundance in faeces increases due to gut microbiota depletion and is linked with patient outcomes. Nat. Microbiol. 9, 1555–1565. doi:10.1038/s41564-024-01680-3
Liu, W. T., and Zhu, L. (2005). Environmental microbiology-on-a-chip and its future impacts. Trends Biotechnol. 23 (4), 174–179. doi:10.1016/j.tibtech.2005.02.004
Lotstedt, B., Strazar, M., Xavier, R., Regev, A., and Vickovic, S. (2023). Spatial host-microbiome sequencing reveals niches in the mouse gut. Nat. Biotechnol. 42, 1394–1403. doi:10.1038/s41587-023-01988-1
Lu, Y. Q., Qiao, H., Tan, X. R., and Liu, N. (2024). Broadening oncological boundaries: the intratumoral microbiota. Trends Microbiol. 32, 807–822. doi:10.1016/j.tim.2024.01.007
Ly, K. L., Luo, X., and Raub, C. B. (2022). Oral mucositis on a chip: modeling induction by chemo- and radiation treatments and recovery. Biofabrication 15 (1), 015007. doi:10.1088/1758-5090/ac933b
Ly, K. L., Rooholghodos, S. A., Rahimi, C., Rahimi, B., Bienek, D. R., Kaufman, G., et al. (2021). An Oral-mucosa-on-a-chip sensitively evaluates cell responses to dental monomers. Biomed. Microdevices 23 (1), 7. doi:10.1007/s10544-021-00543-6
Lynch, S. V., and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375 (24), 2369–2379. doi:10.1056/nejmra1600266
Lythgoe, M. P., Mullish, B. H., Frampton, A. E., and Krell, J. (2022). Polymorphic microbes: a new emerging hallmark of cancer. Trends Microbiol. 30 (12), 1131–1134. doi:10.1016/j.tim.2022.08.004
Martin, A., Woolbright, B. L., Umar, S., Ingersoll, M. A., and Taylor, J. A. (2022). Bladder cancer, inflammageing and microbiomes. Nat. Rev. Urol. 19 (8), 495–509. doi:10.1038/s41585-022-00611-3
Maulana, T. I., Teufel, C., Cipriano, M., Roosz, J., Lazarevski, L., van den Hil, F. E., et al. (2024). Breast cancer-on-chip for patient-specific efficacy and safety testing of CAR-T cells. Cell Stem Cell 31 (7), 989–1002.e9. doi:10.1016/j.stem.2024.04.018
Min, S., Than, N., Shin, Y. C., Hu, G., Shin, W., Ambrosini, Y. M., et al. (2022). Live probiotic bacteria administered in a pathomimetic Leaky Gut Chip ameliorate impaired epithelial barrier and mucosal inflammation. Sci. Rep. 12 (1), 22641. doi:10.1038/s41598-022-27300-w
Mitrofanova, O., Nikolaev, M., Xu, Q., Broguiere, N., Cubela, I., Camp, J. G., et al. (2024). Bioengineered human colon organoids with in vivo-like cellular complexity and function. Cell Stem Cell 31 (8), 1175–1186.e7. doi:10.1016/j.stem.2024.05.007
Moossavi, S., Arrieta, M. C., Sanati-Nezhad, A., and Bishehsari, F. (2022). Gut-on-chip for ecological and causal human gut microbiome research. Trends Microbiol. 30 (8), 710–721. doi:10.1016/j.tim.2022.01.014
Muniraj, G., Tan, R. H. S., Dai, Y., Wu, R., Alberti, M., and Sriram, G. (2023). Microphysiological modeling of gingival tissues and host-material interactions using gingiva-on-chip. Adv. Healthc. Mater 12 (32), e2301472. doi:10.1002/adhm.202301472
Narunsky-Haziza, L., Sepich-Poore, G. D., Livyatan, I., Asraf, O., Martino, C., Nejman, D., et al. (2022). Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 185 (20), 3789–3806.e17. doi:10.1016/j.cell.2022.09.005
Nejman, D., Livyatan, I., Fuks, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368 (6494), 973–980. doi:10.1126/science.aay9189
Neufeld, L., Yeini, E., Pozzi, S., and Satchi-Fainaro, R. (2022). 3D bioprinted cancer models: from basic biology to drug development. Nat. Rev. Cancer 22 (12), 679–692. doi:10.1038/s41568-022-00514-w
Nikolaev, M., Mitrofanova, O., Broguiere, N., Geraldo, S., Dutta, D., Tabata, Y., et al. (2020). Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585 (7826), 574–578. doi:10.1038/s41586-020-2724-8
Ortega Quesada, B. A., Cuccia, J., Coates, R., Nassar, B., Littlefield, E., Martin, E. C., et al. (2024). A modular microfluidic platform to study how fluid shear stress alters estrogen receptor phenotype in ER(+) breast cancer cells. Microsyst. Nanoeng. 10, 25. doi:10.1038/s41378-024-00653-0
Owens, J. A., Saeedi, B. J., Naudin, C. R., Hunter-Chang, S., Barbian, M. E., Eboka, R. U., et al. (2021). Lactobacillus rhamnosus GG orchestrates an antitumor immune response. Cell Mol. Gastroenterol. Hepatol. 12 (4), 1311–1327. doi:10.1016/j.jcmgh.2021.06.001
Penarete-Acosta, D., Stading, R., Emerson, L., Horn, M., Chakraborty, S., Han, A., et al. (2024). A microfluidic co-culture model for investigating colonocytes-microbiota interactions in colorectal cancer. Lab. Chip 24 (15), 3690–3703. doi:10.1039/d4lc00013g
Plaza-Diaz, J., Alvarez-Mercado, A. I., Ruiz-Marin, C. M., Reina-Perez, I., Perez-Alonso, A. J., Sanchez-Andujar, M. B., et al. (2019). Association of breast and gut microbiota dysbiosis and the risk of breast cancer: a case-control clinical study. BMC Cancer 19 (1), 495. doi:10.1186/s12885-019-5660-y
Poceviciute, R., and Ismagilov, R. F. (2019). Human-gut-microbiome on a chip. Nat. Biomed. Eng. 3 (7), 500–501. doi:10.1038/s41551-019-0425-0
Radhakrishnan, J., Varadaraj, S., Dash, S. K., Sharma, A., and Verma, R. S. (2020). Organotypic cancer tissue models for drug screening: 3D constructs, bioprinting and microfluidic chips. Drug Discov. Today 25 (5), 879–890. doi:10.1016/j.drudis.2020.03.002
Rahimi, C., Rahimi, B., Padova, D., Rooholghodos, S. A., Bienek, D. R., Luo, X., et al. (2018). Oral mucosa-on-a-chip to assess layer-specific responses to bacteria and dental materials. Biomicrofluidics 12 (5), 054106. doi:10.1063/1.5048938
Ramirez-Labrada, A. G., Isla, D., Artal, A., Arias, M., Rezusta, A., Pardo, J., et al. (2020). The influence of lung microbiota on lung carcinogenesis, immunity, and immunotherapy. Trends Cancer 6 (2), 86–97. doi:10.1016/j.trecan.2019.12.007
Ramos da Silva, J., Bitencourt Rodrigues, K., Formoso Pelegrin, G., Silva Sales, N., Muramatsu, H., de Oliveira Silva, M., et al. (2023). Single immunizations of self-amplifying or non-replicating mRNA-LNP vaccines control HPV-associated tumors in mice. Sci. Transl. Med. 15 (686), eabn3464. doi:10.1126/scitranslmed.abn3464
Rath, E., and Haller, D. (2022). Intestinal epithelial cell metabolism at the interface of microbial dysbiosis and tissue injury. Mucosal Immunol. 15 (4), 595–604. doi:10.1038/s41385-022-00514-x
Recaldin, T., Steinacher, L., Gjeta, B., Harter, M. F., Adam, L., Kromer, K., et al. (2024). Human organoids with an autologous tissue-resident immune compartment. Nature 633 (8028), 165–173. doi:10.1038/s41586-024-07791-5
Rodrigues, J., Heinrich, M. A., Teixeira, L. M., and Prakash, J. (2021). 3D in vitro model (R)evolution: unveiling tumor-stroma interactions. Trends Cancer 7 (3), 249–264. doi:10.1016/j.trecan.2020.10.009
Sathish, N., Wang, X., and Yuan, Y. (2014). Human papillomavirus (HPV)-associated oral cancers and treatment strategies. J. Dent. Res. 93 (7 Suppl. l), 29S–36S. doi:10.1177/0022034514527969
Sfanos, K. S. (2023). Intratumoral bacteria as mediators of cancer immunotherapy response. Cancer Res. 83 (18), 2985–2986. doi:10.1158/0008-5472.can-23-1857
Shah, P., Fritz, J. V., Glaab, E., Desai, M. S., Greenhalgh, K., Frachet, A., et al. (2016). A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 7, 11535. doi:10.1038/ncomms11535
Shin, Y. C., Than, N., Min, W., Shin, W., and Kim, H. J. (2024). Modelling host–microbiome interactions in organ-on-a-chip platforms. Nat. Rev. Bioeng. 2, 175–191. doi:10.1038/s44222-023-00130-9
Sibilio, S., Mennella, R., Gregorio, V., Rocca, A., Urciuolo, F., Imparato, G., et al. (2024). A novel membrane-on-chip guides morphogenesis for the reconstruction of the intestinal crypt-villus axis. Biofabrication 16 (4), 045019. doi:10.1088/1758-5090/ad6599
Siegel, R. L., Giaquinto, A. N., and Jemal, A. (2024). Cancer statistics, 2024. CA Cancer J. Clin. 74 (1), 12–49. doi:10.3322/caac.21820
Simpson, R. C., Shanahan, E. R., Batten, M., Reijers, I. L. M., Read, M., Silva, I. P., et al. (2022). Diet-driven microbial ecology underpins associations between cancer immunotherapy outcomes and the gut microbiome. Nat. Med. 28 (11), 2344–2352. doi:10.1038/s41591-022-01965-2
Sontheimer-Phelps, A., Hassell, B. A., and Ingber, D. E. (2019). Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 19 (2), 65–81. doi:10.1038/s41568-018-0104-6
Srinivasan, A., Uppuluri, P., Lopez-Ribot, J., and Ramasubramanian, A. K. (2011). Development of a high-throughput Candida albicans biofilm chip. PLoS One 6 (4), e19036. doi:10.1371/journal.pone.0019036
Tabatabaei, F., Moharamzadeh, K., and Tayebi, L. (2020). Three-Dimensional in vitro oral mucosa models of fungal and bacterial infections. Tissue Eng. Part B Rev. 26 (5), 443–460. doi:10.1089/ten.teb.2020.0016
Thacker, V. V., Dhar, N., Sharma, K., Barrile, R., Karalis, K., and McKinney, J. D. (2020). A lung-on-chip model of early Mycobacterium tuberculosis infection reveals an essential role for alveolar epithelial cells in controlling bacterial growth. Elife 9, e59961. doi:10.7554/elife.59961
Tzeng, A., Sangwan, N., Jia, M., Liu, C. C., Keslar, K. S., Downs-Kelly, E., et al. (2021). Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. 13 (1), 60. doi:10.1186/s13073-021-00874-2
Udayasuryan, B., Nguyen, T. T. D., Slade, D. J., and Verbridge, S. S. (2020). Harnessing tissue engineering tools to interrogate host-microbiota crosstalk in cancer. iScience 23 (12), 101878. doi:10.1016/j.isci.2020.101878
Urbaniak, C., Gloor, G. B., Brackstone, M., Scott, L., Tangney, M., and Reid, G. (2016). The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 82 (16), 5039–5048. doi:10.1128/aem.01235-16
Valiei, A., Aminian-Dehkordi, J., and Mofrad, M. R. K. (2023). Gut-on-a-chip models for dissecting the gut microbiology and physiology. Apl. Bioeng. 7 (1), 011502. doi:10.1063/5.0126541
Vitale, S., Calapa, F., Colonna, F., Luongo, F., Biffoni, M., De Maria, R., et al. (2024). Advancements in 3D in vitro models for colorectal cancer. Adv. Sci. (Weinh) 11, e2405084. doi:10.1002/advs.202405084
Voigt, A. Y., Emiola, A., Johnson, J. S., Fleming, E. S., Nguyen, H., Zhou, W., et al. (2022). Skin microbiome variation with cancer progression in human cutaneous squamous cell carcinoma. J. Invest Dermatol 142 (10), 2773–2782.e16. doi:10.1016/j.jid.2022.03.017
Wang, N., and Fang, J. Y. (2023). Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 31 (2), 159–172. doi:10.1016/j.tim.2022.08.010
Wang, X., Wu, S., Wu, W., Zhang, W., Li, L., Liu, Q., et al. (2023a). Candida albicans promotes oral cancer via IL-17a/IL-17ra-macrophage Axis. mBio 14 (3), e0044723. doi:10.1128/mbio.00447-23
Wang, X., Zhang, W., Wu, W., Wu, S., Young, A., and Yan, Z. (2023b). Is Candida albicans a contributor to cancer? A critical review based on the current evidence. Microbiol. Res. 272, 127370. doi:10.1016/j.micres.2023.127370
Wang, Z., Wang, Q., Tao, Y., Chen, J., Yuan, Z., and Wang, P. (2023c). Characterization of immune microenvironment in patients with HPV-positive and negative head and neck cancer. Sci. Data 10 (1), 694. doi:10.1038/s41597-023-02611-3
White, M. T., and Sears, C. L. (2023). The microbial landscape of colorectal cancer. Nat. Rev. Microbiol. 22, 240–254. doi:10.1038/s41579-023-00973-4
Woerner, J., Huang, Y., Hutter, S., Gurnari, C., Sanchez, J. M. H., Wang, J., et al. (2022). Circulating microbial content in myeloid malignancy patients is associated with disease subtypes and patient outcomes. Nat. Commun. 13 (1), 1038. doi:10.1038/s41467-022-28678-x
Wong, C. C., and Yu, J. (2023). Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 20 (7), 429–452. doi:10.1038/s41571-023-00766-x
Wong-Rolle, A., Dong, Q., Zhu, Y., Divakar, P., Hor, J. L., Kedei, N., et al. (2022). Spatial meta-transcriptomics reveal associations of intratumor bacteria burden with lung cancer cells showing a distinct oncogenic signature. J. Immunother. Cancer 10 (7), e004698. doi:10.1136/jitc-2022-004698
World Health Organization (2023a). Breast cancer. Available online at: https://www.who.int/news-room/fact-sheets/detail/breast-cancer.
World Health Organization (2023b). Colorectal cancer. Available online at: https://www.who.int/news-room/fact-sheets/detail/colorectal-cancer.
World Health Organization, I. (2024). Absolute numbers, mortality, in lip, oral cavity, salivary glands, nasopharynx, hypopharynx, and larynx in 2022. Available online at: https://gco.iarc.who.int.
Yang, L., Tu, L., Bisht, S., Mao, Y., Petkovich, D., Thursby, S. J., et al. (2024). Tissue-location-specific transcription programs drive tumor dependencies in colon cancer. Nat. Commun. 15 (1), 1384. doi:10.1038/s41467-024-45605-4
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of MicroRNA-21. Gastroenterology 152 (4), 851–866 e824. doi:10.1053/j.gastro.2016.11.018
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170 (3), 548–563.e16. doi:10.1016/j.cell.2017.07.008
Zepeda-Rivera, M., Minot, S. S., Bouzek, H., Wu, H., Blanco-Miguez, A., Manghi, P., et al. (2024). A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature 628 (8007), 424–432. doi:10.1038/s41586-024-07182-w
Zervantonakis, I. K., Hughes-Alford, S. K., Charest, J. L., Condeelis, J. S., Gertler, F. B., and Kamm, R. D. (2012). Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. U. S. A. 109 (34), 13515–13520. doi:10.1073/pnas.1210182109
Zhang, J., Wang, P., Wang, J., Wei, X., and Wang, M. (2024). Unveiling intratumoral microbiota: an emerging force for colorectal cancer diagnosis and therapy. Pharmacol. Res. 203, 107185. doi:10.1016/j.phrs.2024.107185
Keywords: tumor on-a-chip, host-microbes, microbes, tissue engineering, microfluidic devices
Citation: Sousa MGC, Brasino DSK, Krieger M, Dindar DA, Duhen R, Zhang Z, Franca CM and Bertassoni LE (2025) Host-microbe-cancer interactions on-a-chip. Front. Bioeng. Biotechnol. 13:1505963. doi: 10.3389/fbioe.2025.1505963
Received: 04 October 2024; Accepted: 11 March 2025;
Published: 31 March 2025.
Edited by:
Stefaan Verbruggen, Queen Mary University of London, United KingdomReviewed by:
Revathy Ramachandran, National Institutes of Health (NIH), United StatesCopyright © 2025 Sousa, Brasino, Krieger, Dindar, Duhen, Zhang, Franca and Bertassoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luiz E. Bertassoni, YmVydGFzc29Ab2hzdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.