- 1Department of Biology, Gondar College of Teachers Education, Gondar, Ethiopia
- 2Institute of Biotechnology, University of Gondar, Gondar, Ethiopia
These days, bioethanol research is looking at using non-edible plant materials, called lignocellulosic feedstocks, because they are cheap, plentiful, and renewable. However, these materials are complex and require pretreatment to release fermentable sugars. Saccharomyces cerevisiae, the industrial workhorse for bioethanol production, thrives in sugary environments and can handle high levels of ethanol. However, during lignocellulose fermentation, S. cerevisiae faces challenges like high sugar and ethanol concentrations, elevated temperatures, and even some toxic substances present in the pretreated feedstocks. Also, S. cerevisiae struggles to efficiently convert all the sugars (hexose and pentose) present in lignocellulosic hydrolysates. That’s why scientists are exploring the natural variations within Saccharomyces strains and even figuring out ways to improve them. This review highlights why Saccharomyces cerevisiae remains a crucial player for large-scale bioethanol production from lignocellulose and discusses the potential of genome shuffling to create even more efficient yeast strains.
1 Introduction
The growing energy crisis and rising greenhouse gas concerns are driving the development of bioethanol, a transportation fuel derived from renewable sources. Bioethanol is already a significant contributor to renewable energy in many countries, even being the leading bio-based product globally in terms of volume and economic value (Favaro et al., 2019). However, current bioethanol production often relies on food crops as feedstock. Earlier reports showed that sugarcane and sugar beet account for about 40% of the global bioethanol production, with starch-based feedstocks contributing the remaining 60% (Dhar et al., 2012). This raises concerns about potential competition with food production and indirect land-use change emissions. Additionally, the cost of production can be high, with feedstock costs accounting for up to 70% of the final price (Topaloglu et al., 2023). This highlights the importance of developing bioethanol production processes that utilize readily available and inexpensive resources. In Europe, some pilot and demonstration bioethanol plants from lignocellulosic biomass are either operational or in the process of being commissioned (https://www.etipbioenergy.eu/fact-sheets/ethanol-fact-sheet, accessed on 27 August 2024).
At the global scale, bioethanol is primarily produced from either starch or selected edible agricultural crops like corn, sugar cane, and sugar beet (Busic et al., 2018; Tse et al., 2021). The utilization of agricultural residues and other low-value carbohydrate sources for bioethanol production is of great interest due to economic and environmental factors (Favaro et al., 2019). Lignocellulosic materials, including residues from agriculture and forestry, are gaining prominence in bioethanol studies owing to their abundance, renewable nature, and sustainable characteristics. Furthermore, these materials do not pose a competition with food or fuel crops, rendering them a favorable choice (Tse et al., 2021; Arora et al., 2015; Zhang and Geng, 2012). Lignocellulosic biomass primarily comprises cellulose (40%–60%), hemicellulose (20%–40%), and lignin (10%–25%) (Deng et al., 2023). The breakdown of lignocellulosic biomass through hydrolysis produces hexose sugars like glucose, predominantly from cellulose, and pentose sugars such as xylose and arabinose from hemicellulose (Cunha et al., 2019; Nijland and Driessen, 2020). Therefore, an ideal microorganism should have the capability to ferment both hexose and pentose sugars to enhance the economic viability of bioethanol industry.
Despite the considerable potential it holds, the utilization of lignocellulosic substrates in the production of bioethanol is faced with numerous obstacles. These challenges include the necessity of a pre-treatment phase, inhibitors that arise from the pre-treatment process, and the incapacity of S. cerevisiae to simultaneously ferment glucose and xylose (Deng et al., 2023; Sandri et al., 2023). Industrial strains for lignocellulosic ethanol production need to be ethanol tolerant, temperature-tolerant (typically above 40°C), osmo- and pH-tolerant, and inhibitor-tolerant (Jansen et al., 2017). Therefore, developing an S. cerevisiae strain with high capacity for stress tolerance and co-fermentation of glucose and xylose has been the subject of extensive research in recent years.
Efficient utilization of lignocellulosic biomass for biofuel production requires a proficient microorganism capable of fermenting the feedstocks while tackling the challenges associated with pretreatment and hydrolysis techniques. The target for the industry should be to achieve a minimum of 90% theoretical yields, which equates to around 0.511 g of ethanol per g of glucose consumed (Della-Bianca et al., 2013; Walker and Walker, 2018), where the residual sugar is <2 g/L (Tao et al., 2012). S. cerevisiae emerges as the predominant microorganism utilized in industrial ethanol manufacturing due to its remarkable fermentation capabilities and ability to withstand stress factors (Topaloglu et al., 2023; Wang et al., 2022). In the realm of industrial-scale ethanol production, S. cerevisiae consistently attains a yield exceeding 90% of the theoretical maximum (Della-Bianca et al., 2013). Hence, even a marginal enhancement in ethanol yield could result in hundreds of millions of dollars in extra profits annually (Ruchala et al., 2019).
However, S. cerevisiae encounters various environmental stressors throughout the commercial fermentation process. For example, optimal saccharification typically occurs at temperatures ranging from 45°C to 50°C, implying that S. cerevisiae must withstand elevated temperatures and high ethanol concentrations during the latter stages of the fermentation process. Furthermore, strains intended for very high-gravity fermentation (>250 g/L glucose) must exhibit tolerance to high sugar concentrations (Wang et al., 2022). This review highlights the industrially relevant attributes of S. cerevisiae and the contribution of genome shuffling in achieving these traits.
2 Desired attributes of strains to ferment lignocellulosic biomass
Lignocellulosic biomass represents a vast, renewable resource for biofuel production. The conversion of this biomass into fermentable sugars and subsequently into bioethanol or other valuable chemicals is a promising alternative to fossil fuels. However, the efficient fermentation of lignocellulosic hydrolysates requires robust yeast strains with specific attributes to overcome the challenges posed by the complex nature of the biomass. The essential characteristics of an industrial strain necessary for fermenting lignocellulosic hydrolysate include rapid fermentation kinetics, high ethanol yield, tolerance to ethanol, sugars, and fermentation inhibitors, and efficient utilization of both hexoses and pentose sugars (Zhang and Geng, 2012; Mukherjee et al., 2014; Zhao et al., 2024). However, the conditions encountered during bioethanol fermentation present several challenges, including exceedingly high levels of sugar and ethanol, elevated temperatures, and the presence of toxic compounds. Consequently, the development of an industrial yeast strain with enhanced thermotolerance, osmotolerance, and resistance to secondary metabolites has become imperative for bioethanol production (Wawro, 2018; Kong et al., 2018). Exploring the natural diversity within Saccharomyces is widely perceived as an intriguing approach to discover superior bioethanol strains and gain insights into the challenges faced by yeast cells during bioethanol production (Mukherjee et al., 2014).
Although several bacteria, such as Zymomonas mobilis and genetically modified Escherichia coli, can ferment sugars, S. cerevisiae remains the preferred organism for industrial ethanol production (Dien et al., 2003; Moyses et al., 2016; Techaparin et al., 2017). This preference primarily stems from its high tolerance to ethanol and its ability to ferment under strictly anaerobic conditions. Additionally, S. cerevisiae is resilient to low pH and resistant to bacteriophage infection, all of which are crucial factors in large-scale industrial processes (Moyses et al., 2016; Techaparin et al., 2017). Therefore, a strong multiple stress-tolerance is the desirable characteristic for S. cerevisiae when different feedstocks are used for economical industrial ethanol production. This review examines the desired attributes of yeast strains for fermenting lignocellulosic biomass, focusing on their ability to tolerate elevated temperature and high ethanol concentration, efficiently utilize available sugars, tolerate inhibitory compounds, and perform under very high-gravity fermentation conditions.
2.1 Temperature tolerance
Numerous critical factors must be considered in the production of industrial ethanol. One pivotal environmental factor affecting ethanol production is temperature. The optimal temperature range for yeast growth is typically between 25°C and 35°C. However, heat stress significantly impacts industrial ethanol production (Techaparin et al., 2017). According to Prado et al. (2020), ethanol-producing strains in Brazil ferment within the range of 28°C–33°C, with mills employing water coolers to regulate temperature. Nevertheless, addressing the challenge of ethanol production under high temperatures can be achieved through the utilization of thermotolerant microbes (Prado et al., 2020; Phong et al., 2022). Hence, it is generally imperative to explore the natural diversity of Saccharomyces strains to find superior thermotolerant bioethanol strains from new and less explored areas.
Interest in producing ethanol at high temperatures has been driven by several advantages, including enhanced metabolism, accelerated fermentation rates and yield, decreased media viscosity, reduced energy consumption, and minimized contamination risks (Techaparin et al., 2017; Prado et al., 2020; Phong et al., 2022; Pattanakittivorakul et al., 2019). Moreover, the predominant method for producing lignocellulosic ethanol involves separate hydrolysis and fermentation processes, which increase costs and require distinct vessels for saccharification (40°C–50°C) and fermentation (30°C–35°C) (Yuan et al., 2017; Choudhary et al., 2017). Thus, there is a need to develop a simultaneous saccharification and fermentation process where the average temperature for both steps is approximately 40°C which will allow simultaneous saccharification and fermentation of lignocellulosic biomass (Prado et al., 2020). Mostly, mesophilic yeasts are employed for fermentation; however, a simultaneous saccharification and fermentation approach necessitates thermotolerant yeast capable of withstanding the optimal temperature range of saccharifying enzymes. Consequently, a critical aspect of bioethanol production via simultaneous saccharification and fermentation is the isolation of thermotolerant ethanologenic yeasts. Abrams and Brem (2022) showed that S. cerevisiae maintained viability at temperature 37°C, whereas S. paradoxus became inviable. Novel thermotolerant S. cerevisiae strains obtained by different researchers had been reviewed and presented in Table 1.
Table 1. A list of thermotolerant S. cerevisiae strains that can produce ethanol from different lignocellulosic substrates.
Numerous researchers have identified many thermotolerant strains of S. cerevisiae from various sources, demonstrating their ability to produce ethanol at elevated temperatures (Table 1). In nearly all cases, the optimal temperature for ethanol production was reported to be 40°C. Notably, S. cerevisiae KKU-VN8, S. cerevisiae JRC6, S. cerevisiae TC-5 and S. cerevisiae HG1.1 achieve over 90% of the theoretical ethanol yield at 40°C, meeting the industrial threshold. On the other hand, the residual sugar in many of the strains was not reported though the industrial requirement is less than 2 g/L. Some of the strains like DMKU 3-S087 and TBRC 12151, though thermotolerant, produced ethanol below the industrially significant threshold of 90% theoretical yield. Besides thermotolerance, some strains, such as S. cerevisiae UV-20, tolerate 50% glucose and exhibit remarkable ethanol tolerance of up to 21% (v/v) (Yi et al., 2018). In contrast, S. cerevisiae KKU-VN8 and S. cerevisiae TBRC 12151 resist 13% (v/v) and 12% (v/v) ethanol, respectively (Table 1).
Several studies have been conducted to improve the thermotolerance of S. cerevisiae through different mechanisms. For example, Yang et al. (2023) modified S. cerevisiae’s sterol composition using CRISPR–Cas9 system to boost thermotolerance. Others used adaptive laboratory evolution (Zhang B. et al., 2023), regulating expression of spt23 (Lu et al., 2022), regulation of transcription factors (Gan et al., 2021), manipulation of heat shock protein genes (Zhang M. L. et al., 2023) to enhance yeasts’ response to temperature. Since thermotolerance is just one aspect, it is essential to evaluate additional characteristics of a strain beyond its thermotolerance. Hence genome shuffling can be a powerful tool because it has the potential to improve multiple traits simultaneously by creating a combinatorial effect from beneficial mutations across the genome.
2.2 Ethanol tolerance
Saccharomyces cerevisiae can metabolize sugar to produce ethanol at levels that are harmful to most organisms (Morard et al., 2019; Riles and Fay, 2019). However, there are variations in ethanol tolerance among different strains (Morard et al., 2019; Vamvakas and Kapolos, 2020). During the final stages of bioethanol production, S. cerevisiae strains are exposed to high ethanol concentrations. This ethanol stress often slows down or halts fermentation, resulting in reduced ethanol yield (Snoek et al., 2015; Tao et al., 2012). Therefore, new strains with improved ethanol tolerance are needed to increase efficiency and maximize ethanol production at an industrial scale.
The ethanol tolerance of S. cerevisiae is a complex trait controlled by numerous genes and cannot be easily modified by altering a single gene. Research has shown that improvements in ethanol tolerance result from extensive genomic changes (Tao et al., 2012). Nevertheless, there existed disparities in ethanol tolerance across various yeast strains (Morard et al., 2019; Vamvakas and Kapolos, 2020; Wolf et al., 2023). The severity of a given ethanol concentration can vary between strains. For example, if one strain is tolerant to 5% ethanol and another to 10%, only the first strain will activate stress-specific pathways when both are exposed to 5% ethanol. This makes it challenging to compare their ability to cope with ethanol stress since 5% ethanol is severe only for the first strain (Wolf et al., 2023).
Different researchers employed various strategies to enhance the ethanol tolerance of S. cerevisiae. Morard et al. (2019) identified that polysomy of chromosome III plays a significant role in ethanol tolerance. Strains with an extra copy of this chromosome showed higher ethanol tolerance, which could be reversed by restoring euploidy. Others have shown that the overexpression of certain genes can enhance ethanol tolerance. For instance, Varize et al. (2022) reported that the overexpression of the TRP1 gene (involved in tryptophan biosynthesis) or MSN2 gene (general stress response activator) increased ethanol tolerance up to 14%. Wang et al. (2022) identified genes like ASP3 (encodes L-asparaginase II), ENA5 (encodes a P-type ATPase to reduce cytotoxicity), YOL162W and YOR012W of unknown function, and two transcription factors (Crz1p, Tos8p-that may regulate multiple-tolerant phenotypes) could be used as gene targets to improve multiple stress-tolerance (high ethanol, high temperature, high glucose, high salt) and ethanol production capacities of S. cerevisiae.
Similarly, Samakkarn et al. (2021) reported a transcription factor called Znf1 that plays a central role in ethanol stress response by activating genes for glycerol and fatty acid production to preserve plasma membrane integrity. Comparative genomics has revealed that ethanol-tolerant strains exhibit distinct genetic modifications, where the ethanol tolerance strains are associated with a combination of beneficial alleles across the genome (Wohlbach et al., 2014). To cope with ethanol stress, strains alter the levels of key metabolites, including amino acids, organic acids, and fatty acids, indicating a reprogramming of metabolic fluxes (Ming et al., 2019). In their findings, Haas et al. (2019) suggested that ethanol tolerance is under natural evolutionary fitness-selection for an optimum phenotype that would tend to eliminate alleles of large effect. Despite substantial research endeavors in recent years dedicated to pinpointing evolutionary pathways that could boost ethanol tolerance, this trait remains partially elucidated (Morard et al., 2019; Ming et al., 2019).
Ethanol tolerance is also influenced by temperature. Studies by Riles and Fay (2019) identified that amino acid polymorphisms in the SEC24 gene contribute to ethanol sensitivity at elevated temperatures. Other researchers, like Techaparin et al. (2017) and Kruasuwan et al. (2023), obtained strains that could tolerate up to 12% ethanol at 40°C and up to 13% ethanol at 45°C, respectively. However, there are limited studies reporting on the interplay between thermal and ethanol stress responses.
Natural isolates of S. cerevisiae from various environments exhibit inherent ethanol tolerance. For instance, strains isolated from vineyards in the Western Cape, South Africa displayed high phenotypic diversity including the ability to grow at 45°C and in the presence of 20% (v/v) ethanol (Jansen et al., 2017). It has been found by Zheng and Wang (2015) that isolates from human-associated environments exhibited higher stress tolerance compared to those from forests unaffected by human activity. This may be attributed to evolutionary adaptations to environments influenced by human activities. Dhar et al. (2012) demonstrated that yeasts exhibit adaptive responses to fluctuating stress conditions, characterized by the development of cross-protection and anticipatory gene regulation. Thus, investigating the genetic diversity of such kind of isolates can provide valuable insights into the mechanisms of ethanol tolerance (Minnaar and Haan, 2023).
Therefore, ethanol tolerance in S. cerevisiae is a multifaceted trait governed by genetic, physiological, and environmental factors. Future research should explore the complex interplay between these factors to develop robust yeast strains capable of sustaining high ethanol yields in industrial bioethanol production.
2.3 Tolerance to lignocellulosic-derived inhibitors
Lignocellulosic biomass is composed of cellulose, hemicellulose, and lignin, which are tightly bound together. Pretreatment is necessary to break down this complex structure to enhance the enzymatic hydrolysis of cellulose into fermentable sugars. Common pretreatment methods include acid hydrolysis, steam explosion, and alkaline treatment. Although these methods are effective in breaking down the biomass, they produce various inhibitory byproducts such as acetic acid, formic acid, furfural, 5-hydroxymethylfurfural (HMF), and phenolic compounds (Li et al., 2022; Divate et al., 2022). These inhibitors can negatively impact the growth, metabolism, and fermentation performance of S. cerevisiae, reducing ethanol yield and overall productivity (Cunha et al., 2019; Li et al., 2022; Ko et al., 2019; Brandt et al., 2019). Therefore, understanding yeast responses and adaptations to these stresses is crucial for developing strategies to improve yeast resilience and bioconversion efficiency from lignocellulosic biomass (Cunha et al., 2019).
Enhancing yeast robustness against lignocellulosic-derived inhibitors is crucial for transitioning to a bio-based economy (Moreno et al., 2022; Wang et al., 2015). Engineering yeast to overexpress genes involved in the detoxification of inhibitors has shown to improve tolerance. For example, overexpression of ARI1 (encodes aldehyde reductase) appears to confer higher tolerance to aldehyde inhibitors, thereby increasing the growth rate and ethanol production capacity of S. cerevisiae in an aldehyde-containing environment (Divate et al., 2022). Wang et al. (2015) also demonstrated that overexpressing the key genes PRO1 or INO1, which enhance the synthesis of proline or myo-inositol, significantly increases yeast tolerance to furfural, acetic acid, and phenol (FAP). Ren et al. (2024) demonstrated the effectiveness of a combined X-ray radiation and adaptive evolution approach in creating furfural-resistant yeast mutants. Another strategy involves combining beneficial traits from multiple strains through iterative cycles of DNA recombination and selection, known as genome shuffling, resulting in strains with enhanced tolerance to inhibitors (Cunha et al., 2019).
Natural S. cerevisiae isolates exhibit niche-specific phenotypic and metabolic diversity that has evolved to overcome external stresses, utilize available resources, and thrive in challenging environments (Jansen et al., 2017; Witt et al., 2019). In contrast, industrial and laboratory strains lack these adaptations due to domestication (De Witt et al., 2019). For example, Jansen et al. (2017) identified strains from various vineyards with high tolerance to ethanol (20% v/v), temperature (45°C), pH (2–11), and lignocellulosic inhibitor cocktails (25%). Other studies suggest that a strain’s exposure to specific environments influences its phenotypic resistance to lignocellulosic inhibitors (Narayanan et al., 2017). Pre-cultivating yeast in lignocellulosic hydrolysate containing furfural and HMF induces the expression of NADPH-dependent oxidoreductases, which convert aldehyde groups into less inhibitory furfuryl alcohols. Consequently, pre-exposing cell populations to moderate levels of inhibitors during the pre-cultivation phase can enhance fermentation efficiency of lignocellulosic substrates (Narayanan et al., 2017).
Therefore, natural strains can be a valuable resource for mitigating engineering constraints by studying the molecular mechanisms involved in phenotypic variance and informing future industrial strain improvements for lignocellulosic hydrolysates (Witt et al., 2019). While significant progress has been made through genetic engineering and the use of natural isolates, further research is needed to fully understand the complex interactions between yeast cells and inhibitors.
2.4 Co-fermentation of hexose and pentose sugars
The utilization of all types of sugars obtained from the process of biomass pretreatment, namely hexoses and pentoses, plays a crucial role in the advancement of viable processes within the framework of a bio-refinery. Microorganisms have the ability to convert hexoses into various end products through biochemical pathways. However, a significant obstacle lies in the effective conversion of pentoses, especially when they are present alongside hexoses. The enhancement of sugar conversion during co-fermentation poses a substantial challenge that researchers and industry professionals are actively working to address (Sandri et al., 2023; Jamaluddin et al., 2023). In industrial bioethanol production, S. cerevisiae is commonly utilized due to its rapid digestion of hexose sugars and high tolerance to ethanol compared to other species. However, wild S. cerevisiae strains are unable to ferment pentose sugars, specifically xylose, despite it being the second most abundant sugar in lignocellulosic hydrolysates (Zhang and Geng, 2012; Li et al., 2024; Zhu et al., 2020). In contrast, non-Saccharomyces yeasts such as Kluyveromyces marxianus, Scheffersomyces (Pichia) stipitis, Pachysolen tannophilus, and Candida shehatae are capable of fermenting pentose sugars. Nevertheless, they have not been considered for large-scale production due to their low ethanol yield. Consequently, the majority of research on pentose fermentation in yeast has focused on genetically engineering S. cerevisiae strains to incorporate heterologous xylose metabolic pathways (Cunha et al., 2019; Moyses et al., 2016; Ko et al., 2016; Liu et al., 2018; Nijland et al., 2023).
Efforts to genetically modify S. cerevisiae to incorporate xylose metabolic pathways have had limited success and does not enable yeast to rapidly utilize xylose. Several limitations still need to be addressed, including glucose inhibition and slow xylose transport, cofactor imbalance in the xylose reductase/xylitol dehydrogenase pathway, functional expression of a heterologous xylose isomerase, the low efficiency of downstream pathways and low ethanol production (Hou et al., 2017; Sharma et al., 2021; Ko et al., 2016). To facilitate the conversion of D-xylose into bioethanol, two distinct pathways have been integrated and optimized in S. cerevisiae: the xylose reductase/xylitol dehydrogenase (XR/XDH) pathway and the xylose isomerase (XI) pathway (Cunha et al., 2019; Ko et al., 2016; Liu et al., 2018; Nijland et al., 2023). Regardless of the approach employed in genetic engineering, S. cerevisiae strains designed to metabolize D-xylose rely on the activity of xylulose kinase (Xks1), which catalyzes the conversion of D-xylulose to D-xylulose-5-phosphate at the cost of one ATP molecule (Nijland et al., 2023). Given that this enzymatic reaction represents a crucial step in D-xylose metabolism, it is conceivable that precise control of XKS1 expression may be necessary. Nevertheless, the regulatory mechanisms associated with D-xylose metabolism in S. cerevisiae seem to be absent, possibly due to the fact that this particular sugar is not recognized as a primary carbon source (Sharma et al., 2021; Han et al., 2010). Through combined efforts of reinforcing the pathway of xylose catabolism and elevating the fermentation temperature, Tran et al. (2020) achieved a simultaneous co-fermentation of lignocellulosic hydrolysates, composed of 39.6 g/L glucose and 23.1 g/L xylose, within 24 h yielding 0.48 g/g ethanol (94% theoretical maximum).
Enhancing S. cerevisiae’s ability to efficiently utilize pentose sugars can also be achieved by engineering endogenous hexose transporters (Hxt) to function as pentose transporters. However, these modified transport proteins remain susceptible to glucose-level controlled degradation (Nijland and Driessen, 2020; Nijland et al., 2016). A more successful approach involves engineering the endogenous Hxt transporter family and using evolutionary selection for D-glucose insensitive growth on pentose sugars. This strategy has led to the identification of a critical and conserved asparagine residue in Hxt transporters that, when mutated, reduces D-glucose affinity while maintaining D-xylose affinity (Nijland and Driessen, 2020).
While several xylose-consuming S. cerevisiae strains have been constructed by the heterologous expression of the XR/XDH or the XI pathways, the reported ethanol yields from xylose are still far below the theoretical yield (Hou et al., 2017; Ko et al., 2016; Hoang et al., 2018). On the other hand, there is a lack of consensus in the few studies comparing the use of the different xylose-consuming pathways: while all describe the strains containing the XI pathway as capable of reaching higher ethanol yields than the ones containing the XR/XDH pathway, the XI role in xylose consumption rates and ethanol productivity is not clear (Karhumaa et al., 2007; Kobayashi et al., 2018; Li et al., 2016).
At industrially relevant concentrations, the co-consumption of D-xylose and D-glucose has been realized through transporter engineering when expressed in a strain lacking hxt1-7 (hexose transporters) and gal2 (galactose permease) genes (Nijland and Driessen, 2020). Although this is the main achievement, to translate this method to real industrial conditions will still be a challenge because sugar metabolism flux is distributed over multiple sugars, as the maximal capacity of primary metabolism is exploited. Specifically, D-xylose utilization and growth rates are still significantly lower as compared to D-glucose, and thus a careful balance is necessary throughout fermentation (Nijland and Driessen, 2020). Therefore, in addition to the directed genetic manipulations, random mutational approaches would provide better insights into complex network linked with xylose metabolism. This would assist in the development of an improved strain with mixed sugar utilization/fermentation potential (Sharma et al., 2021).
One of the random mutational approaches to develop an effective strain with improved capability is protoplast fusion (Sharma et al., 2021). It causes random mutations in the genomes of microorganisms without requiring any pre-targeted approach. This is applicable for developing inter-specific, intraspecific, and inter-generic, intra-generic super hybrids with higher capability. It is a significant tool for genetic manipulation as it resolves the barrier to genetic exchange imposed by conventional mating systems. It is particularly useful for industrially important microorganisms (Zhang and Geng, 2012; Sharma et al., 2021; Kahar and Tanaka, 2014).
Several researchers have employed genome shuffling to produce fusants/hybrids with desired traits. For instance, Pasha et al. (2007) constructed a fusant strain, F11, showing a 0.459 g/L ethanol yield with 90% fermentation efficiency. Likewise, Sharma et al. (2021) employed protoplast fusion to derive a hybrid strain from S. cerevisiae LN and Pichia stipitis NCIM 3498. They showed higher ethanol tolerance (10%), enhanced XR, XDH, and XKS activities, and exhibited the ability to ferment glucose in lignocellulosic hydrolysate, with xylose consumption (∼40%) surpassing that of S. cerevisiae LN. Zhang and Geng (2012) obtained a potential recombinant yeast strain, after two rounds of genome shuffling (ScF2), from S. cerevisiae and P. stipites that can utilize a high concentration of xylose (100–250 g/L xylose). This hybrid, ScF2, produced ethanol more rapidly than the naturally occurring xylose-fermenting P. stipitis, with an improved ethanol titer and a much greater increase in xylose tolerance. In another study, the gene shuffling protocol using the highly homologues hexose transporters family provides a powerful tool to enhance the D-xylose affinity of Hxt transporters in S. cerevisiae to increase the D-xylose uptake flux at low D-xylose concentrations (Nijland et al., 2018). Through protoplast fusion and metabolic engineering, Zhao et al. (2024) created a recombinant S. cerevisiae strain, BLH510, capable of co-fermenting glucose, xylose, cellobiose, and xylooligosaccharides while exhibiting tolerance to inhibitors commonly present in lignocellulosic hydrolysates (109). These findings suggest that S. cerevisiae strains can be genetically modified to efficiently utilize a wider range of sugars found in lignocellulosic biomass.
2.5 High- and very-high-gravity fermentation
Ethanol production continues to advance within industrial settings. One notable innovation that has significantly transformed industrial ethanol production is very high-gravity (VHG) fermentation, also known as high-sugar-concentration fermentation (Camargos et al., 2020; Cruz et al., 2020). Generally, sugar concentrations for ethanol production can be divided into normal gravity (<180 g/L total sugars), high gravity (180 - 240 g/L of total sugars), and very high gravity (≥250 g/L of total sugars) (Bai et al., 2008; Deesuth et al., 2015). Both HG and VHG technologies have the potential to reduce distillation costs and yield up to 120 g/L ethanol (Cruz et al., 2020; Liu et al., 2012) and facilitate increased ethanol production without necessitating plant capacity or workforce adjustments. Very high gravity fermentation can achieve more than 15% (v/v) of ethanol, compared to the average of 10%–12% (v/v) that is observed in most distilleries (Puligundla et al., 2011). VHG technology, in particular, holds promise as a highly effective industrial ethanol production method due to its ability to generate high ethanol yields, minimal waste, and cost-effectiveness (Tse et al., 2021; Cruz et al., 2020; Arshad et al., 2017). Compared to submerged and solid-state fermentation, very high gravity fermentation has the greatest potential for high ethanol yields (Tse et al., 2021). However, under VHG fermentation conditions, yeasts undergo multiple stresses due to increased metals and sodium ions, nutrient stresses, increased temperatures, acidic conditions, osmotic stress, and increased ethanol concentrations (Tse et al., 2021; Varize et al., 2022; Puligundla et al., 2011).
Several studies reported ethanol production under HG and VHG fermentation conditions. For instance, Camargos et al. (2020) reported an ethanol concentration of 104.4 g/L from 222 g/L total reducing sugars under fed-batch fermentation conditions with a maximum ethanol productivity of 2.98 g/L.h from 209 g/L total reducing sugars. In a similar study, an ethanol concentration of 135 g/L, with ethanol productivity of 4.42 g/L.h, and a 90% ethanol yield were achieved at a sugar concentration of 300 g/L (Cruz et al., 2020). These findings demonstrate the feasibility of HG and VHG fermentation for commercial-scale ethanol production. However, VHG fermentation is not without drawbacks, including unfermented sugars and the risk of stuck fermentation, leading to prolonged fermentation times (Tao et al., 2012). Osmotic stress, glycerol production, and substrate and ethanol inhibition are among the adverse effects associated with VHG fermentation (Tao et al., 2012; Camargos et al., 2020). Tao et al. (2012) used a combination of metabolic engineering (to delete glycerol gene) and genome shuffling to circumvent the limitations of VHG fermentation and improved the bioethanol production performance of S. cerevisiae. They found that the fusant, SZ3-1, was capable of effectively fermenting 280 g/L glucose within 72 h while simultaneously maintaining a high fermentation rate, enhanced ethanol tolerance, and a low glycerol yield. Similarly, Zheng et al. (2013) obtained isolates with significantly enhanced stress tolerance and ethanol titer under VHG conditions after whole-genome shuffling of the S. cerevisiae ZTW1. They attributed the stress tolerance largely due to copy number variations in large DNA regions.
Some studies claimed that it is difficult to change properties under the control of multiple genes through classical breeding, metabolic engineering, and other genetic manipulation methods with specific genes or pathways as targets (Teixeira et al., 2012; Wang and Hou, 2010). However, whole genome engineering approaches involving yeast cell ploidy manipulation and genome shuffling can be employed to improve the fermentation performances under VHG conditions (Teixeira et al., 2012; Hou et al., 2015). Hou et al. (2015) compared the methods of metabolic pathway modification, cell ploidy manipulation, global transcription machinery engineering, and genome shuffling to improve ethanol production under VHG fermentation. The results suggested that genome shuffling was the most effective way to significantly manipulate yeast strains where by ethanol production was enhanced by up to 11% more than the control, reaching 120 g ethanol/L in 35 h using 300 g glucose/L. The results demonstrated that genome shuffling was valuable for creating yeast strains with desired multiplex traits during VHG fermentations. However, further studies are necessary to verify this finding.
3 Potential of genome shuffling in microbial strain improvement
Microbial strains have been extensively utilized for the production of various valuable commodities across different industries. However, the majority of natural strains cannot be directly used for industrial purposes due to limitations in their productivity, stability, and resistance to environmental stresses. To address these challenges, one effective approach is strain improvement (Chen et al., 2020; Magocha et al., 2018; Sanghavi et al., 2020). Among the techniques available for strain improvement, genome shuffling has emerged as a promising method for rapid enhancement of microbial strains, yielding new strains by combining whole genomes of multiple parental microorganisms using recursive protoplast fusion principles (Chen et al., 2020; Magocha et al., 2018; Hospet et al., 2023). It is a laboratory evolution method involving the combinatorial evolution of complex phenotypes in whole organisms by genome-scale and recursive recombination of mutants (Biot-Pelletier and Martin, 2014). Genome shuffling has brought a major breakthrough in the strain-improvement concept as it is found to be effective and reliable for expressing complex phenotypes (Magocha et al., 2018).
Genome shuffling has some attributes similar to those of classical strain-improvement as both offer genomic diversification and screening for improved strains. In case of random mutagenesis, it generates microbial strains with desired traits and does not necessitate knowledge of the microbe’s genetic makeup, however, it is a labor-intensive process. In contrast, recombinant DNA technology, metabolic engineering, and genome editing techniques are rational genetic engineering methods with a narrow scope of application, as they require a comprehensive understanding of the genetics of the target strain and specialized genetic tools (Chen et al., 2020). To accelerate the enhancement of complex traits and swiftly observe phenotypic improvements in industrially significant strains, genome shuffling has emerged as a more practical and innovative whole-genome improvement technique. Genome shuffling facilitates recombination among multiple parents in each generation, and through repeated fusion of protoplasts, fusants successfully amalgamate genetic traits from diverse parents (Hospet et al., 2023). In conventional protoplast fusion between 2 cells with diverse genetic backgrounds, robust recombinants exhibiting traits from both parents can emerge. Here, the fusion of only two parents in a single generation induces recombination. Conversely, genome shuffling facilitates multiple rounds of genome fusion and the amalgamation of numerous parents from each generation (Zhang and Geng, 2012; Chen et al., 2020; Hospet et al., 2023) without requiring genome sequencing data (Magocha et al., 2018; Biot-Pelletier and Martin, 2014). It can be used in most laboratories without expensive equipment (Gong et al., 2009). Furthermore, genome shuffling allows for the fusion of isolated DNA fragments with a cell’s genome, merging genomes of organisms spanning distant taxonomic groups (Luna-Flores et al., 2016). This approach significantly enhances the likelihood of obtaining a high yield of the strain and unquestionably expands the genetic diversity of the offspring (Wawro, 2018).
4 Application of genome shuffling on Saccharomyces cerevisiae to enhance bioethanol production
Research shows that genome shuffling enhances the phenotypic traits of S. cerevisiae (Table 2). This technique is widely used to improve the desirable performance of microbes, particularly for complex traits that are challenging to achieve through traditional genetic engineering methods (Gong et al., 2009). Chen et al. (2020) summarized the application of genome shuffling in increasing the yield of various microbial metabolites, enhancing strain tolerance, and improving substrate utilization across different microbial strains. This review specifically focuses on applying genome shuffling to improve S. cerevisiae strains for bioethanol production.
Shuffled strains of S. cerevisiae exhibited enhanced traits such as xylose utilization, temperature resilience, ethanol production, and high-gravity fermentation (Table 2). For example, Jetti et al. (2018) developed a fusant strain, S. cerevisiae SP2-18, capable of utilizing 34% xylose and tolerating 15% (v/v) ethanol after two rounds of genome shuffling. Utilizing super-strains with elevated temperature, acidity, and ethanol tolerance has proven economically viable for ethanol production. Benjaphokee et al. (2012) created fusants from thermotolerant and high ethanol-producing S. cerevisiae strains, resulting in strains that could tolerate 41°C and a pH of 3.5, producing an ethanol yield of 0.47 g with 91.2% theoretical efficiency. Similarly, Orosco et al. (2017) obtained a shuffled strain capable of growing at 42°C and in 18% ethanol, yielding 10.83% higher ethanol than parental strain and 15.16% over the industrial strain S. cerevisiae HBY3.
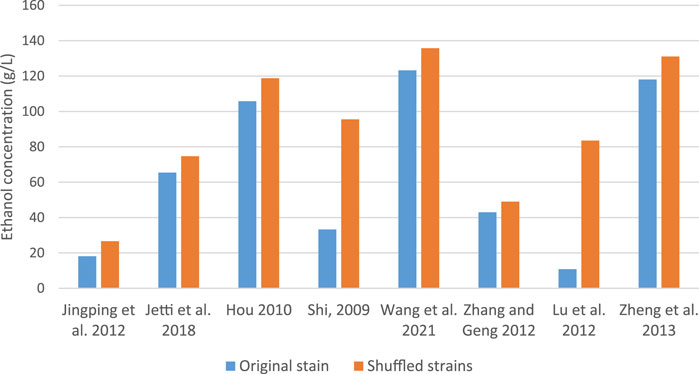
Others like Inokuma et al. (2017) developed a xylose-utilizing S. cerevisiae strain with improved fermentation ability under heat and acid co-stress using genome shuffling. The shuffled strain successfully inherited desirable traits from parental strains and exhibited high xylose fermentation ability under heat and acid co-stress, attributed to the upregulation of genes in Hyb-8 associated with cellular transition metal ion homeostasis. Lu et al. (2012) suggested that trehalose accumulation in recombinant strains contributes to thermotolerance and other stress tolerances. Lin et al. (2019) also created hybrid strains with improved glucose-xylose co-fermentation properties at high temperatures by combining genome shuffling with adaptive evolution. Hou et al. (2015) compared the ethanol production of strains generated by four methods: metabolic pathway modification, cell ploidy manipulation, global transcription machinery engineering and genome shuffling. Notably, ethanol yield in a strain created by genome shuffling enhanced by 11% as compared to the parent strain. As illustrated in Figure 1, shuffled S. cerevisiae strains consistently produced higher ethanol yields compared to their respective parental strains. The more pronounced differences observed in the studies of Shi et al. (2009) and Lu et al. (2012) may be attributed to variations in the specific strains used and the fermentation conditions employed.
Overall, genome shuffling has been employed to confer traits such as thermotolerance, ethanol tolerance, xylose utilization, high-gravity fermentation, and multi-stress tolerance. Studies have demonstrated that just two or three rounds of genome shuffling can generate potential S. cerevisiae hybrids with desirable characteristics. Therefore, employing genome shuffling for strain enhancement serves as a crucial method to address complex phenotypic features and facilitate the rapid evolution of strains with industrial significance.
5 Conclusion and future prospects
Bioethanol derived from lignocellulosic feedstocks presents a compelling option for the development of sustainable biofuels. Saccharomyces cerevisiae stands out as the outstanding microorganism for industrial bioethanol production due to its exceptional fermentation capabilities and tolerance to harsh environments. However, the commercial fermentation process exposes S. cerevisiae to a multitude of stresses. For optimal lignocellulosic bioethanol production, industrial yeast strains require a specific range of traits, including rapid fermentation rates, high ethanol yields, tolerance to fermentation inhibitors and various sugars, and efficient utilization of both hexose and pentose sugars. Unfortunately, most naturally occurring strains lack the requisite productivity or stress tolerance for direct industrial application.
Strain improvement strategies present a promising avenue to overcome these challenges. Wild isolates of S. cerevisiae harbor an extensive reservoir of genetic diversity that can be tapped through genetic modifications to explore the genetic foundations of novel phenotypes. Genome shuffling has emerged as a potent tool for rapidly enhancing complex traits critical to industrial applications. This technique facilitates precise recombination among multiple parental strains in each generation. By conducting repeated protoplast fusions, these “fusants” effectively consolidate the genetic strengths inherited from numerous parents. However, genome shuffling alone may not suffice to achieve strains meeting industrial requirements and should therefore be complemented with other genetic manipulation strategies.
Overall, the combination of lignocellulosic feedstocks, S. cerevisiae, and strain improvement via genome shuffling holds immense promise for the future of sustainable bioethanol production. This approach can potentially yield optimized yeast strains that enhance the efficiency of bioethanol conversion from lignocellulose, thereby advancing the environmental sustainability and economic competitiveness of the biofuel industry. Moreover, future research should consider reporting the theoretical efficiency and residual sugar activity of microbial strains before asserting their industrial applicability.
Author contributions
KT: Conceptualization, Writing–original draft, Writing–review and editing. MA: Writing–review and editing. NB: Supervision, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article..
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrams, M. B., and Brem, R. B. (2022). Temperature-dependent genetics of thermotolerance between yeast species. Front. Ecol. Evol. 10, 859904–859906. doi:10.3389/fevo.2022.859904
Arora, R., Behera, S., Sharma, N. K., and Kumar, S. (2015). A new search for thermotolerant yeasts, its characterization and optimization using response surface methodology for ethanol production. Front. Microbiol. 6 (889), 1–16. doi:10.3389/fmicb.2015.00889
Arshad, M., Hussain, T., Iqbal, M., and Abbas, M. (2017). Enhanced ethanol production at commercial scale from molasses using high gravity technology by mutant S. cerevisiae. Braz J. Microbiol. 48 (3), 403–409. doi:10.1016/j.bjm.2017.02.003
Bai, F. W., Anderson, W. A., and Moo-Young, M. (2008). Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnol. Adv. 26 (1), 89–105. doi:10.1016/j.biotechadv.2007.09.002
Benjaphokee, S., Hasegawa, D., Yokota, D., Asvarak, T., Auesukaree, C., Sugiyama, M., et al. (2012). Highly efficient bioethanol production by a Saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol. N. Biotechnol. 29 (3), 379–386. doi:10.1016/j.nbt.2011.07.002
Biot-Pelletier, D., and Martin, V. J. (2014). Evolutionary engineering by genome shuffling. Appl. Microbiol. Biotechnol. 98 (9), 3877–3887. doi:10.1007/s00253-014-5616-8
Boonchuay, P., Techapun, C., Leksawasdi, N., Seesuriyachan, P., Hanmoungjai, P., Watanabe, M., et al. (2021). Bioethanol production from cellulose-rich corncob residue by the thermotolerant Saccharomyces cerevisiae TC-5. J. Fungi (Basel) 7 (7), 547. doi:10.3390/jof7070547
Brandt, B. A., Jansen, T., Görgens, J. F., and van Zyl, W. H. (2019). Overcoming lignocellulose-derived microbial inhibitors: advancing the Saccharomyces cerevisiae resistance toolbox. Biofuels, Bioprod. Biorefining 13 (6), 1520–1536. doi:10.1002/bbb.2042
Busic, A., Marđetko, N., Kundas, S., Morzak, G., Belskaya, H., Ivančić Šantek, M., et al. (2018). Bioethanol production from renewable raw materials and its separation and purification: a review. Food Technol. Biotechnol. 56 (3), 289–311. doi:10.17113/ftb.56.03.18.5546
Camargos, C. V., Moraes, V. D., de Oliveira, L. M., Guidini, C. Z., Ribeiro, E. J., and Santos, L. D. (2020). High gravity and very high gravity fermentation of sugarcane molasses by flocculating Saccharomyces cerevisiae: experimental investigation and kinetic modeling. Appl. Biochem. Biotechnol. 193 (3), 807–821. doi:10.1007/s12010-020-03466-9
Chen, L., Xin, Q. H., Ma, L. M., Li, R. F., and Bian, K. (2020). Applications and research advance of genome shuffling for industrial microbial strains improvement. World J. Microbiol. Biotechnol. 36 (10), 158. doi:10.1007/s11274-020-02936-w
Cheng, C., Almario, M. P., and Kao, K. C. (2015). Genome shuffling to generate recombinant yeasts for tolerance to inhibitors present in lignocellulosic hydrolysates. Biotechnol. Lett. 37 (11), 2193–2200. doi:10.1007/s10529-015-1895-0
Choudhary, J., Singh, S., and Nain, L. (2017). Bioprospecting thermotolerant ethanologenic yeasts for simultaneous saccharification and fermentation from diverse environments. J. Biosci. Bioeng. 123 (3), 342–346. doi:10.1016/j.jbiosc.2016.10.007
Cruz, M. L., de Resende, M. M., and Ribeiro, E. J. (2020). Improvement of ethanol production in fed-batch fermentation using a mixture of sugarcane juice and molasse under very high-gravity conditions. Bioprocess Biosyst. Eng. 44 (3), 617–625. doi:10.1007/s00449-020-02462-x
Cunha, J. T., Soares, P. O., Romaní, A., Thevelein, J. M., and Domingues, L. (2019). Xylose fermentation efficiency of industrial Saccharomyces cerevisiae yeast with separate or combined xylose reductase/xylitol dehydrogenase and xylose isomerase pathways. Biotechnol. Biofuels 12 (20), 20–14. doi:10.1186/s13068-019-1360-8
Deesuth, O., Laopaiboon, P., Klanrit, P., and Laopaiboon, L. (2015). Improvement of ethanol production from sweet sorghum juice under high gravity and very high gravity conditions: effects of nutrient supplementation and aeration. Industrial Crops Prod. 74, 95–102. doi:10.1016/j.indcrop.2015.04.068
Della-Bianca, B. E., Basso, T. O., Stambuk, B. U., Basso, L. C., and Gombert, A. K. (2013). What do we know about the yeast strains from the Brazilian fuel ethanol industry?. Appl. Microbiol. Biotechnol. 97 (3), 979–991. doi:10.1007/s00253-012-4631-x
Demeke, M. M., Dietz, H., Li, Y., Foulquié-Moreno, M. R., Mutturi, S., Deprez, S., et al. (2013). Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol. Biofuels 6 (89), 89–24. doi:10.1186/1754-6834-6-89
Deng, W., Feng, Y., Fu, J., Guo, H., Guo, Y., Han, B., et al. (2023). Catalytic conversion of lignocellulosic biomass into chemicals and fuels. Green Energy and Environ. 8 (1), 10–114. doi:10.1016/j.gee.2022.07.003
De Witt, R. N., Kroukamp, H., and Volschenk, H. (2019). Proteome response of two natural strains of Saccharomyces cerevisiae with divergent lignocellulosic inhibitor stress tolerance. FEMS Yeast Res. 19 (1). doi:10.1093/femsyr/foy116
Dhar, R., Sagesser, R., Weikert, C., and Wagner, A. (2012). Yeast adapts to a changing stressful environment by evolving cross-protection and anticipatory gene regulation. Mol. Biol. Evol. 30 (3), 573–588. doi:10.1093/molbev/mss253
Dien, B. S., Cotta, M. A., and Jeffries, T. W. (2003). Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63 (3), 258–266. doi:10.1007/s00253-003-1444-y
Divate, N. R., Huang, P. J., Chen, G. H., and Chung, Y. C. (2022). Construction of recombinant Saccharomyces cerevisiae with ethanol and aldehydes tolerance via overexpression of aldehyde reductase. Microorganisms 10 (5), 850. doi:10.3390/microorganisms10050850
Favaro, L., Jansen, T., and Zyl, W. H. v. (2019). Exploring industrial and natural Saccharomyces cerevisiae strains for the bio-based economy from biomass: the case of bioethanol. Crit. Rev. Biotechnol., 1–18. doi:10.1080/07388551.2019.1619157
Gan, Y., Qi, X., Lin, Y., Guo, Y., Zhang, Y., and Wang, Q. (2021). A hierarchical transcriptional regulatory network required for long-term thermal stress tolerance in an industrial Saccharomyces cerevisiae strain. Front. Bioeng. Biotechnol. 9, 826238. doi:10.3389/fbioe.2021.826238
Gong, J., Zheng, H., Wu, Z., Chen, T., and Zhao, X. (2009). Genome shuffling: progress and applications for phenotype improvement. Biotechnol. Adv. 27 (6), 996–1005. doi:10.1016/j.biotechadv.2009.05.016
Haas, R., Horev, G., Lipkin, E., Kesten, I., Portnoy, M., Buhnik-Rosenblau, K., et al. (2019). Mapping ethanol tolerance in budding yeast reveals high genetic variation in a wild isolate. Front. Genet. 10, 998. doi:10.3389/fgene.2019.00998
Han, J. H., Park, J. Y., Kang, H. W., Choi, G. W., Chung, B. W., and Min, J. (2010). Specific expression patterns of xyl1, xyl2 and xyl3 in response to different sugars in Pichia stipitis. J. Microbiol. Biotechnol. 20 (5), 946–949. doi:10.4014/jmb.0912.12028
Hoang, P. T. N., Ko, J. K., Gong, G., Um, Y., and Lee, S. M. (2018). Genomic and phenotypic characterization of a refactored xylose-utilizing Saccharomyces cerevisiae strain for lignocellulosic biofuel production. Biotechnol. Biofuels 11, 268. doi:10.1186/s13068-018-1269-7
Hospet, R., Thangadurai, D., Cruz-Martins, N., Sangeetha, J., Anu Appaiah, K. A., Chowdhury, Z. Z., et al. (2023). Genome shuffling for phenotypic improvement of industrial strains through recursive protoplast fusion technology. Crit. Rev. Food Sci. Nutr. 63 (17), 2960–2969. doi:10.1080/10408398.2021.1983763
Hou, J., Qiu, C., Shen, Y., Li, H., and Bao, X. (2017). Engineering of Saccharomyces cerevisiae for the efficient co-utilization of glucose and xylose. FEMS Yeast Res. 17 (4), 1–14. doi:10.1093/femsyr/fox034
Hou, L. (2009). Novel methods of genome shuffling in Saccharomyces cerevisiae. Biotechnol. Lett. 31 (5), 671–677. doi:10.1007/s10529-009-9916-5
Hou, L. H., Meng, M., Guo, L., and He, J. y. (2015). A comparison of whole cell directed evolution approaches in breeding of industrial strain of Saccharomyces cerevisiae. Biotechnol. Lett. 37 (7), 1393–1398. doi:10.1007/s10529-015-1812-6
Inokuma, K., Iwamoto, R., Bamba, T., Hasunuma, T., and Kondo, A. (2017). Improvement of xylose fermentation ability under heat and acid Co-stress in Saccharomyces cerevisiae using genome shuffling technique. Front. Bioeng. Biotechnol. 5, 81. doi:10.3389/fbioe.2017.00081
Jamaluddin, , Ida Riyanti, E., Rachmania Mubarik, N., and Listanto, E. (2023). Construction of novel yeast strains from Candida tropicalis KBKTI 10.5.1 and Saccharomyces cerevisiae DBY1 to improve the performance of ethanol production using lignocellulosic hydrolysate. Trop. Life Sci. Res. 34 (2), 81–107. doi:10.21315/tlsr2023.34.2.5
Jansen, T., Hoff, J. W., Jolly, N., and van Zyl, W. H. (2017). Mating of natural Saccharomyces cerevisiae strains for improved glucose fermentation and lignocellulosic inhibitor tolerance. Folia Microbiol. 63, 155–168. doi:10.1007/s12223-017-0546-3
Jetti, K. D., Gns, R. R., Garlapati, D., and Nammi, S. K. (2018). Improved ethanol productivity and ethanol tolerance through genome shuffling of Saccharomyces cerevisiae and Pichia stipitis. Int. Microbiol. 22 (2), 247–254. doi:10.1007/s10123-018-00044-2
Jingping, G., Hongbing, S., Gang, S., Hongzhi, L., and Wenxiang, P. (2012). A genome shuffling-generated Saccharomyces cerevisiae isolate that ferments xylose and glucose to produce high levels of ethanol. J. Ind. Microbiol. Biotechnol. 39, 777–787. doi:10.1007/s10295-011-1076-7
Kahar, P., and Tanaka, S. (2014). A xylose-fermenting yeast hybridized by intergeneric fusion between Saccharomyces cerevisiae and Candida intermedia mutants for ethanol production. Sustain. Chem. Process. 2 (17), 17–12. doi:10.1186/s40508-014-0017-y
Karhumaa, K., Sanchez, R. G., Hahn-Hägerdal, B., and Gorwa-Grauslund, M. F. (2007). Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 6, 5. doi:10.1186/1475-2859-6-5
Ko, J. K., Enkh-Amgalan, T., Gong, G., Um, Y., and Lee, S. (2019). Improved bioconversion of lignocellulosic biomass by Saccharomyces cerevisiae engineered for tolerance to acetic acid. GCB Bioenergy 12 (1), 90–100. doi:10.1111/gcbb.12656
Ko, J. K., Um, Y., Woo, H. M., Kim, K. H., and Lee, S. M. (2016). Ethanol production from lignocellulosic hydrolysates using engineered Saccharomyces cerevisiae harboring xylose isomerase-based pathway. Bioresour. Technol. 209, 290–296. doi:10.1016/j.biortech.2016.02.124
Kobayashi, Y., Sahara, T., Ohgiya, S., Kamagata, Y., and Fujimori, K. E. (2018). Systematic optimization of gene expression of pentose phosphate pathway enhances ethanol production from a glucose/xylose mixed medium in a recombinant Saccharomyces cerevisiae. Amb. Express 8 (1), 139. doi:10.1186/s13568-018-0670-8
Kong, I. I., Turner, T. L., Kim, H., Kim, S. R., and Jin, Y. S. (2018). Phenotypic evaluation and characterization of 21 industrial Saccharomyces cerevisiae yeast strains. FEMS Yeast Res. 18 (1). doi:10.1093/femsyr/foy001
Kruasuwan, W., Puseenam, A., Am-in, S., Trakarnpaiboon, S., Sornlek, W., Kocharin, K., et al. (2023). Evaluation of thermotolerant and ethanol-tolerant Saccharomyces cerevisiae as an alternative strain for bioethanol production from industrial feedstocks. 3 Biotech. 13 (23), 23–12. doi:10.1007/s13205-022-03436-4
Li, B., Liu, N., and Zhao, X. (2022). Response mechanisms of Saccharomyces cerevisiae to the stress factors present in lignocellulose hydrolysate and strategies for constructing robust strains. Biotechnol. Biofuels Bioprod. 15 (28), 28–20. doi:10.1186/s13068-022-02127-9
Li, F., Bai, W., Zhang, Y., Zhang, Z., Zhang, D., Shen, N., et al. (2024). Construction of an economical xylose-utilizing Saccharomyces cerevisiae and its ethanol fermentation. FEMS Yeast Res. 24, 1–10. doi:10.1093/femsyr/foae001
Li, X., Park, A., Estrela, R., Kim, S. R., Jin, Y. S., and Cate, J. H. (2016). Comparison of xylose fermentation by two high-performance engineered strains of Saccharomyces cerevisiae. Biotechnol. Rep. 9, 53–56. doi:10.1016/j.btre.2016.01.003
Lin, Y., Cai, Y., Guo, Y., Li, X., Qi, X., Qi, Q., et al. (2019). Development and genomic elucidation of hybrid yeast with improved glucose-xylose co-fermentation at high temperature. FEMS Yeast Res. 19 (3), foz015. doi:10.1093/femsyr/foz015
Liu, C.-G., Wang, N., Lin, Y. H., and Bai, F. W. (2012). Very high gravity ethanol fermentation by flocculating yeast under redox potential-controlled conditions. Biotechnol. Biofuels 5 (61), 61–67. doi:10.1186/1754-6834-5-61
Liu, T., Huang, S., and Geng, A. (2018). Recombinant diploid Saccharomyces cerevisiae strain development for rapid glucose and xylose Co-fermentation. Fermentation 4 (3), 59. doi:10.3390/fermentation4030059
Lu, Y., Cheng, Y. F., He, X. P., Guo, X. N., and Zhang, B. R. (2012). Improvement of robustness and ethanol production of ethanologenic Saccharomyces cerevisiae under co-stress of heat and inhibitors. J. Ind. Microbiol. Biotechnol. 39 (1), 73–80. doi:10.1007/s10295-011-1001-0
Lu, Z., Wu, Y., Chen, Y., Chen, X., Wu, R., Lu, Q., et al. (2022). Role of spt23 in Saccharomyces cerevisiae thermal tolerance. Appl. Microbiol. Biotechnol. 106 (9-10), 3691–3705. doi:10.1007/s00253-022-11920-3
Luna-Flores, C. H., Palfreyman, R. W., Krömer, J. O., Nielsen, L. K., and Marcellin, E. (2016). Improved production of propionic acid using genome shuffling. Biotechnol. J. 12 (2). doi:10.1002/biot.201600120
Magocha, T. A., Zabed, H., Yang, M., Yun, J., Zhang, H., and Qi, X. (2018). Improvement of industrially important microbial strains by genome shuffling: current status and future prospects. Bioresour. Technol. 257, 281–289. doi:10.1016/j.biortech.2018.02.118
Ming, M., Wang, X., Lian, L., Zhang, H., Gao, W., Zhu, B., et al. (2019). Metabolic responses of Saccharomyces cerevisiae to ethanol stress using gas chromatography-mass spectrometry. Mol. Omics 15 (216), 216–221. doi:10.1039/c9mo00055k
Minnaar, L., and Haan, R. d. (2023). Engineering natural isolates of Saccharomyces cerevisiae for consolidated bioprocessing of cellulosic feedstocks. Appl. Microbiol. Biotechnol. 107, 7013–7028. doi:10.1007/s00253-023-12729-4
Morard, M., Macías, L. G., Adam, A. C., Lairón-Peris, M., Pérez-Torrado, R., Toft, C., et al. (2019). Aneuploidy and ethanol tolerance in Saccharomyces cerevisiae. Front. Genet. 10, 1–12. doi:10.3389/fgene.2019.00082
Moreno, A. D., González-Fernández, C., and Tomás-Pejó, E. (2022). Insights into cell robustness against lignocellulosic inhibitors and insoluble solids in bioethanol production processes. Sci. Rep. 12 (557), 557–613. doi:10.1038/s41598-021-04554-4
Moyses, D. N., Reis, V., Almeida, J., Moraes, L., and Torres, F. (2016). Xylose fermentation by Saccharomyces cerevisiae: challenges and prospects. Int. J. Mol. Sci. 17 (3), 207. doi:10.3390/ijms17030207
Mukherjee, V., Steensels, J., Lievens, B., Van de Voorde, I., Verplaetse, A., Aerts, G., et al. (2014). Phenotypic evaluation of natural and industrial Saccharomyces yeasts for different traits desirable in industrial bioethanol production. Appl. Microbiol. Biotechnol. 98 (22), 9483–9498. doi:10.1007/s00253-014-6090-z
Narayanan, V., Schelin, J., Gorwa-Grauslund, M., van Niel, E. W., and Carlquist, M. (2017). Increased lignocellulosic inhibitor tolerance of Saccharomyces cerevisiae cell populations in early stationary phase. Biotechnol. Biofuels 10 (114), 114. doi:10.1186/s13068-017-0794-0
Nijland, J. G., and Driessen, A. J. M. (2020). Engineering of pentose transport in Saccharomyces cerevisiae for biotechnological applications. Front. Bioeng. Biotechnol. 7, 464. doi:10.3389/fbioe.2019.00464
Nijland, J. G., Shin, H., de Waal, P., Klaassen, P., and Driessen, A. (2018). Increased xylose affinity of Hxt2 through gene shuffling of hexose transporters in Saccharomyces cerevisiae. J. Appl. Microbiol. 124 (2), 503–510. doi:10.1111/jam.13670
Nijland, J. G., Vos, E., Shin, H. Y., de Waal, P. P., Klaassen, P., and Driessen, A. J. M. (2016). Improving pentose fermentation by preventing ubiquitination of hexose transporters in Saccharomyces cerevisiae. Biotechnol. Biofuels 9, 158. doi:10.1186/s13068-016-0573-3
Nijland, J. G., Zhang, X., and Driessen, A. J. M. (2023). D-xylose accelerated death of pentose metabolizing Saccharomyces cerevisiae. Biotechnol. Biofuels Bioprod. 16 (1), 67. doi:10.1186/s13068-023-02320-4
Orosco, F. L., Estrada, S. M., Simbahan, J. F., Alcantara, V. A., and Pajares, I. G. (2017). Genome shuffling for improved thermotolerance, ethanol tolerance and ethanol production of Saccharomyces cerevisiae 2013. Philipp. Sci. Lett. 10 (1).
Pasha, C., Kuhad, R. C., and Rao, L. V. (2007). Strain improvement of thermotolerant Saccharomyces cerevisiae VS3 strain for better utilization of lignocellulosic substrates: strain improvement of thermotolerant S. cerevisiae. J. Appl. Microbiol. 103 (5), 1480–1489. doi:10.1111/j.1365-2672.2007.03375.x
Pattanakittivorakul, S., Lertwattanasakul, N., Yamada, M., and Limtong, S. (2019). Selection of thermotolerant Saccharomyces cerevisiae for high temperature ethanol production from molasses and increasing ethanol production by strain improvement. Ant. Van Leeuwenhoek 112 (7), 975–990. doi:10.1007/s10482-019-01230-6
Phong, H. X., Klanrit, P., Dung, N. T. P., Thanonkeo, S., Yamada, M., and Thanonkeo, P. (2022). High-temperature ethanol fermentation from pineapple waste hydrolysate and gene expression analysis of thermotolerant yeast Saccharomyces cerevisiae. Sci. Rep. 12 (1), 13965. doi:10.1038/s41598-022-18212-w
Prado, C. D., Mandrujano, G. P. L., Souza, J. P., Sgobbi, F. B., Novaes, H. R., da Silva, J. P. M. O., et al. (2020). Physiological characterization of a new thermotolerant yeast strain isolated during Brazilian ethanol production, and its application in high-temperature fermentation. Biotechnol. Biofuels 13, 178. doi:10.1186/s13068-020-01817-6
Puligundla, P., Smogrovicova, D., Obulam, V. S. R., and Ko, S. (2011). Very high gravity (VHG) ethanolic brewing and fermentation: a research update. J. Ind. Microbiol. Biotechnol. 38 (9), 1133–1144. doi:10.1007/s10295-011-0999-3
Ren, J., Zhang, M., Guo, X., Zhou, X., Ding, N., Lei, C., et al. (2024). Furfural tolerance of mutant Saccharomyces cerevisiae selected via ionizing radiation combined with adaptive laboratory evolution. Biotechnol. Biofuels Bioprod. 17 (1), 117. doi:10.1186/s13068-024-02562-w
Riles, L., and Fay, J. C. (2019). Genetic basis of variation in heat and ethanol tolerance in Saccharomyces cerevisiae. Genes. genomes.Genet. 9, 179–188. doi:10.1534/g3.118.200566
Ruchala, J., Kurylenko, O. O., Dmytruk, K. V., and Sibirny, A. A. (2019). Construction of advanced producers of first- and second-generation ethanol in Saccharomyces cerevisiae and selected species of non-conventional yeasts (Scheffersomyces stipitis, Ogataea polymorpha). J. Industrial Microbiol. and Biotechnol., 1–24. doi:10.1007/s10295-019-02242-x
Samakkarn, W., Ratanakhanokchai, K., and Soontorngun, N. (2021). Reprogramming of the ethanol stress response in Saccharomyces cerevisiae by the transcription factor Znf1 and its effect on the biosynthesis of glycerol and ethanol. Appl. Environ. Microbiol. 87 (16), e0058821. doi:10.1128/aem.00588-21
Sandri, J. P., Milessi, T. S., Zangirolami, T. C., and Mussatto, S. I. (2023). Screening of yeast co-culture using crude hydrolysate for co-fermentation of pentose and hexose. Biofuels, Bioprod. Bioref 17, 1639–1653. doi:10.1002/bbb.2529
Sanghavi, G., Gupta, P., Singh, R. M., Oza, T., Trivedi, U., and Singh, N. K. (2020). “Microbial strain engineering,” in Engineering of microbial biosynthetic pathways. Editor V. Singh (Singapore: Springer Nature).
Sharma, S., Ghoshal, C., Arora, A., Samar, W., Nain, L., and Paul, D. (2021). Strain improvement of native Saccharomyces cerevisiae LN ITCC 8246 strain through protoplast fusion to enhance its xylose uptake. Appl. Biochem. Biotechnol. 193 (8), 2455–2469. doi:10.1007/s12010-021-03539-3
Shi, D.-j., Wang, C.-l., and Wang, K.-m. (2009). Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 36, 139–147. doi:10.1007/s10295-008-0481-z
Snoek, T., Picca Nicolino, M., Van den Bremt, S., Mertens, S., Saels, V., Verplaetse, A., et al. (2015). Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance. Biotechnol. Biofuels 8 (32), 32–19. doi:10.1186/s13068-015-0216-0
Tao, X., Zheng, D., Liu, T., Wang, P., Zhao, W., Zhu, M., et al. (2012). A novel strategy to construct yeast Saccharomyces cerevisiae strains for very high gravity fermentation. Plos One 7 (2), 312355–e31310. doi:10.1371/journal.pone.0031235
Techaparin, A., Thanonkeo, P., and Klanrit, P. (2017). High-temperature ethanol production using thermotolerant yeast newly isolated from Greater Mekong Subregion. Braz J. Microbiol. 48 (3), 461–475. doi:10.1016/j.bjm.2017.01.006
Teixeira, M. C., Godinho, C. P., Cabrito, T. R., Mira, N. P., and Sá-Correia, I. (2012). Increased expression of the yeast multidrug resistance ABC transporter Pdr18 leads to increased ethanol tolerance and ethanol production in high gravity alcoholic fermentation. Microb. Cell Factories 11 (98), 98–99. doi:10.1186/1475-2859-11-98
Topaloglu, A., Esen, Ö., Turanlı-Yıldız, B., Arslan, M., and Çakar, Z. P. (2023). From Saccharomyces cerevisiae to ethanol: unlocking the power of evolutionary engineering in metabolic engineering applications. J. Fungi 9 (984), 984–1026. doi:10.3390/jof9100984
Tran, P. H. N., Ko, J. K., Gong, G., Um, Y., and Lee, S. M. (2020). Improved simultaneous co-fermentation of glucose and xylose by Saccharomyces cerevisiae for efficient lignocellulosic biorefinery. Biotechnol. Biofuels 13 (12), 1–14. doi:10.1186/s13068-019-1641-2
Tse, T. J., Wiens, D. J., and Reaney, M. J. T. (2021). Production of bioethanol—a review of factors affecting ethanol yield. Fermentation 7 (268), 268–318. doi:10.3390/fermentation7040268
Vamvakas, S. S., and Kapolos, J. (2020). Factors affecting yeast ethanol tolerance and fermentation efficiency. World J. Microbiol. Biotechnol. 36 (114), 114–118. doi:10.1007/s11274-020-02881-8
Varize, C. S., Bücker, A., Lopes, L. D., Christofoleti-Furlan, R. M., Raposo, M. S., Basso, L. C., et al. (2022). Increasing ethanol tolerance and ethanol production in an industrial fuel ethanol Saccharomyces cerevisiae strain. Fermentation 8 (10), 470. doi:10.3390/fermentation8100470
Walker, G. M., and Walker, R. S. K. (2018). Enhancing yeast alcoholic fermentations. Adv. Appl. Microbiol. 105, 87–129. doi:10.1016/bs.aambs.2018.05.003
Wang, H., and Hou, L. (2010). Genome shuffling to improve fermentation properties of top-fermenting yeast by the improvement of stress tolerance. Food Sci. Biotechnol. 19 (1), 145–150. doi:10.1007/s10068-010-0020-3
Wang, L., Li, B., Su, R. R., Wang, S. P., Xia, Z. Y., Xie, C. Y., et al. (2022). Screening novel genes by a comprehensive strategy to construct multiple stress-tolerant industrial Saccharomyces cerevisiae with prominent bioethanol production. Biotechnol. Biofuels Bioprod. 15 (11), 11–19. doi:10.1186/s13068-022-02109-x
Wang, L., Li, B., Wang, S. P., Xia, Z. Y., Gou, M., and Tang, Y. Q. (2021). Improving multiple stress-tolerance of a flocculating industrial Saccharomyces cerevisiae strain by random mutagenesis and hybridization. Process Biochem. 102, 275–285. doi:10.1016/j.procbio.2020.12.022
Wang, X., Bai, X., Chen, D. F., Chen, F. Z., Li, B. Z., and Yuan, Y. J. (2015). Increasing proline and myo-inositol improves tolerance of Saccharomyces cerevisiae to the mixture of multiple lignocellulose-derived inhibitors. Biotechnol. Biofuels 8 (142), 142–213. doi:10.1186/s13068-015-0329-5
Wawro, A. (2018). Genome shuffling as an alternative method of improving the properties of distillery yeast. Postępy Mikrobiologii - Adv. Microbiol. 57 (3), 278–285. doi:10.21307/pm-2018.57.3.278
Witt, R. N. d., Kroukamp, H., Van Zyl, W. H., Paulsen, I. T., and Volschenk, H. (2019). QTL analysis of natural Saccharomyces cerevisiae isolates reveals unique alleles involved in lignocellulosic inhibitor tolerance. FEMS Yeast Res. 19 (5), foz047. doi:10.1093/femsyr/foz047
Wohlbach, D. J., Rovinskiy, N., Lewis, J. A., Sardi, M., Schackwitz, W. S., Martin, J. A., et al. (2014). Comparative genomics of Saccharomyces cerevisiae natural isolates for bioenergy production. Genome Biol. Evol. 6 (9), 2557–2566. doi:10.1093/gbe/evu199
Wolf, I. R., Marques, L. F., de Almeida, L. F., Lázari, L. C., de Moraes, L. N., Cardoso, L. H., et al. (2023). Integrative analysis of the ethanol tolerance of Saccharomyces cerevisiae. Int. J. Mol. Sci. 24 (5646), 5646–5725. doi:10.3390/ijms24065646
Yang, P., Wu, W., Chen, J., Jiang, S., Zheng, Z., Deng, Y., et al. (2023). Thermotolerance improvement of engineered Saccharomyces cerevisiae ERG5 Delta ERG4 Delta ERG3 Delta, molecular mechanism, and its application in corn ethanol production. Biotechnol. Biofuels Bioprod. 16 (1), 66. doi:10.1186/s13068-023-02312-4
Yi, S., Zhang, X., Li, H. x., Du, X. x., Liang, S. w., and Zhao, X. h. (2018). Screening and mutation of Saccharomyces cerevisiae UV-20 with a high yield of second generation bioethanol and high tolerance of temperature, glucose and ethanol. Indian J. Microbiol. 58 (4), 440–447. doi:10.1007/s12088-018-0741-1
Yuan, S. F., Guo, G. L., and Hwang, W. S. (2017). Ethanol production from dilute-acid steam exploded lignocellulosic feedstocks using an isolated multistress-tolerant Pichia kudriavzevii strain. Microb. Biotechnol. 10 (6), 1581–1590. doi:10.1111/1751-7915.12712
Zhang, B., Geberekidan, M., Yan, Z., Yi, X., and Bao, J. (2023a). Very high thermotolerance of an adaptive evolved Saccharomyces cerevisiae in cellulosic ethanol fermentation. Fermentation 9 (4), 393. doi:10.3390/fermentation9040393
Zhang, M. L., Zhang, H., He, Y. x., Wu, Z. h., and Xu, K. (2023b). Improving thermo-tolerance of Saccharomyces cerevisiae by precise regulation of the expression of small HSP. RSC Adv. 13 (51), 36254–36260. doi:10.1039/d3ra05216h
Zhang, W., and Geng, A. (2012). Improved ethanol production by a xylosefermenting recombinant yeast strain constructed through a modified genome shuffling method. Biotechnol. Biofuels 5 (46), 46–11. doi:10.1186/1754-6834-5-46
Zhao, J., Zhao, Y., Wu, L., Yan, N., Yang, S., Xu, L., et al. (2024). Development of a robust Saccharomyces cerevisiae strain for efficient Co-fermentation of mixed sugars and enhanced inhibitor tolerance through protoplast fusion. Microorganisms 12 (8), 1526. doi:10.3390/microorganisms12081526
Zheng, D. Q., Chen, J., Zhang, K., Gao, K. H., Li, O., Wang, P. M., et al. (2013). Genomic structural variations contribute to trait improvement during whole-genome shuffling of yeast. Appl. Microbiol. Biotechnol. 98 (7), 3059–3070. doi:10.1007/s00253-013-5423-7
Zheng, D.-Q., Wu, X. C., Tao, X. L., Wang, P. M., Li, P., Chi, X. Q., et al. (2011). Screening and construction of Saccharomyces cerevisiae strains with improved multi-tolerance and bioethanol fermentation performance. Bioresour. Technol. 102, 3020–3027. doi:10.1016/j.biortech.2010.09.122
Zheng, Y. L., and Wang, S. A. (2015). Stress tolerance variations in Saccharomyces cerevisiae strains from diverse ecological sources and geographical locations. PLoS One 10 (8), e0133889. doi:10.1371/journal.pone.0133889
Keywords: bioethanol, genome shuffling, lignocellulose, Saccharomyces cerevisiae, strain improvement
Citation: Tsegaye KN, Alemnew M and Berhane N (2024) Saccharomyces cerevisiae for lignocellulosic ethanol production: a look at key attributes and genome shuffling. Front. Bioeng. Biotechnol. 12:1466644. doi: 10.3389/fbioe.2024.1466644
Received: 18 July 2024; Accepted: 17 September 2024;
Published: 25 September 2024.
Edited by:
Haifeng Zhao, South China University of Technology, ChinaReviewed by:
Dung Minh Ha-Tran, National Chung Hsing University, TaiwanAna P. Jacobus, São Paulo State University, Brazil
Copyright © 2024 Tsegaye, Alemnew and Berhane. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kindu Nibret Tsegaye, a2luZHV0c2VnYXllMjFAZ21haWwuY29t
 Kindu Nibret Tsegaye
Kindu Nibret Tsegaye Marew Alemnew
Marew Alemnew Nega Berhane
Nega Berhane