
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 13 March 2024
Sec. Biosensors and Biomolecular Electronics
Volume 12 - 2024 | https://doi.org/10.3389/fbioe.2024.1363186
This article is part of the Research Topic Magnetic Particle-Assisted Sensing and Magnetic Biosensors View all 5 articles
 Michael Sveiven1
Michael Sveiven1 Ana K. Serrano2
Ana K. Serrano2 Joshua Rosenberg3
Joshua Rosenberg3 Douglas J. Conrad4
Douglas J. Conrad4 Drew A. Hall1,3*
Drew A. Hall1,3* Anthony J. O’Donoghue5*
Anthony J. O’Donoghue5*Hydrolytic enzymes play crucial roles in cellular processes, and dysregulation of their activities is implicated in various physiological and pathological conditions. These enzymes cleave substrates such as peptide bonds, phosphodiester bonds, glycosidic bonds, and other esters. Detecting aberrant hydrolase activity is vital for understanding disease mechanisms and developing targeted therapeutic interventions. This study introduces a novel approach to measuring hydrolase activity using giant magnetoresistive (GMR) spin valve sensors. These sensors change resistance in response to magnetic fields, and here, they are functionalized with specific substrates for hydrolases conjugated to magnetic nanoparticles (MNPs). When a hydrolase cleaves its substrate, the tethered magnetic nanoparticle detaches, causing a measurable shift in the sensor’s resistance. This design translates hydrolase activity into a real-time, activity-dependent signal. The assay is simple, rapid, and requires no washing steps, making it ideal for point-of-care settings. Unlike fluorescent methods, it avoids issues like autofluorescence and photobleaching, broadening its applicability to diverse biofluids. Furthermore, the sensor array contains 80 individually addressable sensors, allowing for the simultaneous measurement of multiple hydrolases in a single reaction. The versatility of this method is demonstrated with substrates for nucleases, Bcu I and DNase I, and the peptidase, human neutrophil elastase. To demonstrate a clinical application, we show that neutrophil elastase in sputum from cystic fibrosis patients hydrolyze the peptide-GMR substrate, and the cleavage rate strongly correlates with a traditional fluorogenic substrate. This innovative assay addresses challenges associated with traditional enzyme measurement techniques, providing a promising tool for real-time quantification of hydrolase activities in diverse biological contexts.
Enzymes play a pivotal role in various cellular processes, and their activities are tightly regulated to maintain cellular homeostasis. Monitoring enzymes is essential to understand cellular processes and disease mechanisms, and many enzyme-based disease biomarkers have been identified. For example, serum amylase and serum lipase assays are used to diagnose acute pancreatitis (Walkowska et al., 2022), alkaline phosphatase activity in blood is a marker for liver or bone conditions (Cannalire et al., 2023), creatine kinase tests diagnose Duchenne’s Muscle Dystrophy in neonates (de Freitas Nakata et al., 2021), and elevated lactate dehydrogenase in blood is associated with poor prognosis in COVID-19 patients, potentially for use as a triage mechanism (Martha et al., 2022). Among enzymes, hydrolases are crucial in regulating intracellular pathways and, as a result, influence diverse physiological and pathological conditions. Hydrolases break down macromolecules such as lipids, carbohydrates, proteins, and nucleic acids. In humans, these enzymes are essential for food digestion, wound healing, cell signaling, and immune defense (Riise et al., 2019; Chen et al., 2020; Rack et al., 2020; Salhi et al., 2021). When these enzymes become dysfunctional, they become drivers of cancer, neurodegeneration, autoimmune disease, and heart disease (Liu et al., 2016; Mondanelli et al., 2019; Moll et al., 2020; Sama et al., 2020). Therefore, developing techniques to detect aberrant hydrolase activity as a disease biomarker provides an important tool for clinicians and researchers.
Nucleases are hydrolases that cleave the phosphodiester bonds between nucleotides in DNA and RNA (Garcia Gonzalez and Hernandez, 2022) and can be broadly divided into DNases and RNases based on their ability to cleave DNA or RNA, respectively (Santa et al., 2021). Nucleases hold great potential as biomarkers for many cancers (Balian and Hernandez, 2021). For example, high DNase I activity is observed in patients with oral and breast cancers, while FEN I is a nuclease that is over-expressed in lung, prostate, brain, gastric, pancreatic, and breast cancer. In addition, RNase I is linked to pancreatic cancer (Lauková et al., 2020; Balian and Hernandez, 2021). Peptidases (or proteases) are hydrolases that cleave the peptide bonds between amino acids in a protein chain. Peptidases are involved in all biological processes, including food digestion, blood clotting, and immune defense (Armstrong, 2001; Walsh and Ahmad, 2002; Shpacovitch et al., 2008; Kårlund et al., 2021). Many diseases are characterized by dysfunctional peptidase activity, including cancer, arthritis, and Alzheimer’s disease (Eatemadi et al., 2017; Lucena and McDougall, 2021; Lichtenthaler et al., 2022). Considering the vast influence of peptidases and nucleases on human health, measuring their activities in biofluids is of great interest.
Common nuclease measurement techniques include hybridization assays, immunohistochemistry, reverse-transcription polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA), mass spectrometry, and western blots (Balian and Hernandez, 2021). Hybridization assays are qualitative and unsuitable for point-of-care testing (Singh et al., 2008), while immunohistochemistry assays are qualitative and require intensive processing (Wang et al., 2014). Reverse transcription-PCR is semi-quantitative and is usually a measurement of nuclease expression, not activity (Wang et al., 2014). ELISA uses fluorophores or chromogenic substrates that measure only enzyme concentration and can have photobleaching issues or incompatibility problems with sample matrices (Zhang et al., 2016). Many of these assays require a microplate reader to measure absorption or fluorescence (Lauková et al., 2020; Balian and Hernandez, 2021). While effective, they are limited by the size and complexity of the spectrophotometers, confining the assays to centralized labs.
Peptidases are typically quantified by fluorescent or colorimetric assays, where cleavage of a peptide sequence leads to a time-dependent increase in signal (Zhang, 2004; Hao Ong and Yang, 2017; Wei et al., 2019). These enzyme assays are amenable to microplate format and have been used extensively for high-throughput screening but have limitations regarding sensitivity, specificity, and adaptability (Oishi et al., 2008; Nozeret et al., 2019; Hammond and Ferro, 2023). Moreover, a persistent challenge inherent to these assays is their susceptibility to background signals, originating from non-specific interactions, autofluorescence, and photobleaching. This susceptibility can curtail the sensitivity and specificity of the measurements, often necessitating rigorous background correction procedures. Several surface-based (i.e., heterogeneous) peptidase assays have been developed that utilize electrochemical, surface plasmon resonance (SPR), or surface-enhanced Raman spectroscopy (SERS) detection. Electrochemical assays are sensitive and amenable to point-of-care use but susceptible to sample matrix effects (Chen et al., 2015; Menon et al., 2020). SPR sensors are label-free and real-time, but concerns exist over the complexity of the optical equipment required (Wang et al., 2019; Chang, 2021). SERS sensors have issues generating a reproducible colorimetric response, which means that the secondary enzyme will need specific conditions for activity (Ding and Yang, 2014). Overall, the assays described exhibit many desirable characteristics, but there is a need for new assays that overcome their shortcomings, especially regarding ease-of-use concerns and equipment complexity.
This study reports on a new technique that measures real-time hydrolase activity using giant magnetoresistive (GMR) sensors, as illustrated in Figure 1. GMR sensors are elaborately engineered thin-film stacks where the operation is deeply rooted in quantum mechanics; specifically, they exhibit a phenomenon known as spin-dependent scattering. This property makes them very sensitive to changes in the local magnetic field, enabling them to be used as ultrasensitive biosensors (Baselt et al., 1998; Wu et al., 2022). Past research has shown the utility of GMR sensors for measuring antigen levels using an antibody-antigen-antibody sandwich assay. The capture antibody is coupled to the GMR sensor and binds to the antigen. The bound antigen is then quantified using a biotinylated detection antibody as it recruits streptavidin-coated magnetic nanoparticles (MNP) close to the sensor surface (Osterfeld et al., 2008; Gaster et al., 2013; Kim et al., 2013; Klein et al., 2019; Lin et al., 2019; Zhou et al., 2019; Antarnusa et al., 2022; Mostufa et al., 2023; Sveiven et al., 2023). The increase in magnetoresistance directly correlates with the concentration of antigen. Miniaturization of the GMR system has been demonstrated, which allows for greater portability (Olazarra et al., 2022; Gaster et al., 2011; Yao et al., 2022). In the hydrolase assay described here, MNPs are tethered to the sensor substrate via a substrate sequence cleavable by the target enzyme. Therefore, the decrease in magneto resistance over time directly correlates with the enzyme concentration.
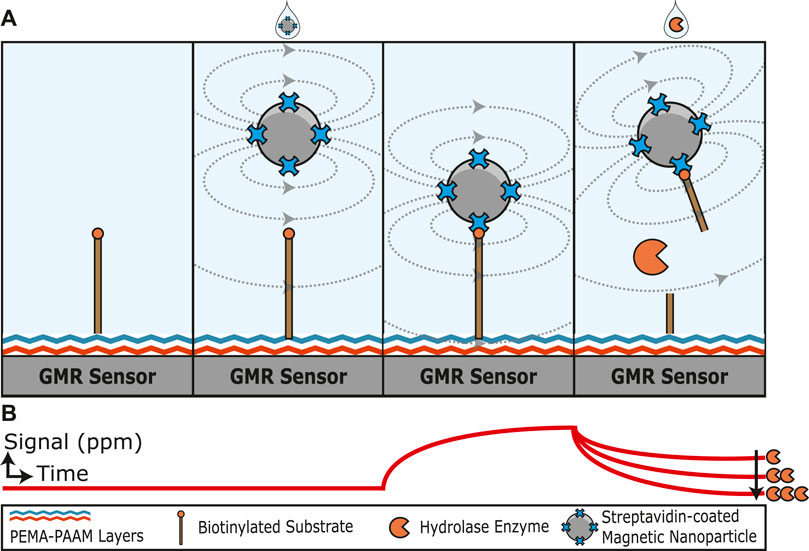
FIGURE 1. Graphical illustration of a hydrolase assay that uses magnetoresistance to quantify substrate cleavage. (A) A biotinylated substrate is covalently attached to the sensor surface through amine coupling. The addition of streptavidin-coated magnetic nanoparticles increases the magnetoresistance signal as the nanoparticles are tethered to the biotin substrate close to the sensor surface. The substrate is cleaved by a hydrolase enzyme and releases the magnetic nanoparticle. (B) Illustration of how the magnetoresistance signal changes with time. In the absence of magnetic nanoparticles, a low signal is detected. Upon the addition of streptavidin-coated magnetic nanoparticles, the signal increases with time as the particles bind to the substrate and are orientated close to the GMR sensor surface. The signal then decreases in proportion to the concentration of active hydrolase enzyme added.
A previous attempt to develop a GMR-based peptidase assay utilized a thioether linkage between a cysteine residue on the substrate and a maleimide-coated surface (Adem et al., 2020). However, the thioether bond was insufficiently stable in aqueous solutions and, therefore, not useful for point-of-care assays that require storage of the assembled substrate on the GMR sensor. We have recently functionalized GMR sensors for covalent binding to antibodies via primary amines and, therefore, used this approach to attach hydrolase substrates to the sensor surface. Prior to the addition of the hydrolase enzyme, the signal for the bound substrate can be quantified using streptavidin-coated MNPs. The signal decreases when the MNPs are released following substrate cleavage, enabling real-time, activity-dependent enzyme monitoring. The substrates consist of an amino acid sequence or a nucleic acid sequence to quantify the activity of a peptidase or nuclease. These substrates are flanked by a biotin molecule to capture streptavidin-coated MNPs and an amine chemical group to bind to the sensor surface. GMR sensors allow for a fast, kinetic, wash-free, portable assay with point-of-care capability. As this assay does not involve optical readout, the common issues of autofluorescence and photobleaching are avoided, enabling higher sensitivity. The GMR sensors are arrayed into 80 individually addressable sensors, creating multiplex ability. This study shows the viability of GMR sensors for quantifying hydrolase activity using specific substrates and highlights the potential of this technology to provide physicians and patients with greater opportunities to detect and monitor diseases.
GMR sensor arrays were purchased from MagArray, Inc (#BZ0078). Each GMR sensor array has 80 sensors arranged in an 8 × 10 matrix, where each sensor is 120 × 120 μm2 on a 280 µm pitch with a nominal resistance (R0) of 1.464 kΩ and a mean magnetoresistance (MR) ratio of 7.99% (Supplementary Figure S1). Each of the 80 sensors can be independently addressed. A custom holder was fabricated from Teflon to create a 100 µL reaction well with an o-ring atop the sensor array (Supplementary Figure S2).
The measurement setup consists of a computer, a power amplifier (Texas Instruments, OPA549), a Helmholtz coil (180 turns of 22 gauge wire per coil, resulting in a 40.5 Oe/A coil constant), and custom readout electronics (Hall et al., 2010a), as shown in Supplementary Figure S3. A double modulation readout scheme rejects 1/f noise from the sensors and electronics, and a temperature compensation technique is used to reduce the temperature drift (Hall et al., 2010b). The computer digitally adjusts the frequency and amplitude of the sensor bias voltage and magnetic field through a National Instruments data acquisition card (PCIe-6351) and a LabVIEW graphical user interface. Specifically, the power amplifier controlled by the computer provides a current to the Helmholtz coil, which creates a homogenous magnetic field (23–34 Oerms based on the sensor MR) for the sensor array. The readout electronics contain 8× transimpedance amplifiers to convert the currents to voltages that the data acquisition card quantizes. Time-multiplexing is applied to read the 8 × 10 sensor array with a 10 s update rate. The measured signal is the change in MR from the initial MR in parts-per-million (ppm).
To functionalize the sensor surface, a thorough cleaning process is initiated by sequential washes with 600 µL of acetone, methanol, and isopropanol. Subsequently, the sensor arrays are subjected to a 10-min treatment in an ultraviolet-ozone cleaner (Uvotech Systems, Helios 500). An optimized protocol consisted of adding 100 µL of 1% Poly (allylamine) in 200 mM MES, pH 6.0 to the sensor wells for 10 min, followed by a rinse with 600 µL of deionized (DI) water. The sensors are then baked at 110°C for 90 min in an oven under atmospheric conditions (Thermo Scientific #PR305225G). Poly (ethylene-alt-maleic anhydride) (PEMA) is rendered aqueous by immersing the microcentrifuge tube containing PEMA in a gently boiling water bath (∼100°C) on a hot plate set to 170°C for 90 min, before adding it to the sensor surface. Then 100 µL of 2% aqueous PEMA in 200 mM MES pH 6.0, filtered through a 0.22 µm filter, is placed onto the sensors for 5 min. The sensors are rinsed with 1 mL DI water and then baked at 160°C for 1 h before spotting. This protocol is adapted from a previous study (Kim et al., 2013).
Peptide, DNA, and PEG substrates are diluted in a printing buffer consisting of PBS, 1 M betaine, and 12.5% 2,3-butanediol. Individual sensors are spotted with the substrate using an iTWO-300P automated spotter (axiVEND, Florida). Twenty droplets of ∼100 pL are spotted on each sensor, sufficient to cover the sensor (Supplementary Figure S4). The automated spotter chamber is then brought to 70% humidity for 1 h, and the sensors are incubated overnight.
To assess the stability of the functionalized sensors, EZ-Link™ Amine-PEG11-Biotin (Thermo Fisher #26136) is coupled to the sensor, as described above. 1% ethanolamine is added and incubated for 30 min at room temperature. The sensor is placed in the GMR readout station, and 50 µL of streptavidin-coated magnetic nanoparticles (Miltenyi Biotec #130–048–101) is added and incubated for 15 min. The sensor is washed with 1 mL of PBS and then incubated for 21 days at 4°C under humid conditions by placing wet Kim wipes in a Petri dish with parafilm to create a seal. The stability evaluation is initiated by returning the assay to the GMR readout station for measurement. Next, the sensors are washed with 1 mL PBS, and streptavidin is blocked by incubating with 100 µL of 1 mM biotin in PBS. Stability is assessed by incubating the sensor array for 5 min each with 0.2, 1, and 5 M NaCl in water. The sensor is then washed briefly with water and sequentially incubated with PBS at 22, 4, and 50°C for 5 min each. For pH stability, the sensor is incubated in PBS adjusted to pH 3 with HCl. It is then incubated in PBS pH 7.2 and PBS adjusted to pH 9 and pH 13.5 with NaOH.
The sensors are functionalized with a substrate (specific to the target enzyme) containing a biotin on one end and an amine group on the other. After overnight incubation with the substrate, the sensors are blocked for 30 min using 1% ethanolamine, followed by a 1 mL wash with PBS. The sensors are placed into the GMR readout station, and 50 µL of MNPs (Miltenyi Biotec #130–048–101) is added. The sensor resistance is measured continuously for 15 min. Then, a 1 mM solution of biotin in PBS is added for 15 min followed by a wash with 1 mL PBS. The enzyme solution is added to the sensors to initiate the magnetic enzyme activity assay, and the MR signal is measured.
The sensors are functionalized with 9.56 ng of double-stranded DNA (5′– CCCCACTAGTAAAAAAAAAAAAAAAAAAAA–biotin–3′, Complementary: 5′– ACTAGTGGGGAAAAAAAAAAAAAAAAAAAA–NH2–3′), where the 3′end of one strand is derivatized with biotin and the 3′end of the other strand is derivatized with an amine group. The Bcu I recognition site (ACTAGT) is underlined. A double-stranded DNA sequence containing a scrambled Bcu I site (TACATG) was also synthesized as a control. The assay setup is described in the magnetic enzyme activity assay section above. To initiate the nuclease assay, 15 µL of the restriction enzyme Bcu I (Thermo Scientific #FD1254) is mixed with 45 µL of fast digest buffer (Thermo Scientific #FD1254) and added onto the sensors. Activity is monitored for 25 min at room temperature. The DNase I is set up under similar conditions, except for 3 units (3 µL) of DNase I solution in 97 µL of DNase I buffer (Thermo Scientific #EN0521). For the sequential Bcu I/DNase I assay, the sensors were functionalized using the Bcu I DNA substrate and the scrambled Bcu I sequence. 6.5 µL of Bcu I solution is combined with 43.5 µL of fast digest buffer for 15 min on the sensor. This is followed by a washing step with 1 mL of PBS before adding 5 units (5 µL) of DNase I in 45 µL of DNase I buffer for 10 min.
The peptidase assay setup follows the protocol described in the nuclease assay, except the sensors are functionalized with a linker sequence consisting of biotin-PEG36-RQPVnWG-PEG36-NH2 or a scrambled version of the same peptide, biotin-PEG36-VWnRQGP-PEG36-NH2. The assay is initiated by adding 100 µL of 677 nM (20 μg/mL) human neutrophil elastase (Athens Research & Technology #16-14–051200) in PBS containing 0.01% Tween-20. To determine repeatability, three independent assays were performed using 100 μL of 500 nM human neutrophil elastase in PBS, 0.01% Tween-20, and the change in MR was recorded after 20 min of incubation. For the enzyme concentration curve assays, the enzyme was serially 2-fold diluted from 125 to 3.9 nM. For inhibition assays, the sensors are functionalized with the human neutrophil elastase substrate or its scrambled counterpart. The inhibitor, sivelestat sodium (VWR #89161–706), is introduced at 0, 4, or 20 μg/mL in a 30 µL PBS, 0.01% Tween-20 onto the sensors for 10 min. Subsequently, 677 nM of HNE is introduced into the inhibitor solution and the change in MR is recorded after 10 min.
Sputum samples were collected from adult cystic fibrosis patients (>18 years) according to a UC San Diego institutional review board-approved protocol for human subject research (#160078) from the UC San Diego Adult Cystic Fibrosis Clinic during routine visits (Quinn et al., 2019). Samples were diluted 1:20 in PBS and stored at −20°C. Before use, samples are thawed, diluted 5-fold in PBS, and then mixed with an equal volume of 10 µM Ala-Ala-Pro-Val-7-amino-4-methylcoumarin (Alfa Aesar) in a black 384-well plate at a final volume of 30 µL such that the final dilution of sputum is 1 in 200. The reaction is incubated at 37°C for 2 h assays in a Synergy HTX microplate reader (BioTek), and readings are obtained in 47 s intervals at excitation and emission wavelengths of 360 and 460 nm, respectively. Enzyme velocity in relative fluorescent units per sec (RFU/s) is calculated using the highest slope recorded for 10 consecutive fluorescent readings, and the mean and standard deviation are determined from three technical replicates.
The cystic fibrosis sputum assay setup follows the protocol described above in the magnetic enzymatic activity assay section. The sensors are functionalized with the human neutrophil elastase substrate. Cystic fibrosis samples are prepared by diluting the frozen stocks 1:10 in PBS containing 0.01% Tween-20, then applying 50 µL of the diluted sample onto the sensors. The final dilution of sputum is 1 in 200.
All data shown are the mean values with one standard deviation as error bars. Sensors that show a signal of more than 117 ppm or less than −117 ppm before MNP addition were excluded. In the hydrolase assays, the sensor was excluded if it did not have sufficient loading after MNP addition (3,300 ppm for human neutrophil elastase substrates and 470 ppm for restriction enzyme substrates). Statistical analysis (Pearson’s coefficient and Deming analysis) is done with custom-written code using NumPy (v1.18.5) and SciPy (v1.6.0) in Python (v3.8). The max negative velocity (-[ΔMR/MR0]/s) for the magnetic neutrophil elastase assays is calculated using LinearRegression from sklearn (v1.0).
The efficacy of the GMR sensor platform hinges upon optimizing surface chemistry to ensure robustness and adaptability. This is crucial as instability of the immobilized substrate can affect the measurement of enzymatic activity and the sensor’s ability to be assembled and stored for use in a point-of-care setting. We previously showed that treating GMR sensors with polyallylamine (PAAM) followed by poly (ethylene-alt-maleic anhydride) (PEMA) yields a surface functionalized with maleic anhydride (Sveiven et al., 2023). Proteins that contain free amines, such as antibodies, can be covalently coupled to the surface for use in an immunoassay. Using this same surface chemistry approach, we coupled an amine-PEG11-biotin linker molecule to a GMR surface via an amine reaction with maleic anhydride (Figure 2A). Upon addition of streptavidin-coated MNPs, an average increase in resistance of 5,380 ppm was quantified on the 22 sensors containing amine-PEG11-biotin, while sensors that lacked this molecule had an average increase in resistance of 304 ppm, revealing that the biotin groups on the PEG linker are responsible for binding to the MNPs (Figure 2B). Following extensive washing with PBS, we obtained no noticeable reduction in signal, indicating that the MNPs are tightly bound to the sensor surface.
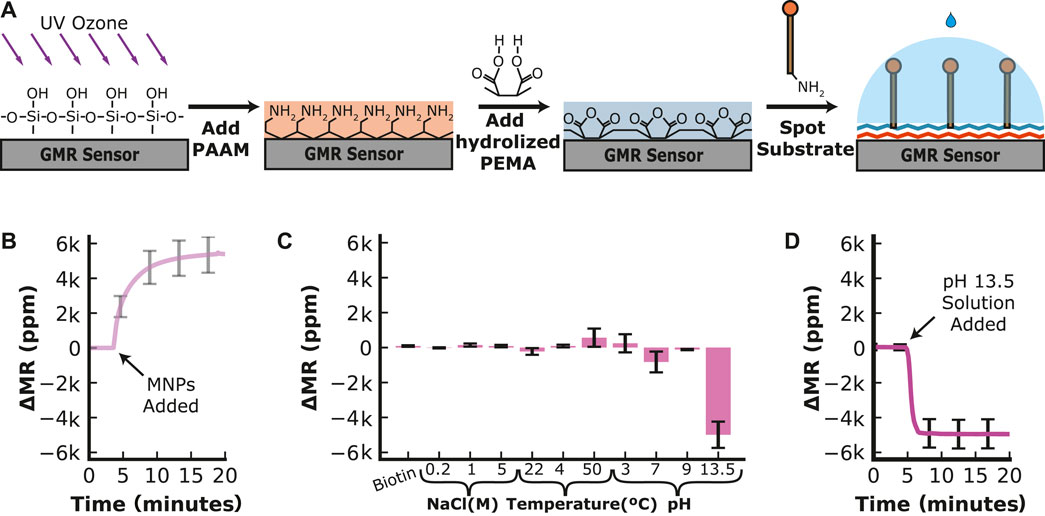
FIGURE 2. Sensor Functionalization and Stability of the Substrate. (A) The sensor surface is cleaned and activated by ultraviolet-ozone treatment before poly (allylamine) (PAAM) is added. The hydrolyzed poly (ethylene-alt-maleic anhydride) (PEMA) is added to create a layer of maleic groups. When the amine-containing substrate is spotted on the sensor surface, the maleic groups form a covalent bond with the amines on the substrate. (B) Time-dependent loading of streptavidin-MNP onto a sensor surface containing PEG11-biotin. (C) Change in magnetoresistance for a fully assembled PEG11-MNP complex that was stored for 21 days at 4°C and then sequentially incubated at various conditions. The change is recorded from one condition to the next. (D) Time-dependent decrease in signal in the presence of an extremely basic reagent (pH 13.5).
The functionalized sensor chip was stored at 4°C for 21 days to evaluate the long-term stability and then placed back in the GMR readout station. We then performed extensive washing steps using a range of buffer conditions to determine if the signal decreased. Little change (<5%) in resistance was quantified in excess biotin, revealing that PEG11-biotin was tightly bound to streptavidin-MNP and could not be competed off. Sequentially adding an increasing concentration of NaCl or changing the temperature from 22°C (room temperature) to 4°C did not alter the signal more than 5% of the saturated signal after MNP addition. When the temperature was changed from 4°C to 50°C, the signal increased by 9.6% of the saturated signal after MNP addition. The sensor was then washed with reagents buffered at various pH values. Changing the pH from neutral to pH 3 increased the signal by 4.9%, then decreased by 15.2% when the buffer was changed back to pH 7. Increasing the pH further to pH 9.0 has little effect on the signal (<5%). However, adding pH 13.5 buffer reduced the signal by 4,970 ppm, corresponding to a 92.5% reduction of the saturated signal after MNP addition (Figure 2C). By monitoring the real-time chemical release of MNPs under extreme alkaline conditions, we showed that the signal decreased rapidly within 3 min and then stabilized (Figure 2D). These data revealed that the sensor chip stored at 4°C for 21 days retained the PEG11 linker sequence and the streptavidin-coated MNPs and that this complex was only broken by treatment with a strong alkaline solution.
From these studies, it was unclear if the decrease in signal was due to the breakage of the amine-maleic anhydride bond or the streptavidin–biotin bond. To determine if the signal reduction was due to the release of the streptavidin–biotin, a reaction was set up whereby the linker-MNP complex was treated with a pH 13.5 solution, washed with PBS at neutral pH, and then incubated with fresh streptavidin-MNPs. The resistance signal decreased by nearly 75% upon treatment with pH 13.5 but then increased to 97% of its original signal upon adding fresh MNPs (Supplementary Figure S5). These studies reveal that the amine–maleic anhydride bond is stable in extreme alkaline conditions, but the biotin–streptavidin bond is broken, most likely due to the denaturation of streptavidin. The stability of the substrate-MNP complex on the sensor surface is compatible with the buffer conditions needed for hydrolase activity assays, as there are no documented human enzymes that require extreme alkaline conditions to be functional.
We next evaluated the ability of hydrolytic enzymes to cleave a substrate sequence and, therefore, replaced the PEG11 linker with double-stranded DNA containing an amine group and biotin on each end (Figure 3A). We chose a sequence containing the restriction site for cleavage by Bcu I that corresponds to A*CTAGT, where * is the cleaved bond. This sequence is flanked by 4 bases on the 5′side and 20 bases on the 3′side and is coupled to the sensor surface as described for the PEG11 linker. Another DNA linker sequence was synthesized with a scrambled restriction site sequence, TACATG, which was expected to resist Bcu I cleavage. Upon cleavage of the DNA substrate, the bound MNPs are predicted to be released into solution, thereby reducing the magnetoresistance signal.
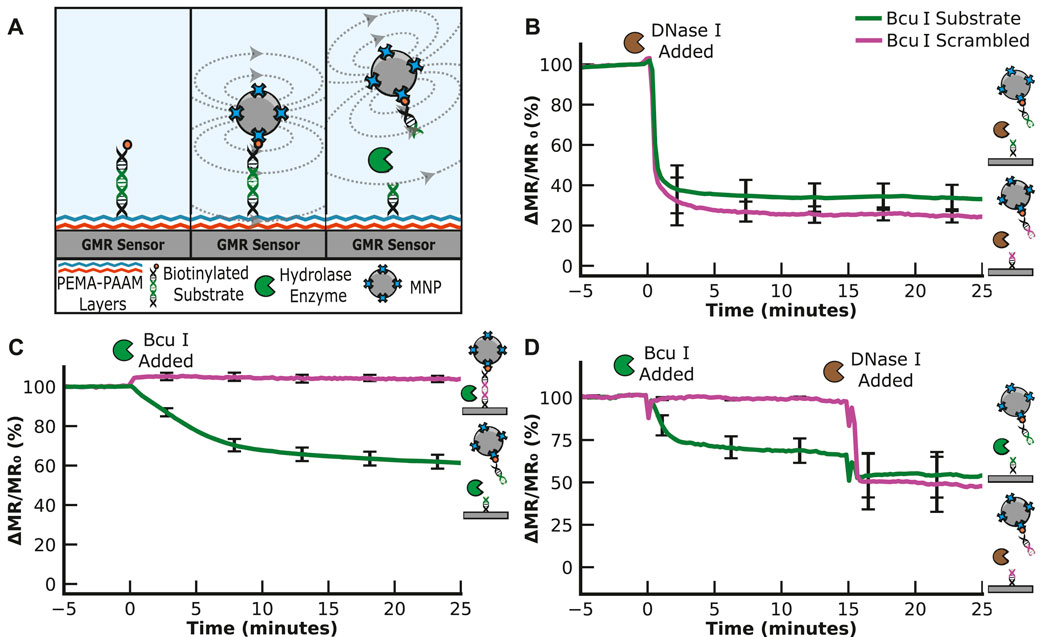
FIGURE 3. Nuclease Assay. (A) Graphical illustration of the nuclease assay. The signal decreases when the nuclease cleaves the substrate. (B) Hydrolysis of the ACTAGT (Bcu I substrate) and TACATG (Bcu I scrambled) sequences by DNase I results in a time-dependent decrease in the MR signal. (C) Hydrolysis of ACTAGT and not TACATG by the restriction enzyme, Bcu I. (D) Sequential addition of Bcu I and DNase I shows that Bcu I specifically cleaves the ACTAGT substrate, while DNase I cleaves the TACATG sequence.
We found that the DNA sequences could be coupled to the sensor surface using the same chemical protocol optimized using the PEG11 linker. Following the wash steps, the signal was evaluated for 5 min to ensure stability. To determine if the two DNA sequences are accessible for nuclease cleavage, we added DNase I, a broad-spectrum nuclease enzyme that non-specifically cleaves the phosphodiester bonds in double-stranded DNA sequences. This enzyme rapidly cleaved both DNA sequences, decreasing the signal by 59.0% for the Bcu I substrate and 64.1% for the scrambled Bcu I substrate after only a 1-min incubation (Figure 3B). The signal stabilized, indicating that all available DNA linker sequences were hydrolyzed. These data confirm that the surface-tethered DNA sequence is accessible for cleavage by a nuclease.
To assess the specificity of the Bcu I substrate over the scrambled sequence, we added Bcu I to a sensor containing both sequences. After incubation for 25 min with the scrambled substrate, no reduction in signal was detected, indicating that this DNA sequence was not cleaved. However, in the adjacent sensors containing the Bcu I substrate, a time-dependent change in signal was detectable, with 22.9% reduction in signal within 5 min and an additional 21.2% reduction over the remaining 20 min (a total of 44.1% reduction in 25 min) (Figure 3C). We next set up an assay where Bcu I was incubated with the DNA sequences for 15 min, and then DNase I was added to the same sensor (Figure 3D). These studies showed that both DNA sequences are cleavable by DNase I, but only the ACTAGT sequence is a substrate for Bcu I.
Knowing that the DNA sequences can be cleaved by nucleases, we evaluated their stability in saliva, a biofluid of interest for use in point-of-care applications. Saliva contains numerous hydrolytic enzymes such as salivary amylase, peptidases, lysozyme, and lipase (Chojnowska et al., 2018; des Gachons and Breslin, 2016; Feng et al., 2019; Mennella et al., 2014). When exposed to saliva, the Bcu I substrate and scrambled DNA sequence had only a 1.3% and 1.6% reduction in signal after 10 min of incubation, respectively (Supplementary Figure S6). When compared to the reductions observed in the presence of Bcu I and DNase I, the signal change caused by saliva is statistically insignificant. These studies reveal that the DNA-MNP complex is stable in a complex biological sample containing numerous hydrolytic enzymes.
After validating the MNP assay with a nuclease, we evaluated the assay format for a peptidase, an alternative hydrolase that cleaves peptide bonds instead of phosphodiester bonds. For these studies, we chose human neutrophil elastase (HNE) as the target enzyme as it has been established as a sputum biomarker for exacerbations associated with chronic obstructive pulmonary disease (COPD) and cystic fibrosis (CF). During neutrophil degranulation, elastase from granules is released and efficiently kills bacteria; however, the excess enzyme also damages lung tissue by degrading extracellular matrix proteins that are important for lung structure and elasticity (Mecham et al., 1997; Kafienah et al., 1998; Saetta et al., 2001; Kawabata et al., 2002). The amount of neutrophil elastase in sputum is directly proportional to the amount of activated neutrophils. We have detected neutrophil elastase activity in the sputum of patients with CF using fluorogenic substrates and revealed that patients with severe disease and more pathogenic bacteria have higher levels of elastase activity (Quinn et al., 2019). While numerous assays have been developed to quantify this enzyme, we were interested in designing a point-of-care peptidase assay using the MNP sensor system.
We first needed to find a peptide efficiently cleaved by HNE. In previous studies by our group, we incubated HNE with 124 different 14-mer peptides, each highly diversified in sequence. The enzyme cleaved 78 of these peptides, and a substrate specificity profile was generated using the most frequently found amino acids in each position surrounding the cleavage site (O’Donoghue et al., 2013). A consensus peptide sequence consisting of Arg-Gln-Pro-Val*Nle-Trp-Gly (RQPVnWG) was developed as a sequence cleaved by HNE, where * is the cleavage site and Nle (n) is norleucine, a non-natural amino acid. In parallel, we identified a scrambled peptide sequence, VWnRQGP, that contains the same seven amino acids but is not cleaved by HNE. These peptides were synthesized with a PEG36 linker on each end. On the N-terminal PEG36, an amine group was included to covalently attach to the sensor surface, while the C-terminal PEG36 contains a biotin group to bind MNPs (Figure 4A). Upon exposure of the sensor to a sample containing HNE, it was predicted that the RQPVnWG peptide would be cleaved between V and n, while the scrambled peptide would not be cleaved.
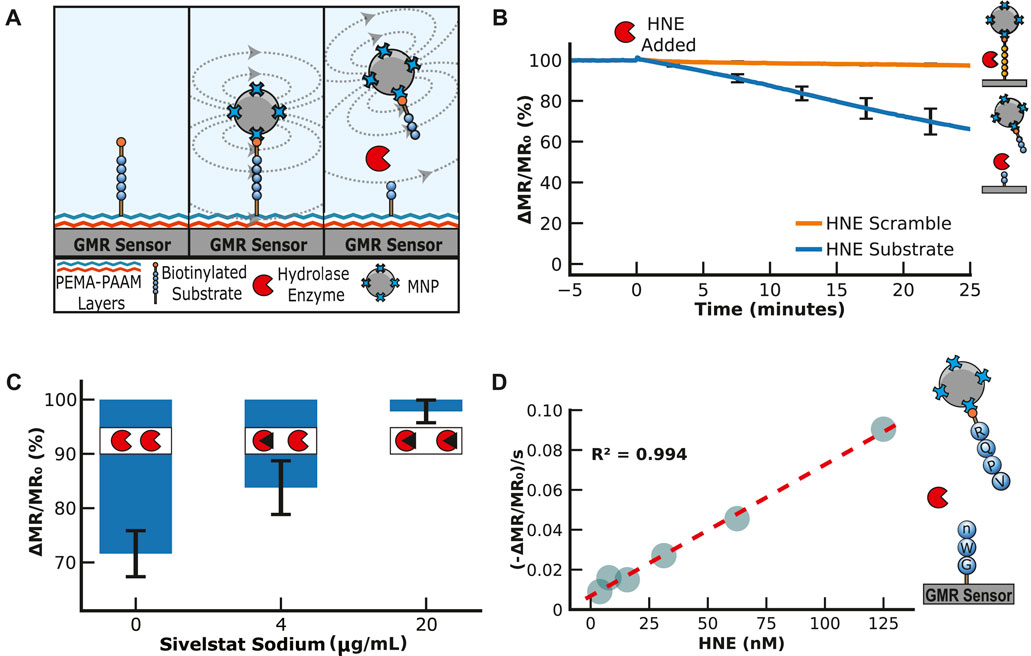
FIGURE 4. Human Neutrophil Elastase Assay. (A) Illustration of the neutrophil elastase assay. The peptide substrate is covalently attached to the sensor surface and then bound to streptavidin-MNPs via a biotin group on the peptide. This results in an increase in magnetoresistance. The addition of human neutrophil elastase cleaves the substrate, decreasing the signal. (B) An example of how the signal is loaded and then reduced by adding human neutrophil elastase. The signal is displayed as a percentage of the loading signal after the magnetic nanoparticle binding has saturated. (C) Human neutrophil elastase assays with inhibition by sivelestat sodium. All three assays have 20 μg/mL of neutrophil elastase, but the inhibitor concentration increases from 0 to 20 μg/mL. (D) Human neutrophil elastase titration serial diluting from 125 to 3.9 nM by a factor of 2 in PBS containing 0.01% Tween-20. The magnetic assay results are shown as the maximum velocity.
Both peptides were coupled to the sensor surface using the protocol outlined previously for PEG11 and DNA, and upon the addition of MNPs, a signal increased by ∼4,000 ppm, which confirmed the interaction between the biotin on the peptide with the streptavidin-coated MNPs. Following a wash step, the signal was monitored for 5 min to ensure stability, and then HNE was added. The peptide containing the substrate sequence was cleaved, releasing the MNPs into the solution in a time-dependent manner (Figure 4B). The signal from the scrambled peptide sequence remained unchanged in the assay. This confirmed that cleavage by HNE was specific for the RQPVnWG substrate and revealed that the enzyme does not cleave the streptavidin protein, which could non-specifically release the MNP due to the breakdown of the streptavidin-biotin interaction. Three independent assays were performed, and the coefficient of variation from all sensors after 20 min of incubation was calculated to be 11% (Supplementary Figure S7).
Next, we evaluated the ability of the assay to distinguish between active and inhibited HNE. Sivelestat sodium is a clinically approved HNE inhibitor for treating acute lung injury or acute respiratory distress syndrome (Pu et al., 2017). HNE was added to RQPVnWG sensors that contained either 4 or 20 μg/mL of sivelestat sodium, and the reactions were monitored for 10 min. The change in magnetoresistance was compared to the HNE digestion assay without the inhibitor. In the absence of sivelestat sodium, the signal decreased by 28.4%, while in the presence of 4 μg/mL, the signal decreased by 16.2%. At 5× higher inhibitor concentration, the signal decreases by only 2.2%, revealing that the enzyme was mostly inactivated under these conditions (Figure 4C). These data showed that the release rate of MNPs correlates with the amount of active enzyme in the assay. We next performed a serial dilution of HNE from 125 to 4 nM and calculated the change in MR signal per second at each concentration. From these studies, a linear concentration curve was calculated with an R2 value of 0.994 (Figure 4D), confirming that the change in MR signal directly correlates with enzyme concentration.
One of the most commonly used fluorogenic peptide substrates for monitoring HNE activity is Ala-Ala-Pro-Val-7-amino-4-methylcoumarin (AAPV-amc), where cleavage between V and amc results in an increase in fluorescence at 460 nm. To directly compare the peptide-MNP assay with a traditional fluorogenic peptide assay, the fluorogenic substrate was also assayed with 125 to 4 nM of HNE. The assay yielded an expected concentration-dependent increase in the reaction velocity. When comparing the velocities of both assays, a Pearson correlation coefficient of 0.974 was calculated, indicating a very strong positive correlation between the surface-based MNP release assay and the traditional solution-based fluorogenic assay (Figure 5A).
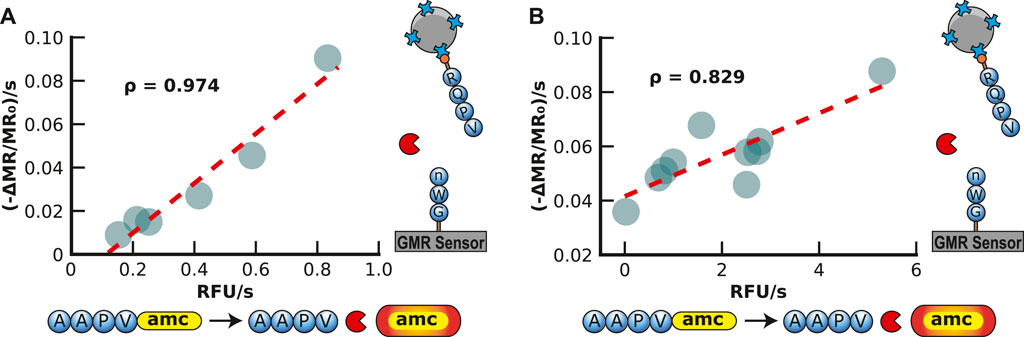
FIGURE 5. Validation of Magnetic Neutrophil Elastase Assay in Buffer and Sputum. (A) Validation of the magnetic human neutrophil elastase assay by comparison to a traditional AMC assay readout with a spectrometer. Each point represents a different concentration of human neutrophil elastase in buffer readout by the GMR readout station (shown in the y-axis) and the spectrometer (shown in the x-axis). The concentration ranges from 3.9 to 125 nM of human neutrophil elastase. (B) Validation of the magnetic human neutrophil elastase assay compared to a traditional amc assay readout with a spectrometer. Each point represents a different human sputum sample of patients with cystic fibrosis measured by the GMR station (shown in the y-axis) and the spectrometer (shown in the x-axis).
To evaluate the peptide-MNP assay using clinically relevant biofluids, we obtained sputum from 10 CF patients, diluted them 200-fold in assay buffer, and incubated it with both the peptide-MNP sensor and the fluorogenic substrate (Figure 5B). Each sputum sample contained sufficient HNE activity to release the MNPs, with the release rate ranging from 0.036 to 0.088 ppm/s. Compared with the fluorescent assay, the data was strongly correlative (Pearson correlation coefficient of 0.829). The velocity of MNP release by HNE in the sputum samples was then compared to the HNE concentration curve, and it was revealed that the amount of enzyme in each sample was between 8.8 and 24.6 μM (Supplementary Table S1). This concentration of HNE in these sputum is comparable to previous studies on CF sputum that used a colorimetric substrate to quantify HNE in CF sputum at a range of 0.47–18.5 μM (Rees et al., 1997; Dittrich et al., 2018). This study shows that the peptide-MNP assay applies to quantifying HNE in patient sputum samples.
The sputum samples used in this study were obtained from different patients; therefore, we could not perform a longitudinal study on how the HNE levels change when the patient is experiencing an exacerbation event. However, a potential application of this assay for patients with CF, COPD, or other lung diseases would be to monitor the HNE levels in sputum daily or weekly, thereby providing the healthcare team with data to monitor neutrophil levels in their lungs. The landscape of enzymes, their functions, and the impact of their dysfunction in disease states is vast. Table 1 lists a subset of these enzymes and the current methodologies used to measure their abundance. Nucleases and peptidases are activity-based markers for lung, inflammatory, and infectious diseases in addition to cancer. Many of these enzymes are currently assayed by ELISA, which cannot distinguish between active and inactive (inhibited enzymes or pro-enzymes). The standard fluorogenic methods for quantifying peptidase activity require a microplate reader and a trained technician. These methods can process many patient samples but are not amenable to point-of-care use. The magnetic enzymatic activity assay described here is designed for single-use in a point-of-care setting where the user (patient or healthcare provider) adds a biofluid sample to a pre-assembled substrate. The adaptability of the magnetic assay for other hydrolase substrates means that it can be readily modified to detect many different enzymes. The magnetic-based enzyme assay could greatly benefit the medical community as it seeks to diagnose patients and monitor their daily health.
In this study, we report on a sensor platform for real-time hydrolase activity monitoring that improves upon some of the limitations of traditional methods. The field of bioengineering has long been accustomed to relying on optical assays to decipher enzymatic processes. However, the constraints of sample matrices, equipment complexity, and issues with autofluorescence and photobleaching have hindered their use. We introduce a novel sensor platform that quantifies hydrolase activity in a wash-free and highly adaptable assay. This adaptability is exemplified by incorporating either DNA or peptide substrates to facilitate the measurement of nucleases and peptidases, respectively. Moreover, the platform’s distinct attribute lies in its readiness for point-of-care diagnostics, a feature that traditional methods often struggle to accommodate. The inherent adaptability of our platform, coupled with its reliance on magnetic nanoparticles, enables the facile development of robust point-of-care assays. The resilience exhibited in complex biological matrices, such as saliva and sputum, further solidifies its applicability in scenarios requiring storage over extended periods.
In summary, the novel sensor platform, presented within the context of this study, emerges as a step forward in enzymatic activity analysis. This platform offers an adaptable and point-of-care capable solution by improving upon the limitations that have tethered traditional methods. With potential applications spanning diverse biomedical domains, this innovation paves the way for precision enzymatic activity analysis, promising to empower researchers and clinicians alike in exploring the complexities of enzymatic processes and their implications in health and disease.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by the UC San Diego institutional review board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
MS: Conceptualization, Formal Analysis, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing. AS: Methodology, Validation, Writing–review and editing. JR: Methodology, Writing–review and editing. DC: Resources, Writing–review and editing. DH: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing–original draft, Writing–review and editing. AD: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing–original draft, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was partly supported by the National Institutes of Health (NIH) (R21HD094359, R21EB028485, and UL1TR001442). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
We thank the volunteers who provided sputum samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2024.1363186/full#supplementary-material
Adem, S., Jain, S., Sveiven, M., Zhou, X., O’Donoghue, A. J., and Hall, D. A. (2020). Giant magnetoresistive biosensors for real-time quantitative detection of protease activity. Sci. Rep. 10, 7941. doi:10.1038/s41598-020-62910-2
Antarnusa, G., Esmawan, A., Dwi Jayanti, P., Rizki Fitriani, S., Suherman, A., Kinarya Palupi, E., et al. (2022). Synthesis of Fe3O4 at different reaction temperatures and investigation of its magnetic properties on giant magnetoresistance (GMR) sensors for bio-detection applications. J. Magn. Magn. Mat. 563, 169903. doi:10.1016/j.jmmm.2022.169903
Armstrong, P. B. (2001). The contribution of proteinase inhibitors to immune defense. Trends Immunol. 22, 47–52. doi:10.1016/S1471-4906(00)01803-2
Balian, A., and Hernandez, F. J. (2021). Nucleases as molecular targets for cancer diagnosis. Biomark. Res. 9, 86. doi:10.1186/s40364-021-00342-4
Bao, Q., Lin, D., Gao, Y., Wu, L., Fu, J., Galaa, K., et al. (2021). Ultrasensitive off-on-off fluorescent nanosensor for protamine and trypsin detection based on inner-filter effect between N,S-CDs and gold nanoparticles. Microchem. J. 168, 106409. doi:10.1016/j.microc.2021.106409
Baselt, D. R., Lee, G. U., Natesan, M., Metzger, S. W., Sheehan, P. E., and Colton, R. J. (1998). A biosensor based on magnetoresistance technology. Biosens. Bioelectron. 13, 731–739. doi:10.1016/S0956-5663(98)00037-2
Bikker, F. J., Nascimento, G. G., Nazmi, K., Silbereisen, A., Belibasakis, G. N., Kaman, W. E., et al. (2019). Salivary total protease activity based on a broad-spectrum fluorescence resonance energy transfer approach to monitor induction and resolution of gingival inflammation. Mol. Diagn. Ther. 23, 667–676. doi:10.1007/s40291-019-00421-1
Cannalire, G., Pilloni, S., Esposito, S., Biasucci, G., Franco, A. D., and Street, M. E. (2023). Alkaline phosphatase in clinical practice in childhood: focus on rickets. Front. Endocrinol. 14, 1111445. doi:10.3389/fendo.2023.1111445
Chan, L. W., Anahtar, M. N., Ong, T.-H., Hern, K. E., Kunz, R. R., and Bhatia, S. N. (2020). Engineering synthetic breath biomarkers for respiratory disease. Nat. Nanotechnol. 15, 792–800. doi:10.1038/s41565-020-0723-4
Chang, C.-C. (2021). Recent advancements in aptamer-based surface plasmon resonance biosensing strategies. Biosensors 11, 233. doi:10.3390/bios11070233
Chen, H., Zhang, J., Gao, Y., Liu, S., Koh, K., Zhu, X., et al. (2015). Sensitive cell apoptosis assay based on caspase-3 activity detection with graphene oxide-assisted electrochemical signal amplification. Biosens. Bioelectron. 68, 777–782. doi:10.1016/j.bios.2015.02.007
Chen, W., Jiang, X., and Yang, Q. (2020). Glycoside hydrolase family 18 chitinases: the known and the unknown. Biotechnol. Adv. 43, 107553. doi:10.1016/j.biotechadv.2020.107553
Chojnowska, S., Baran, T., Wilińska, I., Sienicka, P., Cabaj-Wiater, I., and Knaś, M. (2018). Human saliva as a diagnostic material. Adv. Med. Sci. 63, 185–191. doi:10.1016/j.advms.2017.11.002
de Freitas Nakata, K. C., da Silva Pereira, P. P., and Salgado Riveros, B. (2021). Creatine kinase test diagnostic accuracy in neonatal screening for Duchenne Muscular Dystrophy: a systematic review. Clin. Biochem. 98, 1–9. doi:10.1016/j.clinbiochem.2021.09.010
des Gachons, C. P., and Breslin, P. A. S. (2016). Salivary amylase: digestion and metabolic syndrome. Curr. Diab. Rep. 16, 102. doi:10.1007/s11892-016-0794-7
Ding, X., and Yang, K.-L. (2014). Enzymatic deposition of silver particles for detecting protease activity. Part. Part. Syst. Charact. 31, 1300–1306. doi:10.1002/ppsc.201400107
Dittrich, A. S., Kühbandner, I., Gehrig, S., Rickert-Zacharias, V., Twigg, M., Wege, S., et al. (2018). Elastase activity on sputum neutrophils correlates with severity of lung disease in cystic fibrosis. Eur. Respir. J. 51, 1701910. doi:10.1183/13993003.01910-2017
Eatemadi, A., Aiyelabegan, H. T., Negahdari, B., Mazlomi, M. A., Daraee, H., Daraee, N., et al. (2017). Role of protease and protease inhibitors in cancer pathogenesis and treatment. Biomed. Pharmacother. 86, 221–231. doi:10.1016/j.biopha.2016.12.021
Feng, Y., Li, Q., Chen, J., Yi, P., Xu, X., Fan, Y., et al. (2019). Salivary protease spectrum biomarkers of oral cancer. Int. J. Oral Sci. 11, 7–11. doi:10.1038/s41368-018-0032-z
Flenker, K. S., Burghardt, E. L., Dutta, N., Burns, W. J., Grover, J. M., Kenkel, E. J., et al. (2017). Rapid detection of urinary tract infections via bacterial nuclease activity. Mol. Ther. 25, 1353–1362. doi:10.1016/j.ymthe.2017.03.015
Garcia Gonzalez, J., and Hernandez, F. J. (2022). Nuclease activity: an exploitable biomarker in bacterial infections. Expert Rev. Mol. diagn. 22, 265–294. doi:10.1080/14737159.2022.2049249
Gaster, R. S., Hall, D. A., and Wang, S. (2011). nanoLAB: an ultraportable, handheld diagnostic laboratory for global health. Lab. Chip 11, 950–956. doi:10.1039/C0LC00534G
Gaster, R. S., Hall, D. A., and Wang, S. X. (2013). “Magneto-Nanosensor diagnostic chips,” in Point-of-Care diagnostics on a chip, biological and medical physics, biomedical engineering. Editors D. Issadore, and R. M. Westervelt (Berlin, Heidelberg: Springer), 153–176. doi:10.1007/978-3-642-29268-2_7
Hall, D. A., Gaster, R. S., Lin, T., Osterfeld, S. J., Han, S., Murmann, B., et al. (2010a). GMR biosensor arrays: a system perspective. Biosens. Bioelectron. 25, 2051–2057. doi:10.1016/j.bios.2010.01.038
Hall, D. A., Gaster, R. S., Osterfeld, S. J., Murmann, B., and Wang, S. X. (2010b). GMR biosensor arrays: correction techniques for reproducibility and enhanced sensitivity. Biosens. Bioelectron. 25, 2177–2181. doi:10.1016/j.bios.2010.01.039
Hammond, E., and Ferro, V. (2023). “An enzymatic activity assay for heparanase that is useful for evaluating clinically relevant inhibitors and studying kinetics,” in Proteoglycans: methods and protocols, methods in molecular biology. Editor N. K. Karamanos (New York, NY: Springer US), 227–238. doi:10.1007/978-1-0716-2946-8_16
Hao Ong, I. L., and Yang, K.-L. (2017). Recent developments in protease activity assays and sensors. Analyst 142, 1867–1881. doi:10.1039/C6AN02647H
Hirai, K., Yamaguchi-Tomikawa, T., Eguchi, T., Maeda, H., and Takashiba, S. (2020). Identification and modification of porphyromonas gingivalis cysteine protease, gingipain, ideal for screening periodontitis. Front. Immunol. 11, 1017. doi:10.3389/fimmu.2020.01017
Jung, K., Zachow, J., Lein, M., Brux, B., Sinha, P., Lenk, S., et al. (1999). Rapid detection of elevated prostate-specific antigen levels in blood: performance of various membrane strip tests compared. Urology 53, 155–160. doi:10.1016/S0090-4295(98)00419-1
Kafienah, W., Buttle, D. J., Burnett, D., and Hollander, A. P. (1998). Cleavage of native type I collagen by human neutrophil elastase. Biochem. J. 330, 897–902. doi:10.1042/bj3300897
Kårlund, A., Paukkonen, I., Gómez-Gallego, C., and Kolehmainen, M. (2021). Intestinal exposure to food-derived protease inhibitors: digestion physiology- and gut health-related effects. Healthcare 9, 1002. doi:10.3390/healthcare9081002
Kawabata, K., Hagio, T., and Matsuoka, S. (2002). The role of neutrophil elastase in acute lung injury. Eur. J. Pharmacol. 451, 1–10. doi:10.1016/S0014-2999(02)02182-9
Kim, D., Marchetti, F., Chen, Z., Zaric, S., Wilson, R. J., Hall, D. A., et al. (2013). Nanosensor dosimetry of mouse blood proteins after exposure to ionizing radiation. Sci. Rep. 3, 2234. doi:10.1038/srep02234
Klein, T., Wang, W., Yu, L., Wu, K., Boylan, K. L. M., Vogel, R. I., et al. (2019). Development of a multiplexed giant magnetoresistive biosensor array prototype to quantify ovarian cancer biomarkers. Biosens. Bioelectron. 126, 301–307. doi:10.1016/j.bios.2018.10.046
Lauková, L., Konečná, B., Janovičová, Ľ., Vlková, B., and Celec, P. (2020). Deoxyribonucleases and their applications in biomedicine. Biomolecules 10, 1036. doi:10.3390/biom10071036
Lichtenthaler, S. F., Tschirner, S. K., and Steiner, H. (2022). Secretases in Alzheimer’s disease: novel insights into proteolysis of APP and TREM2. Curr. Opin. Neurobiol. 72, 101–110. doi:10.1016/j.conb.2021.09.003
Lin, C.-W., Chen, J.-M., Lin, Y.-J., Chao, L.-W., Wei, S.-Y., Wu, C.-H., et al. (2019). Magneto-optical characteristics of streptavidin-coated Fe3O4@Au core-shell nanoparticles for potential applications on biomedical assays. Sci. Rep. 9, 16466. doi:10.1038/s41598-019-52773-7
Liu, W.-L., Liu, D., Cheng, K., Liu, Y.-J., Xing, S., Chi, P., et al. (2016). Evaluating the diagnostic and prognostic value of circulating cathepsin S in gastric cancer. Oncotarget 7, 28124–28138. doi:10.18632/oncotarget.8582
Lucena, F., and McDougall, J. J. (2021). Protease activated receptors and arthritis. Int. J. Mol. Sci. 22, 9352. doi:10.3390/ijms22179352
Machado, I., Garrido, V., Hernandez, L. I., Botero, J., Bastida, N., San-Roman, B., et al. (2019). Rapid and specific detection of Salmonella infections using chemically modified nucleic acid probes. Anal. Chim. Acta 1054, 157–166. doi:10.1016/j.aca.2018.12.027
Martha, J. W., Wibowo, A., and Pranata, R. (2022). Prognostic value of elevated lactate dehydrogenase in patients with COVID-19: a systematic review and meta-analysis. Postgrad. Med. J. 98, 422–427. doi:10.1136/postgradmedj-2020-139542
Mecham, R. P., Broekelmann, T. J., Fliszar, C. J., Shapiro, S. D., Welgus, H. G., and Senior, R. M. (1997). Elastin degradation by matrix metalloproteinases: CLEAVAGE site specificity and mechanisms of elastolysis. J. Biol. Chem. 272, 18071–18076. doi:10.1074/jbc.272.29.18071
Mennella, I., Fogliano, V., and Vitaglione, P. (2014). Salivary lipase and α-amylase activities are higher in overweight than in normal weight subjects: influences on dietary behavior. Food Res. Int. 66, 463–468. doi:10.1016/j.foodres.2014.10.008
Menon, S., Mathew, M. R., Sam, S., Keerthi, K., and Kumar, K. G. (2020). Recent advances and challenges in electrochemical biosensors for emerging and re-emerging infectious diseases. J. Electroanal. Chem. Lausanne Switz. 878, 114596. doi:10.1016/j.jelechem.2020.114596
Moll, T., Shaw, P. J., and Cooper-Knock, J. (2020). Disrupted glycosylation of lipids and proteins is a cause of neurodegeneration. Brain 143, 1332–1340. doi:10.1093/brain/awz358
Mondanelli, G., Iacono, A., Carvalho, A., Orabona, C., Volpi, C., Pallotta, M. T., et al. (2019). Amino acid metabolism as drug target in autoimmune diseases. Autoimmun. Rev. 18, 334–348. doi:10.1016/j.autrev.2019.02.004
Mostufa, S., Rezaei, B., Yari, P., Xu, K., Gómez-Pastora, J., Sun, J., et al. (2023). Giant magnetoresistance based biosensors for cancer screening and detection. ACS Appl. Bio Mat. 6, 4042–4059. doi:10.1021/acsabm.3c00592
Nazar Majeed, Z., Philip, K., Alabsi, A. M., Pushparajan, S., and Swaminathan, D. (2016). Identification of gingival crevicular fluid sampling, analytical methods, and oral biomarkers for the diagnosis and monitoring of periodontal diseases: a systematic review. Dis. Markers 2016, 1–23. doi:10.1155/2016/1804727
Newby, P. R., Crossley, D., Crisford, H., Stockley, J. A., Mumford, R. A., Carter, R. I., et al. (2019). A specific proteinase 3 activity footprint in α1-antitrypsin deficiency. ERJ Open Res. 5, 00095–02019. doi:10.1183/23120541.00095-2019
Ni, J., Lin, H., Yang, W., Liao, Y., Wang, Q., Luo, F., et al. (2019). Homogeneous electrochemiluminescence biosensor for the detection of RNase A activity and its inhibitor. Anal. Chem. 91, 14751–14756. doi:10.1021/acs.analchem.9b04194
Nozeret, K., Boucharlat, A., Agou, F., and Buddelmeijer, N. (2019). A sensitive fluorescence-based assay to monitor enzymatic activity of the essential integral membrane protein Apolipoprotein N-acyltransferase (Lnt). Sci. Rep. 9, 15978. doi:10.1038/s41598-019-52106-8
O’Donoghue, A. J., Jin, Y., Knudsen, G. M., Perera, N. C., Jenne, D. E., Murphy, J. E., et al. (2013). Global substrate profiling of proteases in human neutrophil extracellular traps reveals consensus motif predominantly contributed by elastase. PLoS ONE 8, e75141. doi:10.1371/journal.pone.0075141
Oishi, J., Asami, Y., Mori, T., Kang, J.-H., Niidome, T., and Katayama, Y. (2008). Colorimetric enzymatic activity assay based on noncrosslinking aggregation of gold nanoparticles induced by adsorption of substrate peptides. Biomacromolecules 9, 2301–2308. doi:10.1021/bm800192d
Olazarra, A. S. D., Cortade, D. L., and Wang, S. X. (2022). From saliva to SNP: non-invasive, point-of-care genotyping for precision medicine applications using recombinase polymerase amplification and giant magnetoresistive nanosensors. Lab. Chip 22, 2131–2144. doi:10.1039/D2LC00233G
Oriano, M., Terranova, L., Sotgiu, G., Saderi, L., Bellofiore, A., Retucci, M., et al. (2019). Evaluation of active neutrophil elastase in sputum of bronchiectasis and cystic fibrosis patients: a comparison among different techniques. Pulm. Pharmacol. Ther. 59, 101856. doi:10.1016/j.pupt.2019.101856
Osterfeld, S. J., Yu, H., Gaster, R. S., Caramuta, S., Xu, L., Han, S.-J., et al. (2008). Multiplex protein assays based on real-time magnetic nanotag sensing. Proc. Natl. Acad. Sci. 105, 20637–20640. doi:10.1073/pnas.0810822105
Ping, J., Wu, W., Qi, L., Liu, J., Liu, J., Zhao, B., et al. (2021). Hydrogel-assisted paper-based lateral flow sensor for the detection of trypsin in human serum. Biosens. Bioelectron. 192, 113548. doi:10.1016/j.bios.2021.113548
Pu, S., Wang, D., Liu, D., Zhao, Y., Qi, D., He, J., et al. (2017). Effect of sivelestat sodium in patients with acute lung injury or acute respiratory distress syndrome: a meta-analysis of randomized controlled trials. BMC Pulm. Med. 17, 148. doi:10.1186/s12890-017-0498-z
Qing, T., Long, C., Wang, X., Zhang, K., Zhang, P., and Feng, B. (2019). Detection of micrococcal nuclease for identifying Staphylococcus aureus based on DNA templated fluorescent copper nanoclusters. Mikrochim. Acta 186, 248. doi:10.1007/s00604-019-3363-3
Quinn, R. A., Adem, S., Mills, R. H., Comstock, W., DeRight Goldasich, L., Humphrey, G., et al. (2019). Neutrophilic proteolysis in the cystic fibrosis lung correlates with a pathogenic microbiome. Microbiome 7, 23. doi:10.1186/s40168-019-0636-3
Rack, J. G. M., Palazzo, L., and Ahel, I. (2020). (ADP-ribosyl)hydrolases: structure, function, and biology. Genes. Dev. 34, 263–284. doi:10.1101/gad.334631.119
Rees, D. D., Brain, J. D., Wohl, M. E., Humes, J. L., and Mumford, R. A. (1997). Inhibition of neutrophil elastase in CF sputum by L-658,758. J. Pharmacol. Exp. Ther. 283, 1201–1206.
Retout, M., Amer, L., Yim, W., Creyer, M. N., Lam, B., Trujillo, D. F., et al. (2023). A protease-responsive polymer/peptide conjugate and reversible assembly of silver clusters for the detection of porphyromonas gingivalis enzymatic activity. ACS Nano 17, 17308–17319. doi:10.1021/acsnano.3c05268
Riise, R., Odqvist, L., Mattsson, J., Monkley, S., Abdillahi, S. M., Tyrchan, C., et al. (2019). Bleomycin hydrolase regulates the release of chemokines important for inflammation and wound healing by keratinocytes. Sci. Rep. 9, 20407. doi:10.1038/s41598-019-56667-6
Rofael, S., Leboreiro Babe, C., Davrandi, M., Kondratiuk, A. L., Cleaver, L., Ahmed, N., et al. (2023). Antibiotic resistance, bacterial transmission and improved prediction of bacterial infection in patients with antibody deficiency. JAC-Antimicrob. Resist. 5, dlad135. doi:10.1093/jacamr/dlad135
Saetta, M., Turato, G., Maestrelli, P., Mapp, C. E., and Fabbri, L. M. (2001). Cellular and structural bases of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 163, 1304–1309. doi:10.1164/ajrccm.163.6.2009116
Salhi, A., Carriere, F., Grundy, M. M. L., and Aloulou, A. (2021). “Enzymes involved in lipid digestion,” in Bioaccessibility and digestibility of lipids from food. Editors M. M. L. Grundy, and P. J. Wilde (Cham: Springer International Publishing), 3–28. doi:10.1007/978-3-030-56909-9_1
Sama, I. E., Ravera, A., Santema, B. T., van Goor, H., ter Maaten, J. M., Cleland, J. G. F., et al. (2020). Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin–angiotensin–aldosterone inhibitors. Eur. Heart J. 41, 1810–1817. doi:10.1093/eurheartj/ehaa373
Santa, P., Garreau, A., Serpas, L., Ferriere, A., Blanco, P., Soni, C., et al. (2021). The role of nucleases and nucleic acid editing enzymes in the regulation of self-nucleic acid sensing. Front. Immunol. 12, 629922. doi:10.3389/fimmu.2021.629922
Serena, T. E., Bayliff, S. W., Brosnan, P. J., DiMarco, D. T., Doner, B. A., Guthrie, D. A., et al. (2021). Bacterial protease activity as a biomarker to assess the risk of non-healing in chronic wounds: results from a multicentre prospective cohort clinical trial. Wound Repair Regen. Off. Publ. Wound heal. Soc. Eur. Tissue Repair Soc. 29, 752–758. doi:10.1111/wrr.12941
Shpacovitch, V., Feld, M., Hollenberg, M. D., Luger, T. A., and Steinhoff, M. (2008). Role of protease-activated receptors in inflammatory responses, innate and adaptive immunity. J. Leukoc. Biol. 83, 1309–1322. doi:10.1189/jlb.0108001
Singh, P., Yang, M., Dai, H., Yu, D., Huang, Q., Tan, W., et al. (2008). Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol. Cancer Res. 6, 1710–1717. doi:10.1158/1541-7786.MCR-08-0269
Sonawane, S., Khanolkar, V., Namavari, A., Chaudhary, S., Gandhi, S., Tibrewal, S., et al. (2012). Ocular surface extracellular DNA and nuclease activity imbalance: a new paradigm for inflammation in dry eye disease. Invest. Ophthalmol. Vis. Sci. 53, 8253–8263. doi:10.1167/iovs.12-10430
Stanforth, K. J., Wilcox, M. D., Chater, P. I., Brownlee, I. A., Zakhour, M. I., Banecki, K. M. R. M., et al. (2022). Pepsin properties, structure, and its accurate measurement: a narrative review. Ann. Esophagus 5, 31. doi:10.21037/aoe-20-95
Sveiven, M., Gassman, A., Rosenberg, J., Chan, M., Boniface, J., O’Donoghue, A. J., et al. (2023). A dual-binding magnetic immunoassay to predict spontaneous preterm birth. Front. Bioeng. Biotechnol. 11, 1256267. doi:10.3389/fbioe.2023.1256267
Thulborn, S. J., Mistry, V., Brightling, C. E., Moffitt, K. L., Ribeiro, D., and Bafadhel, M. (2019). Neutrophil elastase as a biomarker for bacterial infection in COPD. Respir. Res. 20, 170. doi:10.1186/s12931-019-1145-4
Voynow, J. A., and Shinbashi, M. (2021). Neutrophil elastase and chronic lung disease. Biomolecules 11, 1065. doi:10.3390/biom11081065
Walkowska, J., Zielinska, N., Tubbs, R. S., Podgórski, M., Dłubek-Ruxer, J., and Olewnik, Ł. (2022). Diagnosis and treatment of acute pancreatitis. Diagnostics 12, 1974. doi:10.3390/diagnostics12081974
Walsh, P. N., and Ahmad, S. S. (2002). Proteases in blood clotting. Essays Biochem. 38, 95–111. doi:10.1042/bse0380095
Wang, D., Loo, J. F. C., Chen, J., Yam, Y., Chen, S.-C., He, H., et al. (2019). Recent advances in surface plasmon resonance imaging sensors. Sensors 19, 1266. doi:10.3390/s19061266
Wang, K., Xie, C., and Chen, D. (2014). Flap endonuclease 1 is a promising candidate biomarker in gastric cancer and is involved in cell proliferation and apoptosis. Int. J. Mol. Med. 33, 1268–1274. doi:10.3892/ijmm.2014.1682
Wang, Y.-N., Lee, H.-H., Chou, C.-K., Yang, W.-H., Wei, Y., Chen, C.-T., et al. (2018). Angiogenin/ribonuclease 5 is an EGFR ligand and a serum biomarker for erlotinib sensitivity in pancreatic cancer. Cancer Cell 33, 752–769.e8. doi:10.1016/j.ccell.2018.02.012
Wei, T., Wang, F., Zhang, Z., Qiang, J., Lv, J., Chen, T., et al. (2019). Recent progress in the development of fluorometric chemosensors to detect enzymatic activity. Curr. Med. Chem. 26, 3923–3957. doi:10.2174/0929867325666180214105552
Wu, K., Tonini, D., Liang, S., Saha, R., Chugh, V. K., and Wang, J.-P. (2022). Giant magnetoresistance biosensors in biomedical applications. ACS Appl. Mat. Interfaces 14, 9945–9969. doi:10.1021/acsami.1c20141
Yao, C., Ng, E., and Wang, S. X. (2022). An automated and mobile magnetoresistive biosensor system for early hepatocellular carcinoma diagnosis. Biosens. Bioelectron. 202, 113982. doi:10.1016/j.bios.2022.113982
Zhang, G. (2004). “Protease assays,” in Assay guidance manual. Editors S. Markossian, A. Grossman, K. Brimacombe, M. Arkin, D. Auld, C. Austinet al. (Bethesda, MD: Eli Lilly and Company and the National Center for Advancing Translational Sciences).
Zhang, S., He, L., Dai, N., Guan, W., Shan, J., Yang, X., et al. (2016). Serum APE1 as a predictive marker for platinum-based chemotherapy of non-small cell lung cancer patients. Oncotarget 7, 77482–77494. doi:10.18632/oncotarget.13030
Keywords: disease monitoring, point-of-care testing, DNA substrate, peptide substrate, enzymatic activity, giant magnetoresistive sensor, GMR
Citation: Sveiven M, Serrano AK, Rosenberg J, Conrad DJ, Hall DA and O’Donoghue AJ (2024) A GMR enzymatic assay for quantifying nuclease and peptidase activity. Front. Bioeng. Biotechnol. 12:1363186. doi: 10.3389/fbioe.2024.1363186
Received: 30 December 2023; Accepted: 01 February 2024;
Published: 13 March 2024.
Edited by:
Kai Wu, Texas Tech University, United StatesReviewed by:
Deyuan Lyu, University of Minnesota Twin Cities, United StatesCopyright © 2024 Sveiven, Serrano, Rosenberg, Conrad, Hall and O’Donoghue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Drew A. Hall, ZHJld2hhbGxAdWNzZC5lZHU=; Anthony J. O’Donoghue, YWpvZG9ub2dodWVAaGVhbHRoLnVjc2QuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.