
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol., 27 February 2024
Sec. Industrial Biotechnology
Volume 12 - 2024 | https://doi.org/10.3389/fbioe.2024.1363183
This article is part of the Research TopicMicrobial Biosurfactants: Updates on their Biosynthesis, Production and ApplicationsView all 16 articles
Lipopeptides (LPs) produced by Pseudomonas spp. are specialized metabolites with diverse structures and functions, including powerful biosurfactant and antimicrobial properties. Despite their enormous potential in environmental and industrial biotechnology, low yield and high production cost limit their practical use. While genome mining and functional genomics have identified a multitude of LP biosynthetic gene clusters, the regulatory mechanisms underlying their biosynthesis remain poorly understood. We propose that regulation holds the key to unlocking LP production in Pseudomonas for biotechnology. In this review, we summarize the structure and function of Pseudomonas-derived LPs and describe the molecular basis for their biosynthesis and regulation. We examine the global and specific regulator-driven mechanisms controlling LP synthesis including the influence of environmental signals. Understanding LP regulation is key to modulating production of these valuable compounds, both quantitatively and qualitatively, for industrial and environmental biotechnology.
The climate and environmental challenges we face today are immense. Novel solutions encouraging the development of sustainable processes are urgently needed to support the green transition and contribute to the circular economy. One promising strategy to drive green technologies is the exploitation of microbes and their natural product diversity.
Pseudomonas species represent a large and diverse group of bacteria of significant importance for numerous biotechnological applications owing to unique characteristics of rapid growth, versatile utilization of sustainable carbon sources, high metabolic diversity and tolerance to extreme environments (Wang et al., 2020). Ubiquitous in nature, they perform key functions in complex ecosystems, e.g., plant surfaces, the rhizosphere, water, insects, humans and soils, including those with a history of chemical waste pollution (Nikel et al., 2014). Pseudomonas therefore possess an extensive application potential in some of the most challenging fields of industrial and environmental biotechnology, e.g., environmental restoration, plant growth promotion and protection, and the production of specialized metabolites (Nikel et al., 2014; Wang et al., 2020).
Bacterial specialized metabolites (SM), also called secondary metabolites or natural products, are high-value bioactive compounds with vast biotechnological potential. A significant challenge in the development of these compounds is the activation of SM pathways under laboratory conditions. Synthesis is catalyzed by mega-enzymatic complexes encoded by portions of bacterial genomes known as biosynthetic gene clusters (BGCs). Under standard laboratory conditions BGCs are often expressed at low levels or not at all, termed “silent” (Gram, 2015), so that bacterial genome sequences reveal a larger number of SM gene clusters than indicated by chemical analysis of culture extracts (Sanahuja et al., 2011; Reddy and Saravanan, 2013; Monfil and Casas-Flores, 2014). This complicates the lab-based isolation and characterization of SMs and hinders the development of bioprocesses for their production at scale.
Pseudomonas species are natural producers of a vast number of high-value bioactive compounds including biosurfactants such as rhamnolipids, and linear and cyclic lipopeptides (CLPs). Rhamnolipids and LPs are powerful biosurfactants and antibiotics with enormous potential for applications in medicine (e.g., antibiotics, antitumor, immunosuppressants, and cytotoxic agents acting on cancer cells), food and beverage (e.g., anti-spoilage agents, emulsifiers, foaming agents), cosmetics (e.g., antiaging and moisturizing products), textiles (e.g., preparation of fibers), cleaning products (e.g., household detergents and personal care products), bioremediation (e.g., degradation of xenobiotics, heavy metal removal from polluted soil) and agriculture (e.g., bioprotectants). They are attractive ecofriendly alternatives to chemical surfactants owing to their high specificity, biodegradability, low toxicity and effectiveness at extreme temperatures, pH and salinity (Abdel-Mawgoud et al., 2010). Pseudomonas biosurfactants have so far mainly been used in oil recovery and production, including as dispersants in the bioremediation of oil spills (Banat et al., 2000; De Almeida et al., 2016).
While LPs are produced by other bacteria, e. g., Bacillus species in addition to fungi, Pseudomonas-derived LPs in comparison represent a structurally and functionally large, and diverse group of compounds with broad-spectrum antimicrobial, antitumor, cytotoxic, immunosuppressant and surfactant properties (Raaijmakers et al., 2006; Geudens and Martins, 2018). LPs share a common structural blueprint consisting of a fatty acid tail coupled to the N-terminal of a short oligopeptide. In the case of CLPs, a lactone ring is formed between two amino acids resulting in a cyclic structure (Raaijmakers et al., 2006). The diverse structures and biological activities of linear lipopeptides (LLPs) and CLPs result from differences in fatty acid tail length and modifications in addition to the number, type, order and configuration of amino acids in the peptide moiety and lactone ring. LPs belong to the SM family of non-ribosomal peptides (NRPs) which unlike ribosomal peptides (RPs) are synthesized by enzymes capable of incorporating and subsequently modifying both proteinogenic and so called unusual non-proteinogenic amino acids into the oligopeptide. As a result, LPs display an increased level of diversity and multifunctionality.
High cost of production and low yields are major bottlenecks restricting the development and application of LPs. We propose that unravelling the regulatory networks of global and specific regulators underpinning LP synthesis in Pseudomonas holds the key to unlocking production for biotechnology. While numerous regulatory genes have been identified in several LP-producing strains, knowledge of the regulatory mechanisms and critically, the environmental signals controlling production remains in its infancy.
In this review, we summarize the structure and function of Pseudomonas-LPs and examine the regulatory mechanisms and environmental signals, i.e., biotic and abiotic factors influencing their synthesis. Finally, the challenges and opportunities of exploiting regulation to optimize LP production in Pseudomonas will be discussed. A graphical summary of the review is presented in Figure 1.
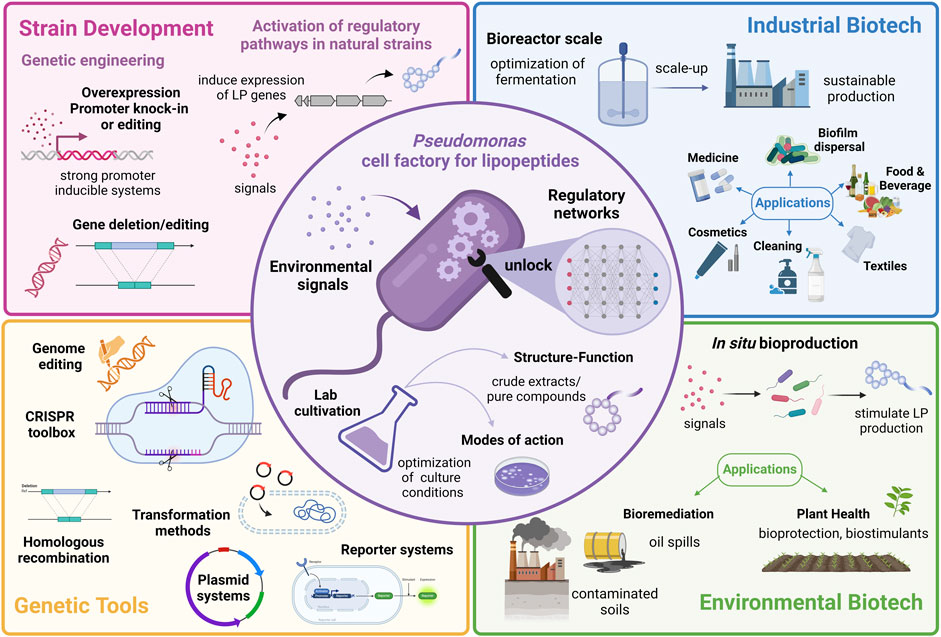
FIGURE 1. Graphical abstract showing strategies to exploit the regulatory pathways controlling lipopeptide production in Pseudomonas for applications in industrial and environmental biotechnology. Created with BioRender.com.
The two main lineages of the genus Pseudomonas (Pseudomonas aeruginosa and Pseudomonas fluorescens lineages) both naturally produce biosurfactants, but strains belonging to the P. aeruginosa lineage produce rhamnolipids whereas members of the P. fluorescens lineage produce LPs. Based on their structures Pseudomonas LPs can be divided into at least 14 different families that differ in oligopeptide length (L) ranging from 8 to 25 amino-acids and macrocycle length (M) ranging from 0 in LLPs to 4 to 9 amino acids in CLPs (Table 1).
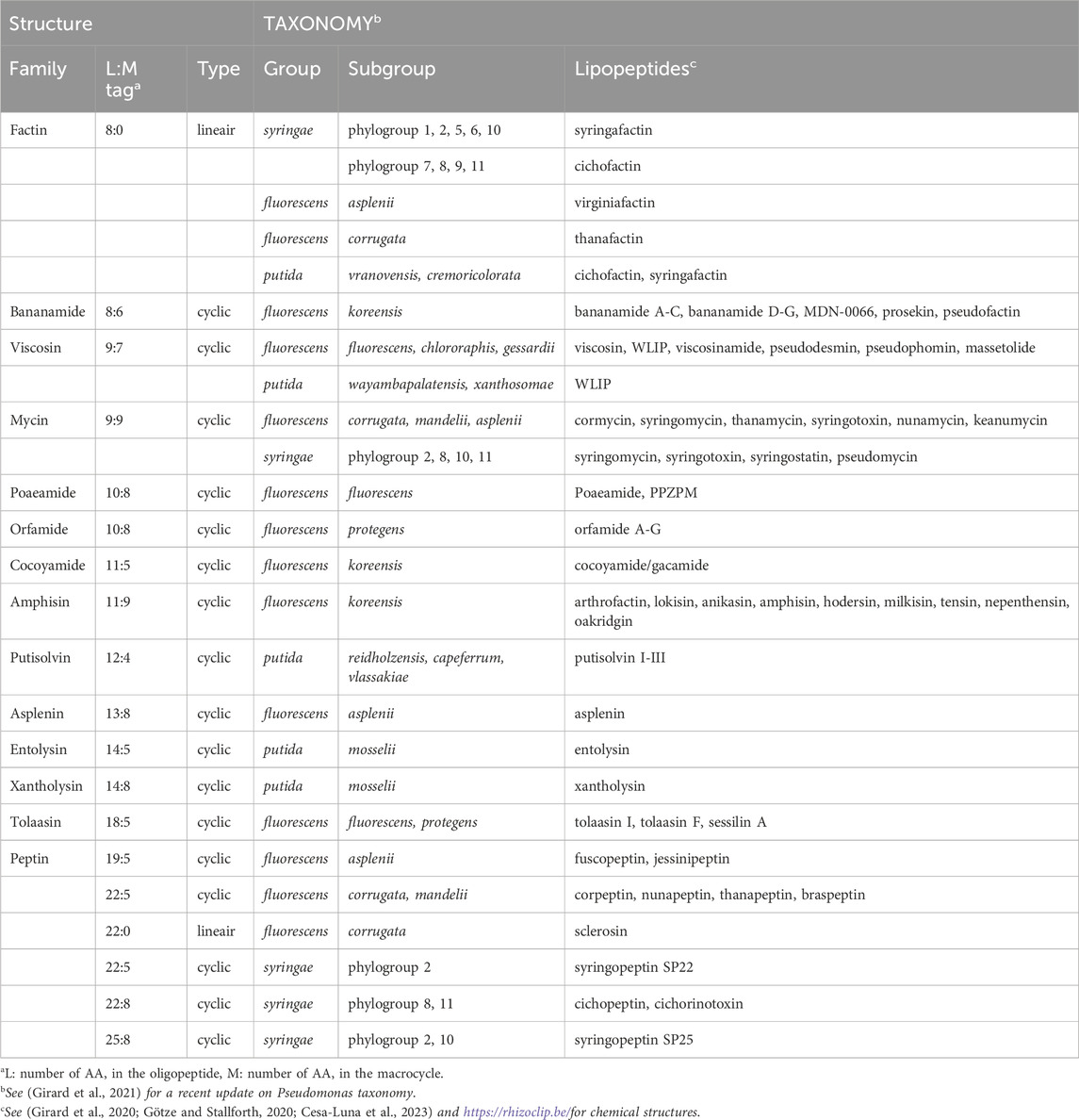
TABLE 1. Structural diversity of Pseudomonas lipopeptides and producing taxonomic groups and subgroups.
Within the P. fluorescens lineage, LP producers reside in the P. fluorescens, Pseudomonas putida and Pseudomonas syringae groups (Cesa-Luna et al., 2023). The P. fluorescens group is further divided into nine major subgroups (P. mandelii, P. jessenii, Pseudomonas koreensis, Pseudomonas corrugata, P. fluorescens, P. gessardii, Pseudomonas chlororaphis, and Pseudomonas protegens) (Girard et al., 2021) comprising both beneficial bacteria and plant pathogens. The P. syringae group, harboring many plant pathogens, is further divided into 13 phylogroups (Berge et al., 2014). LP producers are found in phylogroups 2, 8, 10 and 11 and they typically produce two types of CLPs, respectively from the Mycin and Peptin families, in addition to a LLP from the Factin family. Single Factin producers are found in various other P. syringae phylogroups (Bricout et al., 2022) (Table 1). Strains producing Mycin and Peptin variants are also found in the P. fluorescens group, notably in the P. asplenii, P. mandelii and P. corrugata subgroups (Girard et al., 2020). Most other strains belonging to the P. putida and P. fluorescens group are LP mono-producers, and they produce one or more LP variants that belong to a single LP family. There is a strong correlation between LP type produced and species diversification (Cesa-Luna et al., 2023) with a few notable exceptions, strains that have obtained an LP biosynthetic gene cluster by horizontal transfer. To date, roughly 120 LPs have been described in strains inhabiting diverse environments, but LP producers are often associated with plants.
Pseudomonas LPs display an incredible architectural and chemical diversity and consequently exhibit a range of different biological activities. Here, we briefly present the various biological properties and functions of LPs and discuss their potential biotechnological applications. More detailed information on the natural functions and roles of Pseudomonas LPs is provided in various reviews (D'Aes et al., 2010; Raaijmakers et al., 2010; Geudens and Martins, 2018; Girard et al., 2020; Götze and Stallforth, 2020).
LPs are surface-active molecules also called biosurfactants. Biosurfactants help to condition the producing strain’s environment by supporting various processes including bacterial motility, attachment and colonization of surfaces, biofilm development and access to nutrients and water (Nybroe and Sorensen, 2004; D'Aes et al., 2010; Raaijmakers et al., 2010; Geudens and Martins, 2018; Bricout et al., 2022).
Various lab-based studies show a strong correlation between LP production and swarming motility when comparing wild-type and LP-deficient mutant strains (de Bruijn et al., 2008; de Bruijn and Raaijmakers, 2009; Geudens and Martins, 2018). The ability of LPs to alter surface tension and viscosity is determined by their structural properties (D'Aes et al., 2010). In the amphisin producer Pseudomonas sp. DSS73, amsY and gacS mutant strains are unable to swarm on soft agar and as expected swarming motility is restored by the addition of amphisin but also viscosin, tensin and serrawettin from Serratia liquefaciens (Andersen, 2003). In contrast, synthetic surfactants were unable to complement the non-motile phenotype indicating that unique physiochemical properties relating to the chemical structure of LPs contribute to bacterial movement (Andersen, 2003). The diverse structures and physiochemical properties of LPs make them attractive alternatives to chemical surfactants for numerous industrial applications (Ceresa et al., 2023).
One of the most effective biosurfactants reported to date is surfactin from Bacillus subtilis capable of lowering the surface tension of water from 71 to 27 mN/n at a critical micelle concentration (CMC) of 20 µM (Yeh et al., 2005). Viscosin also shows strong surface activity reducing water surface tension to 25 mN/n at CMC of 4 μg/mL (Nybroe and Sorensen, 2004). Similarly, viscosinamide reduces water surface tension to 27 mN/n however no CMC values are available (Nybroe and Sorensen, 2004). Syringomycin and syringopeptin reduce water surface tension in ranges of 31–35 mN/n comparable to values of 31.5 mN/n and 38 mN/n recorded for putisolvin II and tolaasin, respectively (Hutchison and Johnstone, 1993; Nybroe and Sorensen, 2004; Janek et al., 2010). For most Pseudomonas LPs however information on physiochemical properties, e.g., surface reduction activities and CMC values, foaming capacity, emulsifying activity and compound stability are lacking.
In situ studies support the role of LPs in motility and colonization of specific habitats, e.g., plant material or fungal tissues. Massetolide A contributes to colonization of tomato roots by P. lactis (fluorescens) SS101, viscosin is required for colonizing broccoli florets by a pectolytic strain of P. fluorescens, cichofactin is essential for Pseudomonas cichorii to colonize lettuce leaves, while Pseudomonas sp. DSS73 uses amphisin to colonize sugar beet seeds and roots (Hildebrand et al., 1998; Nielsen and Sorensen, 2003; Tran et al., 2007; Pauwelyn et al., 2013). Other LPs, for example, poaeamide from Pseudomonas poae RE*1-1-13 and putisolvin from P. putida 257 do not positively contribute to rhizosphere competence as no differences in plant root colonization are observed between WT and biosurfactant deficient mutant strains (Kruijt et al., 2009; Zachow et al., 2015).
For many plant-associated LP-producers, biosurfactant-assisted motility is a key determinant for successful rhizosphere and phyllosphere colonization, where in addition to plant material, LPs also mediate interactions with fungi. In the phytopathogen Ralstonia solanacearum, the LP ralsolamycin (RM) facilitates fungal tissue invasion by inducing the formation of fungal survival structures known as chlamydospores in Aspergillus flavus (Spraker et al., 2016). An RM deficient rmyA mutant cannot induce chlamydospore formation and shows reduced hyphal invasion (Spraker et al., 2016). Subsequently (Venkatesh et al., 2022), demonstrated that compared to the rmyA mutant, WT R. solanacearum is internalized in chlamydospores during co-culture with A. flavus and shows increased fitness under starvation and cold stress. Phylogenetically distinct bacteria (not associated with endofungal lifestyles) including the nitrogen-fixing bacterium Herbaspirillium seropedicae were also shown to colonize chlamydospores of A. flavus when treated with WT supernatants further confirming the role of RM in facilitating fungal invasion (Venkatesh et al., 2022). Such knowledge could be translated to stimulate diverse endofungal interactions for improved ecosystem services, e.g., nitrogen fixation (Venkatesh et al., 2022). While a number of other LPs including the Pseudomonas CLPs viscosinamide and tensin can induce fungal survival structures, their role in fungal invasion and endosymbiosis is currently unknown (Nielsen et al., 1999; Nielsen et al., 2000; Venkatesh et al., 2022). More information on interactions with fungi is described in Section 3.2.2.
Biosurfactants also regulate biofilm development with a number of CLPs shown to promote biofilm formation (massetolide A, sessilin and xantholysin) and others involved in biofilm dispersal (arthrofactin, orfamide and putisolvin) (Kuiper et al., 2004; Bonnichsen et al., 2015). In P. sessilinigenes CMR12a, orfamides are indispensable for swarming motility, while sessilin is important for biofilm formation (D’aes et al., 2014). Based on contrasting studies, viscosin is proposed to mediate both biofilm formation and dispersal (de Bruijn et al., 2007; Bonnichsen et al., 2015). Viscosin-mediated biofilm dispersal is dependent on carbon starvation and by microscopic analysis it was observed that cells exhibiting high viscA (required for viscosin biosynthesis) expression levels were leaving biofilms, further supporting the role of LPs in motility. However, information on the mechanisms of LP-mediated biofilm development and dispersal, particularly in situ is limited and could benefit from more temporal studies examining LP function during biofilm lifecycles. For the producing bacteria, the ability to disperse is an important escape function under unfavorable nutrient conditions to support their spread throughout the environment and enable the colonization of new niches. From an industrial perspective, LPs displaying roles in biofilm formation could be used for bulk chemical production using biofilm fermentations whereas dispersal functions could be exploited as surface-coating agents or used in disinfectant formulations (Bonnichsen et al., 2015; Leonov et al., 2021).
Additional biological properties of LPs include the chelation of metal ions and xenobiotic degradation, e.g., petroleum hydrocarbons and pesticides (Nybroe and Sorensen, 2004; Raaijmakers et al., 2010; de Cássia et al., 2014; Raj et al., 2021). Research on biosurfactants as bioremediation agents to clean up contaminated soils is largely limited to rhamnolipids or Pseudomonas-leachates containing uncharacterized LPs (Sekhon Randhawa and Rahman, 2014; Sun et al., 2021). The CLPs viscosin, amphisin, massetolide A and putisolvin can emulsify alkane hydrocarbons such as n-hexadecane (Bak et al., 2015). Viscosin was also shown to stimulate alkane mineralization by a diesel-degrading bacterial consortium however the activity of the CLP was short-lived due to rapid (likely microbial) degradation (Bak et al., 2015). While LPs are proposed to chelate heavy metals and degrade insoluble hydrocarbons to increase their bioavailability and/or detoxify polluted soils for protection against toxicants, roles of LPs in such processes remain unclarified (Raaijmakers et al., 2010; Gutiérrez-Chávez et al., 2021).
Pseudomonas-CLPs display broad-spectrum antimicrobial properties exerting effects against bacteria, fungi, oomycetes and viruses as previously reviewed by (Nielsen et al., 2002; Geudens and Martins, 2018; Girard et al., 2020; Oni et al., 2022).
Pseudomonas-LPs show antagonistic activities against diverse Gram-positive and Gram-negative bacteria including human, plant and animal pathogens (Raaijmakers et al., 2006; Geudens and Martins, 2018). In general, Gram-positive bacteria are more susceptible, for example; viscosin, massetolide A and syringomycins 22A and E inhibit Mycobacteria spp,; amphisin, syringopeptins 22A and 25A in addition to corpeptin and WLIP are active against Bacillus spp.; syringopeptins also display inhibitory effects active against Rhodococcus spp., and Micrococcus spp. (Nybroe and Sorensen, 2004; Geudens and Martins, 2018), while viscosin and tensin are active against Streptomyces scabies (Pacheco-Moreno et al., 2021). Medipeptin A, produced by Pseudomonas mediterranea EDOX is active against Staphylococcus aureus and Bacillus cereus with a MIC of 8 μg/mL and against Micrococcus flavus with a MIC of 2 μg/mL (Zhou et al., 2021). Medipeptin A exerts its activity against S. aureus by binding to the cell wall polymer lipoteichoic acid and the cell wall precursor lipid II and by forming pores in membranes (Zhou et al., 2021). Jessenipeptin and mupirocin (a polyketide antibiotic), co-produced by Pseudomonas sp. QS1027, show synergistic activity against methicillin-resistant S. aureus (Arp et al., 2018).
Fewer LPs show activity against Gram-negative bacteria possibly due to the inability of LPs to access the outer membrane or peptidoglycan layer of Gram-negative cell walls (Nybroe and Sorensen, 2004). For example, syringomycins E and syringopeptin 25A show inhibitory effects against P. syringae but only upon treatment with lysozyme (Fogliano et al., 2002). Tolaasin I and WLIP show low inhibition of Gram-negatives whereas LPs of the xantholysin group are active against various Gram-negative and Gram-positive bacteria (Li et al., 2013). Interestingly, Rainey et al. (1991) reported tolaasin resistant Gram-negative bacteria (Pseudomonas reactans, P. putida and E. coli) become sensitive to the toxin when challenged by tolaasin and polymyxin B, highlighting the importance of synergistic activities of compounds during antagonism.
Extensive research has focused on the inhibitory activities of Pseudomonas-LPs against numerous fungi, oomycetes and yeasts (Nybroe and Sorensen, 2004; Raaijmakers et al., 2006; Geudens and Martins, 2018; Omoboye et al., 2019b; Oni et al., 2022). One of the most active compounds is tolaasin, and 18:5 CLP produced by the mushroom pathogens Pseudomonas tolaasii and P. costantinii (Scherlach et al., 2013). These bacteria cause brown blotch disease characterized by dark brown lesions on the fruiting bodies of various mushroom species including the button mushroom Agaricus bisporus, the oyster mushroom Pleurotus ostreatus and shiitake (Lentinula edodes) (Soler-Rivas et al., 1999; Osdaghi et al., 2019). Tolaasin I is the main virulence factor of P. tolaasii and toxic towards mushrooms (Rainey et al., 1991; Lo Cantore et al., 2006; Andolfi et al., 2008). Tolaasin disrupts the fungal membrane by forming trans-membrane pores, allowing the producing bacteria access to cell nutrients. Tolaasin I is most active and also shows strong antimicrobial activity against other Basidiomycetes (Bassarello et al., 2004; Lo Cantore et al., 2006), a variety of Ascomycetes (Lo Cantore et al., 2006; Andolfi et al., 2008; Ferrarini et al., 2022a), and Oomycetes (Lo Cantore et al., 2006; Andolfi et al., 2008; Ferrarini et al., 2022a), with minimum inhibitory quantities (MIQs) ranging from 0.08 µg for R. solani and A. bisporus to 0.64 µg for some plant pathogenic Ascomycetes. Yeast-like fungi that cause diseases in animals and humans were less sensitive to tolaasin I (Andolfi et al., 2008). Ferrarini et al. (2022a) recently showed that within the Oomycetes P. nicotianae (EC50 = 5.6 µM) is considerably less sensitive to tolaasin I than Pythium myriotylum (EC50 = 0.30 µM in the absence of sterols).
A variant of tolaasin, called sessilin, is made by the well-studied biocontrol strain P. sessilinigenes CMR12a. This strain, isolated from cocoyam roots in Cameroon, also produces the 10:8 CLP orfamide (D’aes et al., 2014). Sessilin and orfamide contribute to the control of the cocoyam root rot disease caused by the Oomycete pathogen P. myriotylum, with sessilin showing the strongest inhibitory activity (Oni et al., 2019b).
Members of the Viscosin, Orfamide, Poaeamide and Putisolvin family (see Cesa-Luna et al. (2023) and Supplementary Table S1 for producing strains) cause immobilization and lysis of zoospores produced by oomycetes at concentrations around 25 µM (Gross et al., 2007; Raaijmakers et al., 2010; Zachow et al., 2015; Ma et al., 2016a). Microscopy studies show that various LPs of the Viscosin, Bananamide and Amphisin family induce morphological changes in fungi and oomycetes; viscosinamide increases branching, hyphal swelling and rosette formation in R. solani in addition to reduced mitochondria activities and changes in mitochondria morphology (Nielsen et al., 1999; Raaijmakers et al., 2006). Comparable microscopic observations have been made for viscosinamide against Pythium ultimum (Nielsen et al., 1999), while P. myriotylum challenged by pseudodesmin, viscosinamide and WLIP at concentrations ranging from 100 nm to 50 µM shows hyphal disintegration with pseudodesmin showing reduced hyphal branching at 1 and 50 µM while viscosinamide distorts fungal hyphae causing lysis at concentrations below 50 µM (Oni et al., 2020a). Increased branching and swelling also occurs for fungal hyphae treated with tensin (Nielsen et al., 2000). Bananamides target P. oryzae causing extensive hyphal branching, leakage and vacuolation (Omoboye et al., 2019a). Entolysin A and B permeabilize the membranes of Pyricularia oryzae and B. cinerea spores and mycelium as revealed by propidium iodide assays, starting at concentrations of 32 μM, with entolysin B being more active than entolysin A (Muangkaew et al., 2023). Antifungal activity of xantholysin has been tested using mutants, suggesting some activity against Ascomycetes, but this remains unconfirmed with pure compounds (Li et al., 2013).
Mycin and Peptin variants produced by specific strains from the P. syringae and the P. fluorescens group show interesting antifungal and anti-oomycete activity (Girard et al., 2020). Activity has mainly been shown by using mutants impaired in LP production. Only a few studies have used pure compounds, reflecting the difficulties in obtaining enough pure compound for biological assays. Keanumycin A, from Pseudomonas sp. QS1027, shows strong antifungal activity against human fungal pathogens including Candida spp. (MIC = 0.86 µM) and was extremely effective against B. cinerea (0.07 µM, 80 μg/L) (Götze et al., 2023). Also syringomycin E, syringotoxin B and syringostatin A, produced by strains of P. syringae pv. syringae, show fungicidal activity against Candida spp. (Sorensen et al., 1996). Nunamycin and nunapeptin are produced by P. nunensis In5, isolated from a potato soil suppressive against R. solani AG3 in southern Greenland (Michelsen and Stougaard, 2011; Ntana et al., 2023). Nunamycin production is required to inhibit R. solani growth in co-culture on agar plates and in a soil microcosm where disease incidence in tomato seedlings was significantly increased in a nunamycin mutant strain compared to the WT (Michelsen et al., 2015b). By using purified CLPs it was shown that nunamycin is more active against R. solani compared to Pythium aphanidermatum which appears more sensitive to nunapeptin. Thanamycin and thanapeptin are produced by Pseudomonas sp. SH-C52, a well-studied biocontrol agent isolated from sugar beet plants grown in a soil naturally suppressive to R. solani (Van Der Voort et al., 2015). Thanamycin has antimicrobial activity against R. solani and a range of other fungi, while some derivatives of thanapeptin have anti-oomycete activity. Sclerosin, a 22:0 LLP from the Peptin family made by Pseudomonas brassicacearum DF41 isolated from canola roots, has activity against the fungal pathogen Sclerotinia sclerotiorum (Berry et al., 2012). Mutant analysis has revealed activity against Botryosphaeria dothidea for braspeptin, made by Pseudomonas sp. 11K1 (Zhao et al., 2019). Other Peptin family members such as syringopeptins show strong activity against various yeasts (Grgurina et al., 1996; Lavermicocca et al., 1997), while Fuscopeptin A and syringopeptin 22-A are toxic to B. cinerea at 20 µM.
Synergistic activities of LP product mixtures and/or with other molecules appear key to fungal antagonism. For example, P. syringae pv. syringae strain B359 secretes syringomycin E and syringomycin 25A in tandem with cell-wall degrading enzymes to inhibit fungal growth (Fogliano et al., 2002). The toxins show inhibitory activity against numerous fungi whereby antifungal activity is enhanced by the addition of purified enzymes and in vivo during co-culture with Trichoderma atroviride. Interestingly, syringomycin 25A is more potent against fungi in the presence of hydrolytic enzymes whereas syringomycin E shows greater inhibition of fungal growth and spore germination without hydrolytic enzymes (Fogliano et al., 2002). Other examples of antifungal synergism include orfamide and sessilin production in P. sessilinigenes CMR12a as well as nunamycin and nunapeptin production in P. nunensis In5 where co-production of the compounds increases inhibition of fungal growth. Crude extracts of P. nunensis In5 show greater inhibition against R. solani and P. aphanidermatum than pure compounds indicating the importance of synergistic activities of CLPs during interactions with pathogens and likely other organisms (Michelsen et al., 2015a). Moreover, sessilin and orfamide act additively in the biological control of the basidiomycete pathogen R. solani in bean and cabbage (Olorunleke et al., 2015). Interestingly, orfamide A and sessilin show no antifungal activity against R. solani when applied individually whereas nunamycin and nunapeptin target R. solani and P. aphanidermatum respectively (Michelsen et al., 2015a; Olorunleke et al., 2015).
More recently, viscosin-like CLPs produced by P. cichorii (identification based on 16S rRNA gene and probably not correct) demonstrated antagonistic activity against the human and vertebrate pathogens Aspergillus fumigatus and Batrachochytrium dendrobatidis (Martin et al., 2019). Using chemical imaging viscosin and massetolide were detected at the fungal inhibition zone suggesting synergistic activities may enhance their antifungal properties. Lab-based assays showing inhibition effects against both pathogens were however only reported for purified viscosin (Martin et al., 2019). In general, the mechanisms underlying synergistic interactions of LPs remain largely unknown.
Antiviral activity is documented for viscosin against bronchitis virus and human-pathogenic viruses but the mechanism of viral inactivation is unknown (Raaijmakers et al., 2006). An analogue of xantholysin (MA026) from Pseudomonas sp. RtlB025 suppresses infectious hematopoietic necrosis virus (IHNV) and displays antiviral activity against hepatitis C virus (HCV) infection (Shimura et al., 2013). Following the global corona pandemic there has been considerable interest in developing diverse antiviral drugs. Xia et al. (2021) modelled the ability of diverse LPs to target coronavirus replication and transcription machinery and found Pseudomonas-derived ferrocin A and iron-chelating ferrocin A to be the best performing molecules (Xia et al., 2021). However, no studies examining the biological activities of Pseudomonas LPs against COVID-19 and SARS-CoV-2 exist.
LPs also possess anti-proliferative activities against different cancer cell lines including viscosin (breast and prostate cancer cell lines) (Siani et al., 2008), xantholysin A (Pascual et al., 2014) and MDN-0066 (Cautain et al., 2015) (kidney tumor cell lines), pseudofactin II (melanoma cell lines) (Janek et al., 2013) and nunamycin/nunapeptin (mantle cell lymphoma, melanoma cell lines, T-cells leukemia) (Michelsen et al., 2015a). Accurate comparison of LP cytotoxic activities is challenged by the lack of standardization across assays. Similar to antimicrobial testing crude extracts over purified compounds are often used making the interpretation of results difficult as the bioactivities observed may derive from other compounds or synergistic activities between compounds. This has been observed in P. nunensis In5, where increased cytotoxic activity was seen when purified nunamycin and nunapeptin are mixed instead of applied individually (Michelsen et al., 2015a).
Pseudomonas LP producers are implicated in both positive and negative interactions with plants. Phytopathogenic strains typically co-produce phytotoxic CLPs from the Mycin and Peptin family, which act as virulence factors and form pores in plant membranes causing electrolyte leakage and necrosis. They usually also co-produce a third LLP or CLP not directly involved in virulence but instead aiding in plant tissue colonization. Phytopathogenic Pseudomonas LP producing strains taxonomically belong to the P. syringae group, or to the P. asplenii and P. corrugata subgroup of the P. fluorescens group (Girard et al., 2020). Phytotoxic CLPs from the Mycin and Peptin family also have strong antimicrobial activity (see above), demonstrating a dual role in pathogenicity and antagonism against competitors. Strains belonging to the P. corrugata subgroup can behave as plant pathogens, causing pit necrosis on tomato and pepper, but also show strong biological control activity against plant pathogens. They are often isolated from the roots and rhizosphere of non-diseased plants and from bulk soil (Catara, 2007; Gislason and de Kievit, 2020). Cormycin and corpeptin produced by P. corrugata double up as phytotoxic compounds and antimicrobial molecules against bacterial and fungal pathogens. Pseudomonas sp. SH-C52 (Mendes et al., 2011) (see above) also belongs to the P. corrugata subgroup and produces Mycin and Peptin-type CLPs with antifungal and anti-oomycete activity, indicating that there is no clear-cut line between plant pathogens and beneficials in these groups (Girard et al., 2020).
Disease suppressive soils are a rich source of LP-producing Pseudomonas strains. The potent biocontrol agents P. nunensis In5 (Michelsen and Stougaard, 2011) and Pseudomonas sp. SH-52 have been obtained from R. solani suppressive soils. Irrigation is known to protect potato tubers against the scab pathogen S. scabies. Microbiome analysis revealed that irrigated potato field had a larger proportion of Pseudomonadales bacteria than a non-irrigated potato field and that the presence of biosynthetic gene clusters encoding CLPs was positively correlated with disease suppression. Tensin, an 11:9 amphisin family CLP proved to be key determinant of in planta inhibition of potato scab in glasshouse trials (Pacheco-Moreno et al., 2021). Likewise, Pseudomonas strains able to produce CLPs belonging to 11 different families are dominant in the rhizosphere of cocoyam plants grown in a tropical soil suppressive to the cocoyam root rot disease caused by P. myriotylum (Oni et al., 2019a; Oni et al., 2020b).
Pseudomonas LP-producers demonstrating fungal and/or oomycete antagonism have gained considerable interest as candidates for controlling plant diseases. A detailed overview of Pseudomonas LP-mediated biocontrol is given by (D'Aes et al., 2010; Raaijmakers et al., 2010; Höfte, 2021; Oni et al., 2022). However, it is important to note that many studies are centered on the collection of data from lab-based experiments with only a handful of reports linking lab data to microcosm- or field-based studies. For example, for the viscosinamide producer P. fluorescens DR54 (Nielsen and Sorensen, 2003) a strong correlation between the ability of DR54 to inhibit growth of R. solani and P. ultimum in co-culture and during colonization of the rhizosphere of germinating sugar beet using plant-soil microcosms was observed. The study highlights the multifunctionality of CLPs with antibiotic and surfactant properties enabling the producing strain to condition its environment for successful rhizosphere colonization.
As wetting agents LPs can increase the solubility of nutrients and hydrophobic substrates for the producing strain (Nybroe and Sorensen, 2004; D'Aes et al., 2010; Mavrodi et al., 2010). For example, Bunster et al. (1989) reported that compared to a non-surface active Pseudomonas strain, surface-active isolates of P. fluorescens and P. putida increased the wetness of wheat leaves. Likewise, syringafactins are strong biosurfactants exerting hygroscopic activities to attract water vapor from the atmosphere increasing water availability and reducing water stress in P. syringae pv. syringae B728a on dry leaves and the apoplast of bean (Burch et al., 2014). Increasing the availability of nutrients and water potentially offers LP-producers a competitive advantage against other microbes including phytopathogens and thus may indirectly contribute to plant disease management by reducing pathogen growth delaying disease onset in the host plant (Nybroe and Sorensen, 2004).
A number of CLPs including massetolide A (Tran et al., 2007), sessilin, orfamide (Ma et al., 2016b; Ma et al., 2017), WLIP, lokisin and entolysin (Omoboye et al., 2019b) are involved in the induction of systemic resistance in plants. This type of resistance is systemically expressed rendering plants less susceptible to subsequent infection with pathogens (Pršić and Ongena, 2020). These studies are typically conducted with CLP mutants and CLP crude extracts with only a few studies demonstrating induction of ISR by pure compounds. Application of purified massetolide A at a concentration of 44 µM to tomato leaves or roots reduced the lesion area caused by Phytopthora infestans, but did not reduce disease incidence (Tran et al., 2007). Pure orfamide triggered ISR against R. solani web blight in bean at concentrations ranging from 0.001 to 0.1 µM (Ma et al., 2016b), while 25 µM orfamide was needed to elicit resistance to Cochliobolus miyabeanus in rice (Ma et al., 2017) suggesting different mechanisms involved.
While protists stimulate beneficial plant-microbe interactions and contribute important functions, e.g., nutrient cycling and pathogen removal they are major bacterial predators (Gao et al., 2019; Bahroun et al., 2021; Guo et al., 2021; Hawxhurst et al., 2023). One predation defense mechanism used by bacteria involves LPs. Using a combination of wild-type and mutant P. fluorescens strains, Mazzola et al. (2009) showed that massetolide and viscosin protect bacteria against predation by the amoeba Naegleria americana, with the predator showing a greater sensitivity to viscosin. LP-producers showed better persistence and protection in soil against the predator but the effect was only temporal (Mazzola et al., 2009). P. nunensis 4Aze was co-isolated with the social amoeba Polyspondyllium pallidum RM1 from forest soil. This strain produces keanumycin D, and nunapeptin B and C with suppressive activities against amoebal predators and the bacterivorous nematode Oscheius myriophilus, highlighting the broad-spectrum activity of LPs and underexplored anti-predator function of these compounds (Pflanze et al., 2023) Also the keanumycins produced by Pseudomonas sp. QS1027 have strong amoebicidal activity (Götze et al., 2023). More information on the influence of predator interactions and predator-derived molecules on LP regulation is needed.
Antiparasitic activities are documented for viscosin against the human parasitic protozoan Trypanosoma cruzi, the causal agent of Chagas disease. A viscosin-like LP from Pseudomonas sp. H6 was active against the fish parasitic ciliate Ichthyophthirius multifiliis and showed inhibitory effects against green algae, crustaceans, cyanobacteria and zebrafish embryos (Raaijmakers et al., 2006; Korbut et al., 2022).
LPs also mediate insect interactions with orfamides, sessilin and viscosin shown to possess insecticidal properties (Geudens and Martins, 2018). Insecticidal activity appears a multifactorial process involving LPs and other metabolites, e.g., Fit toxin, rhizoxin and HCN wherein the role of LPs appears to be strain-specific (Jang et al., 2013; Flury et al., 2017).
The main mode of action of LPs is membrane disruption through pore-formation causing membrane leakage and cell death (Geudens and Martins, 2018). The majority of mechanistic studies rely on model cell membranes that enable simpler and well-controlled experiments however they lack the complexity of real biological membranes. Consequently, the detailed mechanism of action underlying LP pore-formation including membrane selectivity, is largely unknown. As biological organisms are capable of altering their lipid membrane composition in response to external signals in order to adapt to their physical environment (Chwastek et al., 2020), it is likely that LPs have not one but multiple modes of action and display context-dependent activities. This may also contribute to the low resistance towards LPs in the environment despite their ubiquitous nature (Peschel and Sahl, 2006; Steigenberger et al., 2023). Future work should determine the influence of basic membrane parameters, e.g., membrane thickness in addition to physical membrane properties and lipid composition of different membrane types, e.g., bacterial, fungal and mammalian on LP activity. This was recently highlighted by (Ferrarini et al., 2022a) wherein the bioactivities of tolaasin and sessilin were reduced against oomycetes when membrane sterol composition is altered.
In Pseudomonas, LPs are synthesized by large BGCs encoding multi-modular nonribosomal peptide synthetases (NRPSs) (see Götze and Stallforth (2020); Roongsawang et al. (2011) for a detailed overview). These enzymes recognize, activate, modify, and link amino acid intermediates to the product peptide and can synthesize peptides with unusual amino acids including D-amino acids. A typical module comprises a condensation domain, an adenylation domain and a thiolation domain. A specialized condensation starter domain (Cs) with N-acylation activity attaches the fatty acid to the first amino acid. The adenylation (A) domain is responsible for amino acid recognition and adenylation, the thiolation (T) domain binds the adenylated amino acid to a phosphopantetheine carrier. A regular condensation (C) domain catalyzes the formation of a peptide bond between two consecutively bound L-amino acids. A condensation domain with built-in epimerization capacity (C/E domain) located downstream of D-amino acid incorporating modules catalyzes the conversion of L-amino acids to D-isomers. Separate epimerization domains as described in NRPSs of Bacillus and Streptomyces are lacking in Pseudomonas NRPSs (Götze and Stallforth, 2020). Cyclization and release of the peptide are carried out by a tandem of thioesterase (TE) domains associated with the last module. The order of modules is usually co-linear to the peptide sequence. CLPs in mono-producers are usually synthesized by NRPS systems encoded by three large open reading frames that are either organized in one operon or are split, with the first NRPS gene typically composed of two modules, located elsewhere in the genome. The NRPS genes are flanked by one or two luxR-type regulatory genes and three genes encoding a tripartite PleABC export system. LLPs are synthesized by NRPS systems encoding by two large open reading frames (Figure 2).
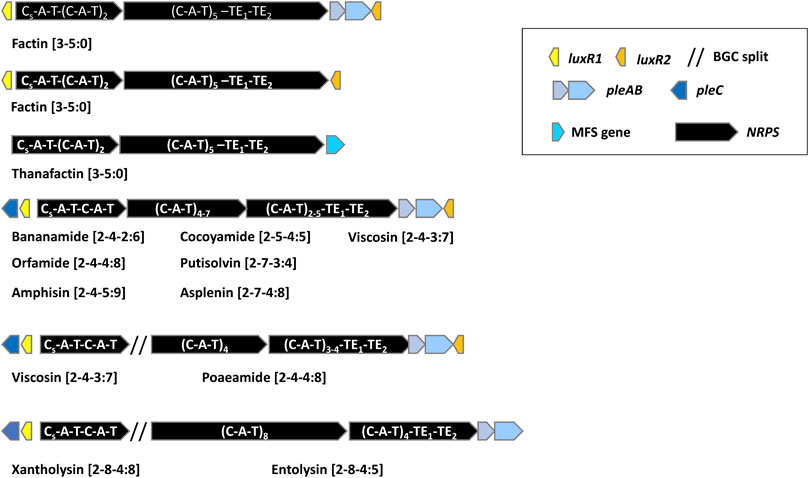
FIGURE 2. General overview of the organization of lipopeptide biosynthetic gene clusters in lipopeptide mono-producers.
The chlorinated 9:9 CLPs belonging to the Mycin family are synthesized by one or two NRPSs and separate enzymes encoded by syrB1, syrB2, syrC and syrP homologues located upstream of the Mycin BGC (Figures 3, 4). All Mycin family members have the unusual amino acids 3-hydroxy-aspartic acid at position 8 and 4-chlorothreonine at position 9 in the peptide (see Girard et al. (2020) for an overview). SyrB1 is a separate enzyme with an A-T module that activates and loads threonine. SyrB2 is a non-heme FeII halogenase that chlorinates threonine. SyrC is an aminoacyltransferase that shuttles the threonyl moiety in trans between the T domain of SyrB1 and the T domain of the last module of the NRPS (Bender et al., 1999; Singh et al., 2007). SyrP is an aspartyl hydroxylase that hydroxylates L-Asp after selection, activation and installation of L-Asp on the T domain of the eighth module of the NRPS cluster (Singh et al., 2008). A pleC transporter gene is usually located downstream of the Mycin BGC preceded by a luxR-type regulatory gene called syrF in P. syringae pv. syringae (Figure 3). CLPs belonging to the Peptin family are usually synthesized by three NRPSs composed of in total 19, 22 or 25 modules. PleAB and pseABC transporter genes and a dab gene are usually located downstream of the Peptin NRPS genes (Figures 3, 4), while A syrD transporter gene (Quigley et al., 1993; Quigley and Gross, 1994) is positioned upstream of the Peptin BGC. DAB is involved in the synthesis of 2,4-diaminobutyric acid, a non-protein amino acid present in all peptins and most mycins (Girard et al., 2020). In Pseudomonas fuscovaginae, the dab gene is located upstream of the asplenin BGC and flanked by two luxR-type regulatory genes (Figure 3). In P. cichorii, pleAB transporter genes and three luxR-type regulatory genes are located downstream of the cichopeptin BGC, but dab and pseABC transporter genes are located upstream of the cichopeptin BGC (Figure 3).
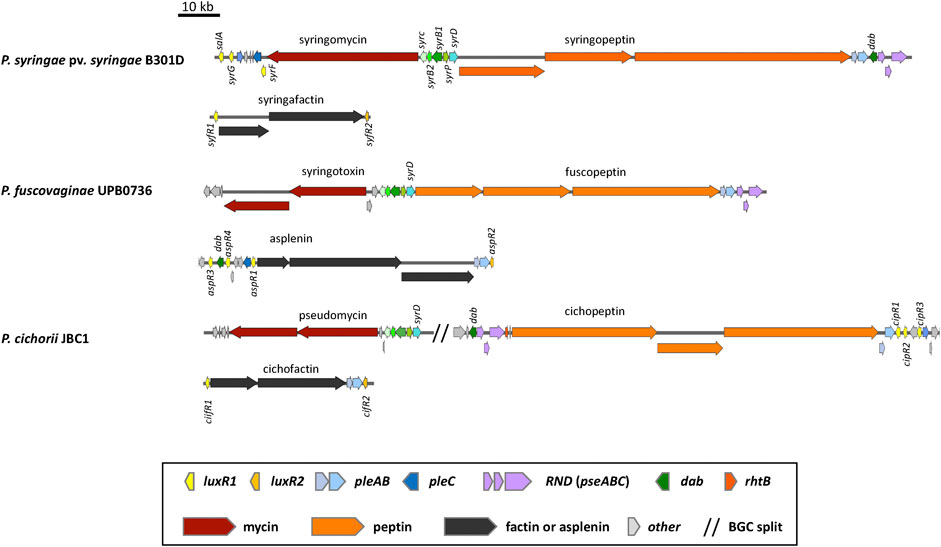
FIGURE 3. Organization of the lipopeptide biosynthetic gene clusters in non-quorum sensing lipopeptide poly-producers. Representative strains are shown. See Supplementary Table S1 for strain and sequence information.
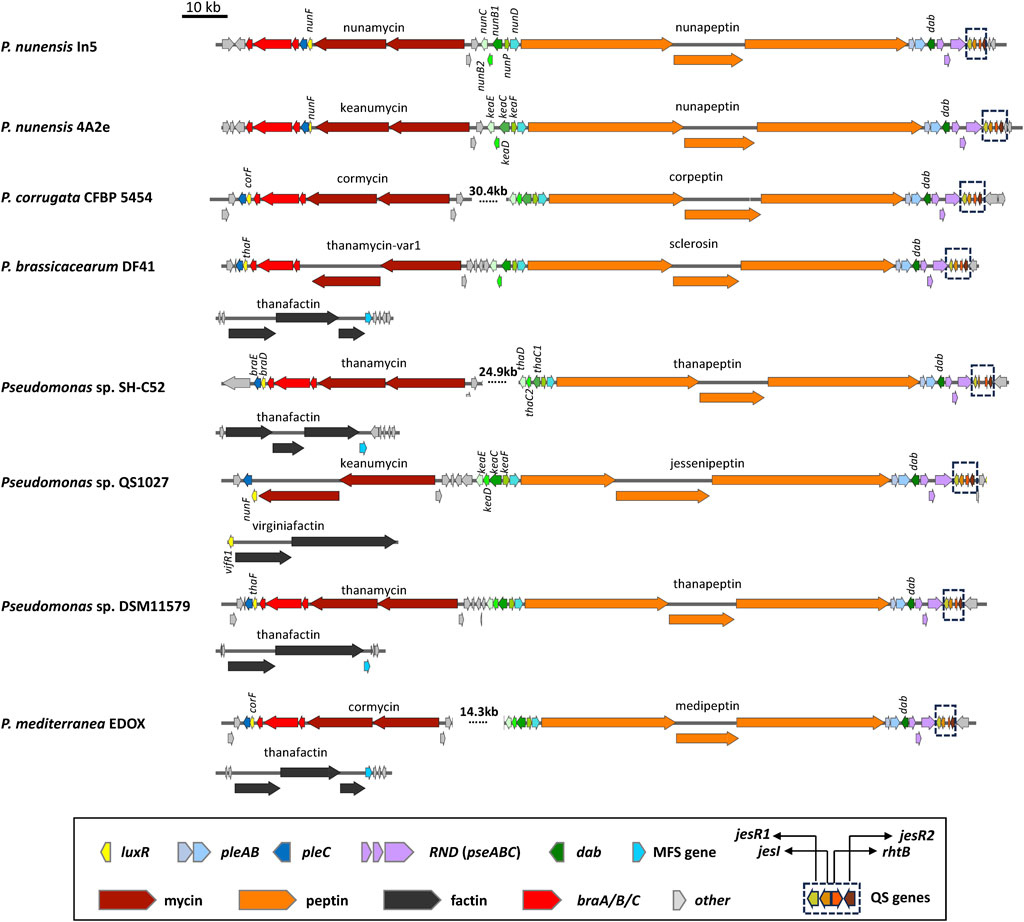
FIGURE 4. Organization of lipopeptide biosynthetic gene clusters in quorum sensing lipopeptide poly-producers. Representative strains are shown. See Supplementary Table S1 for strain and sequence information.
Information on the regulatory mechanisms and environmental signals controlling LPs is lacking but key to modulating LP production both quantitatively and qualitatively in the lab or in the environment for industrial and environmental biotechnology applications. Here, we present what is known so far.
The best studied global regulatory system associated with LP production is the Gac/Rsm signal transduction pathway (Haas and Défago, 2005) (Figure 5). The cascade is initiated by the GacS/GacA two-component system composed of the membrane-bound GacS sensor kinase and the cognate GacA response regulator located in the cytoplasm. The GacS sensor kinase, initially called LemA, was first described in the bean pathogen P. syringae pv. syringae strain B728a and found essential to produce the CLP syringomycin (Hrabak and Willis, 1992; Hrabak and Willis, 1993). At high cell densities autophosphorylation of the GacS sensor kinase is triggered by an unknown chemical signal in the periplasm and the phosphate group is then transferred to the GacA response regulator via a phospho-relay mechanism. Phosphorylated GacA triggers the expression of small RNA genes. The resulting small RNAs (rsmX, rsmY, rsmZ) specifically bind to post-transcriptional repressor proteins (RsmA, RsmE) thereby relieving the translation repression exerted by these proteins at the ribosomal binding site of mRNAs encoding genes (Sonnleitner and Haas, 2011) involved in the biosynthesis or regulation of bioactive molecules including LPs. In most LP-producing strains mutations in gacS or gacA lead to a complete loss of LP production (Koch et al., 2002; de Bruijn et al., 2007; de Bruijn et al., 2008; Song et al., 2014; Olorunleke et al., 2017). Likewise, a rsmXYZ mutant in P. protegens CHA0 is no longer able to produce the CLP orfamide (Sobrero et al., 2017). Mutations in both rsmY and rsmZ resulted in loss of massetolide production in P. lactis SS101, while a double mutation in rsmE and rsmA restored massetolide production in a gacS mutant. In this strain, the most likely target of the RsmE and RsmA repressor proteins is the LuxR-type regulator MassAR (Song et al., 2015b).
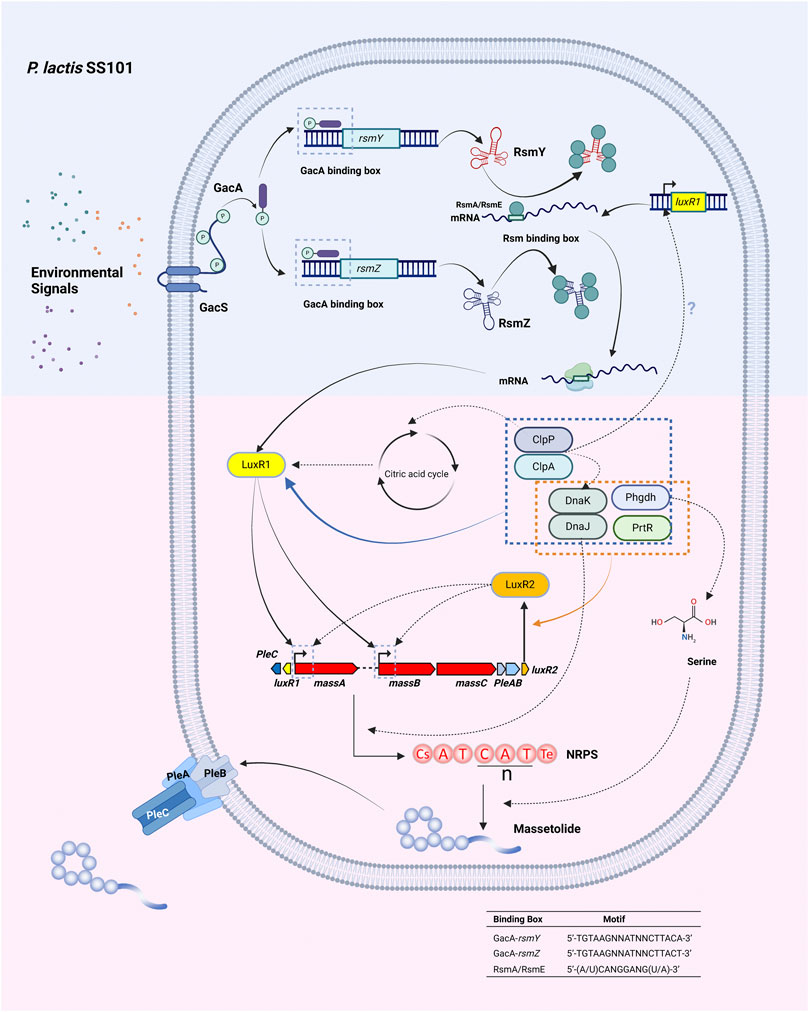
FIGURE 5. Regulation of lipopeptide production in the massetolide mono-producer P. lactis SS101. Massetolide production is under the control of the Gac/Rsm signal transduction pathway. The sensor kinase GacS is triggered by unknown environmental signals in the periplasm resulting in autophosphorylation. The phosphate group is transferred to the GacA response regulator via a phospho-relay system. Phosphorylated GacA binds to the GacA box in the promoter region of the rsmY and rsmZ genes encoding small RNAs. The resulting small RNAs RsmY and RsmZ bind to the repressor proteins RsmA and RsmE, relieving translational repression at the ribosomal binding site of the mRNAs of the luxR1 (massAR) gene. LuxR1 (MassAR) and LuxR2 (MassBCR) activate transcription of the massetolide biosynthetic gene cluster. The serine protease ClpP and its chaperone ClpA additionally regulate massetolide production via LuxR1, the heat shock proteins DnaK and DnaJ, and proteins involved in the citric acid cycle. DnaK and DnaJ may be required for proper folding of LuxR proteins or for assembly of the NRPS complex. Phgdh: D-3 phosphoglycerate dehydrogenase involved in the biosynthesis of the amino acid L-serine. Serine makes up two of the nine amino acids in massetolide. PrtR: antisigma factor which interacts with extra-cytoplasmic function sigma factors and affects the transcription of both luxR1 and luxR2 by an unknown mechanism. Secretion of massetolide occurs by the ABC transporter PleABC. Motifs: GacA-binding box see (Humair et al., 2010), Rsm box, see (Olorunleke et al., 2017). Created with BioRender.com.
Targeting global regulators is an effective strategy to activate silent gene clusters as demonstrated in several Streptomyces studies. In P. fluorescens Pf0-1, a silent gene cluster encoding a novel CLP (gacamide) was identified by genome mining and subsequently activated by repairing a defective gacA through complementation (Jahanshah et al., 2019).
LP mono-producers are found in the P. fluorescens, P. putida and P. syringae group and possess roles in surface motility, biofilm formation or break down, solubilization of nutrients, protection against competitors and predators, induction of systemic resistance in plants, and interactions with insects (D'Aes et al., 2010; Raaijmakers et al., 2010; Götze and Stallforth, 2020; Oni et al., 2022).
The organization of the BGCs encoding CLPs in mono-producers is very well conserved with usually three NRPS genes arranged in either one operon, or in a split configuration in which the first NRPS gene is located elsewhere in the genome (Cesa-Luna et al., 2023) (Figure 6).
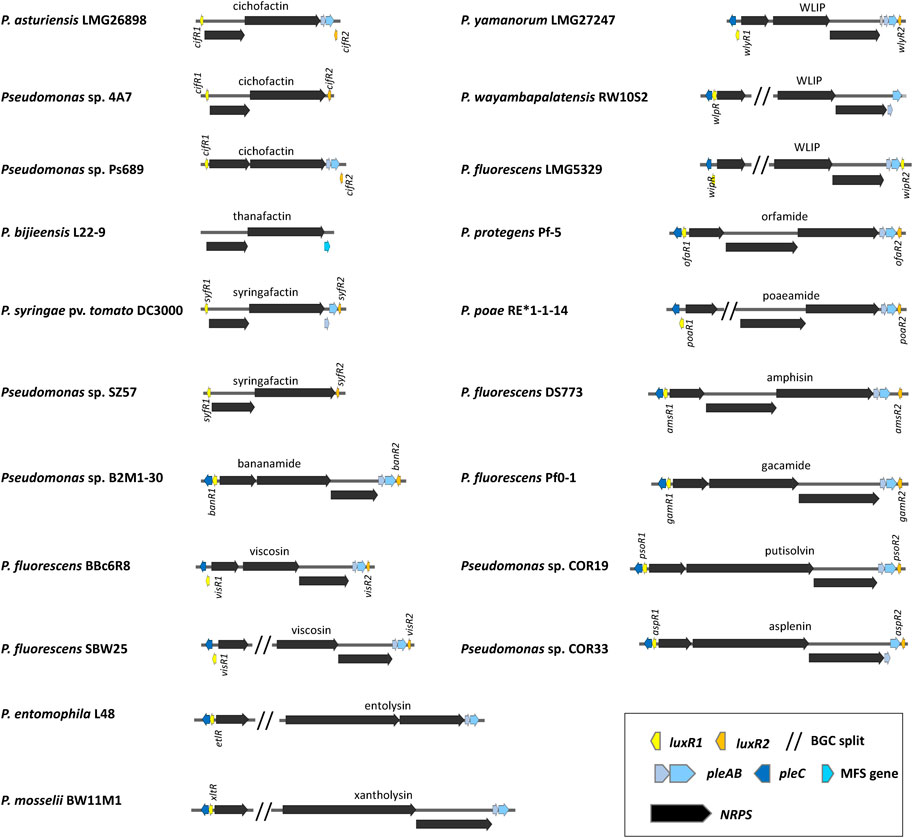
FIGURE 6. Organization of lipopeptide biosynthetic gene clusters in lipopeptide mono-producers. See Supplementary Table S1 for strain and sequence information.
The genomic regions flanking the NRPS gene clusters encoding CLPs typically contain well conserved regulatory and transporter genes. Pseudomonas CLPs are secreted by the PleABC tripartite efflux system, homologous to the MacAB-TolC ABC-type multidrug efflux pump that is found in many Gram-negative bacteria (Fitzpatrick et al., 2017). The PleABC machinery is composed of the inner membrane protein PleB (MacB), the periplasmic adapter PleA (MacA) and the outer membrane protein PleC (TolC) (Girard et al., 2022) (Figure 5). The pleC gene is usually located upstream of the first NRPS gene and preceded by a regulatory gene (luxR1) encoding a protein of the LuxR family. These LuxR proteins contain a typical DNA-binding helix-turn-helix (HTH) motif but lack an N-acylhomoserine lactone (N-AHL)-binding domain. The pleA and pleB genes are positioned downstream of the third NRPS gene and are often, but not always, followed by a luxR-type regulatory gene (luxR2) transcribed in the opposite direction (Figure 6). Recently is was shown that PleB is suitable as a diagnostic sequence for genome mining allowing the detection and/or typing of Pseudomonas LP producers (Girard et al., 2022).
Mutation of either luxR1 (viscAR) or luxR2 (viscBCR) results in loss of viscosin production in P. fluorescens strain SBW25 (de Bruijn and Raaijmakers, 2009). A viscAR mutation in this strain could be complemented with the luxR1 (massAR) regulatory gene of the massetolide producer P. lactis (fluorescens) SS101 (de Bruijn and Raaijmakers, 2009). Mutation of luxR1 (arfF) in Pseudomonas sp. MIS38 likewise leads to loss of arthrofactin production (Washio et al., 2010). Mutation of psoR1 leads to loss of putisolvin production in P. putida PCL1445, while mutation of pleA (macA) or pleB (macB) in this strain leads to reduced putisolvin production (Dubern et al., 2008). In P. protegens CHA0 it has been shown that translation of the LuxR-type regulatory genes orfR1 and orfR2 located up- and downstream of the orfamide BGC are under the direct control of the Gac/Rsm pathway (Sobrero et al., 2017).
LuxR2-type regulatory proteins are lacking in BGCs encoding xantholysin (Li et al., 2013) and entolysin (Vallet-Gely et al., 2010), while the situation is variable for WLIP producers (Figure 6). A luxR2 gene is present downstream of the WLIP BGC in P. yamanorum LMG27247 (wlyR2), P. fluorescens LMG5329 (wipR2) and two WLIP producers from the P. putida group (P. xanthosomae COR54 (wlfR2) and P. fakonensis COW40 (wlfR2)), but absent in P. wayambapalatensis RW10S2 (Rokni-Zadeh et al., 2012) (Figure 6). The LuxR1 regulators XltR, EltR, WlpR and WipR are required for xantholysin, entolysin and WLIP production in the respective strains P. mosselii BW11M1, Pseudomonas entomophila L48, P. wayambapalatensis RW10S2, and P. fluorescens LMG5329 (Vallet-Gely et al., 2010; Rokni-Zadeh et al., 2012; Li et al., 2013; Rokni-Zadeh et al., 2013). Intriguingly, all LP BGCs that lack luxR2 are produced by strains that taxonomically belong to the P. putida group (Girard et al., 2021). The reason for this is unknown but could symbolize a different ecological role.
Mono-producers of the LLPs syringafactin and cichofactin are described in various phylogroups of the P. syringae group (Bricout et al., 2022; Cesa-Luna et al., 2023) and in some isolates of the P. putida group (Cesa-Luna et al., 2023). LLPs are encoded by two NRPS genes arranged in an operon. Homologues of luxR1 and luxR2 are present upstream and downstream of the BGC encoding syringafactin in P. syringae DC3000, and cichofactin in P. putida 4A7, but the pleC transporter gene is absent in P. syringae DC3000 (Berti et al., 2007) and no transporter genes are present in the BGC encoding cichofactin in P. putida 4A7 (Figure 6). Mutation of luxR1 (also called syfR), but not of luxR2 (pspto2833) leads to loss of syringafactin production in P. syringae DC3000 (Berti et al., 2007). P. bijieensis L22-9 is a thanafactin mono-producer. The thanafactin BGC lacks regulatory genes, and a major facilitator superfamily (MFS) transporter is located downstream of the BGC (Figure 6).
Phylogenetic analysis shows that LuxR1 and LuxR2 proteins from CLP and LLP mono-producers form two distinct phylogenetic groups (Figure 7).
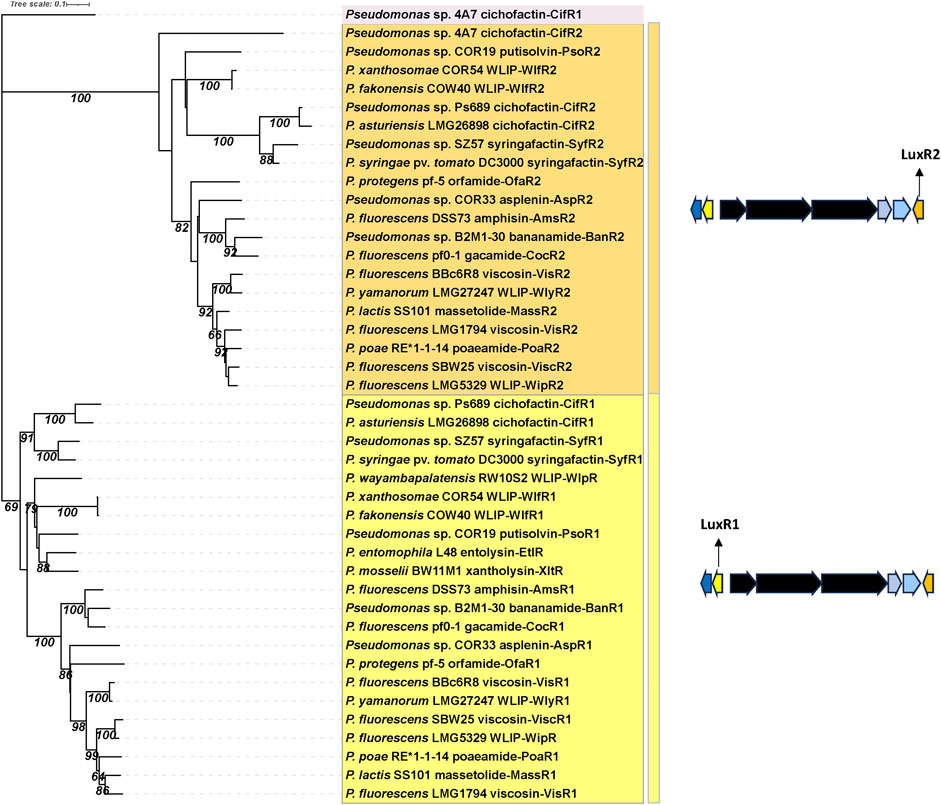
FIGURE 7. Phylogenetic tree of LuxR-type regulatory proteins in lipopeptide mono-producers. Neighbor-Joining phylogenetic tree (JTT model) constructed with MEGA 11 with MUSCLE alignment of LuxR amino acid sequences from LP mono-producers. Bootstrap values (percentage of 1,000 replicates) are shown in the figure. LuxR1 protein encoding genes (yellow) are located upstream of the lipopeptide NRPS genes, LuxR2 protein encoding genes (orange) are located downstream of the lipopeptide NRPS genes. See Supplementary Table S1 for strain and sequence information.
Regulation of LP production in mono-producers in response to cell density by autoinducers, also called quorum sensing regulation (Venturi, 2006), has to our knowledge only been described for the viscosin producer P. fluorescens 5064 (Cui et al., 2005) and in the putisolvin producer P. putida PCL1445. P. fluorescens 5064, an opportunistic soft rot pathogen of broccoli, produces the 9:7 CLP viscosin that is important for plant surface colonization. The N-acyl homoserine lactone (HSL) quorum sensing signal 3-OH-C8-HSL regulates viscosin production in this strain (Cui et al., 2005). P. putida PCL1445 was isolated from grass roots grown in soil polluted with polyaromatic hydrocarbons. The strain produces the 12:4 CLPs putisolvin I and II. These compounds inhibit biofilm formation and break down existing Pseudomonas biofilms (Kuiper et al., 2004). The ppuI-rsaL-ppuR quorum sensing system is involved in putisolvin production and mutants impaired in either ppuI or ppuR show a severe reduction in putisolvin production (Dubern et al., 2006). The quorum-sensing signals 3-oxo-C10-N-acyl homoserine lactone (3-oxo-C10-AHL) or 3-oxo-C12-AHL induce expression of the biosynthesis genes activating production of putisolvin I and II in this strain (Dubern et al., 2006).
Additional compounds involved in LP regulation in mono-producers include heat shock proteins, Clp proteases and enzymes involved in amino acid metabolism (Figure 5). The Hsp70 class heat shock protein DnaK regulates putisolvin production together with DnaJ at low temperature in P. putida PCL1445 (Dubern et al., 2005). DnaK is also involved in the regulation of massetolide in P. lactis SS101 (Song et al., 2014). Mutation of the gene encoding the Hsp90 class heat shock protein HtpG leads to loss of arthrofactin synthesis, while arthrofactin biosynthesis genes are normally expressed suggesting a role in posttranscriptional processes (Washio et al., 2010). Heat shock proteins may be required for the proper folding of positive (LuxR-type?) transcription factors or for assembly of the NRPS complex (Song et al., 2014). In P. putida, DnaK is under the control of the Gacs/GacA regulatory system, but this may not be the case in P. lactis SS101. In P. lactis SS101 the serine protease ClpP and its chaperone ClpA are required for massetolide biosynthesis. ClpP is an ATP-dependent serine protease that associates with different ATPases, including ClpA. ClpA selects target proteins for degradation by ClpP. Transcriptomic and proteomic analyses suggest that the ClpAP complex regulates massetolide biosynthesis via the LuxR1 transcriptional regulator MassAR, the heat shock proteins DnaK and DnaJ and via proteins involved in the citric acid cycle (Song et al., 2015a) (Figure 5). Additional regulators identified in P. lactis SS101 include D-3-phosphoglycerate dehydrogenase (Phgdh) and the antisigma factor PrtR (Song et al., 2014). Phgdh is involved in the biosynthesis of L-serine, an amino acid that makes up two of the nine amino acids in massetolide. PrtR interacts with extra-cytoplasmic function sigma factors of the sigma 70 family and regulates the expression of luxR1 and luxR2 by an unknown mechanism (Figure 5).
Concerning environmental signals that regulate LP production in mono-producers Mazzola et al. (2009) showed that the massetolide and viscosin biosynthesis genes, massABC and viscABC respectively, in P. lactis SS101 and P. fluorescens SBW25 are upregulated upon protozoal exposure conferring protection to each strain against predation. Interestingly, the authors observed that physical contact between prey and predator was not necessary to activate the massABC and viscABC genes. It would be interesting to test extracts from different protists in order to identify specific protist signals that trigger CLP production.
So, for LP mono-producers, quorum sensing regulation is not common, the LuxR1 regulator is always present (with the notable exception of the thanafactin producer P. bijieensis L22-9) and essential for LP production, while LuxR2 is sometimes lacking and when present, not always essential for LP production. LuxR1 and in some cases also LuxR2 are under translational control of the Gac/Rsm regulon. LP production is additionally regulated by heat shock proteins and the ClpAP complex.
LP poly-producers belonging to the P. syringae group, or P. corrugata and P. mandelii subgroup within the P. fluorescens group, typically co-produce CLPs from the Peptin and Mycin family and often produce an additional LLP of the Factin family, while poly-producers of the P. asplenii subgroup produce an additional CLP of the Asplenin family (Girard et al., 2020). Strains that produce a CLP of the Tolaasin family belonging to the P. fluorescens subgroup co-produce a CLP of the Viscosin family, while some members of the P. protegens group co-produce sessilin (a member of the tolaasin family) and orfamide (Cesa-Luna et al., 2023). Regulation of LP production in poly-producers is more complex than in mono-producers and can be quorum sensing dependent or independent.
Production of Mycins and Peptins is independent from QS in various plant pathogenic Pseudomonas bacteria including the closely related bean pathogens P. syringae pv. syringae B301D and B728a, the wide host range pathogens P. cichorii JBC1 and SF1-54, and the rice pathogen P. fuscovaginae UPB0736. These plant pathogenic Pseudomonas strains produce two CLPs simultaneously, one representative from the Peptin family, and one from the Mycin family. These CLPs always seem to co-occur (Figure 3). They function as phytotoxins, are usually co-produced and their secretion involves the same transporters. They can act synergistically to cause disease on plants by forming pores in plant membranes (Bender et al., 1999). Plant pathogenic CLP-producing Pseudomonas strains usually attack above-ground plant parts such as leaves or leaf sheaths. The third LLP produced by plant pathogens such as P. syringae pv. syringae, P. cichorii and P. fuscovaginae is not directly involved in virulence, but is essential for swarming motility and in planta colonization (Girard et al., 2020).
Most of our current knowledge on CLP regulation stems from the phytotoxins syringomycin and syringopeptin produced by P. syringae pv. syringae. Production of these compounds is dependent on the global regulatory GacS/GacA system in addition to the LuxR-type transcription factors SalA, SyrF, and SyrG which combined activate CLP synthesis in response to plant signal molecules. In P. syringae pv. syringae B301D and B728a, syrF, salA and syrG are associated with the syringomycin BGC (Figure 8) (Lu et al., 2002). The resulting proteins all carry a C-terminal HTH DNA binding motif typical for LuxR regulatory genes, but lack an N-terminal AHL domain and belong to the fourth subfamily of the LuxR superfamily (Wang et al., 2006a; Vaughn and Gross, 2016). SalA, SyrF and SyrG were shown to control syringopeptin and syringomycin biosynthesis in a hierarchical organization (Lu et al., 2005; Vaughn and Gross, 2016). The salA gene is positively regulated by the GacS/GacA regulon. SalA positively regulates its own expression (Kitten et al., 1998) but also the expression of both syrG and syrF (Lu et al., 2002; Wang et al., 2006a), while SyrG functions as an transcriptional activator of syrF (Vaughn and Gross, 2016). SyrF activates the syringomycin and syringopeptin biosynthesis and transporter genes by binding to a specific syr-syp box in the promoter region as a dimer (Figure 8). Plants signals that trigger the production of syringomycin and syringopeptin include the phenolic glycoside arbutin and sugars that occur in large quantities in leaf tissues such as D-fructose. GacS, SalA and SyrF transduce the plant signals to activate the syringomycin and syringopeptin BGCs. Sensing of the plant signal molecules probably occurs via GacS (Wang et al., 2006b).
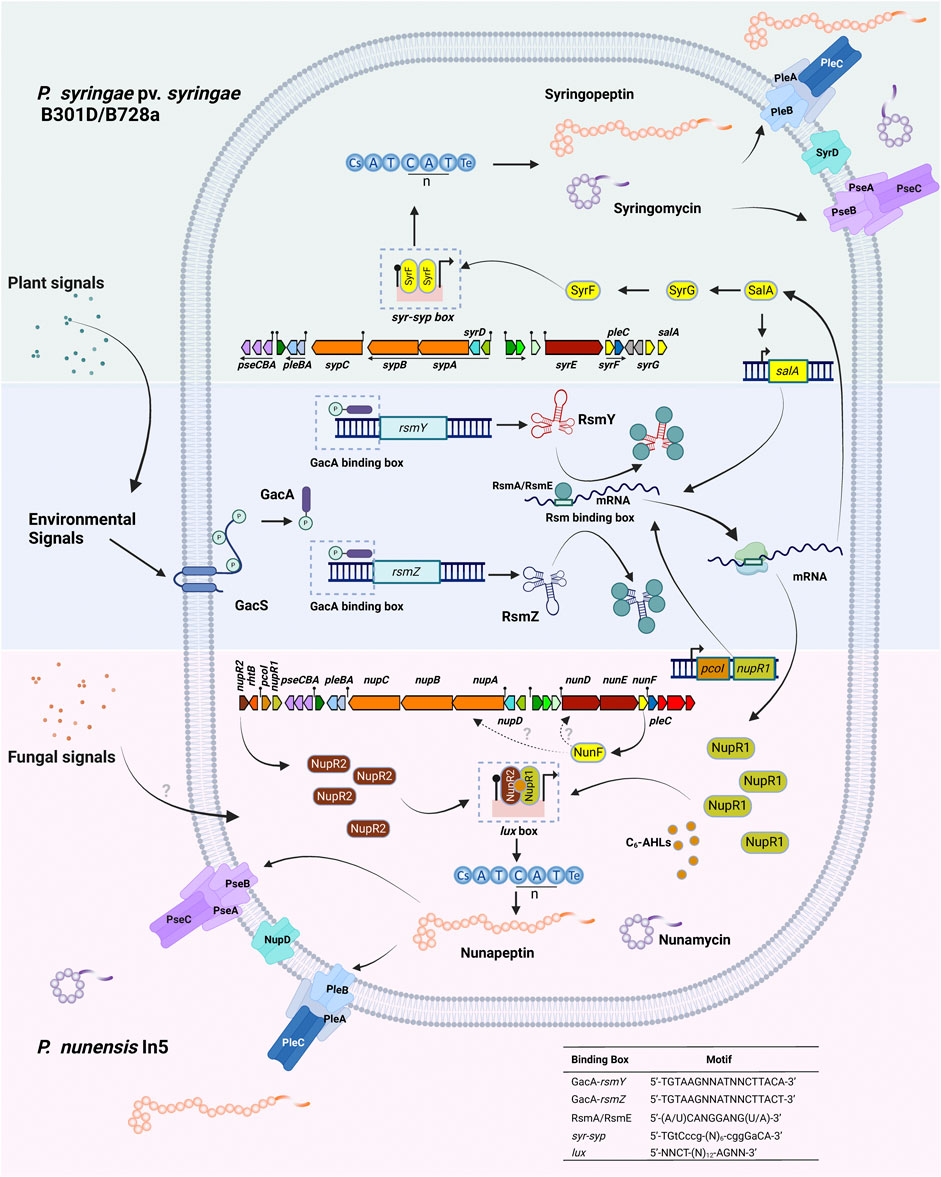
FIGURE 8. Regulation of lipopeptide production in Mycin and Peptin producers. Top: Non-quorum sensing regulation of lipopeptide production in the syringomycin and syringopeptin producers P. syringae pv. syringae B301D and B728a. Syringomycin and Syringopeptin production is under the control of the SalA regulon. SalA is activated by the Gac/Rsm signal transduction pathway in response to plant signals. SalA positively regulates its own transcription and activates the expression of both syrG and syrF. SyrG acts as a transcriptional activator of syrF. SyrF binds as a homodimer to a specific syr-syp box (indicated with a pin) in the promoter region of syringomycin and syringopeptin biosynthesis genes and transporters. Secretion of syringomycin and syringopeptin occurs by the ABC transporter PleABC, the cytoplasmic membrane protein SyrD and the RND transporter PseABC. Operons in the biosynthetic gene cluster are underlined. Bottom: Proposed model of quorum-sensing regulation of lipopeptide production in the nunamycin and nunapeptin producer P. nunensis In5. PcoI is an acyl-homoserine lactone (AHL) synthase encoding the autoinducer N-hexanoyl-L-homoserine lactone (C6-AHL), NupR1 is a LuxR family protein lacking an N-AHL binding domain under the control of the Gac/Rsm regulon, NupR2 is a LuxR protein with an N-AHL binding domain. The promoter region of nunamycin and nunapeptin biosynthesis genes and transporters harbor a specific lux box (indicated with a pin) to which a NupR1-NupR2-C6-AHL complex may bind (not experimentally proven). Production of nunamycin and nunapeptin is triggered by fungal signals that activate nunF. How NunF further activates lipopeptide synthesis and secretion is unknown. Nunamycin and nunapeptin secretion probably occurs by the ABC transporter PleABC, the cytoplasmic membrane protein NupD and the RND transporter PseABC. Motifs: GacA-binding box, see (Humair et al., 2010); Rsm-binding box, see (Olorunleke et al., 2017); syr-syp box, see (Wang et al., 2006a); lux-box, see (Whiteley and Greenberg, 2001). Created with BioRender.com.
P. syringae pv. syringae strains produce a third 8:0 LLP called syringafactin that is needed for swarming motility and enhances fitness on leaf surfaces by attracting moisture and facilitating access to nutrients (Burch et al., 2014). Two luxR1 and luxR2 type regulatory genes named syfR1 and syfR2 are situated up and downstream of the syringafactin BGC in P. syringae pv. syringae strains (Figure 3). Expression of both syfR1 and the surfactin biosynthesis gene syfA are dependent on SalA in P. syringae pv. syringae B728a (Hockett et al., 2013). No pleAB-type transporter genes are associated with the syringafactin BGC in P. syringae pv. syringae B301D (Figure 3) and B728a in contrast to the syringafactin BGC in the mono-producer P. syringae DC3000 (Figure 6).
The rice sheath brown rot pathogen P. fuscovaginae UPB0736 belongs to the P. asplenii subgroup and produces the 19:5 CLP fuscopeptin, the 9:9 CLP syringotoxin and the 13:8 CLP asplenin. Syringotoxin and fuscopeptin act synergistically in inhibiting plant H+-ATPase activity in plant membranes (Batoko et al., 1998) and both CLPs are involved in causing sheath rot symptoms on rice. In addition, syringotoxin is also toxic to the rice sheath blight pathogen Rhizoctonia solani AG1-1A. Asplenin is needed for swarming motility (Ferrarini et al., 2022b). The syringotoxin and fuscopeptin BGCs in P. fuscovaginae UPB0736 are completely devoid of luxR type regulatory genes, while three luxR genes (termed luxR1, luxR2 and luxR3 by (Ferrarini et al., 2022b) and renamed here as asp3, asp4 and asp1) are situated upstream of the asplenin BGC, and one luxR gene (aspR2) downstream of the last NRPS gene of this cluster (Figure 3). Phylogenetic analysis reveals that AspR3 and AspR1 cluster in the same clade as the LuxR1 regulators situated upstream of the BGCs in mono-producers, while AspR2 and AspR4 cluster with the LuxR2 regulators downstream of the BGCs in mono-producers (Figure 9). Intriguingly, aspR1 and aspR2 genes are flanking the asplenin BGC in the asplenin mono-producer Pseudomonas sp. COR33 (Figure 6), but this strain lacks the aspR3 and aspR4 genes. The function of the LuxR regulators in P. fuscovaginae is unknown but it is likely that they co-regulate all three CLPs.
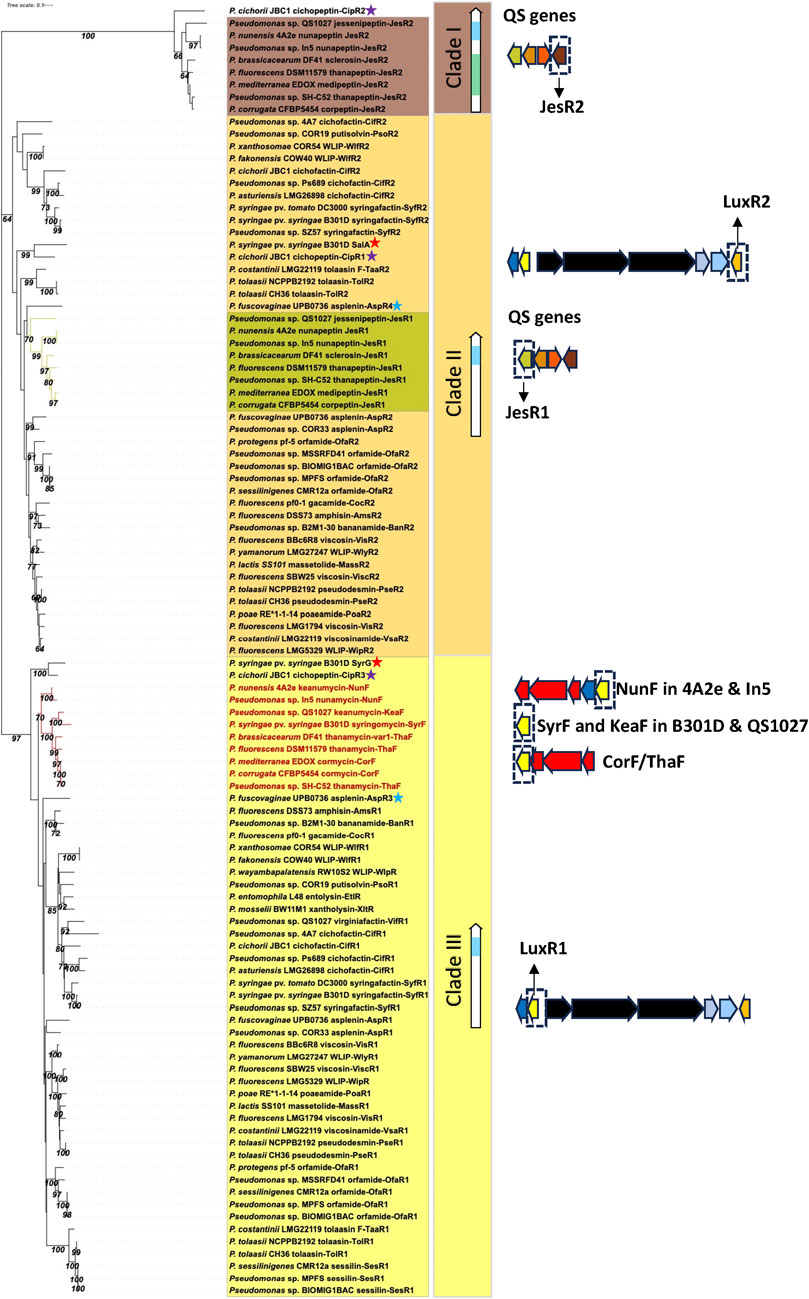
FIGURE 9. Phylogenetic tree of LuxR-type regulatory proteins in lipopeptide mono- and poly-producers. Neighbor-Joining phylogenetic tree (JTT model) constructed with MEGA 11 with MUSCLE alignment of LuxR amino acid sequences from mono- and poly-producers. Bootstrap values (percentage of 1,000 replicates) are shown in the figure. Clade I: JesR2-type regulatory proteins with a helix-turn-helix (HTH, indicated in blue) and an autoinducer binding motif (indicated in green) produced by LP poly-producers. This clade also contains the CipR2 protein from P. cichorii (purple star). Clade II: LuxR2-type regulatory proteins with a HTH motif (indicated in blue) found in LP mono-producers downstream of the LP BGCs. This clade also contains the JesR1 type regulators found in poly-producers with a quorum sensing system (indicated in green) and the SalA (red star), CipR1 (purple star) and AspR4 (blue star) regulatory proteins of P. syringae pv. syringae, P. cichorii and P. fuscovaginae, respectively. Clade III: LuxR1-type regulatory proteins with a HTH motif (in blue) located upstream of the LP BGCs in mono- and poly-producers. This clade also contains the SyrF homologues (indicated in red in the tree) located downstream of the Mycin or Brabantamide BGCs in LP poly-producers. See Supplementary Table S1 for strain and sequence information.
P. cichorii is a broad-host range pathogen that produces the 22:8 CLPs cichopeptin A and B (Huang et al., 2015), the 8:0 LLPs cichofactin A and B (Pauwelyn et al., 2013) and a third 8:8 Mycin-type CLP that is presumably pseudomycin (Girard et al., 2020). Cichopeptins are important virulence factors in the lettuce midrib rot pathogen P. cichorii SF1-54 and cause necrotic symptoms on leaves (Huang et al., 2015). Cichopeptins contain two residues of glycine in their peptide backbone and production is stimulated by glycine betaine. Cichopeptins are produced in planta at early stages of infection (Huang et al., 2015). Cichofactins are needed for swarming motility and a cichofactin-mutant formed significantly more biofilm. They are produced in planta and needed for in planta spread of P. cichorii but are not phytotoxic per se (Pauwelyn et al., 2013). In P. cichorii JBC1 and SF1-54 three luxR-type regulatory genes (cipR1, cipR2, cipR3) are located downstream of the cichopeptin BGC and two luxR-type regulatory genes (cifR1, cifR2) up and downstream of the cichofactin BGC (Figure 3). The cichofactin BGC contains pleAB transporter genes, but pleC is lacking. CipR1 associated with the cichopeptin BGC cluster is similar to SalA (about 60% identity) from P. syringae pv. syringae and clusters in clade II, while CipR3 is similar to SyrG (about 70% identity) and clusters in clade III in a phylogenetic tree in which all LuxR-type regulatory proteins associated with LP BGCs are included (Figure 9). Intriguingly, CipR2 carries an N-AHL binding domain (see further), but the genome of JBC1 does not encode LuxI type proteins involved in AHL synthesis. The role of these regulatory proteins in LP production in P. cichorii has to our knowledge not been studied.
Strains producing Mycin and Peptin variants that are quorum sensing regulated are found within the P. asplenii, P. mandelii and P. corrugata subgroups of the P. fluorescens group or complex (Girard et al., 2020). These strains produce a 19:5 or 22:5 CLP of the Peptin family, a second CLP of the Mycin family that is composed of two NRPSs, and often a third LLP of the Factin family. Many of these strains also harbor a brabantamide BGC downstream of the Mycin cluster. Brabantamides are cyclocarbamate antibiotics with activity against Gram-positive bacteria and Oomycetes (Van Der Voort et al., 2015). Brabantamide genes are co-transcribed with the Mycin and Peptin BGCs and possibly also co-secreted with Mycins and Peptins (Girard et al., 2020). The Mycin, brabantamide and Peptin BGCs are located on a so-called pathogenicity island, designed LPQ (lipopeptide/quorum sensing) island because conserved quorum sensing genes are located downstream of the peptin cluster in all these strains (Melnyk et al., 2019) (Figure 4).
P. nunensis In5 produces the 22:5 CLP nunapeptin and the 9:9 CLP nunamycin but is not known to produce a third LLP (Figure 4; Figure 8). Studies in P. nunensis In5 reported an inter-kingdom communication cascade that upon detection of fungal signals activates a Pseudomonas specific regulator called NunF required for expression of the antifungal CLPs nunamycin and nunapeptin. A nunF mutant is unable to produce nunamycin and nunapeptin (Hennessy et al., 2017a). The nunF promoter showed no induction with plant signal molecules, but was induced during co-culture with Fusarium graminearum, and by unknown components of a Fusarium-derived fungal extract in addition to pure fungal-associated molecules trehalose and glycerol (Hennessy et al., 2017b; Christiansen et al., 2020). NunF is a homologue of SyrF in the plant pathogen P. syringae pv. syringae. The syrF promoter however, showed a lower induction by fungal extract and hardly any induction by trehalose (Christiansen et al., 2020).
Most CLP poly-producers belonging to the P. corrugata, P. asplenii and P. mandelii subgroup carry a homologue of syrF/nunF downstream of the Mycin BGC or Mycin/brabantamide BGCs, followed by a pleC transporter gene (Figure 4) such as nunF associated with nunamycin in P. nunensis In5, and keanumycin in P. nunensis 4A2e and Pseudomonas sp. QS1027, corF associated with cormycin/brabantamide in P. corrugata CFBP5454 and P. mediterranea EDOX, and thaF (braD) associated with thanamycin/brabantamide in Pseudomonas sp. SH-C52. A syrF homologue is also present downstream of the uncharacterized Mycin in P. brassicacearum DF41 (Figure 4). Given that all these strains show strong antifungal activity, it is likely that their NunF/SyrF homologues also react to fungal signals, but this remains to be investigated. The NunF/SyrF-type regulatory proteins form a separate cluster in clade III (indicated in red in Figure 9), a clade that also contains all LuxR1-type regulators located upstream of LP BGCs in mono-producers.
Some strains also encode a third LLP that is either thanafactin or virginiafactin (Figure 4). The thanafactin BGCs lack luxR-type regulatory genes and all contain an MFS transporter downstream of the BGC that it typical for thanafactin producers. The virginiafactin BGC in Pseudomonas sp. QS1027 lacks transporters but carries a luxR1-type regulatory gene (vifR1) upstream of the first NRPS gene. The VifR1 protein clusters with LuxR1-type proteins located upstream of cichofactin and syringafactin in mono- and poly-producers in clade III (Figure 9).
In addition, all these strains harbour a four-gene quorum sensing system composed of jesR1 (rfiA, nupR1), jesI (pcoI, pdfI), rhtB (orf1) and jesR2 (pcoR, nupR2, pdfR) downstream of the Peptin BGC and preceded by an operon (pcoABC) encoding an RND transporter system (Figure 4). The PcoABC efflux system is homologous to the PseABC RND efflux system in P. syringae pv. syringae B301D involved in the secretion of syringomycin and syringopeptin (Kang and Gross, 2005). JesI (PcoI, PdfI) is an AHL synthase, JesR2 (PcoR, NupR2, pdfR) is a LuxR family protein with an N-terminus AHL-binding domain, while JesR1 (RfiA, NupR1) is a LuxR family protein with a helix-turn-helix motif but lacking an N-AHL binding domain. In P. corrugata CFBP 5454 mutants in pcoR, rfiA, or pcoI and rfiA are unable to produce and/or secrete cormycin and corpeptin (Licciardello et al., 2009). In Pseudomonas sp. QS1027 production of jessenipeptin is regulated by a QS system involving the AHL signal hexanoyl homoserine lactone (C6-AHL). Interestingly, the biosynthesis genes for jessenipeptin are located adjacent to those encoding another specialized metabolite mupirocin that works in synergy with the CLP against methicillin-resistant S. aureus (MRSA). However, the AHL signal required to induce production of the CLPs differ. Deletion of jesI, jesR1 or jesR2 led to a complete suppression of jessenipeptin production (Arp et al., 2018). Pflanze et al. (2023) have shown that the regulatory network governing production of CLPs in P. nunensis 4A2 required for protection against predation by amoeba and nematodes involves LuxR-type regulatory genes and the QS signal N-hexanoyl-L-homoserine lactone (C6-AHL). PcoI, nupR1 or nupR2 mutants in P. nunensis 4A2 no longer produce keanumycin or nunapeptin and the authors were able to demonstrate that complementation of knockout mutants with the signaling molecule C6-AHL restored CLP production (Pflanze et al., 2023). C6-AHL also regulates CLP production in other strains and has been detected in chemical extracts of P. nunensis In5 when nunamycin and nunapeptin are produced (Hennessy et al., 2017a). The situation is slightly different in P. brassicacearum DF41 where an AHL deficient strain expressing the AHL lactonase gene aiiA from Bacillus subtilis still produced sclerosin, but the rfiA (jesR1) mutant was strongly reduced in sclerosin production (Berry et al., 2014). In this strain pdfI and rfiA are co-transcribed and positively regulated by the Gac-Rsm network. Recently, it was shown that a QS system is also involved in the regulation of medpeptin by P. mediterranea S58 however the specific AHL signal required is unknown (Gu et al., 2023). A model showing how quorum sensing may regulate Mycin and Peptin production is depicted in Figure 8.
Phylogenetic analysis reveals that the JesR2 homologues in the various strains cluster together in a separate subgroup (clade I) that also contains the CipR2 protein with an N-AHL binding domain associated with the cichopeptin BGC in P. cichorii JBC1. JesR1 homologues form a distinct subgroup (indicated in green) within clade II harboring all LuxR2 proteins associated with LP mono-producers (Figure 9).
Tolaasin is the main virulence factor of the mushroom pathogens P. tolaasii and P. costantinii (Scherlach et al., 2013). P. tolaasii can occur in two reversible phenotypic variants, the pathogenic or smooth phenotype that is opaque, mucoid, non-fluorescent and produces tolaasin, and a non-pathogenic or rough variant that is translucent, non-mucoid, fluorescent and no longer produces tolaasin. Switching between the two phenotypes occurs by a reversible duplication of a 661 bp element in the 5’ end of a regulatory gene called pheN (Grewal et al., 1995) or rtpA (Murata et al., 1998), but that is actually the homologue of gacS (Heeb and Haas, 2001). The duplication introduces a frameshift mutation that results in the loss of part of the sensor domain of GacS (PheN) (Han et al., 1997). Compounds of the P. ostreatus fruiting body activate tolaasin production. Nonpathogenic variants occur at 22°C–30°C but not at 17°C and 20°C or in the presence of Pleurotus extracts (Murata et al., 1998).
P. tolaasii and P. costantinii also produce a second CLP of the viscosin family (pseudodesmin or viscosinamide), required to colonize the mushroom cap (Hermenau et al., 2020; Cesa-Luna et al., 2023). In the tolaasin/pseudodesmin producers P. tolaasii NCPBB 2192T and CH36, and the tolaasin/viscosinamide producer P. costantinii LMG22119 (Cesa-Luna et al., 2023) luxR1 and luxR2-type regulatory genes (tolR1/taaR1 and tolR1/taaR2) are present up and downstream of the tolaasin BGC next to the transporter genes pleC and pleAB. Similar regulators are also present in the split second BGCs encoding pseudodesmin and viscosinamide, respectively. Unlike pseudomodesmin mono-producers, however (Oni et al., 2020a), the pseudodesmin BGCs lack a pleC type transporter (Figure 10).
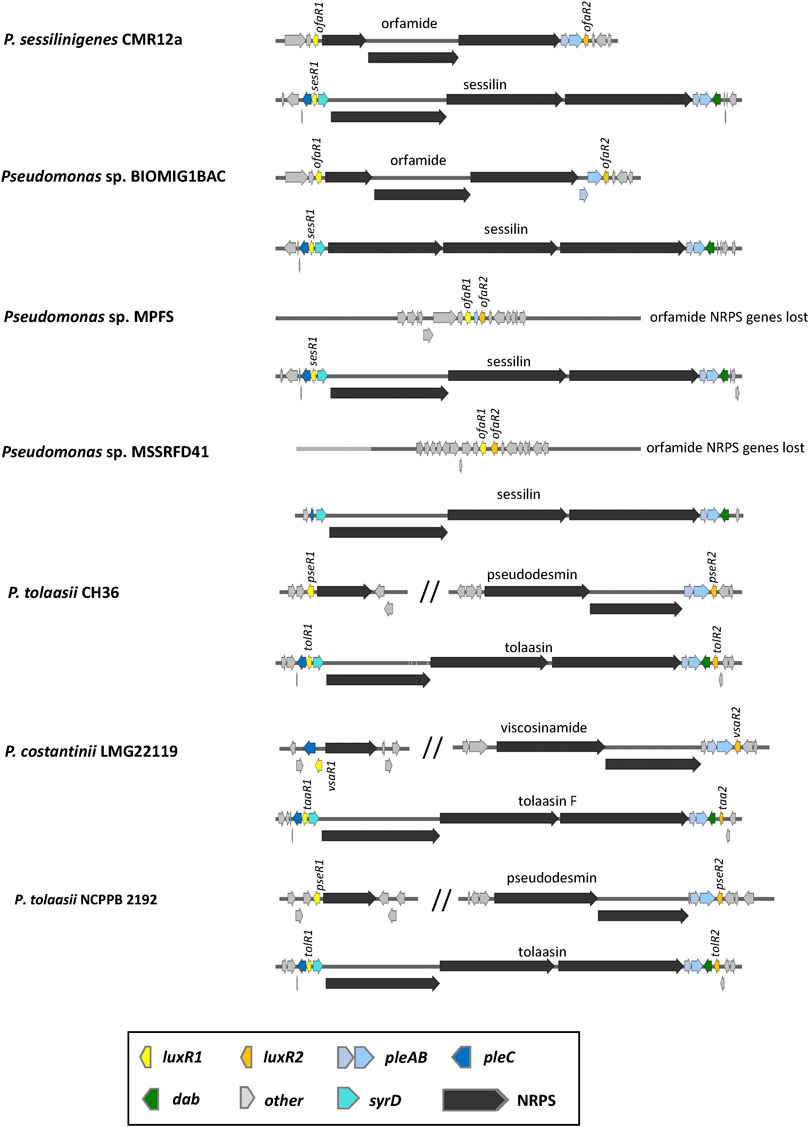
FIGURE 10. Organisation of lipopeptide biosynthetic gene clusters in tolaasin/sessilin producers. See Supplementary Table S1 for strain and sequence information.
A variant of tolaasin, called sessilin, is produced by the well-studied biocontrol strain P. sessilinigenes CMR12a, which also produces the 10:8 CLP orfamide (D’aes et al., 2014). Sessilin/orfamide co-producers have also been obtained from an urban wastewater treatment plant in Turkey (Pseudomonas sp. BIOMIG1BAC) (Altinbag et al., 2020) and from the rhizosphere of banana in Sri Lanka (Pseudomonas aestus BW16M1) (Cesa-Luna et al., 2023). The taxonomically closely related strains Pseudomonas sp. MPFS isolated from the skin of a treefrog in Brazil (Brunetti et al., 2022) and Pseudomonas sp. MSSRFD41 from the rhizosphere of finger millet in India (Sekar et al., 2018) only produce sessilin (Cesa-Luna et al., 2023) (Figure 10). The BGC for sessilin in P. sessilinigenes CMR12a is located on a genomic island acquired by horizontal gene transfer that also contains a phenazine BGC (Biessy et al., 2019).
In P. sessilinigenes CMR12a, typical luxR1 and luxR2-type regulatory genes (ofaR1 and ofaR2) are present up and downstream of the orfamide BGC (Figure 10), like in orfamide mono-producers such as P. protegens CHA0 and Pf-5 (Ma et al., 2016a). The orfamide BGC in CMR12a lacks a pleC transporter gene (Olorunleke et al., 2017), while this gene is present in orfamide mono-producers (Ma et al., 2016a). Only one LuxR-type regulatory gene (sesR1) is found upstream of the sessilin BGC next to a pleC transporter gene in strain CMR12a (Figure 10). Functional analysis of LuxR type regulators in CMR12a has revealed that ofaR1 and ofaR2 mutants are completely abolished in both orfamide and sessilin production, while a sesR1 mutant is still able to produce the two CLPs and has no clear phenotype. Rsm binding sites are located upstream of all three luxR-like genes suggesting regulation by the Gac/Rsm system (Olorunleke et al., 2017). In P. sessilinigenes, phenotypic switching by duplication of a fragment in the gacS gene is not known to happen. Intriguingly, however, spontaneous variants of P. sessilinigenes strains that have lost the genomic island with the sessilin and phenazine BGC occur both in the lab and on plants (Omoboye, 2019). The sessilin/orfamide co-producers Pseudomonas sp. BIOMIG1B and P. aestus BW16M1 have a very similar sessilin and orfamide BGC organization as CMR12a (Figure 10). Pseudomonas sp. MPFS and Pseudomonas sp. MSSRFD41 also carry a sessilin BGC, but strain MSSRFD41 has lost the luxR1-type regulatory gene sesR1 and part of the pleC transporter. Intriguingly, both strains have lost the orfamide BGC, which is considered part of the core genome of P. protegens but retained the ofaR1 and ofaR2 regulatory genes associated with the orfamide BGC in CMR12a and BIOMIG1B (Figure 10). Moreover, strain MPFS still has part of the pleB transporter again suggesting a gene loss.
Phylogenetic analysis reveals that OfaR1/VsaR1/PseR1 and SesR1/TolR1/TaaR1 cluster with other LuxR1 regulators located upstream of LP BGCs in clade III, while OfaR2/VsaR1/PseR2 and TolR2/TaaR2 clusters with the LuxR2 regulators located downstream of the LP BGCs in mono-producers in clade II (Figure 9).
It remains to be investigated whether the LuxR regulators associated with the pseudodesmin/viscosinamide BGC co-regulate tolaasin production in P. tolaasii and P. costantinii and whether the PleAB transporters downstream of the tolaasin and viscosinamide/pseudodesmin BGCs share the outer membrane protein PleCtol for CLP secretion.
As highlighted above, LPs are multifunctional molecules with boundless potential applications in research and industry. However, high production cost and low titers are major bottlenecks in their commercialization. Information on the specific conditions that favor LP production in Pseudomonas is scarce and the factors limiting their production largely unknown. Here, we describe LP production in natural versus controlled lab-scale environments and present strategies to modulate production in vitro and/or in situ.
Owing to the complexity of natural environments, a significant proportion of the Pseudomonas-LP research performed to date is only based on lab-based analyses. Production of several CLPs in the environment notably amphisin, tensin and viscosinamide has been detected, for example, in the sugar beet rhizosphere (Nielsen and Sorensen, 2003). Interestingly in this study no LPs were found in bulk soil signifying production is niche specific (Nielsen and Sorensen, 2003). Production of LPs by other isolates was also quantified on beet seeds and found in ranges of 0.22–0.65 µg CLP per seed. The P. fluorescens strain DSS73 (originally isolated from the sugar beet rhizosphere) was shown to produce amphisin on germinating sugar beet seeds in soil correlating with lab-based findings that unknown components of a sugar beet extract induce amsY expression needed for amphisin production (Koch et al., 2002). The environmental conditions required for amphisin production thus appear to reflect the producing strain’s specific habitat. A similar observation was made for P. syringae pv syringae where conditions required for lab-based production of syringomycin reflect the environmental conditions required for pathogenesis (Gross, 1985). As amphisin has antifungal properties it would be interesting to determine whether components of fungal extracts also induce amsY expression. Interestingly, the concentration of LPs detected over time remained similar in sterile soil whereas levels in non-sterile soil were rapidly reduced suggesting degradation by indigenous microbes (Nielsen and Sorensen, 2003).
While numerous studies have indicated that LPs are susceptible to degradation in the environment, the detailed mechanisms underpinning this process remain obscure. However, a key outcome of such research to date is the finding that microbial degradation of LPs can result in structural changes that alter their biological activities. For example, in Bacillus, degradation of surfactin generates a linear surfactin which can no longer induce systemic resistance (ISR) in tobacco (Rigolet et al., 2023). Linear surfactant also displays a reduced ability to lower surface tension when compared to cyclic surfactin (Liu et al., 2015). In the common button mushroom, protective helper bacteria disarm the causal agent of brown blotch P. tolaasii by enzymatic linearization of the toxin tolaasin and surfactant pseudodesmin, required to colonize the mushroom cap, to yield inactive linear forms of the CLPs (Hermenau et al., 2020). The linearization of CLPs is thought to be a resistance mechanism against competing bacterial species (Hermenau et al., 2020). More recently (Hansen et al., 2023), reported the cooperative degradation of orfamide A by Rhodococcus globerulus D757 and Stenotrophomonas indicatrix D763 to protect orfamide-sensitive members of a synthetic community during co-culture with the orfamide-producer P. protegens. It has been proposed that degradation of CLPs could be a strategy deployed by competitors to prevent CLP producers from performing “critical” functions in their environment such as biofilm formation, enhancing motility or colonization of specific niches (Rigolet et al., 2023). In another recent study (Zhang et al., 2021), present a new role of LPs in mediating bacterial cooperation to evade amoebal predation. Synthesis of syringafactins by Pseudomonas sp. SZ57 induces peptidase production in Paenibacillus sp. SZ31 resulting in partial LP-degradation yielding a mix of modified natural products that become amoebicidal (Zhang et al., 2021). These findings expand current knowledge on the ecological functions of LPs and demonstrate how interactions with other microbes can be exploited to unlock production of new compounds. Additionally, the identification of specific compounds capable of natural product modification could be exploited to select for the synthesis of LPs with specific functions during, for example, lab or large-scale cultivations (see Section 6.4).
Going forward it will be critical to quantify production of LPs in situ and correlate expression of LP-associated genes to LP production. Moreover, information on the stability of LPs in the environment, the mechanisms by which they are degraded and importantly the impact of degradation on LP structure and function is key for developing applications in environmental biotechnology and potentially opens a new avenues for natural product discovery.
LP production is influenced by growth phase in addition to abiotic factors (e.g., temperature, pH, oxygen) and nutritional factors (e.g., carbon, nitrogen and phosphorus sources, trace elements) (Raaijmakers et al., 2006). However, comparable studies are limited and the information available is scattered across a handful of strains (Nybroe and Sorensen, 2004). Optimization of growth conditions required for LP production is best described for syringomycin in P. syringae (Gross and DeVay, 1977; Gross and DeVay, 1977; Gross, 1985; Grgurina et al., 1996) showed higher levels of syringomycin production in still potato dextrose broth cultures (PDB) compared to aerated cultures. Viscosinamide is also produced in still cultures or under carbon, nitrogen or phosphorus starvation (Nybroe and Sorensen, 2004). Whereas syringomycin and syringopeptin are produced in the stationary phase, production of viscosinamide, tensin and amphisin is growth-coupled and occurs in the exponential phase (Nielsen et al., 1999; Nielsen et al., 2000; Koch et al., 2002). Syringomycin is regulated by iron (>2 µM), requires L-histidine as a nitrogen source and is repressed by inorganic phosphate (Gross, 1985). In plant tissues, levels of iron are high whereas concentrations of inorganic phosphate are not sufficient to inhibit the phytotoxin demonstrating that the environmental conditions required to support syringomycin production correlate with those necessary for pathogenesis (Gross, 1985).
While pH had no effect on syringomycin production, temperature was identified as an important factor with optimal production recorded at 24°C (Gross, 1985). In P. nunensis In5, regulation of nunamycin and nunapeptin is also temperature-dependent with optimal production at 15°C. Antifungal activity of In5 decreases with increasing temperature correlating with a reduction in production of both LPs (Michelsen and Stougaard, 2011; Christiansen et al., 2020). Production of putisolvin in P. putida PCL1445 is likewise temperature dependent with the highest production at 11 °C (Dubern et al., 2005). Also, syringafactin production in P. syringae pv. syringae is thermoregulated with much higher production at 20°C than at 30°C (Hockett et al., 2013). The molecular mechanisms underpinning the effects of temperature on CLP production are not well known. It has been suggested that temperature may directly impact synthetase formation and thereby alter iron uptake required for specialized metabolism (Gross, 1985). For P. putida PCL1445 it was shown that low temperature positively regulates putisolvin production during the late exponential phase via the DnaK stress response system (Dubern et al., 2005).
Screening for cultivation conditions inducing nunamycin and nunapeptin production revealed that nunapeptin is produced on a range of media both liquid and agar-based personal communication. In contrast, nunamycin production occurs on select agar-based media or in defined liquid minimal media supplemented with either glucose, glycerol and trehalose as carbon source (Christiansen et al., 2020). Glycerol also supports viscosin production by P. antarctica and has been used as a carbon source for rhamnolipid production (Zhao et al., 2021; Ciurko et al., 2023). Carbon source has been shown to influence tensin production in P. fluorescens, phytotoxin production in P. syringae and fungitoxin production in P. nunensis In5 (Nielsen et al., 2000; Woo et al., 2002; Christiansen et al., 2020). In contrast to nunapeptin, production of nunamycin appears to be tightly regulated occurring only under select conditions at low quantities. Nunamycin is a potent antimicrobial peptide and therefore potentially toxic to In5. Interestingly, production of syringomycin which displays a similar structure to nunamycin is not toxic to P. syringae at concentrations naturally produced (Gross, 1985). Gross (1985) observed that high syringomycin titers did not negatively impact bacterial growth as comparable cell densities were recorded for producers and non-producers. Finally, culture optimization has also been reported for thanamycin production in P. fluorescens SH-C52 resulting in a 3.3-fold product increase sufficient to recover 40 mg/120 L culture for NMR studies however the specific parameters altered were not specified (Johnston et al., 2015).
One major obstacle in natural product research is activating silent BGCs and/or improving the production of those expressed at low levels. For Pseudomonas LPs the challenge typically resides in low titers rather than no production. Several approaches can be used to activate or enhance BGC expression including cultivation-based approaches, molecular based techniques or synthetic biology strategies and combinatorial chemistry (Reen et al., 2015).
Cultivation-based approaches involve optimization of cultivation conditions, e.g., nutrients, carbon source, aeration, pH, temperature or using environmental cues, e.g., chemicals to induce compound production. While some information on optimal conditions needed for LP production exist, more systematic screening of culture conditions and in particular the identification of specific environmental signals that positively (or negatively) regulate expression of LP-associated genes is needed. In the well-studied and prolific producers of SMs actinomycetes, a number of key triggers have been identified to access antibiotic production including chemicals, microbial metabolites, interactions with microbes, environmental factors and enzymes that could serve as a useful starting point for studies in Pseudomonas (Seyedsayamdost, 2014; Zhu et al., 2014; Rosen and Seyedsayamdost, 2017; Zong et al., 2022). Information on the regulatory pathways and environmental signals influencing LPs can then be integrated into cultivation-based approaches and depending on the application, combined with other strategies, e.g., strain engineering to further enhance LP production.
Pseudomonas spp. are becoming increasingly attractive cell factories for the production of high-value chemicals including native and non-native SMs in part due to their capacity to utilize cheap carbon sources (Nikel et al., 2014; Wang et al., 2020). Using renewable resources, e.g., agricultural and food waste would greatly increase the commercialization potential of CLPs making bioprocessing of these compounds more sustainable (Ceresa et al., 2023). Importantly, Pseudomonas spp. are naturally competent and typically well-suited to genetics and molecular research. When selecting strains (particularly environmental isolates) for improved LP production it will be necessary to determine their genetic tractability and consider their origin, e.g., beneficial or pathogen. Select strains can then be engineered using traditional techniques, e.g., homologous recombination or next-generation CRISPR-Cas9 based technologies for increased production of target LPs either by introducing genes that promote LP production, deleting genes that inhibit LP biosynthesis, or overexpressing genes using native or synthetic promoters (Batianis et al., 2020). Depending on the complexity of the regulation, simple single-gene mutations to more complex multi-gene knockouts and/or insertions may be required.
In Pseudomonas LP-producers, genetic manipulation of regulatory genes has been limited to functional genomics studies and not used to improve LP titers. Interestingly, a study investigating the regulation of massetolide and viscosin demonstrated that the massA gene encoding the first NRPS needed for massetolide production can be heterologously expressed to complement a viscA mutant deficient in the first NRPS needed for viscosin production (de Bruijn and Raaijmakers, 2009). However, for the LuxR-type regulators only massAR (luxR1) and not massBCR (luxR2) could restore viscosin production in the viscAR (luxR1) mutant (de Bruijn and Raaijmakers, 2009) indicating that while some LP genes can be exchanged among different Pseudomonas strains, differences in the functionality of structural and regulatory genes may be at play.
Metabolic engineering can be used to increase LP production using strong indigenous or artificial promoters to increase the copy number of biosynthesis and/or regulator genes or alternatively using inducible promoters to overexpress entire gene clusters. For example, the use of promoter systems to improve LP production has been successful in the rhamnolipid producing strains P. aeruginosa, Burkholderia kururiensis and P. chlororaphis (Chong and Li, 2017). Overexpression of the rhlAB operon required for rhamnolipid biosynthesis under the tac promoter in B. kururiensis yielded a mixture of over 50 rhamnolipid congeners (Tavares et al., 2013). Similarly, overexpression of the rhamnolipid biosynthesis gene rhlC in P. chlororaphis resulted in the synthesis of di-rhamnolipids instead of mono-rhamnolipids (Solaiman et al., 2015). These findings demonstrate the potential of metabolic engineering-based approaches not only to increase product titer but equally to alter the structure and function of LPs.
Synthetic biology can serve to overcome challenges in engineering native strains for large scale production, for example, by refactoring BGCs to reduce the complexity of LP regulation (Temme et al., 2012; Wang et al., 2021). However, cloning and heterologous expression of large NRPS gene clusters that can span over 100 Kb (Meleshko et al., 2019) is difficult. Drawbacks include PCR amplification or synthesis of large DNA fragments, decrease in cloning efficiency, stability of vectors and/or successful integration onto the chromosome. Moreover, compared to Pseudomonas spp., classical hosts such as E. coli and yeast are not natural producers of LPs and may, depending on the LP encounter toxicity issues. Furthermore, with the rapid development of advanced gene engineering techniques, e. g., CRISPRi toolbox precise editing of model and natural Pseudomonas genomes for the controllable manipulation of gene expression will greatly facilitate strain engineering strategies for improved production in native hosts or other Pseudomonas strains (Batianis et al., 2020; Wirth et al., 2020).
For pharmaceutical or health-related applications where high purity grade compounds are needed it may be more desirable to synthesize LPs in vitro. While total chemical synthesis of the Pseudomonas CLPs from the Viscosin, Bananamide and Entolysin family has been achieved, the synthesis of larger LP molecules, for example, found in the poly-producers is more challenging (De Vleeschouwer et al., 2014; De Vleeschouwer et al., 2016; De Roo et al., 2022; Ji et al., 2023; Muangkaew et al., 2023). Chemical synthesis is also used to increase the chemical diversity of molecules, for structure-function studies, and to elucidate the stereochemistry of CLPs (Steigenberger et al., 2021; De Roo et al., 2022; Muangkaew et al., 2023).
Limited studies on optimization of the conditions necessary to induce synthesis and improve production of LPs exist. Consequently, no Pseudomonas-derived LPs are currently commercially available with the exception of rhamnolipids used in broad-spectrum applications and produced at industrially viable yields (Soberón-Chávez et al., 2021).
Bioreactor-scale production of LPs has only been reported for pseudofactins (PFs), 8:6 CLPs from the Bananamide family in P. fluorescens BD5 (Biniarz et al., 2018; Biniarz et al., 2020). Optimal production of PFs requires high glycerol (80 g/L) and tryptone (15 g/L) concentrations with high culture aeration (30 L/min) to achieve 7.2 g/30 L yield of PFs (Biniarz et al., 2018). LPs are often produced as a mixture of LPs comprising variants with minor structural changes that can greatly impact their bioactivity (Dufour et al., 2005; Eeman et al., 2006; Biniarz et al., 2020). Biniarz et al. (2018) observed that media supplementation with valine and leucine causes a shift in the ratio of PFs produced and can be used to select structural LP variants. The selective production of LPs will greatly benefit the purification of LPs for structure-function studies and potentially provide opportunities to identify new LP variants with novel biological activities.
Going forward it will be important to determine the limiting factors of LP production across scales (lab-bioreactor-technical scale). Additional parameters influencing growth and metabolic activity to optimize for include temperature, pH, oxygen, agitation, speed or vessel type (Guez et al., 2021). More basic studies on media and culture conditions required for LP production are needed to (i) routinely produce LPs of interest at lab scale for biological and chemical characterization, and (ii) to scale production from lab-to-bioreactor to develop an efficient bioprocess for LP production. Moreover (Gross and DeVay, 1977), observed that syringomycin production varied considerably across different P. syringae strains. Thus, it will be important to determine the specific impact of growth phase, carbon source, nutrients and inducer molecules on LP production in individual strains and to tailor culture conditions accordingly.
LPs are clearly attractive molecules with enormous potential for versatile and eco-friendly applications within biotechnology. However, low titers coupled with high production costs continue to constrain their commercial development.
Targeting key regulators of LP pathways to improve production in Pseudomonas is a promising yet underexplored avenue. To achieve this, future Pseudomonas-LP research efforts should focus on (i) understanding the regulatory mechanisms controlling LP production and integrate knowledge on the influence of environmental signals; (ii) strain engineering for improved production and (iii) media optimization and fermentation conditions for scalable manufacturing.
Moreover, it is becoming increasingly evident that LPs are pivotal to the ecological fitness of Pseudomonads in the environment. Additional information on why and when LPs are produced in natural environments to facilitate versatile Pseudomonas lifestyles in diverse habitats will provide new insights into the ecological functions of these molecules and potentially open new avenues for natural product discovery.
Ultimately, expanding our understanding of the regulatory mechanisms and environmental signals influencing the biosynthesis, structure and function of LPs is key to developing and optimizing industrial-scale production and wide-spread use of these “green compounds” for the future.
LZ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Writing–original draft, Writing–review and editing. MH: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Writing–original draft, Writing–review and editing. RH: Conceptualization, Formal Analysis, Funding acquisition, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Novo Nordisk Foundation grant NNF22OC0074616—Engineering small molecule-mediated regulation in bacteria to boost synthesis and production of bioactive compounds (BoostR) awarded to RH. CLP research in the lab of MH was supported by the concerted research action grant MEMCLiP from Ghent University (GOA 028-19) and the Excellence of Science Grant RhizoCLiP (EOS ID 30650620) from the Research Foundation Flanders (FWO). LZ is supported by a postdoctoral grant from the Research Foundation—Flanders (FWO ID 12AM524N).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2024.1363183/full#supplementary-material
Abdel-Mawgoud, A. M., Lépine, F., and Déziel, E. (2010). Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86 (5), 1323–1336. doi:10.1007/s00253-010-2498-2
Altinbag, R. C., Ertekin, E., and Tezel, U. (2020). Complete genome sequence of Pseudomonas sp. strain BIOMIG1(BAC), which mineralizes benzalkonium chloride disinfectants. Microbiol. Resour. Announc. 9 (20), e00309–e00320. doi:10.1128/MRA.00309-20
Andersen, J. B. (2003). Surface motility in Pseudomonas sp. DSS73 is required for efficient biological containment of the root-pathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiology 149 (1), 37–46. doi:10.1099/mic.0.25859-0
Andolfi, A., Cimmino, A., Cantore, P. L., Iacobellis, N. S., and Evidente, A. (2008). Bioactive and structural metabolites of Pseudomonas and Burkholderia species causal agents of cultivated mushrooms diseases. Perspect. Med. Chem. 2, 1177391X0800200. doi:10.1177/1177391x0800200004
Arp, J., Gotze, S., Mukherji, R., Mattern, D. J., Garcia-Altares, M., Klapper, M., et al. (2018). Synergistic activity of cosecreted natural products from amoebae-associated bacteria. P Natl. Acad. Sci. U. S. A. 115 (15), 3758–3763. doi:10.1073/pnas.1721790115
Bahroun, A., Jousset, A., Mrabet, M., Mhamdi, R., and Mhadhbi, H. (2021). Protists modulate Fusarium root rot suppression by beneficial bacteria. Appl. Soil Ecol. 168, 104158. doi:10.1016/j.apsoil.2021.104158
Bak, F., Bonnichsen, L., Jørgensen, N. O., Nicolaisen, M. H., and Nybroe, O. (2015). The biosurfactant viscosin transiently stimulates n-hexadecane mineralization by a bacterial consortium. Appl. Microbiol. Biotechnol. 99 (3), 1475–1483. doi:10.1007/s00253-014-6054-3
Banat, I. M., Makkar, R. S., and Cameotra, S. S. (2000). Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 53 (5), 495–508. doi:10.1007/s002530051648
Bassarello, C., Lazzaroni, S., Bifulco, G., Lo Cantore, P., Iacobellis, N. S., Riccio, R., et al. (2004). Tolaasins A-E, five new lipodepsipeptides produced by Pseudomonas tolaasii. J. Nat. Prod. 67 (5), 811–816. doi:10.1021/np0303557
Batianis, C., Kozaeva, E., Damalas, S. G., Martín-Pascual, M., Volke, D. C., Nikel, P. I., et al. (2020). An expanded CRISPRi toolbox for tunable control of gene expression in Pseudomonas putida. Microb. Biotechnol. 13 (2), 368–385. doi:10.1111/1751-7915.13533
Batoko, H., d'Exaerde, A. D., Kinet, J. M., Bouharmont, J., Gage, R. A., Maraite, H., et al. (1998). Modulation of plant plasma membrane H+-ATPase by phytotoxic lipodepsipeptides produced by the plant pathogen Pseudomonas fuscovaginae. Biochimica Biophysica Acta-Biomembranes 1372 (2), 216–226. doi:10.1016/S0005-2736(98)00060-1
Bender, C. L., Alarcon-Chaidez, F., and Gross, D. C. (1999). Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63 (2), 266–292. doi:10.1128/MMBR.63.2.266-292.1999
Berge, O., Monteil, C. L., Bartoli, C., Chandeysson, C., Guilbaud, C., Sands, D. C., et al. (2014). A user's guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS ONE 9 (9), e105547. doi:10.1371/journal.pone.0105547
Berry, C. L., Brassinga, A. K. C., Donald, L. J., Fernando, W. G. D., Loewen, P. C., and de Kievit, T. R. (2012). Chemical and biological characterization of sclerosin, an antifungal lipopeptide. Can. J. Microbiol. 58 (8), 1027–1034. doi:10.1139/w2012-079
Berry, C. L., Nandi, M., Manuel, J., Brassinga, A. K. C., Fernando, W. G. D., Loewen, P. C., et al. (2014). Characterization of the Pseudomonas sp DF41 quorum sensing locus and its role in fungal antagonism. Biol. Control 69, 82–89. doi:10.1016/j.biocontrol.2013.11.005
Berti, A. D., Greve, N. J., Christensen, Q. H., and Thomas, M. G. (2007). Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 189 (17), 6312–6323. doi:10.1128/JB.00725-07
Biessy, A., Novinscak, A., Blom, J., Léger, G., Thomashow, L. S., Cazorla, F. M., et al. (2019). Diversity of phytobeneficial traits revealed by whole-genome analysis of worldwide-isolated phenazine-producing Pseudomonas spp. Environ. Microbiol. 21 (1), 437–455. doi:10.1111/1462-2920.14476
Biniarz, P., Coutte, F., Gancel, F., and Łukaszewicz, M. (2018). High-throughput optimization of medium components and culture conditions for the efficient production of a lipopeptide pseudofactin by Pseudomonas fluorescens BD5. Microb. Cell Factories 17 (1), 121. doi:10.1186/s12934-018-0968-x
Biniarz, P., Henkel, M., Hausmann, R., and Łukaszewicz, M. (2020). Development of a bioprocess for the production of cyclic lipopeptides pseudofactins with efficient purification from collected foam. Front. Bioeng. Biotechnol. 8, 565619. doi:10.3389/fbioe.2020.565619
Bonnichsen, L., Bygvraa Svenningsen, N., Rybtke, M., de Bruijn, I., Raaijmakers, J. M., Tolker-Nielsen, T., et al. (2015). Lipopeptide biosurfactant viscosin enhances dispersal of Pseudomonas fluorescens SBW25 biofilms. Microbiology 161 (12), 2289–2297. doi:10.1099/mic.0.000191
Bricout, A., Morris, C. E., Chandeysson, C., Duban, M., Boistel, C., Chataigne, G., et al. (2022). The diversity of lipopeptides in the Pseudomonas syringae complex parallels phylogeny and sheds light on structural diversification during evolutionary history. Microbiol. Spectr. 10 (6), e0145622. doi:10.1128/spectrum.01456-22
Brunetti, A. E., Bunk, B., Lyra, M. L., Fuzo, C. A., Marani, M. M., Spröer, C., et al. (2022). Molecular basis of a bacterial-amphibian symbiosis revealed by comparative genomics, modeling, and functional testing. ISME J. 16 (3), 788–800. doi:10.1038/s41396-021-01121-7
Bunster, L., Fokkema, N. J., and Schippers, B. (1989). Effect of surface-active Pseudomonas spp on leaf wettability. Appl. Environ. Microbiol. 55 (6), 1340–1345. doi:10.1128/Aem.55.6.1340-1345.1989
Burch, A. Y., Zeisler, V., Yokota, K., Schreiber, L., and Lindow, S. E. (2014). The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environ. Microbiol. 16 (7), 2086–2098. doi:10.1111/1462-2920.12437
Catara, V. (2007). Pseudomonas corrugata: plant pathogen and/or biological resource? pathogen profile. Mol. Plant Pathol. 8 (3), 233–244. doi:10.1111/j.1364-3703.2007.00391.x
Cautain, B., De Pedro, N., Schulz, C., Pascual, J., Sousa, T. D. S., Martin, J., et al. (2015). Identification of the lipodepsipeptide MDN-0066, a novel inhibitor of VHL/HIF pathway produced by a new Pseudomonas species. PLoS ONE 10 (5), e0125221. doi:10.1371/journal.pone.0125221
Ceresa, C., Fracchia, L., Sansotera, A. C., De Rienzo, M. A. D., and Banat, I. M. (2023). Harnessing the potential of biosurfactants for biomedical and pharmaceutical applications. Pharmaceutics 15 (8), 2156. doi:10.3390/pharmaceutics15082156
Cesa-Luna, C., Geudens, N., Girard, L., De Roo, V., Maklad, H. R., Martins, J. C., et al. (2023). Charting the lipopeptidome of nonpathogenic Pseudomonas. mSystems 8, e0098822. doi:10.1128/msystems.00988-22
Chong, H. Q., and Li, Q. X. (2017). Microbial production of rhamnolipids: opportunities, challenges and strategies. Microb. Cell Factories 16, 137. doi:10.1186/s12934-017-0753-2
Christiansen, L., Alanin, K. S., Phippen, C. B. W., Olsson, S., Stougaard, P., and Hennessy, R. C. (2020). Fungal-associated molecules induce key genes involved in the biosynthesis of the antifungal secondary metabolites nunamycin and nunapeptin in the biocontrol strain Pseudomonas fluorescens In5. Appl. Environ. Microbiol. 86, 012844. doi:10.1128/AEM.01284-20
Chwastek, G., Surma, M. A., Rizk, S., Grosser, D., Lavrynenko, O., Rucińska, M., et al. (2020). Principles of membrane adaptation revealed through environmentally induced bacterial lipidome remodeling. Cell Rep. 32 (12), 108165. doi:10.1016/j.celrep.2020.108165
Ciurko, D., Chebbi, A., Kruszelnicki, M., Czapor-Irzabek, H., Urbanek, A. K., Polowczyk, I., et al. (2023). Production and characterization of lipopeptide biosurfactant from a new strain of Pseudomonas Antarctica 28E using crude glycerol as a carbon source. RSC Adv. 13 (34), 24129–24139. doi:10.1039/D3RA03408A
Cui, X., Harling, R., Mutch, P., and Darling, D. (2005). Identification of -3-hydroxyoctanoyl-homoserine lactone production in Pseudomonas fluorescens 5064, pathogenic to broccoli, and controlling biosurfactant production by quorum sensing. Eur. J. Plant Pathology 111 (4), 297–308. doi:10.1007/s10658-004-4171-z
D'Aes, J., De Maeyer, K., Pauwelyn, E., and Höfte, M. (2010). Biosurfactants in plant-Pseudomonas interactions and their importance to biocontrol. Environ. Microbiol. Rep. 2 (3), 359–372. doi:10.1111/j.1758-2229.2009.00104.x
D'aes, J., Kieu, N. P., Léclère, V., Tokarski, C., Olorunleke, F. E., De Maeyer, K., et al. (2014). To settle or to move? The interplay between two classes of cyclic lipopeptides in the biocontrol strain Pseudomonas CMR12a. Environ. Microbiol. 16 (7), 2282–2300. doi:10.1111/1462-2920.12462
De Almeida, D. G., Soares Da Silva, R. C., Luna, J. M., Rufino, R. D., Santos, V. A., Banat, I. M., et al. (2016). Biosurfactants: promising molecules for petroleum biotechnology advances. Front. Microbiol. 7, 1718. doi:10.3389/fmicb.2016.01718
de Bruijn, I., de Kock, M. J., de Waard, P., van Beek, T. A., and Raaijmakers, J. M. (2008). Massetolide A biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 190 (8), 2777–2789. doi:10.1128/JB.01563-07
de Bruijn, I., de Kock, M. J., Yang, M., de Waard, P., van Beek, T. A., and Raaijmakers, J. M. (2007). Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 63 (2), 417–428. doi:10.1111/j.1365-2958.2006.05525.x
de Bruijn, I., and Raaijmakers, J. M. (2009). Diversity and functional analysis of LuxR-type transcriptional regulators of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens. Appl. Environ. Microbiol. 75 (14), 4753–4761. doi:10.1128/AEM.00575-09
de Cássia, F. S. S. R., Almeida, D. G., Rufino, R. D., Luna, J. M., Santos, V. A., and Sarubbo, L. A. (2014). Applications of biosurfactants in the petroleum industry and the remediation of oil spills. Int. J. Mol. Sci. 15 (7), 12523–12542. doi:10.3390/ijms150712523
De Roo, V., Verleysen, Y., Kovács, B., Vleeschouwer, M. D., Muangkaew, P., Girard, L., et al. (2022). An nuclear magnetic resonance fingerprint matching approach for the identification and structural Re-evaluation of Pseudomonas lipopeptides. Microbiol. Spectr. 10 (4), e0126122. doi:10.1128/spectrum.01261-22
De Vleeschouwer, M., Martins, J., and Madder, A. (2016). First total synthesis of WLIP: on the importance of correct protecting group choice. J. Peptide Sci. 22 (3), 149–155. doi:10.1002/psc.2852
De Vleeschouwer, M., Sinnaeve, D., den Begin, J., Coenye, T., Martins, J., and Madder, A. (2014). Rapid total synthesis of cyclic lipodepsipeptides as a premise to investigate their self-assembly and biological activity. Chem. - A Eur. J. 20 (25), 7766–7775. doi:10.1002/chem.201402066
Dubern, J. F., Coppoolse, E. R., Stiekema, W. J., and Bloemberg, G. V. (2008). Genetic and functional characterization of the gene cluster directing the biosynthesis of putisolvin I and II in Pseudomonas putida strain PCL1445. Microbiology 154, 2070–2083. doi:10.1099/mic.0.2008/016444-0
Dubern, J. F., Lagendijk, E. L., Lugtenberg, B. J. J., and Bloemberg, G. V. (2005). The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J. Bacteriol. 187 (17), 5967–5976. doi:10.1128/Jb.187.17.5967-5976.2005
Dubern, J. F., Lugtenberg, B. J., and Bloemberg, G. V. (2006). The ppuI-rsaL-ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J. Bacteriol. 188 (8), 2898–2906. doi:10.1128/JB.188.8.2898-2906.2006
Dufour, S., Deleu, M., Nott, K., Wathelet, B., Thonart, P., and Paquot, M. (2005). Hemolytic activity of new linear surfactin analogs in relation to their physico-chemical properties. Biochimica Biophysica Acta (BBA) - General Subj. 1726 (1), 87–95. doi:10.1016/j.bbagen.2005.06.015
Eeman, M., Berquand, A., Dufrêne, Y. F., Paquot, M., Dufour, S., and Deleu, M. (2006). Penetration of surfactin into phospholipid monolayers: nanoscale interfacial organization. Langmuir 22 (26), 11337–11345. doi:10.1021/la061969p
Ferrarini, E., Roo, V. D., Geudens, N., Martins, J. C., and Höfte, M. (2022a). Altering in vivo membrane sterol composition affects the activity of the cyclic lipopeptides tolaasin and sessilin against Pythium. Biochimica Biophysica Acta - Biomembr. 1864, 184008. doi:10.1016/j.bbamem.2022.184008
Ferrarini, E., Spacapan, M., Lam, V. B., McCann, A., Cesa-Luna, C., Marahatta, B., et al. (2022b). Versatile role of Pseudomonas fuscovaginae cyclic lipopeptides in plant and microbial interactions. Front. Plant Sci. 13, 1008980. doi:10.3389/fpls.2022.1008980
Fitzpatrick, A. W. P., Llabres, S., Neuberger, A., Blaza, J. N., Bai, X. C., Okada, U., et al. (2017). Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2, 17070. doi:10.1038/nmicrobiol.2017.70
Flury, P., Vesga, P., Péchy-Tarr, M., Aellen, N., Dennert, F., Hofer, N., et al. (2017). Antimicrobial and insecticidal: cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front. Microbiol. 8 (100), 100. doi:10.3389/fmicb.2017.00100
Fogliano, V., Ballio, A., Gallo, M., Woo, S., Scala, F., and Lorito, M. (2002). Pseudomonas lipodepsipeptides and fungal cell wall-degrading enzymes act synergistically in biological control. Mol. Plant-Microbe Interact. 15 (4), 323–333. doi:10.1094/Mpmi.2002.15.4.323
Gao, Z., Karlsson, I., Geisen, S., Kowalchuk, G., and Jousset, A. (2019). Protists: puppet masters of the rhizosphere microbiome. Trends Plant Sci. 24 (2), 165–176. doi:10.1016/j.tplants.2018.10.011
Geudens, N., and Martins, J. C. (2018). Cyclic lipodepsipeptides from Pseudomonas spp – biological Swiss-army knives. Front. Microbiol. 9, 1867. doi:10.3389/fmicb.2018.01867
Girard, L., Geudens, N., Pauwels, B., Höfte, M., Martins, J. C., and De Mot, R. (2022). Transporter gene-mediated typing for detection and genome mining of lipopeptide-producing Pseudomonas. Appl. Environ. Microbiol. 88 (2), e0186921. doi:10.1128/AEM.01869-21
Girard, L., Höfte, M., and De Mot, R. (2020). Lipopeptide families at the interface between pathogenic and beneficial Pseudomonas-plant interactions. Crit. Rev. Microbiol. 46 (4), 397–419. doi:10.1080/1040841X.2020.1794790
Girard, L., Lood, C., Höfte, M., Vandamme, P., Rokni-Zadeh, H., Van Noort, V., et al. (2021). The ever-expanding Pseudomonas genus: description of 43 new species and partition of the Pseudomonas putida group. Microorganisms 9, 1766. doi:10.3390/microorganisms9081766
Gislason, A. S., and de Kievit, T. R. (2020). Friend or foe? Exploring the fine line between Pseudomonas brassicacearum and phytopathogens. J. Med. Microbiol. 69 (3), 347–360. doi:10.1099/jmm.0.001145
Götze, S., and Stallforth, P. (2020). Structure, properties, and biological functions of nonribosomal lipopeptides from pseudomonads. Nat. Product. Rep. 37 (1), 29–54. doi:10.1039/c9np00022d
Götze, S., Vij, R., Burow, K., Thome, N., Urbat, L., Schlosser, N., et al. (2023). Ecological niche-inspired genome mining leads to the discovery of crop-protecting nonribosomal lipopeptides featuring a transient amino acid building block. J. Am. Chem. Soc. 145 (4), 2342–2353. doi:10.1021/jacs.2c11107
Gram, L. (2015). Silent clusters - speak up. Microb. Biotechnol. 8 (1), 13–14. doi:10.1111/1751-7915.12181
Grewal, S. I. S., Han, B., and Johnstone, K. (1995). Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of Brown blotch disease of Agaricus bisporus. J. Bacteriol. 177 (16), 4658–4668. doi:10.1128/jb.177.16.4658-4668.1995
Grgurina, I., Gross, D. C., Iacobellis, N. S., Lavermicocca, P., Takemoto, J. Y., and Benincasa, M. (1996). Phytotoxin production by Pseudomonas syringae pv. syringae: syringopeptin production by syr mutants defective in biosynthesis or secretion of syringomycin. FEMS Microbiol. Lett. 138 (1), 35–39. doi:10.1111/j.1574-6968.1996.tb08131.x
Gross, D. C. (1985). Regulation of syringomycin synthesis in Pseudomonas syringae pv. syringae and defined conditions for its production. J. Appl. Bacteriol. 58 (2), 167–174. doi:10.1111/j.1365-2672.1985.tb01444.x
Gross, D. C., and DeVay, J. E. (1977). Production and purification of syringomycin, a phytotoxin produced by Pseudomonas syringae. Physiol. Plant Pathol. 11 (1), 13–28. doi:10.1016/0048-4059(77)90083-2
Gross, H., Stockwell, V. O., Henkels, M. D., Nowak-Thompson, B., Loper, J. E., and Gerwick, W. H. (2007). The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 14 (1), 53–63. doi:10.1016/j.chembiol.2006.11.007
Gu, Y.-L., Li, J.-Z., Li, Y., Cong, S., Wang, J., Ma, Y.-N., et al. (2023). Pseudomonas cyclic lipopeptide medpeptin: biosynthesis and modulation of plant immunity. Engineering 28, 153–165. doi:10.1016/j.eng.2023.05.016
Guez, J. S., Vassaux, A., Larroche, C., Jacques, P., and Coutte, F. (2021). New continuous process for the production of lipopeptide biosurfactants in foam overflowing bioreactor. Front. Bioeng. Biotechnol. 9, 678469. doi:10.3389/fbioe.2021.678469
Guo, S., Xiong, W., Hang, X., Gao, Z., Jiao, Z., Liu, H., et al. (2021). Protists as main indicators and determinants of plant performance. Microbiome 9 (1), 64. doi:10.1186/s40168-021-01025-w
Gutiérrez-Chávez, C., Benaud, N., and Ferrari, B. C. (2021). The ecological roles of microbial lipopeptides: where are we going? Comput. Struct. Biotechnol. J. 19, 1400–1413. doi:10.1016/j.csbj.2021.02.017
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3 (4), 307–319. doi:10.1038/nrmicro1129
Han, B., Pain, A., and Johnstone, K. (1997). Spontaneous duplication of a 661 bp element within a two-component sensor regulator gene causes phenotypic switching in colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol. Microbiol. 25 (2), 211–218. doi:10.1046/j.1365-2958.1997.4411811.x
Hansen, M. L., Dénes, Z., Jarmusch, S. A., Wibowo, M., Lozano-Andrade, C. N., Kovács, Á. T., et al. (2023). Resistance towards and biotransformation of Pseudomonas-produced secondary metabolites during community invasion. BioRxiv. doi:10.1101/2023.06.20.545698
Hawxhurst, C. J., Micciulla, J. L., Bridges, C. M., Shor, M., Gage, D. J., and Shor, L. M. (2023). Soil protists can actively redistribute beneficial bacteria along Medicago truncatula roots. Appl. Environ. Microbiol. 89 (3), e0181922. doi:10.1128/aem.01819-22
Heeb, S., and Haas, D. (2001). Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant-Microbe Interact. 14 (12), 1351–1363. doi:10.1094/Mpmi.2001.14.12.1351
Hennessy, R. C., Phippen, C. B. W., Nielsen, K. F., Olsson, S., and Stougaard, P. (2017a). Biosynthesis of the antimicrobial cyclic lipopeptides nunamycin and nunapeptin by Pseudomonas fluorescens strain In5 is regulated by the LuxR-type transcriptional regulator NunF. Microbiologyopen 6 (6), e00516. doi:10.1002/mbo3.516
Hennessy, R. C., Stougaard, P., and Olsson, S. (2017b). A microplate reader-based system for visualizing transcriptional activity during in vivo microbial interactions in space and time. Sci. Rep. 7 (1), 281. doi:10.1038/s41598-017-00296-4
Hermenau, R., Kugel, S., Komor, A. J., and Hertweck, C. (2020). Helper bacteria halt and disarm mushroom pathogens by linearizing structurally diverse cyclolipopeptides. P Natl. Acad. Sci. U. S. A. 117 (38), 23802–23806. doi:10.1073/pnas.2006109117
Hildebrand, P. D., Braun, P. G., McRae, K. B., and Lu, X. (1998). Role of the biosurfactant viscosin in broccoli head rot caused by a pectolytic strain ofPseudomonas fluorescens. Can. J. Plant Pathology 20 (3), 296–303. doi:10.1080/07060669809500396
Hockett, K. L., Burch, A. Y., and Lindow, S. E. (2013). Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. Plos One 8 (3), e59850. doi:10.1371/journal.pone.0059850
Höfte, M. (2021). “The use of Pseudomonas spp. as bacterial biocontrol agents to control plant disease,” in Microbial bioprotectants for plant disease management. Editors J. Köhl,, and W. R. Burleigh Dodds (Cambridge: Burleigh Dodds Science Publishing), 301–374.
Hrabak, E. M., and Willis, D. K. (1992). The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 174 (9), 3011–3020. doi:10.1128/jb.174.9.3011-3020.1992
Hrabak, E. M., and Willis, D. K. (1993). Involvement of the IemA gene in production of syringomycin and protease by Pseudomonas syringae pv syringae. Mol. Plant-Microbe Interact. 6 (3), 368–375. doi:10.1094/Mpmi-6-368
Huang, C. J., Pauwelyn, E., Ongena, M., Debois, D., Leclère, V., Jacques, P., et al. (2015). Characterization of cichopeptins, new phytotoxic cyclic lipodepsipeptides produced by Pseudomonas cichorii SF1-54 and their role in bacterial midrib rot disease of lettuce. Mol. Plant-Microbe Interact. 28 (9), 1009–1022. doi:10.1094/MPMI-03-15-0061-R
Humair, B., Wackwitz, B., and Haas, D. (2010). GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 76 (5), 1497–1506. doi:10.1128/Aem.02014-09
Hutchison, M. L., and Johnstone, K. (1993). Evidence for the involvement of the surface-active properties of the extracellular toxin tolaasin in the manifestation of Brown blotch disease symptoms by Pseudomonas tolaasii on Agaricus bisporus. Physiological Mol. Plant Pathology 42 (5), 373–384. doi:10.1016/S0885-5765(05)80013-X
Jahanshah, G., Yan, Q., Gerhardt, H., Pataj, Z., Lämmerhofer, M., Pianet, I., et al. (2019). Discovery of the cyclic lipopeptide gacamide A by genome mining and repair of the defective GacA regulator in Pseudomonas fluorescens Pf0-1. J. Nat. Prod. 82 (2), 301–308. doi:10.1021/acs.jnatprod.8b00747
Janek, T., Krasowska, A., Radwańska, A., and Łukaszewicz, M. (2013). Lipopeptide biosurfactant pseudofactin II induced apoptosis of melanoma A 375 cells by specific interaction with the plasma membrane. PLoS ONE 8 (3), e57991–e57999. doi:10.1371/journal.pone.0057991
Janek, T., Łukaszewicz, M., Rezanka, T., and Krasowska, A. (2010). Isolation and characterization of two new lipopeptide biosurfactants produced by Pseudomonas fluorescens BD5 isolated from water from the Arctic Archipelago of Svalbard. Bioresour. Technol. 101 (15), 6118–6123. doi:10.1016/j.biortech.2010.02.109
Jang, J. Y., Yang, S. Y., Kim, Y. C., Lee, C. W., Park, M. S., Kim, J. C., et al. (2013). Identification of orfamide A as an insecticidal metabolite produced by Pseudomonas protegens F6. J. Agric. Food Chem. 61 (28), 6786–6791. doi:10.1021/jf401218w
Ji, X. J., Nielsen, A. L., and Heinis, C. (2023). Cyclic peptides for drug development. Angew. Chem. Int. Ed. 63, e202308251. doi:10.1002/anie.202308251
Johnston, C. W., Skinnider, M. A., Wyatt, M. A., Li, X., Ranieri, M. R. M., Yang, L., et al. (2015). An automated Genomes-to-Natural Products platform (GNP) for the discovery of modular natural products. Nat. Commun. 6, 8421. doi:10.1038/ncomms9421
Kang, H. J., and Gross, D. C. (2005). Characterization of a resistance-nodulation-cell division transporter system associated with the syr-syp genomic island of Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 71 (9), 5056–5065. doi:10.1128/Aem.71.9.5056-5065.2005
Kitten, T., Kinscherf, T. G., McEvoy, J. L., and Willis, D. K. (1998). A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 28 (5), 917–929. doi:10.1046/j.1365-2958.1998.00842.x
Koch, B., Nielsen, T. H., Sorensen, D., Andersen, J. B., Christophersen, C., Molin, S., et al. (2002). Lipopeptide production in Pseudomonas sp. strain DSS73 is regulated by components of sugar beet seed exudate via the Gac two-component regulatory system. Appl. Environ. Microbiol. 68 (9), 4509–4516. doi:10.1128/AEM.68.9.4509-4516.2002
Korbut, R., Skjolding, L. M., Mathiessen, H., Jaafar, R., Li, X., Jørgensen, L. V. G., et al. (2022). Toxicity of the antiparasitic lipopeptide biosurfactant SPH6 to green algae, cyanobacteria, crustaceans and zebrafish. Aquat. Toxicol. 243, 106072. doi:10.1016/j.aquatox.2021.106072
Kruijt, M., Tran, H., and Raaijmakers, J. M. (2009). Functional, genetic and chemical characterization of biosurfactants produced by plant growth-promotingPseudomonas putida267. J. Appl. Microbiol. 107 (2), 546–556. doi:10.1111/j.1365-2672.2009.04244.x
Kuiper, I., Lagendijk, E. L., Pickford, R., Derrick, J. P., Lamers, G. E. M., Thomas-Oates, J. E., et al. (2004). Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 51 (1), 97–113. doi:10.1046/j.1365-2958.2003.03751.x
Lavermicocca, P., Iacobellis, N. S., Simmaco, M., and Graniti, A. (1997). Biological properties and spectrum of activity of Pseudomonas syringae pv syringae toxins. Physiological Mol. Plant Pathology 50 (2), 129–140. doi:10.1006/pmpp.1996.0078
Leonov, P. S., Flores-Alsina, X., Gernaey, K. V., and Sternberg, C. (2021). Microbial biofilms in biorefinery - towards a sustainable production of low-value bulk chemicals and fuels. Biotechnol. Adv. 50, 107766. doi:10.1016/j.biotechadv.2021.107766
Li, W., Rokni-Zadeh, H., De Vleeschouwer, M., Ghequire, M. G. K. K., Sinnaeve, D., Xie, G.-L. L., et al. (2013). The antimicrobial compound xantholysin defines a new group of Pseudomonas cyclic lipopeptides. PLoS ONE 8 (5), e62946. doi:10.1371/journal.pone.0062946
Licciardello, G., Bertani, I., Steindler, L., Bella, P., Venturi, V., and Catara, V. (2009). The transcriptional activator rfiA is quorum-sensing regulated by cotranscription with the luxI homolog pcoI and is essential for plant virulence in Pseudomonas corrugata. Mol. Plant-Microbe Interact. 22 (12), 1514–1522. doi:10.1094/MPMI-22-12-1514
Liu, J. F., Mbadinga, S. M., Yang, S. Z., Gu, J. D., and Mu, B. Z. (2015). Chemical structure, property and potential applications of biosurfactants produced by Bacillus subtilis in petroleum recovery and spill mitigation. Int. J. Mol. Sci. 16 (3), 4814–4837. doi:10.3390/ijms16034814
Lo Cantore, P., Lazzaroni, S., Coraiola, M., Dalla Serra, M., Cafarchia, C., Evidente, A., et al. (2006). Biological characterization of white line-inducing principle (WLIP) produced by Pseudomonas reactans NCPPB1311. Mol. Plant Microbe Interact. 19 (10), 1113–1120. doi:10.1094/mpmi-19-1113
Lu, S. E., Scholz-Schroeder, B. K., and Gross, D. C. (2002). Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol. Plant-Microbe Interact. 15 (1), 43–53. doi:10.1094/MPMI.2002.15.1.43
Lu, S. E., Wang, N., Wang, J., Chen, Z. J., and Gross, D. C. (2005). Oligonucleotide microarray analysis of the salA regulon controlling phytotoxin production by Pseudomonas syringae pv. syringae. Mol. Plant-Microbe Interact. 18 (4), 324–333. doi:10.1094/MPMI-18-0324
Ma, Z., Geudens, N., Kieu, N. P., Sinnaeve, D., Ongena, M., Martins, J. C., et al. (2016a). Biosynthesis, chemical structure, and structure-activity relationship of orfamide lipopeptides produced by Pseudomonas protegens and related species. Front. Microbiol. 7, 382. doi:10.3389/fmicb.2016.00382
Ma, Z., Hua, G. K. H., Ongena, M., and Höfte, M. (2016b). Role of phenazines and cyclic lipopeptides produced by Pseudomonas sp. CMR12a in induced systemic resistance on rice and bean. Environ. Microbiol. Rep. 8 (5), 896–904. doi:10.1111/1758-2229.12454
Ma, Z., Ongena, M., and Höfte, M. (2017). The cyclic lipopeptide orfamide induces systemic resistance in rice to Cochliobolus miyabeanus but not to Magnaporthe oryzae. Plant Cell Rep. 36 (11), 1731–1746. doi:10.1007/s00299-017-2187-z
Martin, C., Ibáñez, R., Nothias, L.-F., Boya P, C. A., Reinert, L. K., Rollins-Smith, L. A., et al. (2019). Viscosin-like lipopeptides from frog skin bacteria inhibit Aspergillus fumigatus and Batrachochytrium dendrobatidis detected by imaging mass spectrometry and molecular networking. Sci. Rep. 9 (1), 3019. doi:10.1038/s41598-019-39583-7
Mavrodi, D. V., Peever, T. L., Mavrodi, O. V., Parejko, J. A., Raaijmakers, J. M., Lemanceau, P., et al. (2010). Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 76 (3), 866–879. doi:10.1128/AEM.02009-09
Mazzola, M., de Bruijn, I., Cohen, M. F., and Raaijmakers, J. M. (2009). Protozoan-induced regulation of cyclic lipopeptide biosynthesis is an effective predation defense mechanism for Pseudomonas fluorescens. Appl. Environ. Microbiol. 75 (21), 6804–6811. doi:10.1128/AEM.01272-09
Meleshko, D., Mohimani, H., Tracanna, V., Hajirasouliha, I., Medema, M. H., Korobeynikov, A., et al. (2019). BiosyntheticSPAdes: reconstructing biosynthetic gene clusters from assembly graphs. Genome Res. 29 (8), 1352–1362. doi:10.1101/gr.243477.118
Melnyk, R. A., Hossain, S. S., and Haney, C. H. (2019). Convergent gain and loss of genomic islands drive lifestyle changes in plant-associated Pseudomonas. ISME J. 13 (6), 1575–1588. doi:10.1038/s41396-019-0372-5
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., Van Der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332 (6033), 1097–1100. doi:10.1126/science.1203980
Michelsen, C. F., Jensen, H., Venditto, V. J., Hennessy, R. C., and Stougaard, P. (2015a). Bioactivities by a crude extract from the Greenlandic Pseudomonas sp. In5 involves the nonribosomal peptides, nunamycin and nunapeptin. PeerJ 3, e1476. doi:10.7717/peerj.1476
Michelsen, C. F., and Stougaard, P. (2011). A novel antifungal Pseudomonas fluorescens isolated from potato soils in Greenland. Curr. Microbiol. 62 (4), 1185–1192. doi:10.1007/s00284-010-9846-4
Michelsen, C. F., Watrous, J., Glaring, M. A., Kersten, R., Koyama, N., Dorrestein, P. C., et al. (2015b). Nonribosomal peptides, key biocontrol components for Pseudomonas fluorescens In5, isolated from a Greenlandic suppressive soil. mBio 6 (2), e00079–e00015. doi:10.1128/mBio.00079-15
Monfil, V. O., and Casas-Flores, S. (2014). “Chapter 32 - molecular mechanisms of biocontrol in Trichoderma spp. and their applications in agriculture,” in Biotechnology and biology of Trichoderma. Editors V. K. Gupta, M. Schmoll, A. Herrera-Estrella, R. S. Upadhyay, I. Druzhinina, and M. G. Tuohy (Amsterdam: Elsevier), 429–453.
Muangkaew, P., De Roo, V., Zhou, L., Girard, L., Cesa-Luna, C., Hofte, M., et al. (2023). Stereomeric lipopeptides from a single non-ribosomal peptide synthetase as an additional source of structural and functional diversification in Pseudomonas lipopeptide biosynthesis. Int. J. Mol. Sci. 24 (18), 14302. doi:10.3390/ijms241814302
Murata, H., Tsukamoto, T., and Shirata, A. (1998). rtpA, a gene encoding a bacterial two-component sensor kinase, determines pathogenic traits of Pseudomonas tolaasii, the causal agent of brown blotch disease of a cultivated mushroom. Pleurotus Ostreatus. Mycoscience 39, 261–271. doi:10.1007/bf02464007
Nielsen, T. H., Christophersen, C., Anthoni, U., and Sorensen, J. (1999). Viscosinamide, a new cyclic depsipeptide with surfactant and antifungal properties produced by Pseudomonas fluorescens DR54. J. Appl. Microbiol. 87 (1), 80–90. doi:10.1046/j.1365-2672.1999.00798.x
Nielsen, T. H., Sørensen, D., Tobiasen, C., Andersen, J. B., Christophersen, C., Givskov, M., et al. (2002). Antibiotic and biosurfactant properties of cyclic lipopeptides produced by fluorescent Pseudomonas spp. from the sugar beet rhizosphere. Appl. Environ. Microbiol. 68 (7), 3416–3423. doi:10.1128/aem.68.7.3416-3423.2002
Nielsen, T. H., and Sorensen, J. (2003). Production of cyclic lipopeptides by Pseudomonas fluorescens strains in bulk soil and in the sugar beet rhizosphere. Appl. Environ. Microbiol. 69 (2), 861–868. doi:10.1128/Aem.69.2.861-868.2003
Nielsen, T. H., Thrane, C., Christophersen, C., Anthoni, U., and Sorensen, J. (2000). Structure, production characteristics and fungal antagonism of tensin - a new antifungal cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. J. Appl. Microbiol. 89 (6), 992–1001. doi:10.1046/j.1365-2672.2000.01201.x
Nikel, P. I., Martínez-García, E., and de Lorenzo, V. (2014). Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 12 (5), 368–379. doi:10.1038/nrmicro3253
Ntana, F., Hennessy, R. C., Zervas, A., and Stougaard, P. (2023). Pseudomonas nunensis sp. nov. isolated from a suppressive potato field in Greenland. Int. J. Syst. Evol. Microbiol. 73 (2), 005700. doi:10.1099/ijsem.0.005700
Nybroe, O., and Sorensen, J. (2004). “Production of cyclic lipopeptides by fluorescent pseudomonads,” in Pseudomonas, biosynthesis of macromolecules and molecular metabolism. Editor J.-L. Ramos (Kluwer Academic/Plenum), 147–172.
Olorunleke, F. E., Hua, G. K. H., Kieu, N. P., Ma, Z., and Höfte, M. (2015). Interplay between orfamides, sessilins and phenazines in the control of Rhizoctonia diseases by Pseudomonas sp. CMR12a. Environ. Microbiol. Rep. 7 (5), 774–781. doi:10.1111/1758-2229.12310
Olorunleke, F. E., Kieu, N. P., De Waele, E., Timmerman, M., Ongena, M., and Höfte, M. (2017). Coregulation of the cyclic lipopeptides orfamide and sessilin in the biocontrol strain Pseudomonas sp CMR12a. Microbiologyopen 6 (5), e00499. doi:10.1002/mbo3.499
Omoboye, O. O. (2019). Cyclic lipopeptide diversity and biocontrol versatility of Pseudomonas species associated with the cocoyam rhizospere. PhD Thesis. Ghent: Ghent University.
Omoboye, O. O., Geudens, N., Duban, M., Chevalier, M., Flahaut, C., Martins, J. C., et al. (2019a). Pseudomonas sp. COW3 produces new bananamide-type cyclic lipopeptides with antimicrobial activity against Pythium myriotylum and Pyricularia oryzae. Molecules 24, 4170. doi:10.3390/molecules24224170
Omoboye, O. O., Oni, F. E., Batool, H., Yimer, H. Z., De Mot, R., and Höfte, M. (2019b). Pseudomonas cyclic lipopeptides suppress the rice blast fungus Magnaporthe oryzae by induced resistance and direct antagonism. Front. Plant Sci. 10, 901. doi:10.3389/fpls.2019.00901
Oni, F. E., Esmaeel, Q., Onyeka, J. T., Adeleke, R., Jacquard, C., Clement, C., et al. (2022). Pseudomonas lipopeptide-mediated biocontrol: chemotaxonomy and biological activity. Molecules 27 (2), 372. doi:10.3390/molecules27020372
Oni, F. E., Geudens, N., Adiobo, A., Omoboye, O. O., Enow, E. A., Onyeka, J. T., et al. (2020a). Biosynthesis and antimicrobial activity of pseudodesmin and viscosinamide cyclic lipopeptides produced by pseudomonads associated with the cocoyam rhizosphere. Microorganisms 8 (7), 1079. doi:10.3390/microorganisms8071079
Oni, F. E., Geudens, N., Omoboye, O. O., Bertier, L., Hua, H. G. K., Adiobo, A., et al. (2019a). Fluorescent Pseudomonas and cyclic lipopeptide diversity in the rhizosphere of cocoyam (Xanthosoma sagittifolium). Environ. Microbiol. 21 (3), 1019–1034. doi:10.1111/1462-2920.14520
Oni, F. E., Geudens, N., Onyeka, J. T., Olorunleke, O. F., Salami, A. E., Omoboye, O. O., et al. (2020b). Cyclic lipopeptide-producing Pseudomonas koreensis group strains dominate the cocoyam rhizosphere of a Pythium root rot suppressive soil contrasting with P. putida prominence in conducive soils. Environ. Microbiol. 22 (12), 5137–5155. doi:10.1111/1462-2920.15127
Oni, F. E., Olorunleke, O. F., and Höfte, M. (2019b). Phenazines and cyclic lipopeptides produced by Pseudomonas sp. CMR12a are involved in the biological control of Pythium myriotylum on cocoyam (Xanthosoma sagittifolium). Biol. control 129, 109–114. doi:10.1016/j.biocontrol.2018.10.005
Osdaghi, E., Martins, S. J., Ramos-Sepulveda, L., Vieira, F. R., Pecchia, J. A., Beyer, D. M., et al. (2019). 100 Years since tolaas: bacterial blotch of mushrooms in the 21st century. Plant Dis. 103 (11), 2714–2732. doi:10.1094/Pdis-03-19-0589-Fe
Pacheco-Moreno, A., Stefanato, F. L., Ford, J. J., Trippel, C., Uszkoreit, S., Ferrafiat, L., et al. (2021). Pan-genome analysis identifies intersecting roles for Pseudomonas specialized metabolites in potato pathogen inhibition. eLife 10, e71900. doi:10.7554/eLife.71900
Pascual, J., García-López, M., Carmona, C., Sousa, T. d.S., de Pedro, N., Cautain, B., et al. (2014). Pseudomonas soli sp. nov., a novel producer of xantholysin congeners. Syst. Appl. Microbiol. 37 (6), 412–416. doi:10.1016/j.syapm.2014.07.003
Pauwelyn, E., Huang, C. J., Ongena, M., Leclère, V., Jacques, P., Bleyaert, P., et al. (2013). New linear lipopeptides produced by Pseudomonas cichorii SF1-54 are involved in virulence, swarming motility, and biofilm formation. Mol. Plant-Microbe Interact. 26 (5), 585–598. doi:10.1094/MPMI-11-12-0258-R
Peschel, A., and Sahl, H. G. (2006). The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4 (7), 529–536. doi:10.1038/nrmicro1441
Pflanze, S., Mukherji, R., Ibrahim, A., Gunther, M., Gotze, S., Chowdhury, S., et al. (2023). Nonribosomal peptides protect Pseudomonas nunensis 4A2e from amoebal and nematodal predation. Chem. Sci. 14 (41), 11573–11581. doi:10.1039/d3sc03335j
Pršić, J., and Ongena, M. (2020). Elicitors of plant immunity triggered by beneficial bacteria. Front. Plant Sci. 11, 594530. doi:10.3389/fpls.2020.594530
Quigley, N. B., and Gross, D. C. (1994). Syringomycin production among strains of Pseudomonas syringae pv syringae - conservation of the syrB and syrD genes and activation of phytotoxin production by plant signal molecules. Mol. Plant-Microbe Interact. 7 (1), 78–90. doi:10.1094/Mpmi-7-0078
Quigley, N. B., Mo, Y. Y., and Gross, D. C. (1993). SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP-binding secretion proteins. Mol. Microbiol. 9 (4), 787–801. doi:10.1111/j.1365-2958.1993.tb01738.x
Raaijmakers, J. M., de Bruijn, I., and de Kock, M. J. D. (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant-Microbe Interact. 19 (7), 699–710. doi:10.1094/mpmi-19-0699
Raaijmakers, J. M., de Bruijn, I., Nybroe, O., and Ongena, M. (2010). Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34 (6), 1037–1062. doi:10.1111/j.1574-6976.2010.00221.x
Rainey, P. B., Brodey, C. L., and Johnstone, K. (1991). Biological properties and spectrum of activity of tolaasin, a lipodepsipeptide toxin produced by the mushroom pathogen Pseudomonas tolaasii. Physiological Mol. Plant Pathology 39 (1), 57–70. doi:10.1016/0885-5765(91)90031-C
Raj, A., Kumar, A., and Dames, J. F. (2021). Tapping the role of microbial biosurfactants in pesticide remediation: an eco-friendly approach for environmental sustainability. Front. Microbiol. 12, 791723. doi:10.3389/fmicb.2021.791723
Reddy, C. A., and Saravanan, R. S. (2013). Polymicrobial multi-functional approach for enhancement of crop productivity. Adv. Appl. Microbiol. 82, 53–113. doi:10.1016/b978-0-12-407679-2.00003-x
Reen, F. J., Romano, S., Dobson, A. D., and O'Gara, F. (2015). The sound of silence: activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 13 (8), 4754–4783. doi:10.3390/md13084754
Rigolet, A., Arias, A. A., Anckaert, A., Quinton, L., Rigali, S., Tellatin, D., et al. (2023). The fate of bacterial secondary metabolites in the rhizosphere: Streptomyces degrades and feeds on cyclic lipopeptides produced by competitors. BioRxiv. doi:10.1101/2023.07.27.550914
Rokni-Zadeh, H., Li, W., Sanchez-Rodriguez, A., Sinnaeve, D., Rozenski, J., Martins, J. C., et al. (2012). Genetic and functional characterization of cyclic lipopeptide white-line-inducing principle (WLIP) production by rice rhizosphere isolate Pseudomonas putida RW10S2. Appl. Environ. Microbiol. 78 (14), 4826–4834. doi:10.1128/AEM.00335-12
Rokni-Zadeh, H., Li, W., Yilma, E., Sanchez-Rodriguez, A., and De Mot, R. (2013). Distinct lipopeptide production systems for WLIP (white line-inducing principle) in Pseudomonas fluorescens and Pseudomonas putida. Environ. Microbiol. Rep. 5 (1), 160–169. doi:10.1111/1758-2229.12015
Roongsawang, N., Washio, K., and Morikawa, M. (2011). Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int. J. Mol. Sci. 12 (1), 141–172. doi:10.3390/ijms12010141
Rosen, P. C., and Seyedsayamdost, M. R. (2017). Though much is taken, much abides: finding new antibiotics using old ones. Biochemistry 56 (37), 4925–4926. doi:10.1021/acs.biochem.7b00782
Sanahuja, G., Banakar, R., Twyman, R. M., Capell, T., and Christou, P. (2011). Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol. J. 9 (3), 283–300. doi:10.1111/j.1467-7652.2011.00595.x
Scherlach, K., Lackner, G., Graupner, K., Pidot, S., Bretschneider, T., and Hertweck, C. (2013). Biosynthesis and mass spectrometric imaging of tolaasin, the virulence factor of Brown blotch mushroom disease. Chembiochem 14 (18), 2439–2443. doi:10.1002/cbic.201300553
Sekar, J., Raju, K., Duraisamy, P., and Vaiyapuri, P. R. (2018). Potential of finger millet indigenous rhizobacterium Pseudomonas sp. MSSRFD41 in blast disease management - growth promotion and compatibility with the resident rhizomicrobiome. Front. Microbiol. 9, 1029. doi:10.3389/fmicb.2018.01029
Sekhon Randhawa, K. K., and Rahman, P. K. S. M. (2014). Rhamnolipid biosurfactants-past, present, and future scenario of global market. Front. Microbiol. 5, 454. doi:10.3389/fmicb.2014.00454
Seyedsayamdost, M. R. (2014). High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. 111 (20), 7266–7271. doi:10.1073/pnas.1400019111
Shimura, S., Ishima, M., Nakajima, S., Fujii, T., Himeno, N., Ikeda, K., et al. (2013). Total synthesis and anti-hepatitis C virus activity of MA026. J. Am. Chem. Soc. 135 (50), 18949–18956. doi:10.1021/ja410145x
Siani, H. S., Barragan-Huerta, B. E., Lebron-Paler, A., Pemberton, J. E., Vazquez, R. R., Burns, A. M., et al. (2008). Efficient purification of the biosurfactant viscosin from Pseudomonas libanensis strain M9-3 and its physicochemical and biological properties. J. Nat. Prod. 71 (6), 1011–10115. doi:10.1021/np800069u
Singh, G. M., Fortin, P. D., Koglin, A., and Walsh, C. T. (2008). β-Hydroxylation of the aspartyl residue in the phytotoxin syringomycin E: characterization of two candidate hydroxylases AspH and SyrP in Pseudomonas syringae. Biochemistry 47 (43), 11310–11320. doi:10.1021/bi801322z
Singh, G. M., Vaillancourt, F. H., Yin, J., and Walsh, C. T. (2007). Characterization of SyrC, an aminoacyltransferase shuttling threonyl and chlorothreonyl residues in the syringomycin biosynthetic assembly line. Chem. Biol. 14 (1), 31–40. doi:10.1016/j.chembiol.2006.11.005
Soberón-Chávez, G., González-Valdez, A., Soto-Aceves, M. P., and Cocotl-Yañez, M. (2021). Rhamnolipids produced by Pseudomonas: from molecular genetics to the market. Microb. Biotechnol. 14 (1), 136–146. doi:10.1111/1751-7915.13700
Sobrero, P. M., Muzlera, A., Frescura, J., Jofré, E., and Valverde, C. (2017). A matter of hierarchy: activation of orfamide production by the post-transcriptional Gac-Rsm cascade of Pseudomonas protegens CHA0 through expression upregulation of the two dedicated transcriptional regulators. Environ. Microbiol. Rep. 9 (5), 599–611. doi:10.1111/1758-2229.12566
Solaiman, D. K., Ashby, R. D., Gunther, N. W. t., and Zerkowski, J. A. (2015). Dirhamnose-lipid production by recombinant nonpathogenic bacterium Pseudomonas chlororaphis. Appl. Microbiol. Biotechnol. 99 (10), 4333–4342. doi:10.1007/s00253-015-6433-4
Soler-Rivas, C., Jolivet, S., Arpin, N., Olivier, J. M., and Wichers, H. J. (1999). Biochemical and physiological aspects of brown blotch disease of Agaricus bisporus. FEMS Microbiol. Rev. 23 (5), 591–614. doi:10.1016/S0168-6445(99)00023-6
Song, C., Aundy, K., van de Mortel, J., and Raaijmakers, J. M. (2014). Discovery of new regulatory genes of lipopeptide biosynthesis in Pseudomonas fluorescens. FEMS Microbiol. Lett. 356 (2), 166–175. doi:10.1111/1574-6968.12404
Song, C., Sundqvist, G., Malm, E., de Bruijn, I., Kumar, A., van de Mortel, J., et al. (2015a). Lipopeptide biosynthesis in Pseudomonas fluorescens is regulated by the protease complex ClpAP. BMC Microbiol. 15 (1), 29–367. doi:10.1186/s12866-015-0367-y
Song, C., van der Voort, M., van de Mortel, J., Hassan, K. A., Elbourne, L. D. H., Paulsen, I. T., et al. (2015b). The Rsm regulon of plant growth-promoting Pseudomonas fluorescens SS101: role of small RNAs in regulation of lipopeptide biosynthesis. Microb. Biotechnol. 8 (2), 296–310. doi:10.1111/1751-7915.12190
Sonnleitner, E., and Haas, D. (2011). Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl. Microbiol. Biotechnol. 91 (1), 63–79. doi:10.1007/s00253-011-3332-1
Sorensen, K. N., Kim, K. H., and Takemoto, J. Y. (1996). In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv syringae. Antimicrob. Agents Chemother. 40 (12), 2710–2713. doi:10.1128/Aac.40.12.2710
Spraker, J. E., Sanchez, L. M., Lowe, T. M., Dorrestein, P. C., and Keller, N. P. (2016). Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J. 10 (9), 2317–2330. doi:10.1038/ismej.2016.32
Steigenberger, J., Mergen, C., De Roo, V., Geudens, N., Martins, J. C., and Heerklotz, H. (2023). The effect of membrane thickness on the membrane permeabilizing activity of the cyclic lipopeptide tolaasin II. Front. Mol. Biosci. 9, 1064742. doi:10.3389/fmolb.2022.1064742
Steigenberger, J., Verleysen, Y., Geudens, N., Martins, J. C., and Heerklotz, H. (2021). The optimal lipid chain length of a membrane-permeabilizing lipopeptide results from the balance of membrane partitioning and local damage. Front. Microbiol. 12, 669709–669714. doi:10.3389/fmicb.2021.669709
Sun, W., Zhu, B., Yang, F., Dai, M., Sehar, S., Peng, C., et al. (2021). Optimization of biosurfactant production from Pseudomonas sp. CQ2 and its application for remediation of heavy metal contaminated soil. Chemosphere 265, 129090. doi:10.1016/j.chemosphere.2020.129090
Tavares, L. F., Silva, P. M., Junqueira, M., Mariano, D. C., Nogueira, F. C., Domont, G. B., et al. (2013). Characterization of rhamnolipids produced by wild-type and engineered Burkholderia kururiensis. Appl. Microbiol. Biotechnol. 97 (5), 1909–1921. doi:10.1007/s00253-012-4454-9
Temme, K., Zhao, D., and Voigt, C. A. (2012). Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc. Natl. Acad. Sci. U. S. A. 109 (18), 7085–7090. doi:10.1073/pnas.1120788109
Tran, H., Ficke, A., Asiimwe, T., Höfte, M., and Raaijmakers, J. M. (2007). Role of the cyclic lipopeptide massetolide a in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol. 175 (4), 731–742. doi:10.1111/j.1469-8137.2007.02138.x
Vallet-Gely, I., Novikov, A., Augusto, L., Liehl, P., Bolbach, G., Péchy-Tarr, M., et al. (2010). Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl. Environ. Microbiol. 76 (3), 910–921. doi:10.1128/AEM.02112-09
Van Der Voort, M., Meijer, H. J. G., Schmidt, Y., Watrous, J., Dekkers, E., Mendes, R., et al. (2015). Genome mining and metabolic profiling of the rhizosphere bacterium Pseudomonas sp. SH-C52 for antimicrobial compounds. Front. Microbiol. 6, 693. doi:10.3389/fmicb.2015.00693
Vaughn, V. L., and Gross, D. C. (2016). Characterization of salA, syrF, and syrG genes and attendant regulatory networks involved in plant pathogenesis by Pseudomonas syringae pv. syringae B728a. PLoS One 11 (3), e0150234. doi:10.1371/journal.pone.0150234
Venkatesh, N., Greco, C., Drott, M. T., Koss, M. J., Ludwikoski, I., Keller, N. M., et al. (2022). Bacterial hitchhikers derive benefits from fungal housing. Curr. Biol. 32 (7), 1523–1533.e6. doi:10.1016/j.cub.2022.02.017
Venturi, V. (2006). Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30 (2), 274–291. doi:10.1111/j.1574-6976.2005.00012.x
Wang, N., Lu, S.-E., Records, A. R., and Gross, D. C. (2006a). Characterization of the transcriptional Activators SalA and SyrF, which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 188 (9), 3290–3298. doi:10.1128/JB.188.9.3290-3298.2006
Wang, N., Lu, S. E., Wang, J. L., Chen, Z. J., and Gross, D. C. (2006b). The expression of genes encoding lipodepsipeptide phytotoxins by Pseudomonas syringae pv. syringae is coordinated in response to plant signal molecules. Mol. Plant-Microbe Interact. 19 (3), 257–269. doi:10.1094/Mpmi-19-0257
Wang, S., Cui, J., Bilal, M., Hu, H., Wang, W., and Zhang, X. (2020). Pseudomonas spp. as cell factories (MCFs) for value-added products: from rational design to industrial applications. Crit. Rev. Biotechnol. 40 (8), 1232–1249. doi:10.1080/07388551.2020.1809990
Wang, W., Zheng, G., and Lu, Y. (2021). Recent advances in strategies for the cloning of natural product biosynthetic gene clusters. Front. Bioeng. Biotechnol. 9, 692797. doi:10.3389/fbioe.2021.692797
Washio, K., Lim, S. P., Roongsawang, N., and Morikawa, M. (2010). Identification and characterization of the genes responsible for the production of the cyclic lipopeptide arthrofactin by Pseudomonas sp. MIS38. Biosci. Biotechnol. Biochem. 74 (5), 992–999. doi:10.1271/bbb.90860
Whiteley, M., and Greenberg, E. P. (2001). Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183 (19), 5529–5534. doi:10.1128/Jb.183.19.5529-5534.2001
Wirth, N. T., Kozaeva, E., and Nikel, P. I. (2020). Accelerated genome engineering of Pseudomonas putida by I-SceI-mediated recombination and CRISPR-Cas9 counterselection. Microb. Biotechnol. 13 (1), 233–249. doi:10.1111/1751-7915.13396
Woo, S., Fogliano, V., Scala, F., and Lorito, M. (2002). Synergism between fungal enzymes and bacterial antibiotics may enhance biocontrol. Ant. Van Leeuwenhoek 81 (1-4), 353–356. doi:10.1023/a:1020540818163
Xia, B., Luo, M., Pang, L., Liu, X., and Yi, Y. (2021). Lipopeptides against COVID-19 RNA-dependent RNA polymerase using molecular docking. Biomed. J. 44 (6 Suppl. 1), S15–s24. doi:10.1016/j.bj.2021.11.010
Yeh, M. S., Wei, Y. H., and Chang, J. S. (2005). Enhanced production of surfactin from Bacillus subtilis by addition of solid carriers. Biotechnol. Progr 21 (4), 1329–1334. doi:10.1021/bp050040c
Zachow, C., Jahanshah, G., De Bruijn, I., Song, C., Ianni, F., Pataj, Z., et al. (2015). The novel lipopeptide poaeamide of the endophyte Pseudomonas poae RE-1-1-14 is involved in pathogen suppression and root colonization. Mol. Plant-Microbe Interact. 28 (7), 800–810. doi:10.1094/MPMI-12-14-0406-R
Zhang, S., Mukherji, R., Chowdhury, S., Reimer, L., and Stallforth, P. (2021). Lipopeptide-mediated bacterial interaction enables cooperative predator defense. Proc. Natl. Acad. Sci. 118 (6), e2013759118. doi:10.1073/pnas.2013759118
Zhao, F., Wu, Y., Wang, Q., Zheng, M., and Cui, Q. (2021). Glycerol or crude glycerol as substrates make Pseudomonas aeruginosa achieve anaerobic production of rhamnolipids. Microb. Cell Factories 20 (1), 185. doi:10.1186/s12934-021-01676-2
Zhao, H., Liu, Y. P., and Zhang, L. Q. (2019). In silico and genetic analyses of cyclic lipopeptide synthetic gene clusters in Pseudomonas sp. 11K1. Front. Microbiol. 10, 544–611. doi:10.3389/fmicb.2019.00544
Zhou, L., de Jong, A., Yi, Y. H., and Kuipers, O. P. (2021). Identification, isolation, and characterization of medipeptins, antimicrobial peptides from Pseudomonas mediterranea EDOX. Front. Microbiol. 12, 732771. doi:10.3389/fmicb.2021.732771
Zhu, H., Sandiford, S. K., and van Wezel, G. P. (2014). Triggers and cues that activate antibiotic production by actinomycetes. J. Industrial Microbiol. Biotechnol. 41 (2), 371–386. doi:10.1007/s10295-013-1309-z
Keywords: Pseudomonas, specialized metabolites, lipopeptides, biosurfactants, antibiotics, regulation, bioengineering, bioprocessing
Citation: Zhou L, Höfte M and Hennessy RC (2024) Does regulation hold the key to optimizing lipopeptide production in Pseudomonas for biotechnology?. Front. Bioeng. Biotechnol. 12:1363183. doi: 10.3389/fbioe.2024.1363183
Received: 29 December 2023; Accepted: 12 February 2024;
Published: 27 February 2024.
Edited by:
Eric Déziel, Université du Québec, CanadaReviewed by:
Luis Felipe Muriel-Millán, National Autonomous University of Mexico, MexicoCopyright © 2024 Zhou, Höfte and Hennessy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosanna C. Hennessy, aGVubmVzc3lAcGxlbi5rdS5kaw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.