
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 06 March 2024
Sec. Tissue Engineering and Regenerative Medicine
Volume 12 - 2024 | https://doi.org/10.3389/fbioe.2024.1347159
Background: This study aims to construct a three-dimensional model of skin dermis utilizing continuous tissue sections, with the primary objective of obtaining anatomical structure data for normal human dermal tissues.
Methods: Normal skin tissue specimens were acquired, paraffin-embedded, and subjected to HE staining. Panoramic images of skin sections were captured using a microscope. Tissue section images were aligned using the SIFT and StackReg image alignment methods, with analysis conducted using the OpenCV module. Mimics17 software facilitated the reconstruction of the skin dermal 3D model, enabling the calculation of dermal porosity and the void diameter.
Results: Panoramic skin slices exhibited high-resolution differentiation of dermal fibers and cellular structures. Both SIFT and StackReg image alignment methods yielded similar results, although the SIFT method demonstrated greater robustness. Successful reconstruction of the three-dimensional dermal structure was achieved. Quantitative analysis revealed a dermal porosity of 18.96 ± 4.41% and an average pore diameter of 219.29 ± 34.27 μm. Interestingly, the porosity of the dermis exhibited a gradual increase from the papillary layer to the fourth layer, followed by a transient decrease and then a gradual increase. The distribution of the mean pore diameter mirrored the pattern observed in porosity distribution.
Conclusion: Utilizing the continuous skin tissue slice reconstruction technique, this study successfully reconstructed a high-precision three-dimensional tissue structure of the skin. The quantitative analysis of dermal tissue porosity and average pore diameter provides a standardized dataset for the development of biomimetic tissue-engineered skin.
Autologous dermal grafts and flaps are commonly employed for wound repair; however, they are associated with clinical challenges such as dermal contracture, scar hyperplasia, and impaired appearance and function (Barbara et al., 2022; Feng and Qiang, 2021; Núria and Joan, 2023). The structural integrity of the dermis plays a crucial role in enhancing skin elasticity, flexibility, and cushioning ability, thereby mitigating contracture and scar formation. Consequently, tissue-engineered skin has emerged as a research hotspot in wound repair (Cuesta et al., 2023; Shila et al., 2023; Youngnam et al., 2024).
Tissue-engineered skin represents a significant approach to effectively address wounds, ameliorate skin contractures, reduce scarring, and enhance clinical outcomes. The characterization of tissue-engineered skin is vital as it influences the physicochemical properties of the skin and impacts the proliferation, differentiation, and tissue regeneration of fibroblasts, as well as the growth of blood vessels and nutrient penetration (Maryam and Bita, 2023; Narayanan et al., 2023; Sarah et al., 2023; Taghizadeh et al., 2023). The design of viable tissue-engineered skin necessitates a foundation in the anatomical structure of the human dermis. Tissue-engineered skin that closely mimics the composition and microstructure of normal skin exhibits superior efficacy in promoting wound healing. However, current tissue-engineered skin lacks essential data pertaining to normal skin, such as dermal fiber composition, porosity, pore diameter, and pore wall thickness (Maria et al., 2022; Ewa et al., 2023a). Therefore, a comprehensive quantitative and qualitative analysis of the micro 3D structure of the skin is beneficial for constructing biomimetic tissue-engineered skin.
The optimal spatial distribution of simulated restored skin tissue cells and extracellular matrix serves as the foundation for the regenerative repair of skin defects. Traditional 3D reconstruction methods such as CT and MRI have low resolution for skin tissue structure, making it difficult to recognize the skin epidermis and dermal fiber structure in the images. However, skin tissue section images provide a clear resolution of dermal fiber and cellular structures, thereby representing the microscopic three-dimensional structure of the tissue (Muro and Akita, 2023). This study focused on reconstructing the three-dimensional anatomical structure of skin tissue based on continuous skin tissue sections.
The key to 3D reconstruction of continuous skin tissue slices lies in the acquisition of high-resolution panoramic images of the sections and accurate alignment of the continuous images. The current literature indicates that multiple small-field-of-view images can be acquired on a continuous basis by these images of tissue sections through an optical microscope and then stitching the images together to form a complete panoramic image of the section. However, this method has disadvantages of low stitching accuracy and a heavy workload. In addition, the existing literature describes manual methods that align continuous tissue section images with low alignment accuracy, making it difficult to quickly and accurately align a large amount of image data (Kiemen et al., 2022; Chang et al., 2023). Our study used a new technical method of panoramic imaging and automatic alignment technology to reconstruct the three-dimensional anatomical structure of skin tissues. This new method improves the accuracy rate and work efficiency, and it lays a foundation for the three-dimensional reconstruction of skin tissues.
The study involved the collection of whole skin from the outer thigh of normal adults, ensuring that the collection site was devoid of any visible injuries or skin diseases. Skin tissue specimens were fixed and preserved in a 10% formalin fixative solution. We placed the fixed specimen in an alcohol gradient for dehydration, followed by xylene for clarification. The skin specimen was trimmed into a 1 cm × 1 cm skin tissue block, with depth to subcutaneous, and was then paraffin-embedded and HE-stained. The study protocol was approved by the Institutional Review Board of Guangzhou Red Cross Hospital (APPROVAL NUMBER/2023-009-01) on 15 February 2023.
Each section was scanned using a Motic BA600 Mot-7.5 fully automatic microscope. Panoramic tissue sections were generated, and the resulting images were stored in JPG format. The region of interest was carefully selected for subsequent analysis.
Continuous tissue section images often exhibit deviations such as translation and rotation. To address this, the images of skin tissue sections underwent alignment processing. SIFT and StackReg are image registration plugins used in ImageJ software. These can quickly align a series of image slices. Each image slice is used as a template for aligning the next slice. These two image alignment methods have been successfully used in industrial applications and in the aerospace and medical fields. In this study, continuous tissue section images were imported into ImageJ software, and the SIFT and StackReg plugins were used to quickly and automatically align continuous section images by selecting the “Similarity” mode. Ten randomly selected groups of section images were used to assess the effectiveness of image alignment. The OpenCV tool was used to calculate the root-mean-square error (MSE), structural similarity score (SSIM), and mutual information measure (MI) value of the two image alignment methods. Statistical analysis was then conducted to compare and evaluate the alignment effects of the two methods.
Following HE staining, the skin tissue structure became clearly distinguishable, with the gray value of dermal fibers exhibiting a noticeable contrast against other tissue structures in the gray image. Leveraging the threshold segmentation method, the epidermal and dermal fiber structures of the skin tissue were accurately segmented. Upon importing continuous skin tissue section images into Mimics20.0 software and ensuring precise alignment, the threshold segmentation tool was employed. Grayscale values of the skin tissue section were adjusted until the desired skin tissue structure was selected. Any missed portions were manually edited for segmentation refinement.
Next, skin tissue slices were imported into Mimics20.0 software, and the epidermal and dermal structures were segmented, generating corresponding binary images. The 3D reconstruction tool was then utilized to select the optimal quality 3D reconstruction through contour interpolation. Sequentially, 3D reconstruction based on surface drawing was performed for each skin tissue structure, resulting in the visualization of the 3D model representing the skin tissue structure. A flowchart of the experimental design shows the process of panorama imaging, image registration, and three-dimensional reconstruction (Figure 1).
A total of 100 consecutive skin tissue sections were randomly chosen, and the dermal structure was divided equally into 10 layers. These sections were imported into Mimics20.0 software. Within this software, 60 regions of interest (ROIs) were randomly selected for further analysis. The Tissue Engineering Analysis Module in Mimics20.0 was then employed to perform quantitative analyses on the selected regions of interest: dermal tissue porosity and average pore diameter size were measured, providing valuable insights into the microstructural characteristics of the dermal tissue.
Pair t-testing was used to assess the accuracy of the two methods of image registration. All the data were analyzed using SPSS statistical software, version 27.0 (SPSS, Inc.). The level of significance was p < 0.05 (two-sided).
The panoramic image of the tissue section, following HE staining, exhibits high resolution and provides a detailed view of the epidermis, dermis, hair follicles, and sweat glands. This image accurately outlines the cellular structure of the epidermis and dermis, as well as the dermal fiber structure. These findings serve as the foundation for the subsequent three-dimensional reconstruction of the skin tissue structure (Figure 2).
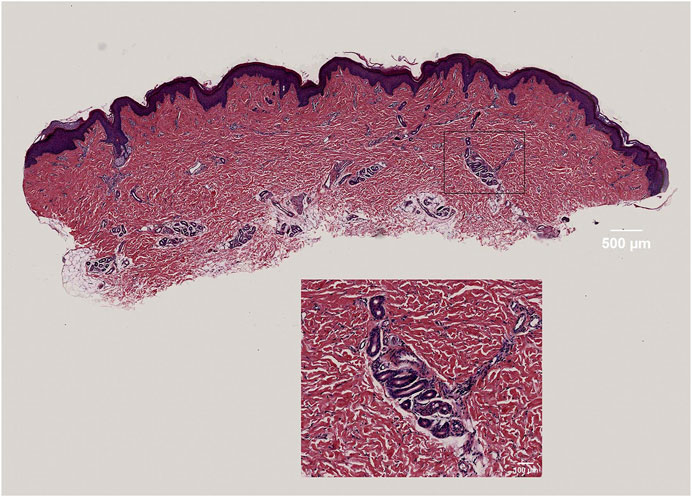
FIGURE 2. High-resolution display of dermal fibers, fibroblasts, and sweat gland structures in skin tissue.
A quantitative analysis of the alignment effects was conducted by statistically evaluating the mean square error, mutual information, and structural similarity scores of two image alignment methods. The results indicate that the mean square error for images aligned using the SIFT method was 66.319, while, for images aligned with StackReg, it was 66.727. The mean mutual information value for SIFT-aligned images was 0.5062054, and for StackReg-aligned images, it was 0.5495679. Additionally, the mean structural similarity score for SIFT-aligned images was 0.454, while, for StackReg-aligned images, it was 0.442. A paired t-test was performed, yielding a p-value greater than 0.05. This suggests that the performance of the two alignment methods is comparable. However, it is noted that the SIFT algorithm demonstrates greater stability than the StackReg-based alignment method (Figure 3).
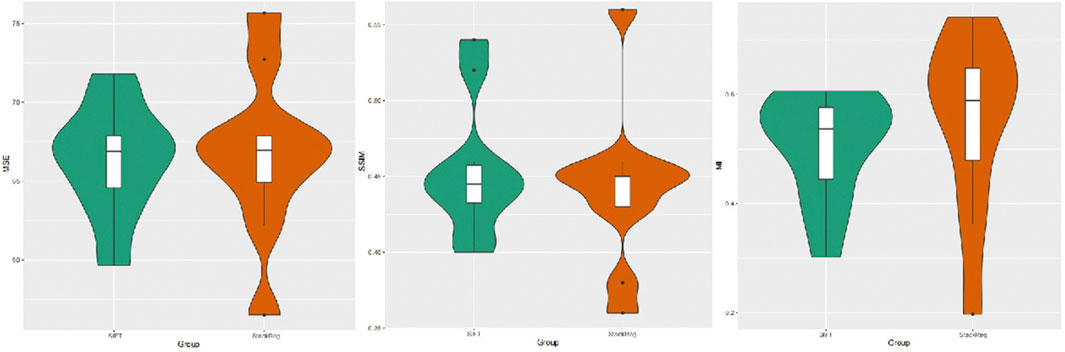
FIGURE 3. Image mean square error, mutual information measure, and structural similarity scores obtained by the SIFT and StackReg alignment methods.
This study successfully identified dermal fibers and fibroblasts, allowing for the clear three-dimensional reconstruction of the dermal tissue structure. This reconstruction provides a vivid representation of the three-dimensional spatial image, illustrating that dermal fibers within the skin’s dermis were scattered. These fibers interweave to form pores of varying sizes, with each pore containing differing numbers of fibroblasts (Figure 4).
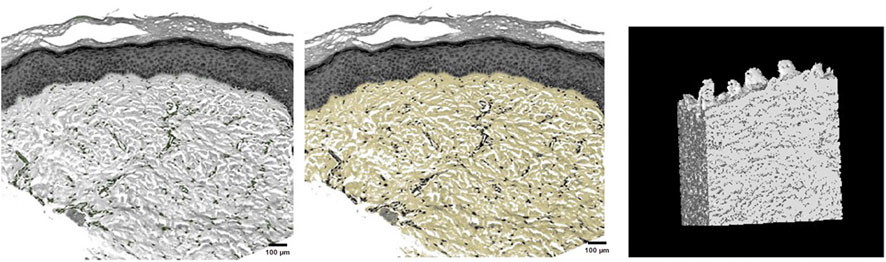
FIGURE 4. Fibroblast distribution in dermal tissue, precise selection of dermal fibers, and three-dimensional reconstruction of dermal tissue.
A longitudinal quantitative analysis of dermal porosity and average pore diameter in human skin tissues was conducted. The results revealed that dermal porosity gradually increased from the papillary layer, decreased in the fourth layer, and then exhibited a gradual increase once again. Furthermore, the distribution of the mean pore diameter mirrored the pattern observed in the distribution of porosity (Figure 5). The site of interest was randomly selected and subjected to statistical analysis, indicating a dermal porosity of 18.96% ± 4.41% and an average pore diameter of 219.29 ± 34.27 μm in human skin tissues.
Tissue-engineered skin has emerged as an optimal method for repairing tissue defects, with current research emphases on scaffold materials, seed cells, and tissue construction (Sorour et al., 2021; Jie et al., 2022; Robab et al., 2022). The construction of tissue-engineered skin for mending skin tissue defects has evolved into a research hotspot, offering innovative possibilities for tissue repair. Tissue-engineered dermal scaffolds play a pivotal role by providing a three-dimensional environment conducive to cell adhesion, growth, and differentiation. Notably, the biomimicry of tissue-engineered dermal scaffolds significantly influences the outcomes of wound repair. Crucial parameters in scaffold materials include skin dermal tissue porosity distribution and average pore diameter. The acquisition of these parameters from normal human skin dermal tissues is instrumental in the development of artificial dermal scaffold materials (Nupur et al., 2021; Yawen et al., 2021; Ewa et al., 2023b). The three-dimensional reconstruction of skin dermis, based on continuous tissue slices, proves to be a valuable approach for obtaining and understanding these essential parameters.
Porosity stands out as a crucial parameter in tissue engineering materials, exerting a notable impact on fluid transport properties within the dermis (Bohumila et al., 2023; Jeyachandran and Cerruti, 2023; Rasha et al., 2023). The three-dimensional structure of decellularized allogeneic dermis, reconstructed using micro-CT, yielded a calculated porosity of 68.3 ± 5.8% (Wang et al., 2015). However, our reconstruction of the 3D structure of normal human dermis through continuous tissue sectioning techniques revealed a dermal porosity of 18.96 ± 4.41%. This discrepancy may be attributed to significant skin shrinkage post-ex vivo, resulting in reduced porosity.
Another pivotal parameter in tissue engineering materials is the mean pore diameter, which influences cell adhesion and migration. It is now understood that fibroblasts in the dermis deposit within pores composed of dermal fibers. Smaller pore diameters in dermal scaffolds pose challenges for fibroblast adhesion and migration in dermal pores (Ghazal et al., 2022; Zohaib et al., 2022; Olevsky et al., 2023). Our reconstruction of the three-dimensional structure of normal human dermis through continuous tissue sectioning techniques yielded an average pore diameter of 219.29 ± 34.27 μm. Therefore, it is imperative that the pore diameter for constructing dermal scaffolds exceeds the diameter of fibroblasts. The reconstruction of the three-dimensional dermal structure using continuous tissue sectioning techniques also revealed that the distribution of dermal scaffold pores lacks an obvious pattern.
Image alignment is pivotal in reconstructing the three-dimensional structural features of human skin dermis, where the quality of alignment directly influences the accuracy of the reconstructed features. Continuous skin tissue slices exhibit variations in grayscale attributes, position (translation and rotation), scale, and nonlinear deformation. Previous studies on image alignment have only paid attention to translation and rotation while ignoring the existence of transformations such as scale expansion and deformation in tissue section images (Liu et al., 2018). In this study, we optimized the global image alignment on the basis of previous studies and used the Similarity alignment mode to reduce the misalignment caused by image deformation. These methods improved the accuracy of the data and produced data that were more representative of the real dermal porosity and pore diameter. The SIFT algorithm, recognized as an excellent feature alignment operator, can be harnessed to address these challenges (Li et al., 2015; Erbing et al., 2023; Meng et al., 2023). Key parameters for evaluating image alignment efficacy include mean square error, mutual information, and structural similarity score. In this study, a statistical analysis of these parameters for two image alignment methods revealed that the SIFT algorithm-based method is comparable in effectiveness to the StackReg image alignment method but demonstrates higher stability.
Current natural and degradable chemically synthesized polymeric materials used in constructing tissue-engineered skin scaffolds fall short of truly mimicking the structural and mechanical properties of the dermis. However, the use of decellularized dermal graft materials has shown excellent clinical results in repairing skin tissue defects. Therefore, understanding the structural features of human skin dermis is crucial for guiding the construction of biomimetic tissue-engineered skin.
The reconstruction of the three-dimensional microstructure of skin dermal tissue, based on serial skin tissue sections, coupled with quantitative analysis at the tissue or cellular level, provides a standard dataset. This dataset is invaluable for advancing the construction of biomimetic tissue-engineered skin and enhances our ability to replicate the intricate features of human skin dermis in engineered substitutes.
By utilizing the continuous skin tissue slice reconstruction technique, this study successfully achieved a high-precision three-dimensional reconstruction of skin tissue structure. The quantitative analysis of dermal tissue porosity and average pore diameter not only enhances our understanding of the microstructural characteristics but also contributes to the establishment of a standard dataset.
The original contributions presented in the study are included in the article/Supplementary material; further inquiries can be directed to the corresponding author.
The studies involving humans were approved by the Institutional Review Board of Guangzhou Red Cross Hospital. The studies were conducted in accordance with local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
PL: Conceptualization, Data curation, Funding acquisition, Methodology, Writing–original draft. TZ: Formal Analysis, Funding acquisition, Methodology, Writing–original draft. YH: Conceptualization, Funding acquisition, Supervision, Visualization, Writing–original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by research grant 202102020481 (PL) from the Guangzhou Science and Technology Plan Project, research grant 202102010074 (TZ) from the Guangzhou Science and Technology Plan Project School (Institute) Joint Funding Project, research grant RHPG05 from the Guangzhou Research Hospital Construction Project, research grant 2023A03J0524 (YH) from the Guangzhou Science and Technology Plan Project School (Institute) Joint Funding Project, and research grant from the 2018 Project Funds of Guangzhou Red Cross Hospital.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Barbara, H., Mounia, S. M., Romain, B., and Jean-Paul, M. (2022). Autologous platelet-rich plasma/thrombin gel combined with split-thickness skin graft to manage postinfectious skin defects: a randomized controlled study. Adv. skin wound care 30 (11). doi:10.1097/01.ASW.0000524399.74460.87
Bohumila, P., Eva, C., Jana, D., Miroslav, Š., Libor, K., Ognen, P. G., et al. (2023). Soft hydrogels with double porosity modified with rgds for tissue engineering. Macromol. Biosci. doi:10.1002/mabi.202300266
Chang, S., Li, L., Hong, B., Jing, L., Yuxuan, X., Keliang, P., et al. (2023). An intelligent workflow for sub-nanoscale 3D reconstruction of intact synapses from serial section electron tomography. BMC Biol. 21 (1). doi:10.1186/s12915-023-01696-x
Cuesta, P. A., Musa, A., Songul, U., Eray, A., Mehmet, M. G., Burak, A., et al. (2023). Fucoidan-loaded electrospun Polyvinyl-alcohol/Chitosan nanofibers with enhanced antibacterial activity for skin tissue engineering. J. Mech. Behav. Biomed. Mater., 148. doi:10.1016/j.jmbbm.2023.106163
Erbing, Y., Fei, C., Meiqing, W., Hang, C., Rong, L., and Patty, H. (2023). Local property of depth information in 3D images and its application in feature matching. Mathematics 11 (5). doi:10.3390/math11051154
Ewa, K., Agnieszka, L., Marcin, G., Anna, N., Anna, Ś. C., Alicja, R. K., et al. (2023a). Correlation between porosity and physicochemical and biological properties of electrospinning PLA/PVA membranes for skin regeneration. Biomater. Adv., 152. doi:10.1016/j.bioadv.2023.213506
Ewa, K., Agnieszka, L., Marcin, G., Anna, N., Anna, Ś-C., Alicja, R. K., et al. (2023b). Correlation between porosity and physicochemical and biological properties of electrospinning PLA/PVA membranes for skin regeneration. Biomater. Adv., 152. doi:10.1016/j.bioadv.2023.213506
Feng, S., and Qiang, H. (2021). Effects of allogenic acellular dermal matrix combined with autologous razor-thin graft on hand appearance and function of patients with extensive burn combined with deep hand burn. Int. Wound J. 18 (3). doi:10.1111/iwj.13532
Ghazal, B., Masoumeh, H. N., and Mehran, S. (2022). Fabrication and evaluation of combined 3D printed/pamidronate-layered double hydroxides enriched electrospun scaffolds for bone tissue engineering applications. Appl. Clay Sci., 225. doi:10.1016/j.clay.2022.106538
Jeyachandran, D., and Cerruti, M. (2023). Glass, ceramic, polymeric, and composite scaffolds with multiscale porosity for bone tissue engineering. Adv. Eng. Mater. 25 (17). doi:10.1002/ADEM.202201743
Jie, X., Huan, F., Ya, S., Yue, K., Deli, X., YuenYee, C., et al. (2022). A 3D bioprinted decellularized extracellular matrix/gelatin/quaternized chitosan scaffold assembling with poly(ionic liquid)s for skin tissue engineering. Int. J. Biol. Macromol., 220. doi:10.1016/j.ijbiomac.2022.08.149
Kiemen, A. L., Braxton, A. M., Grahn, M. P., Kyu, S. H., Jaanvi, M. B., Rebecca, R., et al. (2022). CODA: quantitative 3D reconstruction of large tissues at cellular resolution. Nat. Methods 19 (11). doi:10.1038/s41592-022-01650-9
Liu, P., Zhu, J. Y., Tang, B., and Hu, Z. C. (2018). Three-dimensional digital reconstruction of skin epidermis and dermis. J. Microsc. 270 (2). doi:10.1111/jmi.12671
Maria, E. M., Chian, C., Jeniree, M., Perry, T., and Kyriakos, K. (2022). Novel electrospun polycaprolactone/calcium alginate scaffolds for skin tissue engineering. Materials 16 (1). doi:10.3390/ma16010136
Maryam, K., and Bita, G. (2023). Basic aspects of skin tissue engineering: cells, biomaterials, scaffold fabrication techniques, and signaling factors. J. Med. Biol. Eng. 43 (5). doi:10.1007/s40846-023-00822-y
Muro, S., and Akita, K. (2023). Novel combination method of wide-range serial sectioning and 3D reconstruction visualizing both macro-level dynamics and micro-level interactions in an attempt to analyze the female pelvic floor. Anat. Sci. Int. 98 (3). doi:10.1007/s12565-023-00710-0
Li, L., Guo, S. S., Mei, S. L., and Zhang, N. N (2015). Mosaic and repair method of locust slices based on feature extraction and matching. Editor. Office Trans. Chin. Soc. Agric. Eng. 31 (7). doi:10.3969/j.issn.1002-6819.2015.07.023
Narayanan, K. B., Bhaskar, R., Kim, H., and Sung, S. H. (2023). In vitro cytocompatibility assessment of novel 3D chitin/glucan- and cellulose-based decellularized scaffolds for skin tissue engineering. Sustainability 15 (21). doi:10.3390/su152115618
Nupur, G., Nan, L. Y., Wei, C. J., and Xiao, H. (2021). Highly robust interfacially polymerized PA layer on thermally responsive semi-IPN hydrogel: toward on-demand tuning of porosity and surface charge. ACS Appl. Mater. interfaces 13 (50). doi:10.1021/acsami.1c16639
Núria, B. P., and Joan, B. B (2023). Healing techniques for split-thickness skin grafts donor sites. Umbrella review. Enfermeria Clin. doi:10.1016/j.enfcle.2023.10.004
Olevsky, L. M., Anup, A., Jacques, M., Nadia, K., Eric, P. H., and Katherine, R. H. (2023). Direct integration of 3D printing and cryogel scaffolds for bone tissue engineering. Bioengineering 10 (8). doi:10.3390/bioengineering10080889
Rasha, A. Y., Emad, A. A., and Mohammed, A. T. (2023). Role of porosity in the strength, dielectric properties, and bioactivity of hardystonite ceramic material for use in bone tissue engineering applications. Ceram. Int. 49 (24). doi:10.1016/j.ceramint.2023.10.029
Robab, G., Mehdi, J., Fatemeh, N., Alireza, A., Tizazu, H. M., and Ahmad, R. T. (2022). Fabrication and characterization of bilayer scaffolds - nanocellulosic cryogels - for skin tissue engineering by co-culturing of fibroblasts and keratinocytes. Int. J. Biol. Macromol. 223. doi:10.1016/j.ijbiomac.2022.10.281
Sarah, C. W., Layla, R., and Gymama, S. (2023). Advances in tissue engineering and biofabrication for in vitro skin modeling. Bioprinting, 35. doi:10.1016/j.bprint.2023.e00306
Shila, N., Morteza, E., Hossein, D., Mehdi, J., and Bakhshali, M. (2023). Biomimetic electroactive nanofibrous hydrogel scaffolds based on polythiophene-grafted tragacanth gum and poly(vinyl alcohol) for skin tissue engineering application. Mater. Today Commun., 37. doi:10.1016/j.ijbiomac.2023.126041
Sorour, J., Majid, K., Farid, N., and Hassan, E. D. (2021). Preparation and characterization of sodium alginate-PVA polymeric scaffolds by electrospinning method for skin tissue engineering applications. RSC Adv. 11 (49). doi:10.1039/d1ra04176b
Taghizadeh, M. L., Arash, A., Asadollah, A., and Saber, Z. (2023). Characterization and biocompatibility evaluation of acellular rat skin scaffolds for skin tissue engineering applications. Cell tissue Bank., doi:10.1007/s10561-023-10109-w
Wang, Y., Xu, R., He, W., Zhihui, Y., Haisheng, L., Junyi, Z., et al. (2015). Three-dimensional histological structures of the human dermis. Tissue Eng. Part C Methods 21 (9), 932–944. doi:10.1089/ten.tec.2014.0578
Meng, X., Chen, S., Jun, Z., Zhipeng, W., Zhiwei, R., Stefan, P., et al. (2023). Improved HardNet and stricter outlier filtering to guide reliable matching. Comput. Mater. Continua 75 (3). doi:10.32604/CMC.2023.034053
Yawen, L., Ching-Cheng, H., Yuanyuan, W., Jun, X., GuoDing, W., and XinPeng, B. (2021). Biological evaluations of decellularized extracellular matrix collagen microparticles prepared based on plant enzymes and aqueous two-phase method. Regen. Biomater. 8 (2). doi:10.1093/rb/rbab002
Youngnam, K., Miji, Y., Deniz, D. I., Dino, J. R., Yogendra, P. S., Mecit, A. A, et al. (2024). Intraoperative bioprinting of human adipose-derived stem cells and extra-cellular matrix induces hair follicle-like downgrowths and adipose tissue formation during full-thickness craniomaxillofacial skin reconstruction. Bioact. Mater., 33. doi:10.1016/j.bioactmat.2023.10.034
Keywords: 3D structure, serial histological sections, dermis, image registration, porosity, average pore diameter
Citation: Liu P, Zhang T and Huang Y (2024) Three-dimensional model of normal human dermal tissue using serial tissue sections. Front. Bioeng. Biotechnol. 12:1347159. doi: 10.3389/fbioe.2024.1347159
Received: 01 December 2023; Accepted: 15 February 2024;
Published: 06 March 2024.
Edited by:
Antonella Motta, University of Trento, ItalyReviewed by:
Sônia Maria Malmonge, Federal University of ABC, BrazilCopyright © 2024 Liu, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yihui Huang, aHloMjAyMzI2MDRAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.