- 1Top Runner Incubation Center for Academia-Industry Fusion, Nagaoka University of Technology, Nagaoka, Japan
- 2Department of Mechanical Engineering, Nagaoka University of Technology, Nagaoka, Japan
Gene expression is a fundamental process that regulates diverse biological activities across all life stages. Given its vital role, there is an urgent need to develop innovative methodologies to effectively control gene expression. Light-controlled gene expression is considered a favorable approach because of its ability to provide precise spatiotemporal control. However, current light-controlled technologies rely on photosensitive molecular tags, making their practical use challenging. In this study, we review current technologies for light-controlled gene expression and propose the development of label-free light-controlled technologies using mid-infrared (mid-IR) and terahertz light.
1 Introduction
Light-controlled technologies have enabled researchers to control gene expression with spatiotemporal precision, leading to remarkable advancements in fundamental biological research and medical and pharmaceutical applications (Ryu et al., 2021; Jia and Sletten, 2022). This approach is ideal for studying biology because traditional methods that rely on chemical inducers or genetic modifications have inherent limitations in terms of specificity, reversibility, and control of the timing and location of gene expressions (Khalil, 2020; Ziegler et al., 2022). Photosensitive molecules have been modified into nucleotides, peptides, and small molecules to control in vivo and in vitro gene expression (Hoorens and Szymanski, 2018). Classically, ultraviolet (UV) light has been the main light source for activating photosensitive molecules because of its high energy and wide range. However, UV light exhibits shallow penetration in tissues and damages biological molecules, making it less ideal for biomedical applications (Sharma and Friedman, 2021). The recent development of visible light-reactive molecules has led to improvements in biocompatibility and expanded applications for cells (Dong et al., 2015; Weinstain et al., 2020). Moreover, much weaker light, such as near-infrared light (NIR), is promising for controlling biological activities due to less damage of biological molecules and better light penetration in tissues (Feng et al., 2021; Chen et al., 2020).
Although light-controlled gene expression-modulating technologies have become favorable for the regulation and study of biological systems, there are challenges. One of the challenges in conventional approaches is the requirement for the chemical conjugation of photosensitive molecules to target biological molecules. Specifically, researchers need to examine the target biological molecule structure and select suitable photosensitive molecules for the target process, as well as design a protocol for photosensitive molecule attachment that can yield a sufficient amount of the high-purity product. Another challenge is that the addition of photosensitive molecules for regulating gene expression using cell-free systems or living cells may interfere with enzyme activity, necessitating system optimization. For example, an appropriate laser power has to be chosen because a weak power may not trigger a photoreaction and a strong power can damage biological molecules, potentially leading to undesired photocatalysis. Therefore, optimization of parameters such as the chemical components and incubation conditions is necessary.
The main requirements of light-controlled gene expression within a completely natural and untreated system are a chemical component that can uniquely absorb the excitation light and that the light itself does not damage biological molecules. Considering these, a wavelength in the range of mid-infrared (mid-IR) to terahertz would be desirable because this range of light is absorbed in molecular vibration modes, which are specific to the molecules themselves. In this study, to aid the development of label-free light-controlled gene expression regulation systems, we review the potential of using mid-IR–terahertz wavelengths for regulating gene expression for fundamental research and practical applications. First, we discuss how light can interact with materials as the underlying concept in light-regulated technologies and introduce the common photosensitive molecule modification approaches, including photocages and photoswitches. Second, we outline the use of visible light and the NIR region for activating photosensitive DNA/RNA, proteins, and small molecules for regulating gene expression and discuss the challenges of the current light-controlled technologies, not at the life level, but at the basic conceptual level. Finally, we introduce the recently developed light-controlled technology utilizing mid-IR and terahertz wavelengths and address the challenges of this approach.
2 Photosensitive molecules: photocages and photoswitches
2.1 Role of light and photosensitive molecules in gene expression regulation
Light is an electromagnetic wave that can interact with the molecules in the medium that it traverses (Gutzler et al., 2021). Light can be specifically made to interact with photoactive molecules to modulate gene expression (Hoorens and Szymanski, 2018). Upon the absorption of light, these molecules undergo specific photochemical reactions including cleavage and crosslinking, leading to chemical modifications, bond formation, and changes in their molecular structure (Tavakoli and Min, 2022). Light-controlled technologies provide a non-invasive approach to study and manipulate gene expression with high precision and selectivity. In this approach, a wide range of light can be applied, from UV to NIR and beyond. The photochemical reaction mechanism depends on the wavelength of light: UV and visible light (200–900 nm) leads to electronic transitions, while mid-IR/terahertz light (2.5 µm–3 mm) results in vibrational transitions, affecting intramolecular/intermolecular vibrations (Figure 1A). Two common strategies have been developed using photosensitive molecules: photocages and photoswitches. The following sections describe these strategies. For the ideal design of photosensitive molecules, the following characteristics are required (Singh et al., 2021): first, photosensitive molecules should show high light absorption at wavelengths that are not absorbed by or are not damaging to other biological molecules. In addition, the photoreaction should show high efficiency. Second, photosensitive molecules should have low intrinsic activity and should be stable in the reaction medium before light irradiation. Third, the by-products of the photoreaction should be transparent to the photoreactive trigger light to suppress other competitive reactions that may interfere with the designed photoreaction, and they should not react with molecules in the reaction medium. Furthermore, some photochemical reactions undergo several steps that involve excited/ground-state intermediates; therefore, the detailed mechanism of the photochemical reactions should be studied to understand the correct light irradiation.
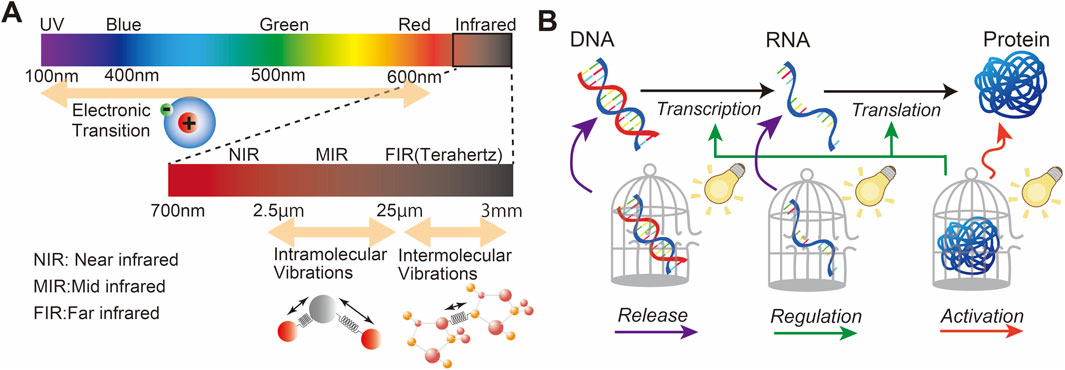
Figure 1. (A) Light wavelength for the excitation of electron transition, intramolecular vibrations, and intermolecular vibrations. (B) Overview of the basic biological process underlying gene expression and possible pathways for light-controlled gene expression.
Gene expression involves various biological processes, including transcription, where the DNA sequence of a gene is copied into complementary RNA, and translation, where messenger RNA (mRNA) serves as a template to assemble amino acids into polypeptide chains to form proteins. Proteins act as enzymes that modulate cellular metabolism and chemical reactions. Although light itself does not play an active role in this process, it can, with the assistance of photosensitive molecules, regulate a wide range of biological processes, including enzymatic activity. Figure 1B shows an example of this concept using photosensitive molecules of DNA, RNA, and proteins for light-controlled gene expression, which is further reviewed in Section 3.
2.2 Photocage
A photocage is a photoremovable protective molecule that blocks the bioactivity of the biological molecule to which it is conjugated. Photocage molecules absorb specific wavelengths of light, typically in the UV or visible range, and undergo a photoreaction that results in their removal or cleavage. This process releases the bioactive molecules and initiates their bioactivity. Popular photocages include boron–dipyrromethene (BODIPY), heptamethine cyanine (Cy7), and coumarin.
BODIPY is a fluorescence dye commonly used in bioimaging for biological studies and medical applications because it has low biotoxicity, is highly stable in various medium conditions, and has outstanding optical properties, such as high quantum yield, high absorption coefficient, narrow fluorescence spectrum, and long fluorescence lifetime (Singh et al., 2021). Additionally, structural modifications of BODIPYs can tune the absorption band to include the NIR region (Liu et al., 2019). However, the photolysis of some BODIPYs occasionally results in reduced photoreactivity (Singh et al., 2021), and real-time monitoring of the released active biomolecules is not feasible (Paul et al., 2019). Figure 2A shows an example of the photochemical reaction of BODIPY, where light irradiation initiates the cleavage of the cargo, resulting in a carbocation intermediate with a solvent-assisted nucleophilic attack (Peterson et al., 2018). A variety of BODIPY derivatives have been discussed in previous reviews (Singh et al., 2021; Cheng et al., 2023; Bobrov et al., 2021). Various types of caging groups can be used to create BODIPY photocages depending on the specific application and desired photorelease mechanisms. Some caging groups that are commonly attached to the BODIPY core include carboxylic acid, amine, alcohol, thiol, halide, xanthane, and phenol (Vohradská et al., 2018; Dockalova et al., 2020; Sitkowska et al., 2018).
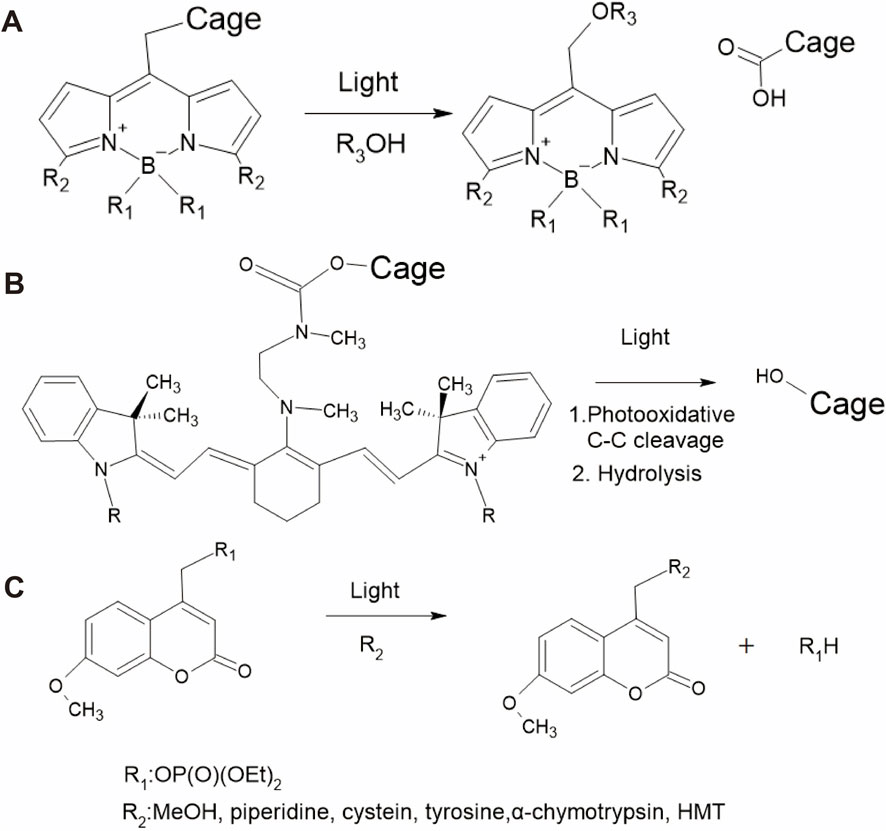
Figure 2. Reaction pathways of different photocage molecules. (A) BODIPY, (B) heptamethine cyanine (Cy7), and (C) 7-methoxycoumarin derivatives.
Cy7 is another dye that is frequently used in bioimaging because it binds to biological molecules, including DNA, RNA, and proteins (Luciano et al., 2019; Rozovsky et al., 2019; Šmidlehner et al., 2018). The advantages of Cy7 are its low toxicity, low background absorbance interference, and high NIR light absorption (Yamamoto et al., 2019). Some Cy7 derivatives have been engineered to function in vivo by developing NIR-absorbing photocage molecules that are less harmful to tissues (Alachouzos et al., 2022; Janeková et al., 2022). The general photoreaction of the Cy7 photocage is shown in Figure 2B, where photooxidative cleavage at the C-C bond and hydrolysis result in the release of the caged molecule (Weinstain et al., 2020; Gorka et al., 2014).
Coumarins are widely used because they can be easily synthesized and rapidly released from substrates. As an example, the reaction of a photoactivatable phosphate-releasing group using 7-methoxycoumarin derivatives is shown in Figure 2C (Tavakoli and Min, 2022; Givens and Matuszewski, 1984). Using structural modifications, the photophysical properties of coumarins such as quantum yield and aqueous solubility can be improved (Bardhan and Deiters, 2019). The advantages of coumarin are its high absorption coefficient, high photoresponse efficiency, fast photolysis kinetics, and suitability for engineering two-photon experiments. Typically, coumarins can be connected to caging groups such as carbonates, alkoxides, carbamates, thiols, sulfonates, azides, halides, phosphates, and carboxylates (Weinstain et al., 2020).
2.3 Photoswitch
Photoswitches are photochromic molecules that undergo reversible conversion between two or more stable states under light irradiation. This photochemical isomerization process involves structural changes that result in distinct differences in the UV–visible absorption spectra of the isomeric states, making them photochromic. Mostly, photoswitch molecules show positive photochromism, with the generated species showing a higher maximum absorption (λmax) than the initial state (Bouas-Laurent and Dürr, 2001). However, when the initial molecules undergo bleaching upon photoisomerization, the photoswitch molecules show negative photochromism. Examples of such photoswitch molecules include azobenzenes, stilbenes, and spiropyrans.
Azobenzene is a well-studied photoswitch that exists in two distinct isomeric states: trans and cis (Axelrod et al., 2022; Volarić et al., 2022). They are widely used in biological applications because of their ease of synthesis, high quantum yield, low photobleaching, high photostationarity, and fast isomerization (Fuentes et al., 2020). Notably, trans-azobenzene is approximately 10 kcal/mol more stable than cis-azobenzene. The UV–visible absorption of trans-azobenzene exhibits two maximum peaks: a strong absorption peak near 320 nm due to a π–π* transition and a weaker peak absorption near 440 nm related to an n−π* transition (Gelabert et al., 2023; Kuntze et al., 2021). cis-azobenzene has a stronger absorption band of the π–π* transition near 400 nm and two shorter absorption bands at 280 and 250 nm (Dias et al., 1992). Absorption near 320 nm induces the rotation of the nitrogen double bond, leading to the formation of the non-polar cis isomer, whereas absorption at 440 nm is associated with trans-to-cis isomerization through various pathways (Giles et al., 2021). Figure 3A shows the structures of trans- and cis-azobenzene (Cattaneo and Persico, 1999). Thermal stimulation or visible light irradiation can induce a switch from the cis to trans isomer, which requires ∼95 kJ mol−1 of activation energy (Garcia-Amorós et al., 2018).
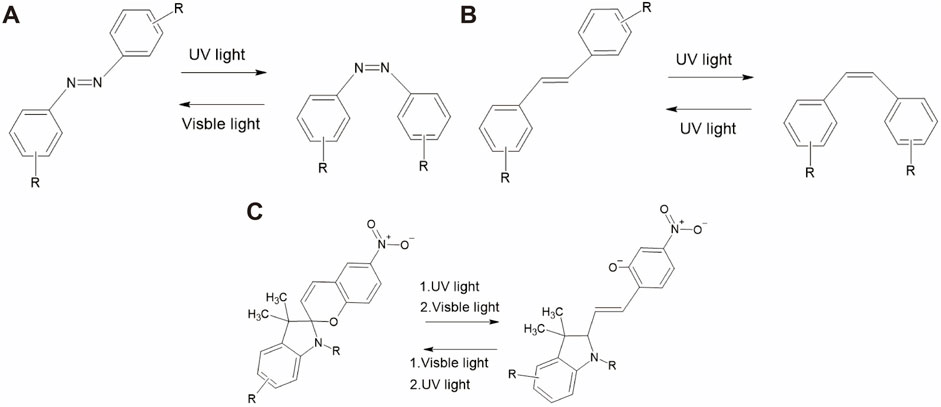
Figure 3. Reaction pathways of different photoswitch molecules. (A) Azobenzene, (B) stilbene, and (C) spiropyran.
Stilbene is another photoswitch molecule that undergoes trans–cis isomerization (Villarón et al., 2021). It is a hydrocarbon comprising two phenyl rings connected by an ethylene (–CH=CH–) bridge that forms a central double bond. The photoisomerization mechanism of stilbene has been reported to be slightly different from that of azobenzene. The main reason is that the excited state of the trans-stilbene, initiated through the π−π* transition, is metastable (Han et al., 2002). Isomerization of unsubstituted stilbene occurs under irradiation near 300 nm, resulting in the formation of the cis isomer. An example of this transition is shown in Figure 3B. The thermal cis-to-trans reisomerization requires 41–46 kcal/mol, which cannot be achieved at room temperature. In terms of biological applications, the disadvantage of stilbene is its irreversible cyclization and oxidization to the cis isomer.
Spiropyran undergoes a photochromic reaction under UV irradiation, which induces heterocyclic cleavage at the C-O bond, leading to the formation of a zwitterionic structure, as shown in Figure 3C, an example of the transition pathway (Kortekaas and Browne, 2019). This isomerization process produces a significant polarity shift (8–15D) (Mukherjee et al., 2022; Shiraishi et al., 2023). More importantly, this process is reversible upon thermal stimulation and photochemical activation under visible light irradiation (>460 nm). The importance of spiropyran is that photoisomerization causes a significant change in polarity, which influences its hydrophilicity/hydrophobicity (Wang et al., 2021). Spiropyran has been reported to strongly interact with certain biological molecules (Keyvan Rad et al., 2022; Ali et al., 2019).
2.4 Challenges in using photosensitive molecules
There are many candidates for photosensitive molecules that can be attached to target molecules. However, the challenge lies in the need to use specific attachment reactions, which limits the number of available pairs of photosensitive molecules and target molecules, or to implement tag modification. For example, prior to attachment, many photoswitch molecules are pre-labeled with protein tags such as SNAP tags, which can fuse with any target protein (Leng et al., 2017). Additionally, the modification process can be quite time-consuming. As an example, we analyzed the duration of photosensitive molecular modification based on protocol journal papers by picking up all related processes, as shown in Figure 4 (Becker et al., 2018; Cheong et al., 2024; Coelho et al., 2020; Conic et al., 2018; Erlendsson et al., 2019; Fields et al., 2019; Forero-Quintero et al., 2022; Gáspár et al., 2018; Horii et al., 2017; Kajimoto and Nakamura, 2018; Klena et al., 2023; Koch et al., 2021; Luo et al., 2023; Mai-Morente et al., 2021; Mazzucco et al., 2020; Mu et al., 2019; Munson and Ganley, 2016; Paweletz et al., 2022; Rawat and Sharma, 2024; Remenyi et al., 2019; Roberts et al., 2018; Sharma et al., 2019; Tanimizu, 2015; Teng et al., 2019; Tronnet and Oswald, 2018; Udupa et al., 2022; Wirth et al., 2020; Xu and Liu, 2019; York and Milush, 2015). The results indicate that the entire process, including preparation and related procedures, typically takes at least a day, with some cases requiring even more time.
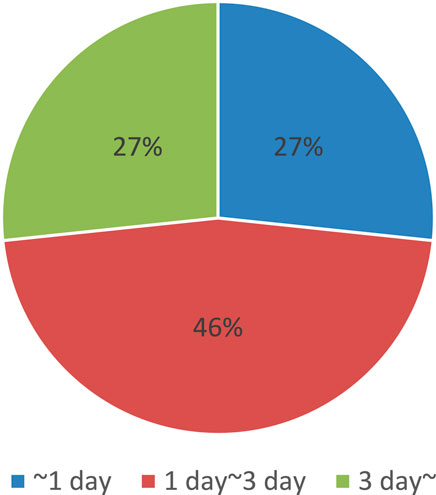
Figure 4. Summary of time required for photosensitive molecular modification, obtained from references (Becker et al., 2018; Cheong et al., 2024; Coelho et al., 2020; Conic et al., 2018; Erlendsson et al., 2019; Fields et al., 2019; Forero-Quintero et al., 2022; Gáspár et al., 2018; Horii et al., 2017; Kajimoto and Nakamura, 2018; Klena et al., 2023; Koch et al., 2021; Luo et al., 2023; Mai-Morente et al., 2021; Mazzucco et al., 2020; Mu et al., 2019; Munson and Ganley, 2016; Paweletz et al., 2022; Rawat and Sharma, 2024; Remenyi et al., 2019; Roberts et al., 2018; Sharma et al., 2019; Tanimizu, 2015; Teng et al., 2019; Tronnet and Oswald, 2018; Udupa et al., 2022; Wirth et al., 2020; Xu and Liu, 2019; York and Milush, 2015).
Additionally, light absorption by photosensitive molecules is limited (Welleman et al., 2020). For practical use, sufficient light penetration through the cells and tissues without causing damage is ideal. Nevertheless, most photosensitive molecules have not yet been engineered to operate at longer wavelengths, such as NIR light (Jia and Sletten, 2022).
Furthermore, some photosensitive molecules show inherent poor water solubility, which might cause biomolecular aggregation and result in interference with gene expression (Deng et al., 2021; Poryvai et al., 2022; Berdnikova, 2019). For instance, traditional BODIPY dyes dissolve only in organic solvents, which is why, to date, tremendous efforts have been made to improve their water solubility by introducing hydrophilic groups such as phosphonates, quaternary ammonium salts, and sulfonates (Koh et al., 2019; Mao et al., 2020; Zhou et al., 2020).
Finally, the development of a new photosensitive molecule design is likely serendipitous (Deng et al., 2021; Dcona et al., 2020). Therefore, laborious efforts are often required to develop photosensitive molecules that are suitable for any untried gene expression system of interest (Buglioni et al., 2022; Roßmann et al., 2023).
3 Current light-controlled gene expression technology
In this section, we review how light irradiation can be used to control gene expression. As shown in Figure 1B, light irradiation can activate photosensitive DNA/RNA, proteins, and small molecules that influence gene expression; these categories are discussed in the following sections.
3.1 DNA/RNA-based light-controlled gene expression
The photocontrol of gene expression using nucleic acids is typically facilitated by photosensitive molecule-modified DNA or RNA, as shown in Figure 1B. There are two main strategies for this approach: light-induced gene activation and light-induced knockdown using photocage molecule-modified oligonucleotides.
Active light control of transcription and translation has been demonstrated by modifying oligonucleotides with photocage molecules. The modifications in the oligonucleotides can be introduced into the phosphate backbone, Watson–Crick face, or nucleotide base. For example, gene expression in cells and synthetic cells has been controlled using photolabile groups on the phosphate backbone of the DNA and RNA modified with photosensitive molecules, including coumarin (Kaufmann et al., 2023; Ando et al., 2001), 2-nitroveratryl bromide (Hartmann and Booth, 2023), and thioether-enol phosphate (Feng et al., 2017; Wang et al., 2016). Other than at the DNA backbone, psoralen cross-linking at the Watson–Crick face of the DNA promoter impedes the unwinding of the double helix, resulting in the blocking of the transcription process (Figure 5A) (Stafforst and Stadler, 2013; Stadler and Stafforst, 2014). Photosensitive molecules can also be introduced at nucleotide bases. Nucleotides modified with photosensitive molecules, such as benzophenones (Anhäuser et al., 2020), diethylaminocoumarin (Menge and Heckel, 2011), 2-nitrobenzyl bromide (Chakrapani et al., 2020), and 6-nitropiperonyloxymethyl group (Lee et al., 2021), have been integrated into DNA or RNA templates for light-controlled transcription and translation. When photoswitch molecules are modified into DNA/RNA, they can reversibly regulate the transcription and translation processes based on light irradiation. For instance, photoswitch molecules, including azobenzene (Tian et al., 2016; Xing et al., 2015; Dudek et al., 2018), stilbenes (O’Hagan et al., 2019; O’Hagan et al., 2020), arylstilbazolium (Czerwinska and Juskowiak, 2012), and 8-pyrenylvinyl deoxyguanosine (Ogasawara, 2018), which contain G-quadruplexes in a hyperstable state, can effectively inhibit transcription and translation processes. Light irradiation induces an unstable state, allowing activity, and additional light irradiation in the unstable state recovers hyperstable G-quadruplexes.
Figure 5. (A) Selective activation of psoralen-cross-linked DNA with UV and blue light (Stafforst and Stadler, 2013). (B) Photoactivation of siRNAs with red light for a non-toxic cellular approach (Meyer and Mokhir, 2014). (C) Spatiotemporal control of gene expression using photocaged T7 RNA polymerase (Hemphill et al., 2013). (D) Light-responsive nanocarrier for precise CRISPR-Cas9-mediated gene editing (Pan et al., 2019). (E) Spatiotemporal control of gene expression using riboswitches and photocaged ligands (Walsh et al., 2014). All figures have been adapted with permission from John Wiley and Sons Copyright (2013) and (2014), American Chemical Society Copyright (2013), and Springer Nature Copyright (2019).
One approach to knock down gene expression is to use photocage-modified antisense oligonucleotides (ASOs), which are short single-stranded nucleotides that specifically bind to target DNA or RNA to stop transcription and translation. Modification of ASOs with 2-nitrobenzyl-caged thymidines has been shown to cause the photoactive knockdown of cancer-related genes (Govan et al., 2013; Tang et al., 2007; Sakamoto et al., 2014) and developmental genes (Tallafuss et al., 2012; Deiters et al., 2010). The inclusion of 2-nitrobenzyl can be achieved in the base-pairing region, nucleobase, and backbone of the complementary strand. Circularized ASOs can also be formed with photocleavable linkers, such as coumarin- (Yamazoe et al., 2014), 2-nitrobenzyl- (Yang et al., 2018), quinoline- (O’Connor et al., 2019), and Ru-BEP (Griepenburg et al., 2015), which allows the control of gene expression using light irradiation. Another approach is to use small interfering RNAs (siRNAs), which are approximately 20 base pairs of dsRNAs, that specifically suppress gene expression by destroying mRNA. By attaching 2-nitrobenzyl on the phosphate backbone and 5′ or 3′phosphate termini of siRNAs, light-controlled knockdown could be achieved in zebrafish (Blidner et al., 2008), mammalian cells (Zhang L. et al., 2018), and HeLa cells (Yu et al., 2018; Kala et al., 2014). When a 9-alkoxyanthracenyl fragment as an O2-sensitive moiety is incorporated in photocage molecules, the selectivity of the activation wavelength can be tuned by attaching photosensitizers to the 3′-terminus (Figure 5B) (Meyer and Mokhir, 2014). The use of light-responsive nanoparticles (NPs) is also a common approach for spatiotemporally manipulating siRNA functions. Typically, irradiation with NIR light triggers the release of thiol-modified siRNAs attached to gold NPs, resulting in the NIR-controlled knockdown of gene expression in cells (Huang et al., 2015; Riley et al., 2018; Braun et al., 2009). In addition, siRNA/mPEG-b-P(APNBMA) NPs containing o-nitrobenzyl moieties have been engineered to initiate siRNA release post-UV light irradiation (Foster et al., 2015).
3.2 Protein-based light-controlled gene expression
Photocontrol of proteins can affect the regulation of DNA/RNA synthesis or activity and the direct activation of protein function, as shown in Figure 1B, using photosensitive molecule-modified proteins.
The specific incorporation of amino acids with photosensitive molecules is required to generate photosensitive molecule-modified proteins in cells (Arbely et al., 2012; Chou and Deiters, 2011). One approach to demonstrate this incorporation is to select an amber stop codon as the insertion site for amino acids with photosensitive molecules (Wang et al., 2012; Neumann, 2012). Examples of this approach include the modification of a nitrobenzyl group on RNA polymerases to allow light-controlled gene expression in cells, as shown in Figure 5C (Hemphill et al., 2013; Chou et al., 2010). Additionally, other light-activated gene-editing tools, such as Cre recombinases (Edwards et al., 2009) and zinc finger (ZF) nucleases (Chou and Deiters, 2011), have been engineered by modifying o-nitrobenzyl on essential residues in catalytic sites, enabling activation upon light irradiation (Chou and Deiters, 2011; Luo et al., 2016). By fusing the GIGANTEA (GI) protein with the ZF protein, blue light irradiation induces heterodimerization between the GI and the light–oxygen–voltage (LOV) domain, which assists the transcriptional activation domain VP16 in initiating the transcription of the gene of interest (Polstein and Gersbach, 2012). Similarly, when the light-sensitive cryptochrome 2 (CRY2) protein is fused with transcription activator-like effector (TALE) proteins, blue light-induced conformational changes in CRY2 recruit a CIB1–effector domain, which exerts the active control of transcription in the same endogenous genome (Konermann et al., 2013). Furthermore, 2-nitrobenzyl-modified tamoxifen is used in light-controlled Cre systems to effectively regulate gene expression in cells (Inlay et al., 2013). Optical dimerization of the CRY2–CIB system has been widely used to reconstitute the split recombinase of Cre and Flp, which enables gene expression control in cells (Cautereels et al., 2024) and mouse brains (Jung et al., 2019).
The CRISPR-Cas9 system is also a well-known gene editing tool that can delete, replace, or insert any part of the genome sequence and has been extensively explored for the light-activated control of gene expression. Light-activated control of the CRISPR-Cas9 system has been achieved by engineering light-responsive Cas9 (Hemphill et al., 2015), gRNA (Moroz-Omori et al., 2020; Jain et al., 2016), and transcription factors (Shao et al., 2018). An example of an engineered Cas9 nuclease is the incorporation of a 2-nitrobenzyl-modified lysine amino acid, which reversibly affects gene function (Hemphill et al., 2015). As shown in Figure 5D, when Cas9 is bound to lanthanide-doped upconversion NPs by a 2-nitrobenzyl photocage, NIR irradiation produces local UV light, resulting in a photocleavage reaction and the release of the CRISPR-Cas9 system (Pan et al., 2019). Other candidate proteins, such as light-activated phosphorylation (Nguyen et al., 2018), dimeric green fluorescent protein (Zhou et al., 2018), and cyclic diguanylate monophosphate signaling cascades (Shao et al., 2018), have also been reported for use in light-activated CRISPR-Cas9 systems.
3.3 Small-molecule-based light-controlled gene expression
In addition to DNA, RNA, and proteins, small-molecule compounds are also general candidates for the photoreactive control of gene expression. Popular small molecules include nucleotides, peptides, and ligands. Nucleotides are the basic building blocks that constitute DNA and RNA; thus, the photocontrol of nucleotides is a straightforward approach for controlling gene expression. In this regard, [7-(diethylamino)coumarin-4-yl]methyl (DEACM)-modified ATP and 2-nitrobenzyl-modified UTP/GTP have been demonstrated as molecules to photocontrol the transcription reaction in vitro (Shao et al., 2018; Pinheiro et al., 2008). The modification of ligands with 2-nitrobenzyl and azobenzene, which effectively interact with DNA/RNA, is a useful approach for controlling gene expression (Walsh et al., 2014; Young et al., 2009; Paul et al., 2021). An example molecule is theophylline, which can specifically bind to the mRNA riboswitch; the photocontrol of gene expression could be achieved by modifying theophylline with 2-nitrobenzyl (Figure 5E) (Walsh et al., 2014). The binding affinity of tamoxifen and cyclophen to the estrogen receptor (ER)-binding domain can be utilized for photoactive regulation by modifying tamoxifen and cyclophen with 2-nitrobenzyl, coumarin, and cyanine derivatives (Zhang W. et al., 2018; Wong et al., 2017; Cruz et al., 2000). The tetracycline (Tet) system in mammalian cells is controlled by nitrobenzyl-modified doxycyclines (Cambridge et al., 2006). Many other small molecules, such as coumarin-modified cAMP-response element-binding protein (CREB) inhibitor (Imoto et al., 2020) and 2-nitrobenzyl-modified ecdysone and nuclear hormones (Link et al., 2004; Lin et al., 2002), have also been engineered to regulate gene expression in cells.
3.4 Challenges with current light-controlled gene expression approaches
To understand the nature of cells, photosensitive molecules can be used to probe fundamental aspects, such as molecular reactions, molecular kinetics, and live cell dynamics. However, the introduction of photosensitive molecules into cells or cell-free systems might influence molecular activity, thereby biasing the outcomes of the study. For example, exogenous photosensitive molecules in live cells can potentially harm normal cellular activities, including unintended effects on cell behavior, signaling pathways, and cellular homeostasis (Chen et al., 2022). Some photosensitive molecules may exhibit cytotoxic effects that affect cell viability and overall cellular health (Dcona et al., 2020). Therefore, researchers must carefully assess the concentration and exposure time required to minimize cytotoxicity.
As mentioned in Section 2.4, the introduction of photosensitive molecules is also a lengthy process. As shown in Figure 6, which summarizes the total time required for molecular introduction into cells using electroporation based on protocol journal papers by picking up all related processes, the process can be done within a day. However, in certain cases, it can take a significantly longer time (Alfastsen et al., 2021; Biase and Schettini, 2024; Chan et al., 2023; Clements et al., 2024; Crestani et al., 2022; Denoth-Lippuner et al., 2022; Egashira et al., 2023; Flumens et al., 2023; Gao et al., 2022; Gesuita et al., 2022; Heinz et al., 2020; Lankford and Hulleman, 2024; Laprie-Sentenac et al., 2023; Lax et al., 2022; Layden et al., 2021; Li et al., 2023; Ling et al., 2022; Meka et al., 2024; Nagahama et al., 2021; Nomura et al., 2020; Sakamoto et al., 2022; Sanketi et al., 2023; Skruber et al., 2021; Turchetto et al., 2020; Wang et al., 2020; Williams and Sauka-Spengler, 2021; Wilson et al., 2020; Wu et al., 2023; Xu et al., 2021; Xu et al., 2020; Zhou et al., 2021).
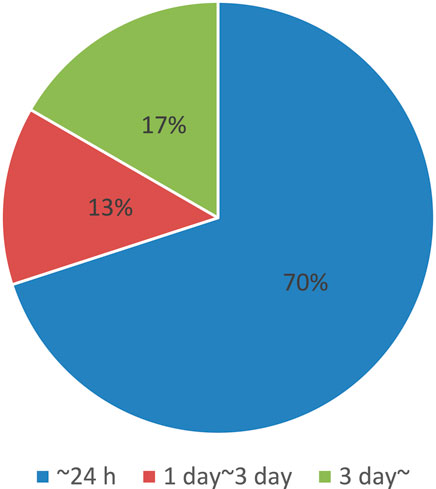
Figure 6. Summary of time required for electroporation, obtained from references (Alfastsen et al., 2021; Biase and Schettini, 2024; Chan et al., 2023; Clements et al., 2024; Crestani et al., 2022; Denoth-Lippuner et al., 2022; Egashira et al., 2023; Flumens et al., 2023; Gao et al., 2022; Gesuita et al., 2022; Heinz et al., 2020; Lankford and Hulleman, 2024; Laprie-Sentenac et al., 2023; Lax et al., 2022; Layden et al., 2021; Li et al., 2023; Ling et al., 2022; Meka et al., 2024; Nagahama et al., 2021; Nomura et al., 2020; Sakamoto et al., 2022; Sanketi et al., 2023; Skruber et al., 2021; Turchetto et al., 2020; Wang et al., 2020; Williams and Sauka-Spengler, 2021; Wilson et al., 2020; Wu et al., 2023; Xu et al., 2021; Xu et al., 2020; Zhou et al., 2021).
Furthermore, achieving high specificity and selectivity for target molecules can be challenging. Ideally, photosensitive molecules should interact only with the intended targets; however, off-target effects may occur, leading to the misinterpretation of the experimental results (Laczi et al., 2022). Moreover, efficient delivery of light-sensitive molecule-tagged biomolecules to specific locations in live cells is required to achieve the desired control of gene expression (Deng et al., 2020).
In addition, light irradiation can induce cellular stress and damage, which is known as phototoxicity. This is particularly relevant in live-cell imaging studies, in which prolonged exposure to light may lead to alterations in cellular behavior (Talone et al., 2021). Different cell types respond differently to photosensitive molecules (Powell et al., 2018). It is important to consider the specific characteristics of the cell type under investigation and validate the applicability of the chosen photosensitive molecule to the cell system of interest. The above discussion highlights that the development of light-regulated translation technologies requires addressing many challenges through enormous efforts, testing, and optimization to improve the effectiveness of the methodologies (Welleman et al., 2020).
4 Use of mid-IR–terahertz light for label-free light-controlled gene expression
Based on the challenges discussed above, we propose that mid-IR and terahertz light can be used as solutions for light-controlled gene expression regulation; this section discusses this proposal in detail.
4.1 Potential for using mid-IR–terahertz light for gene expression regulation
In principle, as described in Section 2.1, mid-IR and terahertz light (2.5 μm–3 mm) has a strong interaction with intramolecular/intermolecular vibrations (López-Lorente et al., 2016; Tonouchi, 2007; Ma et al., 2019; Hoshina et al., 2020). Therefore, mid-IR and terahertz light has been used as light sources for imaging with high special selectivity and sensitivity and molecular detection (Jain et al., 2020). For example, mid-IR photothermal imaging is an emerging technology that can effectively excite the target vibration modes in biological molecules. This results in local property changes, such as the refractive index, molecular volume, and vibration modes, and monitoring these changes allows a high spatial resolution with low photodamage to biological molecules (Samolis et al., 2023; Ishigane et al., 2023).
Since mid-IR and terahertz light can interact with specific molecular vibrations, the irradiation of these light sources can influence biological molecular activity; thus, there is potential to control specific gene expression. The strong light energy at this wavelength can influence molecular activity in a label-free manner (Morichika and Ashihara, 2022; Park et al., 2018; Windhorn et al., 2002). However, to the best of our knowledge, mid-IR and terahertz light has not yet been commonly used for the light control of gene expression. One of the reasons for this is the high light absorption of water in this wavelength range, which leads to an increase in ambient temperature and hampers biomolecular activity (Holmstrom and Nesbitt, 2010; Ashwood et al., 2020). A short-pulse laser can break this fundamental barrier because cyclical ultrashort-term heating and cooling suppress the increase in ambient temperature (Toda et al., 2019; Zhang et al., 2019). Interestingly, when analyzing the energy of mid-IR and terahertz light (2.5 µm–3 mm), which corresponds to 7.95 × 10−20 to 6.62 × 10−23 joule/photon, the energy of biological processes, such as hydrolysis of a peptide bond (−1.39 × 10−20 to −2.78 × 10−20 joule/molecule), ATP hydrolysis (−5.21 × 10−20 joule/molecule), and folding energies of RNA secondary structures (−2.78 × 10−20 to −1.11 × 10−19 joule/molecule), is not very different (Jia et al., 2021). In Figure 7A, Morichika et al. (2019) showed that the irradiation of a plasmonic structure using a mid-IR pulse laser enhanced the localized light field, resulting in CO dissociation of n-hexane mediated by vibrational ladder climbing (Morichika et al., 2019). Other plasmonic studies have showed that strong mid-IR light in a plasmon nanocavity perturbs a few-nm-thick shell of water, predicting the mid-IR and terahertz light interaction with the molecular reaction (Chikkaraddy et al., 2022). Even without plasmonic enhancement, pulsed mid-IR light can selectively induce vibrational excitation, allowing the activation energy barriers to be overcome and facilitating ground-state reactions with minimal heat generation in Figure 7B (Stensitzki et al., 2018). This can result in bimolecular alcoholysis reactions, bidirectional tautomerization of thiotropolone, and the formation of urethane and polyurethane (Stensitzki et al., 2018; Heyne and Kühn, 2019; Nunes et al., 2020).
Figure 7. (A) Plasmonic enhanced mid-IR light field dissociation of CO of n-hexane (Morichika et al., 2019). (B) Selective vibrational excitation-induced ground-state reactions by mid-IR light (Heyne and Kühn, 2019). (C) Terahertz light promoting the hydrophobic hydration around the protein (Sugiyama et al., 2023). All figures have been adapted with permission from American Chemical Society Copyright (2019) and Springer Nature Copyright (2019), (2023).
Although understanding the detailed mechanism of mid-IR and terahertz light irradiation on live cells is complicated, Toyama et al. (2022) showed that strong pulsed-light irradiation of E. coli cells in the mid-IR absorption region can affect their growth rate and survival. Furthermore, the much weaker energy of terahertz light influences the biological molecular activities. Recently, Sugiyama et al. (2023) discovered that the irradiation of pulsed terahertz light on proteins enhances hydrophobic hydration, leading to an increase in the number of hydrogen bonds at the hydration layer in Figure 7C, suggesting that even terahertz light can influence gene expression (Sugiyama et al., 2023). Although previous reports did not show clear findings, some previous studies have already predicted the influence of protein activity and expression (Sun et al., 2021). For example, Tan et al. found the downregulation of SYN expression in primary hippocampal neurons and PSD95 expression in cortical neurons under 0.16–0.17-THz irradiation, with a positive correlation with the exposure time and laser power (Zhi et al., 2019).
4.2 Challenges associated with mid-IR–terahertz light technology
Although some previous studies have shown the effect of mid-IR and terahertz light at the molecular level and presented some molecular reactions, many challenges remain for the active utilization of these wavelengths of light. First, the vibrational mode of molecules is influenced by a variety of factors, such as temperature, humidity, and substrates. Thus, an absorption spectrum shift of target biological molecules can occur in the environment, and researchers may need to calibrate the excitation wavelength. Second, as mentioned above, water absorption of mid-IR and terahertz light is quite high (100–104 cm−1); therefore, limited light penetration might be a critical issue when target samples are a large volume of biological molecules or tissues (Prahl, 2017). Third, instruments and optical elements for the mid-IR and terahertz regions are often expensive and not widely available for commercial purchase, necessitating researchers to develop a setup if cost considerations come into play. Fourth, targeting specific molecules is required to obtain absorption spectrum profiles using Fourier-transform infrared spectroscopy, which requires the preparation of a large volume of purified biological samples.
5 Conclusion
Here, we provide an overview of light-controlled gene expression and propose a label-free light control approach using mid-IR and terahertz light. First, we explain the interaction between light and the materials, offering insights into the selection of appropriate wavelengths. Second, we introduce common techniques for modifying photosensitive molecules, such as photocages and photoswitches, and review previous studies on light-controlled gene expression via the manipulation of photosensitive DNA/RNA, proteins, and small molecules. In addition, we discuss the technological challenges associated with the current technologies utilizing photosensitive molecules for the regulation of gene expression. Looking ahead, we explore the exciting prospect of extending label-free light-controlled technologies using the mid-IR and terahertz wavelength ranges, which are traditionally limited by water absorption, in biological applications. Because it allows the photocontrol of gene expression without any photosensitive molecular tag, this uncharted territory shows potential for groundbreaking innovations in the field of gene expression, contributing to medical and pharmaceutical development.
Author contributions
HY: conceptualization, funding acquisition, writing–original draft, and writing–review and editing. RS: writing–original draft, writing–review and editing. YT: writing–original draft, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. HY acknowledges the support of the Exploratory Research for Advanced Technology (ERATO, JPMJER2002) from the Japan Science and Technology Agency (JST).
Acknowledgments
We appreciate the support of the Exploratory Research for Advanced Technology and editing support of academic papers in English at Nagaoka University of Technology. We acknowledges Editage (https://campaigns.editage.jp/) and ChatGTP(https://chatgpt.com) to improve the language and readability of this paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alachouzos, G., Schulte, A. M., Mondal, A., Szymanski, W., and Feringa, B. L. (2022). Computational design, synthesis, and photochemistry of cy7-PPG, an efficient NIR-activated photolabile protecting group for therapeutic applications. Angew. Chem. Int. Ed. 61, e202201308. doi:10.1002/anie.202201308
Alfastsen, L., Peng, X., and Bhoobalan-Chitty, Y. (2021). Genome editing in archaeal viruses and endogenous viral protein purification. Star. Protoc. 2, 100791. doi:10.1016/j.xpro.2021.100791
Ali, A. A., Kang, M., Kharbash, R., and Kim, Y. (2019). Spiropyran as a potential molecular diagnostic tool for double-stranded RNA detection. BMC Biomed. Eng. 1, 6. doi:10.1186/s42490-019-0008-x
Ando, H., Furuta, T., Tsien, R. Y., and Okamoto, H. (2001). Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat. Genet. 28, 317–325. doi:10.1038/ng583
Anhäuser, L., Klöcker, N., Muttach, F., Mäsing, F., Špaček, P., Studer, A., et al. (2020). A benzophenone-based photocaging strategy for the N7 position of guanosine. Angew. Chem. Int. Ed. 59, 3161–3165. doi:10.1002/anie.201914573
Arbely, E., Torres-Kolbus, J., Deiters, A., and Chin, J. W. (2012). Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J. Am. Chem. Soc. 134, 11912–11915. doi:10.1021/ja3046958
Ashwood, B., Lewis, N. H. C., Sanstead, P. J., and Tokmakoff, A. (2020). Temperature-jump 2D IR spectroscopy with intensity-modulated CW optical heating. J. Phys. Chem. B 124, 8665–8677. doi:10.1021/acs.jpcb.0c07177
Axelrod, S., Shakhnovich, E., and Gómez-Bombarelli, R. (2022). Excited state non-adiabatic dynamics of large photoswitchable molecules using a chemically transferable machine learning potential. Nat. Commun. 13, 3440. doi:10.1038/s41467-022-30999-w
Bardhan, A., and Deiters, A. (2019). Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr. Opin. Struct. Biol. 57, 164–175. doi:10.1016/j.sbi.2019.03.028
Becker, Y., Green, K. A., Scott, B., and Becker, M. (2018). Artificial inoculation of epichloë festucae into lolium perenne, and visualisation of endophytic and epiphyllous fungal growth. Bio-protocol 8, e2990. doi:10.21769/bioprotoc.2990
Berdnikova, D. V. (2019). Design, synthesis and investigation of water-soluble hemi-indigo photoswitches for bioapplications. Beilstein J. Org. Chem. 15, 2822–2829. doi:10.3762/bjoc.15.275
Biase, F. H., and Schettini, G. (2024). Protocol for the electroporation of CRISPR-Cas for DNA and RNA targeting in Bos taurus zygotes. Star. Protoc. 5, 102940. doi:10.1016/j.xpro.2024.102940
Blidner, R. A., Svoboda, K. R., Hammer, R. P., and Monroe, W. T. (2008). Photoinduced RNA interference using DMNPE-caged 2′-deoxy-2′-fluoro substituted nucleic acidsin vitro and in vivo. Mol. Biosyst. 4, 431–440. doi:10.1039/b801532e
Bobrov, A. V., Kishalova, M. V., Merkushev, D. A., and Marfin, Y. S. (2021). BODIPY in matrices: brief review. J. Phys. Conf. Ser. 1822, 012020. doi:10.1088/1742-6596/1822/1/012020
Bouas-Laurent, H., and Dürr, H. (2001). Organic photochromism (IUPAC technical report). Pure Appl. Chem. 73, 639–665. doi:10.1351/pac200173040639
Braun, G. B., Pallaoro, A., Wu, G., Missirlis, D., Zasadzinski, J. A., Tirrell, M., et al. (2009). Laser-activated gene silencing via gold Nanoshell−siRNA conjugates. ACS Nano 3, 2007–2015. doi:10.1021/nn900469q
Buglioni, L., Raymenants, F., Slattery, A., Zondag, S. D. A., and Noël, T. (2022). Technological innovations in photochemistry for organic synthesis: flow chemistry, high-throughput experimentation, scale-up, and photoelectrochemistry. Chem. Rev. 122, 2752–2906. doi:10.1021/acs.chemrev.1c00332
Cambridge, S. B., Geissler, D., Keller, S., and Cürten, B. (2006). A caged doxycycline analogue for photoactivated gene expression. Angew. Chem. Int. Ed. 45, 2229–2231. doi:10.1002/anie.200503339
Cattaneo, P., and Persico, M. (1999). An abinitio study of the photochemistry of azobenzene. Phys. Chem. Chem. Phys. 1, 4739–4743. doi:10.1039/a905055h
Cautereels, C., Smets, J., De Saeger, J., Cool, L., Zhu, Y., Zimmermann, A., et al. (2024). Orthogonal LoxPsym sites allow multiplexed site-specific recombination in prokaryotic and eukaryotic hosts. Nat. Commun. 15, 1113. doi:10.1038/s41467-024-44996-8
Chakrapani, A., Vaňková Hausnerová, V., Ruiz-Larrabeiti, O., Pohl, R., Krásný, L., and Hocek, M. (2020). Photocaged 5-(hydroxymethyl)pyrimidine nucleoside phosphoramidites for specific photoactivatable epigenetic labeling of DNA. Org. Lett. 22, 9081–9085. doi:10.1021/acs.orglett.0c03462
Chan, D. K. H., Collins, S. D., and Buczacki, S. J. A. (2023). Generation and immunofluorescent validation of gene knockouts in adult human colonic organoids using multi-guide RNA CRISPR-Cas9. Star. Protoc. 4, 101978. doi:10.1016/j.xpro.2022.101978
Chen, G., Cao, Y., Tang, Y., Yang, X., Liu, Y., Huang, D., et al. (2020). Advanced near-infrared light for monitoring and modulating the spatiotemporal dynamics of cell functions in living systems. Adv. Sci. 7, 1903783. doi:10.1002/advs.201903783
Chen, W., Li, C., Liang, W., Li, Y., Zou, Z., Xie, Y., et al. (2022). The roles of optogenetics and technology in neurobiology: a review. Front. Aging Neurosci. 14, 867863. doi:10.3389/fnagi.2022.867863
Cheng, H.-B., Cao, X., Zhang, S., Zhang, K., Cheng, Y., Wang, J., et al. (2023). BODIPY as a multifunctional theranostic reagent in biomedicine: self-assembly, properties, and applications. Adv. Mater. 35, 2207546. doi:10.1002/adma.202207546
Cheong, S.-S., Luis, T. C., Hind, M., and Dean, C. H. (2024). A novel method for floxed gene manipulation using TAT-cre recombinase in ex vivo precision-cut lung slices (PCLS). Bio-protocol 14, e4980. doi:10.21769/bioprotoc.4980
Chikkaraddy, R., Xomalis, A., Jakob, L. A., and Baumberg, J. J. (2022). Mid-infrared-perturbed molecular vibrational signatures in plasmonic nanocavities. Light Sci. and Appl. 11, 19. doi:10.1038/s41377-022-00709-8
Chou, C., and Deiters, A. (2011). Light-activated gene editing with a photocaged zinc-finger nuclease. Angew. Chem. Int. Ed. 50, 6839–6842. doi:10.1002/anie.201101157
Chou, C., Young, D. D., and Deiters, A. (2010). Photocaged T7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells. ChemBioChem 11, 972–977. doi:10.1002/cbic.201000041
Clements, M., Simpson Ragdale, H., Garcia-Diaz, C., and Parrinello, S. (2024). Generation of immunocompetent somatic glioblastoma mouse models through in situ transformation of subventricular zone neural stem cells. Star. Protoc. 5, 102928. doi:10.1016/j.xpro.2024.102928
Coelho, C., Vij, R., Smith, D. Q., Brady, N. R., Hamacher-Brady, A., and Casadevall, A. (2020). Study of microbial extracellular vesicles: separation by density gradients, protection assays and labelling for live tracking. Bio-protocol 10, e3502. doi:10.21769/bioprotoc.3502
Conic, S., Desplancq, D., Tora, L., and Weiss, E. (2018). Electroporation of labeled antibodies to visualize endogenous proteins and posttranslational modifications in living metazoan cell types. Bio-protocol 8, e3069. doi:10.21769/bioprotoc.3069
Crestani, M., Dini, T., Gauthier, N. C., and Monzo, P. (2022). Protocol to assess human glioma propagating cell migration on linear micropatterns mimicking brain invasion tracks. Star. Protoc. 3, 101331. doi:10.1016/j.xpro.2022.101331
Cruz, F. G., Koh, J. T., and Link, K. H. (2000). Light-activated gene expression. J. Am. Chem. Soc. 122, 8777–8778. doi:10.1021/ja001804h
Czerwinska, I., and Juskowiak, B. (2012). Photoisomerizable arylstilbazolium ligands recognize parallel and antiparallel structures of G-quadruplexes. Int. J. Biol. Macromol. 51, 576–582. doi:10.1016/j.ijbiomac.2012.06.027
Dcona, M. M., Mitra, K., and Hartman, M. C. T. (2020). Photocontrolled activation of small molecule cancer therapeutics. RSC Med. Chem. 11, 982–1002. doi:10.1039/d0md00107d
Deiters, A., Garner, R. A., Lusic, H., Govan, J. M., Dush, M., Nascone-Yoder, N. M., et al. (2010). Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and Xenopus embryos. J. Am. Chem. Soc. 132, 15644–15650. doi:10.1021/ja1053863
Deng, P., Xiao, F., Wang, Z., and Jin, G. (2021). A novel BODIPY quaternary ammonium salt-based fluorescent probe: synthesis, physical properties, and live-cell imaging. Front. Chem. 9, 650006. doi:10.3389/fchem.2021.650006
Deng, Z., Wang, N., Liu, Y., Xu, Z., Wang, Z., Lau, T.-C., et al. (2020). A photocaged, water-oxidizing, and nucleolus-targeted Pt(IV) complex with a distinct anticancer mechanism. J. Am. Chem. Soc. 142, 7803–7812. doi:10.1021/jacs.0c00221
Denoth-Lippuner, A., Royall, L. N., Gonzalez-Bohorquez, D., Machado, D., and Jessberger, S. (2022). Injection and electroporation of plasmid DNA into human cortical organoids. Star. Protoc. 3, 101129. doi:10.1016/j.xpro.2022.101129
Dias, A. R., Minas Da Piedade, M. E., Martinho Simões, J. A., Simoni, J. A., Teixeira, C., Diogo, H. P., et al. (1992). Enthalpies of formation of cis-azobenzene and trans-azobenzene. J. Chem. Thermodyn. 24, 439–447. doi:10.1016/s0021-9614(05)80161-2
Dockalova, V., Sanchez-Carnerero, E. M., Dunajova, Z., Palao, E., Slanska, M., Buryska, T., et al. (2020). Fluorescent substrates for haloalkane dehalogenases: novel probes for mechanistic studies and protein labeling. Comput. Struct. Biotechnol. J. 18, 922–932. doi:10.1016/j.csbj.2020.03.029
Dong, M., Babalhavaeji, A., Samanta, S., Beharry, A. A., and Woolley, G. A. (2015). Red-shifting azobenzene photoswitches for in vivo use. Accounts Chem. Res. 48, 2662–2670. doi:10.1021/acs.accounts.5b00270
Dudek, M., Deiana, M., Pokladek, Z., Mlynarz, P., Samoc, M., and Matczyszyn, K. (2018). Light-driven chiroptical photoswitchable DNA assemblies mediated by bioinspired photoresponsive molecules. Nanoscale 10, 11302–11306. doi:10.1039/c8nr01784k
Edwards, W. F., Young, D. D., and Deiters, A. (2009). Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem. Biol. 4, 441–445. doi:10.1021/cb900041s
Egashira, T., Nakagawa-Tamagawa, N., Abzhanova, E., Kawae, Y., Kohara, A., Koitabashi, R., et al. (2023). In vivo two-photon calcium imaging of cortical neurons in neonatal mice. Star. Protoc. 4, 102245. doi:10.1016/j.xpro.2023.102245
Erlendsson, S., Thorsen, T. S., and Madsen, K. L. (2019). Supported cell membrane sheets to monitor protein assembly. Bio-protocol 9, e3368. doi:10.21769/bioprotoc.3368
Feng, M., Ruan, Z., Shang, J., Xiao, L., Tong, A., and Xiang, Y. (2017). Photocaged G-quadruplex DNAzyme and aptamer by post-synthetic modification on phosphodiester backbone. Bioconjugate Chem. 28, 549–555. doi:10.1021/acs.bioconjchem.6b00646
Feng, Z., Tang, T., Wu, T., Yu, X., Zhang, Y., Wang, M., et al. (2021). Perfecting and extending the near-infrared imaging window. Light Sci. and Appl. 10, 197. doi:10.1038/s41377-021-00628-0
Fields, C., Sheng, P., Miller, B., Wei, T., and Xie, M. (2019). Northern blot with IR fluorescent probes: strategies for probe preparation. Bio-protocol 9, e3219. doi:10.21769/bioprotoc.3219
Flumens, D., Campillo-Davo, D., Janssens, I., Roex, G., De Waele, J., Anguille, S., et al. (2023). One-step CRISPR-Cas9-mediated knockout of native TCRαβ genes in human T cells using RNA electroporation. Star. Protoc. 4, 102112. doi:10.1016/j.xpro.2023.102112
Forero-Quintero, L. S., Raymond, W., Munsky, B., and Stasevich, T. J. (2022). Visualization, quantification, and modeling of endogenous RNA polymerase II phosphorylation at a single-copy gene in living cells. Bio-protocol 12, e4482. doi:10.21769/bioprotoc.4482
Foster, A. A., Greco, C. T., Green, M. D., Epps, T. H., and Sullivan, M. O. (2015). Light-mediated activation of siRNA release in diblock copolymer assemblies for controlled gene silencing. Adv. Healthc. Mater. 4, 760–770. doi:10.1002/adhm.201400671
Fuentes, E., Gerth, M., Berrocal, J. A., Matera, C., Gorostiza, P., Voets, I. K., et al. (2020). An azobenzene-based single-component supramolecular polymer responsive to multiple stimuli in water. J. Am. Chem. Soc. 142, 10069–10078. doi:10.1021/jacs.0c02067
Gao, L., Wang, F., Wang, Y., Hu, L., and Mao, J. (2022). A protocol for the generation of patient-specific iPSC lines from peripheral blood mononuclear cells. Star. Protoc. 3, 101530. doi:10.1016/j.xpro.2022.101530
Garcia-Amorós, J., Stopa, G., Stochel, G., van Eldik, R., Martínez, M., and Velasco, D. (2018). Activation volumes for cis-to-trans isomerisation reactions of azophenols: a clear mechanistic indicator? Phys. Chem. Chem. Phys. 20, 1286–1292. doi:10.1039/c7cp07349f
Gáspár, I., Wippich, F., and Ephrussi, A. (2018). Terminal deoxynucleotidyl transferase mediated production of labeled probes for single-molecule FISH or RNA capture. Bio-protocol 8, e2750. doi:10.21769/bioprotoc.2750
Gelabert, R., Moreno, M., and Lluch, J. M. (2023). Predicting the electronic absorption band shape of azobenzene photoswitches. Int. J. Mol. Sci. 24, 25. doi:10.3390/ijms24010025
Gesuita, L., Argunsah, A. Ö., and Karayannis, T. (2022). Sparse postnatal labeling and quantification of superficial cortical cell synapses in the mouse neocortex. Star. Protoc. 3, 101837. doi:10.1016/j.xpro.2022.101837
Giles, L. W., Faul, C. F. J., and Tabor, R. F. (2021). Azobenzene isomerization in condensed matter: lessons for the design of efficient light-responsive soft-matter systems. Mater. Adv. 2, 4152–4164. doi:10.1039/d1ma00340b
Givens, R. S., and Matuszewski, B. (1984). Photochemistry of phosphate esters: an efficient method for the generation of electrophiles. J. Am. Chem. Soc. 106, 6860–6861. doi:10.1021/ja00334a075
Gorka, A. P., Nani, R. R., Zhu, J., Mackem, S., and Schnermann, M. J. (2014). A near-IR uncaging strategy based on cyanine photochemistry. J. Am. Chem. Soc. 136, 14153–14159. doi:10.1021/ja5065203
Govan, J. M., Uprety, R., Thomas, M., Lusic, H., Lively, M. O., and Deiters, A. (2013). Cellular delivery and photochemical activation of antisense agents through a nucleobase caging strategy. ACS Chem. Biol. 8, 2272–2282. doi:10.1021/cb400293e
Griepenburg, J. C., Rapp, T. L., Carroll, P. J., Eberwine, J., and Dmochowski, I. J. (2015). Ruthenium-caged antisense morpholinos for regulating gene expression in zebrafish embryos. Chem. Sci. 6, 2342–2346. doi:10.1039/c4sc03990d
Gutzler, R., Garg, M., Ast, C. R., Kuhnke, K., and Kern, K. (2021). Light–matter interaction at atomic scales. Nat. Rev. Phys. 3, 441–453. doi:10.1038/s42254-021-00306-5
Han, W.-G., Lovell, T., Liu, T., and Noodleman, L. (2002). Density functional studies of the ground- and excited-state potential-energy curves of stilbene cis–trans isomerization. ChemPhysChem 3, 167–178. doi:10.1002/1439-7641(20020215)3:2<167::aid-cphc167>3.0.co;2-g
Hartmann, D., and Booth, M. J. (2023). Accessible light-controlled knockdown of cell-free protein synthesis using phosphorothioate-caged antisense oligonucleotides. Commun. Chem. 6, 59. doi:10.1038/s42004-023-00860-2
Heinz, M. C., Oost, K. C., and Snippert, H. J. G. (2020). Introducing the stem cell ASCL2 reporter STAR into intestinal organoids. Star. Protoc. 1, 100126. doi:10.1016/j.xpro.2020.100126
Hemphill, J., Borchardt, E. K., Brown, K., Asokan, A., and Deiters, A. (2015). Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 137, 5642–5645. doi:10.1021/ja512664v
Hemphill, J., Chou, C., Chin, J. W., and Deiters, A. (2013). Genetically encoded light-activated transcription for spatiotemporal control of gene expression and gene silencing in mammalian cells. J. Am. Chem. Soc. 135, 13433–13439. doi:10.1021/ja4051026
Heyne, K., and Kühn, O. (2019). Infrared laser excitation controlled reaction acceleration in the electronic ground state. J. Am. Chem. Soc. 141, 11730–11738. doi:10.1021/jacs.9b02600
Holmstrom, E. D., and Nesbitt, D. J. (2010). Real-time infrared overtone laser control of temperature in picoliter H2O samples: “nanobathtubs” for single molecule microscopy. J. Phys. Chem. Lett. 1, 2264–2268. doi:10.1021/jz100663e
Hoorens, M. W. H., and Szymanski, W. (2018). Reversible, spatial and temporal control over protein activity using light. Trends Biochem. Sci. 43, 567–575. doi:10.1016/j.tibs.2018.05.004
Horii, Y., Matsuda, S., Watari, K., Nagasaka, A., Kurose, H., and Nakaya, M. (2017). An assay to determine phagocytosis of apoptotic cells by cardiac macrophages and cardiac myofibroblasts. Bio-protocol 7, e2553. doi:10.21769/bioprotoc.2553
Hoshina, H., Saito, Y., Furuhashi, T., Shimazaki, T., Sawada, M., Hioki, Y., et al. (2020). Terahertz spectroscopy for characterization of hydrogen bonding and cross-linked structure dynamics in polyurethane. J. Infrared, Millim. Terahertz Waves 41, 265–275. doi:10.1007/s10762-019-00667-0
Huang, X., Hu, Q., Braun, G. B., Pallaoro, A., Morales, D. P., Zasadzinski, J., et al. (2015). Light-activated RNA interference in human embryonic stem cells. Biomaterials 63, 70–79. doi:10.1016/j.biomaterials.2015.06.006
Imoto, T., Kawase, A., Minoshima, M., Yokoyama, T., Bito, H., and Kikuchi, K. (2020). Photolytic release of a caged inhibitor of an endogenous transcription factor enables optochemical control of CREB-mediated gene expression. Org. Lett. 22, 22–25. doi:10.1021/acs.orglett.9b03568
Inlay, M. A., Choe, V., Bharathi, S., Fernhoff, N. B., Baker, J. R., Weissman, I. L., et al. (2013). Synthesis of a photocaged tamoxifen for light-dependent activation of Cre-ER recombinase-driven gene modification. Chem. Commun. 49, 4971–4973. doi:10.1039/c3cc42179a
Ishigane, G., Toda, K., Tamamitsu, M., Shimada, H., Badarla, V. R., and Ideguchi, T. (2023). Label-free mid-infrared photothermal live-cell imaging beyond video rate. Light Sci. and Appl. 12, 174. doi:10.1038/s41377-023-01214-2
Jain, P. K., Ramanan, V., Schepers, A. G., Dalvie, N. S., Panda, A., Fleming, H. E., et al. (2016). Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem. Int. Ed. 55, 12440–12444. doi:10.1002/anie.201606123
Jain, R. K., Hoffman, A. J., Jepsen, P. U., Liu, P. Q., Turchinovich, D., and Vitiello, M. S. (2020). Mid-infrared, long-wave infrared, and terahertz photonics: introduction. Opt. Express 28, 14169–14175. doi:10.1364/oe.395165
Janeková, H., Russo, M., Ziegler, U., and Štacko, P. (2022). Photouncaging of carboxylic acids from cyanine dyes with near-infrared light. Angew. Chem. Int. Ed. 61, e202204391. doi:10.1002/anie.202204391
Jia, B., Wang, T., and Lehmann, J. (2021). Peptidyl transferase center decompaction and structural constraints during early protein elongation on the ribosome. Sci. Rep. 11, 24061. doi:10.1038/s41598-021-02985-7
Jia, S., and Sletten, E. M. (2022). Spatiotemporal control of biology: synthetic photochemistry toolbox with far-red and near-infrared light. ACS Chem. Biol. 17, 3255–3269. doi:10.1021/acschembio.1c00518
Jung, H., Kim, S.-W., Kim, M., Hong, J., Yu, D., Kim, J. H., et al. (2019). Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat. Commun. 10, 314. doi:10.1038/s41467-018-08282-8
Kajimoto, T., and Nakamura, S.-i. (2018). Quantitative analysis of cargo density in single-extracellular vesicles by imaging. Bio-protocol 8, e3111. doi:10.21769/bioprotoc.3111
Kala, A., Jain, P. K., Karunakaran, D., Shah, S., and Friedman, S. H. (2014). The synthesis of tetra-modified RNA for the multidimensional control of gene expression via light-activated RNA interference. Nat. Protoc. 9, 11–20. doi:10.1038/nprot.2013.165
Kaufmann, J., Wolf, J., and Heckel, A. (2023). Extending the palette of green coumarin photocages – oligonucleotide fragmentation and superior 5’-caps. Chem. – A Eur. J. 29, e202300390. doi:10.1002/chem.202300390
Keyvan Rad, J., Balzade, Z., and Mahdavian, A. R. (2022). Spiropyran-based advanced photoswitchable materials: a fascinating pathway to the future stimuli-responsive devices. J. Photochem. Photobiol. C Photochem. Rev. 51, 100487. doi:10.1016/j.jphotochemrev.2022.100487
Khalil, A. M. (2020). The genome editing revolution: review. J. Genet. Eng. Biotechnol. 18, 68. doi:10.1186/s43141-020-00078-y
Klena, N. T., Maltinti, G., Batman, U., Pigino, G., Guichard, P., and Hamel, v. (2023). An in-depth guide to the ultrastructural expansion microscopy (U-ExM) of chlamydomonas reinhardtii. Bio-protocol 13, e4792. doi:10.21769/bioprotoc.4792
Koch, A. L., Morisaki, T., and Stasevich, T. J. (2021). A multi-color bicistronic biosensor to compare the translation dynamics of different open reading frames at single-molecule resolution in live cells. Bio-protocol 11, e4096. doi:10.21769/bioprotoc.4096
Koh, X. Y., Koh, X. H., Hwang, L.-A., Ferrer, F. J., Rahmat, S. A. B., Lama, D., et al. (2019). Therapeutic anti-cancer activity of antibodies targeting sulfhydryl bond constrained epitopes on unglycosylated RON receptor tyrosine kinase. Oncogene 38, 7342–7356. doi:10.1038/s41388-019-0946-8
Konermann, S., Brigham, M. D., Trevino, A. E., Hsu, P. D., Heidenreich, M., Le, C., et al. (2013). Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476. doi:10.1038/nature12466
Kortekaas, L., and Browne, W. R. (2019). The evolution of spiropyran: fundamentals and progress of an extraordinarily versatile photochrome. Chem. Soc. Rev. 48, 3406–3424. doi:10.1039/c9cs00203k
Kuntze, K., Isokuortti, J., Siiskonen, A., Durandin, N., Laaksonen, T., and Priimagi, A. (2021). Azobenzene photoswitching with near-infrared light mediated by molecular oxygen. J. Phys. Chem. B 125, 12568–12573. doi:10.1021/acs.jpcb.1c08012
Laczi, D., Johnstone, M. D., and Fleming, C. L. (2022). Photoresponsive small molecule inhibitors for the remote control of enzyme activity. Chem. – Asian J. 17, e202200200. doi:10.1002/asia.202200200
Lankford, K. P., and Hulleman, J. D. (2024). Protocol for HiBiT tagging endogenous proteins using CRISPR-Cas9 gene editing. Star. Protoc. 5, 103000. doi:10.1016/j.xpro.2024.103000
Laprie-Sentenac, M., Cretet-Rodeschini, C., and Menger, L. (2023). Optimized protocol to generate genome-wide inactivated Cas9-expressing murine T cells. Star. Protoc. 4, 101922. doi:10.1016/j.xpro.2022.101922
Lax, C., Navarro-Mendoza, M. I., Pérez-Arques, C., Navarro, E., Nicolás, F. E., and Garre, V. (2022). Transformation and CRISPR-Cas9-mediated homologous recombination in the fungus Rhizopus microsporus. Star. Protoc. 3, 101237. doi:10.1016/j.xpro.2022.101237
Layden, H. M., Eleuteri, N. A., Hiebert, S. W., and Stengel, K. R. (2021). A protocol for rapid degradation of endogenous transcription factors in mammalian cells and identification of direct regulatory targets. Star. Protoc. 2, 100530. doi:10.1016/j.xpro.2021.100530
Lee, Y.-H., Yu, E., and Park, C.-M. (2021). Programmable site-selective labeling of oligonucleotides based on carbene catalysis. Nat. Commun. 12, 1681. doi:10.1038/s41467-021-21839-4
Leng, S., Qiao, Q.-L., Gao, Y., Miao, L., Deng, W.-G., and Xu, Z.-C. (2017). SNAP-tag fluorogenic probes for wash free protein labeling. Chin. Chem. Lett. 28, 1911–1915. doi:10.1016/j.cclet.2017.03.034
Li, Z., Reint, G., and Haapaniemi, E. M. (2023). Protocol for editing fibroblasts with in vitro transcribed Cas9 mRNA and profile off-target editing by optimized GUIDE-seq. Star. Protoc. 4, 102662. doi:10.1016/j.xpro.2023.102662
Lin, W., Albanese, C., Pestell, R. G., and Lawrence, D. S. (2002). Spatially discrete, light-driven protein expression. Chem. and Biol. 9, 1347–1353. doi:10.1016/s1074-5521(02)00288-0
Ling, X., Chang, L., Chen, H., and Liu, T. (2022). Efficient generation of locus-specific human CAR-T cells with CRISPR/cCas12a. Star. Protoc. 3, 101321. doi:10.1016/j.xpro.2022.101321
Link, K. H., Cruz, F. G., Ye, H.-F., O’Reilly, K. E., Dowdell, S., and Koh, J. T. (2004). Photo-caged agonists of the nuclear receptors RARγ and TRβ provide unique time-dependent gene expression profiles for light-activated gene patterning. Bioorg. and Med. Chem. 12, 5949–5959. doi:10.1016/j.bmc.2004.08.022
Liu, M., Ma, S., She, M., Chen, J., Wang, Z., Liu, P., et al. (2019). Structural modification of BODIPY: improve its applicability. Chin. Chem. Lett. 30, 1815–1824. doi:10.1016/j.cclet.2019.08.028
López-Lorente, Á. I., Wang, P., Sieger, M., Vargas Catalan, E., Karlsson, M., Nikolajeff, F., et al. (2016). Mid-infrared thin-film diamond waveguides combined with tunable quantum cascade lasers for analyzing the secondary structure of proteins. Phys. status solidi (a) 213, 2117–2123. doi:10.1002/pssa.201600134
Luciano, M. P., Crooke, S. N., Nourian, S., Dingle, I., Nani, R. R., Kline, G., et al. (2019). A nonaggregating heptamethine cyanine for building brighter labeled biomolecules. ACS Chem. Biol. 14, 934–940. doi:10.1021/acschembio.9b00122
Luo, H., Zhang, J., Yang, A., Ouyang, W., Long, S., Lin, X., et al. (2023). Large-scale isolation of exosomes derived from NK cells for anti-tumor therapy. Bio-protocol 13, e4693. doi:10.21769/bioprotoc.4693
Luo, J., Arbely, E., Zhang, J., Chou, C., Uprety, R., Chin, J. W., et al. (2016). Genetically encoded optical activation of DNA recombination in human cells. Chem. Commun. 52, 8529–8532. doi:10.1039/c6cc03934k
Ma, J., Qin, Z., Xie, G., Qian, L., and Tang, D. (2019). Review of mid-infrared mode-locked laser sources in the 2.0 μm–3.5 μm spectral region. Appl. Phys. Rev. 6. doi:10.1063/1.5037274
Mai-Morente, S. P., Irigoyen, J. P., Carriquiry, V. M., Marset, V. M., Di Doménico, M., Isasi, E., et al. (2021). Pericyte mapping in cerebral slices with the far-red fluorophore TO-PRO-3. Bio-protocol 11, e4222. doi:10.21769/bioprotoc.4222
Mao, Z., Yang, X., Mizutani, S., Huang, Y., Zhang, Z., Shinmori, H., et al. (2020). Hydrogen sulfide mediates tumor cell resistance to thioredoxin inhibitor. Front. Oncol. 10, 252. doi:10.3389/fonc.2020.00252
Mazzucco, M. R., Vartanian, T., and Linden, J. R. (2020). In vivo blood-brain barrier permeability assays using Clostridium perfringens epsilon toxin. Bio-protocol 10, e3709. doi:10.21769/bioprotoc.3709
Meka, D. P., Richter, M., Rücker, T., Voss, H., Rissiek, A., Krisp, C., et al. (2024). Protocol for differential multi-omic analyses of distinct cell types in the mouse cerebral cortex. Star. Protoc. 5, 102793. doi:10.1016/j.xpro.2023.102793
Menge, C., and Heckel, A. (2011). Coumarin-caged dG for improved wavelength-selective uncaging of DNA. Org. Lett. 13, 4620–4623. doi:10.1021/ol201842x
Meyer, A., and Mokhir, A. (2014). RNA interference controlled by light of variable wavelength. Angew. Chem. Int. Ed. 53, 12840–12843. doi:10.1002/anie.201405885
Morichika, I., and Ashihara, S. (2022). Ultrafast infrared plasmonics advances vibrational spectroscopy. Photonics Rev. 2022. doi:10.11470/photo.220204
Morichika, I., Murata, K., Sakurai, A., Ishii, K., and Ashihara, S. (2019). Molecular ground-state dissociation in the condensed phase employing plasmonic field enhancement of chirped mid-infrared pulses. Nat. Commun. 10, 3893. doi:10.1038/s41467-019-11902-6
Moroz-Omori, E. V., Satyapertiwi, D., Ramel, M.-C., Høgset, H., Sunyovszki, I. K., Liu, Z., et al. (2020). Photoswitchable gRNAs for spatiotemporally controlled CRISPR-cas-based genomic regulation. ACS Central Sci. 6, 695–703. doi:10.1021/acscentsci.9b01093
Mu, W., Homann, S., Hofmann, C., Gorin, A., Huynh, D., Yang, O. O., et al. (2019). A flow cytometric method to determine transfection efficiency. Bio-protocol 9, e3244. doi:10.21769/bioprotoc.3244
Mukherjee, D., Chakraborty, G., Hasan, M. N., Pal, U., Singh, P., Rakshit, T., et al. (2022). Reversible photoswitching of spiropyran in biomolecular interfaces: a combined spectroscopy and computational study. J. Photochem. Photobiol. A Chem. 430, 113958. doi:10.1016/j.jphotochem.2022.113958
Munson, M. J., and Ganley, I. G. (2016). Determination of cellular phosphatidylinositol-3-phosphate (PI3P) levels using a fluorescently labelled selective PI3P binding domain (PX). Bio-protocol 6, e1903. doi:10.21769/bioprotoc.1903
Nagahama, K., Fujino, S., Watanabe, T., Uesaka, N., and Kano, M. (2021). Combining electrophysiology and optogenetics for functional screening of pyramidal neurons in the mouse prefrontal cortex. Star. Protoc. 2, 100469. doi:10.1016/j.xpro.2021.100469
Neumann, H. (2012). Rewiring translation – genetic code expansion and its applications. FEBS Lett. 586, 2057–2064. doi:10.1016/j.febslet.2012.02.002
Nguyen, N. T., He, L., Martinez-Moczygemba, M., Huang, Y., and Zhou, Y. (2018). Rewiring calcium signaling for precise transcriptional reprogramming. ACS Synth. Biol. 7, 814–821. doi:10.1021/acssynbio.7b00467
Nomura, T., Yoshikawa, M., Suzuki, K., and Mochida, K. (2020). Highly efficient CRISPR-associated protein 9 ribonucleoprotein-based genome editing in euglena gracilis. Star. Protoc. 1, 100023. doi:10.1016/j.xpro.2020.100023
Nunes, C. M., Pereira, N. A. M., Reva, I., Amado, P. S. M., Cristiano, M. L. S., and Fausto, R. (2020). Bond-breaking/bond-forming reactions by vibrational excitation: infrared-induced bidirectional tautomerization of matrix-isolated thiotropolone. J. Phys. Chem. Lett. 11, 8034–8039. doi:10.1021/acs.jpclett.0c02272
O’Connor, M. J., Beebe, L. L., Deodato, D., Ball, R. E., Page, A. T., VanLeuven, A. J., et al. (2019). Bypassing glutamic acid decarboxylase 1 (Gad1) induced craniofacial defects with a photoactivatable translation blocker morpholino. ACS Chem. Neurosci. 10, 266–278. doi:10.1021/acschemneuro.8b00231
Ogasawara, S. (2018). Transcription driven by reversible photocontrol of hyperstable G-quadruplexes. ACS Synth. Biol. 7, 2507–2513. doi:10.1021/acssynbio.8b00216
O’Hagan, M. P., Haldar, S., Duchi, M., Oliver, T. A. A., Mulholland, A. J., Morales, J. C., et al. (2019). A photoresponsive stiff-stilbene ligand fuels the reversible unfolding of G-quadruplex DNA. Angew. Chem. Int. Ed. 58, 4334–4338. doi:10.1002/anie.201900740
O’Hagan, M. P., Peñalver, P., Gibson, R. S. L., Morales, J. C., and Galan, M. C. (2020). Stiff-stilbene ligands target G-quadruplex DNA and exhibit selective anticancer and antiparasitic activity. Chem. – A Eur. J. 26, 6224–6233. doi:10.1002/chem.201905753
Pan, Y., Yang, J., Luan, X., Liu, X., Li, X., Yang, J., et al. (2019). Near-infrared upconversion–activated CRISPR-Cas9 system: a remote-controlled gene editing platform. Sci. Adv. 5, eaav7199. doi:10.1126/sciadv.aav7199
Park, W., Lee, Y., Kang, T., Jeong, J., and Kim, D.-S. (2018). Terahertz-driven polymerization of resists in nanoantennas. Sci. Rep. 8, 7762. doi:10.1038/s41598-018-26214-w
Paul, A., Biswas, A., Sinha, S., Shah, S. S., Bera, M., Mandal, M., et al. (2019). Push–pull stilbene: visible light activated photoremovable protecting group for alcohols and carboxylic acids with fluorescence reporting employed for drug delivery. Org. Lett. 21, 2968–2972. doi:10.1021/acs.orglett.9b00124
Paul, A., Huang, J., Han, Y., Yang, X., Vuković, L., Král, P., et al. (2021). Photochemical control of bacterial gene expression based on trans encoded genetic switches. Chem. Sci. 12, 2646–2654. doi:10.1039/d0sc05479h
Paweletz, L. C., Veit, S., and Pomorski, T. G. (2022). A fluorescence-based approach utilizing self-labeling enzyme tags to determine protein orientation in large unilamellar vesicles. Bio-protocol 12, e4542. doi:10.21769/bioprotoc.4542
Peterson, J. A., Wijesooriya, C., Gehrmann, E. J., Mahoney, K. M., Goswami, P. P., Albright, T. R., et al. (2018). Family of BODIPY photocages cleaved by single photons of visible/near-infrared light. J. Am. Chem. Soc. 140, 7343–7346. doi:10.1021/jacs.8b04040
Pinheiro, A. V., Baptista, P., and Lima, J. C. (2008). Light activation of transcription: photocaging of nucleotides for control over RNA polymerization. Nucleic Acids Res. 36, e90. doi:10.1093/nar/gkn415
Polstein, L. R., and Gersbach, C. A. (2012). Light-inducible spatiotemporal control of gene activation by customizable Zinc finger transcription factors. J. Am. Chem. Soc. 134, 16480–16483. doi:10.1021/ja3065667
Poryvai, A., Galkin, M., Shvadchak, V., and Slanina, T. (2022). Red-shifted water-soluble BODIPY photocages for visualisation and controllable cellular delivery of signaling lipids. Angew. Chem. Int. Ed. 61, e202205855. doi:10.1002/anie.202205855
Powell, C. E., Gao, Y., Tan, L., Donovan, K. A., Nowak, R. P., Loehr, A., et al. (2018). Chemically induced degradation of anaplastic lymphoma kinase (ALK). J. Med. Chem. 61, 4249–4255. doi:10.1021/acs.jmedchem.7b01655
Rawat, S., and Sharma, M. (2024). CD8α-CI-M6PR particle motility assay to study the retrograde motion of CI-M6PR receptors in cultured living cells. Bio-protocol 14, e4979. doi:10.21769/bioprotoc.4979
Remenyi, R., Li, R., and Harris, M. (2019). On-demand labeling of SNAP-tagged viral protein for pulse-chase imaging, quench-pulse-chase imaging, and nanoscopy-based inspection of cell lysates. Bio-protocol 9, e3177. doi:10.21769/bioprotoc.3177
Riley, R. S., Dang, M. N., Billingsley, M. M., Abraham, B., Gundlach, L., and Day, E. S. (2018). Evaluating the mechanisms of light-triggered siRNA release from nanoshells for temporal control over gene regulation. Nano Lett. 18, 3565–3570. doi:10.1021/acs.nanolett.8b00681
Roberts, S. K., Hirsch, M., McStea, A., Zanetti-Domingues, L. C., Clarke, D. T., Claus, J., et al. (2018). Cluster analysis of endogenous HER2 and HER3 receptors in SKBR3 cells. Bio-protocol 8, e3096. doi:10.21769/bioprotoc.3096
Rozovsky, A., Patsenker, L., and Gellerman, G. (2019). Bifunctional reactive pentamethine cyanine dyes for biomedical applications. Dyes Pigments 162, 18–25. doi:10.1016/j.dyepig.2018.09.064
Roßmann, K., Gonzalez-Hernandez, A. J., Bhuyan, R., Börjesson, K., Levitz, J., and Broichhagen, J. (2023). Deuteration provides a general strategy to enhance azobenzene-based photopharmacology. Biorxiv. 2023.11.09.566420. doi:10.1101/2023.11.09.566420
Ryu, K. A., Kaszuba, C. M., Bissonnette, N. B., Oslund, R. C., and Fadeyi, O. O. (2021). Interrogating biological systems using visible-light-powered catalysis. Nat. Rev. Chem. 5, 322–337. doi:10.1038/s41570-021-00265-6
Sakamoto, M., Ota, K., Kondo, Y., Okamura, M., Fujii, H., and Bito, H. (2022). In utero electroporation and cranial window implantation for in vivo wide-field two-photon calcium imaging using G-CaMP9a transgenic mice. Star. Protoc. 3, 101421. doi:10.1016/j.xpro.2022.101421
Sakamoto, T., Shigeno, A., Ohtaki, Y., and Fujimoto, K. (2014). Photo-regulation of constitutive gene expression in living cells by using ultrafast photo-cross-linking oligonucleotides. Biomaterials Sci. 2, 1154–1157. doi:10.1039/c4bm00117f
Samolis, P. D., Zhu, X., and Sander, M. Y. (2023). Time-resolved mid-infrared photothermal microscopy for imaging water-embedded axon bundles. Anal. Chem. 95, 16514–16521. doi:10.1021/acs.analchem.3c02352
Sanketi, B. D., Sivakumar, A., and Kurpios, N. A. (2023). Visualizing and manipulating the production and accumulation of hyaluronan for functional assessment in chicken embryos. Star. Protoc. 4, 102200. doi:10.1016/j.xpro.2023.102200
Shao, J., Wang, M., Yu, G., Zhu, S., Yu, Y., Heng, B. C., et al. (2018). Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl. Acad. Sci. 115, E6722–E6730. doi:10.1073/pnas.1802448115
Sharma, M., and Friedman, S. H. (2021). The issue of tissue: approaches and challenges to the light control of drug activity. ChemPhotoChem 5, 611–618. doi:10.1002/cptc.202100001
Sharma, R., Sharma, A., Kumar, A., and Jaganathan, B. G. (2019). Phospho-protein analysis in adherent cells using flow cytometry. Bio-protocol 9, e3395. doi:10.21769/bioprotoc.3395
Shiraishi, Y., Yomo, K., and Hirai, T. (2023). Polarity-Driven isomerization of a hydroxynaphthalimide-containing spiropyran at room temperature. ACS Phys. Chem. Au 3, 290–298. doi:10.1021/acsphyschemau.2c00067
Singh, P. K., Majumdar, P., and Singh, S. P. (2021). Advances in BODIPY photocleavable protecting groups. Coord. Chem. Rev. 449, 214193. doi:10.1016/j.ccr.2021.214193
Sitkowska, K., Feringa, B. L., and Szymański, W. (2018). Green-light-sensitive BODIPY photoprotecting groups for amines. J. Org. Chem. 83, 1819–1827. doi:10.1021/acs.joc.7b02729
Skruber, K., Read, T.-A., and Vitriol, E. A. (2021). Delivering defined amounts of purified protein with high precision into living cells. Star. Protoc. 2, 100272. doi:10.1016/j.xpro.2020.100272
Šmidlehner, T., Kurutos, A., Slade, J., Belužić, R., Ang, D. L., Rodger, A., et al. (2018). Versatile click cyanine amino acid conjugates showing one-atom-influenced recognition of DNA/RNA secondary structure and mitochondrial localisation in living cells. Eur. J. Org. Chem. 2018, 1682–1692. doi:10.1002/ejoc.201701765
Stadler, J. M., and Stafforst, T. (2014). Pyrene chromophores for the photoreversal of psoralen interstrand crosslinks. Org. and Biomol. Chem. 12, 5260–5266. doi:10.1039/c4ob00603h
Stafforst, T., and Stadler, J. M. (2013). Photoactivation of a psoralen-blocked luciferase gene by blue light. Angew. Chem. Int. Ed. 52, 12448–12451. doi:10.1002/anie.201306150
Stensitzki, T., Yang, Y., Kozich, V., Ahmed, A. A., Kössl, F., Kühn, O., et al. (2018). Acceleration of a ground-state reaction by selective femtosecond-infrared-laser-pulse excitation. Nat. Chem. 10, 126–131. doi:10.1038/nchem.2909
Sugiyama, J.-i., Tokunaga, Y., Hishida, M., Tanaka, M., Takeuchi, K., Satoh, D., et al. (2023). Nonthermal acceleration of protein hydration by sub-terahertz irradiation. Nat. Commun. 14, 2825. doi:10.1038/s41467-023-38462-0
Sun, L., Zhao, L., and Peng, R.-Y. (2021). Research progress in the effects of terahertz waves on biomacromolecules. Mil. Med. Res. 8, 28. doi:10.1186/s40779-021-00321-8
Tallafuss, A., Gibson, D., Morcos, P., Li, Y., Seredick, S., Eisen, J., et al. (2012). Turning gene function ON and OFF using sense and antisense photo-morpholinos in zebrafish. Development 139, 1691–1699. doi:10.1242/dev.072702
Talone, B., Bazzarelli, M., Schirato, A., Dello Vicario, F., Viola, D., Jacchetti, E., et al. (2021). Phototoxicity induced in living HeLa cells by focused femtosecond laser pulses: a data-driven approach. Biomed. Opt. Express 12, 7886–7905. doi:10.1364/boe.441225
Tang, X., Swaminathan, J., Gewirtz, A. M., and Dmochowski, I. J. (2007). Regulating gene expression in human leukemia cells using light-activated oligodeoxynucleotides. Nucleic Acids Res. 36, 559–569. doi:10.1093/nar/gkm1029
Tanimizu, N. (2015). Clonal culture of mouse liver progenitor cells. Bio-protocol 5, e1624. doi:10.21769/bioprotoc.1624
Tavakoli, A., and Min, J.-H. (2022). Photochemical modifications for DNA/RNA oligonucleotides. RSC Adv. 12, 6484–6507. doi:10.1039/d1ra05951c
Teng, I. T., Bu, X., and Chung, I. (2019). Conjugation of fab’ fragments with fluorescent dyes for single-molecule tracking on live cells. Bio-protocol 9, e3375. doi:10.21769/bioprotoc.3375
Tian, T., Song, Y., Wang, J., Fu, B., He, Z., Xu, X., et al. (2016). Small-molecule-triggered and light-controlled reversible regulation of enzymatic activity. J. Am. Chem. Soc. 138, 955–961. doi:10.1021/jacs.5b11532
Toda, K., Tamamitsu, M., Nagashima, Y., Horisaki, R., and Ideguchi, T. (2019). Molecular contrast on phase-contrast microscope. Sci. Rep. 9, 9957. doi:10.1038/s41598-019-46383-6
Tonouchi, M. (2007). Cutting-edge terahertz technology. Nat. Photonics 1, 97–105. doi:10.1038/nphoton.2007.3
Toyama, T., Fujioka, J., Watanabe, K., Yoshida, A., Sakuma, T., Inaba, K., et al. (2022). Investigation of bactericidal effect of a mid-infrared free electron laser on Escherichia coli. Sci. Rep. 12, 18111. doi:10.1038/s41598-022-22949-9
Tronnet, S., and Oswald, E. (2018). Quantification of colibactin-associated genotoxicity in HeLa cells by in cell western (ICW) using γ-H2AX as a marker. Bio-protocol 8, e2771. doi:10.21769/bioprotoc.2771
Turchetto, S., Broix, L., and Nguyen, L. (2020). Ex vivo recording of axonal transport dynamics on postnatal organotypic cortical slices. Star. Protoc. 1, 100131. doi:10.1016/j.xpro.2020.100131
Udupa, S., Nagaraja, V., and Karambelkar, S. (2022). Binding affinity quantifications of the bacteriophage Mu DNA modification protein mom using microscale thermophoresis (MST). Bio-protocol 12, e4472. doi:10.21769/bioprotoc.4472
Villarón, D., Duindam, N., and Wezenberg, S. J. (2021). Push-pull stiff-stilbene: proton-gated visible-light photoswitching and acid-catalyzed isomerization. Chem. – A Eur. J. 27, 17346–17350. doi:10.1002/chem.202103052
Vohradská, N., Sánchez-Carnerero, E. M., Pastierik, T., Mazal, C., and Klán, P. (2018). Controlled photorelease of alkynoic acids and their decarboxylative deprotection for copper-catalyzed azide/alkyne cycloaddition. Chem. Commun. 54, 5558–5561. doi:10.1039/c8cc03341b
Volarić, J., Buter, J., Schulte, A. M., van den Berg, K.-O., Santamaría-Aranda, E., Szymanski, W., et al. (2022). Design and synthesis of visible-light-responsive azobenzene building blocks for chemical biology. J. Org. Chem. 87, 14319–14333. doi:10.1021/acs.joc.2c01777
Walsh, S., Gardner, L., Deiters, A., and Williams, G. J. (2014). Intracellular light-activation of riboswitch activity. ChemBioChem 15, 1346–1351. doi:10.1002/cbic.201400024
Wang, K., Schmied, W. H., and Chin, J. W. (2012). Reprogramming the genetic code: from triplet to quadruplet codes. Angew. Chem. Int. Ed. 51, 2288–2297. doi:10.1002/anie.201105016
Wang, Q., Wu, Z., Qin, P., Ji, J., Lai, L., and Yin, M. (2021). Photoregulated morphological transformation of spiropyran derivatives achieving the tunability of interfacial hydrophilicity. Langmuir 37, 11170–11175. doi:10.1021/acs.langmuir.1c02053
Wang, X., Feng, M., Xiao, L., Tong, A., and Xiang, Y. (2016). Postsynthetic modification of DNA phosphodiester backbone for photocaged DNAzyme. ACS Chem. Biol. 11, 444–451. doi:10.1021/acschembio.5b00867
Wang, Y., Wang, Z., and Ji, Q. (2020). CRISPR-Cas9-Based genome editing and cytidine base editing in acinetobacter baumannii. Star. Protoc. 1, 100025. doi:10.1016/j.xpro.2020.100025
Weinstain, R., Slanina, T., Kand, D., and Klán, P. (2020). Visible-to-NIR-Light activated release: from small molecules to nanomaterials. Chem. Rev. 120, 13135–13272. doi:10.1021/acs.chemrev.0c00663
Welleman, I. M., Hoorens, M. W. H., Feringa, B. L., Boersma, H. H., and Szymański, W. (2020). Photoresponsive molecular tools for emerging applications of light in medicine. Chem. Sci. 11, 11672–11691. doi:10.1039/d0sc04187d
Williams, R. M., and Sauka-Spengler, T. (2021). Ex ovo electroporation of early chicken embryos. Star. Protoc. 2, 100424. doi:10.1016/j.xpro.2021.100424
Wilson, C., Rozés-Salvador, V., and Cáceres, A. (2020). Protocol for evaluating neuronal polarity in murine models. Star. Protoc. 1, 100114. doi:10.1016/j.xpro.2020.100114
Windhorn, L., Witte, T., Yeston, J. S., Proch, D., Motzkus, M., Kompa, K. L., et al. (2002). Molecular dissociation by mid-IR femtosecond pulses. Chem. Phys. Lett. 357, 85–90. doi:10.1016/s0009-2614(02)00444-x
Wirth, R., Gao, P., Nienhaus, G. U., Sunbul, M., and Jäschke, A. (2020). Confocal and super-resolution imaging of RNA in live bacteria using a fluorogenic silicon rhodamine-binding aptamer. Bio-protocol 10, e3603. doi:10.21769/bioprotoc.3603
Wong, P. T., Roberts, E. W., Tang, S., Mukherjee, J., Cannon, J., Nip, A. J., et al. (2017). Control of an unusual photo-claisen rearrangement in coumarin caged tamoxifen through an extended spacer. ACS Chem. Biol. 12, 1001–1010. doi:10.1021/acschembio.6b00999
Wu, Y., Sidharta, M., Zhong, A., Persily, B., Li, M., and Zhou, T. (2023). Protocol for the design, conduct, and evaluation of prime editing in human pluripotent stem cells. Star. Protoc. 4, 102583. doi:10.1016/j.xpro.2023.102583
Xing, X., Wang, X., Xu, L., Tai, Y., Dai, L., Zheng, X., et al. (2015). Correction: light-driven conformational regulation of human telomeric G-quadruplex DNA in physiological conditions. Org. and Biomol. Chem. 13, 4613. doi:10.1039/c5ob90054a
Xu, H., Kita, Y., Bang, U., Gee, P., and Hotta, A. (2021). Optimized electroporation of CRISPR-Cas9/gRNA ribonucleoprotein complex for selection-free homologous recombination in human pluripotent stem cells. Star. Protoc. 2, 100965. doi:10.1016/j.xpro.2021.100965
Xu, J., and Liu, Y. (2019). Imaging higher-order chromatin structures in single cells using stochastic optical reconstruction microscopy. Bio-protocol 9, e3160. doi:10.21769/bioprotoc.3160
Xu, Z., Li, Y., and Yan, A. (2020). Repurposing the native type I-F CRISPR-cas system in Pseudomonas aeruginosa for genome editing. Star. Protoc. 1, 100039. doi:10.1016/j.xpro.2020.100039
Yamamoto, T., Caldwell, D. R., Gandioso, A., and Schnermann, M. J. (2019). A cyanine photooxidation/β-elimination sequence enables near-infrared uncaging of aryl amine payloads. Photochem. Photobiol. 95, 951–958. doi:10.1111/php.13090
Yamazoe, S., Liu, Q., McQuade, L. E., Deiters, A., and Chen, J. K. (2014). Sequential gene silencing using wavelength-selective caged morpholino oligonucleotides. Angew. Chem. Int. Ed. 53, 10114–10118. doi:10.1002/anie.201405355
Yang, L., Kim, H. B., Sul, J.-Y., Yeldell, S. B., Eberwine, J. H., and Dmochowski, I. J. (2018). Efficient synthesis of light-triggered circular antisense oligonucleotides targeting cellular protein expression. ChemBioChem 19, 1250–1254. doi:10.1002/cbic.201800012
York, V. A., and Milush, J. M. (2015). Ex vivo human natural killer (NK) cell stimulation and intracellular IFNγ and CD107a cytokine staining. Bio-protocol 5, e1501. doi:10.21769/bioprotoc.1501
Young, D. D., Garner, R. A., Yoder, J. A., and Deiters, A. (2009). Light-activation of gene function in mammalian cells viaribozymes. Chem. Commun., 568–570. doi:10.1039/b819375d
Yu, L., Jing, N., Yang, Z., Zhang, L., and Tang, X. (2018). Caged siRNAs with single folic acid modification of antisense RNA for photomodulation of exogenous and endogenous gene expression in cells. Org. and Biomol. Chem. 16, 7029–7035. doi:10.1039/c8ob01952e
Zhang, D., Lan, L., Bai, Y., Majeed, H., Kandel, M. E., Popescu, G., et al. (2019). Bond-selective transient phase imaging via sensing of the infrared photothermal effect. Light Sci. and Appl. 8, 116. doi:10.1038/s41377-019-0224-0
Zhang, L., Chen, C., Fan, X., and Tang, X. (2018). Photomodulating gene expression by using caged siRNAs with single-aptamer modification. ChemBioChem 19, 1259–1263. doi:10.1002/cbic.201700623
Zhang, W., Hamouri, F., Feng, Z., Aujard, I., Ducos, B., Ye, S., et al. (2018). Control of protein activity and gene expression by cyclofen-OH uncaging. ChemBioChem 19, 1232–1238. doi:10.1002/cbic.201700630
Zhi, T. S., Cheng, T. P., Qing, L. L., Liang, C. Y., Long, -.Y. Z., Long, Z. X., et al. (2019). - Exposure effects of terahertz waves on primary neurons and neuron-like cells under nonthermal conditions. Biomed. Environ. Sci. - 32–739. doi:10.3967/bes2019.094
Zhou, L., Cheng, Z.-Q., Li, N., Ge, Y.-X., Xie, H.-X., Zhu, K., et al. (2020). A highly sensitive endoplasmic reticulum-targeting fluorescent probe for the imaging of endogenous H2S in live cells. Spectrochimica Acta Part A Mol. Biomol. Spectrosc. 240, 118578. doi:10.1016/j.saa.2020.118578
Zhou, X. X., Zou, X., Chung, H. K., Gao, Y., Liu, Y., Qi, L. S., et al. (2018). A single-chain photoswitchable CRISPR-cas9 architecture for light-inducible gene editing and transcription. ACS Chem. Biol. 13, 443–448. doi:10.1021/acschembio.7b00603
Zhou, Z., Ma, L., and Zhang, X. (2021). Protocol for genome-scale CRISPR screening in engineered lineage reporter hPSCs to study cell fate determination. Star. Protoc. 2, 100548. doi:10.1016/j.xpro.2021.100548
Keywords: light-regulated technology, mid-infrared light, gene expression, label-free, photocage, photoswitch, terahertz light
Citation: Yamazaki H, Sugawara R and Takayama Y (2024) Development of label-free light-controlled gene expression technologies using mid-IR and terahertz light. Front. Bioeng. Biotechnol. 12:1324757. doi: 10.3389/fbioe.2024.1324757
Received: 20 October 2023; Accepted: 25 September 2024;
Published: 11 October 2024.
Edited by:
Dipesh Dhakal, University of Florida, United StatesReviewed by:
Tianwen Wang, Anhui Polytechnic University, ChinaRichard Kelwick, Imperial College London, United Kingdom
Copyright © 2024 Yamazaki, Sugawara and Takayama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hirohito Yamazaki, aGlyb2hpdG95YW1hemFraUB2b3MubmFnYW9rYXV0LmFjLmpw
 Hirohito Yamazaki
Hirohito Yamazaki Ryusei Sugawara2
Ryusei Sugawara2