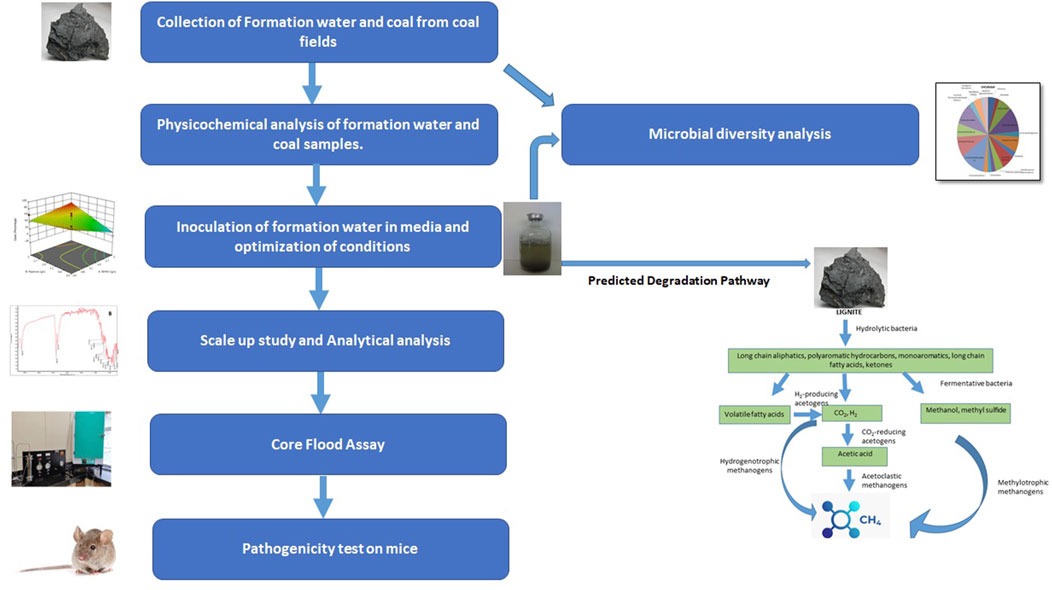
- 1The Energy and Resources Institute (TERI), New Delhi, India
- 2Ongc Energy Centre, Delhi, India
Over the last several decades, coalbed methane (CBM) has emerged as an important energy source in developing nations like India as well as worldwide and is expected to play a significant role in the energy portfolio of the future. The current scenario of rapid exhaustion of fossil fuels is leading to the need to explore alternative and efficient fuel resources. The present study demonstrates enhanced methane production per gram of lignite (lowest-rank coal). Optimization of the bioconversion of lignite to methane revealed 55°C temperature and 1.5 g/L NaCl concentration as ambient conditions for the process. A scale-up study in the optimized condition showed 2,800 mM methane production per 25 g of lignite in anaerobic conditions. Further, Fourier transform Infrared (FTIR) and Gas Chromatography Mass Spectrometry (GCMS) analysis showed bioconversion of lignite into simpler intermediate substrates required for methane production. The results highlighted that the bacterial action first converts lignite into volatile fatty acids, which subsequently get converted into methane. Further, the exploration of indigenous microbial consortia in Tharad well (THAA) mainly comprises the order Methanosarcinales and Methanomicrobiales. The pathogenicity of the microbial consortium THAA was declared safe for use in mice via the oral route by The Energy and Resources Institute (TERI), India. The study demonstrated the development of indigenous consortia (TERI THAA), which can potentially enhance methane production from the lowest coal grade under extreme conditions in Indian coal beds.
1 Introduction
Petroleum-based hydrocarbons are a major energy source used worldwide (Litvinenko, 2020). Coal is the world’s largest and most prevalent conventional energy, mainly categorized into four main types or ranks: anthracite, bituminous, sub-bituminous, and lignite. The lignite and sub-bituminous coals are the low-ranked coal with low commercial and calorific values (Flores, 2014; Schobert, 2017). The combustion of coal leads to the release of toxic and harmful substances, which negatively impacts the human habitat and environment (Goodwin, 2014; Perera, 2017). Therefore, with a constantly increasing demand and finite fossil fuel sources, there is a surging demand to seek an alternate source of hydrocarbons.
The abundance of lignite and sub-bituminous coal accounts for 53% of the total coal resources in the world. According to the report of Ministry of Coal, the all India Production of coal during 2022-23 was 893.19 MT with a positive growth of 14.77% (https://coal.gov.in/en/major-statistics/production-and-supplies). According to the Geological Survey of India, the total geological resource of lignite of the country stands at 45,758.70 million tonne as on 1 April 2019 and major deposits are located in the state of Tamil Nadu, followed by Rajasthan, Gujarat, Kerala, West Bengal, Union Territory of Jammu & Kashmir and, Pondicherry (Fan et al., 2019). Though India has significant lignite reserves, the majority of these are located in environmentally sensitive geographical areas or at a depth of economically unviable options for exploration. Moreover, the high moisture content, low calorific value, and spontaneous combustion of lignite are the factors responsible for its lower economical utilization (Fan et al., 2019). Therefore, it is important to study how to utilize lignite resources (conventional and unconventional) present in India and worldwide to bridge the gap between demand and supply.
Production of biogenic coalbed gas has been reported from coal reserves in many countries (Goraya et al., 2019). This motivates the effective utilization and understanding of environmentally friendly method: biological methane production from coal. Previous studies have shown that “methane producing” consortium extracted from coal was observed to grow on low-volatile bituminous coal as a sole carbon source (Thielemann et al., 2004). A microbial consortium was successfully applied to bioassay methane generating potential in 18 coal samples (Jones et al., 2008).
Biogenic methane production is the consequence of microbial action leading to complex biochemical reactions taking place during the decomposition of organic matter in anoxic environments. It has been noted that all coal types contain gases, with methane typically being the major constituent (90%–97% of all gases), regardless of maturity, organic makeup, or nature of occurrence. Most appropriate for commercial methane extraction are deep-seated seams (depth of roughly 1,000 m or more) having coal of higher rank (carbon content >83.5%, Vitrinite reflectance R0 max 0.7% and above). There are several pathways using which the methanogens produce methane (Boone, 2000). The most prevalent mode of methane production is a hydrogenotrophic pathway which is the hydrogen-mediated reduction of carbon dioxide. Various environmental factors like pH, salinity, temperature and nutritional factors (inorganic and organic) can significantly affect methane production from microbes (Boone, 2000). For instance, methane synthesis is limited in conditions with slow rates of organic matter turnover because the hydrolysis of big polymers typically limits microbial activity in anoxic environments, and low molecular weight intermediates do not accumulate significantly (Menzel et al., 2020; Mulat et al., 2018). Despite being extremely complex and poorly understood, biogenic methane generation pathways are widespread in nature (Basera et al., 2022). It is believed that 20% of the natural gas in the earth is from methanogens, of which two-thirds are from acetate fermentation and one-third from carbon dioxide reduction (Enzmann et al., 2018; Schöne and Rother, 2018). Several studies have reported coal degradation by microbial cultures to produce methane (Iram et al., 2017). Preliminary evidence suggests that lignite can be microbially degraded and biologically converted to methane (biological pathway). Chemical and isotopic data have established that anaerobic lignite breakdown results in the formation of biogenic methane (Yoon et al., 2016). Anaerobic lignite degradation, leading to biogenic methane generation, has been demonstrated using chemical and isotopic evidence (Yoon et al., 2016). Because it is a rich source of complex organic materials, lignite serves as a substrate for microbial decomposition. However, it is a challenging microbiological substrate due to complex aromatic compounds and macromolecules produced from lignin in the coal matrix. The complex process of degrading lignite to methane by microbes involves the initial disintegration of the lignite’s macromolecular matrix and the fermentation of huge intermediates to simpler organic molecules, allowing them to undergo additional transition to produce methane (Wang et al., 2017).
However, all the previous studies on biogenic methane production from lignite conducted in the laboratory were under mesophilic conditions. But, generally, many unminable lignite reserves in India are high in temperature (>50°C). Such thermophilic lignite reserves are a potent candidate for biogenic methane production for Coal bed methane (CBM). Therefore there is a need to direct the research toward biogenic methane production from sub-surface lignite at such high-temperature conditions (55°C–60°C). With the increasing fuel prices and economic forecasting, this technology is expected to significantly contribute towards producing alternative gaseous fuel.
The present study provides important insights into the bioconversion of lignite to methane by exploring the microbial diversity of the consortia involved in methanogenesis. Further optimization of the conditions was also conducted for enhanced recovery of gases. The findings of the study will contribute significantly to paving future directions in the related field.
2 Materials and methods
2.1 Sample collection and onsite inoculation
During the drilling of wells of Mehsana, Ahmedabad, and Patan-Tharad area of Gujarat, fresh lignite core and water samples were collected from the different lignite seams. The first phase of sampling (December 2017) included the collection of formation water samples from different depths of the well located in Tharad (THAA). In the second phase of sampling (January 2018), lignite and formation water samples were collected from different depths of the well in Langnaj (LJAY).
All the samples were inoculated with formation water onsite in specific anaerobic methanogenic media (MPB) broth followed by storing at 4°C and then transported to the laboratory for further analysis. The composition of MPB culture medium (g/l in deionized water) used: K2HPO4—0.3; KH2PO4—0.3; NH4Cl—0.5; MgSO4.6H2O—0.2; NaCl—1.0; yeast extract—1.0; casein peptone—1.0; resazurin—0.001; L-cysteine HCl—0.5 (pH 7.00 ± 0.2) (Basera et al., 2022).
2.2 Physico-chemical analysis of coal and formation water
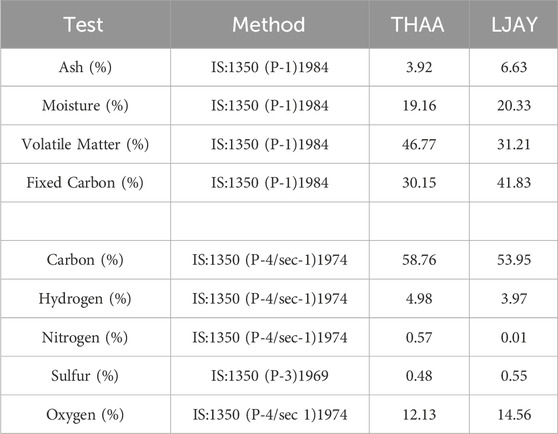
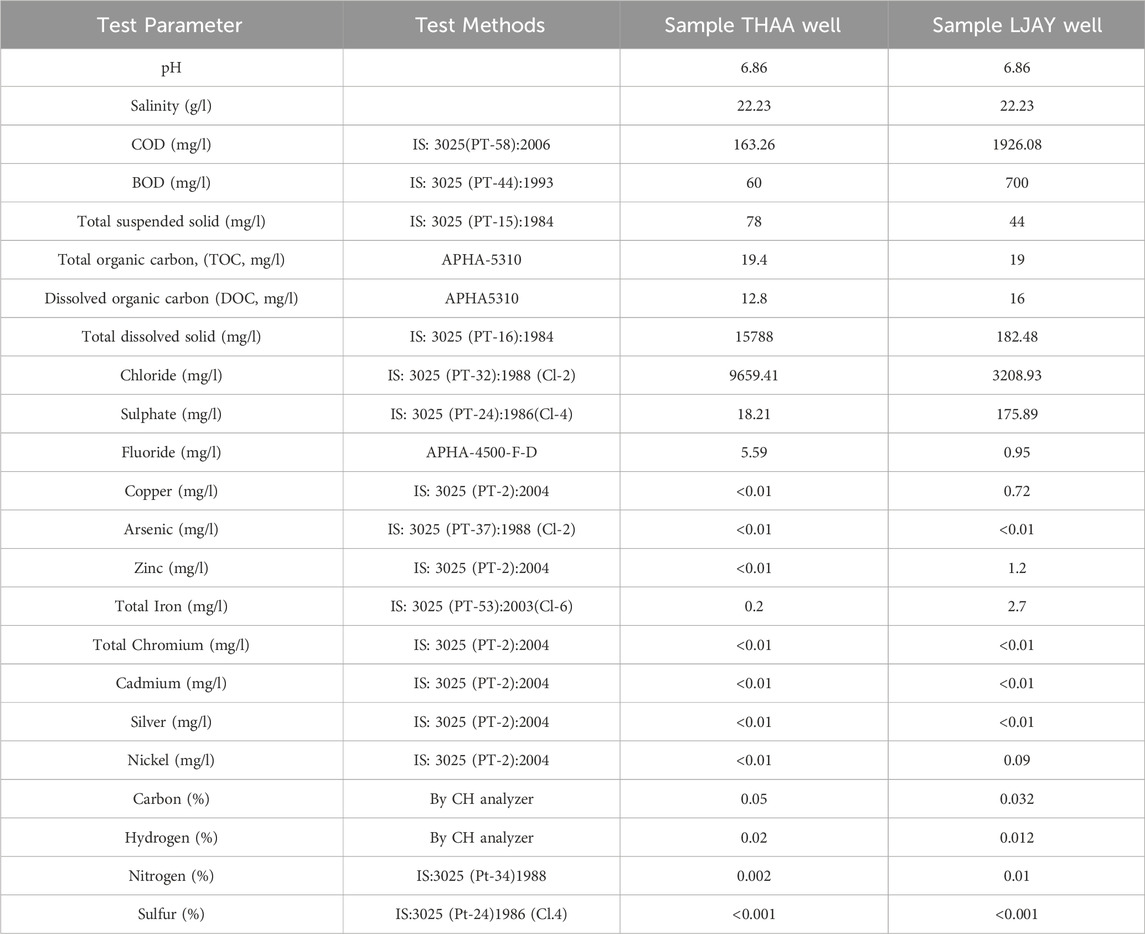
The proximate and ultimate analysis of coal from wells (THAA and LJAY), in the context of ash, moisture, volatile matter, and fixed carbon, along with the specific Carbon, Hydrogen, Nitrogen, Sulphur, and Oxygen (CHNSO) profile, were determined as per the guideline of Bureau of Indian Standards. The method followed and the reference number is mentioned in Test method column of Table 1. The physicochemical analysis of the formation water samples was done. The presence of heavy metals such as Arsenic, Cadmium, Chromium, Copper, Zinc, Copper, and Mercury was also estimated as per the Bureau of Indian Standards and American Public health association standard methods (Rajkovic et al., 2008). The reference of the method followed is described in Table 2.
2.3 Enrichment of indigenous methanogenic consortia
To separate and characterise methanogens from the formation water samples, an enrichment approach was used. A volume of 45 mL of medium was dispensed into each 130 mL serum bottle flushed with O2-free N2. The serum bottles were sealed with butyl rubber stoppers and crimped with aluminum seals. Before inoculation, the sealed, pressurized bottles were sterilized in an autoclave at 121°C temperature for 15 min. At time zero, 45 mL of culture medium was inoculated with 5 mL of the formation fluid/culture using aseptic, strict anaerobic techniques and incubated at 55°C (reservoir’s bottom hole temperature) for 20 days. After the first enrichment cycle was completed, the enriched cultures were transferred to a fresh media (MPB) containing lignite (1% w/v) as a carbon source and incubated for 21 days.
Based on the initial analysis (concerning methane production) of the consortia obtained from formation water, the microbial consortia recovered from the Tharad well were selected for further studies (Supplementary Figure S1). The consortium was also tested for its efficiency in converting coal into methane at different temperatures and salinity (NaCl) conditions.
2.4 Analysis of microbial community in formation water and identification of consortia
With the aim of identifying the microbial community in the formation water, the total genomic DNA was extracted from the formation water, 500 mL of water sample was passed through a membrane filter (90 mm, 0.22 μm). The formation water contained suspended coal particles. To avoid losing those microorganisms attached to coal particles, all that was retained on the filter were then processed for DNA extraction using Power water DNA extraction kit (Mo Bio, United States). DNA was extracted as per the manufacturer’s protocol. Following extraction, DNA samples were quantified and evaluated using Nanodrop. The extracted DNA sample with good quality (A260/A280: 1.8–2.0) and concentrations (more than 50 ng/μL) was taken up further for sequencing.
Polymerase chain reaction (PCR) amplification was done using primers for 16S rRNA (338F: ACTCCTACGGGAGGCAGCA, 806R: GGACTACHVGGGTWTCTAAT, targeting the V3-V4 region) (Caporaso et al., 2010). Amplicon libraries were prepared with high-quality DNA and Nextera Index Kit 16S metagenomic sequencing library preparation protocol. Libraries were sequenced using the Illumina MiSeq platform with 2*300 base-pair chemistry at Medgenome Pvt. Ltd., Bangalore, India.
FASTQC tool v 0.11.8 (Babraham bioinformatics) verified the quality of sequences after demultiplexing and adaptor/primer/barcodes sequence removal from raw reads. Merging of paired-end reads of each sample was carried out using FLASH v 1.2.11 software (Magoč and Salzberg, 2011). Quantitative Insights Into Microbial Ecology (QIIME 2) standard protocol (Bolyen et al., 2019) was followed for quality filtering, library preparation, and Operational Taxonomic Units (OTU) picking. DNA sequences with more than 97% similarity were assembled into the same operational taxonomic units (OTUs). OTU selection and taxonomic assignment were performed according to the SILVA reference database version 138.
2.5 Optimization of lab conditions for enhanced methane production
Physico-chemical parameters greatly impact biogenic methane production (Goswami et al., 2016). Therefore the optimization of laboratory conditions is necessary to study their impact on microbial activity.
The influence of temperature and salt concentrations were analysed by measuring methane production at different conditions. The analysis of the effect of variable concentrations of NaCl was done at different concentrations of 0.5, 1.0, and 1.5 g/L and was checked at selected temperatures of 55°C, 60°C, and 70°C. The biogenic production of methane was analyzed at each condition. The concentration and temperature at which the methane production was found to be highest were chosen for further experiments.
2.6 Analytical methods
2.6.1 Gas analysis and VFA analysis
Production of the gases in the headspace (hydrogen, methane, carbon dioxide, and nitrogen) was quantified by gas chromatography GC 7890A (Agilent Ltd. United States), equipped with a molecular sieve packed stainless steel column (2 m × 2 mm id NUCON, INDIA) and a thermal conductivity detector (TCD) (Rathi et al., 2015). Argon was used as the carrier gas at a flow rate of 1.0 mL/min, and the operating temperatures of the injector, oven, and detector were taken to be at 100, 50, and 150°C, respectively (Rathi et al., 2015). During the enrichment process, methane production in incubated cultures bottle were tested with regular interval of 10 days in each enrichment cycle by taking 0.5 mL of headspace gas samples from the anaerobic serum bottles using gas-tight syringe. The expected headspace gases such as methane, hydrogen, carbon dioxide and nitrogen were quantified by gas chromatography and method was calibrated by injecting standard gases (Sigma) with the range of concentration (0.1 mL, 0.2 mL, 0.3 mL, 0.4 mL, and 0.5 mL). The calibration curve was obtained for all the standards with R2 approximately 0.998. The measured LOD is 0.08 ppm and LOQ is 0.170 ppm.
The water samples were also analyzed for volatile fatty acids (VFAs) and CHNS analysis, The concentration and composition of VFA (formic, acetic, butyric, propionic, valeric, isovaleric, hexanoic and heptanoic acids) in the effluents were analyzed by gas chromatography (GC 6890, Agilent) with a flame ionization detector. The column (CP-Sil 5 CB, 25 m × 0.32 mm × 5 μm, Agilent) operating temperature profiles for 70°C for 0.5 min, then increased to 200°C at 5°C/min and then to 250°C at 10°C/min hold for 1 min. The injector and detector temperature were 250°C. Volatile Free Acid Mix (SupelCo, United States) was used as a VFA standard.
2.6.2 Detection of hydrocarbon
Hydrocarbon were measured in GC 6890 (Agilent Technologies) equipped with HP-5 (5% phenylmethyl siloxane) capillary column (30 m × 320 μm × 0.5 µm) and helium (5 mL min-1) as the carrier gas was used for the analysis of aliphatic and aromatic fractions. The injector and detector (flame ionization detector) temperatures were 280°C and 300°C respectively. The oven was programmed to rise from 80°C to 300°C with an increment of 5°C min-1 and then held for 30 min at this temperature. Authentic standards for aliphatic (C14-C38) and aromatic (naphthalene, anthracene, fluoranthene, pyrene, benzo [a] anthracene, chrysene, benzo [b] fluoranthene, benzo [k] fluoranthene and dibenzo [b] anthracene) were used as reference (Sigma).
2.6.3 Fourier transform infrared spectroscopy (FTIR)
FTIR was carried out to identify the functional groups present in the microbially degraded coal sample. The functional group was characterized by using Fourier Transform Infrared Spectroscopy (Perkin Elmer). All spectra were recorded in an absorbance scale with a mid-measuring region of 400–4,000 cm−1 (mid-infrared range). The resolution was set at 4 cm−1 with 64 scans per spectrum (Sharma et al., 2020).
2.6.4 Scanning electron microscopy (SEM)
The interaction between microbial species and coal was visualized with the help of Scanning Electron Microscopy (Carl Zeiss, Germany). Under aseptic conditions, the sample was absorbed for 2–4 h in a 2.5% glutaraldehyde solution. Phosphate buffer (0.1 M) was used for primary washing, with a pH of 7.2. The sample was dehydrated using ethanol solution in a series of 10%–100% followed by acetone. The overnight air-dried sample was then coated with gold-palladium and visualized through SEM (10 KV in Zeiss EVO MA 10).
2.7 Optimization of the nutrient recipe for enhanced biogenic methane generation from lignite
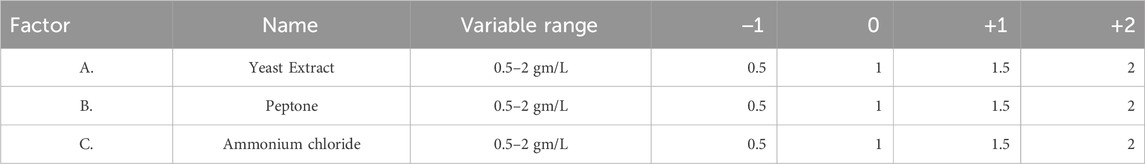
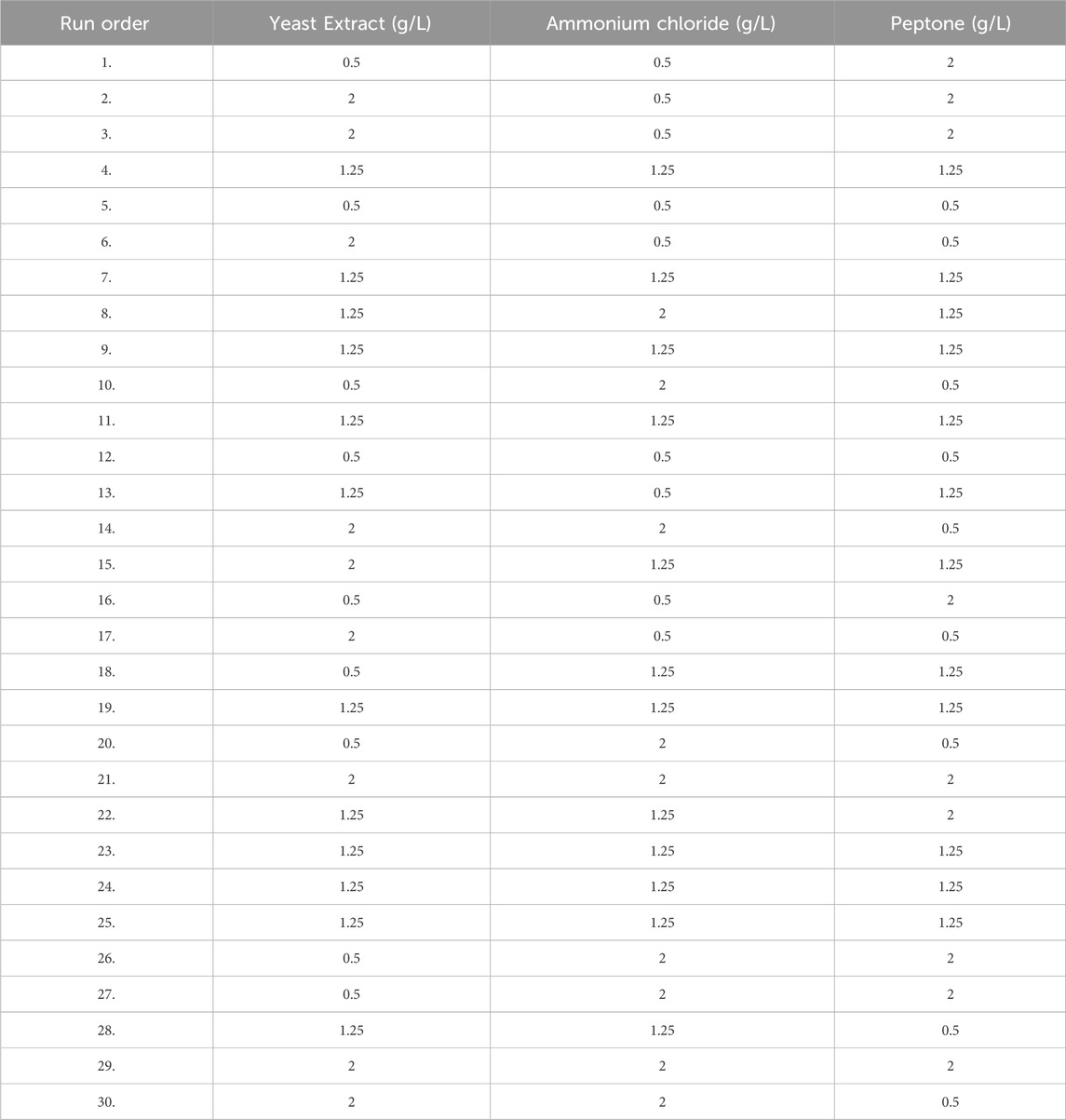
Response surface methodology (RSM) using Box Behnken design was applied to optimize the three important medium components, namely ammonium chloride (0.5–2 g/L; A), casein peptone (0.5–2 g/L; B), and yeast extract (0.5–2 g/L; C) of MPB medium. All experiments were performed in 130 mL serum bottles with a working volume of 45 mL of liquid anaerobic medium with 0.5 g of ground coal and 5 mL of inoculum (enriched media), incubated at 55°C. After 20 days of incubation, methane production was studied as described. All the experiments were performed in triplicate. The data points are the average of the triplicate ±standard deviation (less than 5% of the average), and the calculated significance p-values are ≤0.01.
Utilizing Response Surface Methodology (RSM), optimization studies for increased methane generation from coal were created experimentally (Arslan-Alaton et al., 2010). RSM was used to assess the association between independent variables and biologically improved methane generation (Gopal et al., 2021). The primary composite design model is the one employed in numerical experiments. The model used for numerical experiments is a central composite design. Three major essential nutrients (Yeast Extract, Peptone, and ammonium chloride) were taken in Table 3.
Table 3 describes the range of variable concentrations designed for the optimization of methane production. For the above-mentioned four factors, a total of 30 experiments (Table 4) were designed based on response surface models (Central Composite Design). Two variables, such as yeast extract and concentration of ammonium chloride, have a distinct effect on the production of biological methane and volatile fatty acid production from lignite. For all factors, the interactive effect was identified to be insignificant (p > 0.05). In a polynomial equation, the correlation coefficient (R2) was calculated as 0.9940 for the production of methane, which indicates the variability of the observed response values.
2.8 Scale-up experiment
Scale-up studies were conducted to study the efficacy of Tharad consortia for methane production. The start-up culture was prepared with 1,000 mL of the liquid MPB medium and lignite (1% w/v) as substrate. The sterile nutrient medium was inoculated with 5 mL of the Tharad culture under aseptic conditions and incubated at 55°C for 20 days, after which it was checked for methane production (as described in Section 2.6). After 20th day, 150 mL grown culture was transferred in a 5 L anaerobic bottle containing 2.5 L of liquid MPB medium containing lignite (1% w/v) as the carbon source under strictly anaerobic conditions. The inoculated bottle was then incubated at 55°C for 20 days, post which the methane production was quantified.
2.9 Core flood assay
The core-flood experiment was set up to examine the feasibility of lignite bioconversion to methane using THAA consortia when exposed to temperature and pressure conditions. The study was conducted using the core-flood apparatus (Figure 6A) at a pressure of 900–940 psi and 55°C temperature. Before setting-up up the core-flood study, the internal tubing of the apparatus was flushed with nitrogen gas (99.9% purity) to get rid of atmospheric oxygen. A total of 0.65 gm of lignite core/block was powdered and placed inside the core holder. The core holder was then flooded with ∼650 mL of the medium containing the inoculum. Nitrogen gas was used to maintain the desired pressure and anaerobic condition in the core-flooding apparatus throughout the experiment. The core-flood apparatus was maintained at the described experimental conditions for a period of 21 days. The cell counting was performed with the help of hemocytometer. Cells were stained using Trpan Blue and the total cells were counted using the following formula-
2.10 Pathogenicity of microbial consortia
Since the study’s goal is to deploy lignite bioconversion into methane in the field, it is crucial to carry out the pathogenicity investigation to confirm that the consortiums being employed are suitable for industrial use. The pathogenicity test of THAA was studied by acute oral toxicity under EPA 712-C-96-322 OPPTS 885.3550 guidelines as described by Sharma et al. (2020). Forty mice—20 male and 20 female—were divided into the control and test dose groups in order to evaluate this. 8 mice (4 male and 4 female) were kept as control group. Mice were gavaged with the test substance once (1 mL per mouse). Three to 4 hours before and 2 hours after the test drug was administered, the mice were not given food. The experimental animals that were still alive at the end of the observation period were put to death. Gross necropsy was carried out, and each animal was closely inspected to look for bacterial culture. This study was performed by National Toxicology Centre (APT testing & Research PVT.LTD), Pune under EPA 712-C-96-322 OPPTS 885.3550 guidelines.
2.11 Data availability
All sequence data and metadata information are available through NCBI’s Sequence Read Archive under Bioproject PRJNA970089 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA970089?reviewer=8bjo8u44povj5m32rhkvdu075b).
3 Results and discussion
3.1 Physico-chemical analysis of coal and formation fluid
The ultimate and proximate analysis data of the lignite samples from the well THAA and LJAY have been mentioned in Table 1. The biological conversion of lignite to methane is crucial for securing stable energy resources and sustainable development. Therefore, there have been several physicochemical and biological attempts to improve coal conversion to methane (Piao et al., 2018). Nevertheless, the methane yield depends upon the lignite composition. The data of ultimate and proximate analysis reveals its suitability for anaerobic digestion.
The physicochemical properties of the selected formation water samples (Tharad and Langnaj) were examined (Table 2). All samples showed pH in the range of 6.86–6.94. Total dissolved solids were estimated in all the formation water samples and were found to be in the range of 182–15788 mg/L.
Heavy metal analysis of formation water samples from both wells was done by the standard APHA method, wherein, except for iron, no other heavy metal was detected in the collected formation water samples. Detection of the heavy metals in the formation water is essential because, beyond the threshold levels of toxic ions, the growth of methanogens may be affected, thereby influencing the production of methane (Head et al., 2012). Toxic ions analysis of formation water samples was done by standard APHA 4500 and IS 3025 methods. In these methods, detection limits of the chloride were found at 9,659.41 mg/L in THAA and 3,208.93 mg/L in LJAY. Fluoride concentrations were found 5.59 mg/L in THAA and 0.95 mg/L in LJAY. Sulfate concentrations were found to be 183.21 mg/L in THAA and 175.89 mg/L in LJAY (Table 2). All these concentrations are in the range which doesn’t hinder the growth of methanogens. The physico-chemical analysis of formation water shows absence of heavy metal toxicity and ambient conditions for the growth of microbes (Rathi et al., 2019).
3.2 Enrichment studies
Enrichment studies were conducted with formation water collected from two lignite wells (THAA) to develop the indigenous bacterial consortia for bioconversion of lignite to methane. Due to the significant higher production of methane in the THAA well, the THAA samples were chosen for further assessment (Supplementary Figure S1). The amount of methane generated from the THAA formation water in each subsequent enrichment has been depicted in Figure 1. The first enrichment cycle comprised consideration of THAA cultures for onsite enrichment in two methanogen-specific media MPB containing 1%w/v of coal from the THAA well (lignite). It was in the fourth enrichment cycle where the maximum amount of methane was obtained.
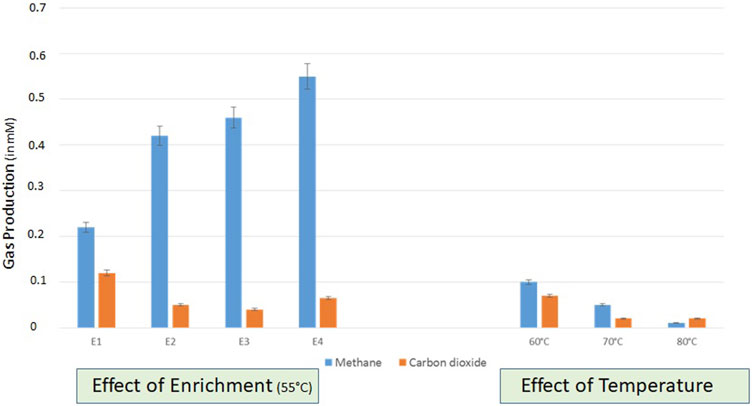
FIGURE 1. Assessment of gas production in different enrichments set uo at 55°C and right panel shows the effect of variable temperatures (60, 70, and 80°C) on the gas production (CH4 and CO2) by indigenous methanogenic consortia. Data was recorded after 20 days of incubation.
The developed consortia from THAA formation water samples were further tested for a range of temperatures and salt concentrations to optimize the bioconversion of lignite to methane. The analysis was conducted at three temperatures (60°C, 70°C, and 80°C), and lignite samples from the THAA well were used with THAA inoculum. Figure 1 illustrates methane production at selected temperatures. From enrichment studies using Tharad inoculum, optimum methane bioconversion was observed at 55°C.
The analysis of the effect of variable concentrations of NaCl (0.5, 1.0, and 1.5 g/L) at selected temperatures (55°C, 60°C, and 70°C) showed the conditions of 55°C temperature and high NaCl concentration (1.5 gm/L) being the most suitable for methane production (Supplementary Figure S2). The optimized conditions supports the fesibility of the process on field for biogenic enhancement in methane production.
3.3 Bacterial community analysis
The microbial communities present in the formation water sample and enrichment culture of the THAA well (Tharad) have been illustrated in Figure 2. The figure shows the rich diversity of bacterial and archaea members in the formation water. The analysis shows the presence of various uncultured members of Methanosaeta, Firmicutes, Clostridia, Archaeon, and, Thermoanaerobacteraceae as the dominant community members in the formation water. Previous studies have reported the presence of Firmicutes, Clostridia, Archaeon, and, Methanosaeta in the formation water extracted from coal beds in various parts of the world. The finding suggests their key role in methane generation and the ability of microbes to survive in extreme environments (Gründger et al., 2015; Beckmann et al., 2019; Rathi et al., 2019). Dictyoglomus thermophillum is a chemoorganotroph and is extremely thermophilic. It plays an essential role in the degradation of organic matter, which is a crucial step for the generation of methane (Coil et al., 2014). Members of the class Clostridia are known to be involved in hydrogen-producing mechanisms (Gomez-Flores et al., 2017; Wang and Yin, 2021). Hydrogen acts as the limiting factor in the hydrogenotrophic methanogenesis pathway; therefore, the presence of these members may immensely impact the gas production in the wells (Kakuk et al., 2021). Members of class Dictyoglomus are abundantly present in producing wells. Members of Dictyoglomus are extremely thermophilic, chemoorganotrophic, and obligate anaerobes (Coil et al., 2014).
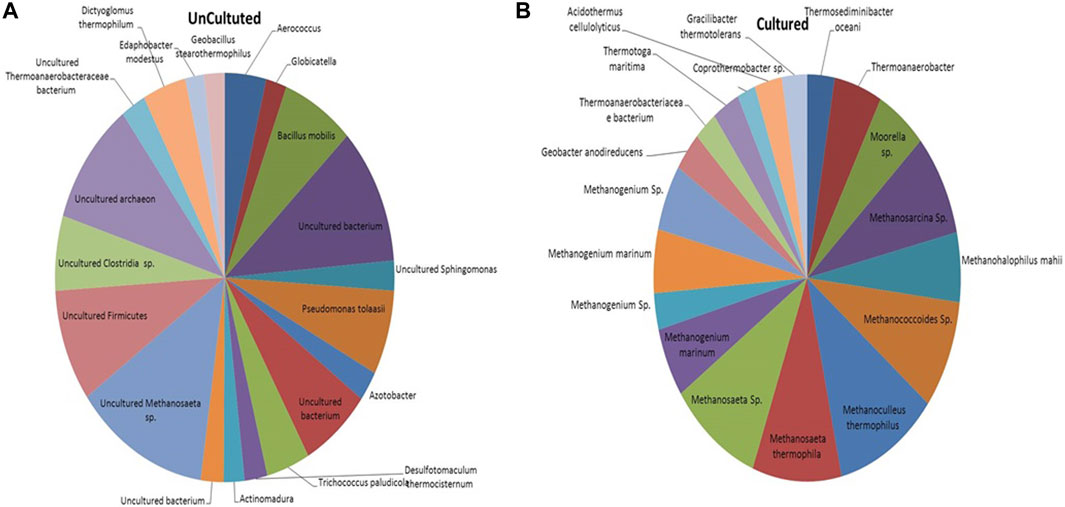
FIGURE 2. Figure illustrating the microbial community composition present in the (A) Formation water sample and (B) Enriched culture sample of THAA well, Tharad.
In the case of cultured adapted microbial consortia, the diversity was high of archaea groups. Dominant members belong to the order Methanosarcinales and Methanomicrobiales. Members like Methanogenium, Methanosaeta, Methanocullens, Methanococcoides, Methanohalophilus, Methanosarcina, and others. The dominance of methanogens in the enriched media shows its suitability for the process. The enriched culture shows presence of Acidothermus cellulyticus, a bacteria known for hydrolysis of organic matter (Shivlata and Tulasi, 2015). Members like Thermoanaerobacteriacea, Thermoanaerobacter, Thermotoga, and Thermosediminibacter are found in the enrichment samples. These members belong to the thermophilic group (Basen et al., 2018; Counts et al., 2017). The enrichment of methanogens and thermophillic microbes shows compatibility of the media to support the growth of microbes in higher temperatures. Various other studies have also supported that the enriched media can support only a subpopulation that dominates in the specific selective conditions (Bonnet et al., 2020).
3.4 Optimized nutrient recipe for enhanced biogenic methane generation
Three-dimensional response surface plots have been designed by keeping a variable at the center and the range of experiments differed with other variables. The effect of interaction for the production of methane gas and volatile fatty acid was analyzed from the three-dimensional surface curve plot. The variable interaction and the calculated response were plotted, and the three-dimension surface curves are shown in Figure 3; Supplementary Figure S3. The maximum response towards the methane gas production 0.897 mM (69.2%) was found to be with the concentration of yeast extract 1.43 g/L, peptone 1.87 g/L and ammonium chloride 1.25 gm/L. Similarly, the maximum response towards the volatile fatty acid production (27.12 mg/L) was found to be with the concentration of yeast extract 1.43 g/L, peptone 1.87 g/L, and ammonium chloride 1.25 g/L. Since the indigenous bacterial community involved in biological methane production are highly sensitive, it requires a specific concentration of each factor for its growth and production of methane, followed by volatile fatty acid. Similar findings with RSM were also shown by (Wu et al., 2017) in terms of a similar methane gas production trend found within the range. For maximal methane gas production, the optimum condition was found to be at 50°C temperature, the concentration of yeast extract 1.43 g/L, peptone 1.87 g/L and ammonium chloride 1.25 gm/L 7.2 pH, substrate concentration, 5 s agitation times.
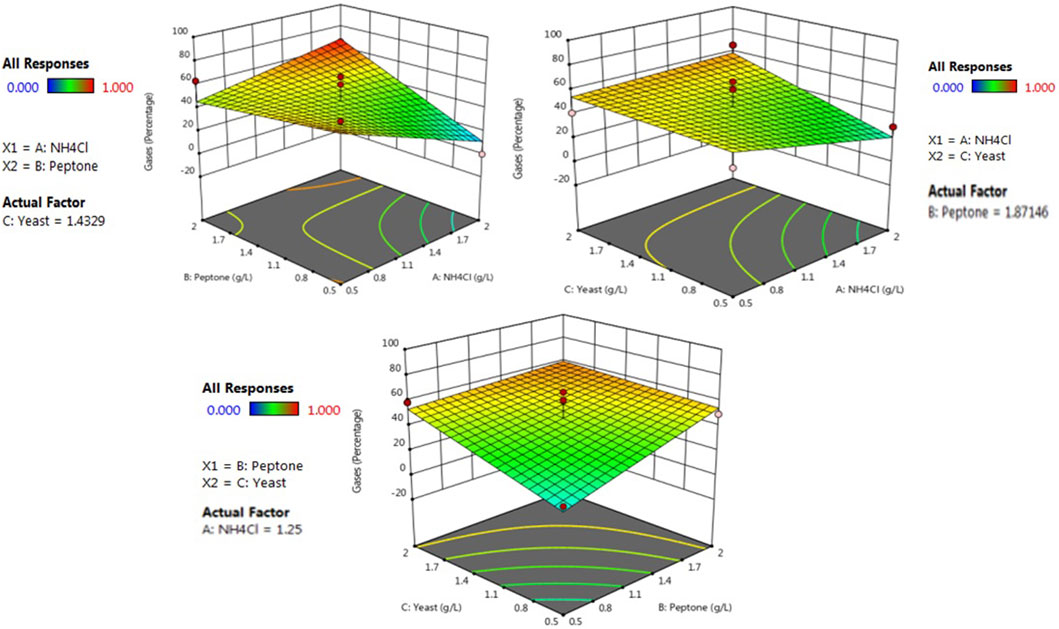
FIGURE 3. RSM contour plots of interactive effects of yeast extract, casein peptone, and ammonium chloride on methane production.
3.5 Scale up study
The bioconversion of lignite to methane was conducted at a large scale (2.5 L) with an optimized nutrient recipe for the THAA consortia and lignite as the sole carbon source. Figure 4 depicts the enhanced methane production by the Tharad consortia after incubation at regular intervals of 7–37 days. Figures 5A, B illustrate the lignite sample’s FTIR spectra before and after the bio consortia interaction. The recorded spectra showed that the peaks range between 2,360–2000 cm−1 are due to aliphatic groups in coal samples and from 1,600 to 1,655 cm−1 corresponds to aromatic C=C structure. The presence of C=O and C-O-R is revealed by the spectra obtained between the 1800–1,000 cm−1 region. In the treated condition, the aromatic C=C stretching indicates the presence of a high amount of carbon. The possibility is due to the reduction of oxygen through the transformation of C=O to CH2 or by decarboxylation of the matrix, which leads to an improved, calorific value. The results depicted that the bacterial treatment biologically converts the lignite into volatile compounds which can then be easily converted into methane.
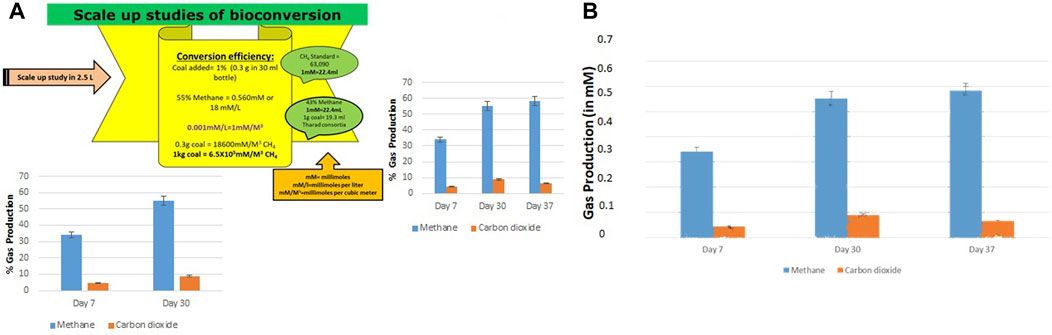
FIGURE 4. (A) Conversion efficiency calculation of scale up study and percentage production of methane and carbon dioxide during the scale-up (2.5 L) studies. (B) Gas production in scale up study in mM on Day 7, 30, and 37.
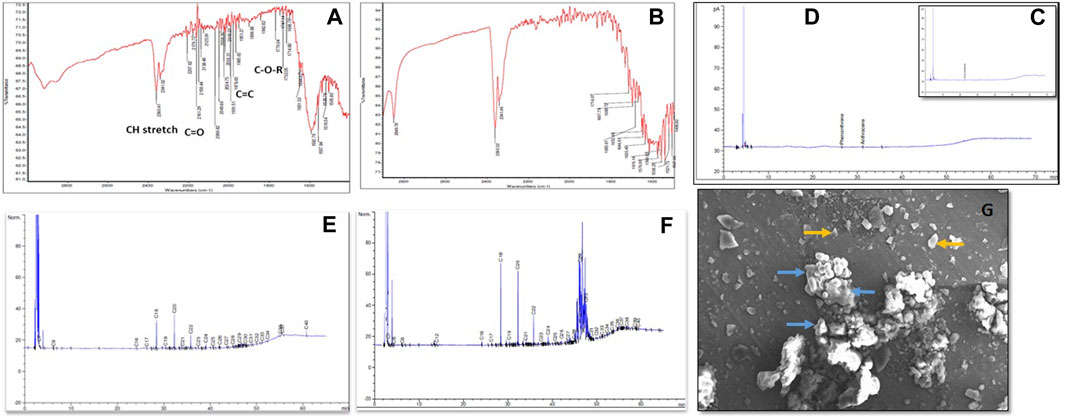
FIGURE 5. (A) FTIR spectra of residual Lignite (Before bioconversion process) (B) FTIR spectra of residual lignite (After the bioconversion process) C and D depicted the GC chromatograms representing aromatic hydrocarbon content in the coal where, (C) Before the bacterial treatment and (D) After the bacterial treatment. (E) Represents the aliphatic hydrocarbon in control, (F) Aliphatic hydrocarbons profile after bacterial treatment. (G) SEM depicted the interaction between lignite and bacteria at 920X magnification, where yellow arrows the coal particles, and blue arrows indicate microbial cells.
Hydrocarbon degradation studies of the lignite sample have been represented in Figures 5C, D, which shows the alkane groups present in the lignite before and after the bacterial treatment. By observing the fragment pattern, a carbon chain from C7-C40 was detected. Major peaks were recorded for C18, C20, C22, C29, and C30. The results indicate partial depolymerization of geopolymers. A characterized THAA bacterial consortium was chosen for studying the interaction between lignite particles and the bacterial inoculum using scanning electron microscopy (SEM). Figure 5E reveals the morphology of bacteria as being coccus shaped, besides the interaction between the bacteria and lignite particles can also be seen.
3.6 Core flood
After an incubation period of 21 days, the spent medium was collected in a sealed N2-flushed 1 L bottle. Under set experimental conditions, the consortium produced 249.51 ± 1.51 mL methane per gram of lignite. VFAs were also produced and detected in the collected spent medium sample. The total VFA content was 9.851 ± 0.367 mM, with Acetic Acid being the most abundant fatty acid. Acetic acid and propionic acids were 6.656 ± 0.169 mM and 1.170 ± 0.169 mM, respectively, along with fewer quantities of butyric acid and i-Valoric acid (Figure 6B). The pH of the spent medium collected was 5.82 ± 0.2, with the total cell count being 3.2 ×
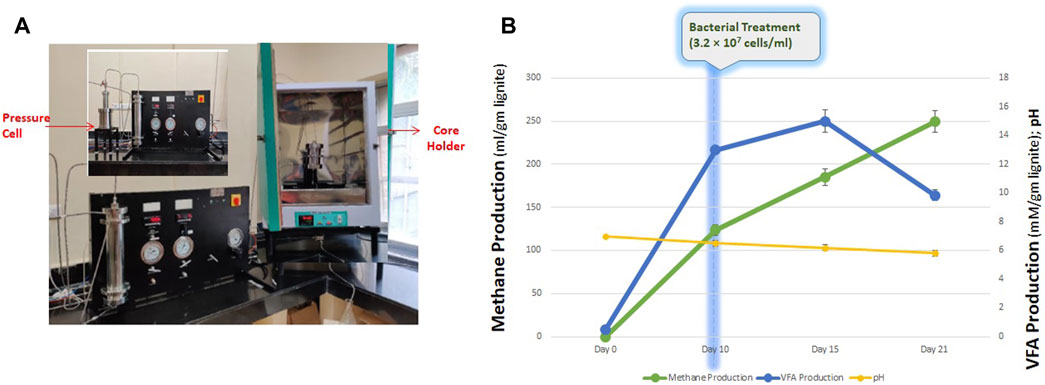
FIGURE 6. (A) Schematic diagram of core flood apparatus. (B) Production of methane, VFA and final pH of the core flood study.
3.7 Pathogenicity of culture
Pathogenicity of microbial consortia obtained from formation water sample THAA, Tharad did not cause any mortality when administered in mice by oral route at the dose of 1.0 mL containing 1×
4 Conclusion
Lignite is abundantly found in various parts of the world, but its low calorific value has less economic significance than other forms of coal. Therefore studies focusing on the effective bioconversion of lignite to methane carry great ecological importance. The present study showed around 0.897 mM production of methane gas through the bioconversion of lignite. The optimization study shows maximum methane production at the temperature of 50°C. The microbial diversity analysis showed diverse groups of bacteria and archaea present in the formation water. The enriched media showed their role in the effective bioconversion of lignite to methane. The analytical analysis shows the partial depolymerization of coal geopolymer. The SEM analysis showed the interaction of the microbes with the lignite surface.
After the optimization of conditions, the scale-up study revealed methane production as 2,800 mM/25 gm of coal, indicating the effective bioconversion of lignite to methane.
The results clearly show how this study is a step forward in the bioproduction of methane, as it utilizes the large lignite reserves, thus enhancing its ecological importance. In addition, the study was carried out successfully at high-temperature conditions, above 50°C, and at depth, too far down from the earth’s surface, compared to otherwise moderate conditions at the oil/coal wells situated in different regions of the world.
Besides, it uses indigenous nonpathogenic microbial consortia for bioconversion of lignite to methane, thus making it a viable choice for similar lignite-bearing oil wells across the country and ensuring there’s caused.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/ the accession numbers are SRR24475124 and SRR24475123.
Ethics statement
The animal studies were approved by Dr. K. G. Apte, APT testing Research Private Limited. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
PB: Formal Analysis, Investigation, Data curation, Writing–original draft. ML: Formal Analysis, Investigation, Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing–review and editing. NS: Funding acquisition, Project administration, Supervision, Writing–review and editing. BL: Project administration, Resources, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2024.1308308/full#supplementary-material
References
Arslan-Alaton, I., Ayten, N., and Olmez-Hanci, T. (2010). Photo-Fenton-like treatment of the commercially important H-acid: process optimization by factorial design and effects of photocatalytic treatment on activated sludge inhibition. Appl. Catal. B-environmental - Appl. Catal. B-ENVIRON 96, 208–217. doi:10.1016/j.apcatb.2010.02.023
Basen, M., Geiger, I., Henke, L., and Müller, V. (2018). A genetic system for the thermophilic acetogenic bacterium thermoanaerobacter kivui. Appl. Environ. Microbiol. 84, e02210-17. doi:10.1128/AEM.02210-17
Basera, P., Lavania, M., Shinde, O., Sarkar, S. K., and Lal, B. (2022). An operative laboratory investigation of bioconversion route from waste coal to natural energy. Ann. Microbiol. 72, 13. doi:10.1186/s13213-021-01659-z
Beckmann, S., Luk, A. W. S., Gutierrez-Zamora, M.-L., Chong, N. H. H., Thomas, T., Lee, M., et al. (2019). Long-term succession in a coal seam microbiome during in situ biostimulation of coalbed-methane generation. ISME J. 13, 632–650. doi:10.1038/s41396-018-0296-5
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 37 (8), 852–857. doi:10.1038/s41587-019-0209-9
Bonnet, M., Lagier, J. C., Raoult, D., and Khelaifia, S. (2020). Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New microbes new Infect. 34, 100622. doi:10.1016/j.nmni.2019.100622
Boone, D. R. (2000). in Biological Formation and consumption of methane BT - atmospheric methane: its role in the global environment. Editor M A K Khalil (Berlin, Heidelberg: Springer), 42–62. doi:10.1007/978-3-662-04145-1_4
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–336. doi:10.1038/nmeth.f.303
Coil, D. A., Badger, J. H., Forberger, H. C., Riggs, F., Madupu, R., Fedorova, N., et al. (2014). Complete genome sequence of the extreme thermophile Dictyoglomus thermophilum H-6-12. Genome announc. 2, e00109-14. doi:10.1128/genomeA.00109-14
Counts, J. A., Zeldes, B. M., Lee, L. L., Straub, C. T., Adams, M. W. W., and Kelly, R. M. (2017). Physiological, metabolic and biotechnological features of extremely thermophilic microorganisms. WIREs Syst. Biol. Med. 9, e1377. doi:10.1002/wsbm.1377
Enzmann, F., Mayer, F., Rother, M., and Holtmann, D. (2018). Methanogens: biochemical background and biotechnological applications. Amb. Express 8, 1. doi:10.1186/s13568-017-0531-x
Fan, J., Wang, G., and Zhang, J. (2019). Study on spontaneous combustion tendency of coals with different metamorphic grade at low moisture content based on TPO-DSC. Energies 12, 3890. doi:10.3390/en12203890
Flores, R. M. (2014). Coal composition and reservoir characterization. Coal Coalbed Gas. 2014, 235–299. doi:10.1016/B978-0-12-396972-9.00005-7
Gomez-Flores, M., Nakhla, G., and Hafez, H. (2017). Hydrogen production and microbial kinetics of Clostridium termitidis in mono-culture and co-culture with Clostridium beijerinckii on cellulose. Amb. Express 7, 84. doi:10.1186/s13568-016-0256-2
Goodwin, R. W. (2014). Coal combustion residue disposal options. Combust. Ash Residue Manag. 2014, 105–113. doi:10.1016/B978-0-12-420038-8.00008-2
Gopal, L. C., Govindarajan, M., Kavipriya, M. R., Mahboob, S., Al-Ghanim, K. A., Virik, P., et al. (2021). Optimization strategies for improved biogas production by recycling of waste through response surface methodology and artificial neural network: sustainable energy perspective research. J. King Saud. Univ. - Sci. 33, 101241. doi:10.1016/j.jksus.2020.101241
Goraya, N. S., Rajpoot, N., and Marriyappan Sivagnanam, B. (2019). Coal bed methane enhancement techniques: a review. ChemistrySelect 4, 3585–3601. doi:10.1002/slct.201803633
Goswami, R., Chattopadhyay, P., Shome, A., Banerjee, S. N., Chakraborty, A. K., Mathew, A. K., et al. (2016). An overview of physico-chemical mechanisms of biogas production by microbial communities: a step towards sustainable waste management. 3 Biotech. 6, 72. doi:10.1007/s13205-016-0395-9
Gründger, F., Jiménez, N., Thielemann, T., Straaten, N., Lüders, T., Richnow, H.-H., et al. (2015). Microbial methane formation in deep aquifers of a coal-bearing sedimentary basin, Germany. Front. Microbiol. 6, 200. doi:10.3389/fmicb.2015.00200
Iram, A., Akhtar, K., and Ghauri, M. (2017). Coal methanogenesis: a review of the need of complex microbial consortia and culture conditions for the effective bioconversion of coal into methane. Ann. Microbiol. 67, 275–286. doi:10.1007/s13213-017-1255-5
Jones, E., Voytek, M., Warwick, P., Corum, M., Cohn, A., Bunnell, J., et al. (2008). Bioassay for estimating the biogenic methane-generating potential of coal samples. Int. J. Coal Geol. 76, 138–150. doi:10.1016/j.coal.2008.05.011
Kakuk, B., Wirth, R., Maróti, G., Szuhaj, M., Rakhely, G., Laczi, K., et al. (2021). Early response of methanogenic archaea to H2 as evaluated by metagenomics and metatranscriptomics. Microb. Cell Fact. 20, 127. doi:10.1186/s12934-021-01618-y
Litvinenko, V. (2020). The role of hydrocarbons in the global energy agenda: the focus on liquefied natural gas. Resources 9, 59. doi:10.3390/resources9050059
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27, 2957–2963. doi:10.1093/bioinformatics/btr507
Menzel, T., Neubauer, P., and Junne, S. (2020). Role of microbial hydrolysis in anaerobic digestion. Energies 13, 5555. doi:10.3390/en13215555
Mulat, D. G., Dibdiakova, J., and Horn, S. J. (2018). Microbial biogas production from hydrolysis lignin: insight into lignin structural changes. Biotechnol. Biofuels 11, 61. doi:10.1186/s13068-018-1054-7
Perera, F. (2017). Pollution from fossil-fuel combustion is the leading environmental threat to global pediatric health and equity: solutions exist. Int. J. Environ. Res. Public Health 15, 16. doi:10.3390/ijerph15010016
Piao, D.-M., Song, Y.-C., and Kim, D.-H. (2018). Bioelectrochemical enhancement of biogenic methane conversion of coal. Energies 11, 2577. doi:10.3390/en11102577
Rajkovic, M. B., Lacnjevac, C. M., Ralevic, N. R., Stojanović, M. D., Tosković, D. V., Pantelic, G. K., et al. (2008). Identification of metals (heavy and radioactive) in drinking water by an indirect analysis method based on scale tests. Sensors (Basel) 8, 2188–2207. doi:10.3390/s8042188
Rathi, R., Lavania, M., Sawale, M., Kukreti, V., Kumar, S., and Lal, B. (2015). Stimulation of an indigenous thermophillic anaerobic bacterial consortium for enhanced oil recovery. RSC Adv. 5, 88115–88124. doi:10.1039/C5RA10489K
Rathi, R., Lavania, M., Singh, N., Sarma, P. M., Kishore, P., Hajra, P., et al. (2019). Evaluating indigenous diversity and its potential for microbial methane generation from thermogenic coal bed methane reservoir. Fuel 250, 362–372. doi:10.1016/j.fuel.2019.03.125
Schobert, H. (2017). “Introduction to low-rank coals: Types, resources, and current utilization,” in Low-Rank Coals for Power Generation, Fuel and Chemical Production. ISBN 9780081008959, 3–21. doi:10.1016/B978-0-08-100895-9.00001-2
Schöne, C., and Rother, M. (2018). in Methanogenesis from carbon monoxide BT - biogenesis of hydrocarbons. Editors A J M Stams, and D Sousa (Cham: Springer International Publishing), 1–29. doi:10.1007/978-3-319-53114-4_4-1
Sharma, N., Lavania, M., Kukreti, V., and Lal, B. (2020). Instigation of indigenous thermophilic bacterial consortia for enhanced oil recovery from high temperature oil reservoirs. PLoS One 15, e0229889. doi:10.1371/journal.pone.0229889
Shivlata, L., and Tulasi, S. (2015). Thermophilic and alkaliphilic Actinobacteria: biology and potential applications. Front. Microbiol. 6, 1014. doi:10.3389/fmicb.2015.01014
Thielemann, T., Cramer, B., and Schippers, A. (2004). Coalbed methane in the Ruhr Basin, Germany: a renewable energy resource? Org. Geochem. 35, 1537–1549. doi:10.1016/j.orggeochem.2004.05.004
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi:10.1093/nar/22.22.4673
Wang, B., Tai, C., Wu, L., Chen, L., Liu, J., Hu, B., et al. (2017). Methane production from lignite through the combined effects of exogenous aerobic and anaerobic microflora. Int. J. Coal Geol. 173, 84–93. doi:10.1016/j.coal.2017.02.012
Wang, J., and Yin, Y. (2021). Clostridium species for fermentative hydrogen production: an overview. Int. J. Hydrogen Energy 46, 34599–34625. doi:10.1016/j.ijhydene.2021.08.052
Keywords: lignite, volatile fatty acids, methane, anaerobic scale up, coreflood experiment
Citation: Basera P, Lavania M, Singh N and Lal B (2024) Laboratory investigation and core flood demonstration of enhanced biogenic methane generation from lignite. Front. Bioeng. Biotechnol. 12:1308308. doi: 10.3389/fbioe.2024.1308308
Received: 06 October 2023; Accepted: 10 January 2024;
Published: 19 February 2024.
Edited by:
Balaram Mohapatra, Gujarat Biotechnology Research Centre (GBRC), IndiaReviewed by:
Preeti Solanki, Chandigarh University, IndiaDhiraj Paul, National Centre for Cell Science, India
Copyright © 2024 Basera, Lavania, Singh and Lal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meeta Lavania, bWVldGFsQHRlcmkucmVzLmlu
 Priyanka Basera
Priyanka Basera Meeta Lavania
Meeta Lavania Nimmi Singh2
Nimmi Singh2