
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 01 August 2023
Sec. Bioprocess Engineering
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1227619
This article is part of the Research Topic Plants and Environmental Threats View all 6 articles
 Pei Liu1,2†
Pei Liu1,2† Haiyu Long1,2†
Haiyu Long1,2† Han Cheng1,2
Han Cheng1,2 Mengdi Liang1,2
Mengdi Liang1,2 Zhengwei Liu1,2
Zhengwei Liu1,2 Zhenlian Han3
Zhenlian Han3 Zhen Guo1,2
Zhen Guo1,2 Hao Shi1,2
Hao Shi1,2 Min Sun4
Min Sun4 Shuai He1,2*
Shuai He1,2*Introduction: Bacillus species are known for their ability to produce nanoparticles with various potential applications.
Methods: In this study, we present a facile approach for the green synthesis of selenium nanoparticles (Se NPs) using the biogenic selenate-reducing bacterium Bacillus paramycoides 24522. We optimized the growth conditions and sodium selenite reduction efficiency (SSRE) of B. paramycoides 24522 using a response surface approach.
Results: Se NPs were synthesized by reducing selenite ions with B. paramycoides 24522 at 37 °C, pH 6, and 140 r/min, resulting in stable red-colored Se NPs and maximal SSRE (99.12%). The synthesized Se NPs demonstrated lethality against Staphylococcus aureus and Escherichia coli with MICs of 400 and 600 μg/mL, and MBCs of 600 and 800 μg/mL, respectively, indicating the potential of Se NPs as antibacterial agents. Furthermore, the Se NPs showed promising antioxidant capabilities through scavenging DPPH radicals and reducing power.
Discussion: This study highlights the environmentally friendly production of Se NPs using B. paramycoides 24522 and their possible applications in addressing selenium pollution, as well as in the fields of environment and biotechnology.
Selenium (Se) plays a vital role in the construction of plants and biological systems at lower concentrations (Al-Hagar et al., 2021). As shown in Figure 1 Se is present in different oxidation states, such as soluble Se oxyanions (SeO42− and SeO32−), which are released by geologic and anthropogenic sources (volcanos and mining) into the environment, such as lakes and rivers (Song et al., 2017). These Se oxyanions can easily be assimilated and bioaccumulated by fish, shrimp, or some other aquatic animals from contaminated rivers, lakes, or oceans, adversely affecting the health of human beings (Shi et al., 2021). Therefore, Se contamination of water resources has become an increasingly severe health risk issue (Jadhav et al., 2022). Due to the toxicity of Se oxyanions, the maximum acceptable content in drinking water assigned by the World Health Organization (WHO) is 40 μg/L and 10 μg/L in the Bureau of Indian Standards (BIS) guidelines (Sonkeshariya et al., 2020). Additionally, the Ministry of Environment and Climate Change Strategy in Canada assigned a policy for background Se concentrations of 2 μg/L in the water columns and sediment (English et al., 2022). In this regard, eliminating and/or reducing the adverse effects caused by high concentrations of Se oxyanions has become increasingly imminent.
Selenium nanoparticles (Se NPs), however, exhibit low or no cytotoxicity when compared with Se oxyanions (Xu et al., 2018; He et al., 2021). Recently, Se NPs have received widespread attention because of their unique properties, such as antioxidant (Azimi et al., 2021; Brar et al., 2022), photocatalytic (Velayati et al., 2022), antibacterial and anticancer activities (Han et al., 2020; Rana, 2021). Therefore, the development of green, efficient and sustainable biotechnological synthetic routes for transforming toxic Se oxyanions into nontoxic Se NPs is of great significance.
There are numerous techniques available for the production of Se NPs, including chemical reduction, physical processes, and biological approaches. Nonetheless, these methods are associated with drawbacks such as high expenses, limited safety, significant toxicity, and environmental contamination (Shreyash et al., 2021). Microbial cells possess a distinctive metabolic pathway that allows them to generate unique metabolites with special characteristics, including enzymes and bioactive substances. As living organisms, they offer great potential for exploring new avenues in reducing the soluble toxic selenium oxides to the insoluble hypotoxic selenium element (Se0), which in turn forms Se NPs (Zhang et al., 2018). Bacillus strains, such as Bacillus mycoides SeITE01 (Lampis et al., 2014), Bacillus subtilis 168 (Jia et al., 2022), and Bacillus niabensis OAB2 (Al-Hagar et al., 2021), have shown promise as nano-factories capable of producing a range of Se NPs, when culturing these bacteria in media containing nutrients and the precursor selenite. The reaction mixture changes from colorless to reddish color, generally indicating successful synthesis of SeNPs. Some Gram-negative bacteria, such as Stenotrophomonas maltophilia SeITE02 (Wadhwani et al., 2016), Shewanella sp. 023S (Staicu et al., 2022), Pseudomonas aeruginosa ATCC 27853 (Kora and Rastogi, 2016), Escherichia coli ATCC 35218 (Kora and Rastogi, 2017), Enterobacter cloacae Z0206 (Song et al., 2017), and Lactobacillus casei ATCC 393 (Qiao et al., 2020) have also been reported to synthesize Se NPs.
Among the microorganisms that have been reported, B. paramycoides is a valuable microbe that can efficiently degrade organic matter and eliminate pollutants. This bacterium possesses excellent reduction abilities and can reduce selected metal ions, transforming them into nanoparticles (Yadav et al., 2023). In a study by (Dharmaraj et al., 2021), zinc oxide nanoparticles (ZnO NPs) were synthesized using the cell-free supernatant of B. paramycoides. Additionally, the culture supernatant of this bacterium was utilized to produce silver oxide nanoparticles (Ag2O NPs), which were evaluated for their potential antibiofilm activity and cytotoxic effects. These findings suggest that B. paramycoides could be a promising organism for the synthesis of nanoparticles with potential environmental remediation and waste management applications. Its potential should be further explored to unlock its full benefits for society.
In this study, an aquatic-derived B. paramycoides 24522 with remarkable reduction efficiency of selenite was isolated from selenium-rich aquaculture pond sediment in China. Factors affecting B. paramycoides 24522 growth and sodium selenite reduction efficiency (SSRE) were optimized using response surface methodology. The Se NPs were characterized using dynamic light scattering (DLS), X-ray diffraction (XRD), Fourier transform infrared (FT-IR) spectroscopy, and scanning electron microscopy-energy dispersive spectrometry (SEM-EDS). The antimicrobial and antioxidant capabilities of Se NPs were also evaluated.
Sediment samples were collected from the Hongze Lake aquaculture pond (33°02′N, 118°28′E) in Huai’an, China. To generate selenium-rich aquaculture pond sediment, sodium selenite (Na2SeO3) powder was added to the sediment and cultivated for 30 days, sprayed with water and stirred thoroughly 5–6 times every day. Na2SeO3 and all the other analytical grade chemicals were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
One gram of selenium-rich aquaculture pond sediment was dispensed into 100 mL of LB medium (5 g/L peptone, 3 g/L beef extract, 5 g/L NaCl) and incubated at 37°C with shaking at 200 rpm for 1 week. After that, 1 mL of the upper liquid was added into 9 mL sterilized deionized water, and this was a 10−1 dilution. Following a similar procedure, we prepared 10−2–10−6 dilutions. To screen Se NP-producing microorganisms, 5 mM Na2SeO3 was first added to solid LB plates, and 0.2 mL of each dilution was then added, spread and placed at 37 °C for 1–7 days. The isolated microorganisms were purified by streaking on a plate many times. Single colonies were picked out and stored at 4°C for further experiments.
For screening efficient selenate-reducing microbes, 0.1 mL of the single isolated colonies were inoculated in 100 mL LB liquid medium with 5 mM Na2SeO3 and cultured at 37°C for 24 h. The fermentation broth turned reddish in 24 h, indicating that the corresponding microbes possess a relatively high conversion ability of Na2SeO3. The most efficient selenate-reducing isolate, 1805, was identified based on its physiological/biochemical characteristics with the help of 16S rDNA sequencing. PCR products of isolate 1805 were viewed on an agarose gel (1% w/v), sequenced, and BLAST searched through the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/). The phylogenetic tree of strain 1805 was plotted using MEGA 11 software (Tamura et al., 2021).
The growth curve of B. paramycoides 24522 was plotted by culturing it in LB broth at 37°C and 200 rpm for 48 h. The effects of temperature (20, 25, 30, 37, 40, 42°C), pH (4, 5, 6, 7, 8, 9, 10, 11) and rotation speed (0, 50, 100, 150, 200, 250, 300, 350 rpm) on the growth of B. paramycoides 24522 were determined.
The sodium selenite concentration was determined by the Na2S chromogenic method following the published protocols of (Biswas et al., 2011). In brief, a standard curve (Supplementary Figure S1) of elemental selenium was first plotted by accurately adding 1, 2, 4, 6, 8, 10, 12, 14, 16, 18, and 20 mg of elemental selenium powder into 30 mL of sodium sulfide nonahydrate solution (1 M) and mixing thoroughly. The absorbances were measured at a wavelength of 500 nm (OD500), taking sodium sulfide nonahydrate solution without selenium powder as a control.
After that, the content of synthetic elemental selenium in the culture medium was determined by inoculating 1% of B. paramycoides 24522 into LB medium with 2 mM Na2SeO3, which was prepared as a 1 M stock solution and filter sterilized. The cells were cultured at 37°C and 200 rpm for 24 h. The mixture was centrifuged at 5,000×g and 4°C for 20 min, the supernatant was discarded, and the sample was washed and centrifuged three times with 1 M NaCl (5,000×g, 20 min, resuspended in 30 mL of 1 M sodium sulfide nonahydrate solution for 30 min. The sample was centrifuged again at 10,000×g for 30 min. The absorbance at OD500 of the supernatant liquid was measured using B. paramycoides 24522 cultures for 24 h without adding Na2SeO3 and under the same operation as the control. The content of synthetic elemental selenium was calculated by a standard curve.
SSRE was calculated as follows:
The effects of temperature (20, 25, 30, 35, 37, 40, 42, 45°C), pH (2, 4, 6, 8, 10, 12), and rotation speed (0, 50, 100, 150, 200, 250, 300, 350 rpm) on SSRE of B. paramycoides 24522 were studied in the presence of 2 mM Na2SeO3. SSRE under different conditions was measured and calculated as described in Section 2.4.1.
Response surface methodology was used to optimize the conditions for SSRE of B. paramycoides 24522. The experiment was conducted using Box-Behnken design (BBD) with Design-Expert version 12 software. The pH (A), temperature (B), and rotation speed (C) were detected as independent variables according to the results of the above single-factor experiments. SSRE was used as a response value. The factors and levels of these independent variables are presented in Supplementary Table S2. BBD was applied to obtain the second-order response surface.
To determine the location and distribution of the biosynthesized Se NPs, B. paramycoides 24522 was inoculated into LB medium supplemented with 2 mM Na2SeO3. A negative control was set up with the isolate cultured in only LB medium. After incubation at 37°C for 24 h, the bacterial cultures were harvested through gentle centrifugation (5000×g for 10 min at 4°C). The strain was then subjected to cell fragmentation for the extraction of proteins and polysaccharides from each site. To begin, the supernatant and precipitate were collected. The supernatant contained exocytosis protein (ECP), while the precipitate was washed using 1×PBS buffer (pH7) and resuspended via centrifugation. Next, we centrifuged the supernatant at 10,000×g for 30 min to obtain periplasmic space protein (PSP). The precipitate from the previous step was dissolved in 1×PBS buffer (pH7) and treated with Dnase Ⅰ (125 mg/mL), which enzymatically cleaves phosphodiester bonds between DNA bases. Ultrasound was then used to further break up the sample, which was then centrifuged at 5000 g for 10 min to discard the precipitate and collect the supernatant, containing cytoplasmic protein (CPP). This precipitate was dissolved in 1×PBS buffer (pH7) to obtain cell membrane and cell wall protein (MWP). To obtain intracellular polysaccharides (IPS), 50 mL of ECP was filtered through a 0.22 um membrane. The resulting sample was then treated with an equal volume of ice ethanol and refrigerated overnight at −20°C. Finally, we centrifuged the sample at 12,000×g for 30 min at 4°C to obtain IPS.The isolated proteins and polysaccharides were incubated with sodium selenite, and changes in color activity were observed in EP tubules. All samples were kept on ice or at 4°C during the operation to prevent damage from external factors.
To purify Se NPs from the culture broth, a 24 h culture of B. paramycoides 24522 was grown in LB medium containing 2 mM Na2SeO3. The culture was then collected by centrifugation at 5000 g for 20 min and washed three times with 0.1 M phosphate buffered saline (PBS). The resulting pellets were subjected to an ultrasonication treatment using 500 W for 50 min of 2 s of sonication with 8 s of rest in between. The Se NPs were then harvested by centrifugation at 11,000×g for 20 min, following the method outlined in (Huang et al., 2021). This approach was successful in recovering Se NPs from the B. paramycoides 24522 culture, providing a potential avenue for further study of Se NPs and their applications.
To provide additional details on the properties of Se NPs, UV-Vis spectroscopy (MD SpectraMax Plus384, United States), DLS (Brookhaven Instruments NanoBrook 90Plus Zeta with Particle Solutions Ver. 3.5), XRD (Bruker, United States), FT-IR spectroscopy, and SEM-EDS were carried out as described in our previous work (Liu et al., 2022; Nie et al., 2022).
In this study, the antibacterial properties of the as-prepared Se NPs at a concentration of 1 mg/mL were evaluated using the well diffusion method (Liu et al., 2022). Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922 were subcultured on nutrient agar and evenly coated on each plate. Then, 8 mm diameter wells containing the prepared Se NPs were added to the nutrient agar, along with controls of B. paramycoides 24522 culture (without Na2SeO3, 200 μL) and Na2SeO3 (200 μL, 2 mM). The plates were then incubated at 37°C for 24 h, after which the size of the inhibition zone was measured.
The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values of the biosynthesized Se NPs were determined according to a protocol published in our previous work (Nie et al., 2022). Different concentrations of Se NPs ranging from 100 to 1,000 μg/mL were added to the primary bacterial culture and incubated at 180 rpm and 37°C for 24 h. Bacterial growth was monitored using spectrophotometry at a wavelength of 600 nm (OD). The bacterial inhibition rate was determined using the formula below:
The MIC was determined as the concentration that hindered the normal growth of the test pathogens, and the MBC was defined as the concentration that completely suppressed the normal growth of the test pathogens.
The abilities of Se NPs, butyl hydroxy anisole (BHA), and butylated hydroxytoluene (BHT) as scavengers of 1,1-diphenyl-2-picrylhydrazyl (DPPH) radicals were evaluated using spectrophotometric techniques (Liu et al., 2022). A solution containing 6 mL of DPPH at a concentration of 0.06 mM was mixed with Se NPs (100 μg/mL) and then incubated in darkness for 30 min. Absorbance was measured at 517 nm against a blank. The scavenging capacity of DPPH was calculated as follows:
To evaluate the reducing power of the as-prepared Se NPs, we modified the method proposed by Mao (Mao et al., 2013). We prepared reaction mixtures by adding varying amounts of Se NPs (5, 10, 20, 30, 40, and 50 μg) into 2.5 mL of phosphate buffer (0.2 mol/L, pH 6.6) and 2.5 mL of potassium ferricyanide (1%). To start the experiment, the mixture was incubated in a water bath at 50°C for 20 min. Afterwards, 2.5 mL of 10% trichloroacetic acid (TCA) were added to each solution and then centrifuged at 1,000 g for 10 min. After separation, 2.5 mL of the supernatant was mixed with 2.5 mL of distilled water and 0.5 mL of FeCl3 (0.1%). The mixture was then allowed to react for 10 min, and the absorbance was measured at 700 nm. As control experiments, 0.1 g/L BHA and 0.1 g/L BHT were used.
The data were analyzed using Origin Pro 9 software and reported as the mean ± SD. To determine statistical significance (p < 0.05), one-way ANOVA was performed on the experimental results.
In this study, we investigated 162 isolates capable of converting Na2SeO3 into Se NPs within 48 h. These isolates were obtained from LB agar plates supplemented with 5 mM Na2SeO3. Six bacterial isolates (0903, 1801, 1802, 1804, 1805 and 2505) that could synthesize SeNPs within 24 h were further isolated from LB liquid medium with different concentrations of Na2SeO3. Of these strains, isolate 1805 exhibited the highest sodium selenite reduction efficiency (SSRE) and the greatest resistance to Na2SeO3 in the isolation medium (Figure 2). The bacteria capable of reducing Na2SeO3 appeared as reddish bacterial fluid in the culture medium, indicating the accumulation of Se NPs (Al-Hagar et al., 2021).
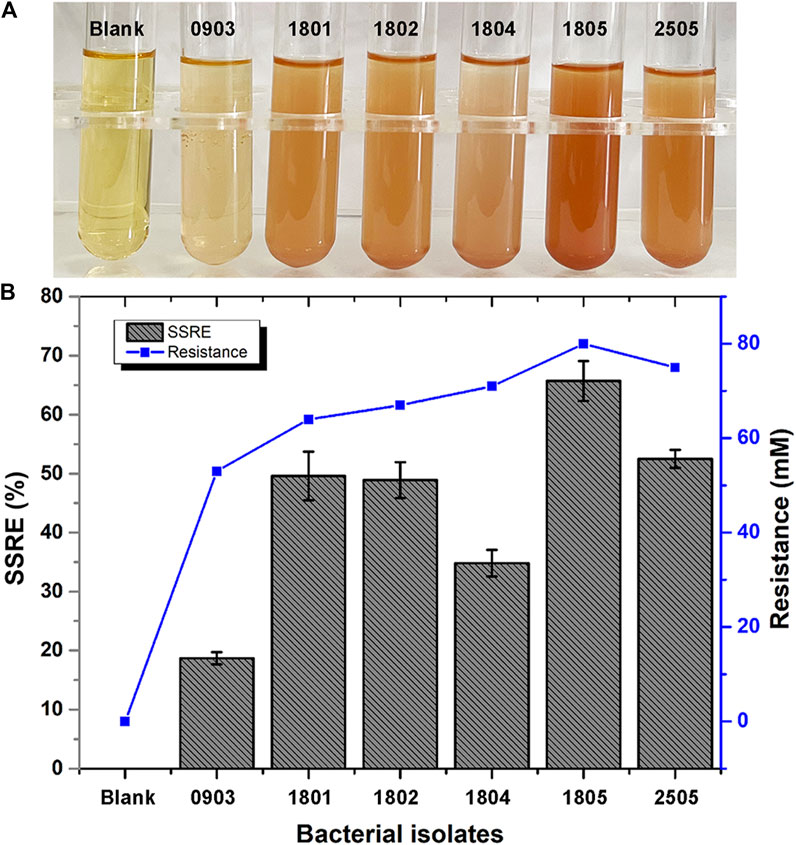
FIGURE 2. (A) Color changes during Se NPs biosynthesis (Blank: LB broth before synthesis, 0903, 1801, 1802, 1804, 1805, 2505: After synthesis with isolates 0903, 1801, 1802, 1804, 1805 and 2505). (B) Sodium selenite reduction efficiency (SSRE) of 6 strains after 24 h.
As shown in Figures 3B, C, isolate 1805 was milky white, opaque, viscous, and slightly protruding on the LB plate without Na2SeO3 but changed to dark red with supplementation of 5 mM Na2SeO3. Physiological and biochemical results indicated that strain 1805 was a Gram-positive bacterium that had contact enzyme activity to liquefy gelatin and peptonize milk and could also reduce nitrate and hydrolyze starch (Supplementary Table S1). The pH and temperature for isolate 1805 growth were 4–10 and 20°C–45°C, respectively, (Supplementary Table S1).
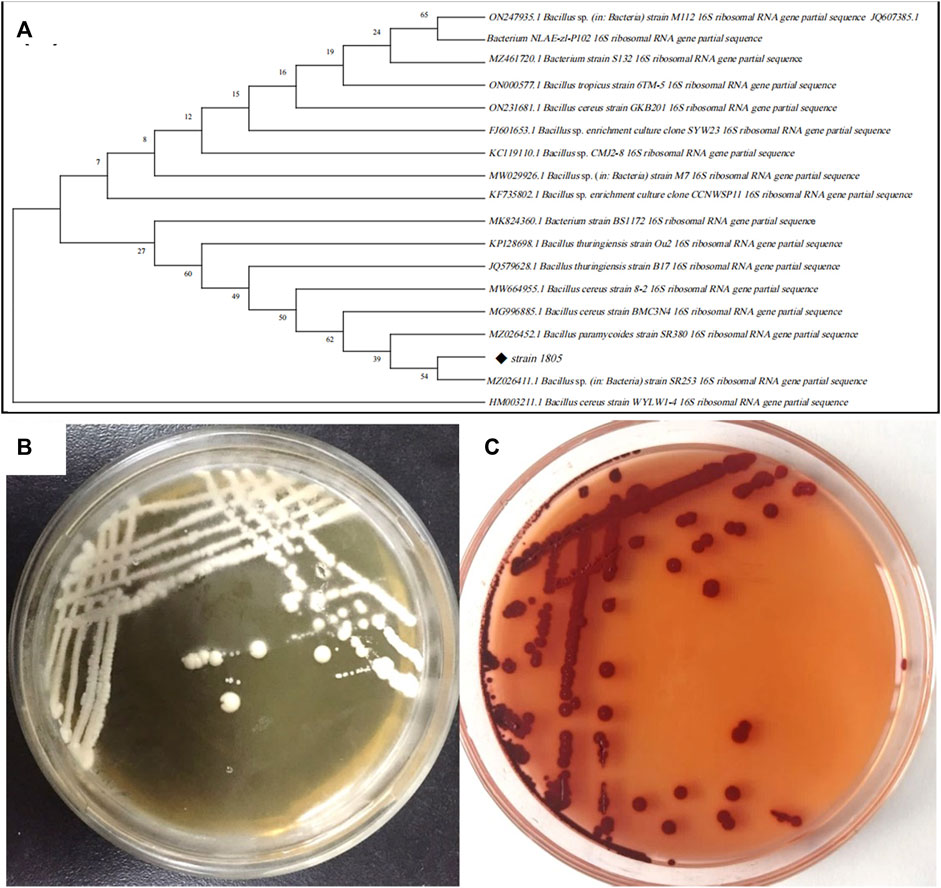
FIGURE 3. (A) Phylogenetic tree of strain 1805 and its related sequences from the NCBI database, (B) Growth of strain 1805 in solid LB medium, (C) Growth of strain 1805 in the presence of 5 mmol/L sodium selenite.
The best match model species of the 16S rDNA gene sequences to the isolate 1805 in NCBI GenBank are listed in Figure 3A, which revealed that this strain belonged to the genus Bacillus and had the highest similarities to the same species B. paramycoides, which scored 99.79%. Phylogenetic analysis using the neighbor-joining method (Abol-Fotouh et al., 2021) indicated that strain 1805 fell in the same clusters as B. paramycoides (MZ026452.1) (Figure 3A).
According to the appearance of the isolated colony, physiological/biochemical and sequencing results, the isolated strain was identified as B. paramycoides. This strain was preserved in the China General Microbiological Culture Collection Center (CGMCC, NO.24522). Therefore, this isolate was named B. paramycoides 24522.
The growth state of the inoculated bacteria is crucial for Se NPs biosynthesis. Therefore, we first studied the growth profile of B. paramycoides 24522 and observed that it grew rapidly from 4 h, entered a stable phase at 18 h, and declined after 24 h (Supplementary Figure S2). Therefore, it is necessary to select the optimal growth period for activation before 24 h, which provides a preliminary basis for optimizing the growth and SSRE conditions of B. paramycoides 24522.
Optimum growth and fermentation conditions, such as temperature, pH, and rotation speed, of B. paramycoides 24522 for production as a biofactory are required but have not been found in the current literature. In this study, the effects of temperature (20, 25, 30, 35, 37, 40, 42, and 45°C), pH (2, 4, 6, 8, 10, and 12), and rotation speed (0, 50, 100, 150, 200, 250, 300, and 350 rpm) on the SSRE of B. paramycoides 24522 were studied in the presence of 2 mM Na2SeO3. As shown in Figures 4A–C, B. paramycoides 24522 grew best at 30°C, pH 6 and 200 rpm. Concerning the growth feature of B. paramycoides 24522 (Supplementary Figure S2), it could be perceived that the strain cultured at 30°C, pH 6, and 200 rpm for 24 h can be used as the optimal inoculum for subsequent SSRE optimization.
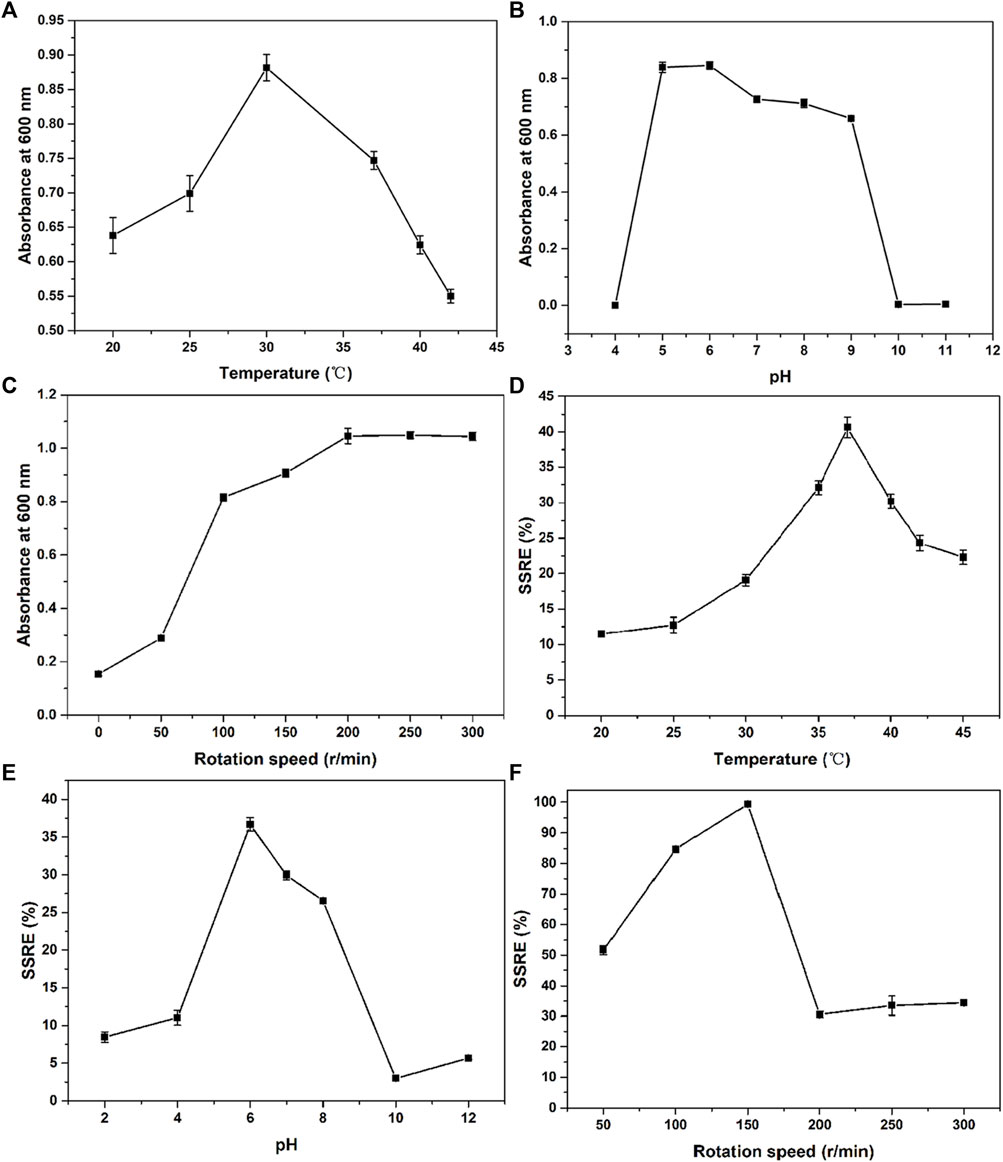
FIGURE 4. Effect of temperature (A), pH (B) and rotation speed (C) on the growth and SSRE (D–F) of B. paramycoides 24522.
The ideal temperature, pH, and rotation speed for Se NPs production by microorganisms differ from species to species (Ikram et al., 2021; Romano et al., 2022). Herein, one-factor approach experiments were applied to explore these conditions on the SSRE of B. paramycoides 24522. The maximum SSRE was perceived at 37°C, recording 40.61%, as displayed in Figure 4D. The initial pH value affects the chargeability and permeability of the bacterial cell membrane and has an essential influence on the growth of the bacteria and the oxidation potential of Na2SeO3, which further affects the rate of Na2SeO3 reduction. The optimal pH for SSRE of B. paramycoides 24522 was observed at pH 6 (Figure 4E), which was similar to Lactobacillus paracasei HM1 (El-Saadony et al., 2021). Rotation speed is one of the critical factors affecting the dissolved oxygen level of the medium during cultivation (You et al., 2021). Therefore, it was speculated that rotation speed might be one of the crucial factors affecting the SSRE of B. paramycoides 24522, which is a strictly aerobic strain. As demonstrated in Figure 4F, the SSRE was gradually augmented with increasing rotation speed, and the maximum SSRE (99.39%) was achieved at 150 rpm but decreased significantly when the rotation speed was between 200 and 300 rpm.
Based on the results of one-factor optimization, BBD was used to optimize the conditions for SSRE of B. paramycoides 24522. The results are displayed in Table 1. Quadratic equations indicating the linear relationship between response (SSRE) and independent variables [pH (A), temperature (B), and rotation speed (C)] were:
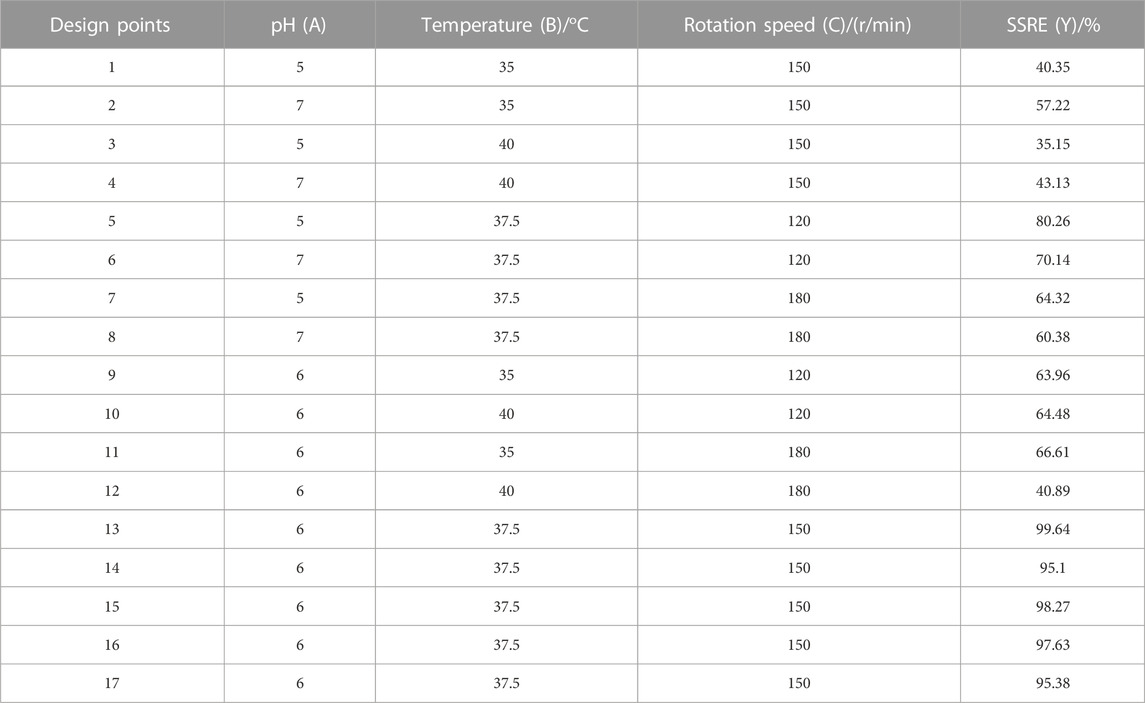
TABLE 1. Experimental design with the corresponding values observed by studying the combined effect of pH (A), temperature (B) and rotation speed (C) on the SSRE of B. paramycoides 24522.
The ANOVA of SSRE is shown in Table 2. The p-value of the model was 0.0001, meaning high significance (95% confidence) and indicating that the built quadratic equation was relatively credible for evaluation of the Na2SeO3 reducing ability of B. paramycoides 24522. In addition, all the coefficients of the equation to determine SSRE were significant (p≦0.05), except pH (A, p = 0.5100) and the combined effect of temperature and rotation speed (AC, p = 0.5915). The quadratic effects of both pH and temperature (p < 0.0001) were very significant.
The optimum pH, temperature, and rotation speed for SSRE of B. paramycoides 24522 were 6.018, 37.383°C and 137.681 rpm, respectively. Under these conditions, the predicted responses were 98.541% (Figure 5). Considering the operational feasibility, the optimal conditions for SSRE of B. paramycoides 24522 were set to pH 6, 37°C, and 140 r/min. Under optimal conditions, we conducted experiments and found that the SSRE was 99.12%, which was in line with the prediction (98.541%). This indicated that the simulation model could predict the relationship between each factor and SSRE well.
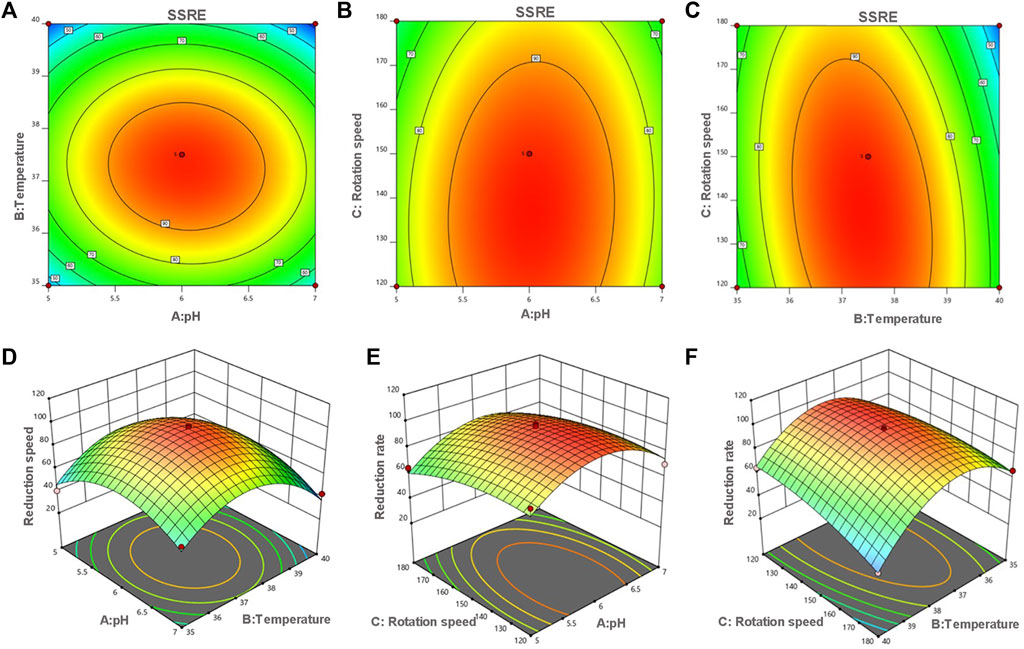
FIGURE 5. Contour (A–C) and response surface plots (D–F) of the reduction rate as a function of pH, temperature and rotation speed. The flags indicate the optimum reduction rate predicted values by the models. Red points indicate the design points above the model. Pink points indicate the design points below the model.
SEM images of SeNPs synthesized by B. paramycoides 24522 with 2 mM Na2SeO3-supplemented LB broth at 24 h are given in Figure 6A. The as-prepared SeNPs were well dispersed with fairly regular spherical shapes. The particle size varied from 100 to 180 nm, and the mean size was ∼150 nm in the DLS analysis (Figure 6B). The EDS spectra (Figure 6C) displayed a strong signal in the Se area, indicating the presence of Se in the SeNPs with a weight of 73.7% (insert of Figure 6C). The SeNPs displayed an absorption peak at ∼1.4 keV, which was consistent with previous reports (Xu et al., 2018).
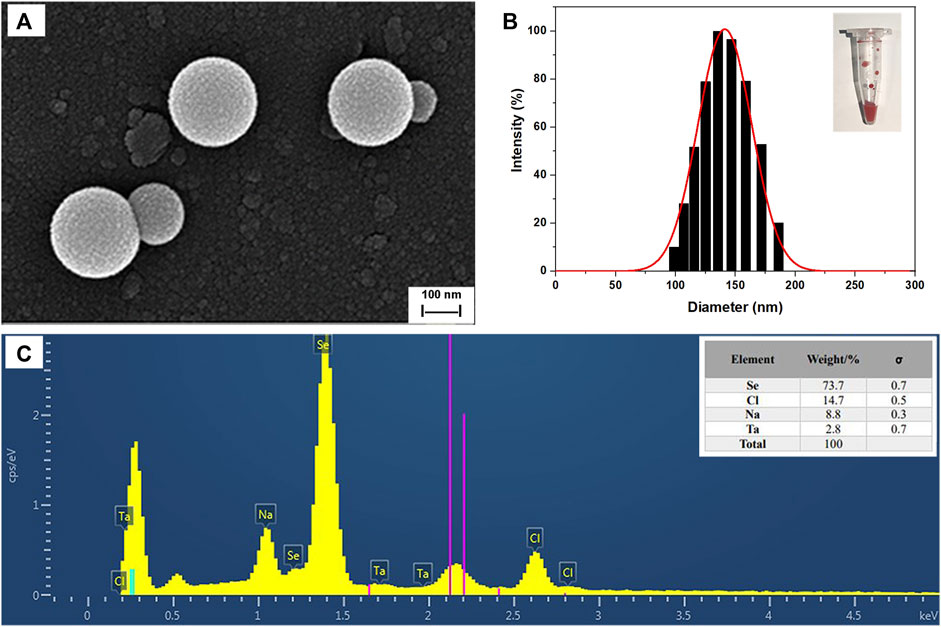
FIGURE 6. Characterization of SeNPs synthesized by B. paramycoides 24522. (A) SEM image and (B) size distribution histograms of SeNPs. The insets show the corresponding photographs of SeNPs. (C) EDS analysis of the contents of selenium (Se), chlorine (Cl), and sodium (Na) in the area of (A).
The crystal composition and average crystal size of biosynthesized Se NPs were analyzed using XRD, as it provides information on the state of observed atoms. XRD studies, shown in Figure 7A, were conducted on Se NPs synthesized by B. paramycoides 24522. The results indicated that the Se NPs had a mix of crystalline and amorphous compositions (Velayati et al., 2022). Additionally, the XRD analysis of the biosynthesized Se NPs, shown in Figure 7A, revealed diffraction peaks at 2ɵ values of 23.875°, 30.140°, 41.998°, 44.281°, 46.08°, 52.549° and 83.084°, corresponding to Bragg’s reflections at (100), (101), (110), (012), (111), (201), and (104), respectively.
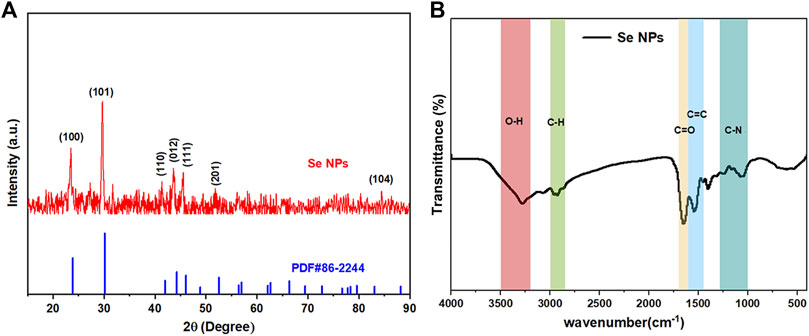
FIGURE 7. Crystallinity, surface bonding, and functional group analysis of the biosynthesized Se NPs. (A) XRD spectrum of the prepared Se NPs; (B) FT-IR spectrum of the prepared Se NPs.
Figure 7B displays the FT-IR spectrum of Se NPs synthesized using B. paramycoides 24522. The spectrum reveals five clear peaks at 3274.34, 2928.78, 1653.31, 1543.26, and 1042.90. The band at 3274.34 cm-1 indicates the–O– vibrations of aliphatic amines. The IR bands at 2928.78 and 1,543.26 cm−1 correspond to the characteristics of C–H and C=O stretching, respectively. Additionally, the peaks at 1,653.31 cm−1 and 1,042.90 cm−1 suggest the presence of C=O and N–H groups, including primary and secondary amines and amides (Fan et al., 2020). These findings suggest that the protein molecule functions as a stabilizing agent to prevent and reduce the presence of agents in forming Se NPs, thereby suggesting that the synthesized protein is a key component of the final product.
The Se NPs showed a significant inhibitory effect against S. aureus and E. coli, with inhibition zones of 16.01 ± 0.51 mm and 14.12 ± 0.89 mm, respectively. In comparison, the Na2SeO3 solution (2 mM) exhibited inhibition zones of 15.75 ± 0.77 mm and 8.04 ± 1.01 mm for S. aureus and E. coli, respectively. These findings suggest that Se NPs are more effective at inhibiting bacterial growth than Na2SeO3 solution. The B. paramycoides 24522 culture, however, did not show any zone of inhibition for either S. aureus or E. coli. Additionally, S. aureus was inhibited (MIC) at 400 μg/mL and killed (MBC) at 600 μg/mL, while E. coli was inhibited at 600 μg/mL and killed at 800 μg/mL (Table 3). These results demonstrate the potential of Se NPs as antibacterial agents, and further research is necessary to explore their broader applications in healthcare and environmental settings.
This study also investigated the potential antioxidant activity of biosynthesized Se NPs compared to the synthetic antioxidants BHA and BHT. As shown in Figure 8A, the Se NPs showed a DPPH radical scavenging activity of 72.79% at a concentration of 100 μg/mL, which was comparable with the standards, BHA and BHT. This finding is significant because it suggests that Se NPs may have potential applications in the food and pharmaceutical industries as antioxidants. The reducing capacity of all tested samples increased with increasing amounts, and the Se NPs showed the highest reducing capacity, followed by BHA and BHT (Figure 8B). This result was consistent with the DPPH free radical scavenging activity, which also demonstrated that the biosynthesized Se NPs had strong antioxidant activity. These findings have important implications for the development of eco-friendly, sustainable and natural antioxidants.
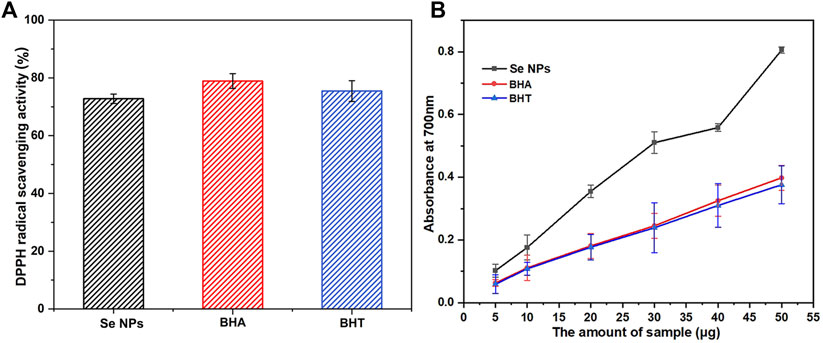
FIGURE 8. The scavenging ability on 1,1-diphenyl-2-picrylhydrazil (DPPH) radicals (A) and reducing capacity (B) of the bio-synthesized Se NPs, butyl hydroxy anisole (BHA) and butylated hydroxytoluene (BHT).
The aim of the present study was to synthesize Se NPs using the biogenic selenate-reducing bacterium B. paramycoides 24522 and investigate their antibacterial and antioxidant activities. Our results showed that B. paramycoides 24522 efficiently reduced 2 mM Na2SeO3 with a maximal SSRE of 99.12% at 37°C, pH 6, and 140 r/min within 24 h. The synthesized Se NPs exhibited a stable red color and demonstrated significant antibacterial activity against S. aureus and E. coli, with MICs ranging from 400 μg/mL to 600 μg/mL and MBCs ranging from 600 μg/mL to 800 μg/mL. Additionally, the Se NPs showed promising antioxidant capabilities through scavenging DPPH radicals and reducing capacity.
Our findings suggest that that protein molecules function as a stabilizing agent in the biosynthesis of Se NPs by B. paramycoides 24522. Furthermore, five different proteins exocytosis protein (ECP), periplasmic space protein (PSP), cytoplasmic protein (CPP), cell membrane and cell wall protein (MWP), and intracellular polysaccharides (IPS), were isolated from the bacteria using ultrasonic fragmentation (Supplementary Figure S3). And CPP from the cytoplasmic protein played a pivotal role in the synthesis of Se NPs. FT-IR spectroscopy results also demonstrated the critical role of proteins in facilitating the biosynthesis of Se NPs, providing insights into the mechanisms involved in synthesizing these nanoparticles through biological means. However, which protein plays the crucial role in sodium selenite reduction and Se NPs synthesis from MWP remains to be investigated. The research also highlights the potential utility of Se NPs as an environmentally friendly solution to selenium pollution and in various applications such as new drug delivery systems, sensors, and photocatalysts.
Compared to previous studies, our optimized biosynthesis method taking only 24 h is time-efficient (Table 4). For instance, Kora et al. (Kora and Rastogi, 2016; Kora, 2018) found that both Pseudomonas aeruginosa ATCC 27853 and Bacillus cereus AJ3 take 24–72 h to prepare SeNPs; Khoei et al. (Khoei et al., 2017) and Song et al. (Song et al., 2017) reported that Burkholderia fungorum DBT1, Burkholderia fungorum 95, and Enterobacter cloacae Z0206 take 96 h to complete Se NPs synthesis. Meanwhile, B. paramycoides, isolated in this work, is a highly beneficial bacteria that can effectively break down organic matter and remove pollutants, and has great potential for improving environmental sustainability and reducing the harmful impact of waste on our planet.
It is noteworthy that Borah et al. (Borah et al., 2021) reported that B. paramycoides SP3, which was isolated from the leachate of coal mine overburden, is capable of synthesizing Se NPs within 72 h. However, our isolate, B. paramycoides 24522, differs in two aspects from the aforementioned isolate: (1) it was obtained from sediments of Se-enriched culture ponds, and (2) it can accomplish synthesis within 24 h with a remarkable sodium selenite reduction efficiency of 99.12%.
Moreover, the biogenic Se NPs have been recently discovered to possess an exceptional ability to combat bacterial infections, making them a highly promising and potent agent in the field of antibacterial research. Studies have shown that Se NPs have broad-spectrum antibacterial effects that target both gram-positive and gram-negative bacteria (Indhira et al., 2023). The antibacterial activity of Se NPs is attributed to several mechanisms, including disintegration of the bacterial cell wall and membrane, hampering bacterial metabolism and impeding bacterial DNA replication (Abadi et al., 2022). When compared to traditional antibiotics, Se NPs have been shown to have advantages such as a smaller size, higher surface area-to-volume ratio, and greater ability to penetrate bacterial cell walls, which make them effective antimicrobial agents. In addition, while the antimicrobial activity of Se NPs may not be as strong as that of Ag NPs against certain microorganisms (Davoodbasha et al., 2016), they have been shown to be effective against a wider range of microorganisms. Therefore, Se NPs may be a promising alternative to traditional antibiotics and other antimicrobial agents. Se NPs also possess potent antioxidant activity, which can effectively scavenge free radicals and reactive oxygen species (ROS) in cells, thereby protecting against diseases such as cancer, diabetes, and neurodegenerative disorders (Hashem et al., 2023). Due to their outstanding antioxidant properties, Se NPs have been identified as promising therapeutic candidates for treating oxidative stress-related diseases (Fan et al., 2020).
In conclusion, the present study demonstrates the efficient biosynthesis of Se NPs using B. paramycoides 24522 and their potential applications in addressing selenium pollution, as well as in the fields of environment and biotechnology. The synthesized Se NPs exhibited significant antibacterial and antioxidant activities, indicating their potential use as antibacterial agents and therapeutic candidates for treating oxidative stress-related diseases. Further research is needed to identify the specific protein responsible for Se NP synthesis and to investigate their potential toxicity and efficacy in vivo.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
PL, HL, and ZG conceptualized and designed the research project. HL and HC oversaw data management and organization. ML and ZL conducted statistical analyses. PL, HL, and SH drafted and revised the manuscript. HS, ZH, and MS contributed to manuscript revisions. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Grant numbers 32202158, 22278171); the Natural Science Foundation of Jiangsu Province of China (Grant number BK20220703); and the Agricultural Science and Technology Independent Innovation Project of Jiangsu Province, China [Grant number CX(22)3078].
The authors would like to thank Newacademic for the English language review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1227619/full#supplementary-material
Abadi, B., Hosseinalipour, S., Nikzad, S., Pourshaikhali, S., Fathalipour-Rayeni, H., Shafiei, G., et al. (2022). Capping agents for selenium nanoparticles in biomedical applications. J. Clust. Sci. 34, 1669–1690. doi:10.1007/s10876-022-02341-3
Abol-Fotouh, D., AlHagar, O. E. A., and Hassan, M. A. (2021). Optimization, purification, and biochemical characterization of thermoalkaliphilic lipase from a novel Geobacillus stearothermophilus FMR12 for detergent formulations. Int. J. Biol. Macromol. 181, 125–135. doi:10.1016/j.ijbiomac.2021.03.111
Al-Hagar, O. E. A., Abol-Fotouh, D., Abdelkhalek, E. S., Abo Elsoud, M. M., and Sidkey, N. M. (2021). Bacillus niabensis OAB2: Outstanding bio-factory of selenium nanoparticles. Mat. Chem. Phys. 273, 125147. doi:10.1016/j.matchemphys.2021.125147
Azimi, F., Oraei, M., Gohari, G., Panahirad, S., and Farmarzi, A. (2021). Chitosan-selenium nanoparticles (Cs–Se NPs) modulate the photosynthesis parameters, antioxidant enzymes activities and essential oils in Dracocephalum moldavica L. under cadmium toxicity stress. Plant Physiol. bioch. 167, 257–268. doi:10.1016/j.plaphy.2021.08.013
Biswas, K. C., Barton, L. L., Tsui, W. L., Shuman, K., Gillespie, J., and Eze, C. S. (2011). A novel method for the measurement of elemental selenium produced by bacterial reduction of selenite. J. Microbiol. Meth. 86, 140–144. doi:10.1016/j.mimet.2011.04.009
Borah, S. N., Goswami, L., Sen, S., Sachan, D., Sarma, H., Montes, M., et al. (2021). Selenite bioreduction and biosynthesis of selenium nanoparticles by Bacillus paramycoides SP3 isolated from coal mine overburden leachate. Environ. Pollut. 285, 117519. doi:10.1016/j.envpol.2021.117519
Brar, K. K., Magdouli, S., Othmani, A., Ghanei, J., Narisetty, V., Sindhu, R., et al. (2022). Green route for recycling of low-cost waste resources for the biosynthesis of nanoparticles (NPs) and nanomaterials (NMs)-A review. Environ. Res. 207, 112202. doi:10.1016/j.envres.2021.112202
Davoodbasha, M., Kim, S.-C., Lee, S.-Y., and Kim, J.-W. (2016). The facile synthesis of chitosan-based silver nano-biocomposites via a solution plasma process and their potential antimicrobial efficacy. Arch.Biochem. Biophys. 605, 49–58. doi:10.1016/j.abb.2016.01.013
Dharmaraj, D., Krishnamoorthy, M., Rajendran, K., Karuppiah, K., Jeyaraman, R., and Ethiraj, K. (2021). Protein leakage induced marine antibiofouling activity of biosynthesized zinc oxide nanoparticles. J. Clust. Sci. 32, 643–650. doi:10.1007/s10876-020-01827-2
El-Saadony, M. T., Saad, A. M., Taha, T. F., Najjar, A. A., Zabermawi, N. M., Nader, M. M., et al. (2021). Selenium nanoparticles from Lactobacillus paracasei HM1 capable of antagonizing animal pathogenic fungi as a new source from human breast milk. Saudi J. Biol. Sci. 28, 6782–6794. doi:10.1016/j.sjbs.2021.07.059
English, S. G., Hess, H., Bishop, C. A., Porter, E., Cheng, K. M., and Elliott, J. E. (2022). Bioaccumulation and effects of selenium from surface coal mining in an aquatic songbird. Environ. Res. 208, 112702. doi:10.1016/j.envres.2022.112702
Fan, D., Li, L., Li, Z., Zhang, Y., Ma, X., Wu, L., et al. (2020). Biosynthesis of selenium nanoparticles and their protective, antioxidative effects in streptozotocin induced diabetic rats. Sci. Technol. Adv. Mat. 21, 505–514. doi:10.1080/14686996.2020.1788907
Faramarzi, S., Anzabi, Y., and Jafarizadeh-Malmiri, H. (2020). Nanobiotechnology approach in intracellular selenium nanoparticle synthesis using Saccharomyces cerevisiae—Fabrication and characterization. Archives Microbiol. 202, 1203–1209. doi:10.1007/s00203-020-01831-0
Han, W., Liu, X., Wang, L., and Zhou, X. (2020). Engineering of lipid microbubbles-coated copper and selenium nanoparticles: Ultrasound-stimulated radiation of anticancer activity ian human ovarian cancer cells. Process Biochem. 98, 113–121. doi:10.1016/j.procbio.2020.07.013
Hashem, A. H., Saied, E., Ali, O. M., Selim, S., Al Jaouni, S. K., Elkady, F. M., et al. (2023). Pomegranate peel extract stabilized selenium nanoparticles synthesis: Promising antimicrobial potential, antioxidant activity, biocompatibility, and hemocompatibility. Appl. Biochem. Biotech. doi:10.1007/s12010-023-04326-y
He, L., Zhao, J., Wang, L., Liu, Q., Fan, Y., Li, B., et al. (2021). Using nano-selenium to combat coronavirus disease 2019 (COVID-19)? Nano Today 36, 101037. doi:10.1016/j.nantod.2020.101037
Huang, S., Wang, Y., Tang, C., Jia, H., and Wu, L. (2021). Speeding up selenite bioremediation using the highly selenite-tolerant strain Providencia rettgeri HF16-A novel mechanism of selenite reduction based on proteomic analysis. J. Hazard. Mat. 406, 124690. doi:10.1016/j.jhazmat.2020.124690
Ikram, M., Javed, B., Raja, N. I., and Mashwani, Z. U. R. (2021). Biomedical potential of plant-based selenium nanoparticles: A comprehensive review on therapeutic and mechanistic aspects. Int. J. Nanomedicine 16, 249–268. doi:10.2147/IJN.S295053
Indhira, D., Aruna, A., Manikandan, K., Albeshr, M. F., Alrefaei, A. F., Vinayagam, R., et al. (2023). Antimicrobial and photocatalytic activities of selenium nanoparticles synthesized from Elaeagnus indica leaf extract. Processes 11, 1107. doi:10.3390/pr11041107
Islam, S. N., Naqvi, S. M. A., Raza, A., Jaiswal, A., Singh, A. K., Dixit, M., et al. (2022). Mycosynthesis of highly fluorescent selenium nanoparticles from Fusarium oxysporum, their antifungal activity against black fungus Aspergillus niger, and in-vivo biodistribution studies. 3 Biotech. 12, 309. doi:10.1007/s13205-022-03383-0
Jadhav, A. S., Ramteke, P., Singh, S. K., and Labhasetwar, N. K. (2022). Sustainable selenium remediation from water using aluminium–iron mixed oxide: Batch and column adsorption studies. J. Water Process Eng. 48, 102824. doi:10.1016/j.jwpe.2022.102824
Jia, H., Huang, S., Cheng, S., Zhang, X., Chen, X., Zhang, Y., et al. (2022). Novel mechanisms of selenite reduction in Bacillus subtilis 168:Confirmation of multiple-pathway mediated remediation based on transcriptome analysis. J. Hazard. Mat. 433, 128834. doi:10.1016/j.jhazmat.2022.128834
Khoei, N. S., Lampis, S., Zonaro, E., Yrjälä, K., Bernardi, P., and Vallini, G. (2017). Insights into selenite reduction and biogenesis of elemental selenium nanoparticles by two environmental isolates of Burkholderia fungorum. New Biotech. 34, 1–11. doi:10.1016/j.nbt.2016.10.002
Kora, A. J. (2018). Bacillus cereus, selenite-reducing bacterium from contaminated lake of an industrial area: A renewable nanofactory for the synthesis of selenium nanoparticles. Bioresour. Bioprocess. 5, 30. doi:10.1186/s40643-018-0217-5
Kora, A. J., and Rastogi, L. (2017). Bacteriogenic synthesis of selenium nanoparticles by Escherichia coli ATCC 35218 and its structural characterisation. IET nanobiotechnol 11, 179–184. doi:10.1049/iet-nbt.2016.0011
Kora, A. J., and Rastogi, L. (2016). Biomimetic synthesis of selenium nanoparticles by Pseudomonas aeruginosa ATCC 27853: An approach for conversion of selenite. J. Environ. Manage. 181, 231–236. doi:10.1016/j.jenvman.2016.06.029
Lampis, S., Zonaro, E., Bertolini, C., Bernardi, P., Butler, C. S., and Vallini, G. (2014). Delayed formation of zero-valent selenium nanoparticles by Bacillus mycoides SeITE01 as a consequence of selenite reduction under aerobic conditions. Microb. Cell Fact. 13, 35. doi:10.1186/1475-2859-13-35
Liu, P., Zhao, W.-H., Song, Y.-P., Wang, Q., Kan, Y.-F., Wang, S.-Y., et al. (2022). Characterization, antimicrobial, and antioxidant potentialities of first-time isolated silver nanoparticles synthesizing protein secreted by Lysinibacillus sphaericus. Process Biochem. 122, 230–237. doi:10.1016/j.procbio.2022.08.032
Mao, X., Liu, P., He, S., Xie, J., Kan, F., Yu, C., et al. (2013). Antioxidant properties of bio-active substances from shrimp head fermented by Bacillus licheniformis OPL-007. Appl. Biochem. Biotechnol. 171, 1240–1252. doi:10.1007/s12010-013-0217-z
Nie, X., Xing, Y., Li, Q., Gao, F., Wang, S., Liu, P., et al. (2022). ARTP mutagenesis promotes selenium accumulation in Saccharomyces boulardii. LWT 168, 113916. doi:10.1016/j.lwt.2022.113916
Qiao, L., Dou, X., Yan, S., Zhang, B., and Xu, C. (2020). Biogenic selenium nanoparticles synthesized by Lactobacillus casei ATCC 393 alleviate diquat-induced intestinal barrier dysfunction in C57BL/6 mice through their antioxidant activity. Food Funct. 11, 3020–3031. doi:10.1039/D0FO00132E
Rana, T. (2021). Prospects and future perspectives of selenium nanoparticles: An insight of growth promoter, antioxidant and anti-bacterial potentials in productivity of poultry. J. Trace Elem. Med. Bio. 68, 126862. doi:10.1016/j.jtemb.2021.126862
Romano, I., Vitiello, G., Gallucci, N., Di Girolamo, R., Cattaneo, A., Poli, A., et al. (2022). Extremophilic microorganisms for the green synthesis of antibacterial nanoparticles. Microorganisms 10, 1885. doi:10.3390/microorganisms10101885
Samanta, P., Dey, S., Ghosh, A. R., and Kim, J. K. (2022). Nanoparticle nutraceuticals in aquaculture: A recent advances. Aquaculture 560, 738494. doi:10.1016/j.aquaculture.2022.738494
Shi, X.-D., Tian, Y.-Q., Wu, J.-L., and Wang, S.-Y. (2021). Synthesis, characterization, and biological activity of selenium nanoparticles conjugated with polysaccharides. Crit. Rev. Food Sci. 61, 2225–2236. doi:10.1080/10408398.2020.1774497
Shreyash, N., Bajpai, S., Khan, Mohd. A., Vijay, Y., Tiwary, S. K., and Sonker, M. (2021). Green synthesis of nanoparticles and their biomedical applications: A review. ACS Appl. Nano Mat. 4, 11428–11457. doi:10.1021/acsanm.1c02946
Song, D., Li, X., Cheng, Y., Xiao, X., Lu, Z., Wang, Y., et al. (2017). Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci. Rep-UK 7, 3239. doi:10.1038/s41598-017-03558-3
Sonkeshariya, H., Shakya, A. K., and Ghosh, P. K. (2020). Development of a sulfidogenic bioreactor system for removal of co-existent selenium, iron and nitrate from drinking water sources. J. Environ. Manage. 254, 109757. doi:10.1016/j.jenvman.2019.109757
Staicu, L. C., Wójtowicz, P. J., Molnár, Z., Ruiz-Agudo, E., Gallego, J. L. R., Baragaño, D., et al. (2022). Interplay between arsenic and selenium biomineralization in Shewanella sp. O23S. Environ. Pollut. 306, 119451. doi:10.1016/j.envpol.2022.119451
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi:10.1093/molbev/msab120
Velayati, M., Hassani, H., Sabouri, Z., Mostafapour, A., and Darroudi, M. (2022). Green-based biosynthesis of Se nanorods in chitosan and assessment of their photocatalytic and cytotoxicity effects. Environ. Technol. Inno. 27, 102610. doi:10.1016/j.eti.2022.102610
Wadhwani, S. A., Shedbalkar, U. U., Singh, R., and Chopade, B. A. (2016). Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biot. 100, 2555–2566. doi:10.1007/s00253-016-7300-7
Xu, C., Qiao, L., Guo, Y., Ma, L., and Cheng, Y. (2018). Preparation, characteristics and antioxidant activity of polysaccharides and proteins-capped selenium nanoparticles synthesized by Lactobacillus casei ATCC 393. Carbohyd. Polym. 195, 576–585. doi:10.1016/j.carbpol.2018.04.110
Yadav, P., Pandey, S., and Dubey, S. K. (2023). Selenite bioreduction with concomitant green synthesis of selenium nanoparticles by a selenite resistant EPS and siderophore producing terrestrial bacterium. BioMetals. doi:10.1007/s10534-023-00503-y
You, J., Yang, C., Pan, X., Hu, M., Du, Y., Osire, T., et al. (2021). Metabolic engineering of Bacillus subtilis for enhancing riboflavin production by alleviating dissolved oxygen limitation. Bioresour. Technol. 333, 125228. doi:10.1016/j.biortech.2021.125228
Zhang, H., Zhou, H., Bai, J., Li, Y., Yang, J., Ma, Q., et al. (2019). Biosynthesis of selenium nanoparticles mediated by fungus Mariannaea sp. HJ and their characterization. Colloid. Surf. A 571, 9–16. doi:10.1016/j.colsurfa.2019.02.070
Keywords: selenium nanoparticles, Bacillus paramycoides, antibacterial activity, antioxidant activity, green synthesis
Citation: Liu P, Long H, Cheng H, Liang M, Liu Z, Han Z, Guo Z, Shi H, Sun M and He S (2023) Highly-efficient synthesis of biogenic selenium nanoparticles by Bacillus paramycoides and their antibacterial and antioxidant activities. Front. Bioeng. Biotechnol. 11:1227619. doi: 10.3389/fbioe.2023.1227619
Received: 23 May 2023; Accepted: 21 July 2023;
Published: 01 August 2023.
Edited by:
Li Zhou, Shanghai Academy of Agricultural Sciences, ChinaReviewed by:
Muhammad Ikram, Pir Mehr Ali Shah Arid Agriculture University, PakistanCopyright © 2023 Liu, Long, Cheng, Liang, Liu, Han, Guo, Shi, Sun and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai He, aGVzaHVhaUBoeWl0LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.