
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Bioeng. Biotechnol. , 05 July 2023
Sec. Synthetic Biology
Volume 11 - 2023 | https://doi.org/10.3389/fbioe.2023.1208784
Phosphoramidite chemical DNA synthesis technology is utilized for creating de novo ssDNA building blocks and is widely used by commercial vendors. Recent advances in enzymatic DNA synthesis (EDS), including engineered enzymes and reversibly terminated nucleotides, bring EDS technology into competition with traditional chemical methods. In this short study, we evaluate oligos produced using a benchtop EDS instrument alongside chemically produced commercial oligonucleotides to assemble a synthetic gene encoding green fluorescent protein (GFP). While enzymatic synthesis produced lower concentrations of individual oligonucleotides, these were available in half the time of commercially produced oligonucleotides and were sufficient to assemble functional GFP sequences without producing hazardous organic chemical waste.
Phosphoramidite solid-phase chemical DNA synthesis (CDS) (Beaucage and Caruthers, 1981) is the standard method for producing de novo single-stranded DNA (ssDNA) and is used on a large scale by commercial vendors to produce oligonucleotides (oligos) for PCR, cloning, and increasingly for de novo construction of genes and plasmids (Kosuri and Church, 2014; Song et al., 2021). CDS methods have evolved to maximize product length while minimizing overall cost and errors, but they are limited to production of oligos of 200 or fewer nucleotides (nt) and require the use of hazardous chemicals such as acetonitrile, tetrahydrofuran, and dichloromethane (Hughes et al., 2011; Kosuri and Church, 2014; Song et al., 2021).
Controlled enzymatic DNA synthesis (EDS) was first developed in the 1950s–1970s (Grunberg-Manago et al., 1955; Bollum, 1962; Chang and Bollum, 1971) with the discovery of the terminal deoxynucleotidyl transferase (TdT) enzyme that adds dNTPs to the 3′-hydroxyl group of DNA. Recent breakthroughs have addressed historical limitations of TdT (Palluk et al., 2018; Song et al., 2021) by evolving more thermally stable TdT enzymes (Chua et al., 2020; Lu et al., 2022), developing reversible 3′-dNTP protecting groups that can be accommodated by TdT (Mathews et al., 2016; Mathews et al., 2017; Hoff et al., 2020), and improving enzymatic coupling efficiency to enable a cyclic, solid-phase-templated extension of DNA (Palluk et al., 2018; Barthel et al., 2020; Hoff et al., 2020). EDS has the added benefit of eliminating the use of hazardous organic chemicals by utilizing a primarily aqueous workflow (Song et al., 2021).
Several EDS-focused companies were launched within the last decade, including Molecular Assemblies and Nuclera (2013), DNA Script (2014), Camena Bioscience (2016), Ansa Biotechnologies (2018), and Kern Systems (2019) (Eisenstein, 2020). Camena Bioscience and DNA Script report greater than 99% coupling efficiency while producing 300- and 280-mer oligos, respectively (Eisenstein, 2020), elevating this technology to a competitive level with chemical synthesis (Masaki et al., 2022). More recently, Ansa Biotechnologies, Inc., reported the enzymatic synthesis of the longest de novo oligonucleotide ever produced in a single synthesis at 1,005 nucleotides long (Ansa Biotechnologies, 2023), and Camena Bioscience reported the production of a whole 2.7-kb plasmid using multi-enzymatic de novo DNA synthesis and gene assembly (Camenabio, 2020). However, EDS products are not readily available yet for purchase from commercial sources, and interested users are limited to early access programs (Ansa and Molecular Assemblies) or purchasing a benchtop EDS instrument (DNA Script).
The Syntax-100™ from DNA Script is the first commercially available benchtop EDS instrument. It is marketed to have a user-friendly platform and essentially aqueous waste stream and can synthesize 96 × 60 mers in 13 hours, including cleanup and quantitation. Here, we evaluate the Syntax-100™ to produce oligos using EDS and compare them to commercial CDS oligos to assemble a synthetic gene encoding green fluorescent protein (GFP). In this brief study, we consider the time of oligo synthesis and gene assembly, cost per oligo/base, sequence accuracy, and waste production.
Primerize (Tian and Das, 2017) was used to design overlapping oligos for a GFP-coding sequence (Snapgene, 2023) with 20 nt overlap, 60 or 90 nt maximum length, and a minimum annealing temperature of 60°C. A set of twelve (90 nt, 100 nmol) or twenty (60 nt, 25 nmol) commercial CDS oligos was purchased from Integrated DNA Technologies (IDT, USA). Each set of CDS oligos was used for independent assembly experiments. The Syntax-100™ enzymatic DNA synthesizer was set up according to the user instructions, with reagents and consumables provided for a 60-mer kit. A set of twenty oligos (60 nt max) and Gibson assembly primers were synthesized in two or more replicates per plate on the Syntax-100™ at 0.3 nmol per oligo, normalized to a final concentration of 5 μM. After completion of each run (13 hours), the oligos were immediately stored at −20°C. Two individual plates of EDS oligos were produced and used for separate assembly experiments. Supplementary Table S1 lists all oligos used in this study.
Method modified from Dormitzer et al. (2013). A set of twelve (90 nt) CDS oligos, twenty (60 nt) CDS oligos, or twenty (60 nt) EDS oligos was pooled by combining 2 µL of each 5 µM oligo. A volume of 10 μL of ×2 Gibson Assembly Master Mix (New England Biosciences, #E2611) was combined with 10 µL of pooled oligos and incubated at 50°C/30 min. Next, 5 µL of this mixture was added to 12.5 µL of ×2 Q5 Master Mix (NEB, # M0492S), 2.5 µL each end oligo, and H2O to reach a final concentration of 25 µL. The PCR cycles were run as follows: 98°C/60 s, 98°C/10 s, 60°C/30 s, and 72°C/1.5 min, return to step 2 for 24 cycles, followed by 72°C/5 min and 10°C hold. An enrichment PCR using 2 µL of PCR1 and 2.5 µL of 5 µM end oligos was performed under the aforementioned conditions to obtain a full-length gfp product.
Following gel confirmation of the assembled gene product, 1 µL of Taq polymerase (NEB, #M0273S) was added to PCR to add a single deoxyadenosine (A) to the 3′-ends. TOPO cloning and transformation were performed using a TOPO® TA Cloning® Kit (Invitrogen #K457540) (Thermo Fisher Scientific, 2023). Transformants were screened by PCR, and plasmids were isolated via a Miniprep Kit (Zymo, #D4211) and were sequenced (Eurofins).
Confirmed, correct gfp sequences from each assembly were cloned into an expression vector using Gibson assembly primers (Supplementary Table S1). Separate PCRs were run with 2 μL of the template (pET28a (+) or PCR4_GFP), 25 μL of ×2 Q5 Master Mix, 2.5 μL of each 5 μM forward or reverse primer, and H2O to 50 μL using the online Q5 PCR protocol (New England Biolabs, 2023). Vector and insert sequences were gel-extracted and cloned in a Gibson assembly reaction according to the online protocol (New England Biolabs, 2023) and transformed into NEB5α competent cells (NEB, #C2987). Sequence-confirmed plasmids were transformed into BL21 DE3 competent cells (Thermo Fisher Scientific, #EC0114) and plated on LB/kanamycin plates. Cells transformed with pET28-GFP from each assembly were sub-cultured onto fresh LB/kanamycin plates with or without the addition of 10 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), incubated overnight at 37°C, and imaged under UV light to confirm the expression of fluorescent GFP.
In this study, we sought to compare CDS- and EDS-sourced oligonucleotides for use in the assembly of the gene encoding GFP. We used the application Primerize (Tian and Das, 2017) to divide a GFP coding sequence (Snapgene, 2023) into overlapping oligos of 60 nt or 90 nt maximum length, a 20 nt overlap, and a minimum annealing temperature of 60°C that were amenable to assembly using previously established polymerase cycling assembly (PCA) methods (Supplementary Table S1) (Stemmer et al., 1995; Dormitzer et al., 2013). The 60 nt oligos provided the maximum length available from our EDS benchtop instrument, and the 90 nt oligos provided a longer yet cost-effective option for CDS oligos that could be used to compare the effects of oligo length on gene assembly. We synthesized replicates of twenty 60 nt oligos via EDS in two independent runs on the Syntax-100™. The total run time for each plate was thirteen hours, including sample cleanup and quantitation. A set of twenty (60 nt max, 25 nmol each) and twelve (90 nt max, 100 nmol each) commercial CDS oligos was ordered and delivered in 2 days.
Our adapted gene assembly approach (Dormitzer et al., 2013) included an initial isothermal assembly of pooled oligos, followed by two rounds of PCR: PCR1 for PCA and PCR2 to enrich the final product (Figure 1A). The CDS 90 and 60 nt oligo assemblies showed a faint band at the expected size (717 bp) after PCR1 and a clear band after PCR2, whereas the two EDS 60 nt oligo sets did not produce a visible full-length product until PCR2 (Figure 1B). Full gene products assembled from each oligo set were TOPO-cloned into pCR4 vectors (Thermo Fisher Scientific, 2023) and sequenced after transformation and colony screening. A minimum of ten transformants were screened and sequenced for each independent assembly (Table 1). We observed that the designed gfp sequence was successfully assembled using each of the four oligo sets (Table 1). Notably, the longer 90 nt CDS oligos did produce a higher percentage of correct sequences (40%) compared to the 60 nt oligo sets (27%, CDS and 24%, EDS). Sequence-validated gfp gene products from each oligo set were sub-cloned into an expression vector, transformed into competent E. coli cells, and analyzed for GFP protein expression and fluorescence. EDS- and CDS-sourced DNA sequences successfully produced functional gene products with confirmed expression and fluorescence of GFP (Figure 1C).
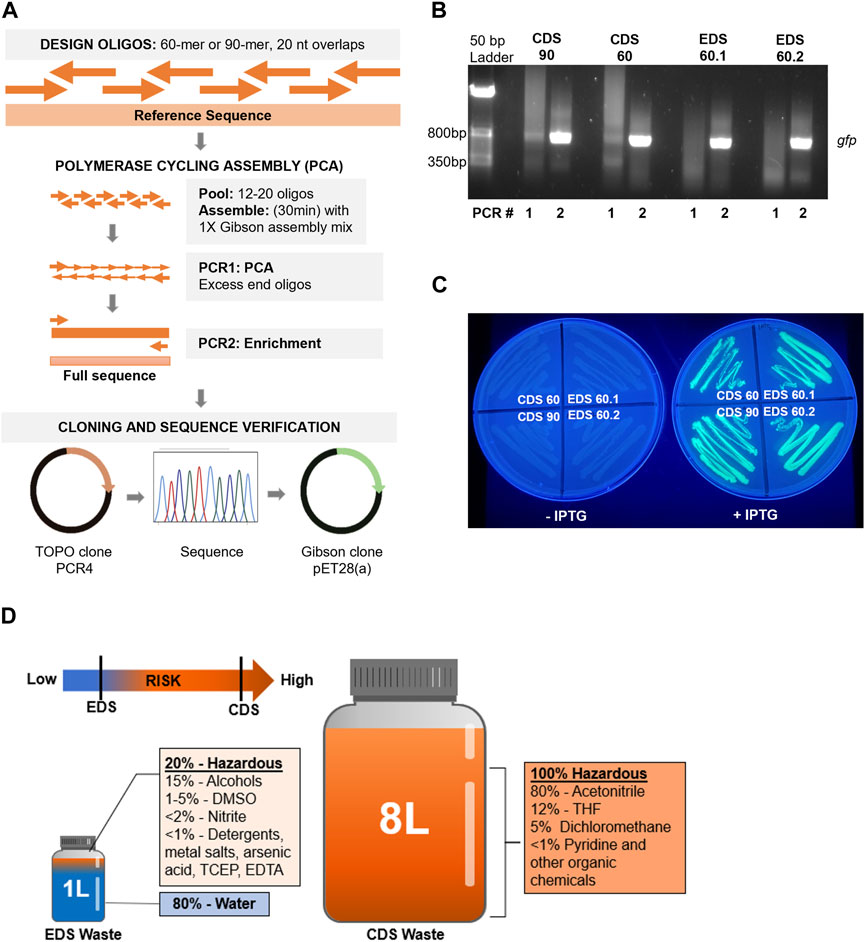
FIGURE 1. (A) Workflow for oligo design, gene assembly, cloning, sequence verification, and sub-cloning of synthetic gfp sequences, created with BioRender.com. (B) Pooled EDS (60 nt) and CDS (90 nt or 60 nt) oligos were assembled into gfp using an isothermal incubation step, followed by PCR1 for polymerase cycling assembly and PCR2 to enrich the final product (717 bp). (C) Sequence-verified GFP-encoding sequences assembled from each oligo set were cloned into a pET28 vector and transformed into BL21 DE3 E. coli that were grown on LB/kanamycin plates in the presence or absence of 10 mM IPTG; plates were imaged under UV light to demonstrate GFP expression in induced samples (right) compared to uninduced samples (left). (D) Analysis of waste stream for a benchtop EDS instrument (Syntax-100™) and CDS instrument (MerMade 192e) with the hazardous nature of waste indicated by color (non-hazardous = blue; hazardous = orange). Note: components and volumes are from production of one set of 96 × 60-mers per instrument.
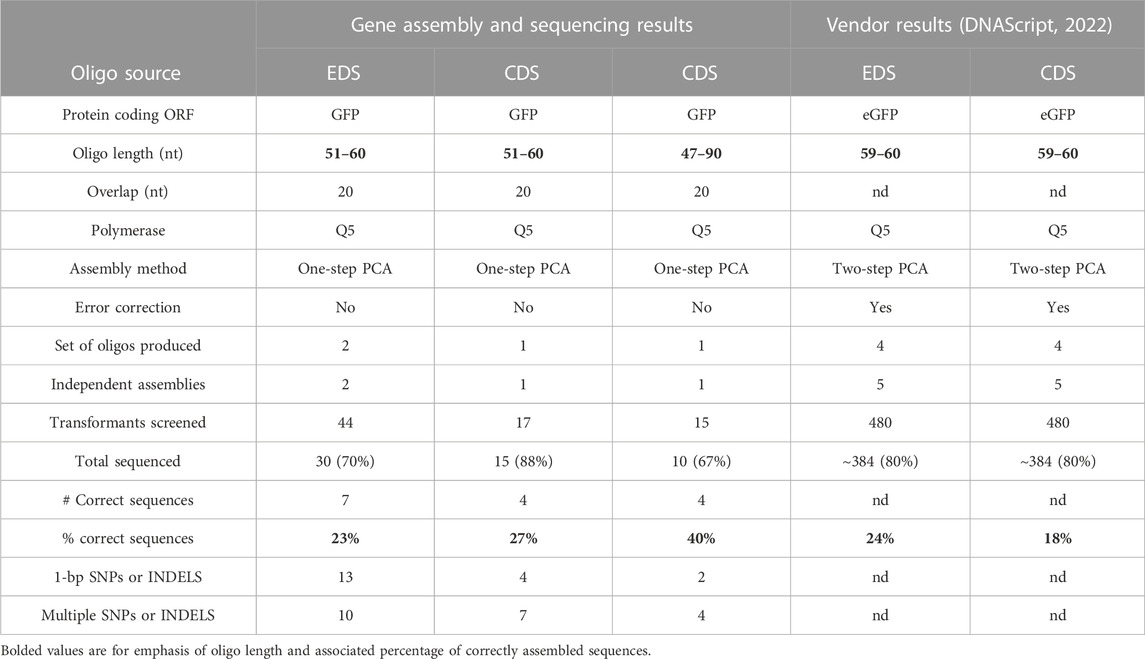
TABLE 1. Summary of gene assembly and sequence results from enzymatically or chemically synthesized oligos (EDS or CDS) and comparison with published results from the vendor.
While this work was being completed, the vendor for the Syntax-100™ published an application note on their website detailing a similar set of experiments (DNA Script, 2022). We have summarized those findings in Table 1 along with our independent validation of EDS oligos compared to commercially prepared CDS oligos. Using an analogous gene assembly method, we obtained similar percentages of correctly assembled sequences using EDS oligos (23%) compared to the vendor (24%). However, our results for CDS 60 nt oligos differed, with 27% of our sequenced transformants having the designed sequence compared to only 18% reported by the vendor. The vendor does not state the source of their CDS oligos.
Currently, the most cost-effective way to utilize the Syntax-100™ is to produce 96 × 60-mers (or 80-mers and 120-mers that have recently become available), which is comparable to ordering the equivalent number, length, and quality of oligos from IDT (Table 2) at approximately $0.20–$0.40 per base, if labor and the initial cost of the instrument are not included. The yield for EDS oligos is much lower (0.3 nmol per oligo per well), but in our experiments, EDS oligos were of sufficient quality and quantity to assemble a functional gfp gene. It is worth noting that purchasing a CDS synthetic gfp gene from commercial sources currently costs approximately $280, which is still substantially cheaper than purchasing or producing the assembly oligos by EDS or CDS methods. Finally, we were interested in analyzing the waste generated in our laboratory on a benchtop CDS instrument (MerMade 192E™, LGC Limited) compared to the Syntax-100™ after production of 96 × 60-mers. Our analysis indicated a clear reduction in hazardous chemical waste production in both volume and individual waste components when using the benchtop EDS method (Figure 1D).
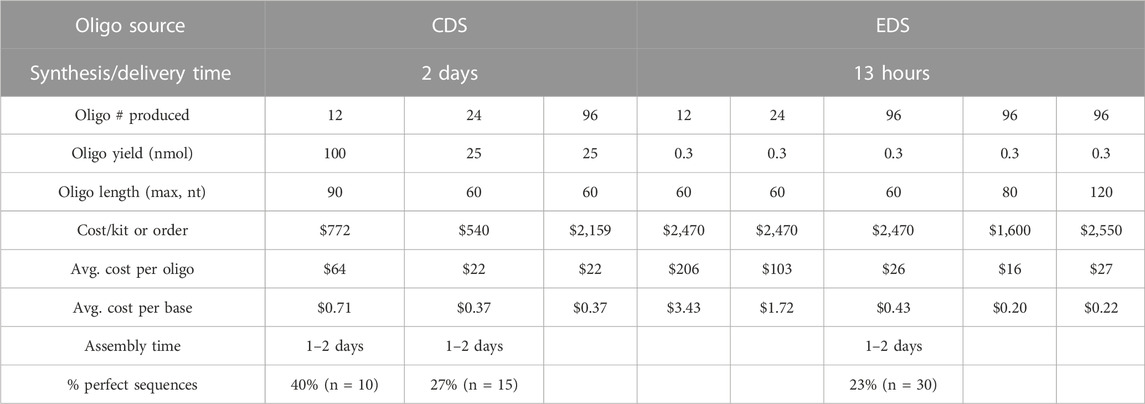
TABLE 2. Analysis of cost, time, and accuracy of GFP sequences assembled from oligos produced via enzymatic (EDS) or chemical (CDS) DNA synthesis.
This brief study provides an independent validation that EDS oligonucleotides are suitable for use in gene synthesis workflows, with 23% of assembled samples containing the designed gfp sequence (Table 1). Our findings are similar to those described by the vendor in their recently published application note (DNA Script, 2022). Notably, we obtained similar results without adding a correction step in our PCA protocol. Our results for CDS gene assembly indicate a higher rate of correct sequences assembled (40% and 27% compared to 23% for EDS oligos) that may be due to longer starting oligo length (in the case of CDS 90 nt oligos) or the higher reported error rate for EDS (0.4%) versus CDS (0.1%) (Masaki et al., 2022). Further experiments are required to differentiate between errors introduced during DNA synthesis or gene assembly. Both EDS- and CDS-produced oligos used in gene assembly resulted in functional GFP-coding sequences.
Although commercial CDS sources remain the most cost-effective solution for obtaining oligos, it is worth noting that benchtop DNA synthesis instruments (CDS and EDS) provide users with improved access to on-demand DNA synthesis in mobile or austere laboratory settings. In our studies, EDS produced substantially less hazardous waste than a CDS instrument, suggesting this technology could provide a safer and more environmentally friendly alternative for large-scale DNA synthesis. Finally, the recent report of kilobase-length DNA by EDS methods (Ansa Biotechnologies, 2023) suggests that this technology may overcome the inherent length limitations faced by CDS methods. Future studies may include testing additional EDS kits, offering oligo modifications such as the addition of biotin, fluorophores, and quenchers for a variety of downstream applications.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
NR and NM designed the experiments. BS performed the main experiments and data analysis. NR and BS prepared the first draft, and NM reviewed and edited the manuscript. NR and NM supervised this project. All authors contributed to the article and approved the submitted version.
This work was funded by the Defense Threat Reduction Agency Joint Science and Technology Office (DTRA-JSTO). The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of Defense or the U.S. government.
The authors would like to thank Kelley D. Betts for her contribution to designing Figure 1D.
NR was employed by the company Excet, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1208784/full#supplementary-material
CDS, chemical DNA synthesis; EDS, enzymatic DNA synthesis; NT, nucleotide; TdT, terminal nucleotidyl transferase; dNTPs, dinucleotide triphosphates; IDT, Integrated DNA Technologies; DNA, deoxyribonucleic acid; GFP, green fluorescence protein; SNP, single-nucleotide polymorphism; INDEL, insertion/deletion; IPTG, isopropyl β- d-1-thiogalactopyranoside.
Ansa Biotechnologies, I. (2023). Ansa Biotechnologies Announces Successful de novo Synthesis of World’s Longest Oligonucleotide at 1005 Bases. Available at: https://www.businesswire.com/news/home/20230309005124/en/Ansa-Biotechnologies-Announces-Successful-de-novo-Synthesis-of-World%E2%80%99s-Longest-Oligonucleotide-at-1005-Bases.
Barthel, S., Palluk, S., Hillson, N. J., Keasling, J. D., and Arlow, D. H. (2020). Enhancing terminal deoxynucleotidyl transferase activity on substrates with 3 ' terminal structures for enzymatic de novo DNA synthesis. Genes-Basel 11 (1), 102. doi:10.3390/genes11010102
Beaucage, S. L., and Caruthers, M. H. (1981). Deoxynucleoside phosphoramidites - a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 22 (20), 1859–1862. doi:10.1016/s0040-4039(01)90461-7
Bollum, F. J. (1962). Oligodeoxyribonucleotide-primed reactions catalyzed by calf thymus polymerase. J. Biol. Chem. 237, 1945–1949. doi:10.1016/s0021-9258(19)73964-7
Camenabio (2020). Camena Bioscience uses gSynth™ to synthesise a 2.7kb plasmid [press release]. Available at: https://www.camenabio.com/gSynth-production-long-DNA-molecules.
Chang, L. M. S., and Bollum, F. J. (1971). Deoxynucleotide-polymerizing enzymes of calf thymus gland. VI. Enzymic synthesis of oligodeoxynucleotides. Biochemistry-Us 10 (3), 536–542. doi:10.1021/bi00779a029
Chua, J. P. S., Go, M. K., Osothprarop, T., Mcdonald, S., Karabadzhak, A. G., Yew, W. S., et al. (2020). Evolving a thermostable terminal deoxynucleotidyl transferase. Acs Synth. Biol. 9 (7), 1725–1735. doi:10.1021/acssynbio.0c00078
DNAScript (2022). Application note: Gene assembly. Available at: https://www.dnascript.com/wp-content/uploads/2022/09/DNAScript-Gene-Assembly_AppNote_version1.0.pdf. p1-12 (Accessed April 18, 2023).
Dormitzer, P. R., Suphaphiphat, P., Gibson, D. G., Stockwell, T. B., Algire, M. A., Alperovich, N., et al. (2013). Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci. Transl. Med. 5 (185), 185ra68. doi:10.1126/scitranslmed.3006368
Eisenstein, M. (2020). Enzymatic DNA synthesis enters new phase. Nat. Biotechnol. 38 (10), 1113–1115. doi:10.1038/s41587-020-0695-9
Grunberg-Manago, M., Oritz, P. J., and Ochoa, S. (1955). Enzymatic synthesis of nucleic acidlike polynucleotides. Science 122 (3176), 907–910. doi:10.1126/science.122.3176.907
Hoff, K., Halpain, M., Garbagnati, G., Edwards, J. S., and Zhou, W. (2020). Enzymatic synthesis of designer DNA using cyclic reversible termination and a universal template. Acs Synth. Biol. 9 (2), 283–293. doi:10.1021/acssynbio.9b00315
Hughes, R. A., Miklos, A. E., and Ellington, A. D. (2011). Gene synthesis: Methods and applications. Methods Enzymol. 498, 277–309. doi:10.1016/B978-0-12-385120-8.00012-7
Kosuri, S., and Church, G. M. (2014). Large-scale de novo DNA synthesis: Technologies and applications. Nat. Methods 11 (5), 499–507. doi:10.1038/nmeth.2918
Lu, X. Y., Li, J. L., Li, C. Y., Lou, Q. Q., Peng, K., Cai, B. J., et al. (2022). Enzymatic DNA synthesis by engineering terminal deoxynucleotidyl transferase. Acs Catal. 12 (5), 2988–2997. doi:10.1021/acscatal.1c04879
Masaki, Y., Onishi, Y., and Seio, K. (2022). Quantification of synthetic errors during chemical synthesis of DNA and its suppression by non-canonical nucleosides. Sci. Rep. 12 (1), 12095. doi:10.1038/s41598-022-16222-2
Mathews, A. S., Yang, H., and Montemagno, C. (2017). 3'-O-Caged 2'-deoxynucleoside triphosphates for light-mediated, enzyme-catalyzed, template-independent DNA synthesis. Curr. Protoc. Nucleic Acid. Chem. 71, 1–38. doi:10.1002/cpnc.41
Mathews, A. S., Yang, H. K., and Montemagno, C. (2016). Photo-cleavable nucleotides for primer free enzyme mediated DNA synthesis. Org. Biomol. Chem. 14 (35), 8278–8288. doi:10.1039/c6ob01371f
New England Biolabs (2023). Protocol for Q5® high-fidelity 2X master Mix 2023. Available at: https://www.neb.com/protocols/2012/12/07/protocol-for-q5-high-fidelity-2x-master-mix-m0492.
Palluk, S., Arlow, D. H., de Rond, T., Barthel, S., Kang, J. S., Bector, R., et al. (2018). De novo DNA synthesis using polymerase-nucleotide conjugates. Nat. Biotechnol. 36 (7), 645–650. doi:10.1038/nbt.4173
Snapgene (2023). Aequorea victoria green fluorescent protein. Available at: https://www.snapgene.com/plasmids/fluorescent_protein_genes_and_plasmids/GFP.
Song, L. F., Deng, Z. H., Gong, Z. Y., Li, L. L., and Li, B. Z. (2021). Large-Scale de novo Oligonucleotide Synthesis for Whole-Genome Synthesis and Data Storage: Challenges and Opportunities. Front. Bioeng. Biotechnol. 9, 689797. doi:10.3389/fbioe.2021.689797
Stemmer, W. P., Crameri, A., Ha, K. D., Brennan, T. M., and Heyneker, H. L. (1995). Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164 (1), 49–53. doi:10.1016/0378-1119(95)00511-4
ThermoFisherScientific (2023). TOPO TA for sequencing. Available at: https://www.thermofisher.com/order/catalog/product/450071?ICID=search-4500712023.
Keywords: enzymatic DNA synthesis, de novo DNA, gene assembly, chemical DNA synthesis, benchtop DNA synthesis
Citation: Simmons BL, McDonald ND and Robinett NG (2023) Assessment of enzymatically synthesized DNA for gene assembly. Front. Bioeng. Biotechnol. 11:1208784. doi: 10.3389/fbioe.2023.1208784
Received: 19 April 2023; Accepted: 20 June 2023;
Published: 05 July 2023.
Edited by:
Patrick O’Donoghue, Western University, CanadaReviewed by:
Noah Reynolds, University of Illinois at Springfield, United StatesCopyright © 2023 Simmons, McDonald and Robinett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalie G. Robinett, bmF0YWxpZS5nLnJvYmluZXR0LmN0ckBhcm15Lm1pbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.